Novel lytic phages isolated from various environmental settings were systematically characterized for their critical genetic traits, morphology structures, host ranges against laboratory strains and clinical multidrug-resistant (MDR) Pseudomonas aeruginosa isolates, and antibacterial capacity both in vitro and in mouse models. First, we characterized the genetic traits and compared them with those of other existing phages.
KEYWORDS: bacteriophage, antibiotic resistance, Pseudomonas aeruginosa, acute and chronic pneumonia, antibacterial efficacy, phage therapy
ABSTRACT
With the fast emergence of serious antibiotic resistance and the lagging discovery of novel antibacterial drugs, phage therapy for pathogenic bacterial infections has acquired great attention in clinics. However, the development of therapeutic phages also faces tough challenges such as the laborious screening and time to generate effective phage drugs since each phage may lyse only a narrow scope of bacterial strains. Identifying highly effective phages with broad host ranges is crucial for improving phage therapy. Here, we isolated and characterized several lytic phages from various environments specific for Pseudomonas aeruginosa by testing their growth, invasion, host ranges, and potential for killing targeted bacteria. Importantly, we identified several therapeutic phages (HX1, MYY9, and TH15) with broad host ranges to lyse laboratory strains and clinical isolates of P. aeruginosa with multidrug resistance (MDR) both in vitro and in mouse models. In addition, we analyzed critical genetic traits related to high-level broad-host-range coverages by genome sequencing and subsequent computational analysis against known phages. Collectively, our findings establish that these novel phages may have potential for further development as therapeutic options for patients who fail to respond to conventional treatments.
IMPORTANCE Novel lytic phages isolated from various environmental settings were systematically characterized for their critical genetic traits, morphology structures, host ranges against laboratory strains and clinical multidrug-resistant (MDR) Pseudomonas aeruginosa isolates, and antibacterial capacity both in vitro and in mouse models. First, we characterized the genetic traits and compared them with those of other existing phages. Furthermore, we utilized acute pneumonia induced by laboratory strain PAO1 and strain W19, an MDR clinical isolate, and chronic pneumonia by agar beads laden with FRD1, a mucoid-phenotype strain isolated from the sputum of a cystic fibrosis (CF) patient. Consequently, we found that these phages not only suppress bacteria in vitro but also significantly reduce infection symptoms and disease progression in vivo, including lowered bug burdens, inflammatory responses, and lung injury in mice, suggesting that they may be further developed as therapeutic agents against MDR P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa, a significant Gram-negative pathogen, is commonly related to hospital-acquired pneumonia (HAP), particularly in individuals with cancer, cystic fibrosis (CF), ventilator-associated pneumonia, or other infectious diseases (1). A study described that P. aeruginosa was the second most common cause of HAP in the United States, accounting for 14% to 16% (2). Similarly, according to the China Antimicrobial Surveillance Network (CHINET) (http://www.chinets.com/), in 2019, P. aeruginosa was recorded as the third most common pathogen isolated from specimens of the airway tract and the fourth most common bacterium from skin infectious wounds in hospitals. In addition, P. aeruginosa is also the third main contributor to urinary tract infections in patients who had frequently received broad-spectrum antibiotics (3).
Critically, P. aeruginosa, especially multidrug-resistant (MDR) strains, defined as being resistant to ≥3 agents of different antimicrobial classes, has become a life-threatening pathogen (4, 5). According to the U.S. Centers for Disease Control and Prevention (CDC) (https://www.cdc.gov/), at least 2.8 million Americans were infected with antibiotic-resistant (AR) bacteria or fungi, causing more than 35,000 deaths each year. Carbapenem antibiotics, including imipenem and meropenem, are the last resort for the treatment of severe Gram-negative bacterial infection. However, in 2019, clinically isolated P. aeruginosa from Chinese tertiary hospitals showed high-level resistance to imipenem (27.5%), next to Acinetobacter and Klebsiella, and meropenem (23.5%), ranking only second to Acinetobacter, in accordance with CHINET surveillance data (6, 7). Meanwhile, according to the CDC, the average incidence of carbapenem-resistant P. aeruginosa in the United States was 19.2%, which is also next to Acinetobacter (49.5%) among health care-associated infections (HAIs), in 2014 (8). Hence, it is of high urgency to develop novel effective drugs to control refractory infections caused by MDR Pseudomonas.
Bacteriophages (here phages) are viruses coexisting and coevolving with bacteria and other microorganisms, which are omnipresent and extraordinarily abundant in the microbial ecosystems of water, soil, and sediment, at ∼1031 individual particles (9, 10). Defined as natural predators of bacteria, fungi, or archaea, phages can specifically invade and destroy their hosts (7). At present, comparative genomic analysis of novel lytic phages is one of the key approaches for determining their potential for clinical applications. Being the most abundant biont on earth and owing to the ability to lyse host populations, phages have long been considered to have potential utility as favorable therapeutic agents. Consequently, phage therapy is deemed an innovative, alternative, and nonantibiotic treatment to control severe infections by MDR bacteria, against which the currently available antibiotics have lost their efficacy.
Although phage therapy was popular in eastern European countries before the 1940s, it has been forsaken for years in the Western world because of the advent of antibiotics and their inherent disadvantages, such as the lack of control of quality, narrow host ranges, and so on (1, 11). It has also been proven that phages can shape the microbial ecology and evolution of pathogens by the means of adaptive immune systems, CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR associated), and anti-CRISPR proteins (Acrs) that counteract CRISPR-Cas (12, 13). Recent accumulating research from academic communities and pharmaceutical companies has acclaimed phage therapy as a viable alternative treatment due to the limited side effects, low drug resistance, and effective pharmacodynamics in vitro and in vivo. For instance, a recent report revealed that a 15-year-old patient suffering from CF and an unmanageable Mycobacterium abscessus infection was challenged with a three-lytic-phage cocktail after a bilateral lung transplantation operation, which showed great tolerance to phages and an inspiring improvement in clinical symptoms (14). Hence, phage therapy has regained great attention to treat serious infections caused by MDR pathogenic microorganisms due to the widespread improper application and abuse of antibiotics.
In order to search for effective and broad-target therapeutic phages, we set out to isolate and purify P. aeruginosa phages from community and hospital sewage as well as feces from farms using an efficient double-agar-layer method. In addition, aiming to further identify the characteristics of new lytic phages, we analyzed their genomes, morphology structures, and host ranges. We next identified the phage candidates that can effectively kill clinical isolates of MDR P. aeruginosa that cause refractory infection. Next, we employed acute and chronic bronchopulmonary infection mouse models through airway delivery of P. aeruginosa. Two strains were employed to establish acute pneumonia, including laboratory strain PAO1 and W19, an MDR clinical isolate. The cell aggregates used for the development of chronic pneumonia are based on agar beads laden with clinical isolate FRD1, a mucoid-phenotype strain that was noted for its high mucus yield with a mucA mutation separated from a CF patient’s sputum, which mimics the pathogenesis of human inflammatory disease (15–18). As expected, several highly effective phages that target relatively broad-range hosts were identified as potential killers of MDR bacterial strains. Indeed, we validated that these phages exhibit strong antibacterial effects on acute and chronic pneumonic mice infected with P. aeruginosa, which may have promise to treat clinical patients with severe infections as compassionate therapies.
RESULTS
Isolation of phages from different environments.
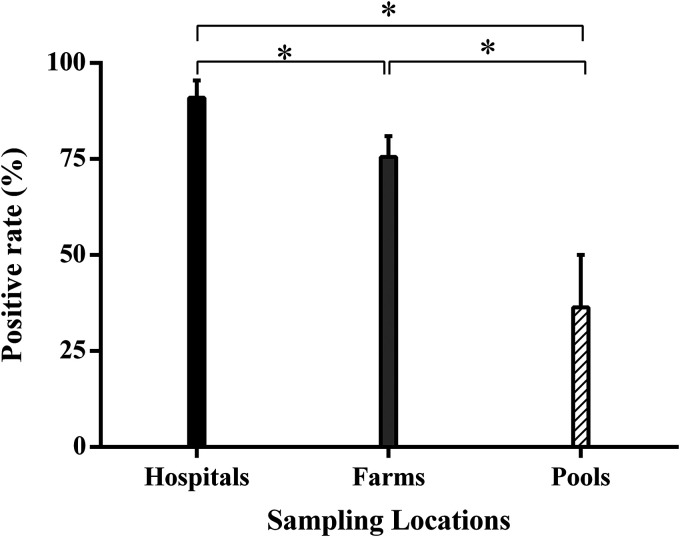
Considering sites for collecting phages, sampling from hospital effluent was found to be optimal because of the characteristics of the ecological microenvironment in hospitals. Here, to increase the diversity of phage species, we extensively sampled from wild environments, not only in hospital sewage but also in poultry farms and stagnant pools in the community. By comparing phages sampled from different sources, we observed that the positive rate of phage detection and isolation from farms was lower than in hospitals (90.9%), despite remaining at high levels (up to 72.2%), as shown in Fig. 1.
FIG 1.
Positive targeting rate of phage detection and isolation from the different types of sampling locations using P. aeruginosa laboratory strains PAO1, ATCC 27853, and PA14 as the hosts. The data are presented as means ± standard deviations (SD), and unpaired Student’s t test was applied to compare differences between the two groups. “*” indicates a P value of <0.05.
Morphological and structural characteristics of phages.
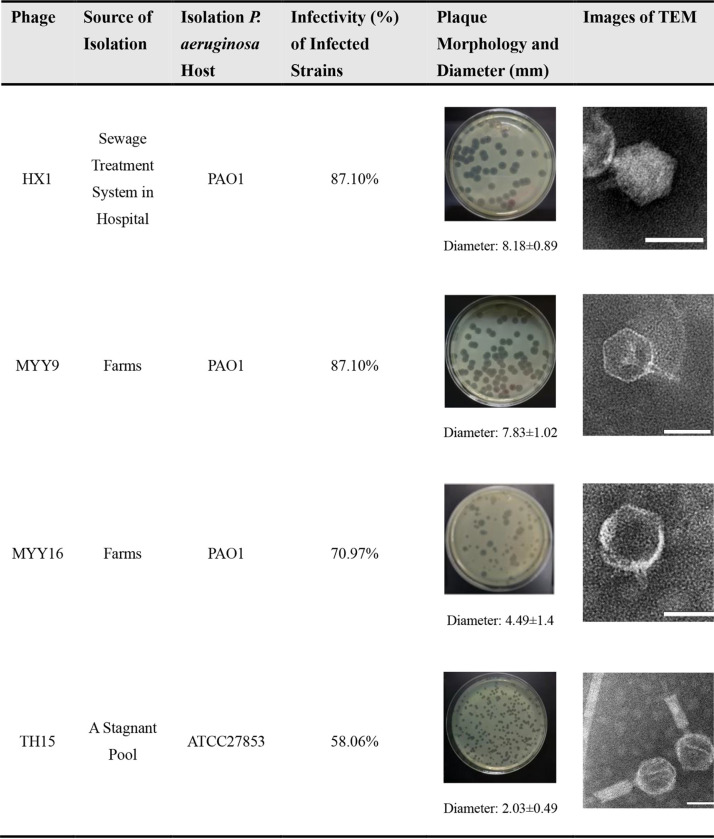
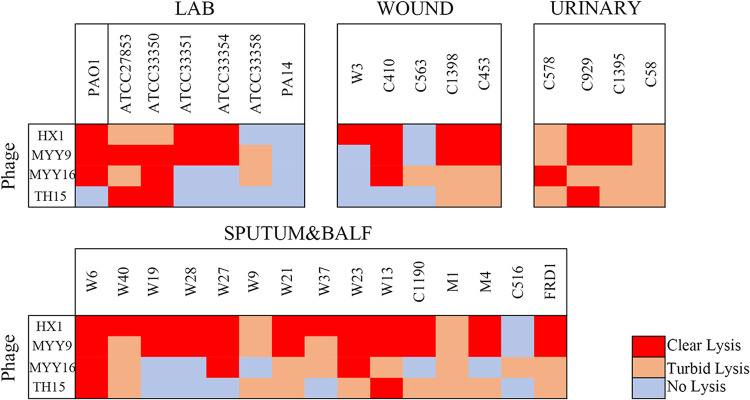
Using PAO1 and ATCC 27853 as host strains, we successfully isolated several phages, named HX1, MYY9, MYY16, and TH15. All of these phages are capable of forming clear plaques on a lawn of their host cells, indicating that they may be lytic phages. Table 1 summarizes details of all identified phages, including sources of isolation, host bacteria, plaque morphologies, plaque diameters, infectivities (percentages) of and laboratory strains clinical isolates of MDR P. aeruginosa, as well as transmission electron microscopy (TEM) images. Although MYY16 infected more strains (70.97%) than TH15 (58.06%), MYY16 exhibited weaker bactericidal effects and less clear plaques than TH15 and other phages.
TABLE 1.
Host ranges and morphologies of the four phages identified in this studya
Phages were isolated from various environments: sewage treatment systems in hospitals, feces of poultry, and stagnant pools. The infectivity (percent) of the phages was calculated based on the host ranges (Fig. 4). Phages exhibited clear plaque morphologies with different diameters in double-agar plates with the P. aeruginosa host. TEM images of phages that were negatively stained by 2% phosphotungstic acid are also shown. Bars at the bottom right, 50 nm.
Using TEM analysis, we observed that all phages had icosahedral heads. In addition, TH15 had a contractile tail, suggesting that it belongs to the order Caudovirales and the family Myoviridae. Other phages, MYY9, HX1, and MYY16, possessed a short tail, belonging to the order Caudovirales and the family Podoviridae according to the International Committee on Taxonomy of Viruses (ICTV). In addition, TH15 had a capsid symmetry length of approximately 68.38 nm and contractile tails of approximately 99.7 nm in length. Furthermore, the intact HX1 particles also contained an ∼51.5-nm-diameter icosahedral head and a short tail of ∼15.0 nm in length, while MYY9 and MYY16 had ∼54.7- and ∼57.5-nm-diameter icosahedral heads, with short tails of 28.1 and 17.2 nm in length, respectively, as shown in Table 1. Finally, Table 2 summarizes the morphological features of phages from the TEM analysis.
TABLE 2.
Summarized information on phage morphological characteristics for the TEM imagesa
| Phage | Host | Mean capsid diam (nm) ± SD | Mean tail length (nm) ± SD | Family | Order |
|---|---|---|---|---|---|
| HX1 | PAO1 | 51.46 ± 2.89 | 15.00 ± 1.42 | Podoviridae | Caudovirales |
| MYY9 | PAO1 | 54.70 ± 2.17 | 28.10 ± 0.98 | Podoviridae | Caudovirales |
| MYY16 | PAO1 | 57.50 ± 3.52 | 17.17 ± 1.57 | Podoviridae | Caudovirales |
| TH15 | ATCC 27853 | 68.38 ± 3.31 | 99.72 ± 3.81 (contractile) | Myoviridae | Caudovirales |
The diameter of each phage icosahedral head and the length of the tail were measured using ImageJ software. The orders and families to which the phages belong are indicated.
Sequencing analysis of lytic phage genomes.
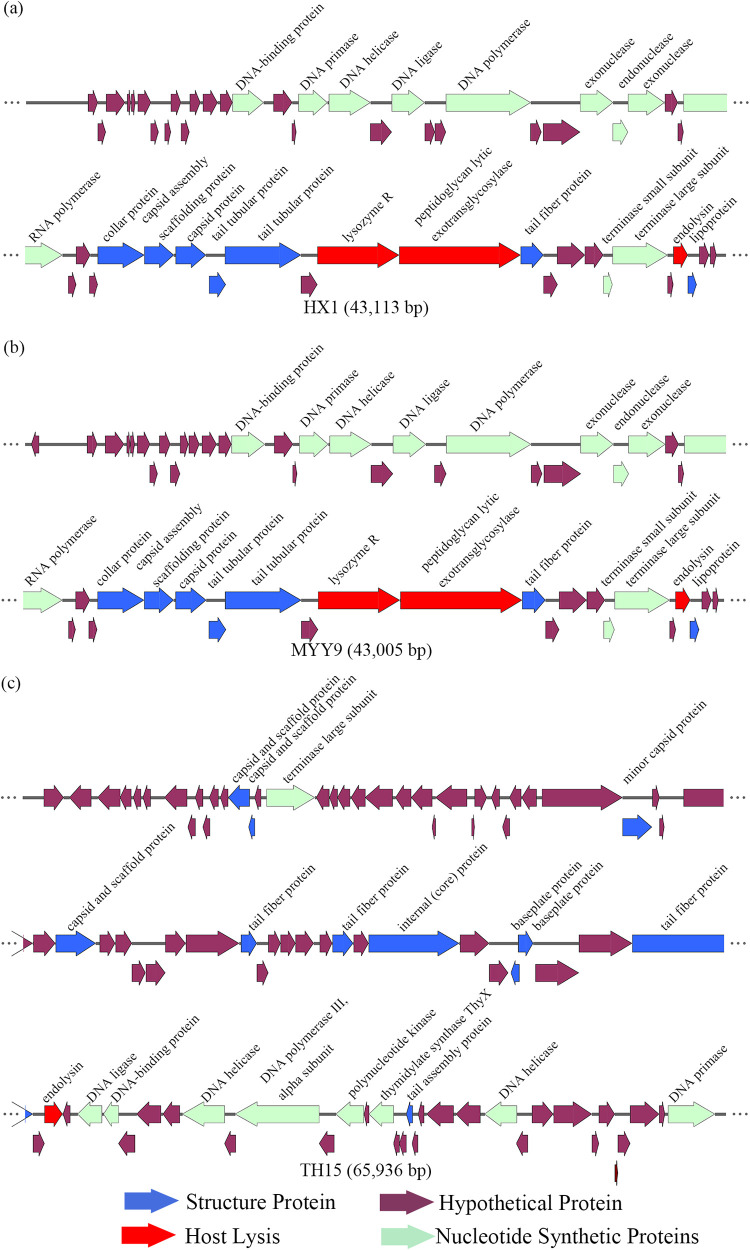
To gain an understanding of the genetic traits of the new phages, we chose three of the four phages to perform genome sequencing analysis based on the infectivity of lysed cell types, levels of clear-plaque formation, and the capacity for modulating the growth status of their hosts. As a result, HX1, MYY9, and TH15 qualified for additional assessments and showed a strong cleavage ability or a broader host range, making them potential therapeutic candidates. The whole genomes of phages were detected by Illumina NovaSeq (Tsingke, Beijing, China), and analysis results are summarized in Fig. 2 and Table 3.
FIG 2.
Functional gene maps of phages HX1 (a), MYY9 (b), and TH15 (c) prepared using SnapGene software. The different-colored arrows show the CDSs of the phages and their transcription direction. The putative products of each CDS were predicted and divided into four modules, including putative structural proteins, nucleotide metabolism proteins, putative lytic enzymes, and proteins with hypothetical function.
TABLE 3.
Summarized data on phage genome features and comparison of three phagesa
| Phage | Host | Total length (bp) | GC content (%) | No. of restriction endonucleases | No. of CDSs |
|||
|---|---|---|---|---|---|---|---|---|
| Structure proteins | Nucleotide synthesis | Lytic enzymes | Hypothetical function | |||||
| HX1 | PAO1 | 43,113 | 62.24 | 7 | 7 | 11 | 3 | 32 |
| MYY9 | PAO1 | 43,005 | 62.21 | 8 | 7 | 11 | 3 | 31 |
| TH15 | ATCC 27853 | 65,936 | 55.57 | 9 | 11 | 9 | 1 | 72 |
The putative coding sequences (CDSs) were identified and divided into four modules using bioinformatics analysis. The numbers of CDSs in each module are listed.
Analysis of the features of phage HX1 and MYY9 genomes.
We noted that MYY9 and HX1, belonging to the same family, Podoviridae, shared highly similar features: they had 97.84% identical sequences by primary amino acid sequencing analysis and were closely related to a lytic Pseudomonas phage, PAXYB1, a “phiKMV-like” phage by BLASTn analysis, which was isolated from wastewater samples (GenBank accession number NC_047952.1) (19). First, HX1 and MYY9 consisted of circular double-stranded DNA (dsDNA) molecules of 43,113 bp and 43,005 bp accompanied by GC contents of 62.24% and 62.21%, respectively. Second, eight different restriction endonucleases showed the ability to digest MYY9 DNA, while seven showed the ability to digest HX1. Bioinformatics analysis of the phage genomes identified 52 putative coding sequences (CDSs) in MYY9. Also, 51 of the CDSs were transcribed in a forward orientation, but 1 was transcribed in reverse, while 53 CDSs were identified in HX1, and all CDSs had a forward orientation. Third, in MYY9, 44 CDSs can start translation with an AUG start codon, 8 CDSs can start with GUG, and none can start with UUG, while 47 CDSs initiate with an AUG, 6 CDSs initiate with GUG, and none initiate with UUG in HX1 (Fig. 2 and Table 3).
According to the amino acid sequences and homology to functional domains aligned with known phages, we predicted the putative products of the CDSs and categorized them into four modules, including structural proteins (blue), such as tail fiber components/proteins and capsids and scaffold proteins; nucleotide synthesis proteins (turquoise), such as the coding DNA polymerase III alpha subunit, DNA helicase, and so on; putative lytic enzymes (red); and hypothetical proteins (brown), as shown in Fig. 2. Finally, none of the tRNA genes were detected in both genomes using the tRNAscan-SE program, indicating that the isolated phages possibly utilize the host tRNA machinery to synthesize proteins (20). No CDSs that are associated with drug resistance, human virulence genes, or lysogenization, such as site-specific integrases or repressors, were noticed, which may benefit drug development.
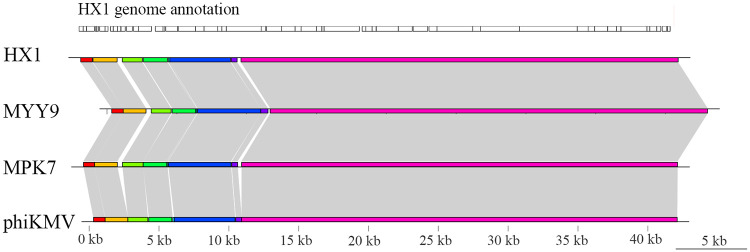
We considered that HX1 and MYY9 may be new members of the “phiKMV-like” phage genus using genomic analysis. Briefly, we compared HX1 and MYY9 sequences with those of two members of this genus, MPK7 and phiKMV, and found that all genomic architectures exhibited high similarity in base pair content and length, comprising a conservative colinear area, while the less conserved regions were located in the early middle cluster, sharing a few discrepancies (Fig. 3). Overall, these analyses supported that horizontal gene transfer (HGT) and host gene integration events might rarely occur in this genus, which is in line with previous studies (21, 22).
FIG 3.
Comparative genome analysis reveals that HX1 and MYY9 are new members of the “phiKMV-like” genus. The analysis was performed using Mauve and genoPlotR software. The genomes of MPK7 and phiKMV were available in the GenBank database. The CDSs from HX1 are shown as boxes in parallel approximately to scale at the top. Colored bars indicated nucleotide syntenic blocks, while white spaces between colored bars are the missing regions. Gray shading indicates homologous regions.
Features of the phage TH15 genome.
TH15 was characterized as having a circular dsDNA molecule, and the genome had a size of 65,936 bp and a GC content of 55.57%. TH15 contained nine different restriction endonucleases. TH15 was found to be closely related to a lytic Pseudomonas phage, PB1-like lytic phage vB_PaeM_SCUT-S1, which was also assigned to the Myoviridae family (GenBank accession number NC_048745.1) (20). Next, a total of 93 CDSs were identified, 50 of which had a leftward orientation and 43 of which had a rightward orientation. Additionally, the majority of all CDSs can initiate translation with an AUG, 8 CDSs started with GUG, and only one started with UUG. The putative products predicted for each CDS are shown in Fig. 2. Finally, no tRNA genes or any risk genes were detected in its genome.
Identification of broad-host-range phages.
We next evaluated these phages’ host spectra by using spot plaque testing with an undiluted phage stock (approximately 109 PFU/ml). The bacteria that were used include 24 clinically isolated P. aeruginosa strains that were resistant to meropenem and/or aztreonam and 7 laboratory strains belonging to different serotypes (see Table S1 in the supplemental material). The results are referred to as three categories: clear zones (red), turbid zones (orange), or no zones (gray) (Fig. 4). We defined clear and turbid zones as having effects on bacterial growth, indicating different levels of bacterial lysis. As a result, we measured the efficiency of each of the phages infected with the different bacteria that are listed in Table 1. We found that phages HX1 (87.10%), MYY9 (87.10%), and MYY16 (70.97%) have relatively broad host ranges and may have potential for future therapeutic applications.
FIG 4.
Host ranges of each phage against laboratory and clinical P. aeruginosa strains (see Table S1 in the supplemental material). A spot test was performed with a 5-μl undiluted phage stock (approximately 109 PFU/ml). Results were evaluated as “not clear” when no inhibition zone was observed, while they were evaluated as “clear” and “turbid” when completely clear and turbid inhibition zones were observed, respectively. The experiment was performed at least three times.
Bactericidal effects of the phages in vitro.
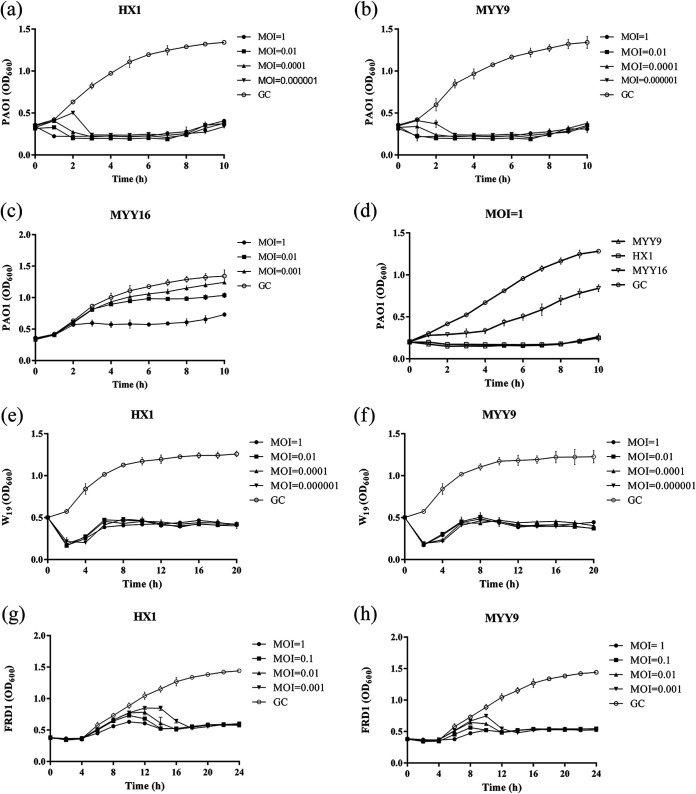
The bacteriolytic activities of three phages (HX1, MYY9, and MYY16) were evaluated using PAO1 and clinical isolates W19 and FRD1 at different multiplicities of infection (MOIs) in vitro (Fig. 5). The infection assays of phages HX1, MYY9, and MYY16 (used as a negative control) against PAO1 that were used to isolate them from the environment were performed at 37°C (Fig. 5a to c). Our results showed that even at an MOI of 0.000001, both HX1 and MYY9 could effectively inhibit the growth of PAO1, keeping the optical density at 600 nm (OD600) at approximately 0.3 within 10 h (Fig. 5a and b). In contrast, Fig. 5c indicates that when the MOI was ≤1, MYY16 had no overt impact on the growth of PAO1. In addition, when the MOI was 0.01, there were no differences between the growth control (GC) group and the MYY16-treated group (Fig. 5c). Compared to MYY16, MYY9 and HX1 exhibited strong abilities to modulate PAO1 growth when the MOI was 1 (Fig. 5d). Next, the inhibitory effects of HX1 and MYY9 against W19 (Fig. 5e and f) and FRD1 (Fig. 5g and h) revealed the strong potential to impede bacterial growth but to different extents. These data demonstrated that HX1 and MYY9 may be potential candidates for phage therapy of clinical diseases.
FIG 5.
Infection assay of the phages against P. aeruginosa in vitro. (a to c) Bactericidal effects of HX1, MYY9, and MYY16 independently against their host strain PAO1 at different multiplicities of infection (MOIs). (d) Comparison of antibacterial activities of phages HX1, MYY9, and MYY16 against host strain PAO1 at an MOI of 1. (e to h) Inhibition of bacterial growth by HX1 and MYY9 against clinical isolates with the multidrug resistance P. aeruginosa strain W19 (e and f) and high-mucus-yield strain FRD1 (g and h). The experiment with the growth control (GC) group was conducted by adding equal volumes of SM buffer. The optical density (OD600) was recorded every 10 min. Data are calculated as the means (±SD). The experiment was repeated three times.
Determination of endotoxins.
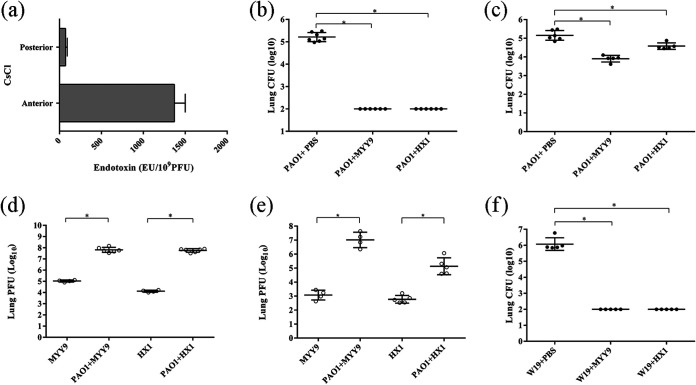
Endotoxin, or lipopolysaccharide (LPS), is a component of the Gram-negative bacterial outer membrane and is widespread in crude phage lysates. Endotoxin may induce immune responses, in particular innate immunity (23). Here, we tested the pyrogen effects of the crude phages and found that the levels of endotoxin were exceptionally high, up to 1,370.3 ± 130.0 endotoxin units (EU)/109 PFU. However, we made purifications of the phages, leading to a significant reduction of the LPS quantity, ∼71.5 ± 20.2 EU/109 PFU (Fig. 6a). Hence, the level of pyrogen carried by the phages may not cause an obvious adverse response.
FIG 6.
Antibacterial efficacy of phages HX1 and MYY9 against P. aeruginosa infections in mouse acute pneumonia models. (a) The levels of endotoxin were determined in crude phage lysates and preparations after the purification procedures. (b and c) Phage HX1 and MYY9 therapy via intratracheal instillation or tail vein injection reduced bacterial loads (CFU) in infected mouse lungs 24 h after challenging the host with strain PAO1. (d and e) The phage titers (PFU) in infected lungs of mice treated by intratracheal instillation (d) and tail vein injection (e) were enumerated. (f) Phage HX1 and MYY9 therapy via intratracheal instillation reduced bacterial loads in infected mouse lungs 24 h after challenge with the multidrug-resistant P. aeruginosa clinical isolate W19. The lower limit of detection is 2 log10 units. Each group contained at least three mice. The data are presented as means (±SD), and unpaired Student’s t test was applied to compare differences between two groups. “*” indicates a P value of <0.05.
Phage therapy reduces bacterial loads in lungs of mice with acute pneumonia.
Intratracheal instillation of P. aeruginosa was performed to induce mouse acute pneumonia. The challenge dose of PAO1, approximately 5 × 106 CFU, was proven to be an effective dose since high bacterial loads were normally achieved, and it caused systemic dissemination within 24 h. The phage-treated groups with intratracheal instillation exhibited significant decreases in bacterial counts by approximately 3.21 log units for MYY9 and HX1, whereas small decreases of approximately 1.25 log units for MYY9 and 0.57 log units for HX1 were observed in groups treated via tail vein injection. (Fig. 6b and c). To examine whether phage-mediated bacterial lysis may occur in the lung, we determined the phage titers (PFU per milliliter) in infected lungs (Fig. 6d and e). As expected, the phage loads obviously increased compared to those of controls without infection by PAO1. We successfully established acute pneumonia models utilizing clinical isolate W19, whose effective dose was approximately 1 × 106 CFU, and observed that phage therapy also reduced the bacterial colonies in infected lungs (approximately 4.07 log units for MYY9 and HX1) (Fig. 6f).
Phage therapy alleviates bacterial loads and inflammatory responses in chronic pneumonia mouse models.
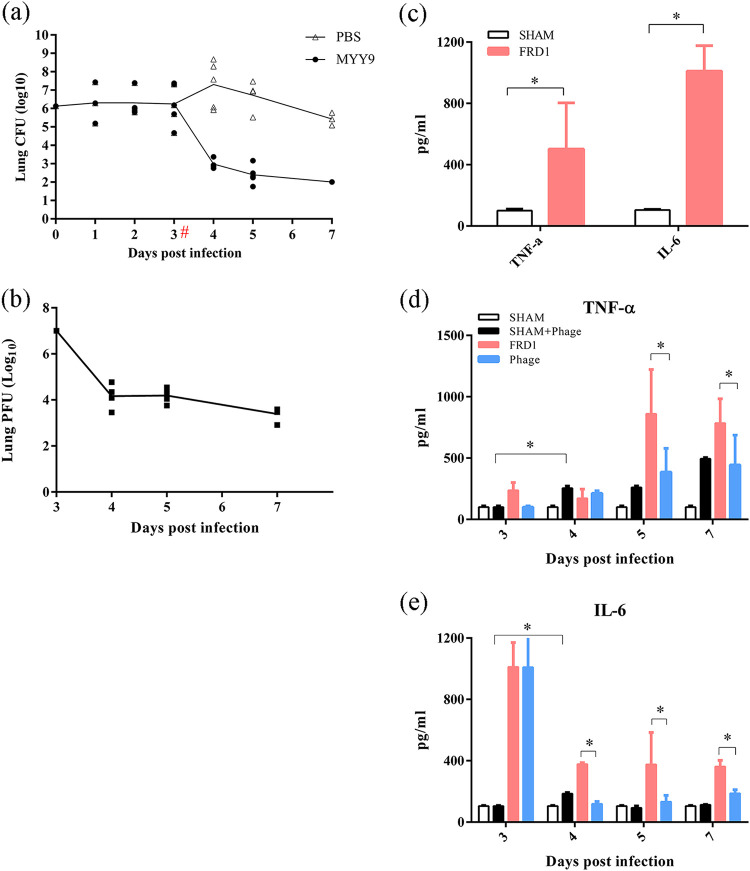
The challenge dose that ensured an effective colony under the condition of no mortality was determined by repeated experimental exploration. Bacterial loads (CFU) were monitored over 7 days, as shown in Fig. 7a. While at 3 days postinfection (dpi), all mice were stably infected by FRD1, some mice cleared the bacteria, leading to a CFU decrease to 4.67 log units, but the majority developed a stable chronic infection after this time point. Moreover, bacterial loads increased (approximately 1.06 log units) at 4 dpi because of the possible influence of the anesthesia operation. After 3 dpi, compared to the controls, a statistically significant reduction in bacterial loads was observed in the phage-treated groups at all time points. Specifically, the bacterial count in the treatment group decreased by 4.735 log units at 4 dpi and by 3.743 log units at 5 dpi. At 7 dpi, no bacteria were detected in the treatment group (Fig. 7a).
FIG 7.
Therapeutic efficacy of phage MYY9 against high-mucus-yield P. aeruginosa clinical isolate FRD1 infections in chronic pneumonia mouse models. (a) Curves of bacterial loads (CFU) in infected lungs over time in the model group (PBS) and the treatment group (MYY9) when the MOI is 1. (b) Curves of bacteriophage titers (PFU) in infected lungs over time in the treatment group (MYY9). (c) Cytokine concentrations in BALF in chronic pneumonia models. (d and e) Concentration dynamics of selected inflammatory biomarkers (TNF-α and IL-6) in BALF over time. “#” indicates the time point of phage treatment, while “*” indicates a P value of <0.05. The lower limit of detection is 2 log10 units. Each group contained at least three mice. For each panel, data represent the means (±SD).
Curves of phage titers (PFU) in infected lungs over time in the treatment group revealed dynamic changes in vivo. The phage titer decreased after 24 h (4 dpi), which indicated that all of the phages entering the body could not be totally utilized. Moreover, PFU decreased with the elimination of bacteria at 5 dpi and 7 dpi (Fig. 7b).
Lung inflammation has been shown to be closely related to increased cytokine levels. The cytokine profile that we selected in bronchoalveolar lavage fluid (BALF) of FRD1-infected mice was enormously different from that of the uninfected controls (Fig. 7c). The levels of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) were significantly increased, by approximately 401.90 pg/ml and 906.25 pg/ml, respectively, in FRD1-infected mice compared to sham controls. It was found that TNF-α levels increased mildly in the group of uninfected animals treated with phages (sham plus phage) from 4 dpi to 7 dpi, while IL-6 levels increased at 4 dpi only, suggesting that TNF-α and IL-6 responses were slightly induced by phages. Furthermore, the synthesis and release of TNF-α and IL-6 were decreased at all of the time points in the phage-treated group (MYY9), indicating that phage therapy can alleviate inflammatory reactions due to the subdued infection (Fig. 7d and e).
Histopathological analysis of lungs in chronic and acute pneumonia.
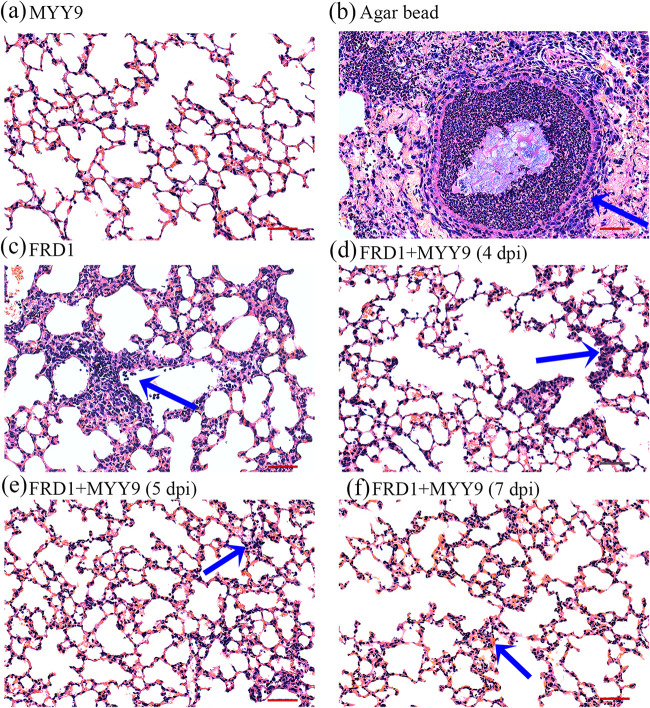
The histopathology of mouse lungs persistently infected with the FRD1 strain, as determined by light microscopy, showed that the infection is plurifocal. First, there was no obvious pathological damage in mice that received only phage therapy compared with normal ones, suggesting that phages entering the body do not induce an overt immune response (Fig. 8a). However, an agar bead could be visualized in the bronchial lumen; microcolonies of the bacterium existed in it, which induced strong inflammatory cell responses, and the bead was filled with massive neutrophils (Fig. 8b). Figure 8c shows that FRD1 had a huge potential to stimulate a diffuse inflammatory cell infiltrate, which contained macrophages, lymphocytes, and neutrophils, at 3 dpi. The phage therapy group showed milder inflammatory responses with few cell infiltrations and weaker tissue damage signs whether in the lung mesenchyme or parenchyma at 24 h (4 dpi), 48 h (5 dpi), and 96 h (7 dpi) posttherapy (Fig. 8d to f).
FIG 8.
Histopathological analyses (H&E staining) of inflammatory responses and lung injury following infection with FRD1. (a) The MYY9 group, which was challenged with the therapeutic dose of phage MYY9 alone and no bacterial infection, exhibited no differences compared with normal controls. (b) P. aeruginosa FRD1-laden agar beads colonized the respiratory tract, leading to massive inflammatory cell responses (blue arrow). (c) Phage-untreated group 3 days after infection by FRD1 (3 dpi). (d to f) Lung tissues from the phage-treated group at different time points: 4 dpi (after 24 h of phage therapy) (d), 5 dpi (after 48 h of therapy) (e), as well as 7 dpi (after 96 h of therapy) (f). Compared to the untreated group, the treatment group shown greatly attenuated inflammatory cell infiltration in the perivascular and peribronchial areas. All photos were taken at a magnification of ×200 by an optical microscope. Bars, 50 μm.
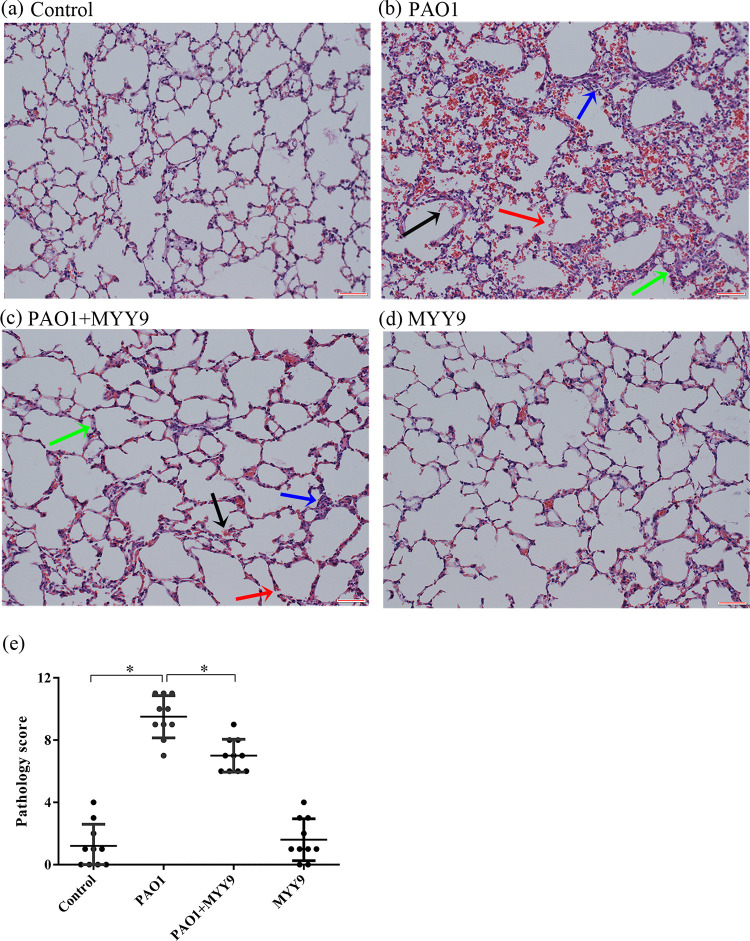
As for acute pneumonia models, in normal lungs (instillation of phosphate-buffered saline [PBS]) as controls, mice infected with PAO1 suffered more serious lung damage, including diffuse inflammatory cell infiltration, edema, and severe hemorrhage, as well as integrity loss (Fig. 9a and b). However, tissue damage signs in the phage therapy groups improved significantly (Fig. 9c). Finally, to eliminate the interference of endotoxin, we set up a group that received only phages, which showed no difference from the normal lung tissue (Fig. 9d). The total pathology scores, representing individual grades of disease, were assessed in a double-blind manner, suggesting that bacterial infection induces significant tissue injury and that phage therapy effectively alleviated inflammation-induced tissue damage (Fig. 9e).
FIG 9.
Histopathological analyses of inflammatory responses and lung injury in mouse acute pneumonia models and therapeutic efficacy with phage MYY9 via airway administration. (a) Histological morphology in a normal mouse. (b) Representative mouse lung infected with PAO1 showing diffuse inflammatory cell infiltration (blue arrow), edema (green arrow), serious hemorrhage (black arrow), and integrity loss (red arrow). (c) Phage MYY9-treated mouse lung 2 h after infection with P. aeruginosa (MOI = 1). (d) Phage MYY9 alone was also used to test the effect of the endotoxin’s potential tissue-damaging and inflammatory responses. (e) The total pathology scores were assessed in a double-blind manner by different researchers (0 to 4, mild pneumonia; 5 to 7, moderate pneumonia; 8 to 10, severe pneumonia). All of these photos were taken at a magnification of ×200 by an optical microscope. Bars, 50 μm.
DISCUSSION
P. aeruginosa is an important antibiotic-resistant (AR) priority pathogen since it possesses three major mechanisms to counter antibiotic attack: intrinsic resistance, adaptive resistance, and immune evasion (5, 20, 24, 25). Importantly, nearly all acute or chronic hospital-acquired infections and pneumonias caused by P. aeruginosa are associated with AR (26). With emerging AR, an increasing number of researchers are refocusing on phage therapy against a “superbacterium.” To this end, we isolated and characterized four lytic phages with broad host ranges targeting clinical isolates of MDR P. aeruginosa using the laboratory strains PAO1 and ATCC 27853 as the hosts. Specifically, our study analyzed phage morphologies, genome architectures, host ranges, safety, and antibacterial capacity both in vitro and in vivo, which characterized several potential therapeutic phages for future development.
Traditionally, spot tests and plaque assays are viable approaches to isolate and purify phages from the biosphere (27, 28). In addition, virion structure observation via TEM is still the most widely used criterion for phage classification (29). The order Caudovirales containing the tailed phages is comprised of three families: Myoviridae, Siphoviridae, and Podoviridae (30). Thus far, characterized phages for P. aeruginosa have covered all families of the Caudovirales, and the families Myoviridae and Podoviridae were found to constitute the majority (1, 30). TEM analyses indicated that HX1, MYY9, and MYY16 belong to the Podoviridae family, whereas TH15 belongs to the Myoviridae family, consistent with previous studies.
To investigate whether these phages are bona fide therapeutic candidates with high efficacy, we performed genome sequencing and analyzed their genetic features. This ensured that there is no potential risk involving toxins, virulence factor-coding genes, lysogenic genes, AR genes, and the ability to integrate into the host genome. Indeed, analysis of genetic features revealed that all phages not only lacked these hazardous genes but also had putative lytic-enzyme-coding genes (31). Particularly, MYY9 and HX1 belonged to the “phiKMV-like” genus, which is dependent on functional type IV pili (TFP) for infections (32, 33). Moreover, this genus was highly inclined to develop phage drugs because of its broad host ranges, even across some other genera, and the generation of a lysis cassette and enzymes that interfere with targeted bacterial metabolism or regulate cell lysis potency (22, 34, 35).
Meropenem and aztreonam, belonging to the carbapenem and β-lactam antibiotic classes, respectively, are the most common drugs to treat Gram-negative MDR bacterial infection. Therefore, we employed clinical Pseudomonas strains resistant to these antibiotics isolated from infectious wounds, the urinary tract, sputum, and BALF to test phage host spectra since they exhibited different levels of toxins and adhesins associated with virulence, colonization ability, and biofilm formation. In addition, we tested laboratory strains that belong to the most prevalent serotypes seen in clinics since serotyping is a critical method for P. aeruginosa typing (36–38). As expected, all the phage candidates shared relatively broad host coverages and may be useful for therapeutic applications. We used spot tests rather than efficiency-of-plating (EOP) assays in this study because the ratios of EOP values were positively correlated with plaque clarity in spot tests, and both methods were suggested to be complementarily applied for testing bacterial coverage (39).
With respect to antibacterial efficacy in vitro, three phages showed a strong ability to prevent host growth in a dose-dependent manner. Moreover, we tested the inhibitory effects of HX1 and MYY9 on the W19 and FRD1 clinical strains that were intended to induce acute and chronic lung infections, respectively. Remarkably, these data indicated that both phages had significant inhibitory effects on the growth of W19 and FRD1 and may be tested as promising antimicrobial agents for P. aeruginosa.
Previous studies about the therapeutic potential of lytic phages often used mouse models with various pathogens, such as Klebsiella pneumoniae, Staphylococcus aureus, Elizabethkingia anophelis, and Acinetobacter baumannii (40–43). Moreover, the majority of these infection models are acute pneumonia, while relatively less is done in skin infection models, sepsis, and hemorrhagic pneumonia, etc. To date, there have been limited reports regarding phage application in experimental chronic infections such as chronic pneumonia (39, 43–45); however, P. aeruginosa is closely associated with hospital-acquired acute or chronic infectious diseases. Consequently, we conducted an analysis of bacterial loads in mouse models of acute pneumonia induced by PAO1 and W19 and chronic pneumonia based on the FRD1 strain.
We performed intratracheal instillation to induce mouse acute pneumonia models. There are two methods of bacterial inoculation into the lung through the airway according to previous studies: intratracheal inoculation and intranasal administration. One advantage of intranasal administration is that airway integrity was not compromised, while one of the merits of intratracheal inoculation is its ability to cause illness with reproducible clinical characteristics and similar pathological features (46). Additionally, there are several routes for applying phages, such as intranasal, intraperitoneal, intramuscular, and intravenous administration, to achieve therapeutic goals. However, the mode of drug administration closely affects the phage dose that reaches the nidus, having the given indications and usage. For instance, intranasal inhalation, the most common form, is appropriate for acute or chronic respiratory tract infections or pneumonia, and intravenous administration applies to hematogenous infection or sepsis, while the oral delivery of phage has focused on acute gastrointestinal infections (4, 47–49). As a consequence, two routes of phage delivery, intravenous injection and intratracheal instillation, were chosen to evaluate in vivo anti-P. aeruginosa efficacy in this study. One reason why tail vein injection was adopted is that high bacterial loads achieved in infected mouse lungs may cause systemic dissemination within 24 h. Another consideration is that intravenous injection is one of the most efficient routes of administration for antibacterial drug delivery to the lung. Since intratracheal instillation reflects local drug delivery, it would be important to test its antibacterial efficacies to compare with other administration approaches in lung infection models. In this study, intratracheal administration was found to be superior to intravenous administration. A pivotal factor is that the phage titers in the lungs varied under the same therapeutic dose. Nevertheless, regardless of the routes, phages could decrease bacterial loads and reduce histological damage in infected lungs in our study.
Concerning chronic pneumonia, previous studies have been restricted to a few preclinical evaluations involving chronic infections of otitis and burn wounds (45). For this reason, we conducted an evaluation of MYY9 against chronic pneumonia caused by FRD1-laden agar beads that can result in comparatively low mortality, high bacterial colonies, severe lung lesions, and high chronic infection under the challenge dose. In addition, previous studies stated that the individual P. aeruginosa agar bead preparations are considered optimal when the average diameter of the bead is approximately 150 μm and its bacterial loads are about 2 × 107 CFU/ml (15). We explored a minimal lethal dose (MLD) of FRD1 (5 × 106 CFU/ml). Analyzing a combination of parameters, we studied the cytokine levels in BALF and performed histopathological analyses of inflammatory responses and lung injury. We observed strong protection in the phage-treated group, such as no death, overt decreases in bacterial burdens, and obvious improvement in histopathology.
Phages target bacteria via diversified host-associated receptors such as proteins, sugars, and other cell surface structures (36), but we did not study the host receptor that our phages can recognize, which may be a next topic. In addition, we focused on only the therapeutic efficacy of phage therapy using naturally isolated monophage, but recently, an increasing number of studies have concentrated on assessments of the combination of therapeutic phage application with antibiotics, phage enzymes, phage cocktails, or engineered phage and have proven that the combinations or gene-modified phages are superior to natural or single phages (9, 10, 50). Therefore, potential combination therapy and gene modification may show an even better therapeutic efficacy, which warrants further investigation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All research protocols were performed according to standard guidelines. The P. aeruginosa strains used in our study, which were obtained from a lab in the State Key Laboratory of Biotherapy of Sichuan University and from patients in Chongqing General Hospital and Sichuan Provincial People’s Hospital, China, are shown in Table S1 in the supplemental material. Tryptic soy broth (TSB) (Becton, Dickinson and Company, Franklin Lakes, NJ) and tryptic soy agar (TSA) (Becton, Dickinson and Company, Franklin Lakes, NJ) media were applied to culture Pseudomonas strains throughout this study. Double-concentration TSB (2× TSB) was prepared by weighing twice as much dry ingredients of single-concentration TSB in the procedure for phage separation. The double-layer plate for the procedure for purifying the bacteriophages was prepared with solid medium that contained TSA and soft-agar overlays, which contained 0.6% agar in TSB.
Animal maintenance.
C57BL/6 mice (6 to 8 weeks of age) were purchased from Beijing HFK Bioscience Co. Ltd., Beijing, China. The mice were kept under specific-pathogen-free (SPF) and temperature- and light-controlled conditions and were fed with a standard laboratory diet during the entire experiment. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Sichuan University.
Isolation and identification of phages by spot tests.
P. aeruginosa phages were separated from the effluent of West China Hospital, Sichuan University, Chengdu; feces from farms in Mianyang City, Sichuan Province; and stagnant pools in Chengdu suburb areas, Sichuan Province, by using a traditional method described previously, with some modifications (1). In short, samples were collected into a 50-ml centrifuge tube and centrifuged at 3,000 × g for 15 min to discard the precipitate. Next, the supernatant was filtered using Millipore filters (0.22-μm pore size). The collected filtrate (3 ml) was mixed with 3 ml 2× TSB containing late-exponential/early-stationary-phase P. aeruginosa host strain ATCC 27853, PAO1, and PA14 bacteria. The mixture was incubated in a shaking incubator at 220 rpm at 37°C for 12 h or until it became clear.
After incubation, with the aim of removing bacteria and their debris, the cultures were centrifuged at 3,000 × g for 2 min, and the supernatant was filtered using Millipore filters (0.22 μm) and collected in a sterile 5-ml centrifuge tube. The bacterial lysate containing target phages was obtained.
The presence of the target phages was identified by a spot test. In brief, 5 μl of the bacterial lysate was spotted onto the individual host bacterial lawns and cultured at 37°C for 16 h.
Phage enumeration by single-plaque assays.
In order to accurately calculate the concentrations of phages by single-plaque assays, in which the numbers of plaques were between 30 and 300, the solution was diluted with sodium chloride and magnesium sulfate (SM) buffer (100 mM NaCl, 8 mM MgSO4, 2% gelatin, 50 mM Tris-HCl [pH 7.5]) in a series of 10-fold dilutions (i.e., 100 μl phages plus 900 μl SM buffer), and the optimal dilution multiples for performing single-plaque assays were ascertained.
Propagation and purification of phages.
The procedures for the purification of the target phages were confirmed by a single-plaque assay. In brief, the mixture containing 90 μl of late-exponential-phase bacteria (optical density [OD] = 1) and 30 μl of the phage solution was incubated at 37°C for 15 min to ensure phage adsorption before vortexing. Next, the mixture was added to 4 ml soft-agar broth and promptly poured onto the bottom plate containing TSA medium. After solidification, the plate was placed at 37°C for culturing at a constant temperature for 16 h. After observing the formation of plaques, single plaques of different shapes were picked using 1 ml of a pipette nozzle and dissolved in 500 μl SM buffer. These steps were repeated until the plaques formed with the same size on the plate (repeated at least 3 times in general).
The phage solution was treated with NaCl and polyethylene glycol 8000 (PEG 8000) (Sigma-Aldrich, St. Louis, MO) at final concentrations of 1 M and 10%, respectively, and incubated for 12 h. The concentrated phages were collected by ultracentrifugation at 14,000 × g for 20 min at 4°C and resuspended with sterilized SM buffer (1 ml). The phage concentrations were measured using single-plaque assays (39).
A concentrated phage preparation was obtained by using a cesium chloride (CsCl) (Sigma-Aldrich, St. Louis, MO) gradient, which consists of three variable densities of 1.30, 1.50, and 1.7. The different concentrations were transferred into a 36.5-ml ultracentrifuge tube (Beckman Coulter, Seton Scientific, United Kingdom). After ultracentrifugation for 3 h (240,000 × g at 4°C), the phage was collected. Next, these phage collections were dialyzed with 500 volumes of SM buffer at 4°C for 30 min to remove residual CsCl. This procedure was repeated at least 3 times (51).
Transmission electron microscopy.
Purified phage solutions (approximately 109 PFU/ml) were laid onto a carbon-coated copper grid and negatively stained with 2% (wt/vol) potassium phosphotungstate (pH 7.2) for 3 min. Prepared phage samples were viewed using a transmission electron microscope (JEM-2100 Plus; Japan) at 80 kV (52).
Determination of host ranges of the phages by spot tests.
Host range determinations were similarly performed according to a previous publication (47). In brief, 5 μl of phage droplets (approximately 109 PFU/ml) was titrated on a fresh lawn containing the indicated bacteria, and the plate was incubated at 37°C for 12 h to evaluate the lytic spots. The clear plaques generated demonstrated high host sensitivity, turbid plaques indicated low host sensitivity, and no plaques indicated no host sensitivity. Details of bacterial strains used in the study are listed in Table S1. Antimicrobial susceptibility testing of bacteria was performed by measuring MICs in a 96-well plate. “R” stands for resistance, and “S” stands for sensitivity, while “I” stands for intermediate, between sensitivity and resistance.
DNA preparation and sequencing analysis of phage genomes.
Bacteriophage DNA extraction was performed using the phenol-chloroform-isoamyl alcohol (25:24:1) extraction protocol after it was enriched with PEG 8000. Next, the DNA was precipitated by adding anhydrous ethanol as previously described, with some modifications (53).
Global genome sequencing was performed by Tsingke Biological Technology using Illumina NovaSeq. The genome sequences were assembled with the SPAde package (version 3.13.0; parameter -k127) by using 2G clean data (54). Sequence analysis and gene structure prediction were performed using GeneMarkS (version 4.28) (55). The map of the phage’s genome was made with SnapGene3.2.1 (20, 56, 57). Potential coding sequences (CDSs) were predicted and analyzed by BLAST and GeneMarkS. Genes encoding tRNA were identified by tRNAscan-SE. Comparative genome analysis was performed using Mauve (version 2.2.5) and the genoPlotR package (21).
Bacteriolytic activity in vitro.
The in vitro bacteriolytic activity of phages was determined by optical densitometry (OD600) (40, 41). Briefly, the phages were added to bacterial cultures in the exponential growth phase at different MOIs (multiplicities of infection) (ratios of phages to bacteria) (SM buffer was added to the culture as a control) in a unique honeycomb in sterile plates using Bioscreen C (Bioscreen, Finland). The mixture was incubated with Bioscreen C at 37°C for 24 h. The OD600 values were measured at 10-min intervals.
Determination of endotoxins by an EndoLISA.
Endotoxin levels were evaluated by an EndoLISA (enzyme-linked immunosorbent assay [ELISA]-based endotoxin detection assay; Hyglos, Germany) in accordance with the recommendations of the manufacturer. The fluorescent signal was detected immediately in a fluorescence microplate reader. The value was defined as endotoxin units per phage titer (EU/PFU) (23). All experimental samples conformed to international standards (≤5 EU/kg of body weight).
In vivo anti-P. aeruginosa infection efficacy of phages MYY9 and HX1 in therapy of murine pneumonia models.
Mice were randomly divided into six groups (five mice per group): bacteria only, MYY9 therapy, HX1 therapy, MYY9 only, HX1 only, and the normal control. To establish the acute infection models, P. aeruginosa PAO1 and W19 strains were used to set up acute pneumonia by the intratracheal administration of approximately 1 × 108 and 2 × 107 CFU in 50 μl, respectively, under anesthetized conditions (58). After 2 h of bacterial infection, the phages were administered to achieve therapeutic aims by tail vein injection (system delivery) and intratracheal instillation (topical application) in mice (47, 48).
Making P. aeruginosa FRD1-laden agar beads.
The procedures for making FRD1-laden agar beads were adapted from the protocol described previously by Marcella Facchini et al. (15). P. aeruginosa FRD1 was grown to log phase in a shaking incubator at 37°C. Next, the bacterial pellet was collected by centrifugation at 2,700 × g for 15 min at 4°C and resuspended in 1 ml sterile phosphate-buffered saline (PBS). One milliliter of the bacterial suspension was mixed with 9 ml of liquid TSA (TSB plus 1.5% agar) preequilibrated at 50°C. The TSA-FRD1 mixture was added to heavy mineral oil that was prewarmed at 50°C, rapidly stirred for 6 min at room temperature, slowly stirred for 35 min at 4°C, and rested on ice over 10 min in turn. The agar beads were washed with sterile PBS 4 to 6 times to remove mineral oil by centrifugation at 2,700 × g for 15 min at 4°C. Quantitative bacteriology was assessed after the aseptically homogenized agar beads were collected. The agar beads were stored overnight at 4°C.
P. aeruginosa FRD1 infection and MYY9 therapy in mice.
To induce chronic pneumonia in mouse models, mice were challenged with agar beads that were embedded with 1 × 107 CFU of mucous strain FRD1 in 50 μl directly by intratracheal instillation. The clinical symptoms and body weights of mice were observed and monitored every day. At day 3 after FRD1 administration, MYY9, at a concentration of 1 × 107 PFU/ml, was given via intratracheal instillation in a volume of 50 μl (MOI = 1). Moreover, mice were sacrificed at six different time points.
Quantitative culture.
At each time point, mouse lungs were removed aseptically, weighed, and homogenized in sterile PBS (pH 7.2). Serial dilutions of the homogenate were plated on TSA to evaluate bacterial loads (CFU counts) and phage titers (PFU counts) using an agar overlay technique as mentioned above (50).
Bronchoalveolar lavage fluid collection and determination of inflammatory biomarkers.
The mouse lung and trachea were fully exposed under anesthetized conditions at a particular point. The trachea was excised and intubated with a catheter. The lavage fluid was collected via instilling and aspirating 1 ml of sterile PBS repeatedly (58). Concentrations of the inflammatory biomarkers interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) were measured via an ELISA kit (Neobioscreen, China) according to the recommendations of the manufacturer.
Histopathological analysis.
After sacrifice, the lungs were placed in 4% (wt/vol) paraformaldehyde for 48 h and routinely processed in paraffin sections, involving sequential infiltration of increasing concentrations of ethanol and xylene. The fixed tissues were sectioned and stained with hematoxylin and eosin (H&E). The histopathological scores were recorded as previously described (46).
Statistical analysis.
All statistical analyses, unless otherwise stated, were executed using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). Error bars in graphs show the standard errors of the means. The unpaired t test or one-way analysis of variance (ANOVA) with Tukey’s post hoc analysis was applied to compare differences between two groups. Most assays were performed three times in triplicate, and the data were expressed as means from three independent experiments. Statistical significance was considered a P value of <0.05.
Data availability.
The relevant data sets in this study can be found under GenBank accession numbers MW406974 to MW406976.
ACKNOWLEDGMENTS
This work is supported by the National Major Scientific and Technological Special Project for Significant New Drug Development (2019ZX09721001-004-004) as well as funds from Southwest Medical University and West China Hospital, Sichuan University, to Z.W. This work was also supported by National Institutes of Health grants R01 AI109317-01A1, R01 AI138203, P20 GM113123, and AI097532-01A1 to M.W.
We thank the farm staff for their support and assistance and are grateful to the staff of Chongqing General Hospital and Sichuan Provincial People’s Hospital for providing bacterial strains.
F.C., Z.W., and M.W. conceived and designed experiments. F.C. performed most of the experiments with help from others (X.C., J.L., X.Y., X.H., M.L., W.L., T.H., Y.X., J.L., P.G., and X.W.). All authors analyzed the data and wrote and approved the manuscript.
We declare we have no conflict of interest.
REFERENCES
- 1.Alemayehu D, Casey PG, McAuliffe O, Guinane CM, Martin JG, Shanahan F, Coffey A, Ross RP, Hill C. 2012. Bacteriophages phiMR299-2 and phiNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. mBio 3:e00029-12. doi: 10.1128/mBio.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu YY, Cao JM, Yang Q, Chen S, Lv HY, Zhou HW, Wu Z, Zhang R. 2019. Risk factors for carbapenem-resistant Pseudomonas aeruginosa, Zhejiang Province, China. Emerg Infect Dis 25:1861–1867. doi: 10.3201/eid2510.181699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomila A, Shaw E, Carratala J, Leibovici L, Tebe C, Wiegand I, Vallejo-Torres L, Vigo JM, Morris S, Stoddart M, Grier S, Vank C, Cuperus N, Van den Heuvel L, Eliakim-Raz N, Vuong C, MacGowan A, Addy I, Pujol M, COMBACTE-MAGNET WP5-RESCUING Study. 2018. Predictive factors for multidrug-resistant gram-negative bacteria among hospitalised patients with complicated urinary tract infections. Antimicrob Resist Infect Control 7:111. doi: 10.1186/s13756-018-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heo YJ, Lee YR, Jung HH, Lee J, Ko G, Cho YH. 2009. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob Agents Chemother 53:2469–2474. doi: 10.1128/AAC.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazares A, Moore MP, Hall JPJ, Wright LL, Grimes M, Emond-Rheault JG, Pongchaikul P, Santanirand P, Levesque RC, Fothergill JL, Winstanley C. 2020. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat Commun 11:1370. doi: 10.1038/s41467-020-15081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basalla J, Chatterjee P, Burgess E, Khan M, Verbrugge E, Wiegmann DD, LiPuma J, Wildschutte H. 2019. The identification of loci that encode potentially active compounds against drug resistant pathogens amidst a decreasing pool of novel antibiotics. Appl Environ Microbiol 85:e01438-19. doi: 10.1128/AEM.01438-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao K, Song S, Zhao Z, Liu Z, Ji Y, Gu P, Fan X, Li Q. 2019. The complete genome sequence of Escherichia phage SRT7, a novel T7-like phage. Arch Virol 164:1217–1219. doi: 10.1007/s00705-019-04182-4. [DOI] [PubMed] [Google Scholar]
- 8.Walters MS, Grass JE, Bulens SN, Hancock EB, Phipps EC, Muleta D, Mounsey J, Kainer MA, Concannon C, Dumyati G, Bower C, Jacob J, Cassidy PM, Beldavs Z, Culbreath K, Phillips WE, Jr, Hardy DJ, Vargas RL, Oethinger M, Ansari U, Stanton R, Albrecht V, Halpin AL, Karlsson M, Rasheed JK, Kallen A. 2019. Carbapenem-resistant Pseudomonas aeruginosa at US emerging infections program sites, 2015. Emerg Infect Dis 25:1281–1288. doi: 10.3201/eid2507.181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalewska-Piatek B, Piatek R. 2020. Phage therapy as a novel strategy in the treatment of urinary tract infections caused by E. coli. Antibiotics (Basel) 9:304. doi: 10.3390/antibiotics9060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pires DP, Costa AR, Pinto G, Meneses L, Azeredo J. 2020. Current challenges and future opportunities of phage therapy. FEMS Microbiol Rev 44:684–700. doi: 10.1093/femsre/fuaa017. [DOI] [PubMed] [Google Scholar]
- 11.Wei S, Chelliah R, Rubab M, Oh DH, Uddin MJ, Ahn J. 2019. Bacteriophages as potential tools for detection and control of Salmonella spp. in food systems. Microorganisms 7:570. doi: 10.3390/microorganisms7110570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, Fineran PC. 2020. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol 5:48–55. doi: 10.1038/s41564-019-0612-5. [DOI] [PubMed] [Google Scholar]
- 13.Chevallereau A, Meaden S, Fradet O, Landsberger M, Maestri A, Biswas A, Gandon S, van Houte S, Westra ER. 2020. Exploitation of the cooperative behaviors of anti-CRISPR phages. Cell Host Microbe 27:189–198.e6. doi: 10.1016/j.chom.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facchini M, De Fino I, Riva C, Bragonzi A. 2014. Long term chronic Pseudomonas aeruginosa airway infection in mice. J Vis Exp 2014:51019. doi: 10.3791/51019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutone A, Lepanto MS, Rosa L, Scotti MJ, Rossi A, Ranucci S, De Fino I, Bragonzi A, Valenti P, Musci G, Berlutti F. 2019. Aerosolized bovine lactoferrin counteracts infection, inflammation and iron dysbalance in a cystic fibrosis mouse model of Pseudomonas aeruginosa chronic lung infection. Int J Mol Sci 20:2128. doi: 10.3390/ijms20092128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silo-Suh LA, Suh S-J, Ohman DE, Wozniak DJ, Pridgeon JW. 2015. Complete genome sequence of Pseudomonas aeruginosa mucoid strain FRD1, isolated from a cystic fibrosis patient. Genome Announc 3:e00153-15. doi: 10.1128/genomeA.00153-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren H, Liu Y, Zhou J, Long Y, Liu C, Xia B, Shi J, Fan Z, Liang Y, Chen S, Xu J, Wang P, Zhang Y, Zhu G, Liu H, Jin Y, Bai F, Cheng Z, Jin S, Wu W. 2019. Combination of azithromycin and gentamicin for efficient treatment of Pseudomonas aeruginosa infections. J Infect Dis 220:1667–1678. doi: 10.1093/infdis/jiz341. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Xu Y, Gu Y, Zhu Y, Liu X. 2017. Characterization and genomic study of “phiKMV-like” phage PAXYB1 infecting Pseudomonas aeruginosa. Sci Rep 7:13068. doi: 10.1038/s41598-017-13363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Chen P, Lin Z, Wang T. 2019. Characterization of two Pseudomonas aeruginosa viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses 11:318. doi: 10.3390/v11040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Melo ACC, da Mata Gomes A, Melo FL, Ardisson-Araújo DMP, de Vargas APC, Ely VL, Kitajima EW, Ribeiro BM, Wolff JLC. 2019. Characterization of a bacteriophage with broad host range against strains of Pseudomonas aeruginosa isolated from domestic animals. BMC Microbiol 19:134. doi: 10.1186/s12866-019-1481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briers Y, Peeters LM, Volckaert G, Lavigne R. 2011. The lysis cassette of bacteriophage varphiKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 1:25–30. doi: 10.4161/bact.1.1.14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szermer-Olearnik B, Boratyński J. 2015. Removal of endotoxins from bacteriophage preparations by extraction with organic solvents. PLoS One 10:e0122672. doi: 10.1371/journal.pone.0122672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curran CS, Bolig T, Torabi-Parizi P. 2018. Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am J Respir Crit Care Med 197:708–727. doi: 10.1164/rccm.201705-1043SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang Z, Raudonis R, Glick BR, Lin T-J, Cheng Z. 2019. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Botelho J, Grosso F, Peixe L. 2019. Antibiotic resistance in Pseudomonas aeruginosa—mechanisms, epidemiology and evolution. Drug Resist Updat 44:100640. doi: 10.1016/j.drup.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Karumidze N, Thomas JA, Kvatadze N, Goderdzishvili M, Hakala KW, Weintraub ST, Alavidze Z, Hardies SC. 2012. Characterization of lytic Pseudomonas aeruginosa bacteriophages via biological properties and genomic sequences. Appl Microbiol Biotechnol 94:1609–1617. doi: 10.1007/s00253-012-4119-8. [DOI] [PubMed] [Google Scholar]
- 28.Gu J, Li X, Yang M, Du C, Cui Z, Gong P, Xia F, Song J, Zhang L, Li J, Yu C, Sun C, Feng X, Lei L, Han W. 2016. Therapeutic effect of Pseudomonas aeruginosa phage YH30 on mink hemorrhagic pneumonia. Vet Microbiol 190:5–11. doi: 10.1016/j.vetmic.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Gao S, Dong Y, Lu C, Liu Y. 2020. Isolation and characterization of bacteriophages against virulent Aeromonas hydrophila. BMC Microbiol 20:141. doi: 10.1186/s12866-020-01811-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garbe J, Bunk B, Rohde M, Schobert M. 2011. Sequencing and characterization of Pseudomonas aeruginosa phage JG004. BMC Microbiol 11:102. doi: 10.1186/1471-2180-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holguin AV, Rangel G, Clavijo V, Prada C, Mantilla M, Gomez MC, Kutter E, Taylor C, Fineran PC, Barrios AFG, Vives MJ. 2015. Phage PhiPan70, a putative temperate phage, controls Pseudomonas aeruginosa in planktonic, biofilm and burn mouse model assays. Viruses 7:4602–4623. doi: 10.3390/v7082835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae H-W, Cho Y-H. 2013. Complete genome sequence of Pseudomonas aeruginosa podophage MPK7, which requires type IV pili for infection. Genome Announc 1:e00744-13. doi: 10.1128/genomeA.00744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chibeu A, Ceyssens PJ, Hertveldt K, Volckaert G, Cornelis P, Matthijs S, Lavigne R. 2009. The adsorption of Pseudomonas aeruginosa bacteriophage phiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol Lett 296:210–218. doi: 10.1111/j.1574-6968.2009.01640.x. [DOI] [PubMed] [Google Scholar]
- 34.Ceyssens PJ, De Smet J, Wagemans J, Akulenko N, Klimuk E, Hedge S, Voet M, Hendrix H, Paeshuyse J, Landuyt B, Xu H, Blanchard J, Severinov K, Lavigne R. 2020. The phage-encoded N-acetyltransferase Rac mediates inactivation of Pseudomonas aeruginosa transcription by cleavage of the RNA polymerase alpha subunit. Viruses 12:976. doi: 10.3390/v12090976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Essoh C, Vernadet J-P, Vergnaud G, Coulibaly A, Kakou-N’Douba A, N’Guetta AS-P, Ouassa T, Pourcel C. 2020. Characterization of sixteen Achromobacter xylosoxidans phages from Abidjan, Côte d’Ivoire, isolated on a single clinical strain. Arch Virol 165:725–730. doi: 10.1007/s00705-019-04511-7. [DOI] [PubMed] [Google Scholar]
- 36.Bertozzi Silva J, Storms Z, Sauvageau D. 2016. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, Huang K, Yang X, Sun L, You J, Pan X, Cui X, Yang H. 2018. Characterization of a novel lytic podovirus O4 of Pseudomonas aeruginosa. Arch Virol 163:2377–2383. doi: 10.1007/s00705-018-3866-y. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Du Y, Qian C, Li L, Jiang L, Jiang X, Cao H, Guo X, Liu B. 2018. Establishment of a suspension array for Pseudomonas aeruginosa O-antigen serotyping. J Microbiol Methods 155:59–64. doi: 10.1016/j.mimet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Jeon J, Yong D. 2019. Two novel bacteriophages improve survival in Galleria mellonella infection and mouse acute pneumonia models infected with extensively drug-resistant Pseudomonas aeruginosa. Appl Environ Microbiol 85:e02900-18. doi: 10.1128/AEM.02900-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji J, Liu Q, Wang R, Luo T, Guo X, Xu M, Yin Q, Wang X, Zhou M, Li M, He P. 2020. Identification of a novel phage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Microb Pathog 139:104317. doi: 10.1016/j.micpath.2020.104317. [DOI] [PubMed] [Google Scholar]
- 41.Peng SY, Chen LK, Wu WJ, Paramita P, Yang PW, Li YZ, Lai MJ, Chang KC. 2020. Isolation and characterization of a new phage infecting Elizabethkingia anophelis and evaluation of its therapeutic efficacy in vitro and in vivo. Front Microbiol 11:728. doi: 10.3389/fmicb.2020.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Can K, Aksu U, Yenen OS. 2018. Investigation of PhiKZ phage therapy against Pseudomonas aeruginosa in mouse pneumonia model. Turk J Med Sci 48:670–678. doi: 10.3906/sag-1711-22. [DOI] [PubMed] [Google Scholar]
- 43.Jeon J, Park JH, Yong D. 2019. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol 19:70. doi: 10.1186/s12866-019-1443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Z, Zhang J, Niu YD, Cui N, Ma Y, Cao F, Jin L, Li Z, Xu Y. 2015. Isolation and characterization of a “phiKMV-like” bacteriophage and its therapeutic effect on mink hemorrhagic pneumonia. PLoS One 10:e0116571. doi: 10.1371/journal.pone.0116571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Waters EM, Neill DR, Kaman B, Sahota JS, Clokie MRJ, Winstanley C, Kadioglu A. 2017. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 72:666–667. doi: 10.1136/thoraxjnl-2016-209265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis 198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 47.Yang M, Du C, Gong P, Xia F, Sun C, Feng X, Lei L, Song J, Zhang L, Wang B, Xiao F, Yan X, Cui Z, Li X, Gu J, Han W. 2015. Therapeutic effect of the YH6 phage in a murine hemorrhagic pneumonia model. Res Microbiol 166:633–643. doi: 10.1016/j.resmic.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Prazak J, Iten M, Cameron DR, Save J, Grandgirard D, Resch G, Goepfert C, Leib SL, Takala J, Jakob SM, Que Y-A, Haenggi M. 2019. Bacteriophages improve outcome in experimental Staphylococcus aureus ventilator-associated pneumonia. Am J Respir Crit Care Med 200:1126–1133. doi: 10.1164/rccm.201812-2372OC. [DOI] [PubMed] [Google Scholar]
- 49.Malik DJ, Sokolov IJ, Vinner GK, Mancuso F, Cinquerrui S, Vladisavljevic GT, Clokie MRJ, Garton NJ, Stapley AGF, Kirpichnikova A. 2017. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci 249:100–133. doi: 10.1016/j.cis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 50.Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MT, de la Fuente-Nunez C, Lu TK. 2019. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179:459–469.e9. doi: 10.1016/j.cell.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins AT. 2014. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol 80:6694–6703. doi: 10.1128/AEM.01789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu M, Pasula R, Smith PA, Martin WJ, II. 2003. Mapping alveolar binding sites in vivo using phage peptide libraries. Gene Ther 10:1429–1436. doi: 10.1038/sj.gt.3302009. [DOI] [PubMed] [Google Scholar]
- 53.Lavelle K, Martinez I, Neve H, Lugli GA, Franz C, Ventura M, Bello FD, Sinderen DV, Mahony J. 2018. Biodiversity of Streptococcus thermophilus phages in global dairy fermentations. Viruses 10:577. doi: 10.3390/v10100577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng T, Li Q, Liu Z, Li X, Liu Z, Liu H, Liu F, Xie L, Wang H, Zhang L, Wu D, Chen M, Li Y, Ji A. 2019. Characterization and genome analysis of novel Klebsiella phage Henu1 with lytic activity against clinical strains of Klebsiella pneumoniae. Arch Virol 164:2389–2393. doi: 10.1007/s00705-019-04321-x. [DOI] [PubMed] [Google Scholar]
- 57.Tabassum R, Shafique M, Khawaja KA, Alvi IA, Rehman Y, Sheik CS, Abbas Z, Rehman SU. 2018. Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae. Sci Rep 8:17904. doi: 10.1038/s41598-018-36229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venuprasad K, Theivanthiran B, Cantarel B. 2016. Intra-tracheal administration of Haemophilus influenzae in mouse models to study airway inflammation. J Vis Exp 2016:53964. doi: 10.3791/53964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The relevant data sets in this study can be found under GenBank accession numbers MW406974 to MW406976.