Abstract
We reassess progress in the field of biomolecular modeling and simulation, following up on our perspective published in 2011. By reviewing metrics for the field’s productivity and providing examples of success, we underscore the productive phase of the field, whose short-term expectations were overestimated and long-term effects underestimated. Such successes include prediction of structures and mechanisms; generation of new insights into biomolecular activity; and thriving collaborations between modeling and experimentation, including experiments driven by modeling. We also discuss the impact of field exercises and web games on the field’s progress. Overall, we note tremendous success by the biomolecular modeling community in utilization of computer power; improvement in force fields; and development and application of new algorithms, notably machine learning and artificial intelligence. The combined advances are enhancing the accuracy and scope of modeling and simulation, establishing an exemplary discipline where experiment and theory or simulations are full partners.
Keywords: biomolecular modeling, biomolecular simulation, biomolecular dynamics, structure prediction, protein folding, DNA folding, RNA folding, multiscale modeling, citizen science projects, machine learning, artificial intelligence
Not all those who wander are lost.
—J.R.R. Tolkien, The Fellowship of the Ring
On the mountains of truth you can never climb in vain: either you will reach a point higher up today, or you will be training your powers so that you will be able to climb higher tomorrow.
—Friedrich Nietzsche, Human, All Too Human
1. INTRODUCTION
1.1. Background
Several years ago, my students and I from New York University (NYU) published a perspective article on the field of biomolecular modeling and simulation (171). We sought to trace the field’s trajectory from its early days to recent developments and applications. Our trajectory traced emerging simulation techniques by Alder & Wainwright (3), by Rahman & Stillinger (154), and by Stillinger & Rahman (188); consistent force field developments by the groups of Lifson (18, 106), Scheraga (129, 164), and Allinger (4, 5); and pioneering applications using energy minimization for structure determination, crystal structure refinement (71), enzyme reactions (199), and molecular dynamics (MD) simulations (115). We were particularly interested in charting objective measures of the field’s evolution, assessing the field’s fulfillment of its early high expectations, evaluating interactions between modelers and experimentalists, describing notable examples of success and failure, and pinpointing areas for future growth. Overall, we probed how the fundamental problems of force-field imperfections and limited conformational sampling were being addressed, and what the field’s prospects for the future were. Our final verdict of a “field coming of age” (171, p. 191), despite early inflated expectations and unrealistic goals, was summarized in a field expectation curve projecting steady progress onto 2020. This view is reproduced in Figure 1 with new accompanying images.
Figure 1.
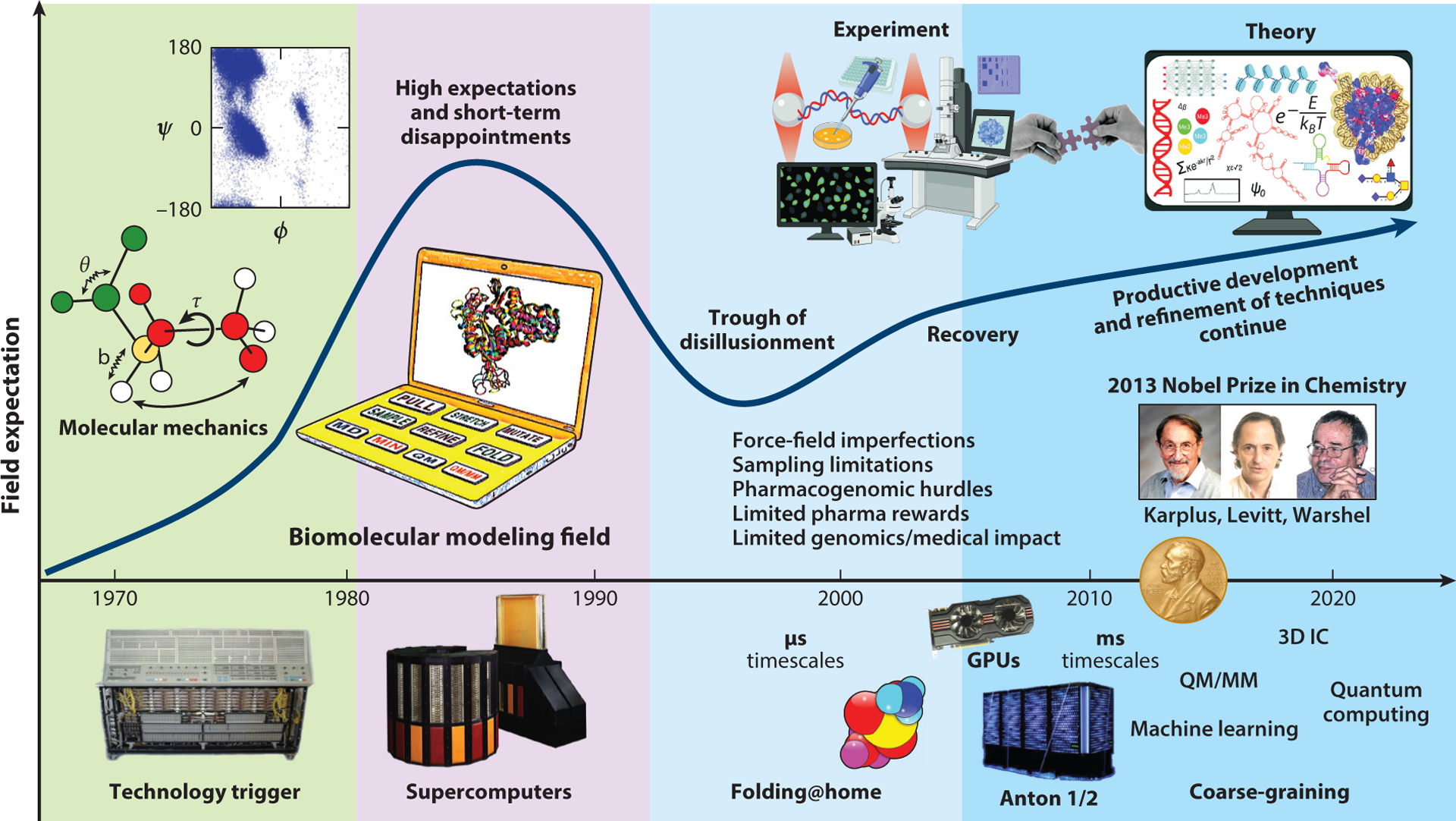
Expectation curve for the field of biomolecular modeling and simulation. The field started with comprehensive molecular mechanics efforts, and it took off with the increasing availability of fast workstations and, later, supercomputers. Following unrealistically high short-term expectations and disappointments concerning the limited medical impact of modeling and genomic research on human disease treatment, better collaborations between theory and experiment have ushered the field to its current productive stage. Problems realized in the decade between 2000 and 2010 and later addressed include force field imperfections, conformational sampling limitations, some pharmacogenomics hurdles, and limited medical impact of genomics-based therapeutics for human diseases. Technological innovations that have helped drive the field include distributed computations and the advent of the use of graphic processing units for biomolecular computations. The molecular-dynamics-specialized supercomputer Anton made it possible in 2009 to reach the millisecond timescale for explicit-solvent all-atom simulations. The 2013 Nobel Prize in Chemistry awarded to Levitt, Karplus, and Warshel helped validate a field that lagged behind experiment and propel its trajectory. Abbreviation: QM/MM, quantum mechanics/molecular mechanics.
1.2. Field Expectation Curve
Such an expectation curve was first described by computer scientist James Bezdek (17) for new technologies. It begins with a technology trigger (in this case, the advent and rising availability of supercomputers) and often displays an early peak where unrealistic, inflated short-term expectations accompany the initial euphoria upon introduction of the new product. When obstacles and disappointments emerge for the new technology—in this case, resulting from the inaccuracies of force fields and limited conformational sampling that became apparent at the end of the 20th century—most new technologies disappear. The field of biomolecular modeling and simulation also suffered from disappointments realized in the pharmaceutical industry using drug design initiatives (126) and in our ability to immediately utilize information from the Human Genome Project for improved diagnosis and treatment of human diseases. Although most newly introduced technologies disappear, those that survive in the long term demonstrate steady progress, when deficiencies are carefully addressed and notable advances are made, as illustrated in our revised expectation curve of biomolecular modeling and simulation (Figure 1).
1.3. Progress
Much has happened in the field in the intervening years since our perspective (171). Most notably, the 2013 Nobel Prize in Chemistry was awarded to Martin Karplus, Michael Levitt, and Arieh Warshel for pioneering work in computational biology and chemistry, including multiscale computations and MD applied to enzyme structure and mechanisms. This prize is significant because it has validated a field that historically lagged behind experimentation (168). Now, scientists studying chemical and biological systems routinely employ computations using graphic processing units and cloud-based computing, in combination with experiments, to probe structures, energetics, kinetics, mechanisms, and functions of these systems. With visionary leaders and open source programs, like NAMD and GROMACS, modern bioinformatics and computational biology tools have opened our eyes to biomolecules in action, much like the light microscope did for biology in the 17th century. Simulations of millions (44) or billions (83) of atoms are now possible, and millisecond time frames are easily approachable, especially when using enhanced sampling methods and coarse-grained or multiscale models. Biomolecular modeling and simulation applications have allowed us to pose and answer new questions and pursue difficult challenges in both basic and applied research. Problems range from unraveling the folding pathways of proteins and identification of new therapeutic targets for common human diseases to design of novel materials and pharmaceuticals.
A spectacular demonstration of the ability of molecular modelers to exploit state-of-the-art software and collaborate productively emerged recently, with the rise of the COVID-19 pandemic. Rapidly, the fastest supercomputers were recruited to simulate viral proteins and explore potential binding of drugs to them by in silico screening of large databases. Consortia like the COVID-19 HPC and BioExcel were established to bring together government, industry, and academic leaders to support COVID-19 research by providing modelers worldwide access to the fastest supercomputers. Public science projects like Foldit, Eterna, and others also recruited the community to address specific subprojects related to SARS-CoV-2.
1.4. Update
In this updated perspective, we follow up on some of these objective measures of the field’s trajectory, report on exciting recent examples of success and instructive failures, pinpoint emerging challenges and subfields, and discuss the role of community exercises on the field. Some of our discussions are based on community responses to the questionnaire developed in our NYU course on Biomolecular Modeling (see the Supplemental Appendix).
Overall, we find that the field of biomolecular modeling and simulation is not only thriving in this era of rapidly evolving genomics sciences and technology, but also a truly exemplary multidisciplinary field that exploits, integrates, and applies numerous elements from science, mathematics, technology, and engineering to solve fundamental scientific problems that are impacting human health.
In the next section, we present metrics of the field’s rise in popularity, as reflected by publication records and computer power progress. Sections 3 and 4 discuss, in turn, examples of success and failure. Modeling-inspired experiments and experimental–modeling collaborations are noted in Section 5. We then discuss, in Section 6, the impact of community-wide initiatives like the Critical Assessment of protein Structure Prediction (CASP), RNA-Puzzles, Foldit, and Eterna on the field’s evolution. We summarize our general findings in Section 7 and offer recommendations to accelerate the field’s progress in Section 8.
In a separate review, Schlick & Portillo-Ledesma (172) expand on aspects of technology advances in the field, such as knowledge-based (or data-mining) versus physics-based approaches and the role of hardware and software development in the field’s evolution. A recent review by Dill and colleagues (47) further discusses the field’s progress, focusing on the role of computational molecular physics in advancing protein modeling on high-performance computing platforms, the importance of enhanced sampling methods, and contributions made by community-wide exercises.
2. METRICS OF THE FIELD’S RISE IN POPULARITY
As discussed in our previous perspective (171), the rise in popularity of the molecular modeling and simulation field is evident from the steady increase in the number of scientific publications since 1970. Earlier, we noted an exponential increase in publications since 1990, commensurate with the advent of supercomputers, and a sharper slope since 2005.
2.1. Publication Volume
As shown in Figure 2a, the increase in publication volume has been sustained. To obtain these data, we surveyed the Scopus database for peer-reviewed articles related to biomolecular modeling. Figure 2b shows the 20 journals with the highest numbers of biomolecular modeling articles from this search. An overall growth of modeling articles is also seen for these journals, with a particular increase in more diverse journals that do not specialize in biomolecular modeling. Figure 2c shows the trend of modeling articles across high-impact journals. A similar growth trend is seen, demonstrating continued outreach and impact of modeling applications into medicine and biotechnology.
Figure 2.
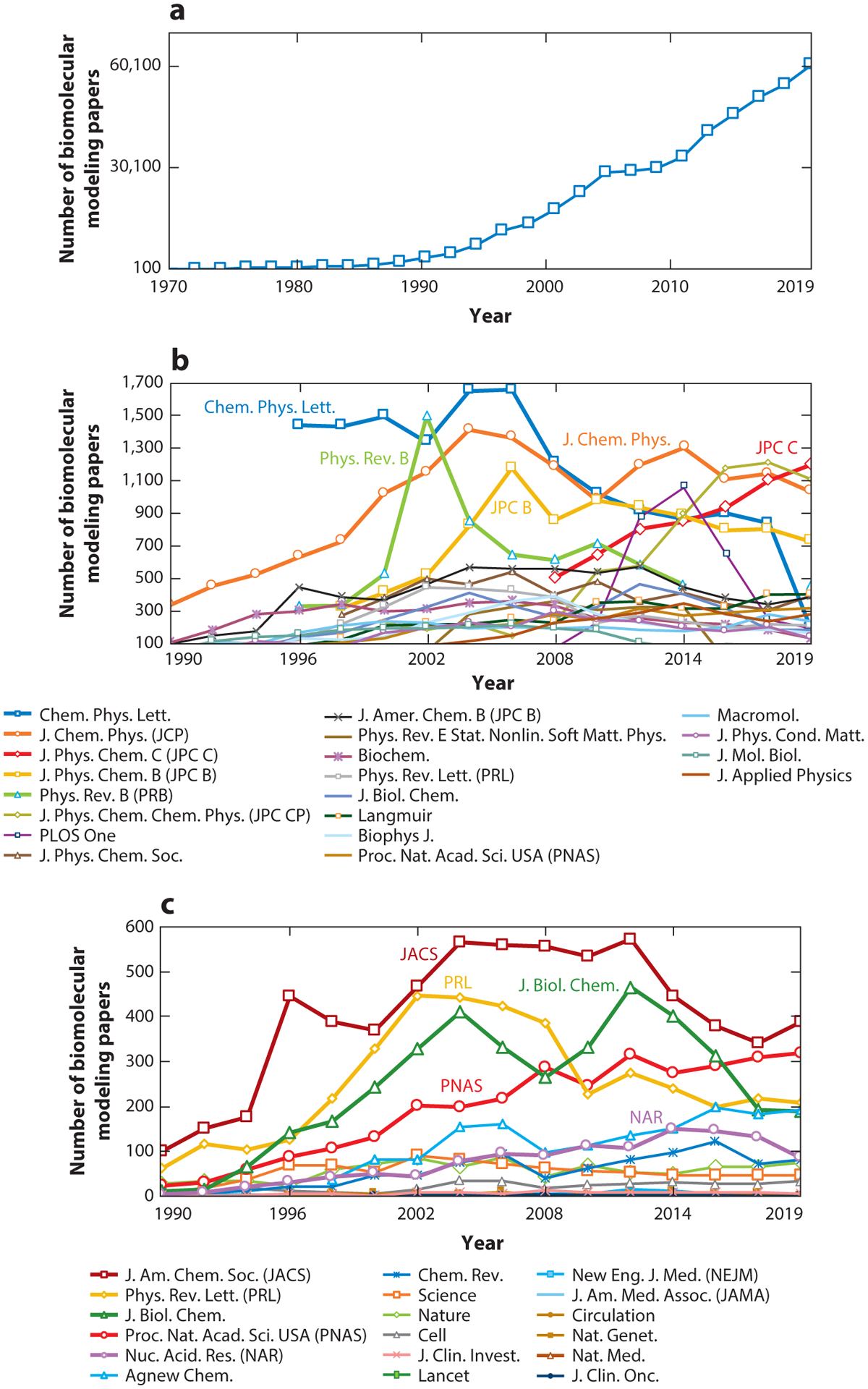
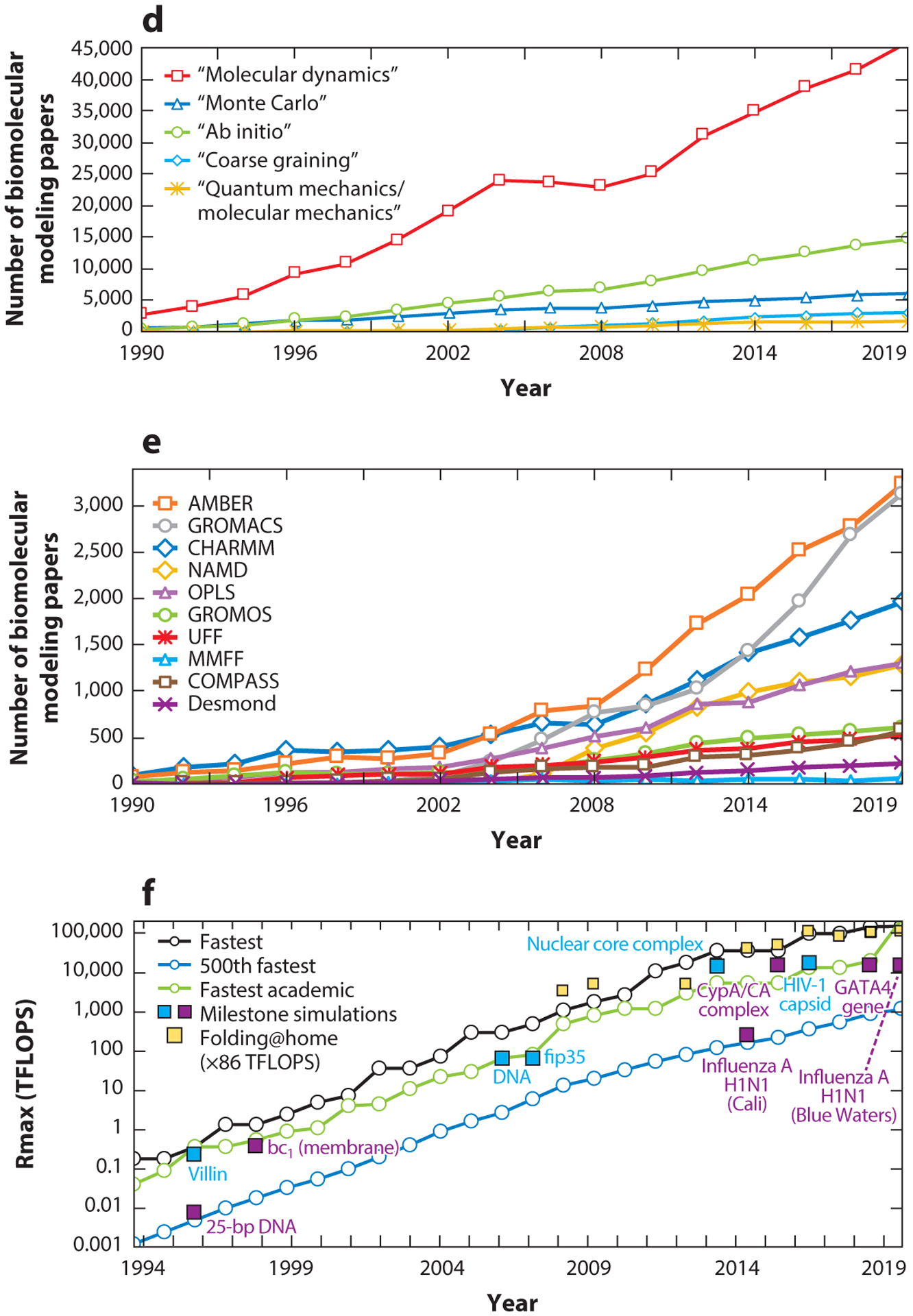
Metrics of the field’s rise in popularity and the evolution of computational performance. (a) Biomolecular modeling papers per year in peer-reviewed journals, as found in Scopus using the query search: “molecular dynamics” OR “biomolecular simulation” OR “molecular modeling” OR “molecular simulation” OR “biomolecular modeling”. (b) Biomolecular modeling papers from panel a in the 20 journals with the most numbers of modeling papers, rank-ordered according to the average number of modeling papers across the years sampled. (c) Biomolecular modeling papers from panel a appearing in high-impact-factor journals, rank-ordered by the SCImago Journal Rank (SJR) h-index. (d) Biomolecular modeling papers from panel a decomposed by method using the query search: ((“molecular modeling” OR “molecular simulation” OR “biomolecular modeling” OR ‘biomolecular simulation”) AND (“method name”)), where method name is “molecular dynamics”, “Monte Carlo”, “ab initio,” “coarse graining”, or “quantum mechanics/molecular mechanics”. (e) Biomolecular modeling papers from panel a decomposed by use of seven popular molecular dynamics packages and force fields using the query search: ((“molecular modeling” OR “molecular simulation” OR “biomolecular modeling” OR ‘biomolecular simulation”) AND (“package/force field”)), where package/force field is Amber (140), CHARMM (24), GROMACS (16), NAMD (145), OPLS (81), GROMOS (57), UFF (157), COMPASS (191), MMFF (60), and Desmond (22). (f) Ranked overall and academic computational systems as reported according to the LINPACK benchmark, as assembled in the Top500 supercomputer lists (www.top500.org). The estimated total speed for the Folding@home distributed computing project is shown in x86 TFLOPS for direct comparison with LINPACK speeds. Biomolecular modeling milestones are dated assuming the computations were performed about a year prior to publication, except for the two 1998 publications, which we associate with computations started in 1996. These include the 25-base-pair DNA system using NCSA SGI machines (204); villin using the Cray T3E900 (43); the bc1 membrane complex using the Cray T3E900 (70); the B-DNA dodecamer using MareNostrum/Barcelona (141); the fip35 protein run on NCSA Abe clusters (49); influenza A H1N1 using the Jade supercomputer (158); the HIV capsid using the Titan Cray XK7 (143); the GATA4 gene using the Trinity Phase 2 (83); and three simulations on Blue Waters, the nuclear core complex (50), the CypA/CA complex (111), and influenza A H1N1 (44). For the simulations in Blue Waters, which has opted out of the Top500 benchmark since 2012, we use estimates of sustained system performance/sustained petascale performance from 2012 and 2020 (14).
2.2. Simulation Techniques and Programs
In Figure 2d, we further decompose the modeling papers from Figure 2a according to simulation techniques. Evaluation by technique shows that MD dominates, followed by quantum mechanics (QM), Monte Carlo (MC) simulations, coarse-grained (CG) approaches, and quantum mechanics/molecular mechanics (QM/MM) calculations. Figure 2e shows papers from Figure 2a decomposed by force field and software package. We see that Amber and CHARMM continue to show increasing usage, as do open source packages like GROMACS and NAMD due to their ready availability as well as suitability for parallelized computer architectures.
2.3. Computational Power
The dramatic increase in computational power since 1990 has certainly helped fuel the field, but how do the computers used in biomolecular modeling compare to the fastest computer systems available today? We assess this feature in Figure 2f by comparing computer power used in landmark simulations (see Supplemental Table 1), namely those notable for large system size or long simulation span, to the fastest systems available using the biannual Top500 ranking (see Supplemental Table 2). In Top500, each computer is ranked according to its maximal-achieved performance (Rmax) measured by the LINPACK Benchmark, a test to solve a dense system of linear equations.
For the landmark simulations performed on Blue Waters, which opted out of the Top500, we approximate the speed by using estimates of sustained system performance/sustained petascale performance reported in 2012 (14) and the peak performance reported in 2020 (http://www.ncsa.illinois.edu/enabling/bluewaters). We also include computational speed for the Folding@home distributed computing network in x86 TFLOPS, an estimate of an x86 class CPU (15). Overall, we see that landmark biomolecular simulations have remained on par, even exceeding the world’s fastest computers over the periods 2008–2011 and 2014–2016. Technology has clearly helped fuel the field and will undoubtedly continue to do so (172).
2.4. Landmark Simulations
Figure 3 highlights these milestone simulations. Early important simulations in 1996 included the ns simulation of the 25-base-pair DNA (204) and μs simulation of villin (43), and the short simulation of the huge bc1 membrane complex (70) in 1998. Later on, μs timescales were achieved for much larger systems, such as the B-DNA dodecamer in 2006 (141). The 2007 long 10 μs simulation of the fast-folding Fip35 protein (49), and the subsequent misfolding into an α-helical structure in spite of the sufficiently long sampling, helped reveal limitations in state-of-the-art force fields.
Figure 3.
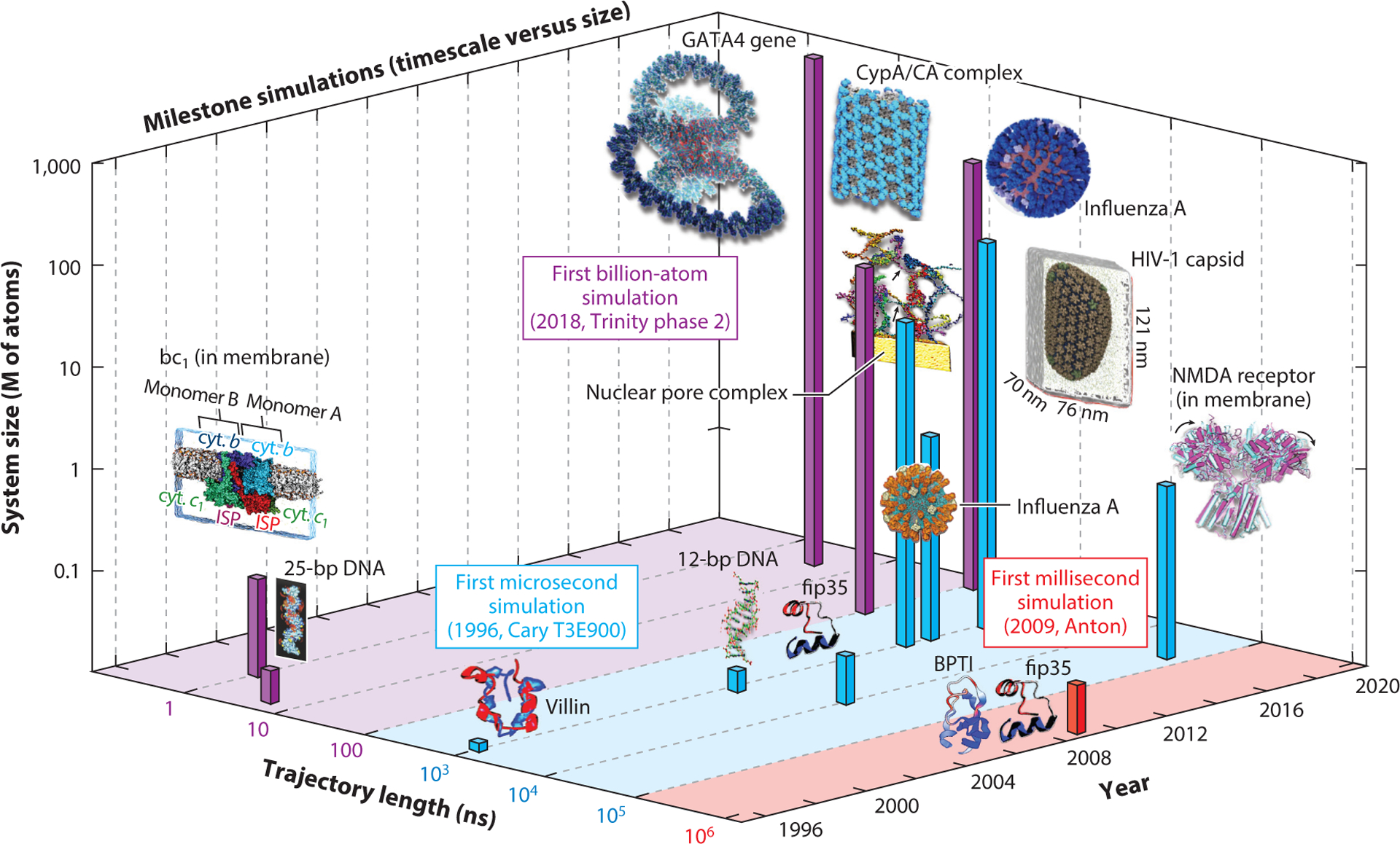
Milestone simulations in biomolecular modeling showing evolution in molecular dynamics timescale and system size. Consistent with Figure 2f, we assume that computations were performed one year before publication, except for the publications in 1998, for which the calculations were performed in 1996. The simulated systems in temporal order are: 25-base-pair DNA (5 ns and ~21k atoms) (204), villin protein (1 μs and 12k atoms) (43), bc1 membrane complex (1 ns and ~91k atoms) (70), B-DNA dodecamer (1.2 μs and ~16k atoms) (141), Fip35 protein (10 μs and ~30k atoms) (49), Fip35 and BPTI proteins (100 μs for Flip35 and 1 ms for BPTI, and ~ 13k atoms) (182), nuclear pore complex (1 μs and 15.5M atoms) (50), influenza A virus (1 μs and >1M atoms) (158), NMDA receptor in membrane (60 μs and ~507k atoms) (187), tubular CypA/CA complexes (100 ns and 25.6M atoms) (111), HIV-1 fully solvated empty capsid (1 μs and 64M atoms) (143), GATA4 gene (1 ns and 1B atoms) (83), and influenza A virus H1N1 (121 ns and ~160M atoms) (44).
The ms simulations of small proteins in 2009 (182) were made possibly by the specialized hardware of the Anton supercomputer (180), with architecture and software optimized for efficient parallelization of nonbonded interactions (22). Large explicit-solvent ms simulations are now possible for proteins (182), membrane complexes (187), and other systems.
Several simulations from the Schulten group on the Blue Waters supercomputer have greatly advanced both the size and trajectory lengths of biomolecular systems. These include the μs simulations of the 15.5-million-particle nuclear pore complex in 2013 (50) and CG models of influenza A (≥1M particles) in 2014 (158). In 2015, the Schulten group simulated a huge 25.6 million–atom antiviral complex for 100 ns (111) and, in 2016, the enormous 64.4 million–atom HIV-1 capsid for 1.2 μs (143). Recently, the Amaro group extended these limits by simulating on Blue Waters the first explicitly solvated viral lipid envelope of the influenza H1N1 virus of approximately 160M atoms (44).
Specialized software, like the cellPACK software by Olson and colleagues (80), has also helped expand the system size and trajectory length limits. Recently, the first billion-atom biomolecular simulation of the entire GATA4 gene was achieved based on our nucleosome-resolution GATA4 model using the state-of-the-art MD program GENESIS performed on the Trinity phase 2 supercomputer at Los Alamos (83).
2.5. Overall Simulation Trends
Overall, tremendous progress is evident from the publication volume, varied techniques, and force fields and simulation packages, as well as simulation trends in system size, trajectory lengths, and high-speed computers. A bright future can certainly be anticipated on this basis.
It has already been stated (195) that the computational biology community has realized an increase of three orders of magnitude in simulation scope per decade. The trends that we illustrate in Figures 2 and 3 suggest that biomolecular researchers have utilized well and will continue to exploit ever more powerful machines with combined software and hardware advances.
Across the world, technology corporations like IBM, Google, Intel, Macintosh, and others are racing to build quantum computing machines for cloud services. While such technological developments have been fueled by international competition or smart phone and video game markets, biomolecular modelers continue to exploit state-of-the-art resources to study biologically important questions with improved algorithms. These combined advances will undoubtedly continue to drive the field forward and push its frontiers in terms of larger systems, longer simulations, and biophysical insights, and it will ultimately close the gap between experimental and computational timescales.
3. MODELING AND SIMULATION SUCCESSES
Because computations in biology allow researchers to follow the dynamics of biomolecular systems, connect static experimental structures to pathways, or explore mechanistic questions, numerous success stories can be collected from labs worldwide pursuing such problems, as detailed in our prior perspective (171). In this section, we highlight general areas of notable success that are likely to impact future research in modeling and its application to biomedicine. We focus on protein folding, biomolecular design, machine learning (ML) and artificial intelligence, prediction of protein flexibility, and force field polarization, although there are many more.
3.1. Equilibrium Simulations of Protein Folding
Since Anfinsen (9) first proposed his thermodynamic hypothesis, protein folding has been a central model problem in biomolecular modeling. Due to the enormity of the conformational space, finding explicit solvent trajectories with sufficient sampling and resolution to capture the full folding and unfolding pathways of proteins has remained a computationally demanding challenge even for small peptides, despite advances in computational speed and power. Physics-based models, which describe molecular behavior based on molecular mechanics principles (167), have been successful since the 1960s, as they can provide us a mechanistic understanding of the pathways, structures, and energetics involved. Although knowledge-based approaches using Google’s α-fold were shown recently to be very powerful for folding proteins, there is no doubt that force-field-based models will continue to make fundamental impacts (for a separate review on this issue, see 172).
One of the landmark simulations in this context is the reliable folding and unfolding of a β-heptapeptide polymer (36). The relevant pathways were possible to capture decades ago because the frequency of the interchange between the folded and unfolded conformations is high relative to the simulation timescale.
As state-of-the-art examples, Piana and colleagues characterized the thermodynamics and kinetics of the folding and unfolding of several proteins near their melting temperature by μs to ms atomistic simulations (110) on Anton (180, 181). Recently, they reported the first simulation of protein folding inside the cavity of a chaperonin protein (148). The chaperonin strongly interacts with the unfolded protein, stabilizing it and slowing the folding process compared to the rate in solution. Such interactions could help substrates escape kinetic traps along the pathway associated with compact, misfolded states (Figure 4a).
Figure 4.
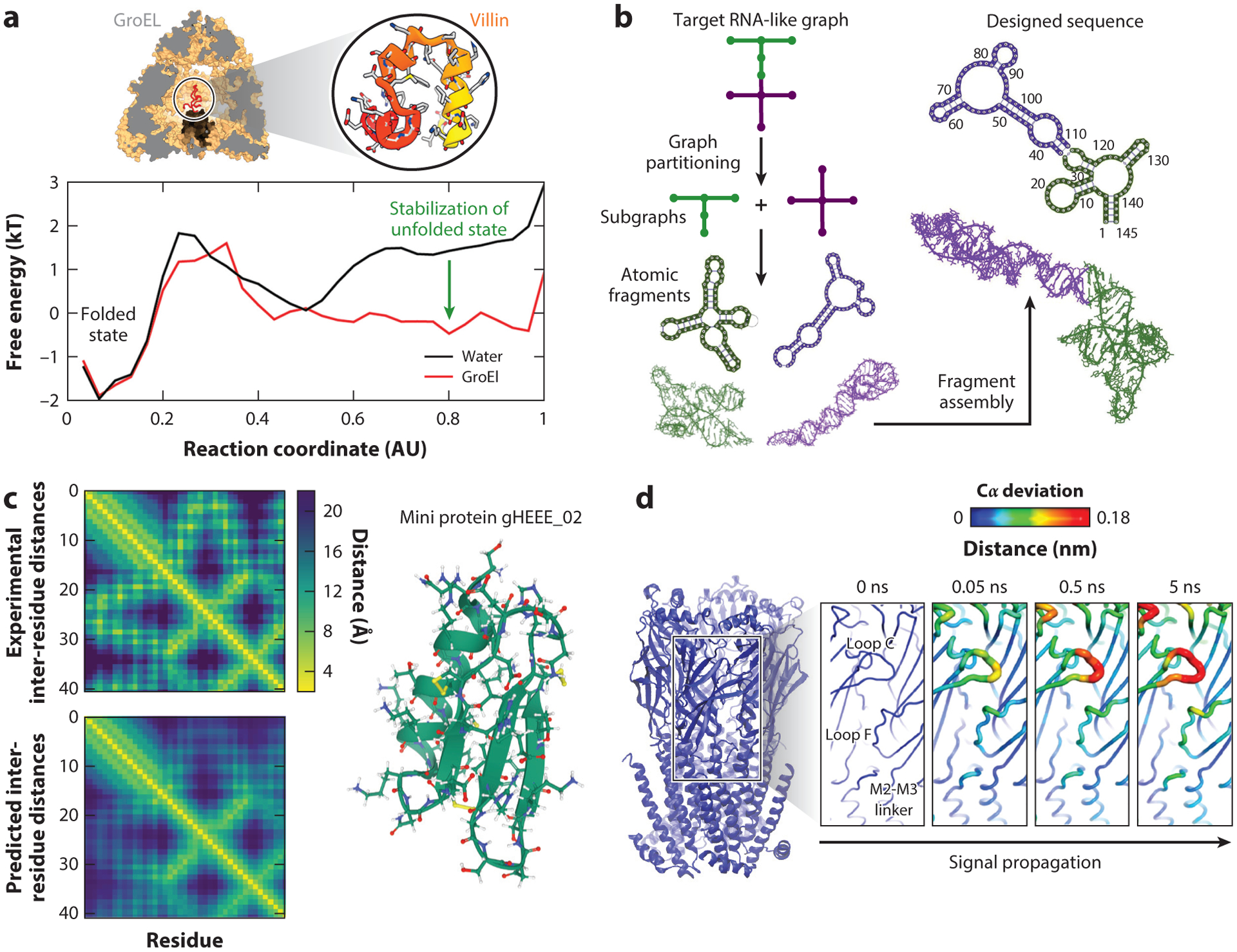
Examples of modeling successes. (a) Protein folding. The folding of a small protein (top right), villin, inside the GroEL protein cavity (top left) is compared to its folding in bulk. The corresponding energy profiles of folding show significant differences in the shape of the unfolded basin, indicating the role of the chaperonin protein in stabilizing unfolded states. Panel adapted with permission from Reference 148. (b) RNA novel motif design. Shown is a flowchart of the computational pipeline for design of RNA-like tree graph topologies (73, 75). In these tree graph representations of RNA secondary structures, edges denote stems, and vertices denote loops, bulges, and junctions (169). We use graph partitioning to segment the target RNA-like graph into subgraphs, extract the corresponding atomic fragments from our RAG-3D database, construct a new sequence or structure using fragment assembly, and screen the top-scoring sequences using RNA 2D structure prediction programs to produce successful sequences that will fold onto the target RNA-like topology (73–75, 117). (c) AlphaFold performance on prediction of inter-residue distances. Inter-residue distance distributions are obtained from the experimental structure (top) and predicted structure (bottom) of the miniprotein gHEEE02 (right), showing good agreement. Distance maps were obtained from https://deepmind.com. (d) Cloud computing to accelerate molecular dynamics. Extensive nonequilibrium simulations of nicotine unbinding from the nicotinic acetylcholine receptor are shown. The Cα fluctuation levels (colored on a scale from blue to red as fluctuations increase) show the sequence of structural changes coupled to the unbinding of the nicotine from the receptor that leads to its activation through a conformational change. Panel adapted with permission from Reference 135.
Better sampling (54, 165, 166), better force fields, and innovative computational approaches are generally needed to attack the problem broadly using available experimental information.
For example, Markov state models were used to characterize folding and misfolding mechanisms of a dimeric protein (183), resulting in the finding that folded and misfolded states can be reached from both pathways. Molecular fragment replacement (19) can fold proteins from extended states by ensuring consistency between local structures and experimental nuclear magnetic resonance (NMR) chemical shifts. Replica exchange MD and accelerated MD simulations have been used to study the effects of denaturing and stabilizing agents on folding processes (2) and the folding of helical proteins (42). CG models have been used to study the folding of intrinsically disordered proteins (IDPs) involved in neurodegenerative diseases (150, 155).
For many specific cases where experimental data are available, protein folding can now be addressed with advanced sampling combined with state-of-the-art force fields.
3.2. Protein and Nucleic Acid Design
Computational tools and algorithms to design novel sequences and structures have played an important role in the field of biomolecular structure, as they apply our growing knowledge of structures and mechanisms to new potential therapeutic and technological designs.
One of the most attractive outcomes in biomolecular modeling is the engineering of proteins with specific folds and binding partners based on peptide sequence, theoretical principles, and computational methods. The 2002 engineering of the Trp-cage peptide with a novel fold and fast folding kinetics (128) helped launch miniprotein design (11), potentially offering useful scaffolds for various applications like biomolecular modeling or catalysis. Rational design based on known rules and fragment-based design was used to create other intriguing assemblies of secondary structure motifs (e.g., 12, 34, 105), providing insights into stabilizing tertiary structure interactions.
Various high-throughput miniprotein design techniques, such as the massively parallel de novo protein design platform of the Baker group (29), can design proteins with customized shapes to bind therapeutic targets. Thousands of miniproteins can be designed by generating scaffold libraries of hundreds of backbone geometries and docking them onto targets, followed by high-affinity optimization.
Further advances are also coming from expanding rotamer libraries derived from experimental data, such as that created by the Daggett group by MD sampling (27). The modular design of protein binding pockets based on known structures (66) has also advanced the field.
Specific therapeutic or biophysical targets and their binding can now be addressed through protein design. Reminiscent of the predictions that we quoted in 2011 (171) from The Economist in 1998 “that most chemical experiments [may one day be] conducted inside the silicon of chips instead of in the glassware of laboratories,” recent protein blacksmithing (109) techniques apply mechanical deformations along collective modes to encourage equilibration. While it was unrealistic 20 years ago, protein engineering could become routine in the next decade.
Computational design of nucleic acids is also an active and attractive area in the field, with biophysical, biomedical, and industrial applications (173, 176). A pioneering notion in DNA nanotechnology developed by Seeman (175) in 1981 to demonstrate how to generate DNA sequences that fold into junction topologies has been followed by the development of computational tools that combine thermodynamics, energy minimization, stochastic search, evolutionary information, and experimental data for nucleic acid design (7, 73).
DNA design tools (7) guide DNA strands into tertiary structures that are then optimized. DNA origami (161) has been used to assemble inorganic nanostructures and proteins (176). Short designed oligonucleotides labeled with fluorophores (DNA-PAINT) that bind to specific genome regions are used for genome visualization (84). Designed DNA molecules are also used to create nanoparticles with biomedical applications, such as drug delivery (201).
RNA design tools (7) use thermodynamic properties, such as melting temperatures and Gibbs free energies, stochastic searches, or graph theory elements, to optimize construction of novel RNA motifs. In our group’s pipeline (Figure 4b), we apply a graph theory–based approach with graph partitioning and fragment assembly tools to design RNA sequences that fold onto novel RNA folds predicted by our RNA clustering analysis of RNA’s theoretical motif universe (73). This computational pipeline has recently been adapted to identify SARS-CoV-2 viral drug targets by destroying the pseudoknot of the frame-shifting element (174).
RNA design has been successful in nanomedicine, for example, in the design of RNAs for therapeutic diagnostics (23). Engineered riboswitch elements are used, for example, to regulate gene expression, to control metabolic flux, and as fluorescence biosensors (61).
There are many challenges involved in the design of biomolecules, including imperfections in large-structure predictions and modeling of RNA molecules that interact with proteins and other macromolecular complexes (173). Because the amount of biological sequence data is growing faster than the development of algorithms and increase of computer power, there is a continuous need for faster and parallel algorithms for filtering the designs, assembling larger structures hierarchically, and dealing with multiple strands or complex designs (39).
3.3. Machine Learning and Artificial Intelligence Applications
The increased usage of ML algorithms in biology has great potential to discern patterns and extract salient features from large and complex data sets, such as brain neural networks, genomic data, or protein–protein complexes.
In biology, ML approaches have for a long time helped generate comparative structural and functional predictions for proteins using sequence–structure similarity and inference of function from evolutionary relationships. Other important applications of known structural databases are fragment assembly and design and virtual screening of active compounds for drug discovery. In applications where approximations made by physics-based methods fail, or where the use of experimental constraints are crucial, knowledge-based methods can be the methods of choice (for recent reviews, see 59, 132). The sequence and structural databases that have been curated since the 1970s provide a gold mine of data, driving the biomolecular modeling field.
Neural network models can also be particularly successful for classification and prediction problems. Such approaches are based on mathematical models of the brain. Community exercises such as the Tox21 toxicology prediction challenge (114) and the CASP (179) have demonstrated their potential. Notably, the 2018 CASP experiment highlighted Google’s AlphaFold system for de novo protein structure prediction; this approach outperformed all competitors in its category (94, 178). AlphaFold uses three neural networks trained to predict the distribution of distances between every pair of residues within a target protein (Figure 4c), estimate the accuracy of the candidate structures, and generate protein structures (178). This approach led to dramatic progress in de novo structure prediction due to better contact and inter-residue distance prediction (94).
Another important recent application involves the use of neural networks to accelerate conformation-dependent electronic structure calculation for organic semiconductors (72). Such problems are computationally expensive: CG simulations are required for conformational sampling, mapping thousands of structures onto their atomistic representation, and performing QM calculations to obtain orbital energies. By using artificial neural networks trained to approximate electronic structure from CG configurations, the time-consuming components of mapping from CG to atomic coordinates and QM calculations can be eliminated.
In the area of genome modeling, neural networks have been used to model 3D chromosome conformations at 50-kb resolution. Di Pierro and coworkers (149) trained neural networks with Chip-seq data of epigenetic marks to produce chromatin-type sequences. These sequences were then used as input to a physical model for chromatin folding to obtain conformational ensembles of specific chromosomes. With this approach, genome folding based on epigenetic marks can be predicted.
Importantly, the quality and robustness of ML approaches are highly dependent on the training data set, and rigorous testing and validation are needed to avoid false positives, biases, or overfitting. Nonetheless, ML- and artificial intelligence–based methods, in conjunction with force field methods, can make a huge impact on our ability to predict, simulate, and understand biomolecular systems.
3.4. Modeling Target Flexibility in Drug Design Studies
The prediction of protein flexibility is important to account for allosteric effects in targeted drug design. Exploring the large set of conformations available to a drug target helps improve design accuracy. G protein–coupled receptors (GPCRs), for example, are highly flexible and configurationally complex (97), so computations can help narrow down their target binding areas.
A wide range of novel MD simulation methods are currently being applied to investigate the interactions between GPCRs and ligand molecules (192). For example, accelerated MD developed by the McCammon group was used to engineer the target flexibility in virtual screening for drug-like compounds (121, 122). This modeling approach successfully discovered new allosteric modulators of the M2 muscarinic acetylcholine receptor, a potential target to treat heart diseases. Later, μs atomic simulations on Anton helped determine binding modes and drug–receptor interactions for multiple allosteric modulators of this receptor (40).
In another study, simulations on Anton elucidated the mechanism by which GPCRs activate heteromeric G proteins by enhancing guanosine diphosphate release (41). Results revealed that GPCRs accelerate nucleotide release by favoring a structural rearrangement on the G protein that weakens its nucleotide affinity.
Recently, extensive equilibrium and nonequilibrium MD simulations were performed to understand how the agonist nicotine affects the nicotinic acetylcholine receptor, an ion channel that modulates synaptic signaling of a wide range of neurotransmitters (135) (Figure 4d). The Oracle Cloud infrastructure made possible 450 simulations of 5 ns in 5 days, notably accelerating the speed of discovery compared to shared local high-performance computing resources. By analyzing the Cα displacement, researchers were able to describe the receptor response to nicotine unbinding, delineating a signal propagation pathway that may be relevant to ion channels.
Other important areas of GPCR research include study of membrane cholesterol effects on structure and function (177) and free energy predictions of experimental mutagenesis and ligand modifications (20). High-throughput methods for engineering GPCR flexibility have now been developed, such as the GPCR-ModSim web server (48).
More general methods for predicting protein flexibility combine MD simulations with experimental data. For example, the popular PredyFlexy method combines root mean square deviations (RMSDs) of MD trajectories with crystallographic B-factors to define flexibility classes through protein sequence predictions (37). Methods like FLEXc, which utilize neural networks to combine evolutionary sequence information with amino acid flexibility statistics, can also perform well in terms of predicting secondary structures, solvent accessibility, and amino acid properties (203). A clever alternative approach is the Site Identification by Ligand Competitive Saturation (SILCS) developed by the MacKerell group (58): The protein target is immersed in an aqueous solution with multiple drug-like ligands, and the system is sampled by MD or MC simulations to allow competitive binding to a flexible target.
3.5. Polarization
Fourth-generation force fields now include polarizability effects to better treat induced electronic polarization, improving on effective empirical fixed charges. Several types of classical polarization models exist: Drude oscillators, the fluctuating charges model, and inducible dipoles. Some of the latest polarizable force fields are AMBER, AMOEBA, and SIBFA, which use induced dipole models; CHARMM-Drude, GROMOS, and OPLS, which use the Drude oscillator model; and CHARMM-FQ and ABEEMσπ, which rely on the fluctuating charge model (for recent reviews, see 10, 77, 116). Since reparametrization of the entire force field is required to generate a polarizable force field, the effort is substantial, and there is a time lag until users switch force fields.
Initially parameterized for small molecules, polarizable force fields are increasingly being extended to include more classes of macromolecules. For example, the AMOEBA and CHARMM polarizable force fields have recently been extended to RNA (101, 209), and several polarizable force fields are applicable to lipids (31), carbohydrates (63, 138, 139), and organic molecules (108).
Lipid-parameterized force fields are more accurate for studying phospholipid bilayer membrane systems because they provide better descriptions of the dipole potential across the water–lipid interface (104). Polarizable water models, such as the six-site SWM6 model, better describe local hydrogen bonding structures (206). Similarly, hydrogen bonds formed between water molecules and protein residues, and within protein residues, are more accurate when treated with polarizable force fields (130).
For protein simulations, the accuracy of polarizable force fields has been evaluated for structure refinement, IDPs, and protein folding (197). Due to a better description of the protein–water interactions compared to additive force fields, polarization was found to improve protein structure refinement and conformational sampling of IDPs. However, polarizable force fields can fail in finding native structures of proteins due to overstabilization of the open structures by protein–water interactions. Updated polarizable force fields for proteins, such as Drude-2019 (107), further improve the treatment of hydrogen bonds and backbone and side chain parameters to remedy this problem.
For protein–metal interactions, Ren and colleagues have shown that the inclusion of manybody polarization effects is necessary to properly describe metal selectivity (78) and metalloprotein structures and energies (79). To model the interaction between ATP and Mg2 + correctly, polarizable force fields are needed (196). For protein–ligand interactions, the AMOEBA polarizable force field has demonstrated excellent performance in predicting binding affinities in the 2019 Statistical Assessment of the Modeling of Proteins and Ligands (SAMPL) challenge (6).
In nucleic acid modeling, polarizable force fields have been successful in describing π–π and ion molecular interactions, where polarization and charge redistribution are important. For example, the MacKerell group reported that the ionic distribution and ion-dependent DNA conformational dynamics are in better agreement with experiments when using polarizable force fields compared with additive force fields (162). Polarization was also important for capturing base flipping (102). Similarly, in the development of AMOEBA for nucleic acids and aromatic molecules, considering the crucial π-electron polarizability was found to be important for correctly describing the liquid structure of benzene (208).
Some biomolecular applications apply polarizable force fields in QM/MM schemes to account for electronic polarization in the MM portion (21, 113). Thiel’s group has suggested that polarization only modestly affects computed activation and reaction energies (21, 51).
Importantly, most force fields only work well for the classes of molecules that they were designed to work for, such as globular proteins. Better force fields and broader classes of systems are now continuously becoming available. Further applications and developments are discussed in References 10, 77, and 68.
4. INSTRUCTIVE MODELING AND SIMULATION FAILURES
Many general failures and limitations of biomolecular modeling applications regarding force fields or sampling algorithms are well known. This is especially true as our molecular subjects become more complex and ambitious, deviating from the simpler chemical systems and relatively short time frames utilized in parameterizing general-purpose force fields.
Experienced modelers often train young scientists so that they learn from incorrect models, protocols, and/or parameters. Well-trained practitioners learn quickly about the importance of ample statistics, tests for robustness, convergence limitations, and so on.
A common concern is force field parameters for new applications. Known vulnerabilities involve poor convergence in replica-exchange MD, normal-mode analysis, and various path sampling approaches.
As the field matures, and molecular modeling simulations are applied increasingly by users rather than by developers of modeling packages, old problems resurface, and new ones emerge. Failures are instructive to novices and experts alike and help us learn, diagnose, and fix errors and deficiencies in our models and algorithms, some expected and many surprising.
4.1. Force Field Limitations
Areas of force field weaknesses include modeling of disordered proteins (159), RNAs (124), and nucleic acid–protein complexes (86, 91).
IDPs, which are important for many biomolecular functions, are poorly modeled by modern force fields, as these force fields were originally developed for folded proteins (146, 147). In general, they do not capture well representative ensembles of structures and associated transitions.
Several groups have developed modifications for reliable studies of disordered proteins. One example is the a99SB-disp force field (159), parameterized by an iterative process in which variables are adjusted until the simulations reproduce experimental data. Another example is the AMBER ff14IDPs force field, containing revised ϕ/ψ dihedral terms to correct the dihedral angle distributions of some residues associated with disorder (185). Force fields can also fail in describing ordered proteins; common limitations are the bias toward helix conformations and the difficulty in predicting β-hairpin folding motifs (32).
For RNA, improvements in torsion and van der Waals parameters can help remedy some problems that emerge in reproduction of experimental structures. However, recent work suggests that RNA force fields may still be lacking, even for reproducing tetraloop folding in comparison with NMR and X-ray data (95). Moreover, some RNA force fields produce structure instability during the simulation trajectory, leading to the prediction of incorrect structures (13, 124) (Figure 5a). The protocols used may also be a factor.
Figure 5.
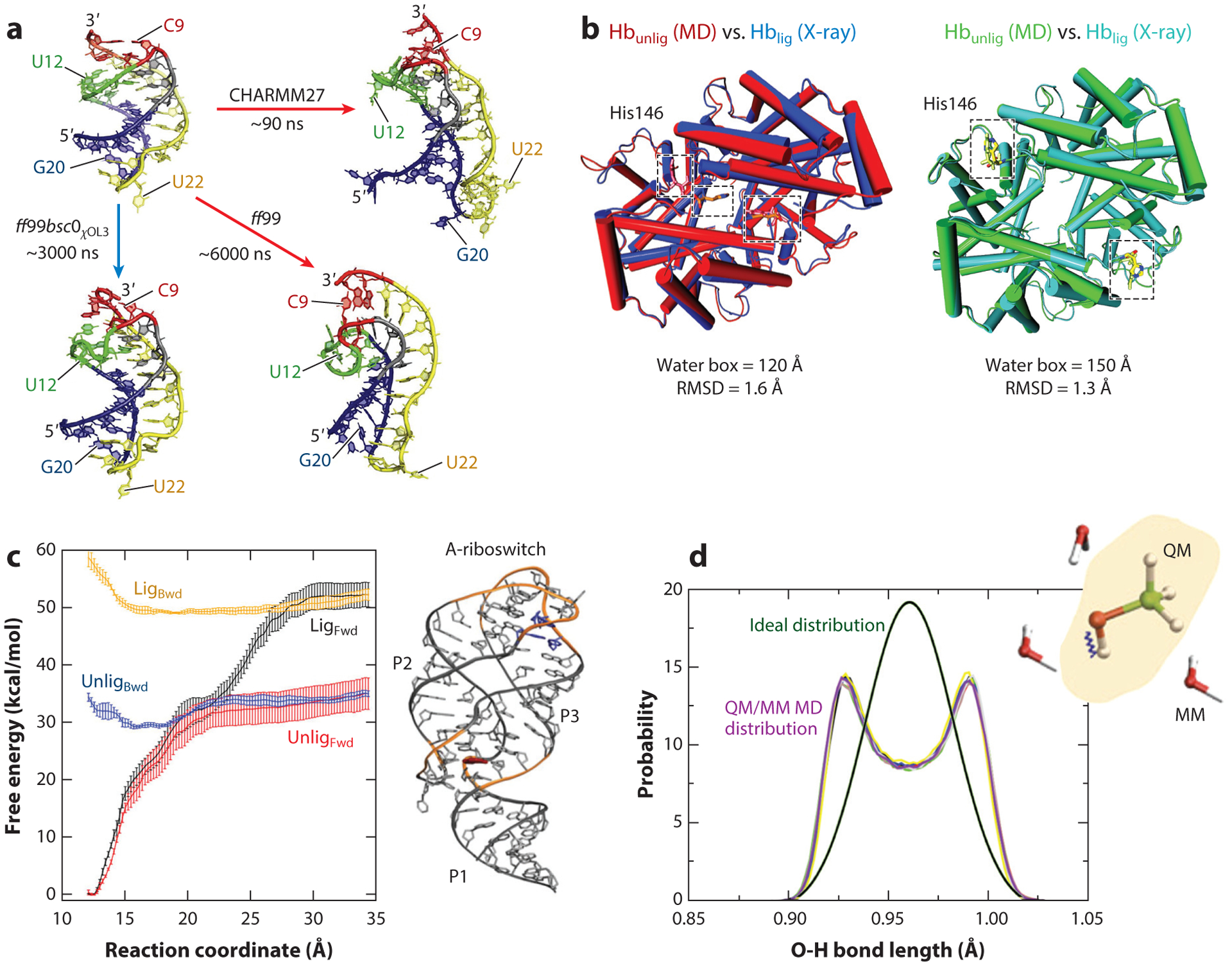
Examples of modeling failures. (a) RNA force fields. Riboswitch aptamer simulations with different force fields indicate that structural details and stabilities are force field dependent. With the CHARMM and ff99 force fields, the aptamer pseudoknotted fold is distorted compared to the X-ray structure, whereas with the ff99bsc0χOL3 force field, the aptamer fold is maintained. Panel adapted with permission from Reference 13. (b) Configurational sampling. A large water box is required to stabilize the unliganded tetramer of the hemoglobin. The unliganded structure obtained with molecular dynamics simulations using a water box of 150Å aligns well with the corresponding experimental structure (right), whereas the unliganded structure obtained with a water box of 120Å is instead similar to the experimental structure of the liganded conformation (left). Panel adapted with permission from Reference 45. (c) Advanced sampling techniques. Different free energy profiles are obtained with umbrella sampling for the forward (Fwd) and backward (Bwd) processes of a conformational change for a riboswitch in the presence (Lig) and absence (Unlig) of a ligand, a consequence of poor convergence of the simulations. Panel adapted with permission from Reference 38. (d) QM/MM simulations. Simulations of solutes treated at the quantum-mechanical level embedded in a rigid water model treated by classical molecular mechanics can lead to poor sampling due to insufficient coupling between the two regions. Shown is the incorrect distribution of the methanol O-H bond length obtained from QM/MM simulations (violet curve) compared to the ideal distribution (green curve). Panel adapted with permission from Reference 65. Abbreviations: MD, molecular dynamics; QM/MM, quantum mechanics/molecular mechanics.
Current force fields also struggle to describe complexes between nucleic acids and proteins. For example, μs simulations of several RNA–protein complexes showed that some systems can progressively deviate from the experimental structures during the course of a simulation due to force field imbalances, incorrect starting experimental structures, and/or poor treatment of the system flexibility due to insufficient sampling (91). Similarly, DNA–protein interactions and binding free energies are still notoriously difficult to predict due to insufficient description of DNA–protein interface relaxation (86).
4.2. Molecular Dynamics and Configurational Sampling Limitations
Problems involved in the numerical integration of the equations of motion in MD—concerning stability, resonance, and accuracy—are well recognized (167). In addition, numerous other details in the simulations, from boundary conditions to equilibration protocols, can lead to artifacts due to the inherent chaotic nature of biomolecular simulations (167).
A recent example related to hemoglobin reported by Karplus and colleagues (45) shows the consequences of the water box dimension that surrounds a complex biomolecular system (Figure 5b). The unliganded tetramer of hemoglobin was found to be stable in solution only when the water box contained 10 times more water molecules than the standard size for such a system. The standard size in this case does not account for the hydrophobic effect, crucial for the stabilization of the unliganded conformation and agreement with experiments. This report stirred some follow-up discussions (46, 52, 53) that suggested that other simulation aspects, like the extent of statistical sampling, might also influence simulation results.
Insufficient sampling can lead to problems in the calculation of theoretical spectra. Usually, spectroscopic maps computed from MD simulations are used to calculate theoretical amide I spectra by connecting observables in the MD simulations to quantum spectroscopic variables (202). However, large errors in the frequencies are noted when theoretical and experimental spectra are compared. Such errors have been attributed to improper configurational sampling during the MD simulation when studying complex systems (26).
Many enhanced sampling methods were developed to address the sampling limitations of traditional integration methods (e.g., 165, 166). However, such methods are vulnerable on many fronts. A study on a conformational change in a riboswitch in the presence and absence of ligand binding (38) showed that the energy profiles calculated with umbrella sampling for the forward and backward processes were different (Figure 5c). This poor convergence points to the need to use more complex collective variables to approximate reaction coordinates and obtain accurate free energies. Furthermore, the widely used free energy methods to estimate protein–protein binding often yield large errors, in the order of 6–9 kJ/mol. This is due to the irreversibility of the binding process, hysteresis, or insufficient sampling of the phase space (144).
A particular area of high failure concerns the modeling of membrane systems. Free energy simulations of solute–bilayer systems are prone to sampling errors due to the presence of large free energy barriers not associated with the reaction coordinate considered (127). That is, metastable states are trapped, and the answers are not reliable. In addition, the free energy calculations strongly depend on the force field used and the resolution of the molecular model (all-atom versus CG) (127, 190).
4.3. Other General Failures
In general, protein–protein docking scores and prediction of interfaces have low accuracies (118, 189). However, some tangible progress has been made using better integration of different modeling tools with docking procedures, as evident in the latest edition of the Critical Assessment of Predicted Interactions (CAPRI) (103).
In particular, ML algorithms for protein–protein hot-spot prediction (112) are vulnerable to overfitting due to the small number of samples available for training. They also suffer from imbalances due to the much larger number of non-hot-spots compared to hot-spots, which can alter the associated clustering optimization. To obtain reliable results, the training database should contain not only positive examples, but also negative data, which are difficult to define. Finally, the best prediction method depends highly on the sequence similarity between the training data and the target system (62).
Similarly, secondary structure prediction methods for large RNAs are often imperfect, as they are based on minimum free energy algorithms that assume a simple relationship between RNA structure and free energy. Large RNAs deviate from the minimum free energy status due to their complex internal environment (173).
Problems have also been recognized in QM/MM simulations, especially concerning boundary treatments. For example, simulations of small solutes treated at a QM level embedded in a rigid solvent model can lead to poor sampling (Figure 5d). It was shown recently that insufficient coupling between the high-frequency vibrations of the QM system and the MM rigid water molecules affects the efficient energy exchange between the two parts of the system (65). As a result, the solute molecule does not achieve thermal equilibration and leads to incorrect bond-length distributions. Thus, no universal protocols for QM/MM simulations exist, and tailoring is often needed.
5. MODELING-INSPIRED EXPERIMENTS AND EXPERIMENTAL–MODELING COLLABORATIONS
The prediction of structures, functions, and/or mechanisms from biomolecular simulations is a frequent goal. However, modelers are often hesitant to publish predictions without experimental validation, as recently discussed by Karplus & Lavery (85). Historically, experimentalists may have viewed predictions with suspicion (168), but the climate has changed quickly. Collaborations involving side-by-side predictions and experimental validation are now common. In this section, we describe different categories of modeling predictions with representative examples: experiments inspired by simulations or theory, theoretical predictions independently confirmed by experiments, and concurrent experimental and modeling studies.
5.1. Experiments Inspired By Simulation or Theory
New experiments that were motivated by modeling or theoretical computations represent an exciting category.
MD simulations of the sodium-coupled betaine transporter BetP predicted the position of a second sodium binding site, previously unidentified (87). These results inspired X-ray crystallographic studies that confirmed the coordinating groups in the second sodium binding site (142).
Mechanistic predictions from modeling followed by experiments have also helped elucidate protein membrane insertion mechanisms. Microsecond simulations of BamA, a bacterial β-barrel membrane protein, suggested a dynamic mechanism of membrane permeation: Destabilization of the interaction between two strands (β1 and β16) leads to a lateral opening of the barrel (134). Subsequent cross-linking experiments in which artificial disulfide bonds between both β strands were inserted revealed loss of activity (133), consistent with the predicted mechanism (Figure 6a).
Figure 6.
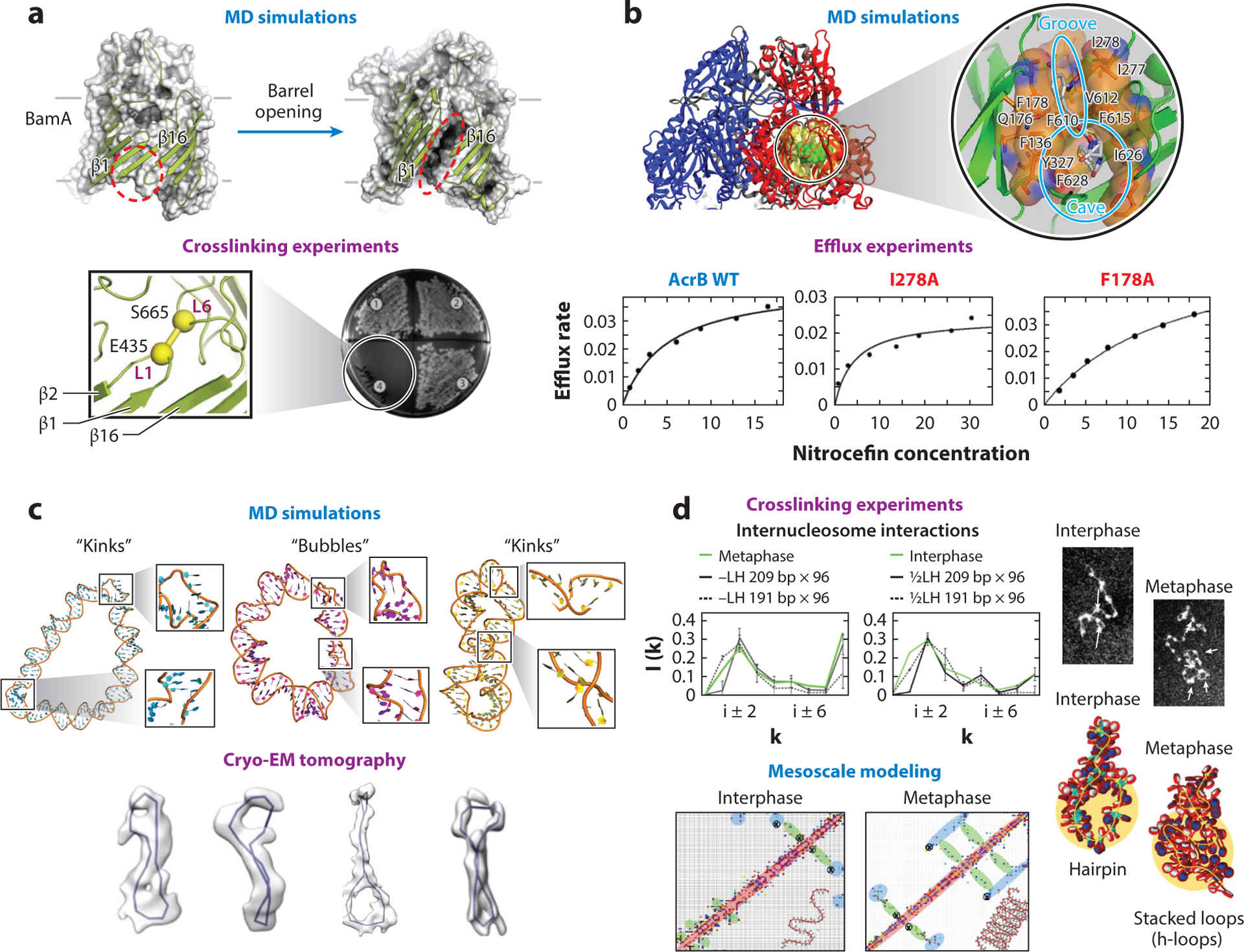
Interplay modeling experiment. (a) Molecular dynamics (MD) simulations of the bacterial protein BamA predicted a membrane insertion mechanism by lateral opening of the β barrel that involves the strands β1 and β16 (top). Crosslinking experiments created artificial disulfide bonds between loops L1 and L6 that connect both β strands and confirmed that BamA function was inhibited (bottom). Panel adapted with permission from Reference 133. (b) MD simulations of the AcrB multidrug transporter with the drug nitrocefin predicted a drug-binding pocket that includes residues such as I278 and F178 (top). Mutagenesis and biophysics experiments, which measured the efflux rate at different nitrocefin concentrations, confirmed the role of these residues in the binding of the drugs (bottom). Panel adapted with permission from References 194 and 89. (c) MD simulations of DNA minicircles predicted the formation of bubbles and kinks under torsional stress (top). Electron cryo-tomography experiments confirmed the formation of such geometries in DNA minicircles (bottom). Panel adapted with permission from Reference 69. (d) Chromatin crosslinking experiments show increased long-range internucleosome contacts without loss of zigzag short-range contacts for metaphase chromatin, compared to interphase chromatin (top; interaction patterns in green). Mesoscale model of fibers typical of interphase [0.5 LH/nucleosome and nucleosome repeat length (NRL) = 191 bp] and metaphase (no LH and NRL = 209 bp) chromatin provide the folding mechanism of hierarchical looping (or stacked loops in 3D) to explain such increases in long-range contacts with maintenance of short-range contacts. This can be observed in the computed contact maps and fiber structures (bottom), and in the interaction patterns for fibers with NRL = 191 or 209 bp (top, black solid and dashed lines) without LH (left) and with 0.5 LH/nulceosome (right) (56).
In the area of nucleic acid structure prediction, theoretical and experimental work have inspired new experiments. For example, several years of predictions from mesoscale chromatin simulations (151) and high-resolution structural data (163, 186) inspired researchers to conduct single-molecule fluorescence resonance energy transfer (FRET) experiments (88) to study the modulation of chromatin dynamics by the heterochromatin protein 1α (HP1α), a typical component of silenced genes. Results revealed that HP1α modulates chromatin dynamics by transiently binding and stabilizing stacked nucleosomes.
Motivated by a desire for improved accuracy in the field, single-molecule tweezer experiments (30) have been designed to test how well base pair–level models (BPLMs) (136) predict the flexibility of double stranded DNA and RNA. Overall, the results showed that some predictions, such as those for persistence lengths of double-stranded RNA, were accurate, while others, such as those for torsional properties, suffered from inaccuracies, revealing a crucial area for further improvement.
5.2. Theoretical Predictions Independently Confirmed by Experiment
More generally, predictions are often made in computational works to motivate future experiments. Below are several examples in which theoretical predictions were eventually confirmed experimentally.
Nikaido and colleagues, using all-atom MD simulations, predicted that some residues in the bacterial multidrug transporter AcrB, a membrane protein involved in antibiotic resistance, are important for the binding of the drug nitrocefin (194). Three years later, some of these predictions were confirmed using fluorescent efflux assays of AcrB mutants (89) (Figure 6b). These experiments further showed that interactions with the drugs doxorubicin and minocycline were consistent with previous simulations from 2011 (193) and 2013 (160). Together, these studies shed light into the way substrates are bound to the cavity of AcrB and then extruded by a conformational change.
The Whitford group, using structure-based MD simulations, predicted a novel tilting motion of the small ribosomal head subunit (30S) during mRNA–tRNA translocation (131). Independently, the Blanchard group performed single-molecule FRET experiments to image the complete translocation mechanism (200). The experimental results revealed an exaggerated motion of the 30S subunit head, verifying the predicted tilt motion. Taken together, these studies revealed how the motion of the 30S subunit facilitates the movement of the tRNA into its final posttranslocation position.
In the area of nucleic acid structure, the sequence-dependent behavior of torsionally stressed supercoiled DNA and DNA minicircles has long been the subject of experiments and theoretical work. For example, MD simulations of DNA minicircles predicted spontaneous formation of noncanonical structures such as kinks, local openings of the double helix, and wrinkles, formed to relieve bending and torsional stress (96, 123). Recently developed cryo-electron tomography techniques have confirmed the existence of such 3D conformations in circular DNA structures (69, 198) (Figure 6c).
5.3. Concurrent Experimental and Modeling Studies
Collaborations between experimentalists and modelers have become common. In this section, we illustrate such synergies in elucidating transport mechanisms in channels, catalytic mechanisms in enzymes, study of protein structure, and chromatin folding.
From long (29.5 μs) MD simulations of potassium channels, the Roux group deduced a kinetic model that predicted the effect of buried water molecules occupancy on the rate of conversion of the channel from its inactive to its conductive state (137). This model gained support from experimental measurements of the channel conversion rate at high osmotic stress. Under these conditions, water occupancy is reduced, and the conversion rate is accelerated, according to the predicted mechanism.
Recently, in another channel study, a combination of MD simulations and in vivo experiments demonstrated that a single residue in the plant aquaporin PIP2 is responsible for water blockage (25). The residue position at the entrance of the channel serves as a crucial steric gate. Similarly, mutagenesis studies and MD simulations were used to identify lipid-binding sites in a protein channel that transports chloride ions (205). Results revealed multiple binding sites, indicating that the channel gating is allosterically regulated.
Combined QM/MM approaches provide mechanistic insights, with atomic and electronic detail, into enzyme kinetics experiments. In a computer-aided enzyme design study, the Warshel group performed QM/MM calculations of several mutants of a dehalogenase enzyme to determine the maximal catalytic improvement that could be achieved by residue substitution (76). Based on the computational prediction, several mutants were constructed and characterized by kinetic assays, confirming some predictions and validating the computational strategy. In another example, the Mulholland group integrated QM/MM simulations with site-directed mutagenesis experiments to gain insight into the influence of active site residues on the product outcome of a monoterpene synthase enzyme (99). The simulations revealed the residues responsible for guiding the product outcome, which might be important in the design of altered enzymes to produce clean products.
In the area of protein structure, Chen & Hub (28) calculated wide-angle X-ray scattering (WAXS) profiles from MD simulations of several proteins to validate the use of protein dynamics to interpret WAXS experiments. They showed that the water and protein force fields have a minor effect on the calculated profiles. Further incorporation of atomic fluctuations significantly increases the agreement between computed and experimental WAXS curves. In a recent study, long all-atom simulations were combined with rapid pressure-drop experiments to analyze how water squeezes out of the hydrophobic core as proteins fold (152). Results demonstrated that, for some proteins, the dehydratation and folding processes occur at different times, and several desolvated states are visited before the protein reaches the native conformation, while in others, the drying and folding processes occur together, and water is excluded from the core as proteins fold.
Finally, the mesoscale modeling of chromatin in the work of Gigoryev et al. (56) has helped interpret puzzling cross-linking experimental data that showed different interaction patterns for interphase and metaphase chromosomes. By modeling interphase and metaphase chromatin, they suggested hierarchical looping as the mechanism that explains the increased long-range internucleosome contacts without the loss of the zigzag motifs observed in the cross-linking experiments (Figure 6d). These findings were recently supported by chromosome conformation maps (micro-C) that revealed interdigitated zigzag fibers in mammal cells (64, 92). In another study, by simulating chromatin fibers containing segments with acetylated and wild-type histone tails, Rao et al. (156) suggested an epigenetic mechanism of segregation induced by acetylated domains on the kilobase level, commensurate with patterns observed in experimental contact maps on a larger scale. Recently, the structural effects of different modeled linker histone protein densities bound to chromatin fibers were also used to help interpret different chromatin architectures in lymphoma cells (207).
6. THE IMPACT OF COMMUNITY EXERCISES AND WEB PROGRAMS
Community exercises and games that bring together researchers with common goals help propel the field of biomolecular modeling. Such initiatives reveal what works and what fails, and also heighten interest in scientific problems and solutions, often helping recruit fresh talent.
6.1. CASP and RNA-Puzzles
The community-wide CASP (125) has helped advance methods for predicting protein structure from amino acid sequences since 1994. CASP exercises occur biannually and provide researchers worldwide with the opportunity to test their predictions for established targets that will soon be resolved experimentally. In this way, the exercise illuminates the current state-of-the-art techniques and software for protein structure prediction and refinement.
The number of CASP participants increased continuously from 1994 to 2005, after which they gradually decreased (see Supplemental Figure 1). Similar trends are observed in the number of predictions, but with a delay; the maximum number of predictions was reached in 2010. Unlike the participants trend, the number of predictions increased in 2018 with respect to 2016. With recent exciting contributions from artificial intelligence, interest in CASP may be revitalized.
CASP results have taught us many things. The quality of template-based modeling predictions has increased dramatically since 1994 (93). The ab initio modeling category (prediction of targets with no obvious templates) has yielded a more moderate improvement (1), until recently, when Google’s AphaFold system shined (179).
CASP has also highlighted that structure refinement is challenged by random errors in force fields and poor sampling, prompting force field improvements, new sampling strategies, and knowledge-based methods to bias simulations (55).
Similar to CASP, RNA-Puzzles, a collective experiment for blind de novo RNA tertiary structure prediction (35) launched in 2012, has encouraged the community to improve current tools and develop new approaches by determining the capability and limitations of methods for RNA tertiary structure prediction from sequence.
There have been RNA-Puzzles rounds focused on the prediction of small and medium RNA structures (35), large RNA structures (120), and riboswitches and ribozymes (119), with an increasing number of participants and puzzles solved (see Supplemental Figure 1).
Overall, RNA-Puzzles results have highlighted that template-based and homology-based structure predictions can achieve a high level of accuracy but have emphasized that the prediction of large RNA structures remains challenging, as does the prediction of non-Watson-Crick interactions. As discussed by Pyle & Schlick (153) and Schlick & Pyle (173), current challenges include clustering of predicted secondary structure candidates to determine alternative low-energy states, annotation of RNA motifs and updating of structural databases, quality check of deposited structures solved by experimental techniques, use of experimental data for proper structural and functional interpretation, RNA force field inaccuracies for all-atom simulations, generation of atomic models from CG structures, and modeling of large RNA where few experimental data are available. With the COVID-19 pandemic, a resurgence of RNA modeling and improvement in handling large RNAs have been realized.
6.2. Foldit and Eterna, Citizen Science Projects
Other excellent examples of community exercises are Foldit (33) and Eterna (98), which are online programs that challenge players to fold proteins and RNA molecules, respectively, from their constituent residues. These citizen science projects encourage nonexperts to participate, increasing the general public’s interest in scientific problems in the biomolecular field. They aim to find the functional 3D structure of a protein or RNA from its sequence using force field–based calculations as well as human intuition. By modifying the positions of the backbone and side chains to change interresidue and residue–solvent interactions, players manipulate the molecules to seek structures with low energies. The lower is the energy, the higher is the participant score.
Foldit participants are challenged to design stable folded proteins de novo and create the lowest free energy model (33). Since launched in 2008 by the Rosetta group, Foldit has attracted more than 800,000 users, who have solved more than 1,800 puzzles (see Supplemental Figure 1). A total of 56 designs of soluble proteins were created by 36 different players, representing 20 different folds, including a new fold (90). Foldit predictions were found to substantially outperform automated algorithms, such as Rosetta (33). By using human creativity and instinct guided by scientific understanding, Foldit has helped advance the de novo protein design problem. Moreover, it has demonstrated the importance of using human intelligence combined with computational algorithms to solve structure prediction problems (90).
Eterna players are challenged to design RNA molecules from an initial sequence by changing, adding, or deleting nucleotide residues to obtain a target conformation (98). Eterna also went one step further: The best designs will be synthesized in the laboratory. Since launched in 2011, 119,032 users have registered, solving at least one puzzle each (see Supplemental Figure 1). Of these players, 4,366 have participated in lab challenges. Of the 365,843 designs submitted (see Supplemental Figure 1), 167,730 have been synthesized, and improvements have been invited. From these efforts, EternaBot, a ML algorithm for determining RNA sequences that fold onto target structures, was developed. The Eternacon annual meeting brings together players, scientists, and developers to Stanford University to discuss puzzle solving, scientific advances, and future challenges.
Similar to Foldit, the Eterna community, as well as the EternaBot algorithm, offers alternatives (8) to automated algorithms. Overall, Eterna has helped accelerate the progress of in vitro RNA design by generating hundreds of designs and creating a data set of approximately 100,000 potential RNA designs, some of which were subsequently tested.
6.3. Response of the Biomolecular Modeling Community to the COVID-19 Outbreak
The importance of community collaborations in science has been demonstrated recently during the emergence of the COVID-19 pandemic. Besides worldwide collaborations among the high-performance communities (e.g., BioExcel, https://covid.bioexcel.eu, and the COVID-19 HPC Consortium, https://covid19-hpc-consortium.org) and many others, specific initiatives have been launched to help develop drugs and vaccines.
CASP has launched a SARS-CoV-2 structure modeling initiative for predicting the structure of viral proteins. In the first round, approximately 1,600 models were predicted from 52 groups. These predicted structures aid in the development of vaccines and drugs to help fight this horrific disease, for example, by recruiting the SUMMIT supercomputer to screen drug databases for compatible COVID-19 protein target residues (184).
Individual groups have also predicted the structures of many viral proteins. For example, the DeepMind group from Google has deployed AlphaFold to predict the structure of the membrane protein and other viral proteins (82). Similarly, many groups are predicting structures of the spike protein bound to various inhibitors (67).
Foldit and Eterna have both launched challenges related to COVID-19 to help design proteins to aid in the immune system response upon infection, bind the spike protein of the virus, and/or design mRNA-based vectors for vaccines.
6.4. Community Progress
Overall, these community exercises and citizen science projects have had a positive impact on the biomolecular modeling field, as they increase general interest in the field, highlight its importance, recruit fresh talent, lead to new approaches, stimulate discussions, and increase data generation and products. The new communities established by CASP or Eterna attract millennials, bring new energy into these important scientific efforts, and extend research efforts into the arena of social networks. Other communities, like the one established by more than 500 developers of the suite Rosetta for macromolecular modeling and design (100), are revolutionizing and accelerating the field by encouraging strong collaborations and discouraging competition.
New initiatives will undoubtedly continue to drive the field forward and train a new generation of science, technology, engineering, and mathematics researchers. As the COVID-19 pandemic has shown us, scientists are able to come together for the common good quickly and successfully.
7. SUMMARY
Our reassessment of the progress in the field of biomolecular modeling and simulation highlights how far the field has come from its early days at the dawn of digital computers. The skepticism that enveloped early molecular computers, replaced by inflated and unrealistic expectations with the advent of supercomputers and high-speed human genome sequencing, has advanced to a productive stage where computations and instrumentation are hand-in-hand partners, as well as effective methods in their own right to explore molecular structures, functions, and mechanisms (Figure 1). Improved force fields, better sampling techniques, usage of available information from structural and functional databases, emerging community exercises and games, emphasis on merging scales, infusion of clever ideas from many areas of science and engineering, and ever-expanding technologies have been utilized well and applied successfully to solve and advance many scientific and medical problems with wide-ranging societal and health impacts. The expeditious responses by the high-performance computing community to the COVID-19 pandemic illustrate how unified the goals are and how far the technology has evolved to impact medicine and human health.
Gone are the days where biomolecular scientists worked in isolation. Labs, teams, and nations are collaborating as never before to address pressing problems, from pollution to energy to pandemics. With gene editing approaches, dazzling improvement in structural determination, and increasing reliability of computational predictions, scientists are well positioned to address many important problems in science, health, and industry. Despite numerous technical and ethical challenges that lie ahead, the foundations are firm, and the trajectory of the field is guaranteed to take us into a bright future.
8. RECOMMENDATIONS
To exploit the field’s great potential for continued impact on society and human health, we make the following general recommendations.
8.1. Important Algorithmic Directions
The promising CG and multiscale models that bridge multiple scales associated with complex biomolecular systems (see 172) require better direction and assessment to be effective at large and applied generally. The many approaches available have not been unified or tested in any consistent way, as the classical force fields have been. More effort is required to share such approaches, apply them systematically, and make programs available to the community at large. Similarly, artificial intelligence and ML approaches can be better shared, organized, and applied in key areas like structure prediction, ligand binding, or biomolecular interactions.
8.2. Community Initiatives
Community exercises like CASP and its many descendants or web programs like Eterna and Foldit have been instrumental in highlighting problems in the field, recruiting a community of scientists and citizens, and improving state-of-the-art force fields. While some initiatives retire, others will replace them to engage interest and advance emerging subfields. Perhaps more public funding could help in encouraging scientists to develop and support such exercises, computer games, and initiatives. Special societal subsections, such as the multiscale genome organization subsection just added to the Biophysical Society or the Molecular Sciences Sustainable Software Institute, can go a long way toward promoting and advancing public interest in subfields of biomolecular modeling.
8.3. Public Engagement
Biomolecular scientists have done well at engaging public interest and participation in modeling challenges (e.g., Folding@home, Human Proteome Folding Project), but more could be done. Introducing molecular modeling and simulation early into the high school curriculum could help children understand that the molecules of life are not static and that there is much more to explore related to the activities of these molecules. Problem-based learning in molecular simulation could help youngsters bridge many fields and encourage innovation and exploration (see the advice column to science, technology, engineering, and mathematics students in 170). As our scientific capabilities advance, for example, in the area of gene editing, we will need a public that understands both the scientific ethical dilemmas and potential benefits of these technologies. The earlier such an education begins, the better informed our society can become. The COVID-19 pandemic provides an excellent ground for introducing many relevant scientific topics to young students, from infection spread models to phylogenetic trees of the viral genome to drug–target simulations and vaccine development.
8.4. Multidisciplinarity, Diversity, and Education
Undoubtedly, biomolecular modeling is an exemplary field for merging together many scientific and engineering disciplines with the goal of solving scientific problems with state-of-the-art technologies (172). However, more could be done to enhance diversity and outreach. This general problem in science may be easier to address in our field, where large teams often work together, and new talent can help look at each problem with a fresh perspective. Against recent discriminatory or biased trends in the world, biomolecular scientists could take the lead in establishing new programs to include and bring together minorities and accommodate disabilities, starting from the elementary school level. Curricular changes, experience in research, moving scientific exhibits, and other educational models could help recruit talent from all corners of the world. Young people today have taken the lead in making statements about gun control, environmental damage, and education for all. We still have much work to do to educate better, recruit, and retain such young talent further. We are better together.
Supplementary Material
ACKNOWLEDGMENTS
Support from the National Institutes of Health, the National Institute of General Medical Sciences award R35-GM122562, the National Science Foundation RAPID Award (2030377) from the Division of Mathematical Sciences and the Division of Chemistry, and Philip-Morris USA Inc. and Philips-Morris International to T.S. is gratefully acknowledged. We thank the following questionnaire respondents for their time and opinions: Elena Akhmatskaya, Russ B. Altman, Nir Ben Tal, Giovanni Bussi, Qiang Cui, Mauricio Esguerra, James Gumbart, Jonathan Ipsaro, G. Ali Mansoori, Andy McCammon, Mihaly Mezei, Stephen Neidle, Chris Oostenbrink, Ognjen Perisic, Stefano Piana-Agnostinetti, Benoit Roux, Robert Skeel, James Skinner, Paul Whitford, Celerino Abad Zapatero, and Yingkai Zhang. We also thank David Baker and Brian Koepnick for providing information on Foldit participants and solved puzzles, and Rhiju Das and Jonathan Romano for providing information on Eterna participants and solved puzzles. We thank David Case, Ron Elber, Alex MacKerell, and Pengyu Ren for their thoughtful comments on polarizable force fields. We apologize in advance to the many authors of excellent biomolecular papers who we could not cite due to page limits.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abriata LA, Tamo GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. 2018. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment-based contact prediction methods. Proteins 86(Suppl. 1):97–112 [DOI] [PubMed] [Google Scholar]
- 2.Adamczak B, Kogut M, Czub J. 2018. Effect of osmolytes on the thermal stability of proteins: replica exchange simulations of Trp-cage in urea and betaine solutions. Phys. Chem. Chem. Phys 20(16):11174–82 [DOI] [PubMed] [Google Scholar]
- 3.Alder BJ, Wainwright TE. 1959. Studies in molecular dynamics. I. General method. J. Chem. Phys 31(2):459–66 [Google Scholar]
- 4.Allinger NL. 1976. Calculation of molecular structure and energy by force-field methods. In Advances in Physical Organic Chemistry, Vol. 13, ed. Gold V, Bethell D, pp. 1–82. Cambridge, MA: Academic [Google Scholar]
- 5.Allinger NL, Miller MA, Van Catledge FA, Hirsch JA. 1967. Conformational analysis. LVII. The calculation of the conformational structures of hydrocarbons by the Westheimer-Hendrickson-Wiberg method. J. Am. Chem. Soc 89(17):4345–57 [Google Scholar]
- 6.Amezcua M, Mobley D. 2020. SAMPL7 challenge overview: assessing the reliability of polarizable and non-polarizable methods for host-guest binding free energy calculations. ChemRxiv 12768353. 10.26434/chemrxiv.12768353,v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen ES. 2010. Prediction and design of DNA and RNA structures. New Biotechnol. 27(3):184–93 [DOI] [PubMed] [Google Scholar]
- 8.Anderson-Lee J, Fisker E, Kosaraju V, Wu M, Kong J, et al. 2016. Principles for predicting RNA secondary structure design difficulty. J. Mol. Biol 428(5A):748–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science 181(4096):223–30 [DOI] [PubMed] [Google Scholar]
- 10.Baker CM. 2015. Polarizable force fields for molecular dynamics simulations of biomolecules. WIREs Comput. Mol. Sci 5(2):241–54 [Google Scholar]
- 11.Baker EG, Bartlett GJ, Porter Goff KL, Woolfson DN. 2017. Miniprotein design: past, present, and prospects. Acc. Chem. Res 50(9):2085–92 [DOI] [PubMed] [Google Scholar]
- 12.Baker EG, Williams C, Hudson KL, Bartlett GJ, Heal JW, et al. 2017. Engineering protein stability with atomic precision in a monomeric miniprotein. Nat. Chem. Biol 13(7):764–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banáš P, Sklenovský P, Wedekind JE, Šponer J, Otyepka M. 2012. Molecular mechanism of preQ1 riboswitch action: a molecular dynamics study. J. Phys. Chem. B 116(42):12721–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer G, Hoefler T, Kramer W, Fiedler R. 2012. Analyses and modeling of applications used to demonstrate sustained petascale performance on blue waters. Paper presented at CUG 2012, Stuttgart, Germany [Google Scholar]
- 15.Beberg AL, Ensign DL, Jayachandran G, Khaliq S, Pande VS. 2009. Folding@home: lessons from eight years of volunteer distributed computing. In 2009 IEEE International Symposium on Parallel Distributed Processing, pp. 1–8. Piscataway, NJ: IEEE [Google Scholar]
- 16.Berendsen HJC, van der Spoel D, van Drunen R. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun 91(1):43–56 [Google Scholar]
- 17.Bezdek JC. 1993. Fuzzy models—what are they, and why? [Editorial]. IEEE Trans. Fuzzy Syst 1(1):1–6 [Google Scholar]
- 18.Bixon M, Lifson S. 1967. Potential functions and conformations in cycloalkanes. Tetrahedron 23(2):769–84 [Google Scholar]
- 19.Boomsma W, Tian P, Frellsen J, Ferkinghoff-Borg J, Hamelryck T, et al. 2014. Equilibrium simulations of proteins using molecular fragment replacement and NMR chemical shifts. PNAS 111(38):13852–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boukharta L, Gutiérrez-de Terán H, Åqvist J. 2014. Computational prediction of alanine scanning and ligand binding energetics in G-protein coupled receptors. PLOS Comput. Biol 10(4):e1003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulanger E, Thiel W. 2014. Toward QM/MM simulation of enzymatic reactions with the Drude oscillator polarizable force field. J. Chem. Theory Comput 10(4):1795–809 [DOI] [PubMed] [Google Scholar]
- 22.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, et al. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC ‘06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, pp. 43–43. New York: ACM [Google Scholar]
- 23.Bramsen JB, Kjems J. 2012. Development of therapeutic-grade small interfering RNAs by chemical engineering. Front. Genet 3:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem 4(2):187–217 [Google Scholar]
- 25.Canessa Fortuna A, Zerbetto De Palma G, Aliperti Car L, Armentia L, Vitali V, et al. 2019. Gating in plant plasma membrane aquaporins: the involvement of leucine in the formation of a pore constriction in the closed state. FEBS J. 286(17):3473–87 [DOI] [PubMed] [Google Scholar]
- 26.Carr JK, Zabuga AV, Roy S, Rizzo TR, Skinner JL. 2014. Assessment of amide I spectroscopic maps for a gas-phase peptide using IR-UV double-resonance spectroscopy and density functional theory calculations. J. Chem. Phys 140(22):224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter Childers M, Daggett V. 2017. Insights from molecular dynamics simulations for computational protein design. Mol. Syst. Des. Eng 2(1):9–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P-C, Hub JS. 2014. Validating solution ensembles from molecular dynamics simulation by wide-angle X-ray scattering data. Biophys. J 107(2):435–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chevalier A, Silva D-A, Rocklin GJ, Hicks DR, Vergara R, et al. 2017. Massively parallel de novo protein design for targeted therapeutics. Nature 550(7674):74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou F-C, Lipfert J, Das R. 2014. Blind predictions of DNA and RNA tweezers experiments with force and torque. PLOS Comput. Biol 10(8):e1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowdhary J, Harder E, Lopes PEM, Huang L, MacKerell AD, Roux B. 2013. A polarizable force field of dipalmitoylphosphatidylcholine based on the classical Drude model for molecular dynamics simulations of lipids. J. Phys. Chem. B 117(31):9142–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cino EA, Choy W-Y, Karttunen M. 2012. Comparison of secondary structure formation using 10 different force fields in microsecond molecular dynamics simulations. J. Chem. Theory Comput 8(8):2725–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, et al. 2010. Predicting protein structures with a multiplayer online game. Nature 466:756–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craven TW, Cho M-K, Traaseth NJ, Bonneau R, Kirshenbaum K. 2016. A miniature protein stabilized by a cation-μ interaction network core. J. Am. Chem. Soc 138(5):1543–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz JA, Blanchet M-F, Boniecki M, Bujnicki JM, Chen S-J, et al. 2012. RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. RNA 18(4):610–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daura X, Jaun B, Seebach D, van Gunsteren WF, Mark AE. 1998. Reversible peptide folding in solution by molecular dynamics simulation. J. Mol. Biol 280(5):925–32 [DOI] [PubMed] [Google Scholar]
- 37.de Brevern AG, Bornot A, Craveur P, Etchebest C, Gelly J-C. 2012. PredyFlexy: flexibility and local structure prediction from sequence. Nucleic Acids Res. 40:W317–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Palma F, Bottaro S, Bussi G. 2015. Kissing loop interaction in adenine riboswitch: insights from umbrella sampling simulations. BMC Bioinformat. 16(Suppl. 9):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dirks RM, Lin M, Winfree E, Pierce NA. 2004. Paradigms for computational nucleic acid design. Nucleic Acids Res. 32(4):1392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, et al. 2013. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature 503(7475):295–99 [DOI] [PubMed] [Google Scholar]
- 41.Dror RO, Mildorf TJ, Hilger D, Manglik A, Borhani DW, et al. 2015. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348(6241):1361–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan L, Guo X, Cong Y, Feng G, Li Y, Zhang JZH. 2019. Accelerated molecular dynamics simulation for helical proteins folding in explicit water. Front. Chem 7:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Y, Kollman PA. 1998. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science 282(5389):740–44 [DOI] [PubMed] [Google Scholar]
- 44.Durrant JD, Kochanek SE, Casalino L, Ieong PU, Dommer AC, Amaro RE. 2020. Mesoscale all-atom influenza virus simulations suggest new substrate binding mechanism. ACS Central Sci. 6(2):189–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hage K, Hédin F, Gupta PK, Meuwly M, Karplus M. 2018. Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size. eLife 7:e35560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Hage K, Hédin F, Gupta PK, Meuwly M, Karplus M. 2019. Response to comment on “Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size”. eLife 8:e45318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emiliano B, Simmerling C, Dill K. 2020. Protein storytelling through physics. Science. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esguerra M, Siretskiy A, Bello X, Sallander J, Gutiérrez-de Terán H. 2016. GPCR-ModSim: a comprehensive web based solution for modeling G-protein coupled receptors. Nucleic Acids Res. 44(W1):W455–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freddolino PL, Liu F, Gruebele M, Schulten K. 2008. Ten-microsecond molecular dynamics simulation of a fast-folding WW domain. Biophys. J 94(10):L75–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamini R, Han W, Stone JE, Schulten K. 2014. Assembly of Nsp1 nucleoporins provides insight into nuclear pore complex gating. PLOS Comput. Biol 10(3):e1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganguly A, Boulanger E, Thiel W. 2017. Importance of MM polarization in QM/MM studies of enzymatic reactions: assessment of the QM/MM Drude oscillator model. J. Chem. Theory Comput 13(6):2954–61 [DOI] [PubMed] [Google Scholar]
- 52.Gapsys V, de Groot BL. 2019. Comment on “Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size”. eLife 8:e44718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gapsys V, de Groot BL. 2020. On the importance of statistics in molecular simulations for thermodynamics, kinetics and simulation box size. eLife 9:e57589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genheden S, Ryde U. 2012. Will molecular dynamics simulations of proteins ever reach equilibrium? Phys. Chem. Chem. Phys 14(24):8662–77 [DOI] [PubMed] [Google Scholar]
- 55.Gniewek P, Kolinski A, Jernigan RL, Kloczkowski A. 2012. How noise in force fields can affect the structural refinement of protein models? Proteins 80(2):335–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T. 2016. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. PNAS 113(5):1238–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunsteren WFV. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Zürich: Biomos [Google Scholar]
- 58.Guvench O, MacKerell AD Jr. 2009. Computational fragment-based binding site identification by ligand competitive saturation. PLOS Comput. Biol 5(7):e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haghighatlari M, Hachmann J. 2019. Advances of machine learning in molecular modeling and simulation. Curr. Opin. Chem. Eng 23:51–57 [Google Scholar]
- 60.Halgren TA. 1996. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem 17(5–6):490–519 [Google Scholar]
- 61.Hallberg ZF, Su Y, Kitto RZ, Hammond MC. 2017. Engineering and in vivo applications of riboswitches. Annu. Rev. Biochem 86:515–39 [DOI] [PubMed] [Google Scholar]
- 62.Hamp T, Rost B. 2015. More challenges for machine-learning protein interactions. Bioinformatics 31(10):1521–25 [DOI] [PubMed] [Google Scholar]
- 63.He X, Lopes PEM, MacKerell AD. 2013. Polarizable empirical force field for acyclic polyalcohols based on the classical Drude oscillator. Biopolymers 99(10):724–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsieh T-HS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, et al. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78(3):539–53.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu H, Liu H. 2013. Pitfall in quantum mechanical/molecular mechanical molecular dynamics simulation of small solutes in solution. J. Phys. Chem. B 117(21):6505–11 [DOI] [PubMed] [Google Scholar]
- 66.Huang P-S, Boyken SE, Baker D. 2016. The coming of age of de novo protein design. Nature 537(7620):320–27 [DOI] [PubMed] [Google Scholar]
- 67.Huang X, Pearce R, Zhang Y. 2020. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging 12(12):11263–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inakollu VSS, Geerke DP, Rowley CN, Yu H. 2020. Polarisable force fields: What do they add in biomolecular simulations? Curr. Opin. Struct. Biol 61:182–90 [DOI] [PubMed] [Google Scholar]
- 69.Irobalieva RN, Fogg JM, Catanese DJ Jr., Sutthibutpong T, Chen M, et al. 2015. Structural diversity of supercoiled DNA. Nat. Commun 6:8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Izrailev S, Crofts AR, Berry EA, Schulten K. 1999. Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc1 complex. Biophys. J 77(4):1753–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jack A, Levitt M. 1978. Refinement of large structures by simultaneous minimization of energy and R factor. Acta Crystallogr. A 34(6):931–35 [Google Scholar]
- 72.Jackson NE, Bowen AS, Antony LW, Webb MA, Vishwanath V, de Pablo JJ. 2019. Electronic structure at coarse-grained resolutions from supervised machine learning. Sci. Adv 5(3):eaav1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain S, Laederach A, Ramos SBV, Schlick T. 2018. A pipeline for computational design of novel RNA-like topologies. Nucleic Acids Res. 46(14):7040–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jain S, Schlick T. 2017. F-rag: generating atomic coordinates from RNA graphs by fragment assembly. J. Mol. Biol 429(23):3587–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jain S, Zhu Q, Paz AS, Schlick T. 2020. Identification of novel RNA design candidates by clustering the extended RNA-As-Graphs library. Biochim. Biophys. Acta Gen. Subj 1864(6):129534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jindal G, Slanska K, Kolev V, Damborsky J, Prokop Z, Warshel A. 2019. Exploring the challenges of computational enzyme design by rebuilding the active site of a dehalogenase. PNAS 116(2):389–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jing Z, Liu C, Cheng SY, Qi R, Walker BD, et al. 2019. Polarizable force fields for biomolecular simulations: recent advances and applications. Annu. Rev. Biophys 48:371–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jing Z, Liu C, Qi R, Ren P. 2018. Many-body effect determines the selectivity for Ca2+ and Mg2+ in proteins. PNAS 115(32):E7495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jing Z, Qi R, Liu C, Ren P. 2017. Study of interactions between metal ions and protein model compounds by energy decomposition analyses and the AMOEBA force field. J. Chem. Phys 147(16):161733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 2014. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discuss. 169(0):23–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jorgensen WL, Madura JD, Swenson CJ. 1984. Optimized intermolecular potential functions for liquid hydrocarbons. J. Am. Chem. Soc 106(22):6638–46 [Google Scholar]
- 82.Jumper J, Tunyasuvunakool K, Kohlim P, Hassabis D, Team A. 2020. Computational predictions of protein structures associated with COVID-19. Rep., DeepMind, London. https://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19 [Google Scholar]
- 83.Jung J, Nishima W, Daniels M, Bascom G, Kobayashi C, et al. 2019. Scaling molecular dynamics beyond 100,000 processor cores for large-scale biophysical simulations. J. Comput. Chem 40(21):1919–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. 2014. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11:313–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karplus M, Lavery R. 2014. Significance of molecular dynamics simulations for life sciences. Isr. J. Chem 54(8–9):1042–51 [Google Scholar]
- 86.Khabiri M, Freddolino PL. 2017. Deficiencies in molecular dynamics simulation-based prediction of protein–DNA binding free energy landscapes. J. Phys. Chem. B 121(20):5151–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khafizov K, Perez C, Koshy C, Quick M, Fendler K, et al. 2012. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. PNAS 109(44):E3035–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kilic S, Felekyan S, Doroshenko O, Boichenko I, Dimura M, et al. 2018. Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1α. Nat. Commun 9(1):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kinana AD, Vargiu AV, Nikaido H. 2016. Effect of site-directed mutations in multidrug efflux pump AcrB examined by quantitative efflux assays. Biochem. Biophys. Res. Commun 480(4):552–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koepnick B, Flatten J, Husain T, Ford A, Silva D-A, et al. 2019. De novo protein design by citizen scientists. Nature 570(7761):390–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krepl M, Havrila M, Stadlbauer P, Banas P, Otyepka M, et al. 2015. Can we execute stable microsecondscale atomistic simulations of protein–RNA complexes? J. Chem. Theory Comput 11(3):1220–43 [DOI] [PubMed] [Google Scholar]
- 92.Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, et al. 2020. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78(3):554–65.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kryshtafovych A, Monastyrskyy B, Fidelis K, Moult J, Schwede T, Tramontano A. 2018. Evaluation of the template-based modeling in CASP12. Proteins 86(Suppl. 1):321–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. 2019. Critical assessment of methods of protein structure prediction (CASP)—round XIII. Proteins 87(12):1011–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kührová P, Best RB, Bottaro S, Bussi G, Šponer J, et al. 2016. Computer folding of RNA tetraloops: identification of key force field deficiencies. J. Chem. Theory Comput 12(9):4534–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lankaš F, Lavery R, Maddocks JH. 2006. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure 14(10):1527–34 [DOI] [PubMed] [Google Scholar]
- 97.Latorraca NR, Venkatakrishnan AJ, Dror RO. 2017. GPCR dynamics: structures in motion. Chem. Rev 117(1):139–55 [DOI] [PubMed] [Google Scholar]
- 98.Lee J, Kladwang W, Lee M, Cantu D, Azizyan M, et al. 2014. RNA design rules from a massive open laboratory. PNAS 111(6):2122–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leferink NGH, Ranaghan KE, Karuppiah V, Currin A, van der Kamp MW, et al. 2018. Experiment and simulation reveal how mutations in functional plasticity regions guide plant monoterpene synthase product outcome. ACS Catal. 8(5):3780–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, et al. 2020. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods 17(7):665–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lemkul JA, MacKerell AD. 2018. Polarizable force field for RNA based on the classical Drude oscillator. J. Comput. Chem 39(32):2624–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemkul JA, Savelyev A, MacKerell AD Jr. 2014. Induced polarization influences the fundamental forces in DNA base flipping. J. Phys. Chem. Lett 5(12):2077–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lensink MF, Velankar S, Wodak SJ. 2017. Modeling protein-protein and protein-peptide complexes: CAPRI 6th edition. Proteins 85(3):359–77 [DOI] [PubMed] [Google Scholar]
- 104.Leonard AN, Wang E, Monje-Galvan V, Klauda JB. 2019. Developing and testing of lipid force fields with applications to modeling cellular membranes. Chem. Rev 119(9):6227–69 [DOI] [PubMed] [Google Scholar]
- 105.Liang H, Chen H, Fan K, Wei P, Guo X, et al. 2009. De novo design of a beta alpha beta motif. Angew. Chem 48(18):3301–3 [DOI] [PubMed] [Google Scholar]
- 106.Lifson S 1986. Theoretical foundation for the empirical force field method. Gazz. Chim. Ital 116(12):687–92 [Google Scholar]
- 107.Lin F-Y, Huang J, Pandey P, Rupakheti C, Li J, et al. 2020. Further optimization and validation of the classical Drude polarizable protein force field. J. Chem. Theory Comput 16(5):3221–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin F-Y, MacKerell ADJ. 2019. Force fields for small molecules. Methods Mol. Biol 2022:21–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin X, Schafer NP, Lu W, Jin S, Chen X, et al. 2019. Forging tools for refining predicted protein structures. PNAS 116(19):9400–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. 2011. How fast-folding proteins fold. Science 334(6055):517–20 [DOI] [PubMed] [Google Scholar]
- 111.Liu C, Perilla JR, Ning J, Lu M, Hou G, et al. 2016. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun 7:10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu S, Liu C, Deng L. 2018. Machine learning approaches for protein-protein interaction hot spot prediction: progress and comparative assessment. Molecules 23(10):2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Loco D, Lagardère L, Cisneros GA, Scalmani G, Frisch M, et al. 2019. Towards large scale hybrid QM/MM dynamics of complex systems with advanced point dipole polarizable embeddings. Chem. Sci 10(30):7200–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mayr A, Klambauer G, Unterthiner T, Hochreiter S. 2016. Deeptox: toxicity prediction using deep learning. Front. Environ. Sci 3:80 [Google Scholar]
- 115.McCammon JA, Gelin BR, Karplus M. 1977. Dynamics of folded proteins. Nature 267(5612):585–90 [DOI] [PubMed] [Google Scholar]
- 116.Melcr J, Piquemal J-P. 2019. Accurate biomolecular simulations account for electronic polarization. Front. Mol. Biosci 10.3389/fmolb.2019.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meng G, Tariq M, Jain S, Elmetwaly S, Schlick T. 2019. RAG-Web: RNA structure prediction/design using RNA-As-Graphs. Bioinformatics 36(2):647–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mezei M 2017. Rescore protein–protein docked ensembles with an interface contact statistics. Proteins 85(2):235–41 [DOI] [PubMed] [Google Scholar]
- 119.Miao Z, Adamiak RW, Antczak M, Batey RT, Becka AJ, et al. 2017. RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA 23(5):655–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miao Z, Adamiak RW, Blanchet M-F, Boniecki M, Bujnicki JM, et al. 2015. RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA 21(6):1066–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miao Y, Goldfeld DA, Moo EV, Sexton PM, Christopoulos A, et al. 2016. Accelerated structure-based design of chemically diverse allosteric modulators of a muscarinic G protein-coupled receptor. PNAS 113(38):E5675–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miao Y, Huang Y-M, Walker RC, McCammon JA, Chang C-EA. 2018. Ligand binding pathways and conformational transitions of the HIV protease. Biochemistry 57(9):1533–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mitchell JS, Laughton CA, Harris SA. 2011. Atomistic simulations reveal bubbles, kinks and wrinkles in supercoiled DNA. Nucleic Acids Res. 39(9):3928–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mlýnský V, Banás P, Hollas D, Réblová K, Walter NG, et al. 2010. Extensive molecular dynamics simulations showing that canonical G8 and protonated A38H+ forms are most consistent with crystal structures of hairpin ribozyme. J. Phys. Chem. B 114(19):6642–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moult J, Pedersen JT, Judson R, Fidelis K. 1995. A large-scale experiment to assess protein structure prediction methods. Proteins 23(3):2–4 [DOI] [PubMed] [Google Scholar]
- 126.Munos B 2009. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov 8(12):959–68 [DOI] [PubMed] [Google Scholar]
- 127.Neale C, Pomès R. 2016. Sampling errors in free energy simulations of small molecules in lipid bilayers. Biochim. Biophys. Acta Biomembranes 1858(10):2539–48 [DOI] [PubMed] [Google Scholar]
- 128.Neidigh JW, Fesinmeyer RM, Andersen NH. 2002. Designing a 20-residue protein. Nat. Struct. Mol. Biol 9(6):425–30 [DOI] [PubMed] [Google Scholar]
- 129.Némethy G, Scheraga HA. 1965. Theoretical determination of sterically allowed conformations of a polypeptide chain by a computer method. Biopolymers 3(2):155–84 [Google Scholar]
- 130.Ngo VA, Fanning JK, Noskov SY. 2019. Comparative analysis of protein hydration from MD simulations with additive and polarizable force fields. Adv. Theory Simul 2(2):1800106 [Google Scholar]
- 131.Nguyen K, Whitford PC. 2016. Steric interactions lead to collective tilting motion in the ribosome during mRNA–tRNA translocation. Nat. Commun 7:10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Noé F, Tkatchenko A, Müller K-R, Clementi C. 2020. Machine learning for molecular simulation. Annu. Rev. Phys. Chem 71:361–90 [DOI] [PubMed] [Google Scholar]
- 133.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22(7):1055–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, et al. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501(7467):385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oliveira ASF, Edsall CJ, Woods CJ, Bates P, Nunez GV, et al. 2019. A general mechanism for signal propagation in the nicotinic acetylcholine receptor family. J. Am. Chem. Soc 141(51):19953–58 [DOI] [PubMed] [Google Scholar]
- 136.Olson WK, Colasanti AV, Czapla L, Zheng G. 2008. Insights into the sequence-dependent macromolecular properties of DNA from base-pair level modeling: coarse-graining of condensed phase and biomolecular systems. In Coarse-Graining of Condensed Phase and Biomolecular Systems, ed. Voth GA, pp. 205–23. Boca Raton, FL: CRC Press [Google Scholar]
- 137.Ostmeyer J, Chakrapani S, Pan AC, Perozo E, Roux B. 2013. Recovery from slow inactivation in K+ channels is controlled by water molecules. Nature 501(7465):121–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pandey P, Mallajosyula SS. 2016. Influence of polarization on carbohydrate hydration: a comparative study using additive and polarizable force fields. J. Phys. Chem. B 120(27):6621–33 [DOI] [PubMed] [Google Scholar]
- 139.Patel DS, He X, MacKerell AD. 2015. Polarizable empirical force field for hexopyranose monosaccharides based on the classical Drude oscillator. J. Phys. Chem. B 119(3):637–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, et al. 1995. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun 91(1):1–41 [Google Scholar]
- 141.Pérez A, Luque FJ, Orozco M. 2007. Dynamics of B-DNA on the microsecond time scale. J. Am. Chem. Soc 129(47):14739–45 [DOI] [PubMed] [Google Scholar]
- 142.Perez C, Faust B, Mehdipour AR, Francesconi KA, Forrest LR, Ziegler C. 2014. Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling. Nat. Commun 5(1):4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perilla JR, Schulten K. 2017. Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat. Commun 8:15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perthold JW, Oostenbrink C. 2017. Simulation of reversible protein–protein binding and calculation of binding free energies using perturbed distance restraints. J. Chem. Theory Comput 13(11):5697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem 26(16):1781–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Piana S, Donchev AG, Robustelli P, Shaw DE. 2015. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 119(16):5113–23 [DOI] [PubMed] [Google Scholar]
- 147.Piana S, Klepeis JL, Shaw DE. 2014. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr. Opin. Struct. Biol 24:98–105 [DOI] [PubMed] [Google Scholar]
- 148.Piana S, Shaw DE. 2018. Atomic-level description of protein folding inside the GroEL cavity. J. Phys. Chem. B 122(49):11440–49 [DOI] [PubMed] [Google Scholar]
- 149.Pierro MD, Cheng RR, Aiden EL, Wolynes PG, Onuchic JN. 2017. De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. PNAS 114(46):12126–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Poma AB, Guzman HV, Li MS, Theodorakis PE. 2019. Mechanical and thermodynamic properties of αβ42, αβ40, and α-synuclein fibrils: a coarse-grained method to complement experimental studies. Beilstein J. Nanotechnol 10:500–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Portillo-Ledesma S, Schlick T. 2020. Bridging chromatin structure and function over a range of experimental and spatial temporal scales by molecular modeling. WIREs Comput. Mol. Sci 10:e1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Prigozhin MB, Zhang Y, Schulten K, Gruebele M, Pogorelov TV. 2019. Fast pressure-jump all-atom simulations and experiments reveal site-specific protein dehydration-folding dynamics. PNAS 116(12):5356–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pyle AM, Schlick T. 2016. Challenges in RNA structural modeling and design. J. Mol. Biol 428(5A):733–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rahman A, Stillinger FH. 1971. Molecular dynamics study of liquid water. J. Chem. Phys 55(7):3336–59 [Google Scholar]
- 155.Ramis R, Ortega-Castro J, Casasnovas R, Mariño L, Vilanova B, et al. 2019. A coarse-grained molecular dynamics approach to the study of the intrinsically disordered protein α-synuclein. J. Chem. Inf. Model 59(4):1458–71 [DOI] [PubMed] [Google Scholar]
- 156.Rao SSP, Huang S-C, Glenn St. Hilaire B, Engreitz JM, Perez EM, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171(2):305–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rapp AK, Casewit CJ, Colwell KS, Goddard WA, Skiff WM. 1992. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc 114(25):10024–35 [Google Scholar]
- 158.Reddy T, Shorthouse D, Parton DL, Jefferys E, Fowler PW, et al. 2015. Nothing to sneeze at: a dynamic and integrative computational model of an influenza A virion. Structure 23(3):584–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Robustelli P, Piana S, Shaw DE. 2018. Developing a molecular dynamics force field for both folded and disordered protein states. PNAS 115(21):E4758–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ruggerone P, Murakami S, Pos KM, Vargiu AV. 2013. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem 13(24):3079–100 [DOI] [PubMed] [Google Scholar]
- 161.Rothemund PWK. 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440(7082):297–302 [DOI] [PubMed] [Google Scholar]
- 162.Savelyev A, MacKerell AD. 2015. Competition among Li+, Na+, K+, and Rb+ monovalent ions for DNA in molecular dynamics simulations using the additive CHARMM36 and Drude polarizable force fields. J. Phys. Chem. B 119(12):4428–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schalch T, Duda S, Sargent DF, Richmond TJ. 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436(7047):138–41 [DOI] [PubMed] [Google Scholar]
- 164.Scheraga HA. 2011. Respice, Adspice, and Prospice. Annu. Rev. Biophys 40:1–39 [DOI] [PubMed] [Google Scholar]
- 165.Schlick T 2009. Molecular dynamics-based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules. F1000 Biol. Rep 1:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schlick T 2009. Monte Carlo, harmonic approximation, and coarse-graining approaches for enhanced sampling of biomolecular structure. F1000 Biol. Rep 1:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schlick T 2010. Molecular Modeling and Simulation: An Interdisciplinary Guide. Berlin: Springer. 2nd ed. [Google Scholar]
- 168.Schlick T 2013. The 2013 Nobel Prize in Chemistry celebrates computations in chemistry and biology. SIAM News 46:1–4 [Google Scholar]
- 169.Schlick T 2018. Adventures with RNA graphs. Methods 143:16–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schlick T 2020. Eight suggestions for future leaders of science and technology. Biophysicist 1(1):1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schlick T, Collepardo-Guevara R, Halvorsen LA, Jung S, Xiao X. 2011. Biomolecular modeling and simulation: a field coming of age. Q. Rev. Biophys 44(2):191–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schlick T, Portillo-Ledesma S. 2020. Biomolecular modeling thrives in the age of technology. Nat. Comput. Sci In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schlick T, Pyle AM. 2017. Opportunities and challenges in RNA structural modeling and design. Biophys. J 113(2):225–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schlick T, Zhu Q, Jain S, Yan S. 2020. Structure-altering mutations of the SARS-CoV-2 frame shifting RNA element. Biophys. J In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Seeman NC. 1982. Nucleic acid junctions and lattices. J. Theor. Biol 99(2):237–47 [DOI] [PubMed] [Google Scholar]
- 176.Seeman NC, Sleiman HF. 2017. DNA nanotechnology. Nat. Rev. Mater 3(1):17068 [Google Scholar]
- 177.Sengupta D, Chattopadhyay A. 2015. Molecular dynamics simulations of GPCR–cholesterol interaction: an emerging paradigm. Biochim. Biophys. Acta Biomembr 1848(9):1775–82 [DOI] [PubMed] [Google Scholar]
- 178.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, et al. 2019. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins 87(12):1141–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, et al. 2020. Improved protein structure prediction using potentials from deep learning. Nature 577(7792):706–10 [DOI] [PubMed] [Google Scholar]
- 180.Shaw DE, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, et al. 2008. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM 51(7):91–97 [Google Scholar]
- 181.Shaw DE, Grossman JP, Bank JA, Batson B, Butts JA, et al. 2014. Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. In SC14: International Conference for High Performance Computing, Networking, Storage and Analysis, pp. 41–53. Piscataway, NJ: IEEE [Google Scholar]
- 182.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, et al. 2010. Atomic-level characterization of the structural dynamics of proteins. Science 330(6002):341–46 [DOI] [PubMed] [Google Scholar]
- 183.Sirur A, De Sancho D, Best RB. 2016. Markov state models of protein misfolding. J. Chem. Phys 144(7):075101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Smith MD, Smith JC. 2020. Repurposing therapeutics for COVID-19: supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv 11871402. https://doi.org/10.26434.chemrxiv.11871402.v4 [Google Scholar]
- 185.Song D, Wang W, Ye W, Ji D, Luo R, Chen H-F. 2017. ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins. Chem. Biol. Drug Des 89(1):5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Song F, Chen P, Sun D, Wang M, Dong L, et al. 2014. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344(6182):376–80 [DOI] [PubMed] [Google Scholar]
- 187.Song X, Jensen MØ, Jogini V, Stein RA, Lee C-H, et al. 2018. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 556(7702):515–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stillinger FH, Rahman A. 1974. Improved simulation of liquid water by molecular dynamics. J. Chem. Phys 60(4):1545–57 [Google Scholar]
- 189.Stranges PB, Kuhlman B. 2013. A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Sci. 22(1):74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sun D, Forsman J, Woodward CE. 2015. Evaluating force fields for the computational prediction of ionized arginine and lysine side-chains partitioning into lipid bilayers and octanol. J. Chem. Theory Comp 11(4):1775–91 [DOI] [PubMed] [Google Scholar]
- 191.Sun H 1998. COMPASS: an ab initio force-field optimized for condensed-phase applications—overview with details on alkane and benzene compounds. J. Phys. Chem. B 102(38):7338–64 [Google Scholar]
- 192.Tautermann CS, Seeliger D, Kriegl JM. 2015. What can we learn from molecular dynamics simulations for GPCR drug design? Comput. Struct. Biotechnol. J 13:111–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vargiu AV, Collu F, Schulz R, Pos KM, Zacharias M, et al. 2011. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J. Am. Chem. Soc 133(28):10704–7 [DOI] [PubMed] [Google Scholar]
- 194.Vargiu AV, Nikaido H. 2012. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. PNAS 109(50):20637–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vendruscolo M, Dobson CM. 2011. Protein dynamics: Moore’s law in molecular biology. Curr. Biol 21(2):R68–70 [DOI] [PubMed] [Google Scholar]
- 196.Walker B, Jing Z, Ren P. 2020. Molecular dynamics free energy simulations of ATP:Mg2+ and ADP:Mg2+ using the polarisable force field AMOEBA. Mol. Simul In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang A, Zhang Z, Li G. 2018. Higher accuracy achieved in the simulations of protein structure refinement, protein folding, and intrinsically disordered proteins using polarizable force fields. J. Phys. Chem. Lett 9(24):7110–16 [DOI] [PubMed] [Google Scholar]
- 198.Wang Q, Irobalieva RN, Chiu W, Schmid MF, Fogg JM, et al. 2017. Influence of DNA sequence on the structure of minicircles under torsional stress. Nucleic Acids Res. 45(13):7633–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Warshel A, Levitt M. 1976. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol 103(2):227–49 [DOI] [PubMed] [Google Scholar]
- 200.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. 2016. Multiperspective smFRET reveals ratedetermining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol 23(4):333–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Williford J-M, Santos JL, Shyam R, Mao H-Q. 2015. Shape control in engineering of polymeric nanoparticles for therapeutic delivery. Biomater. Sci 3:894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Woys AM, Almeida AM, Wang L, Chiu C-C, McGovern M, et al. 2012. Parallel β-sheet vibrational couplings revealed by 2D IR spectroscopy of an isotopically labeled macrocycle: quantitative benchmark for the interpretation of amyloid and protein infrared spectra. J. Am. Chem. Soc 134(46):19118–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yaseen A, Nijim M, Williams B, Qian L, Li M, et al. 2016. FLEXc: protein flexibility prediction using context-based statistics, predicted structural features, and sequence information. BMC Bioinformat. 17(8):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Young MA, Beveridge DL. 1998. Molecular dynamics simulations of an oligonucleotide duplex with adenine tracts phased by a full helix turn. J. Mol. Biol 281(4):675–87 [DOI] [PubMed] [Google Scholar]
- 205.Yu K, Jiang T, Cui Y, Tajkhorshid E, Hartzell HC. 2019. A network of phosphatidylinositol 4,5-bisphosphate binding sites regulates gating of the Ca2+-activated Cl- channel ANO1 (TMEM16A). PNAS 116(40):19952–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yu W, Lopes PEM, Roux B, MacKerell AD. 2013. Six-site polarizable model of water based on the classical Drude oscillator. J. Chem. Phys 138(3):034508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yusufova N, Soshnev AA, Kloetgen A, Teater M, Osunsade A, et al. 2020. H1 deficiency drives lymphoma through disruption of 3D chromatin architecture. Nature. In press [Google Scholar]
- 208.Zhang C, Bell D, Harger M, Ren P. 2017. Polarizable multipole-based force field for aromatic molecules and nucleobases. J. Chem. Theory Comput 13(2):666–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang C, Lu C, Jing Z, Wu C, Piquemal J-P, et al. 2018. AMOEBA polarizable atomic multipole force field for nucleic acids. J. Chem. Theory Comp 14(4):2084–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


