Yang and Wang review the mechanisms regulating lysosome biogenesis and functions during the cellular response to diverse environmental cues and in organismal development and aging.
Abstract
Lysosomes are degradation centers and signaling hubs in cells and play important roles in cellular homeostasis, development, and aging. Changes in lysosome function are essential to support cellular adaptation to multiple signals and stimuli. Therefore, lysosome biogenesis and activity are regulated by a wide variety of intra- and extracellular cues. Here, we summarize current knowledge of the regulatory mechanisms of lysosome biogenesis, including synthesis of lysosomal proteins and their delivery via the endosome–lysosome pathway, reformation of lysosomes from degradative vesicles, and transcriptional regulation of lysosomal genes. We survey the regulation of lysosome biogenesis in response to nutrient and nonnutrient signals, the cell cycle, stem cell quiescence, and cell fate determination. Finally, we discuss lysosome biogenesis and functions in the context of organismal development and aging.
Introduction
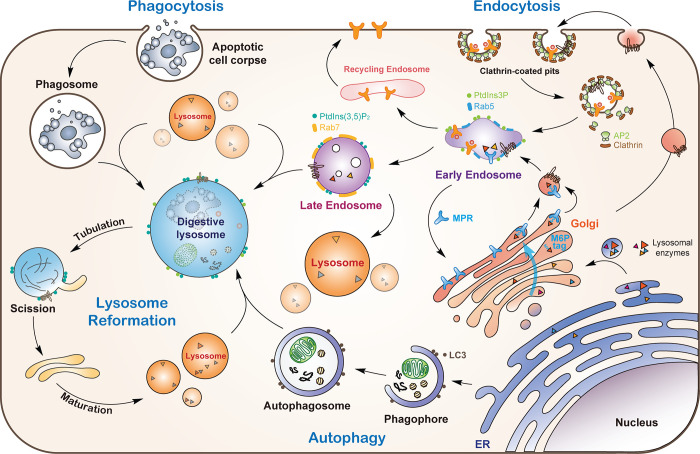
The Nobel Prize–winning discovery by de Duve in the 1950s established lysosomes as central degradative and metabolic organelles in cells (de Duve, 2005). Lysosomes are single membrane–limited, dynamic, heterogeneous organelles, which vary in their positioning, morphology, size, enzyme content, and substrates. Lysosomal membranes contain hundreds of integral and peripheral membrane proteins, including diverse transporters and ion channels (Bagshaw et al., 2005; Chapel et al., 2013). The acidic lysosomal lumen is maintained by the lysosomal multi-subunit V-ATPase. This low pH of 4.5–5.5 enables the activation of >50 intralysosomal hydrolases, which digest macromolecules including proteins, nucleic acids, lipids, and carbohydrates. Lysosomes receive and digest materials generated by endocytosis of small molecules and cell surface proteins, phagocytosis of large particles such as apoptotic cell corpses and pathogenic bacteria, or autophagy of cytoplasmic contents including damaged mitochondria, ER, and lysosomes (Bright et al., 2016; Chen et al., 2010; Cinque et al., 2020; Khaminets et al., 2015; Liu et al., 2012; Maejima et al., 2013; Pickles et al., 2018; Xu et al., 2019; Fig. 1). The digestion products are exported and reused as building blocks to maintain cellular homeostasis. Consequently, lysosomes have long been regarded as the cell’s recycling bins. Lysosome dysfunction causes a wide variety of human disorders such as lysosomal storage diseases (LSDs) and neurodegenerative diseases (Appelqvist et al., 2013; Bajaj et al., 2019; Cox and Cachón-González, 2012; Platt et al., 2012).
Figure 1.
Lysosomes receive proteins and cargos from multiple pathways. Lysosomal hydrolases are synthesized and modified by linkage with oligosaccharides in the ER and transported to the Golgi apparatus. Following recognition of the mannose residues in the oligosaccharide chain by MPR, the hydrolase–MPR complexes are delivered to early endosomes. Newly synthesized lysosomal membrane proteins are either sorted at the TGN and delivered to endosomes (direct pathway) or first delivered to the plasma membrane and then endocytosed to reach early endosomes (indirect pathway). Receptors not destined for lysosomes are recycled back to the plasma membrane or Golgi. Early endosomes undergo a conversion to late endosomes, which then fuse with lysosomes. Phagocytosed cargos are enclosed in phagosomes, which undergo a maturation process and fuse with lysosomes. Autophagic cargos are delivered to lysosomes by fusion of autophagosomes with lysosomes. Lysosomes reform from digestive lysosomes (endo-, phago-, and autolysosomes) by tubulation and scission to form protolysosomes, which mature into functional lysosomes.
Lysosomes play important roles in numerous cellular processes rather than functioning merely as recycling bins. They serve as signaling hubs that are critical for energy and amino acid sensing, signal transduction, and autophagy regulation (Perera and Zoncu, 2016). In addition, lysosomes interact with other intracellular organelles (e.g., mitochondria, ER) for mutual homeostatic regulation (Bonifacino and Neefjes, 2017; Wong et al., 2019). Lysosomes are involved in secretion and plasma membrane repair (Andrews et al., 2014). They also play important roles in tumorigenesis, immune responses, and many other physiological or pathological processes (Hipolito et al., 2018; Perera et al., 2019). Because of their essential roles in cellular homeostasis, lysosomes must adapt to extra- and intracellular cues to maintain their own homeostasis. Lysosome biogenesis is one of the most important mechanisms for lysosomal adaptation. It increases lysosome numbers to meet different cellular demands such as starvation-induced autophagy and the dispensation of lysosomes to daughter cells during cell division. In this review, we summarize our current understanding of lysosome biogenesis, including synthesis of lysosomal proteins and their endosomal-lysosomal delivery, transcriptional regulation, and the role of lysosome biogenesis in the context of cellular and organismal development and aging, particularly in mammals and the model organism Caenorhabditis elegans.
Lysosomal protein delivery and lysosomal fusion
Lysosome biogenesis requires the coordination of lysosomal protein biosynthesis and endosome-lysosome trafficking (Fig. 1). Lysosomal hydrolases are first synthesized and modified by linkage with oligosaccharides in the ER. They are then transported to the Golgi apparatus, where mannose residues in the oligosaccharide chain on most lysosomal hydrolases are phosphorylated and recognized by the mannose 6-P receptor (MPR). The hydrolase–MPR complex is then enclosed in clathrin-coated vesicles at the TGN and delivered to early endosomes. Some hydrolases are sent to endosomes in an MPR-independent manner (Braulke and Bonifacino, 2009; Luzio et al., 2014). Lysosomal delivery of newly synthesized lysosomal membrane proteins occurs through both direct and indirect pathways. In the direct pathway, lysosomal membrane proteins are sorted at the TGN and delivered directly to endosomes, while the indirect pathway involves delivery to the plasma membrane before undergoing endocytosis to reach early endosomes (Braulke and Bonifacino, 2009; Luzio et al., 2014; Saftig and Klumperman, 2009). To deliver endosomal proteins to lysosomes, early endosomes undergo conversion to late endosomes, which is marked by switching of the early endosome–specific Rab5 and phosphatidylinositol 3-phosphate (PtdIns3P) to late endosome-specific Rab7 and phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2; Gillooly et al., 2000; Huotari and Helenius, 2011; Schink et al., 2016). Proteins not destined for lysosomes are recycled back to the plasma membrane or Golgi via retromer-dependent and -independent pathways (McNally and Cullen, 2018). The Rab switch involves Rab5 inactivation by TBC-2 and Rab7 activation by the Sand1–Ccz1 complex (Chotard et al., 2010; Li et al., 2009; Nordmann et al., 2010; Poteryaev et al., 2010; Rink et al., 2005). Endosomal PtdIns3P synthesis is suppressed by the interaction of WDR91/WDR81 with the Vps34 complex, allowing the production of PtdIns(3,5)P2 from PtdIns3P accompanied by degradation of PtdIns3P (Liu et al., 2016; Liu et al., 2017; Rapiteanu et al., 2016). WDR91 serves as a Rab7 effector to couple the endosomal Rab switch with PtdIns3P down-regulation during endosome conversion (Liu et al., 2017). For late endosome–lysosome fusion, the Rab7 small GTPase, homotypic fusion and protein sorting (HOPS) multi-subunit tethering complex, and SNARE proteins are generally required, and sometimes the kiss-and-run mechanism is also involved (Lürick et al., 2018; Luzio et al., 2010).
Phagosomes containing apoptotic cell corpses or other particles undergo a maturation process, in which phagocytic receptors are recycled and phagosomal lipids and proteins are changed, so as to fuse with lysosomes for degradation of phagosomal contents (Wang and Yang, 2016; Fig. 1). Autophagosomes similarly undergo maturation for fusion with late endosomes/lysosomes, essentially using similar sets of proteins (Rab7/HOPS/SNAREs) as in endosome–lysosome fusion (Fig. 1). Autophagosome-specific proteins are also involved, including LC3/Atg8, ATG14, and autophagosome-related SNARE proteins (Zhao and Zhang, 2019). Notably, fusion of autophagosomes with endosomes/lysosomes is regulated by nutrients. In C. elegans and mammalian cells, O-linked β-N-acetylglucosamine (O-GlcNAc) transferase senses nutrient signals and mediates O-GlcNAcylation of SNAP29 (Guo et al., 2014; Hanover et al., 2010). This modification reduces the interaction of SNAP29 with Stx17 and VAMP8, thus inhibiting SNARE complex assembly and autolysosome formation.
Lysosome reformation
Cargo degradation rapidly consumes the pool of lysosomes in the cell. Thus, lysosomal regeneration after degradation is a critical homeostatic mechanism for maintaining the lysosome pool (Fig. 1). Defective lysosome reformation is observed in cells from patients with LSDs (Bright et al., 1997; Durchfort et al., 2012; Lloyd-Evans et al., 2008; Luzio et al., 2014; Schmid et al., 1999) and is implicated in Parkinson’s disease (Magalhaes et al., 2016).
Endocytic lysosome reformation (ELR)
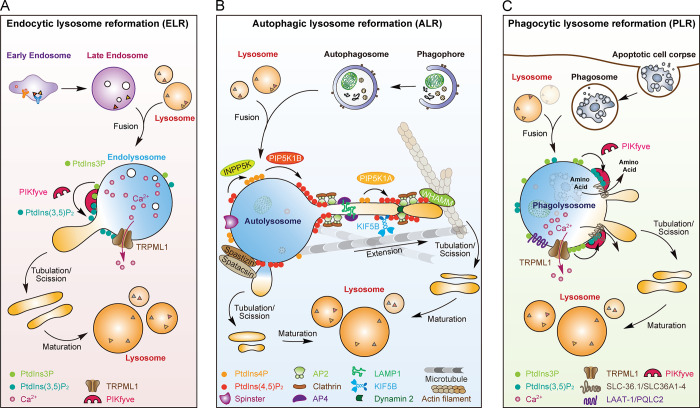
Earlier work indicated that ELR is an ATP-dependent process (Bright et al., 2005; Pryor et al., 2000). The requirement for ATP is likely to support the maintenance of a proton gradient, as inhibition of lysosomal acidification blocked lysosome reformation. Intralysosomal Ca2+ is also essential (Pryor et al., 2000). Later, it was found that the PtdIns3P 5-kinase PIKfyve and the lysosomal calcium channel TRPML1 are both required for ELR (Treusch et al., 2004; Nicot et al., 2006; Miller et al., 2015; Bissig et al., 2017). PIKfyve generates PtdIns(3,5)P2, which activates TRPML1 to control lysosomal Ca2+ efflux (Dong et al., 2010; McCartney et al., 2014; Fig. 2 A). PIKfyve, TRPML1, and mechanistic target of rapamycin (mTOR) also regulate phagosome and entotic vacuole shrinkage (Krajcovic et al., 2013; Krishna et al., 2016), which suggests that these factors are generally required for lysosome regeneration.
Figure 2.
Graphic summary of lysosome reformation. (A) ELR is regulated by PIKfyve, TRPML1, and Ca2+. PIKfyve converts PtdIns3P to PtdIns(3,5)P2, which activates TRPML1 to regulate lysosomal Ca2+ efflux required for lysosomal tubulation. (B) ALR is achieved by PtdIns(4,5)P2-, clathrin-, and AP2-mediated membrane budding on autolysosomes; KIF5B-driven elongation of membrane tubules along microtubules; dynamin 2–dependent protolysosome scission; and finally protolysosome maturation. AP4 enriches lysosomal membrane proteins for tubulation. WHAMM promotes lysosome tubulation by binding to PtdIns(4,5)P2. The sugar transporter Spinster is also involved in ALR. The generation of PtdIns(4,5)P2 from PtdIns4P is controlled by PtdIns4P 5-kinase 1B on autolysosome membranes and PtdIns4P 5-kinase 1A on protolysosomal tubules. The balance between PtdIns4P and PtdIns(4,5)P2 is also regulated by inositol polyphosphate-5-phosphatase K. Spastizin and spatacsin form a complex to promote tubule initiation on the autolysosome. (C) PLR is regulated by PIKfyve, TRPML1, and amino acid transporters (e.g., SLC-36.1/SLC36A1-4 and LAAT-1/PQLC2). PIKfyve probably regulates the activity of these lysosomal transporters through PtdIns(3,5)P2 to promote lysosome tubulation from phagolysosomes.
Autophagic lysosome reformation (ALR)
Lysosome reformation from autolysosomes, also referred to as ALR, was initially identified in starvation-induced autophagy (Yu et al., 2010). In the early stage of starvation-induced autophagy, mTOR, a major kinase regulating cell growth, is inhibited, and all lysosomes are essentially converted into autolysosomes. With prolonged starvation, mTOR is reactivated by lysosomal degradation of macromolecules. This allows autolysosomes to generate lysosomal tubules and form protolysosomes devoid of lysosomal contents. Protolysosomes subsequently mature into functional lysosomes, but the mechanisms remain to be elucidated. ALR is thus important for restoring the lysosome pool during autophagy (Yu et al., 2010). Fibroblasts derived from patients with several different LSDs were defective in mTOR reactivation and ALR.
Mechanistically, ALR is achieved by the coordinated actions of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), clathrin, and the adaptor protein complexes AP2 and AP4 (Rong et al., 2012; Fig. 2 B). On certain microdomains of the autolysosome, PtdIns(4,5)P2 is produced from the membrane-ubiquitous phosphatidylinositol 4-phosphate (PtdIns4P) by PtdIns4P 5-kinase 1B. PtdIns(4,5)P2 further recruits clathrin through the AP2 complex. On these microdomains, lysosomal membrane proteins (e.g., LAMP1) are enriched by interacting with AP4. Clathrin recruitment then facilitates the formation of PtdIns(4,5)P2-enriched microdomains, where protolysosomal tubulation initiates (Du et al., 2016; Rong et al., 2012). The kinesin heavy chain KIF5B is recruited to the microdomains by interacting with PtdIns(4,5)P2 and drives protolysosomal tubule extension along microtubules (Du et al., 2016). PtdIns(4,5)P2 also binds to WHAMM, a WASP family member that activates Arp2/3 for actin nucleation. WHAMM promotes branched actin network formation on autolysosomes, facilitating protolysosome tubulation (Dai et al., 2019). On the protolysosomal tubules, another PtdIns4P 5-kinase, PIP5K1A, generates PtdIns(4,5)P2 to trigger another round of clathrin recruitment, leading to dynamin 2–dependent protolysosome scission (Rong et al., 2012; Fig. 2 B). The balance between PtdIns4P and PtdIns(4,5)P2 on autolysosomal microdomains is critical for ALR. Loss of inositol polyphosphate-5-phosphatase K, an inositol polyphosphate 5-phosphatase that hydrolyses PtdIns(4,5)P2 to PtdIns4P, suppresses lysosome repopulation by impairing ALR, leading to muscular dystrophy (McGrath et al., 2021). ALR probably involves additional factors. For example, spastizin/SPG15 and spatacsin/SPG11, the gene products responsible for two common autosomal-recessive hereditary forms of spastic paraplegia, form a complex and localize to lysosomes through the spastizin FYVE domain, which promotes tubule initiation on the autolysosome (Chang et al., 2014; Fig. 2 B). How the spastizin–spatacsin complex cooperates with the PtdIns(4,5)P2-clathrin axis to regulate ALR remains unclear.
Phagocytic lysosome reformation (PLR)
Phagocytosis of apoptotic or living cells leads to formation of phagosomes that ultimately fuse with lysosomes (Wang and Yang, 2016). In bone marrow–derived macrophages, it was observed that degradation of phagocytosed contents (e.g., erythrocytes) permitted the shrinkage of phagolysosomes concomitant with reformation of the tubuloreticular network of lysosomes, while nondegradable contents (e.g., latex beads) prevented reformation of the tubular network (Knapp and Swanson, 1990). Studies in mammalian cells and C. elegans revealed that PIKfyve, TRPML1, and SLC-36.1 are essential regulators of PLR (Fig. 2 C). In mammalian cells, inactivation of PIKfyve impairs the shrinkage of phagolysosomes and entotic vacuoles but does not affect events upstream of phagolysosome formation (Krishna et al., 2016). Inhibiting the lysosomal Ca2+ channel TRPML1 has a similar effect on phagolysosome shrinkage. TRPML1 overexpression bypasses the defective phagolysosome shrinkage induced by loss of PIKfyve. Thus, PIKfyve likely functions in part through TRPML1 to regulate phagolysosome shrinkage (Krishna et al., 2016). In C. elegans, genetic screening identified PPK-3, the worm orthologue of PIKfyve, and SLC-36.1, the homologue of the mammalian neutral amino acid transporters SLC36A1-4/PAT1-4, as essential regulators of PLR (Gan et al., 2019). In wild-type embryos, tubulation occurs on cell corpse–containing phagolysosomes. However, ppk-3 and slc-36.1 mutants accumulate phagolysosomal vacuoles that fail to generate lysosomal tubules. SLC-36.1 localizes to lysosomes and forms a complex with PPK-3 to regulate PLR. In ppk-3 and slc-36.1 mutant embryos, there are fewer free lysosomes in the engulfing cells, which suggests that PLR is important for maintaining the regular number of lysosomes. In addition, PIKfyve/PPK-3 and SLC-36.1 are required for ALR in C. elegans adult hypodermis, as their loss leads to accumulation of many enlarged autolysosomes that fail to generate lysosome reformation tubules (Gan et al., 2019).
The identification of PIKfyve, TRMPL1, and SLC-36.1/SLC30A1-4 suggests that the mechanisms of lysosome reformation from different types of digestive lysosomes likely share common regulators (Figs. 1 and 2). Notably, export of amino acids from phago- and autolysosomes is not only important for lysosome reformation but also essential for embryonic development and cell survival (Gan et al., 2019; Krishna et al., 2016; Liu et al., 2012). The promotion of nutrient export by PIKfyve from phagolysosomes concomitant with lysosome reformation is likely achieved through lysosomal transporters, such as the neutral amino acid transporter SLC-36.1/SLC30A1-4, the basic amino acid exporter LAAT-1/PQLC2 (Liu et al., 2012), and the lysosomal sugar transporter Spinster (Rong et al., 2011; Fig. 2, B and C). It remains to be investigated whether PIKfyve regulates these lysosomal transporters through PtdIns(3,5)P2 and whether additional lysosomal exporters are involved in lysosome reformation.
Transcriptional regulation of lysosome biogenesis
To meet the need for cellular degradation, lysosomes increase their numbers and sizes by transcriptional activation of lysosomal and autophagy genes. This is mainly achieved by coordinated actions of transcription factors, transcription repressors, and epigenetic regulators.
TFEB/TFE3 and coordinated expression and regulation of lysosomal genes
In mammalian cells, the promoter regions of many lysosomal and autophagic genes share one or more 10–base pair sequence (5′-GTCACGTGAC-3′), which was named the CLEAR (coordinated lysosomal expression and regulation) element (Sardiello et al., 2009). The CLEAR consensus sequence contains the E-box sequence (5′-CACGTG-3′) that is targeted by members of the MiT/TFE (microphthalmia/transcription factor E) family of basic helix-loop-helix transcription factors. This family comprises microphthalmia-associated transcription factor, TFEB, TFE3, and TFEC (Hemesath et al., 1994). Among them, TFEB and TFE3 bind to CLEAR elements to activate lysosomal gene expression, thus promoting lysosome biogenesis (Martina et al., 2014; Palmieri et al., 2011; Sardiello et al., 2009). Microphthalmia-associated transcription factor was also reported to promote endolysosome biogenesis in melanoma cells (Ploper et al., 2015). Importantly, TFEB/TFE3 transcriptionally activate genes involved in autophagosome biogenesis and autophagosome–lysosome fusion, thereby coupling autophagy with lysosome biogenesis (Martina et al., 2014; Settembre et al., 2011). Reinforced expression of TFEB/TFE3 enhances the degradation of long-lived proteins (Settembre et al., 2011), lipid droplets, and damaged mitochondria (Nezich et al., 2015; Settembre et al., 2013). TFEB/TFE3 also facilitate lysosome exocytosis (Martina et al., 2014; Medina et al., 2011), which mediates plasma membrane repair during cell injury.
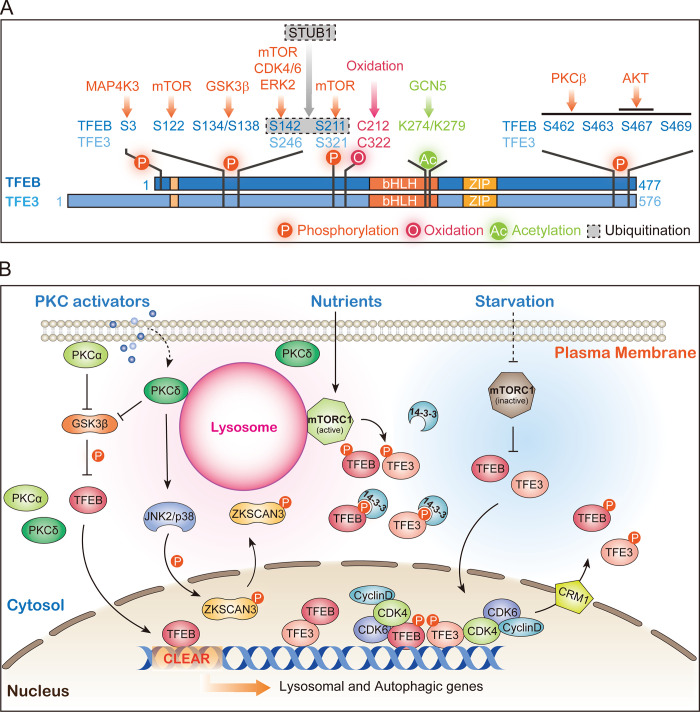
TFEB/TFE3 activity is regulated by their nuclear-cytoplasmic shuttling (Fig. 3). Under normal conditions, TFEB/TFE3 are phosphorylated by multiple kinases, including mTOR, ERK, GSK3β, Akt, and PKC, which likely mediate different extra- or intracellular signals (Puertollano et al., 2018; Fig. 3 A). Phosphorylated TFEB/TFE3 can be dephosphorylated by the calcineurin phosphatase or protein phosphatase 2A (Chen et al., 2017; Martina and Puertollano, 2018; Medina et al., 2015). Dephosphorylated TFEB/TFE3 translocate into the nucleus and activate expression of autophagy and lysosomal genes (Fig. 3 B). GSK3β specifically phosphorylates TFEB but not TFE3 in the PKC-GSK3β signaling cascade, which distinguishes the role of TFEB from TFE3 in lysosome biogenesis (Li et al., 2016). Nuclear TFEB/TFE3 can be rephosphorylated and exported to the cytoplasm in a manner dependent on the exportin CRM1 (Li et al., 2018; Napolitano et al., 2018; Yin et al., 2020; Fig. 3 B). Thus, the phosphorylation–dephosphorylation balance regulates TFEB/TFE3 activities by controlling their subcellular localizations. TFEB is also regulated by ubiquitination and acetylation (Fig. 3 A). STUB1, a chaperone-dependent E3 ubiquitin ligase, targets phosphorylated TFEB for degradation through the ubiquitin-proteasome pathway, thereby regulating the steady state of phosphorylated TFEB (Sha et al., 2017). The histone acetyltransferase GCN5 acetylates TFEB at Lys274 and Lys279, disrupting TFEB dimerization and binding to target genes (Wang et al., 2020b). TFEB/TFE3 also undergo oxidation at Cys212 (TFEB) or Cys322 (TFE3) in their 14-3-3–binding motifs in response to changes in redox balance (Martina et al., 2021; Wang et al., 2020a). More recently, TFEB was found to act through liquid-liquid phase separation (LLPS) in the nucleus, forming transcriptional condensates containing the Mediator complex and TFEB target gene mRNAs (Chen et al., 2020). IPMK1, an inositol polyphosphate multikinase, negatively regulates lysosomal biogenesis and autophagy by interacting with and inhibiting LLPS of TFEB, leading to dissolution of TFEB condensates. TFE3 similarly undergoes LLPS but is not regulated by IMPK1 (Chen et al., 2020). Thus, like GSK3β, IMPK1 regulation of lysosome biogenesis is specific to TFEB, which suggests that TFEB and TFE3 can be regulated differently despite their highly similar amino acid sequences.
Figure 3.
TFEB/TFE3-dependent lysosome biogenesis. (A) Regulatory modifications of TFEB/TFE3. TFEB/TFE3 are phosphorylated (orange arrows), acetylated (green arrow), and ubiquitinated (gray arrow) by the indicated enzymes. Direct oxidation by reactive oxygen species can also occur (red arrow). (B) mTOR-dependent and PKC-dependent lysosome biogenesis pathways. Under nutrient-rich conditions, TFEB/TFE3 is phosphorylated by lysosome-localized mTOR. Phosphorylated TFEB/TFE3 bind to 14-3-3 proteins and are sequestered in the cytoplasm. With starvation, mTOR is inactivated and unable to phosphorylate TFEB/TFE3, leading to their nuclear translocation and activation. Activation of PKC leads to phosphorylation and inactivation of GSK3β, which then fails to phosphorylate TFEB, thereby inducing nuclear translocation of TFEB. Activated PKC induces JNK2/p38 activation, which phosphorylates ZKSCAN3 and leads to its cytoplasmic translocation and de-repression of lysosomal genes. Nuclear TFEB/TFE3 are rephosphorylated by CDK4/6 (and probably also by mTOR and GSK3β) and relocate back to the cytoplasm in a CRM1-dependent manner.
STAT3 and lysosomal hydrolases
Lysosomes contain three major types of proteolytic hydrolases: papain-like cysteine proteases (e.g., cathepsin B and L), pepsin-related aspartyl proteases (e.g., cathepsin D and E), and asparaginyl endopeptidase. Deficiency in these key lysosomal proteases or lysosomal protein overload leads to generation of lysosomal reactive oxygen species and oxidative stress, which activates STAT3 rather than TFEB. Activated STAT3, which is phosphorylated, binds to the promoters of the genes encoding these three families of lysosomal proteases to trigger their expression (Martínez-Fábregas et al., 2018). Oxidative stress–induced activation of STAT3 does not trigger expression of autophagy genes, which suggests that autophagy is not involved when elevated lysosomal capacity is required for digesting accumulated substrates received from endocytosis (micropinocytosis) or resulting from deficiency of key lysosomal proteases. STAT3-dependent activation of lysosomal proteases also contributes to lysosome-mediated and caspase-independent cell death. In postlactational regression of the mammary gland, STAT3 up-regulates cathepsins B and L but down-regulates their inhibitor Spi2A. This probably increases lysosomal membrane permeability, which, together with the decreased expression of LAMP1 and LAMP2, leads to lysosomal cell death (Kreuzaler et al., 2011). Thus, STAT3-mediated transcription of lysosomal proteases regulates lysosomal functions in a manner that is subject to stress or developmental context. Intriguingly, STAT3 was reported to interact with V-ATPase to enhance its activity upon cytosolic acidification, which suggests that STAT3 regulates intracellular and lysosomal pH independently of its transcriptional activity (Liu et al., 2018).
MYC and ZKSCAN3 are transcriptional repressors of lysosomal genes
The fact that the CLEAR element contains the E-box consensus sequence suggests the possibility that E-box–binding transcription factors might be involved in TFEB/TFE3-mediated lysosome biogenesis. Indeed, MYC, a basic helix-loop-helix transcription factor important for cell proliferation and many other cellular processes, antagonizes TFEB/TFE3 transcriptional activities (Annunziata et al., 2019). MYC represses lysosome biogenesis by occupying the promoters of lysosomal genes, including TFEB and TFE3. Down-regulating MYC allows TFEB/TFE3 to bind to lysosomal gene promoters. MYC also interacts with histone deacetylases (HDACs) on the promoters of lysosomal genes, which epigenetically regulates lysosomal gene expression. The antagonistic effect of MYC-HDACs on TFEB/TFE3 likely contributes to cell fate determination. In neoplastic cells expressing nuclear MYC and HDAC2, TFE3 is more frequently cytoplasmic. In human induced pluripotent stem cells (iPSCs), MYC and HDAC2 levels are increased, whereas TFEB levels are reduced. In contrast, TFEB/TFE3-activated lysosome biogenesis is essential for maintenance of the differentiated state of cells (Annunziata et al., 2019). Thus, MYC probably helps to maintain lysosome activity within a given range that favors a context-specific cell state. ZKSCAN3 (ZNF306) is a DNA-binding protein with tandem zinc fingers, a KRAB domain that represses gene expression, and a SCAN3 domain. ZKSCAN3 is normally nuclear, and its inactivation increases the transcription of a subset of autophagy and lysosome genes. The promoters of many of these genes contain multiple ZKSCAN3-binding sites (KRDGGG, K: G/T, R: A/G, D: A/G/T; Yang et al., 2008). Furthermore, silencing ZKSCAN3 enhances TFEB-dependent lysosomal gene expression. Thus, ZKSCAN3 probably acts in conjunction with TFEB to negatively regulate the expression of lysosomal and autophagy genes (Chauhan et al., 2013).
Epigenetic regulation of lysosomal genes
The expression of lysosomal and autophagy genes involves other epigenetic regulators in addition to the aforementioned MYC-HDAC complexes. Nuclear TFEB activity is enhanced by elevated levels of CARM1, a protein arginine methyltransferase that catalyzes dimethylation of histone H3 at Arg17 (Shin et al., 2016). Glucose starvation leads to an increase in genome-wide H3 Arg17 dimethylation associated with the nuclear translocation of CARM1. CARM1 interacts with TFEB and serves as a coactivator (Shin et al., 2016). Glucose starvation also activates ACSS2 by AMPK-mediated phosphorylation. Phosphorylated ACSS2 translocates into the nucleus and interacts with TFEB at the promoter regions of lysosomal and autophagy genes, where ACSS2 locally produces acetyl-CoA for histone acetylation (Li et al., 2017). In contrast to CARM1 and ACSS2, BRD4 negatively regulates lysosomal and autophagy genes independently of TFEB/TFE3 (Sakamaki et al., 2017). BRD4 binds to acetylated histones at the promoters of autophagy and lysosome genes and recruits the histone methyltransferase G9a to catalyze mono- and dimethylation of histone H3 at Lys9 (H3K9), thereby repressing lysosomal and autophagy gene expression.
mTOR-dependent nutrient sensing and regulation of lysosome biogenesis
The lysosomal surface is a residential site of mTOR, which suggests that lysosomes are important signaling hubs in cells (Perera and Zoncu, 2016; Sancak et al., 2010; Sancak et al., 2008). mTORC1 is recruited to lysosomes by a heterodimeric complex consisting of GTP-bound RagA/B and GDP-bound RagC/D, where it is activated by the Rheb GTPase (Angarola and Ferguson, 2019; Kim et al., 2008; Menon et al., 2014; Sancak et al., 2010; Sancak et al., 2008). The multi-subunit complex Ragulator and GATOR1 were respectively identified as the guanine exchange factor and GTPase activating protein (GAP) of RagA/B (Bar-Peled et al., 2013; Bar-Peled et al., 2012). While the guanine exchange factor of RagC/D is not known, the Folliculin-FNIP1/2 complex functions as the GAP of RagC/D (Petit et al., 2013; Tsun et al., 2013). Current studies revealed that formation of the active mTORC1-recruiting RagA/B-RagC/D complex is regulated by amino acids, allowing mTOR to sense changes in cytoplasmic or lysosomal amino acid levels. Sensing of cytoplasmic amino acids is achieved by a negative protein interaction cascade that directs GAP activity toward RagA/B. In the absence of nutrients, Sestrin2 and CASTOR1/2 inhibit GATOR2. GATOR2 is an inhibitor of GATOR1, and therefore nutrient deprivation activates the GAP function of GATOR1, which in turn inactivates RagA/B and blocks mTORC1 recruitment to the lysosome. Thus, mTOR is inactivated. In nutrient-rich cells, the inhibitory role of Sestrin2 and CASTOR1/2 on GATOR2 is abolished by leucine and arginine, respectively. GATOR2 then inhibits the GAP activity of GATOR1, allowing RagA/B in the GTP-loaded active form to interact with GDP-loaded RagC/D, thus recruiting and activating mTORC1 on the lysosome (Chantranupong et al., 2016; Wolfson et al., 2016). The lysosome transporter SLC38A9 functions as a lysosomal arginine sensor. It interacts with the V-ATPase complex and Ragulator, allowing RagA/B activation and consequently mTOR recruitment and activation on lysosomes (Rebsamen et al., 2015; Wang et al., 2015a; Zoncu et al., 2011).
When amino acids are available, TFEB/TFE3 are recruited to lysosomes by interacting with active Rag GTPase complexes (Martina and Puertollano, 2013). On lysosomes in nutrient-rich cells, mTOR phosphorylates TFEB at Ser142 and Ser211 and TFE3 at Ser321 (Fig. 3). Phosphorylated TFEB/TFE3 are released into the cytosol and bind to 14-3-3 proteins. TFEB/TFE3 are thus sequestered from the nucleus and are inactive (Martina et al., 2012; Martina et al., 2014; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). In addition, MAP4K3 phosphorylates TFEB at Ser3 to promote its interaction with the mTORC1–Ragulator complex and its subsequent phosphorylation at Ser211 (Hsu et al., 2018). mTOR may also phosphorylate TFEB at Ser122 (Vega-Rubin-de-Celis et al., 2017). In nutrient-deficient cells, the Rag GTPase complex is inactivated and fails to recruit either mTORC1 or TFEB/TFE3 onto the lysosomal surface. Thus, mTOR is inactive and unable to phosphorylate TFEB/TFE3. Dephosphorylated TFEB/TFE3 translocate into the nucleus and activate the expression of lysosomal and autophagy genes (Fig. 3 B). This “lysosome-to-nucleus” signaling thus regulates lysosome biogenesis and autophagy in response to nutrient signals. Additionally, nutrient deprivation induces lysosomal Ca2+ release through TRPML1/MCOLN1, which activates the Ca2+-dependent phosphatase calcineurin. Calcineurin then dephosphorylates TFEB and induces its nuclear translocation (Medina et al., 2015; Wang et al., 2015b).
mTOR-independent regulation of lysosome biogenesis
In many cases, lysosome biogenesis occurs even when mTORC1 is active. A PKC-dependent and mTOR-independent mechanism for lysosome biogenesis was identified by screening lysosome-inducing compounds (Li et al., 2016). PKC-activating diterpenes act through PKCα and PKCδ to induce phosphorylation and inactivation of GSK3β, leading to dephosphorylation and nuclear translocation of TFEB (Fig. 3). GSK3β phosphorylates TFEB, but not TFE3, on Ser138 and Ser134. Thus, PKC activation distinguishes TFEB from TFE3 for lysosome biogenesis. In parallel to GSK3β inactivation and TFEB nuclear translocation, PKC activation leads to phosphorylation and activation of the MAPKs JNK2 and p38, which in turn phosphorylate ZKSCAN3 on Thr153 in a putative nuclear export signal. This induces cytoplasmic relocation of ZKSCAN3 and consequently alleviates the transcriptional repression of lysosomal genes. Thus, PKC switches on two parallel signaling pathways to activate lysosome biogenesis (Li et al., 2016; Fig. 3 B). Because of the central roles of PKC in signal transduction across the plasma membrane, this PKC-dependent and mTOR-independent mechanism probably regulates lysosome biogenesis in response to many extracellular signals. In addition, AKT was found to phosphorylate TFEB at Ser467 to repress its nuclear translocation, while trehalose decreases AKT activity to activate TFEB in an mTORC1-independent manner (Palmieri et al., 2017).
In melanoma with the most prevalent BRAF(V600E) mutation, TFEB, but not TFE3, is phosphorylated and inactivated by ERK independently of mTOR. Inhibition of BRAF leads to TFEB activation via ERK inhibition. This synergizes with phosphorylation and inactivation of ZKSCAN3 by JNK2/p38 to induce lysosome biogenesis (Li et al., 2019). In osteoclasts, PKCβ directly phosphorylates TFEB at three serine residues in the C terminus, thus stabilizing TFEB to promote its activity (Ferron et al., 2013). The cellular response to ER stress involves TFEB/TFE3 translocation to the nucleus in a manner that depends on protein kinase RNA-like ER kinase and calcineurin, but not mTORC1, to up-regulate ATF4 and other unfolded protein response genes (Martina et al., 2016). During mitophagy, translocation of TFEB/TFE3 into the nucleus depends on Parkin and Pink1 but not mTORC1. In this case, nuclear translocation of TFEB/TFE3 requires the autophagy regulators Atg5 and Atg9, but the molecular mechanisms remain elusive (Nezich et al., 2015). More recently, it was found that defective endocytosis or osmotic stress activates TFEB-dependent lysosome biogenesis without perturbing mTORC1 activity. Inhibition of endocytosis leads to accumulation of NHE7, a Na+/H+ exchanger, on the cell surface, which causes influx of Na+ and elevation of cytosolic pH. This in turn leads to elevation of Ca2+ levels, mediated by the Na+/Ca2+ exchanger NCX1, and consequently activation of calcineurin, which dephosphorylates TFEB and promotes TFEB nuclear translocation and activation (López-Hernández et al., 2020).
mTOR-independent regulation of TFEB/TFE3 also occurs in cancers. In translocation-renal cell carcinoma and alveolar soft part sarcoma, the TFE3 gene is translocated to other chromosomal loci, resulting in fusion gene products that retain the transcriptional activity of TFE3 (Perera et al., 2019). These TFE3 fusion proteins usually have much higher expression levels than wild-type TFE3 and constitutively localize in the nucleus. In an alveolar soft part sarcoma mouse model, fusion of TFE3 with ASPSCR1 up-regulated lysosomal and autophagy genes, which increased lysosomal abundance (Tanaka et al., 2017). In pancreatic ductal adenocarcinoma cells, MiT/TFE proteins constitutively localize to the nucleus regardless of nutrient availability, leading to a strong increase in lysosomes (Perera et al., 2015). Constitutively active MiT/TFE factors promote mTORC1 signaling by enhancing the expression of RagD (Di Malta et al., 2017; Napolitano et al., 2020).
Lysosome biogenesis and function in the cell cycle
Mitotic cells increase their numbers through the cell cycle, a process involving accurate DNA replication followed by division of the nucleus and partitioning of cytoplasmic contents including lysosomes. In budding yeast, vacuoles/lysosomes are transmitted from mother to daughter cells during cell division. If this fails, new vacuoles can be generated in daughter cells; indeed, a functional vacuole is required for the new daughter cell to initiate cell cycle progression through the TORC1-SCH9 pathway (Jin and Weisman, 2015). In mammals, the requirement for lysosomes in cell cycle progression is not clear. Moreover, it is poorly understood how lysosomes increase their numbers in mother cells before dispensation to daughter cells. A recent study revealed that CDK4 and 6, the cyclin D–dependent kinases that drive G1-S transition, regulate lysosome biogenesis in the cell cycle (Yin et al., 2020). Chemical inhibition and genetic inactivation of CDK4/6, but not other major cyclin-dependent kinases, leads to nuclear retention and activation of TFEB and TFE3, which enhance lysosome biogenesis and autophagy. The cyclin D–CDK4/6 complexes interact with and phosphorylate TFEB (at Ser142) and TFE3 (at Ser246) in the nucleus, promoting their CRM1-dependent nuclear export and inactivation (Fig. 3). In S, G2, and M phases, Ser142 phosphorylation of TFEB is markedly reduced, and nuclear retention of TFEB and lysosome biogenesis are greatly increased. Thus, the high activities of CDK4/6 suppress lysosome biogenesis at the G1 phase, while their low activities enhance lysosome biogenesis in the S-M phases by relieving the inhibition of TFEB and TFE3 (Yin et al., 2020). This allows the cell to coordinate DNA replication with an increased abundance of lysosomes, which can then be distributed to daughter cells by cell division. The findings that CDK4/6 inhibition induces lysosome biogenesis and autophagy have implications for the use of CDK4/6 inhibitors in cancer therapy. For example, simultaneous inhibition of CDK4/6 and autophagy/lysosome functions has synergistic effects in treatment of breast cancer and other solid tumors (Fassl et al., 2020; Vijayaraghavan et al., 2017).
Lysosome biogenesis and function in stem cell quiescence
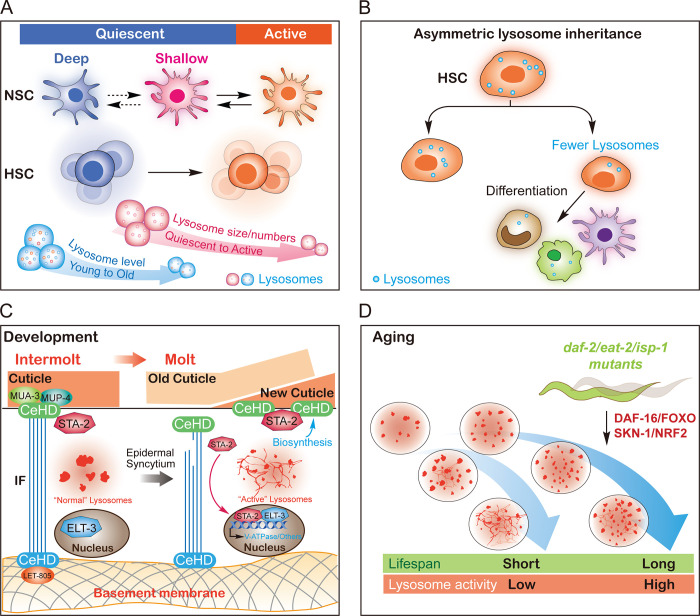
Quiescence is an important strategy to maintain somatic stem cell pools. In mouse brain, quiescent and active neural stem cells (NSCs) derived from the subventricular zone of the lateral ventricle exhibit high expression of lysosome- and proteosome-associated genes, respectively (Leeman et al., 2018). In quiescent NSCs (qNSCs), lysosomes are larger and more abundant and accumulate more protein aggregates. Impairing lysosome function causes further accumulation of protein aggregates and decreases qNSC activation in response to growth factors. In contrast, enhancing lysosome function reduces protein aggregates and enhances qNSC activation. Moreover, qNSCs exhibit age-dependent lysosomal decrease, protein aggregate increase, and decline in activation (Fig. 4 A). Reinforced TFEB expression reduces protein aggregates and promotes the activation of old qNSCs. Thus, lysosomal degradation of protein aggregates is restricted and declines with age in qNSCs, which can be triggered to promote qNSC activation (Leeman et al., 2018; Fig. 4 A). Like qNSCs, quiescent hematopoietic stem cells (qHSCs) display increased lysosomal gene expression and contain abundant enlarged lysosomes. Reducing lysosomal activity enhances the quiescence of HSCs and the in vivo potency of primed HSCs (Liang et al., 2020; Fig. 4 A).
Figure 4.
Role of lysosome biogenesis and function in development and aging. (A) The correlation between lysosomes and the different states of NSCs or HSCs. qNSCs and qHSCs have larger and more abundant lysosomes with lower degradation activity (indicated in pink). qNSCs exhibit an age-dependent decrease in lysosome levels (indicated in blue). (B) The correlation between lysosomes and the asymmetric division and differentiation of HSCs. Lysosomes are asymmetrically distributed to daughter cells, and the daughter cells with low levels of lysosomes are metabolically active and induced to differentiate. (C) STA-2– and ELT-3–dependent lysosome biogenesis in ECM remodeling during C. elegans molt. During molt, the cuticle-epidermis attachments are damaged and detected by STA-2. STA-2 translocates to the nucleus and functions together with ELT-3 to activate the expression of lysosomal V-ATPase genes. This accelerates lysosome maturation at molt and facilitates the ECM remodeling required for larval development. (D) The correlation between lysosome biogenesis and lifespan in C. elegans. In aging C. elegans, lysosomes show reduced vesicular morphology; increased tubular morphology; increased mean and total volume; and decreased acidity, motility, and degradation activity. In long-lived mutants from three different longevity pathways, lysosomal gene expression is up-regulated, which requires DAF-16/FOXO and SKN-1/NRF2. Lysosome morphology and activity are well maintained during aging.
It is currently unclear why lysosomes exhibit reduced cargo degradation in qNSCs and qHSCs and whether this leads to a feedback increase in lysosomal gene expression. In another study, however, higher lysosome protease activity was observed in different types of qNSCs in vitro (Kobayashi et al., 2019). TFEB activation decreases NSC proliferation in vitro and induces quiescence of NSCs in the hippocampal dentate gyrus of adult mouse brain. Tfeb knockout enhances the levels of activated EGFR and NotchR and increases the number of active NSCs in the dentate gyrus (Kobayashi et al., 2019). In this case, higher lysosome activity is important for adaptation and maintenance of NSC quiescence, probably by removing unwanted cellular signals. It seems paradoxical that lysosome function is required for both maintenance and exit of NSC quiescence. This may reflect the complex regulation and function of lysosomes in these processes. Accordingly, a study performed in rat embryonic fibroblasts suggests that lysosome function modulates quiescence depth as a “dimmer switch” (Fujimaki et al., 2019). Lysosomal gene expression is significantly up-regulated as quiescence deepens, whereas lysosomal degradation declines. Reducing lysosome function drives cells progressively deeper into quiescence, while increasing lysosome function pushes cells into shallower quiescence. Interestingly, expression of Mitf and Tfe3 but not Tfeb increases significantly in deep quiescence, while Mitf but not Tfe3 or Tfeb shows a high degree of coexpression with lysosomal genes in quiescence. This suggests a unique role of Mitf in regulating lysosome biogenesis in quiescent rat embryonic fibroblasts (Fujimaki et al., 2019).
Lysosome biogenesis and activity in cell fate determination
Lysosome biogenesis and activity are linked to stem cell fate determination and transition. Hyperactivation of mTORC1 drives aberrant lineage differentiation, while reprogramming of mouse and human somatic cells to iPSCs requires repression of the mTORC1 pathway (Julian and Stanford, 2020). In HSCs, lysosomes are asymmetrically distributed to daughter cells, which affects the fate of the recipient cells (Loeffler et al., 2019). The daughter cells with low levels of lysosomes are metabolically active and induced to differentiate (Fig. 4 B). The asymmetric inheritance of lysosomes may regulate mitochondrial clearance, autophagy, and Notch signaling, thus contributing to HSC fate determination (Loeffler et al., 2019). A lysosome-based signaling pathway has also been identified as a driver of mouse embryonic stem cell differentiation. Mechanistically, Ragulator and Folliculin recruit and activate RagC/D on the lysosomal surface, leading to cytoplasmic retention and inactivation of TFE3 (Villegas et al., 2019). TFE3 activation drives expression of genes related to lysosome biogenesis and metabolic signaling and represses transcriptional programs associated with periimplantation development and neural lineage differentiation. Consistent with this, perturbing delivery and maturation of lysosomal enzymes induces nuclear translocation of TFE3 and impairs cell exit from the self-renewal circuitry (Villegas et al., 2019). Together with the aforementioned antagonistic effect of MYC on TFE3 in neoplastic cells and human iPSCs (Annunziata et al., 2019), these findings suggest that regulation of lysosome biogenesis is important for the self-renewal of stem cells or neoplastic cells. In addition, lysosome-related metabolic signaling pathways are found to regulate cell-type specification in adipocytes and during embryoid body formation (Wada et al., 2016; Young et al., 2016). In the latter case, TFEB and proper lysosome function are required for endodermal specification, probably by regulating canonical Wnt signaling (Young et al., 2016). These studies suggest that lysosomes affect cell fate determination by serving as both metabolic signaling hubs and degradative compartments.
Lysosome biogenesis and animal development
It is largely unknown how lysosome biogenesis contributes to development. Recently, lysosome biogenesis and activity were reported to be up-regulated during C. elegans molt, to promote ECM remodeling for larval development (Miao et al., 2020). C. elegans larvae undergo four molts to reach adulthood. In each molt, the larva sheds and resynthesizes apical ECM/cuticle to accommodate growth. At molt, lysosomal properties alter significantly in the big multinuclear epidermal syncytium, including formation of tubular lysosomal structures and increases in lysosome abundance, motility, and degradation activity. When worms exit from molt, lysosome morphology and properties return to the typical intermolt pattern. Impairing lysosome function affects endocytic cargo degradation and cuticle collagen turnover, blunts the increase in protein synthesis at molt, and causes molting defects. Thus, lysosomes are activated specifically at molt in the epidermis to maintain tissue homeostasis by degrading old cuticle and recycling the resulting catabolites for new cuticle synthesis. Interestingly, lysosome activation is triggered when the cuticle-epidermis attachments are disrupted, and this structural damage is detected by STA-2, a signal-activated STAT family transcription factor (Zhang et al., 2015), and ELT-3, a tissue-specific generic GATA transcription factor. At molt, STA-2 and ELT-3 become more enriched in the nuclei to enhance the expression of V-ATPase genes. This accelerates lysosome maturation at molt. Thus, STA-2 and ELT-3 cooperate to provide temporal (molt) and spatial (epidermis) lysosomal activation, which facilitates the ECM remodeling required for larval development (Fig. 4 C).
The C. elegans anchor cell is a specialized uterine cell that invades the vulval tissue to initiate uterine–vulval connection during hermaphrodite development (Hagedorn and Sherwood, 2011). It is reported that lysosomes are modulated to promote basement membrane breach during anchor cell invasion. In this process, UNC-6/netrin, the guidance factor secreted by the vulval tissue, interacts with the receptor UNC-40/DCC at the basement membrane breach site to direct the formation of a transient lysosome-derived membranous protrusion for cell invasion (Naegeli et al., 2017). During meiotic maturation, sperm signaling triggers degradation of the major translational repressor GLD-1 and releases its repression of V-ATPase synthesis, leading to lysosome acidification and activation in proximal oocytes (Bohnert and Kenyon, 2017). Lysosome activation promotes clearance of protein aggregates before fertilization, thus preventing transmission of damaged materials to the next generation and providing raw materials to developing oocytes. Similar lysosome acidification is observed during Xenopus laevis oocyte maturation, suggestive of an evolutionarily conserved process that enhances oocyte proteostasis in multiple species (Bohnert and Kenyon, 2017).
Regulation of lysosome biogenesis and function in maintenance of longevity
In C. elegans, the TFEB homologue HLH-30 promotes lifespan by enhancing lysosomal lipolysis (O’Rourke and Ruvkun, 2013). HLH-30 is required for lifespan extension in C. elegans defective in multiple pathways that depend on autophagy-lysosome functions, including food intake, mitochondrial respiration, insulin/IGF-1 signaling (IIS), TOR signaling, and germline signaling (Lapierre et al., 2013; Nakamura et al., 2016). Nevertheless, further investigation is required to determine whether C. elegans HLH-30 functions similarly to TFEB/TFE3. Interestingly, HLH-30 acts in complex with the DAF-16/FOXO transcription factor to extend the lifespan under oxidative stress conditions (Lin et al., 2018). In addition, a recent study suggests that HLH-30 regulates adult reproductive diapause, a long-lived quiescent state that enables survival without food (Gerisch et al., 2020). HLH-30 is essential for the entry, survival, and recovery of adult reproductive diapause, but it is unclear whether and how lysosome biogenesis plays a role in these processes.
While lysosome activity declines with age, age-related increases in the number and size of lysosomes have been observed in various species such as Paramecium, nematodes, and different human cell lines (Brandes et al., 1972; Lipetz and Cristofalo, 1972; Sundararaman and Cummings, 1976). In C. elegans aging, lysosomes undergo a series of age-associated changes, including reduced vesicular but increased tubular morphology, increased mean and total volume, and decreased acidity, motility, and degradation activity (Sun et al., 2020). Moreover, 43 lysosomal genes exhibited reduced expression with age. Among them were genes encoding the two main classes of lysosomal proteins important for lysosomal acidity and degradation, i.e., the cathepsin proteases (17 genes) and V-ATPase subunits (15 genes). In long-lived mutants representing three different longevity pathways (IIS, caloric restriction, and impaired mitochondria function), lysosome gene expression is up-regulated, and this requires DAF-16/FOXO and SKN-1/NRF2 transcription factors (Fig. 4 D). Notably, the IIS and caloric restriction pathways seem to target lysosomal hydrolases and V-ATPase components, respectively. The genes up-regulated in the isp-1 mutant, which has dysfunctional mitochondria, are mostly shared with the IIS pathway.
Removing germ cells significantly increases lifespan in both C. elegans and Drosophila melanogaster (Kenyon, 2010). It was reported recently that lysosome activity in the C. elegans intestine is regulated by steroid signaling from the gonad. The DAF-12/DAF-9 steroid hormone signaling pathway potentiates the nuclear localization of DAF-16/FOXO in the intestine, which leads to increased expression of the V-ATPase for maintenance of lysosome acidity in reproductive worms. Moreover, neuronal expression of XBP-1s, the spliced and active form of the UPRER transcription factor, increases expression of lysosomal genes in the intestine. This enhances lysosome acidity and activity required for XBP-1s to increase proteostasis and longevity (Imanikia et al., 2019). It remains unclear how lysosomal transcripts are selectively regulated by different pathways and how this contributes to lysosome functionality in different long-lived animals.
Lysosomes can release signaling molecules to modulate longevity. Overexpression of the lysosomal acid lipase LIPA-4 promotes C. elegans longevity. LIPA-4 induces nuclear translocation of the fatty acid–binding protein LBP-8 from the lysosomal surface. Nuclear LBP-8 in turn presents oleoylethanolamine to its nuclear receptor NHR-80, which cooperates with NHR-49 to induce target gene expression and thus extend the lifespan (Folick et al., 2015). Thus, lysosomal lipid metabolism may signal to nuclear transcription to modulate longevity.
Conclusion and perspective
As reviewed here, the biogenesis of lysosomes requires integration of multiple pathways involving biosynthesis, endosome-lysosome trafficking, lysosomal degradation and reformation, and transcription regulation. Our knowledge of lysosomes and their critical roles in cellular homeostasis, development, and aging has greatly advanced in recent years. Nonetheless, many outstanding questions remain to be addressed. These include, but are not limited to, the following: (1) Although stress-responsive lysosome biogenesis is mainly regulated through TFEB/TFE3, it is unclear how lysosomal genes are regulated at the basal level. Are additional transcriptional regulators engaged in context-specific lysosome biogenesis? (2) How does the interaction of lysosomes with other intracellular organelles affect lysosome biogenesis and activity? How do the signals from organelles such as the ER, mitochondria, or nucleus affect lysosome biogenesis? (3) How do lysosome biogenesis and function contribute to animal development, especially in stem cell self-renewal and differentiation and in tissue and organ development? (4) How are lysosome biogenesis and function regulated in animal aging?
Answering these questions will provide a deeper and more thorough view of lysosome biogenesis and functions under diverse physiological and pathological conditions. In particular, the application of model animals (e.g., C. elegans and rodents) is anticipated to contribute enormously to dissecting the regulation of lysosome homeostasis at an organismal level in normal and diseased states.
Acknowledgments
The authors thank Dr. Q. Yin for help with figures and Dr. I. Hanson for proofreading the manuscript.
Research in the laboratories of X. Wang and C. Yang is supported by grants from the National Natural Science Foundation of China (91754203 and 31630018 to X. Wang; 91954204 and 31730053 to C. Yang), grants from the Ministry of Science and Technology of China (2016YFA0500203 to X. Wang; 2017YFA0503403 to C. Yang), and grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000 to X. Wang) and Yunnan Province Science and Technology Bureau (202001BB050077 to C. Yang).
The authors declare no competing financial interests.
References
- Andrews, N.W., Almeida P.E., and Corrotte M.. 2014. Damage control: Cellular mechanisms of plasma membrane repair. Trends Cell Biol. 24:734–742. 10.1016/j.tcb.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angarola, B., and Ferguson S.M.. 2019. Weak membrane interactions allow Rheb to activate mTORC1 signaling without major lysosome enrichment. Mol. Biol. Cell. 30:2750–2760. 10.1091/mbc.E19-03-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata, I., van de Vlekkert D., Wolf E., Finkelstein D., Neale G., Machado E., Mosca R., Campos Y., Tillman H., Roussel M.F., et al. 2019. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat. Commun. 10:3623. 10.1038/s41467-019-11568-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelqvist, H., Wäster P., Kågedal K., and Öllinger K.. 2013. The lysosome: From waste bag to potential therapeutic target. J. Mol. Cell Biol. 5:214–226. 10.1093/jmcb/mjt022 [DOI] [PubMed] [Google Scholar]
- Bagshaw, R.D., Mahuran D.J., and Callahan J.W.. 2005. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol. Cell. Proteomics. 4:133–143. 10.1074/mcp.M400128-MCP200 [DOI] [PubMed] [Google Scholar]
- Bajaj, L., Lotfi P., Pal R., Ronza A.D., Sharma J., and Sardiello M.. 2019. Lysosome biogenesis in health and disease. J. Neurochem. 148:573–589. 10.1111/jnc.14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled, L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., and Sabatini D.M.. 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340:1100–1106. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled, L., Schweitzer L.D., Zoncu R., and Sabatini D.M.. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig, C., Hurbain I., Raposo G., and van Niel G.. 2017. PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic. 18:747–757. 10.1111/tra.12525 [DOI] [PubMed] [Google Scholar]
- Bohnert, K.A., and Kenyon C.. 2017. A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature. 551:629–633. 10.1038/nature24620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Neefjes J.. 2017. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 47:1–8. 10.1016/j.ceb.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes, D., Murphy D.G., Anton E.B., and Barnard S.. 1972. Ultrastructural and cytochemical changes in cultured human lung cells. J. Ultrastruct. Res. 39:465–483. 10.1016/S0022-5320(72)90114-1 [DOI] [PubMed] [Google Scholar]
- Braulke, T., and Bonifacino J.S.. 2009. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 1793:605–614. 10.1016/j.bbamcr.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Bright, N.A., Davis L.J., and Luzio J.P.. 2016. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr. Biol. 26:2233–2245. 10.1016/j.cub.2016.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright, N.A., Gratian M.J., and Luzio J.P.. 2005. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15:360–365. 10.1016/j.cub.2005.01.049 [DOI] [PubMed] [Google Scholar]
- Bright, N.A., Reaves B.J., Mullock B.M., and Luzio J.P.. 1997. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J. Cell Sci. 110:2027–2040. 10.1242/jcs.110.17.2027 [DOI] [PubMed] [Google Scholar]
- Chang, J., Lee S., and Blackstone C.. 2014. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J. Clin. Invest. 124:5249–5262. 10.1172/JCI77598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong, L., Scaria S.M., Saxton R.A., Gygi M.P., Shen K., Wyant G.A., Wang T., Harper J.W., Gygi S.P., and Sabatini D.M.. 2016. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 165:153–164. 10.1016/j.cell.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapel, A., Kieffer-Jaquinod S., Sagné C., Verdon Q., Ivaldi C., Mellal M., Thirion J., Jadot M., Bruley C., Garin J., et al. 2013. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell. Proteomics. 12:1572–1588. 10.1074/mcp.M112.021980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, S., Goodwin J.G., Chauhan S., Manyam G., Wang J., Kamat A.M., and Boyd D.D.. 2013. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell. 50:16–28. 10.1016/j.molcel.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Wang Z., Zhao Y.G., Zheng H., Zhao H., Liu N., and Zhang H.. 2020. Inositol polyphosphate multikinase inhibits liquid-liquid phase separation of TFEB to negatively regulate autophagy activity. Dev. Cell. 55:588–602.e7. 10.1016/j.devcel.2020.10.010 [DOI] [PubMed] [Google Scholar]
- Chen, D., Xiao H., Zhang K., Wang B., Gao Z., Jian Y., Qi X., Sun J., Miao L., and Yang C.. 2010. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 327:1261–1264. 10.1126/science.1184840 [DOI] [PubMed] [Google Scholar]
- Chen, L., Wang K., Long A., Jia L., Zhang Y., Deng H., Li Y., Han J., and Wang Y.. 2017. Fasting-induced hormonal regulation of lysosomal function. Cell Res. 27:748–763. 10.1038/cr.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard, L., Mishra A.K., Sylvain M.A., Tuck S., Lambright D.G., and Rocheleau C.E.. 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 21:2285–2296. 10.1091/mbc.e09-11-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinque, L., De Leonibus C., Iavazzo M., Krahmer N., Intartaglia D., Salierno F.G., De Cegli R., Di Malta C., Svelto M., Lanzara C., et al. 2020. MiT/TFE factors control ER-phagy via transcriptional regulation of FAM134B. EMBO J. 39:e105696. 10.15252/embj.2020105696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, T.M., and Cachón-González M.B.. 2012. The cellular pathology of lysosomal diseases. J. Pathol. 226:241–254. 10.1002/path.3021 [DOI] [PubMed] [Google Scholar]
- Dai, A., Yu L., and Wang H.W.. 2019. WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat. Commun. 10:3699. 10.1038/s41467-019-11694-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve, C. 2005. The lysosome turns fifty. Nat. Cell Biol. 7:847–849. 10.1038/ncb0905-847 [DOI] [PubMed] [Google Scholar]
- Di Malta, C., Siciliano D., Calcagni A., Monfregola J., Punzi S., Pastore N., Eastes A.N., Davis O., De Cegli R., Zampelli A., et al. 2017. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 356:1188–1192. 10.1126/science.aag2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M., et al. 2010. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 1:38. 10.1038/ncomms1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, W., Su Q.P., Chen Y., Zhu Y., Jiang D., Rong Y., Zhang S., Zhang Y., Ren H., Zhang C., et al. 2016. Kinesin 1 drives autolysosome tubulation. Dev. Cell. 37:326–336. 10.1016/j.devcel.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Durchfort, N., Verhoef S., Vaughn M.B., Shrestha R., Adam D., Kaplan J., and Ward D.M.. 2012. The enlarged lysosomes in beige j cells result from decreased lysosome fission and not increased lysosome fusion. Traffic. 13:108–119. 10.1111/j.1600-0854.2011.01300.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassl, A., Brain C., Abu-Remaileh M., Stukan I., Butter D., Stepien P., Feit A.S., Bergholz J., Michowski W., Otto T., et al. 2020. Increased lysosomal biomass is responsible for the resistance of triple-negative breast cancers to CDK4/6 inhibition. Sci. Adv. 6:eabb2210. 10.1126/sciadv.abb2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron, M., Settembre C., Shimazu J., Lacombe J., Kato S., Rawlings D.J., Ballabio A., and Karsenty G.. 2013. A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 27:955–969. 10.1101/gad.213827.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folick, A., Oakley H.D., Yu Y., Armstrong E.H., Kumari M., Sanor L., Moore D.D., Ortlund E.A., Zechner R., and Wang M.C.. 2015. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science. 347:83–86. 10.1126/science.1258857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimaki, K., Li R., Chen H., Della Croce K., Zhang H.H., Xing J., Bai F., and Yao G.. 2019. Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Proc. Natl. Acad. Sci. USA. 116:22624–22634. 10.1073/pnas.1915905116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Q., Wang X., Zhang Q., Yin Q., Jian Y., Liu Y., Xuan N., Li J., Zhou J., Liu K., et al. 2019. The amino acid transporter SLC-36.1 cooperates with PtdIns3P 5-kinase to control phagocytic lysosome reformation. J. Cell Biol. 218:2619–2637. 10.1083/jcb.201901074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch, B., Tharyan R.G., Mak J., Denzel S.I., Popkes-van Oepen T., Henn N., and Antebi A.. 2020. HLH-30/TFEB is a master regulator of reproductive quiescence. Dev. Cell. 53:316–329.e5. 10.1016/j.devcel.2020.03.014 [DOI] [PubMed] [Google Scholar]
- Gillooly, D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. 2014. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16:1215–1226. 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- Hagedorn, E.J., and Sherwood D.R.. 2011. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr. Opin. Cell Biol. 23:589–596. 10.1016/j.ceb.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover, J.A., Krause M.W., and Love D.C.. 2010. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta. 1800:80–95. 10.1016/j.bbagen.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath, T.J., Steingrímsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., and Fisher D.E.. 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8:2770–2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Hipolito, V.E.B., Ospina-Escobar E., and Botelho R.J.. 2018. Lysosome remodelling and adaptation during phagocyte activation. Cell. Microbiol. 20:e12824. 10.1111/cmi.12824 [DOI] [PubMed] [Google Scholar]
- Hsu, C.L., Lee E.X., Gordon K.L., Paz E.A., Shen W.C., Ohnishi K., Meisenhelder J., Hunter T., and La Spada A.R.. 2018. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat. Commun. 9:942. 10.1038/s41467-018-03340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari, J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanikia, S., Özbey N.P., Krueger C., Casanueva M.O., and Taylor R.C.. 2019. Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans. Curr. Biol. 29:2322–2338.e7. 10.1016/j.cub.2019.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y., and Weisman L.S.. 2015. The vacuole/lysosome is required for cell-cycle progression. eLife. 4:e08160. 10.7554/eLife.08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian, L.M., and Stanford W.L.. 2020. Organelle cooperation in stem cell fate: Lysosomes as emerging regulators of cell identity. Front. Cell Dev. Biol. 8:591. 10.3389/fcell.2020.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C.J. 2010. The genetics of ageing. Nature. 464:504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Khaminets, A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522:354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Kim, E., Goraksha-Hicks P., Li L., Neufeld T.P., and Guan K.L.. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10:935–945. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, P.E., and Swanson J.A.. 1990. Plasticity of the tubular lysosomal compartment in macrophages. J. Cell Sci. 95:433–439. 10.1242/jcs.95.3.433 [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Piao W., Takamura T., Kori H., Miyachi H., Kitano S., Iwamoto Y., Yamada M., Imayoshi I., Shioda S., et al. 2019. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat. Commun. 10:5446. 10.1038/s41467-019-13203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic, M., Krishna S., Akkari L., Joyce J.A., and Overholtzer M.. 2013. mTOR regulates phagosome and entotic vacuole fission. Mol. Biol. Cell. 24:3736–3745. 10.1091/mbc.e13-07-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzaler, P.A., Staniszewska A.D., Li W., Omidvar N., Kedjouar B., Turkson J., Poli V., Flavell R.A., Clarkson R.W., and Watson C.J.. 2011. Stat3 controls lysosomal-mediated cell death in vivo. Nat. Cell Biol. 13:303–309. 10.1038/ncb2171 [DOI] [PubMed] [Google Scholar]
- Krishna, S., Palm W., Lee Y., Yang W., Bandyopadhyay U., Xu H., Florey O., Thompson C.B., and Overholtzer M.. 2016. PIKfyve regulates vacuole maturation and nutrient recovery following engulfment. Dev. Cell. 38:536–547. 10.1016/j.devcel.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre, L.R., De Magalhaes Filho C.D., McQuary P.R., Chu C.C., Visvikis O., Chang J.T., Gelino S., Ong B., Davis A.E., Irazoqui J.E., et al. 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4:2267. 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman, D.S., Hebestreit K., Ruetz T., Webb A.E., McKay A., Pollina E.A., Dulken B.W., Zhao X., Yeo R.W., Ho T.T., et al. 2018. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science. 359:1277–1283. 10.1126/science.aag3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Friedrichsen H.J., Andrews S., Picaud S., Volpon L., Ngeow K., Berridge G., Fischer R., Borden K.L.B., Filippakopoulos P., and Goding C.R.. 2018. A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat. Commun. 9:2685. 10.1038/s41467-018-04849-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Song Y., Quach C., Guo H., Jang G.B., Maazi H., Zhao S., Sands N.A., Liu Q., In G.K., et al. 2019. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 10:1693. 10.1038/s41467-019-09634-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., and Wang X.. 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 136:2445–2455. 10.1242/dev.035949 [DOI] [PubMed] [Google Scholar]
- Li, X., Yu W., Qian X., Xia Y., Zheng Y., Lee J.H., Li W., Lyu J., Rao G., Zhang X., et al. 2017. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol. Cell. 66:684–697.e9. 10.1016/j.molcel.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Xu M., Ding X., Yan C., Song Z., Chen L., Huang X., Wang X., Jian Y., Tang G., et al. 2016. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 18:1065–1077. 10.1038/ncb3407 [DOI] [PubMed] [Google Scholar]
- Liang, R., Arif T., Kalmykova S., Kasianov A., Lin M., Menon V., Qiu J., Bernitz J.M., Moore K., Lin F., et al. 2020. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 26:359–376.e7. 10.1016/j.stem.2020.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X.X., Sen I., Janssens G.E., Zhou X., Fonslow B.R., Edgar D., Stroustrup N., Swoboda P., Yates J.R. III, Ruvkun G., and Riedel C.G.. 2018. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat. Commun. 9:4400. 10.1038/s41467-018-06624-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipetz, J., and Cristofalo V.J.. 1972. Ultrastructural changes accompanying the aging of human diploid cells in culture. J. Ultrastruct. Res. 39:43–56. 10.1016/S0022-5320(72)80005-4 [DOI] [PubMed] [Google Scholar]
- Liu, B., Du H., Rutkowski R., Gartner A., and Wang X.. 2012. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 337:351–354. 10.1126/science.1220281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Palmfeldt J., Lin L., Colaço A., Clemmensen K.K.B., Huang J., Xu F., Liu X., Maeda K., Luo Y., and Jäättelä M.. 2018. STAT3 associates with vacuolar H+-ATPase and regulates cytosolic and lysosomal pH. Cell Res. 28:996–1012. 10.1038/s41422-018-0080-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Jian Y., Sun X., Yang C., Gao Z., Zhang Z., Liu X., Li Y., Xu J., Jing Y., et al. 2016. Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. J. Cell Biol. 212:181–198. 10.1083/jcb.201506081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Xing R., Jian Y., Gao Z., Ma X., Sun X., Li Y., Xu M., Wang X., Jing Y., et al. 2017. WDR91 is a Rab7 effector required for neuronal development. J. Cell Biol. 216:3307–3321. 10.1083/jcb.201705151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans, E., Morgan A.J., He X., Smith D.A., Elliot-Smith E., Sillence D.J., Churchill G.C., Schuchman E.H., Galione A., and Platt F.M.. 2008. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14:1247–1255. 10.1038/nm.1876 [DOI] [PubMed] [Google Scholar]
- Loeffler, D., Wehling A., Schneiter F., Zhang Y., Müller-Bötticher N., Hoppe P.S., Hilsenbeck O., Kokkaliaris K.D., Endele M., and Schroeder T.. 2019. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature. 573:426–429. 10.1038/s41586-019-1531-6 [DOI] [PubMed] [Google Scholar]
- López-Hernández, T., Puchkov D., Krause E., Maritzen T., and Haucke V.. 2020. Endocytic regulation of cellular ion homeostasis controls lysosome biogenesis. Nat. Cell Biol. 22:815–827. 10.1038/s41556-020-0535-7 [DOI] [PubMed] [Google Scholar]
- Lürick, A., Kümmel D., and Ungermann C.. 2018. Multisubunit tethers in membrane fusion. Curr. Biol. 28:R417–R420. 10.1016/j.cub.2017.12.012 [DOI] [PubMed] [Google Scholar]
- Luzio, J.P., Gray S.R., and Bright N.A.. 2010. Endosome-lysosome fusion. Biochem. Soc. Trans. 38:1413–1416. 10.1042/BST0381413 [DOI] [PubMed] [Google Scholar]
- Luzio, J.P., Hackmann Y., Dieckmann N.M., and Griffiths G.M.. 2014. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6:a016840. 10.1101/cshperspect.a016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima, I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T.. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes, J., Gegg M.E., Migdalska-Richards A., Doherty M.K., Whitfield P.D., and Schapira A.H.. 2016. Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson disease. Hum. Mol. Genet. 25:3432–3445. 10.1093/hmg/ddw185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., and Puertollano R.. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200:475–491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., and Puertollano R.. 2018. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J. Biol. Chem. 293:12525–12534. 10.1074/jbc.RA118.003471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., Chen Y., Gucek M., and Puertollano R.. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., and Puertollano R.. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7:ra9. 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., Diab H.I., Brady O.A., and Puertollano R.. 2016. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35:479–495. 10.15252/embj.201593428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, J.A., Guerrero-Gómez D., Gómez-Orte E., Antonio Bárcena J., Cabello J., Miranda-Vizuete A., and Puertollano R.. 2021. A conserved cysteine-based redox mechanism sustains TFEB/HLH-30 activity under persistent stress. EMBO J. 40:e105793. 10.15252/embj.2020105793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Fábregas, J., Prescott A., van Kasteren S., Pedrioli D.L., McLean I., Moles A., Reinheckel T., Poli V., and Watts C.. 2018. Lysosomal protease deficiency or substrate overload induces an oxidative-stress mediated STAT3-dependent pathway of lysosomal homeostasis. Nat. Commun. 9:5343. 10.1038/s41467-018-07741-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, A.J., Zhang Y., and Weisman L.S.. 2014. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays. 36:52–64. 10.1002/bies.201300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, M.J., Eramo M.J., Gurung R., Sriratana A., Gehrig S.M., Lynch G.S., Lourdes S.R., Koentgen F., Feeney S.J., Lazarou M., et al. 2021. Defective lysosome reformation during autophagy causes skeletal muscle disease. J. Clin. Invest. 131:e135124. 10.1172/JCI135124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.E., and Cullen P.J.. 2018. Endosomal retrieval of cargo: Retromer is not alone. Trends Cell Biol. 28:807–822. 10.1016/j.tcb.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Medina, D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17:288–299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M., et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21:421–430. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., Cantley L.C., and Manning B.D.. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 156:771–785. 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, R., Li M., Zhang Q., Yang C., and Wang X.. 2020. An ECM-to-Nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev. Cell. 52:21–37.e5. 10.1016/j.devcel.2019.10.020 [DOI] [PubMed] [Google Scholar]
- Miller, A., Schafer J., Upchurch C., Spooner E., Huynh J., Hernandez S., McLaughlin B., Oden L., and Fares H.. 2015. Mucolipidosis type IV protein TRPML1-dependent lysosome formation. Traffic. 16:284–297. 10.1111/tra.12249 [DOI] [PubMed] [Google Scholar]
- Naegeli, K.M., Hastie E., Garde A., Wang Z., Keeley D.P., Gordon K.L., Pani A.M., Kelley L.C., Morrissey M.A., Chi Q., et al. 2017. Cell invasion in vivo via rapid exocytosis of a transient lysosome-derived membrane domain. Dev. Cell. 43:403–417.e10. 10.1016/j.devcel.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, S., Karalay Ö., Jäger P.S., Horikawa M., Klein C., Nakamura K., Latza C., Templer S.E., Dieterich C., and Antebi A.. 2016. Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7:10944. 10.1038/ncomms10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano, G., Di Malta C., Esposito A., de Araujo M.E.G., Pece S., Bertalot G., Matarese M., Benedetti V., Zampelli A., Stasyk T., et al. 2020. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature. 585:597–602. 10.1038/s41586-020-2444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano, G., Esposito A., Choi H., Matarese M., Benedetti V., Di Malta C., Monfregola J., Medina D.L., Lippincott-Schwartz J., and Ballabio A.. 2018. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat. Commun. 9:3312. 10.1038/s41467-018-05862-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich, C.L., Wang C., Fogel A.I., and Youle R.J.. 2015. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210:435–450. 10.1083/jcb.201501002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot, A.S., Fares H., Payrastre B., Chisholm A.D., Labouesse M., and Laporte J.. 2006. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol. Biol. Cell. 17:3062–3074. 10.1091/mbc.e05-12-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann, M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- O’Rourke, E.J., and Ruvkun G.. 2013. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15:668–676. 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Palmieri, M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L., Chaudhury A., Bajaj L., Bondar V.V., Bremner L., et al. 2017. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8:14338. 10.1038/ncomms14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R.M., and Zoncu R.. 2016. The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 32:223–253. 10.1146/annurev-cellbio-111315-125125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R.M., Di Malta C., and Ballabio A.. 2019. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu. Rev. Cancer Biol. 3:203–222. 10.1146/annurev-cancerbio-030518-055835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R.M., Stoykova S., Nicolay B.N., Ross K.N., Fitamant J., Boukhali M., Lengrand J., Deshpande V., Selig M.K., Ferrone C.R., et al. 2015. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 524:361–365. 10.1038/nature14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, C.S., Roczniak-Ferguson A., and Ferguson S.M.. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202:1107–1122. 10.1083/jcb.201307084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles, S., Vigié P., and Youle R.J.. 2018. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, F.M., Boland B., and van der Spoel A.C.. 2012. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J. Cell Biol. 199:723–734. 10.1083/jcb.201208152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper, D., Taelman V.F., Robert L., Perez B.S., Titz B., Chen H.W., Graeber T.G., von Euw E., Ribas A., and De Robertis E.M.. 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA. 112:E420–E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev, D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141:497–508. 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Pryor, P.R., Mullock B.M., Bright N.A., Gray S.R., and Luzio J.P.. 2000. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J. Cell Biol. 149:1053–1062. 10.1083/jcb.149.5.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano, R., Ferguson S.M., Brugarolas J., and Ballabio A.. 2018. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37:e98804. 10.15252/embj.201798804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiteanu, R., Davis L.J., Williamson J.C., Timms R.T., Paul Luzio J., and Lehner P.J.. 2016. A genetic screen identifies a critical role for the WDR81-WDR91 complex in the trafficking and degradation of tetherin. Traffic. 17:940–958. 10.1111/tra.12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen, M., Pochini L., Stasyk T., de Araújo M.E., Galluccio M., Kandasamy R.K., Snijder B., Fauster A., Rudashevskaya E.L., Bruckner M., et al. 2015. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 519:477–481. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink, J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson, A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42. 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C., et al. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14:924–934. 10.1038/ncb2557 [DOI] [PubMed] [Google Scholar]
- Rong, Y., McPhee C.K., Deng S., Huang L., Chen L., Liu M., Tracy K., Baehrecke E.H., Yu L., and Lenardo M.J.. 2011. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. USA. 108:7826–7831. 10.1073/pnas.1013800108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig, P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Sakamaki, J.I., Wilkinson S., Hahn M., Tasdemir N., O’Prey J., Clark W., Hedley A., Nixon C., Long J.S., New M., et al. 2017. Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell. 66:517–532.e9. 10.1016/j.molcel.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak, Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak, Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello, M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Schink, K.O., Tan K.W., and Stenmark H.. 2016. Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32:143–171. 10.1146/annurev-cellbio-111315-125349 [DOI] [PubMed] [Google Scholar]
- Schmid, J.A., Mach L., Paschke E., and Glössl J.. 1999. Accumulation of sialic acid in endocytic compartments interferes with the formation of mature lysosomes. Impaired proteolytic processing of cathepsin B in fibroblasts of patients with lysosomal sialic acid storage disease. J. Biol. Chem. 274:19063–19071. 10.1074/jbc.274.27.19063 [DOI] [PubMed] [Google Scholar]
- Settembre, C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., et al. 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15:647–658. 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, Y., Rao L., Settembre C., Ballabio A., and Eissa N.T.. 2017. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 36:2544–2552. 10.15252/embj.201796699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H.J., Kim H., Oh S., Lee J.G., Kee M., Ko H.J., Kweon M.N., Won K.J., and Baek S.H.. 2016. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 534:553–557. 10.1038/nature18014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Li M., Zhao D., Li X., Yang C., and Wang X.. 2020. Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. eLife. 9:e55745. 10.7554/eLife.55745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman, V., and Cummings D.J.. 1976. Morphological changes in aging cell lines of Paramecium aurella. I. Alterations in the cytoplasm. Mech. Ageing Dev. 5:139–154. 10.1016/0047-6374(76)90014-2 [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Homme M., Yamazaki Y., Shimizu R., Takazawa Y., and Nakamura T.. 2017. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 77:897–907. 10.1158/0008-5472.CAN-16-2486 [DOI] [PubMed] [Google Scholar]
- Treusch, S., Knuth S., Slaugenhaupt S.A., Goldin E., Grant B.D., and Fares H.. 2004. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 101:4483–4488. 10.1073/pnas.0400709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun, Z.Y., Bar-Peled L., Chantranupong L., Zoncu R., Wang T., Kim C., Spooner E., and Sabatini D.M.. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell. 52:495–505. 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rubin-de-Celis, S., Peña-Llopis S., Konda M., and Brugarolas J.. 2017. Multistep regulation of TFEB by MTORC1. Autophagy. 13:464–472. 10.1080/15548627.2016.1271514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan, S., Karakas C., Doostan I., Chen X., Bui T., Yi M., Raghavendra A.S., Zhao Y., Bashour S.I., Ibrahim N.K., et al. 2017. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat. Commun. 8:15916. 10.1038/ncomms15916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas, F., Lehalle D., Mayer D., Rittirsch M., Stadler M.B., Zinner M., Olivieri D., Vabres P., Duplomb-Jego L., De Bont E.S.J.M., et al. 2019. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell. 24:257–270.e8. 10.1016/j.stem.2018.11.021 [DOI] [PubMed] [Google Scholar]
- Wada, S., Neinast M., Jang C., Ibrahim Y.H., Lee G., Babu A., Li J., Hoshino A., Rowe G.C., Rhee J., et al. 2016. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 30:2551–2564. 10.1101/gad.287953.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Wang N., Xu D., Ma Q., Chen Y., Xu S., Xia Q., Zhang Y., Prehn J.H.M., Wang G., and Ying Z.. 2020a. Oxidation of multiple MiT/TFE transcription factors links oxidative stress to transcriptional control of autophagy and lysosome biogenesis. Autophagy. 16:1683–1696. 10.1080/15548627.2019.1704104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D., Chantranupong L., Comb W., et al. 2015a. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 347:188–194. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Gao Q., Yang M., Zhang X., Yu L., Lawas M., Li X., Bryant-Genevier M., Southall N.T., Marugan J., et al. 2015b. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. USA. 112:E1373–E1381. 10.1073/pnas.1419669112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and Yang C.. 2016. Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell. Mol. Life Sci. 73:2221–2236. 10.1007/s00018-016-2196-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Huang Y., Liu J., Zhang J., Xu M., You Z., Peng C., Gong Z., and Liu W.. 2020b. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 21:e48335. 10.15252/embr.201948335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson, R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R., and Sabatini D.M.. 2016. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 351:43–48. 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.C., Kim S., Peng W., and Krainc D.. 2019. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29:500–513. 10.1016/j.tcb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Zhou P., Cheng S., Lu Q., Nowak K., Hopp A.K., Li L., Shi X., Zhou Z., Gao W., et al. 2019. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 178:552–566.e20. 10.1016/j.cell.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Yang, L., Zhang L., Wu Q., and Boyd D.D.. 2008. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J. Biol. Chem. 283:35295–35304. 10.1074/jbc.M806965200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Q., Jian Y., Xu M., Huang X., Wang N., Liu Z., Li Q., Li J., Zhou H., Xu L., et al. 2020. CDK4/6 regulate lysosome biogenesis through TFEB/TFE3. J. Cell Biol. 219:e201911036. 10.1083/jcb.201911036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N.P., Kamireddy A., Van Nostrand J.L., Eichner L.J., Shokhirev M.N., Dayn Y., and Shaw R.J.. 2016. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 30:535–552. 10.1101/gad.274142.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946. 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Li W., Li L., Li Y., Fu R., Zhu Y., Li J., Zhou Y., Xiong S., and Zhang H.. 2015. Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity. 42:309–320. 10.1016/j.immuni.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Zhao, Y.G., and Zhang H.. 2019. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 218:757–770. 10.1083/jcb.201810099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu, R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]