Abstract
Background
Since the outbreak of coronavirus disease 2019 (COVID-19), the pattern of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA shedding has not been well characterized.
Methods
In our study, 652 patients in Wuhan Designated Hospital were recruited, and their clinical and laboratory findings were extracted and analyzed.
Results
The median duration of SARS-CoV-2 RNA detection was 23 days [interquartile range (IQR), 18 days] from symptom onset. Compared to patients with early viral RNA clearance (<23 days after illness onset), we found that patients with late viral RNA clearance (≥23 days) had a higher proportion of clinical features, as follows: symptoms, including fever, dry cough, and sputum production; comorbidities, including hypertension, chronic kidney disease, uremia, chronic liver disease, anemia, hyperlipidemia, and bilateral lung involvement; complications, such as liver injury; delayed admission to hospital; laboratory parameters at baseline, including higher eosinophils, uric acid, cholesterol, triglycerides, and lower hemoglobin; and less treatment with arbidol, chloroquine, or any antivirals. After generalized linear regression, prolonged SARS-CoV-2 RNA shedding was independently associated with younger age; delayed admission to hospital; symptoms including fever, shivering, and sputum production; comorbidities including hypertension, diabetes, cardiovascular disease, anemia, hyperlipidemia, uremia, and lung involvement; and higher alanine aminotransferase (ALT), uric acid, and cholesterol levels at baseline.
Conclusions
In conclusion, the factors mentioned above are associated with the negative conversion of SARS-CoV-2 RNA. A deeper insight into virological dynamics will be helpful for establishing patient discharge and quarantine release criteria.
Keywords: Coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), viral RNA clearance, negative conversion, viral shedding
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a type of beta-coronavirus that crossed species barriers and induced coronavirus disease 2019 (COVID-19) (1,2). At the time of writing the manuscript, the virus has infected 73 million patients and killed 1.62 million people worldwide. Unlike SARS or MERS (3,4), which occurred in the last decade with a higher mortality rate and were caused by the other 2 coronaviruses [human severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV)], COVID-19 reportedly has a lower fatality rate, while its contagion has caused more casualties than either SARS or MERS. Several studies have reported that the median time of viral RNA shedding is 12-23 days, and the longest carrying history was 118 days in a study of 38 patients with COVID-19 (5-8). Further, it was proved that the long-term carriers still pose a transmission risk (8). The prolonged viral RNA shedding means a longer contagious period, requiring numerous tests and a longer isolation time, making it challenging to establish patient discharge and quarantine criteria.
The previous study showed outpatients who were old, had severe illness, and had severe underlying diseases had high viral RNA shedding (9). A recent study (10) suggested that male patients, delayed admission to hospital after illness onset, and invasive mechanical ventilation during hospitalization were associated with prolonged SARS-CoV-2 RNA shedding. Fu et al. (11) reported that patients with coronary heart disease (CHD) comorbidity, decreased albumin levels, and delayed antiviral therapy experienced delays in SARS-CoV-2 RNA clearance. Another study included 251 patients showed patients with cough and/or fever at the time of laboratory-confirmed COVID-19 disease are more likely to have a persistent SARS-CoV-2 PCR test result (7). Those with asthma or receiving immunosuppression are less likely to achieve cessation of viral RNA shedding (7). However, in these studies, the data was particularly focused on the correlation between the duration of viral RNA shedding and different severity, or symptoms alone, or comorbidities alone, or in outpatients alone, or in mild patients alone. The pattern of SARS-CoV-2 RNA shedding during treatment has not been well characterized. There is limited information to predict which patients infected with SARS-CoV-2 RNA will need a longer time of viral RNA shedding.
Here, we aim to determine the persistence and clearance time of SARS-CoV-2 RNA and describe the correlation of clinical features and candidate molecular markers with the negative conversion of SARS-CoV-2 RNA in a large patient cohort enrolled 652 COVID-19 patients with different severity. All patients were in the same center and all tests were performed according to the unified requirements and standardized methods, and nasopharyngeal swabs were collected on average every 3 to 7 days (serial time points). We take a more comprehensive view from the following aspects, including comorbidities, symptoms, laboratory tests, treatments, complications, and outcome data to evaluate negative conversion of SARS-CoV-2 RNA. We hope that our findings may be helpful for identifying the clinical characteristics of patients with a longer duration of viral RNA detection and inspire further clinical research on SARS-CoV-2 RNA infection.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-445).
Methods
Study design and participants
A total of 684 laboratory-confirmed symptomatic COVID-19 patients were recruited in this study from February 13 to April 6, 2020, in Wuhan Designated Hospital. All hospitalized patients received a standard diagnosis and treatment protocol based on the “Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7)” (12). Among the 684 SARS-CoV-2 RNA positive patients, 652 were analyzed while the other 32 were excluded, as 20 were PCR negative and only specific IgM and IgG positive, and 12 had only 1 positive sample and died before further samples were collected (Figure 1). Ethics approval (No. 2020-LS04) was obtained from the Institutional Review Board of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. All patients provided written informed consent. The study followed Declaration of Helsinki (as revised in 2013).
Figure 1.

Flow diagram for patients with confirmed coronavirus disease 2019 (COVID-19) included in the study.
According to the “Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7)” (12), patients could be classified into 4 levels of severity: mild cases (mild clinical symptoms with no abnormal radiological findings), moderate cases [fever, cough, and other symptoms with pneumonia on chest computed tomography (CT)], severe cases (severe pneumonia with respiratory distress, oxygen saturation in room air at rest ≤93%, or partial pressure of oxygen in arterial blood/fraction of inspired oxygen ≤300 mmHg), and critical cases (respiratory failure, shock, and other organ failure with intensive care or mechanical ventilation). We combined mild cases and moderate cases and categorized our COVID-19 patients into 3 groups: moderate, severe, and critical.
Demographic characteristics, comorbidities, symptoms, laboratory tests, treatments, complications, and outcome data were extracted using electronic medical records. Nasopharyngeal swabs were collected on average every 3 to 7 days (serial time points). The definitions and descriptions related to the results are as follows:
Symptom onset was defined as the day when initial symptoms were noticed;
Diagnosis time was defined as the days from symptom onset (DFSO) to the first positive detection of viral RNA in respiratory tract specimens;
Viral persistence time was defined as the DFSO to the last positive detection of respiratory tract specimens;
According to the “Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7)” (9), clinically recovered COVID-19 patients may be released after 2 negative real-time reverse transcription PCR (rRT-PCR) tests from respiratory specimens after a 24-hour interval. Viral shedding time was defined as the DFSO to the first negative day of 2 persistent negative detections.
Detection of SARS-CoV-2 RNA
Nasopharyngeal and oropharyngeal samples were collected according to the manufacturer’s protocol, and SARS-CoV-2 RNA was detected using rRT-PCR. The criteria for diagnosis followed the recommendation of the National Institute for Viral Disease Control and Prevention (China) (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html).
Statistical analysis
Statistical analysis was performed using SPSS 25 (IBM, USA) and R software (Version 3.6.0). Continuous variables were expressed as median with interquartile range (IQR), and categorical variables were expressed as number (%). The Kruskal-Wallis H test or Mann-Whitney test were used to compare the differences between groups. Categorical variables were compared using the Chi-square (χ2) test or Fisher’s exact test (if more than 20% of the cells had an expected count <5). The data were analyzed through Spearman’s bivariate correlations to evaluate the covariation among viral shedding time, symptom alleviation time, and chest CT image improvement time. Negative conversion of viral RNA, as time-to-event data, was the outcome measure, and was presented with Kaplan-Meier curves. Significant risk factors identified on univariate analyses were further analyzed by the generalized linear regression (GLM) model to identify the independent risk factors associated with the prolonged duration of SARS-CoV-2 shedding. The significance level of the hypothesis tests was set at 0.05 (two-sided).
Results
Clinical characteristics
The median age of the 652 patients was 58 years (IQR, 19), and 52.1% were female (Table S1). The median time from symptom onset to hospital admission was 5 days (IQR, 8) and the median diagnosis time was 9 days (IQR, 12). The median viral persistence time was 13 days (IQR, 15), while the median duration of viral shedding was 23 days (IQR, 18). Nearly 75% showed a negative PCR for viral RNA within 31 days, while the longest time was 69 days. The PCR negative conversion rate showed no differences across the different severity groups (data not shown). Furthermore, this cohort was divided into 2 groups: patients with viral RNA clearance <23 days (EC, n=312), and those with prolonged viral RNA shedding ≥23 days (LC, n=340). The median duration of viral RNA shedding was 15 days (IQR, 8) in the EC group, and 32 days (IQR, 14) in the LC group (Table S1).
Symptoms and related comorbidities
Fever (68.7%) and dry cough (68.4%) were the most common symptoms. Other symptoms included fatigue, breathlessness, myalgia, shivering, and sputum production, amongst others. The percentage of patients with fever, dry cough, and sputum production in the LC group was significantly higher than the EC group (P<0.05, Table S1). After stratification, the percentage of patients with fever, shivering or dry cough at diagnosis in moderate group was higher in the LC group than in the EC group (P<0.05, Table S1), indicating patients with these symptoms are more difficult to achieve virus clearance. In severe group, the proportion of patients with sputum production, sore throat, thoracic tightness or fatigue at diagnosis was higher in the LC group than in the EC group (P<0.05, Table S1). In critical group, the proportion of patients with fever or fatigue was higher in the LC group than in the EC group (P<0.05, Table S1). In the moderate and critical groups, patients with fever were more likely with late viral RNA clearance.
The common comorbidities included hypertension (230 cases), diabetes (99 cases), cardiovascular disease (63 cases), anemia (50 cases), chronic kidney disease (40 cases, 18 of them were uremia), chronic liver disease (34 cases), chronic obstructive pulmonary disease (29 cases), hyperlipidemia (26 cases), and cerebrovascular disease (20 cases), amongst others. The percentage of hypertension, chronic kidney disease, uremia, chronic liver disease, anemia, and hyperlipidemia was higher in the LC group than in the EC group (40.6% vs. 29.5%, 9.7% vs. 2.2%, 4.7% vs. 0.6%, 7.4% vs. 2.9%, 12.9% vs. 1.9%, 5.9% vs. 1.9%, P<0.05, respectively, Table S1). After stratification, in moderate group the percentage of hypertension and anemia was higher in the LC group than in the EC group (P<0.05, Table S1). In severe group, the percentage of clinical factors, including hypertension, chronic kidney disease, uremia, chronic liver disease, anemia, hyperlipidemia, and bilateral lung involvement was increased in LC group than in EC group (P<0.05, Table S1). Moreover, in critical group the percentage of clinical factors, including hypertension, chronic kidney disease, chronic liver disease, and anemia was lower in the EC group than in the LC group (P<0.05, Table S1).
The percentage of patients treated with an angiotensin II receptor blocker (ARB) or angiotensin-converting enzyme inhibitors (ACEI) in the LC group was higher than in the EC group (P<0.05, Table S1).
Laboratory testing and CT findings on admission
The analysis of laboratory parameters on admission day showed that the level of hemoglobin was lower in the LC group than in the EC group (P<0.05). Other parameters, including eosinophils, uric acid, troponin I, albumin, cholesterol, and triglycerides were much higher in the LC group (P<0.05). Lung condition was assessed by chest CT scan. The LC group had a higher percentage of bilateral lung involvement than the EC group (P<0.05). After stratification, eosinophils, albumin and uric acid was higher in the LC group than in the EC group (P<0.05, Table S1). In severe group, eosinophils, uric acid at baseline and the percentage of bilateral lung involvement was higher in the LC group (P<0.05, Table S1). In critical group, urea and d-Dimer was higher in the LC group than in the EC group (P<0.05).
Treatment, complications, and outcomes
A total of 393 patients (60.3%) were treated with antiviral drugs, including chloroquine, lopinavir and ritonavir, arbidol, ribavirin, and interferon. Furthermore, 313 patients (48.0%) received antibacterial therapy, including moxifloxacin, ceftriaxone, and azithromycin. The percentage of patients treated with arbidol, chloroquine or any antivirals was higher in the EC group than in the LC group (P<0.05). After stratification, there was no significant difference in treatment plan except that the percentage of patients treated with chloroquine was higher in the EC group than in the LC group (P<0.05, Table S1). In severe group the proportion of people receiving arbidol or any antivirals in LC group was lower than that in EC group (P<0.05, respectively, Table S1), while in critical group, the proportion of patients treated with any antivirals was higher in the EC group than in the LC group (P<0.05, Table S1). There was no difference in the percentage of 42 patients treated with corticosteroids (prednisone equivalent >40 mg) between the 2 groups (P>0.05).
The LC group had a longer time to symptom alleviation and chest CT imaging improvement. Viral RNA shedding was positively correlated with symptom alleviation time and chest CT image improvement time (r=0.588, P<0.001; r=0.605, P<0.001, respectively).
We evaluated the proportion of major complications such as acute respiratory distress syndrome (ARDS), acute cardiac injury (ACI), liver injury, acute kidney injury (AKI), shock, and disseminated intravascular coagulation (DIC). The results showed that the percentage of patients with liver injury in the LC group was higher than in the EC group (P<0.05).
As of April 6, 2020, 624 (95.7%) patients had been discharged, 14 (2.1%) had died, and 14 (2.2%) were still hospitalized for complications or hypoxia with viral RNA clearance. The proportion of outcomes in the two groups was not significantly different (P>0.05).
Negative conversion of SARS-CoV-2 RNA
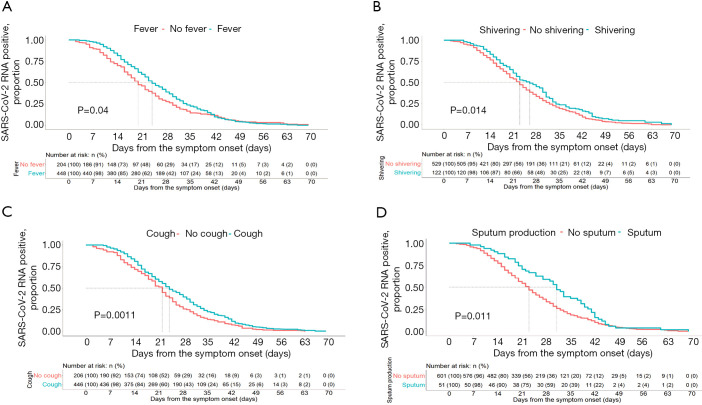
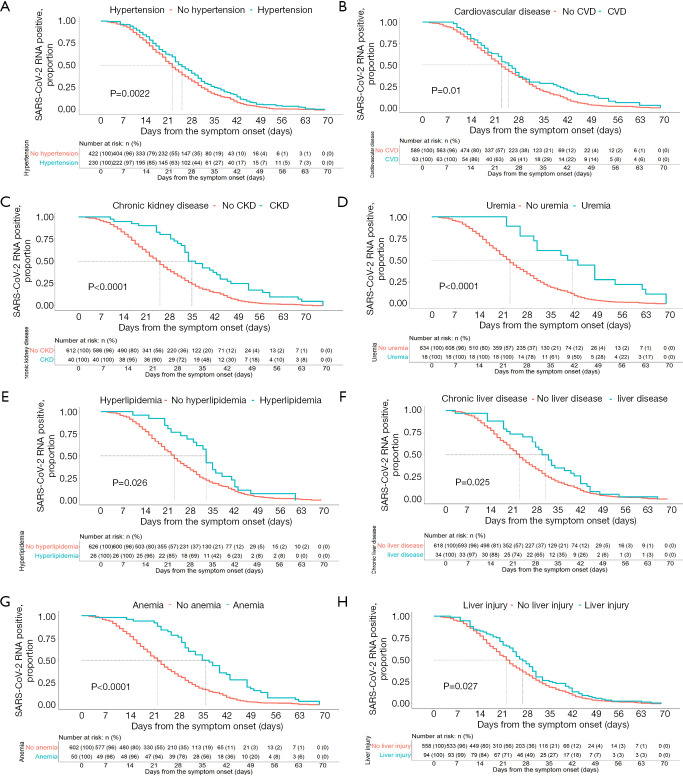
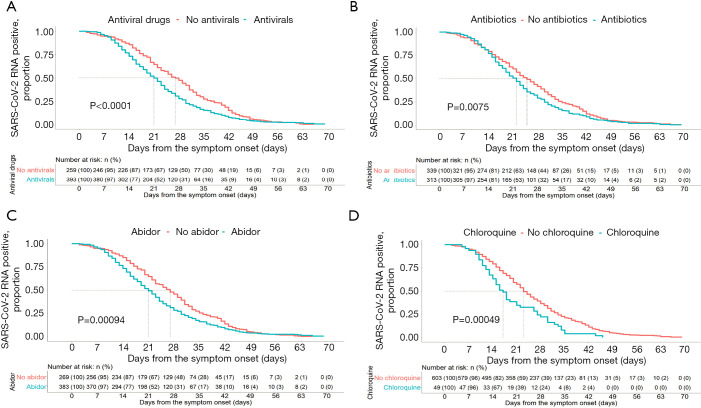
From the Kaplan-Meier curves, the RNA positive proportional changes within 1–70 days among patients with different symptoms, comorbidities, treatments, and complications are shown in Figures 2-4. The RNA negative conversion rate showed no significant difference between the 3 different severity groups (data not shown). For the patients with fever, shivering, dry cough, and sputum production, it took more days to clear SARS-CoV-2 RNA than for those without these symptoms (P<0.05, Figure 2A,B,C,D). SARS-CoV-2 RNA clearance was significantly delayed in patients with comorbidities, including hypertension, cardiovascular disease, chronic kidney disease, uremia, hyperlipidemia, chronic liver disease, and anemia (Figure 3A,B,C,D,E,F,G). Patients with complications, including liver injury, required a longer time for viral RNA clearance (P<0.05, Figure 3H). Patients treated with antivirals, including arbidol and chloroquine, and antibiotics had faster viral RNA clearance (P<0.05, Figure 4A,B,C,D).
Figure 2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA shedding curves for coronavirus disease 2019 (COVID-19) patients according to symptom predictors. (A) Cumulative proportion of patients with detectable SARS-CoV-2 RNA by day from symptom onset between patients with and without fever (log-rank P=0.04). (B) Cumulative proportion of patients with detectable SARS-CoV-2 RNA by day from symptom onset between patients with and without shivering (log-rank P=0.014). (C) Cumulative proportion of patients with detectable SARS-CoV-2 RNA by day from symptom onset between patients with and without cough (log-rank P=0.0011). (D) Cumulative proportion of patients with detectable SARS-CoV-2 RNA by day from symptom onset between patients with and without sputum production (log-rank P=0.0011).
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA shedding curves for coronavirus disease 2019 (COVID-19) patients according to predictors of comorbidities and complications. Cumulative proportion of patients with detectable SARS-CoV-2 RNA by day from symptom onset between patients with and without hypertension (A, log-rank P=0.0022), cardiovascular disease (B, log-rank P=0.0022), chronic kidney disease (C, log-rank P<0.001), uremia (D, log-rank P<0.001), hyperlipidemia (E, log-rank P=0.026), chronic liver disease (F, log-rank P=0.025), anemia (G, log-rank P<0.001), and liver injury (H, log-rank P=0.027).
Figure 4.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA shedding curves for coronavirus disease 2019 (COVID-19) patients according to predictors of treatments. Patients treated with antivirals, including arbidol and chloroquine, and antibiotics had faster viral RNA clearance than patients without treatment. (A) Antiviral drugs (log-rank P<0.001); (B) antibiotics (log-rank P=0.075); (C) arbidol (log-rank P=0.00094); (D) chloroquine (log-rank P=0.00049).
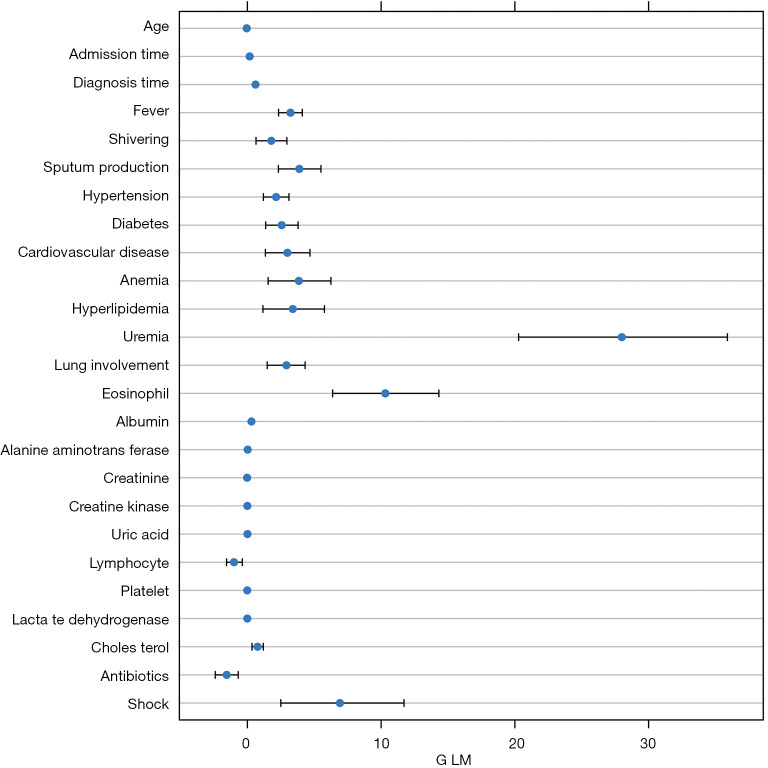
Risk factors for negative conversion of SARS-CoV-2 RNA
The effect of each factor on prolonged viral RNA shedding was evaluated through a GLM model. Variables, including age, sex, smoking, symptoms, comorbidities, use of antivirals and corticosteroids, duration from illness onset to hospitalization, diagnosis time, major complications, and laboratory parameters at baseline, were tested. We found that younger age was associated with delayed virological clearance (P<0.05, Table 1, Figure 5). GLM analysis indicated that both the time from illness onset to admission and the diagnosis time were independent factors associated with SARS-CoV-2 RNA shedding time (P<0.05, Table 1, Figure 5). Symptoms including fever, shivering, sputum production, and lung involvement on chest CT scan were significantly related to prolonged viral RNA shedding (P<0.05, Table 1, Figure 5). Comorbidities including hypertension, diabetes, cardiovascular disease, anemia, hyperlipidemia, and uremia delayed virus clearance (P<0.05, Table 1, Figure 5). Higher expression levels of laboratory parameters including eosinophils, alanine aminotransferase (ALT), uric acid, and cholesterol at baseline were associated with prolonged viral RNA shedding (P<0.05).
Table 1. Results for the generalized linear model (GLM).
| Coefficients | Estimate | Std. error | z value | Pr(>|z|) | P |
|---|---|---|---|---|---|
| Age | −0.05 | 0.01 | −3.50 | 0.0005 | <0.001 |
| Admission time | 0.17 | 0.04 | 4.26 | 2.02e-05 | <0.001 |
| Diagnosis time | 0.61 | 0.03 | 18.04 | <2e-16 | <0.001 |
| Fever | 3.22 | 0.45 | 7.16 | 8.30e-13 | <0.001 |
| Shivering | 1.79 | 0.59 | 3.03 | 0.002 | <0.01 |
| Sputum production | 3.88 | 0.80 | 4.85 | 1.26e-06 | <0.001 |
| Hypertension | 2.15 | 0.49 | 4.43 | 9.60e-06 | <0.001 |
| Diabetes | 2.57 | 0.62 | 4.12 | 3.84e-05 | <0.001 |
| Cardiovascular disease | 2.99 | 0.84 | 3.58 | 0.0003 | <0.001 |
| Anemia | 3.85 | 1.15 | 3.34 | 0.0008 | <0.001 |
| Hyperlipidemia | 3.40 | 1.18 | 2.89 | 0.004 | <0.01 |
| Uremia | 28.00 | 3.67 | 7.64 | 2.24e-14 | <0.001 |
| Bilateral lung involvement | 2.92 | 0.69 | 4.22 | 2.40e-05 | <0.001 |
| EOS count at baseline | 10.31 | 1.97 | 5.23 | 1.70e-07 | <0.001 |
| Albumin at baseline | 0.31 | 0.04 | 8.57 | <2e-16 | <0.001 |
| ALT at baseline | 0.02 | 0.006 | 3.96 | 7.63e-05 | <0.001 |
| Creatinine at baseline | −0.02 | 0.005 | −4.07 | 4.77e-05 | <0.01 |
| CK at baseline | −0.002 | 0.0003 | −5.20 | 2.03e-07 | <0.001 |
| Uric acid at baseline | 0.008 | 0.002 | 3.08 | 0.002 | <0.01 |
| Lymphocytes at baseline | −1.00 | 0.29 | −3.45 | 0.0006 | <0.001 |
| PLT at baseline | −0.01 | 0.002 | −4.66 | 3.10e-06 | <0.001 |
| Lactate dehydrogenase at baseline | −0.004 | 0.002 | −2.58 | 0.01 | <0.01 |
| Cholesterol at baseline | 0.77 | 0.21 | 3.63 | 0.0003 | <0.001 |
| Antibiotics treatment | −1.54 | 0.43 | −3.59 | 0.0003 | <0.001 |
| Shock | 6.92 | 2.30 | 3.01 | 0.003 | <0.01 |
Figure 5.
Results for the generalized linear model (GLM). GLM indicated that the time from illness onset to admission and diagnosis were independent factors associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA shedding time (P<0.05). Symptoms, including fever, shivering, sputum production, and lung involvement on chest CT scan, were significantly related to longer viral RNA conversion time (P<0.05). Comorbidities including hypertension, diabetes, cardiovascular disease, anemia, hyperlipidemia, and uremia could delay virus clearance (P<0.05). Higher expression levels of laboratory parameters, including eosinophils, alanine aminotransferase (ALT), uric acid, and cholesterol, were associated with prolonged viral RNA shedding time (P<0.05).
Discussion
There have been few studies on the correlations between clinical features and the duration of SARS-CoV-2 RNA shedding. A better understanding of the viral RNA shedding profile is crucial to establish a standard quarantine and discharge plan.
In our study, the median time from symptom onset to viral RNA shedding was 23 days, which was similar to findings by Zhou et al. (13), who reported that the median time was 20 days for 137 cases, and Fu et al., who reported a median time of 19 days for 410 cases (11).
Generally, patients with prolonged viral RNA clearance were younger (p<0.05). The findings here are consistent with a recent study from Portugal, which stated that younger patients with mild disease severity need more time for viral RNA clearance (14). Furthermore, there was no sex-related difference in viral clearance time, which is inconsistent with the study by Li et al. (10) who showed that male patients usually had longer viral RNA shedding. This may be due to the study population, in that all patients in Li et al.’s study were mild cases.
By comparing clinical symptoms between the EC and LC groups, we found that fever, dry cough, and sputum production were correlated with prolonged viral RNA shedding. Patients with fever, dry cough, shivering and sputum production at diagnosis were more likely to have a persistently positive SARS-CoV-2 PCR result if tested within 3 weeks of symptom onset from the Kaplan-Meier curves. However, dry cough was not a significant factor after GLM. We hypothesize that these patients are more severely affected by the SARS-CoV-2 infection in the lungs. After we scored all the lung imaging of these patients, we found that bilateral lung involvement was higher in the LC group. Our results also suggest that viral RNA shedding time was positively correlated with symptom alleviation time and chest CT image improvement time.
Delayed hospital admission, delayed diagnosis time, and comorbidities, including hypertension, cardiovascular disease, diabetes, hyperlipidemia, uremia, and anemia, were independent risk factors for prolonged SARS-CoV-2 RNA shedding.
Our findings showed delayed hospital admission and diagnosis time were relevant to longer viral RNA clearance, suggesting that symptomatic patients should be admitted to hospital as soon as possible if they have SARS-CoV-2 contact history. Although no specific antiviral drug for SARS-CoV-2 has been proven effective (15,16), our study showed that chloroquine and arbidor could improve virus clearance in the EC group, while chloroquine and arbidor were not significant risk factors after the GLM model. After early hospital admission, general supportive treatment, including oxygen inhalation and nutritional support, may help in early viral clearance.
As previously reported (10), the proportion of patients with hypertension was higher in the longer viral RNA shedding population. We also found a higher proportion of patients with hypertension in the LC group, and in the GLM model hypertension was also a significant risk factor. Cardiovascular disease was also proven to be an independent risk factor for prolonged viral RNA shedding, which is consistent with the findings of Fu et al. (11).
Uremia has been proven to be a risk factor for viral RNA clearance. A recent study (17) reported that ACE2 receptor expression in kidney tissue is higher compared to that in lung tissue. ACE2 and TMPRSS genes are potential targets for SARS-CoV-2, and are co-expressed on podocytes and proximal convoluted tubule cells. There is growing concern that the use of renin-angiotensin-aldosterone system (RAAS) inhibitors may alter ACE2 expression and play a role in disease virulence (18). We found that the proportion of patients who previously used ACEIs or ARBs in the LC group was higher. This change could partially explain why SARS-CoV-2 tends to infect patients with chronic kidney disease and is harder to remove, though further studies are required to clarify the mechanisms. Another possible explanation of these findings is that patients with kidney dysfunction have deficits in immune cell populations (19), leading to increased lung inflammation.
We also found that virus clearance was delayed in hyperlipidemia patients, which is consistent with the result that high cholesterol levels on admission were associated with prolonged viral RNA shedding time. The reason we took this into consideration is due to the role of cholesterol during viral RNA infection. The entry of flaviviruses and coronaviruses into human cells is via lipids and cholesterol-rich membrane microdomains, which is facilitated by the interaction between the surface glycoprotein S of SARS-CoV and the cellular receptor ACE2 (20). Moreover, after cellular entry, a high amount of intracellular cholesterol and fatty acids are required for the formation of the replication complex of the RNA virus (21,22). The high level of cholesterol may exacerbate cardiovascular risk, which was proven to be an independent risk factor of prolonged viral RNA shedding, consistent with Fu et al. (11). Further studies are needed to evaluate the impact of these lipids in hyperlipidemia patients infected with SARS-CoV-2.
Patients with anemia had more difficulty achieving viral clearance, and lower hemoglobin was more common in the LC group. As yet, there is no solid evidence that SARS-CoV-2 attacks blood cells. However, patients with hemoglobin disorders are generally more prone to viral and bacterial infections, including the current global health situation (23-25). Two potential pathophysiological mechanisms are as follows:
SARS-CoV-2 interacts with the hemoglobin molecule through CD147, CD26, and other receptors located on erythrocytes;
Hepcidin-mimetic action of a viral spike protein, inducing ferroportin blockage.
A hemoglobinopathy, hypoxia, and cell iron overload might play additional roles. Oxygen-deprived blood disease should also be considered. Anemia and hypertension are common in patients with chronic kidney disease. These factors may partially explain why uremic patients had delayed virus clearance.
COVID-19 patients with diabetes had greater difficulty achieving viral clearance in our study. This is similar to the finding that diabetes was related to prolonged viral clearance in patients with SARS or influenza A (26,27).
Several studies have reported that the use of corticosteroids is linked to persistent viral RNA shedding in patients with avian influenza A (H7N9), MERS, and SARS (28-30). However, there was no statistical difference in the proportion of patients using corticosteroids in the 2 groups (7.4% vs. 6.8%, P=0.544). Thus, we cannot conclude that the use of corticosteroids is associated with the negative conversion of SARS-CoV-2 RNA.
ALT elevation was a risk factor after GLM. Liver injury in COVID-19 patients could be the result of direct viral hepatitis or the immune-mediated inflammatory response. ACE2 receptors are present on cholangiocytes and hepatocytes, providing a plausible mechanism of SARS-CoV-2 related hepatotoxicity (31). Additionally, etiologies including drug toxicity, sepsis, and congestion cannot be ruled out. SARS-CoV-2 related viral hepatitis and its corresponding immune-mediated inflammatory response are possible reasons for slower viral clearance (32,33). However, further in-depth studies are needed to understand liver injury-related prolonged viral RNA shedding.
One limitation of our study is that the virus was only detected by rRT-PCR without virus culture. The rRT-PCR test is used to amplify the virus gene for detection, and a positive result does not mean the virus is alive. It is not known whether individuals with persistent positive results are transmissible. Even though the intervals between specimen collection were fixed, the virological clearance time could not be exactly identical to viral conversion. Additionally, this study was performed at a single center. Therefore, the results must be confirmed in other settings or in a prospective study.
In conclusion, prolonged SARS-CoV-2 RNA shedding is independently associated with delayed admission; symptoms including fever, shivering, and sputum production; and comorbidities such as hypertension, cardiovascular disease, diabetes, hyperlipidemia, uremia, and anemia. Higher ALT, uric acid, and cholesterol were also associated with the negative conversion of SARS-CoV-2 RNA. A more in-depth insight into virological dynamics will be helpful for managing patients with COVID-19.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank Yingqi Hua, Xiaojian Zhou, Yinyan He, Yan Wang, Kai Li, Ning Li, Songwen Chen, Wei Bao, Yuhang Ma, Yi Lin, Jiangfeng Hu, Linyuan Zhang, Dezhi Liu, Jun Wang, Xiaoyun Feng, Wenlan Sun, Xinxin Xia, Runwei Tang, Hao Xu, Chuan Liu, Mengdan Xu, Yuting Yao, Zhijian Zhang, Ruihua Chen, Xiaojin Zhang, Yanling Zhao, Chongmei Huang, Dafan Chen, Qi Su, Xiangdong Meng, Xiaobo Yu, Ruijie Chen, Xiaomiao Zhang, Weidong Wu, Fang Wang, and Jieshuang Jia of Shanghai General Hospital for their assistance in the data collection.
Funding: This work was supported by National Natural Science Foundation of China (Grant No. 81873402; Grant No. 81900016), Appropriate Technology Application Program of Shanghai Municipal Health System (Grant No.2019SY042), Program of Shanghai Municipal Health System (Grant No. 201740039), First People’s Hospital of Shanghai research project “No.3 Sub-center Study on Early Identification and Severe Early Warning of Acute Respiratory Infectious Diseases” (Program No. 20Z11900903), Shanghai Jiao Tong University scientific research fund for COVID-19 prevention and control (Grant No. YG2020YQ22), and Zhejiang University special scientific research fund for COVID-19 prevention and control (Grant No. 2020XGZX009).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics approval (No. 2020-LS04) was obtained from the Institutional Review Board of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. All patients provided written informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-445
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-445
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-445). The authors have no conflicts of interest to declare.
References
- 1.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cevik M, Bamford C, Ho A. COVID-19 pandemic- A focused review for clinicians. Clin Microbiol Infect 2020;26:842-7. 10.1016/j.cmi.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, van Doremalen N, Falzarano D, et al. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523-34. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang S, Xia S, Ying T, et al. A novel coronavirus (2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell Mol Immunol 2020;17:554. 10.1038/s41423-020-0372-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian GQ, Chen XQ, Lv DF, et al. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis (Lond) 2020;52:511-2. 10.1080/23744235.2020.1748705 [DOI] [PubMed] [Google Scholar]
- 6.Li J, Zhang L, Liu B, et al. Case report: viral shedding for 60 days in a woman with novel coronavirus disease (COVID-19). Am J Trop Med Hyg 2020;102:1210-3. 10.4269/ajtmh.20-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corsini Campioli C, Cano Cevallos E, Assi M, et al. Clinical predictors and timing of cessation of viral RNA shedding in patients with COVID-19. J Clin Virol 2020;130:104577. 10.1016/j.jcv.2020.104577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Zheng XS, Shen XR, et al. Prolonged shedding of severe acute respiratory syndrome coronavirus 2 in patients with COVID-19. Emerg Microbes Infect 2020;9:2571-7. 10.1080/22221751.2020.1852058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao TT, Yin ZR, Xu JJ, et al. The Correlation Between Clinical Features and Viral RNA Shedding in Outpatients With COVID-19. Open Forum Infect Dis 2020;7:ofaa331. [DOI] [PMC free article] [PubMed]
- 10.Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clin Infect Dis 2020;71:799-806. 10.1093/cid/ciaa351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Han P, Zhu R, et al. Risk Factors for Viral RNA Shedding in COVID-19 Patients. Eur Respir J 2020;56:2001190. 10.1183/13993003.01190-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Health Commission & National Administration of Traditional Chinese Medicine . Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7). Chin Med J (Engl) 2020;133:1087-95. 10.1097/CM9.0000000000000819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054-62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmo A, Pereira-Vaz J, Mota V, et al. Clearance and Persistence of SARS-CoV-2 RNA in COVID-19 patients. J Med Virol 2020;92:2227-31. 10.1002/jmv.26103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan JF, Yao YF, Yeung ML, et al. Treatment With Lopinavir/Ritonavir or Interferon-beta1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J Infect Dis 2015;212:1904-13. 10.1093/infdis/jiv392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao B, Wang Y, Wen D, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med 2020;382:1787-99. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan XW, Xu D, Zhang H, et al. Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: a study based on single-cell transcriptome analysis. Intensive Care Med 2020;46:1114-6. 10.1007/s00134-020-06026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adapa S, Chenna A, Balla M, et al. COVID-19 Pandemic Causing Acute Kidney Injury and Impact on Patients With Chronic Kidney Disease and Renal Transplantation Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. J Clin Med Res 2020;12:352-61. 10.14740/jocmr4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betjes MG. Immune cell dysfunction and inflammation in end-stage renal disease. Nat Rev Nephrol 2013;9:255-65. 10.1038/nrneph.2013.44 [DOI] [PubMed] [Google Scholar]
- 20.Neufeldt CJ, Cortese M, Acosta EG, et al. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 2018;16:125-42. 10.1038/nrmicro.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glende J, Schwegmann-Wessels C, Al-Falah M, et al. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology 2008;381:215-21. 10.1016/j.virol.2008.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto-Acosta R, Mosso C, Cervantes-Salazar M, et al. The increase in cholesterol levels at early stages after dengue virus infection correlates with an augment in LDL particle uptake and HMG-CoA reductase activity. Virology 2013;442:132-47. 10.1016/j.virol.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Chen W, Zhou Y, et al. SARS CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020;10. doi: 10.1101/2020.05.14.090332. [DOI]
- 24.Radzikowska U, Ding M, Tan G, et al. Distribution of ACE2, CD147, cyclophilins, CD26 and other SARSCoV-2 associated molecules in human tissues and immune cells in health and disease. Allergy 2020;75:2829-45. 10.1111/all.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhang Z, Yang L, et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience 2020;23:101160. 10.1016/j.isci.2020.101160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Tang F, Fontanet A, et al. Long-term SARS coronavirus excretion from patient cohort, China. Emerg Infect Dis 2004;10:1841-3. 10.3201/eid1010.040297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee N, Chan PK, Hui DS, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009;200:492-500. 10.1086/600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Guo Q, Yan Z, et al. Factors Associated With Prolonged Viral Shedding in Patients With Avian Influenza A(H7N9) Virus Infection. J Infect Dis 2018;217:1708-17. 10.1093/infdis/jiy115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arabi YM, Mandourah Y, Al-Hameed F, et al. Corticosteroid Therapy for Critically Ill Patients with Middle East Respiratory Syndrome. Am J Respir Crit Care Med 2018;197:757-67. 10.1164/rccm.201706-1172OC [DOI] [PubMed] [Google Scholar]
- 30.Lee N, Allen Chan KC, Hui DS, et al. Effects of early corticosteroid treatment on plasma SARS-associated Coronavirus RNA concentrations in adult patients. J Clin Virol 2004;31:304-9. 10.1016/j.jcv.2004.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv 2020;13. doi: 10.1101/2020.02.03.931766. [DOI]
- 32.Zhang C, Shi L, Wang FS. Liver injury in COVID-19: Management and challenges. Lancet Gastroenterol Hepatol 2020;5:428-30. 10.1016/S2468-1253(20)30057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Aghemo A, Forner A, et al. COVID-19 and liver disease. Liver Int 2020;40:1278-81. 10.1111/liv.14470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as