Significance
Oncogenic Ras isoforms are the subject of intense study due to the difficulty in targeting these biomedically important yet “undruggable” proteins. Recent success in covalent targeting of a Ras mutant illustrates avenues for ligand design; however, many mutant Ras forms do not feature appropriately-placed nucleophiles, suggesting that strategies for noncovalent engagement of Ras are required. We report the design of a conformationally-defined proteomimetic that reproduces a key binding surface of Sos, a well-characterized effector of Ras. The proteomimetic binds wild-type and various mutant forms of Ras and modulates downstream signaling. Significantly, the compound shows enhanced internalization and selective toxicity toward cancer cells that up-regulate macropinocytosis. We anticipate these studies will foster new therapeutic modalities to engage mutant Ras.
Keywords: Ras, proteomimetic, peptide, Sos
Abstract
Aberrant Ras signaling is linked to a wide spectrum of hyperproliferative diseases, and components of the signaling pathway, including Ras, have been the subject of intense and ongoing drug discovery efforts. The cellular activity of Ras is modulated by its association with the guanine nucleotide exchange factor Son of sevenless (Sos), and the high-resolution crystal structure of the Ras–Sos complex provides a basis for the rational design of orthosteric Ras ligands. We constructed a synthetic Sos protein mimic that engages the wild-type and oncogenic forms of nucleotide-bound Ras and modulates downstream kinase signaling. The Sos mimic was designed to capture the conformation of the Sos helix–loop–helix motif that makes critical contacts with Ras in its switch region. Chemoproteomic studies illustrate that the proteomimetic engages Ras and other cellular GTPases. The synthetic proteomimetic resists proteolytic degradation and enters cells through macropinocytosis. As such, it is selectively toxic to cancer cells with up-regulated macropinocytosis, including those that feature oncogenic Ras mutations.
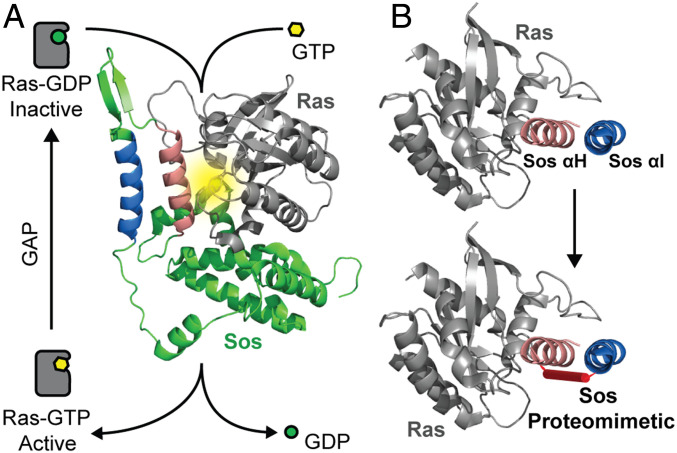
The Ras-specific guanine nucleotide exchange factor Son of sevenless (Sos) mediates the conversion of Ras from its inactive GDP-bound form to the active GTP-bound state (1). Sos catalyzes nucleotide exchange via insertion of a critical helical segment (αH) between the conformationally dynamic Switch I and II regions that flank the Ras nucleotide binding pocket leading to disruption of water-mediated and direct interactions between the protein and the cofactor (Fig. 1A) (2). Given the biomedical importance of the conformationally dynamic Sos-binding interface of Ras, several rational design and screening strategies have been attempted to develop ligands for this interface (3, 4). Recent efforts to engage the Ras G12C (5, 6) and the G12D isoforms (7, 8) suggest that targeted screens may afford small molecules and peptide macrocycles as potential leads. The structure of the Ras–Sos complex provides a basis for the rational design of Sos helix mimics that engage the Ras switch regions. Our group has previously developed a conformationally stabilized α-helix mimic to target the Ras–Sos protein–protein interaction (PPI) (9, 10). The stabilized α-helix was shown to bind Ras at the orthosteric binding site and inhibit Sos-mediated nucleotide exchange, Ras activation, and phosphorylation of the downstream effector protein ERK (11), a well-characterized kinase implicated in cell proliferation and differentiation. However, this compound preferred to bind Ras in its nucleotide-free form, suggesting that a single Sos helix is likely insufficient to properly engage the dynamic Ras interface in its nucleotide-bound form. While wild-type Ras toggles between its two nucleotide-bound forms (Fig. 1A), the oncogenic forms of Ras remain activated in their GTP-bound states (3). Therefore, a compound that preferentially engages the nucleotide-free form of Ras may have limited biological utility.
Fig. 1.
Overview of the Ras activation cycle and design of a Sos-based proteomimetic. (A) The cellular activity of Ras is tightly controlled as part of a balanced feedback loop. Oncogenic mutations shift this balance and increase the cellular concentration of Ras-GTP leading to aberrant downstream signaling. The molecular model shows the complex between Ras (gray ribbon) and its guanine exchange factor Sos (green). Sos inserts a helical hairpin (pink and blue helices) into the nucleotide binding pocket of Ras to mediate nucleotide exchange. The Ras nucleotide binding pocket is highlighted in yellow. Segments of Sos are not shown to highlight interactions of the helical hairpin with Ras (PDB code: 1NVW). (B) The molecular models depict critical Sos helices and the design of a constrained Sos proteomimetic as a Ras inhibitor. GDP, guanosine 5′-diphosphate; GTP, guanosine 5′-triphosphate; GAP, GTPase activating protein; GEF, guanine nucleotide exchange factor.
Our prior results with the Sos helix mimic encapsulate a critical challenge in developing minimal protein secondary-structure mimics. Although mimics of protein secondary structures have proven to be a potent class of PPI inhibitors (12–15), many protein interfaces feature binding epitope complexity beyond what can be captured by reproduction of minimal elements of protein structure (16–19). We hypothesized that the introduction of additional contact residues from Sos may allow engagement of nucleotide-bound Ras (Fig. 1B). Sos inserts αH into the switch region of Ras, but analysis of the complex shows that several other residues from Sos also interact with Ras (SI Appendix, Fig. S1). The conformation of the αH helix, itself, is controlled by the αI domain as part of a hairpin helix organization. The αI helix makes important electrostatic contacts with the Ras effector loop in the Switch I region.
We sought to develop a tertiary-structure mimic of Sos that encompasses critical binding residues from the helix–loop–helix motif to determine if the additional contacts allow engagement of nucleotide-bound Ras (Fig. 1B). We utilized a recently described synthetic approach from our group to capture the conformation of the Sos αH and αI helices. In prior efforts, we learned that helix dimers may be stabilized by judicious substitution of a surface salt bridge with a covalent bond and appropriate sculpting of the dimeric interface to coerce knob-into-hole helix packing (20, 21). These stabilized proteomimetics are termed crosslinked helix dimers or CHDs. Here, we show that Sos CHDs are proteolytically stable, selectively cell permeable, and engage the Sos-binding surface of nucleotide-bound Ras with high specificity in biochemical and cellular contexts. We utilized a combination of rational design principles and computational modeling to exploit previously unexplored and underutilized pockets at the target interface (22, 23). The optimized proteomimetic binds wild-type and mutant Ras forms with nanomolar to low micromolar affinities, modulates nucleotide exchange, engages Ras and other Ras subfamily GTPases as demonstrated by chemoproteomic assays, inhibits downstream activation of the Ras-mediated signaling cascade, and is selectively toxic to cancer cells with oncogenic Ras mutations.
Results
Design of Sos Tertiary Helix Mimics.
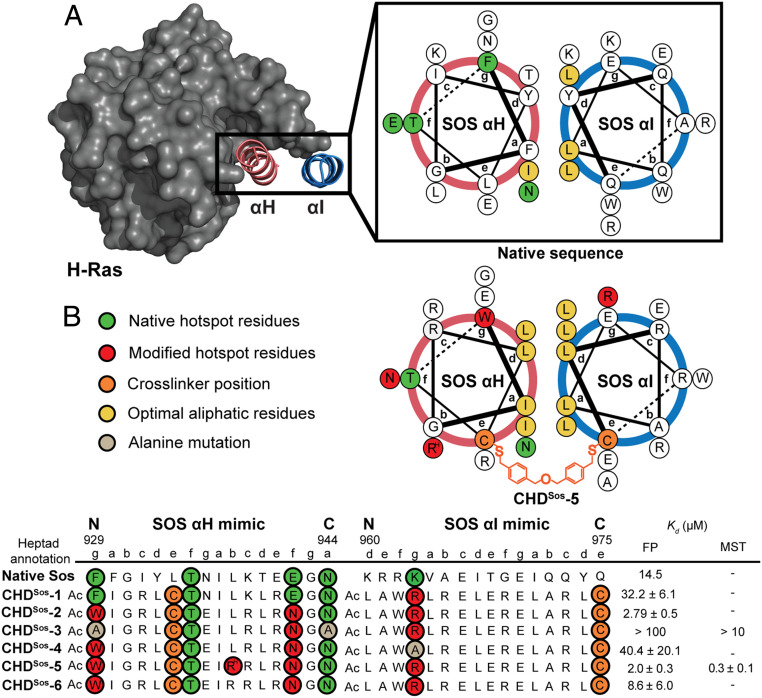
High-resolution structures of the Ras/Sos complex (Protein Data Bank [PDB]: 1NVW) show the catalytic region of Sos binding to Ras as a helical hairpin consisting of the αH and αI domains (Fig. 2A). Computational alanine scanning mutagenesis suggests that the critical Ras-contacting residues, or the hot-spot residues, are populated on the αH helix of the Sos hairpin (F929, T935, E942, and N944), with the αI helix potentially engaging an ionic patch on the Ras Switch I loop (SI Appendix, Table S1) (10, 24). We sought to develop CHDs that capture the conformation of the αH and αI helix–loop–helix tertiary structure. Synthesis of CHDs requires an appropriately placed crosslinker at the solvent-exposed surface of the two helical segments in addition to enhanced intramolecular contacts at the dimer interface. To mimic the αH/αI helix dimer, the crosslinker is placed at the “e” position of the antiparallel helix construct. In prior efforts, we optimized the synthesis of the CHDs by placing cysteine residues at the “e” positions followed by alkylation of the thiol group with a dibenzylether crosslinker (Fig. 2B) (20). The native αH/αI interface features an unoptimized knob-into-hole packing; the aromatic residues at the “a” and “d” positions were mutated to enhance conformational stability in the minimal coiled-coil mimics (18, 21, 25–27). Other noninteracting residues were also mutated to increase intra- and interstrand salt bridge interactions and the conformational stability of the helix dimer (28). The designed sequences are listed in Fig. 2B and SI Appendix, Fig. S12 and Table S2. We rationally designed Sos derivatives with the aid of two Rosetta-based computational webservers, Robetta (24) and Rosetta Backrub (22). The Robetta server provides alanine scanning mutagenesis analysis using static models of protein interfaces, and Backrub suggests potential residue substitutions to improve scores. Beyond these computational webservers, we utilized the Chimera visualization software (29) to predict nonnatural side chains, particularly to engage ionic patches on the protein surface. Modeling of the Ras–Sos complex and apo-Ras crystal structures suggest a potential ionic interaction between K963 residue of the αI helix and a negative patch on the Ras Switch I loop; the Sos lysine residue is sandwiched between E31 and D33 of Ras. We hypothesized that the guanidine functionality of an arginine group may engage both Ras ionic residues better than the primary amine group of a lysine side chain. Based on this hypothesis, K963R substitution was incorporated in the designed CHDs.
Fig. 2.
Rational design of Sos proteomimetics as Ras ligands. Antiparallel helix wheel diagrams depicting native (Top) Sos helical hairpin and (Bottom) the optimized constrained helix dimer. (A) Sos αH (pink) and αI (blue) helical domains make direct contacts with Sos, with many of the energetically important Ras contacting residues, termed hot-spot residues, positioned on the αH helix. (B) We designed and synthesized constrained Sos mimics with a hydrophobic interface and nonnative residues on both helices to enhance binding interactions with Ras. A dibenzyl ether crosslinker is placed at the “e” position of each helix to enhance conformational stability. The binding affinity of the Sos derivatives for Ras was measured by an FP assay; the binding affinity of the lead derivative CHDSos-5 and alanine control CHDSos-3 was further confirmed by MST. RH, L-homoarginine.
CHDSos-1 most closely mimics the native Sos hairpin sequence; the fluorescently-labeled derivative of this proteomimetic binds H-Ras with a binding affinity, Kd = 32 ± 6 μM, as measured in a fluorescence polarization (FP) assay with fluorescein-derivatized CHDs (SI Appendix, Fig. S2). In the cellular context, the binding affinity of Sos protein for Ras is highly dependent on the membrane localization of two proteins. The reported Kd value for the in-solution interaction of nucleotide-bound Ras with the catalytic domain of Sos is 14.5 μM (30). Therefore, the binding affinity of CHDSos-1 for Ras is in range of what is expected of a Sos fragment in the biochemical context in the absence of membrane localization. Computational analysis performed by Rosetta Backrub indicated that two contact residues (F929 and E942) were not optimally engaged within their respective subpockets on the Ras surface. The analysis suggested substitutions of F929W and E942N. Substitution of these two residues in CHDSos-2 provided a 10-fold enhancement in binding affinity (Kd = 2.8 ± 0.5 μM). CHDSos-3 (F929A and N944A) was designed as a negative control by substituting computationally predicted critical αH binding residues with alanine. As expected, the alanine double mutant bound with a significantly reduced affinity (Kd > 100 μM). To analyze the importance of the ionic interactions between αI and the Ras Switch I loop, we designed CHDSos-4 in which the single cationic arginine residue (K963R) in CHDSos-2 is substituted with an alanine. Binding analysis suggests that the ionic interactions between αI and Ras are critical for the overall complex formation as the substitution of the arginine group reduced the binding affinity to Kd = 40 ± 20 μM. Lastly, we recognized that L938 on αH is situated close to a negatively charged groove in Ras; we mutated this residue to a noncanonical homoarginine residue to potentially gain an ionic contact in CHDSos-5. CHDSos-5 binds H-Ras with a slightly improved affinity, Kd = 2.0 ± 0.3 μM, over CHDSos-2. The extra cationic residue in CHDSos-5 also improves its aqueous solubility as compared to CHDSos-2. CHDSos-6, which contains an arginine residue at position 938, bound Ras with a fourfold reduced affinity (Kd = 8.6 ± 6.0 μM) than CHDSos-5, illustrating the need for the longer homoarginine side chain at this position.
We further analyzed the binding affinity of the lead proteomimetic CHDSos-5 and the negative control CHDSos-3 with microscale thermophoresis (MST). In the MST assay, CHDSos-5 binds fluorescently-labeled Ras with nanomolar binding affinity (Kd = 0.3 ± 0.1 μM, Fig. 2B, SI Appendix, Fig. S3); as in with the polarization assay, the alanine mutant CHDSos-3 binds poorly to the protein target (Kd > 100 μM). The improved binding affinities observed with MST suggest that the dye placement on the CHDs may not be optimal for the FP assay (31).
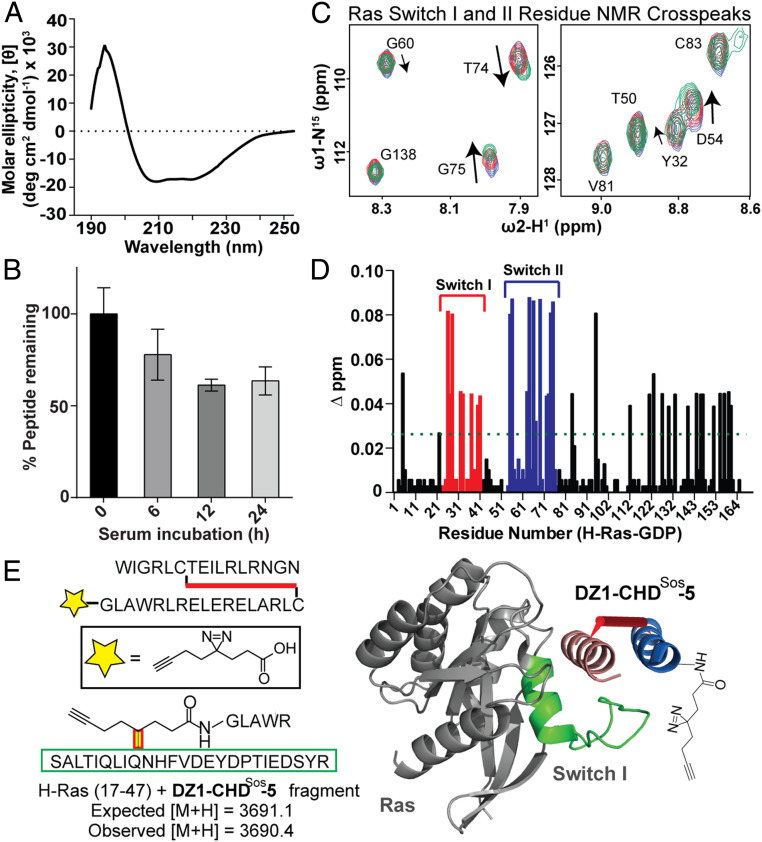
Sos Proteomimetic CHDSos-5 Is Conformationally and Proteolytically Stable and Binds Ras in its Dynamic Switch Region.
We assessed the conformational stability of the Sos proteomimetic CHDSos-5 with circular dichroism (CD) spectroscopy. CD provides a unique signature for α-helices with a local maximum at 195 nm and local minima at 208 and 222 nm (32, 33). CD experiments indicate that CHDSos-5 has a well-defined helical character (Fig. 3A). In contrast, the individual peptides corresponding to the Sos αH and αI display a random coil-like signature (SI Appendix, Fig. S4). Equimolar mixtures of the unlinked peptides also did not reveal a defined structure, while the crosslinked CHDSos-2 displayed prominent helical characteristics.
Fig. 3.
Biophysical characterization, proteolytic stability, and the Ras-binding site analysis of CHDSos-5. (A) CD spectrum of CHDSos-5. The CD study was conducted in 50 mM aqueous potassium fluoride buffer (pH 7.5) at 20 μM peptide concentration. (B) Proteolytic stability of CHDSos-5 in 25% FBS was analyzed in an HPLC assay as discussed in the Materials and Methods. Error bars are mean ± SD of biological replicates. (C) 1H-15N HSQC titration spectra of uniformly 15N-labeled GDP-loaded wild-type H-Ras. Examples of specific Ras residues that shift upon titration with increasing equivalents (1, 2.5, and 5) of CHDSos-5. (D) Bar graph shows mean chemical shift changes observed for the 15N-labeled H-Ras upon titrations with increasing amounts of CHDSos-5. (E) The CHDSos-5 binding site on Ras was further confirmed by a proximity-guided protein crosslinking reaction. DZ1-CHDSos-5 contains a photoactivable diazirine group that reacts with proximal residues on Ras. A fragment with mass corresponding to DZ1-CHDSos-5 crosslinked to Ras Switch I loop was identified. The identified fragment corresponds to the Switch I region and is depicted as a green ribbon in the molecular model.
Enzymatic proteolysis is a critical factor limiting the potential of peptide therapeutics. The CHDs have shown resistance to proteolytic degradation because of their conformational stability (18). We analyzed the proteolytic stability of CHDSos-5 in serum. The rate of proteolysis of CHDSos-5 was determined in a high performance liquid chromatography (HPLC)-based time-course assay under ex vivo conditions. CHDSos-5 exhibits considerable tolerance to proteolysis by serum proteases with a calculated t1/2 > 24 h. Approximately 60% of the proteomimetic remains intact after 24 h (Fig. 3B).
The MST and FP binding data indicate that CHDSos-5 binds Ras with high nanomolar to low micromolar binding affinity. To determine if the proteomimetic engaged Ras at the Sos-binding region, as designed, we conducted a titration heteronuclear single quantum coherence NMR spectroscopy (HSQC) experiment, which monitors chemical shift changes of specific protein residues upon ligand binding or conformation change. The addition of increasing concentrations (2.5 and 5 eq.) of CHDSos-5 to 15N-labeled H-Ras (100 μM) led to changes in peak shifts that correspond to residues within the Ras Switch I and II regions (Fig. 3C and SI Appendix, Fig. S5). Fig. 3D displays the observed chemical shift changes after CHDSos-5 titration (5 eq.) as a bar graph.
To further confirm the binding site occupancy, we elaborated CHDSos-5 with a diazirine moiety as a phototriggered crosslinking group at the N terminus of the αI helix (Fig. 3E). We hypothesized that upon irradiation of the diazirine, the resultant reactive carbene would covalently label the Ras hinge region if the association occurred at the Sos binding site. H-Ras was incubated with 5 eq. DZ1-CHDSos-5, and the complex was exposed to ultraviolet (UV) light. Agarose gel electrophoresis on protein-peptide mixture revealed a distinct monolabeled protein band, which was treated with trypsin and analyzed by mass spectroscopy to identify a crosslinked Ras fragment. We observed a fragment mass of digested DZ1-CHDSos-5 photocrosslinked with the Ras hinge region (Fig. 3E and SI Appendix, Fig. S6). Both the titration HSQC and the chemical crosslinking results are consistent with our prediction that the designed proteomimetic binds to the Sos binding surface of Ras.
CHDSos-5 Exhibits Selective Cellular Penetration in Mutant Ras Cancer Cells.
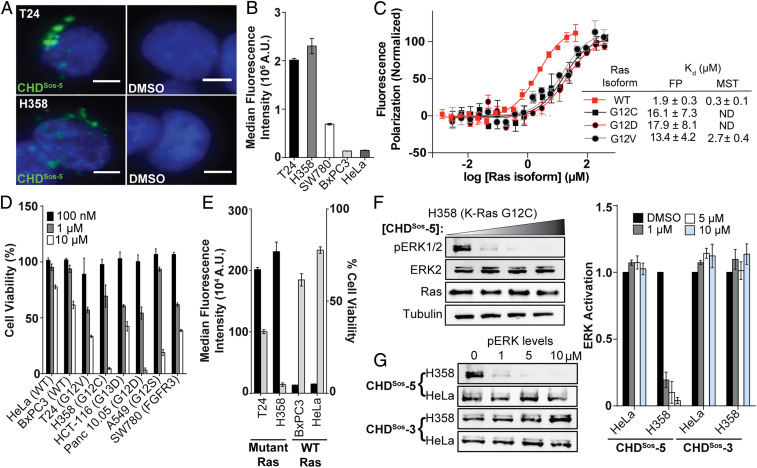
The in vitro results suggest that the designed CHDs may modulate Ras signaling. Effective cellular modulation requires efficient uptake and cytoplasmic localization of the compounds into the cell. We recently comprehensively analyzed mechanisms of peptidomimetic transport into cancer cells, and observed that efficient uptake of conformationally constrained peptidomimetics is directly correlated with the macropinocytic activity of each cell line regardless of size, charge, and conformation of the peptidomimetic (34, 35). In particular, CHDs exhibited high levels of cellular uptake and endosomal escape into the cytoplasm in macropinocytic cells despite their higher molecular weight—the uptake of CHDs was similar to that of Tat, a polycationic, cell-penetrating peptide known for high cellular internalization (36, 37). Macropinocytosis may be up-regulated in cells with activating mutations in Ras or other endemic mutations within the Ras pathway (34, 35). In keeping with these earlier analyses, live-cell fluorescence microscopy showed significant cellular uptake of fluorescein-labeled CHDSos-5 into the cytosol in the Ras mutant T24 (H-Ras G12V) bladder and H358 (K-Ras G12C) lung cancer cells (Fig. 4A). The intracellular intensity of fluorescent CHDSos-5 is similar to that of fluorescently-labeled Tat peptide (SI Appendix, Fig. S7A). To support the microscopy studies, we quantified peptide uptake using flow cytometry analysis and observed similar results for CHDSos-5 and Tat. Negative controls CHDSos-2 and CHDSos-3 exhibited lower uptake than CHDSos-5 presumably due to the removal of an arginine residue; this result supports the hypothesis that macropinocytic uptake is aided by cationic residues (Fig. 4B and SI Appendix, Fig. S7B). As expected, low cellular uptake was observed for peptides in HeLa and BxPC3 cells, both of which have a low macropinocytosis activity. When compared to cells that were incubated at 37 °C, cold-treated T24 and H358 cells showed >90% reduction in uptake for fluorescently-tagged CHDSos-2 and CHDSos-5 (SI Appendix, Fig. S7 C and D)—this result is expected because macropinocytosis is an energy-dependent uptake pathway. Enhanced macropinocytic activity is not limited to cells with Ras mutations, and certain other mutations are also known to up-regulate this activity. For example, CHDSos-5 is also permeable in SW780 cells, which contain oncogenic FGFR3 fusions (Fig. 4B). Overall, reliance of cellular permeability on certain cancer mutations suggests that certain cancers may be more amenable to therapeutic proteomimetics.
Fig. 4.
Cellular internalization and efficacy of Sos proteomimetics is modulated with oncogenic Ras mutations. (A) Live-cell fluorescence imaging of Hoechst-stained Ras mutant T24 and H358 cells incubated with fluorescently labeled CHDSos-5 or DMSO for 4 h at 40× magnification. (Scale bar, 5 μm.) (B) Flow cytometry analysis of fluorescently labeled CHDSos-5 (1 μM) in T24, H358, SW780, BxPC3, and HeLa cells after 1 h treatment. (C) FP and MST analyses were performed to determine the binding affinity of CHDSos-5 and GDP-loaded H-Ras wild-type and mutant isoforms. (D) The cellular toxicity of CHDSos-5 was analyzed in an MTT cell viability assay. Bar graph shows viability of Ras wild-type and mutant cell lines treated with increasing concentrations of CHDSos-5. The results from the MTT assay were confirmed in the CellTiter-Glo luminescent cell viability assay (SI Appendix). (E) Double y-axis graph shows correlation of CHDSos-5 cellular uptake and toxicity. Results from cellular uptake studies (left axis) with 1 μM fluorescent analog and MTT cell viability assay (right axis) at 10 μM concentration are shown. (F) Representative Western blot showing ERK phosphorylation levels in H358 cells upon treatment with 0, 1, 5, and 10 μM CHDSos-5. (G) Western blots showing ERK phosphorylation levels in HeLa and H358 cells upon treatment with CHDSos-5 or negative control CHDSos-3. Bar graphs compare ERK phosphorylation in HeLa and H358 cells posttreatment with CHDSos-3 and CHDSos-5. Error bars are mean ± SD of biological duplicates.
CHDSos−5 Binds Wild-Type and Mutant Isoforms of Ras.
The enhanced cellular uptake of CHDSos-5 in cancer cells with mutated forms of Ras suggests that the Sos proteomimetic may be selectively toxic to these cells. Based on this premise, we explored the potential of CHDSos-5 to engage the mutant forms of H-Ras, specifically where G12 is mutated to cysteine, aspartic acid, and valine (G12C, G12D, and G12V). Substitution of the glycine residue with polar and β-branched residues modulates the conformation of the dynamic switch regions between the “open” and “closed” Ras forms, and these mutations are often observed in the oncogenic Ras isoforms (38). We expressed the mutants in the H-Ras construct for in vitro binding analyses. Activating mutations are more often found in the K-Ras isoform in human cancers, but the Sos-binding surface of H-Ras and K-Ras are fully conserved with differences largely localized to the C-terminal hypervariable region, which is implicated in membrane anchoring of Ras proteins (39). Titration of fluorescein-derivatized CHDSos-5 with wild-type and G12X mutant H-Ras proteins reveals that the proteomimetic binds all Ras proteins within a similar low micromolar affinity range—albeit with a preference for the wild-type form (Fig. 4C). We anticipate that the binding affinity of CHDSos-5 for wild-type, G12C, G12D, and G12V H-Ras proteins would translate to wild type and mutant forms of K-Ras.
CHDSos-5 Is Selectively Toxic to Mutant Ras Cancer Cells by Down-Regulating Ras Signaling.
Encouraged by the in vitro results, which showed that CHDSos-5 can bind wild-type and mutant Ras forms with high nanomolar to low micromolar affinity and that CHDSos-5 has selectively high permeability in Ras mutant cell lines, we evaluated the potential of the Sos proteomimetic to inhibit Ras signaling in cells. We began by assessing the toxicity of the Sos proteomimetic using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell viability assay. In preliminary studies, we learned that CHDSos-5 can function in the presence of serum. This result is important because cells often need to be treated with peptides under artificial serum-free conditions to assess their cellular potential because peptides may be retained in the medium by the hydrophobic components present in serum. As expected from the cell-permeability results, CHDSos-2 and CHDSos-5 exhibited concentration-dependent toxicity against cell lines containing oncogenic Ras mutations in comparison to the wild-type Ras HeLa control cell line (Fig. 4D and SI Appendix, Fig. S8A). Cell viability was shown to be inversely correlated to the inherent macropinocytic uptake level between different cell lines (Fig. 4E). The results suggest that exploitation of up-regulated macropinocytosis presents a potentially unexploited advantage for delivering therapeutics to mutant cancer cells. Importantly, the alanine control CHDSos-3 displayed little to no effect on the viability of the tested cell lines, suggesting that targeting of Ras is leading to cellular toxicity (SI Appendix, Fig. S8B). The MTT assay results were validated with a second cell viability assay based on the CellTiter-Glo Luminescent system, which confirmed the selective toxicity of CHDSos-5 against a mutant Ras (H358) cell line relative to the wild-type Ras control (HeLa) (SI Appendix, Fig. S8C).
To demonstrate that the cellular toxicity of CHDSos-5 is correlated with down-regulation of Ras signaling, we probed the impact of the Sos proteomimetic on the cellular concentrations of GTP-bound Ras and phosphorylated ERK. We first determined if treatment of cells with CHDSos-5 modulates Ras activation. Cellular levels of Ras-GTP were assessed with a Raf1 Ras-binding domain (RBD) pulldown assay (40). The extent of Ras phosphorylation was significantly reduced in the presence of 10 μM CHDSos-5 in H358 cells (SI Appendix, Fig. S9). Ras is a critical mediator of multiple signal transduction pathways, and ERK activation is a well-studied node in the Ras effector pathway (41). To determine if the impact of CHDSos-5 on Ras-GTP levels leads to the intended decrease in ERK phosphorylation, we treated H358 and HeLa cells in complete medium with increasing concentrations of CHDSos-3 or CHDSos-5. The resulting lysates were blotted for phosphorylated ERK. CHDSos-5 significantly reduced ERK activation levels (42), IC50 < 1 μM, in a concentration-dependent manner in the K-Ras G12C mutant cell line while exerting little effect in HeLa cells. This differential activity is consistent with the cellular permeability differences of the proteomimetic between the two cell lines (Fig. 4 F–G and SI Appendix, Fig. S10A). As expected, the alanine control CHDSos-3 did not suppress ERK phosphorylation in either cell line (Fig. 4G and SI Appendix, Fig. S10B). The observed down-regulation of activated ERK with CHDSos-5 treatment is consistent with the intracellular engagement of Ras by the proteomimetic.
Chemoproteomic Analysis Reveals Cellular Targets of CHDSos-5.
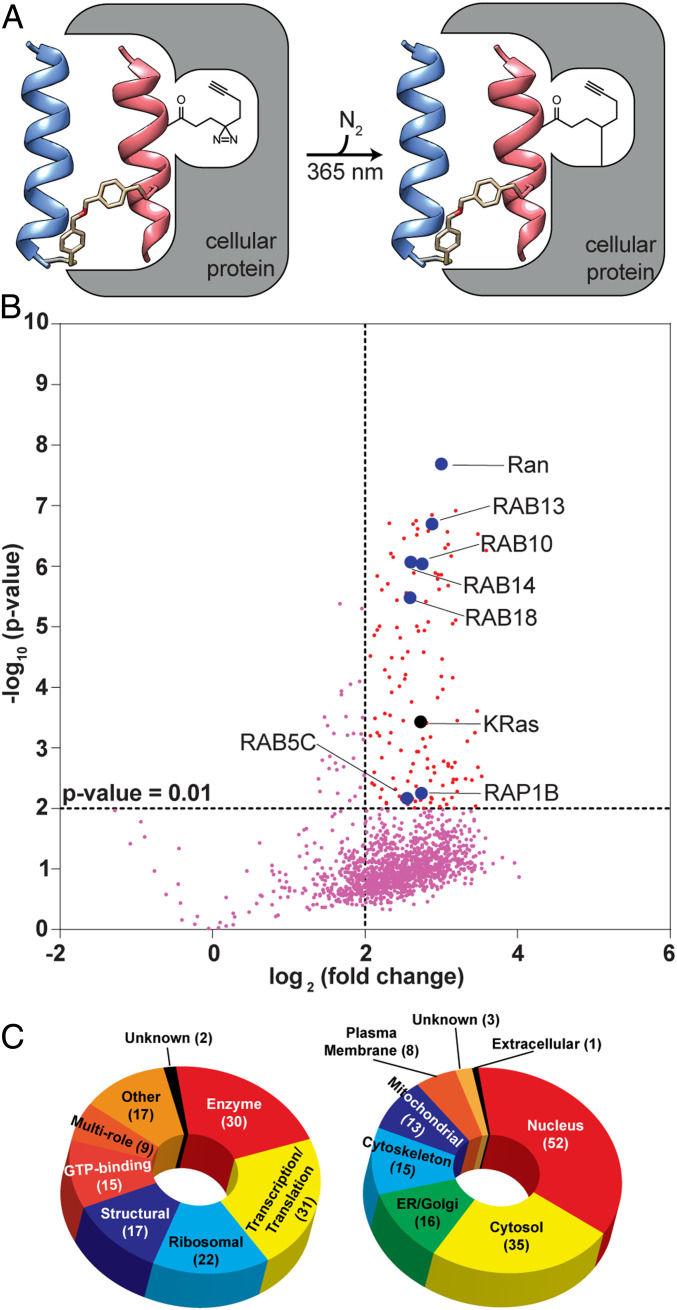
The Ras superfamily of small GTPases consists of over 150 members and includes the Ras, Rho, Rab, Arf, and Ran subfamilies (43). The superfamily has high structural and sequence conservation in the GTP/GDP nucleotide-binding domain which is engaged by various structurally conserved GEFs. The Ras–Sos complex formation is promoted by membrane localization of both proteins, but it is not known how many Ras family members Sos may engage if membrane recruitment was not a determining factor. The designed Sos proteomimetic, CHDSos-5, mimics a portion of the Sos nucleotide-binding domain and does not contain a membrane anchor. We hypothesized that CHDSos-5 would likely have multiple cellular partners, and a chemoproteomic analysis may reveal its major targets. To identify putative binding partners of CHDSos-5, we subjected H358 lung cancer cells to photoaffinity labeling followed by enrichment of the interactors and their characterization by established mass spectroscopy-based proteomics protocols (44). Labeling was accomplished using a variant of CHDSos-5 with a photolabile diazirine crosslinker attached in place of the nonnatural homoarginine residue (DZ2-CHDSos-5, Fig. 5A).
Fig. 5.
Analysis of Sos proteomimetic cellular interacting partners by quantitative MS-based proteomics. (A) Schematic depicting proximity-driven photocrosslinking of DZ2-CHDSos-5 with cellular proteins. (B) Volcano plot reveals statistically significant (P < 0.01) enriched proteins in H358 cells upon treatment with DZ2-CHDSos-5 (10 μM) for 4 h. Enriched protein targets that are members of the Ras GTPase superfamily are labeled. (C) Pie charts outlining functional diversity (Left) and cellular localization (Right) of the enriched protein targets.
Separate populations of cells were treated with DZ2-CHDSos-5 (10 μM) alongside a previously described negative control diazirine probe (CP-2-66, SI Appendix, Table S3) (45) and then exposed to UV light (365 nm) to induce photocrosslinking of DZ2-CHDSos-5–bound protein targets. Cells were lysed, and the probe-labeled proteins were conjugated to biotin azide via CuAAC, enriched with streptavidin-coated resin, and trypsinized as previously described (46). Subsequently, tryptic peptides from each population were labeled with isobaric tandem mass tags (TMT) (47) to enable quantitative comparisons and analyzed by mass spectrometry (MS)-based proteomics. Proteins were considered targets if they were enriched by an average value of >fourfold across biological duplicate experiments (P < 0.01; SI Appendix, Table S3). Overall, 143 protein targets were identified; among these, K-Ras was shown to be enriched by DZ2-CHDSos-5 along with seven other members of the Ras GTPase superfamily: the GTP-binding nuclear protein (RAN) and several Ras-related proteins (RAB13, RAB10, RAB14, RAB18, RAB5C, and RAP1B) (Fig. 5B). SI Appendix, Table S4 compares sequences of Ras-related GTPases and other G proteins enriched from proteomics analysis to human K-Ras. Each sequence was aligned and compared to K-Ras within the G-domain (aa 1 to 166) and the Sos αH/αI hairpin binding region (aa 10 to 40, 56 to 75). Sequence identity assesses the degree of fully conserved residues within the indicated regions, while sequence similarity refers to variable residue substitutions with similar chemical properties according to the Gonnet PAM 250 matrix (48). The analysis suggests that there is a high degree of sequence similarity (50 to 70%) between the enriched Ras-related and other G proteins in the Sos helix hairpin binding region (49).
The enriched targets identified by the photoaffinity labeling method include proteins with a wide array of functions (Fig. 5C). A majority of these interactors is localized within the intracellular compartment of the cell, which supports the hypothesis that CHDSos-5 avoids endosomal entrapment upon internalization (Fig. 5C). The biological impact of targeting other GTPases, beyond Ras, and non-Ras family proteins with the Sos proteomimetic remains to be determined.
Discussion
Ras remains an intractable target for traditional drug discovery. Multiple strategies to develop lead compounds have been described (3, 4, 10, 50–56), but with the exception of covalent G12C inhibitors, these leads have not advanced into clinical trials (5, 57). Ras-targeting compounds should ideally be able to engage specific mutant isoforms because pan-Ras inhibitors are expected to present unwanted side effects. Here, we describe a Sos proteomimetic that binds the nucleotide binding pocket of wild-type and mutant Ras forms but is selectively toxic to oncogenic Ras cells because the cellular internalization of the proteomimetic is governed by macropinocytosis that is up-regulated in the mutant Ras cells.
The synthetic derivative mimics a Sos helical hairpin that mediates nucleotide exchange and activation of Ras. The design and synthesis of the proteomimetic are based on a recently described strategy from our group to develop conformationally-defined minimal mimics of helical tertiary structures. Our strategy leads to CHDs whose conformational stability requires optimal knob-into-hole helix packing and an appropriately placed covalent crosslinker. We utilized computational modeling and rational design principles to incorporate noncanonical residues to enhance binding interactions of the Sos derivatives with Ras. Several design and sequence iterations were needed to develop a lead CHD with the requisite aqueous solubility and strong binding affinity for wild-type and mutant Ras isoforms. Titration 1H-15N-HSQC NMR and photoaffinity labeling studies reveal that the lead derivative, CHDSos-5, engages Ras at its nucleotide binding surface.
CHDSos-5 proved to be cell permeable and exhibited similar uptake as the well-characterized cell-penetrating Tat peptide. Significantly, CHDSos-5 displayed higher cellular uptake in Ras mutant cells relative to those with only the wild-type variant. This superior cellular internalization result in mutant Ras cells was expected based on our recent characterization of the uptake pathways for peptidomimetics—we learned that macropinocytosis is a key mechanism utilized by medium-sized peptidic compounds (34). The up-regulation of macropinocytosis in cell lines carrying mutations in Ras serves as a selectivity filter for the Sos proteomimetic to specifically engage oncogenic Ras even though its binding affinity is slightly better for wild-type Ras in biochemical experiments. We assayed the potential of CHDSos-5 to modulate phosphorylation of ERK—a well-documented downstream kinase impacted by Ras activation. CHDSos-5 proved to be a potent inhibitor of ERK with IC50 < 1 μM in the mutant Ras cell line but not in the cell line expressing wild-type Ras. Significantly, the alanine control CHDSos-3 did not modulate ERK phosphorylation at the concentrations tested. This result suggests that the efficacy of the lead proteomimetic is sequence specific. Lastly, we employed photoaffinity labeling integrated with quantitative MS-based proteomics to map interactors for CHDSos-5 in an oncogenic Ras cell line and identified K-Ras and other Ras family members. The fact that CHDSos-5 engages proteins other than Ras is not surprising; the result may reflect unknown natural partners of Sos in the absence of its membrane recruitment. While there have been many successful proteomics studies utilizing small molecules and peptides to interrogate PPIs (45, 58, 59), the use of protein tertiary mimics to map binding partners of therapeutically important protein domains has not been previously described. The biological relevance of targeting previously unknown binding partners of the Sos helical hairpin remains to be deciphered. In summary, we describe a pan-Ras ligand that is selectively toxic to cells that express mutant Ras isoforms and up-regulate macropinocytosis as a nutrient uptake pathway. The selective uptake of the Sos proteomimetic suggests a therapeutic strategy for targeting oncogenic Ras without the requirement for ligands that specifically engage its various mutations. Our preliminary studies focused on the evaluation of a handful of model mutant Ras cell lines with the lead proteomimetic. Further modifications of the lead proteomimetic are required to improve its binding affinity for Ras for advanced in vitro and in vivo applications.
Materials and Methods
Synthesis of CHD Peptides.
Peptide synthesis was performed using standard Fmoc solid-phase peptide synthesis on Rink Amide resin unless otherwise specified. After final Fmoc deprotection, the peptides were acetylated at the N terminus using a solution of 0.5 M acetic anhydride and 5% diisopropylethylamine (DIEA) in NMP for 30 min prior to cleavage from resin. The two desired peptide strands were cleaved from resin, purified via reversed phase (RP)-HPLC, and characterized by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometry. The two strands are crosslinked as previously described (SI Appendix, Scheme S1) (18, 20, 21). Briefly, each strand contains a single cysteine residue strategically placed for addition of the dibenzylether crosslinker. The crosslinker was synthesized according to previously published protocols (20). A purified strand was initially dissolved in 20 mM NH4CO3 buffer (pH 8.1) and subsequently added to a solution of the dibenzylether crosslinker (5 to 10 eq.) in acetonitrile. The reaction mixture was stirred for 90 min at 20 °C followed by purification via RP-HPLC (25 to 65% acetonitrile gradient in water with 0.1% trifluoroacetic acid (TFA) over 40 min) on a C18 reverse-phase column. The purified product was subsequently lyophilized. The lyophilized monocysteine alkylated peptide was then dissolved in 1:1 acetonitrile:20 mM aqueous NH4CO3 solution (pH 8.1). An excess amount of the second strand with a free cysteine (1.5 eq.) was added, and the resulting mixture was stirred for 1 h at 20 °C. The reaction mixture was subjected to RP-HPLC (25 to 65% acetonitrile gradient in water with 0.1% TFA over 40 min) on a C18 reverse-phase column) and characterized by analytical HPLC and MALDI-TOF spectroscopy (SI Appendix, Figs. S11 and S12 and Table S2)
Synthesis of Fluorescein-Labeled Peptides.
The peptide sequences (SI Appendix, Table S2) were synthesized as previously described with β-alanine added to the N terminus to serve as a linker prior to coupling with fluorescein isothiocyanate (3 eq.) and DIEA (3 eq.) overnight and protected from light. The fluorophore-conjugated peptides were then cleaved from resin, purified via RP-HPLC, and characterized with MALDI-TOF spectrometry.
Synthesis of Diazirine Photocrosslinker Peptides.
The peptide sequences (SI Appendix, Fig. S12 and Table S2) were synthesized as previously described with L-glycine added to the N terminus as a linker prior to coupling of 3-(3-(but-3-ynyl)-3H-diazirin-3-yl)propanoic acid (Sigma-Aldrich) under standard peptide coupling conditions overnight and protected from light. The diazirine-conjugated peptides were then cleaved from resin, purified via RP-HPLC, and characterized with analytical liquid chromatography mass spectrometry (LCMS).
Circular Dichroism Spectroscopy.
CD experiments were conducted on an Jasco J-1500 Circular Dichroism spectrometer equipped with a temperature controller using 1 mm length cells and a scan speed of 4.0 nm/min at 298 K. The generated spectra were averaged over five scans with baseline subtraction. Raw values were normalized to molar residue ellipticity. The samples were prepared in a buffer containing 50 mM potassium fluoride in water (pH 7.4) to a final peptide concentration of 20 μM, unless otherwise mentioned. The concentration of each sample was monitored via the UV absorbance at 280 nm of a tryptophan residue.
Protein Purification.
Wild-type and mutant N-terminal His6-H-Ras (residues 1 to 166) in pProEx HTb expression vector were expressed in Escherichia coli (BL21). Cells were grown at 37 °C to an absorbance of 0.7 at 600 nm. Protein expression was induced with 500 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 6 h at 30 °C or overnight at 18 °C. The bacterial cells were pelleted via centrifugation at 4,500 rpm for 15 min. Bacterial pellets were resuspended in a lysis buffer containing 20 mM Tris pH 7.4, 300 mM NaCl, 2 mM 2-mercaptoethanol, and a complete, EDTA-free protease inhibitor mixture (Roche). The lysis, elution, and dialysis buffers were also supplemented with 2.5 mM MgCl2. The resuspended pellets were sonicated using a Branson Cell Disrupter 200. Clarified lysates were formed upon centrifugation at 13,000 rpm for 20 min at 4 °C and incubated with charged Ni-NTA resin (Invitrogen) at 4 °C for 1 h. The resin beads were washed five times with resuspension buffer containing 5 mM imidazole. The His6-tagged proteins were eluted via gravity flow with buffer containing 200 mM imidazole in buffer containing 20 mM Tris, 300 mM NaCl, pH 7.4. Eluted proteins were dialyzed twice against buffer containing 20 mM Tris, 300 mM NaCl, 2.5 mM MgCl2, pH 7.4 for H-Ras. For HSQC and photoaffinity labeling studies, the polyhistidine tag was cleaved using Tobacco etch virus (TEV) protease before proceeding to nucleotide loading. Concentrated eluates were subjected to nucleotide loading and size-exclusion chromatography (GE Life Sciences) at 4 °C with 25 mM Tris, 50 mM NaCl, pH 7.5 buffer, and the desired monomeric peaks were collected. The eluted protein samples were concentrated with 3,000 Da molecular weight cutoff Amicon centrifugal columns (Millipore) in dialysis buffer containing 10% glycerol (v/v) before being snap frozen in liquid N2 and stored at −80 °C until further use.
Nucleotide Loading of Ras.
Nucleotide loading for FP, HSQC, and nucleotide displacement studies were conducted as previously described (60). Purified H-Ras protein was incubated with GDP or GTP (10 eq.) in loading buffer (20 mM Tris, 300 mM NaCl, 5 mM EDTA, pH 7.4) for 90 min on ice. Reactions were then quenched with 12 mM MgCl2 and incubated for 30 min on ice. Depending on the subsequent assay, the protein was subjected to size-exclusion chromatography via fast protein liquid chromatography or preequilibrated NAP-5 (GE Life Sciences) columns to remove excess free nucleotide and additional purification.
Fluorescence Polarization Binding Assay.
The relative binding affinities of CHDs for wild-type and mutant His6-tagged H-Ras (1 to 166) preloaded with GDP were determined using direct FP assays with fluorescein-tagged CHD peptides (Flu-αHBS and Flu-α3βHBS). These experiments were conducted with a DTX 880 Multimode Detector (Beckman) at 25 °C with excitation and emission wavelengths set to 485 and 525 nm, respectively. The addition of increasing concentrations of His6-H-Ras-GDP to a 15 nM solution of fluorescein-labeled CHD in 50 mM Tris, 100 mM NaCl, 0.1% pluronic acid, pH 7.4 was carried out in black, U-bottom 96-well plates (Brand). The generated binding-affinity (KD) values for each compound are from biological triplicate studies and were determined via fitting to a sigmoidal dose–response nonlinear regression model on Graphpad Prism 6.
where RT is the total concentration of H-Ras (1-166), LST is the total concentration of fluorescence peptide, and FSB is the fraction of bound fluorescence peptide.
Microscale Thermophoresis Binding Assay.
Fluorescent labeling of the H-Ras protein was performed using the Monolith Protein Labeling Kit RED-MALEIMIDE second Generation from NanoTemper (MO-L014) or AFDye 647 maleimide fluoroprobes (1122-1). Briefly, 20 nM of H-Ras protein was labeled with 1.5 equivalents of cysteine reactive dye in 1× phosphate buffered saline (PBS) under dark for 90 min. Labeled Ras proteins were then purified via the kit-provided column, and the degree of label (DOL) was calculated by using the dye’s absorbance and protein’s original absorbance. The observed DOL value after the labeling for wild-type H-Ras-GDP was 0.43 and G12V H-Ras-GDP was 0.98.
MST binding assays were conducted with NanoTemper Monolith NT.115 Pico. Assay conditions were optimized with a premium coated capillary tube from NanoTemper to avoid random adsorption. Measurement was performed at 25 °C using 10 to 15% laser excitation power for 20 s. The assay buffer was composed of 50 mM Tris, 100 mM NaCl, and 0.1% pluronic acid. Before the measurements, 5 to 10 nM of the dye-labeled protein was treated with serial dilutions of indicated CHDs and the resulting solution incubated for 90 mins at room temperature. Error bars are generated from triplicates. Binding affinities were obtained from the MO analysis software in NT.115 Pico with the following equation:
where Unbd is the response value of unbound state, Bd is the response value of bound state, and TC is the final concentration of fluorescent molecule.
Serum Stability Assay.
Proteolytic stability of CHDSos-5 was assessed in 25% serum by using fetal bovine serum (FBS, Innovative Research) in Roswell Park Memorial Institute (RPMI)-1640 medium. A 45 µL mixture of 33% FBS (v/v) in serum-free medium was prepared for each sample. The addition of 15 µL indicated peptide in serum-free medium (25% FBS final) per each experiment was recorded as the starting time point. Time points of 5 min, 6 h, 12 h, and 24 h were analyzed in triplicate. Each experiment was quenched at the determined time point by adding 30 μL 100% EtOH, which was prepared at −70 °C. After cooling on ice for 10 min, each sample was pelleted at 14,000 × g for 5 min. A total of 30 µL supernatant was isolated, and 3 µL 500 µM L-Tryptophan was titrated into the sample to be used as an internal standard. The resulting mixture was then immediately subjected to RP-HPLC with a C18 3.5 μm 2.1 × 150 mm analytical column and monitored at 280 nm. Eluting peaks were collected, and the mass of each peak was determined using MALDI-TOF spectroscopy. Integrated peak area of nondegraded peptides was used to determine the percent surviving in each given condition.
Photoaffinity Labeling and Visualization of the Conjugated CHD-Ras Complex.
For each experiment, a sample of 250 μM diazirine-labeled CHD peptide was incubated with 50 μM purified H-Ras for 90 min at room temperature (100 μL total volume) in binding buffer (50 mM Tris, 100 mM NaCl, 0.1% pluronic acid, pH 7.4). Incubated samples were then photoirradiated for 5 min at 24 °C. Photoconjugated samples were then mixed with 1× sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 5% 2-mercaptoethanol (200 μL final volume) and heated at 95 °C for 5 min. Heated samples were then separated via SDS-PAGE gel electrophoresis on a 1.0 mm 15% polyacrylamide gel. The resulting gel was stained with Coomassie Brilliant Dye and subsequently destained (40% MeOH/ 10% acetic acid/ 50% water).
In-Solution Tryptic Digest MS.
In-gel tryptic digest was conducted with the In-Gel Tryptic Digestion Kit (Thermo Scientific) according to the manufacturer’s instructions. Upon photoaffinity labeling, the conjugated band was isolated as a 1 × 1 mm segment for in-gel trypsin digest. The isolated gel band was then incubated with 200 μL Coomassie destaining solution (25 mM ammonium bicarbonate in 1:1 μL trapure water:acetonitrile) for 30 min at 37 °C. After incubation, destaining buffer was removed, and the destaining step was repeated twice more. The destained gel band was treated with reducing buffer (25 mM ammonium bicarbonate, 50 mM TCEP) for 10 min at 60 °C. After discarding the reducing buffer, the band was treated with 200 μL iodoacetamide alkylation solution (25 mM ammonium bicarbonate, 100 mM iodoacetamide) for 30 min at 37 °C while shaking in the dark. After alkylation, the gel piece was again washed with 100 μL destaining buffer twice before treatment with 100 μL acetonitrile for 15 min at room temperature. The gel band was then air dried for 15 min after removal of the acetonitrile. Trypsin solution was comprised of 5 μg trypsin protease (Pierce, MS grade) dissolved in 100 μL 25 mM ammonium bicarbonate buffer, pH 8.0. The dried gel was treated with 100 μL trypsin solution overnight at 30 °C while shaking. After digestion, the buffer was removed, and the digested peptide was extracted from the gel with 25 μL 1% trifluoroacetic acid. Extracted samples in 1% trifluoroacetic acid were then diluted with 25 μL ultrapure water and submitted for MALDI-TOF MS analysis.
HSQC Titration NMR Analysis.
The purification procedure is nearly identical to the previously described protocol with a couple exceptions. The E. coli (BL21) cells (4 L) containing the His6-H-Ras construct were grown at 37 °C in fully supplemented Luria broth until optical density (O.D.) 0.8 was reached. The cells were pelleted and resuspended in minimal M9 medium (1 L) before being supplemented with 20% glucose and 15NH4Cl as the sole nitrogen source (61). Protein expression was induced with 1 mM IPTG at O.D. 0.8 overnight at 18 °C. Protein purification and subsequent concentration were performed as described above. The His6-tag was cleaved off upon incubation with recombinant His6-tagged TEV protease (Invitrogen) overnight at 4 °C according to the manufacturer’s protocol. The resulting protein mixture was loaded onto a charged Ni-NTA column (Invitrogen), and the tagless protein was collected in the flow-through fraction. 15N-labeled H-Ras was then preloaded with GDP according to the previously mentioned protocol. Uniformly, 15N-labeled H-Ras-GDP underwent buffer exchange into NMR buffer (20 mM Na2HPO4-NaH2PO4, pH 5.5, 150 mM NaCl, 10 mM MgCl2) concentrated with an Amicon Ultra centrifugal filter (Millipore) and was supplemented with 10% D2O prior to analysis. Data collection was conducted on a 600 MHz Bruker 4-channel NMR system equipped with a 5 mm TCI cryogenic probe at 25 °C using a standard pulse sequence and analyzed by TopSpin 4.0.6 (Bruker). 15N-1H-HSQC spectra of 15N-labeled H-Ras-GDP (100 μM) alone was collected, and peaks were assigned based on published data. For peptide titration experiments, 2.5 and 5 equivalents of CHDSos-5 were dissolved in matching buffer and incubated with 15N-1H-Ras (100 μM). The mean chemical shift difference (∆δ-NH) observed for 1H and 15N nuclei of various resonances was calculated according to the following equation with α = 0.14:
Cell Culture.
T24, HCT-116, and HeLa cells were maintained in Dulbecco’s modified eagle medium (DMEM, Corning Cellgro) supplemented with 10% FBS (Innovative Research), 1× penicillin/streptomycin (EMD Millipore), and 10 mM Hepes buffer (Sigma-Aldrich). H358, Panc 10.05, A549, and BxPC3 cells were maintained in RPMI-1640 medium supplemented with 10% FBS (Innovative Research), 1× penicillin/streptomycin, 10 mM Hepes buffer, and 1× sodium pyruvate (Sigma-Aldrich). All cells were kept in a humidified incubator at 37 °C and 5% CO2.
Live-Cell Fluorescence Microscopy.
The indicated cell lines were seeded at 5 × 105 cells/well in poly-D-lysine–coated 35 mm plates (MatTek) and incubated overnight. The growth medium is aspirated and washed with serum-free medium. The cells are then incubated with 1 μM (final) of fluorescein-conjugated peptides dissolved in serum-free medium (0.4% DMSO v/v) for 4 h at 37 °C while protected from light. All compounds are dissolved as concentrated stocks in DMSO. After the specified incubation time, each plate was aspirated and treated with Hoechst dye solution (Hoechst 33342, ThermoFisher Scientific) for 10 min to stain cell nuclei. The Hoechst dye stock solution (10 mg/mL) was diluted 1:2,000 in PBS to form the working mixture. The dye solution was removed, and the plate was gently washed 3× with PBS. DMEM (high glucose, Hepes, no phenol red, 20% FBS) was added to each plate and used as the imaging solution. Fluorescence images were acquired on an Eclipse Ti Fluorescence Microscope (Nikon) equipped with NIS-Elements imaging software and using a 40× objective lens.
Flow Cytometry.
The indicated cell lines were seeded at 1 × 105 cells/well in clear polystyrene 24-well plates (Corning) and incubated overnight. The initial growth medium is replaced with serum-free medium and incubated for 2 h at 37 °C. Upon aspiration, the cells are incubated in serum-free medium containing 0.4% DMSO (v/v for 1 h. The cells were then treated with 1 μM (final) fluorescein-conjugated peptides in serum-free medium for another hour while protected from light. All compounds are dissolved as concentrated stocks in DMSO. Each well was aspirated and treated with 1× trypsin (0.25% trypsin, 2.21 mM EDTA, Corning Cellgro) for 10 min at 37 °C. After trypsinization, the resulting solution was mixed with cold serum-free medium and collected. The samples are centrifuged at 500 rpm for 5 min at 4 °C. The supernatant was removed, and the cell pellets were resuspended with cold PBS before being placed on ice. Each sample was treated with 10% trypan blue (v/v) immediately before analysis by flow cytometry on a Becton Dickinson Accuri flow cytometer. The presented data consist of the median fluorescence intensities for at least 10,000 cells/sample and were processed using FlowJo (Tree Star Inc.).
Cell Viability Assay.
Cell viability was monitored using the MTT (Sigma-Aldrich, M2128) or CellTiter-Glo Luminescent cell viability assay (Promega).
MTT Cell Viability Assay.
Cell viability was monitored using the MTT (Sigma-Aldrich, M2128) Luminescent cell viability assay. Cells were plated in clear 96-well plates at 2,000 cells/well and allowed to affix overnight at 37 °C. Each well is gently aspirated and washed with serum-free medium before introducing peptide inhibitors dissolved in complete medium with 0.5% DMSO (v/v final) to desired concentrations (90 μL/well). The cells are incubated in the presence of peptide for 72 h at 37 °C. MTT reagent solution is composed of Thiazolyl Blue Tetrazolium Bromide dissolved in Dulbecco’s Phosphate Buffered Saline (DPBS), pH 7.4 to a concentration of 5 mg/mL and subsequently sterile filtered into a light-protected container. MTT reagent is added to each well (0.45 mg/mL/well final). The cells are incubated at 37 °C for an additional 4 h. Upon careful removal of the supernatant, 150 μL DMSO (solubilization solution) is added to each well and mixed to ensure complete solubilization and release of the insoluble purple formazan precipitate into solution. Absorbance values are recorded with a Synergy HT Multi-Detection Microplate Reader (BioTek) at 570 nm.
CellTiter-Glo Luminescent Cell Viability Assay.
Cell viability was monitored using the CellTiter-Glo Luminescent cell viability assay (Promega). Cells were plated in white, opaque 96-well plates at 2,000 cells/well and allowed to affix overnight at 37 °C. Each well is gently aspirated and washed with serum-free medium before introducing peptide inhibitors dissolved in complete medium (DMSO is not needed to dissolve CHDSos-5) to desired concentrations (90 μL/well). The cells are incubated in the presence of peptide for 72 h at 37 °C. The plate and its contents are then equilibrated to room temperature for ∼30 min after peptide treatment. Upon gentle removal of the well supernatant, 50 μL 1× PBS is added to each well. An equivalent volume (50 μL) of CellTiter-Glo 2.0 reagent is then added to each well with the resulting contents mixed for 2 min on an orbital shaker to induce cell lysis. The plate is allowed to incubate at room temperature for 10 min before recording the luminescent signal (0.25 to 1 s integration time) with a DTX 880 Multimode Detector (Beckman). Each plate included triplicate wells of a positive death control (cells with 10% DMSO), a negative control (cells with 0.05% DMSO), and a blank (no cells with 0.05% DMSO).
Ras Activation Assay.
H358 cells were initially seeded in 6-well plates (1 × 106 cells/well) and allowed to attach overnight. The cells were treated with indicated peptides dissolved in complete medium at indicated concentrations for 6 h at 37 °C. Ras activity was determined by the Active Ras Pull-Down and Detection Kit (Thermo Scientific, catalog No. 16117) according to the manufacturer’s instructions. In brief, cells were lysed with 250 μL lysis buffer and scraped off; the resulting lysate was centrifuged at 13,000 rpm for 10 min at 4 °C. Precleared lysates were subsequently added to 80 μg GST-tagged RBD and prewashed glutathione agarose beads for 1 h at 4 °C under constant rocking. The beads were then pelleted, washed three times with buffer, and eluted for Western blotting with 50 μL 2× reducing sample buffer.
ERK Activation Assay.
H358 or HeLa cells were initially seeded in 6-well plates (1 × 106 cells/well) and allowed to attach overnight. The cells were then incubated with indicated peptides dissolved in complete medium at specified concentrations for 6 h at 37 °C. The cells were washed twice with ice-cold PBS and then lysed in cold radioimmunoprecipitation assay (RIPA) buffer (200 μL/well) containing 25 mM Tris HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium doexycholic acid, 0.1% SDS, Roche Complete Protease inhibitor mixture (1×, Sigma-Aldrich, P2714), and Roche PhosSTOP phosphatase inhibitor mixture tablets (1×, Sigma-Aldrich, 4906845001). The cells are kept on ice for 5 min with occasional swirling for uniform spreading. Cell lysates are generated with a cell scraper, transferred to a microcentrifuge tube, and centrifuged at 13,000 rpm for 10 min at 4 °C. The clarified supernatant was collected, and the total protein concentration was measured by Pierce BCA protein assay. The lysates were subjected to SDS-PAGE (10 μg/lane loading) and Western blot analysis via immunoblotting. Primary antibodies include Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling 4370), p44/42 MAPK (Erk1/2) (Cell Signaling 4695), Ras (Abcam ab108602), and α-tubulin (Cell Signaling 2144). Secondary antibodies include anti-rabbit IgG, HRP-linked antibody (Cell Signaling 7074P2, 1:2,000). After primary antibody incubation, blotted membranes were probed with secondary antibodies and SignalFire ECL Reagent (Cell Signaling 6883P3) and imaged using a ChemiDoc Imaging System (Bio-Rad). Comparative blot densitometry was performed with ImageJ (NIH) and normalized to tubulin expression.
Preparation and Enrichment of Sample Lysates for Proteomics.
H358 cells were grown to 80 to 95% confluence in 10 cm plates with FBS-supplemented RPMI-1640. The growth medium was aspirated, and the cells were washed with DPBS. The cells were then incubated with serum-free medium containing probes DZ2-CHDSos-5 (20 µM in RPMI-1640 with 0.4% DMSO, two replicates) or control probe CP-2-66 (20 µM in RPMI-1640 with 0.4% DMSO, four replicates) for 4 h at 37 °C under an atmosphere of 5% CO2 before being irradiated under UV light (365 nm, 30 min, 4 °C). The cells were washed with DPBS, scraped and transferred to 1.5 mL Eppendorf tubes, pelleted, and stored at −80 °C until the next step.
Cell pellets were resuspended in 500 μL DPBS and lysed by sonication (15 ms on, 40 ms off, 15% amplitude, 1 s total on × 2). Protein concentrations were normalized (1 to 2 mg/mL; final volume of 500 μL with DPBS) using the Lowry Protein Assay (Pierce). To each sample was added solutions of Tris((1-benzyl-4-triazolyl)methyl)amine (30 µl, 1.7 mM in DMSO/t-BuOH 1:4 v/v), Tris(2-carboxyethyl)phosphine (10 μL, 50 mM), biotin-PEG3-azide (5 μL, 100 μM), and CuSO4 (10 μL, 50 mM), and the samples were shaken at room temperature for 1 h. A cold MeOH/CHCl3 mixture (2.5 mL, 4:1 v/v) was added to each lysate, followed by 1 mL cold DPBS. The resulting mixture was vortexed and centrifuged (4,700 × g, 10 min, 4 °C). The organic and aqueous layers were aspirated, and the remaining protein disk was further washed via sonication in cold MeOH/CHCl3 solution (2 mL, 4:1) and pelleted by centrifugation (4,700 × g, 10 min, 4 °C). The protein pellet was aspirated and combined with freshly prepared urea solution (500 μL, 6 M in DPBS) and a solution of SDS (with 10 μL 10% w/v) by sonication. A freshly prepared 1:1 solution (50 μL) of TCEP (200 mM in DPBS) and K2CO3 (600 mM in DPBS) was added, and the mixture was incubated for 30 min at 37 °C with shaking. A solution of freshly prepared iodoacetamide (70 μL, 400 mM in DPBS) was added, and the mixture was incubated at 25 oC in the absence of light. Each sample was combined with a solution of SDS (130 μL, 10% in DPBS w/v) followed by DPBS (5.5 mL) and incubated with preequilibrated streptavidin-agarose beads (100 μL 50% slurry; Pierce) for 1.5 h at RT while rotating. The streptavidin beads were pelleted by centrifugation (750 g, 2 min, 4 °C) and sequentially washed with SDS solution (1 × 5 mL, 0.2% in DPBS), DPBS (2 × 5 mL), and triethylammonium bicarbonate buffer (1 × 5 mL, TEAB, 100 mM, pH 8.4). The beads were resuspended in TEAB (0.5 mL, 100 mM pH 8.5) and transferred into LoBind microcentrifuge tubes. The tube was washed once more with TEAB (0.5 mL, 100 mM pH 8.5) to ensure complete transfer of the beads, which were then pelleted by centrifugation (750 × g, 2 min, 4 °C) and the supernatant aspirated. Each sample of beads was combined with a solution of CaCl2 (2 μL, 100 mM) and a solution of sequencing-grade porcine trypsin (2 μg, Promega in 200 μL TEAB, 100 mM, pH 8.4) and incubated for 14 h at 37 °C with shaking. The beads were pelleted by centrifugation (750 × g, 2 min, 4 °C), and the supernatants were transferred to new LoBind microcentrifuge tubes. Each digested sample was TMT 10plex labeled (Thermo Fisher Scientific): for each sample, a stock solution of TMT reagent (8 μL, 20 μg/μL) was added along with dry MS-grade acetonitrile (final acetonitrile concentration 30% v/v), followed by incubation at RT for 1 h. The reaction was quenched by adding hydroxylamine (6 μL) and left to stand for 15 min, followed by the addition of formic acid (5 μL). Each TMT-labeled sample was dried via vacuum centrifugation, and the samples were combined by redissolving one sample in a solution of TFA (200 μL, 0.1% in water) and transferring the solution into each sample tube until all samples were redissolved. The process was repeated with a further volume of TFA solution (100 μL, in water, final volume 300 μL), and the combined sample was dried via vacuum centrifugation. The samples were fractionated using a fractionation kit (Pierce High pH Reversed-Phase Peptide Fractionation Kit) according to the manufacturer’s instructions. Briefly, the peptide fractions were eluted from reversed-phase spin columns with consecutive solutions of 0.1% triethylamine combined with MeCN (5, 7.5, 10, 12.5, 15, 17.5, 20, 22.5, 25, 30, 50, and 95% MeCN). The fractions were combined pairwise (fraction 1 with fraction 7, fraction 2 with fraction 8, etc.), dried by vacuum centrifugation, and stored at −80 °C until ready for injection.
LC-MS Analysis for Proteomics.
TMT samples were analyzed using a Thermo Fisher Scientific Orbitrap Fusion Lumos mass spectrometer equipped with an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific) following previously reported procedures (46, 62). Briefly, dissolved samples (20 µL; 0.1%, v/v formic acid in water) were injected (3 µL/injection) onto an Acclaim PepMap RSLC analytical column (75 μm × 15 cm) equipped with an Acclaim PepMap 100 trap column (75 μm × 20 mm) and eluted using the following gradient (300 µL/min, column temperature 35 °C): 2% buffer B (0.1% formic acid in acetonitrile) and 98% buffer A (0.1% formic acid in water) for 10 min; buffer B increased to 30% over 192 min, then to 60% over 6 min, followed by an increase to 95% over 1 min and held steady for 5 min; buffer B was decreased to 2% over 1 min where it remained for 6 min. The voltage applied to the nano-LC electrospray source was 2.0 kV, and MS1 spectra were acquired at a resolution of 120,000 with an automatic gain control (AGC) target value of 1 × 106 ions and a maximum injection time of 50 ms. A data-dependent acquisition mode was used (repeat count 1, duration 20 s), with a scan range of 375 to 1,500 m/z. Collision-induced dissociation (CID) was performed for MS2 peptide fragmentation (quadrupole ion trap analysis, AGC 1.8 × 104, CID collision energy 30%, maximum injection time 120 ms, isolation window 1.6), and the MS3 precursor was fragmented through high-energy CID (collision energy 65%). Synchronous precursor selection was enabled to include up to 10 MS2 fragment ions for the MS3 spectrum, detected with the Orbitrap (resolution of 50,000, AGC target value of 1.5 × 105, maximum injection time of 120 ms).
Proteomics Data Analysis.
Data processing was performed as in a previously reported procedure (62). Briefly, the Proteome Discoverer 2.4 software package (Thermo Fisher Scientific) was used to determine peptide sequences using the Homo sapiens proteome database (42,358 sequences) via SEQUEST HT algorithm (10 ppm precursor mass tolerance, 0.6 Da fragment ion mass tolerance, one missed cleavage allowed, target false discovery rate 1% [Percolator]). Static modifications were set for arbamidomethyl (C, +57.02146) and the TMT-tag (K and N-terminal, +229.1629), while oxidation (M, +15.994915) as a variable modification. Peptide quantitation was performed on the MS3 level with reporter ion mass tolerance set to 20 ppm. Subsequent statistical analysis was performed in Python 3.6.5. TMT abundances and ratios obtained from Proteome Discoverer were transformed with log2(x), and P values were obtained using t tests with two biological replicates. Additionally, identified proteins were required to have a minimum of three unique peptides. Quantitative data are listed in SI Appendix, Table S3. The mass spectrometric proteomics datasets have been deposited to the MassIVE repository with the dataset identifier MSV000086972.
Supplementary Material
Acknowledgments
We thank Dafna Bar-Sagi's Laboratory (NYU School of Medicine) for the Ras mutant cancer cell lines and plasmids for the expression of recombinant H-Ras and their associated mutant isoforms. P.S.A. thanks the NIH (R35GM130333) for a grant in support of this work. S.H.H. is supported by the Ted Keusseff Fellowship. K.C.R.-C. thanks the New York University Dean’s Undergraduate Research Fund.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.-A.S. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101027118/-/DCSupplemental.
Data Availability
Proteomics data have been deposited in MassIVE Repository (MSV000086972). All other study data are included in the article and/or SI Appendix.
References
- 1.Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G., Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J., The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Moore A. R., Rosenberg S. C., McCormick F., Malek S., RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Smaill J. B., Liu T., Ding K., Lu X., Small-molecule inhibitors directly targeting KRAS as anticancer therapeutics. J. Med. Chem. 63, 14404–14424 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Canon J., et al., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., et al., GTP-state-selective cyclic peptide ligands of K-Ras(G12D) block its interaction with Raf. ACS Cent. Sci. 6, 1753–1761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakamoto K., Masutani T., Hirokawa T., Generation of KS-58 as the first K-Ras(G12D)-inhibitory peptide presenting anti-cancer activity in vivo. Sci. Rep. 10, 21671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo D. Y., Hauser A. D., Joy S. T., Bar-Sagi D., Arora P. S., Covalent targeting of Ras G12C by rationally designed peptidomimetics. ACS Chem. Biol. 15, 1604–1612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patgiri A., Yadav K. K., Arora P. S., Bar-Sagi D., An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 7, 585–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kholodenko B. N., Hancock J. F., Kolch W., Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol. 11, 414–426 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y. S., et al., Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 110, E3445–E3454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kushal S., et al., Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 15602–15607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., Cheloha R. W., Watanabe T., Gardella T. J., Gellman S. H., Receptor selectivity from minimal backbone modification of a polypeptide agonist. Proc. Natl. Acad. Sci. U.S.A. 115, 12383–12388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelay-Gimeno M., Glas A., Koch O., Grossmann T. N., Structure-based design of inhibitors of protein-protein interactions: Mimicking peptide binding epitopes. Angew. Chem. Int. Ed. Engl. 54, 8896–8927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horne W. S., Grossmann T. N., Proteomimetics as protein-inspired scaffolds with defined tertiary folding patterns. Nat. Chem. 12, 331–337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adihou H., et al., A protein tertiary structure mimetic modulator of the Hippo signalling pathway. Nat. Commun. 11, 5425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadek J., et al., Modulation of virus-induced NF-κB signaling by NEMO coiled coil mimics. Nat. Commun. 11, 1786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins A. M., Wuo M. G., Arora P. S., Protein-protein interactions mediated by helical tertiary structure motifs. J. Am. Chem. Soc. 137, 11622–11630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuo M. G., Hong S. H., Singh A., Arora P. S., Synthetic control of tertiary helical structures in short peptides. J. Am. Chem. Soc. 140, 16284–16290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuo M. G., Mahon A. B., Arora P. S., An effective strategy for stabilizing minimal coiled coil mimetics. J. Am. Chem. Soc. 137, 11618–11621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauck F., Smith C. A., Friedland G. F., Humphris E. L., Kortemme T., RosettaBackrub–A web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 38, W569–W575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rooklin D., et al., Targeting unoccupied surfaces on protein-protein interfaces. J. Am. Chem. Soc. 139, 15560–15563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kortemme T., Kim D. E., Baker D., Computational alanine scanning of protein-protein interfaces. Sci. STKE 2004, pl2 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Steinkruger J. D., et al., The d’–d–d’ vertical triad is less discriminating than the a′–a–a′ vertical triad in the antiparallel coiled-coil dimer motif. J. Am. Chem. Soc. 134, 2626–2633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadley E. B., Testa O. D., Woolfson D. N., Gellman S. H., Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proc. Natl. Acad. Sci. U.S.A. 105, 530–535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher J. M., et al., A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 1, 240–250 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Lavigne P., Sönnichsen F. D., Kay C. M., Hodges R. S., Interhelical salt bridges, coiled-coil stability, and specificity of dimerization. Science 271, 1136–1138 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Sondermann H., et al., Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Moerke N. J., Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr. Protoc. Chem. Biol. 1, 1–15 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Kallenbach N. R., Lyu P. C., Zhou H. X., “CD spectroscopy and the helix-coil transition in peptides and polypeptides” in Circular Dichroism and the Conformational Analysis of Biomolecules, Fasman G. D., Ed. (Plenum Press, New York, 1996), pp. 201–259. [Google Scholar]
- 33.Manning M. C., Woody R. W., Theoretical CD studies of polypeptide helices: Examination of important electronic and geometric factors. Biopolymers 31, 569–586 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Yoo D. Y., et al., Macropinocytosis as a key determinant of peptidomimetic uptake in cancer cells. J. Am. Chem. Soc. 142, 14461–14471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Commisso C., et al., Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lönn P., et al., Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 6, 32301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lönn P., Dowdy S. F., Cationic PTD/CPP-mediated macromolecular delivery: Charging into the cell. Expert Opin. Drug Deliv. 12, 1627–1636 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Lu S., Jang H., Nussinov R., Zhang J., The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci. Rep. 6, 21949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castellano E., Santos E., Functional specificity of ras isoforms: So similar but so different. Genes Cancer 2, 216–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor S. J., Resnick R. J., Shalloway D., “Nonradioactive determination of Ras-GTP levels using activated ras interaction assay” in Methods in Enzymology (Academic Press, 2001), vol. 333, pp. 333–342. [DOI] [PubMed] [Google Scholar]
- 41.García-Gómez R., Bustelo X. R., Crespo P., Protein-protein interactions: Emerging oncotargets in the RAS-ERK pathway. Trends Cancer 4, 616–633 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Samatar A. A., Poulikakos P. I., Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 13, 928–942 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Vigil D., Cherfils J., Rossman K. L., Der C. J., Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker C. G., Pratt M. R., Click chemistry in proteomic investigations. Cell 180, 605–632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker C. G., et al., Ligand and target discovery by fragment-based screening in human cells. Cell 168, 527–541.e29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., et al., Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs. Nat. Chem. 11, 1113–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McAlister G. C., et al., MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonnet G. H., Cohen M. A., Benner S. A., Exhaustive matching of the entire protein sequence database. Science 256, 1443–1445 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Colicelli J., Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer-Smith R., et al., Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welsch M. E., et al., Multivalent small-molecule pan-RAS inhibitors. Cell 168, 878–889.e29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer T., et al., Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 109, 5299–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Q., et al., Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 51, 6140–6143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostrem J. M., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Downward J., Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Stephen A. G., Esposito D., Bagni R. K., McCormick F., Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Lanman B. A., et al., Discovery of a covalent inhibitor of KRASG12C (AMG 510) for the treatment of solid tumors. J. Med. Chem. 63, 52–65 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Kleiner R. E., Hang L. E., Molloy K. R., Chait B. T., Kapoor T. M., A chemical proteomics approach to reveal direct protein-protein interactions in living cells. Cell Chem. Biol. 25, 110–120.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murale D. P., Hong S. C., Haque M. M., Lee J.-S., Photo-affinity labeling (PAL) in chemical proteomics: A handy tool to investigate protein-protein interactions (PPIs). Proteome Sci. 15, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margarit S. M., et al., Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Ito Y., et al., Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 36, 9109–9119 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Joeh E., et al., Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. U.S.A. 117, 27329–27338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited in MassIVE Repository (MSV000086972). All other study data are included in the article and/or SI Appendix.