Significance
We report a methodology for the pooled construction of mutants bearing precise genomic sequence variations and multiplex phenotypic characterization of these mutants using next-generation sequencing (NGS). Unlike existing techniques depending on CRISPR-Cas–directed genomic breaks for genome editing, this strategy instead uses single-stranded DNA produced by a retron element for recombineering. This enables libraries of millions of elements to be constructed and offers relaxed design constraints which permit natural DNA or random variation to be used as inputs.
Keywords: genetic engineering, synthetic biology, functional genomics, antibiotic resistance, retron
Abstract
Creating and characterizing individual genetic variants remains limited in scale, compared to the tremendous variation both existing in nature and envisioned by genome engineers. Here we introduce retron library recombineering (RLR), a methodology for high-throughput functional screens that surpasses the scale and specificity of CRISPR-Cas methods. We use the targeted reverse-transcription activity of retrons to produce single-stranded DNA (ssDNA) in vivo, incorporating edits at >90% efficiency and enabling multiplexed applications. RLR simultaneously introduces many genomic variants, producing pooled and barcoded variant libraries addressable by targeted deep sequencing. We use RLR for pooled phenotyping of synthesized antibiotic resistance alleles, demonstrating quantitative measurement of relative growth rates. We also perform RLR using the sheared genomic DNA of an evolved bacterium, experimentally querying millions of sequences for causal variants, demonstrating that RLR is uniquely suited to utilize large pools of natural variation. Using ssDNA produced in vivo for pooled experiments presents avenues for exploring variation across the genome.
Constructing genotypes of interest and observing their effect on phenotype is central to our modern understanding of genetics and genome function. As methods for editing genomes have progressed, this “reverse genetics” approach has expanded in breadth and scale, from knockout libraries (1) to refactored genomes (2, 3). These experiments can now be performed within multiplexed pools, which allow an ever greater number of mutations to be explored across varied conditions. Critically, both creating genotypes and observing phenotypes within pools has necessitated development of new techniques. Transposon insertions (4), marked integrations (5), and CRISPR inhibition (6, 7) can create thousands of variants simultaneously within pooled experiments, and targeted sequencing of these elements enables pooled measurement of variant phenotypes. These advancements in experimental scale have fundamentally transformed our understanding of genome function (8).
However, these current high-throughput genetic techniques remain limited, in that they typically introduce, ablate, or regulate kilobases of DNA to create variation. This contrasts with point mutations, which are ubiquitous in natural variation (9), and are indispensable for engineering proteins (10) and metabolic pathways (11). While modifying kilobases of DNA can add and subtract functional elements such as genes and regulatory sequences from the genome, point mutations can alter the function of these elements, accessing a larger phenotypic landscape.
Point mutations and other precision edits can be performed by oligonucleotide recombineering, which creates precise genomic changes in many bacteria without incorporating selective markers or other large DNA sequences (12, 13) (Fig. 1B). This technique enables multiple variants to be created simultaneously in a pooled experiment (14), but provides no means for determining the phenotypes of individual mutations within the pool. Therefore, genome-wide recombineering requires individual mutant clones to be isolated, genotyped, and phenotyped for causality to be established, severely limiting throughput.
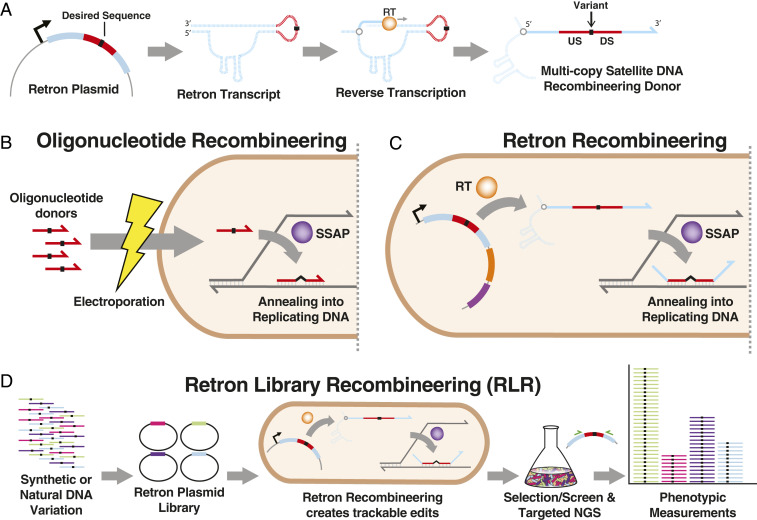
Fig. 1.
Overview of retron recombineering. (A) The retron msr/msd region undergoes transcription, then targeted reverse transcription catalyzed by retron reverse transciptase (RT), producing multicopy satellite DNA. When a sequence (red) containing homology upstream (US) and downstream (DS) of a sequence alteration (black) is introduced, this DNA functions as a recombineering donor. Dashed lines depict RNA, and solid lines depict DNA. (B) In oligonucleotide recombineering, synthetic oligonucleotide donors introduced into bacteria anneal to replicating DNA, directed by a single-stranded annealing protein (SSAP). This introduces desired sequence alterations (black) into the genome. (C) Retron recombineering uses RT-derived DNA as a donor, rather than transformed oligonucleotides, but similarly incorporates these into replicating DNA using an SSAP. (D) Libraries of synthetic or natural DNA variants can be incorporated into retrons to perform recombineering. This produces mutant cells bearing a retron plasmid, available for targeted amplicon sequencing using complementary primers (green) to measure mutant abundance in multiplex.
We reasoned that it would be possible to transform recombineering into a method for pooled, high-throughput genome-wide phenotypic measurement by using bacterial retroelements known as retrons. Retrons are prokaryotic retroelements that undergo targeted reverse transcription, producing single-stranded multicopy satellite DNA (msDNA) (15, 16). Recently, retrons were discovered to participate in antiphage defense systems, addressing the decades-long mystery of their function (17, 18). Retron msDNA are just one component of such systems, but this component can produce functioning recombineering donors within cells, creating specified genomic edits (16, 19). This is accomplished by altering the retron sequence to produce single-stranded DNA (ssDNA) containing a mutation of interest surrounded by homology to a targeted genomic locus (Fig. 1 A and C). Retron-based recombineering has been previously demonstrated for recording stimuli using genome modification (19), but, because the editing rates were far lower than recombineering using electroporated DNA (14, 20), it was impractical for studying mutants of physiological interest.
Results
Here, we combine several modifications to the genome and retron construct which improve editing efficiency, and demonstrate the use of the retron sequence itself as a “barcode” to identify mutants within mixed pools, enabling pooled screens of precisely created mutant strains. This system allows for simultaneous, pooled characterization of precision mutations across the genome, and offers advantages to similar CRISPR-Cas techniques.
The Effect of Endogenous Factors on Retron Recombineering.
To measure editing performance, retron plasmids were constructed to confer drug resistance variants within the essential genes rpoB and gyrA, and coexpressed with Red. The fraction of cells edited after growth and induction of this system in batch culture for approximately 20 generations was measured by deep sequencing the targeted locus (Fig. 2 A and B), and additionally confirmed by plating efficiency on the relevant antibiotic. Initially, less than 0.1% of Escherichia coli bearing the retron recombineering system incorporated the desired mutation, often below the limit of confident quantification in this assay. Previous results establish editing in this range (19), which is likely too low to link a mutant to its retron plasmid for pooled screens.
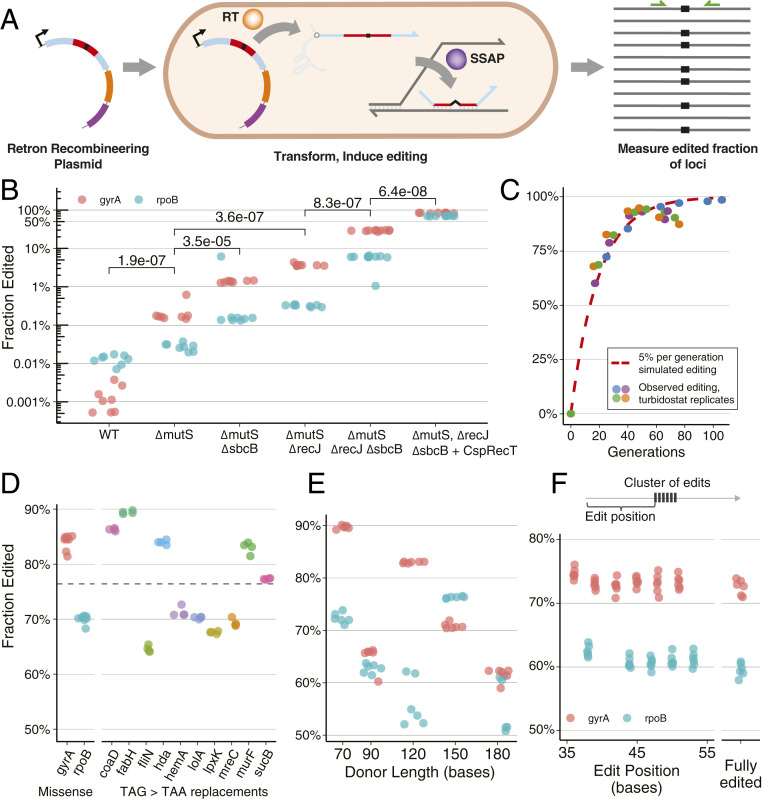
Fig. 2.
Characterization and optimization of retron recombineering. (A) A retron plasmid creates multicopy ssDNA via a retron RT. This DNA is incorporated into the genome via a coexpressed SSAP. Editing is observed by amplicon NGS, facilitated by primers targeting the locus (green). (B) Optimization of RLR. Colors differentiate gyrA and rpoB edits. Biological replicates are indicated with dots. Two-sided, unpaired parametric t tests were performed between genotypes indicated with brackets, and P values are displayed. (C) Editing of cultures in a turbidostat, with continuous growth and induction. mutS sbcB recJ genotype was used, transformed with gyrA-editing plasmid, expressing Red as an SSAP. These data are shown alongside a simulated editing trajectory of an allele with neutral fitness effect, editing at 5% per generation. (D) Edited fraction observed across alleles, after approximately 20 generations of induction and batch growth. A retron plasmid containing CspRecT as an SSAP was expressed in mutS recJ sbcB in all cases. Results of individual replicates are shown as dots. Results for gyrA and rpoB missense alleles (in B) are shown again for comparison alongside TAG > TAA stop codon editing for 10 essential genes. The mean edited fraction achieved across the 12 loci, 76.4%, is indicated by the dashed line. (E) The effect of retron length on editing. Six replicates for each experiment are indicated with dots colored by locus. (F) Integration of a cluster of mismatched bases in a retron donor. Editing of each position across eight replicates is shown for gyrA and rpoB alleles. Observations having all mutations are reported as fully edited.
Inactivating mismatch repair with a mutS genotype improved editing by approximately 150-fold and twofold for gyrA and rpoB donors, respectively (Fig. 2B), likely by preventing repair of genomic mismatches, which are necessary recombineering intermediates (12, 21) (Fig. 2A and SI Appendix, Fig. S1A). Mismatch repair was therefore inactivated in subsequent experiments to improve editing efficiency. Exonuclease inactivation is known to improve oligonucleotide recombineering by up to threefold (22, 23), and there is evidence that this is generalizable to retron recombineering (24, 25). Inactivation of exonuclease genes recJ or sbcB (also known as xonA) individually provided benefit, and inactivation of both together increased the edited fraction by 131- and 201-fold for the gyrA and rpoB donors, respectively (Fig. 2B and SI Appendix, Fig. S1C). This large improvement came as a surprise, given the modest effect that exonuclease inactivation has on oligonucleotide recombineering efficiency. We hypothesize that the high concentrations of transformed donor DNA introduced during electroporation overwhelm native exonucleases and decrease this effect for oligonucleotide recombineering.
In contrast to oligonucleotide recombineering methods (14) and CRISPR-Cas methods (26–29), retron recombineering is expected to be a continuous rather than discrete process, with each generation resulting in additional edited progeny. We observed these dynamics during continuous growth and induction in the eVOLVER turbidostat (30). Seventy-three hours of growth and induction across four replicate experiments produced edited fractions that increased rapidly and then slowed, in a manner consistent with a simulation of 5% editing of unedited cells per generation (SI Appendix, Fig. S5). This process ultimately produced edited fractions as high as 99%, with a mean of 92% (Fig. 2C and SI Appendix, Fig. S1D). Extended induction can thus lead to nearly complete editing in the population, producing nearly direct correspondence between a mutant and the corresponding retron “barcode.”
Optimization of Exogenous Retron Library Recombineering Components.
Improved editing per generation is expected to enhance retron library recombineering (RLR) by improving the correspondence between mutant and barcode. The 5% editing per generation estimated in the turbidostat is only one-fifth the theoretical maximum of 25% possible because of semiconservative replication during recombineering (21), indicating further improvements were possible. To understand the impact of possible improvements, we built a population genetics model simulating editing and the growth of edited strains, to explore the impact of editing efficiency on multiplexed experiments (SI Appendix, Fig. S5A). We predict that beneficial phenotypes like antibiotic resistance can be observed and quantified accurately with modest editing efficiency, because edited alleles will rapidly outcompete their nonedited counterparts (SI Appendix, Fig. S2C). Detrimental alleles, however, require more potent editing to observe even lethal effects (SI Appendix, Fig. S5D). Detrimental phenotypes also require persistent induction to be observed, whereas beneficial alleles can be observed by following an initial pulse of induction with selection (SI Appendix, Fig. S5B).
These considerations led us to seek further improvements in editing rate by exploring other SSAPs, the proteins which catalyze recombineering by recruiting and annealing ssDNA to the replication fork (21). While Red is the canonical SSAP used in E. coli, its distant relative CspRecT was recently found to improve recombineering efficiency in E. coli (31). Replacing Red with CspRecT further increased the edited fraction observed in batch growth by nearly threefold to more than 12-fold for gyrA and rpoB donors, respectively (Fig. 2B and SI Appendix, Fig. S6B). Notably, requirements for inactivating mismatch repair and exonucleases are also somewhat relaxed when using CspRecT (SI Appendix, Fig. S6A).
To characterize improved editing across a sample of sequences and genomic regions, we altered TAG “amber” stop codons to TAA “ochre” variants within essential genes, a focus of genome recoding projects (32). Altering 10 such stop codons resulted in 65 to 89% editing after 20 generations, with a mean of 76% (Fig. 2D). This is, to our knowledge, the highest efficiency achieved using oligonucleotide recombineering, and appears effective across a range of target sequences.
To alleviate the requirement for the mutagenic mutS genotype for efficient editing, we explored expressing a dominant-negative mutL allele (mutL*) as an alternative (13). Cultures transiently expressing mutL* during induction produced edited fractions equivalent to that observed with mutS (SI Appendix, Fig. S2B). Whole genome sequencing revealed that mutL*-expressing cultures accrue five random mutations, on average, throughout the editing process, fewer than mutS (SI Appendix, Fig. S4), but approximately 15 times the rate expected with mismatch repair intact (33). Mutation outside the editing loci occurred randomly, with no evidence of systematic off-target editing due to retrons (Dataset S2). Dominant-negative mismatch repair suppression is a viable strategy for improving recombineering across multiple prokaryotes (13) and eukaryotes (34), providing a possible route for extending this strategy to new organisms.
Increasing donor length would presumably provide more homology for annealing and integration at higher rates, but, surprisingly, we find that shorter donors are more effective, as a general rule, with 70-bp donors outperforming 90-bp sequences used previously (Fig. 2E). This stands in contrast to CRISPR-Cas methods in which hundreds of bases of homology are typically used (35, 36). Notably, while efficiency exceeded 50% for all lengths attempted, donors of similar length can behave quite differently (Fig. 2E). We hypothesize that this is due to variation in structure and folding; further investigation here may yield improvements in efficiency and consistency.
To characterize the effect of introducing multiple changes, a cluster of six synonymous mismatch edits were incorporated in a 16-bp window in the center of a 90-bp donor (Fig. 2F). Overall, any edited strain was likely to have all mutations incorporated, and the cluster of edits functioned at a similar efficiency to a single mismatch edit. Insertion of C-terminal tags onto essential genes was also successful, at a lower efficiency (SI Appendix, Fig. S3A).
In summary, inactivation of a subset of E. coli exonucleases and transient suppression of mismatch repair improves retron recombineering thousands-fold, and can result in edited fractions greater than 90% after continuous editing. This enables a retron to reliably create and identify its corresponding mutant strain within pooled experiments. Characterizing deleterious mutations poses additional challenges, and is expected to be very sensitive to variation in editing rate across donor sequences.
Detection of Phenotypes Using RLR, Barcoded Mutant Pools.
With efficient retron recombineering established, targeted sequencing of retron cassettes can be used as a measure of mutant abundance in a population, and therefore its phenotype within a pooled assay (Fig. 3). Antibiotic resistance is a growth phenotype of clinical importance, so we first sought to investigate antibiotic resistance mutations using RLR. We constructed a retron library conferring known E. coli rifampicin resistance mutations, known Mycobacterium tuberculosis rifampicin resistance mutations, and mutations affecting resistance to other antibiotics, and neutral, deleterious, and lethal control mutations (Fig. 3A).
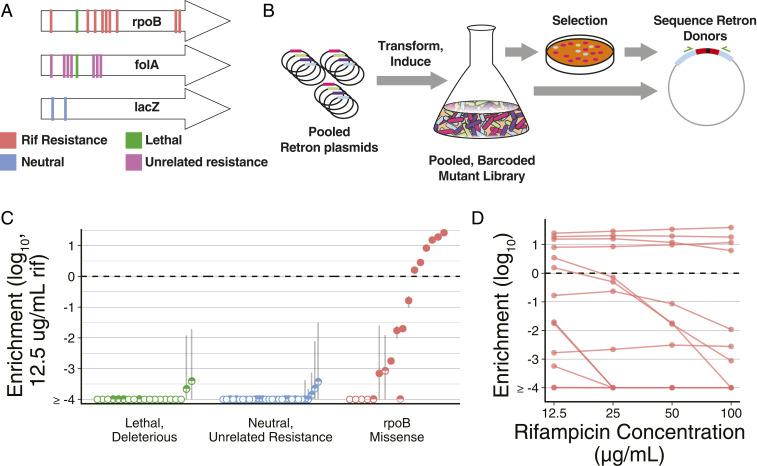
Fig. 3.
Pooled measurement of phenotypes using RLR. (A) The rpoB mutations, including known rifampicin resistance alleles, were specified, as well as resistance alleles for unrelated drugs, and control alleles expected to be neutral, lethal, or deleterious (SI Appendix, Table S3). (B) A pool of Retron plasmids conferring these alleles are transformed into cells. Transformants are induced, and editing produces a pooled, barcoded mutant library. A selection is performed, and frequencies of retron donors are compared before and after treatment for each allele. (C) RLR enrichment values observed with rifampicin treatment. The median of three replicates is indicated with a dot, and error bars are the SE of the mean. Pseudocounts of one are given to alleles not detected after treatment, such that frequencies are a lower limit of detection in these cases. Unfilled points indicate alleles not detected among any replicates after rifampicin treatment, and half-filled points indicate alleles detected in a subset of replicates. An enrichment value of zero is marked with a horizontal dashed line, indicating identical relative abundance before and after selection. (D) For rpoB mutation, allelic enrichment across concentration of rifampicin is displayed. The median of three independent experiments is indicated with a dot, and lines connect an allele across concentrations of rifampicin.
mutS recJ sbcB E. coli were transformed with this retron plasmid library, induced in batch growth to acquire the desired mutations, and plated on solid medium with rifampicin. Sequencing retron donors from these samples before and after selection correctly identified known rifampicin resistance mutations (37) by enrichment, while resistance alleles to unrelated drugs and other control alleles were depleted (Fig. 3C).
Mutations observed in rifampicin-resistant M. tuberculosis (38) did not confer detectable resistance in E. coli, with the exception of alleles altering the H526 residue of RpoB previously implicated in E. coli rifampicin resistance. This suggests some context dependence of rifampicin resistance mutations across differing rpoB sequences.
A range of enrichment values was observed across resistance alleles, reflecting variation in the ability to grow under selection. Selection across rifampicin concentrations establishes inhibition curves for mutants within the pool (Fig. 3D). In this way, RLR enables high-throughput, pooled identification of antibiotic-resistant alleles and facile characterization of their relative effects across a range of conditions. Red, 90-bp donors, and mutS were used in these experiments because the benefit of CspRecT, shorter donors, and mutL* had not yet been determined. The improved efficiency and lower rate of mutagenesis associated with these changes are expected to further improve and refine these types of screens.
Quantitative Characterization of Growth Rate.
Many phenotypes of interest produce small changes in fitness, rather than binary growth/death phenotypes (9, 39). Mutations producing small fitness improvements at low antibiotic concentrations can lead to the development of high-level antibiotic resistance (40). We thus used growth in subinhibitory concentrations of rifampicin as a model phenotype for measuring the subtle growth rates of mutants. Critically, neutral mutation controls remain viable in this system, and normalizing to these mutants enables a quantitative measurement indicative of relative growth rates.
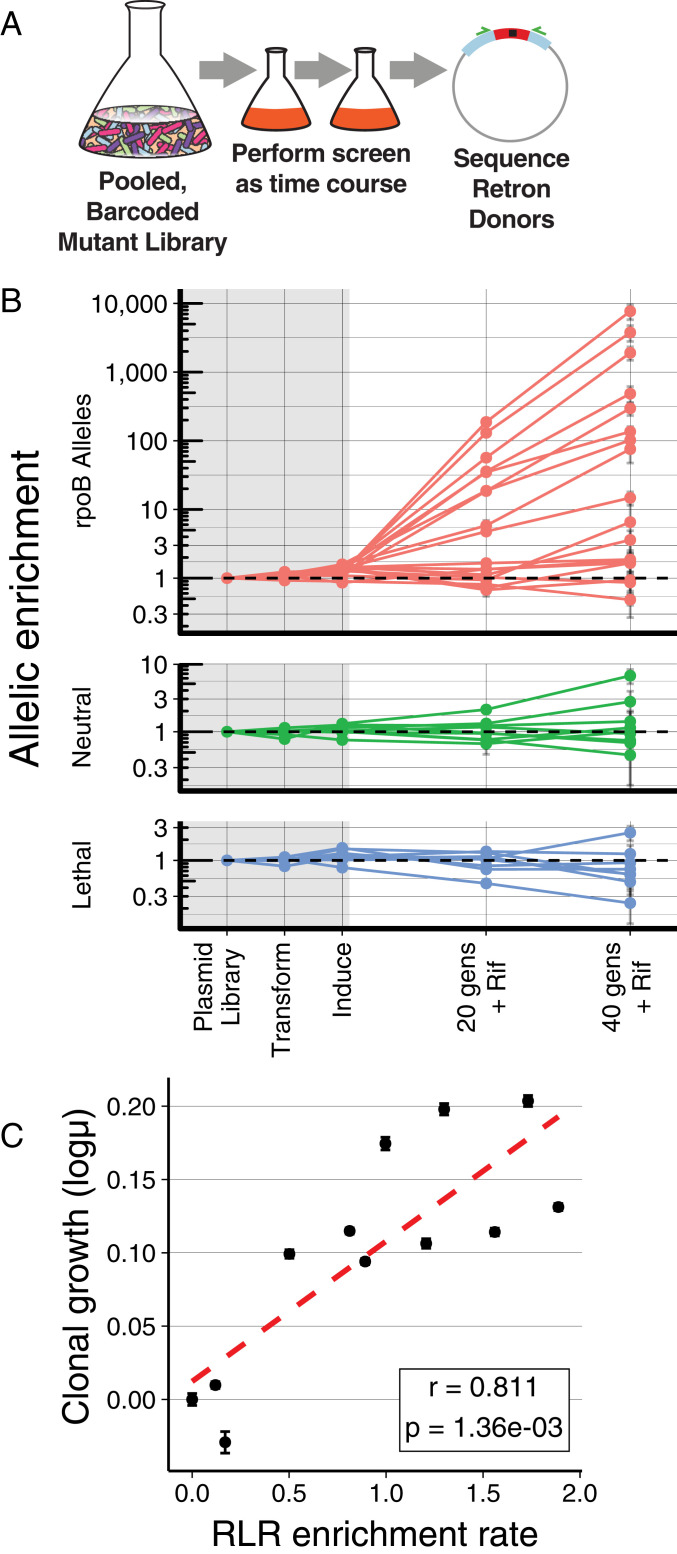
The pooled, barcoded mutant library constructed previously by RLR was grown at a subinhibitory concentration of rifampicin (5 g/mL), and samples were collected across multiple time points. Relative abundance of plasmids remained stable during transformation and induction of editing, but began to diverge once subinhibitory rifampicin was applied (Fig. 4B). The degree to which resistant mutants outpace neutral controls is a quantitative measure of growth rate which correlates well to mutations when their growth is measured individually (Fig. 4C and SI Appendix, Fig. S7A). RLR, therefore, affords quantitative growth metrics for alleles within a pool that is comparable to testing all mutants individually, but is inherently more scalable.
Fig. 4.
Quantitative measurements using RLR. (A) Graphical representation of quantitative RLR during a time course. The relative abundance of pooled, barcoded mutants is measured over time by NGS. (B) Relative abundance of donors during an experiment. Frequencies of each allele are normalized to the median of neutral controls within each time point, and to their starting abundance in the plasmid pool. Measurements within the gray rectangle occur during construction, transformation, and induction of the library, whereas those in the white area occur during growth in subinhibitory rifampicin (5 g/mL). The mean of three replicates is indicated with a dot; error bars are the standard error of the mean. Horizontal dashed lines indicate no change in frequency. (C) Growth rate of all mutants in the pool can be determined from the observed trajectories (RLR enrichment rate). Growth rate was measured individually for 11 resistance mutants using classical methods, and plotted against the log10 of RLR enrichment rate. Error bars depict standard error of these measurements; r is the Pearson’s correlation coefficient between these two measures, and the P is the probability of these results given the null hypothesis of no correlation.
RLR Detects Causal Variants, Using Libraries Prepared from Evolved Genomic DNA.
Directed evolution can produce antibiotic resistance, producing mutant strains able to grow in thousands-fold more concentrated antibiotic than their ancestors (41). We reasoned that RLR could be used to determine the causal mutations leading to resistance in such isolates, by using random fragments of their genomic DNA to construct an RLR library. Most fragments containing a variant should be capable of editing, because we found little effect of mutation position within donor oligonucleotides (SI Appendix, Fig. S6C).
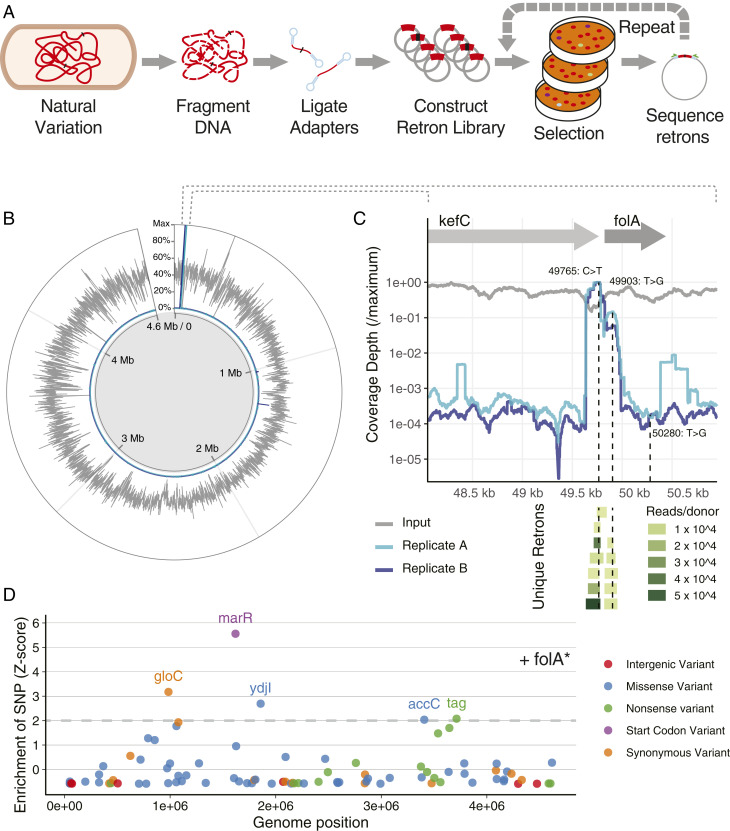
Genomic DNA from an E. coli strain highly resistant to TMP (41) was acoustically sheared into fragments, ligated to custom adapter sequences, and used to construct RLR libraries containing tens of millions of members (Fig. 5A) in Retron plasmids expressing Red as an SSAP. Retron donors comprised sequences averaging approximately 100 bp in length, providing over 50-fold coverage of the genome in unique fragments (Fig. 5B and SI Appendix, Fig. S9), ensuring that variants present in the evolved genome are well represented in the genomic RLR library. This library was then introduced into a mutS recJ sbcB strain.
Fig. 5.
Detecting causal variants in genomic DNA using retron libraries. (A) Graphical summary of an RLR experiment using genomic DNA (gDNA) as input. Adapters ligated to randomly fragmented gDNA enable pooled cloning, creating a multimillion member library of retron plasmids. Induction results in editing, and selection enriches for relevant mutants, whose retrons are sequenced as a pool via amplicon NGS. (B) Results of genomic DNA screen. Deduplicated genomic coverage contained in the retron library is displayed, showing the mean coverage across 1-kb base pair windows, normalizing to maximum coverage (gray; see SI Appendix, Fig. S9B for additional detail). After induction and selection of TMP, coverage depth of variants is depicted for two replicates of the library, again normalized to maximum coverage (blue, light blue). (C) The folA locus is displayed, with genomic position on the x axis. Sequence coverage observed for each base is plotted on the y axis for the input library and two replicates postselection, normalized to maximum coverage. Below, retron donor sequences observed more than 1,000 times in replicate A are depicted as a “pileup” aligned to the genome, and are colored by their abundance in postselection sequencing. Detected SNPs are indicated by vertical dashed lines. (D) Optionally, mutants surviving selection can be transformed by the pool again, screening for additional mutations and combinatorial effects. Transforming the library into a strain already bearing detected folA mutations (folA*) exposes additional candidate variants adaptive in additional TMP. Z scores for each allele describe their deviation from mean allele coverage depth. Variants with Z scores over two have been labeled by the gene in which they occur, colored by their relationship to coding sequences.
Induction of RLR and selection with TMP dramatically increased the abundance of retron donors containing variants at the folA locus (Fig. 5 B and C), which encodes the protein target of TMP (41). Within RLR donors mapping to this region, coverage of two single nucleotide polymorphisms (SNPs) are highly enriched, indicating they individually increase TMP resistance (Fig. 5C). Multiple retron donor sequences independently contribute to the enrichment of both alleles (Fig. 5C). The more highly enriched allele lies upstream of the folA coding sequence and likely increases it’s transcription, a well-described route to TMP resistance (42). The other is known to increase the catalytic rate of FolA, leading to TMP resistance (42). A third SNP within folA is not enriched, suggesting little or no effect individually at this concentration of TMP. Mutations at this third position are known to interact with folA mutations at the active site but not confer resistance on their own (43). The ability to individually measure mutation effects, especially for mutations near each other in the genome, distinguishes this method from P1 transduction, which transfers 100 kb of contiguous variation, and requires subsequent sequencing and deconvolutions of hits for interpretation (44).
To identify alleles outside the folA locus providing additional resistance, we performed a subsequent transformation of the genomic library into a strain bearing all folA mutations, induced editing, and selected with increased TMP. Sequencing of retron plasmids identified several enriched variants (Fig. 5D), including variants within known resistance determinants such as the marR regulator of the multiple antibiotic resistance operon (45). In this manner, causal alleles can be determined repeatedly from evolved or environmentally derived pools of variation, in order to deconvolute phenotypes requiring multiple mutations.
Discussion
Here we show that retron recombineering is a flexible, generalizable tool for genome editing which surpasses the editing frequencies achieved by other markerless editing tools like oligonucleotide recombineering. Pooled, barcoded mutant libraries can be prepared in this way and used for multiplexed characterization of natural and synthesized allelic variants, a process we call RLR. RLR enables millions of experiments to be performed simultaneously, observing the effects of mutations across the genome.
RLR is an alternative to CRISPR-based methods which also perform pooled phenotypic measurements of mutant sequences. These CRISPR-based methods create edits by using a guide RNA to direct targeted breakage, and a plasmid-borne donor DNA to repair these breaks with the desired sequence, with phenotypic tracking permitted by amplicon sequencing of these components (26–29). RLR’s “donor only” nature distinguishes it from these “guide + donor” methods in several key ways. RLR eliminates the requirement for ablating CRISPR targeting, allowing single-base pair changes to be characterized without requiring additional protospacer-adjacent motif (PAM)-ablating mutations to be incorporated (26–28, 36). RLR overcomes the requirement for targeting a suitable PAM altogether, whereas CRISPR “guide + donor” methods decrease in performance as the distance to a PAM increases (26–28). Intriguingly, production of a retron ssDNA donor appears to improve CRISPR “guide + donor” methods in Saccharomyces cerevisiae (26) and E. coli (46), presumably by making the recombination donor more abundant or accessible.
RLR’s lack of guides simplifies and streamlines RLR elements. In contrast to the two unique elements required for “guide + donor” strategies, or the three required for efficient, “PE3” prime editing (47), RLR’s sole requirement is a unique, short donor sequence within the retron. This relaxed design constraint enables RLR using nondesigned variation. We demonstrate this using genomic DNA fragments as an input, but we expect this flexibility will facilitate numerous applications.
Because RLR does not require CRISPR, perhaps it can be combined with engineered selections/screens of which CRISPR is already a component. The lack of CRISPR activity may also enable use in systems for which CRISPR-Cas is toxic, as is often observed (48, 49). Nontoxic editing methods like RLR may enable new applications otherwise hampered by expression of deleterious components.
Certain pitfalls and challenges remain, however. The continuous, replication-dependent nature of RLR over multiple generations is instrumental to its effectiveness, but may limit its utility in certain organisms where this is undesirable. RLR also relies on inactivating mismatch repair if short mismatches are to be incorporated, increasing the mutation rate. More work remains be done to increase editing rates, and decrease variation in efficiency between alleles. Improving both will likely be required for accurate pooled measurement of deleterious mutations. RLR’s simple, streamlined nature may lend itself to applications incorporating combinatorial sets of mutations, and strategies remain to be developed in this area. It remains unexplored whether RLR could facilitate alterations extending outside the donor itself, such as inversions, duplications, and rearrangements.
Here RLR measures beneficial growth and selective phenotypes, but nongrowth phenotypes could also be made accessible to pooled measurement by fluorescent reporters (50, 51), biosensors (50), single-cell transcriptomics (52), and a myriad of other screens (8). Retron recombineering may even find use as a method of producing barcoded variation in constructs cloned into E. coli, for use in other organisms. Given that recombineering is possible in a range of organisms (53–55), and retrons occur across a range of organisms (16), RLR should not be limited to use in E. coli, and development of RLR in other genetic systems is an exciting future area for which this work helps to establish a road map. We foresee a wide array of possible future technologies using the flexible and simple editing and barcoding framework to accomplish pooled mutant screens.
Materials and Methods
Preparation of Strains and Plasmids.
Strain ECNR1 (14) was modified to replace the bla ampicillin resistance cassette and the tetR repressor at the lambda prophage locus with the tetA tetracycline resistance cassette, using double-stranded recombineering as per Datsenko and Wanner (56). All strains were also modified by inactivation of the araBAD operon using this method, conferring arabinose auxotrophy and ensuring consistent induction with arabinose. Other genes were likewise inactivated, and the antibiotic resistance markers were removed using FLP recombinase (56). Retron plasmid pFF745 (Addgene 61450) was a generous gift from F.F. and T.K.L. New retron donor sequences were introduced by Kinase, Ligase, DpnI (KLD) mutagenesis (New England Biolabs [NEB]), or Gibson Assembly (NEB). Plasmids were modified to contain a low-copy origin under stringent replication control (SC101) and a tightly regulated promoter (AraC-pBAD) to minimize variability in growth and plasmid titer among retron plasmid-bearing populations, and enable tight repression of the editing system. Expression in this system produced an effect on growth comparable to expressing GFP, an improvement on pFF745-derived COLE1-pL_lacO expression systems reported previously (19, 24) which had larger impacts on growth (SI Appendix, Fig. S8). Red recombinase was replaced by CspRecT, and mutL-E32K was added in relevant plasmids by Gibson Assembly (NEB). See SI Appendix, Table S1 for oligonucleotides used to perform these alterations, and see Dataset S1 for representative plasmid maps.
Measurement of Editing.
To perform editing, strains were transformed with retron plasmids via electroporation (56), and plated to lysogeny broth (LB; Lennox formulation: 10 g of tryptone, 5 g of yeast extract, 5 g of sodium chloride per 1 L of distilled, deionized water [ddH2O]) with 25 g/mL chloramphenicol (LB-CM25) with 1.5% agar added. After 18 h to 24 h of growth at 34 °C, colonies were picked into 100 L of LB-CM25 in a 96-well plate and allowed to grow 6 h to 8 h, reaching confluence. These uninduced precultures were diluted 1,000-fold into 1 mL of LB-CM25 containing 0.2% L-arabinose (LB-CM25ara) in a 96-well plate, and allowed to grow for 24 h at 34 °C with shaking at 900 rpm, reaching confluence. This 1,000-fold dilution and growth was repeated once more for all cultures. Assuming density at confluence to be consistent, the number of generations experienced with induction across both 1:1,000 batch cultures is approximately 20, because is equal to 1,024; 120 L of these saturated cultures (optical density at 600 nm [OD600] approximately 3.0) were sampled to measure editing via amplicon sequencing as described below. When growth in a turbidostat is indicated, strains were grown in an eVOLVER instrument (30) (FynchBio). Turbidostat vials were maintained between OD600 of 0.2 and 0.4, in LB-CM25ara with 0.05% tween added to prevent biofilm formation, growing at 34 C with a stir rate of eight. One-milliliter samples were sampled over 73 h to measure editing via amplicon sequencing as described below. The number of generations experienced by a given time point was inferred from the turbidostat growth record using a custom Matlab script, and the edited fraction was determined by gyrA amplicon next-generation sequencing (NGS) for each sample. To measure editing by amplicon sequencing, samples were centrifuged for 5 min at 4,800 g, and the pellet was resuspended in 10 mM NaOH solution with 0.01% Triton-100 and incubated at 95 °C for 10 min in a thermocycler. The resulting lysates were centrifuged for 10 min at 4,800 g at 4 °C, and supernatants were stored at −20 °C. PCR amplification of the genomic region was performed in reactions containing 10 L of Q5 2× Mastermix (NEB), 2 L of lysate supernatant, 0.2 L of 50-M primer mixture for the locus (SI Appendix), 1 L of Evagreen dye (Biotium), and 6.8 L of ddH2O. Amplification was monitored in the SYBR channel on an Eppendorf Realplex4 real-time PCR system until several cycles of productive amplification were observed (typically less than 10 cycles), and the resulting amplicons were indexed for sequencing as described below. Source data and summarized data for Fig. 2B can be found in SI Appendix, Table S2; data and analysis scripts for all figures are available at https://github.com/churchlab/rlr (57).
Preparation of Genome-Derived Retron Plasmid Libraries.
TMP-adapted strains s100 and s102 were a kind gift from Michael Baym, Department of Biomedical Informatics, Harvard Medical School, MG1655 was obtained from the Coli Genetic Stock Center (Yale University). Genomic DNA was isolated from 1.5 mL of LB cultures using the Promega Wizard Genomic DNA purification kit (Promega), and quantified using the Qubit double-stranded DNA (dsDNA) HS reagent (Thermo Fisher). Four micrograms of DNA was placed in a Covaris 520045 tube, and sheared for 1,200 s in a Covaris M220 using 50-W peak incident power, 20% duty factor, and 200 cycles per burst. Fragmented DNA was visualized with a Bioanalyzer (Agilent), using a DNA 1000 chip (Agilent). The resulting fragments were repaired, dA-tailed, and ligated to a custom adapter sequence (SI Appendix) using the NEBnext Ultra II DNA library prep kit for Illumina (NEB). The resulting purified products were amplified in a reaction containing 25 L of Q5 2× Mastermix (NEB), 2 L of template, 0.5 L of 50-M primer mixture (SI Appendix), 2.5 L of Evagreen dye (Biotium), and 22.5 L of ddH2O. Amplification was monitored in the SYBR channel on an Eppendorf Realplex4 real-time PCR system until several cycles of productive amplification were observed, typically less than 10 cycles, and products were purified using DNA-binding magnetic beads (58). Adapter dimer fragments containing no ligated insert were removed by digestion with BsaI enzyme and size-selective purification using DNA-binding magnetic beads (58). After this purification, adapter dimer fragments were no longer detectable by gel electrophoresis, indicating their rarification in the pooled library. A vector for Type-IIs “Golden Gate” assembly was prepared via PCR, using a pBAD-SC101 retron plasmid (with Red or CspRecT, as indicated) as a template, and the resulting amplicon was purified using DNA-binding magnetic beads (58). Vector and insert were ligated together in a Type-IIs “Golden Gate” assembly, with restriction and ligation occurring in one reaction mixture. This was performed in a reaction containing approximately 0.3 pmol of vector and 1 pmol of inserts, 2 L of BsaI-v2, and 2 L of concentrated T4 DNA ligase (M202 T, NEB). Reactions were incubated for 5 min at 37 C, then for 30 cycles of 37 C and 20 C for 4 min each, then finally for 60 C for 10 min, in a thermocycler.
Preparation of Synthetic Retron Plasmid Libraries.
Recombineering donors were designed using the Mage Oligo Design Tool (MODEST) web tool (59). Donors were assembled from individual oligonucleotides (Integrated DNA Technologies) using Primerize (60). These donors were assembled, by Hifi DNA assembly (NEB), into a compatible vector prepared by PCR; see SI Appendix, Table S1 for oligonucleotide sequences used to create vectors for this assembly. For more details on the alleles specified, as well as source and summary data of their performance in the qualitative rifampicin selection, see SI Appendix, Table S3.
Transforming Strains with Plasmid Libraries, Induction and Editing.
Plasmid library assembly reactions were purified by Ethanol precipitation, eluted into 2 L of TE buffer, and chilled on ice. Fifty microliters of thawed electrocompetent cells (Lucigen ELITE 10G) were introduced, and electroporated using the EC1 setting on a Bio-Rad MicroPulser. After recovery in Lucigen Recovery medium for 1 h at 37 C with shaking, 10 mL of LB with 25 g/mL chloramphenicol was added for overnight growth at 30 C with shaking. Glycerol stocks were prepared at this point, and plasmids were isolated by midi-prep (Qiagen). Electrocompetent cells were prepared by growing 50 mL of mutS recJ sbcB in LB at 30 C until an OD600 of 0.5 to 0.8 was achieved. These cultures were chilled on ice; pelleted, resuspended, and rinsed twice with 50 mL of chilled 10% glycerol; then resuspended in 2 mL of chilled glycerol for a final centrifugation. This pellet was resuspended in 150 L of 10% glycerol, and 50-L aliquots were used for electroporation and recovery as described above. This recovery was washed with phosphate-buffered saline (PBS) and resuspended into LB, and a 1/5 dilution of this recovery was performed into 10 mL of LB-CM25Ara and grown for 6 h to 8 h at 34 C to saturate the culture and dilute out nontransformed and/or dead cells from the recovery. Editing was performed by diluting 1/1,000 into 10 mL (synthetic libraries) or 50 mL (genomic libraries) of LB-CM25Ara and grown to saturation at 34 C overnight. This process was repeated once more to achieve greater than 20 generations of growth and induction.
Qualitative Drug Resistance Screens.
Barcoded synthetic mutant libraries prepared as above were pelleted and rinsed with PBS, before resuspending in PBS. Diluted samples were plated onto 100-mm Petri dishes containing LB with relevant concentrations of rifampicin and 1 g/mL chloramphenicol, to determine the colony-forming unit (CFU) per milliliter plating efficiency across different conditions when grown overnight at 34 C. Not fewer than six petri dishes per condition were plated using a dilution targeting 1,000 colonies per plate, and grown overnight at 34 C. The resulting colonies were scraped from the plates, rinsed, and resuspended in PBS, and plasmids were isolated by Mini-prep (Qiagen). For genomic libraries, the identical procedure was performed, except 150-mm Petri plates containing cation-adjusted Miller–Hinton Broth containing 1 g/mL TMP and 1 g/mL chloramphenicol were used, plates were incubated for 2 d at 34 C, and plasmids were isolated by Midi-prep (Qiagen). For the secondary screen in the folA* strain background, TMP was used at 1 mg/mL, necessitating 1% dimethyl sulfoxide in the medium for solubility.
Quantitative Drug Resistance Screens (Time Course).
Barcoded synthetic mutant libraries prepared as above were diluted 1/1,000 into 50 mL of LB medium containing 10 g/mL chloramphenicol and 5 g/mL rifampicin. After growth overnight at 34 C, a 5-mL sample was taken, and this dilution was repeated for a second time point. Samples of the initial plasmid library, the library when transformed into mutS recJ sbcB cells, and at time points, were obtained for plasmid purification and amplicon sequencing.
Deep Sequencing of Retron Donors.
E. coli containing retron plasmids were obtained from the relevant liquid or solid growth condition, and washed once with Dulbecco’s PBS, and their plasmids were purified via Miniprep or Midiprep (Qiagen). PCR amplification of retron donor sequences was performed in a reaction containing 20 L of q5 2× Mastermix (NEB), 2 L of purified plasmid DNA, 0.4 L of 50-M primer mixture (SI Appendix, Table S1), 1 L of Evagreen dye (Biotium), and 15.6 L of ddH2O. Amplification was monitored in the SYBR channel on an Eppendorf Realplex4 real-time PCR system until several cycles of productive amplification were observed, typically less than 10 cycles. Products were indexed for sequencing as described below.
Amplicon Sequencing.
Amplicons from previous steps were prepared for Illumina sequencing by first removing oligonucleotides via treatment with Exonuclease I (NEB), then performing PCR using primers adding Indexes 1 and 2 for Illumina Paired-end sequencing (SI Appendix, Table S1). PCR was performed in a reaction containing 10 L of Q5 2× Mastermix (NEB), 1 L of PCR product, 0.2 L of 50-M primer mixture, 1 L of Evagreen dye (Biotium), and 7.8 L of ddH2O. Amplicons were purified using DNA-binding beads (58) and quantified using the Qubit dsDNA HS reagent (Thermo Fisher). The resulting DNA were pooled and sequenced on MiSeq, NextSeq, or HiSeq Illumina instruments, producing paired-end, 150-bp reads.
Sequence Analysis: Editing.
No fewer than 10,000 paired-end reads per replicate were merged using Paired-End read merger (PEAR) (61), and adapter and primer sequences were trimmed using Cutadapt (62). Sequences surrounding the edited region were trimmed again with Cutadapt, and counts of identical sequences were determined. Any sequence occurring fewer than 20 times was assumed to be a rare sequencing error, and was discarded at this stage. The edited fraction was calculated as the fraction of edited sequences divided by the number of edited and wild-type reference sequences detected within a sample. To minimize the impact of sequencing error on conclusions, additional synonymous mutations were incorporated along with the desired missense gyrA and rpoB alleles for Fig. 2B, where edited events are often rare. This more unambiguously links an edited sequence to an editing event, and not to mutation or errors during PCR or sequencing. A subset of these results were additionally confirmed by a phenotypic test—the efficiency of plating on antibiotic (SI Appendix, Fig. S1C). The fraction of all observed sequences which were either edited or wild type was monitored, and exceeded 93% for the results in Fig. 2B, and exceeded 95% for other panels in Fig. 2D. Scripts are available at https://github.com/churchlab/rlr (57).
Culture-Based Confirmation of Editing Results.
In parallel with amplicon sequencing to measure editing, serial dilutions of samples bearing the RpoB donor were plated to LB with 25 g/mL chloramphenicol, and LB with 25 g of Rifampicin. CFU per milliliter was determined by counting colonies, and the proportion of Rifampicin-resistant CFUs was used as a proxy for the edited fraction of cells.
Sequence Analysis: Synthetic DNA Experiments.
Paired-end reads were merged using PEAR (61), adapter and primer sequences were trimmed using Cutadapt (62), and counts of identical sequences were determined. Donor sequences not matching expected sequences were discarded. The fraction of sequences discarded correlated with estimated sequencing error rate and quality scores, and was never 25% of sequences. The frequency of each donor in the resulting dataset was then determined. For time courses, these frequencies are normalized to the median frequency of the neutral allele retron pool within each replicate and time point, and initialized to their relative frequency in the first time point prior to plotting. For calculating relative growth rates, an exponential curve was fit to noninitialized trajectories using a linear model. Scripts are available at https://github.com/churchlab/rlr (57).
Sequence Analysis: Natural DNA Experiments.
For determining the length of retron donor regions in cloned gDNA libraries, reads were first aligned to the MG1655 reference genome using Burrows–Wheeler Aligner (BWA) (63). The length of the region between aligned read pairs, the template length or Tlen column, was extracted from this alignment and visualized as a histogram (SI Appendix, Fig. S9A). Template lengths longer than 500 bp were assumed to be the result of erroneous alignment, and were discarded. For other analyses, paired-end reads were merged using PEAR (61), and adapter and primer sequences were trimmed off using Cutadapt (62). The resulting sequences were aligned to the MG1655 reference genome using BWA (63). To determine coverage of the reference genome, depth was determined using Bedtools genomecov (64), and visualized by plotting the mean coverage of 1,000-bp sliding windows (Fig. 5B). Alternatively, to examine nonredundant coverage conferred by unique donors, sequences with identical start and end alignment positions were collapsed into a single sequence before determining depth, visualized by plotting the mean coverage of 500-bp windows (SI Appendix, Fig. S9B). When examining a subset of the genome, coverage at every base was used, rather than using sliding windows (Fig. 5C). While mean coverage of genomic positions was sufficient, coverage at some loci was zero, or was unusually high. These loci are summarized in SI Appendix, and are interpreted as alignment artifacts in all cases. Zero coverage can result from differences between the MG1655 reference genome and that of the BW25113:lacA ancestor of the evolved strain, which is missing some genomic regions. Likewise, sequences from the circular genome may not align correctly near the termini of the linear reference sequence, resulting in zero coverage. Unusually high coverage results from incorrect mapping of short reads to high-copy sequences such as insertion elements and ribosomal RNA. To determine the abundance of retron donors conferring SNPs, reads from amplicons obtained after selection were aligned to the MG1655 reference genome, and SNPs were inferred using Millstone (65). Depth of SNP coverage in the resulting output reflects abundance in the pool, and was used for visualization and analysis.
Individual Growth Rate Measurements.
Colonies were picked into 100 L of LB medium in 96-well plates and grown for 4 h to 6 h at 34 °C. Ten microliters of a 10-fold dilution of this “preculture” was used to inoculate 190 L of LB medium with relevant additives in a Nunclon 96-well microwell plate (Thermo Scientific). These plates were cultured in a shaking plate reader at 34 °C, measuring OD600 every 15 min. The resulting data were analyzed using the analyze_growth.m script to derive growth rates for all wells. Scripts are available at https://github.com/churchlab/rlr (57).
Population Genetics Model.
A simulation was written in the Matlab scripting language, in which, each generation, a fraction “r” of unedited cells become edited, for a population bearing a particular retron. Once edited, that population reproduces at a rate of “f,” which may be more or less than the parental rate of one. Simulations can be performed with various r and f and initial conditions. The model makes implicit assumptions that populations experience constant exponential growth and are of an arbitrarily large size, for simplicity. Scripts are available at https://github.com/churchlab/rlr (57).
Supplementary Material
Acknowledgments
This work was supported by the US Department of Energy (Grant DE-FG02-02ER63445), the National Defense Science and Engineering Graduate Fellowship, and the Harvard Consortium for Space Genetics. We thank Harvard Biopolymers for next-generation sequencing services. M.G.S. thanks professors T. Bernhardt and M. Springer for support and advice.
Footnotes
Competing interest statement: M.G.S., T.M.W., F.F., T.K.L., S.L.S., and G.M.C. have filed patent applications based on related work. T.K.L. is a cofounder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, and Eligo Biosciences, in which he has related financial interests. T.K.L. also holds financial interests in nest.bio, Ampliphi, IndieBio, MedicusTek, Quark Biosciences, and Personal Genomics. G.M.C. is a cofounder of companies in which he has related financial interests: EnEvolv and 64-x. For a complete list of G.M.C.’s financial interests, see https://arep.med.harvard.edu/gmc/tech.html.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018181118/-/DCSupplemental.
Data Availability
Read counts, read depths, all data used for creating figures, and code used to create them have been deposited in GitHub (https://github.com/churchlab/rlr) (57).
References
- 1.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson S. M., et al. , Design of a synthetic yeast genome. Science 355, 1040–1044 (2017). [DOI] [PubMed] [Google Scholar]
- 3.C. A. Hutchison, 3rd, et al. , Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016). [DOI] [PubMed] [Google Scholar]
- 4.van Opijnen T., Camilli A., Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner J. R., Reeder P. J., Karimpour-Fard A., Woodruff L. B. A., Gill R. T., Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 28, 856–862 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Gilbert L. A., et al. , Genome-Scale CRISPR-Mediated control of gene repression and activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters J. M., et al. , A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray A. N., et al. , High-throughput bacterial functional genomics in the sequencing era. Curr. Opin. Microbiol. 27, 86–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenaillon O., et al. , Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heim R., Tsien R. Y., Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Hong K.-K., Nielsen J., Recovery of phenotypes obtained by adaptive evolution through inverse metabolic engineering. Appl. Environ. Microbiol. 78, 7579–7586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costantino N., Court D. L., Enhanced levels of lambda red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 15748–15753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyerges Á., et al. , A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl. Acad. Sci. U.S.A. 113, 2502–2507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H. H, et al. , Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lampson B. C., Inouye M., Inouye S., Retrons, msDNA, and the bacterial genome. Cytogenet. Genome Res. 110, 491–499 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Simon A. J., Ellington A. D., Finkelstein I. J., Retrons and their applications in genome engineering. Nucleic Acids Res. 47, 11007–11019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millman A., et al. , Bacterial retrons function in anti-phage defense. Cell 183, 1551–1561.e12 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Bobonis J., et al. , Phage proteins block and trigger retron toxin/antitoxin systems. bioRxiv [Preprint] (2020). 10.1101/2020.06.22.160242 (Accessed 22 June 2020). [DOI] [Google Scholar]
- 19.Farzadfard F., Lu T. K., Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lajoie M. J., Gregg C. J., Mosberg J. A., Washington G. C., Church G. M., Manipulating replisome dynamics to enhance lambda red-mediated multiplex genome engineering. Nucleic Acids Res. 40, e170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosberg J. A., Lajoie M. J., Church G. M., Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics 186, 791–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawitzke J. A., et al. , Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J. Mol. Biol. 407, 45–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosberg J. A., Gregg C. J., Lajoie M. J., Wang H. H., Church G. M.. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. Plos One 7, e44638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anna J. S., Morrow B. R., Ellington A. D., Retroelement-based genome editing and evolution. ACS Synth. Biol. 7, 2600–2611 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Farzadfard F., Gharaei N., Citorik R. J., Lu T. K., Efficient retroelement-mediated DNA writing in bacteria. bioRxiv [Preprint] (2020). 10.1101/2020.02.21.958983 (Accessed 22 February 2020). [DOI] [PubMed] [Google Scholar]
- 26.Sharon E., et al. , Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garst A. D., et al. , Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Roy K. R., et al. , Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat. Biotechnol. 36, 512–520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X., et al. , High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR–Cas9 in yeast. Nat. Biotechnol. 36, 540–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong B. G., Mancuso C. P., Kiriakov S., Bashor C. J., Khalil A. S., Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 36, 614–623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wannier T. M., et al. , Improved bacterial recombineering by parallelized protein discovery. Proc. Natl. Acad. Sci. U.S.A. 117, 13689–13698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lajoie M. J., et al. , Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jee J., et al. , Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature 534, 693–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang Z., Metzner E., Isaacs F. J., Advanced eMAGE for highly efficient combinatorial editing of a stable genome. bioRxiv [Preprint] (2020). 10.1101/2020.08.30.256743 (Accessed 31 August 2020). [DOI] [Google Scholar]
- 35.Chavez A., et al. , Precise Cas9 targeting enables genomic mutation prevention. Proc. Natl. Acad. Sci. U.S.A. 115, 3669–3673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhury A., et al. , CRISPR/Cas9 recombineering-mediated deep mutational scanning of essential genes in Escherichia coli. Mol. Syst. Biol. 16, e9265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrick J. E., Kauth M. R., Strelioff C. C., Lenski R. E., Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27, 1338–1347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pozzi G., et al. , RpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J. Clin. Microbiol. 37, 1197–1199 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenaillon O., et al. , The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Kepler T. B., Perelson A. S., Drug concentration heterogeneity facilitates the evolution of drug resistance. Proc. Natl. Acad. Sci. U.S.A. 95, 11514–11519 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baym M., et al. , Spatiotemporal microbial evolution on antibiotic landscapes. Science 353, 1147–1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamer Y. T., et al. , High-Order epistasis in catalytic power of dihydrofolate reductase gives rise to a rugged fitness landscape in the presence of trimethoprim selection. Mol. Biol. Evol. 36, 1533–1550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dion A., Linn C. E., Bradrick T. D., Georghiou S., Howell E. E., How do mutations at phenylalanine-153 and isoleucine-155 partially suppress the effects of the aspartate-27serine mutation in Escherichia coli dihydrofolate reductase? Biochemistry 32, 3479–3487 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Thomason L. C., Costantino N., Court D. L., E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 79, 1.17.1–1.17.8 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Sulavik M. C., Gambino L. F., Miller P. F., The MarR repressor of the multiple antibiotic resistance (MAR) operon in Escherichia coli: Prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1, 436 (1995). [PMC free article] [PubMed] [Google Scholar]
- 46.Lim H., et al. , Multiplex generation, tracking, and functional screening of substitution mutants using a CRISPR/retron system. ACS Synth. Biol. 15, 1003–1009 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Anzalone A. V., et al. , Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rock J. M., et al. , Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2, 16274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho S., et al. , High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 7, 1085–1094 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Rogers J. K., et al. , Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 43, 7648–7660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosuri S., et al. , Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 14024–14029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixit A., et al. , Perturb-seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy K. C., Recombination and recombineering. EcoSal Plus, 10.1128/ecosalplus.ESP-0011-2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbieri E. M., Muir P., Akhuetie-Oni B. O., Yellman C. M., Isaacs F. J., Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171, 1453–1467.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filsinger G. T., et al. , Characterizing the portability of phage-encoded homologous recombination proteins. Nat. Chem. Biol. 17, 394–402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert M., Goodman D.. RLR Documentation, Code, and Figures. GitHub. https://github.com/churchlab/rlr. Accessed 15 March 2021. [Google Scholar]
- 58.Oberacker P., et al. , Bio-On-Magnetic-Beads (BOMB): Open platform for high-throughput nucleic acid extraction and manipulation. PLoS Biol. 17, e3000107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonde M. T., et al. , MODEST: A web-based design tool for oligonucleotide-mediated genome engineering and recombineering. Nucleic Acids Res. 42, W408–W415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian S., Das R., Primerize-2D: Automated primer design for RNA multidimensional chemical mapping. Bioinformatics 33, 1405–1406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J., Kobert K., Flouri T., Stamatakis A., PEAR: A fast and accurate illumina paired-end read merger. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 1 (2011). [Google Scholar]
- 63.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997 (Accessed 26 May 2013). [Google Scholar]
- 64.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodman D. B., et al. , Millstone: Software for multiplex microbial genome analysis and engineering. Genome Biol. 18, 101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Read counts, read depths, all data used for creating figures, and code used to create them have been deposited in GitHub (https://github.com/churchlab/rlr) (57).