Abstract
Background
NAC (NAM, ATAF1/2, and CUC2) transcription factors play an important role in plant growth and development. However, in tumorous stem mustard (Brassica juncea var. tumida), one of the economically important crops cultivated in southwest China and some southeast Asian countries, reports on the identification of NAC family genes are lacking. In this study, we conducted a genome-wide investigation of the NAC family genes in B. juncea var. tumida, based on its recently published genome sequence data.
Methods
The NAC genes were identified in B. juncea var. tumida using the bioinformatics approach on the whole genome level. Additionally, the expression of BjuNAC genes was analyzed under high- and low-temperature stresses by quantitative real-time PCR (qRT-PCR).
Results
A total of 300 BjuNAC genes were identified, of which 278 were mapped to specific chromosomes. Phylogenetic analysis of B. juncea var. tumida, Brassica rapa, Brassica nigra, rice and Arabidopsis thaliana NAC proteins revealed that all NAC genes were divided into 18 subgroups. Furthermore, gene structure analysis showed that most of the NAC genes contained two or three exons. Conserved motif analysis revealed that BjuNAC genes contain a conserved NAM domain. Additionally, qRT-PCR data indicated that thirteen BjuNAC genes with a varying degree of up-regulation during high-temperature stress. Conversely, four BjuNAC genes (BjuNAC006, BjuNAC083, BjuNAC170 and BjuNAC223) were up-regulated and two BjuNAC genes (BjuNAC074 and BjuNAC295) down-regulated under low temperature, respectively. Together, the results of this study provide a strong foundation for future investigation of the biological function of NAC genes in B. juncea var. tumida.
Keywords: Brassica juncea var. Tumida, NAC gene family, Transcription factors, Bioinformatics
Introduction
Plant transcription factors are a group of regulators that inhibit the transcription rate of target genes by regulating their expression during plant growth and development to facilitate the response to various biotic and abiotic stresses (Latchman, 1997; Jin et al., 2017). Transcription factors form a transcriptional complex of regulatory genes by binding to DNA sequences or specifically interacting with other proteins (Welner et al., 2015; Kim, Nam & Lim, 2016). The NAC (NAM, ATAF1/2, and CUC2) transcription factor family is one of the largest families of transcription factors in plants. NAC transcription factors are named after no apical meristem (NAM) proteins found in Petunia hybrids (Souer et al., 1996), Arabidopsis transcription activation factor (ATAF1/2) proteins, and cup-shaped cotyledon (CUC2) proteins of Arabidopsis thaliana (Aida et al., 1997). The N-terminal of NAC proteins contains a conserved domain comprising 150–160 amino acid, which is further divided into five subdomains (A–E), whereas the C-terminal is a height diversity transcriptional regulatory region (Welner et al., 2015). Additionally, a group of NAC proteins, named NTL proteins (NAC with transmembrane motif 1-like), have been identified, which contain transmembrane regions with α-helical transmembrane motifs (Kim et al., 2006; Seo & Park, 2010).
Genes encoding NAC transcription factors have been identified in many plant species, including Arabidopsis, rice (Oryza sativa), radish (Raphanus sativus), melon (Cucumis melo), Brassica napus, Tartary buckwheat (Fagopyrum tataricum), cacao (Theobroma cacao), and sesame (Sesamum indicum) (Ooka et al., 2003; Nuruzzaman et al., 2010; Wang et al., 2015; Wei et al., 2016; Karanja et al., 2017; Zhang et al., 2018b; Liu et al., 2019; Shen et al., 2019). In some plant species, the NAC transcription factors were known to play diverse roles. The first NAC genes identified in Petunia were shown to regulate the formation of embryos and flowers (Souer et al., 1996). Subsequent studies have shown that NAC transcription factors are involved in the response to many biotic and abiotic stresses (Nakashima et al., 2012; Lv et al., 2016), such as drought (Wu, Wang & Wang, 2016; Hussain et al., 2017), salt stress (Wang et al., 2017; Alshareef et al., 2019), floral morphogenesis (Hendelman et al., 2013), lateral root development (He et al., 2005), leaf senescence (Guo & Gan, 2006; Kim, Nam & Lim, 2016), embryo development (Dalman et al., 2017), hormone signaling (Wang, Rashotte & Dane, 2014), and xylogenesis, fiber development, and wood formation in vascular plants (Ouyang et al., 2016; Sun et al., 2018; Zhang et al., 2018a; Yao et al., 2020).
Currently, various NAC TFs are reportedly induced by temperature stress and participate in plant responses to cold and hot stress. Reports have shown that the expression of NAC TFs is significantly increased in tomato (Liu et al., 2012) and chickpeas (Ha et al., 2014) due to high and low temperatures. Overexpression of GmNAC20 enhances salt and freezing tolerance in transgenic Arabidopsis plants. GmNAC20 mediates stress tolerance by regulating the expression of COR (cold-responsive) genes, DREB1A/CBF3 (Dehydration Responsive Element-Binding Protein/C-repeat Binding Factor 3), and DREB1C/CBF2. GmNAC20 can directly bind to the promoters of DREB1A/CBF3 and DREB1C/CBF2, thereby causing increased transcription of CBF3 and decreased transcription of CBF2. MaNAC1 acts as a target gene of MaICE1 (Musa acuminata inducer of CBF expression 1), wherein MaNAC1 protein interacts with MaCBF1 to regulate the cold tolerance of banana fruit (Hao et al., 2011). The overexpression of a Malus baccata NAC transcription factor gene MbNAC25 also increases cold and salt tolerances of Arabidopsis (Han et al., 2020). NAC transcription factors play an essential role in high temperature stress. Arabidopsis plants with NAC019 overexpression are more heat-tolerant than the wild-type, and heat shock factors (HSFs) binding to the promoter element of heat shock proteins are the primary mechanisms for heat shock protein accumulation under heat stress. Heat stress can also regulate the expression of RCF2 (regulators of C-REPEAT BINDING FACTOR gene expression 2) (Guan et al., 2014), which interacts with and dephosphorylates NAC019; both proteins are necessary for heat induction of HSFs and thermotolerance. High temperature causes accumulation of H2O2 in plant cells, whereas positive feedback induces NTL4 gene expression in Arabidopsis. Reactive oxygen species (ROS) induce NTL4 gene transcription and NTL4 protein processing, wherein NTL4 gene and ROS constitute a positive feedback loop. High temperatures cause rapid accumulation of ROS in Arabidopsis and recycling of nutrients and metabolites in damaged tissues to meristems or newly formed leaves, thereby causing local programmed cell death and enhancing plants’ survival rate (Lee et al., 2014).
Tumorous stem mustard (Brassica juncea var. tumida) is an economically important crop widely cultivated in China. B. juncea is an allotetraploid (AABB, 2n = 36) that originated from hybridization between Brassica rapa (AA, 2n = 20) and Brassica nigra (BB, 2n = 16), followed by chromosome doubling. The enlarged stem of B. juncea var. tumida is used as an important seasoning, such as Fuling mustard. The growth of B. juncea var. tumida is affected by temperature, humidity, and light (Liu, 1996). The current research on B. juncea var. tumida is mainly focused on increasing yield via genetic breeding; however, few studies have been conducted on its internal molecular regulation mechanism of NAC genes. The genome data of Brassica juncea var.tumida has been published recently, which made it possible for us to find more information (Yang et al., 2016).
In this study, we used the HMMsearch software (Potter et al., 2018) to identify the NAC transcription factors in B. juncea var. tumida cultivar “Yongan xiaoye”. The physiological and biochemical characteristics of BjuNAC transcription factors were analyzed, and a phylogenetic tree of B. juncea var. tumida, Brassica rapa, Brassica nigra, rice and Arabidopsis thaliana NAC proteins was constructed. Additionally, the expression of temperature-sensitive BjuNAC genes was analyzed at high and low temperatures, and the yield and quality of mustard tubers was investigated at high temperature (39 °C), low temperature (5 °C), and room temperature (25 °C; control). Overall, our results provide a theoretical basis of heat and cold stress tolerance in mustard.
Materials & Methods
Identification of NAC transcription factors
The genome sequence of B. juncea var. tumida was downloaded from the Brassica database (http://brassicadb.cn/#/Download/). The Hidden Markov Model (HMM) profile (Eddy, 1998) of the NAC protein (PF02365), which was downloaded from the Pfam database (http://pfam.xfam.org/) (Finn et al., 2014), was used as a query to identify the NAC genes in the B. juncea var. tumida genome using the local HMMER3.1 software, with E-value <1E-10 (Potter et al., 2018). Then, a species-specific HMM file of B. juncea var. tumida was generated using the HMMbuild protocol, and a new HMMsearch was performed. All putative NAC genes were identified using the NCBI CDD search (https://www.ncbi.nlm.nih.gov/cdd) (Marchler-Bauer & Bryant, 2004); the conserved NAM domain was identified using SMART (http://smart.embl-heidelberg.de/).
Multiple sequence alignment and phylogenetic analysis
The AtNAC protein sequences were downloaded from The Arabidopsis Information Resource database (https://www.arabidopsis.org/) (Garcia-Hernandez et al., 2002). Furthermore, the NAC protein sequences in rice were obtained from Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) (Kawahara et al., 2013). The NAC protein sequences of Brassica nigra were obtained using HMMsearch method above. And NAC proteins in Brassica rapa were downloaded from the Brassica database (http://brassicadb.cn/#/SearchTranscriptionFactorGene/?subfamily=NAC). All amino acid sequences of BjuNAC, AtNAC, Brassica rapa, Brassica nigra and rice proteins were aligned using Muscle alignment (Edgar, 2004), and a neighbor-joining phylogenetic tree was constructed using MEGA X, with 1,000 bootstrap replications (Kumar et al., 2018).
Characterization of BjuNAC proteins and BjuNAC genes
The physicochemical properties (such as molecular weight and isoelectric point) of all putative BjuNAC proteins were determined using the ExPASy website (https://web.expasy.org) (Artimo et al., 2012). Conserved motifs were identified using MEME (http://meme-suite.org/) (Bailey et al., 2009), with the following parameters: maximum number of motifs = 15; search model = zero or one occurrence per sequence; motif length = 6–50 amino acids; and default settings for all other parameters. Information on the structure of all identified NAC genes was obtained from the GFF file, and the exon–intron map of NAC genes was constructed using the Gene Structure Display Server (GSDS) website (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). Phylogenetic trees, motifs, and gene structure maps were drawn using the TBtools software (Chen et al., 2020).
Chromosomal distribution cis-element and evolutionary analysis of BjuNAC genes
Information on the chromosomal location of BjuNAC genes was obtained from the GFF file using a Perl script (Supplemental 1) and then compiled manually. The chromosomal map of BjuNAC genes was drawn using the Mapchart software (Voorrips, 2002). MCSCANX (Wang et al., 2012) was used to analyze the collinearity and duplication events of BjuNAC genes. The synonymous substitution ratio (Ka/Ks) value of segmental and tandem duplication genes was used as the protocol sample Ka/Ks calculator in TBtools. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) was used to perform cis-element analysis of the upstream regions (−1,500 bp) of BjuNAC genes.
RNA extraction and qRT-PCR analysis
The expression pattern of BjuNAC genes in young leaves was studied under high and low temperatures. Seeds were sown in meteorite: soil (2:1) mix, incubated at 25 °C under 12-h light/12-h dark photoperiod in a growth chamber. After 5 weeks of seed culture, seedlings with the same growth conditions were placed at high temperature (39 °C) and low temperature (5 °C) and cultivated for 0, 6, 12, 24, and 48 h; whole seedlings were used as the samples. After harvest, all samples were immediately frozen in liquid nitrogen and stored at −70 °C until needed for RNA isolation.
Total RNA was isolated from whole seedlings of B. juncea var. tumida using the Plant MiniBESTRNA Extraction Kit (TaKaRa Biotechnology, Dalian, China), and cDNA was synthesized using the PrimeScript™ RT Kit and gDNA Eraser (TaKaRa Biotechnology, Dalian, China). Then, quantitative real-time PCR (qRT-PCR) analysis was performed on the Bio-Rad iQ5 Real-Time PCR Detection System using the TBGreen® PremixEx Taq II Kit (Tli RNaseH Plus) RR820A (TaKaRa, Japan) and 14 pairs of gene-specific primers (Table S1), under the following thermocycling conditions: 95 °C for 30 s; 40 cycles at 95 °C for 5 s and 55 °C for 30 s; and 72 °C for 30 s. The BjuActin gene (BjuB012485) was used as an internal reference. Three biological replicates were performed for each gene, with each replicate containing three technical repeats. To analyze the expression change under different stress, the 2−ΔΔCt method was applied (Livak & Schmittgen, 2001; Rao et al., 2013).
Results
Genome-wide identification of BjuNAC genes
The HMM model file (PF02365) of NAC was downloaded from the Pfam database. A total of 336 BjuNAC genes were identified in the genome sequence data of B. juncea var. tumida using the HMMER 3.0 software. The redundant and non-NAM domain-containing genes were removed by domain identification using the SMART and NCBI websites, and 300 BjuNAC genes were extracted for further analysis. According to their chromosomal locations, 278 of 300 BjuNAC genes were named as BjuNAC 001–278, whereas the remaining 22 genes belonging to the scaffold or contig data were named as BjuNAC 279–300. Proteins encoded by BjuNAC genes varied in length from 164 amino acids (aa; BjuNAC037) to 1,432 aa (BjuNAC236) (Table S2), and their isoelectric point ranged from 4.19 (BjuNAC012) to 10.03 (BjuNAC058), with an average value of 6.50. The molecular weight (MW) of BjuNAC proteins ranged from 18.74 kDa (BjuNAC037) to 161 kDa (BjuNAC236), with an average molecular weight of 38.45 kDa. Among the NAC TFs studied, only 3 NAC proteins have MW >100 kDa and 36 NAC proteins are between 50 and 100 kDa. The MW of most NAC proteins is 20–50 kDa. The physiological and biochemical properties of all BjuNAC proteins are listed in Table S2.
Phylogenetic analysis of BjuNAC proteins
We constructed a phylogenetic tree using the NJ method, with 1,000 bootstrap replications. On the basis of the classification of NAC proteins in other species (Ooka et al., 2003), all 300 BjuNAC proteins were divided into 18 subfamilies (Fig. S1). The results showed that BjuNAC proteins are as diverse as AtNAC and rice proteins, and 300 BjuNACs were unevenly distributed in 18 subgroups. In our analysis, no Brassica NAC members from the subgroups ONAC001 and OsNAC3 were identified. The largest branch (NAC 2) contained 45 NAC proteins, whereas the smallest branch (TIP) contained only one NAC protein. Each subgroup contained different numbers of proteins, probably due to different genomic events during the evolution of B. juncea var. tumida and other species.
Analysis of chromosomal distribution and duplication of BjuNAC genes
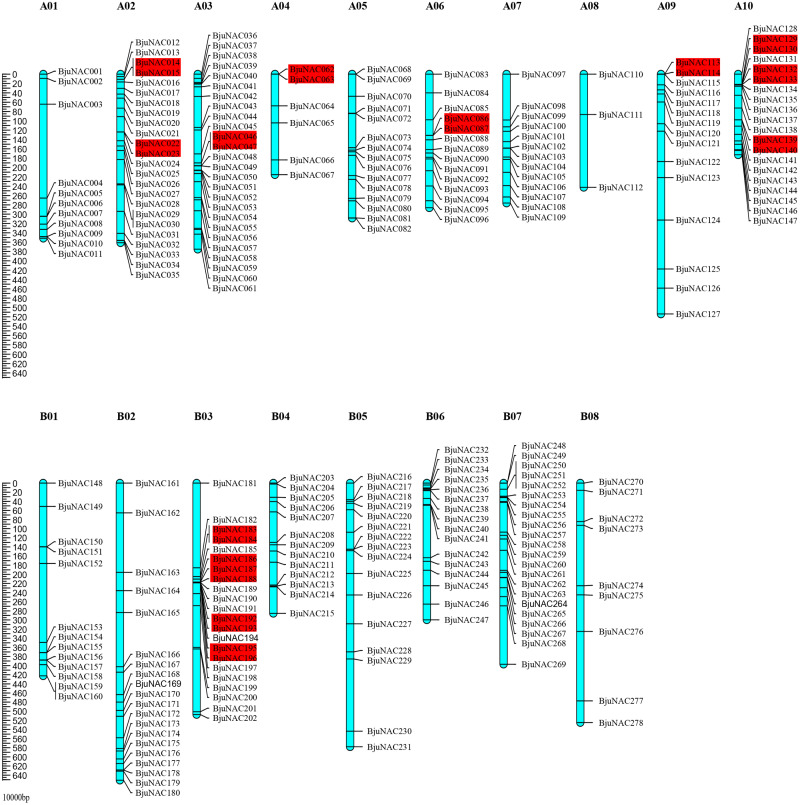
Out of 300 BjuNAC genes identified in this study, 278 were mapped to 18 chromosomes (A01–A10 and B01–B08). The highest number of BjuNAC genes (26) was mapped to chromosome A03; this was followed by 24, 22, 22, and 20 genes on chromosomes A02, B03, B07, and A10, respectively, and only 3 genes on chromosome A08 (Fig. 1). Distribution of BjuNAC genes on the chromosome is similar to NAC genes of Brassica rapa (BrNAC). It contains more NAC genes on chromosome A03 and A10, most of which are at the telomeric ends or near the centromere (Liu et al., 2014).
Figure 1. Chromosomal distribution of BjuNAC genes on the genetic map.
Graphical representation of locations for putative BjuNAC genes on each chromosome. The tandem duplicated genes were colored with red backgrounds. A01∼A10, B01∼B08 shows the chromosome numbers. Other 22 BjuNAC genes information belongs to scaffold or contig data were shown in Table S2.
Collinearity analysis revealed that the mapped 220 BjuNAC genes formed 288 collinear pairs, including 14 tandem repeat pairs (Fig. S2). Additionally, 14 collinear pairs belonged to tandemly duplicated genes, including BjuNAC14/15, BjuNAC22/23, BjuNAC46/47, BjuNAC62/63, BjuNAC86/87, BjuNAC113/114, BjuNAC129/130, BjuNAC132/133, BjuNAC139/140, BjuNAC183/184, BjuNAC186/187/188 (three tandem repeats), BjuNAC192/193, BjuNAC195/196, and BjuNAC280/282 (on the Contig11_2007833_2318034).
Exon–intron gene structure and conserved motif analysis
To investigate the function of BjuNAC genes, we determined their exon–intron structure using the GSDS (Fig. S3). We also constructed a phylogenetic tree of all BjuNAC genes (Fig. S3A). The results showed that most of the BjuNAC genes (289 out of 300) contained introns, whereas the remaining 11 genes were intronless. Approximately half of the BjuNAC genes (159 of 300) contained two introns. BjuNAC186 and BjuNAC236 contained 18 introns, the highest number of introns among all BjuNAC genes (Fig. S3C).
To further analyze the structural diversity of BjuNAC proteins, conserved motifs were searched using the MEME program (Fig. S4). The distribution of motifs is shown in (Fig. S3B). Generally, NAC genes clustered within the same subgroup shared a similar motif composition (Fig. S5). Most of the NAC proteins included motif1 (representing subdomain A), motif4 (representing subdomain B), motif5 and motif3 (corresponding to subdomain C), motif2 and motif6 (corresponding to subdomain D), and motif7 (represented subdomain E). However, some of the conserved motifs were partially lost in proteins belonging to certain subgroups. For example, there were three BjuNAC proteins lost motif7 and three of them also lost motif 2, motif 3, motif 5 and motif6 in group VI. Half of the BjuNAC proteins in group IV; contained only motif1 and motif4. In group VII and VIII, almost all of BjuNAC proteins contained motif 8, especially in group VII, only BjuNAC276 lost motif 1 and motif 12 (Fig. S3B).
Cis-acting element analysis
Cis-acting elements usually located within 1,500 bp sequence upstream of the transcription start site play a key role in regulating gene expression. Studies have shown that the expression of many genes depends on the presence or absence of specific cis-acting elements. To predict the function of BjuNAC genes, we analyzed the upstream regulatory sequence (−1,500 to +1 bp) of BjuNAC genes using PlantCARE. The results showed that the upstream sequences of most of the 300 BjuNAC genes contained cis-acting elements (Fig. S6) that respond to light, including the 3-AF1 binding site, AAAC-motif, ACA-motif, ACE, AT1-motif ATC-motif, ATCT-motif, Box 4, chs-CMA1a, GA-motif, GATA-motif, G-Box, and GT1-motif. Additionally, 77% of the BjuNAC genes contained anaerobic induction-responsive element and abscisic acid-responsive element (ABRE), 70% contained methyl jasmonate-responsive element (CGTCA-motif/TGACG-motif), 39% contained auxin-responsive element (TGA-box), 34% contained drought stress response element (MBS) and salicylic acid-responsive elements (TCA-element), 35% contained defense and stress response element (TC-rich repeats), and 36% contained low-temperature response element (LTR). The upstream sequences of only 22% and 11% of the BjuNAC genes contained circadian control regulatory elements (circadian) and seed development-specific elements (RY-element), respectively. A total of 110 genes have one or more LTR cis-elements involved in low-temperature responsiveness. Four abiotic stress response elements, including ABRE, DRE, LTR, and TC-rich repeats, lay the foundation for further research on the potential regulatory mechanism of NAC genes in response to abiotic stress.
Gene expression analysis under temperature stress
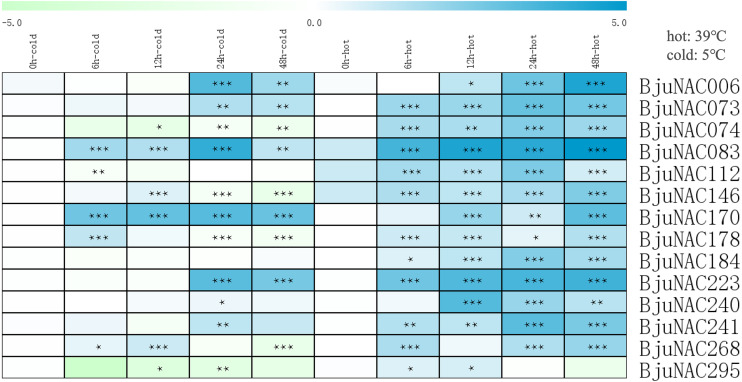
To further study the response of BjuNAC genes to abiotic stress, 14 BjuNAC genes enriched in the “temperature stress response” Gene Ontology term were selected for expression analysis under cold and heat stress treatments by qRT-PCR. Heat maps showing the expression of these 14 genes under the two stresses are shown in Fig. 2, the expression results are listed in Table S5. Most of the genes were up-regulated under the heat treatment, whereas some genes were induced by low-temperature stress. BjuNAC006, BjuNAC083, BjuNAC170 and BjuNAC223 were highly up-regulated under low-temperature conditions. Under the cold stress, most of the genes were up-regulated at 24 or 48 h, but some genes showed down-regulation after 48 h of cold treatment, for example, BjuNAC295 was down-regulated under both cold and heat treatments.
Figure 2. Expression levels of BjuNACs under cold and hot treatment by qRT PCR.
The number represented the treatment times (hours). The colour scales represent relative expression data. Note: *** p < 0.001; ** p < 0.01; * p < 0.05.
Discussion
Tumorous stem mustard is an economically important food crop; however, its yield is affected by many abiotic stresses (Cai et al., 2019; Li et al., 2019). As one of the largest transcription factor families in plant species, the NAC family plays an important role in plant growth, development, and biotic and abiotic stress responses (Seo & Park, 2010; Nakashima et al., 2012). NAC transcription factors have been characterized at the genome level in many plant species (Ling et al., 2017; Ahmad et al., 2018; Zhang et al., 2018b; Liu et al., 2019). In this study, we conducted a comprehensive analysis of BjuNAC genes and the encoded transcription factors, including analysis of the gene structure, evolutionary relationships, conserved motifs, chromosomal distribution, duplication events, and expression patterns under high and low temperatures.
A total of 300 NAC genes were identified in the B. juncea var. tumida genome, which is far greater than the number of NAC genes identified in other plant species. This maybe because the B. juncea var. tumida has a larger genome, which is an allotetraploid (AABB), resulting from the hybridization between diploid ancestors of B. rapa (AA) and B. nigra (BB), followed by spontaneous chromosome doubling (He et al., 2020). Therefore, the number of NAC genes in B. juncea var. tumida doubled. The number of NAC proteins in mustard tubers is more than that in Brassica rapa (204 members) (Liu et al., 2014) but less than that in NAC proteins in Brassica napus (410 members) (Mohanta et al., 2020), which indicates that the number of NAC proteins is positively correlated to the size of the Brassica genome. NAC family members were identified in Brassica juncea var. tumida (300 members) and Brassica nigra (212 members) (Table S3), using the HMMsearch method. Phylogenetic tree of NAC transcription factors in Brassica juncea var. tumida, Brassica nigra, Brassica rapa, Arabidopsis and rice, and all the NAC members were divided into 18 subfamilies. In the distribution of each subfamily, the number of Brassica nigra and Brassica rapa NAC members in subgroups was the same, but it was more in Brassica juncea than that of diploid. The copy number change of NAC family in Brassica juncea is not a simple doubling of the two diploids, but the loss or increase of genes between different subfamilies compared with the number of NAC in Brassica nigra and Brassica rapa. The isoelectric point of BjuNAC proteins varied from 4.19 to 10.03, with an average value of 6.50, which is consistent with most NAC TFs in Brassica napus that are acidic proteins. More than 68.3% of the BjuNAC proteins (205 out of 300) were acidic in nature, which may contribute to the acidic subcellular environment. Different NAC protein subgroups have different number of amino acids, protein MW, and isoelectric points. The versatility of NAC function may be the primary reason for the wide MW ranges of NAC proteins. Phylogenetic analysis revealed that all BjuNAC genes were divided into 18 subgroups. NAC proteins are involved in a variety of plant growth and developmental processes, such as seed germination, cell division, secondary cell wall biosynthesis, organ boundary and meristem formation, lateral root development, flowering, senescence, and iron balance regulation. Phylogenetic analysis can be used to predict gene function, which is very important for further analysis of gene function. NAM subgroups are involved in the formation of organ borders and meristem formation of plant, whereas ATAF1 subgroups are related to plant senescence and leaf curling. The overexpression of OsNAC1 significantly enhanced drought resistance of transgenic rice in the field without phenotypic changes or yield reduction under severe drought stress conditions during the growth period (Hu et al., 2006). Although the expression level of OsNAC7 in rice is extremely low or negligible in leaves, embryos, and callus, high expression levels are detected in stems and young panicles (Kikuchi et al., 2000). Expression of OsNAC022 was induced by drought, high salinity, and abscisic acid (Hong et al., 2016). Salicylic acid and abscisic acid had no significant effect on TaNAC08 gene expression (Xia et al., 2010). The TIP subfamily contains BjuNAC074, whose protein sequence is homologous to Arabidopsis thaliana TIP mRNA, wherein AtTIP gene is resistant to virus invasion in Arabidopsis thaliana (Donze et al., 2014). That implies NAC protein is involved in the response to virus infection during plant vegetative development. AtNAP gene in the subfamily of NAC transcription factors is related to leaf senescence, and overexpression of the AtNAP gene causes premature senescence in Arabidopsis thaliana. According to the results of phylogenetic analysis, the AtNAC3, ATAF, and NAP subgroup proteins share a close relationship, and most of the published stress-related NAC family members are included in these three subgroups. Arabidopsis contains three proteins belonging to the AtNAC3 subgroup (ANAC019, ANAC055, and ANAC072/RD26) and four proteins belonging to the ATAF subgroup (ATAF1/ANAC002, ATAF2/ANAC081, ANAC032, and ANAC102), all of which are NAC proteins. NAC proteins participate in the response to various abiotic and biotic stresses. In rice, the ANAC063 subgroup contained the highest number of NAC proteins, whereas the OsNAC8 subgroup was the smallest, with only three BjuNAC genes in B. juncea var. tumida is higher than that in other species, indicating that BjuNAC genes are more important for plant growth and development under various abiotic and biotic stresses.
Whole genome duplication has played an important role during plant genome evolution process, and segmental duplication is primarily responsible for the expansion of the NAC gene family. Collinearity analysis showed that 223 BjuNAC genes resulted from duplication, including 1 singleton, 27 dispersed, 9 proximal, 248 segmental, and 15 tandem duplications. These genes formed a total of 288 duplicate gene pairs, 50% of which originated from segmental duplication, indicating that segmental duplication represents the main mode of expansion of the BjuNAC gene family. At the same time, we found that 15 gene pairs were tandem repeats. Additionally, non-synonymous to synonymous substitution ratio (Ka/Ks) of segmental and tandem duplications was <1, indicating that the BjuNAC genes may have undergone purifying selection.
We also analyzed the exon–intron structure of all BjuNAC genes and conserved motifs in the encoded proteins. Molecular characterization of BjuNAC genes revealed the motif composition of the NAC transcription factor and is dispersed in a subgroup of gene structures that are not used. The BjuNAC genes clustered within the same subgroup generally shared a similar motif composition. The number of introns in BjuNAC genes varied from 0 to 18; only 11 BjuNAC genes were intronless, and approximately 50% of all BjuNAC genes contained two introns. These molecular features of BjuNAC genes are similar to the NAC genes of other plant species. Variation in the structure of BjuNAC genes indicates that the evolution of environmental stress response in these genes occurred through the acquisition or loss of introns.
We compared the NAC protein sequences of Brassica rapa, Brassica nigra and Brassica juncea respectively by the blast. Through the similarity of greater than 90%, we assumed that the two genes have homology. Finally, 151 BniNACs and 181 BraNACs were filtered. According to the analysis of chromosome distribution, genes deriving from chromosomes of Brassica nigra and Brassica rapa might have caused differences in the distribution of NACs in Brassica juncea var. tumida due to chromosomal rearrangements and structural variations within evolution. Among them, on chromosomes B1, B2 and B7 of Brassica nigra, there were 32, 20, 24 genes homologous to BjuNAC genes. On chromosome of Brassica rapa, 35 genes on chromosome A03 were homologous to BjuNACs, followed by 28 genes on chromosome A02, and the least only 2 genes (Table S4) on A08. BniNACs chromosome distribution was shown in Fig. S7.
B. juncea var. tumida is sown from September to early October; however, late sowing significantly affects its effective tumorous stem yield. A temperature range of 15–20 °C is optimal for the germination of B. juncea var. tumida seeds. At the germination and seedling stages, changes in environmental conditions greatly influence the crop yield; sometimes only 3–5 days of difference on sowing date can cause 30% of the yield reduction of tumorous stem mustard (Liu, 2014). The region of Chongqing in southwest China experiences extreme weather in September and October, which has a greater impact on the germination of mustard seeds and the growth of seedlings. In Chongqing Fuling area, temperatures as high as 35 °C and 30 °C lasted for >5 days in September 2018 and 2019 and for 13 days in September 2019, respectively. Continuously high temperature decreases not only the seed germination rate but also the survival rate of seedlings.
The growth phase of mustard seedlings is from mid-October to late November. After 60 days of the growing season, mustard seedlings enter the stem expansion stage, which generally lasts from early December to mid-February (approximately 100 days) and is the most important growth phase of mustard plants. The optimal temperature for the stem expansion stage is 8 °C–15 °C, and the number of leaves is 25–45. At temperature <4 °C, the stem expansion of the mustard tuber is suppressed, which may lead to growth arrest. At −4 °C, frost damage is lethal for the mustard tuber (Li, Dai & Zhan, 2015). Long-term exposure to low temperature induces the plants to transition from vegetative growth to reproductive growth, resulting in bolting and consequently reduced yield. In 2008, the overall output of tumorous stem mustard was reduced by 30% because of excessive snow and ice (Jian, 2019).
The results of qRT-PCR showed that the expression levels of BjuNAC006, BjuNAC073, BjuNAC083, BjuNAC223, BjuNAC241, and BjuNAC170 increased significantly over time, regardless of the heat or cold stress, indicating that these genes may be involved in response to temperature stimuli. BjuNAC073 and BjuNAC006 are homologous to BnaNAC103 and BnaNAC55, respectively. BnaNAC103 (Niu et al., 2014) and BnaNAC55 (Niu et al., 2016) respond to multiple signals, including heat, cold, salicylic acid, and a fungal pathogen, Sclerotinia sclerotiorum. Simultaneously, they play a role in activating the promoter activity of five genes encoding ROS-scavenging enzymes or mediating defense responses. Expression of BjuNAC083 is consistent with NAC gene expression (similar sequence with Bra029201) under heat and cold stress in Brassica rapa (Liu et al., 2014). BjuNAC006 (sequence similar to ANAC055) and BjuNAC083 (sequence similar to ANAC019) transcription factors bind specifically to 5′-CATGTG-3′ motif and respond to water shortage at high temperatures (Tran et al., 2004). The expression level of BjuNAC178 reached a peak at 6 h, then declined under cold stress and gradually increased under heat stress, which indicated the role of BjuNAC178 in temperature stress response. The expression level of BjuNAC240 reached a peak at 24 h, but did not vary much under cold stress. BLAST (basic local alignment search tool) comparison showed that BjuNAC240 is homologous to BnaNAC60, positively modulating programmed cell death and age-triggered leaf senescence (Yan et al., 2020). The expression of BjuNAC112 and BjuNAC184 was the highest at 24 h under heat stress, but did not change significantly under cold stress, indicating that these genes may be involved in the high temperature stress response. The expression level of two BjuNAC genes (BjuNAC074 and BjuNAC295) was down-regulated under low temperature, and they may respond to cold stress. Nonetheless, the specific biological functions of BjuNAC genes need to be verified experimentally.
Conclusions
In this study, we identified 300 NAC genes in B. juncea var. tumida and characterized their gene structure, chromosomal distribution, phylogenetic relationship, conserved motifs, and expression pattern in response to abiotic stress. BjuNAC genes showed differential expression patterns under high- and low-temperature stresses. Overall, our findings provide a strong basis for further investigation of the biological functions of BjuNAC genes. This information could be used to increase the yield of B. juncea var. tumida, thus increasing its food value and economic importance.
Supplemental Information
Multiple sequence alignment of full-length NAC proteins was done using muscle method and the phylogenetic tree was constructed using MEGAX by the neighbor-joining method with bootstrap 1000 replicates.
Collinear blocks represented by grey background. 288 BjuNAC duplication pairs are linked with green lines, and 15 tandem duplication pairs are linked with red lines.
(a) A phylogenetic tree using the neighbor-joining method by MEGAX, with bootstrap values of 1,000. (b) The conserved motifs were predicted using Multiple Em for Motif Elicitation (MEME) (http://alternate.meme-suite.org/). (c) The box length indicates the number of amino acids in the motif. The coding sequences (CDS) of exons are represented by purple boxes, the introns are represented by lines.
The height of each amino acid code in the sequence logo of each motif represents the degree of conservation.
All domain sequences of BjuNACs divided into five subdomains A-E.
The promoter sequences (1500 bp) upstream of genes were chosen for cis-regulatory element analysis using the PlantCARE online tool (http://www.dna.affrc.go.jp/PLACE/). Each color indicates a cis-regulatory element.
Graphical representation of locations for putative BniNAC genes on each chromosome. B1∼B8 represented the chromosome numbers. Other 25 BniNAC genes information belongs to scaffold data were shown in Table S3.
Funding Statement
This work was supported by grants from the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN201800609) and Natural Sciences Foundation of Chongqing (Grant No. cstc2015jcyjA0752). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Changshu Luo, Email: luochangshu7812@sina.com.
Xiaohong He, Email: hexh@cqupt.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Longxing Jiang performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Quan Sun conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Yu Wang performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Pingan Chang conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Haohuan Kong performed the experiments, prepared figures and/or tables, and approved the final draft.
Changshu Luo analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Xiaohong He conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.
References
- Ahmad et al. (2018).Ahmad M, Yan X, Li J, Yang Q, Jamil W, Teng Y, Bai S. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biology. 2018;18:214. doi: 10.1186/s12870-018-1427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida et al. (1997).Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. The Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshareef et al. (2019).Alshareef NO, Wang JY, Ali S, Al-Babili S, Tester M, Schmöckel SM. Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato. Plant Physiology and Biochemistry. 2019;140:113–121. doi: 10.1016/j.plaphy.2019.04.038. [DOI] [PubMed] [Google Scholar]
- Artimo et al. (2012).Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey et al. (2009).Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2019).Cai Z, Zeng DE, Liao J, Cheng C, Sahito ZA, Xiang M, Fu M, Chen Y, Wang D. Genome-wide analysis of auxin receptor family genes in Brassica juncea var. tumida. Gene. 2019;10(2):165. doi: 10.3390/genes10020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2020).Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Dalman et al. (2017).Dalman K, Wind JJ, Nemesio-Gorriz M, Hammerbacher A, Lundén K, Ezcurra I, Elfstrand M. Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biology. 2017;17:6. doi: 10.1186/s12870-016-0952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze et al. (2014).Donze T, Qu F, Twigg P, Morris TJ. Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology. 2014;449:207–214. doi: 10.1016/j.virol.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy (1998).Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar (2004).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn et al. (2014).Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez et al. (2002).Garcia-Hernandez M, Berardini TZ, Chen GH, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, Mundodi S, Reiser L, Rhee SY, Scholl R, Tacklind J, Weems DC, Wu YH, Xu L, Yoo D, Yoon JW, Zhang PF. TAIR: a resource for integrated Arabidopsis data. Functional & Integrative Genomics. 2002;2:239–253. doi: 10.1007/s10142-002-0077-z. [DOI] [PubMed] [Google Scholar]
- Guan et al. (2014).Guan Q, Yue X, Zeng H, Zhu J. The protein phosphatase RCF2 and its interacting partner nac019 are critical for heat stress–responsive gene regulation and thermotolerance in Arabidopsis. The Plant Cell. 2014;26:438–453. doi: 10.1105/tpc.113.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo & Gan (2006).Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal: for Cell and Molecular Biology. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- Ha et al. (2014).Ha CV, Esfahani MN, Watanabe Y, Tran UT, Sulieman S, Mochida K, Nguyen DV, Tran L-SP. Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLOS ONE. 2014;9:e114107. doi: 10.1371/journal.pone.0114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han et al. (2020).Han D, Du M, Zhou Z, Wang S, Li T, Han J, Xu T, Yang G. Overexpression of a Malus baccata NAC transcription factor gene MbNAC25 increases cold and salinity tolerance in Arabidopsis. International Journal of Molecular Sciences. 2020;21:1198. doi: 10.3390/ijms21041198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao et al. (2011).Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. The Plant Journal: for Cell and Molecular Biology. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- He et al. (2020).He J, He XH, Chang PA, Jiang HZ, Gong D, Sun Q. Genome-wide identification and characterization of TCP family genes in Brassica juncea var. tumida. PeerJ. 2020;8:e9130. doi: 10.7717/peerj.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2005).He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. The Plant Journal: for Cell and Molecular Biology. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- Hendelman et al. (2013).Hendelman A, Stav R, Zemach H, Arazi T. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. Journal of Experimental Botany. 2013;64:5497–5507. doi: 10.1093/jxb/ert324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong et al. (2016).Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Frontiers in Plant Science. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2015).Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2006).Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain et al. (2017).Hussain RM, Ali M, Feng X, Li X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biology. 2017;17:55. doi: 10.1186/s12870-017-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian (2019).Jian Y. E-commerce headlines. 2019. https://m.sohu.com/a/338012352_222128. [01 September 2019]. https://m.sohu.com/a/338012352_222128
- Jin et al. (2017).Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja et al. (2017).Karanja BK, Xu L, Wang Y, Muleke EMM, Jabir BM, Xie Y, Zhu X, Cheng W, Liu L. Genome-wide characterization and expression profiling of transcription factor genes under abiotic stresses in radish (Raphanus sativus L.) PeerJ. 2017;5:e4172. doi: 10.7717/peerj.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara et al. (2013).Kawahara Y, Bastide MDL, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG, Childs KL, Davidson RM, Lin HN, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi et al. (2000).Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY. Molecular analysis of the NAC gene family in rice. Molecular & General Genetics. 2000;262:1047–1051. doi: 10.1007/PL00008647. [DOI] [PubMed] [Google Scholar]
- Kim, Nam & Lim (2016).Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Current Opinion in Plant Biology. 2016;33:48–56. doi: 10.1016/j.pbi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2006).Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. The Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2018).Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman (1997).Latchman DS. Transcription factors: an overview. The International Journal of Biochemistry & Cell Biology. 1997;29:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2014).Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM. The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Science: an International Journal of Experimental Plant Biology. 2014;227:76–83. doi: 10.1016/j.plantsci.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Lescot et al. (2002).Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2019).Li M, Sun B, Xie F, Gong R, Luo Y, Zhang F, Yan Z, Tang H. Identification of the GRAS gene family in the genome provides insight into its role in stem swelling in stem mustard. PeerJ. 2019;7:e6682. doi: 10.7717/peerj.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Dai & Zhan (2015).Li S, Dai XD, Zhan KH. Influence of extreme climate events on the cultivation of mustard tuber in Fuling District, Chongqing. Chinese Agricultural Science Bulletin. 2015;29:174–180. [Google Scholar]
- Ling et al. (2017).Ling L, Song L, Wang Y, Guo C. Genome-wide analysis and expression patterns of the NAC transcription factor family in Medicago truncatula. Physiology and Molecular Biology of Plants: an International Journal of Functional Plant Biology. 2017;23:343–356. doi: 10.1007/s12298-017-0421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2012).Liu H, Ouyang B, Zhang J, Wang T, Li H, Zhang Y, Yu C, Ye Z. Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLOS ONE. 2012;7:e50785. doi: 10.1371/journal.pone.0050785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2019).Liu M, Ma Z, Sun W, Huang L, Wu Q, Tang Z, Bu T, Li C, Chen H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum) BMC Genomics. 2019;20:113. doi: 10.1186/s12864-019-5500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu (1996).Liu PY. Origin, evolution, classification and distribution of Chinese mustard. In: Liu PY, editor. Chinese mustard. China Agriculture Press; Beijing: 1996. pp. 1–26. [Google Scholar]
- Liu et al. (2014).Liu T, Song X, Duan W, Huang Z, Gaofeng L, Li Y, Hou X. Genome-wide analysis and expression patterns of nac transcription factor family under different developmental stages and abiotic stresses in chinese cabbage. Plant Molecular Biology Reporter. 2014;32:1041–1056. doi: 10.1007/s11105-014-0712-6. [DOI] [Google Scholar]
- Liu (2014).Liu YX. Seed Encyclopedia. 2014. http://bk.3456.tv/zhongzibk/13428. [5 May 2014]. http://bk.3456.tv/zhongzibk/13428
- Livak & Schmittgen (2001).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lv et al. (2016).Lv X, Lan S, Guy KM, Yang J, Zhang M, Hu Z. Global expressions landscape of NAC transcription factor family and their responses to abiotic stresses in Citrullus lanatus. Scientific Reports. 2016;6:30574. doi: 10.1038/srep30574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer & Bryant (2004).Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta et al. (2020).Mohanta TK, Yadav D, Khan A, Hashem A, Tabassum B, Khan AL, Abd Allah EF, Al-Harrasi A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLOS ONE. 2020;15:e0231425. doi: 10.1371/journal.pone.0231425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima et al. (2012).Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Niu et al. (2014).Niu F, Wang B, Wu F, Yan J, Li L, Wang C, Wang Y, Yang B, Jiang YQ. Canola (Brassica napus L.) NAC103 transcription factor gene is a novel player inducing reactive oxygen species accumulation and cell death in plants. Biochemical and Biophysical Research Communications. 2014;454:30–35. doi: 10.1016/j.bbrc.2014.10.057. [DOI] [PubMed] [Google Scholar]
- Niu et al. (2016).Niu F, Wang C, Yan J, Guo X, Wu F, Yang B, Deyholos MK, Jiang YQ. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Molecular Biology. 2016;92:89–104. doi: 10.1007/s11103-016-0502-7. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman et al. (2010).Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Ooka et al. (2003).Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research: an International Journal for Rapid Publication of Reports on Genes and Genomes. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Ouyang et al. (2016).Ouyang K, Li J, Zhao X, Que Q, Li P, Huang H, Deng X, Singh SK, Wu AM, Chen X. Transcriptomic analysis of multipurpose timber yielding tree Neolamarckia cadamba during xylogenesis using RNA-Seq. PLOS ONE. 2016;11:e0159407. doi: 10.1371/journal.pone.0159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter et al. (2018).Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Research. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao et al. (2013).Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2(−ΔΔCT) method for quantitative real-time polymerase chain reaction data analysis. Biostatistics, Bioinformatics and Biomathematics. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- Seo & Park (2010).Seo PJ, Park CM. A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signaling & Behavior. 2010;5:481–483. doi: 10.4161/psb.11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2019).Shen S, Zhang Q, Shi Y, Sun Z, Zhang Q, Hou S, Wu R, Jiang L, Zhao X, Guo Y. Genome-wide analysis of the NAC domain transcription factor gene family in Theobroma cacao. Gene. 2019;11(1):35. doi: 10.3390/genes11010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer et al. (1996).Souer E, Van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2018).Sun H, Hu M, Li J, Chen L, Li M, Zhang S, Zhang X, Yang X. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biology. 2018;18:150. doi: 10.1186/s12870-018-1367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran et al. (2004).Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips (2002).Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. The Journal of heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang B, Guo X, Wang C, Ma J, Niu F, Zhang H, Yang B, Liang W, Han F, Jiang YQ. Identification and characterization of plant-specific NAC gene family in canola (Brassica napus L.) reveal novel members involved in cell death. Plant Molecular Biology. 2015;87:395–411. doi: 10.1007/s11103-015-0286-1. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang K, Zhong M, Wu YH, Bai ZY, Liang QY, Liu QL, Pan YZ, Zhang L, Jiang BB, Jia Y, Liu GL. Overexpression of a chrysanthemum transcription factor gene Dg NAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Reports. 2017;36:571–581. doi: 10.1007/s00299-017-2103-6. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2012).Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Rashotte & Dane (2014).Wang Z, Rashotte AM, Dane F. Citrullus colocynthis NAC transcription factors Cc NAC1 and Cc NAC2 are involved in light and auxin signaling. Plant Cell Reports. 2014;33:1673–1686. doi: 10.1007/s00299-014-1646-z. [DOI] [PubMed] [Google Scholar]
- Wei et al. (2016).Wei S, Gao L, Zhang Y, Zhang F, Yang X, Huang D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Reports. 2016;35:1827–1839. doi: 10.1007/s00299-016-1997-8. [DOI] [PubMed] [Google Scholar]
- Welner et al. (2015).Welner D, Deeba F, Lo Leggio L, Skriver K. NAC transcription factors: from structure to function in stress-associated networks. In: Gonzalez DH, editor. Plant transcription factors. Academic Press; Boston: 2015. pp. 199–212. [Google Scholar]
- Wu, Wang & Wang (2016).Wu J, Wang L, Wang S. Comprehensive analysis and discovery of drought-related NAC transcription factors in common bean. BMC Plant Biology. 2016;16:193. doi: 10.1186/s12870-016-0882-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia et al. (2010).Xia N, Zhang G, Sun YF, Zhu L, Xu LS, Chen XM, Liu B, Yu YT, Wang XJ, Huang LL, Kang ZS. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiological and Molecular Plant Pathology. 2010;74:394–402. doi: 10.1016/j.pmpp.2010.06.005. [DOI] [Google Scholar]
- Yan et al. (2020).Yan J, Chen Q, Cui X, Zhao P, Gao S, Yang B, Liu JX, Tong T, Deyholos MK, Jiang YQ. Ectopic overexpression of a membrane-tethered transcription factor gene NAC60 from oilseed rape positively modulates programmed cell death and age-triggered leaf senescence. The Plant Journal. 2020;105(3):600–618. doi: 10.1111/tpj.15057. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2016).Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, Yao P, Li X, Xie K, Zhang J, Wang J, Liu F, Ma W, Shopan J, Zheng H, Mackenzie SA, Zhang M. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nature Genetics. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- Yao et al. (2020).Yao W, Zhang D, Zhou B, Wang J, Li R, Jiang T. Over-expression of poplar NAC15 gene enhances wood formation in transgenic tobacco. BMC Plant Biology. 2020;20:12. doi: 10.1186/s12870-019-2191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018a).Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, Hu S, Li XB. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. The New Phytologist. 2018a;217:625–640. doi: 10.1111/nph.14864. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2018b).Zhang Y, Li D, Wang Y, Zhou R, Wang L, Zhang Y, Yu J, Gong H, You J, Zhang X. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLOS ONE. 2018b;13:e0199262. doi: 10.1371/journal.pone.0199262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of full-length NAC proteins was done using muscle method and the phylogenetic tree was constructed using MEGAX by the neighbor-joining method with bootstrap 1000 replicates.
Collinear blocks represented by grey background. 288 BjuNAC duplication pairs are linked with green lines, and 15 tandem duplication pairs are linked with red lines.
(a) A phylogenetic tree using the neighbor-joining method by MEGAX, with bootstrap values of 1,000. (b) The conserved motifs were predicted using Multiple Em for Motif Elicitation (MEME) (http://alternate.meme-suite.org/). (c) The box length indicates the number of amino acids in the motif. The coding sequences (CDS) of exons are represented by purple boxes, the introns are represented by lines.
The height of each amino acid code in the sequence logo of each motif represents the degree of conservation.
All domain sequences of BjuNACs divided into five subdomains A-E.
The promoter sequences (1500 bp) upstream of genes were chosen for cis-regulatory element analysis using the PlantCARE online tool (http://www.dna.affrc.go.jp/PLACE/). Each color indicates a cis-regulatory element.
Graphical representation of locations for putative BniNAC genes on each chromosome. B1∼B8 represented the chromosome numbers. Other 25 BniNAC genes information belongs to scaffold data were shown in Table S3.
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.