Abstract
Parkinson's disease (PD) is a progressive neurodegenerative disorder that affects around 2% of individuals over 60 years old. It is characterised by the loss of dopaminergic neurons in the substantia nigra pars compacta of the midbrain, which is thought to account for the major clinical symptoms such as tremor, slowness of movement and muscle stiffness. Its aetiology is poorly understood as the physiological and molecular mechanisms leading to this neuronal loss are currently unclear. However, mitochondrial and lysosomal dysfunction seem to play a central role in this disease. In recent years, defective mitochondrial elimination through autophagy, termed mitophagy, has emerged as a potential contributing factor to disease pathology. PINK1 and Parkin, two proteins mutated in familial PD, were found to eliminate mitochondria under distinct mitochondrial depolarisation-induced stress. However, PINK1 and Parkin are not essential for all types of mitophagy and such pathways occur in most cell types and tissues in vivo, even in the absence of overt mitochondrial stress — so-called basal mitophagy. The most common mutation in PD, that of glycine at position 2019 to serine in the protein kinase LRRK2, results in increased activity and this was recently shown to disrupt basal mitophagy in vivo. Thus, different modalities of mitophagy are affected by distinct proteins implicated in PD, suggesting impaired mitophagy may be a common denominator for the disease. In this short review, we discuss the current knowledge about the link between PD pathogenic mutations and mitophagy, with a particular focus on LRRK2.
Keywords: autophagy, leucine-rich repeat kinase, mitophagy, Parkinsons disease
Introduction
A recent report from the United Nations projects that the number of people aged 65 or older, estimated at about 703 million in 2019, will reach almost 1.5 billion in 2050 [1]. As age is the biggest risk factor for neurodegeneration, this maturing of the world population will inevitably lead to a significant increase in the number of patients with neurodegenerative disorders. Parkinson's disease (PD) is a chronic progressive disease and is the second most common neurodegenerative disorder in the world [2]. It affects about 2% of the population over 60 years old, which corresponds today to over 7 million patients. Although symptoms can be relieved by dopamine replacement, there are a lack of treatments that actually alter the course of neurodegeneration [3]. Symptoms were first described in 1817 by James Parkinson in ‘An essay on shaking palsy’ [4], and are characterised by rigidity, bradykinesia, tremor, and postural instability. These symptoms are thought to be due to a loss of striatal dopaminergic (DA) neurons originating in the substantia nigra pars compacta (SNpc), and the accumulation of aggregates of α-synuclein, which are a major constituent of Lewy bodies [5,6]. However, the physiological and molecular mechanisms leading to these phenotypes are still largely unknown. PD is a multifactorial and complex disease that can be subclassified in familial and apparently sporadic PD. While over 90% of cases are sporadic, 22 genes have been identified from familial cases [7]. Many of these PD loci are either directly or indirectly associated with mitochondrial dysfunction [8,9].
Mitochondria are not only in charge of cellular energy production, but also display numerous functions including the production and regulation of reactive oxygen species (ROS), controlling cell death through apoptosis, the regulation of calcium homeostasis, the biosynthesis and metabolism of lipids, the biosynthesis of heme, the regulation of pH within the cell, and much more. These highly specialised and complex organelles form a dynamic network, and their mass is regulated through mitochondrial biogenesis [10] and turnover [11]. In addition to genetics, environmental factors such as exposure to toxins such as MPTP, paraquat, or rotenone (which are all mitochondrial Complex I inhibitors) can lead to the acquisition of Parkinsonian phenotypes [12–15]. Together, this highlights the central role of mitochondria in PD.
Another organelle that has strong links to PD is the lysosome. The lysosome is the end point of many intracellular trafficking pathways and plays a critical function in degrading and recycling a wide variety of cellular components, ranging from single polypeptides to whole organelles, including mitochondria [16]. A large body of work is now emerging that links lysosomal dysfunction to neurodegeneration and in particular PD [17]. Large scale GWAS studies have identified multiple lysosome-related proteins that pose as risk loci for PD [18,19]. Indeed, many lysosomal storage disorders display pronounced neurodegeneration and of relevance here, GBA1, the gene that encodes the lysosomal enzyme glucocerebrosidase and is mutated in Gaucher's disease, is a major genetic risk factor for PD [20,21]. Furthermore, rare variants in other lysosomal genes such as ATP13A2, TMEM175, and VPS13C have also been associated with PD [22]. ATP13A2 (PARK9) is a lysosomal P-type ATPase and its loss-of-function mutation causes Kufor–Rakeb syndrome, a juvenile early onset parkinsonism [23]. Transmembrane protein 175 (TMEM175) is a lysosomal potassium channel located in late endosomes and lysosomes, identified in a PD GWAS [24] and its loss-of-function appears to impair autophagy-mediated degradation of α-synuclein [25]. Vacuolar protein sorting-associated protein 13C (VPS13C), is a protein involved in ER-late endosome/lysosome contact sites [26] and its mutations are a monogenic cause of early onset parkinsonism [27].
If mitochondrial or lysosomal dysfunction can lead to PD, then a pathway that involves both these organelles is a prime candidate for further investigation. One such pathway is mitophagy, or the autophagy of mitochondria, which delivers damaged, dysfunctional or superfluous mitochondria to the lysosome for degradation [11]. Multiple mitophagy pathways have been identified that may operate under distinct scenarios. In most tissues, mitophagy occurs under basal conditions as part of normal cellular physiology, the levels of which varies between cell types independently of their mitochondrial content, suggesting the existence of distinct regulatory mechanisms [28–30]. Mitophagy can also occur as a programmed event, as is the case in the late stages of red blood cell maturation [31], cardiomyocyte maturation [32–34], retinal ganglion cell differentiation [35], and in the elimination sperm-derived mitochondria after fertilisation of the egg [36]. Finally, mitophagy can occur in response to a chemical, genetic, or physiologically induced stress and it is these stress pathways that have previously been linked to PD. Our current knowledge on the regulation of mitophagy has been extensively reviewed [5,7,11,37] and here we will focus on the links between pathogenic PD mutations and mitophagy.
Mitophagy defects in PD
Currently, no core autophagy-essential genes (such as the ATG genes) have been directly implicated in PD. However, several PD-linked genes have been identified that modulate mitophagy, which suggests this pathway may be more relevant to PD pathology compared with autophagy in general. As we will describe in the following paragraphs, these PD-related genes could potentially affect mitophagy at different steps and act independently of one another.
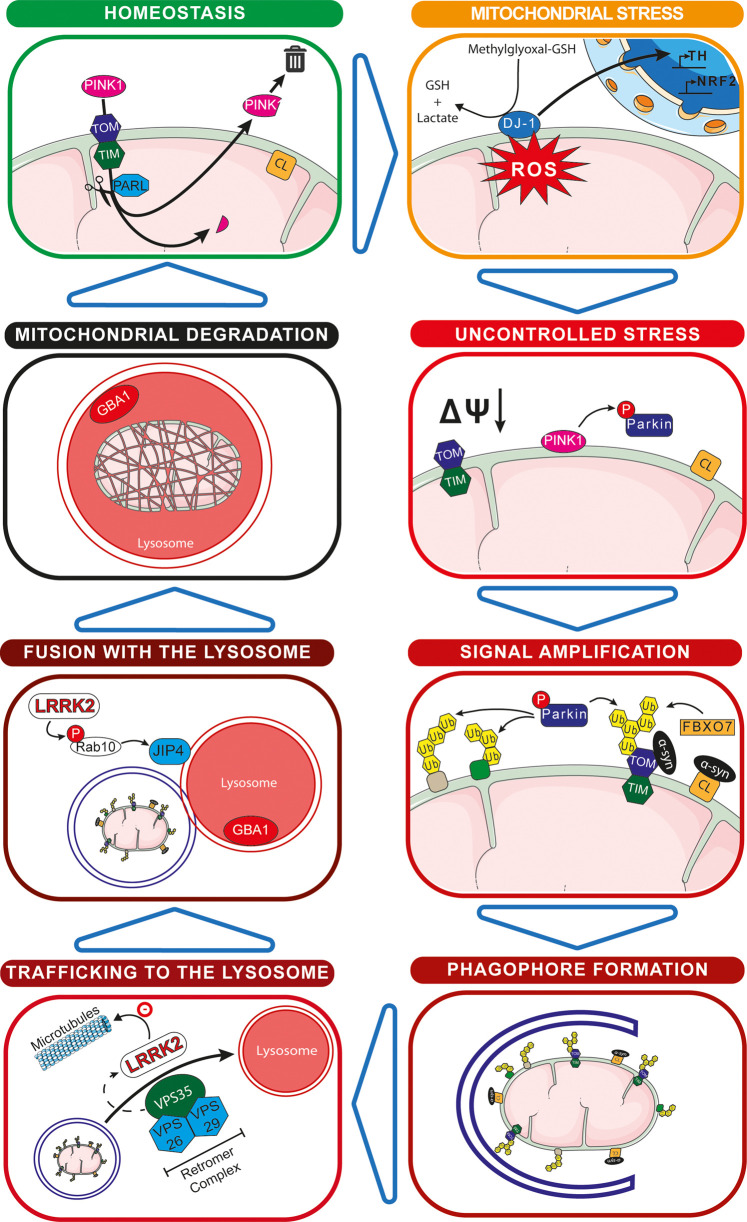
Most of our current knowledge about mitophagy regulation comes from the comprehensive study of the ubiquitin-dependant pathway of mitophagy that relies on the PD-related PINK1 and Parkin proteins. PTEN induced kinase 1 (PINK1, encoded by the PARK6 gene) and Parkin (encoded by the PARK2 gene) are at the heart of the ubiquitin-dependent pathway of mitophagy [3,5,7] and mutation of either results in autosomal recessive forms of PD [38]. PARK2 was the first gene discovered that directly linked mitophagy and PD in 2008 [39], and PARK6 was identified soon after, aided by the fact that these two genes are believed to operate in the same pathway [40–44]. This pathway appears to be activated upon extreme mitochondrial stress, with extensive mitochondrial depolarisation being a key trigger. Briefly, PINK1 is a serine/threonine kinase that in normal physiological conditions (Figure 1 — homeostasis), is imported from the cytosol to the mitochondrial matrix thanks to its mitochondria targeting sequence via the TOM/TIM complexes [45,46]. There it is cleaved and inactivated by the protease PARL (presenilin-associated rhomboid-like protein), and the cleaved are then degraded in the cytosol by the proteasome via a n-end rule pathway [3,7,37,47]. Upon mitochondrial damage and loss of mitochondrial membrane potential (Figure 1 — uncontrolled stress), PINK1 acts as a damage sensor and accumulates on the cytosolic side of the outer mitochondrial membrane (OMM) [44]. There, it phosphorylates both ubiquitin and Parkin at their respective Serine 65 residues [48,49]. Sequentially, ubiquitin is first phosphorylated, as the binding of Parkin to pre-existing phospho-ubiquitin has been shown to be required for its phosphorylation [50,51]. Activated Parkin then ubiquitylates multiple OMM targets, leading to an amplification of the ubiquitin signal [11,52] (Figure 1 — signal amplification). This signal leads to the recruitment of the autophagy machinery, via ubiquitin-binding autophagy receptors such as optineurin and NDP52 [53], and the engulfment of the mitochondrial fragment into a mitophagosome. Concomitant fusion with a lysosome results in mitochondrial degradation and recycling of its constituent components [54]. The PINK1/Parkin pathway has been extensively studied in vitro, often with harsh chemical uncouplers that are at odds with normal physiological conditions. Thus, it has not been clear as to when this pathway may be activated in vivo. Indeed, it was recently shown that this pathway does not seem to affect basal mitophagy in both mice and Drosophila [55–57]. Hence, the prevailing idea is that the PINK1/Parkin pathway constitutes a stress pathway in response to distinct mitochondrial toxins and pathological conditions, the identity of which are currently unclear.
Figure 1. Parkinson's disease-linked genes independently implicated in mitophagy.
Homeostasis: PINK1 is imported within the mitochondria where it is cleaved and is then exported for degradation. Cardiolipin is present at the internal mitochondrial membrane where it interacts with the Complex I–Complex III–Complex IV supercomplex. Mitochondrial stress: Oxidized DJ-1 is translocated to the nucleus where it acts as a transcription factor for genes involved in ROS detoxication. Uncontrolled stress: Following loss of mitochondrial membrane potential, PINK1 is stabilized at the surface of the outer mitochondrial membrane where it phosphorylates Parkin. Cardiolipin is translocated to the cytosolic side of the outer mitochondrial membrane. Signal amplification: Activated Parkin and FBXO7 ubiquitylate proteins on the outer mitochondrial membrane. Cardiolipin interacts with α-synuclein to recruit LC3. Phagophore Formation: The amplificated signal recruits the autophagosome machinery that initiates the formation of the phagophore. Trafficking to the lysosome: The newly formed phagosome is trafficked to the lysosome with the help of the retromer complex (VPS35, VPS26, and VPS29). VPS35 enhances the kinase activity of LRRK2. Fusion with the lysosome: LRRK2 phosphorylates Rab10, which in turn promote the translocation of JIP4 to the lysosome to form sensing tubules, enabling the sorting of vesicles. Mitochondrial degradation: The defective mitochondria is degraded in the mitolysosome that contains GBA1.
It should be noted that aside from mitophagy, PINK1 and Parkin have also been involved in additional modes of mitochondrial turnover that may also be relevant for PD. For example, PINK1/Parkin-mediated proteasomal turnover of OMM proteins in Drosophila not only stimulates mitophagy, but also promotes the selective turnover of a subset of mitochondrial electron transport chain subunits [58]. Additionally, in mammalian cells, the PINK1/Parkin pathway has been involved in the generation of mitochondria-derived vesicles (MDVs) in response to oxidative stress, triggered by uncouplers such as CCCP or oligomycin and antimycin A, which are selectively delivered to late endosomes/lysosomes independently of the mitophagy pathway [59,60]. In a more physiological setting, heat stress in PINK1 knockout primary bone-marrow-derived macrophages increases the mitochondrial antigen presentation (MitAP) of 2-oxoglutarate dehydrogenase through MDVs [61]. Of importance to PD, it was recently shown that intestinal bacterial infection in PINK1 knockout mice induced MitAP and provoked a CD8+ T cell infiltration into the brain that lead to reduced DA neuron axon varicosities and motor impairments [62]. These data highlight a potential cross-talk between mitochondrial quality control and autoimmune mechanisms in the aetiology of PD, an area where LRRK2 has also been implicated [63].
Despite PINK1 and Parkin dominating the mitophagy field, other genes involved in PD have recently been described that influence mitochondria and mitophagy (see Figure 1). Mutations in DJ-1, encoded by the PARK7 gene, cause recessive forms of PD [64] and it acts as a redox sensor that localises to mitochondria [7]. Oxidation of DJ-1 on Cysteine 106 leads to its translocation into the nucleus where it functions as a transcriptional co-activator and corepressor [65,66] (Figure 1 — mitochondrial stress). Notably, it positively regulates the expression of tyrosine hydroxylase, the enzyme responsible for the conversion of tyrosine to L-DOPA, and it also regulates the expression of NRF2, a master regulator of the anti-oxidative stress response [65]. In vitro, DJ-1 showed a methylglyoxal-adduct hydrolase activity, which could protect low-molecular thiols such as Coenzyme A, highlighting its role in redox homeostasis [67]. Although the localisation of this protein is controversial, pathogenic DJ-1 mutants have been reported to translocate from the cytosol to the mitochondrial matrix in vitro [68]. Also, DJ-1 deficiency is associated with the age-dependant relocalisation of OMM hexokinase 1 to the cytosol, which in turn inhibits the PINK1/Parkin pathway [69]. Importantly, loss of DJ-1 leads to oxidative stress within the cell, mitochondrial fragmentation, and potentially impaired mitophagy [64,70].
As mentioned above, one of the histopathological hallmarks of PD is the presence of Lewy bodies [6], a major constituent of which is aggregates of α-synuclein. α-Synuclein, encoded by the PARK1 gene, is a small soluble protein mostly expressed in the brain. Though the function of this protein is not clear, it may play a role in neurotransmitter release [71]. Pathogenic α-synuclein has been shown to bind preferentially to mitochondria [72], in particular to Translocase of the outer mitochondrial membrane 20 (TOMM20), which inhibits mitochondrial protein import leading to mitochondrial dysfunction [73]. Evidence also suggests α-synuclein may indirectly affect mitophagy by interacting with Cardiolipin [74], or up-regulating the expression of Miro, an OMM GTPase regulating mitochondrial movement [75]. Of relevance, it was recently shown that when cardiolipin translocates from the inner mitochondrial membrane to the OMM, it interacts with α-synuclein and can recruit LC3 to the mitochondria to trigger mitophagy [76] (Figure 1 — signal amplification).
Mutations in the FBXO7 gene (PARK15) encoding for F-box only protein 7 (FBXO7) causes autosomal recessive early onset PD. FBXO7 has been shown to ubiquitinate TOMM20 [77] (Figure 1 — signal amplification), and to act in a common mitophagy pathway with PINK1 and Parkin, with overexpression of human FBXO7 in Drosophila rescuing a Parkin loss-of-function phenotype [78].
Mutation in the VPS35 gene (PARK17), encoding for the Vacuolar protein sorting-associated protein 35 (VPS35), causes autosomal dominant late-onset PD [7]. VPS35 is a key component of the retromer cargo recognition complex and thus regulates endocytic membrane trafficking, which can influence autophagy in general [79] (Figure 1 — trafficking to the lysosome). Relevantly, it has been reported to be a Parkin substrate and in turn, its ubiquitylation regulates retromer-mediated endocytic sorting, including that of the autophagy-essential protein ATG9A [80]. Furthermore, the PD-related D620N mutation of VPS35 was recently shown, in patient-derived neurons, to cause α-synuclein accumulation and mitochondrial defects, which was attributed to impaired autophagy [81]. Of note, The D620N mutation has been shown in both mouse and human to be an upstream regulator of LRRK2 kinase activity [82].
As previously mentioned, homozygous mutations in the GBA1 gene cause the lysosomal storage disorder Gaucher disease [83]. However, heterozygous GBA1 mutations are also risk factors for developing PD [20], increasing the risk of disease development by about 21-fold [84]. While it is estimated that between 7% and 12% of patients with PD carry a mutation in the GBA1 gene, only a minority of GBA1 mutation carriers actually develop PD symptoms [20]. Given the lysosomal-related function of GBA1, mutations provoke general autophagy defects that would also be predicted to disrupt mitophagy [85]. Additionally, mice with the GBA1 knock-in mutation L444P have been reported to have impaired mitochondrial function and mitophagy [86] (Figure 1 — mitochondrial degradation).
LRRK2 and basal mitophagy
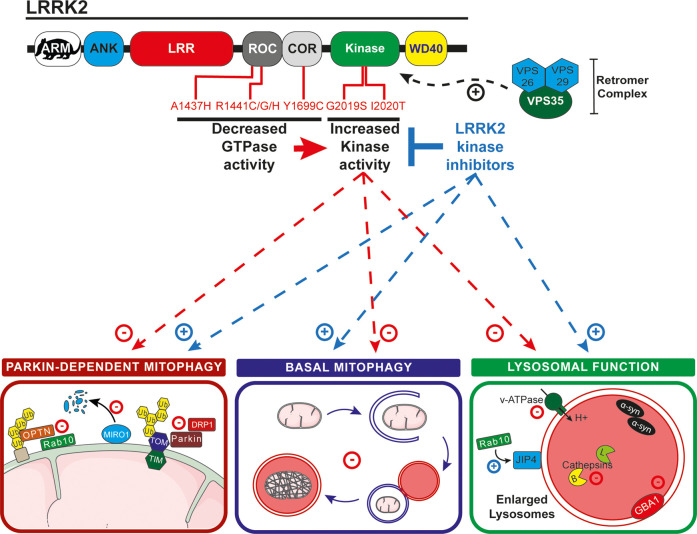
Coding variants in the leucine-rich repeat kinase 2 (LRRK2, PARK8) gene cause an autosomal dominant form of PD and are responsible for the majority of familial cases [87]. LRRK2 is a large (286 kDa) multidomain protein consisting of an armadillo (ARM) domain, an ankyrin domain (ANK), a leucine-rich repeat domain (LRR), a Ras of complex GTPase domain (ROC), a C-terminal of ROC domain (COR), a kinase domain, and a WD40 domain (Figure 2). Coding variants in LRRK2 leading to PD segregate to its catalytic core, such as in the GTPase domain (R1441C/G/H or Y1699C) and in its kinase domain (G2019S and I2020T). The G2019S mutation being the most frequent genetic cause of PD, representing 4–5% of familial cases and about 1% of sporadic cases [88]. All the known pathogenic mutations of LRRK2 lead to an increased kinase activity [89]. Given the track record of small molecule kinase inhibitors as effective drugs for other diseases, LRRK2 kinase inhibitors offer considerable and obvious potential. Indeed, small molecule LRRK2 kinase inhibitors, as well as an antisense LRRK2 oligo, are currently in clinical trials for PD indications [90]. In addition, other types of inhibitors have been developed, either targeting the G-protein cycle by inhibiting its GTPase activity or by inhibiting LRRK2's protein–protein interactions (such as with 14-3-3) [91]. The proliferation in LRRK2 inhibitor development highlights that pharmacologically targeting LRRK2 could be a highly potent solution to diminish pathogenic LRRK2 effects.
Figure 2. Roles of LRRK2 in mitophagy.
Schematic of LRRK2 domain structure is shown at the top and pathogenic mutations in the catalytic domains are noted. VPS35, a retromer subunit, enhances LRRK2 kinase activity. Parkin-dependent mitophagy: Increased LRRK2 activity negatively impacts PINK1/Parkin-dependent mitophagy by reducing PARKIN interaction with the TOMM complex and DRP1, as well as reducing Rab10 recruitment to mitochondria and the interaction between optineurin (OPTN) with ubiquitylated OMM proteins. Basal mitophagy: LRRK2 kinase activity also impairs basal mitophagy and lysosomal function (which impacts the end point of mitophagy). Lysosomal function: At the lysosome LRRK2 regulates sorting via Rab10 phosphorylation and JIP4 and it negatively regulates the function of GBA1. It also impacts the vacuolar ATPase, leading to a higher lysosomal pH, which in turn impacts cathepsin-mediated degradation of α-synuclein. LRRK2 kinase inhibitors attenuate many of these effects, for example, inhibitors such as GSK3357679A, rescues the mitophagy defects in mice carrying the pathogenic G2019S LRRK2 mutation.
The fact that LRRK2 is a large multidomain protein suggests that it has numerous functions within the cell. However, its physiological roles are still poorly understood, as are the aspects of its functions that are relevant for PD. In-line with the hypothesis that mitochondrial and lysosomal dysfunction underlie PD, an emerging body of data supports a key role for LRRK2 at these organelles. Work has shown that patient-derived fibroblasts from G2019S carriers display abnormal mitochondrial morphology and function [92]. Likewise, this same LRRK2 mutation has been shown to increase mitochondrial DNA damage in human-derived cells [88,93]. The abnormal mitochondrial phenotype is also present in mice with aged G2019S knock-in animals displaying hallmarks of impaired fission and altered dynamics [94,95]. Mutations of LRRK2 have also been shown to alter lysosomal function (Figure 2 — lysosomal function). In various in vitro and in vivo models of the G2019S mutation, enlarged lysosomes were reported, suggesting a strong link between LRRK2 and lysosomal function [96]. A recent in vitro study with neuronal primary cultures identified LRRK2 as an interactor with the a1 subunit of the Vacuolar ATPase [97]. In that article, the authors show that the R1441C mutation, but not the G2019S, leads to an increased lysosomal pH and consequently a decreased lysosomal function, though this effect appears independent of the kinase activity of LRRK2. In contrast, it was also reported that the G2019S mutation modestly alters lysosomal morphology and acidification, and that this effect is dependent on the kinase activity of LRRK2 [98]. Here, the defect in lysosomal function disrupted Cathepsin B and L mediated turnover and led to an accumulation of insoluble α-synuclein. Furthermore, LRRK2 kinase activity has also been reported to reduce GBA1 activity in neurons derived from patients, through its substrate Rab10, and this effect was dependent on LRRK2 kinase activity [99]. Additionally, LRRK2 kinase activity plays a key role in phagosome maturation and lysosomal trafficking [100] and recent work has shown LRRK2 interacts with the motor protein adaptor JIP4 at lysosomes to regulate lysosomal tubulation in a process termed LYTL (lysosomal tubulation/sorting driven by LRRK2 [101]) (Figure 1 — fusion with the lysosome, and Figure 2). In relation to intracellular trafficking and the cytoskeleton, LRRK2 PD mutants have previously been shown in vitro to display a higher microtubule association compared with WT LRRK2 [102]. This was recently confirmed with the high-resolution structure of the catalytic portion of LRRK2, showing microtubule binding through its WD40 domain [103]. Lastly, microtubule-based signalling and ciliogenesis was disrupted in LRRK2 R1441C knock-in mice, which displayed decreased ciliation in cholinergic neurons in the striatum [104]. Taken together, these data suggest an important link between LRRK2 and microtubules, which in turn could have consequences for mitochondria and lysosomes.
Given the published evidence of pathogenic LRRK2 playing a role in both mitochondrial and lysosomal dysfunction, it follows that recent work also implicates impaired mitophagy. Research has linked the LRRK2 pathway with that of PINK1/Parkin-dependent mitophagy [105–108] (Figure 2 — Parkin-dependent mitophagy). For example, LRRK2 was shown to form a complex with Miro, which is required for its efficient removal during PINK1/Parkin-dependent mitophagy [108]. Impaired mitochondrial dynamics were also observed in cell lines co-expressing Parkin and LRRK2. Expression of LRRK2 G2019S disrupted Parkin-dependent mitophagy, potentially via reducing Parkin's interaction with OMM proteins, including the fission regulating GTPase DRP-1 [106]. Work has also shown in PD-derived patient fibroblasts that Rab10, a downstream substrate of LRRK2, could recruit the mitophagy receptor optineurin [105]. Impaired mitophagy was also observed in fibroblasts derived from patients with the G2019S mutation [107]. These data are summarised in Figure 2.
The above evidence suggests that impaired stress-induced Parkin-dependent mitophagy could be a contributing factor to PD pathology. However, we have previously shown that mitophagy occurs in vivo in the absence of overt stress, under so-called basal conditions, in a manner that is independent of PINK1 and Parkin [55,56]. It is reasonable to assume that both impaired stress-induced mitophagy and impaired basal mitophagy can lead to the accumulation of dysfunctional mitochondria and reduced cellular health. If so, then compromised basal mitophagy could also be an important contributing factor for PD development. This may well be the case, as we recently showed that LRRK2 kinase activity inversely correlates with basal levels of mitophagy in specific organs and cell types in vivo (Figure 2 — basal mitophagy). Using mitophagy (mito-QC) and autophagy (auto-QC) reporter mice we were able to show that the common pathogenic G2019S LRRK2 mutation, which results in increased kinase activity, reduced basal mitophagy in cells and tissues of clinical relevance, including midbrain DA neurons and microglial cells [109]. In contrast, the absence of LRRK2 resulted in increased basal mitophagy in these same cell types. Interestingly, this phenomenon was not observed in all cell types, implying cell, and tissue-specific consequences for LRRK2 activity. Importantly though, using a next-generation LRRK2 inhibitor, GSK3357679A, we were able to correct the mitophagy defect in LRRK2 G2019S-expressing mice and restore mitophagy to wild type levels in both DA neurons and microglia. The effect of LRRK2 on mitophagy potentially appears to be selective, as general macroautophagy seems to be largely unaffected both in vitro [105] and in vivo [109]. This suggests a significant disruption of lysosomal function is not occurring.
A major question remaining concerns how LRRK2 kinase activity regulates basal mitophagy (and indeed how it regulates other cellular processes). It is not yet clear whether LRRK2 activity plays a direct mitophagic role via regulating autophagy machinery, or more indirect through disruption of mitochondrial movement/function or membrane trafficking in general. However, a subset of Rab GTPases were recently identified to be key substrates of the LRRK2 kinase [110]. Rab GTPases are master regulators of vesicular trafficking and thus ideally placed to regulate and influence auto-lysosomal pathways [87]. Their interaction with LRRK2 has been extensively reviewed recently [89,111,112]. Notably, LRRK2 has been shown to phosphorylate Rab12 [110,113] and SQSTM1/p62 in vitro [114]. Rab12 is a positive regulator for autophagy that regulates mTORC1 activity, a sensor for nutrient availability and key inhibitor of starvation-induced autophagy [115,116]. Additionally, Rab12 has also been shown to directly interact with autophagosomes and influence their trafficking [117,118]. p62 was the first autophagy cargo receptor described in mammals and is thought to be important in regulating the incorporation of cellular components into forming autophagosomes [119,120]. Thus, LRRK2 substrates are known to regulate autophagy but further work is needed to determine if, and how, their regulation by LRRK2 impacts this process.
Conclusions
PD is a complex neurodegenerative disorder and, as with many such diseases, its pathophysiology and molecular mechanisms are still poorly defined. However, these molecular details are slowly being revealed and there is a gathering optimism within the field. Though several pathogenic gene mutations have been associated with defects in mitochondrial elimination, whether mitophagy itself plays an active role in PD pathology remains to be proven. The emergence of the discovery of new links between PD-associated mutations and mitophagy defects imply that impaired mitochondrial elimination might indeed be a key event in PD aetiology. Though a better appreciation of these pathways is still required, enhancing mitophagy pharmacologically could hold great potential for PD treatment.
Perspectives
Ageing of the population will lead to the emergence of neurodegenerative disorders such as PD.
Altered mitophagy appears to be a common impairment between many pathogenic PD mutations, suggesting a potential key role.
A better understanding of mitophagy pathways might be key in the future of understanding the aetiology of Parkinson's disease and its potential treatment.
Acknowledgements
Figures were realised using Servier Medical Art templates (CC BY 3.0). We would like to thank Dr Alastair Reith (GSK) for the critical reading of this manuscript.
Abbreviations
- DA
dopaminergic
- FBXO7
F-box only protein 7
- LRR
leucine-rich repeat domain
- MDVs
mitochondria-derived vesicles
- MitAP
mitochondrial antigen presentation
- OMM
outer mitochondrial membrane
- PD
Parkinson's disease
- PINK1
PTEN induced kinase 1
- TOMM20
translocase of the outer mitochondrial membrane 20
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was funded by a grant from the Medical Research Council, U.K. (IGG; MC_UU_00018/2) and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (Boehringer Ingelheim, GSK and Merck Serono).
Authors Contribution
F.S. and I.G.G. wrote and edited the manuscript.
References
- 1.United Nations - Department of Economic and Social Affairs. World Population Ageing 2019 [Internet]. 2019 [cited 2020 Nov 9]. Available from: https://www.un.org/development/desa/pd/sites/www.un.org.development.desa.pd/files/files/documents/2020/Jan/un_2019_worldpopulationageing_report.pdf
- 2.Coppedè, F. (2012) Genetics and epigenetics of Parkinson's disease. Sci. World J. 2012, 1–12 10.1100/2012/489830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasner, K., Grünewal, A. and Klein, C. (2020) Parkin-linked Parkinson‘s disease: from clinical insights to pathogenic mechanisms and novel therapeutic approaches. Neurosci. Res. 159, 34–39 10.1016/j.neures.2020.09.001 [DOI] [PubMed] [Google Scholar]
- 4.Parkinson, J. (2002) An essay on the shaking palsy. 1817. J. Neuropsychiatry Clin. Neurosci. 14, 223–236; discussion 222 10.1176/jnp.14.2.223 [DOI] [PubMed] [Google Scholar]
- 5.Gao, F., Yang, J., Wang, D., Li, C., Fu, Y., Wang, H.et al. (2017) Mitophagy in Parkinson's disease: pathogenic and therapeutic implications. Front. Neurol. 8, 527 10.3389/fneur.2017.00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holdorff, B., Rodrigues e Silva, A.M. and Dodel, R. (2013) Centenary of Lewy bodies (1912–2012). J. Neural. Transm. 120, 509–516 10.1007/s00702-013-0984-2 [DOI] [PubMed] [Google Scholar]
- 7.Liu, J., Liu, W., Li, R. and Yang, H. (2019) Mitophagy in Parkinson's disease: from pathogenesis to treatment. Cells 8, 712 10.3390/cells8070712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park, J.S., Davis, R.L. and Sue, C.M. (2018) Mitochondrial dysfunction in Parkinson's disease: new mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. Rep. 18, 21 10.1007/s11910-018-0829-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser, D.N. and Hastings, T.G. (2013) Mitochondrial dysfunction and oxidative stress in Parkinson's disease and monogenic parkinsonism. Neurobiol. Dis. 51, 35–42 10.1016/j.nbd.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljubicic, V., Joseph, A.-M., Saleem, A., Uguccioni, G., Collu-Marchese, M., Lai, R.Y.J.et al. (2010) Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging. Biochim. Biophys. Acta 1800, 223–234 10.1016/j.bbagen.2009.07.031 [DOI] [PubMed] [Google Scholar]
- 11.Montava-Garriga, L. and Ganley, I.G. (2020) Outstanding questions in mitophagy: what we do and do not know. J. Mol. Biol. 432, 206–230 10.1016/j.jmb.2019.06.032 [DOI] [PubMed] [Google Scholar]
- 12.Hancock, D.B., Martin, E.R., Mayhew, G.M., Stajich, J.M., Jewett, R., Stacy, M.A.et al. (2008) Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol. 8, 6 10.1186/1471-2377-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose, A. and Beal, M.F. (2016) Mitochondrial dysfunction in Parkinson's disease. J. Neurochem. 139, 216–231 10.1111/jnc.13731 [DOI] [PubMed] [Google Scholar]
- 14.Langston, J.W., Ballard, P., Tetrud, J.W. and Irwin, I. (1983) Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219, 979–980 10.1126/science.6823561 [DOI] [PubMed] [Google Scholar]
- 15.Langston, J.W. (2017) The MPTP story. J. Parkinsons Dis. 7, S11–S19 10.3233/JPD-179006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inpanathan, S. and Botelho, R.J. (2019) The lysosome signaling platform: adapting with the times. Front. Cell Dev. Biol. 7, 113 10.3389/fcell.2019.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherubini, M. and Wade-Martins, R. (2018) Convergent pathways in Parkinson's disease. Cell Tissue Res. 37, 79–90 10.1007/s00441-017-2700-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, D., Nalls, M.A., Hallgrímsdóttir, I.B., Hunkapiller, J., van der Brug, M., Cai, F.et al. (2017) A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 49, 1511–1516 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y.I., Wong, G., Humphrey, J. and Raj, T. (2019) Prioritizing Parkinson's disease genes using population-scale transcriptomic data. Nat. Commun. 10, 994 10.1038/s41467-019-08912-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do, J., McKinney, C., Sharma, P. and Sidransky, E. (2019) Glucocerebrosidase and its relevance to Parkinson disease. Mol. Neurodegener. 14, 36 10.1186/s13024-019-0336-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Migdalska-Richards, A. and Schapira AH, V. (2016) The relationship between glucocerebrosidase mutations and Parkinson disease. J. Neurochem. 139, 77–90 10.1111/jnc.13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopfner, F., Mueller, S.H., Szymczak, S., Junge, O., Tittmann, L., May, S.et al. (2020) Rare variants in specific lysosomal genes are associated with Parkinson's disease. Mov. Disord. 35, 1245–1248 10.1002/mds.28037 [DOI] [PubMed] [Google Scholar]
- 23.Usenovic, M., Tresse, E., Mazzulli, J.R., Taylor, J.P. and Krainc, D. (2012) Deficiency of ATP13A2 leads to lysosomal dysfunction, -synuclein accumulation, and neurotoxicity. J. Neurosci. 32, 4240–4246 10.1523/JNEUROSCI.5575-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nalls, M.A., Pankratz, N., Lill, C.M., Do, C.B., Hernandez, D.G., Saad, M.et al. (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 46, 989–993 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinn, S., Drolet, R.E., Cramer, P.E., Wong, A.H.-K., Toolan, D.M., Gretzula, C.A.et al. (2017) TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc. Natl Acad. Sci. U.S.A. 114, 2389–2394 10.1073/pnas.1616332114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F.A., Li, P., Lees, J.A.et al. (2018) VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesage, S., Drouet, V., Majounie, E., Deramecourt, V., Jacoupy, M., Nicolas, A.et al. (2016) Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 98, 500–513 10.1016/j.ajhg.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montava-Garriga, L., Singh, F., Ball, G. and Ganley, I.G. (2020) Semi-automated quantitation of mitophagy in cells and tissues. Mech. Ageing Dev. 185, 111196 10.1016/j.mad.2019.111196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWilliams, T.G., Prescott, A.R., Allen, G.F.G., Tamjar, J., Munson, M.J., Thomson, C.et al. (2016) . mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McWilliams, T.G., Prescott, A.R., Villarejo-Zori, B., Ball, G., Boya, P. and Ganley, I.G. (2019) A comparative map of macroautophagy and mitophagy in the vertebrate eye. Autophagy 15, 1296–1308 10.1080/15548627.2019.1580509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barde, I., Rauwel, B., Marin-Florez, R.M., Corsinotti, A., Laurenti, E., Verp, S.et al. (2013) A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science 340, 350–353 10.1126/science.1232398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, J.-F., Rodger, C.E., Allen, G.F.G., Weidlich, S. and Ganley, I.G. (2020) HIF1α-dependent mitophagy facilitates cardiomyoblast differentiation. Cell Stress 4, 99–113 10.15698/cst2020.05.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong, G., Song, M., Csordas, G., Kelly, D.P., Matkovich, S.J., Dorn, G.W.et al. (2015) Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 350, aad2459 10.1126/science.aad2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lampert, M.A., Orogo, A.M., Najor, R.H., Hammerling, B.C., Leon, L.J., Wang, B.J.et al. (2019) BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 15, 1182–1198 10.1080/15548627.2019.1580095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteban-Martínez, L., Sierra-Filardi, E., McGreal, R.S., Salazar-Roa, M., Mariño, G., Seco, E.et al. (2017) Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 36, 1688–1706 10.15252/embj.201695916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio-Peña, K., Al Rawi, S., Husson, F., Lam, F., Merlet, J. and Galy, V. (2021) Mitophagy of polarized sperm-derived mitochondria after fertilization. iScience 24, 102029 10.1016/j.isci.2020.102029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan, B.J., Hoek, S., Fon, E.A. and Wade-Martins, R. (2015) Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem. Sci. 40, 200–210 10.1016/j.tibs.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 38.Miller, S. and Muqit, M.M.K. (2019) Therapeutic approaches to enhance PINK1/Parkin mediated mitophagy for the treatment of Parkinson's disease. Neurosci. Lett. 705, 7–13 10.1016/j.neulet.2019.04.029 [DOI] [PubMed] [Google Scholar]
- 39.Narendra, D., Tanaka, A., Suen, D.F. and Youle, R.J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda, N., Sato, S., Shiba, K., Okatsu, K., Saisho, K., Gautier, C.A.et al. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisler, S., Holmström, K.M., Skujat, D., Fiesel, F.C., Rothfuss, O.C., Kahle, P.J.et al. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 42.Geisler, S., Holmström, K.M., Treis, A., Skujat, D., Weber, S.S., Fiesel, F.C.et al. (2010) The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 6, 871–878 10.4161/auto.6.7.13286 [DOI] [PubMed] [Google Scholar]
- 43.Vives-Bauza, C., Zhou, C., Huang, Y., Cui, M., de Vries, R.L.A., Kim, J.et al. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. U.S.A. 107, 378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narendra, D.P., Jin, S.M., Tanaka, A., Suen, D.-F., Gautier, C.A., Shen, J.et al. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene, A.W., Grenier, K., Aguileta, M.A., Muise, S., Farazifard, R., Haque, M.E.et al. (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 10.1038/embor.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deas, E., Plun-Favreau, H., Gandhi, S., Desmond, H., Kjaer, S., Loh, S.H.Y.et al. (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20, 867–879 10.1093/hmg/ddq526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamano, K. and Youle, R.J. (2013) PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 10.4161/auto.24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M.et al. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- 49.Kazlauskaite, A., Kondapalli, C., Gourlay, R., Campbell, D.G., Ritorto, M.S., Hofmann, K.et al. (2014) Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang, M.Y., Vranas, M., Krahn, A.I., Pundlik, S., Trempe, J.-F. and Fon, E.A. (2017) Structure-guided mutagenesis reveals a hierarchical mechanism of Parkin activation. Nat. Commun. 8, 14697 10.1038/ncomms14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ordureau, A., Sarraf, S.A., Duda, D.M., Heo, J.-M., Jedrychowski, M.P., Sviderskiy, V.O.et al. (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 10.1016/j.molcel.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiba-Fukushima, K., Imai, Y., Yoshida, S., Ishihama, Y., Kanao, T., Sato, S.et al. (2012) PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2, 1002 10.1038/srep01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazarou, M., Sliter, D.A., Kane, L.A., Sarraf, S.A., Wang, C., Burman, J.L.et al. (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodger, C.E., McWilliams, T.G. and Ganley, I.G. (2018) Mammalian mitophagy – from in vitro molecules to in vivo models. FEBS J. 285, 1185–1202 10.1111/febs.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McWilliams, T.G., Prescott, A.R., Montava-Garriga, L., Ball, G., Singh, F., Barini, E.et al. (2018) Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449.e5 10.1016/j.cmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McWilliams, T.G., Barini, E., Pohjolan-Pirhonen, R., Brooks, S.P., Singh, F., Burel, S.et al. (2018) Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Open Biol. 8, 180108 10.1098/rsob.180108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee, J.J., Sanchez-Martinez, A., Zarate, A.M., Benincá, C., Mayor, U., Clague, M.J.et al. (2018) Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217, 1613–1622 10.1083/jcb.201801044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincow, E.S., Merrihew, G., Thomas, R.E., Shulman, N.J., Beyer, R.P., MacCoss, M.J.et al. ) The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl Acad. Sci. U.S.A. 110, 6400–6405 10.1073/pnas.1221132110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLelland, G.-L., Soubannier, V., Chen, C.X., McBride, H.M. and Fon, E.A. (2014) Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33, 282–295 10.1002/embj.201385902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLelland, G.-L., Lee, S.A., McBride, H.M. and Fon, E.A. (2016) Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214, 275–291 10.1083/jcb.201603105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matheoud, D., Sugiura, A., Bellemare-Pelletier, A., Laplante, A., Rondeau, C., Chemali, M.et al. (2016) Parkinson's disease-related proteins PINK1 and parkin repress mitochondrial antigen presentation. Cell 166, 314–327 10.1016/j.cell.2016.05.039 [DOI] [PubMed] [Google Scholar]
- 62.Matheoud, D., Cannon, T., Voisin, A., Penttinen, A.-M., Ramet, L., Fahmy, A.M.et al. (2019) Intestinal infection triggers Parkinson's disease-like symptoms in Pink1−/− mice. Nature 571, 565–569 10.1038/s41586-019-1405-y [DOI] [PubMed] [Google Scholar]
- 63.Dzamko, N. and Halliday, G.M. (2012) An emerging role for LRRK2 in the immune system. Biochem. Soc. Trans. 40, 1134–1139 10.1042/BST20120119 [DOI] [PubMed] [Google Scholar]
- 64.Thomas, K.J., McCoy, M.K., Blackinton, J., Beilina, A., van der Brug, M., Sandebring, A.et al. (2011) DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 20, 40–50 10.1093/hmg/ddq430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ariga, H., Takahashi-Niki, K., Kato, I., Maita, H., Niki, T. and Iguchi-Ariga, S.M.M. (2013) Neuroprotective function of DJ-1 in Parkinson's disease. Oxid. Med. Cell Longev. 2013, 1–9 10.1155/2013/683920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi-Niki, K., Niki, T., Iguchi-Ariga, S.M.M. and Ariga, H. (2017) Transcriptional regulation of DJ-1. Adv. Exp. Med. Biol. 1037, 89–95 10.1007/978-981-10-6583-5_7 [DOI] [PubMed] [Google Scholar]
- 67.Matsuda, N., Kimura, M., Queliconi, B.B., Kojima, W., Mishima, M., Takagi, K.et al. (2017) Parkinson's disease-related DJ-1 functions in thiol quality control against aldehyde attack in vitro. Sci. Rep. 7, 12816 10.1038/s41598-017-13146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kojima, W., Kujuro, Y., Okatsu, K., Bruno, Q., Koyano, F., Kimura, M.et al. (2016) Unexpected mitochondrial matrix localization of Parkinson's disease-related DJ-1 mutants but not wild-type DJ-1. Genes Cells 21, 772–788 10.1111/gtc.12382 [DOI] [PubMed] [Google Scholar]
- 69.Hauser, D.N., Mamais, A., Conti, M.M., Primiani, C.T., Kumaran, R., Dillman, A.A.et al. (2017) Hexokinases link DJ-1 to the PINK1/parkin pathway. Mol. Neurodegener. 12, 70 10.1186/s13024-017-0212-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krebiehl, G., Ruckerbauer, S., Burbulla, L.F., Kieper, N., Maurer, B., Waak, J.et al. (2010) Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson's disease-associated protein DJ-1. PLoS One 5, e9367 10.1371/journal.pone.0009367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devoto VM, P. and Falzone, T.L. (2017) Mitochondrial dynamics in Parkinson's disease: a role for α-synuclein? Dis. Model Mech. 10, 1075–1087 10.1242/dmm.026294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, X., Becker, K., Levine, N., Zhang, M., Lieberman, A.P., Moore, D.J.et al. (2019) Pathogenic alpha-synuclein aggregates preferentially bind to mitochondria and affect cellular respiration. Acta Neuropathol. Commun. 7, 41 10.1186/s40478-019-0696-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Maio, R., Barrett, P.J., Hoffman, E.K., Barrett, C.W., Zharikov, A., Borah, A.et al. (2016) α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Sci. Transl. Med. 8, 342ra78 10.1126/scitranslmed.aaf3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rocha, E.M., De Miranda, B. and Sanders, L.H. (2018) Alpha-synuclein: pathology, mitochondrial dysfunction and neuroinflammation in Parkinson's disease. Neurobiol. Dis. 109, 249–257 10.1016/j.nbd.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 75.Shaltouki, A., Hsieh, C.-H., Kim, M.J. and Wang, X. (2018) Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson's models. Acta Neuropathol. 136, 607–620 10.1007/s00401-018-1873-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryan, T., Bamm V, V., Stykel, M.G., Coackley, C.L., Humphries, K.M., Jamieson-Williams, R.et al. (2018) Cardiolipin exposure on the outer mitochondrial membrane modulates α-synuclein. Nat. Commun. 9, 817 10.1038/s41467-018-03241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teixeira, F.R., Randle, S.J., Patel, S.P., Mevissen, T.E.T., Zenkeviciute, G., Koide, T.et al. (2016) Gsk3β and Tomm20 are substrates of the SCFFbxo7/PARK15 ubiquitin ligase associated with Parkinson's disease. Biochem. J. 473, 3563–3580 10.1042/BCJ20160387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burchell, V.S., Nelson, D.E., Sanchez-Martinez, A., Delgado-Camprubi, M., Ivatt, R.M., Pogson, J.H.et al. (2013) The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 16, 1257–1265 10.1038/nn.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cutillo, G., Simon, D.K. and Eleuteri, S. (2020) VPS35 and the mitochondria: connecting the dots in Parkinson's disease pathophysiology. Neurobiol. Dis. 145, 105056 10.1016/j.nbd.2020.105056 [DOI] [PubMed] [Google Scholar]
- 80.Williams, E.T., Glauser, L., Tsika, E., Jiang, H., Islam, S. and Moore, D.J. (2018) Parkin mediates the ubiquitination of VPS35 and modulates retromer-dependent endosomal sorting. Hum. Mol. Genet. 27, 3189–3205 10.1093/hmg/ddy224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanss, Z., Larsen, S.B., Antony, P., Mencke, P., Massart, F., Jarazo, J.et al. (2020) Mitochondrial and clearance impairment in p.D620N VPS35 patient-derived neurons. Mov. Disord. online ahead of print 10.1002/mds.28365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mir, R., Tonelli, F., Lis, P., Macartney, T., Polinski, N.K., Martinez, T.N.et al. (2018) The Parkinson's disease VPS35[D620N] mutation enhances LRRK2-mediated Rab protein phosphorylation in mouse and human. Biochem. J. 475, 1861–1883 10.1042/BCJ20180248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gegg, M.E. and Schapira AH, V. (2016) Mitochondrial dysfunction associated with glucocerebrosidase deficiency. Neurobiol. Dis. 90, 43–50 10.1016/j.nbd.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bultron, G., Kacena, K., Pearson, D., Boxer, M., Yang, R., Sathe, S.et al. (2010) The risk of Parkinson's disease in type 1 Gaucher disease. J. Inherit. Metab. Dis. 33, 167 10.1007/s10545-010-9055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stoker, T.B., Torsney, K.M. and Barker, R.A. (2018) Pathological mechanisms and clinical aspects of GBA1 mutation-associated Parkinson's disease. In Parkinson's Disease: Pathogenesis and Clinical Aspects. Codon Publications (Stoker, T.B. and Greenland, J.C. eds), Brisbane (AU): Codon Publications, [cited 2020 Nov 11]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30702840 [PubMed] [Google Scholar]
- 86.Osellame, L.D., Rahim, A.A., Hargreaves, I.P., Gegg, M.E., Richard-Londt, A., Brandner, S.et al. (2013) Mitochondria and quality control defects in a mouse model of Gaucher disease–links to Parkinson's disease. Cell Metab. 17, 941–953 10.1016/j.cmet.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfeffer, S.R. (2017) Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 28, 712–715 10.1091/mbc.e16-10-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delcambre, S., Ghelfi, J., Ouzren, N., Grandmougin, L., Delbrouck, C., Seibler, P.et al. (2020) Mitochondrial mechanisms of LRRK2 G2019S penetrance. Front. Neurol. 11, 881 10.3389/fneur.2020.00881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alessi, D.R. and Sammler, E. (2018) LRRK2 kinase in Parkinson's disease. Science 360, 36–37 10.1126/science.aar5683 [DOI] [PubMed] [Google Scholar]
- 90.Tolosa, E., Vila, M., Klein, C. and Rascol, O. (2020) LRRK2 in Parkinson disease: challenges of clinical trials. Nat. Rev. Neurol. 16, 97–107 10.1038/s41582-019-0301-2 [DOI] [PubMed] [Google Scholar]
- 91.Soliman, A., Cankara, F.N. and Kortholt, A. (2020) Allosteric inhibition of LRRK2, where are we now. Biochem. Soc. Trans. 48, 2185–2194 10.1042/BST20200424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mortiboys, H., Johansen, K.K., Aasly, J.O. and Bandmann, O. (2010) Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology 75, 2017–2020 10.1212/WNL.0b013e3181ff9685 [DOI] [PubMed] [Google Scholar]
- 93.Sanders, L.H., Laganière, J., Cooper, O., Mak, S.K., Vu, B.J., Huang, Y.A.et al. (2014) LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson's disease patients: reversal by gene correction. Neurobiol. Dis. 62, 381–386 10.1016/j.nbd.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ou, H.C., Song, T.Y., Yeh, Y.C., Huang, C.Y., Yang, S.F., Chiu, T.H.et al. (2010) EGCG protects against oxidized LDL-induced endothelial dysfunction by inhibiting LOX-1-mediated signaling. J. Appl. Physiol. 108, 1745–1756 10.1152/japplphysiol.00879.2009 [DOI] [PubMed] [Google Scholar]
- 95.Yue, M., Hinkle, K.M.M., Davies, P., Trushina, E., Fiesel, F.C.C., Christenson, T.A.A.et al. (2015) Progressive dopaminergic alterations and mitochondrial abnormalities in LRRK2 G2019S knock-in mice. Neurobiol. Dis. 78, 172–195 10.1016/j.nbd.2015.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madureira, M., Connor-Robson, N. and Wade-Martins, R. (2020) LRRK2: autophagy and lysosomal activity. Front. Neurosci. 14, 498 10.3389/fnins.2020.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wallings, R., Connor-Robson, N. and Wade-Martins, R. (2019) LRRK2 interacts with the vacuolar-type H+-ATPase pump a1 subunit to regulate lysosomal function. Hum. Mol. Genet. 28, 2696–2710 10.1093/hmg/ddz088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schapansky, J., Khasnavis, S., DeAndrade, M.P., Nardozzi, J.D., Falkson, S.R., Boyd, J.D.et al. (2018) Familial knockin mutation of LRRK2 causes lysosomal dysfunction and accumulation of endogenous insoluble α-synuclein in neurons. Neurobiol. Dis. 111, 26–35 10.1016/j.nbd.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ysselstein, D., Nguyen, M., Young, T.J., Severino, A., Schwake, M., Merchant, K.et al. (2019) LRRK2 kinase activity regulates lysosomal glucocerebrosidase in neurons derived from Parkinson's disease patients. Nat. Commun. 10, 5570 10.1038/s41467-019-13413-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Härtlova, A., Herbst, S., Peltier, J., Rodgers, A., Bilkei-Gorzo, O., Fearns, A.et al. (2018) LRRK2 is a negative regulator of Mycobacterium tuberculosis phagosome maturation in macrophages. EMBO J. 37, e98694 10.15252/embj.201798694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bonet-Ponce, L., Beilina, A., Williamson, C.D., Lindberg, E., Kluss, J.H., Saez-Atienzar, S.et al. (2020) LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci. Adv. 6, eabb2454 10.1126/sciadv.abb2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kett, L.R., Boassa, D., Ho, C.C.-Y., Rideout, H.J., Hu, J., Terada, M.et al. (2012) LRRK2 parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 21, 890–899 10.1093/hmg/ddr526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deniston, C.K., Salogiannis, J., Mathea, S., Snead, D.M., Lahiri, I., Matyszewski, M.et al. (2020) Structure of LRRK2 in Parkinson's disease and model for microtubule interaction. Nature 588, 344–349 10.1038/s41586-020-2673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dhekne, H.S., Yanatori, I., Gomez, R.C., Tonelli, F., Diez, F., Schüle, B.et al. (2018) A pathway for Parkinson's disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife 7, e40202 10.7554/eLife.40202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wauters, F., Cornelissen, T., Imberechts, D., Martin, S., Koentjoro, B., Sue, C.et al. (2020) LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy 16, 203–222 10.1080/15548627.2019.1603548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bonello, F., Hassoun, S.M., Mouton-Liger, F., Shin, Y.S., Muscat, A., Tesson, C.et al. (2019) LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson's disease. Hum. Mol. Genet. 28, 1645–1660 10.1093/hmg/ddz004 [DOI] [PubMed] [Google Scholar]
- 107.Korecka, J.A., Thomas, R., Christensen, D.P., Hinrich, A.J., Ferrari, E.J., Levy, S.A.et al. (2019) Mitochondrial clearance and maturation of autophagosomes are compromised in LRRK2 G2019S familial Parkinson's disease patient fibroblasts. Hum. Mol. Genet. 28, 3232–3243 10.1093/hmg/ddz126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsieh, C.-H., Shaltouki, A., Gonzalez, A.E., Bettencourt da Cruz, A., Burbulla, L.F., Lawrence, E.S.et al. (2016) Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson's disease. Cell Stem Cell 19, 709 10.1016/j.stem.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Singh, F., Prescott, A.R., Ball, G. and Ganley, I.G. (2020) Pharmacological rescue of impaired mitophagy in Parkinson's disease-related LRRK2 G2019S knock-in mice. bioRxiv 10.1101/2020.12.07.414359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Steger, M., Tonelli, F., Ito, G., Davies, P., Trost, M., Vetter, M.et al. (2016) Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh, P.K. and Muqit, M.M.K. (2020) Parkinson's: a disease of aberrant vesicle trafficking. Annu. Rev. Cell Dev. Biol. 36, 237–264 10.1146/annurev-cellbio-100818-125512 [DOI] [PubMed] [Google Scholar]
- 112.Pfeffer, S.R. (2018) LRRK2 and Rab GTPases. Biochem. Soc. Trans. 46, 1707–1712 10.1042/BST20180470 [DOI] [PubMed] [Google Scholar]
- 113.Thirstrup, K., Dächsel, J.C., Oppermann, F.S., Williamson, D.S., Smith, G.P., Fog, K.et al. (2017) Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci. Rep. 7, 10300 10.1038/s41598-017-10501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kalogeropulou, A.F., Zhao, J., Bolliger, M.F., Memou, A., Narasimha, S., Molitor, T.P.et al. (2018) P62/SQSTM1 is a novel leucine-rich repeat kinase 2 (LRRK2) substrate that enhances neuronal toxicity. Biochem. J. 475, 1271–1293 10.1042/BCJ20170699 [DOI] [PubMed] [Google Scholar]
- 115.Matsui, T. and Fukuda, M. (2013) Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 14, 450–457 10.1038/embor.2013.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Matsui, T., Noguchi, K. and Fukuda, M. (2014) Dennd3 functions as a guanine nucleotide exchange factor for small GTPase Rab12 in mouse embryonic fibroblasts. J. Biol. Chem. 289, 13986–13995 10.1074/jbc.M113.546689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu, J., Fotouhi, M. and McPherson, P.S. (2015) Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 16, 709–718 10.15252/embr.201440006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu, J., Kozlov, G., McPherson, P.S. and Gehring, K. (2018) A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy. J. Biol. Chem. 293, 4566–4574 10.1074/jbc.RA117.001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bjørkøy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Øvervatn, A.et al. (2005) P62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lamark, T., Svenning, S. and Johansen, T. (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 61, 609–624 10.1042/EBC20170035 [DOI] [PubMed] [Google Scholar]