Abstract
Background:
RBMS3 (RNA-binding motif, single-stranded-intervacting protein 3) acts as a tumor-suppressive gene in a number of human cancers, however, its role in breast cancer is not fully understood. This study aimed to investigate the expression and clinicopathological significance of RBMS3 in breast cancer.
Methods:
A total of 998 breast cancer tissue samples in The Cancer Genome Atlas (TCGA) database with survival outcomes were divided into high RBMS3 expression and low expression groups using the median as the cutoff. Clinicopathological characteristics and prognosis were compared between the 2 groups.
Results:
TCGA showed that RBMS3 mRNA was downregulated in breast cancer tissues, and RBMS3 downregulation was correlated with poor prognosis. Immunohistochemistry staining of 127 paraffin-embedded breast cancer tissues showed that RBMS3 protein was localized in the cytoplasm and nucleus; however, nuclear staining was present in 90.0% of normal breast tissues but only 28.3% of breast cancer tissues. Decreased RBMS3 protein expression was significantly correlated with estrogen receptor (ER)-negative status and death at final follow-up. Patients with lower RBMS3 protein expression had substantially shorter survival than those with higher RBMS3 expression. Univariate and multivariate analysis indicated that the combination of RBMS3 expression and ER status (a variable designated as “cofactor”) was an independent prognostic factor in patients with breast cancer (hazard ratio [HR] = 0.420, 95% confidence interval [CI]: 0.223-0.791, P = 0.007).
Conclusion:
RBMS3 downregulation was correlated with poor prognosis in breast cancer patients, and the combination of RBMS3 expression and ER status was an independent prognostic factor.
Keywords: breast cancer, RBMS3, expression, prognosis, tumor suppressor
Introduction
Breast cancer is the most frequently diagnosed cancer in women, and is the leading cause of cancer death among women worldwide, accounting for 25% of all cancer cases and 15% of all cancer deaths among women.1 Due to the highly heterogeneous nature of breast cancer, the effects of treatment and the prognosis of women with breast cancer with disease of the same stage may differ greatly.2,3 Therefore, it is particularly important to identify specific indicators related to breast cancer treatment and prognosis.
Since the identification of the erb-b2 receptor tyrosine kinase 2 (HER2) gene and the development of targeted therapy in the 1980s,4-6 gene markers have played an important role in breast cancer diagnosis and treatment. Since that time, gene expression profiling has divided breast cancer into 4 types,7,8 and multi-gene assays have been developed for assessing the risk of recurrence and benefit from chemotherapy.9-11 It is important to identify tumor markers for breast cancer diagnosis and treatment.
Deletion of chromosome 3p is one of the most common mutations in many solid tumors.12 There are several candidate tumor-suppressor genes (TSGs) on 3p, including VHL3 on 3p2513; RAR-b on 3p2414; FHIT on 3p14.215; and RASSF1A,16 CACNA2D2,17 and DLC11 18 on 3p21.3. RBMS3 (RNA-binding motif, single-stranded-interacting protein 3) is a TSG located in the p23-p24 region of human chromosome 3, and belongs to the family of c-Myc single-strand binding proteins (MSSP).19 RBMS3 expression is normal in many tissues, but is significantly reduced in a variety of malignancies, such as nasopharyngeal carcinoma,20,21 esophageal squamous cell carcinoma,22 lung squamous cell carcinoma,23 and gastric cancer.24 RBMS3 may inhibit cell proliferation and angiogenesis, and promotes apoptosis by regulating gene transcription or RNA metabolism.20 Li et al 22 showed that RBMS3 suppresses esophageal squamous cell carcinoma by downregulating c-Myc and cyclin-dependent kinase 4 (CDK4), thereby inhibiting retinoblastoma protein (Rb) phosphorylation. Chen et al 20 have demstrated that RBMS3 upregulates p53 and p21, and downregulates cyclin E and CDK2, thereby inhibiting Rb Ser780.
Only a few studies have examined the role of RBMS3 in breast cancer. Yang et al 25 found that mRNA and protein expression was downregulated in breast cancer tissue and cell lines, while RBMS3 overexpression suppressed breast cancer cell proliferation, migration, and invasion in vitro, and decreased tumor growth in vivo. Zhu et al 26 also reported that RBMS3 was downregulated in breast cancer and ectopic RBMS3 expression inhibited cell migration and invasion in vitro, and inhibited lung metastasis in vivo. In addition, RBMS3/Twsit1/matrix metalloproteinase 2 (MMP2) axis plays a role in the regulation of invasion and metastasis of breast cancer.26 However, the prognostic value of RBMS3 in patients with breast cancer remains unknown.
Thus, the purpose of the present study was to investigate the clinical significance of RBMS3 in breast cancer patients, and its prognostic value.
Patients and Methods
Patients and Samples
Eight paired fresh breast cancer tissues and the normal tissue adjacent to the tumor (NAT) from surgeries performed in 2015 were obtained from the Department of Thyroid and Breast Surgery of the First Affiliated Hospital of Sun Yat-sen University, and were used for quantitative reverse transcription-PCR (qRT-PCR). In addition, 127 paraffin-embedded breast cancer tissues were obtained from the Department of Thyroid and Breast Surgery, the First Affiliated Hospital of Sun Yat-sen University from surgeries performed from 2001 to 2004. Ten paraffin-embedded normal breast tissues were obtained from the Department of Plastic Surgery, the First Affiliated Hospital of Sun Yat-sen University from surgeries performed in 2015, for immunohistochemistry (IHC) analysis.
The Ethics Committee of our hospital approved this study. Prior written informed consent was obtained from each patient. All breast cancer tissues were pathologically diagnosed as breast invasive ductal carcinoma. The tissues were staged according to the seventh edition of the American Joint Committee on Cancer (AJCC) cancer staging system.27,28 The clinical and pathological characteristics of the 127 patients are summarized in Table 1.
Table 1.
Clinical and Pathological Characteristics of 127 Patients With Breast Cancer.
| Age (years) | |
| ≥ 51 | 67 (52.8) |
| < 51 | 60 (47.2) |
| ER status | |
| Positive | 80 (63.0) |
| Negative | 47 (37.0) |
| Her2 status | |
| Positive | 43 (33.9) |
| Negative | 84 (66.1) |
| Stage | |
| I | 10 (7.9) |
| II | 73 (57.5) |
| III | 44 (34.6) |
| IV | 0 (0.0) |
| T stage | |
| T1 | 26 (20.5) |
| T2 | 88 (69.3) |
| T3 | 13 (10.2) |
| T4 | 0 (0.0) |
| N stage | |
| N0 | 47 (37.0) |
| N1 | 40 (31.5) |
| N2 | 32 (25.2) |
| N3 | 8 (6.3) |
| M stage | |
| M0 | 127 (100.0) |
| M1 | 0 (0.0) |
| Status at follow-up | |
| Alive | 87 (68.5) |
| Dead | 40 (31.5) |
| RBMS3 | |
| Low | 91 (71.7) |
| High | 36 (28.3) |
Abbreviations: ER, estrogen receptor; Her2, human epidermal growth factor receptor 2.
Data are presented as count (percentage).
The Cancer Genome Atlas (TCGA)
RBMS3 mRNA expression and clinical data of 1,092 patients with breast cancer were downloaded from TCGA (http://cancergenome.nih.gov/). The RBMS3 mRNA expressions were compared between cancerous tissue and the adjacent normal tissue. Next, patients were divided into RBMS3 high expression and RBMS3 low expression groups using the median as the cutoff. The association between RBMS3 mRNA expression (high or low) and overall survival (OS) was evaluated using Kaplan–Meier analysis and the log-rank test.
IHC
The paraffin-embedded breast cancer tissues and normal breast tissues were deparaffinized, and then incubated for 30 min with goat serum at room temperature to block endogenous antibodies. Next, the tissues were incubated with rabbit anti-RBMS3 antibody (1:100, Novus, USA) overnight at 4°C, followed by incubation with horseradish peroxidase–linked secondary antibody for 30 min at room temperature. The slides were then stained with diaminobenzidine, and counterstained with hematoxylin. Two investigators blinded to patient clinicopathological data viewed and scored the degree of immunostaining independently. Any disagreements were resolved by discussion. RBMS3 protein expression was scored according to the degree of immunostaining as follows: absent (total absence of staining), very weak (faint staining in < 25% of tumor cells), moderate (moderate staining in 25% to < 75% of tumor cells, or strong staining in < 25% of tumor cells), or strong (moderate staining in >75% of tumor cells, or strong staining in >25% of tumor cells). Moderate/strong staining indicated high RBMS3 expression (RBMS3_high); absent/very weak staining indicated low RBMS3 expression (RBMS3_low) 22 (Supplementary Figure 1).
qRT-PCR
The 8 paired breast cancer tissues and adjacent non-tumorous tissues were lysed with TRIzol to extract the total RNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal PCR control. RBMS3-specific primers (RBMS3-F [forward]: 5′-GCACAGAAAGCGGTAGCATC-3′; RBMS3-R [reverse]: 5′-TGTCCAAAGGGTTTCAGCATA-3′) were purchased from GeneCopoeia (Germantown, MD, USA). One-step SYBR Green I–based semiquantitative RT-PCR (SQRT-PCR) was performed to detect RBMS3 mRNA levels in the tissues (One Step SYBR RT-PCR kit; TaKaRa, Dalian, China). The qRT-PCR results were analyzed using Rotor-Gene Real-Time Analysis Software 6.0 (Corbett Robotics, Brisbane, Australia).
Statistical Analysis
Continuous data were reported as mean ± standard deviation and categorical data as count (percentage). Student’s t-test was used to compare RBMS3 expression between breast cancer tissues and adjacent non-tumorous tissues in the qRT-PCR experiment. The chi-square test was used to analyze the relations between RBMS3 expression and clinical and pathological data. Survival curves were generated using the Kaplan–Meier method, and compared with the log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazards regression model. Statistical analyses were performed with the SPSS statistical software package version 24.0 (IBM, USA). Values of P < 0.05 were considered as statistical significance.
Results
RBMS3 Expression Was Decreased in Breast Cancer Tissue, Which Was Associated With Decreased Survival
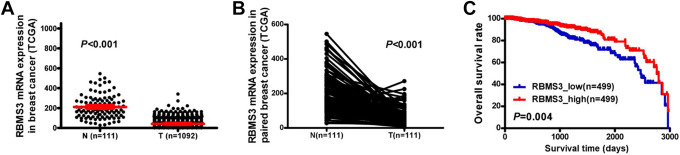
Comparison of 1,092 breast cancer tissues and 111 NAT from TCGA database showed significantly lower RBMS3 mRNA expression in breast cancer tissue than in the NAT (P < 0.001, Figure 1a). RBMS3 expression was significantly downregulated at the mRNA level in the 111 breast cancer tissues compared with the NAT (P < 0.001, Figure 1b). A total 998 breast cancer tissue samples in TCGA database with OS data were divided into high expression (top 50%) and low expression (bottom 50%) groups using the median RBMS3 mRNA expression level as the cutoff. OS was significantly higher in the high expression group as compared to the low expression group (P = 0.004, Figure 1c).
Figure 1.
RBMS3 expression in breast cancer tissues (T) and adjacent non-tumorous (N) tissues from The Cancer Genome Atlas (TCGA) database, and the relations between RBMS3 expression and overall survival (OS) rate. A) RBMS3 mRNA expression in 1,092 breast cancer tissue specimens was lower than that in 111 NAT (P < 0.001). The red lines represent the mean and standard deviation. B) RBMS3 mRNA expression was lower in 111 breast cancer tissue specimens than in paired NAT (P < 0.001). C) Kaplan–Meier survival curve showing a higher OS rate in the RBMS3 high expression group as compared to the RBMS3 low expression group (P = 0.004).
RBMS3 Was Downregulated at the mRNA and Protein Level in Breast Cancer Tissue
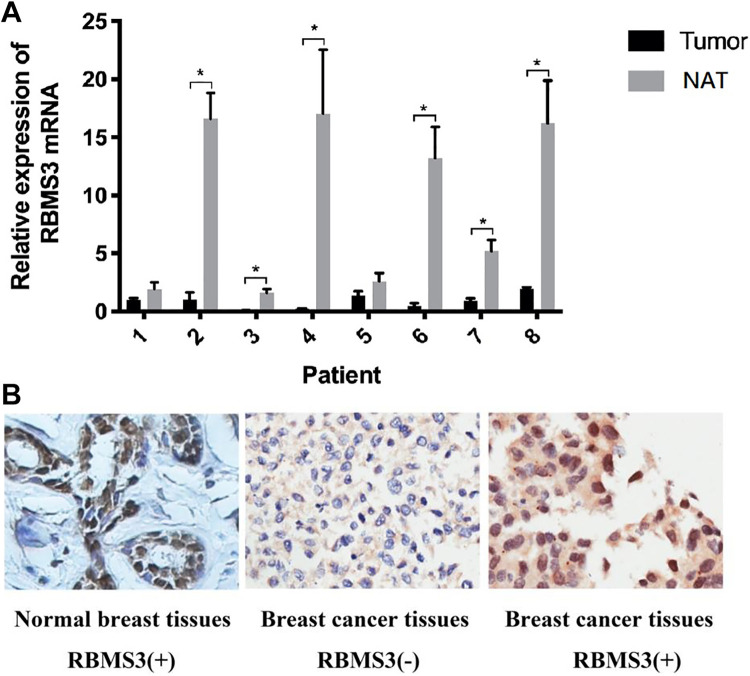
RBMS3 mRNA expression was detected using qRT-PCR in the 8 paired fresh breast cancer and NAT. The results indicated that RBMS3 mRNA expression was lower in breast cancer tissues than in NAT in 6 pairs of samples (75%, Figure 2a).
Figure 2.
RBMS3 expression in breast cancer tissue and NAT. A) qRT-PCR showing lower RBMS3 mRNA expression in breast cancer tissues compared to NAT (*P < 0.05). RBMS3 expression was normalized to the internal control GAPDH. B) Representative immunohistochemical (IHC) staining images of RBMS3 expression and location (brown nuclear staining) in breast cancer tissue and normal breast tissue specimens (×200 magnification).
IHC staining was used to investigate RBMS3 protein expression and localization in 127 paraffin-embedded breast cancer tissues and 10 paraffin-embedded normal breast tissues. RBMS3 protein was localized in the cytoplasm and nucleus (Figure 2b). The nuclear staining was 90.0% (9/10) in normal breast tissue, and 28.3% (36/127) in breast cancer tissue.
Relations of RBMS3 Downregulation With Clinicopathological Characteristics of Patients With Breast Cancer
RBMS3 protein expression downregulation was not significantly related to age, clinical disease stage, T stage, or N stage (all, P > 0.05). However, downregulation was significantly correlated with negative estrogen receptor (ER) status (P = 0.010), and death at the final follow-up (P = 0.024) (Table 2).
Table 2.
Relations Between RBMS3 Expression Level and Clinical and Pathological Data.
| Characteristic | Total | RBMS3 | Chi-square P-value | Fisher’s exact P-value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | |||||
| ≥ 51 | 67 | 52 (77.6) | 15 (22.4) | 0.115 | 0.167 |
| < 51 | 60 | 39 (65.0) | 23 (35.0) | ||
| Stage | |||||
| I-II | 83 | 56 (67.5) | 27 (32.5) | 0.214 | 0.214 |
| III | 44 | 35 (79.5) | 9 (20.5) | ||
| T stage | |||||
| T1-T2 | 114 | 81 (71.1) | 33 (28.9) | 0.757 | 0.757 |
| T3 | 13 | 10 (76.9) | 3 (23.1) | ||
| N stage | |||||
| N0 | 47 | 31 (66.0) | 16 (34.0) | 0.311 | 0.311 |
| N1-N3 | 80 | 60 (75.0) | 20 (25.0) | ||
| ER | |||||
| Positive | 80 | 51 (63.8) | 29 (36.3) | 0.010* | 0.014* |
| Negative | 47 | 40 (85.1) | 7 (14.9) | ||
| Her2 | |||||
| Positive | 43 | 31 (72.1) | 12 (27.9) | 1.000 | 1.000 |
| Negative | 84 | 60 (71.4) | 24 (28.6) | ||
| Status at follow-up | |||||
| Alive | 87 | 57 (65.5) | 30 (34.5) | 0.024* | 0.033* |
| Dead | 40 | 34 (85.0) | 6 (15.0) | ||
Abbreviations: ER, estrogen receptor; Her2, human epidermal growth factor receptor 2.
RBMS3 low and high expression data are reported as count (percentage).
* P < 0.05, indicates statistical significance.
RBMS3 Protein Downregulation Was Associated With a Poor Prognosis
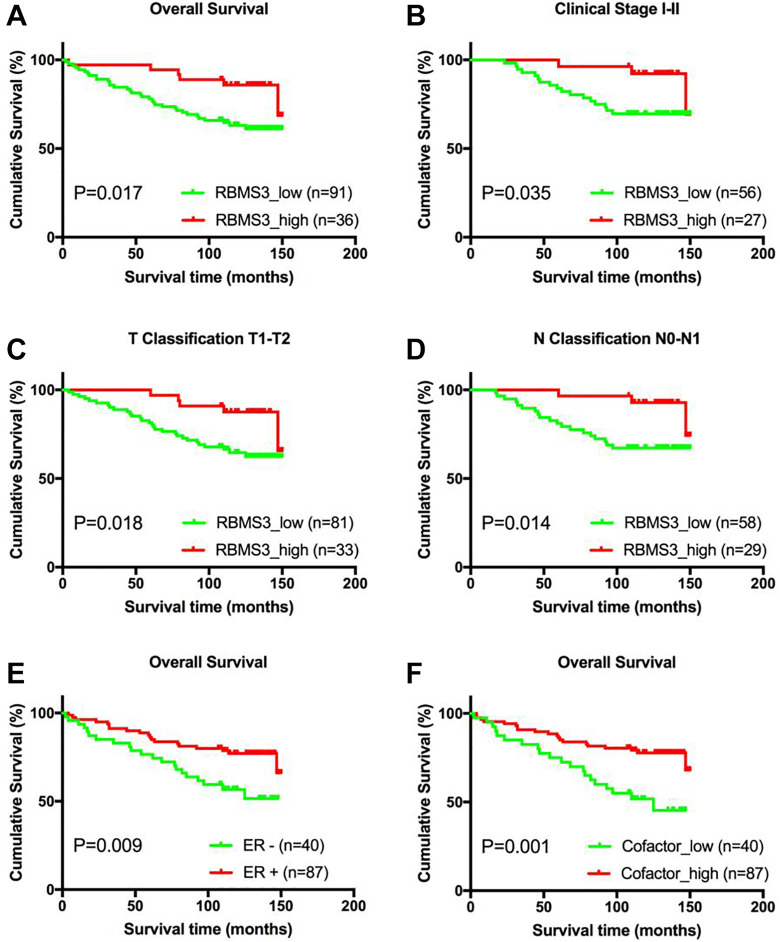
Kaplan–Meier analysis and the log-rank test were used to analyze survival data to assess the clinical significance of downregulated RBMS3 protein expression in patients with breast cancer. The analysis showed that the 5-year OS rate was 94.4% in patients with high RBMS3 expression, and 78% in those with low expression, indicating that low RBMS3 expression was associated with shorter OS (P = 0.017, Figure 3a). In addition, RBMS3 downregulation was associated with shorter OS in patients with early stage (stage I/II disease) (P = 0.035; Figure 3b). Similarly, patients with T1/T2 disease with lower RBMS3 expression had significantly shorter OS (P = 0.018, Figure 3c), as did patients with N0/N1 disease (P = 0.014, Figure 3d). However, no statistically significant differences were found between RBMS3 expression and survival time in the subsets of patients with clinical stage III, T3, or N2/N3 disease, which might be due to the limited number of patients in each subset.
Figure 3.
Kaplan–Meier analysis of overall survival (OS) in patients with breast cancer based on RBMS3 and cofactor expression. A) OS rate of RBMS3 high versus RBMS3 low expression in all patients (P = 0.017). B) OS rate of patients with American Joint Committee on Cancer (AJCC) stage I/II disease (P = 0.035). C) OS rate of patients with T1/T2 disease (P = 0.018). D) OS rate of patients with N0/N1 disease (P = 0.014). E) OS rate of cofactor_high versus cofactor_low expression in all patients (P = 0.001).
Correlation analysis showed that RBMS3 protein downregulation was correlated with negative ER status. As shown in Figure 3e, ER-negative patients had significantly shorter OS (P = 0.009). Accordingly, a variable termed “cofactor” that combined the expression of RBMS3 and ER was created. The samples were classified as cofactor_high (RBMS3_high and/or ER-positive, n = 87) or cofactor_low (RBMS3_low and ER-negative, n = 40). OS was lower in the cofactor_low group as compared to the cofactor_high group (5-year OS: 75.0% and 86.2%, respectively; P = 0.001; Figure 3f).
Univariate analysis showed that clinical disease stage (hazard ratio [HR] = 0.435, 95% confidence interval [CI]: 0.234-0.808, P = 0.008), ER status (HR = 0.444, 95% CI: 0.238-0.828, P = 0.011), RBMS3 expression (HR = 0.360, CI: 0.151-0.861, P = 0.022), and cofactor expression (HR = 0.372, CI: 0.199-0.695, P = 0.002) were significantly correlated with prognosis. Multivariate analysis showed that clinical disease stage (HR = 0.514, CI: 0.273-0.968, P = 0.039) was the independent predictor of overall survival (Table 3).
Table 3.
Univariate and Multivariate Analysis of Variables and Breast Cancer Prognosis.
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% Cl) | P-value | HR (95% Cl) | P-value | |
| Age | 0.645 (0.340, 1.224) | 0.18 | ||
| (≥ 51 vs. < 51) | ||||
| Stage | 0.435 (0.234, 0.808) | 0.008* | 0.514 (0.273, 0.968) | 0.039* |
| (I/II vs. III) | ||||
| T stage | 0.827 (0.365, 1.875) | 0.65 | ||
| (T1 vs. T2/T3) | ||||
| N stage | 0.636 (0.323, 1.252) | 0.19 | ||
| (N0 vs. N1-3) | ||||
| ER status | 0.444 (0.238, 0.828) | 0.011* | 0.828 (0.096, 7.136) | 0.864 |
| (+ vs. −) | ||||
| Her2 status | 1.149 (0.583, 2.262) | 0.688 | ||
| (+ vs. −) | ||||
| RBMS3 expression | 0.360 (0.151, 0.861) | 0.022* | 0.570 (0.204, 1.589) | 0.282 |
| (high vs. low) | ||||
| Cofactor | 0.372 (0.199, 0.695) | 0.002* | 0.430 (0.045, 4.109) | 0.464 |
| (high vs. low) | ||||
Abbreviations: CI, confidence interval; ER, estrogen receptor; Her2, human epidermal growth factor receptor 2; HR, hazard ratio.
* P < 0.05, indicates statistical significance.
However, due to the “cofactor” variable was the combination of RBMS3 expression and ER status, significant collinearity between cofactor and RBMS3/ER were observed (rcofactor, RBMS3 = 0.426, rcofactor, ER = 0.885; both P < 0.001). Therefore, a multivariate model which includes only stage and cofactor was carried out. The results indicate that both clinical disease stage (HR = 0.502, CI: 0.267-0.943, P = 0.032) and cofactor expression (HR = 0.420, CI: 0.223-0.791, P = 0.007) were independent predictors of overall survival.
Discussion
RBMS3 expression is significantly reduced in nasopharyngeal carcinoma,20,21 lung squamous cell carcinoma,23 esophageal squamous cell carcinoma,22,29 and gastric cancer.24 However, the role of RBMS3 in breast cancer development remains not fully understood. In the present study, we found that RBMS3 was significantly downregulated in breast cancer tissues, and was significantly correlated with ER status and mortality. Kaplan–Meier survival analysis showed that lower RBMS3 expression was associated with shorter OS; hence, a poorer prognosis. Univariate and multivariate Cox regression analysis suggested that the cofactor_low (RBMS3_low and ER-negative) was an independent predictor of poorer prognosis (shorter OS) in patients with breast cancer (HR = 0.420, 95% CI: 0.223-0.791, P = 0.007).
Penkov et al 19 first identified RBMS3 when screening fibroblast complementary DNA (cDNA) libraries. The authors found that RBMS3 protein was mainly located in the cytoplasm and can bind to the poly(A/U) region of RNA to regulated RNA metabolism. In a study of liver fibrosis development, Fritz et al 30 found that RBMS3 can directly bind to the Prx1 mRNA 3′ untranslated region, and stabilize the Prx1 mRNA. Jayasena et al 31 reported that RBMS3 can stabilize Smad2 transcripts by binding to the non-translated regions of SMAD2 mRNA. Lu et al 32 reported that RBMS3 binds to the 3′ non-transcribed region of pancreas transcription factor 1 alpha subunit (Ptf1a) mRNA, and regulated PTF1A protein expression and promotes pancreatic exocrine gland and acinar cell differentiation. These findings all indicate that RBMS3, similar to most RNA-binding proteins, localizes in the cytoplasm and can directly bind mRNA, thereby regulating RNA metabolism.
However, Chen et al 20 found that RBMS3 was mainly expressed in the nucleus and inhibited RNA transcription in nasopharyngeal carcinoma.20 Li et al 22 reported that in esophageal squamous cell carcinoma RBMS3 is mainly expressed in the nucleus, and can directly bind to the DNA replication initiation region about 2 kb upstream of the c-MYC gene, and thus regulate c-MYC gene expression. The results of the current study showed that RBMS3 protein was localized in the cytoplasm and nucleus, and only nuclear RBMS3 protein was significantly downregulated in breast cancer, which is similar to the finding of Liang et al.22 This suggests that in breast cancer cells, RBMS3 may play a role in regulating gene transcription, rather than regulating RNA metabolism.
Breast cancer is a highly heterogeneous disease,2,3 and its treatment and prognosis are highly dependent on tumor stage and type. However, the best treatments for patients with different disease stages and tumor types have not been determined. In 2007, an international web-based forum on priorities in translational breast cancer research determined that it is the highest priority to identify molecular signatures to select patients who could be spared chemotherapy.33 Our study showed that RBMS3 protein expression in patients with stage I/II (early clinical stage), T1/T2 (small tumor size), and N0/N1 (less lymph node metastasis) disease was associated with prognosis. This suggests that RBMS3 may be useful for predicting prognosis and assessing the need for chemotherapy in patients with breast cancer. Some patients with early-stage disease and high RBMS3 expression may be spared chemotherapy, while those with low RBMS3 expression may require more intensive therapy to obtain better outcomes.
In the present study, RBMS3 expression was significantly correlated with ER status (P = 0.010, Table 2). Moreover, cofactor (ER and RBMS3) was an independent prognostic factor. It is well-known that c-MYC is a breast cancer oncogene that is involved in breast cancer development and progression.34,35 RBMS3 can bind directly to the upstream initiation region of the c-MYC gene, thereby downregulating c-MYC gene expression.22 c-MYC is amplified in ER-negative breast cancer.36-39 Hence, it is worth to further investigate whether c-Myc is involved in regulating RBMS3 and ER.
The mechanism by which RBMS3 inhibits the development and progression of breast cancer is unknown, but prior studies have suggested possibilities. As in our study, Yang et al 25 found that RBMS3 mRNA and protein expression were significantly downregulated in breast cancer tissue. Interestingly, the authors also observed that RBMS3 inhibited β-catenin, cyclin D1, and c-Myc protein expression in breast cancer cells. The authors concluded that the inhibition effect of RBMS3 on proliferation and tumorigenesis of breast cancer cells was involved in blockage of the Wnt/β-catenin signaling pathway. Zhu et al found that RBMS3 negatively regulated Twsit1 expression by binding to the 3′-UTR of Twist1 mRNA,26 in turn resulting in decreased Twist1-induced expression of MMP2. The study also found that cell migration, invasion, and lung metastasis induced by Twist1 was reversed by upregulation of RBMS3.
Conclusions
In summary, the results of this study suggest that RBMS3 downregulation was correlated with poor prognosis in breast cancer patients. RBMS3 expression was correlated with ER status in breast cancer tissue, and cofactor (RBMS3 and ER) was a significant prognostic factor of OS. Further study is warranted to determine the underlying molecular mechanism.
Supplemental Material
Supplemental Material, sj-jpg-1-tct-10.1177_15330338211004921 for Tumor Suppressor Effect of RBMS3 in Breast Cancer by Chunyang Wang, Yidan Wu, Yunqi Liu, Fushun Pan, Huijuan Zeng, Xiaoxi Li and Liang Yu in Technology in Cancer Research & Treatment
Acknowledgments
We thank all the participants who took part in this study.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CDK4
cyclin-dependent kinase 4
- cDNA
complementary DNA
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- IHC
immunohistochemistry
- MMP2
matrix metalloproteinase 2
- MSSP
Myc single-strand binding proteins
- OS
overall survival
- qRT-PCR
quantitative reverse transcription-PCR
- TCGA
The Cancer Genome Atla
- TSGs
tumor-suppressor genes
Authors’ Note: Chunyang Wang, Yidan Wu, and Yunqi Liu contributed equally to this work. This study was approved by the ethics committee of the First Affiliated Hospital of Sun Yat-sen University [NO. 2012182].
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The present study was supported by the Guangdong Province Natural Science Foundation [2015A030313141] and the Guangdong Province Science and Technology Project [2016A040403113].
ORCID iD: Liang Yu, MD, PhD  https://orcid.org/0000-0002-3265-3802
https://orcid.org/0000-0002-3265-3802
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Spitale A, Mazzola P, Soldini D, Mazzucchelli L, Bordoni A. Breast cancer classification according to immunohistochemical markers: clinicopathologic features and short-term survival analysis in a population-based study from the South of Switzerland. Ann Oncol. 2009;20(4):628–635. [DOI] [PubMed] [Google Scholar]
- 3. Tang P, Wang J, Bourne P. Molecular classifications of breast carcinoma with similar terminology and different definitions: are they the same? Hum Pathol. 2008;39(4):506–513. [DOI] [PubMed] [Google Scholar]
- 4. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/NEU oncogene. Science. 1987;235(4785):177–182. [DOI] [PubMed] [Google Scholar]
- 5. Goldenberg MM. Trastuzumab, a recombinant DNA-derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin Ther. 1999;21(2):309–318. [DOI] [PubMed] [Google Scholar]
- 6. Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45(11):1033–1040. [DOI] [PubMed] [Google Scholar]
- 7. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. [DOI] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albain KS, Barlow WE, Shak S, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. [DOI] [PubMed] [Google Scholar]
- 11. Sparano JA, Gray RJ, Makower DF, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373(21):2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26(52):7283–7301. [DOI] [PubMed] [Google Scholar]
- 13. Kuzmin I, Duh FM, Latif F, Geil L, Zbar B, Lerman MI. Identification of the promoter of the human von Hippel-Lindau disease tumor suppressor gene. Oncogene. 1995;10(11):2185–2194. [PubMed] [Google Scholar]
- 14. de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343(6254):177–180. [DOI] [PubMed] [Google Scholar]
- 15. Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated (3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84(4):587–597. [DOI] [PubMed] [Google Scholar]
- 16. Horiguchi K, Tomizawa Y, Tosaka M, et al. Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene. 2003;22(49):7862–7865. [DOI] [PubMed] [Google Scholar]
- 17. Ji L, Nishizaki M, Gao B, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62(9):2715–2720. [PMC free article] [PubMed] [Google Scholar]
- 18. Daigo Y, Nishiwaki T, Kawasoe T, Tamari M, Tsuchiya E, Nakamura Y. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res. 1999;59(8):1966–1972. [PubMed] [Google Scholar]
- 19. Penkov D, Ni R, Else C, Pinol-Roma S, Ramirez F, Tanaka S. Cloning of a human gene closely related to the genes coding for the c-myc single-strand binding proteins. Gene. 2000;243(1-2):27–36. [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Kwong DL, Zhu CL, et al. RBMS3 at 3p24 inhibits nasopharyngeal carcinoma development via inhibiting cell proliferation, angiogenesis, and inducing apoptosis. PLoS One. 2012;7(9):e44636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J, Fu L, Zhang LY, Kwong DL, Yan L, Guan XY. Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma. Chin J Cancer. 2012;31(5):215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Chen L, Nie CJ, et al. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 2011;71(19):6106–6115. [DOI] [PubMed] [Google Scholar]
- 23. Liang YN, Liu Y, Meng Q, et al. RBMS3 is a tumor suppressor gene that acts as a favorable prognostic marker in lung squamous cell carcinoma. Med Oncol. 2015;32(2):459. [DOI] [PubMed] [Google Scholar]
- 24. Zhang T, Wu Y, Fang Z, et al. Low expression of RBMS3 and SFRP1 are associated with poor prognosis in patients with gastric cancer. Am J Cancer Res. 2016;6(11):2679–2689. [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y, Quan L, Ling Y. RBMS3 inhibits the proliferation and metastasis of breast cancer cells. Oncol Res. 2018;26(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu L, Xi PW, Li XX, et al. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. J Exp Clin Cancer Res. 2019;38(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. [DOI] [PubMed] [Google Scholar]
- 28. Edge SB, Compton CC, Fritz AG, Greene FL, Trotti A, eds. AJCC Cancer Staging Manual. 7th ed. Springer; 2010. [Google Scholar]
- 29. Qin YR, Fu L, Sham PC, et al. Single-nucleotide polymorphism-mass array reveals commonly deleted regions at 3p22 and 3p14.2 associate with poor clinical outcome in esophageal squamous cell carcinoma. Int J Cancer. 2008;123(4):826–830. [DOI] [PubMed] [Google Scholar]
- 30. Fritz D, Stefanovic B. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J Mol Biol. 2007;371(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jayasena CS, Bronner ME. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. J Cell Biol. 2012;199(3):453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu CK, Lai YC, Chen HR, Chiang MK. Rbms3, an RNA-binding protein, mediates the expression of Ptf1a by binding to its 3’UTR during mouse pancreas development. DNA Cell Biol. 2012;31(7):1245–1251. [DOI] [PubMed] [Google Scholar]
- 33. Dowsett M, Goldhirsch A, Hayes DF, Senn HJ, Wood W, Viale G. International Web-based consultation on priorities for translational breast cancer research. Breast Cancer Res. 2007;9(6):R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shiu RP, Watson PH, Dubik D. c-myc oncogene expression in estrogen-dependent and -independent breast cancer. Clin Chem. 1993;39(2):353–355. [PubMed] [Google Scholar]
- 35. Pietilainen T, Lipponen P, Aaltomaa S, Eskelinen M, Kosma VM, Syrjanen K. Expression of c-myc proteins in breast cancer as related to established prognostic factors and survival. Anticancer Res. 1995;15(3):959–964. [PubMed] [Google Scholar]
- 36. Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–4367. [PubMed] [Google Scholar]
- 37. Cuny M, Kramar A, Courjal F, et al. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60(4):1077–1083. [PubMed] [Google Scholar]
- 38. Persons DL, Borelli KA, Hsu PH. Quantitation of HER-2/NEU and c-myc gene amplification in breast carcinoma using fluorescence in situ hybridization. Mod Pathol. 1997;10(7):720–727. [PubMed] [Google Scholar]
- 39. Yasojima H, Shimomura A, Naoi Y, et al. Association between c-myc amplification and pathological complete response to neoadjuvant chemotherapy in breast cancer. Eur J Cancer. 2011;47(12):1779–1788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-jpg-1-tct-10.1177_15330338211004921 for Tumor Suppressor Effect of RBMS3 in Breast Cancer by Chunyang Wang, Yidan Wu, Yunqi Liu, Fushun Pan, Huijuan Zeng, Xiaoxi Li and Liang Yu in Technology in Cancer Research & Treatment