Abstract
For the biogenesis of mitochondria, hundreds of proteins need to be targeted from the cytosol into the various compartments of this organelle. The intramitochondrial targeting routes these proteins take to reach their respective location in the organelle are well understood. However, the early targeting processes, from cytosolic ribosomes to the membrane of the organelle, are still largely unknown. In this study, we present evidence that an integral membrane protein of the endoplasmic reticulum (ER), Ema19, plays a role in this process. Mutants lacking Ema19 show an increased stability of mitochondrial precursor proteins, indicating that Ema19 promotes the proteolytic degradation of nonproductive precursors. The deletion of Ema19 improves the growth of respiration-deficient cells, suggesting that Ema19-mediated degradation can compete with productive protein import into mitochondria. Ema19 is the yeast representative of a conserved protein family. The human Ema19 homologue is known as sigma 2 receptor or TMEM97. Though its molecular function is not known, previous studies suggested a role of the sigma 2 receptor as a quality control factor in the ER, compatible with our observations about Ema19. More globally, our data provide an additional demonstration of the important role of the ER in mitochondrial protein targeting.
INTRODUCTION
Mitochondria house 800 to 1500 different proteins (Calvo et al., 2016; Morgenstern et al., 2017). With the exception of a very small number of mitochondrial translation products, all these proteins are synthesized on cytosolic ribosomes and need to be imported into the mitochondria. Mitochondrial targeting signals allow the specific binding of these proteins to mitochondrial surface receptors. Such receptors are part of the translocase of the outer membrane, or TOM complex (Shiota et al., 2015; Araiso et al., 2019), which serves as the entry gate for almost all mitochondrial protein precursors. Following translocation through the TOM complex, proteins are sorted by several additional complexes in the outer and inner membrane to their respective mitochondrial subcompartment. The mechanisms by which the mitochondrial protein import systems mediates these protein translocation reactions were analyzed in detail over the past three decades and are described in several comprehensive reviews (Chacinska et al., 2009; Endo et al., 2011; Callegari et al., 2020; Drwesh and Rapaport, 2020; Edwards et al., 2020; Mokranjac, 2020; Schneider, 2020).
The early steps of the import process which include all reactions prior to precursor binding to the TOM receptors are less well understood (Avendano-Monsalve et al., 2020; Bykov et al., 2020). Some precursors presumably associate with the mitochondrial surface before ribosomes complete their synthesis (Marc et al., 2002; Williams et al., 2014; Golani-Armon and Arava, 2016). However, in contrast to the situation for secretory proteins destined for the endoplasmic reticulum (ER), there is little evidence for a mechanistic coupling of synthesis and translocation of mitochondrial precursors. Owing to their posttranslational targeting, most mitochondrial precursor proteins presumably explore the cytosol and the cytosol-exposed surface of other compartments before binding the TOM receptors (Gamerdinger et al., 2015; Costa et al., 2018).
Precursor proteins that accumulate in the cytosol can be highly toxic (Wang and Chen, 2015; Wrobel et al., 2015) and elicit response programs to counteract these deleterious consequences (Nargund et al., 2012; Weidberg and Amon, 2018; Boos et al., 2019, 2020; Song et al., 2021; Zöller et al., 2020). The stability of many precursor proteins in the cytosol is low thanks to surveillance of the ubiquitin/proteasome system for uninserted precursors (Bragoszewski et al., 2017; Kowalski et al., 2018; Paasch et al., 2018; Saladi et al., 2020; Shakya et al., 2021). The association of mitochondrial precursors to the ER surface can retard their degradation and facilitate their productive targeting to the TOM complex by a process referred to as ER-SURF (Hansen et al., 2018; Xiao et al., 2020).
A recent screen in the yeast Saccharomyces cerevisiae (from hereon referred to as yeast) for factors that determine the cytosolic stability of mitochondrial precursor proteins identified the so-far uncharacterized protein Ema19 (Hansen et al., 2018). In this study, we elucidated the relevance of Ema19 for mitochondrial biogenesis in more detail. We observe that the absence of Ema19 increases the abundance and stability of mitochondrial proteins, including those associated with the ER surface. These data suggest that Ema19 plays a role, directly or indirectly, as a quality control factor during the early, targeting steps of the mitochondrial import process.
RESULTS
Cells lacking Ema19 show higher levels of nonimported precursor proteins
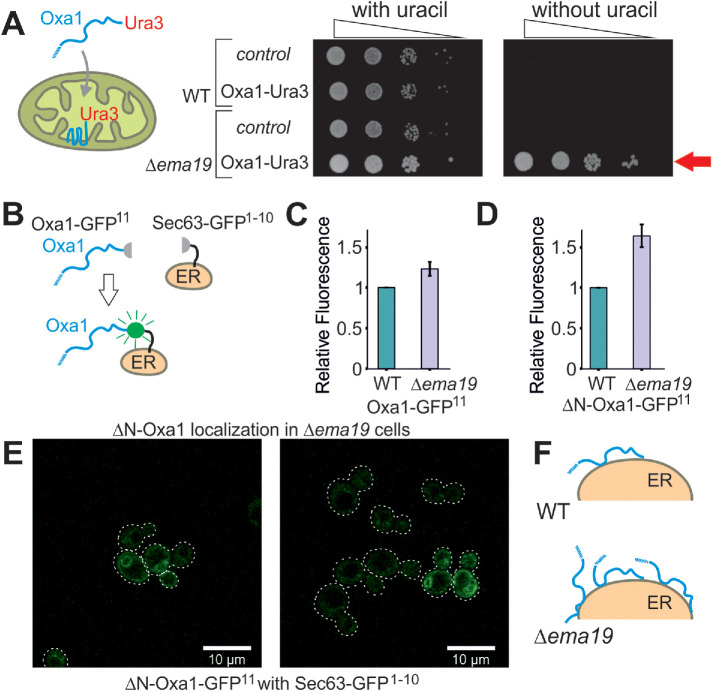
We recently developed a genetic screen, which allows us to measure the efficiency of mitochondrial protein import on the basis of a simple growth assay (Hansen et al., 2018). The assay relies on expressing a fusion between the mitochondrial inner membrane protein Oxa1 and the enzyme Ura3 (orotidine-phosphate decarboxylase) on the background of Δura3 cells (Figure 1A). Efficient mitochondrial import depleted cytosolic pools of the Oxa1-Ura3 fusion protein causing uracil-deficiency, whereas the cytosolic accumulation of the fusion protein (for example, when its mitochondrial presequence was deleted) suppressed the uracil-dependent growth of these cells (Supplemental Figure S1A). Wild-type (WT) cells expressing the Oxa1-Ura3 protein were unable to grow without uracil, whereas Oxa1-Ura3 expression in cells lacking EMA19 grew very efficiently without uracil (Figure 1A). Thus, the absence of Ema19 apparently leads to increased cytosolic levels of the Oxa1-Ura3 fusion protein. This effect was seen very reliably in several genetic backgrounds (YPH499, W303) (Supplemental Figure S1B). This Oxa1–Ura3-mediated growth in the absence of uracil was only found in a small number of yeast mutants, including strains with defects in the ER-associated cochaperone Djp1 or the import component Tim50 (Hansen et al., 2018).
FIGURE 1:
Deletion of Ema19 leads to increased levels of nonimported mitochondrial precursor proteins. (A) A schematic representation of the Oxa1-Ura3 reporter assay is shown on the left. WT and Δema19 cells were transformed with empty (control) or Oxa1-Ura3 expression plasmids and grown on uracil-containing medium to mid-log phase. Serial 10-fold deletions were dropped onto glucose plates which contained or lacked uracil. The arrow depicts the efficient uracil-independent growth induced by Oxa1-Ura3 in Δema19 cells. (B) Schematic representation of the split-GFP assay. (C, D) WT and ∆ema19 cells expressing Sec63-GFP1-10 and either Oxa1-GFP11 or ΔN-Oxa1-GFP11 were grown to mid-log phase before GFP-mediated fluorescence was measured. Values show mean and SD values from three measurements. (E) Microscopy pictures from the cells analyzed in D. Note that the GFP signal shows the characteristic perinuclear and cortical pattern of ER proteins in yeast cells. (F) Model for the accumulation of extramitochondrial precursors in Δema19 cells.
As an independent proof for the cytosolic accumulation of Oxa1 in Δema19 cells, we employed a fluorescence-based screen using a self-complementing split version of superfolder GFP (Pedelacq et al., 2006; Smoyer et al., 2016). We fused one part of GFP to the C-terminus of Oxa1 (Oxa1-GFP11) and the other to the ER surface protein Sec63 (Sec63-GFP1-10) (Figure 1B). Using the precursor form of Oxa1, we only detected very low fluorescent levels in the WT but these were slightly increased in Δema19 cells (Figure 1C; Supplemental Figure S1C). Consistent with previous reports, our data indicate that under normal conditions Oxa1 precursors associate with the ER surface only very transiently (Hansen et al., 2018; Shakya et al., 2020; Xiao et al., 2020), and that Ema19 has a role in regulating the amount or extent of this interaction. When we used an Oxa1 version that lacked its N-terminal mitochondrial targeting sequence (ΔN-Oxa1-GFP11), the fluorescence signal was much higher and significantly enhanced in the Δema19 mutant (Figure 1D). This signal now clearly showed the characteristic pattern of an ER staining (Figure 1E). From this we conclude that the absence of Ema19 does not lead to an initial mislocalization of mitochondrial proteins yet affects their association time with the ER or stability once they reach the ER surface (Figure 1F).
Ema19 is highly conserved but dispensable at normal growth conditions
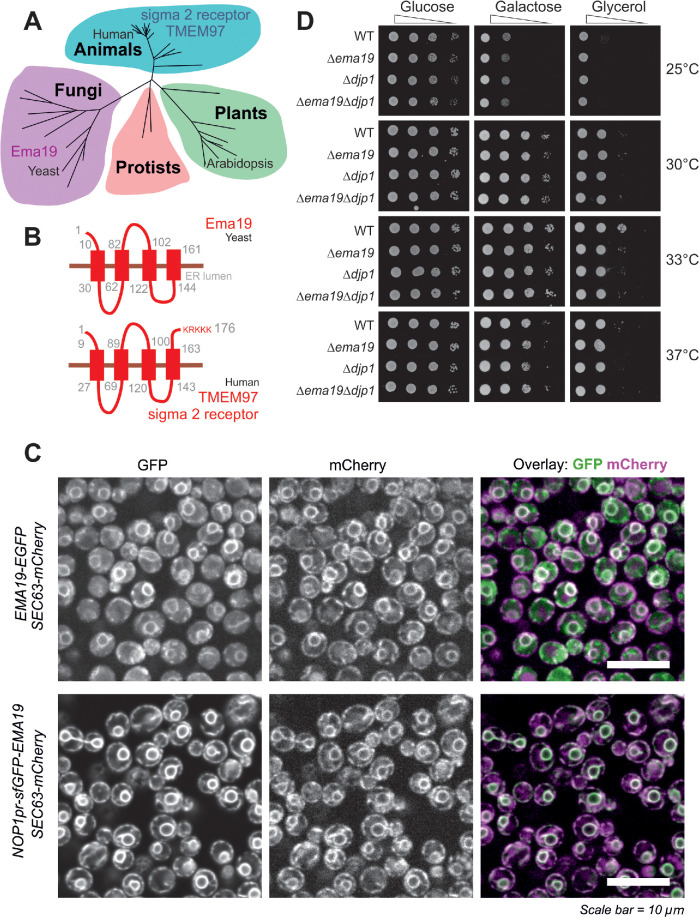
In an attempt to understand the role of Ema19 in mitochondrial precursor biogenesis we performed a BLAST search. Our search identified Ema19 homologues in many eukaryotic species, suggesting that Ema19 is a member of a protein family that is ubiquitously present in fungi and animals, including humans (Figure 2A; Supplemental Figure S2). Consistently, four transmembrane domains can be predicted for these homologues indicating a conserved membrane topology. These hydrophobic regions show many highly conserved positions and seem to be the functionally relevant segments of the Ema19 proteins (Figure 2B; Supplemental Figure S3). N-terminal signal sequences are absent and the human Ema19 homologue, named TMEM97 or sigma 2 receptor, ends with a C-terminal KRKKK sequence that is similar to the canonical KKXX/KXKXX di-lysine ER retrieval signal found in many resident membrane proteins of the ER (Gaynor et al., 1994; Ma and Goldberg, 2013). Fusing Ema19 to GFP indeed confirmed a stable ER localization of Ema19 similar to previously published data on the human protein (Bartz et al., 2009; Alon et al., 2017; Hansen et al., 2018) (Figure 2C). Despite the high conservation of the protein, yeast mutants lacking Ema19 did not show any obvious growth defects on fermentable (glucose, galactose) or nonfermentable (glycerol) carbon sources (Figure 2D).
FIGURE 2:
Ema19 belongs to a conserved family of ER proteins. (A) Phylogenetic analysis of members of the Ema19/TMEM97 family (see Supplemental Figure S4 and Table S4 for sequence details). (B) A model for the orientation of the Ema19 and TMEM97 is shown on the right. Transmembrane domains were predicted using TMpred (Hofmann and Stoffel, 1993). (C) Cells expressing Ema19-EGFP and sfGFP-Ema19 were analyzed by fluorescence microscopy. Sec63-mCherry served as ER marker. The intense perinuclear staining of the GFP signals indicate the localization of Ema19 in perinuclear ER membranes. (D) The respective strains were grown in galactose medium and dropped on plates containing different carbon sources and grown at the indicated temperatures.
Cells lacking Ema19 show morphological changes on respiratory growth conditions
We next used electron microscopy to visualize the ultrastructure of Δema19 mutants (Figure 3, A and B; Supplemental Figure S4). WT and Δema19 cells grown on glucose did not show any obvious differences. However, on respiration-inducing growth conditions on glycerol, we observed the accumulation of intracellular structures in the Δema19 cells, which were not observed in WT cells. These structures were not observed by the lipid-staining reagent BODIPY 493/503 (Supplemental Figure S5). Moreover, when we performed lipidomics under the same growth conditions (growth on glycerol), we did not observe any major changes in phospholipids (Figure 3, C and D). From this we conclude that Ema19 protects cells from defects that are observed specifically when cells grow on respiratory media, thus at conditions which induce the biogenesis of mitochondria.
FIGURE 3:
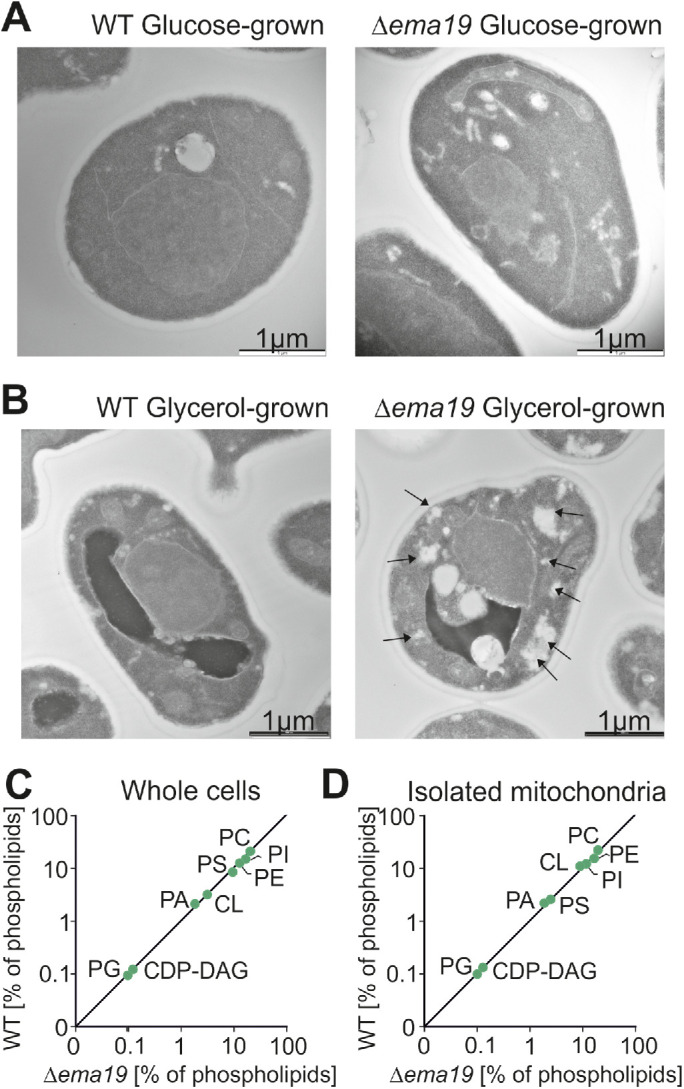
Δema19 cells show morphological changes on growth on glycerol. (A, B) Yeast cells were grown to log phase in media containing glucose or glycerol as carbon source. Cells were fixed in 2% glutaraldehyde, 3% formaldehyde overnight at 4°C and treated as described (Prescianotto-Baschong and Riezman, 2002). Sections were analyzed by transmission electron microscope. Arrows point at the morphological structures observed in Δema19 cells. More examples and enlarged images are shown in Supplemental Figure S4. (C, D) The content of lipids in whole cell or mitochondrial extracts was analyzed by mass spectrometry and is shown here as the percentage of total lipid mass. CDP-DAG, cytidine diphosphate diacylglycerol; CL, cardiolipin; PA, phosphatidic acid, PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine.
The absence of Ema19 leads to increased intracellular levels of some mitochondrial proteins
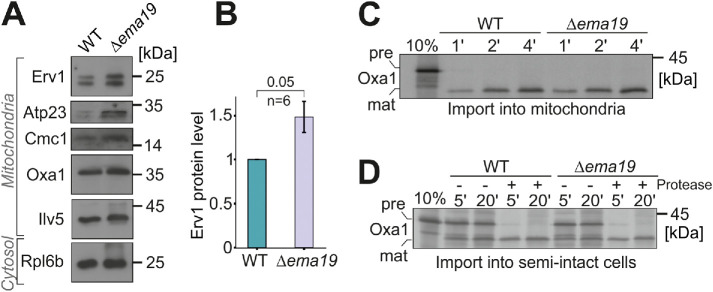
Since Ema19 loss affected Oxa1 fusion protein accumulation in the cytosol, we explored the effect of losing Ema19 on the levels of several mitochondrial proteins from various subcompartments (Figure 4A). We observed that the abundance of most tested mitochondrial proteins was unchanged. However, the levels of the IMS protein Erv1 were considerably increased by about 50% (Figure 4B; Supplemental Figure S6A). Unfortunately, we were not able to test whether Ema19 influences the import reaction of Erv1, as in our hands, in vitro synthesized Erv1 was not imported into isolated mitochondria. However, we tested the import reaction of several other proteins, including Oxa1, Mrpl40, Cox19, or Cmc1, using isolated mitochondria or semi-intact cells in in vitro import reactions (Hansen et al., 2018; Laborenz et al., 2019). WT and Δema19 mitochondria imported these proteins with the same efficiency (Figure 4, C and D; Supplemental Figure S6, B–G). This suggests that Ema19 has no direct relevance for the protein import into mitochondria, but influences mitochondrial protein biogenesis on another step.
FIGURE 4:
Δema19 cells show increased levels of Erv1. (A, B) The levels of the indicated proteins in whole cell extracts were analyzed by Western blotting. (C, D) Radiolabeled Oxa1 was incubated with isolated mitochondria or semi-intact cells for the times indicated. Samples were treated with protease and analyzed by SDS–PAGE and autoradiography; pre, precursor; mat, mature.
Ema19 interacts with many membrane proteins of the ER, vacuole, and mitochondria
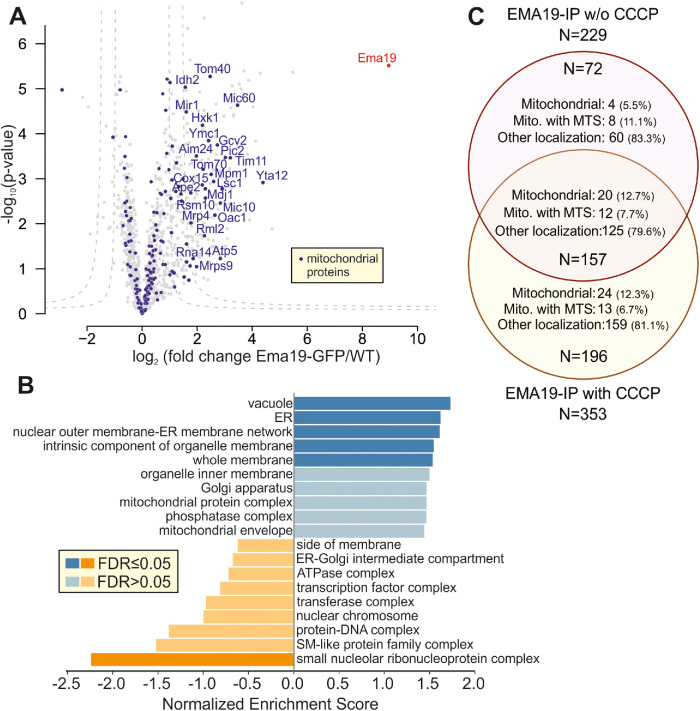
Is Ema19 monomeric or part of a larger complex? To address this question, we made use of the Ema19-GFP strain in which we had observed the ER localization of the protein. WT and Ema19-GFP cells were grown on glucose and lysed with the nonionic detergent NP40. Ema19-GFP was isolated using magnetic GFP-Trap beads and recovered proteins were identified by mass spectrometry (Figure 5A). Ema19 was strongly enriched with the beads confirming the successful affinity purification procedure. No other proteins were similarly enriched, suggesting that Ema19 is not part of a stable heterooligomeric complex. However, many membrane proteins were pulled down with Ema19-GFP (in comparison to the WT extracts), many of which were constituents of the ER, vacuole, and mitochondria (Figure 5B; Supplemental Table S5). The number of mitochondrial proteins that were copurified with Ema19-GFP even further increased when the mitochondrial membrane potential had been dissipated by treatment with the uncoupler carbonylcyanid-m-chlorphenylhydrazon (CCCP) prior to cell lysis (Figure 5C; Supplemental Figure S7). This indicates that Ema19 might particularly interact with those precursor proteins that failed to be imported into mitochondria. Interestingly, the mitochondrial proteins that were recovered with Ema19-GFP included almost exclusively transmembrane proteins such as Tom70, Mic60, Yta12, Tom40, Cox15, Ymc1, Oac1, Pic2, Tim11, Mic10, and Fmp10. This suggests that Ema19 is an interactor of many cellular membrane proteins, including proteins which are not permanent residents of the ER.
FIGURE 5:
Ema19 interacts with many membrane proteins of the ER, vacuole, and mitochondria. (A) Ema19-GFP and WT cells were grown on glucose medium to mid-log phase, harvested and washed. Cells were lysed with glass bead lysis using NP40 as mild detergent. Extracts were cleared by centrifugation and incubated with GFP-Trap beads before bound proteins were analyzed by mass spectrometry. Data are based on four biological replicates for each strain. Enriched proteins are found in the upper right corner of the graph. Significant enrichment was tested with a two-sided t test with permutation-based FDR cut-off to correct for multiple testing (S0 = 4, FDR < 0.05 or FDR < 0.01). (B) Ema19-GFP interactors were analyzed using the WEB-based GEne SeT AnaLysis Toolkit algorithm (Liao et al., 2019) and gene ontology groups are shown which are frequently found with Ema19 (in blue) or which are underrepresented (in orange). (C) The mitochondrial membrane potential influences Ema19 interactions. Cells were treated for 1 h with the uncoupler CCCP to dissipate the mitochondrial membrane potential before cells were harvested and lysed with NP40. Ema19-GFP was isolated and interactors were determined by mass spectrometry. The full dataset is shown in Supplemental Table S5. Venn diagram showing proteins that were significantly enriched in the Ema19-GFP pull-down under the different conditions (S0 = 4, FDR<0.05). Mitochondrial localization according to Morgenstern et al. (Morgenstern et al., 2017); MTS, mitochondrial targeting sequence.
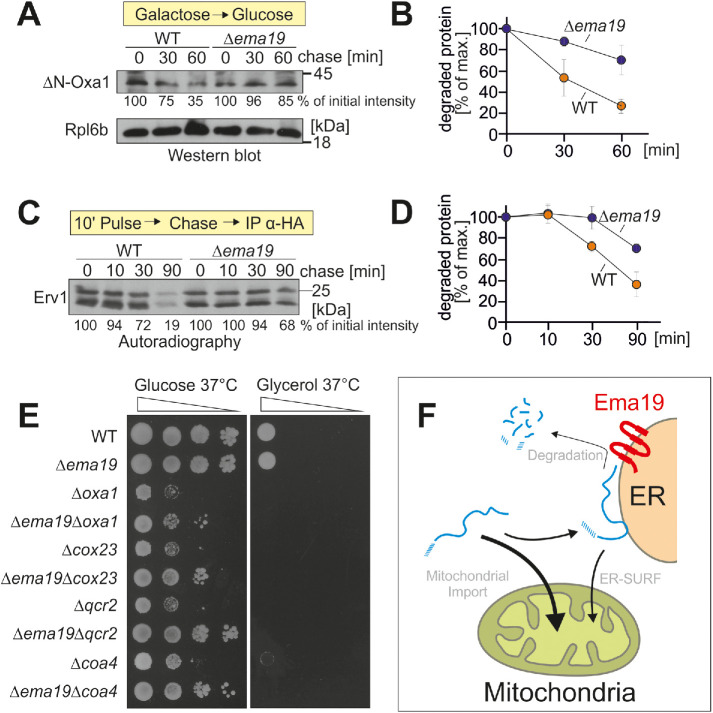
Ema19 promotes the degradation of extramitochondrial precursor proteins
Ema19 might influence the stability of mitochondrial precursor proteins at a stage before their uptake into mitochondria. To test this, we used the established model substrate for an ER-bound precursor protein ΔN-Oxa1 (Hansen et al., 2018). We expressed this protein from a regulatable GAL promoter in WT and Δema19 cells in galactose-containing medium. Then we transferred the cells to glucose-containing medium to switch off its expression and monitored the degradation of the ΔN-Oxa1 over time (Figure 6, A and B). In WT cells, about two-thirds of the ΔN-Oxa1 protein were degraded within 1 h, whereas only a minor fraction of ΔN-Oxa1 was degraded in Δema19 cells. Thus, Ema19 apparently facilitates the recognition or degradation of this nonimportable model protein.
FIGURE 6:
Δema19 cells show reduced protein degradation. (A) The nonimportable ΔN-Oxa1 was expressed in WT and Δema19 cells. Expression was stopped by shifting cells from galactose to glucose for the times indicated (chase). Cells were isolated and the levels of ΔN-Oxa1 were analyzed by Western blotting and quantified. The ribosomal protein Rpl6b served as loading control. (B) The results from three biological replicates were quantified. Mean and standard deviations are shown. (C) Cells expressing HA-tagged Erv1 were grown to log phase. Radiolabeled methionine was added for 10 min (pulse). Cells were incubated in the presence of nonradioactive methionine (chase) for the times indicated. Cells were isolated, lysed, and used for immunoprecipitation with HA-specific antibodies (IP α-HA). The levels of the radiolabeled Erv1-HA protein were detected by autoradiography. (D) The results from three biological replicates were quantified. Mean and standard deviations are shown. (E) Cells of the indicated strains were grown on galactose and dropped onto glucose- or glycerol-containing plates. (F) Model for the role of Ema19 in the degradation of ER-associated mitochondrial precursor proteins.
Next, we asked whether a similar stabilizing effect can also be seen for Erv1. To this end, we expressed Erv1 with a hemagglutinin (HA) tag in WT and Δema19 cells and radiolabeled newly synthesized proteins with 35S methionine for 10 min. Then, cells were washed and transferred to nonradioactive medium.
Erv1 was isolated by immunoprecipitation and its levels were visualized by autoradiography (Figure 6, C and D). Again, we observed a considerably lower turnover in Δema19 cells than in WT cells.
On the basis of systematic genetic screens, it was proposed that EMA19 interacts genetically with many genes coding for subunits of the respiratory chain (Costanzo et al., 2010). This observation inspired us to analyze the growth of double mutants lacking EMA19 and genes required for respiration, such as OXA1, COX23, QCR2, or COA4 (Figure 6E). None of these mutants were able to grow on the nonfermentable carbon source glycerol. However, very surprisingly, we observed that the absence of Ema19 improved growth of all of these double mutants on glucose, particularly at 37°C. Thus, the absence of Ema19 apparently suppresses some of the defects in these respiration-incompetent strains, suggesting that it may allow a longer window of opportunity for precursors to enter mitochondria under limiting conditions.
DISCUSSION
The molecular reactions by which mitochondrial proteins are recognized on the mitochondrial surface and threaded through the protein-conducting channels of the mitochondrial protein translocases were elucidated in great detail. These fascinating mechanistic insights were obtained from extremely powerful in vitro assays with isolated mitochondria to which radiolabeled precursor proteins were mixed. The posttranslational mode of the import reaction, the stability of mitochondrial membranes, the efficiency of the reaction for most mitochondrial proteins tested, and the possibility to regulate and stage the reaction by changing the energetic conditions made this in vitro assay superior over other approaches. However, unfortunately, this assay is largely blind to the initial steps that occur in between protein synthesis and precursor binding to the TOM complex under physiological in vivo conditions where other cellular membranes and structures are crowding the cell.
A number of recent studies reported alternative approaches to follow the import reaction, which resulted in surprising observations: 1) ribosome profiling revealed that cytosolic chaperones and the signal recognition particle play crucial roles in distinguishing mitochondrial and secretory proteins already at very early steps in their synthesis (Schibich et al., 2016; Doring et al., 2017; Costa et al., 2018); 2) proximity labeling suggested that some mitochondrial proteins, in particular hydrophobic inner membrane proteins, explore the mitochondrial surface already during their synthesis (Jan et al., 2014; Williams et al., 2014; Vardi-Oknin and Arava, 2019; Wang et al., 2019) and that, in vivo, many (if not most) mitochondrial surface proteins are in direct proximity to the ER (Hung et al., 2017; Cho et al., 2020); 3) systematic screens of GFP-tagged protein libraries showed that many mitochondrial proteins are prone to accumulate in nonmitochondrial locations under certain growth conditions, in particular on the ER and within the nucleus (Vitali et al., 2018; Backes et al., 2020; Saladi et al., 2020; Shakya et al., 2021; Xiao et al., 2020) and, maybe even more surprising, observed nonmitochondrial residents in mitochondria (Ruan et al., 2017; Bader et al., 2020); and 4) genetic screens reported a very close cooperation of the mitochondrial and ER surface in protein biogenesis (Kornmann et al., 2009; Papic et al., 2013; Okreglak and Walter, 2014; Gamerdinger et al., 2015; Wohlever et al., 2017; Hansen et al., 2018; Vitali et al., 2018; Dederer et al., 2019; Matsumoto et al., 2019). Thus, in vivo, the surfaces of the ER and of mitochondria apparently vividly cooperate to sort proteins to the correct intracellular location.
Ema19 was identified in such a genetic screen for proteins which prevent the cytosolic accumulation of the Oxa1-Ura3 fusion protein (Hansen et al., 2018). The data shown in this study suggest that Ema19 is a protein of the ER membrane that is critical for the degradation of mitochondrial precursor proteins (Figure 6F). It is not clear whether Ema19 interacts with these precursors directly. However, the large number of mitochondrial proteins that were copurified with Ema19-GFP, in particular after dissipation of the mitochondrial membrane potential which blocks mitochondrial import, indicates that Ema19 serves as ER receptor for these stranded proteins. At least three pathways are known which lead to the degradation of ER-associated mitochondrial precursor proteins: 1) the components of the ER-associated degradation system as well as Cdc48 (VCP/p97 in mammals) are known to facilitate the proteasomal degradation of these proteins (Dederer et al., 2019; Matsumoto et al., 2019). 2) Autophagy was shown to play an important role in clearing off nonproductive proteins from the ER surface (Loi et al., 2019; Schäfer et al., 2020). 3) Two recent studies discovered the ER protein Spf1 (P5A-ATPase in mammals) as a transmembrane helix dislocase that extracts missorted mitochondrial tail-anchored proteins from the ER membrane (McKenna et al., 2020; Qin et al., 2020). Interestingly, Spf1 was among the proteins that we coisolated with Ema19-GFP (see Figure 5A) and Ema19 might support the Spf1-mediated extraction of mitochondrial precursor proteins.
Substrates of the Mia40-dependent import pathway are more slowly imported than matrix proteins and their cytosolic forms are often rather stable (Glerum et al., 1996; Fischer et al., 2013; Kowalski et al., 2018; Habich et al., 2019; Mohanraj et al., 2019; Murschall et al., 2020). This might explain why we found higher levels of the Mia40 substrate Erv1 (Kallergi et al., 2012) but not of other mitochondrial proteins we tested. The mitochondrial import machinery and the cytosolic degradation system compete for these Mia40 substrates (Bragoszewski et al., 2017; Kowalski et al., 2018; Mohanraj et al., 2019). Consistent with this idea, we observed that in the context of respiration-incompetent mutants, where the low membrane potential renders protein import inefficient, the deletion of Ema19 increases the growth. Apparently, reducing the degradation of mitochondrial precursors increases the chance of these proteins to be imported.
Ema19 belongs to a protein family which is conserved among eukaryotes. The human ortholog TMEM97 or sigma 2 receptor has been implicated to play critical roles in multiple cellular dysfunctions including tumor formation, inflammation, and neurodegeneration (Tesei et al., 2018; Schmidt and Kruse, 2019; Zeng et al., 2020) potentially caused by disturbed cholesterol homeostasis (Bartz et al., 2009).
Sigma receptors were identified as binding sites for a number of pharmaceuticals, but physiological ligands are not known. Although the anti-proliferating effect of these pharmaceuticals and their potential as anticancer drugs raised much interest, the physiological role of sigma factors is not clear. The sigma 1 receptor is a single-spanning membrane protein that forms homotrimers in the ER (Schmidt et al., 2016). It is enriched at the mitochondria-associated membrane fraction of the ER and was proposed to serve as “membrane chaperone,” though molecular details about its molecular activity are unknown (Hayashi et al., 2009; Fukunaga et al., 2015; Delprat et al., 2020). Even less is known about the sigma 2 receptor, which has no structural similarity with sigma 1 receptors but is targeted by the same pharmaceuticals. Its molecular nature was only unraveled a few years ago and found to be identical to the ER protein TMEM97 (Alon et al., 2017). Several molecular processes were linked to sigma 2 receptor activity, including sterol transport, apoptosis, mitochondrial ROS production, membrane protein trafficking, and the binding and sequestration of misfolded proteins (Bartz et al., 2009; Schmidt and Kruse, 2019). Due to the latter activity, the sigma receptors were also referred to as “ER stress gatekeepers” (Tesei et al., 2018). This function is also supported by our observations in this study, which suggest that the yeast homologue of the sigma 2 receptor, Ema19, is critical for the removal of mitochondrial precursor proteins on the ER surface. Like TMEM97, the mitoprotein extractor of the ER, Spf1/P5A-ATPAse, was found to influence sterol homeostasis and its molecular function was elusive until very recently (Cronin et al., 2000; Sorensen et al., 2015, 2019). It will be exciting to use yeast as a model to elucidate the function of the Ema19/TMEM97/sigma 2 receptor protein family in more detail. More globally, our studies provide another example of the importance of ER membrane proteins in ensuring the fidelity of mitochondrial protein import.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used in this study are based on the WT strains YPH499 (MATa ura3 lys2 ade2 trp1 his3 leu2). However, for some experiments, BY4742 (MATα his3 leu2 lys2 ura3) or W303 (MATa ura3 ade2 trp1 his3 leu2) were used (see Supplemental Tables S1, S2). The plasmids with the superfolder splitGFP sequences were cloned by Gibson assembly (Gibson et al., 2009). To generate the plasmids, the coding regions of Oxa1 (residues 1–402) or ΔN-Oxa1 (residues 43–402) were amplified from genomic DNA introducing the sequence for GFP11 (Smoyer et al., 2016) into the downstream primer and ligated into the EcoR1 and Sma1 sites of a pYX142 vector. We had obtained the sequence for the GFP11 part from the information to the pRS315pr-GFP11-mCherry-PUS1 plasmid on the Addgene webpage. To generate the Sec63-GFP1-10 the coding region of Sec63 was amplified from genomic DNA and fused upstream of the sequence for GFP1-10 that was amplified from pSJ2039 into the EcoR1 and Sma1 sites of a pYX122 plasmid. pSJ2039 (pRS316-NOP1pr-GFP1-10-SCS2TM) was a gift from Sue Jaspersen.
Strains were grown in yeast complete medium (1% yeast extract, 2% peptone), containing 2% of the carbon sources galactose, glucose, or glycerol as indicated. Strains containing plasmids were grown at 30°C in minimal synthetic medium containing 0.67% yeast nitrogen base and 2% glucose or 2% lactate as carbon source.
Superfolder split-GFP assay
Cells containing either the plasmids pYX142-Oxa1-GFP11 and pYX122-Sec63-GFP10 or the plasmids pYX142-ΔN-Oxa1-GFP11 and pYX122-Sec63-GFP10 were grown in selective medium containing 2% glucose to mid-log phase; 3 OD600 were harvested, resuspended in 100 µl selective medium containing 2% glucose, transferred into a black 96 well plate, and centrifuged (5 min at 30 × g). The fluorescence was measured at 480 nm with a fluorescence microplate reader (Clariostar, BMG LABTECH).
∆N-Oxa1 degradation assay
Yeast strains containing a pYX223-∆N-Oxa1-HA plasmid were grown in selective media containing 2% lactate. Expression of ∆N-Oxa1-HA was induced with addition of 0.5% galactose for 4 h. The cells were then shifted to selective medium containing 2% glucose. Whole-cell lysates were taken over time and visualized by Western blotting (see Table S3 for antibodies).
Preparation of semi-intact cells
The protocol for the preparation of semi-intact cells was previously described in Laborenz et al. (2019). Cells were grown in full medium containing 2% galactose at 30°C and grown to mid-log phase. Cells were harvested (700 g, 7 min, RT). The pellets were resuspended in 25 ml SP1 buffer (10 mM dithiothreitol [DTT], 100 mM Tris pH unadjusted) and incubated for 10 min at 30°C. After centrifugation (5 min at 1000 × g), cells were resuspended in 6 ml SP2 buffer (0.6 M sorbitol, 1× YP, 0.2% glucose, 50 mM KPi, pH 7.4, 3 mg/g wet weight zymolyase) and incubated at 30°C for 30–60 min. Spheroplast formation was monitored every 15 min. After generation of the spheroplast, they were centrifuged and resuspended in 40 ml SP3 buffer (1× YP, 1% glucose, 0.7 M sorbitol) and again incubated for 20 min at 30°C. The samples were centrifuged (5 min at 1000 × g) and washed two times with 20 ml of ice-cold permeabilization buffer (20 mM HEPES, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2, 0.4 M sorbitol). The pellet was resuspended in permeabilization buffer containing 0.5 mM EGTA. Aliquots were slowly frozen over liquid nitrogen for 30 min and then stored at –80°C.
Protein import into semi-intact cells
The protocol for the import of radiolabeled proteins into semi-intact cells was previously described in Laborenz et al. (2019). The cells were thawed on ice and the OD600 was measured. An OD600 0.5 per reaction was used. To prepare radiolabeled (35S methionine) proteins for import experiments, the TNT Quick Coupled Transcription/Translation Kit from Promega was used. Semi-intact cells were resuspended in B88 buffer (20 mM HEPES, pH 6.8, 250 mM sorbitol, 150 mM KOAc, 5 mM Mg(OAc)2) containing 2 mM ATP, 2 mM NADH, 5 mM creatine phosphate, 100 µg/ml creatine phosphatase, and 1 µl per reaction radiolabeled lysate. The mixture was incubated for 10 min on ice to allow the cells to take up the lysate. The samples were then incubated at 30°C and samples were taken over time. These samples were directly added into 900 µl B88 buffer containing 50 µM CCCP to stop import and then treated with or without protease for 30 min on ice. Protein digestion was stopped by adding 2 mM phenylmethylsulfonyl fluoride (PMSF). The samples were centrifuged (5 min, 4000 × g, 4°C) and washes with B88 containing 2 mM PMSF. After a last centrifugation step (10 min, 25,000 × g, 4°C), the samples were resuspended in laemmli containing 50 mM DTT. After boiling at 96°C for 3 min, the samples were loaded on an SDS–PAGE, and the gel was blotted onto a nitrocellulose membrane and visualized by autoradiography.
Pulse-chase labeling of cells and immunoprecipitation
For in vivo labeling of translated proteins, cells containing pYX232-Erv1-HA were grown in selective medium without methionine, cysteine, and tryptophane containing 2% galactose to mid-log phase. After washing, the cells with selective medium without amino acids containing 2% galactose, cells were resuspended in the same medium. Amino acids and 1 µl per reaction radiolabeled methionine were added and incubated for 10 min at 30°C. After chasing the radiolabeled methionine with ice-cold methionine and cysteine, samples were taken after different time points. To stop all reactions, the samples were centrifuged and the pellet stored on ice. Trichloracidic acid (TCA) was used for protein precipitation. The pellets were supplemented with 12% TCA and incubated at –80°C for 2 h. The frozen samples were then thawed at room temperature and centrifuged (20 min at 30,000 × g). The pellet was washed with ice-cold 100% acetone and again centrifuged (20 min at 30,000 × g). The pellet was dried 5 min at 30°C and resuspended in lysis buffer (2.5% Triton X-100, 30 mM Tris, pH 8, 100 mM NaCl). The lysate was incubated on ice for 30 min and afterward centrifuged (60 min at 30,000 × g). For immunoprecipitation the supernatant was added to 30 µl equilibrated Sepharose A beads and 3 µl HA antibody and tumbled end-over-end overnight at 4°C. The beads were centrifuged (1 min at 2000 × g) and the supernatant was kept and stored. The beads were washed twice with lysis buffer, twice with wash buffer I (2.5% Triton X-100, 30 mM Tris, pH 8, 100 mM NaCl, 1 M urea), and twice with wash buffer II (30 mM Tris, pH 8, 100 mM NaCl). The beads were resuspended in laemmli buffer containing 50 mM DTT and boiled for 3 min at 96°C.
Identification of Ema19-associated proteins
For co-immunoprecipitation and mass spectrometry, cells were grown in full medium containing 2% glucose to mid-log phase. For samples in which the mitochondrial membrane potential was dissipated, 100 µM CCCP was added for 1 h; 10 OD600 were harvested and washed with water. The supernatant was removed and the pellet resuspended in lysis buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40) and lysed with glass bead lysis using a FastPrep-24 5 G homogenizer (MP Biomedicals) with 3 cycles of 30 s, speed 8.0 m s–1, 120-s breaks. The lysate was centrifuged (2 min at 13,000 × g, 4°C). The supernatant was transferred to a precooled tube and 300 µl dilution buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) were added; 25 µl equilibrated magnetic GFP-Trap A beads were added to the lysate and tumbled end-over-end for 1 h at 4°C. Afterward, the supernatant was discarded. The beads were then washed three times with 800 µl wash buffer I (150 mM NaCl, 50 mM Tris, pH 7.5, 5% glycerol, 0.05% NP40). After adding wash buffer I for the first time, the beads were transferred to a new tub to get rid of the detergent. Then the beads were washed with 500 µl wash buffer II (150 mM NaCl, 50 mM Tris, pH 7.5, 5% glycerol). After wash buffer II was removed completely, 50 µl elution buffer I (2 M urea, 50 mM Tris, pH 7.5, 1 mM DTT, 5 ng/µl trypsin) was added to the beads for an on-bead digest and incubated for 1 h at room temperature; 15 ng/µl fresh trypsin was added for another 10 min at room temperature. Afterward, the supernatant containing the peptides was transferred to a fresh tube; 50 µl elution buffer II (2 M urea, 50 mM Tris, pH 7.5, 5 mM chloroacetamide) was added for alkylating the peptides overnight in the dark at room temperature. For purification of peptides on C18 stage tips, the stage tips were washed with 100 µl methanol (5 min at 500 × g), 100 µl buffer B (0.1% formic acid, 80% acetonitrile in MS grade water), and then with 100 µl buffer A (0.1% formic acid in MS grade water). To acidify the samples, they were treated with TFA to get a pH < 2. The acidified peptides were then added onto the stage tips and centrifuged (5 min at 500 g). After a last wash step with 100 µl buffer A, the peptides were eluted with 60 µl buffer B (5 min, 500 × g). The samples were dried in a speed vac. Then, 9 µl buffer A and 1 µl buffer A* (0.1% formic acid, 0.1% TFA in MS grade water) were added and the samples were used for mass spectrometry.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD022660.
FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2021/03/PXD022660
Miscellaneous
The following methods were used as described: lipid analysis of yeast cells (Aaltonen et al., 2016; Velazquez et al., 2016); electron microscopy, isolation of mitochondria, and import of radiolabeled proteins (Laborenz et al., 2019); mass spectrometry (Backes et al., 2020; Saladi et al., 2020).
Supplementary Material
Acknowledgments
We thank Sabine Knaus and Lena Krämer for technical assistance, Katja Hansen for discussion and advice, and Sue Jasperson for the plasmids and protocols for the splitGFP screen. This project was funded by grants from the Deutsche Forschungsgemeinschaft (DIP MitoBalance to M.S., T.L., and J.M.H. and IRTG1830 to J.M.H.) and the Landesschwerpunkt BioComp (to Z.S. and J.M.H.). Y.S.B. was supported by an EMBO long-term fellowship. M.S. is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Abbreviations used:
- CCCP
carbonylcyanid-m-chlorphenylhydrazon
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- HA
hemagglutinin
- PMSF
phenylmethylsulfonyl fluoride
- TCA
trichloroacetic acid
- TOM
translocase of the outer membrane
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-11-0748) on February 17, 2021.
REFERENCES
- Aaltonen MJ, Friedman JR, Osman C, Salin B, di Rago JP, Nunnari J, Langer T, Tatsuta T (2016). MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria. J Cell Biol 213, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon A, Schmidt HR, Wood MD, Sahn JJ, Martin SF, Kruse AC (2017). Identification of the gene that codes for the sigma2 receptor. Proc Natl Acad Sci USA 114, 7160–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araiso Y, Tsutsumi A, Qiu J, Imai K, Shiota T, Song J, Lindau C, Wenz LS, Sakaue H, Yunoki K, et al. (2019). Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575, 395–401. [DOI] [PubMed] [Google Scholar]
- Avendano-Monsalve MC, Ponce-Rojas JC, Funes S (2020). From cytosol to mitochondria: the beginning of a protein journey. Biol Chem 401, 645–661. [DOI] [PubMed] [Google Scholar]
- Backes S, Bykov YS, Räschle M, Zhou J, Lenhard S, Krämer L, Mühlhaus T, Bibi C, Jann C, Smith JD, et al. (2020). The mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. bioRxiv, https//.org/10.1101/2020.09.14.296194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G, Enkler L, Araiso Y, Hemmerle M, Binko K, Baranowska E, De Craene JO, Ruer-Laventie J, Pieters J, Tribouillard-Tanvier D, et al. (2020). Assigning mitochondrial localization of dual localized proteins using a yeast bi-genomic mitochondrial-split-GFP. Elife 9, e56649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, Erfle H, Muckenthaler M, Pepperkok R, Runz H (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab 10, 63–75. [DOI] [PubMed] [Google Scholar]
- Boos F, Kramer L, Groh C, Jung F, Haberkant P, Stein F, Wollweber F, Gackstatter A, Zoller E, van der Laan M, et al. (2019). Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat Cell Biol 21, 442–451. [DOI] [PubMed] [Google Scholar]
- Boos F, Labbadia J, Herrmann JM (2020). How the mitoprotein-induced stress response safeguards the cytosol: a unified view. Trends Cell Biol 30, 241–254. [DOI] [PubMed] [Google Scholar]
- Bragoszewski P, Turek M, Chacinska A (2017). Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol 7, 170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov YS, Rapaport D, Herrmann JM, Schuldiner M (2020). Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem Sci 45, 650–667. [DOI] [PubMed] [Google Scholar]
- Callegari S, Cruz-Zaragoza LD, Rehling P (2020). From TOM to the TIM23 complex - handing over of a precursor. Biol Chem 401, 709–721. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK (2016). MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44, D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee HW, Lee IK, Carr SA, Ting AY (2020). Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci USA 117, 12143–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa EA, Subramanian K, Nunnari J, Weissman JS (2018). Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. (2010). The genetic landscape of a cell. Science 327, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Khoury A, Ferry DK, Hampton RY (2000). Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol 148, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dederer V, Khmelinskii A, Huhn AG, Okreglak V, Knop M, Lemberg MK (2019). Cooperation of mitochondrial and ER factors in quality control of tail-anchored proteins. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprat B, Crouzier L, Su TP, Maurice T. (2020). At the crossing of ER stress and MAMs: A key role of sigma-1 receptor? Adv Exp Med Biol 1131, 699–718. [DOI] [PubMed] [Google Scholar]
- Doring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O’Brien EP, Kramer G, Bukau B (2017). Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170, 298–311 e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drwesh L, Rapaport D. (2020). Biogenesis pathways of alpha-helical mitochondrial outer membrane proteins. Biol Chem 401, 677–686. [DOI] [PubMed] [Google Scholar]
- Edwards R, Gerlich S, Tokatlidis K (2020). The biogenesis of mitochondrial intermembrane space proteins. Biol Chem 401, 737–747. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S (2011). Structural insight into the mitochondrial protein import system. Biochim Biophys Acta 1808, 955–970. [DOI] [PubMed] [Google Scholar]
- Fischer M, Horn S, Belkacemi A, Kojer K, Petrungaro C, Habich M, Ali M, Kuttner V, Bien M, Kauff F, et al. (2013). Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol Biol Cell 24, 2160–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Shinoda Y, Tagashira H (2015). The role of SIGMAR1 gene mutation and mitochondrial dysfunction in amyotrophic lateral sclerosis. J Pharmacol Sci 127, 36–41. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E (2015). The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348, 201–207. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD (1994). Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol 127, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A (1996). Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 271, 14504–14509. [DOI] [PubMed] [Google Scholar]
- Golani-Armon A, Arava Y (2016). Localization of nuclear-encoded mRNAs to mitochondria outer surface. Biochemistry (Mosc) 81, 1038–1043. [DOI] [PubMed] [Google Scholar]
- Habich M, Salscheider SL, Murschall LM, Hoehne MN, Fischer M, Schorn F, Petrungaro C, Ali M, Erdogan AJ, Abou-Eid S, et al. (2019). Vectorial import via a metastable disulfide-linked complex allows for a quality control step and import by the mitochondrial disulfide relay. Cell Rep 26, 759–774 e755. [DOI] [PubMed] [Google Scholar]
- Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, Schuldiner M, Herrmann JM (2018). An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361, 1118–1122. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP (2009). MAM: more than just a housekeeper. Trends Cell Biol 19, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993). TMbase - A database of membrane spanning proteins segments. Biol Chem 374, 166. [Google Scholar]
- Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, Ting AY (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6, e24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS (2014). Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallergi E, Andreadaki M, Kritsiligkou P, Katrakili N, Pozidis C, Tokatlidis K, Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, et al. (2012). Targeting and maturation of Erv1/ALR in the mitochondrial intermembrane space. ACS Chem Biol 7, 707–714. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski L, Bragoszewski P, Khmelinskii A, Glow E, Knop M, Chacinska A (2018). Determinants of the cytosolic turnover of mitochondrial intermembrane space proteins. BMC Biol 16, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborenz J, Hansen K, Prescianotto-Baschong C, Spang A, Herrmann JM (2019). In vitro import experiments with semi-intact cells suggest a role of the Sec61 paralog Ssh1 in mitochondrial biogenesis. Biol Chem 400, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi M, Raimondi A, Morone D, Molinari M (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun 10, 5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Goldberg J (2013). Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J 32, 926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C (2002). Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Nakatsukasa K, Kakuta C, Tamura Y, Esaki M, Endo T (2019). Msp1 clears mistargeted proteins by facilitating their transfer from mitochondria to the ER. Mol Cell 76, 191–205. [DOI] [PubMed] [Google Scholar]
- McKenna MJ, Sim SI, Ordureau A, Wei L, Harper JW, Shao S, Park E (2020). The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369, eabc5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K, Wasilewski M, Beninca C, Cysewski D, Poznanski J, Sakowska P, Bugajska Z, Deckers M, Dennerlein S, Fernandez-Vizarra E, et al. (2019). Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7. EMBO Mol Med 11, e9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D (2020). How to get to the other side of the mitochondrial inner membrane - the protein import motor. Biol Chem 401, 723–736. [DOI] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lubbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Hoss P, Feuerstein R, Gebert M, et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19, 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murschall LM, Gerhards A, MacVicar T, Peker E, Hasberg L, Wawra S, Langer T, Riemer J (2020). The C-terminal region of the oxidoreductase MIA40 stabilizes its cytosolic precursor during mitochondrial import. BMC Biol 18, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Walter P (2014). The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci USA 111, 8019–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paasch F, den Brave F, Psakhye I, Pfander B, Jentsch S (2018). Failed mitochondrial import and impaired proteostasis trigger SUMOylation of mitochondrial proteins. J Biol Chem 293, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic D, Elbaz-Alon Y, Koerdt SN, Leopold K, Worm D, Jung M, Schuldiner M, Rapaport D. (2013). The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol Cell Biol 33, 4083–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong C, Riezman H (2002). Ordering of compartments in the yeast endocytic pathway. Traffic 3, 37–49. [DOI] [PubMed] [Google Scholar]
- Qin Q, Zhao T, Zou W, Shen K, Wang X. (2020). An endoplasmic reticulum ATPase safeguards endoplasmic reticulum identity by removing ectopically localized mitochondrial proteins. Cell Rep 33, 108363. [DOI] [PubMed] [Google Scholar]
- Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, Li R (2017). Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladi S, Boos F, Poglitsch M, Meyer H, Sommer F, Muhlhaus T, Schroda M, Schuldiner M, Madeo F, Herrmann JM (2020). The NADH dehydrogenase Nde1 executes cell death after integrating signals from metabolism and proteostasis on the mitochondrial surface. Mol Cell 77, 189–202 e186. [DOI] [PubMed] [Google Scholar]
- Schäfer JA, Schessner JP, Bircham PW, Tsuji T, Funaya C, Pajonk O, Schaeff K, Ruffini G, Papagiannidis D, Knop M, et al. (2020). ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J 39, e102586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibich D, Gloge F, Pohner I, Bjorkholm P, Wade RC, von Heijne G, Bukau B, Kramer G (2016). Global profiling of SRP interaction with nascent polypeptides. Nature 536, 219–223. [DOI] [PubMed] [Google Scholar]
- Schmidt HR, Kruse AC (2019). The molecular function of sigma receptors: past, present, and future. Trends Pharmacol Sci 40, 636–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, Kruse AC (2016). Crystal structure of the human sigma1 receptor. Nature 532, 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A (2020). Evolution of mitochondrial protein import - lessons from trypanosomes. Biol Chem 401, 663–676. [DOI] [PubMed] [Google Scholar]
- Shakya VP, Barbeau WA, Xiao T, Knutson CS, Schuler MH, Hughes AL. (2021). A nuclear-based quality control pathway for non-imported mitochondrial proteins. Elife 10, e61230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, et al. (2015). Molecular architecture of the active mitochondrial protein gate. Science 349, 1544–1548. [DOI] [PubMed] [Google Scholar]
- Smoyer CJ, Katta SS, Gardner JM, Stoltz L, McCroskey S, Bradford WD, McClain M, Smith SE, Slaughter BD, Unruh JR, Jaspersen SL (2016). Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J Cell Biol 215, 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Herrmann JM, Becker T (2021). Quality control of the mitochondrial proteome. Nat Rev Mol Cell Biol, 22, 54–70. [DOI] [PubMed] [Google Scholar]
- Sorensen DM, Holen HW, Holemans T, Vangheluwe P, Palmgren MG (2015). Towards defining the substrate of orphan P5A-ATPases. Biochim Biophys Acta 1850, 524–535. [DOI] [PubMed] [Google Scholar]
- Sorensen DM, Holen HW, Pedersen JT, Martens HJ, Silvestro D, Stanchev LD, Costa SR, Gunther Pomorski T, Lopez-Marques RL, Palmgren M (2019). The P5A ATPase Spf1p is stimulated by phosphatidylinositol 4-phosphate and influences cellular sterol homeostasis. Mol Biol Cell 30, 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesei A, Cortesi M, Zamagni A, Arienti C, Pignatta S, Zanoni M, Paolillo M, Curti D, Rui M, Rossi D, Collina S (2018). Sigma receptors as endoplasmic reticulum stress “gatekeepers” and their modulators as emerging new weapons in the fight against cancer. Front Pharmacol 9, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi-Oknin D, Arava Y (2019). Characterization of factors involved in localized translation near mitochondria by ribosome-proximity labeling. Front Cell Dev Biol 7, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M (2016). Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol 212, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali DG, Sinzel M, Bulthuis EP, Kolb A, Zabel S, Mehlhorn DG, Figueiredo Costa B, Farkas A, Clancy A, Schuldiner M, et al. (2018). The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- Wang P, Tang W, Li Z, Zou Z, Zhou Y, Li R, Xiong T, Wang J, Zou P (2019). Mapping spatial transcriptome with light-activated proximity-dependent RNA labeling. Nat Chem Biol 15, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen XJ (2015). A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. (2018). MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 360, eaan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS (2014). Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlever ML, Mateja A, McGilvray PT, Day KJ, Keenan RJ (2017). Msp1 is a membrane protein dislocase for tail-anchored proteins. Mol Cell 67, 194–202 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. (2015). Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488. [DOI] [PubMed] [Google Scholar]
- Xiao T, Shakya VP, Hughes AL. (2021). ER targeting of non-imported mitochondrial carrier proteins is dependent on the GET pathway. Life Sci Alliance 4, e202000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Riad A, Mach RH (2020). The Biological Function of Sigma-2 Receptor/TMEM97 and Its Utility in PET Imaging Studies in Cancer. Cancers (Basel) 12, 1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller E, Laborenz J, Kramer L, Boos F, Raschle M, Alexander RT, Herrmann JM (2020). The intermembrane space protein Mix23 is a novel stress-induced mitochondrial import factor. J Biol Chem 295, 14686–14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD022660.
FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2021/03/PXD022660