Abstract
Genetic and chemical perturbations impact diverse cellular phenotypes, including multiple indicators of cell health. These readouts reveal toxicity and antitumorigenic effects relevant to drug discovery and personalized medicine. We developed two customized microscopy assays, one using four targeted reagents and the other three targeted reagents, to collectively measure 70 specific cell health phenotypes including proliferation, apoptosis, reactive oxygen species, DNA damage, and cell cycle stage. We then tested an approach to predict multiple cell health phenotypes using Cell Painting, an inexpensive and scalable image-based morphology assay. In matched CRISPR perturbations of three cancer cell lines, we collected both Cell Painting and cell health data. We found that simple machine learning algorithms can predict many cell health readouts directly from Cell Painting images, at less than half the cost. We hypothesized that these models can be applied to accurately predict cell health assay outcomes for any future or existing Cell Painting dataset. For Cell Painting images from a set of 1500+ compound perturbations across multiple doses, we validated predictions by orthogonal assay readouts. We provide a web app to browse predictions: http://broad.io/cell-health-app. Our approach can be used to add cell health annotations to Cell Painting datasets.
INTRODUCTION
Perturbing cells with specific genetic and chemical reagents in different environmental contexts impacts cells in various ways (Kitano, 2002). For example, certain perturbations impact cell health by stalling cells in specific cell cycle stages, increasing or decreasing proliferation rate, or inducing cell death via specific pathways (Markowetz, 2010; Szalai et al., 2019). Cell health is normally assessed by eye or measured by specifically targeted reagents, which are either focused on a single cell health parameter (e.g. ATP assays) or multiple in combination via FACS-based or image-based analyses, which involves a manual gating approach, complicated staining procedures, and significant reagent cost. These traditional approaches limit the ability to scale to large perturbation libraries such as candidate compounds in academic and pharmaceutical screening centers.
Image-based profiling assays are increasingly being used to quantitatively study the morphological impact of chemical and genetic perturbations in various cell contexts (Caicedo et al., 2016; Scheeder et al., 2018). One unbiased assay, called Cell Painting, stains for various cellular compartments and organelles using nonspecific and inexpensive reagents (Gustafsdottir et al., 2013). Cell Painting has been used to identify small-molecule mechanisms of action, study the impact of overexpressing cancer mutations, and discover new bioactive mechanisms, among many other applications (Wawer et al., 2014; Rohban et al., 2017; Caicedo et al., 2018; Simm et al., 2018; Christoforow et al., 2019; Pahl and Sievers, 2019; Hughes et al., 2020). Additionally, Cell Painting can predict overall mammalian toxicity levels for environmental chemicals (Nyffeler et al., 2020) and some of its derived morphology measurements are readily interpreted by cell biologists and relate to cell health (Bray et al., 2016). However, no single, inexpensive assay enables discovery of fine-grained cell health readouts that would provide researchers with a more complete understanding of perturbation mechanisms.
We hypothesized that we could predict many cell health readouts directly from the Cell Painting data, which are already available for hundreds of thousands of perturbations. This would enable the rapid and interpretable annotation of small molecules or genetic perturbations. To do this, we first developed two customized microscopy assays, which collectively report on 70 different cell health indicators via a total of seven reagents applied in two reagent panels. Collectively, we call these assays “Cell Health.”
To demonstrate proof of concept, we collected a small pilot dataset of 119 clustered regularly interspersed short palindromic repeats (CRISPR) knockout perturbations in three different cell lines using Cell Painting and Cell Health. We used the Cell Painting morphology readouts to train 70 different regression models to predict each Cell Health indicator independently. We used simple machine learning methods instead of a deep learning approach because of our limited sample size of 119 perturbations and the inability to increase the sample size by linking single cell measurements across assays. We predicted certain readouts, such as the number of S phase cells, with high performance, while performance on other readouts, such as DNA damage in G2 phase cells, was low. We applied and validated these models on a separate set of existing Cell Painting images acquired from 1571 compound perturbations measured across six different doses from the Drug Repurposing Hub project (Corsello et al., 2017). We provide all predictions in an intuitive web-based application at http://broad.io/cell-health-app so that others can extend our work and explore cell health impacts of specific compounds.
RESULTS AND DISCUSSION
We collected Cell Painting images and targeted Cell Health readouts in three different cell lines (A549, ES2, and HCC44), each treated with 119 CRISPR perturbations targeting 59 genes and controls (Supplemental Table S1). We selected these genes to span multiple biological pathways and induce different morphological states. The seven reagents we included in the two Cell Health panels (Supplemental Table S2) include specific stains and antibodies such as Caspase 3/7 dye to target apoptotic cells and γH2Ax antibodies to measure DNA damage.
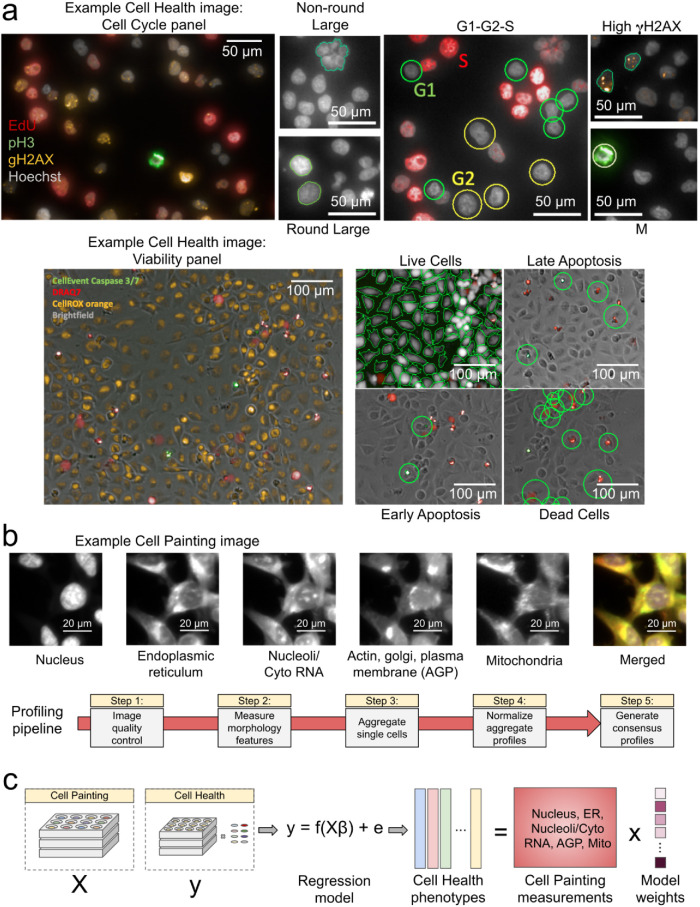
Applying biological knowledge of cell health-related phenotypes and several manual gating strategies, we defined 70 different Cell Health readouts (Supplemental Table S3) based on signals from the seven reagents, plus nucleus morphology measurements from digital phase contrast (DPC) (Figure 1A; Supplemental Figures S1 and S2). While these readouts are relatively easy to interpret, running two separate assays is not ideal for large-scale perturbation screening experiments.
FIGURE 1:
Data processing and modeling approach. (a) Example images and workflow from the Cell Health assays. We apply a series of manual gating strategies (see Materials and Methods) to isolate cell subpopulations and to generate cell health readouts for each perturbation. (Top) In the “Cell Cycle” panel, in each nucleus we measure Hoechst, EdU, PH3, and gH2AX. (Bottom) In the “Viability” panel, we capture DPC images, measure Caspase 3/7, DRAQ7, and CellROX. (b) Example Cell Painting image across five channels, plus a merged representation across channels. The image is cropped from a larger image and shows ES2 cells. Scale bars are 20 µm. Below are the steps applied in an image-based profiling pipeline, after features have been extracted from each cell’s image. (c) Modeling approach where we fit 70 different regression models using CellProfiler features derived from Cell Painting images to predict Cell Health readouts. Model weights refer to the coefficients derived from each regression model.
We developed and applied a bioinformatics pipeline to process features extracted from Cell Painting images by CellProfiler software (Carpenter et al., 2006). The pipeline yields image-based profiles representing gene and guide perturbation signatures (Caicedo et al., 2017) (Figure 1B). We observed that 63% of guide profile replicates were distinguishable from negative controls; that is, they had stronger pairwise correlations than 95% of a null distribution defined by nonreplicate correlations (Supplemental Figure S3). This rate is consistent with previous Cell Painting studies of genetic perturbations (Rohban et al., 2017).
We developed an approach to use the inexpensive reagents from the multiplexed, high-throughput Cell Painting assay to predict Cell Health readouts (Figure 1C). We generated a single “consensus” signature for each guide perturbation across cell lines, producing 357 signatures (3 cell lines × 119 CRISPR guides) with 952 morphology measurements. We independently optimized 70 different elastic net linear regression models using consensus morphology profiles of Cell Painting data to predict each of the 70 Cell Health readouts independently (Figure 1C). The actual identity of the CRISPR guides was not relevant during training.
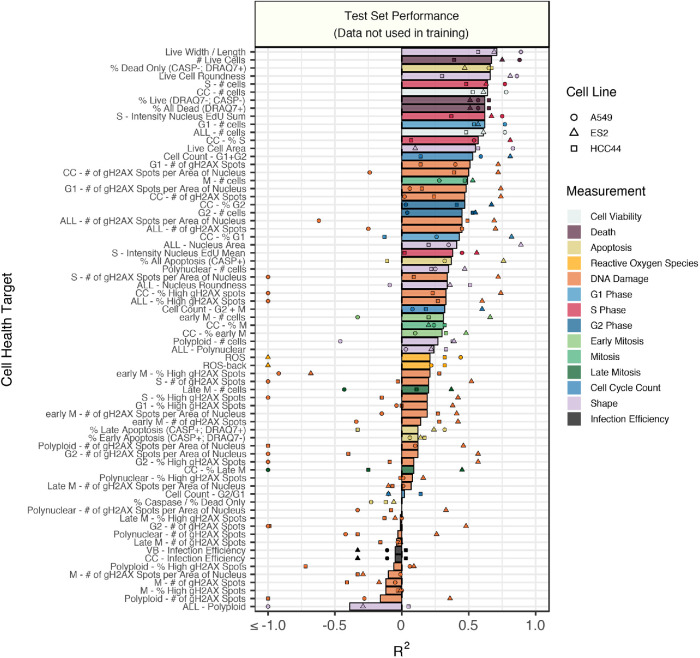
Predictive performance in a held-out test set (a balanced 15% of profiles not used in training) indicates high expected generalizability for many models (Figure 2; Supplemental Figure S4). Performance was better for nearly every model when trained with real data compared with shuffled data, thus beating a random chance baseline (Supplemental Figure S5).
FIGURE 2:
Test set model performance of predicting 70 Cell Health readouts with independent regression models. Performance for each phenotype is shown, sorted by decreasing R2 performance. The bars are colored based on the primary measurement metadata (see Supplemental Table S3), and they represent performance aggregated across the three cell lines. The points represent cell line specific performance. Points falling below –1 are truncated to –1 on the x-axis. See Supplemental Figure S4 for a full depiction.
Many Cell Health readouts were predicted very well, including percentage of dead cells, number of S-phase cells, DNA damage in G1-phase cells, and percentage of apoptotic cells (Supplemental Figure S6A). However, other readouts such as DNA damage in polynuclear cells and percentage of cells in late mitosis could not be predicted better than random (Supplemental Figure S6B). Models derived from different combinations of Cell Health reagents had variable performance, with DRAQ7, shape, and EdU models performing the best (Supplemental Figure S7). Performance differences might result from random technical variation, small sample sizes for training models, different numbers of cells in certain Cell Health subpopulations (e.g., mitosis or polynuclear cells), fewer cells collected in the viability panel (see Materials and Methods), or the inability of Cell Painting reagents to capture certain phenotypes. We observed overall better predictivity in ES2 cells, which had the highest CRISPR infection efficiency (Supplemental Figure S8), suggesting that stronger perturbations provide better information for training and that training on additional data should provide further benefit.
Using a linear model for predictions enables interpretability. For example, inspecting the model for the Cell Health readout Live Cell Area reveals that it relies on cell and cytoplasm shape features from Cell Painting (Supplemental Figure S9). This is expected given that the Live Cell Area readout is derived from cell boundary measurements from the DPC channel. In our approach, each regression model uses a combination of interpretable morphology features to make Cell Health phenotype predictions, unlike so-called “black box” deep learning feature extractors. Therefore, the specific combination of Cell Painting features provides a potentially interpretable morphology signature representing the underlying cell health state.
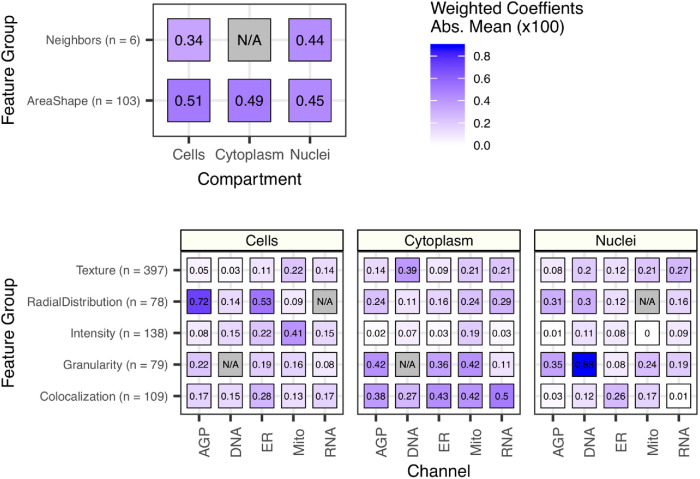
Overall, many different feature classes were important for accurate predictions (Figure 3; Supplemental Figure S10). Some features tended to strongly contribute across multiple Cell Health readouts. For example, particularly informative features include the radial distribution of the actin, golgi, and plasma membrane (AGP) channel in cells and DNA granularity in nuclei. This demonstrates that the Cell Painting assay captures complex cell health phenotypes using a rich variety of morphology feature types.
FIGURE 3:
The importance of each class of Cell Painting features in predicting 70 Cell Health readouts. Each square represents the mean absolute value of model coefficients weighted by test set R2 across every model. The features are broken down by compartment (Cells, Cytoplasm, and Nuclei), channel (AGP, Nucleus, ER, Mito, Nucleolus/Cyto RNA), and feature group (AreaShape, Neighbors, Channel Colocalization, Texture, Radial Distribution, Intensity, and Granularity). The number of features in each group, across all channels, is indicated. For a complete description of all features, see the handbook: http://cellprofiler-manual.s3.amazonaws.com/CellProfiler-3.0.0/index.html. Dark gray squares indicate “not applicable,” meaning either that there are no features in the class or that the features did not survive an initial preprocessing step. Note that for improved visualization we multiplied the actual model coefficient value by 100.
We performed a series of analyses to determine certain parameters and options that are likely to improve models in the future. First, we performed a “cell line holdout” analysis, in which we trained models on two of three cell lines and predicted cell health readouts on the held out cell line. We observed that certain models including those based on viability, S phase, early mitotic, and death phenotypes could be moderately predicted in cell lines agnostic to training (Supplemental Figure S11). Not surprisingly, shape-based phenotypes could not be predicted in holdout cell lines, which emphasizes the limitations of transferring certain cell line intrinsic measurements across cell lines. We also performed a systematic feature removal analysis, in which we retrained cell health models after dropping features that are measured from specific groups, compartments, and channels. We observed that many models were robust to dropping entire feature classes during training (Supplemental Figure S12). This result demonstrates that many Cell Painting features are highly correlated, which might permit prediction “rescue” even if the directly implicated morphology features are not measured. Because of this, we urge caution when generating hypotheses regarding causal relationships between phenotypes and individual Cell Painting features. Last, we performed a sample size titration analysis in which we randomly removed an increasing number of samples from training. For the high- and mid-performing models we observed a consistent performance drop, suggesting that increasing sample size would result in better overall performance (Supplemental Figure S13).
Predictive models of cell health would be most useful if they could be trained once and successfully applied to datasets collected separately from the experiment used for training. Otherwise one could not annotate existing datasets that lack parallel Cell Health results, and Cell Health assays would have to be run alongside each new dataset. We therefore applied our trained models to a large, publicly available Cell Painting dataset collected as part of the Drug Repurposing Hub project (Corsello et al., 2017). The data derive from A549 lung cancer cells treated with 1571 compound perturbations measured in six doses.
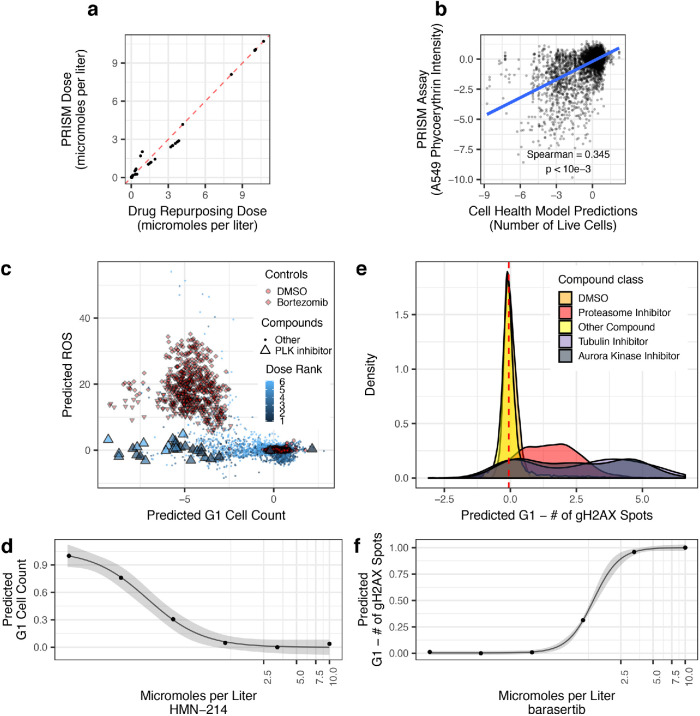
We first chose a high-performing model to validate. The number of live cells model captures the number of cells that are unstained by DRAQ7. We compared model predictions to orthogonal viability readouts from a third dataset: Publicly available PRISM assay readouts, which count barcoded cells after an incubation period (Yu et al., 2016). Despite measuring perturbations with slightly different doses and fundamentally different ways to count live cells (Figure 4A), the predictions correlated with the assay readout (Spearman’s Rho = 0.35, p < 1 × 10–3; Figure 4B).
FIGURE 4:
Validating Cell Health models to Cell Painting data from The Drug Repurposing Hub. The models were not trained using the Drug Repurposing Hub data. (a) The results of the dose alignment between the PRISM assay and the Drug Repurposing Hub data. This view indicates that there was not a one-to-one matching between perturbation doses. (b) Comparing viability estimates from the PRISM assay to the predicted number of live cells in the Drug Repurposing Hub. The PRISM assay estimates viability by measuring barcoded A549 cells after an incubation period. (c) Drug Repurposing Hub profiles stratified by G1 cell count and ROS predictions. Bortezomib and MG-132 are proteasome inhibitors and are used as positive controls in the Drug Repurposing Hub set; DMSO is a negative control. We also highlight all PLK inhibitors in the dataset. (d) HMN-214 is an example of a PLK inhibitor that shows strong dose response for G1 cell count predictions. (e) Tubulin and aurora kinase inhibitors are predicted to have a high number of gH2AX spots in G1 cells compared to other compounds and controls. (f) Barasertib (AZD1152) is an aurora kinase inhibitor that is predicted to have a strong dose response for the number of gH2AX spots in G1 cells predictions.
We also chose to validate three additional models: ROS, G1 cell count, and Number of gH2AX spots in G1 cells. We observed that the two proteasome inhibitors (bortezomib and MG-132) in the Drug Repurposing Hub set yielded high ROS predictions (OR = 76.7; p < 1 × 10–15) (Figure 4C). Proteasome inhibitors are known to induce ROS (Ling et al., 2003; Han and Park, 2010). As well, PLK inhibitors yielded low G1 cell counts (OR = 0.035; p = 3.9 × 10–8) (Figure 4C). The PLK inhibitor HM-214 showed an appropriate dose response (Figure 4D). PLK inhibitors block mitotic progression, thus reducing entry into the G1 cell cycle phase (Lee et al., 2014). Last, we observed that aurora kinase and tubulin inhibitors yielded a high Number of gH2AX spots in G1 cells predictions (OR = 11.3; p < 1 × 10–15) (Figure 4E). In particular, we observed a strong dose response for the aurora kinase inhibitor barasertib (AZD1152) (Figure 4F). Aurora kinase and tubulin inhibitors cause prolonged mitotic arrest, which can lead to mitotic slippage, G1 arrest, DNA damage, and senescence (Orth et al., 2011; Cheng and Crasta, 2017; Tsuda et al., 2017).
We applied uniform manifold approximation (UMAP) to observe the underlying structure of the samples as captured by morphology data (McInnes et al., 2018). We observed that the UMAP space captures gradients in predicted G1 cell count (Supplemental Figure S14A) and in predicted ROS (Supplemental Figure S14B). We also observed similar gradients in the ground truth cell health readouts in the CRISPR Cell Painting profiles used for training cell health models (Supplemental Figure S15). Gradients in our data suggest that cell health phenotypes manifest in a continuum rather than in discrete states.
Last, we observed moderate technical artifacts in the Drug Repurposing Hub profiles, indicated by high DMSO profile dispersion in the Cell Painting UMAP space (Supplemental Figure S14C); this represents an opportunity to improve model predictions with new batch effect correction tools. Additionally, it is important to note that the expected performance of each Cell Health model can only be as good as the performance observed in the original test set (see Figure 2), and that all predictions require further experimental validation.
CONCLUSIONS
We have demonstrated feasibility that information in Cell Painting images can predict many different Cell Health indicators even when trained on a relatively small dataset. The results motivate collecting larger datasets for training, with more perturbations and multiple cell lines. These new datasets would enable the development of more expressive models, based on deep learning, that can be applied to single cells. Including orthogonal imaging markers of CRISPR infection would also enable us to isolate cells with expected morphologies. More data and better models would improve the performance and generalizability of Cell Health models and enable annotation of new and existing large-scale Cell Painting datasets with important mechanisms of cell health and toxicity.
MATERIALS AND METHODS
CRISPR constructs used for knockout
We performed a CRISPR and CRISPR-associated protein 9 (Cas9) knockout experiment to perturb cells (CRISPR-Cas9). We designed guides to target 59 different genes with an average of two guides per gene (Supplemental Table S1). All sgRNAs were selected from the Avana library (Doench et al., 2016; Meyers et al., 2017) or by using CRISPick (https://broad.io/crispick; Hanna and Doench, 2020). Nevertheless, it is important to note that the identity of the CRISPR gene target is not used in training the machine learning models.
Cell lines
We performed CRISPR knockout in three different cell lines (A549, ES2, and HCC44). All cell lines used were stably expressing Cas9 and were part of the Achilles project (Meyers et al., 2017). Prior to data collection, we confirmed cell line identity using single nucleotide polymorphism (SNP) profiling. We confirmed that all cell lines were mycoplasma negative by using MycoAlert Mycoplasma detection kits (Lonza, Walkersville, MD).
Lentiviral infection and plating
Virus was prepared in 96-well plates according to the published protocol (https://portals.broadinstitute.org/gpp/public/resources/protocols). Before initiating the screen, we optimized the number of cells per well and polybrene concentration for each Cas9-expressing cell line. Ultimately, we plated A549, ES2, and HCC44 cells with starting densities of 350, 375, and 150 cells per well, respectively, in 384-well black-wall, clear-bottom plates (Corning Costar). We also optimized sgRNA lentivirus volume to achieve 100% infection while maintaining low toxicity. For the screen, we spin-infected cells with 4 µg/ml polybrene concentration at the optimized density and virus volume (Aguirre et al., 2016). Three parallel plates were seeded per cell line. On one plate, cells were treated with or without 2 µg/ml puromycin 24 h postinfection, and cell viability was determined using CellTiterGlo (Promega) after 96 h of selection to determine infection efficiency. The second and third plates were used for the Cell Health assays (cell cycle and viability).
Cell Painting: cell staining and image acquisition
We followed the traditional Cell Painting protocol to acquire the readouts (Bray et al., 2016). We treated the cells with CRISPR guide perturbations and incubated for 4 d. Following the incubation period, we fixed cells with 10 µl of 16% (wt/vol) methanol-free paraformaldehyde for a final concentration of 3.2% (vol/vol). We imaged cells with a PerkinElmer Opera Phenix confocal HCI microscope at 20× magnification. We applied the standard panel of Cell Painting dyes to mark various cellular compartments: Hoechst 33342 to mark DNA; Concanavalin A/Alexa 488 to mark endoplasmic reticulum (ER); SYTO 14 to mark the nucleoli; cytoplasmic RNA, Phalloidin/Alexa 568, and wheat-germ agglutinin/Alexa 555 to mark actin cytoskeleton, golgi, and plasma membrane (AGP); and MitoTracker Deep Red/Alexa 647 to mark mitochondria (Bray et al., 2016). We collected nine images per well in five different channels for these different unbiased stains. In total, we collected 138,226 pictures after quality control filtering, which includes five channels per site, nine sites per well, across nine 384-well plates. In total, this represents about 2 TB of images. We deposited raw and illumination-corrected images to the Image Data Resource (https://idr.openmicroscopy.org) under accession number idr0080 (Williams et al., 2017).
Cell Painting: image processing
The next step in a Cell Painting protocol is to extract morphology features from the images that can be used as an unbiased systems biology measurement to describe how each perturbation impacts various cellular compartments in the assay. We built a CellProfiler image analysis and illumination correction pipeline (version 2.2.0) pipeline to extract these image-based features (McQuin et al., 2018). We include the CellProfiler pipelines in our github repository. Using the CellProfiler pipeline, we first performed several adjustments to account for potential confounding factors such as background intensity and illumination correction. Next, we used our pipeline to segment cells, distinguish between nuclei and cytoplasm, and then measure specific features related to the various channels captured. We measured the fluorescence intensity, texture, granularity, density, location, and various other measurements for each single cell (see http://cellprofiler-manual.s3.amazonaws.com/CellProfiler-3.0.0/index.html for more details). Following the image analysis pipeline, we obtain 8,964,210 cells and 1785 feature measurements across 9 different plates. We provide the raw output single-cell profiles as extracted from our CellProfiler pipeline on figshare (Way et al., 2019).
Cell Painting: image-based profiling
After the image analysis pipeline, the next step is to process the single-cell image-based features that are output of the CellProfiler pipeline. We used a standard approach (Caicedo et al., 2017) to process the single-cell profiles. First, we aggregated all single cells grouped by perturbation (effectively, by well) by computing the median value per morphology feature. This process takes all single cells and computes a single perturbation profile that is used to compare all perturbations against each other downstream. Next, using the median and median absolute deviation of feature values from empty wells as the center and scale parameters respectively, we normalized all perturbation profiles by subtracting the center and dividing by the scale and did so for each plate independently. This normalization procedure transforms all features to exist on the same scale and enables the perturbation profiles to be compared across plates and batches.
We then applied a feature selection procedure to reduce noisy and retain the most informative features. We removed features with missing values in any profile, features with low variance, features with extreme outlier values, and blocklisted features. Extreme outlier features are defined by having measurements greater than 15 SD following normalization. The blocklisted features are generally unreliable features that are known to be noisy and have caused numerical issues in previous experiments (Way, 2019). We used pycytominer (https://github.com/cytomining/pycytominer) to perform the profiling pipeline, which can be reproduced at https://github.com/broadinstitute/cell-health.
Following these procedures we derived profiles for 357 perturbations representing 119 guides measured across the three different cell lines. We also computed the perturbation consensus signatures of the Cell Painting data (see Materials and Methods: forming consensus signatures). Our final Cell Painting dataset had 357 consensus profiles measured by 952 morphology features (357 × 952). These data are available on github https://github.com/broadinstitute/cell-health/tree/master/1.generate-profiles/data/profiles.
Cell Health assays: cell staining and image acquisition
We treated all cells with a panel of specific reagents, each measuring a different aspect of cell health (see Supplemental Table S2). The seven reagents include unbiased dyes, click chemistry, and specific antibody treatments. The reagents measure various aspects of cell health including proliferation, mitosis, DNA damage, reactive oxygen species (ROS), and apoptosis timing. We collected a minimum of four replicates per treatment. Because many reagents fluoresce in different emission spectra, we applied the reagents in parallel. We applied a series of semimanual gating strategies to isolate specific cell health phenotypes in specific cell subsets (Supplemental Table S3). Together, we refer to the collection of measurements as the “Cell Health” assays.
More specifically, we collect the Cell Health assay data in a series of two distinct panels: cell cycle and viability. In the first panel we measure Hoechst, EdU, PH3, and gH2AX and use these measurements to quantify cell cycle and DNA damage in specific cell cycle subsets. In the second panel, we measure viability phenotypes using Caspase 3/7, DRAQ7, CellRox, and DPC nucleus morphology measurements.
We acquired all cell images using an Opera Phenix HCI Instrument (PerkinElmer) with a 20× water objective (a numerical aperture of 1.0) in confocal mode. We acquired images in four channels using default excitation/emission combinations: for the blue channel (Hoechst) 405/435–480; for the green channel (Alexa 488 and CellEvent), 488/500–550; for the orange channel (Alexa 568 and CellRox Orange), 561/570–630; and for the far red channel (Alexa 647 and DRAQ7), 640/650–760. We applied the Cell Health reagents for cell viability and for cell cycle in two separate plates.
The first set of plates (n = 3 replicate plates) measures cell cycle. We added 5-ethynyl-2′-deoxyuridine (EdU) in live cells for S phase cells to integrate. We then fixed the cells with 4% formaldehyde using standard approaches and detected EdU using Click-iT EdU Alexa Fluor 647 HCS Assay (ThermoFisher C10357) according to the vendor protocol. We then performed standard immunofluorescence staining with two antibodies: one targeting phosphohistone H3 (PH3) to measure cells undergoing mitosis and one to identify DNA damage foci in nuclei via γH2Ax. We followed these PH3 and γH2Ax antibody treatments by secondary antibodies conjugated with Alexa 488 and Alexa 568, respectively. We added Hoecsht 33342 dye to stain nuclear DNA. For the cell cycle plate, we acquired nine fields of view per well.
The second set of plates (n = 3 replicate plates) measures cell viability. We added CellEvent Caspase-3/7 Green Detection Reagent (ThermoFisher), DRAQ7, and CellROX Orange Reagent (ThermoFisher) dyes to measure apoptotic cells, dead cells, and ROS, respectively. We acquired one field of view per well using green, orange, and far red fluorescence channels as well as brightfield and DPC channels. The cells were incubated at 37°C.
Cell Health assays: image analysis
We developed and ran two distinct image analysis pipelines in Harmony software (version 4.1; PerkinElmer) for each of the Cell Health plates. Individually for each cell line assayed, and for both cell cycle and cell viability plates, we established a series of manual gating strategies to identify distinct cell line subpopulations (see Figure 1A).
For the cell cycle plate, we performed nucleus segmentation using the Hoechst channel and discarded all nuclei that were at the field border. We identified cells in specific cell cycle stages using Hoechst, Alexa 488 (pH3), and Alexa 647 (EdU) intensities. We identified γH2AX spots within nuclei based on the Alexa 568 channel. These strategies are standard in the field (Aguirre et al., 2016). More specifically, we identified subpopulations based on the respective channel intensities and morphological properties for each nucleus as specified:
We stratified populations “polyploid,” “polynuclear (large not round nuclei),” and “cells selected for cell cycle” based on total intensity of the Hoechst channel (DNA content) and nucleus “roundness” measurements as output from the PerkinElmer Harmony software.
- We identified four subpopulations within the “cells selected for cell cycle” population as follows:
- a) “G1 cells:” selected based on low total Hoechst intensity and low green (pH3) and far red (EdU) channels. We excluded outlier nuclei with unusually high intensity of Hoechst max.
- b) “G2 cells:” selected based on high total Hoechst intensity and low green (pH3) and far red (EdU) channels. We excluded outlier nuclei with unusually high intensity of Hoechst max.
- c) “G2/M cells”: selected based on the same criteria as for G2, except we included nuclei with high green (pH3) mean.
- d) “M cells”: selected based on high green (pH3) mean.
- e) “S cells”: selected based on high mean far red (EdU) channel intensity.
We counted orange spots (γH2AX) representing DNA damage loci in each of the cell cycle subpopulations. We determined high γH2AX activity if there were more than three spots per nucleus.
For the cell viability plate, we performed cell segmentation based on the cumulative DPC channels. Again, we identified specific subpopulations based on the following channel intensities:
- We separated “Dead Cells” and “Live Cells” based on max intensity far red channel (DRAQ7).
- a) We identified a “Dead Only Cells” subpopulation within the “Dead Cells” population by isolating cells without green (Caspase 3/7) signal.
- We identified “Caspase Positive Cells” based on green (Caspase 3/7) channel max intensity.
- a) We distinguished two subpopulations in the “Caspase Positive Cells” named “Early Apoptotic Cells” and “Late Apoptotic Cells” based on low and high far red (DRAQ7) max signal intensity, respectively.
- We used the mean intensity of the CellROX Orange signal to measure ROS.
- a) We excluded edge wells in the ROS analysis because of consistent poor signal quality.
Additionally, we set these gates for each cell subpopulation using a set of random wells from each cell line and experiment independently. We observed that the intensity measurements used to form the gates were consistent across wells and plates and generally formed distinct cell subpopulation clusters. After using the random wells to set the gates, we used the Harmony microscope software to apply the gates to the remaining wells and plates.
We also used CRISPR infection efficiency, which is measured in a separate assay for both cell cycle and viability imaging assays, as negative control features. In total, considering both plates and all cell subpopulations, we measured 70 different variables in the Cell Health Assay. To standardize plate-level differences, we normalized Cell Health readouts per plate by subtracting median values and dividing by the SD.
Forming consensus signatures
After acquiring the images and processing the data, we prepared the data further before input into our machine learning framework. We generated consensus signatures for each perturbation using a moderated z-score (MODZ) procedure (Subramanian et al., 2017). Briefly, we calculated pairwise Spearman correlations between all replicates of a single perturbation and then combined profiles by weighting their signature contribution by mean pairwise correlation to all other replicates. We applied this transformation to both Cell Painting profiles and Cell Health assay measurements. We collected more replicates of the Cell Painting data than the Cell Health data. In total, we collected assay readouts from 357 common perturbations (119 CRISPR guides across three cell lines). In the Cell Painting data, we filtered and collapsed 3456 morphology profiles to the common set of 357 consensus profiles. In the Cell Health assays, we filtered and collapsed 2302 well profile readouts to the common set of 357 consensus readouts. We generated consensus signatures because there is no way to match replicate-level information across the assays. We applied UMAP (McInnes et al., 2018) to the consensus profiles and visualized patterns of ground truth cell health measurements.
Machine learning framework
We randomly split 15% of the consensus signatures into a separate test set. We balanced this stratification by cell line. We then used the remaining 85% to train all 70 cell health models. In total, we used 303 samples as the training set and 54 as the test set.
We elected to train elastic net regression models using sklearn (version 0.20.3) (Pedregosa et al., 2011; Zou and Hastie, 2005). We chose this model because it is quick to train, is easily interpretable, and will induce sparsity in selecting model features. We also trained classification models and binarized training and testing data by using > 1.5 SD away from the mean as positive examples. However, because the classification approach was unstable and sensitive to low sample sizes, we elected to move forward using only the regression models (see https://github.com/broadinstitute/cell-health/issues/78). To identify optimal alpha and elastic net mixing parameters, we performed a grid search and fivefold cross-validation using the training set only. For each model independently, we observed cross-validation performance across 9 different alpha parameters ([0.01, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8]) and 11 different elastic net mixing parameters ([0.1, 0.12, 0.14, 0.16, 0.2, 0.3, 0.4, 0.5, 0.7, 0.8, 0.9]). Alpha controls the regularization penalty term for all features, and the elastic net mixing parameter controls the trade-off between L1 and L2 regression where 0 = L1 and 1 = L2. Therefore, the closer the elastic net mixing parameter is to 0, the sparser the model. We optimized and trained 70 different elastic net regression models for each of the 70 Cell Health assay readouts independently.
We repeated this procedure and independently optimized 70 additional models using randomly shuffled data. For the shuffling procedure, we randomly shuffled the Cell Painting features independently per column before training. We use the shuffled model performance as a suitable baseline to compare real model performance.
Machine learning evaluation
We evaluated each of the 70 Cell Health regression models independently using R-squared statistics from sklearn version 0.20.3 (Pedregosa et al., 2011). We calculated R-squared for the full training and testing partitions, in shuffled training and testing partitions, and for each cell line independently for all 70 models. The measurement can be interpreted as how well the models could predict the real Cell Health readout with values approaching one as perfect fits. It is best to compare test set performance in real versus shuffled data. The test set performance in real data simulates how models are expected to perform in data not used for model training. The shuffled performance indicates if there is any expected performance inflation.
Machine learning robustness: investigating the impact of sample size
We performed an analysis in which we randomly dropped an increasing amount of samples from the training set before model training. After dropping the predefined number of samples, we retrained all 70 cell health models and assessed performance on the original holdout test set. We performed this procedure 10 times with 10 unique random seeds to mirror a more realistic scenario of new data collection and to reduce the impact of outlier samples on model training.
Machine learning robustness: systematically removing feature classes
We performed an analysis in which we systematically dropped features measured in specific compartments (Nuclei, Cells, and Cytoplasm), specific channels (RNA, Mito, ER, DNA, and AGP), and specific feature groups (Texture, Radial Distribution, Neighbors, Intensity, Granularity, Correlation, and Area Shape) and retrained all models. We omitted one feature class and then independently optimized all 70 cell health models as described in the Machine learning framework results section above. We repeated this procedure once per feature class.
Drug Repurposing Hub Cell Painting data: image-based profiling
A subset of the Drug Repurposing Hub compounds (n = 1571) (Corsello et al., 2017) were profiled using the Cell Painting assay across about six doses per compound. We processed this dataset using a standard image-based profiling pipeline to extract consensus profiles per treatment. See https://github.com/broadinstitute/lincs-cell-painting for complete details and instructions on how to reproduce. Briefly, we applied a CellProfiler image analysis pipeline to segment cells, adjusted for background intensity, and measured morphology features for three compartments: cells, cytoplasm, and nuclei. The output of this procedure was 136 SQLite files (one for each plate) representing unnormalized single-cell profiles. Next, we developed and applied an image-based profiling bioinformatics pipeline to generate treatment consensus profiles from the single-cell measurements (Caicedo et al., 2017). The same image analysis pipeline and bioinformatics pipeline were used to process all the plates in the experiment.
In the pipeline, we first median-aggregated the single cells by feature to form well profiles and then, using the median and median absolute deviation of feature values from DMSO as the center and scale parameters, respectively, we normalized all perturbation profiles by subtracting the center and dividing by the scale and did so for each plate independently.
The plates in this dataset have 24 DMSO-treated wells and therefore represented a good alignment control to adjust for plate level differences. Each plate also has two positive controls (BRD-K50691590 [Bortezomib] and BRD-K60230970 [MG-132]) at 20 mmol/l with 12 replicates each for all plates. We visualized positive and negative control profiles in our UMAP space to determine the extent of technical artifacts present in our data. Following the z-score normalization, we combined all treatment replicates (∼6 per compound and dose pair) using MODZ consensus signatures. We generated consensus profiles for control replicates by well across plate maps. In total, this procedure resulted in 10,752 different treatment profiles and 1788 normalized CellProfiler morphology features.
Drug Repurposing Hub Cell Painting data: predicting cell health readouts
We applied all Cell Health models to the 10,752 Drug Repurposing Hub consensus Cell Painting profiles. We simply applied the Cell Health trained models using the sklearn model.predict() method. Every feature measured in the CRISPR perturbation Cell Painting profiles were also measured in the Drug Repurposing Hub output. The result of the model application was 70 Cell Health readouts for all 10,752 treatments. We used these predictions for model validation with external data and for visualization in the web app scatter plots.
Assessing generalizability of cell health models applied to Drug Repurposing Hub data
We used our cell health webapp (https://broad.io/cell-health-app) to identify compounds with high predictions for three models with high or intermediate performance: ROS, Number of G1 cells, and Number of gH2AX spots in G1 cells. For each model, we identified classes of compounds with consistently high scores, then tested for statistical enrichment: for proteasome inhibitors in the ROS model, PLK inhibitors in the Number of G1 cells model, and aurora kinase and tubulin inhibitors in the Number of gH2AX spots in G1 cells model. We used one-sided Fisher’s exact tests to quantify differences in expected proportions between high and low model predictions. For each case, we determined high and low predictions based on the 50% quantile threshold for each model independently.
Drug Repurposing Hub Cell Painting data: visualization
We also applied UMAP to the 10,752 Drug Repurposing Hub Cell Painting profiles and extracted two lower dimensional representations. UMAP reduces the Cell Painting profiles to two features that capture the global structure of the input data. Prior to UMAP transformation, we applied a feature selection procedure to the Drug Repurposing Hub profiles. We removed features with low variance, features with missing values in any consensus profile, blocklisted features (Way, 2019), and features with extreme outlier values defined by greater than 15 SD following normalization. This procedure reduced the feature dimension from 1788 to 572. By reducing the number of features, we can be more confident that the major sources of variation are not biased by CellProfiler feature redundancies or by technical artifacts of sample processing.
Drug Repurposing Hub Cell Painting data: dose-response analysis
To model dose, we fit Hill equations (4 parameter log-logistic model) to all 1571 Drug Repurposing compounds consensus signatures transformed into each of the 70 different Cell Health model predictions. Before input into the model, we zero-one transformed Cell Health predictions across doses for each compound independently. For most compounds, this normalization procedure happens for six data points (representing six doses per compound consensus signature) at a time. The zero-one procedure assigns a value of zero to the lowest value, one to the highest value, and scales each intermediate value accordingly. This procedure results in 109,970 different model fits. We used the drc R package (version 3.0-1) to fit all models (Ritz et al., 2015). We present all precomputed dose fit models to be explored at https://broad.io/cell-health-app.
Comparing viability predictions to an orthogonal readout
We downloaded the PRISM assay results (version 19Q4) from the Cancer Dependency Map website at https://depmap.org/portal/download/ (Corsello et al., 2017). The PRISM assay measures viability of multiple cell lines in a pooled format and deconvolutes results based on barcoded readouts (Yu et al., 2016). We focused on the A549 cell line and compounds that were measured in both the PRISM assay and the Drug Repurposing collection. The PRISM assay profiled 1382 of the 1571 Drug Repurposing Compounds. The PRISM assay also used slightly different doses than the Drug Repurposing Hub collection procedure. Therefore, to align doses, we converted doses into dose ranks and report Spearman correlations between the two datasets (see Figure 4A).
Code and data availability
All data and code are publicly available. Analysis software to reproduce the full paper is available at https://github.com/broadinstitute/cell-health/tree/v2.0 (Way et al., 2020). Raw and illumination-corrected Cell Painting images are available in the Image Data Resource (accession number idr0080). Single cell morphology profiles derived from these images are available at the National Institutes of Health (NIH) Figshare at https://doi.org/10.35092/yhjc.9995672.v5. Processed Cell Painting profiles and raw and processed Cell Health readouts are also available at https://github.com/broadinstitute/cell-health/tree/v2.0. Processing code and data for the Cell Painting Drug Repurposing Hub data is available at https://github.com/broadinstitute/lincs-cell-painting/tree/v0.1. Cell Health predictions for the Drug Repurposing Hub compounds are available to explore at https://broad.io/cell-health-app.
Supplementary Material
Acknowledgments
We thank Kyle Karhohs for executing the image analysis pipeline; Nasim Jamali, David Stirling, and Beth Cimini for insightful discussions about CellProfiler features and cell images; Kate Hartland for inputs on staining protocols; and the LINCS data generators in the Broad Institute’s Connectivity Map Team for producing the Drug Repurposing Hub Cell Painting data. This work was funded in part by the NIH (MIRA R35 GM122547 to A.E.C and NCI U01 CA176058 to W.C.H.) and The Slim Initiative in Genomic Medicine for the Americas (SIGMA), a joint U.S.–Mexico project funded by the Carlos Slim Foundation (F.V.). T.B. was supported by the Deutsche Forschungsgemeinschaft Research Fellowship (No. 328668586).
Abbreviations used:
- AGP
actin, golgi, and plasma membrane
- CRISPR
clustered regularly interspersed short palindromic repeats
- DPC
Digital Phase Contrast
- EdU
5-ethynyl-2′-deoxyuridine
- ER
endoplasmic reticulum
- MODZ
moderated z-score
- PH3
phosphohistone H3
- ROS
reactive oxygen species
- UMAP
uniform manifold approximation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-12-0784) on February 3, 2021.
REFERENCES
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang C-Z, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. (2016). Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov 6, 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M-A, Singh S, Han H, Davis CT, Borgeson B, Hartland C, Kost-Alimova M, Gustafsdottir SM, Gibson CC, Carpenter AE (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc 11, 1757–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C, Vasilevich AS, Barry JD, Bansal HS, Kraus O, et al. (2017). Data-analysis strategies for image-based cell profiling. Nat Methods 14, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo JC, McQuin C, Goodman A, Singh S, Carpenter AE (2018). Weakly Supervised Learning of Single-Cell Feature Embeddings. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2018, 9309–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo JC, Singh S, Carpenter AE (2016). Applications in image-based profiling of perturbations. Curr Opin Biotechnol 39, 134–142. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Crasta K (2017). Consequences of mitotic slippage for antimicrotubule drug therapy. Endocr Relat Cancer 24, T97–T106. [DOI] [PubMed] [Google Scholar]
- Christoforow A, Wilke J, Binici A, Pahl A, Ostermann C, Sievers S, Waldmann H (2019). Design, synthesis, and phenotypic profiling of pyrano-furo-pyridone pseudo natural products. Angew Chem Int Ed 58, 14715–14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, Johnston SE, Vrcic A, Wong B, Khan M, et al. (2017). The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med 23, 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsdottir SM, Ljosa V, Sokolnicki KL, Anthony Wilson J, Walpita D, Kemp MM, Petri Seiler K, Carrel HA, Golub TR, Schreiber SL, et al. (2013). Multiplex cytological profiling assay to measure diverse cellular states. PLoS One 8, e80999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RE, Doench JG (2020). Design and analysis of CRISPR-Cas experiments. Nat Biotechnol. 10.1038/s41587-020-0490-7 [DOI] [PubMed] [Google Scholar]
- Han YH, Park WH (2010). The changes of reactive oxygen species and glutathione by MG132, a proteasome inhibitor affect As4.1 juxtaglomerular cell growth and death. Chem Biol Interact 184, 319–327. [DOI] [PubMed] [Google Scholar]
- Hughes RE, Elliott RJR, Munro AF, Makda A, Robert O’Neill J, Hupp T, Carragher NO (2020). High content phenotypic profiling in oesophageal adenocarcinoma identifies selectively active pharmacological classes of drugs for repurposing and chemical starting points for novel drug discovery. SLAS Discovery: Adv Life Sci R & D 25, 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2002). Computational systems biology. Nature 420, 206–210. [DOI] [PubMed] [Google Scholar]
- Lee S-Y, Jang C, Lee K-A (2014). Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev Reprod 18, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y-H, Liebes L, Zou Y, Perez-Soler R (2003). Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J Biol Chem 278, 33714–33723. [DOI] [PubMed] [Google Scholar]
- Markowetz F (2010). How to understand the cell by breaking it: network analysis of gene perturbation screens. PLoS Comput Biol 6, e1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML] http://arxiv.org/abs/1802.03426. [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyffeler J, Willis C, Lougee R, Richard A, Paul-Friedman K, Harrill JA (2020). Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol Appl Pharmacol 389, 114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ (2011). Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res 71, 4608–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl A, Sievers S (2019). The cell painting assay as a screening tool for the discovery of bioactivities in new chemical matter. Methods Mol Biol 1888, 115–126. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: machine learning in Python. J Mach Learn Res 12, 2825–2830. [Google Scholar]
- Ritz C, Baty F, Streibig JC, Gerhard D (2015). Dose-Response Analysis Using R. PLoS One 10, e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohban MH, Singh S, Wu X, Berthet JB, Bray M-A, Shrestha Y, Varelas X, Boehm JS, Carpenter AE (2017). Systematic morphological profiling of human gene and allele function via Cell Painting. Elife 6. 10.7554/eLife.24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeder C, Heigwer F, Boutros M (2018). Machine learning and image-based profiling in drug discovery. Curr Opin Syst Biol 10, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm J, Klambauer G, Arany A, Steijaert M, Wegner JK, Gustin E, Chupakhin V, Chong YT, Vialard J, Buijnsters P, et al. (2018). Repurposing high-throughput image assays enables biological activity prediction for drug discovery. Cell Chem Biol 25, 611–618.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. (2017). A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai B, Subramanian V, Holland CH, Alföldi R, Puskás LG, Saez-Rodriguez J (2019). Signatures of cell death and proliferation in perturbation transcriptomics data-from confounding factor to effective prediction. Nucleic Acids Res 47, 10010–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda Y, Iimori M, Nakashima Y, Nakanishi R, Ando K, Ohgaki K, Kitao H, Saeki H, Oki E, Maehara Y (2017). Mitotic slippage and the subsequent cell fates after inhibition of Aurora B during tubulin-binding agent-induced mitotic arrest. Sci Rep 7, 16762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray M-A, Kemp MM, Winchester E, et al. (2014). Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proc Natl Acad Sci USA 111, 10911–10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way G (2019). Blocklist Features—Cell Profiler. Dataset https//.org/10.6084/m9.figshare.10255811.v3 [Google Scholar]
- Way G, Kost-Alimova M, Shibue T, Harrington W, Gill S, Becker T, Hahn WC, Carpenter A, Vazquez F, Singh S (2019). Cell health—cell painting single cell profile. NIH Figshare Archive, https//.org/10.35092/yhjc.9995672.v5 [Google Scholar]
- Way G, Becker T, Carpenter, A, Singh S (2020). broadinstitute/cell-health: response to reviewers. Zenodo. 10.5281/ZENODO.4262789 [Google Scholar]
- Williams E, Moore J, Li SW, Rustici G, Tarkowska A, Chessel A, Leo S, Antal B, Ferguson RK, Sarkans U, et al. (2017). The Image Data Resource: A bioimage data integration and publication platform. Nat Methods 14, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Mannan AM, Yvone GM, Ross KN, Zhang Y-L, Marton MA, Taylor BR, Crenshaw A, Gould JZ, Tamayo P, et al. (2016). High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol 34, 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T (2005). Regularization and Variable Selection via the Elastic Net. J R Stat Soc Series B Stat Methodol 67, 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code are publicly available. Analysis software to reproduce the full paper is available at https://github.com/broadinstitute/cell-health/tree/v2.0 (Way et al., 2020). Raw and illumination-corrected Cell Painting images are available in the Image Data Resource (accession number idr0080). Single cell morphology profiles derived from these images are available at the National Institutes of Health (NIH) Figshare at https://doi.org/10.35092/yhjc.9995672.v5. Processed Cell Painting profiles and raw and processed Cell Health readouts are also available at https://github.com/broadinstitute/cell-health/tree/v2.0. Processing code and data for the Cell Painting Drug Repurposing Hub data is available at https://github.com/broadinstitute/lincs-cell-painting/tree/v0.1. Cell Health predictions for the Drug Repurposing Hub compounds are available to explore at https://broad.io/cell-health-app.