Abstract
Endocytic trafficking controls the density of molecules at the plasma membrane and by doing so, the cell surface profile, which in turn determines how cells interact with their environment. A full apprehension of any cellular process necessitates understanding how proteins associated with the plasma membrane are endocytosed, how they are sorted after internalization, and if and how they are recycled to the plasma membrane. To date, it is still difficult to experimentally gain access to this information, even more to do it in a quantitative way. Here we present a toolset based on photoactivation of fluorescent proteins that enabled us to generate quantitative information on endocytosis, incorporation into sorting and recycling endosomes, delivery from endosomes to the plasma membrane, and on the type of vesicles performing intracellular transport. We illustrate these approaches by revealing striking differences in the endocytic trafficking of T-cell receptor and CD4, which bind to the same molecule at the surface of antigen-presenting cells during T-cell activation.
INTRODUCTION
Individual cells are part of a whole and cannot function without interactions. Availability of surface proteins at the plasma membrane is one of the main factors controlling interactions between cells and regulating their response to extracellular signals. Density of receptors ready to engage ligands is a key determinant of the sensitivity and strength of a cell response to a given stimulus (Achour et al., 2008). Similarly, density of adhesion molecules (Ballestrem et al., 2001), ion channels (Fox et al., 2013), or availability of transporters (Duelli and Kuschinsky, 2001) dictate how tightly cells can bind and how efficiently they can regulate their membrane potential and metabolism.
While gene expression contributes to shape the cell surface profile, the availability of most key proteins required to convey immediate cellular responses is primarily determined by endocytic trafficking (Cullen and Steinberg, 2018; Spang and Mayor, 2018). Residency time at the plasma membrane is tightly and dynamically regulated by rates of endocytosis and recycling. This is why much effort is put into understanding the mechanisms that determine when and how proteins at the plasma membrane are internalized, how they are sorted in endosomal populations, and if and how they are returned to the plasma membrane through recycling or retrograde transport (Hsu et al., 2012; Redpath et al., 2020).
Flow cytometry is a powerful tool to get a general overview of endocytic trafficking by providing access to parameters such as expression of cell surface proteins, their removal from the cell surface (Rossatti et al., 2019), and their recycling through antibody feeding assays (Compeer et al., 2018). A finer view of endocytic trafficking can be obtained through pulse width analysis (Chia et al., 2014), although distinction between different intracellular staining patterns remains limited. Flow cytometry-based approaches to study intracellular trafficking have the strength of high-throughput and unbiased quantification. However, these bulk measurements are not suited to reveal details about the mechanisms underlying endocytic processes. They also fall short when it comes to investigating how targeted endocytic recycling can maintain functional membrane polarity by returning internalized surface proteins to a restricted area of the plasma membrane.
Microscopy approaches are more suitable to gain a detailed understanding of the machinery at play during various steps of endocytic trafficking. Study of endocytosis in particular benefits from the high signal-to-noise ratio of fluorophores at the plasma membrane generated by a 100–150 nm deep evanescent wave in total internal reflection fluorescence (TIRF) microscopy. Hence, TIRF allows visualization and automated quantification of the assembly of endocytic-coated pits at the plasma membrane. This type of approach has been used to understand how the adaptor AP-2 is recruited to clathrin-coated pits (Cocucci et al., 2012) to assemble functional endocytic vesicles (Hong et al., 2015; Kadlecova et al., 2017) or how cargoes are incorporated into clathrin-coated pits (Weigel et al., 2013). Alternating between the narrow TIRF and a deeper epifluorescence illumination further allows following the first molecular events associated with scission and departure from the plasma membrane, for instance, to elucidate the role of actin and actin-binding proteins in this process (Merrifield et al., 2005). While TIRF-based imaging approaches have provided exquisite details about the mechanisms of endocytosis, they are limited to the first and last steps of the endocytic journey (Mattheyses et al., 2010; Redpath et al., 2019).
Fluorescent timers—fluorescent proteins that change color over time–can be used to investigate the intracellular path that brings proteins from the ER to their final destination. This strategy has been used to show that the lysosomal protein LAMP-2A visits the plasma membrane, the early and recycling endosomes before it reaches late endosomes and lysosomes (Subach et al., 2009b). However, the relatively slow change in color and the lack of ability to trigger the color change at a location of interest makes fluorescent timers unsuitable to investigate endocytic recycling. Similarly, bimolecular fluorescence complementation (BiFC) can reveal in which intracellular compartments interactions take place (Dirk et al., 2016), but there is no way to experimentally control when the two halves of the fluorescent proteins are available for complementation. Additionally, once reconstituted, BiFC fluorescent proteins do not dissociate and will be detected in all compartments downstream of the location of complementation.
Fluorescent ligands have been used since the 1980s to follow the endocytic trafficking of surface proteins. Key insights into endocytosis, sorting, and recycling of surface receptors and lipids have been obtained in pulse-chase experiments using fluorescently labeled molecules. These experiments have, for instance, revealed that sorting endosomes process transferrin and low-density lipoprotein differently, rapidly exporting the former and concentrating the later. Results obtained with fluorescently labeled lipids have also demonstrated that internalized lipids are mostly recycled and that this recycling proceeds through the same endocytic compartments than surface receptors (Mukherjee et al., 1997; Maxfield and McGraw, 2004). This approach is not limited to receptors for which ligands can be functionally coupled to a fluorophore, as fluorescently labeled antibodies against surface proteins can be internalized and followed through their endocytic journey.
Typically, antibody-feeding assays represent a robust and conceptually simple way of visualizing endocytic trafficking. Cell surface proteins are labeled with specific antibodies and allowed to be internalized. They can be revealed with fluorescently labeled secondary antibodies or streptavidin at any given time point after internalization (Boucrot et al., 2015) or when they are returned to the cell surface (Fletcher-Jones et al., 2019). A recent study reports a Halo-tag variation of the feeding assay in which the dyes are fluorescent only when bound to the Halo-tag allowing live reporting of the kinetics of recycling (Jonker et al., 2020). While feeding assays enable following the endocytic journey of a given cell surface protein, they are limited to proteins bearing a substantial extracellular domain and provide little information on the regulatory mechanisms of recycling processes.
Recently, photoactivatable or photoswitchable proteins have been widely used for photoactivatable localization microscopy (PALM; Betzig et al., 2006). However, these fluorescent proteins were initially designed to investigate protein dynamics (Patterson and Lippincott-Schwartz, 2002). Here we present a method based on photoactivation of fluorescent proteins to visualize and quantify endocytic trafficking of T-cell surface receptors, similar to what has been performed for integrin trafficking in cancer cells (Caswell et al., 2007). Membrane proteins of interest are expressed fused to photoactivatable fluorescent proteins, such as PA-mCherry, or photoswitchable fluorescent proteins, such as PS-CFP2. To investigate endocytosis, restricted areas of the plasma membrane are illuminated with a 405-nm light to trigger localized photoactivation of the fluorescent proteins. When endocytosis occurs, the PA-mCherry signal can be seen in endocytic vesicles, which can then be counted and followed as they are incorporated into intracellular compartments identified by expression of various Rab small GTPases. We further show that spatially restricted photoconversion within intracellular compartments by two-photon illumination enables us to follow membrane proteins as they travel within transport vesicles from endosomes to be incorporated in the plasma membrane.
We apply these approaches to investigate endocytic trafficking taking place during T-cell activation. We focus on the T-cell receptor (TCR), which we have investigated previously (Compeer et al., 2018; Redpath et al., 2019), and on the coreceptor CD4. Both TCR and CD4 bind to the same peptide-loaded MHCII protein at the surface of antigen presenting cells during T-cell activation. TCR has no intrinsic catalytic activity and TCR triggering and phosphorylation of intracellular domains relies on the kinase Lck, which can engage TCR through binding to CD4 (Veillette et al., 1989; Rossy et al., 2012). We aimed to determine the potential convergences and divergences of the endocytic trafficking of TCR and CD4 to understand if binding to the same molecule on the plasma membrane leads to identical or divergent endocytic trafficking fates.
RESULTS AND DISCUSSION
Photoactivation to determine the endocytic route into the cell
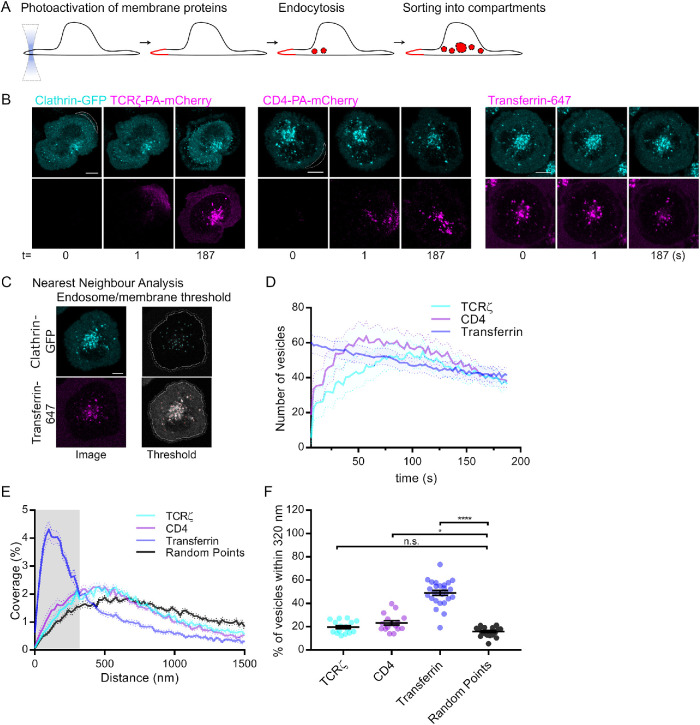
To quantitatively investigate cargo endocytosis, we utilize PA-mCherry fused to proteins of interest (CD4 and TCRζ in this study). PA-mCherry is relatively nonfluorescent until it is exposed to 350–400 nm light (Subach et al., 2009a), allowing specific investigation of the attached protein cargoes by illuminating the cell periphery with ultraviolet light (Figure 1A). The cell periphery was selected so essentially plasma membrane was exposed to the ultraviolet light, ensuring no pre-existing cargo-containing endosomes were photoactivated. By coexpressing the PA-mCherry-fused cargo with clathrin fused to EGFP in the same cells, we were able to determine whether this cargo was internalized in clathrin-positive endocytic vesicles.
FIGURE 1:
Real time endocytosis of plasma membrane receptors. (A) Schematic depicting photoactivation to investigate endocytosis of proteins. (B) Representative confocal images of activated Jurkat T-cells expressing TCRζ-PA-mCherry, CD4-PA-mCherry or added transferrin-Alexa 647 coexpressing Clathrin-EGFP. Cells were photoactivated every three frames for 30 cycles by 405-nm laser on the membrane region of interest (white dashed line) and imaged for 200 s. (C) Example of custom-made MATLAB vesicle tracking and cross-channel nearest-neighbor distance evaluation software showing the identification by threshold of endosomal compartments. (D) Number of PA–mCherry-positive vesicles detected at any given point during 200 s acquisition. (E) Cross-channel nearest neighbor distance between vesicles defined by Clathrin-EGFP and TCRζ-PA-mCherry, CD4-PA-mCherry, or transferrin-Alexa 647. (F) Percentage of vesicles within 320 nm of clathrin-positive vesicles; n.s., not significant; *p < 0.05, ****p < 0.0001 from Student’s t test of means of three independent experiments of four to nine cells. Error bars/lines = mean ± SEM. All images were acquired on a Zeiss 880 confocal microscope. Scale bars = 5 µm.
CD4 is well established as a clathrin cargo when coupled to the human immunodeficiency virus (HIV) protein Nef (Burtey et al., 2007; Pereira and daSilva, 2016; Manrique et al., 2017). However, few studies have investigated CD4 endocytosis in the absence of Nef in T-cells (Pelchen-Matthews et al., 1992, 1993; Foti et al., 1997), and no studies have investigated this process using live-cell imaging. To determine if CD4 undergoes endocytosis in clathrin-coated vesicles, we cotransfected Jurkat T-cells with CD4-PA-mCherry and clathrin light chain fused to GFP (clathrin-GFP). We also cotransfected Jurkat T-cells with TCRζ-PA-mCherry and clathrin-GFP or added transferrin647 to clathrin-GFP transfected T-cells to compare CD4 endocytosis to bona fide clathrin-independent and -dependent cargoes, respectively (Figure 1B) (Hanover et al., 1984; Compeer et al., 2018). Peripheral membrane regions were then exposed to 405 nm light repeatedly over 30 s, after which no further exposure occurred. Vesicles were observed for a further 150 s following photoactivation to quantify their co-occurrence with clathrin over time (Figure 1B, dotted outlines). Labeled transferrin was added to the cellular media and cells imaged for 180 s.
To quantify the occurrence of vesicles positive for both PA-mCherry or transferrin647 and clathrin-EGFP, we used a custom MATLAB routine based on a nearest neighbor approach (Figure 1C). This analysis uses intensity-based thresholds set to levels that identify vesicles in each channel, while preventing identification of peripheral membrane from the analysis to prevent the photoactivation region from contaminating the results. The distance from the center of each vesicle in each channel is then compared with the other channel to identify the nearest neighboring vesicle. Two vesicles localized in alternate channels are considered to be the same if they are within 320 nm. This value is the typical value where the distribution of cross-channel nearest neighbor distances for colocalized vesicles crosses that of the randomized control drawn from the same dataset. This value approximates the 60–500 nm diameter of a typical endocytic vesicle (Klumperman and Raposo, 2014) and resolution of a confocal microscope with a 100×/1.4 NA objective as used in this study.
Photoactivation of TCRζ and CD4 resulted in identification of a rapid influx of endosomes within the cell (Figure 1D), peaking at 50 s (CD4) and 100 s (TCRζ). Both photoactivated proteins formed similar numbers of vesicles to each other. Transferrin is a canonical clathrin-mediated endocytosis cargo and accordingly 50% of transferrin vesicles were within 320 nm of clathrin-positive vesicles over the time course of the experiment compared with 18% for the random control (Figure 1, E and F). Of note, the same experiment performed using heterodimer-forming protein, flotillin 1 and flotillin 2 labeled with PA-mCherry and EGFP, respectively, resulted in 54% of vesicles with a nearest neighbor within 320 nm (Compeer et al., 2018). In contrast, only 20% of TCRζ vesicles were within 320 nm of clathrin-positive vesicles, a nonsignificant level compared with the control and consistent with our previous report of TCRζ being a cargo internalized through a clathrin-independent endocytic pathway (Compeer et al., 2018). Twenty-two percent of CD4-positive endocytic vesicles were clathrin-positive, which was significantly higher than the random control, yet closer to TCRζ (22–20%) than transferrin (22–50%). Together, these results indicate that when expressed alone in T-cells, CD4 endocytosis occurs predominantly independent of clathrin.
These results represent the first quantitative investigation of CD4 endocytosis in live T-cells. Older studies performed in other cell types (HeLa, Chinese hamster ovary, and 3T3) have described CD4 endocytosis as being predominantly clathrin dependent (Marsh et al., 1990; Keller et al., 1992; Pitcher et al., 1999). However, despite the association of CD4 with clathrin-coated pits observed in electron microscopy, early kinetic measurements of CD4 endocytosis suggest that CD4 uptake occurs by relatively nonspecific membrane invagination as opposed to strict clathrin-dependent endocytosis (Marsh et al., 1990). Furthermore, a more recent proteomics study showed that CD4 interacts with beta-integrin, which undergoes both clathrin-dependent and -independent endocytosis and flotillin-1, which consistently label clathrin-independent endocytic pathways (Raposo et al., 2011). The results generated by our approach are consistent with these kinetic and proteomic studies, showing that CD4 likely undergoes both clathrin-independent and -dependent endocytosis. Importantly, these data illustrate that photoactivation of membrane proteins allows to discriminate between endocytic pathways and thus to provide critical information on the mechanism regulating surface expression of receptors.
Photoactivation to determine endocytic sorting
Investigation of the sorting of newly endocytosed cargoes into endocytic pathways normally requires a fluorescently labeled marker that can be fed to the cell exogenously. This can be receptor ligands such as transferrin and epidermal growth factor (Leonard et al., 2008) or antibodies against the extracellular domain of surface proteins (Fletcher-Jones et al., 2019). The use of photoactivatable fluorescent proteins offers a distinct advantage over feeding of fluorescently labeled makers. The start and the end of the visualization of the incorporation of cargoes into endocytic vesicles is accurately timed by exposure to 405 nm light. Thus, the number of endocytic vesicles visualized during the course of a given experiment can be controlled, thereby greatly facilitating the quantification of their progress through endocytic pathways.
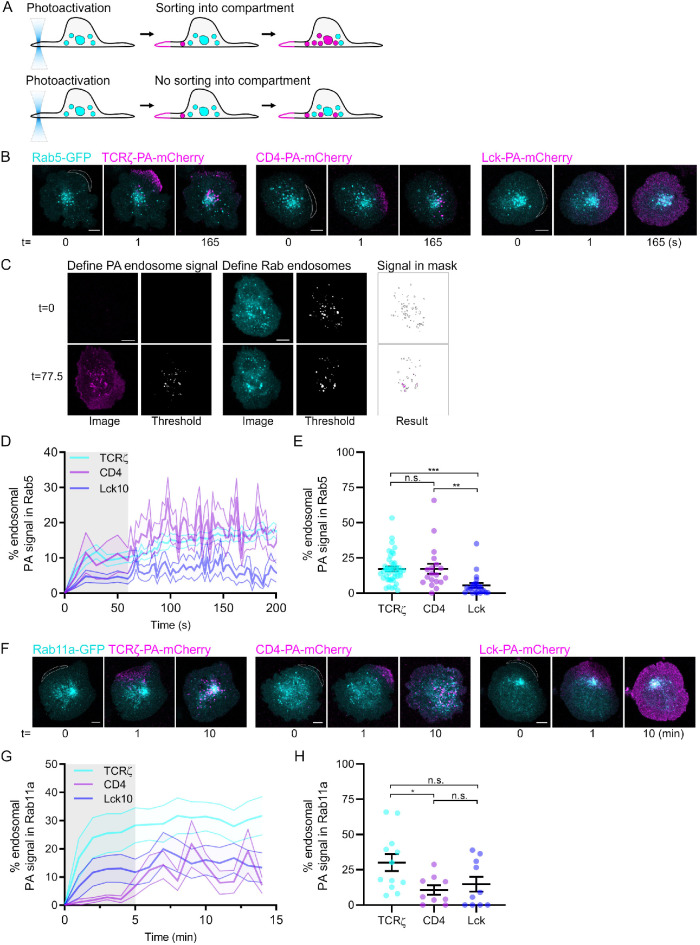
To visualize which endosomes surface proteins are sorted after internalization, we employed a similar approach as when studying endocytosis in cells expressing the protein of interest fused to PA-mCherry and endocytic markers fused to EGFP. We measured, after photoactivation at the plasma membrane, the amount of PA-mCherry fluorescence contained within endosomes demarked by the EGFP signal (Figure 2A). We compared entry of CD4 into Rab5-labeled early endosomes to that of TCRζ, which we know to be rapidly and transiently associated with these endosomes (Redpath et al., 2019). As a negative control we used the membrane-binding region of Lck (Lck10-PA-mCherry), which is not endocytosed at the time scale revealed by our photoactivation approach (Figure 2B).
FIGURE 2:
Real time sorting of receptors into intracellular compartments. (A) Schematic depicting photoactivation to investigate sorting. Top: sorting of photoactivated endosomes (magenta) into the Rab compartment (cyan); bottom: photoactivated endosomes not sorting into the Rab compartment. Dashed lines: quantified compartment mask. (B) Confocal images of activated Jurkat T-cells expressing TCRζ-PA-mCherry, CD4-PA-mCherry, or Lck10-PA-mCherry coexpressing Rab5-GFP. Cells were activated on surfaces, photoactivated for five frames with 5-s intervals by 405-nm laser on the cell periphery (white dashed line), and imaged for 200 s. (C) Example of custom-made automated Fiji-based quantification analysis displaying top: the identification by threshold of photoactivated endosomes and Rab endosomes at 0 s; bottom: the identification by threshold of photoactivated endosomes and Rab endosomes at 77.5 s after photoactivation. (D) Percentage of TCRζ-PA-mCherry, CD4-PA-mCherry, and Lck10-PA-mCherry in GFP–Rab5-positive compartments compared with the cytosol. Grayed region = time of photoactivation. (E) Percentage of TCRζ-PA-mCherry, CD4-PA-mCherry, and Lck10-PA-mCherry in GFP–Rab5-positive compartments at 200 s. (F) Confocal images of activated Jurkat T-cells expressing TCRζ-PA-mCherry, CD4-PA-mCherry, or Lck10-PA-mCherry coexpressing GFP-Rab11a. Cells were activated on surfaces and photoactivated for five frames with 5-s intervals by 405-nm laser on the membrane region of interest (white dashed line), and an image was acquired every 60 s for 10 min. (G) Percentage of TCRζ-PA-mCherry, CD4-PA-mCherry, and Lck-A-mCherry in Rab11a–GFP-positive compartments compared with the cytosol. Grayed region = time of photoactivation. (H) Average percentage of TCRζ-PA-mCherry, CD4-PA-mCherry, and Lck10-PA-mCherry in GFP–Rab11a-positive compartments at 200 s; n.s., not significant; *p < 0.05, **p < 0.01, ***p < 0.001 from Student’s t test of means of at least three independent experiments of three to six cells. Error bars/lines = mean ± SEM. All images were acquired on a Zeiss 880 confocal microscope. Scale bars = 5 µm.
To quantify the entry of photoactivated receptor endosomes into the endocytic compartment of interest, we have established a Fiji/ImageJ macro (Sorting analysis.ijm, Figure 2C). This analysis first segregates the photoactivated signal present in endosomes from the plasma membrane. In the initial frame prior to photoactivation, a threshold is set by the user in the photoactivated channel to ensure only endosomal structures appearing after photoactivation are identified. This is achieved by setting the threshold in order that no signal in the frame prior to photoactivation can be identified (Define PA endosome signal). Throughout the following frames of the time series, this threshold is adjusted by the user so that only the photoactivated signal coming from the endosomes of interest—and not the signal at the plasma membrane—is retained for analysis (Signal in mask). A threshold is then set for the Rab endocytic marker channel to ensure only the endosomal signal is retained for analysis from the Rab channel (Define Rab endosomes). Finally, the intensity of signal of the photoactivated channel present in the Rab endosomes (Signal in mask) is divided by the total endosomal photoactivated signal and plotted as the percentage of total photoactivated signal in endosomes (Figure 2D).
We found that internalized CD4 was incorporated into Rab5-positive endosomes at identical levels to TCRζ (17.14% vs. 17.25% of photoactivated signal in endosomes respectively, Figure 2, D and E). In contrast, Lck10 showed little association with Rab5 endocytic compartments (5.46%). To assess CD4, TCRζ and Lck10 sorting into endocytic compartments downstream of Rab5, we performed the same experiments in Jurkat T-cells expressing a recycling endosomal marker, Rab11a, fused to EGFP. Cells were photoactivated repeatedly on the cell periphery over 30 s, then imaged once per minute for 14 min to limit photobleaching of the photoactivated signal (Figure 2F). This allowed longer-term imaging required to detect trafficking into Rab11-positive recycling endosomes. TCRζ showed robust entry into Rab11a-positive recycling endosomes (Figure 2, G and H), consistent with our previous report (Redpath et al., 2019). By contrast, CD4 and Lck10 entered into Rab11a-positive recycling endosomes at significantly lower levels than TCRζ (10.62 and 14.74% compared with 30.07%, respectively), indicating that neither are typically Rab11a-dependent recycling cargoes.
To ensure that the different incorporations into Rab-positive compartments that we observed were due to genuine biological differences and not differences in photoactivation efficiency, we quantified the mean intensity of TCRζ, CD4, and Lck10-PA-mCherry following photoactivation (Supplemental Figure S1, A and B). In Rab5 sorting experiments, TCR-PA-mCherry was photoactivated to higher levels than CD4 and Lck10-PA-mCherry, which were photoactivated to similar levels. In Rab11 sorting experiments, TCRζ and Lck10-PA-mCherry were photoactivated to near identical levels, while CD4-PA-mCherry photoactivation was lower. Hence, the amount of membrane proteins revealed by photoactivation did not correlate with the amount of the same proteins reaching endosomes containing Rab5 or Rab11. Together, our data strongly support the notion that photoactivation of fluorescent proteins is a valid method to reveal the intracellular fate of internalized membrane proteins.
TCRζ and CD4, unlike Lck10, both reach Rab5-positive endosomes very quickly after endocytosis. This is consistent with the fact that a Rab5-positive compartment represents a sorting hub where freshly internalized membrane proteins are routed for degradation or recycling (Woodman, 2000; Redpath et al., 2020). This method further provides essential information on the kinetics of endocytic trafficking. TCRζ and CD4 were observed in Rab5-positive endosomes seconds after endocytosis, while it took minutes for TCRζ to reach Rab11-positive endosomes. Because of the relatively fast bleaching time of PA-mCherry, these differences also imply that different time intervals between imaging frames have to be used to visualize endocytic processes with different kinetics or cargo delivery into different subcellular compartments. Our data further show that sorting at the level of Rab5 endosomes sends TCRζ for a quick return to the plasma membrane via Rab11-postitive recycling endosomes, while CD4 takes a different route.
Two-photon photoactivation to quantify cargo recycling
Imaging the return to the plasma membrane of surface receptor from intracellular compartments is technically challenging. Constrained photoactivation of photoactivatable fluorescent proteins in specific intracellular compartments using two-photon illumination was an early predicted use of photoactivatable fluorescent proteins (Lippincott-Schwartz and Patterson, 2003). Yet only a single report has utilized two-photon photoactivation to visualize cargo sorting from intracellular compartments (Luo et al., 2006). As such, we established an assay using two-photon photoactivation of cargoes in intracellular compartments to quantify the kinetics of receptor recycling and determine the endosomal populations from which recycling occurs.
In establishing this assay, we found PA-mCherry too rapidly photobleached to obtain meaningful data (data not shown). We settled on the photoswitchable version of CFP, PSCFP2 (Chudakov et al., 2004; photoswitching is referred to as photoactivation hereafter). PSCFP2 exhibited excellent signal and resistance to photobleaching following photoactivation, allowing us to reliably visualize trafficking from intracellular compartments.
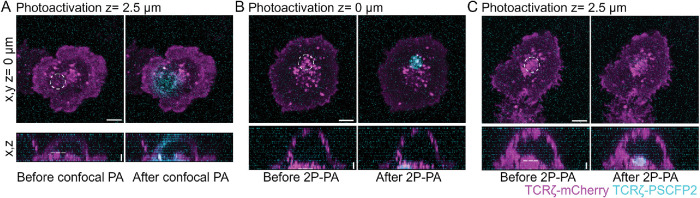
We first validated the two-photon photoactivation conditions in fixed Jurkat T-cells expressing TCRζ-mCherry and TCRζ-PSCFP2. Photoactivation of TCRζ-mCherry compartments 2.5 µm within the cell using a conventional 405-nm laser, resulted in significant photoactivation of the plasma membrane above and below the focal plane (Figure 3A, dashed line). Photoactivation at the plasma membrane (z = 0 µm) using 800 nm light generated by a two-photon laser resulted in constrained photoactivation to within 1 µm of the photoactivation plane, indicating that two-photon photoactivation occurs in a ∼1-µm radius of the focal plane (Figure 3B). To ensure photoactivation occurred without any plasma membrane contamination, we next photoactivated 2.5 µm within the cell (Figure 3C, dashed line). This resulted in robust intracellular photoactivated signal with no detectable signal at the plasma membrane, indicating specific activation of intracellular PSCFP2 was achieved.
FIGURE 3:
Two-photon photoactivation in fixed cells. (A) Representative images of WT Jurkat T-cells expressing TCRζ-mCherry (magenta) and TCRζ-PSCFP2 (cyan), activated on cover glass, and fixed 20 min postactivation. Z-stacks were taken using confocal microscopy of cells before (left) and after (right) single photon photoactivation at 2.5 μm z-depth. (B) Representative images of cells photoactivated with a two-photon (800 nm) laser at Z = 0 µM. Preparation of cells was performed as in A. (C) Representative images of cells photoactivated with a two-photon (800 nm) laser at Z = 2.5 µM. Preparation of cells was performed as in A. Dashed white line indicates photoactivated region. Images were acquired on a Zeiss 880 confocal microscope. Horizontal error bars = 5 μm, vertical = 2 μm.
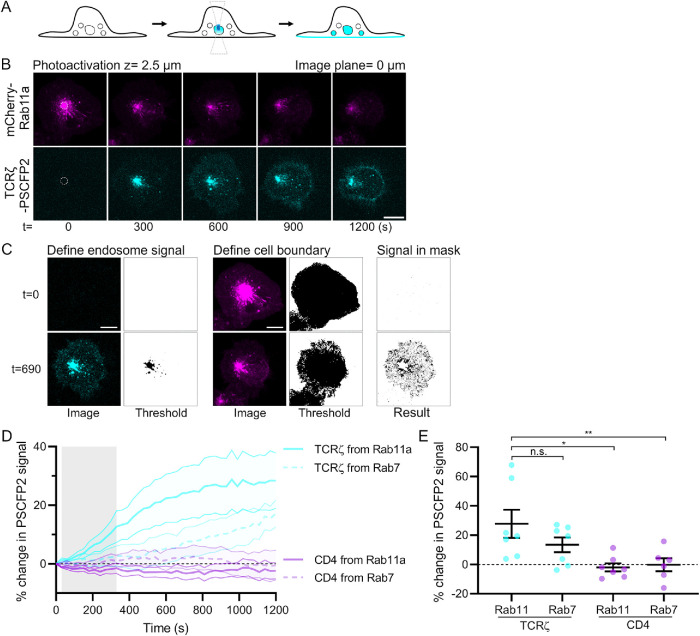
To probe receptor recycling from endosomal compartments of interest, we used Jurkat T-cells expressing TCRζ-PSCFP2 or CD4-PSCFP2 and mCherry-Rab11a or mCherry-Rab7, a marker for late endosomes. In live cells, Rab11a- or Rab7-positive compartments concentrated at the perinuclear region of the cell were photoactivated 2.5 µm within the cell every 30 s for 10 frames, following which cells were imaged for a further 900 s (Figure 4, A and B, dashed circle = photoactivation region). Cells were imaged at the plasma membrane (z = 0 µm) and 1 and 2 µm above the membrane to capture recycling to the plasma membrane and transport vesicles leaving the photoactivated compartment.
FIGURE 4:
Two-photon photoactivation to measure recycling in real time. (A) Schematic displaying two-photon photoactivation in intracellular compartment and recycling after photoactivation. (B) Representative confocal images of WT Jurkat T-cells expressing TCRζ-PSCFP2 (cyan) and mCherry-Rab11a (magenta). Cells were photoactivated with 800 nm light from a 2-Photon laser, 2.5 µm within the cell. Dashed circle = photoactivation region. Image plane = 0 µm. (C) Example of custom-made automated Fiji-based quantification analysis displaying top: the identification by threshold of endosomes and cell boundary at 0 s and bottom: at 690 s after photoactivation. (D) Percentage of change in PSCFP2 signal of TCRζ-PSCFP2 or CD4-PSCFP2 photoactivated from Rab11a- or Rab7-positive endosomes. (E) Average percentage of change in PSCFP2 signal of TCRζ-PSCFP2 or CD4-PSCFP2 photoactivated from Rab11a- or Rab7-positive endosomes at 1200 s; n.s., not significant; *p < 0.05, **p < 0.01 from Student’s t test of means four independent experiments of one to two cells. Error bars/lines = mean ± SEM. All images were acquired on a Zeiss 880 confocal microscope. Scale bars = 5 µm.
These data were quantified using a custom Fiji/ImageJ macro (recycling analysis.ijm, Figure 4C). Initially, a threshold is set by the user so that no PSCFP2 signal present at background levels prior to photoactivation is included in the analysis. This threshold is then checked throughout the image time series to ensure all endosomal signal in the PSCPF2 channel is detected (Define endosome signal). This endosomal signal is subsequently removed from the analysis to ensure only changes in plasma membrane signal are quantified. The boundary of the area of the cell plasma membrane in contact with the activating surface is then user-defined, creating a mask based on the mCherry channel at z = 0 µm (Define cell boundary). Finally, the signal resulting from photoactivation and detected in the plasma membrane mask (Signal in mask) is quantified to determine the level of receptor recycling from the photoactivated compartment. This is then normalized to the background signal at time = 0 to determine the percentage increase in PSCFP2 signal over time.
Photoactivation of TCRζ-PSCFP2 in Rab11a-positive endosomes resulted in a pronounced increase in TCRζ signal at the plasma membrane, while CD4-PSCFP2 photoactivation in Rab11a-positive endosomes resulted in no increase in CD4 plasma membrane signal whatsoever. When TCRζ-PSCFP2 was photoactivated in Rab7-positive endosomes, a modest increase in plasma membrane TCRζ signal was also observed, and again no increase in CD4-PSCFP2 signal was noted following photoactivation from Rab7-positive endosomes. Averaging the percentage increase of the final 300 s of each condition reveals that TCRζ recycled from Rab11a-positive endosomes significantly more than CD4 from either Rab11a- or Rab7-positive endosomes (Figure 4E, 27.73% compared with –2.059 and –0.233%, respectively). Surprisingly, no significant difference was observed between TCRζ recycling from Rab11a and Rab7-positive endosomes (27.73% compared with 13.37%, respectively).
Thus, our photoactivation approach suggests that there are at least two pathways to recycle TCRζ to the plasma membrane in activated Jurkat T-cells: 1) a fast-track route, where TCRζ gets to Rab11-positive recycling endosome after a quick sorting step in Rab5-positive early endosomes, and from there, directly to the plasma membrane (Redpath et al., 2019); 2) a longer way along which TCRζ progresses through endocytic compartments until it reaches Rab7-positive late endosomes, where Rab7 can promote recycling to the plasma membrane via the trans-Golgi network and retrograde transport (Seaman, 2012; Sapmaz et al., 2019). Although it has not been demonstrated for TCRζ, retrograde transport recycles Lat, a key adaptor protein in T-cell activation (Carpier et al., 2018). Additionally, indirect evidence suggests that TCR recycling depends on syntaxin-7 and Rab7 in cytotoxic T-cells (Pattu et al., 2011). In contrast to TCRζ, our results show that CD4 is not transported to the plasma membrane, either from Rab11- or from Rab7-positive endosomes. In summary, these results illustrate the power of our approach. They reveal two different recycling paths for a surface protein, TCRζ, while showing that another receptor, CD4, despite binding to the same molecule—MHCII—than TCR at the surface of antigen-presenting cells, does not recycle via any of these routes.
Two-photon photoactivation to quantify vesicle trafficking from intracellular compartments
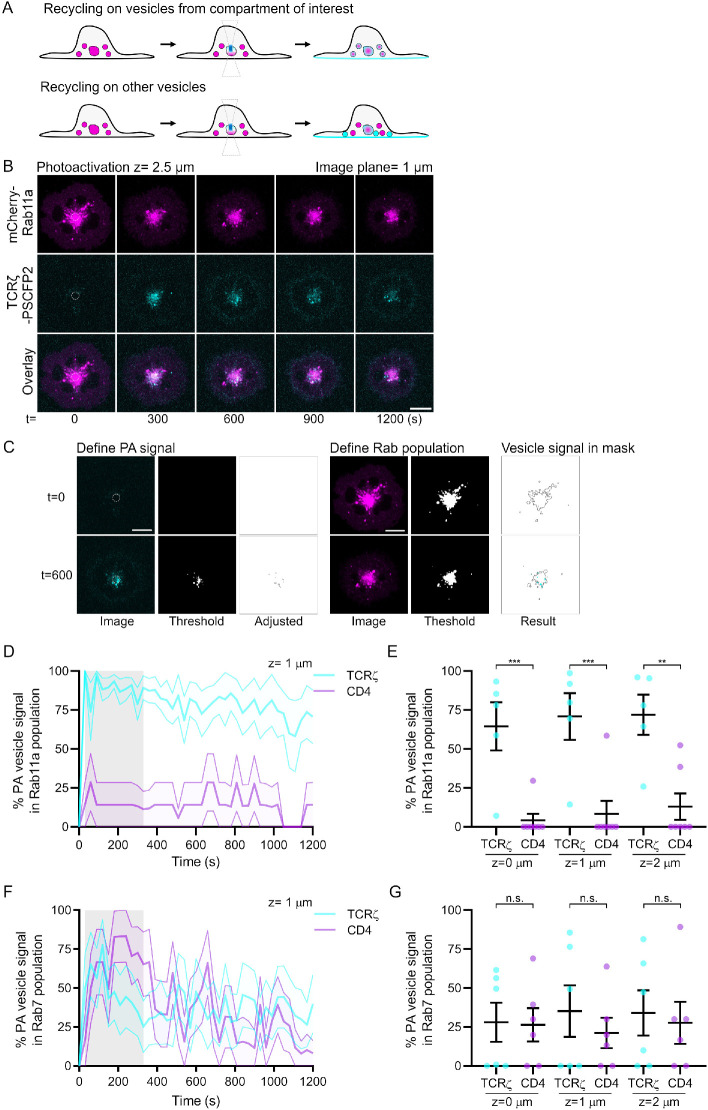
Capturing images from our two-photon membrane delivery assay at multiple z-planes allowed us to quantify vesicular trafficking events occurring from the Rab compartment where photoactivation was performed (Figure 5A). Within the intracellular focal planes, colocalization between the cargo and the endosomal marker was consistently observed over time within and beyond the photoactivated intracellular compartments (Figure 5B, dashed circle). Endocytic trafficking is the sum of highly dynamic fusion, fission, and transport events. Thus, membrane proteins revealed by photoactivation at the surface of a given endosome will eventually leave this endosome in transport vesicles. By excluding the region of endosomes that has been photoactivated from the analysis, we could identify the vesicular population that transports the receptor from this endosomal compartment.
FIGURE 5:
Two-photon photoactivation to identify transport vesicles in real time. (A) Schematic depicting recycling of vesicles from photoactivated compartment of interest, bottom: recycling from other vesicles. (B) Representative confocal images of WT Jurkat T-cells expressing TCRζ-PSCFP2 (cyan) and mCherry-Rab11a (magenta). Cells were activated on surfaces and photoactivated with 800 nm light from a 2-Photon laser, 2.5 µm within the cell. Dashed circle = photoactivation region. Image plane = 1 µm. (C) Example of custom-made automated Fiji-based quantification analysis showing top: the identification by threshold of photoactivated endosome signal and Rab endosomes at 0 s; bottom: the identification by threshold of photoactivated endosome signal and Rab endosomes at 600 s after photoactivation. (D) Percentage of photoactivated TCRζ-PSCFP2 signal that is present in Rab11a endosomal compartments at z = 0. (E) Average percentage of photoactivated TCRζ-PSCFP2 signal that is present in Rab11a endosomal compartments at different z-planes (0, 1, and 2 µm). (F) Percentage of photoactivated TCRζ-PSCFP2 signal that is present in Rab7 endosomal compartments at z = 0. (G) Average percentage of photoactivated TCRζ-PSCFP2 signal that is present in Rab7 endosomal compartments at different z-planes (0, 1, and 2 µm); n.s., not significant; **p < 0.01, ***p < 0.001 from Student’s t test of means of four independent experiments of one to two cells. Error bars/lines = mean ± SEM. All images were acquired on a Zeiss 880 confocal microscope. Scale bars = 5 µm.
To do so, we modified the sorting analysis used in Figure 2 (trafficking analysis for 2PA.ijm). Initially, a user-defined threshold is set to identify the photoactivated signal in the cell. This threshold is set to a level so all signal in the frame prior to photoactivation is not identified to ensure only photoactivated endosomal signal is quantified (Figure 5C) (Define PA signal). A size exclusion is then automatically applied that removes large endosomes where photoactivation was initially performed (as the photoactivated signal within the photoactivated region consistently remains as large endosomal structures) and extremely small regions of photoactivated signal (as the photoactivated signal returning to the plasma membrane is present in very small [less than 5 pixels] clusters; Adjusted). A user-defined threshold is set to identify the Rab population of interest (Define Rab population) and the photoactivated receptor signal outside of the initial photoactivation region and present within the Rab endosomal population of interest quantified (Signal in mask). The endosomal photoactivated signal present in the Rab compartment is then expressed as a percentage of the total endosomal photoactivated signal detected outside of the photoactivated region.
TCRζ was present on Rab11a-positive vesicles outside of the photoactivated endosomal region of the cell at significantly higher levels than CD4 at each z plane of the image (Figure 5, D and E). CD4 was rarely present on Rab11a-positive vesicles, with only two cells having any CD4 signal in Rab11a-positive vesicles across all experiments. In contrast, TCRζ and CD4 were present equally on Rab7-positive vesicles outside of the endosomal photoactivated regions at each z plane of the image (Figure 5, F and G), indicating both receptors are capable of trafficking on Rab7-positive vesicles within the cell.
Quantification of the PSCFP2 intensity following photoactivation revealed that no CD4-PSCFP2 could be photoactivated on Rab11a-positive intracellular compartments (Supplemental Figure S1C), confirming the data in Figure 2, which showed that CD4 does not reach Rab11-positive endosomes after endocytosis. This quantification also revealed that the amount of a given protein photoactivated in a given endosome does not influence the identification of transport vesicles. We detected a similar number of Rab7-positive vesicles containing TCRζ-or CD4, even though PSCFP2 photoactivation occurred at higher levels on Rab7-positive endosomes compared with CD4 (Supplemental Figure S1D).
This quantification of the two-photon photoactivation data confirms that TCRζ is recycled to the plasma membrane from Rab11-positive intracellular compartments within Rab11-positive transport vesicles. Similarly, it shows that TCRζ is transported from Rab7-positive endosomes by Rab7-positive vesicles to be eventually recycled to the cell surface. CD4, which is not returned to the plasma membrane from Rab7-positive endosomes, is nevertheless transported by Rab7-positive vesicles. This raises the question of the destination of CD4 when it leaves late endosomes. HIV infection of T-cells and the resulting expression of the protein Nef lead to CD4 down-regulation from the cell surface by enhanced endocytosis and degradation in lysosomes (Amorim et al., 2014). A Nef-interacting protein, b-COP, directs CD4, via Rab7-positive vesicles, toward late endosomes and lysosomes for degradation in T-cells infected by HIV (Schaefer et al., 2008). Similarly, HIV infection stimulates the expression of the small GTPase HRES-1 which inhibits CD4 recycling and targets it for degradation (Nagy et al., 2006). Our study, which addresses the endocytic trafficking of CD4 in Jurkat T-cells, could not identify a clear return of internalized CD4 to the cell surface from Rab11- or from Rab7-positive compartments in the absence of an HIV infection. Of note, the reported half-life of CD4 is ∼24 h in the absence of the viral Nef protein (Rhee and Marsh, 1994). Because of this long half-life, it is possible that there was not enough time for degradation of CD4 in the time frames our experiments were undertaken. The presence of CD4 in Rab7-positive compartments could mean that CD4 is predominantly targeted for degradation in the lysosome or that it is returned to the cell surface independently of Rab11 or Rab7, or from Rab7 endosomes but at a rate too slow to be measured with our photoactivation approach.
CONCLUSION
The photoactivation assays we present here represent the first comprehensive approach to visually dissect every step of the endocytic recycling pathway in live cells. Through a single methodological concept—photoactivation of fluorescent proteins—we were able to generate quantitative information about mechanisms of endocytosis at the plasma membrane (Figure 1), incorporation into sorting and recycling endosomes (Figure 2), delivery from endosomes to the plasma membrane (Figures 3 and 4), and finally nature of vesicles carrying out intracellular transport (Figure 5).
We illustrate the possibilities of this approach in a system, T-cell activation, which relies greatly on intracellular trafficking (Onnis and Baldari, 2019). Despite the inherent narrow cytoplasmic volume of these cells and the resulting crowding of their endosomes, we were able to observe distinct, if not opposite, endocytic behaviors of two receptors, TCRζ and CD4, that are involved in the same cellular process.
Our data indicate that TCRζ is quickly returned to the plasma membrane after clathrin-independent endocytosis through a fast Rab5-Rab11 recycling pathway as recently published (Redpath et al., 2019). They also reveal that there is at least another recycling route for TCRζ that goes through Rab7-positive endosome and transport vesicles, most likely involving retromers. Why TCRζ is recycled through two distinct routes in the same cells in the same conditions remains to be elucidated, although it may be to facilitate signaling in endosomes (Benzing et al., 2013; Saveanu et al., 2019).
This study further represents the first investigation of CD4 endocytic trafficking in live T-cells. Photoactivation at the plasma membrane revealed that CD4 is only marginally internalized by clathrin-mediated endocytosis, against the current dogma. It is, however, possible that a slow, clathrin-dependent endocytic process occurs in parallel to the fast internalization we observed. Determining if this is the case would require increasing the time interval between imaging frames in order to avoid bleaching PA-mCherry and allow the detection of endocytic vesicles minutes after the initial photoactivation instead of seconds. Using photoactivation of intracellular compartments, we also could not detect recycling of CD4 and observed internalized CD4 in Rab7-positive transport vesicles. Determining why there are two potential endocytic pathways for CD4 and whether its incorporation into Rab7-positive endosome leads to degradation in lysosomes or return to the cell surface via an alternative recycling pathway, perhaps at a very slow rate, will require further investigation.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Plasmids
TCRζ-PSCFP2 and Lck10-PA-mCherry were provided by K. Gaus (University of New South Wales). PA-mCherry expression backbone was obtained from Clontech. TCRζ-PA-mCherry and CD4-PA-mCherry were generated by inserting TCRζ or CD4 into the PA-mCherry backbone with EcoRI and AgeI. Clathrin-EGFP was kindly provided by T. Kirchausen (Harvard Medical School). mCherry-Rab5 was a gift from Gia Voeltz (Addgene plasmid #49201). mCherry-Rab7a (Addgene plasmid #55127) and mCherry-Rab11a (Addgene plasmid #55124) were gifts from Michael Davidson.
Cell culture
Jurkat T-cells (ATCC Cat# TIB-152, RRID:CVCL_0367) were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Invitrogen). T-cells were transfected with 1 µg DNA per 200,000 cells, 12–24 h prior to imaging using the Neon electroporation kit (Invitrogen).
Before imaging, cells were incubated for 10 min at 37°C on 18-mm glass-coated surfaces (Marienfeld) that were prepared by incubating with poly-l-lysine (Sigma) for 30 min at room temperature. Afterward coverslips were washed once with phosphate-buffered saline and incubated with 1 µM anti-CD3ε (16-0037; eBioscience) and anti-CD28 (16-0289; eBioscience) antibodies overnight at 4°C for T-cell activation. Cells were imaged from 10 to 40 min after their deposition on the coverslips.
For transferrin internalization, transfected Jurkat T-cells were activated as above and placed under the microscope. Alexa 647–conjugated transferrin (Jackson ImmunoResearch, USA) was added to T-cells 5 min after activation at 25 µg/ml (5 µl/ml), incubated for a further 10 min, and imaged.
Microscopy
Live-cell confocal microscopy was performed on a Zeiss LSM880 laser-scanning confocal microscope (Zeiss, Germany) equipped with an argon laser (405, 488 nm), a diode pump solid state laser (561, 647 nm), an Airy disk, and a live-cell incubation chamber (Pecon) as well as an Airyscan detector and a Mai Tai Insight DeepSee tuneable multi-photon laser.
GFP constructs and photoactivated TCRζ-PSCFP2 were excited using the 488-nm line of the argon laser source, while PA-mCherry and mCherry tagged proteins were excited with the 561-nm laser line. Images were acquired using a 100× 1.4 NA DIC M27 Apo-Plan oil immersion objective (Zeiss, Germany) and GaAsP-PMTs in simultaneous, bidirectional scanning mode. For each channel, the pinhole was set to 1 Airy Unit.
Confocal photoactivation was achieved by illuminating a region of the cell outer membrane with a 7.2 µW 405-nm laser pulse with 12.24 µs pixel dwell time. Two-photon photoactivation was achieved by illuminating PSCFP2 at a focal point 2.5 μm from the coverslip with a 58 mW 800-nm laser pulse with 65.54 µs pixel dwell time every 30 s for 10 consecutive cycles.
Statistical analysis
All statistics were performed using GraphPad Prism version 8. Statistical significance between datasets was determined by performing two-tailed, unpaired Student’s t tests or one-way ANOVA with Sidak correction for multiple comparisons, where stipulated. Graphs show mean values, and error bars represent the SEM. In statistical analysis, p > 0.05 is indicated as not significant (n.s.), whereas statistically significant values are indicated by asterisks as follows: *p ≤ 0.05, **p < 0.01, ***p < 0.001.
Image analysis and code availability
Image analysis was performed using custom MATLAB analyses for the nearest neighbor distances. Vesicles were identified and localized by intensity-based thresholding from the background of each image of the time series after filtering by a spatial band-pass of 2–9 pixels. Cross-channel nearest-neighbor distances were calculated based on the vesicle localization. The randomized control distribution was generated by retaining the spatial information from the localization measurements and scrambling the temporal information before regenerating the cross-channel nearest-neighbor distance distribution. Repeating this 100 times allowed confidence intervals to this randomized control. Fiji (Schindelin et al., 2012) routines were used as detailed in the main text for incorporation into endosomes, delivery to the plasma membrane, and identification of transport vesicles. Fiji macros are included as a supplementary download for this publication. MATLAB codes are available from Philip R. Nicovich GitHub repo [https://github.com/ PRNicovich/PAVesT.git].
Supplementary Material
Acknowledgments
We thank the staff of the BioMedical Imaging Facility of the University of New South Wales. We thank the following funding bodies: National Health, Medical Research Council (APP1102730), the Swiss National Science Foundation (SNSF 31003A_172969), the Thurgauische Stiftung für Wissenschaft und Forschung, the State Secretariat for Education, Research and Innovation, and the Deutsche Forschungsgemeinschaft (RO 6238/1-1).
Abbreviations used:
- BiFC
bimolecular fluorescence complementation
- HIV
human immunodeficiency virus
- TCR
T-cell receptor
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-10-0669) on February 3, 2021.
REFERENCES
- Achour L, Labbé-Jullié C, Scott MGH, Marullo S (2008). An escort for GPCRs: implications for regulation of receptor density at the cell surface. Trends Pharmacol Sci 29, 528–535. [DOI] [PubMed] [Google Scholar]
- Amorim NA, Da Silva EML, De Castro RO, Da Silva-Januário ME, Mendonça LM, Bonifacino JS, DaCosta LJ, DaSilva LLP (2014). Interaction of HIV-1 nef protein with the host protein Alix promotes lysosomal targeting of cd4 receptor. J Biol Chem 289, 27744–27756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B (2001). Marching at the front and dragging behind: Differential αVβ3-integrin turnover regulates focal adhesion behavior. J Cell Biol 155, 1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing C, Rossy J, Gaus K (2013). Do signalling endosomes play a role in T cell activation? FEBS J 280, 5164–5176. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science (80-) 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira APA, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT (2015). Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465. [DOI] [PubMed] [Google Scholar]
- Burtey A, Rappoport JZ, Bouchet J, Basmaciogullari S, Guatelli J, Simon SM, Benichou S, Benmerah A (2007). Dynamic interaction of HIV-1 Nef with the clathrin-mediated endocytic pathway at the plasma membrane. Traffic 8, 61–76. [DOI] [PubMed] [Google Scholar]
- Carpier J-M, Zucchetti A, Bataille L, Dogniaux S, Shafaq-Zadah M, Bardin S, Lucchino M, Maurin M, Joannes L, Magalhaes J, et al. (2018). Rab6-dependent retrograde traffic of LAT controls immune synapse formation and T cell activation. J Exp Med 215, 1245–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson K, et al. (2007). Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell 13, 496–510. [DOI] [PubMed] [Google Scholar]
- Chia PZC, Ramdzan YM, Houghton FJ, Hatters DM, Gleeson PA (2014). High-throughput quantitation of intracellular trafficking and organelle disruption by flow cytometry. Traffic 15, 572–582. [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA (2004). Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol 22, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Aguet F, Boulant S, Kirchhausen T (2012). The first five seconds in the life of a clathrin-coated pit. Cell 150, 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeer EB, Kraus F, Ecker M, Redpath G, Amiezer M, Rother N, Nicovich P, Kapoor-Kaushik N, Deng Q, Samson G, et al. (2018). A mobile endocytic network connects clathrin-independent receptor endocytosis to recycling and promotes T cell activation. Nat Commun 9, 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Steinberg F (2018). To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat Rev Mol Cell Biol 19, 679–696. [DOI] [PubMed] [Google Scholar]
- Dirk BS, Pawlak EN, Johnson AL, Van Nynatten LR, Jacob RA, Heit B, Dikeakos JD (2016). HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci Rep 6, 37021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli R, Kuschinsky W (2001). Brain glucose transporters: relationship to local energy demand. Physiology 16, 71–76. [DOI] [PubMed] [Google Scholar]
- Fletcher-Jones A, Hildick KL, Evans AJ, Nakamura Y, Wilkinson KA, Henley JM (2019). The C-Terminal helix 9 motif in rat cannabinoid receptor type 1 regulates axonal trafficking and surface expression. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Carpentier JL, Aiken C, Trono D, Lew DP, Krause KH (1997). Second-messenger regulation of receptor association with clathrin-coated pits: a novel and selective mechanism in the control of CD4 endocytosis. Mol Biol Cell 8, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PD, Loftus RJ, Tamkun MM (2013). Regulation of Kv2.1 K+ conductance by cell surface channel density. J Neurosci 33, 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Willingham MC, Pastan I (1984). Kinetics of transit of transferrin and epidermal growth factor through clathrin-coated membranes. Cell 39, 283–293. [DOI] [PubMed] [Google Scholar]
- Hong SH, Cortesio CL, Drubin DG (2015). Machine-learning-based analysis in genome-edited cells reveals the efficiency of clathrin-mediated endocytosis. Cell Rep 12, 2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu VW, Bai M, Li J (2012). Getting active: Protein sorting in endocytic recycling. Nat Rev Mol Cell Biol 13, 323–328. [DOI] [PubMed] [Google Scholar]
- Jonker CTH, Deo C, Zager PJ, Tkachuk AN, Weinstein AM, Rodriguez-Boulan E, Lavis LD, Schreiner R (2020). Accurate measurement of fast endocytic recycling kinetics in real time. J Cell Sci 133, jcs231225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlecova Z, Spielman SJ, Loerke D, Mohanakrishnan A, Reed DK, Schmid SL (2017). Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J Cell Biol 216, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller GA, Siegel MW, Caras IW (1992). Endocytosis of glycophospholipid-anchored and transmembrane forms of CD4 by different endocytic pathways. EMBO J 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Raposo G (2014). The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol 6, a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard D, Hayakawa A, Lawe D, Lambright D, Bellve KD, Standley C, Lifshitz LM, Fogarty KE, Corvera S (2008). Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J Cell Sci 121, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH (2003). Development and use of fluorescent protein markers in living cells. Science (80-) 300, 87–91. [DOI] [PubMed] [Google Scholar]
- Luo H, Nakatsu F, Furuno A, Kato H, Yamamoto A, Ohno H (2006). Visualization of the post-golgi trafficking of multiphoton photoactivated transferrin receptors. Cell Struct Funct 31, 63–75. [DOI] [PubMed] [Google Scholar]
- Manrique S, Sauter D, Horenkamp FA, Lülf S, Yu H, Hotter D, Anand K, Kirchhoff F, Geyer M (2017). Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and. Nat Commun 8, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M, Armes JE, Pelchen-Matthews A (1990). Endocytosis and recycling of CD4. In: Biochemical Society Transactions, Portland Press, 139–143. [DOI] [PubMed] [Google Scholar]
- Mattheyses AL, Simon SM, Rappoport JZ (2010). Imaging with total internal reflection fluorescence microscopy for the cell biologist. J Cell Sci 123, 3621–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE (2004). Endocytic recycling. Nat Rev Mol Cell Biol 5, 121–132. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D (2005). Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell 121, 593–606. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Ghosh RN, Maxfield FR (1997). Endocytosis. Physiol Rev 77, 759–803. [DOI] [PubMed] [Google Scholar]
- Nagy G, Ward J, Mosser DD, Koncz A, Gergeley P Jr., Stancato C, Qian Y, Fernandez D, Niland B, Grossman CE, et al. (2006). Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem 281, 34574–34591. [DOI] [PubMed] [Google Scholar]
- Onnis A, Baldari CT (2019). Orchestration of immunological synapse assembly by vesicular trafficking. Front Cell Dev Biol 7, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J (2002). A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877. [DOI] [PubMed] [Google Scholar]
- Pattu V, Qu B, Marshall M, Becherer U, Junker C, Matti U, Schwarz EC, Krause E, Hoth M, Rettig J (2011). Syntaxin7 is required for lytic granule release from cytotoxic T lymphocytes. Traffic 12, 890–901. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Boulet I, Littman DR, Fagard R, Marsh M (1992). The protein tyrosine kinase p56lck inhibits CD4 endocytosis by preventing entry of CD4 into coated pits. J Cell Biol 117, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Parsons IJ, Marsh M (1993). Phorbol ester-induced downregulation of CD4 is a multistep process involving dissociation from p56lck, increased association with clathrin-coated pits, and altered endosomal sorting. J Exp Med 178, 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira EA, daSilva LLP (2016). HIV-1 Nef: taking control of protein trafficking. Traffic 17, 976–996. [DOI] [PubMed] [Google Scholar]
- Pitcher C, Höning S, Fingerhut A, Bowers K, Marsh M (1999). Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol Cell Biol 10, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo RAS, Thomas B, Ridlova G, James W (2011). Proteomic-based identification of cd4-interacting proteins in human primary macrophages. PLoS One 6, e18690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath GMI, Betzler VM, Rossatti P, Rossy J (2020). Membrane heterogeneity controls cellular endocytic trafficking. Front Cell Dev Biol 8, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath GMI, Ecker M, Kapoor-Kaushik N, Vartoukian H, Carnell M, Kempe D, Biro M, Ariotti N, Rossy J (2019). Flotillins promote T cell receptor sorting through a fast Rab5–Rab11 endocytic recycling axis. Nat Commun 10, 4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S, Marsh J (1994). Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J Virol 68, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossatti P, Ziegler L, Schregle R, Betzler VM, Ecker M, Rossy J (2019). Cdc42 couples T cell receptor endocytosis to GRAF1-mediated tubular invaginations of the plasma membrane. Cells 8, 1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossy J, Williamson DJ, Gaus K (2012). How does the kinase Lck phosphorylate the T cell receptor? Spatial organization as a regulatory mechanism. Front Immunol 3, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapmaz A, Berlin I, Bos E, Wijdeven RH, Janssen H, Konietzny R, Akkermans JJ, Erson-Bensan AE, Koning RI, Kessler BM, et al. (2019). USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7. Nat Commun 10, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu L, Zucchetti AE, Evnouchidou I, Ardouin L, Hivroz C (2019). Is there a place and role for endocytic TCR signaling? Immunol Rev 291, 57–74. [DOI] [PubMed] [Google Scholar]
- Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL (2008). HIV-1 Nef targets MHC-I and CD4 for degradation via a final common β-cop-dependent pathway in T cells. PLoS Pathog 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaying V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfield S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ (2012). The retromer complex - endosomal protein recycling and beyond. J Cell Sci 4693–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Mayor S (2018). Editorial Overview: Membranes and organelles: rethinking membrane structure, function and compartments. Curr Opin Cell Biol 53, A1–A3. [DOI] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV (2009a). Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods 6, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV (2009b). Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat Chem Biol 5, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Bookman MA, Horak EM, Samelson LE, Bolen JB (1989). Signal transduction through the CD4 receptor involves the activation of the internal membrane tyrosine-protein kinase p56 lck. Nature 338, 257–259. [DOI] [PubMed] [Google Scholar]
- Weigel AV, Tamkun MM, Krapf D (2013). Quantifying the dynamic interactions between a clathrin-coated pit and cargo molecules. Proc Natl Acad Sci USA 110, E4591–E4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman PG (2000). Biogenesis of the sorting endosome: The role of Rab5. Traffic 1, 695–701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.