Abstract
The recent outbreak of SARS-CoV-2 has had a profound effect on the world. Similar to that in SARS-CoV, the entry receptor of SARS-CoV-2 is ACE2. The binding of SARS-CoV-2 spike protein to ACE2 is the critical to the virus infection. Recently multiple species (human, Chinese chrysanthemum, Malay pangolin and cat) have been reported to be susceptible to the virus infection. However, the binding capacity and the detailed binding mechanism of SARS-CoV-2 spike protein to ACE2 of these species remains unexplored. Herein free energy calculations with MM-GBSA and Potential of Mean Forces together reveal that the Human-SARS-CoV-2 has a higher stability tendency than Human-SARS-CoV. Meanwhile, we uncover that SARS-CoV-2 has an enhanced ability to bind with the ACE2 in humans, pangolins and cats compared to that in bats. Analysis of key residues with energy decomposition and residue contact maps reveal several important consensus sites in ACE2s among the studied species, and determined the more favorable specified residues among the different types of amino acids. These results provide important implications for understanding SARS-CoV-2 host range which will make it possible to control the spread of the virus and use of animal models, targeted drug screening and vaccine candidates against SARS-CoV-2.
Keywords: SARS-CoV-2, ACE2, Molecular simulation, Binding energy, Animal hosts
1. Introduction
The recent outbreak of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has had a profound impact on the world. So far, the virus has spread to over 220 countries and caused more than 2 million deaths. Therefore, development of vaccines/drugs is critical to the control of the pandemic. However, no effective antiviral drugs are currently available although a number of drug candidates are presently under development [[1], [2], [3], [4]].
According to genome comparison studies, SARS-CoV-2, like SARS-CoV, belongs to the genus beta-coronavirus. The Receptor Binding Domain (RBD) of the SARS-CoV-2 spike protein has 73% sequence identity with the SARS-CoV RBD. SARS-CoV-2 enters the host cell through the interaction of its RBD and the Angiotensin-converting Enzyme 2 (ACE2) [5,6]. in vitro studies showed that the purified recombinant human ACE2 protein is capable of attracting coronaviruses and preventing SARS-CoV-2 from accessing the cells. However, the murine recombinant soluble ACE2 (mrsACE2) was unable to bind directly to viruses and did not significantly affect the infection of cells [7]. Similarly, based on the naive llama single-domain antibody library and PCR, the generated related nanobodies were reported to bind tightly to the RBD and block its interaction with ACE2 [8]. Therefore, ACE2 and the RBD protein are considered as potential targets for novel antiviral drug discovery and several studies have confirmed that ACE2 qualifies as a therapeutic target.
Given the importance of the interaction between ACE2 and spike-RBD, the structures of the Human-SARS-CoV-2 were recently resolved through cryo-electron microscopy (cryo-EM) and X-ray [[9], [10], [11]]. However, despite the intuitive and credible interface formed by ACE2 and RBD on the resolved structure, the protein-protein interactions at the interface are often nonspecific, resulting in structural diversification. In addition, these different structures resolved by cryo-EM and X-ray have different conformations, giving rise to a slightly different interface. More importantly, with increasing research on SARS-CoV-2, several other species have been reported to be susceptible to infection by the virus although bats and pangolins are believed to be the original and intermediate hosts, respectively. Interestingly, the virus was recently detected in domestic cats [12,13]. Therefore, exploring both similarity and difference involved in the interface between ACE2 and RBD, gathering information on key biological processes of species interfaces for understanding potential natural animal-to-human transmission, will guide rational molecular design strategies.
Molecular dynamics (MD) simulation has been proved a powerful technique to investigate protein interactions at the atomic level [14,15]. However, in previous studies MD simulations were mainly used to examine the mechanism involved in the single Human-SARS-CoV-2 system or explore the interaction between small molecule inhibitors and proteins [[16], [17], [18]]. At the beginning of the outbreak, Hao [19] showed that the binding free energy of Human-SARS-CoV (−78.6 kcal⋅mol−1) is higher than that of Human-SARS-CoV-2 (−50.6 kcal⋅mol−1) by MD simulations in MOE2019. Subsequently, an additional study [20] showed similar findings based on a protein-protein docking method. They reported that the receptor-binding affinity between SARS-CoV-2 and human ACE2 was 73% of that of SARS-CoV. On the contrary, opposite results were obtained in several other MD simulation studies. Researchers [[21], [22], [23]] reported that SARS-CoV-2 has the larger interaction area involving human ACE2 contacts and also has the larger correlation in Pearson based matrices analyses. These results supported the stronger binding of SARS-CoV-2 RBD to ACE2 than SARS-CoV. Recently, based on their own developed simulation method, Zhu group [24] confirmed that the CoV-2-S-RBD could maintain higher affinity binding to human-ACE2 than the CoV–S in all fluctuating conformations. Consequently, it is important to conduct MD simulations with a variety of appropriate analytical methods to compare the stability of the Human-SARS-CoV-2 and Human-SARS-CoV complexes, and to extend these methods to explore the detail interaction between SARS-CoV-2 and the ACE2 of reported species.
In this study, focus was directed to the four species previously reported to be susceptible to infection with SARS-CoV-2, including humans, the Malayan pangolin, Cats and Chinese chrysanthemum. The RBD-ACE2 interacted interfaces of these species were revealed by a molecular modeling study through homology modeling, molecular dynamics (MD) simulation, steered molecular dynamics (SMD) simulation and Potential Mean Force (PMF) calculations. First, molecular modeling was used to construct the unknown complexes of the Pangolin-SARS-CoV-2, Cat-SARS-CoV-2 and Bat-SARS-CoV-2. Based on the structural models, MD simulations were performed to obtain information on the molecular interactions between spike-RBD and different ACE2 proteins. Moreover, energy decomposition analysis and PMF calculations were used to determine the critical sites at the interacted interface of all four species and to analyze differences in binding capacity among the species. The obtained results will help in understanding the binding mechanism between virus and multi-species, and provide useful insights on drug and vaccine development.
2. Materials and methods
2.1. Homology modeling and quality assessment
The study used Swiss Model (https://www.swissmodel.expasy.org) and template pdb (ID: 2AJF) to construct the complexes of the ACE2 and SARS-CoV-2-RBD proteins of human (Homo sapiens NP_001358344.1), pangolin (Manis javanica XP_017505746.1), cat (Felis catus AAX59005.1) and bat (Rhinolophus sinicus ADN93475.1) [25]. Sequences of mice (Mus musculus NP_081562.2) was also considered because recent structural studies suggest that mouse ACE2 is likely poor receptors for SARS-CoV-2 [26]. In addition, the online SAVES webserver (https://servicesn.mbi.ucla.edu/SAVES/) was used to assess the quality of the prediction model [27].
2.2. MD simulation
The GROMACS (2018) software with amber14sb force field was used as the MD simulation tool to optimize the structures of the homology model. In addition, TIP3P waters was added to all the systems with the counter-ions Cl- and Na+. Notably, periodic boundary conditions were considered in order to avoid possible problems from the boundary effect. Additionally, the Particle Mesh Ewald (PME) algorithm and a 1.2 nm distance cutoff were applied for the long-range and short-range electrostatic interactions, respectively. The Nose–Hoover algorithm and Parrinello-Rahman were also used to maintain the temperature and pressure at 300 K and 1 atm, receptivity. Moreover, the steepest descent method was considered for energy minimization in all the MD simulation systems and each system was equilibrated under the NVT-ensemble and NPT-ensemble states. The coupling time constant was 2.0ps. Finally, a 500ns simulation was performed on each system to obtain a stable conformation of the complex in the system [28,29].
2.3. Binding free energy calculation/MM-GBSA
The binding free energies of the protein–protein systems were calculated through the MM-GBSA procedure encoded in the AMBER 18.0 software. Generally, the binding free energy (ΔGbind) was calculated as follows:
where was the molecular mechanics free energy, while ΔGsol and TΔS were the solvation free energy and entropy contribution, respectively. Furthermore, the value included the van der Waals energy (ΔGvdw) and the electrostatic (ΔGele) energy:
here, the electrostatic solvation ΔGsol represented the polar solvation free energy (ΔGele,sol) and the nonpolar solvation free energy (ΔGnonpol,sol):
The ΔGele,sol was determined using the Generalized Born (GB) equation with the dielectric constants of the solute and solvent set at 1.0 and 80.0, respectively. The ΔGnonpol,sol represented non-electrostatic contributions to the solvation energy [30,31].
2.4. SMD simulation
SMD simulations were initiated using coordinates and velocities from equilibrium MD simulations to facilitate the unbinding of the spike protein from the ACE2 protein [32,33]. At the beginning of SMD simulations, the 8 nm × 8 nm × 28 nm (x,y,z) box was constructed using the gmx editconf module of the GROMACS software, taking into account the periodic distance and the centroid of the ACE2-RBD complex was placed on one side of the box (4 nm × 4 nm × 4 nm). Thereafter, about 55,000 water molecules and 150 mmol counter NaCl were included to neutralize the resulting system. Notably, the ACE2 receptor was set as a fixed reference during the SMD simulations and external steering forces were applied to the spike-RBD protein to gradually separate it from the complex system along the predefined direction. Moreover, the spring constant of 1000 kJ⋅mol−1 nm−2 and pulling speed of 0.01 nm/ps were applied to ensure the spike-RBD pulled away from the composite along the z-axis within 500 ps. Both coordinates and steering forces were saved after every 1 ps for further analysis [34,35].
2.5. Umbrella sampling and determination of PMF
The PMF profile was constructed from a series of umbrella sampling windows. For each system, 53 to 55 asymmetric windows with a sampling distance of 0.5 Å∼1 Å were selected for the umbrella sampling simulations from the SMD simulation. Each window was then equilibrated for 10ns with a force constant of 1000 kJ/mol/nm2. The total number of sampling windows for the seven systems was 378 and the overall simulation time was ∼3.78 μs. In addition, based on outputs from the above US, the PMF was calculated using the weighted histogram analysis method provided by the gmx wham tool [36,37].
3. Results
3.1. Structure prediction
It is noteworthy that at the time this study began, the complex structure of the human ACE2 and spike-RBD protein had not been experimentally resolved (this was uncovered in a later study). Notably, there exists high homology in the sequences of mammalian ACE2 and there is also a high similarity between SARS-CoV and SARS-CoV-2. There is 80% sequence identity in the ACE2 of humans, pangolins, cats, bats and mice. In addition, the RDB protein is 85% similar in the sequences between SARS-CoV and SARS-CoV-2. Based on the sequence similarities, the homology models of the five species were generated by using the homology modeling program Swiss Model.
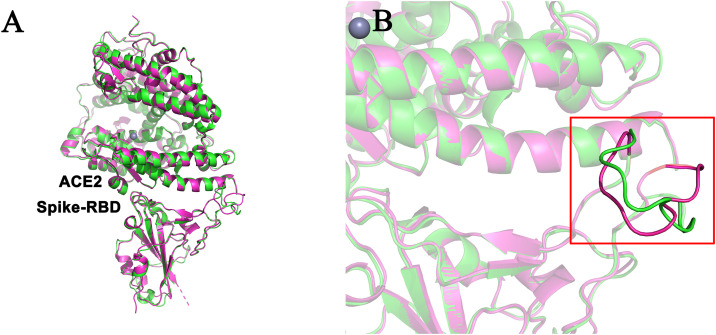
To estimate the quality of a protein model, model quality assessment methods can be used. Notably, each VERIFY 3D score of the predicted structures was greater than 95%, and almost all the ERRAT scores were 90% while the PROVE warning parts were not more than 5%. In addition, analysis of the Ramachandran plot indicated that more than 98% of the residues fell under the favored region, suggesting the reliability of predicted structures (see TableS1). Moreover, the predicted structure of the human system and the currently resolved cryo-EM structure were further analyzed. The two structures were exceptionally similar with a backbone Root Mean Squared Deviation (RMSD) of 0.462 Å (Fig. 1 ). Particularly, two interfaces of structures agree well, even though there is only a minor deviation in a small loop part because of their inherent high flexibilities. Overall, due to the high levels of sequence similarity across the species and the high sequence homology between SARS-CoV and SARS-CoV-2, the predicted results of homology modeling were credible.
Fig. 1.
Comparison between the predicted structure (green) and resolved structure (magenta) of complex formed by human ACE2-RBD complex. The two structures superimpose very well to each other and the interface areas almost all the same.
3.2. The interactions between ACE2 and RBD proteins
Based on the results of homology modeling, all the five complexes of SARS-CoV-2 in the different species, as well as the human-SARS-CoV and mice-SARS-CoV complexes were selected for further MD simulations. The template complex of SARS-CoV and human-ACE2 (PDB id: 2AJF) was simulated as a reference. Simulations for each system covered a minimum of 200 ns, resulting in a total simulation time of 1.4 μs. The Root-mean-square Fluctuation (RMSF) was computed for each MD-simulated system to highlight dynamic behaviours of individual amino acid residues within a system. The RMSF plots indicated that similar residue fluctuation profile for all investigated complexes remained in the range of 0.1–1 nm, which could be inferred that all the virus-receptor complexes remain in stable configurations during MD simulations. Figure S1 also showed that a vast majority of important catalytic residues present at the protein-protein interacted interface had an RMSF value of not more than 0.2 nm, except for N-terminal regions with the inherent flexibility. Although residues involved in the binding sites in the spike protein were partially located in the flexibility loop regions, the low RMSF values of these residues at the corresponding yellow locations were attributed to the stabilizing-effect of the binding proteins.
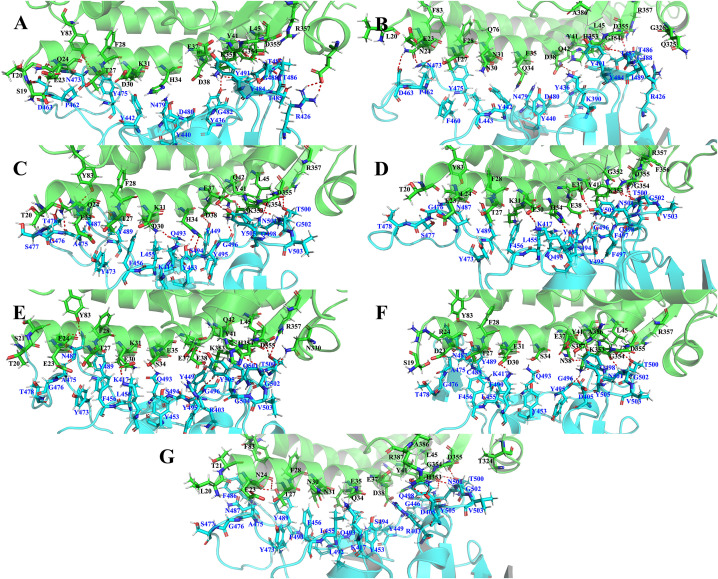
The most stable conformation was uncovered through MD simulations. In terms of human-SARS-CoV complex, the residues at the interface were mainly concentrated at 19, 20, 23–24, 26–28, 30–31, 34–38, 41–42, 45, 79, 82–83, 324–326, 330, 353–357, 386–387 and 393 of the ACE2 protein. Additionally, the involved residues at the interface of the SARS-CoV RBD protein were 403–404, 426, 431–443, 460–463, 470–476 and 479–492. The results also showed that hydrogen bonds were mainly formed by S19/Q24/H34/Y41/Q42/Y83/E329/K353 (ACE2) and R426/Y436/Y440/P462/N473/Y484/T486/G488 (RBD). Furthermore, the salt bridge and π-π interaction were formed by E329(ACE2)-R426(spike) and Y41(ACE2)-Y484(spike), respectively (Fig. 3 A). The results were in general accordance with the resolved structure of Human-SARS-CoV. As to Mice-SARS-CoV, the involved residues at the interface were basically similar to Human-SARS-CoV system, and the hydrogen bonds were mainly formed by N24/Y41/Q42/H353/D355 (ACE2) and Y436/D463/N473/T486/G488 (RBD). However, a remarkable phenomenon is noted that the hydrogen bond between F83 and the spike structure is missing in the Mice-SARS-CoV (Fig. 3B).
Fig. 3.
The interface between ACE2 (in green) and spike-RBD (in blue). Ã G, the complexes of Human-SARS-CoV, Mice-SARS-CoV, Human-SARS-CoV-2, Cat-SARS-CoV-2, Pangolin-SARS-CoV-2, Bat-SARS-CoV-2, Mice-SARS-CoV-2, respectively. Representative structures were extracted from simulation trajectories.
With regard to the SARS-CoV-2 systems, the MD results revealed that the final conformation of the complexes was generally similar, most likely due to the high degree of sequence identity in ACE2 among different species. In addition, the stable conformation was observed to be in line with corresponding resolved structures (if there was one). Importantly, the residues at the interface (in all five complexes) were mainly concentrated at 19–50, 75–85, 325–330, 350–355 and 385–393 of the ACE2 protein (shown in Fig. 2 with the black box). Nonetheless, significant differences were observed in the binding interfaces due to variations in the two spike residues (Fig. 5 ). With regard to the human-ACE2 and SARS-CoV-2 complex, the main residues of the ACE2 protein involved at the protein-protein interaction site were 20, 23–24, 27–31, 34–38, 41–42, 45, 79, 82–83, 324–326, 330, 353–357, 386–387 and 393 (Fig. 3C). On the other hand, the corresponding parts of the spike protein were 403–405, 417, 421, 445–446, 449, 453–456, 473–478 and 485–505. Notably, hydrogen bonding is essential in maintaining the stability and function of complex formed by ACE2 and RBD protein. In the finally obtained major conformation of the ACE2-SARS-CoV-2 system, the main chain oxygen atom of K353 was observed to form a 2.84 Å hydrogen bond with the N atom of G502 in the RBD structure. In addition to forming hydrogen bonds with the backbone and backbone chain, the numerous side chains such as Q24/K31/H34/E37/D38/Y41/Q42/Y83/K353/D355 in ACE2 and Y449/Y453/A475/T478/N487/Q493/S494/Y495/in RBD also had the ability to form hydrogen bonds. These results were roughly consistent with those of the experimentally resolved structures. Similar to hydrogen bonds, salt bridges are also important in maintaining the protein complex structure. Herein, the results revealed the presence of two salt bridges in the human-SARS-CoV-2 system, one of which was formed between D30 (in ACE2) and K417 (in RBD) while the other was formed between E37 (in ACE2) and the R403 (in RBD). Similarly, the π-π interaction formed by Y83 (ACE2) and F486 (RBD) could also contribute to favorable binding. More detailed results are listed in Table S2.
Fig. 2.
ACE2 receptor sequence comparison among different species. The ACE2 protein sequence is very homologous among different species. The corresponding binding site has been marked with black box at residues 19–50, 75–85, 325–330, 350–355 and 385–393.
Fig. 5.
Comparison of the protein sequence of SARS-CoV-2-RBD and SARS-CoV-RBD. The region of action with human ACE2 receptor protein has been marked with a black box in the corresponding region. It can be seen from the figure that the main residue regions of the two types of S proteins involved in the action of ACE2 are similar, but compared with SARS virus, SARS-CoV-2-S protein has more additional residues involved in receptor interaction.
In the cat-SARS-CoV-2 complex (Fig. 3D), comparison with the above human-related results showed that similar residues were involved (20–24, 27–31, 34–38, 41–42, 45, 79, 82–83, 324–326, 330, 353–357, 386–387 and 393 of the ACE2 protein as well as 403–405, 408, 417, 446, 449, 453–458, 473–478, 487–505 of the spike protein) although they had different degrees of interaction. After comparing the two complexes, the results showed that the hydrogen bonds in the cat complex were similar to those of the human-SARS-CoV-2 complex, although they were fewer in number. It is noteworthy that the D38E in the cat ACE2 protein could also form stable hydrogen bonds with relevant residues in the spike protein. Additionally, E23 showed the ability form a salt bridge with K458 of the spike protein, in addition to the salt bridges mentioned earlier (previous paragraph). However, the weak association in cats was mainly caused by the π-π interaction between F28(in ACE2)-Y489(in spike) and the cation-π action composition of K31(ACE2)-Y489(spike)/K353(ACE2)-Y505(spike). Moreover, the structural similarity could be extended to the bat and pangolin systems, which had almost the same set of residues in the interface region but with slightly diverse hydrogen-bonds (Fig. 3E and F). As a reference, Mice-SARS-CoV-2 and Human-SARS-CoV-2 share similar protein-protein interface. Of concern is that the hydrogen bond formed by E30/E31/F83 is missing because of the different substituents in the side chains (Fig. 3G).
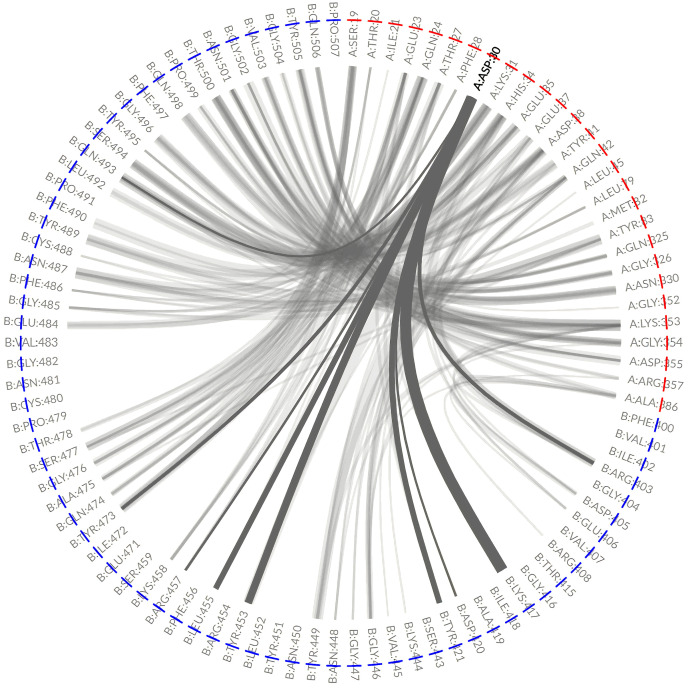
In real environments, there exists inherent differences in the degree of fluctuation and flexibility of protein. Therefore, the analysis of this single stable conformation does not cover protein-protein interactions of different conformations, whether the stable conformation was obtained from simulation snapshots or from resolved structures (Figs. 3 and 4). However, the MD can resolve this deficiency by monitoring innate dynamic fluctuation. In order to exhaustively analyze the dynamic non-covalent contact networks in proteins throughout MD simulation, the GetContacts tool was used to explore all the possible hydrogen bonding interactions by extracting from the MD trajectories. Taking the D30 of ACE2 in human-SARS-CoV-2 complex as an example, the two resolved structures had strikingly different hydrogen bonding interactions, which may correspond to different conformations of the same molecule. Interestingly, analysis of H-bonds in D30 from the MD process provided all the potential hydrogen bonds (Fig. 6 ). The figure shows that nine different residues, including R403, K417, D420, Y421, Y453, L455, R457, Y473 and Q493 in the spike protein, had the potential to form hydrogen bonds with D30. In addition, a thicker line in the figure indicates that a H-bond was highly likely formed between D30 and K417, consistent with the resolved structure (see Fig. 4). Meanwhile, by comparing all resolved structures, D30 forms one hydrogen bond with the Q493 (PDB ID:7KJ4) offered another evidence (Figure S2). In a word, the results of dynamic analysis not only were consistent with resolved structures, but also can predict possible yet unknown conformations. More information on the hydrogen bonding interactions at the protein-protein interface of the other four susceptible systems is provided in Figure S3. Moreover, all the original data is provided in the supplementary material and can be uploaded and visualized on this website; https://gpcrviz.github.io/flareplot/?p=create.
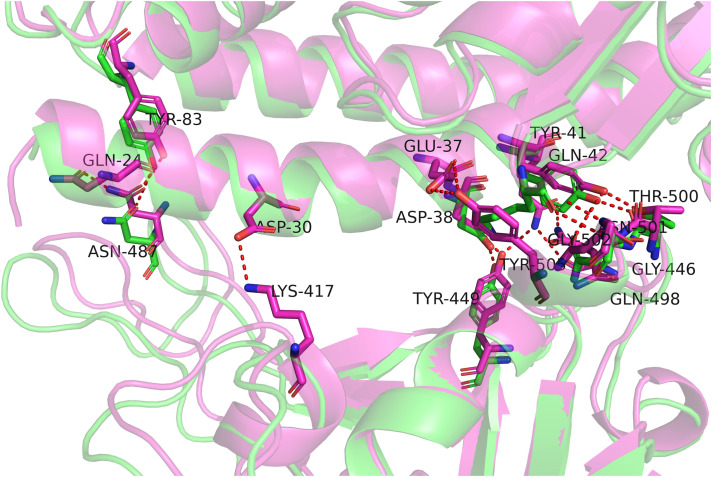
Fig. 4.
The interface structural comparison of two resolved human-SARS-cov2 structures. Both entry 6LZG (magenta) and 6M17 (green) are drawn from the Protein Data Bank (PDB) database. As an overall perspective, these two structures solved by cryo-electron microscopy are well agreement with each other. Some minor yet notable differences existed, nevertheless. Take hydrogen bonding interactions as an example, five additional hydrogen bonds exist in magenta structure, namely Q24-N487, D30-K417, E37-Y498, Q42-Q498 and Q42-G446, respectively. This suggests that even though the human-SARS-cov2 structures has been resolved, a single conformation is not provided adequate information because of the inherent structural fluctuation.
Fig. 6.
The visualization of hydrogen bonding interactions between D30 (in ACE2 with red) and other residues (in spike with blue). The hydrogen bonding interactions for D30 are highlighted by black curves while the other hydrogen-bonding residues represented in the background as a grey curve. The thicker line, the higher the proportion of hydrogen bond made between the residues.
3.3. The stability of the ACE2-RBD complex
Next, we explored the similarities and differences in interaction between the spike protein and the ACE2-RBD of the five different species. Binding free energy calculations were conducted using the MM-GBSA method. As listed in Table 1 , the results showed that the human-SARS-CoV-2 and cat-SARS-CoV-2 systems had the highest binding energy of about −54 kcal⋅mol− 1, followed by the pangolin system (−45.72 kcal/mol). While the bat-SARS-CoV-2, mice-SARS-CoV-2, human-SASRS-CoV-2 and human-SARS-CoV systems with binding energies of −33.97 kcal/mol, −29.97 kcal/mol, −30.36 kcal/mol and −23.16 kcal/mol, respectively. Additionally, electrostatic energy (ΔGele) was shown to play a dominant role in the binding reactions of all the seven systems. Moreover, van der Waals interactions (ΔGvdw) and the nonpolar solvation free energy (ΔGnonpol,sol) were shown to be favorable for binding while the polar solvation free energy (ΔGele,sol) conferred unfavorable effects on binding, in all the complexes. It is noteworthy that the unfavorable ΔGele,sol in the human-SARS-CoV system was significantly higher than in the other groups, which was the major factor associated with low-energy. Although the seven systems had different binding free energy figures, the corresponding large error values of ±7 kcal⋅mol−1 could potentially change the ordering of energy in the systems, with less marked differences. Such include the cross-state range of energy in human-SARS-CoV-2, pangolin-SARS-CoV-2 and cat-SARS-CoV-2 as well as the little distinction between bat-SARS-CoV-2 and human-SARS-CoV-2.
Table 1.
The binding free energy of the seven systems (kcal·mol−1).
| System | polar contributions |
nonpolar contributions |
ΔGbind | |||
|---|---|---|---|---|---|---|
| ΔGele | ΔGele,sol | ΔGvdw | ΔGnonpol,sol | |||
| Human-ACE2-SARS-CoV-2 | −648.15 | 692.62 | −86.30 | −12.19 | −54.03 ± 7.50 | |
| Pangolin-ACE2-SARS-CoV-2 | −512.95 | 566.44 | −87.06 | −12.16 | −45.72 ± 7.20 | |
| Cat-ACE2-SARS-CoV-2 | −502.39 | 546.52 | −86.29 | −12.12 | −54.27 ± 7.20 | |
| Bat-ACE2-SARS-CoV-2 | −606.99 | 662.16 | −78.37 | −10.78 | −33.97 ± 7.10 | |
| Mice-ACE2-SARS-CoV-2 | −471.14 | 539.30 | −86.32 | −11.80 | −29.97 ± 5.60 | |
| Human-ACE2-SARS-CoV | −760.63 | 822.78 | −80.95 | −11.56 | −30.36 ± 7.30 | |
| Mice-ACE2-SARS-CoV | −396.97 | 458.50 | −74.85 | −9.83 | −23.16 ± 3.96 | |
Considering the large uncertainty obtained in the MM-GBSA results, the additional binding energy method with SMD simulations were performed. The seven systems were selected to examine the ability of these spike-RBD proteins to dissociation from the different ACE2 proteins. Notably, it was necessary to select the reasonable values of a spring constant (K) and velocity (V) before the SMD studies, in order to obtain measurable dissociation. Thereafter, a series combination tests were conducted with different values of K and V, to identify the optimal solutions. These combination tests included a total of seven K values ranging from 500 to 2000 kJ/mol/nm2 and four different velocities from 0.005 nm/ps to 0.015 nm/ps (Figure S4). The pre-test results showed that K changed over time as the distance between the protein centroids increased. Therefore, the lower V and K values, the lower the maximum pull force required. Generally, all the position curves were smooth during the separation of complexes although there were some fluctuations at the end of the force curves when the rake speed value (or K) was too high. This indicated that the K and V values greater than 1500 kJ/mol/nm2 and 0.015 nm/ps, respectively, would not be suitable. Finally, the moderate K value of 1000 kJ/mol/nm2 and a V of 0.01 nm/ps were selected for all the SMD studies.
Based on the equilibrium structure and previous tensile tests, SMD simulation with 0.5 ns were performed on all the seven systems by applying a pulling force on the RBD part to disassociate it from the receptor (Figure S5). The results on Figure S4 suggest that all the spike proteins dissociated from the ACE2-spike complex are through a similar process. In addition, the whole RBD structure remained basically stable with a slow dissociation rate from the beginning to the end of the process. This illustrated that reasonable values of K and V were selected without any structural damages in the simulation process. At the initial phase of SMD, the ACE2 and RBD proteins were in their fully bound state and the original external pulling force may not be enough to contend the strong binding forces. Therefore, the complexes did not show large conformational changes and the distances between the ACE2 and spike proteins increased slowly. However, as the pulling force gradually increased, the external force finally offset the binding force in a duration of about 150–180 ps. By comparing the different systems, there were small changes in the conformation of the centroid-centroid distances of the complexes. Nevertheless, these forces played important roles in damaging the conformation of protein-protein interaction sites, particularly, the breakage of hydrogen-bonds. Notably, when the hydrogen bond was gradually destroyed by an increase in external forces, the binding forces of complexes subsequently decreased. Therefore, the required pulling force between ACE2 and RBD was also gradually reduced in about 180–270 ps. At this stage, the most striking feature of change in the complexes was the RBD section being pulled away from the ACE2 protein at higher rate. Moreover, ACE2 and RBD were completely isolated from each other after 270 ps, leading to a further reduction in the required external forces. Consequently, the rate of separation slightly decreased at a constant speed until the end of the process. Overall, there was a similar trend in change in all the seven systems during the stretching process, likely due to the high sequence similarity between different species.
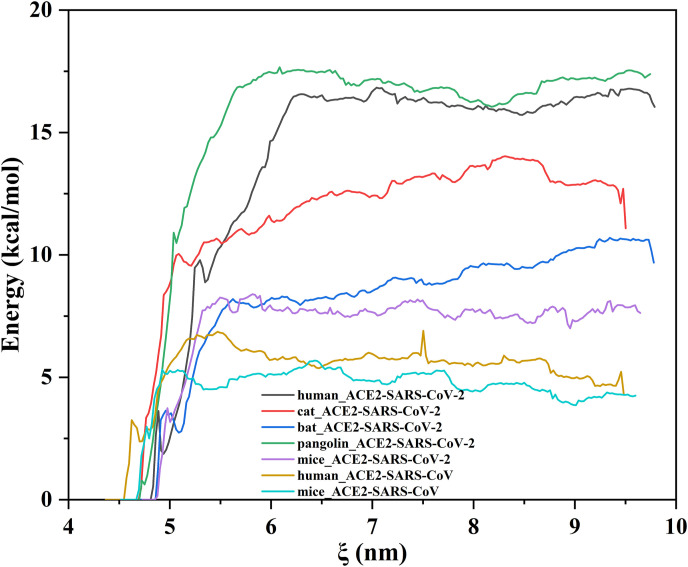
A series of MD simulations combined with umbrella sampling provides the binding energy ΔG along the specific reaction coordinate path. In this study, each complex system produced a series of coordinate conformations along the z-axis during the stretching process and about 53–55 windows were selected for independent MD at a sampling distance of ∼1 Å. Afterwards, the free energy surfaces were obtained by reweighing the umbrella sampling histograms through a weighted histogram analysis method (Fig. 7 ). From the PMF curves, the seven curves differed tremendously from each other. The pangolin-ACE2 bound to SARS-CoV-2 to give the highest unbinding free energy of −16.8 kcal/mol among all the systems studied, followed by the Human-SARS-CoV-2 complex which reduced by only 0.5 kcal/mol. It was implied that the unbinding free energy in both the pangolin and human systems were almost the same. Moreover, the calculated energy had marked differences in three other SARS-CoV-2 systems, with cat-SARS-CoV-2 having −13.1 kcal/mol, bat-SARS-CoV-2 recording −10.1 kcal/mol and mice-SARS-CoV-2 sharing −7.6 kcal/mol. Corresponding to the above SARS-CoV-2 systems, human-SARS-CoV and mice-SARS-CoV with low unbinding free energy of −6.6 kcal/mol and −5.1 kcal/mol, respectively. ACE2 was shown to have the lowest unbinding free energy. Finally, the PMF results presented above show that SARS-CoV-2 may to some extent display a stronger affinity towards the ACE2 proteins than SARS-CoV. Furthermore, the affinity of SARS-CoV-2 towards the intermediate hosts was found to be stronger than that towards the original host.
Fig. 7.
The average potential curve of each composite system in umbrella sampling. The curve reflects the binding ability of ACE2 and Spike-RBD in each complex. From the figure, it can be found that the binding ability of ACE2 and SARS-CoV-2 in the pangolin system is the highest, while the difference between the human host and the pangolin is very small, followed by Cat and Chinese chrysanthemum bat ACE2. As a reference host, mice ACE2 has the low binding force with SARS-CoV/SARS-CoV-2, respectively.
3.4. The key residues of ACE2 in different species
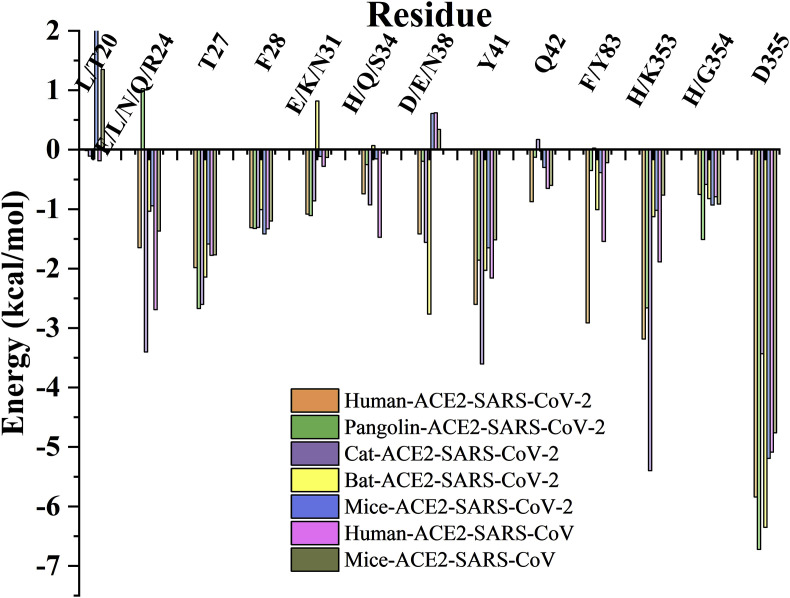
Further, energy decomposition analysis was conducted with the binding free energies of the seven systems before being broken down into individual residues located within the protein-protein interface. The results are presented in Table S3. Generally, residues involved in the protein-protein interface site, such as 19–28, 30–31, 34–38, 41–42, 45, 79, 82–83, 325–326, 330, 352–357 and 386 of ACE2, represented different levels of binding interactions. Out of those, residues 24, 27–28, 31, 34, 38, 41–42, 83 and 353–355 played crucial roles in ACE2-spike interactions (Fig. 8 ). Additionally, the different types of residues were observed to play distinct roles. For instance, residue N/Q/L/R24 had beneficial effects in four systems (range from −3.403 to −0.945 kcal/mol) while the unfavorable effect of E24 was present in the pangolin-SARS-CoV-2 complex. The change from the amino to hydroxy group in Q24E was shown to potentially result in the alteration of electrostatic interactions with the surrounding residues A475/T478/N487 and solvent waters. Furthermore, the conserved amino acids such as T27 and F28, had similar energy contributions in the seven systems, ranging from −1.586 to −2.671 and −1.008 to −1.417 kcal/mol, respectively.
Fig. 8.
Free energy decomposition plots for the seven systems.
Generally, the overall energy contribution of residue K31 in all the seven systems was higher than E/N31 while H34 had a higher input than S/Q34. This is possibly because both K31 and H34 were more likely to form hydrogen bonds with Q493 and Y453/S494/Y495 in the spike protein, respectively. In addition, residue 38 consisting of aspartic, glutamic and asparagine was beneficial to the binding interactions in all the four SARS-CoV-2 related systems although aspartic-38 was unfavorable to the SARS-CoV complex. This may be because the different sequences of residues, 493–499/479-485 in the spikes of SARS-CoV-2 and SARS-CoV changed the spatial orientation of the residues, especially Q498 in SRAS-CoV-2 and the corresponding Y484 in SRAS-CoV. Notably, Y41 had huge energy contributions in all the seven systems while residue 42 with glutamine contributed more to the human-SARS-CoV-2 compared to the other systems. This is likely due to the high probability of the formation of hydrogen bonds between Q42 and Q498. However, the same residue Y83 displayed different energy values in all corresponding systems because of loop fluctuations in residues 81–84, while the absence of a favorable hydrogen-bonding group may impede the energy contributions originate from F83. Moreover, the most important residues, 353–355 were essential in maintaining the binding force between ACE2 and RBD. Compared to H353 in mice, K353 had a great ability to form hydrogen bonds with Y495/G496/Q498/G502 and weak interactions with Y505. Furthermore, residue 354 with histidine was able to form an additional H-bond with D405, indicating a stronger energy contribution than that with glycine. Additionally, D355 had the highest contribution in all the systems probably because of the ease of generating static electricity with T500(SARS-CoV-2)/486(SARS-CoV).
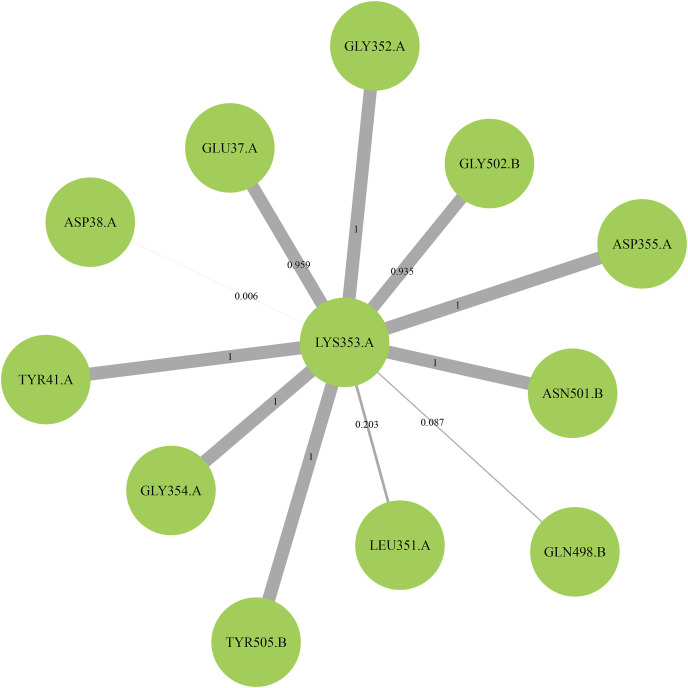
To reveal that what influences the conformation of these key residues, the residue contact maps were used in monitoring the interactions of specific residues throughout the simulation. This was based on the key residues obtained from the binding free energy calculations and energy decomposition analysis [38]. The final plot had the residue of interest at the center and the associated residues around it. Thickness of the line represented the frequency of interactions between two residues from the whole process. As an example, the key residue K353 contact map in the human-SARS-CoV-2 system is shown in Fig. 9 , which indicates that eight residues could form close associations with K353. Among these residues, E37, Y41, G352, G354 and D355 had coefficient values of above 0.9 in the ACE2 protein. These had a great impact in maintaining the conformation of K353 in ACE2 as well as the closely associated residues N501, G502 and Y505 in the spike protein. Additionally, any changes in the type of residue, including the center and neighboring residues, could more or less alter their position. All the key residue contact maps were calculated and the extracted results were shown in Table S4.
Fig. 9.
The residue contact map of K353 in human-SARS-CoV-2 system. Labels with A and B represent the different chains of ACE2 and spike-RBD protein, respectively, while the lines with values stand for the correlation of interactions.
From the comparison, the study found that each type of residue had some special characteristics. Given the same kind of key residues and surrounding residues in all the reported susceptible four species, T27, F28, Y41, Q42, K353 and D355 showed no significant differences between systems. In contrast, the same residue Y83 were affected by the different kinds of residue 21 in the above systems and caused minor effects on the residue contact maps. As for remaining non-conserved sites, a few of these sites had little effect on residue localization (e.g., E/K31 and D/E/N38), but more sites exhibit highly diverse contact maps with different types of residues. The obvious differences in residue 24 were seen on the residue contact maps. On one hand, there were different types of residue 24 while on the other hand, the multi-surrounding residues were not all the same, such as I/S/T21 and E/D23 in the different groups. Moreover, there wasn't a direct link between the energy contributions and residue contact maps, although the significant changes in maps may have had an effect on energy (residue 24 in Fig. 8). With regard to H/S34, similar H34 residue contact maps were observed in the Human and Cat-SARS-CoV-2 systems while there was no significant difference in the S34 residue contact maps of the Pangolin/Bat-SARS-CoV-2 groups. Moreover, a comparison of the residue contact maps showed that a change from H34 to S34 could definitely increase contact with E/D30 in ACE2 and L455 in the spike protein. As to human-SARS-CoV system, despite having the same H34 in the same ACE2, D30 (in ACE2) and Y442 (in SARS-CoV spike) exhibited more differences because of the variation between TYR442 and the corresponding residue L455 (in SARS-CoV-2). Additionally, H354 in Pangolin-SARS-CoV-2 could make a slightly stronger association with G504 in spike and weak association with F356 in ACE2, as opposed to G354 in the other systems. Considering the multiple surrounding residues showed a tight linkage to key residues with the diversified maps in the all five systems. Not only mutations at key sites, but also changes in surrounding residues may have had a large impact on the contact interactions.
4. Discussion
Homology modeling and MD simulations revealed that the seven complexes had high structural similarity. Two methods (MM-GBSA and PMF) were used to obtain the relative binding energy between different ACE2s and spike proteins, in order to explore the stability of complexes formed by SARS-CoV-2 and the ACE2 of different species. The MM-GBSA calculations revealed that electrostatic interaction was the dominant force involved in maintaining the conformation of the complexes. Comparing the results from MM-GBSA and PMF also showed that the displayed trends were consistent. Of these, the SARS-CoV-2-related systems were more stable than the Human-SARS-CoV complex in both calculation methods. Meanwhile, the two mice-SARS-CoV-2 and mice-SARS-CoV-with the low energy in SARS-CoV-2 and SARS-CoV related systems, which is consistent with the reported experimental result that mice were not susceptible to SARS-CoV/SARS-CoV-2 infection [26]. Notably, the Human-SARS-CoV-2 complex exhibited the highest stability in all the five SARS-CoV-2 groups. In the case of stability of Human-SARS-CoV-2 and Human-SARS-CoV systems, this observation is at variance with the results of Hao et al. [19,20], but is in accordance with previously released reports. Structural studies have shown the stronger binding affinity of the CoV-2-S-hACE2 as compared to that of CoV–S-hACE2 [39,40]. Together, active pharyngeal viral shedding findings suggest a more efficient transmission of SARS-CoV-2 than SARS-CoV [41]. Nonetheless, contradictory findings were obtained between MM-GBSA and PMF with regard to Cat/Human-SARS-CoV-2 and Human/Pangolin-SARS-CoV-2. MM-GBSA showed that Cat/Human-SARS-CoV-2 had the highest binding energy while PMF revealed that the Human/Pangolin-SARS-CoV-2 had the highest unbinding energy. This inconsistency may have been due to the large error range of ±7 kcal⋅mol−1 obtained in MM-GBSA or because MM-GBSA and PMF use different calculation approaches.
Moreover, stability of the complexes was determined by relevant residues in the protein-protein interaction sites. Given the conservative nature of ACE2 protein sequences, the residues primarily involved in protein-protein interaction sites in all the systems were 19–24, 27–31, 34–38, 41–42, 45, 79, 82–83, 324–326, 330, 353–357, 386–387 and 393. However, the key residues were Q/E/L/R24, T27, F28, D/E30, K/E31, H/S34, D/E/N38, Y41, Q42, Y83, K353, H/G354, D355 out of which residues 353–355 were the most critical.
In addition, analysis of the hydrogen bonding network in key residues complemented the weaknesses of single conformation in resolved structures. Notably, the same residue in different ACE2 proteins could form similar hydrogen bonds with the spike protein, but at different positions, in the five SARS-CoV-2 systems. As for the different kinds of residues, the hydrogen bonding effect is more different. The L24E showed less distant contact with N487 is predicted to be disruptive. Meanwhile, the S34 could form three hydrogen bonds with residues Y453/Q493/G496 while H34 could more associate with spike R403/G496. Similarly, G354 mainly interacted with T500, unlike H354 which associated with multiple residues, D405/V503/G502/G504/T505. Overall, the specified residues L24/K31/H34/N38/H354 in ACE2 are more favorable for binding with SARS-CoV-2 among four species. In both Human complexes, SARS-CoV-2 and SARS-CoV were present in similar sites but at different frequencies, despite sequence variability between them. Hotspots K31/D38/Y83/K353 contribute to the difference in the stability of the two systems. In corroboration with findings from previous studies, it was reported that, ACE2 of mice exhibited the lowest receptor activity, the E30 N, H34/Q, Y83F and K353H substitutions are all predicted to disrupt interactions in mice ACE2. Results of our reanalysis are generally consistent with those reported in the literature [42].
The hydrogen-bonding interactions were not only determined by the self-residue type but also by the spatial conformation of residues, influenced by surrounding residues. In addition, the residue contact maps could discriminate the close interactions around specific residues. In most cases, the same kind of key residue in four species has similar results residue contact maps (e.g., T27, F28, Y41, Q42, K353 and D355). However, due to the mutated surrounding residues in different species, the conformation and energy contribution of the same type of residues are not exactly the same. The most dramatic example is Y83, which is strongly affected by immediately adjacent residues such as different types residues 82 and 84 in four species. This results in Y83 conformational diversification ultimately leading to energy contribution differences (Fig. 7). As to non-conserved sites I/S/T21, E/D23, H/S34 and H/G354, the role of surrounding residues is more diverse. This clearly shows that when extended to other more species, it is necessary to focus not only on hot spot residues itself, but also on adjacent residue types. The residue contact maps also revealed similar residue contacts among four species and this information might provide a mechanistic basis for further understanding interspecies transmission. Such sites could potentially be helpful in designing novel peptide-based entry inhibitors against specific the RBD of SARS-CoV-2 to disrupt SARS-CoV-2 initiates its entry into human cells by binding to ACE2.
5. Conclusions
In this study, homology modeling and MD simulation were used to construct complexes of SARS-CoV-2-RBD and ACE2 in five species. The MM-GBSA and PMF methods not only confirmed the receptor-binding affinity between SARS-CoV-2-RBD and human ACE2 was stronger than that of SARS-CoV-RBD, but also revealed the binding affinity between the SARS-CoV-2-RBD and ACE2 in five species. The Energy decomposition analysis indicated the key residues in five species. As a supplement, the network of important hydrogen bonding interaction between ACE2s and spikes can make up for the lack of resolved structures. Additionally, the residue contact maps help to understanding species-related differences in affinity. In summary, conducted in the backdrop of an unabated SARS-CoV-2 epidemic, the obtained results will help in understanding the mechanism of infection between species and provide useful insights on novel potent drug development.
Authors’ contribution
Kunqian Yu conceived the project; Kunqian Yu and Shaojie Ma designed the research; Shaojie Ma performed MD simulations; Hui Li performed sequence alignments; Shaojie Ma wrote the paper with help from Kunqian Yu and Jun Yang; All authors reviewed the results and approved the manuscript.
Acknowledgements
This work was supported by the National Key R & D Program of China (2020YFC0841400, 2016YFA0501200 and 2017YFA0505400), the National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711003-003-005), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDC01040100) and the National Natural Science Foundation of China (31770798 and 21921004).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biochi.2021.05.001.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kandimalla R., John A., Abburi C., Vallamkondu J., Reddy P.H. Current status of multiple drug molecules, and vaccines: an update in SARS-CoV-2 therapeutics. Mol. Neurobiol. 2020;57:4106–4116. doi: 10.1007/s12035-020-02022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Sig. Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 Spike protein. Biochimie. 2021;180:143–148. doi: 10.1016/j.biochi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 11.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9:529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:2196–2203. doi: 10.1016/j.cub.2020.05.023. e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhammed M.T., Aki-Yalcin E. Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019;93:12–20. doi: 10.1111/cbdd.13388. [DOI] [PubMed] [Google Scholar]
- 15.Makarova T.M., Bogdanov A.A. Allosteric regulation of the ribosomal A site revealed by molecular dynamics simulations. Biochimie. 2019;167:179–186. doi: 10.1016/j.biochi.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Wang J., Xu X., Zhou X., Chen P., Liang H., Li X., Zhong W., Hao P. Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. J. Gen. Virol. 2020;101:921–924. doi: 10.1099/jgv.0.001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkhivker G.M. Molecular simulations and network modeling reveal an allosteric signaling in the SARS-CoV-2 spike proteins. J. Proteome Res. 2020;19:4587–4608. doi: 10.1021/acs.jproteome.0c00654. [DOI] [PubMed] [Google Scholar]
- 18.Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C., van Zandbergen G., Landry J., de Azevedo N.T.D., Mosalaganti S., Schwarz A., Covino R., Mühlebach M.D., Hummer G., Krijnse Locker J., Beck M. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Q., Herrmann A. bioRxiv; 2020. Fast Assessment of Human Receptor-Binding Capability of 2019 Novel Coronavirus (2019-nCoV) 2020.2002.2001.930537. [DOI] [Google Scholar]
- 21.Lai H.T.T., Nguyen L.H., Kranjc A., Nguyen T.T., Nguyen-Manh D. bioRxiv; 2020. Elucidating the Differences in the Molecular Mechanism of Receptor Binding between 2019-nCoV and the SARS-CoV Viruses Using Computational Tools. 2020.2004.2021.053009. [DOI] [Google Scholar]
- 22.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 24.Peng C., Zhu Z., Shi Y., Wang X., Mu K., Yang Y., Zhang X., Xu Z., Zhu W. Computational insights into the conformational accessibility and binding strength of SARS-CoV-2 spike protein to human angiotensin-converting enzyme 2. J. Phys. Chem. Lett. 2020;11:10482–10488. doi: 10.1021/acs.jpclett.0c02958. [DOI] [PubMed] [Google Scholar]
- 25.Brooke G.N., Prischi F. Structural and functional modelling of SARS-CoV-2 entry in animal models. Sci. Rep. 2020;10:15917. doi: 10.1038/s41598-020-72528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preziuso S. Severe Acute respiratory Syndrome coronavirus 2 (SARS-CoV-2) exhibits high predicted binding affinity to ACE2 from lagomorphs (rabbits and pikas) Animals. 2020;10:1460. doi: 10.3390/ani10091460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham Peele K., Srihansa T., Krupanidhi S., Ayyagari V.S., Venkateswarulu T.C. Design of multi-epitope vaccine candidate against SARS-CoV-2: a in-silico study. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1770127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad Gajula M.N.V., Vogel K.P., Rai A., Dietrich F., Steinhoff H.J. How far in-silico computing meets real experiments. A study on the structure and dynamics of spin labeled vinculin tail protein by molecular dynamics simulations and EPR spectroscopy. BMC Genom. 2013;14:S4. doi: 10.1186/1471-2164-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontiggia F., Pachov D.V., Clarkson M.W., Villali J., Hagan M.F., Pande V.S., Kern D. Free energy landscape of activation in a signalling protein at atomic resolution. Nat. Commun. 2015;6:7284. doi: 10.1038/ncomms828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ylilauri M., Pentikäinen O.T. MMGBSA as a tool to understand the binding affinities of filamin–peptide interactions. J. Chem. Inf. Model. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 31.Maffucci I., Contini A. Improved computation of protein-protein relative binding energies with the nwat-MMGBSA method. J. Chem. Inf. Model. 2016;56:1692–1704. doi: 10.1021/acs.jcim.6b00196. [DOI] [PubMed] [Google Scholar]
- 32.Cuendet M.A., Michielin O. Protein-protein interaction investigated by steered molecular dynamics: the TCR-pMHC complex. Biophys. J. 2008;95:3575–3590. doi: 10.1529/biophysj.108.131383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautieri A., Vesentini S., Montevecchi F.M., Redaelli A. Mechanical properties of physiological and pathological models of collagen peptides investigated via steered molecular dynamics simulations. J. Biomech. 2008;41:3073–3077. doi: 10.1016/j.jbiomech.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 34.Lemkul J.A., Bevan D.R. Assessing the stability of alzheimer's amyloid protofibrils using molecular dynamics. J. Phys. Chem. B. 2010;114:1652–1660. doi: 10.1021/jp9110794. [DOI] [PubMed] [Google Scholar]
- 35.Lesitha Jeeva Kumari J., Jesu Jaya Sudan R., Sudandiradoss C. Evaluation of peptide designing strategy against subunit reassociation in mucin 1: a steered molecular dynamics approach. PloS One. 2017;12 doi: 10.1371/journal.pone.0183041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hub J.S., de Groot B.L., van der Spoel D. g_wham-A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010;6:3713–3720. doi: 10.1021/ct100494z. [DOI] [Google Scholar]
- 37.Domański J., Hedger G., Best R.B., Stansfeld P.J., Sansom M.S.P. Convergence and sampling in determining free energy landscapes for membrane protein association. J. Phys. Chem. B. 2017;121:3364–3375. doi: 10.1021/acs.jpcb.6b08445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown D.K., Penkler D.L., Sheik Amamuddy O., Ross C., Atilgan A.R., Atilgan C., Tastan Ö. Bishop, MD-TASK: a software suite for analyzing molecular dynamics trajectories. Bioinformatics. 2017;33:2768–2771. doi: 10.1093/bioinformatics/btx349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu J., Sun P.D. bioRxiv : the preprint server for biology; 2020. High Affinity Binding of SARS-CoV-2 Spike Protein Enhances ACE2 Carboxypeptidase Activity. 2020.2007.2001.182659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P., Zheng S., Li X., Song C., Li R., Guo J.-T., Junop M., Zeng H., Lin H. Broad and differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.00940-20. e00940-00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.