Serological markers are important for the diagnosis of hepatitis E virus (HEV) infection. This study aims to compare the diagnostic performance of the anti-HEV IgM and the HEV antigen (Ag) assays and establish a multifactorial model to improve the diagnosis of current HEV infection when HEV RNA detection is not available.
KEYWORDS: hepatitis E, diagnosis, dynamics, IgM, HEV Ag, logistic regression
ABSTRACT
Serological markers are important for the diagnosis of hepatitis E virus (HEV) infection. This study aims to compare the diagnostic performance of the anti-HEV IgM and the HEV antigen (Ag) assays and establish a multifactorial model to improve the diagnosis of current HEV infection when HEV RNA detection is not available. A total of 809 serum samples, including 325 anti-HEV IgM-positive and 484 anti-HEV IgM-negative samples, were tested for HEV RNA. The anti-HEV IgM assay had very high sensitivity (99.4%) but moderate accuracy (79.2%) and specificity (74.3%). By retrospective follow-up of 58 patients with sequential samples (n = 143) tested for anti-HEV antibodies, we found anti-HEV IgM remained positive for more than 10 months in some HEV-infected patients, when HEV RNA was already undetectable; thus, decision solely based on anti-HEV IgM may lead to misdiagnosis. In contrast, the HEV Ag assay had very high specificity (100%). However, the detection efficiency of HEV Ag greatly diminished when the HEV RNA level was low or the anti-HEV IgG level was high. By logistic regression, a model integrating anti-HEV IgM, alanine aminotransferase, and HEV Ag was proposed, and the cutoff value was determined based on the testing results of the 143 sequential samples. The model was further evaluated with 67 randomly selected IgM-positive samples from single-visit patients. Overall, the model outperformed the anti-HEV IgM or the HEV Ag assay in the diagnosis of current HEV infection (sensitivity/specificity/accuracy, 89.5%/95.2%/91.9%). The area under the receiver operating characteristics curve of the model was greater than 0.97.
INTRODUCTION
Hepatitis E virus (HEV) infection is a public health concern and recognized as the most common cause of acute viral hepatitis worldwide. It is estimated that 20 million HEV infections occur annually, with more than 3 million symptomatic cases and ∼60,000 deaths (1). It can cause not only endemic hepatitis in developing countries with poor sanitation and limited medical resources but also sporadic hepatitis in developed countries due to contact with HEV-contaminated meat (2) or blood transfusion (3). The prognosis of HEV infection is generally good. However, it is a concern in patients with underlying chronic liver disease (4) and women in their second or third trimester of pregnancy (5). More recently, it was reported that chronic HEV infections have become a significant problem in immunocompromised patients (6).
HEV is a positive-stranded RNA virus (7). It has extensive genetic diversity. Genotypes 1 and 2 can infect only humans, while genotypes 3 and 4 can infect both humans and swine, which makes it hard to eliminate due to the existence of the animal reservoir (8).
A large proportion of hepatitis E cases go unrecognized or are frequently misdiagnosed as drug-induced liver injury (9, 10). Lack of knowledge and awareness of the disease among clinicians is part of the reason. Meanwhile, the accurate diagnosis of hepatitis E is still challenging.
After HEV infection, anti-HEV IgM antibody is induced after an incubation period ranging from 15 to 60 days, and it disappears early in the convalescent period. Anti-HEV IgG may remain detectable for many years (11). Acute hepatitis E is commonly diagnosed by detecting anti-HEV IgM and/or rising IgG levels in the serum (12). In some immunosuppressed patients with chronic HEV infection, anti-HEV antibodies remained negative (13). Therefore, molecular diagnosis of HEV RNA is highly recommended (14). Viral RNA can be detected in the serum and/or feces during the incubation period or early acute phase of disease by reverse transcription-PCR (RT-PCR). It provides a specific and highly sensitive approach to the diagnosis of HEV infection. Because RT-PCR is relatively expensive and technically challenging, the assay is not easily accessible, especially in developing countries where HEV is hyperendemic. Detection of HEV antigen (Ag) has been suggested as a convenient and cost-efficient alternative to RT-PCR, since HEV Ag production may parallel that of HEV RNA. It was reported that the method of HEV Ag detection had good concordance with HEV RNA detection and could serve as a useful tool in early diagnosis of infection (15–18).
The current study is dedicated to comparing the diagnostic performance of the Wantai anti-HEV IgM assay and the HEV Ag assay with the HEV RNA detection assay as a reference. We first evaluated the anti-HEV IgM assay and found it had high sensitivity (99.4%) but poor specificity (74.3%). By retrospective follow-up of samples collected serially from HEV-infected patients, we found the positivity of anti-HEV IgM may last for a long period, which contributed to the low positive predictive value (PPV) of this assay (48.6%). Meanwhile, the diagnostic accuracy of the HEV Ag assay was evaluated, and factors affecting its detection efficiency were explored. Finally, we used regression modeling techniques and receiver operating characteristics (ROC) curve analysis to propose a multifactorial model, which was shown to outperform the anti-HEV IgM or the HEV Ag assay in the diagnosis of current HEV infection when HEV RNA detection is not available.
MATERIALS AND METHODS
Study design.
This noninterventional study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki 1975 and approved by the Human Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Informed consent was waived, since the research involved no more than minimal risk to the subjects. All medical information was coded and kept confidential as required.
Serum samples from subjects who presented with symptoms of hepatitis or came for disease screening purposes were sent to the laboratory for routine testing of anti-HEV antibodies. From August 2018 to December 2019, ∼8,700 samples were tested for anti-HEV antibodies and 565 samples were positive for anti-HEV IgM. A total of 809 samples were subsequently tested for HEV RNA based on sample availability, including 325 anti-HEV IgM-positive samples and 484 anti-HEV IgM-negative samples. Most (479/484) of the anti-HEV IgM-negative samples were included because their alanine aminotransferase (ALT) levels were above the upper limit of normal (ULN; 64 IU/ml). The other 5 anti-HEV IgM-negative samples with ALT levels within the normal range were included because they were the follow-up samples of anti-HEV IgM-positive subjects. Available leftover specimens were collected and stored at −20°C until analyzed. Subjects with HEV RNA detected in serum were defined as current HEV infection. Diagnostic performance evaluation of different assays was based on comparison with HEV RNA.
To explore the duration of the anti-HEV antibody response, we retrospectively identified patients with sequential specimens from the entire set of 809 samples. A total of 143 samples, collected from 58 patients with multiple testing, were available. The kinetics of anti-HEV antibodies and the diagnosis performance of the HEV Ag assay were studied with these serial samples. When we noticed the flaws of the anti-HEV IgM and the HEV Ag assay in the diagnosis of current HEV infection, a model combining anti-HEV IgM, HEV Ag, and ALT was developed. Another set of 67 IgM-positive single-visit samples with available record of ALT level and enough volume (>0.5 ml) for HEV testing then were randomly selected from the entire set, and the performance characteristics of the model were tested. Since the results of these 143 sequential samples are the basis of the proposed model, they are defined as the training set. The other 67 single-visit samples are defined as the test set.
Laboratory methods.
HEV antibodies were detected in 50 μl of serum samples using the chemiluminescence microparticle immunoassay (CMIA), developed by Wantai BioPharm (Beijing, China). It uses a recombinant antigen corresponding to 394 to 606 amino acid residues of the HEV capsid protein (19). The test was performed and interpreted in accordance with the manufacturer's instructions. The results were expressed as cutoff index (COI). A COI of ≥1 and a COI of <1 were defined as a positive result and a negative result, respectively. The CMIA was approved for the diagnostic testing of anti-HEV antibodies by the National Medical Products Administration (NMPA) of China.
HEV Ag was measured with the Wantai HEV Ag enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions. In this system, goat polyclonal anticapsid antibodies are used for antigen capture, and enzyme-linked monoclonal antibodies against the capsid protein are used for detection. Briefly, 100 μl of serum sample was added to each well of the microplate, which was precoated with anti-HEV capsid polyclonal antibodies. The microplate was sealed and incubated at 37°C for 60 min. After washing the wells with washing buffer five times, a second horseradish peroxidase-conjugated monoclonal anti-HEV capsid antibody was added, followed by further incubation at 37°C for 30 min and washing five times with the washing solution again. Subsequently, 100 μl of tetramethylbenzidine substrate solution was added and incubated for 15 min at 37°C. The reaction was stopped by addition of the stop solution, and the absorbance was immediately measured using dual-wavelength detection (450 and 630 nm) with a microplate reader (Multiskan FC microplate reader; Thermo Fisher Scientific). The cutoff value was 0.12 plus the mean absorbance of three negative controls, as recommended by the manufacturer. The absorbance ratio of each specimen to the cutoff value (S/CO) was used to indicate HEV Ag status, with positivity defined as a ratio of ≥1.0. The HEV Ag ELISA kit is intended for research use only.
The level of HEV RNA in clinical samples was determined by a laboratory-developed test (LDT) of RT-PCR. HEV RNA was extracted from 140 μl of serum samples using a TIANamp virus RNA kit (Tiangen, Beijing, China) according to the manufacturer's instructions, and 50 μl of RNA solution was eluted for each sample. The one-step RT-PCR was performed with a HiScript II one-step quantitative RT-PCR probe kit (Vazyme, Nanjing, China). Each reaction mixture included 8 μl of extracted RNA, 10 μl of 2× one-step Q probe mix, 1 μl of one-step probe enzyme mix, 0.1 μM fluorescence probe, and 0.2 μM each primer. RT-PCR conditions included an RT step of 50°C for 15 min, a predenature step of 95°C for 30 s, followed by 45 cycles of 95°C for 10 s and 60°C for 30 s. The sequences of the primers and the probe were described and validated previously (20–22). The forward and reverse primers were 5′-GGT GGT TTC TGG GGT GAC-3′ and 5′-AGG GGT TGG TTG GAT GAA-3′, respectively, and the probe was 5′-FAM-TGA TTC TCA GCC CTT CGC-TAMRA-3′. The amplification was carried out on a ViiA 7 real-time PCR system (Thermo Fisher Scientific) according to the manufacturer’s instructions. HEV RNA was in vitro transcribed from pGEM-7Zf(-)-TW6196E (23) by T7 RNA polymerase to obtain a positive strand as the RNA standard for quantitative RT-PCR (23). The RNA standard was titrated by measuring the optical density at 260 nm with a spectrophotometer. A standard curve was generated from serial dilutions of the standard. The threshold cycle values were plotted as a function of the input HEV RNA viral copy numbers. The LDT was validated against the commercial promoter HEV RNA detection kit (ACON, Hangzhou, China), which was approved for the diagnostic testing of HEV RNA by the NMPA of China to provide a qualitative dichotomous (positive/negative) result. The linearity (see Table S1 in the supplemental material), limit of detection (Table S2), and agreement across specimens (Table S3) were compared between the LDT and the promoter assay.
Samples with positive anti-HEV IgM were tested for HEV RNA individually, and samples with negative anti-HEV IgM were pooled (4 to 5 specimens were mixed into 1 specimen) and tested for HEV RNA. When positive HEV RNA was detected from a pooled sample testing, the subsequent testing of the individual samples from the pool was required.
To test the effect of the anti-HEV IgG on the detection of HEV Ag, serum samples that were either positive or negative for anti-HEV IgG but negative for both anti-HEV IgM and HEV RNA were collected and pooled separately as dilution sera to a final volume of ∼5 ml each. Two serum samples collected from HEV RNA-positive patients (A and B) were 10-fold serially diluted with these pooled anti-HEV IgG-positive or IgG-negative sera to generate 1/10, 1/100, and 1/1,000 dilutions. The original samples and the serially diluted samples were then subjected to HEV Ag detection by ELISA or HEV RNA detection by RT-PCR in duplicates.
Statistical analysis.
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc, Cary, NC) and MedCalc statistical software version 19.3 (MedCalc Software, Ostend, Belgium). Graphs were generated using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA).
Categorical variables were described as proportion (percent), and continuous variables were described as means ± standard errors of the means (SEM) or median (range). Comparisons between two groups were performed using t test, Mann–Whitney U test for continuous variables, and chi-square test for categorical variables as appropriate. Correlation analyses were performed with the Spearman's method. A two-tailed P value of <0.05 was considered statistically significant.
Diagnostic performance was determined with ROC curve analyses, followed by calculation of 95% confidence intervals (CI) using the binomial exact method. Area under the ROC curve (AUROC) of different diagnostic methods was compared using the method of DeLong et al. (24). Detailed diagnostic values were evaluated by calculating the sensitivity, specificity, PPV, negative predictive value (NPV), and accuracy. The optimum cutoff value for diagnosis is selected by maximizing the sum of sensitivity and specificity.
Model estimation was done with the LOGISTIC procedure of SAS. Data of the 143 sequential samples were used to build a model, whereas data of the 67 single-visit patients were used to test the model. Variables of age, gender, ALT/ULN, anti-HEV IgM, anti-HEV IgG, and HEV Ag were included in the logistic regression analysis. Logarithmic transformation of anti-HEV IgM, IgG, and HEV Ag levels and square root transformation of ALT/ULN levels were applied to improve the normality of the distribution. Variables that were significantly different between subjects with and without HEV RNA detected by univariate analysis (P ≤ 0.05) were included in the stepwise multiple logistic regression analysis to identify independent variables associated with current HEV infection. Variables with a P value of <0.05 by multivariate analysis were kept, and the estimate coefficient output by SAS was used to construct a model. The goodness of fit of the model was assessed using the Akaike information criterion (AIC), and the discrimination ability was assessed by AUROC.
RESULTS
Performance of the anti-HEV IgM assay in the diagnosis of current HEV infection.
A total of 809 samples were tested for HEV RNA. Among them, 158 out of the 325 anti-HEV IgM-positive samples and 1 out of the 484 anti-HEV IgM-negative samples had HEV RNA detected. The performance of the anti-HEV IgM assay in diagnosing current HEV infection was summarized in Table 1. The sensitivity and the NPV was quite good (∼99%), followed by accuracy (∼79%) and specificity (∼74%). The main flaw of the anti-HEV IgM assay was its low PPV, only ∼49%, suggesting that half of the cases with positive anti-HEV IgM are not current HEV infection. The only case who was negative for anti-HEV IgM but positive for HEV RNA had another sequential sample collected 1 day later. He was found to have anti-HEV IgM seroconversion and a rising level of anti-HEV IgG at the later time point. HEV RNA was detected in both samples, and acute HEV infection was confirmed.
TABLE 1.
Diagnostic performance of anti-HEV IgM, HEV Ag, and logistic regression models for current HEV infectiona
| Assay/model | Sample set | Cutoff | Sensitivity (%) | Specificity (%) | Accuracy (%) | PPV (%) | NPV (%) | AUROC, 95% CI |
|---|---|---|---|---|---|---|---|---|
| Anti-HEV IgM | Entire set (n = 809) | ≥1 | 99.4 | 74.3 | 79.2 | 48.6 | 99.8 | 0.96, 0.95–0.98 |
| HEV Ag | Training set | ≥1 | 64.8 | 100.0 | 82.5 | 100.0 | 74.2 | 0.88, 0.81–0.94 |
| Test set | 58.7 | 100.0 | 71.6 | 100.0 | 52.5 | 0.87, 0.78–0.95 | ||
| [IgM+Ag+ALT] | Training set | >7.741 | 91.2 | 95.2 | 93.1 | 95.4 | 90.8 | 0.98, 0.96–1.00 |
| Test set | 87.0 | 95.2 | 89.6 | 97.6 | 76.9 | 0.97, 0.94–1.00 | ||
| [IgM+ALT] simplified | Training set | >3.921 | 94.1 | 85.5 | 90.0 | 87.7 | 93.0 | 0.96, 0.94–0.99 |
| Test set | 84.8 | 85.7 | 85.1 | 92.9 | 72.0 | 0.95, 0.90–1.01 |
PPV, positive predictive value; NPV, negative predictive value; AUROC, area under the receiver operating characteristics curve; CI, confidence interval; Ag, antigen; ALT, alanine aminotransferase.
Kinetics of HEV antibodies.
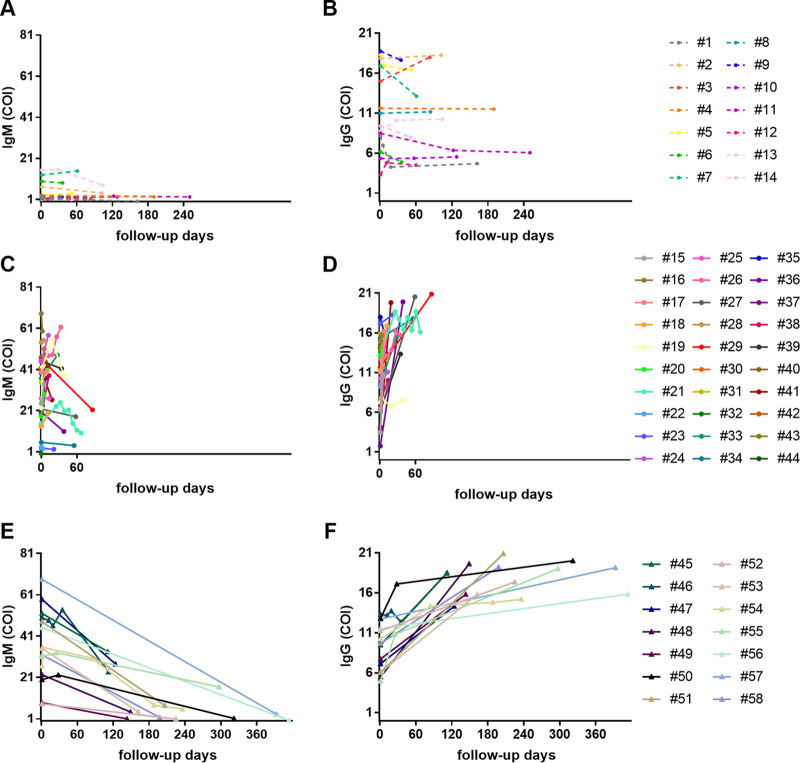
To understand why the anti-HEV IgM assay gives low PPV, we performed a retrospective follow-up study: 58 subjects who had serial sera collected and positive anti-HEV IgM specimen(s) at least once were identified from the entire set of 809 samples. Among them, 14 subjects (1 to 14) remained negative for HEV RNA throughout the observation period, while the other 44 patients (15 to 58) were confirmed as acute HEV infection by detecting positive HEV RNA in the first or following samples collected initially. A total of 143 samples were collected from these 58 subjects, with, on average, 2.5 samples per subject and a median follow-up of 3 months. Twenty-five of the 44 patients with acute HEV infection cleared the virus (19, 21, 23, 24, 26, 27, 33, 34, 36, 39, and 44 to 58), while the other 19 patients had follow-up duration of less than 3 months and remained positive for HEV RNA at the last visit. Patients 15 to 44 had a midterm follow-up period within 3 months, while patients 45 to 58 had long-term follow-up lasting from 90 days to 414 days. The anti-HEV IgM and IgG levels were plotted against the follow-up days (Fig. 1). For the patients who remained negative for HEV RNA (1 to 14), the levels of their anti-HEV IgM were relatively low, and most of them showed neither large increases nor large decreases (Fig. 1A and B). These 14 patients were probably in the convalescent period from recent infection or had nonspecific cross-reactive antibodies to HEV due to unknown reasons. In contrast, for the patients with acute HEV infection (15 to 58), the levels of anti-HEV IgM increased rapidly at the beginning but decreased slowly during the convalescent period, which remained positive for as long as 0.5 to 1 year (Fig. 1C and E). For example, patients 51 and 58 remained positive for 6 months, patients 54 and 55 were positive for 8 months, and 57 remained positive for more than 1 year. The levels of anti-HEV IgG of these 44 patients increased gradually and remained positive throughout the period of observation (Fig. 1D and F). The unexpected long period of anti-HEV IgM positivity interfered with the diagnosis of current HEV infection and explained the low PPV of this assay.
FIG 1.
Dynamics of HEV antibodies during the progression of illness. The levels of anti-HEV IgM (A, C, and E) and IgG (B, D, and F) are plotted against the follow-up days and shown with colored points and connecting lines for each patient. Subjects 1 to 14 were HEV RNA negative throughout the study period, and all others had HEV RNA detected at least once during the study period.
Performance of the HEV Ag assay in the diagnosis of current HEV infection.
HEV Ag has been reported as a useful serological marker for diagnosing HEV infection. Next, we used the 143 sequential samples from the antibody kinetics study to study the diagnostic efficacy of the HEV Ag assay. Of the 143 samples, 72 samples were negative for both HEV RNA and HEV Ag. Only 46 out of the 71 HEV RNA-positive samples were positive for HEV Ag. The sensitivity of the HEV Ag assay was 64.8%. Both the specificity and the PPV were 100%. The accuracy was ∼83%, and the NPV was ∼74% (Table 1).
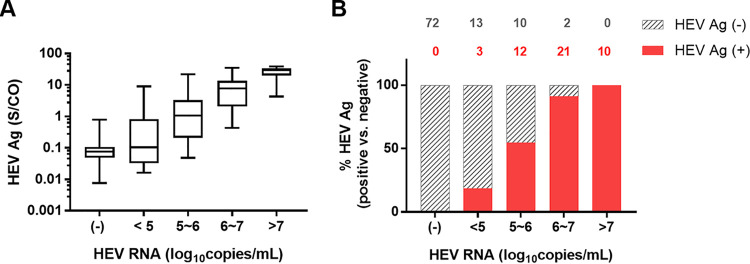
To understand why some samples showed discordant HEV RNA and HEV Ag results, we categorized the samples according to their HEV RNA levels. HEV Ag levels increased with HEV RNA levels (Fig. 2A). The discordance was mainly found in the HEV RNA low viral load groups (Fig. 2B). Among the 71 HEV RNA-positive samples, HEV Ag was detected in 3/16 (18.75%) samples in the HEV RNA range below 105 copies/ml, 12/22 (54.5%) samples in the range of 105 to 106 copies/ml, 21/23 (91.3%) samples in the range of 106 to 107 copies/ml, and 10/10 (100%) samples in the range above 107 copies/ml.
FIG 2.
Comparison between HEV Ag and HEV RNA levels in 72 HEV RNA-negative sera and 71 HEV RNA-positive sera. (A) Box plot of HEV Ag levels in samples at various levels of HEV RNA. The HEV Ag detection results for each group are shown as the range (whiskers), interquartile range (boxes), and median (line within boxes). (B) The proportion of samples with detectable HEV Ag at various HEV RNA levels. The number of samples with positive HEV Ag results is shown in red, and the number of samples with negative HEV Ag results is shown in gray on top of each column.
Factors affecting the detection of HEV Ag.
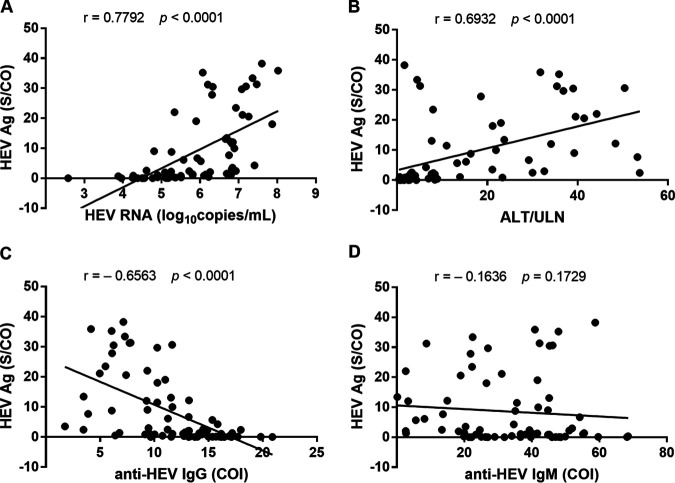
To explore other factors that may affect the detection efficiency of HEV Ag, we determined the correlation between the levels of HEV Ag and HEV RNA, anti-HEV IgG, IgM, or ALT/ULN in the 71 HEV RNA-positive samples (Fig. 3). The Spearman’s correlation coefficient (r) was 0.7792 between HEV Ag and HEV RNA (P < 0.0001), 0.6932 between HEV Ag and ALT/ULN (P < 0.0001), −0.6563 between HEV Ag and anti-HEV IgG (P < 0.0001), and −0.1636 between HEV Ag and anti-HEV IgM (P = 0.1729), respectively. Thus, both HEV RNA level and ALT/ULN had strong positive linear association with HEV Ag level, while anti-HEV IgG level showed a moderate to strong negative linear relationship to HEV Ag level and anti-HEV IgM level had no obvious linear relationship to HEV Ag level.
FIG 3.
Correlation between levels of HEV Ag and HEV RNA (A), ALT/ULN (B), anti-HEV IgG (C) or anti-HEV IgM (D) in 71 HEV RNA-positive sera. P values were calculated using the Spearman’s correlation method.
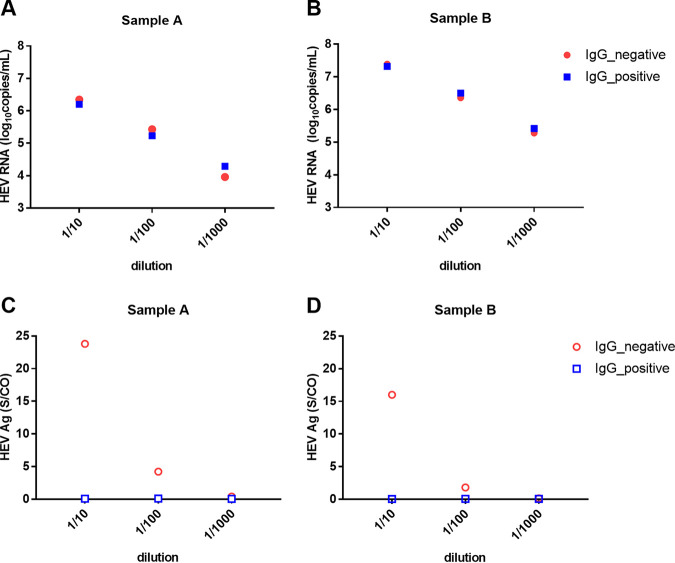
With that said, we hypothesized that the presence of anti-HEV IgG interferes with the detection of HEV Ag. To test this idea, we serially diluted two HEV RNA-positive serum samples (A and B) with either anti-HEV IgG-positive or IgG-negative serum (Table S3). The original samples and the diluted samples were then subjected to HEV Ag detection by ELISA or HEV RNA detection by RT-PCR (Fig. 4). The HEV RNA levels in serial dilutions by either positive or negative anti-HEV IgG serum were comparable (Fig. 4A and B). However, the HEV Ag level was much higher in dilutions with negative anti-HEV IgG serum (Fig. 4C and D), suggesting that the presence of anti-HEV IgG interfered with the detection of HEV Ag.
FIG 4.
HEV RNA (A and B) or HEV Ag (C and D) detection in samples diluted serially with anti-HEV IgG-positive or anti-HEV IgG-negative serum. The mean value from 2 experiments is shown.
Improved diagnostic accuracy of a model combining anti-HEV IgM, HEV Ag and ALT/ULN.
Considering the high seropositivity of anti-HEV IgG in areas where HEV is endemic, we reason that the HEV Ag assay alone is not good enough to diagnose current HEV infection, since it will cause misdiagnosis when the HEV RNA level is low or the anti-HEV IgG level is high. Thus, we decided to establish a model combining the key factors that differentiate current HEV infection from past infection or other liver diseases. Multivariate analyses showed that the levels of HEV Ag, anti-HEV IgM, and ALT/ULN were significantly independent variables associated with current HEV infection (Table S4). By stepwise multiple logistic regression analysis, a model for predicting current HEV infection was proposed. The model combining the 3 key factors in parameter estimates of 3.08 (lgAg), 5.14 (lgIgM), and 2.64, respectively, was identified with the highest AUROC (referred to as [IgM+Ag+ALT]). The model was shown as
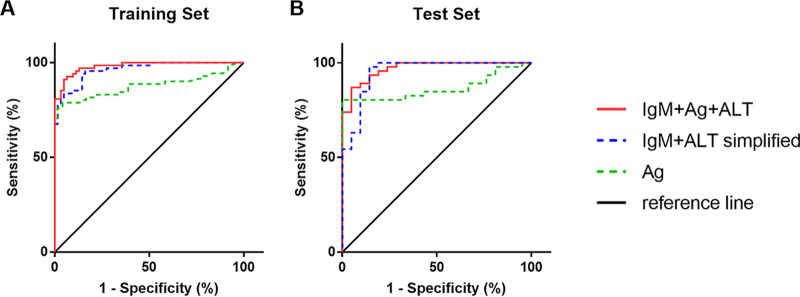
In the above-mentioned 143 sequential samples, 3 samples in the HEV RNA-positive group and 10 samples in the HEV RNA-negative group did not have records of ALT levels, so they were not included. A weighted sum score of 7.741 of the parameter estimates was associated with the highest sum of sensitivity and specificity and determined as the cutoff value. Sixty-two out of the 68 HEV RNA-positive samples had values above the cutoff, and 59 out of the 62 HEV RNA-negative samples had values below the cutoff. The diagnostic performance of the model was visualized by ROC curve (Fig. 5A) and summarized in Table 1. It achieved a sensitivity of 91.2% and a specificity of 95.2% with AUROC of 0.98. Both the PPV and the NPV were above 90%.
FIG 5.
ROC curves for diagnosis of current HEV infection with the HEV Ag assay, the [IgM+Ag+ALT] model, or the simplified [IgM+ALT] model in the training set (A) and the test set (B). The reference line indicates when false-positive results are produced at the same rate as true-positive results.
The diagnostic performance of the HEV Ag assay and the model was repeatedly evaluated in the 67 randomly selected anti-HEV IgM-positive samples. Results confirmed that the diagnostic performance of the model (AUROC and 95% CI, 0.97, 0.94 to 1.00) was better than that of HEV Ag (AUROC and 95% CI, 0.87, 0.78 to 0.95) (Table 1 and Fig. 5B).
If the HEV Ag assay is not available, a simplified model combining only anti-HEV IgM and ALT/ULN level was proposed and shown as follows. Parameters were scaled down and rounded for the ease of clinical application.
A weighted sum score of 3.921 of the parameter estimates was determined as the cutoff value. It achieved a sensitivity of 94.1% and a specificity of 85.5% with an AUROC of 0.96 in the training set. The sensitivity, specificity, and AUROC was 84.8%, 85.7%, and 0.95 in the test set, respectively (Fig. 5B and Table 1).
DISCUSSION
In this study, we evaluated the diagnostic performance of HEV serological markers for current HEV infection. Our findings show that anti-HEV IgM and HEV Ag are on the opposite ends in the diagnosis of HEV infection. A positive anti-HEV IgM result does not necessarily mean there is current HEV infection, while a negative anti-HEV IgM result basically excludes the possibility of HEV infection, except for very rare cases when patients seek medical care during the window period before anti-HEV IgM appears or patients are immunosuppressed and unable to produce anti-HEV IgM. In contrast, a positive result of HEV Ag confirms current HEV infection, while a negative result of HEV Ag does not guarantee there is no HEV infection, especially when the patient has a low viral RNA level or high anti-HEV IgG level.
The concordance between HEV Ag and HEV RNA is 118/143 (82.5%) in the training set and 48/67 (71.6%) in the test set. The HEV Ag assay gave correct diagnosis on all HEV RNA-negative samples but only correctly diagnosed 46/71 (64.5%) HEV RNA-positive samples in the training set and 27/46 (58.7%) in the test set.
It is understandable to have less HEV Ag detected at low levels of HEV RNA, since the HEV Ag assay was reported to be less sensitive than RT-PCR (17, 25, 26). The low efficiency of HEV Ag detection at the presence of anti-HEV IgG is somewhat unexpected. By revisiting the literature, we found the phenomenon was described previously. Zhao et al. used an in-house-developed HEV Ag detection assay and found that the sensitivity of the HEV Ag assay was inversely proportional to the concentration of anti-HEV antibodies in serum (17). In our speculation, epitope competition might lead to the reduced detection efficiency of HEV Ag in the presence of anti-HEV IgG. The anti-HEV IgG antibodies in serum may compete with the anti-capsid polyclonal antibodies used in the ELISA kit to bind with the capsid protein. An analysis of serial samples from HEV-infected monkeys showed that HEV Ag could be detected prior to the appearance of anti-HEV antibodies, almost simultaneously with the fecal RNA, but HEV Ag disappeared 2 to 3 weeks earlier than the RNA, when anti-HEV IgG levels increased (15). Trémeaux et al. showed the HEV Ag assay gave more frequently positive results in immunocompromised patients at the acute phase (26). Behrendt et al. found significantly higher levels of HEV Ag in chronically infected individuals than acutely infected patients (25). These results have several things in common. The easier detection of HEV Ag in either immunocompromised patients at the acute phase or chronic patients probably benefits from the lower level of anti-HEV IgG in these subjects. Another interesting result was shown by Geng et al. (27). They found that HEV Ag was detected more frequently in urine than HEV RNA, and the ratio of HEV Ag to HEV RNA in the urine was significantly higher than in sera and feces. It is possible that since less anti-HEV IgG is secreted into urine than sera (since IgGs are normally too large to pass through the tubules of kidney), it is easier to obtain HEV Ag detection results. These findings indicate that the accuracy of the HEV Ag assay will be affected especially in regions where HEV is endemic and where the anti-HEV IgG seroprevalence is high (28, 29). It is necessary to improve the HEV Ag detection assay in the future to make it less susceptible to concomitant substances in the serum.
The exact duration of anti-HEV antibody response remains uncertain. Dawson et al. reported IgM antibodies disappeared in specimen collected 3.5 years later, and IgG antibodies continued to be detected 4.5 years after the acute-phase infection (11). Myint et al. showed that in acute hepatitis E patients, anti-HEV IgM can persist for an average of 5 months (30). Riveiro-Barciela et al. reported an unexpectedly long persistence of anti-HEV IgM: 4 out of 5 (80%) subjects were positive for anti-HEV IgM with the Wantai assay during the second year after acute hepatitis E, and 2 out of 12 (17%) patients were still positive by the Wantai assay after 3 years (31). In our retrospective follow-up analysis, most patients with confirmed HEV infection experienced a rapid increase of anti-HEV IgM shortly after the acute phase and a slow decrease of anti-HEV IgM thereafter. Four patients were positive for anti-HEV IgM during the 3- to 6-month follow-up (45 to 48), 6 were positive during the 6- to 12-month follow-up (50, 51, 53, 54, 55, and 58), and 1 was positive after a year (57). One patient turned negative after ∼5 months, one turned negative after ∼8 months, and another one turned negative after 13 months. Generally, patients with higher levels of anti-HEV IgM had longer durations of being positive. We currently do not know why the levels of anti-HEV IgM varied among the patients. It may be related to the immune status of the patient per se but not to the previous exposure to HEV, since the patients who had relatively low levels of anti-HEV IgM at presentation were all positive for anti-HEV IgG (22, 23, 32, and 34).
The study has several limitations. First, HEV RNA and HEV Ag were analyzed retrospectively. RNA may degrade in stored sera over time, and the detection accuracy may be impaired. Second, the follow-up duration was not evenly distributed. For some patients, the intervals between sequential samples were too short to describe the long-term trend. For other patients, their samples were collected from 2 time points, and we can only make a rough estimation of their anti-HEV IgM seroconversion. Third, further modification or validation may be necessary before the multifactorial model can be applied to testing results from other products, considering the sensitivity and specificity difference between them (32). It also may not be readily applied to immunosuppressed patients. Fourth, the model is still complicated. To ease the application in clinical settings, an algorithm is proposed. When an immunocompetent person has elevated levels of liver enzymes and is suspected of HEV infection, anti-HEV IgM should be tested first, since it has very high sensitivity. When the anti-HEV IgM testing gives a positive result, HEV Ag testing is recommended. Positive results of HEV Ag can confirm HEV infection. If the HEV Ag assay gives a negative result, diagnosis based on the [IgM+Ag+ALT] model is needed. If the HEV Ag assay is not available, the simplified [IgM+ALT] model could correctly diagnose ∼88% of the cases at a cutoff value greater than 3.921.
In conclusion, we explored the merits and demerits of the anti-HEV IgM and HEV Ag assay and developed a model with satisfactory performance in the diagnosis of current HEV infection when HEV RNA detection is not available. It is a useful tool in clinical decision making, especially in developing countries where HEV is endemic.
Supplementary Material
ACKNOWLEDGMENTS
J.L. and Y.H. conceived and designed the study; J.L., P.W., Q.L., and Z.L. performed the experiments and collected the clinical data; J.L., Y.H., and J.J. conducted the analysis and prepared the manuscript; Q.G., H.G., and Q.X. contributed to the sample collection and data interpretation. All authors approved the final version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (no. 81501733) and the Shanghai Municipal Health Commission [Shanghai Municipal Key Clinical Specialty (shslczdzk01103)].
We do not have any disclosures to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nimgaonkar I, Ding Q, Schwartz RE, Ploss A. 2018. Hepatitis E virus: advances and challenges. Nat Rev Gastroenterol Hepatol 15:96–110. doi: 10.1038/nrgastro.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, Okamoto H. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol 84:2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- 3.Khuroo MS, Kamili S, Yattoo GN. 2004. Hepatitis E virus infection may be transmitted through blood transfusions in an endemic area. J Gastroenterol Hepatol 19:778–784. doi: 10.1111/j.1440-1746.2004.03437.x. [DOI] [PubMed] [Google Scholar]
- 4.Hamid SS, Atiq M, Shehzad F, Yasmeen A, Nissa T, Salam A, Siddiqui A, Jafri W. 2002. Hepatitis E virus superinfection in patients with chronic liver disease. Hepatology 36:474–478. doi: 10.1053/jhep.2002.34856. [DOI] [PubMed] [Google Scholar]
- 5.Khuroo MS, Teli MR, Skidmore S, Sofi MA, Khuroo MI. 1981. Incidence and severity of viral hepatitis in pregnancy. Am J Med 70:252–255. doi: 10.1016/0002-9343(81)90758-0. [DOI] [PubMed] [Google Scholar]
- 6.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 7.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlauder GG, Mushahwar IK. 2001. Genetic heterogeneity of hepatitis E virus. J Med Virol 65:282–292. doi: 10.1002/jmv.2031. [DOI] [PubMed] [Google Scholar]
- 9.Webb GW, Dalton HR. 2019. Hepatitis E: an underestimated emerging threat. Ther Adv Infect Dis 6:2049936119837162. doi: 10.1177/2049936119837162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalton HR, Fellows HJ, Stableforth W, Joseph M, Thurairajah PH, Warshow U, Hazeldine S, Remnarace R, Ijaz S, Hussaini SH, Bendall RP. 2007. The role of hepatitis E virus testing in drug-induced liver injury. Aliment Pharmacol Ther 26:1429–1435. doi: 10.1111/j.1365-2036.2007.03504.x. [DOI] [PubMed] [Google Scholar]
- 11.Dawson GJ, Mushahwar IK, Chau KH, Gitnick GL. 1992. Detection of long-lasting antibody to hepatitis E virus in a US traveller to Pakistan. Lancet 340:426–427. doi: 10.1016/0140-6736(92)91507-5. [DOI] [PubMed] [Google Scholar]
- 12.Dalton HR, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 13.Kamar N, Garrouste C, Haagsma EB, Garrigue V, Pischke S, Chauvet C, Dumortier J, Cannesson A, Cassuto-Viguier E, Thervet E, Conti F, Lebray P, Dalton HR, Santella R, Kanaan N, Essig M, Mousson C, Radenne S, Roque-Afonso AM, Izopet J, Rostaing L. 2011. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140:1481–1489. doi: 10.1053/j.gastro.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 14.European Association of the Study of Liver. 2018. EASL clinical practice guidelines on hepatitis E virus infection. J Hepatol 68:1256–1271. doi: 10.1016/j.jhep.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F, Li X, Li Z, Harrison TJ, Chong H, Qiao S, Huang W, Zhang H, Zhuang H, Wang Y. 2006. Detection of HEV antigen as a novel marker for the diagnosis of hepatitis E. J Med Virol 78:1441–1448. doi: 10.1002/jmv.20717. [DOI] [PubMed] [Google Scholar]
- 16.Wen GP, Tang ZM, Yang F, Zhang K, Ji WF, Cai W, Huang SJ, Wu T, Zhang J, Zheng ZZ, Xia NS. 2015. A valuable antigen detection method for diagnosis of acute hepatitis E. J Clin Microbiol 53:782–788. doi: 10.1128/JCM.01853-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Geng Y, Harrison TJ, Huang W, Song A, Wang Y. 2015. Evaluation of an antigen-capture EIA for the diagnosis of hepatitis E virus infection. J Viral Hepat 22:957–963. doi: 10.1111/jvh.12397. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Rao H, Wang Y, Wang J, Kong X, Ji Y, Zhu L, Liu Y, Fang J, Yang M, Luo B, Wang Z, Shi Y, Wang Y, Wang H, Zhao J, Wei L. 2019. Evaluation of an antigen assay for diagnosing acute and chronic hepatitis E genotype 4 infection. J Gastroenterol Hepatol 34:458–465. doi: 10.1111/jgh.14405. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Ge SX, Huang GY, Li SW, He ZQ, Wang YB, Zheng YJ, Gu Y, Ng MH, Xia NS. 2003. Evaluation of antibody-based and nucleic acid-based assays for diagnosis of hepatitis E virus infection in a rhesus monkey model. J Med Virol 71:518–526. doi: 10.1002/jmv.10523. [DOI] [PubMed] [Google Scholar]
- 20.Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy J-M, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J Clin Microbiol 50:897–902. doi: 10.1128/JCM.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicot F, Cazabat M, Lhomme S, Marion O, Sauné K, Chiabrando J, Dubois M, Kamar N, Abravanel F, Izopet J. 2016. Quantification of HEV RNA by droplet digital PCR. Viruses 8:233. doi: 10.3390/v8080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Ju X, Xiang G, Gong M, Yang R, Qin J, Li Y, Nan Y, Yang Y, Zhang QC, Ding Q. 2020. Identification of functional cis-acting RNA elements in the hepatitis E virus genome required for viral replication. PLoS Pathog 16:e1008488. doi: 10.1371/journal.ppat.1008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong ER, DeLong DM, Clarke-Pearson DL. 1988. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 25.Behrendt P, Bremer B, Todt D, Brown RJ, Heim A, Manns MP, Steinmann E, Wedemeyer H. 2016. Hepatitis E virus (HEV) ORF2 antigen levels differentiate between acute and chronic HEV infection. J Infect Dis 214:361–368. doi: 10.1093/infdis/jiw161. [DOI] [PubMed] [Google Scholar]
- 26.Trémeaux P, Lhomme S, Chapuy-Regaud S, Peron J-M, Alric L, Kamar N, Izopet J, Abravanel F. 2016. Performance of an antigen assay for diagnosing acute hepatitis E virus genotype 3 infection. J Clin Virol 79:1–5. doi: 10.1016/j.jcv.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Geng Y, Zhao C, Huang W, Harrison TJ, Zhang H, Geng K, Wang Y. 2016. Detection and assessment of infectivity of hepatitis E virus in urine. J Hepatol 64:37–43. doi: 10.1016/j.jhep.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 28.Li RC, Ge SX, Li YP, Zheng YJ, Nong Y, Guo QS, Zhang J, Ng MH, Xia NS. 2006. Seroprevalence of hepatitis E virus infection, rural southern People's Republic of China. Emerg Infect Dis 12:1682–1688. doi: 10.3201/eid1211.060332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu F-C, Huang S-J, Wu T, Zhang X-F, Wang Z-Z, Ai X, Yan Q, Yang C-L, Cai J-P, Jiang H-M, Wang Y-J, Ng M-H, Zhang J, Xia N-S. 2014. Epidemiology of zoonotic hepatitis E: a community-based surveillance study in a rural population in China. PLoS One 9:e87154. doi: 10.1371/journal.pone.0087154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myint KSA, Endy TP, Shrestha MP, Shrestha SK, Vaughn DW, Innis BL, Gibbons RV, Kuschner RA, Seriwatana J, Scott RM. 2006. Hepatitis E antibody kinetics in Nepalese patients. Trans R Soc Trop Med Hyg 100:938–941. doi: 10.1016/j.trstmh.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Riveiro-Barciela M, Rando-Segura A, Barreira-Díaz A, Bes M, Ruzo SP, Piron M, Quer J, Sauleda S, Rodríguez-Frías F, Esteban R, Buti M. 2020. Unexpected long-lasting anti-HEV IgM positivity: is HEV antigen a better serological marker for hepatitis E infection diagnosis? J Viral Hepat 27:747–753. doi: 10.1111/jvh.13285. [DOI] [PubMed] [Google Scholar]
- 32.Pas SD, Streefkerk RHRA, Pronk M, de Man RA, Beersma MF, Osterhaus ADME, van der Eijk AA. 2013. Diagnostic performance of selected commercial HEV IgM and IgG ELISAs for immunocompromised and immunocompetent patients. J Clin Virol 58:629–634. doi: 10.1016/j.jcv.2013.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.