Numerous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapid serological tests have been developed, but their accuracy has usually been assessed using very few samples, and rigorous comparisons between these tests are scarce. In this study, we evaluated and compared 10 commercially available SARS-CoV-2 rapid serological tests using the STARD (Standards for Reporting of Diagnostic Accuracy Studies) methodology.
KEYWORDS: RDT, IgG, IgM, antibodies, COVID-19, analytical performances, LFIA, SARS-CoV-2, immunoassays, serology
ABSTRACT
Numerous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapid serological tests have been developed, but their accuracy has usually been assessed using very few samples, and rigorous comparisons between these tests are scarce. In this study, we evaluated and compared 10 commercially available SARS-CoV-2 rapid serological tests using the STARD (Standards for Reporting of Diagnostic Accuracy Studies) methodology. Two hundred fifty serum samples from 159 PCR-confirmed SARS-CoV-2 patients (collected 0 to 32 days after the onset of symptoms) were tested with rapid serological tests. Control serum samples (n = 254) were retrieved from pre-coronavirus disease (COVID) periods from patients with other coronavirus infections (n = 11), positivity for rheumatoid factors (n = 3), IgG/IgM hyperglobulinemia (n = 9), malaria (n = 5), or no documented viral infection (n = 226). All samples were tested using rapid lateral flow immunoassays (LFIAs) from 10 manufacturers. Only four tests achieved ≥98% specificity, with the specificities ranging from 75.7% to 99.2%. The sensitivities varied by the day of sample collection after the onset of symptoms, from 31.7% to 55.4% (days 0 to 9), 65.9% to 92.9% (days 10 to 14), and 81.0% to 95.2% (>14 days). Only three of the tests evaluated met French health authorities’ thresholds for SARS-CoV-2 serological tests (≥90% sensitivity and ≥98% specificity). Overall, the performances varied greatly between tests, with only one-third meeting acceptable specificity and sensitivity thresholds. Knowledge of the analytical performances of these tests will allow clinicians and, most importantly, laboratorians to use them with more confidence; could help determine the general population’s immunological status; and may help diagnose some patients with false-negative real-time reverse transcription-PCR (RT-PCR) results.
INTRODUCTION
Asymptomatic carriage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been estimated to be as high as 86% in some studies (1). Others posit that it may be responsible for up to two-thirds of viral transmission (1–4). As the world increasingly acknowledges the challenges that this poses to disease containment, reliable testing has become central to monitoring the coronavirus disease 2019 (COVID-19) pandemic, informing health policy, rapidly responding to events as they evolve, and mitigating disease transmission (5, 6).
Yet real-time reverse transcription-PCR (RT-PCR), the gold standard for SARS-CoV-2 detection, has substantial limitations. PCR requires specialized, expensive laboratory equipment, which is often located only in laboratories with biosafety level ≥2, and may be affected by sample transport and testing delays of 2 to 3 days, during which time suspected COVID-19 cases may further expose other patients and health workers (7–9). For SARS-CoV-2, RT-PCR also uses nasopharyngeal swab samples that can be complex to obtain, pose considerable risks to health care workers with insufficient personal protective equipment (PPE), and have produced false-negative results in up to 30 confirmed COVID-19 patients (10–12). Chest radiography (CXR) and computed tomography (CT) scans are currently used to overcome the lack of sensitivity PCRs but also require expensive equipment (11, 13). These challenges limit the ability of current molecular and imaging approaches to be scaled up in epidemic settings where rapid, reliable, and easy population screening is needed.
Thus, serological confirmation of COVID-19 antibodies (Abs) could provide an important complementary tool to PCR testing by identifying previously exposed individuals (8, 12). SARS-CoV-2 seroconversion occurs 7 to 14 days after the onset of symptoms (8, 14–16). Classical ELISAs (enzyme-linked immunosorbent assays) are currently available, but considerable effort has been made by manufacturers to offer faster answers with rapid diagnostic tests (RDTs) (17). According to the Foundation for Innovative New Diagnostics (FIND), 177 SARS-CoV-2 antibody RDTs were commercially available on 15 June 2020 (18). Most information was directly submitted by test suppliers or obtained from publicly available sources and was not independently verified. Neither their analytical performances nor their usefulness in a clinical setting has yet been rigorously evaluated with a sufficient panel of samples (19, 20). In addition, validation criteria seem to be different from one country to another (21–23).
We carried out a retrospective clinical evaluation of 10 commercially available RDTs, comparing their performances according to the time between the onset of symptoms and sampling, severity of the disease, and usability of the tests. Our study was designed using the 2015 Standards for Reporting of Diagnostic Accuracy Studies (STARD) (24). We aim to provide accurate clinical performance data to assess the utility of the RDTs and their ability to be integrated into adapted diagnostic algorithms across health systems and epidemiological contexts, especially in areas with limited resources (24).
MATERIALS AND METHODS
Study design.
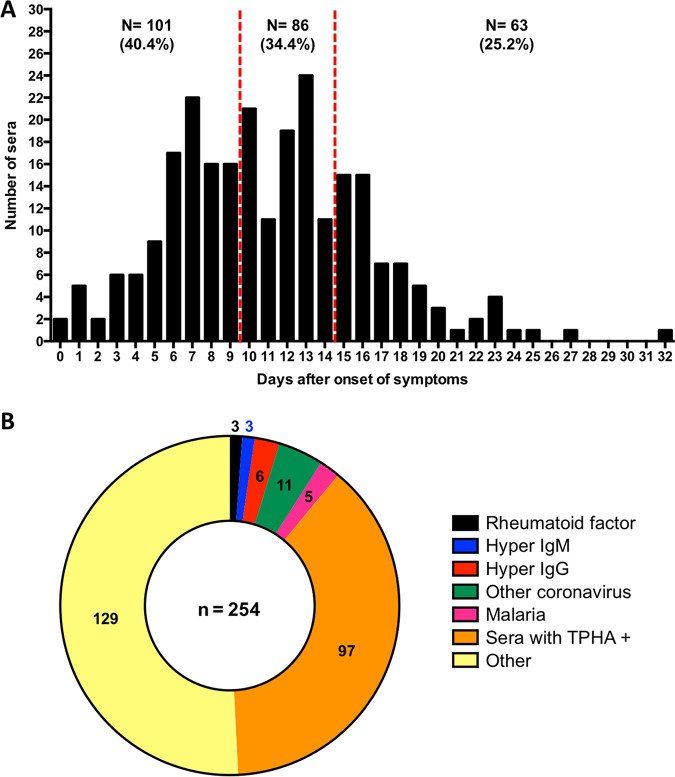
We conducted a retrospective study on 250 serum samples collected between 11 March and 3 April 2020 from 159 patients with documented RT-PCR-positive results for SARS-CoV-2 using nasopharyngeal swabs (eSwabs-Virocult; Copan, Italy). Real-time RT-PCRs targeting the RNA-dependent RNA polymerase and E genes were used to detect the presence of SARS-CoV-2 as described previously by Corman et al. (7). All patients were from 2 university hospitals located in the south of Paris (Bicêtre and Paul Brousse Hospitals) and provided between one and four serum samples. Serum samples from COVID-19 patients were randomly selected and grouped according to the time between the onset of symptoms and patient blood sampling (0 to 9 days, 10 to 14 days, and >14 days) (Fig. 1A).
FIG 1.
Serum collection used for the evaluation. (A) Distribution of 250 serum samples from COVID-positive patients according to the number of days after the onset of symptoms. (B) Distribution of the 254 control serum samples.
To assess specificity, an additional 254 serum samples collected prior to December 2019 were selected, which had previously tested positive for a separate agent or pathology that could potentially interfere with SARS-CoV-2 testing results, either another coronavirus (n = 11), other viral and parasitic infections (including Epstein-Barr virus [EBV], cytomegalovirus [CMV], rubeola virus, and toxoplasma) (n = 129), a rheumatoid factor (n = 3), IgG (n = 6) and IgM (n = 3) hyperglobulinemia, malaria (n = 5), or a positive Treponema pallidum hemagglutination assay (TPHA) (n = 97) (Fig. 1B).
Each RDT was evaluated on the same collection of serum samples. The minimum sample size was calculated assuming an expected sensitivity of 90% (with 5% accuracy) and a specificity of 98% (with 2% accuracy), amounting to 250 true-positive samples and 254 true-negative samples (power, 0.80; alpha, 0.05).
Sample preparation.
Selected serum samples were randomly placed in working boxes so as not to bias the technicians’ interpretation of results. Two sets of these boxes were prepared and stored at 4°C prior to being used.
Selected tests.
Diagnostic tests were selected based on supply, expected performances (based on published literature), and commercial brochures. Ten RDTs that either could detect all antibodies or specifically identified IgG or IgM (in blood, serum, or plasma) were evaluated: NG-Test IgG-IgM COVID-19 (NG-Biotech, Guipry, France) (RDT 1), an anti-SARS-CoV-2 rapid test (Autobio Diagnostic Co., Zhengzhou, China) (RDT 2), a novel coronavirus 2019 (2019-nCoV) antibody IgG/IgM test (Avioq Bio-Tech Co., Yantai, China) (RDT 3), the Nadal COVID-19 IgG/IgM test (Nal Von Minden GmbH, Regensburg, Germany) (RDT 4), Biosynex COVID-19 BSS (Biosynex, Illkirch-Graffenstaden, France) (RDT 5), a 2019-nCoV Ab test (Innovita Biological Technology Co., Qian’an, China) (RDT 6), a 2019-nCoV IgG/IgM test (Biolidics, Mapex, Singapore) (RDT 7), COVID-19-Check-1 (Veda Lab, Alençon, France) (RDT 8), the Finecare SARS-CoV2 antibody test (Guangzhou Wondfo Biotech, Guangzhou, China) (RDT 9), and the Wondfo SARS-CoV2 antibody test (Guangzhou Wondfo Biotech, Guangzhou, China) (RDT 10). The characteristics of these RDTs are summarized in Table S1 in the supplemental material. Tests were performed at room temperature by trained laboratory technicians. All tests were performed according to the manufacturers’ instructions using strict biosecurity measures and good microbiological practices and procedures (8).
The intensity of the reaction line was recorded in 3 gradations: no signal (0), very weak but definitively positive (1), and medium to high intensity (2). Values were not recorded when a control line did not appear, and tests were subsequently repeated (Fig. S1A and B).
Visual test interpretation was conducted independently by two separate readers and recorded on data collection sheets. Readings were determined based on two of three readers’ interpretations. In cases where all three interpretations were different, results were registered as unknown.
Data analysis.
The sensitivity and specificity of each RDT were calculated with their respective 95% confidence intervals (CI95) using VassarStats (http://vassarstats.net/).
The cumulative positivity at different points of illness (from symptom appearance until day 31 after symptom appearance) was determined as follows: (i) a positive result on day N was followed by subsequent positive results on days N + 1, N + 2, and N + n, etc., and (ii) a negative result on day N was preceded by negative results on days N − 1, N − 2, and N − n, etc. Details of the calculation are presented in Fig. S2.
Cumulative curves were fitted to an asymmetrical (five-parameter) logistic equation using GraphPad Prism v6 (25). For comparative purposes, the point at which 50% cumulative positivity was reached was calculated for all RDTs and expressed as the number of days after symptom onset (Table 1; Fig. S3).
TABLE 1.
Median times for SARS-CoV-2 seroconversion using 10 commercially available RDTs in Paris, France, in June 2020
| RDT | Time to seroconversion (days after symptom onset) |
|
|---|---|---|
| Median | CI95a | |
| 1 | 8.3 | 8.2–8.4 |
| 2 | 7.4 | 7.3–7.6 |
| 3 | 7.0 | 6.8–7.1 |
| 4 | 7.2 | 7.0–7.3 |
| 5 | 7.8 | 7.6–7.9 |
| 6 | 9.6 | 9.5–9.7 |
| 7 | 8.2 | 8.1–8.4 |
| 8 | 7.5 | 7.4–7.7 |
| 9 | 7.0 | 6.8–7.1 |
| 10 | 7.0 | 6.8–7.1 |
CI95, 95% confidence interval.
The positive predictive value (PPV) and negative predictive value (NPV) were calculated as follows: PPV = (sensitivity × prevalence)/{(sensitivity × prevalence) + [(1 − specificity) × (1 − prevalence)]}, and NPV = [specificity × (1 − prevalence)]/{[specificity × (1 − prevalence)] + [(1 − sensitivity) × prevalence]}.
Usability evaluation.
A self-administered user experience questionnaire using the Osgood scale was used for all tests and focused on the clarity of the instructions for the test user, the test’s technical complexity, the ease of test result interpretation, and access to legal information (26).
Ethics.
All samples were from a biobank (BIOCOVID-19) after having received ethical clearance from the Patient Protection Committee (PPC) of the Ile-de-France VII (no. 2009-965). Blood samples from patients infected with SARS-CoV-2 who were subjected to routine testing as part of clinical management but whose serum samples had not been entirely used for clinical purposes were approved for use in this study. The biobank is stored at CRB Paris South (BRIF, BB-0033-00089). The planning, conduct, and reporting of studies were in line with the principles of the Declaration of Helsinki.
RESULTS
Clinical characteristics of COVID-19 patients.
Overall, 250 serum samples collected from 159 COVID-19 patients were selected from the BIOCOVID-19 biobank. The distribution of the tested serum samples was as follows: 1 serum sample for 93 patients, 2 serum samples for 42 patients, 3 serum samples for 23 patients, and 4 serum samples for 1 patient. The median age was 62.9 years (range, 12.8 to 97.6 years), and the male/female ratio was 1.69 (100/59). Among these individuals, 4.4% (7/159) were discharged after their initial visit to the emergency room (ER), and 95.6% (152/159) were hospitalized. Over the study period, 44.1% (67/152) of patients required intensive care unit (ICU) care while hospitalized. The overall death rate among hospitalized patients was 19.1% (29/152): 10.5% (9/85) among non-ICU patients and 29.9% (20/67) among ICU patients. Most serum samples were obtained on days 0 to 15 (85.5%; 219/256) after symptoms appeared, although serum samples from later dates (up to day 31) were also available (Fig. 1A).
Test performances.
The cumulative positivity rate rose with time, reaching 100% 20 days after symptom onset for all RDTs (Fig. 2). More than 50% of SARS-CoV-2-infected patients had detectable antibodies 7 to 10 days after symptoms appeared (Fig. 2). The time needed to reach >95% sensitivity varied between 14 days (for half of the RDTs tested) and 18 days (for RDT 6) (Fig. 2). Asymmetrical (five-parameter) logistic analysis demonstrated that the 50% cumulative positivity (or the median time for seroconversion) varied from 7.0 to 9.6 days (Table 1). Failures in migration, as observed by the absence of a control line, were observed once for RDT 2 and RDT 6 and three times for RDT 8. For RDT 1, a weak control line was observed once. After retesting, all RDTs gave correct control lines (see Fig. S1B in the supplemental material).
FIG 2.
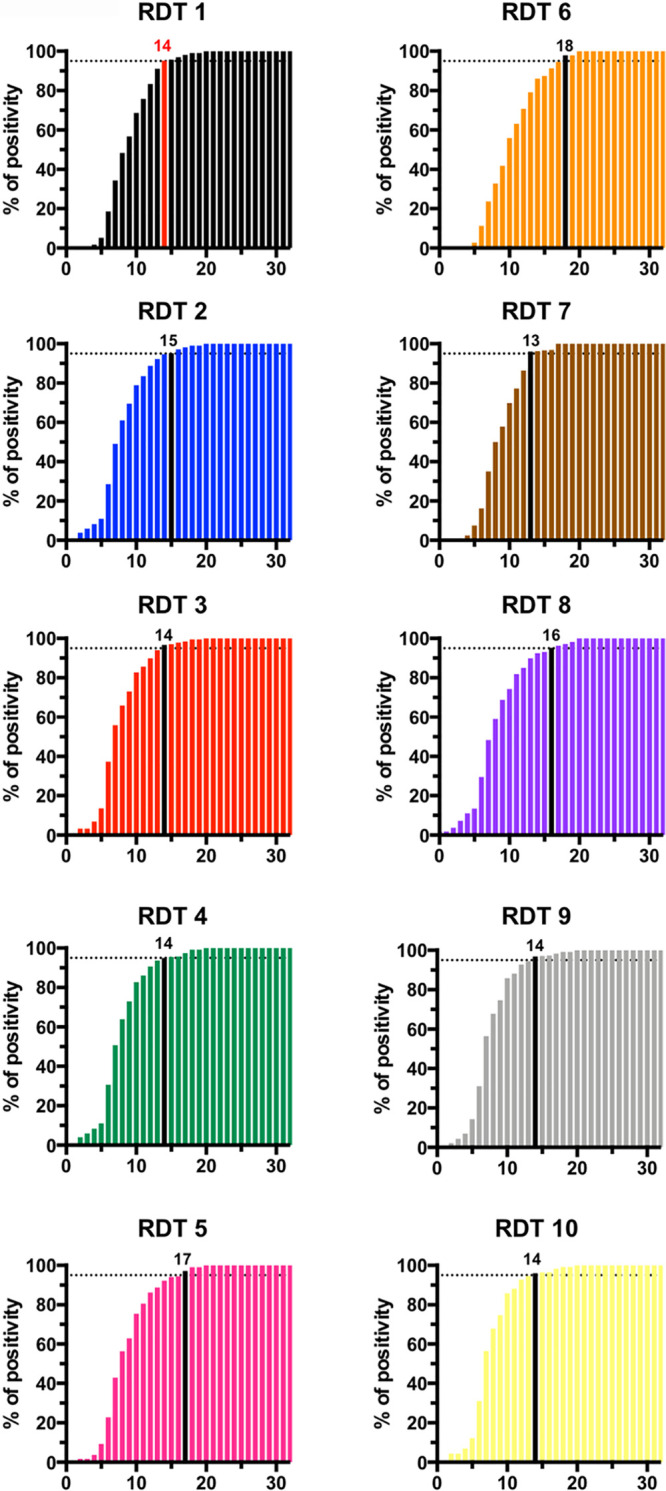
Cumulative positivity rate obtained with 10 RDTs in serum samples from COVID-19 patients stratified by the number of days after the appearance of symptoms. The day after symptom appearance with >95% positivity is indicated by a colored bar (red for RDT 1 and black for the other tests). The abscissas correspond to days after symptoms.
As expected, the overall test sensitivity was highest 15 days after the appearance of symptoms (Table 2), with all RDTs reaching >90% sensitivity at that point, except for RDT 6 and RDT 8 (81.0% and 88.5%, respectively). For the 8 RDTs able to differentiate between IgM and IgG, combined detection significantly increased the overall test sensitivity, with the exception of RDT 1, RDT 4, and RDT 5 (for which IgM detection seemed to be nearly as sensitive as IgM and IgG detection) (Table 2).
TABLE 2.
Performances of 10 rapid serological tests for SARS-CoV-2 antibodies in Paris, France, in June 2020
| Test | Total no. of samples tested | No of test results that were not interpretable | Sensitivity |
Specificity |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of samplesa | Ig type | Sensitivity by time elapsed after symptom onset (%) (CI95) |
No. of samples testedb | Ig type | Specificity (%) (CI95) | |||||
| 0–9 days | 10–14 days | >14 days | ||||||||
| RDT 1 (IgM/IgG) | 499 | 0 | 247 | IgM or IgG | 42.0 (32.3–52.3) | 75.0 (64.1–83.5) | 93.7 (83.7–97.9) | 252 | IgM or IgG | 99.2 (96.9–99.9) |
| IgM | 42.0 (32.3–52.3) | 75.0 (64.1–83.5) | 93.7 (83.7–97.9) | IgM | 99.6 (97.5–100.0) | |||||
| IgG | 33.0 (24.1–43.2) | 70.2 (59.1–79.5) | 85.7 (74.1–92.9) | IgG | 99.2 (96.9–99.9) | |||||
| RDT 2 (IgM/IgG) | 500 | 0 | 247 | IgM or IgG | 52.0 (41.8–62.0) | 87.1 (77.6–93.1) | 90.3 (79.5–96.0) | 253 | IgM or IgG | 94.5 (90.7–96.8) |
| IgM | 46.0 (36.1–56.2) | 81.2 (70.9–88.5) | 82.3 (70.0–90.4) | IgM | 96.0 (92.6–98.0) | |||||
| IgG | 44.0 (34.2–54.3) | 83.5 (73.6–90.4) | 83.9 (71.9–91.6) | IgG | 97.6 (94.7–99.0) | |||||
| RDT 3 (IgM/IgG) | 482 | 1 | 243 | IgM or IgG | 46.5 (36.6–56.7) | 76.5 (65.6–84.9) | 91.8 (81.2–96.9) | 238 | IgM or IgG | 94.1 (90.1–96.6) |
| IgM | 42.6 (32.9–52.8) | 75.3 (64.3–83.9) | 86.9 (75.2–93.8) | IgM | 95.4 (91.7–97.6) | |||||
| IgG | 45.5 (35.7–55.7) | 75.3 (64.3–83.9) | 91.8 (81.2–96.9) | IgG | 95.8 (92.2–97.9) | |||||
| RDT 4 (IgM/IgG) | 503 | 0 | 249 | IgM or IgG | 55.4 (45.2–65.2) | 90.6 (81.8–95.6) | 92.1 (81.7–97.0) | 254 | IgM or IgG | 99.2 (96.9–99.9) |
| IgM | 54.5 (44.3–64.3) | 88.2 (79.0–93.9) | 90.5 (79.8–96.1) | IgM | 100.0 (98.1–100) | |||||
| IgG | 18.8 (12.0–28.1) | 54.1 (43.0–64.9) | 90.5 (79.8–96.1) | IgG | 99.2 (96.9–99.9) | |||||
| RDT 5 (IgM/IgG) | 495 | 0 | 246 | IgM or IgG | 48.0 (38.0–58.2) | 84.3 (74.3–91.1) | 90.5 (79.8–96.1) | 249 | IgM or IgG | 92.4 (83.6–96.9) |
| IgM | 48.0 (38.0–58.2) | 80.7 (70.3–88.3) | 90.5 (79.8–96.1) | IgM | 97.5 (90.3–99.6) | |||||
| IgG | 22.0 (14.6–31.6) | 69.9 (58.7–79.2) | 77.8 (65.2–86.9) | IgG | 94.9 (86.9–98.4) | |||||
| RDT 6 (IgM/IgG) | 502 | 0 | 249 | IgM or IgG | 31.7 (23.0–41.8) | 65.9 (54.7–75.6) | 81.0 (68.7–89.4) | 253 | IgM or IgG | 98.4 (95.7–99.5) |
| IgM | 22.8 (15.3–32.4) | 54.1 (43.0–64.9) | 61.9 (48.8–73.6) | IgM | 99.2 (96.9–99.9) | |||||
| IgG | 21.8 (14.4–31.3) | 60.0 (48.8–70.3) | 71.4 (58.5–81.8) | IgG | 98.8 (96.3–99.7) | |||||
| RDT 7 (IgM/IgG)c | 246 | 0 | 167 | IgM or IgG | 35.7 (24.9–48.1) | 78.8 (64.9–88.5) | 93.3 (80.7–98.3) | 79 | IgM or IgG | 92.4 (83.6–96.7) |
| IgM | 20.0 (11.7–31.6) | 32.7 (20.7–47.3) | 53.3 (38.0–68.1) | IgM | 97.5 (90.3–99.6) | |||||
| IgG | 32.9 (22.4–45.2) | 76.9 (62.8–87.0) | 93.3 (80.7–98.3) | IgG | 94.9 (86.9–98.4) | |||||
| RDT 8 (IgM/IgG) | 488 | 3 | 238 | IgM or IgG | 55.7 (45.2–65.6) | 81.3 (70.6–88.8) | 88.5 (77.2–94.9) | 247 | IgM or IgG | 75.7 (69.8–80.8) |
| IgM | 42.3 (32.4–52.7) | 70.0 (58.6–79.5) | 65.6 (52.2–77.0) | IgM | 79.8 (74.1–84.5) | |||||
| IgG | 46.4 (36.3–56.8) | 71.3 (59.9–80.5) | 85.2 (73.3–92.6) | IgG | 87.9 (83.0–91.5) | |||||
| RDT 9 (total Ig) | 500 | 0 | 249 | Total Ig | 55.4 (45.2–65.2) | 92.9 (84.7–97.1) | 95.2 (85.8–98.8) | 251 | Total Ig | 98.4 (95.7–99.4) |
| RDT 10 (total Ig) | 503 | 0 | 249 | Total Ig | 55.4 (45.2–65.2) | 92.9 (84.7–97.1) | 92.1 (81.7–97.0) | 254 | Total Ig | 96.5 (93.2–98.3) |
Number of tested serum samples from COVID-positive patients.
Number of tested serum samples from COVID-negative patient.
RDT 7 was evaluated on only half of the total serum samples collection (only 250 tests received).
Specificities, calculated with serum samples recovered from patients between 2017 and early 2019, ranged from 75.7% to 99.2%. Only four tests (RDT 1, RDT 4, RDT 5, and RDT 9), reached the >98% threshold recommended by the French health authorities for serological diagnostic tests (Table 2) (23). The presence of a rheumatoid factor did not induce false-positive results except in the case of RDT 3, which systematically gave a positive IgM (3/3) and/or IgG (1/3) signal. Among the 11 serum samples with a non-SARS-CoV-2 agent (other coronaviruses), four tests produced one false-positive result, and one test produced two false-positive results. Notably, the false-positive results occurring in non-SARS-CoV-2 agent samples corresponded to one serum sample recovered from the same patient. No other patterns were detected for other false-positive results (Table S2). The concordance between all tests varied from 77.0% to 96.4% except in the case of RDT 8, which had lower concordance with other RDTs (<80%). Other RDTs gave concordant results (usually ∼90% to 95%) (Fig. 3).
FIG 3.
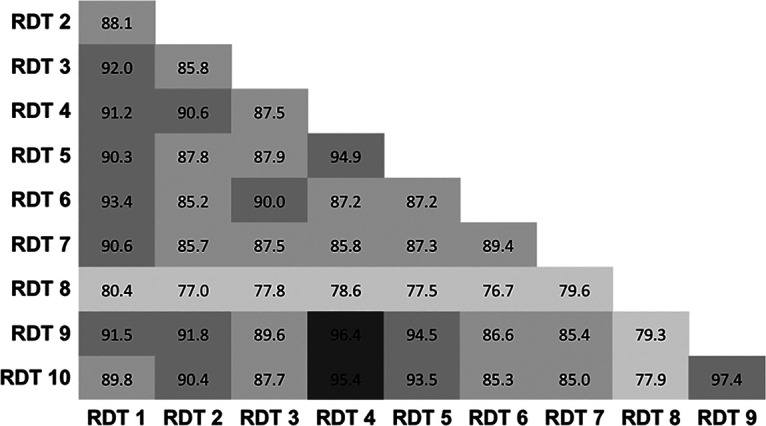
Agreement of results between RDTs. Percent agreement is indicated across all RDT combinations. RDTs were considered positive if any IgG and/or IgM was detected.
The positive and negative predictive values (PPV and NPV, respectively) describe the performances of a diagnostic test. A high value can be interpreted as indicating the accuracy of such a test. The PPV and NPV are not intrinsic to the test (as the true-positive rate and true-negative rate are), but they also depend on the prevalence. As the prevalence increases, the PPV also increases, but the NPV decreases. Similarly, as the prevalence decreases, the PPV decreases, while the NPV increases. As a consequence, having both the NPV and PPV above a certain value can be quite challenging. Among the 10 RDTs evaluated, only 3 presented PPVs and NPVs above 95% over a large window of population prevalence (RDT 1, RDT 4, and RDT 9) (Fig. S4). In France, depending on the region, the seroprevalence was estimated to be around 5% in June 2020, and local estimates now report values ranging from 5% to 15%, depending on regions more or less impacted by the virus (27). Thus, considering a 5% to 15% prevalence range, the PPV (5 to 15%) for RDT 1, RDT 4, and RDT 9 would be 86 to 95.4%, 85.8 to 95.3%, and 75.8 to 91.3%, respectively, and the NPV (5 to 15%) would be 99.7 to 98.9%, 99.5 to 98.6%, and 99.7 to 99.2%, respectively. Overall, the 3 RDTs perform equally well, with a slight advantage for RDT 1.
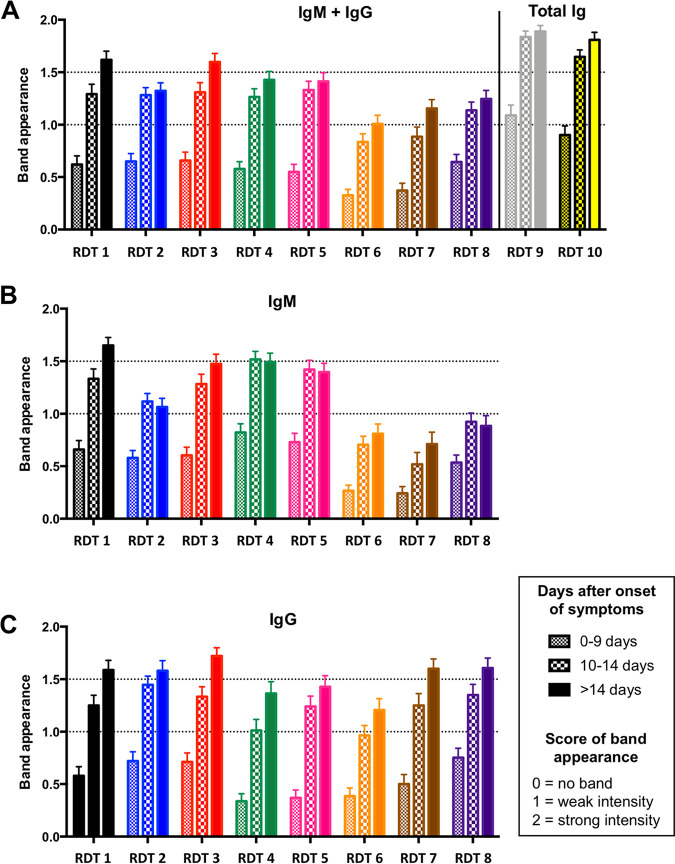
Band intensity.
To compare the ease of reading of the banded results of the RDTs, the intensity of the reaction line was recorded according to 3 gradations: no signal (0), very weak but definitively positive (1), and medium to high intensity (2). As shown in Fig. 4, the overall ease of reading was highest for serum samples recovered >14 days after the appearance of symptoms. The band intensity was most prominent in tests with combined antibody detection (i.e., both IgM and IgG detection [RDT 9 and RDT 10]) (Fig. 4A). Among the eight RDTs that differentiated between antibody types, the IgM band intensity was most pronounced with RDT 1 (Fig. 4B), with RDT 3, RDT 4, and RDT 5 closely following. Conversely, IgM bands obtained with RDT 6, RDT 7, RDT 8, and, to a lesser extent, RDT 2 were significantly less pronounced (Fig. 4B). For IgG tests, the bands produced by RDT 1, RDT 2, RDT 3, RDT 7, and RDT 8 were more prominent than those produced by RDT 4, RDT 5, and RDT 6 (Fig. 4C).
FIG 4.
Result (visible band) intensity for IgM-plus-IgG (A), IgM-only (B), and IgG-only (C) tests.
Ease of use.
All the tests were in cassette form, and nearly all devices used standard colloidal gold antigen-conjugated particles (Table S1). One test (RDT 9) used fluorescent antigen-conjugated particles for visualization using a specific reader. Ease of use could vary from one test to another, and all tests contained instructions for use (IFU) manuals that were in all cases considered easy to understand (Table S3). Only RDT 9’s IFU manual did not provide figures explaining the methods or interpretation of results. Most IFU manuals (6/10) contained figures explaining their methods and interpretation of results, and 3/10 IFU manuals contained figures explaining the interpretation of results (Table S3). No users reported difficulty using the RDTs, although RDT 2 provided a dropper with no clear instruction as to how many drops should be used. Buffer for RDT 9 was included with every test tube. Fewer than half of the RDTs (RDT 1, RDT 3, RDT 4, and RDT 5) included single-use plastic pipettes or similar devices for transferring samples into the test wells. No users reported difficulties in identifying sample and buffer wells. Interpretation of results for all tests, with the exception of RDT 6, was considered easy. The recommended time to reading of results ranged from 10 to 20 min (Table S1). From a packaging and legal point of view, all manufacturers except those of RDT 6 respected the CE-IVD (Conformité Européenne [CE] marking for all in vitro diagnostic [IVD] devices) regulation to describe the needed storage conditions in the IFU, on test packaging, and in product references. RDT 6’s reference test was not found on the box or within the IFU. All tests were in a single, sealed package and included a desiccant pouch.
DISCUSSION
With no curative medications currently available for COVID-19 and vaccines in early stages of development, physical distancing and widespread testing have become the primary tools available to control an unprecedented global health crisis. Serological assays and RDTs are being increasingly used across the world to address other tests’ limitations, but most commercially available RDTs have had their accuracy verified on only a small number of serum samples without including negative samples to evaluate cross-reactivity. Moreover, their usefulness for patient management in active hospital settings and among the general public has almost never been rigorously evaluated (28, 29). By demonstrating the feasibility and accuracy of rapid serological immunoassays with a substantially more robust sample size than what has previously been described, we add depth to the evolving conversation surrounding SARS-CoV-2 testing strategies. We hope that by knowing the analytical performances of nearly a dozen commercially available tests, and by providing comparative details, we will allow clinicians to select and use these tests with more confidence and certainty.
This study is, to our knowledge, the first to compare diagnostic performances and times to seropositivity of nearly a dozen SAR-CoV-2 RDTs using a large sample size (250 selected samples each for specificity and sensitivity, more than double those of other peer-reviewed, published RDT evaluations). Other studies evaluating antibody tests have also not included samples from patients with non-SARS-CoV-2 infections to evaluate specificity.
Overall, after the appearance of symptoms, seroconversion occurred on days 7 to 9 for 50 COVID-positive patients (Table 1), with >95% seroconverting after 14 days using RDT 1, RDT3, RDT 4, RDT 9, and RDT 10 and after 18 days for RDT 6 (Fig. 2). The specificities ranged from 94.5 to 99.2%, except for RDT 8 (75.7%). Notably, RDT 3 produced systematic false-positive results with serum samples of patients who had a high level of rheumatoid factor (see Table S2 in the supplemental material).
Thresholds for sensitivity and specificity for RDTs have been set by many national health authorities (21–23). For diagnosis in symptomatic patients, high sensitivity is required (generally ≥90%), while specificity is less critical, as some false-positive results may be tolerated as other potential diagnoses are considered in parallel (RT-PCR and/or CT scans). However, if lateral flow immunoassays (LFIAs) were deployed as an individual-level approach to inform release from quarantine or immune protection, then high specificity (>98) would be essential, as false-positive results return nonimmune individuals to risk of exposure (23). Using the French health authorities’ (21) acceptable limits for SARS-CoV-2 serological tests (≥90% sensitivity and ≥98% specificity), our evaluation validated only three RDTs for clinical use, namely, NG-Test IgG-IgM COVID-19 (RDT 1) (NG-Biotech), the Nadal COVID-19 IgG/IgM test (RDT 4) (Nal Von Minden GmbH), and the Finecare SARS-CoV2 antibody test (RDT 9) (Guangzhou Wondfo Biotech).
Appraisals of test performances should also consider the influence of population prevalence, as it may change over time and geography and within different population groups. The potential risk of a test providing false-positive results is crucial for release from lockdown of nonimmune individuals. Among the 10 RDTs evaluated, only 3 presented PPVs and NPVs above 95% over a large window of population prevalence (RDT 1, RDT 4, and RDT 9).
These serological tests were able to independently diagnose COVID-19, especially in those with ≥2 weeks of symptoms, and could be used to triangulate unclear or false-negative results from PCR and CT testing. They could also be used to monitor the status of medical and nonmedical frontline workers and, over the longer term, to establish population-level immunity as countries’ social restrictions ease. In the United States (Santa Clara County, CA), rapid antibody tests were used to evaluate the population prevalence of antibodies (ranging from 2.49 to 4.16%) and helped authorities to understand that infection was far more widespread (55-fold) than indicated by the number of confirmed cases. These data are crucial to calibrate epidemic and mortality projections (30).
Among the three RDTs fulfilling the French health authorities’ criteria, only NG-Test IgG-IgM COVID-19 (NG-Biotech) might be considered a self-test since it includes all materials needed for self-puncture and capillary blood recovery. Nevertheless, we authenticated this using serum only, since its use has been previously established in capillary whole blood, and our results in serum confirm those of the initial study (31), namely, that this bedside finger-prick test confirmed infection in <15 min and could be performed by a medical practitioner without specialized training or a pathology laboratory (31).
Our study is limited in the following ways. (i) RT-PCR detection was based on the use of upper respiratory tract specimens from patients with severe symptoms. None were asymptomatic patients (who did not access care). (ii) Most study participants’ diagnoses were based on positive findings from an RT-PCR test using respiratory samples. Patients with negative RT-PCR results but with chest imaging compatible with COVID-19 were not included. (iii) Because the epidemic situation in France was very recent at the time of the study, samples were collected during the acute phase of illness. Accordingly, we do not yet have serum samples from later stages to evaluate antibody persistence. (iv) Only 10 out of more than 170 available RDTs have been evaluated.
The COVID-19 pandemic has revealed gaps in our diagnostic arsenal and highlights the essential role of serodiagnostics in public health responses (32). With the use of carefully verified assays, appropriately designed serological studies will help characterize transmission dynamics, refine disease burden estimates, diagnose suspected cases, and confirm clinically diagnosed patients without access to RT-PCR testing.
Although this assessment demonstrates varied analytical performances across a sample of current SARS-CoV-2 RDTs, they nevertheless hold real utility as tools for establishing population-level exposure: many people have been exposed more than 3 weeks prior to antibody testing and would benefit from the nearly 100% sensitivity (in all tests evaluated) after 3 weeks. However, highly sensitive (as early as 7 days) and specific tests are needed, both to achieve sufficiently high positive predictive values since population prevalence is often estimated to be low (≤5%) and to be clinically useful as an initial diagnostic assay and a complement to direct RNA testing. Only three of the evaluated assays met the thresholds needed (sensitivity of >90% 14 days after symptom appearance and >98% specificity).
Serological assays are simple, inexpensive, rapid, easy to interpret, and practical (can be stored at room temperature). They detect IgM, IgG, or both and can be performed directly at a patient’s bedside, at a general physician’s office, or when triaging in an emergency department, as most have been validated using whole blood.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflict of interest.
This research was supported by Assistance Publique-Hôpitaux de Paris (APHP), Médecins Sans Frontières (MSF), and a grant from the French Defense Innovation Agency (AID).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. 2020. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, Wang M. 2020. Presumed asymptomatic carrier transmission of COVID-19. JAMA 323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizumoto K, Kagaya K, Zarebski A, Chowell G. 2020. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong ZD, Tang A, Li KF, Li P, Wang HL, Yi JP, Zhang YL, Yan JB. 2020. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis 26:1052–1054. doi: 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia Z, Lu Z. 2020. Modelling COVID-19 transmission: from data to intervention. Lancet Infect Dis 20:757–758. doi: 10.1016/S1473-3099(20)30258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patrick K, Stanbrook MB, Laupacis A. 2020. Social distancing to combat COVID-19: we are all on the front line. CMAJ 192:E516–E517. doi: 10.1503/cmaj.200606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang YW, Schmitz JE, Persing DH, Stratton CW. 2020. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol 58:e00512-20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. 2020. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. World Health Organization, Geneva, Switzerland. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117. [Google Scholar]
- 10.Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang F-S, Xie L, Sharma L, Dela Cruz CS, Qin E. 2020. Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med 201:1150–1152. doi: 10.1164/rccm.202003-0524LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. 2020. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. 2020. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IBK, Anderson DJ, Kong C, Altes T, Bush A, Desai SR, Goldin O, Goo JM, Humbert M, Inoue Y, Kauczor HU, Luo F, Mazzone PJ, Prokop M, Remy-Jardin M, Richeldi L, Schaefer-Prokop CM, Tomiyama N, Wells AU, Leung AN. 2020. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology 296:172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, Wang Y, Cui H, Pan K, Xu A. 2020. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis 94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, He X, Qian C, Sun Q, Hu Q, Liu H, Ye S, Xiang X, Zhou Y, Zhang W, Guo Y, Wang X-H, He W, Wan X, Sun F, Wei Q, Chen C, Pan G, Xia J, Mao Q, Chen Y, Deng G. 2020. Viral kinetics and antibody responses in patients with COVID-19. medRxiv 10.1101/2020.03.24.20042382. [DOI]
- 16.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez J, Shelton W, Manuel D-V, Rodriguez-Castellanos VE, Zuluaga JDH, Chamorro DF, Arroyo-Ariza D. 2020. Immunological assays for SARS-CoV-2: an analysis of available commercial tests to measure antigen and antibodies. medRxiv 10.1101/2020.04.10.20061150. [DOI]
- 18.Foundation for Innovative New Diagnostics. 2020. SARS-CoV-2 diagnostic pipeline. Foundation for Innovative New Diagnostics, Geneva, Switzerland. https://www.finddx.org/covid-19/pipeline/?avance=Commercialized&type=Rapid+diagnostic+tests&test_target=Antibody&status=all§ion=show-all&action=default. Accessed 15 June 2020. [Google Scholar]
- 19.Lassaunière R, Frische A, Harboe ZB, Nielsen ACY, Fomsgaard A, Krogfelt KA, Jørgensen CS. 2020. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv 10.1101/2020.04.09.20056325. [DOI]
- 20.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma SA, Lyons AM, Li S, Wong AW, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, II, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington LA, Loudermilk R, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv 10.1101/2020.04.25.20074856. [DOI] [PMC free article] [PubMed]
- 21.Adams ER, Ainsworth M, Ainsworth R, Andersson MI, Auckland K, Baillie JK, Barnes E, Beer S, Bell J, Berry T, Bibi S, Carroll M, Chinnakannan S, Clutterbuck E, Cornall RJ, Crook DW, De Silva T, Dejnirattisai W, Dingle KE, Dold C, Espinosa A, Eyre DW, Farmer H, Fernandez Mendoza M, Georgiou D, Hoosdally SJ, Hunter A, Jeffrey K, Klenerman P, Knight J, Knowles C, Kwok AJ, Leuschner U, Levin R, Liu C, Lopez-Camacho C, Martinez Garrido JC, Matthews PC, McGivern H, Mentzer AJ, Milton J, Mongkolsapaya J, Moore SC, Oliveira MS, Pereira F, Perez Lopez E, Peto T, Ploeg RJ, Pollard A, Prince T, et al. 2020. Antibody testing for COVID-19: a report from the National COVID Scientific Advisory Panel. medRxiv 10.1101/2020.04.15.20066407. [DOI] [PMC free article] [PubMed]
- 22.Food and Drug Administration. 2020. Policy for coronavirus disease-2019 tests during the public health emergency (revised). Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/media/135659/download. [Google Scholar]
- 23.Haute Autorité de Santé. 2020. Cahier des charges définissant les modalités d’évaluation des performances des tests sérologiques détectant les anticorps dirigés contre le SARS-CoV-2. Haute Autorité de Santé, Saint-Denis, France. https://www.has-sante.fr/upload/docs/application/pdf/2020-04/cahier_des_charges_test_serologique_covid19.pdf. Accessed April 16, 2020. [Google Scholar]
- 24.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HC, Bossuyt PM. 2016. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 6:e012799. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giraldo J, Vivas NM, Vila E, Badia A. 2002. Assessing the (a)symmetry of concentration-effect curves: empirical versus mechanistic models. Pharmacol Ther 95:21–45. doi: 10.1016/s0163-7258(02)00223-1. [DOI] [PubMed] [Google Scholar]
- 26.Osgood CE. 1962. Studies on the generality of affective meaning systems. Am Psychol 17:10–28. doi: 10.1037/h0045146. [DOI] [PubMed] [Google Scholar]
- 27.INSERM. 2020. Premiers résultats des enquêtes de santé publique de l’Inserm sur la Covid-19: facteurs de risque individuels et sociaux. INSERM, Paris, France. https://presse.inserm.fr/wp-content/uploads/2020/10/2020-10-09_CP_SAPRIS_EPICOV-1.pdf. [Google Scholar]
- 28.Tuaillon E, Bollore K, Pisoni A, Debiesse S, Renault C, Marie S, Groc S, Niels C, Pansu N, Dupuy AM, Morquin D, Foulongne V, Bourdin A, Le Moing V, Van de Perre P. 2020. Detection of SARS-CoV-2 antibodies using commercial assays and seroconversion patterns in hospitalized patients. J Infect 81:e39–e45. doi: 10.1016/j.jinf.2020.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Elslande J, Houben E, Depypere M, Brackenier A, Desmet S, Andre E, Van Ranst M, Lagrou K, Vermeersch P. 2020. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect 26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendavid E, Mulaney B, Sood N, Shah S, Ling E, Bromley-Dulfano R, Lai C, Weissberg Z, Saavedra-Walker R, Tedrow J, Tversky D, Bogan A, Kupiec T, Eichner D, Gupta R, Ioannidis J, Bhattacharya J. 2020. COVID-19 antibody seroprevalence in Santa Clara County, California. medRxiv 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed]
- 31.Dortet L, Emeraud C, Vauloup-Fellous C, Khecharem M, Ronat J-B, Fortineau N, Roque-Afonso A-M, Naas T. 2020. Rapid determination of SARS-CoV-2 antibodies using a bedside, point-of-care, serological test (4/20/2020). SSRN https://ssrn.com/abstract=3582814. [DOI] [PMC free article] [PubMed]
- 32.Cheng MP, Yansouni CP, Basta NE, Desjardins M, Kanjilal S, Paquette K, Caya C, Semret M, Quach C, Libman M, Mazzola L, Sacks JA, Dittrich S, Papenburg J. 2020. Serodiagnostics for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Ann Intern Med 173:450–460. doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.