The coronavirus disease 2019 (COVID-19) pandemic has highlighted the challenges inherent to the serological detection of a novel pathogen such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Serological tests can be used diagnostically and for surveillance, but their usefulness depends on their throughput, sensitivity, and specificity. Here, we describe a multiplex fluorescent microsphere-based assay, 3Flex, that can detect antibodies to three major SARS-CoV-2 antigens—spike (S) protein, the spike ACE2 receptor-binding domain (RBD), and nucleocapsid (NP).
KEYWORDS: COVID-19, IgG, SARS-CoV-2, coronavirus, fluorescence assays, immunoassays, microsphere, neutralizing antibodies, serology
ABSTRACT
The coronavirus disease 2019 (COVID-19) pandemic has highlighted the challenges inherent to the serological detection of a novel pathogen such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Serological tests can be used diagnostically and for surveillance, but their usefulness depends on their throughput, sensitivity, and specificity. Here, we describe a multiplex fluorescent microsphere-based assay, 3Flex, that can detect antibodies to three major SARS-CoV-2 antigens—spike (S) protein, the spike ACE2 receptor-binding domain (RBD), and nucleocapsid (NP). Specificity was assessed using 213 prepandemic samples. Sensitivity was measured and compared to that of the Abbott Architect SARS-CoV-2 IgG assay using serum samples from 125 unique patients equally binned (n = 25) into 5 time intervals (≤5, 6 to 10, 11 to 15, 16 to 20, and ≥21 days from symptom onset). With samples obtained at ≤5 days from symptom onset, the 3Flex assay was more sensitive (48.0% versus 32.0%), but the two assays performed comparably using serum obtained ≥21 days from symptom onset. A larger collection (n = 534) of discarded sera was profiled from patients (n = 140) whose COVID-19 course was characterized through chart review. This revealed the relative rise, peak (S, 23.8; RBD, 23.6; NP, 16.7 [in days from symptom onset]), and decline of the antibody response. Considerable interperson variation was observed with a subset of extensively sampled intensive care unit (ICU) patients. Using soluble ACE2, inhibition of antibody binding was demonstrated for S and RBD, and not for NP. Taking the data together, this study described the performance of an assay built on a flexible and high-throughput serological platform that proved adaptable to the emergence of a novel infectious agent.
INTRODUCTION
The betacoronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic. As SARS-CoV-2 continues to move through the global population, serological testing is vitally important to understand both the degree of spread (seroprevalence) and the characteristics of the immune response in terms of antibody (Ab) kinetics, dominant antigens, and immunological significance.
A number of antibody tests have been granted emergency use authorization (EUA) by the FDA (1). These include rapid lateral flow assay (LFA) devices, enzyme-linked immunosorbent assays (ELISAs), chemiluminescent microparticle immunoassays (CMIAs), and fluorescence microsphere immunoassays (FMIA). The majority of these tests are qualitative and typically target either the SARS-CoV-2 spike (S) protein or the nucleocapsid (NP) protein. Likewise, these same targets were of interest in the development of serological assays for SARS-CoV-1 (2–6).
The S protein is an ∼180-kDa glycosylated homotrimer that extends from the viral surface and initiates host cell entry (7). The extracellular region of S is organized into S1 and S2, with S1 comprising the outermost region and containing the receptor-binding domain (RBD) for the human ACE2 receptor (8–11). Anti-S antibodies can neutralize SARS-CoV-2 in cell culture (7, 8, 12–15), as can anti-S antibodies against SARS-CoV-1 (3). Thus, S and RBD are of great interest as targets for both immunoassays and vaccines due to their interactions with the human host (16). As a component of serological assays, S is incorporated as the full extracellular domain (ECD) or the outermost S1 domain or the receptor-binding domain (RBD). Additionally, the abundance and antigenicity of NP have made it an attractive target for both diagnostic and vaccine work (17–19). NP has roles in viral replication, transcription, and packaging of the viral genome (20, 21).
Beyond exposure, issues surrounding the serological response to SARS-CoV-2 are becoming more nuanced and include long-term antibody kinetics, measuring anamnestic responses to reinfection or vaccination, and assessment of plasma donors. For these, both qualitative and quantitative/semiquantitative serological assays that target multiple antigens and/or have the ability to assess the neutralization potential of SARS-CoV-2 antibodies are of great interest. To date, only a few assays with FDA EUA incorporate both the NP and S antigens, including technologies such as LFAs (e.g., Assure COVID-19 IgG/IgM rapid test device, CareStart COVID-19 IgM/IgG, Cellex qSARS-CoV-2 IgG/IgM rapid test), CMIAs (e.g., Diazyme DZ-Lite SARS-CoV-2 IgM/IgG, Vibrant COVID-19 Ab assay), and FMIAs (Luminex Inc. xMAP SARS-CoV-2 multiantigen IgG assay) (1). None of the current EUA assays incorporate an assessment of antibody-mediated neutralization of SARS-CoV-2 S protein binding to the ACE2 receptor.
In the current study, we utilized a laboratory-developed FMIA designated SARS-CoV-2 IgG 3Flex (3Flex) built on the Luminex Flexmap three-dimensional (3D) system for the simultaneous detection of antibodies to the N, S, and RBD antigens. We assessed its performance using 534 serum samples from 140 SARS-CoV-2-infected individuals from both inpatient and outpatient settings. In addition, we followed the serological response in a subset of individuals over time and demonstrated longitudinal patterns of antibody levels to each antigen alongside an assessment of neutralization at different time points over the course of disease.
MATERIALS AND METHODS
Clinical laboratory setting.
The University of Rochester (UR) Medicine Labs clinical microbiology laboratory in Rochester, NY (serving several area hospitals, practices, nursing homes, and urgent care providers), provides diagnostic services for institutionalized and ambulatory patients. During the ongoing COVID-19 global pandemic, analyses diagnostic for SARS-CoV-2 infections were provided for an expanded patient population in western NY. This study was approved by the University of Rochester Institutional Review Board (RSRB STUDY00004836).
Sample collection.
Residual serum specimens (n = 534) were collected at convenience in April 2020 from 140 unique patients (including outpatients, nursing home residents, and inpatients). All serum specimens were obtained from symptomatic patients positive for SARS-CoV-2 nucleic acid tested with real-time PCR (RT-PCR)-based assays (cobas SARS-CoV-2 test, Simplexa COVID-19 Direct, and Xpert Xpress SARS-CoV-2). Serum was held at 4°C until discard (7 days) or −80°C for long-term storage. Additionally, pre-COVID-19 sera (n = 125) from 2019 were used for specificity testing and validation and included specimens positive for antibodies raised against agents of bacterial and viral diseases (Lyme disease, syphilis, cytomegalovirus [CMV] infection, Epstein-Barr virus [EBV] infection, and other respiratory illnesses) and autoimmune markers (antinuclear antigen [ANA] and rheumatoid factor).
Chart review.
Patient charts were reviewed by at least two people from a four-person team, and discrepancies were resolved by a third person. Information collected included patient demographics (sex and age), clinical course (including the estimated date of symptom onset, days of hospitalization, intensive care unit [ICU] admission, and death), reported symptoms (including fever, cough, and shortness of breath), and comorbidities (including history of smoking, coronary artery disease, chronic obstructive pulmonary disease [COPD], and diabetes). Given the need to assess for the serological response within a known time frame (days from symptom onset), all patients in this study had symptomatic manifestations of COVID-19. Thus, values representing days from symptom onset were calculated using the collection date for each serum sample. Where possible, RT-PCR cycle threshold (CT) values from multiple RT-PCR tests were obtained and assessed alongside the serological response for the patients included in this study.
Multiplex SARS-CoV-2 IgG microsphere immunoassay development. (i) Antigens.
The immunoassay was constructed using full-length SARS-CoV-2 NP protein (catalog no. Z03480-100), the extracellular domain (ECD) of the S protein (catalog no. Z03481-100), and the RBD (catalog no. Z03483-100), all purchased from GenScript Biotech Corporation, Piscataway, NJ. The S protein (135 kDa; His tag and FLAG tag) was expressed in Sf9 insect cells, NP protein (46 kDa; His tag) was expressed in Escherichia coli, and RBD (30 kDa; His tag) was obtained from expression in human cells. The SARS-CoV-2 spike ECD and RBD are both reported to “bind with human ACE2 in functional ELISA” by the manufacturer. In addition to the viral antigens, two additional control beads were included, i.e., an internal control bead on region 45 (IC45) and a bead coupled with human IgG, as described in the next section.
(ii) Microsphere-antigen coupling.
Protein coupling to magnetic microsphere beads was performed according to the manufacturer’s recommendations using an xMAP antibody coupling (AbC) kit (Luminex, Austin, TX). The S protein was coupled at 10 pM, while the NP and RBD were coupled at 100 pM (i.e., equimolar coupling). Beads were also coated with purified human IgG (catalog no. I2511; Sigma-Aldrich, Inc.) at 0.1 μg/ml. Coupled beads corresponded to bead regions 18 (S), 33 (RBD), 73 (NP), 45 (internal control; IC45), and 66 (human IgG). IgG coupling confirmation was performed using serial dilutions of biotin-SP (long spacer)-conjugated Fcγ fragment-specific goat anti-human IgG (catalog no. 109-065-098; Jackson ImmunoResearch Laboratories, Inc.) and streptavidin-R–phycoerythrin (PE) conjugate (SAPE) (catalog no. S21388; Thermo Fisher Scientific). SARS-CoV-2 antigen coupling was confirmed by testing serial dilutions of rabbit-derived antibodies directed against S (catalog no. NB100-56047) and NP (catalog no. NB100-56049; Novus Biologicals, LLC, Centennial, CO) and detection with PE-labeled goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody (catalog no. P2771MP; Thermo Fisher Scientific).
(iii) Assay procedure.
Serum was diluted to 1:2,000 in PBS-TBN (phosphate-buffered saline [PBS], 0.02%; Tween, 0.1%; bovine serum albumin [BSA], 1%; azide, 0.05%). The assay was performed by incubating a 5-plex bead mix delivering 2,500 beads for each bead region (IC45, each CoV antigen bead, and the IgG-coupled bead) in a 50-μl volume of PBS-TBN with 50 μl of the diluted patient serum for 15 min at 37°C. Beads were washed twice in 150 μl of PBS-TBN with shaking for 2 min at 37°C using magnetic separation. Washed beads were incubated with a mixture of 1:4,500 biotin-SP-conjugated Fcγ fragment-specific goat anti-human IgG and 1 μg/ml SAPE for 15 min at 37°C. Beads were again washed twice in 150 μl of PBS-TBN with shaking for 2 min at 37°C and separated magnetically. Beads were resuspended in 100 μl of PBS-TBN and analyzed on a Flexmap 3D instrument (Luminex, Austin, TX), which provides a measurement of the median fluorescence intensity (MFI) for each analyte on each bead region. For each 96-well plate, patient sera were tested alongside three PBS-TBN blank wells, an IgG-stripped serum control (catalog no. IIGGDS100ML; Innovative Research, Inc., Novi, MI), and two sera from patients positive for SARS-CoV-2 by RT-PCR known to be of “low” and “high” positivity with respect to SARS-CoV-2 IgG serology (as determined by the Abbott SARS-CoV-2 IgG assay). If any of the 3Flex markers (S, RBD, or NP) gave a positive result, then the overall interpretation of the assay was classified as representing positivity.
Comparator SARS-CoV-2 IgG testing.
Sera used in the validation of the 3Flex assay were tested in parallel according to the package insert for the FDA Emergency Use Authorization (EUA) SARS-CoV-2 IgG assay on the Abbott Architect i2000SR platform. This assay detects serological responses to SARS-CoV-2 NP.
Study design and analysis. (i) Multiplex SARS-CoV-2 IgG microsphere immunoassay validation.
To establish the required specificity of the assay, and for cross-reactivity testing, MFI cutoff values were determined by testing a total of 218 “pre-COVID-19” sera. These comprised sera from individuals positive for ANA (n = 20) or rheumatoid factor (n = 10) or from those serologically diagnosed with Lyme disease (n = 10), syphilis (n = 10), CMV infection (n = 10), EBV infection (n = 10), or non-SARS-CoV-2 respiratory illnesses (n = 55) and other negative samples (n = 93). To examine assay performance, sera (n = 125) from unique patients (i.e., n = 125) positive for SARS-CoV-2 nucleic acid and with a known date of symptom onset were tested. These samples were randomly selected and binned into 5 time intervals corresponding to ≤5, 6 to 10, 11 to 15, 16 to 20, and ≥21 days from symptom onset. Results were interpreted qualitatively, and concordance was established with respect to the Abbott SARS-CoV-2 IgG assay. Assay reproducibility was established by testing 5 replicates each of 3 serum samples testing positive by the Abbott Architect SARS-CoV-2 IgG assay (see Table S2 in the supplemental material).
(ii) Serological profiling of SARS-CoV-2 infection.
MFI values for each antigen from all tests (n = 534) were assessed with respect to days from symptom onset. MFI values and corresponding CT values were plotted in GraphPad Prism 8 with a smoothed curve (GraphPad Software, San Diego, CA). Peak values for IgG responses were determined by area under the curve (AUC) analyses. Because some patients were highly represented in the larger data set, a subset of randomly selected serum specimens was used for MFI comparisons by time and for comparisons between selected patient populations (i.e., between ICU-admitted and other patients). Where possible, this subset included no more than 1 serum specimen assayed from a unique patient for each of the 5 time intervals. This allowed 231 serological tests from 140 unique patients to be examined. Additionally, sera from 9 extensively sampled patients were tested to explore interperson (i.e., between-patient) IgG responses and the precision of the assay with repeated measures.
(iii) ACE2 inhibition.
As a proxy for the detection of “neutralizing” titers of antibodies to SARS-CoV-2, the 5-plex bead mix was incubated with soluble recombinant human angiotensin-converting enzyme 2 (ACE2) (catalog no. 0192-30; AdipoGen Corporation, San Diego, CA) for 2 min at 37°C with shaking prior to the addition of sera. A doubling dilution series of ACE2 was used to optimize the concentration needed to produce an ∼50% loss of MFI value for sera tested with and without ACE2. A concentration of 2 μg/ml was selected because MFI values for the spike RBD were reduced by 50% in the majority of SARS-CoV-2-positive samples tested. ACE2 inhibition was also performed for the 9 extensively characterized patients, but only 1 randomly selected serum sample from each of the 5 time intervals was tested if possible. Inhibition values—given as the residual MFI plus ACE2 (percent)—are calculated as the percentage of the MFI value in the presence of ACE2 over the MFI value without ACE2.
Statistical and graphical analysis.
Statistical calculations and plotting were performed in Prism 8 (GraphPad Software, San Diego, CA). Fisher’s exact test was used for patient population comparisons. Unless otherwise indicated, error bars indicate means ± standard deviations (SD).
RESULTS
Multiplex SARS-CoV-2 IgG microsphere immunoassay validation. (i) Specificity.
Specificity was assessed using a set of 218 pre-COVID-19 sera. Serum samples were submitted to the diagnostic laboratory between 1 October 2019 and 1 February 2020 and included those that were negative for all analytes tested, as well as samples that were serologically positive for syphilis, CMV infection, EBV infection/mononucleosis (EBV/Mono), rheumatoid factor, and Lyme disease (Table 1). This also included 55 samples taken from patients within 60 days of an acute respiratory infection. All pre-COVID-19 sera were designated “negative” and formed the basis of cutoff values to establish the positive/negative thresholds used to interpret subsequent testing. The MFI cutoff values (≤) for S, RBD, and NP were 583, 182, and 2,455, respectively. These values were chosen to give 100% specificity and exceeded 6 standard deviations of the means of all negative samples included in this specificity assessment.
TABLE 1.
3Flex MFI values of pre-COVID-19 serum, including serum positive for potential cross-reactivitya
| Sample group | n | MFI avg (95% CI) |
||
|---|---|---|---|---|
| Spike | RBD | Nucleocapsid | ||
| Otherb | 93 | 140 (127–153) | 91 (88–93) | 224 (159–288) |
| Lyme disease | 10 | 148 (138–157) | 97 (92–102) | 564 (58–1,069) |
| Syphilis | 10 | 165 (148–182) | 105 (98–113) | 214 (3–284) |
| CMV | 10 | 180 (113–247) | 96 (91–102) | 230 (99–361) |
| EBV/Mono | 10 | 187 (134–241) | 109 (102–117) | 265 (165–365) |
| Acute viral respiratory illness | 55 | 136 (121–151) | 91 (90–93) | 165 (107–223) |
| ANA | 20 | 178 (153–202) | 101 (97–104) | 348 (162–535) |
| Rheumatoid factor | 10 | 149 (134–164) | 99 (94–104) | 476 (80–873) |
Abbreviations: 95% CI, 95% confidence interval; ANA, antinuclear antigen; CMV, cytomegalovirus; EBV/Mono, Epstein-Barr virus mononucleosis; MFI, median fluorescent intensity.
Samples with no serological diagnosis.
(ii) Sensitivity.
Serum samples from patients with confirmed COVID-19 were categorized by days from symptom onset (≤5, 6 to 10, 11 to 15, 16 to 20, and ≥21 days; n = 25 each) and evaluated with both the 3Flex assay and the Abbott Architect SARS-CoV-2 IgG assay (EUA) (Table 2). With serum obtained ≤5 days from symptom onset, the 3Flex assay demonstrated a 48.0% (12/25) positivity rate, compared to 32.0% for the Architect SARS-CoV-2 IgG assay (8/25) (Table 2). In 3/4 of the discrepant samples, the 3Flex assay was positive due to the S target alone, while in 1/4, all three targets were positive (see Table S1 in the supplemental material). By 16 to 20 days from symptom onset, the two assays showed comparable positivity rates (92%) which were also observed for samples obtained ≥21 days from symptom onset. Although the 3Flex assay is reported with a positive or negative qualitative result, MFI values increased from averages of 2,305.88 (S), 1,168.40 (RBD), and 3,380.28 (NP) in the ≤5 days from symptom onset category to 24,859.64 (S), 14,963.76 (RBD), and 20,801.24 (NP) in the ≥21 days from symptom onset category (Table 2).
TABLE 2.
Percent positivity and average MFIa values of 3Flex versus Abbott Architect SARS-CoV-2 IgG serological tests by days from symptom onset for 125 unique patients
| No. of days from symptom onset | Total no. (%) of tests | Abbott Architect SARS-CoV-2 IgG | 3Flex |
|||
|---|---|---|---|---|---|---|
| No. (%) of positive test results | No. (%) of positive test results | MFI avg (95% CI) |
||||
| Spike | RBD | Nucleocapsid | ||||
| ≤5 | 25 (100.0) | 8 (32.0) | 12 (48.0) | 4,637 (719–8,554) | 2,337 (355–4,319) | 6,578 (2,823–10,333) |
| 6–10 | 25 (100.0) | 13 (54.2)b | 17 (68.0) | 11,688 (5,053–18,324) | 6,283 (1,700–10,867) | 14,779 (6,111–23,477) |
| 11–15 | 25 (100.0) | 20 (80.0) | 21 (84.0) | 24,654 (15,358–33,950) | 14,378 (6,202–22,554) | 19,592 (12,242–26,941) |
| 16–20 | 25 (100.0) | 22 (91.6)b | 23 (92.0) | 23,202 (17,044–29,359) | 14,222 (9,185–19,258) | 25,534 (18,859–32,209) |
| ≥21 | 25 (100.0) | 23 (92.0) | 23 (92.0) | 27,010 (20,231–33,789) | 16,258 (10,727–21,789) | 22,591 (13,629–31,554) |
MFI, median fluorescent intensity.
One test had insufficient volume for the Abbott Architect SARS-CoV-2 IgG assay.
Serological profiling of SARS-CoV-2 infection. (i) Characteristics of patient population.
All 140 unique patients profiled were positive for SARS-CoV-2 nucleic acid by RT-PCR. Of these, 65/140 (46.4%) were women and 75/140 (53.6%) were men (Table 3). Sixty-six patients were admitted to an ICU, and there were 20 mortalities—with 16/20 (80%) being men. Nursing home residents comprised 37/140 (26.4%) of the tested patients, and the average ages of the nursing home residents were 82.6 and 76.0 years for women and men, respectively, compared to 63.4 and 64.1 years for the other patient types (inpatient, outpatient) combined. The most frequently reported symptoms were fever (women: 32/65, 49.2%; men: 51/75, 68.0%) and shortness of breath (women: 34/65, 52.3%; men: 44/75; 58.7%). Gastrointestinal symptoms (diarrhea/vomiting) were noted for 42/140 (30.0%) of the patients (women: 16/65, 24.6%; men: 26/75, 34.7%). Ventilated patients comprised 47/140 (33.6%) of the population (women: 22/65, 33.8%; men: 25/75, 33.3%). The most frequently reported comorbidities included hypertension (women: 45/65, 69.2%; men: 55/75, 73.3%), a history of smoking (women: 33/65, 50.8%; men: 44/75, 58.7%), and diabetes (women: 23/65, 35.4%; men: 33/75, 44.0%).
TABLE 3.
Demographics, symptoms, and comorbidities of 140 patients positive for SARS-CoV-2 by RT-PCR and tested for IgG in this study
| Parameter | Value (%) |
|---|---|
| Demographics | |
| No. of female | 65 (46.4) |
| No. of males | 75 (53.6) |
| No. of nursing home residents | 37 (26.4) |
| Age (avg) in yrs | 68.0 |
| Symptoms | |
| No. with fever | 83 (59.3) |
| No. with cough | 64 (45.7) |
| No. with headache | 12 (8.6) |
| No. with sore throat | 10 (7.1) |
| No. with shortness of breath | 78 (55.7) |
| No. with altered mental status | 45 (32.1) |
| No. with malaise | 36 (25.7) |
| No. with diarrhea/vomiting | 42 (30.0) |
| Course of disease/complications | |
| No. of days of hospitalization (avg) | 13.8 |
| No. with ICU admission | 66 (47.1) |
| No. with low oxygen saturation | 75 (53.6) |
| No. with ventilation | 47 (33.6) |
| No. with septic shock/vasopressor | 28 (20.0) |
| No. with bacterial pneumonia | 28 (20.0) |
| No. deceased | 20 (14.3) |
| Comorbidities | |
| No. with asthma | 15 (10.7) |
| No. with COPD | 13 (9.3) |
| No. with diabetes | 56 (40.0) |
| No. with hypertension | 100 (71.4) |
| No. with obstructive sleep apnea | 24 (17.1) |
| No. with coronary artery disease | 19 (13.6) |
| No. with cerebrovascular disease | 25 (17.9) |
| No. with immunodeficiency/immunosuppression | 11 (7.9) |
| No. with history of smoking | 77 (55.0) |
| Total | 140 (100) |
(ii) Serological response to SARS-CoV-2 S, RBD, and NP antigens.
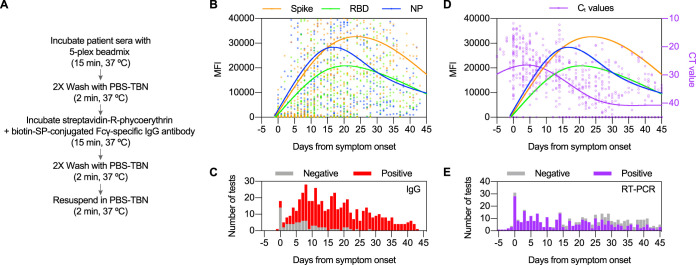
MFI values corresponding to each antigen were measured for all sera tested (n = 534) and analyzed with respect to days from symptom onset (Fig. 1B; see also Table 4). The peak IgG responses to S and RBD antigens occurred 23.8 days and 23.6 days from symptom onset, respectively. The peak IgG response to NP was observed at 16.7 days from symptom onset. No test was qualitatively interpreted as negative at >28 days from symptom onset. Although MFIs for all antigens decreased following the peak response, results for all seroconverted patients with at least one test performed following a positive test result (n = 59) were interpreted as positive on subsequent tests with the exception of three patients with discrepant findings. Two of these three patients represented mortalities with only limited sera available (i.e., two samples) from a narrow time frame (<11 days from symptom onset). The serological response of the third discrepant patient (patient H) was profiled later (see Fig. 4).
FIG 1.
Utility of the 3Flex multiplex microsphere-based immunological assay for SARS-CoV-2 diagnostics and immunological research. (A) Simplified workflow and illustration of the 3Flex assay. (B) Total composite profile of IgG immune reactivity in SARS-CoV-2-infected patients by days from symptom. Data represented are median MFI values for each viral antigen (Spike, RBD, and NP) from 534 serological tests (1,602 data points) of 140 patients. The tests represented in the figure were performed between −5 and 45 days from symptom onset, and the results include those found to be qualitatively negative or positive. Data points outside the axis limits (n = 273) are not shown. Fitted curve lines show smoothed splines with 4 knots. (C) Histogram showing number of serological tests by day from symptom onset and their qualitative positivity. (D) Composite profile of cycle threshold (CT) values from the same SARS-CoV-2-infected patients overlaid with the serological profile. CT values are plotted in reverse on the right axis (purple). Results from a total of 521 RT-PCR tests are depicted, and each has 2 data points plotted. Undetected targets or negative test results were assigned a CT value of 45. Fitted curve lines show smoothed splines with 4 knots. (E) Histogram showing number of RT-PCR (molecular) tests by day from symptom onset and their qualitative positivity.
TABLE 4.
Percent positivity and average MFIa values of all 3Flex serological tests by days from symptom onset for all patients testedb
| No. of days from symptom onset |
Total no. (%) of tests |
No. (%) of negative test results |
No. (%) of positive test results |
MFI avg (95% CI) |
||
|---|---|---|---|---|---|---|
| Spike | RBD | Nucleocapsid | ||||
| ≤5 | 67 (100.0) | 33 (49.3) | 34 (50.7) | 11,565 (5,718–17,412) | 8,723 (3,295–14,150) | 13,794 (8,721–18,866) |
| 6–10 | 106 (100.0) | 21 (19.3) | 85 (80.2) | 23,051 (18,955–27,147) | 16,172 (12,323–20,023) | 27,028 (21,749–32,308) |
| 11–15 | 85 (100.0) | 9 (10.6) | 76 (89.4) | 29,545 (25,190–33,901) | 19,876 (15,534–24,217) | 28,598 (23,978–33,218) |
| 16–20 | 88 (100.0) | 5 (5.7) | 83 (94.3) | 31,006 (26,406–35,606) | 20,694 (16,539–24,848) | 28,474 (24,134–32,814) |
| ≥21 | 188 (100.0) | 5 (2.7) | 183 (97.3) | 31,751 (28,774–34,727) | 19,516 (16,889–22,142) | 20,459 (17,574–23,343) |
MFI, median fluorescent intensity.
Includes multiple serological tests from nonunique patients.
FIG 4.
Interperson variation in longitudinal IgG immunological responses to SARS-CoV-2 in intensively hospitalized patients. Longitudinal serum samples from ICU-admitted patients (n = 9) were tested for IgG to SARS-CoV-2 S, RBD, and NP antigens. Curve lines are fitted for each antigen (smoothed splines with 4 knots). Corresponding RT-PCR CT values are plotted in reverse on the right axis (purple) and connected by straight lines through the mean CT value of each test. A single sample was randomly selected for each time interval bin (≤5, 6 to 10, 11 to 15, 16 to 20, and ≥21 days from symptom onset) to assay for ACE2 inhibition. Values shown in boxes above each plot are percent residual MFI values for each antigen (percentage of MFI detected with ACE2 addition compared to a no-ACE2 control). Box size indicates time interval bins. NEG, negative; POS, positive.
Overall, 61/534 test results were interpreted as negative and 473/534 test results were interpreted as positive (Fig. 1C; see also Table S1). Twelve of 534 test results were interpreted as positive based on one antigen MFI value exceeding the threshold for positivity (S: 9/12, 75%; RBD: 3/12, 25%; NP: 0/12, 0.0%). For 57 test results interpreted as positive based on analysis of two antigens, the frequencies of antigen pairs were 52/57 (91.2%) for S and RBD, 4/57 (7.0%) for S and NP, and 1/57 (1.8%) for RBD and NP. Single-antigen and double-antigen positive results occurred at medians of 5.5 and 8.0 days from symptom onset, respectively. Allowing for only one test (n = 231) per unique patient (n = 140) in each time interval, the IgG positivity rate was 53.4% when serum was collected ≤5 days from symptom onset and was 96.1% when serum was collected ≥21 days from symptom onset (Table 3).
CT values (n = 1,042; 2 CT values per RT-PCR test) were obtained from RT-PCR detection of SARS-CoV-2 nucleic acid and compared to the serological response to S, RBD, and NP (Fig. 1D). Nucleic acid detection peaked (i.e., showed the lowest average CT values) at 4.6 days from symptom onset. Among the tests conducted <21 days from symptom onset, 85.2% were positive, with a mean CT value of 26.3 ± 0.4 (standard errors of the means [SEM]) (Fig. 1E). In contrast, 47.8% of the results of tests conducted ≥21 days from symptom onset were positive, with a mean CT value of 33.4 ± 0.4 (SEM).
(iii) Serological response to SARS-CoV-2 in ICU-admitted patients.
Patient populations were assessed for differences in MFI values obtained for each SARS-CoV-2 antigen. Higher MFI values were observed for patients aged <65 (versus ≥65, not shown) and for those admitted to an ICU (Fig. 2). ICU-admitted patients demonstrated significantly higher MFI values for all SARS-CoV-2 antigens at >21 days from symptom onset. ICU-admitted patients (versus not-admitted patients) were not appreciably different with respect to age (67.0 years versus 68.9 years); sex (49.5% female versus 46.9% female); or histories of smoking, cerebrovascular disease, coronary artery disease, hypertension, diabetes, or asthma. Patients with histories of chronic obstructive pulmonary disease (10/66 versus 3/74, P = 0.0383, Fisher's exact test) and obstructive sleep apnea (16/66 versus 8/74, P = 0.0438) were more frequent in the ICU-admitted patient population than in that of the nonadmitted individuals.
FIG 2.

Violin plot comparison of mean MFI values for patients admitted to the ICU (n = 66) versus all other SARS-CoV-2-infected patients (n = 74). Data shown are for serological test results interpreted as positive only. Only 1 test for each unique patient was randomly selected for inclusion in each time interval bin (≤5, 6 to 10, 11 to 15, 16 to 20, and ≥21 days from symptom onset). Lines indicate means. Differences between violin plots are calculated using an unpaired t test: *, P ≤ 0.05; **, P ≤ 0.01.
(iv) Inhibition of SARS-CoV-2 antibody binding to S and RBD using ACE2.
To assess whether 3Flex could assay for antibodies that neutralize receptor binding, titrated concentrations (0.0625 to 4 μg/ml) of ACE2 were incubated with the 5-plex bead mix prior to addition of serum. The addition of ACE2 resulted in loss of detection of S and RBD antibodies but not loss of detection of NP (Fig. 3). The 2 μg/ml concentration of ACE2 was selected for the following studies because it was capable of producing a >50% drop in anti-RBD IgG detection. The residual MFI values detected at 2 μg/ml of ACE2 were 53.1% ± 8.0%, 43.5% ± 9.72%, and 98.6% ± 4.7%, for S, RBD, and NP, respectively (means ± SD).
FIG 3.

Addition of ACE2 inhibits detection and binding of antibodies to S and RBD, but not to NP. Percent residual (i.e., compared to no ACE2 added) mean MFI values with standard deviation are shown at increasing ACE2 concentrations (x axis log2 scale). Sera tested (n = 11) were from patients positive for SARS-CoV-2 IgG.
(v) Longitudinal serological response to SARS-CoV-2 in extensively sampled individuals.
Nine patients (6 women, 3 men) were extensively sampled due to extended hospitalization (all admitted to an ICU) for COVID-19. The IgG response to SARS-CoV-2 was explored in each of these, alongside ACE2 inhibition testing (Fig. 4). The average age of these patients was 67.3 years (range, 47.0 to 80.0 years). All of these patients survived their illness. All patients were ventilated except for patient I. There were wide-ranging prevalences of comorbidities and COVID-19 complications present in this group. These patients had histories notable for hypertension (9/9), diabetes (7/9), smoking (5/9), and kidney disease (7/9). Where available, CT values typically decreased or were undetectable following a peak IgG response. Highly variable (MFI magnitude and kinetics) serological responses were observed for these individuals. In addition, ACE2 inhibition was demonstrated to differing degrees for each patient, generally with the greatest “neutralization” observed for sera tested ≥21 days from symptom onset. Neutralization was exclusive to S and RBD, with NP showing no significant inhibition. Despite evidence of an IgG response and having tested negative by RT-PCR, 4/9 patients subsequently gave RT-PCR-positive test results for SARS-CoV-2 on later follow-up testing.
DISCUSSION
In this work, we validated 3Flex as a multiplex fluorescent microsphere immunoassay to qualitatively assess the IgG response to infection by SARS-CoV-2. We found that this assay compared favorably with the Abbott Architect SARS-CoV-2 IgG assay in testing serum from a diverse group of individuals with a spectrum of COVID-19 clinical presentations. The overall profile of the IgG serological response in this collection revealed the initial and peak responses for each SARS-CoV-2 antigen, timing of the decline in antibody levels, and correlation with decline in CT values (a proxy for viral load).
Through the early stages of the pandemic, the kinetics of the antibody response to SARS-CoV-2 were described using a variety of methodologies and antigenic targets. Although it is challenging to directly compare results, the overall picture which emerges from the literature is that most individuals develop IgM, IgA, and IgG responses at approximately 5 to 14 days after symptom onset, though some can take up to 14 to 28 days (22–25). In this study, we found similar kinetics. Across 534 tests from 140 individuals, 48% of those with reported symptom onset within the previous 5 days had detectable IgG, but that number rose to 68% by days 6 to 10 and to 92% by days 16 to 20. Further, as the pandemic enters its ninth month since the first cases were identified in the United States, some studies have raised concerns over declining levels of SARS-CoV-2 antibodies and the potential ramifications for natural immunity and vaccine response (26). Among the nine patients for whom we had longitudinal samples, none of the individuals became negative according to the qualitative interpretation of the assay. In addition, several individuals who were tested at later time points (>50 days after symptom onset) also demonstrated lower but still qualitatively positive results (data not shown). These findings are consistent with serological data from Iceland which indicated that infected individuals remained seropositive up to 4 months postinfection (27).
Whereas many serological assays for COVID-19 detect a single antigen, this assay incorporates detection of S, RBD, and NP. This feature may prove useful as it has the added flexibility to potentially discriminate between the immune responses of infected or vaccinated people (albeit its applicability is entirely dependent on the ultimate composition of upcoming SARS-CoV-2 vaccines). This multitarget approach may also improve the sensitivity of the assay, since a positive result for any of the antigens is interpreted as positive overall. We observed that ∼15% of samples were positive for one antigen but not another, though after 28 days from symptom onset the majority of the patients were positive for all antigens. Here, detection of S and RBD increased the sensitivity of the 3Flex assay among some of the earlier samples (≤5 days from symptom onset) compared to that of the NP-only Abbott Architect SARS-CoV-2 IgG assay. Currently, there are mixed reports concerning the antigen-specific dynamics of the antibody response, i.e., the relative timing and duration of responses to one viral antigen versus another. Despite similar findings reported by Liu et al. (28), others have reported greater sensitivity at earlier time points using single-target NP-based assays (29).
The kinetics of the antibody response were not appreciably different by demographic measures such as age or sex. Higher MFI values were observed in patients admitted to the ICU as part of their COVID-19 disease course. This association of higher antibody levels with disease severity has been reported by others (30, 31), though our data suggested that antigen-specific antibody kinetics did not differ significantly between the two groups—i.e., in aggregate, peak responses were observed at the same day from symptom onset.
A limitation of this study was its dependence on retrospective and remnant serum samples, which hindered longitudinal sampling. However, for a subset of nine individuals admitted to the ICU, we had multiple samples that enabled us to track their antibody and viral load data over time. This allowed us to look beyond the composite data (see Fig. 1B) for a depiction of individual responses (Fig. 4). The longitudinal MFI values of these individual patients suggested a high degree of interperson variation in terms of both kinetics and relative antigen responses. For most of these nine patients, the MFIs for anti-S antibodies were higher. This is despite the fact that S protein was coupled to beads at only 10% of the molarity of RBD or NP. Thus, anti-S antibodies potentially comprise a larger proportion of anti-SARS-CoV-2 antibodies. We observed interperson-dependent and time (from symptom onset)-dependent variability in the ability of soluble ACE2 to block the binding of anti-S and anti-RBD antibodies, an indirect measure of neutralizing capability. In general, a trend toward greater neutralization over time from symptom onset was observed. The ability to measure inhibition of signal with ACE2 may prove useful to measure the effectiveness of, and to discriminate between, the immune responses of infected or vaccinated individuals. However, an important caveat is that it is unclear how in vitro neutralization performed in this manner translates to in vivo immunity.
Another important limitation of this study was the relatively small number of samples (218) used to establish specificity. Another caveat is that the criterion used for positivity in this assay (requiring a positive result for only 1 of 3 anti-SARS-CoV-2 antibodies) may decrease the specificity of the assay. In our validation, this criterion was chosen because it increased sensitivity without diminishing specificity. Going forward, it will be important to refine the specificity of this assay using a larger set of contemporary (i.e., COVID-19 pandemic) specimens testing negative for SARS-CoV-2 antibodies by another test with EUA. Additionally, the positivity criteria used here may be reevaluated as experience with the assay performance accumulates. Other limitations included the use of a single-target comparator assay that detects only NP. Thus, the relative performance criteria may be different from those used for assays that also detect S or RBD. This assay requires a high-complexity laboratory and access to a Luminex Flexmap 3D system or a comparable system. Due to the variability in reagent cost, particularly with respect to the supplier of recombinant SARS-CoV-2 proteins, the costs per test may differ between laboratories. Despite these logistical limitations, the assay has a fairly short turnaround time (∼90 min to result) and can accommodate >90 patient serum samples per run.
In summary, the 3Flex assay described here is a versatile and robust method to measure antibodies to the major antigens of SARS-CoV-2. Beyond the current pandemic, the xMAP microsphere technology enables rapid and flexible assay design, as well as straightforward implementation. This provides a serological platform capable of responding to new and emerging pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the UR Medicine Central Laboratory of Clinical Microbiology for specimen collection and serological testing.
A.C. and N.D.P. designed the study, selected sera, and performed statistical analyses. S.A. provided technical expertise with assay design, reagent selection, and assay optimization. S.A., A.C., and N.D.P. interpreted criteria for assay validation. M.T.C. provided sera from confirmed respiratory disease infections for interference testing. J.W. and M.S.Z. provided technical expertise and additional reagents. A.C., C.A.P., J.L.B., A.B.C., and N.D.P. reviewed patient charts for demographic information and COVID-19 histories. L.D.B., Z.P., A.C., L.R.-M., R.A.S., N.D.P., and S.A. performed serological testing. L.R.-M. and R.A.S. contributed to assay validation and were responsible for project supervision, administration, and specimen curation. D.J.H. and N.D.P. provided laboratory resources. A.C. and N.D.P. analyzed data/results and wrote the first draft of the manuscript. All of us participated in editing and reviewing the manuscript and approved the final manuscript. All of us contributed to the article and approved the submitted version.
Funding from the Department of Pathology and Laboratory Medicine, University of Rochester Medical Center, supported this study. This study was also partially supported by the National Institutes of Health Institute of Allergy, Immunology, and Infectious Diseases (grants R21 AI138500 and R01 AI129518) (M.S.Z., J.W.) and University of Rochester Clinical and Translational Science Award UL1 TR002001 from the National Center for Advancing Translational Sciences of the National Institutes of Health (M.S.Z.). The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health. None of the funders named above had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. S.A. is an employee of Luminex Corporation (Austin, TX). Luminex had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. N.D.P. received support from Luminex Corporation for the purchase of reagents and supplies.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.FDA. EUA authorized serology test performance. 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance. Accessed 31 August 2020.
- 2.Meyer B, Drosten C, Muller MA. 2014. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res 194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu M, Shi Y, Guo Z, Chen Z, He R, Chen R, Zhou D, Dai E, Wang X, Si B, Song Y, Li J, Yang L, Wang J, Wang H, Pang X, Zhai J, Du Z, Liu Y, Zhang Y, Li L, Wang J, Sun B, Yang R. 2005. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect 7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. 2020. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma S, Lyons AM, Li S, Wong AW-y, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington L, Loudermilk R, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv 10.1101/2020.04.25.20074856. [DOI] [PMC free article] [PubMed]
- 6.Krammer F, Simon V. 2020. Serology assays to manage COVID-19. Science 368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 7.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. 2009. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol 7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904 e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, Gong Y, Xiao H, Fan Z, Tan S, Wu G, Tan W, Lu X, Fan C, Wang Q, Liu Y, Zhang C, Qi J, Gao GF, Gao F, Liu L. 2020. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai W, Zhang X, He Y, Jiang S, Du L. 2020. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res 179:104820. doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, VIB-CMB COVID-19 Response Team, Hoffmann M, Pöhlmann S, Graham BS, Callewaert N, Schepens B, Saelens X, McLellan JS. 2020. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus A, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch BJ. 2020. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaimes JA, Andre NM, Chappie JS, Millet JK, Whittaker GR. 2020. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Che XY, Hao W, Wang Y, Di B, Yin K, Xu YC, Feng CS, Wan ZY, Cheng VC, Yuen KY. 2004. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis 10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed SF, Quadeer AA, McKay MR. 2020. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu SJ, Leng CH, Lien SP, Chi HY, Huang CY, Lin CL, Lian WC, Chen CJ, Hsieh SL, Chong P. 2006. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surjit M, Lal SK. 2008. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol 8:397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M, Mohammed A, Zhao C, Yang Y, Xie J, Ding C, Ma X, Weng J, Gao Y, He H, Jin T. 2020. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang G, Nie S, Zhang Z, Zhang Z. 2020. Longitudinal change of severe acute respiratory syndrome coronavirus 2 antibodies in patients with coronavirus disease 2019. J Infect Dis 222:183–188. doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Ren L, Yang S, Xiao M, Chang Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang Q, Xu S, Zhu H, Xu Y, Jin Q, Sharma L, Wang L, Wang J. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma WL. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. 2020. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 27.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, Le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. 2020. Evaluation of nucleocapsid and Spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58:e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI. 2020. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. J Infect Dis doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis 26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grzelak L, Temmam S, Planchais C, Demeret C, Tondeur L, Huon C, Guivel-Benhassine F, Staropoli I, Chazal M, Dufloo J, Planas D, Buchrieser J, Rajah MM, Robinot R, Porrot F, Albert M, Chen KY, Crescenzo-Chaigne B, Donati F, Anna F, Souque P, Gransagne M, Bellalou J, Nowakowski M, Backovic M, Bouadma L, Le Fevre L, Le Hingrat Q, Descamps D, Pourbaix A, Laouenan C, Ghosn J, Yazdanpanah Y, Besombes C, Jolly N, Pellerin-Fernandes S, Cheny O, Ungeheuer MN, Mellon G, Morel P, Rolland S, Rey FA, Behillil S, Enouf V, Lemaitre A, Creach MA, Petres S, Escriou N, Charneau P, Fontanet A, et al. 2020. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med 12:eabc3103. doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.