Abstract
Signaling through the CD19-CD81 co-receptor complex, in association with the B cell receptor, is a critical determinant of B cell development and activation. It is unknown how CD81 engages CD19 to enable co-receptor function. Here, we report a 3.8 Å structure of the CD19-CD81 complex bound to a therapeutic Fab, determined by cryo-electron microscopy. The structure includes both the extracellular domains and the transmembrane helices of the complex, revealing a contact interface between the ectodomains that drives complex formation. Upon binding to CD19, CD81 opens its ectodomain to expose a hydrophobic CD19-binding surface and reorganizes its transmembrane helices to occlude a cholesterol binding pocket present in the apoprotein. Our data reveal the structural basis for CD19-CD81 complex assembly, providing a foundation for rational design of therapies for B cell dysfunction.
One Sentence Summary:
The structure of the B cell co-receptor complex reveals a striking conformational reorganization of the tetraspanin CD81.
B cells, a critical component of the adaptive immune system, enable development of long-lasting immunity following infection (1). Upon activation, B cells mature into plasma cells, which circulate in the bloodstream and secrete antibodies that can specifically bind and eliminate the foreign antigen that stimulated their production. Activated B cells also mature into long-lived memory B cells, providing durable immunity following infection. In cell-mediated immunity, B cells also serve as antigen-presenting cells that capture and present antigen to T cells.
B cell activation and maturation are initiated when an antigen is recognized by a cell surface B cell receptor (BCR). Signaling through the BCR leads to B cell maturation and contributes to stimulation of somatic hypermutation and heavy-chain class switching events associated with affinity maturation and antibody secretion. BCR signaling is also frequently subverted by B-cell malignancies to drive their proliferation, growth, and survival (2, 3).
The BCR functions in association with a co-receptor complex that contains the complement receptor CD21, the signaling protein CD19, and the tetraspanin CD81 (4). Analogous to the CD4/CD8 co-receptors of T cells, the B cell co-receptor complex boosts the response to antigen by lowering the signaling threshold for activation by approximately 1000-fold (5). CD 19, a single pass transmembrane protein belonging to the immunoglobulin superfamily, is the key signaling subunit of this complex, acting as a docking and recruitment site for various kinases and signaling components within the B cell signaling pathway (6, 7). Patients with homozygous mutations in the CD19 gene develop Common Variable Immune Deficiency (CVID), an immune disorder characterized by recurrent and severe infections, primarily of the respiratory tract, ears, and sinuses (8), highlighting the importance of CD19 in immune cell function. The restriction of CD19 expression to the B cell lineage has also made it an attractive target of cell-based therapies for cancer and autoimmune diseases (9–15).
Complex formation between CD19 and the tetraspanin CD81 is critical for proper B cell function (16). Tetraspanins are a highly-conserved family of four-transmembrane proteins that interact with each other and with a variety of partner proteins to regulate many signal transduction pathways (17). Through their interactions with partner proteins, tetraspanins play critical roles in numerous cellular functions, including protein trafficking, adhesion, cell signaling, and cell migration (18–22). CD81 chaperones CD19 through the secretory pathway, allowing for cell surface expression of mature, properly folded CD19 (23, 24). On the B cell membrane, the CD19-CD81 interaction is dynamically regulated upon B cell activation (25). CD81 may regulate the diffusion of CD19 by immobilizing CD19 and associated signaling scaffolds in distinct locations in the membrane, thereby regulating the engagement of CD19 with the BCR, for example, to prevent high-level constitutive tonic signaling (21).
Our structure of apo-CD81 revealed a cholesterol binding pocket within its intramembrane cavity (27). However, there is no structural information showing the molecular details of how tetraspanins mediate complex formation with partner proteins to regulate their trafficking and signaling. Molecular dynamics simulations suggested that CD81 exists in both a closed and open conformation, and that cholesterol binding may favor a closed state with lower affinity for CD19 (27). Thus, cholesterol binding by CD81 may regulate its association with CD19 and subsequently, B cell signaling. To better understand how these two proteins function together, we characterize their interaction in detail through a combination of cryo-electron microscopy (cryo-EM) and biochemical methods.
To produce CD19 in complex with CD81 for cryo-EM studies, we developed and validated a fusion protein approach (25). The fusion protein contains the full extracellular and transmembrane domains of CD19 and the first 15 amino acids of its cytoplasmic tail (Figure S1A). Because the cytoplasmic region of CD19 is predicted to be disordered and is not required for CD19 to bind to CD81 (28), we removed the majority of this segment to facilitate structural studies and linked CD19 to full-length CD81 with a Gly-Gly-Ser (GGS)x4 sequence.
To facilitate structure determination by cryo-EM, CD19-CD81 (62 kDa) was bound to a therapeutic anti-CD19 Fab (Coltuximab), increasing the molecular weight to approximately 110 kDa (29, 30). The purified complex contained CD19-CD81 and the Fab in stoichiometric quantities (Figure S1B). Cryo-EM imaging of this complex revealed well-dispersed, single particles. Two-dimensional (2D) class averages showed clear secondary structure features in both the transmembrane region and the ectodomain (Figure S2). After 2D and 3D classifications, a homogenous data set of approximately 245,000 particles was used to produce a final density map at an overall nominal resolution of 3.8 Å (Figure S2; Table S1).
The structure of the CD19-CD81 complex is elongated, with the Fab-bound CD19 ectodomain resting directly on top of the CD81 ectodomain and the transmembrane helices arranged in a five-helix bundle (Figure 1A). The crystal structures of the CD19 ectodomain (31), Coltuximab (32), and full-length CD81 (27) were docked into the cryo-EM density map. Whereas the CD19 ectodomain and Fab fit easily into the density, CD81 did not, and the EC2, TM1-TM2, and TM3-TM4 regions of CD81 were instead separately docked into the density. The quality of the density was sufficient for sequence-specific assignment for most of the ectodomains of both CD19 and CD81, including the contact interface, and the variable domain of the Fab (Figure 1B; Figure S3). Secondary structure is visible in the transmembrane domains, but this region is less well-resolved than the ectodomains, precluding the modeling of side chains within this region (Figure 1B; Figure S3). The final model of the CD19-CD81 complex contains the heavy and light chain of Coltuximab, the complete ectodomain and transmembrane helix (modeled as poly-alanine) of CD19, and full-length CD81, including the small extracellular loop (EC1) which was not resolved in the CD81 crystal structure (Figures 1C, 2A).
Fig. 1. 3D reconstruction and atomic model of the human CD19-CD81 complex bound to Coltuximab.
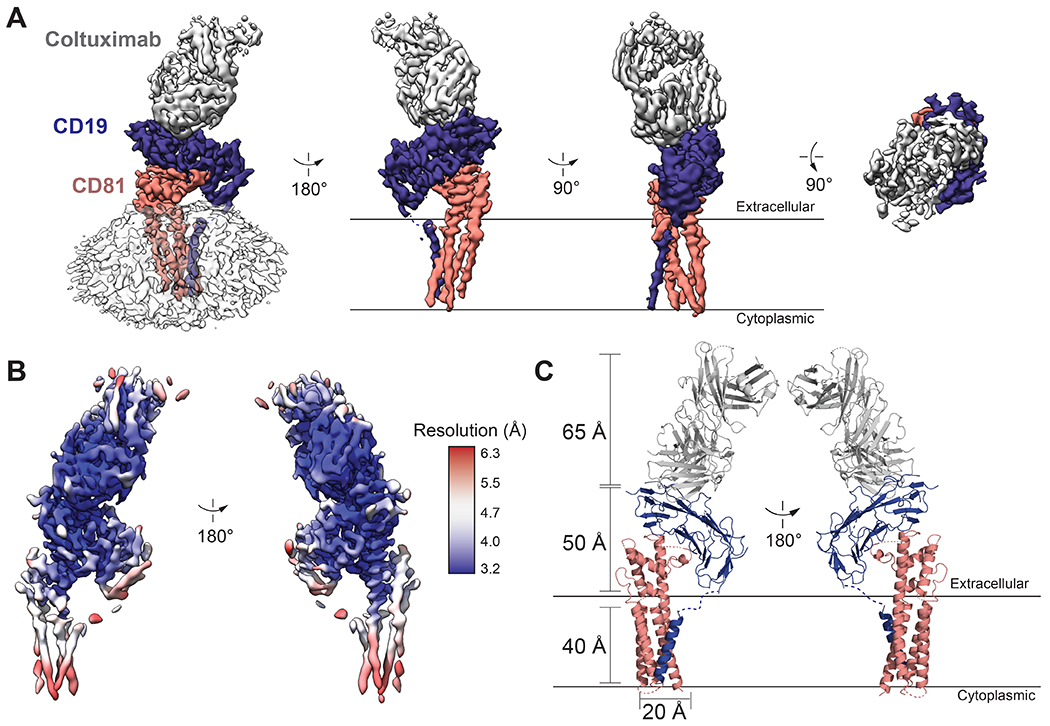
(A) Different views of the cryo-EM map of the CD19-CD81-Coltuximab complex colored by subunit. Density associated with the detergent micelle is rendered transparently in white in the left panel. (B) Locally filtered cryo-EM map of the CD19-CD81-Coltuximab complex colored according to local resolution, calculated in cryoSPARC. (C) Ribbon representation of the structure of the complex. CD19, CD81, and Coltuximab are colored as in A.
Fig. 2. Comparison of apo-CD81 and CD19-CD81 complex structures.
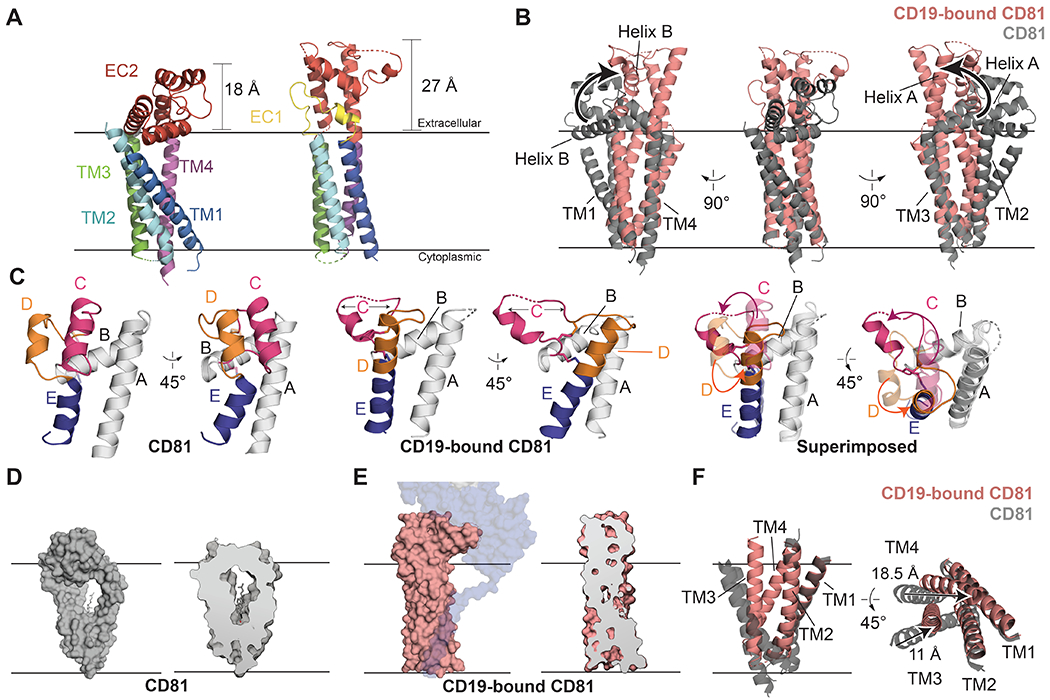
(A) Comparison of apo-CD81 (left) with complexed CD81 (right). The structures are aligned on the TM3/TM4 helices and colored by domain. (B) Overlay of apo-CD81 on complexed CD81. Structures were aligned on the TM3/4 helices. Helices A and B of the large extracellular loop (EC2) are highlighted to show the hinge-opening movement of the ectodomain. (C) Comparison of the large extracellular loops of apo-CD81 (left) and complexed CD81 (middle), and superposition of the two conformations (right). Helices C-E are colored. In the superposition, the large extracellular loop of complexed CD81 is solid, and apo-CD81 is transparent. (D) Surface representation of apo-CD81 (left) and cross-section highlighting cholesterol binding pocket (right). (E) Surface representation of complexed CD81 (left) and cross-section highlighting the occlusion of the cholesterol binding pocket (right). CD81 was modeled was side chains in the transmembrane domain for Panel E. (F) Overlay of the transmembrane region of apo-CD81 on complexed CD81, aligned on the TM1/2 helices. Movements within the transmembrane domain are indicated with black arrows. See also Supplementary Video S1.
The most striking feature of the CD19-CD81 structure is the large-scale conformational change within the tetraspanin CD81 (Figure 2). The transmembrane helices of apo-CD81 resemble a cone, with two pairs of closely associated helices, TM1/TM2 and TM3/TM4, converging near the cytoplasmic side of the membrane (Figure 2A) (27). These helices surround a large central cavity, enclosing a total volume of 3,300 Å3 (27). The five helices that make up the large extracellular loop (EC2) contain a three-helix “stalk” comprised of helices A, B, and E and a “top” face consisting of helices C and D. Upon complex formation, the large extracellular loop (EC2) of CD81 extends into an open conformation, with the A, B and E helices swinging as a rigid body approximately 60° relative to the membrane plane (Figure 2B), positioning helices A and E to be near-continuous with helices TM3 and TM4, respectively. The C and D helices also rearrange dramatically, with helices D and E merging to become one continuous helix, comparable in length to helix A (Figure 2C), while helix C partially unravels. The conformation of helix C is buttressed by a conserved Cys-Cys-Gly (CCG) motif, which uses two disulfide bonds to stabilize its N-terminal end. The opening and reorganization of the ectodomain is also associated with inward movement of the TM1/TM2 and TM3/TM4 pairs of helices, decreasing the distance between TM1 and TM4 by about 15 Å, virtually eliminating the central cavity and thus preventing cholesterol binding (Figure 2D–F).
In the crystal structure of CD81, no electron density was visible for the small extracellular loop (EC1), suggesting that this region is disordered in the apoprotein. Upon complex formation with CD19, however, the movement of Helix B away from the membrane allows interactions between EC1 and EC2, consistent with a role for EC1 in stabilizing the open conformation of the ectodomain (Figure 2A). In contrast to CD81, the conformation of the CD19 ectodomain within the complex is nearly identical to that of the isolated ectodomain in the absence of CD81 (Figure S4).
The CD19-CD81 complex buries a total of approximately 700 Å2 at an interface between the ectodomain of CD19 and the EC2 of CD81 (Figure 3A). The observation that the ectodomains constitute the primary interaction site is consistent with our previous results from domain swap experiments (25), and is inconsistent with a prior report that the primary interaction site lies within the transmembrane region (24). The interface between the CD19 and CD81 ectodomains is highly hydrophobic, with aliphatic hydrophobic residues on helices C and D of CD81 surrounding a patch of exposed tryptophan, phenylalanine and histidine residues on CD19 (Figure 3B). Several polar residues nearby approach within hydrogen bonding distance, providing specificity to this otherwise hydrophobic contact site. The CCG motif anchoring the base of the variable C helix is a defining feature of the tetraspanin family (17), and the overall three-helix bundle architecture of the EC2, capped by the variable and conformationally dynamic C/D helix region, is conserved (33), suggesting that the use of the C and D helices for recognition of partner proteins will be a general feature of the tetraspanin family.
Fig. 3. CD19-CD81 interface.
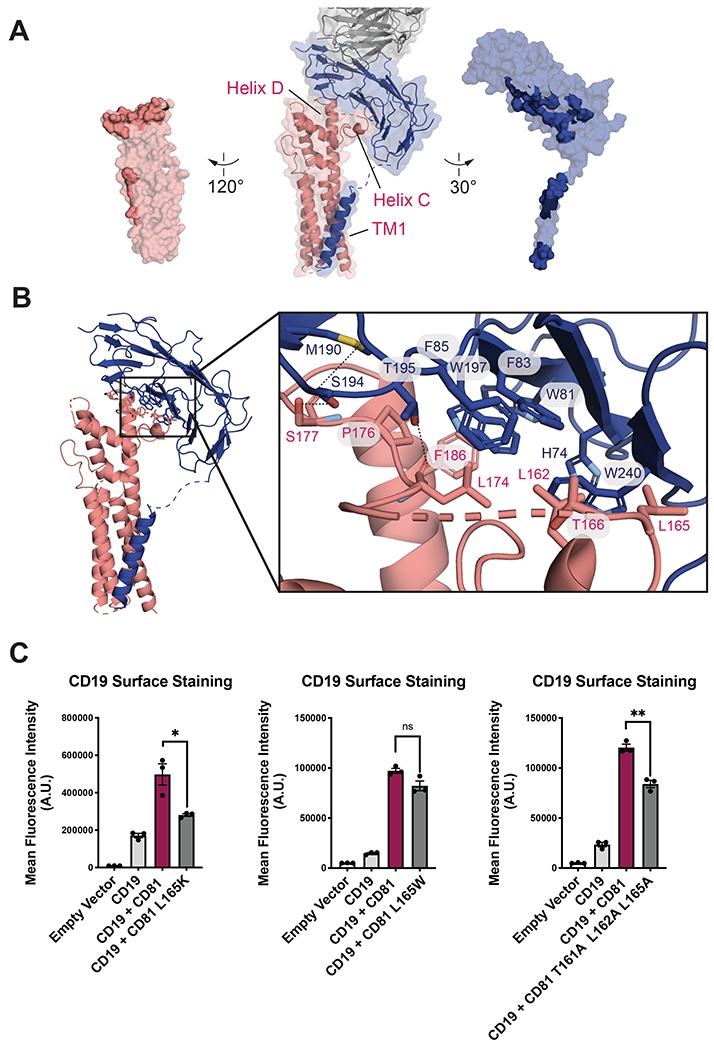
(A) Open book representation of the CD19-CD81 interface. CD81 is salmon, CD19 is blue, and the Fab is grey. Residues at the binding interface are colored in a darker shade. (B) CD19-CD81 binding interface. Zoomed in views show hydrogen bonds and van der Waals interactions with dotted lines. (C) Effect of CD81 interface mutations on CD19 export. Surface CD19 was detected by flow cytometry using an Alexa 488-coupled anti-CD19 antibody. Expression of CD81 mutants was confirmed by flow cytometry (Figure S7). Error bars represent mean ± SEM of three independent experiments. Statistical analysis was performed in GraphPad Prism using an unpaired t test. *p ≤0.05; **p ≤ 0.01; ***p ≤ 0.001.
Our structure also reveals that the therapeutic Fab Coltuximab binds to the upper face of CD19 (Figure 1A). Coltuximab binds to the same region as the therapeutic anti-CD19 antibody B43, and also competes for binding with two other therapeutic antibodies, Inebilizumab and Denintuzumab, suggesting that current antibody selection strategies converge on a dominant CD19 epitope (Figure S5).
Previous work uncovered a human CD81 truncation mutation that results in type VI immunodeficiency (34). Whereas wild-type CD81 rescues export of CD19 to the cell surface in CD81-knockout HEK293T cells, the CD81 disease-associated mutation does not allow CD19 surface delivery in these cells (Figure S6), consistent with prior observations that it fails to traffic CD19 to the cell surface in B cells (34) and in other non-B cell lines (35). To assess whether the observed CD81-CD19 interface is required for the function of CD81 in CD19 surface export, we mutated residues on CD81 at the observed interface and tested the ability of these mutants to traffic CD19 to the cell surface (Figure 3C). The mutants we tested did not affect cell surface abundance of CD81 (Figure S7). L165K, which would place a charged residue in the hydrophobic interface, nearly abolishes the ability of CD81 to traffic CD19 to the surface, whereas an L165W mutation does not interfere with trafficking. A triple mutant, T161A/L162A/L165A also disrupts trafficking. These results are consistent with the hydrophobic character of the CD19-CD81 interface. Though our structure also reveals contact between the transmembrane domain of CD19 and TM1 of CD81, the density in this region is more poorly resolved (Figure 1B) and our prior experiments showed that this interaction is not necessary for complex formation or for export of CD19 to the cell surface by CD81 (25).
CD9 is the closest human homologue of tetraspanin CD81, sharing 45% amino acid sequence identity, yet it does not export CD19 to the cell surface (Figure 4A) (24). Its overall structure is highly similar to CD81 (RMSD 1.35 Å), with minor structural differences only in the C-D helix region of EC2 (33). An accompanying low-resolution structure of CD9 in complex with EWI-2 only made it possible to roughly fit an EWI-2 homology model and the CD9 crystal structure into the cryo-EM map, which has a very different overall architecture from the CD19-CD81 complex (33). To understand the basis of the specificity of CD81 for CD19, we simulated the “open,” cholesterol-free conformation of CD9 with a homology model of the structure of CD19-bound CD81 (36). The comparison shows that the hydrophobic loop of CD81 that engages CD19 is highly polar in the model of CD9, providing an explanation for the CD19 selectivity of CD81 (Figure 4B).
Fig. 4. Structural comparison of CD81 to its tetraspanin CD9 homologue explains CD19-binding specificity.
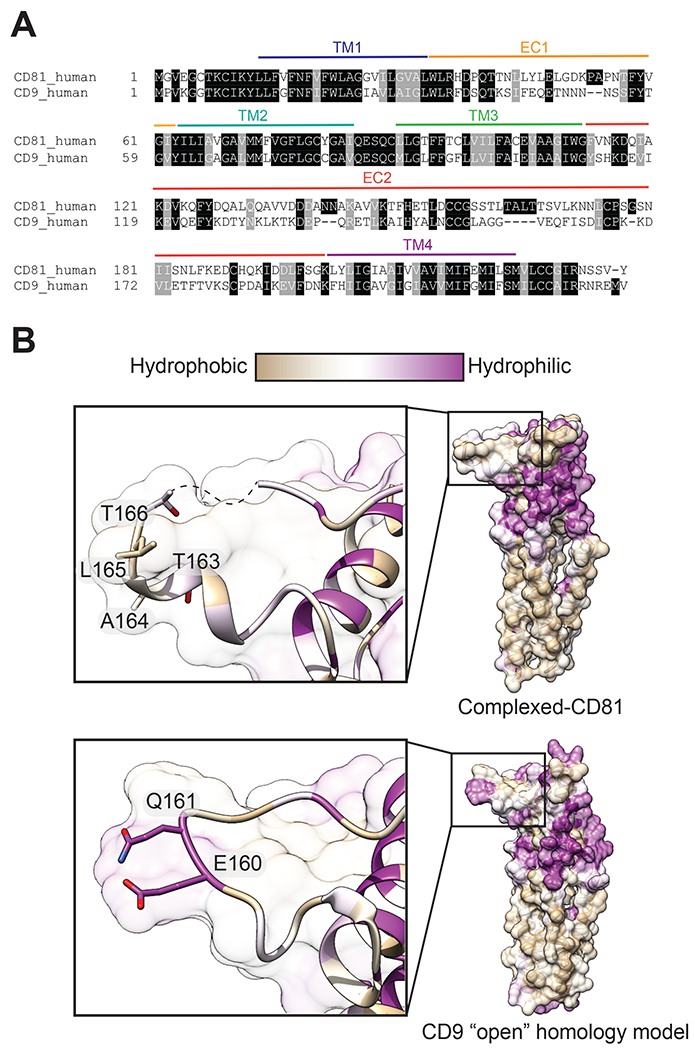
(A) Sequence alignment of human CD81 and human CD9. (B) Homology model of “open” CD9 and CD19 complexed-CD81 with surfaces colored by amino acid hydrophobicity on the Kyte-Doolittle scale. Zoomed in panels show the key hydrophobic “TALT” sequence that is necessary for CD81 to recognize CD19, which is replaced a hydrophilic region in the analogous position on CD9.
The structural and biochemical results reported here reveal molecular details about the B cell coreceptor complex that have eluded previous definition (4). The CD19-CD81 complex is assembled in the endoplasmic reticulum (ER), where cholesterol accounts for only 5% of lipids (compared to up to 50% of plasma membrane lipids) (37). The low cholesterol concentration could favor CD19-CD81 complex formation in the ER, where it then travels through the secretory pathway and is inserted into the B cell membrane in the cholesterol free, “open” conformation. Upon B cell activation, the BCR and co-receptor complex have been shown to partition into cholesterol rich lipid rafts, and CD81 is necessary for this movement (38, 39). Thus, cholesterol binding may allow CD81 to release CD19 in response to B cell activation, allowing CD19 to interact with the BCR and carry out signal amplification during antigen recognition. The switch that triggers the transition from cholesterol-free to cholesterol-bound CD81 remains an area of further investigation, but some studies suggest the palmitoylation of CD81 modulates cholesterol binding (40, 41).
The mechanism of conformational regulation in response to lipid binding is likely a general feature of the tetraspanin family. The functional relationship between cholesterol and numerous tetraspanins has been discussed (42–45), although never directly shown, and analysis of sequence conservation for the 33 tetraspanin paralogs within humans highlights tight evolutionary constraints within the transmembrane region (27). Residue N18, which forms a hydrogen bond to cholesterol in the apo-CD81 structure, is conserved in 27 of 33 human tetraspanins (27). Indeed, the structure of CD9 also revealed electron density consistent with a bound lipid within its intramembrane cavity (33). Our structure of the CD19-CD81 complex confirms that there are at least two distinct conformations of a tetraspanin and suggests that tetraspanins regulate the subcellular localization of their partner proteins based on differences in cholesterol concentration.
We previously showed that the anti-CD81 antibody 5A6 recognizes a conformational epitope that is masked when CD81 is in complex with CD19, but becomes accessible upon B cell activation (25). This antibody is competitive with CD19 for binding to CD81, and the angle of approach of the 5A6 Fab is nearly identical to that of CD19 (Figure S8) (25). 5A6 inhibits B cell lymphoma growth in a xenograft model as effectively as rituximab, a standard treatment for this disease (46). However, unlike rituximab, 5A6 selectively kills human lymphoma cells while sparing normal cells (46), suggesting that conformationally-selective antibodies that distinguish unbound CD81 on chronically active malignant cells from the CD19-CD81 complex on resting B cells show therapeutic promise.
In addition to its function as a signaling subunit of the B cell co-receptor, CD19 is also a target of CAR-T cells, which have revolutionized the treatment of B cell malignancies and show promise for treatment of autoimmune disorders (14, 15, 47). It is currently not possible to selectively target resting or chronically activated, malignant B cells based on differences in CD19 conformation, leading to depletion of all B cells and recurrent infections due to loss of humoral immunity. It should be possible to select antibodies that discriminate between CD81-complexed CD 19 and free CD19 by raising antibodies that recognize a composite epitope on our CD19-CD81 fusion protein. Such an antibody could be used to trap CD19 in complex with CD81, raising the threshold for B cell activation in autoimmune diseases. Alternatively, an antibody that binds to the region of the CD19 ectodomain that interacts with CD81 on resting B cells could selectively deplete chronically activated, malignant B cells, preventing the undesired complication of destroying all B cells with current generation CAR-T approaches.
Supplementary Material
Acknowledgements:
We thank members of the Blacklow and Kruse laboratories for helpful discussions and Marie Bao for critical reading of the manuscript. Cryo-EM data were collected at The Harvard Cryo-EM Center for Structural Biology at Harvard Medical School. We thank Sarah Sterling and Richard Walsh for helpful comments and advice during grid screening and data collection.
Funding:
Financial support for this work was provided by NIH grants R35 CA220340 (S.C.B.), F31 HL147459 (K.J.S.) and DP5 OD021345 (A.C.K.).
Competing Interests:
S.C.B. receives funding for an unrelated project from Novartis, is on the scientific advisory board for Erasca, Inc., is an advisor to MPM Capital, and is a consultant for IFM, Scorpion Therapeutics and Ayala Pharmaceuticals for unrelated projects. A.C.K. is a co-founder and advisor on unrelated projects for the Institute for Protein Innovation, a non-profit research institute and a founder of Tectonic Therapeutic, a for-profit biotechnology company.
Footnotes
Supplementary Materials:
Data and Materials Availability:
Materials will be made available upon request. Accession numbers of the CD19-CD81-Coltuximab Fab model and map are Protein Data Bank (PDB) 7JIC and EMD-22344, respectively.
References and Notes:
- 1.LeBien TW, Tedder TF, B lymphocytes: how they develop and function. Blood. 112, 1570–1580 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, Xu W, Shaffer AL, Wright G, Xiao W, Powell J, Jiang J, Thomas CJ, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Johnson NA, Rimsza LM, Campo E, Jaffe ES, Wilson WH, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pierce SK, Staudt LM, Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 463, 88–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bojarczuk K, Bobrowicz M, Dwojak M, Miazek N, Zapala P, Bunes A, Siernicka M, Rozanska M, Winiarska M, B-cell receptor signaling in the pathogenesis of lymphoid malignancies. Blood Cells. Mol. Dis 55, 255–265 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bradbury LE, Kansas GS, Levy S, Evans RL, Tedder TF, The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. Baltim. Md 1950 149, 2841–2850 (1992). [PubMed] [Google Scholar]
- 5.Carter RH, Fearon DT, CD19: Lowering the Threshold for Antigen Receptor Stimulation of B Lymphocytes. Science. 256, 105–107 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto M, Poe JC, Jansen PJ, Sato S, Tedder TF, CD19 Amplifies B Lymphocyte Signal Transduction by Regulating Src-Family Protein Tyrosine Kinase Activation. J. Immunol 162, 7088–7094 (1999). [PubMed] [Google Scholar]
- 7.Tuveson DA, Carter RH, Soltoff SP, Fearon DT, CD19 of B cells as a surrogate kinase insert region to bind phosphatidylinositol 3-kinase. Science. 260, 986–989 (1993). [DOI] [PubMed] [Google Scholar]
- 8.van Zelm MC, Reisli I, van der Burg M, Castaño D, van Noesel CJM, van Tol MJD, Woellner C, Grimbacher B, Patiño PJ, van Dongen JJM, Franco JL, An Antibody-Deficiency Syndrome Due to Mutations in the CD19 Gene. N. Engl. J. Med 354, 1901–1912 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, Borquez-Ojeda O, Qu J, Wasielewska T, He Q, Bernal Y, Rijo IV, Hedvat C, Kobos R, Curran K, Steinherz P, Jurcic J, Rosenblat T, Maslak P, Frattini M, Sadelain M, CD19-Targeted T Cells Rapidly Induce Molecular Remissions in Adults with Chemotherapy-Refractory Acute Lymphoblastic Leukemia. Sci. Transl. Med 5, 177ra38–177ra38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, Milone MC, Levine BL, June CH, Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med 368, 1509–1518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH, T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med 3, 95ra73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochenderfer JN, Rosenberg SA, Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol 10, 267–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter DL, Levine BL, Kalos M, Bagg A, June CH, Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kansal R, Richardson N, Neeli I, Khawaja S, Chamberlain D, Ghani M, Ghani Q, Balazs L, Beranova-Giorgianni S, Giorgianni F, Kochenderfer JN, Marion T, Albritton LM, Radic M, Sustained B cell depletion by CD19-targeted CAR T cells is a highly effective treatment for murine lupus. Sci. Transl. Med 11 (2019), doi: 10.1126/scitranslmed.aav1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yazawa N, Hamaguchi Y, Poe JC, Tedder TF, Immunotherapy using unconjugated CD19 monoclonal antibodies in animal models for B lymphocyte malignancies and autoimmune disease. Proc. Natl. Acad. Sci. U. S. A 102, 15178–15183 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Zelm MC, Smet J, Adams B, Mascart F, Schandené L, Janssen F, Ferster A, Kuo C-C, Levy S, van Dongen JJM, van der Burg M, CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest 120, 1265–1274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemler ME, Targeting of tetraspanin proteins — potential benefits and strategies. Nat. Rev. Drug Discov 7, 747 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berditchevski F, Odintsova E, Tetraspanins as Regulators of Protein Trafficking. Traffic. 8, 89–96 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Levy S, Todd SC, Maecker HT, CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol 16, 89–109 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME, Tetraspanin CD151 regulates α6β1 integrin adhesion strengthening. Proc. Natl. Acad. Sci 100, 7616–7621 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD, The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 38, 461–474 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Liu L, He B, Liu WM, Zhou D, Cox JV, Zhang XA, Tetraspanin CD151 Promotes Cell Migration by Regulating Integrin Trafficking. J. Biol. Chem 282, 31631–31642 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Braig F, Brandt A, Goebeler M, Tony H-P, Kurze A-K, Nollau P, Bumm T, Bottcher S, Bargou RC, Binder M, Blood, in press, doi: 10.1182/blood-2016-05-718395. [DOI] [PubMed] [Google Scholar]
- 24.Shoham T, Rajapaksa R, Boucheix C, Rubinstein E, Poe JC, Tedder TF, Levy S, The Tetraspanin CD81 Regulates the Expression of CD19 During B Cell Development in a Postendoplasmic Reticulum Compartment. J. Immunol 171, 4062–4072 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Susa KJ, Seegar TC, Blacklow SC, Kruse AC, A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking. eLife. 9, e52337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artac H, Reisli I, Kara R, Pico-Knijnenburg I, Adin-Çinar S, Pekcan S, der Zijde CMJ, van Tol MJD, Bakker-Jonges LE, van Dongen JJM, van der Burg M, van Zelm MC, B-cell maturation and antibody responses in individuals carrying a mutated CD19 allele. Genes Immun. 11, 523 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC, Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell. 167, 1041–1051.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbury LE, Goldmacher VS, Tedder TF, The CD19 signal transduction complex of B lymphocytes. Deletion of the CD19 cytoplasmic domain alters signal transduction but not complex formation with TAPA-1 and Leu 13. J. Immunol. Baltim. Md 1950 151, 2915–2927 (1993). [PubMed] [Google Scholar]
- 29.Hong EE, Erickson H, Lutz RJ, Whiteman KR, Jones G, Kovtun Y, Blanc V, Lambert JM, Design of Coltuximab Ravtansine, a CD19-Targeting Antibody–Drug Conjugate (ADC) for the Treatment of B-Cell Malignancies: Structure–Activity Relationships and Preclinical Evaluation. Mol. Pharm 12, 1703–1716 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kantarjian HM, Lioure B, Kim SK, Atallah E, Leguay T, Kelly K, Marolleau J-P, Escoffre-Barbe M, Thomas XG, Cortes J, Jabbour E, O’Brien S, Bories P, Oprea C, Hatteville L, Dombret H, A Phase II Study of Coltuximab Ravtansine (SAR3419) Monotherapy in Patients With Relapsed or Refractory Acute Lymphoblastic Leukemia. Clin. Lymphoma Myeloma Leuk 16, 139–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teplyakov A, Obmolova G, Luo J, Gilliland GL, Crystal structure of B-cell co-receptor CD19 in complex with antibody B43 reveals an unexpected fold. Proteins Struct. Funct. Bioinforma 86, 495–500 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Ereño-Orbea J, Sicard T, Cui H, Carson J, Hermans P, Julien J-P, Structural Basis of Enhanced Crystallizability Induced by a Molecular Chaperone for Antibody Antigen-Binding Fragments. J. Mol. Biol 430, 322–336 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Umeda R, Satouh Y, Takemoto M, Nakada-Nakura Y, Liu K, Yokoyama T, Shirouzu M, Iwata S, Nomura N, Sato K, Ikawa M, Nishizawa T, Nureki O, Structural insights into tetraspanin CD9 function. Nat. Commun 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zelm MC, Smet J, Adams B, Mascart F, Schandené L, Janssen F, Ferster A, Kuo C-C, Levy S, van Dongen JJM, van der Burg M, CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest 120, 1265–1274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vences-Catalán F, Kuo C-C, Sagi Y, Chen H, Kela-Madar N, van Zelm MC, van Dongen JJM, Levy S, A mutation in the human tetraspanin CD81 gene is expressed as a truncated protein but does not enable CD19 maturation and cell surface expression. J. Clin. Immunol 35, 254–263 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS, Switch-like Control of SREBP-2 Transport Triggered by Small Changes in ER Cholesterol: A Delicate Balance. Cell Metab. 8, 512–521 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherukuri A, Cheng PC, Sohn HW, Pierce SK, The CD19/CD21 Complex Functions to Prolong B Cell Antigen Receptor Signaling from Lipid Rafts. Immunity. 14, 169–179 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Cherukuri A, Shoham T, Sohn HW, Levy S, Brooks S, Carter R, Pierce SK, The Tetraspanin CD81 Is Necessary for Partitioning of Coligated CD19/CD21-B Cell Antigen Receptor Complexes into Signaling-Active Lipid Rafts. J. Immunol 172, 370–380 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Cherukuri A, Carter RH, Brooks S, Bornmann W, Finn R, Dowd CS, Pierce SK, B Cell Signaling Is Regulated by Induced Palmitoylation of CD81. J. Biol. Chem 279, 31973–31982 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Delandre C, Penabaz TR, Passarelli AL, Chapes SK, Clem RJ, Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Exp. Cell Res 315, 1953–1963 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charrin S, Manié S, Thiele C, Billard M, Gerlier D, Boucheix C, Rubinstein E, A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol 33, 2479–2489 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Buschiazzo J, Ialy-Radio C, Auer J, Wolf J-P, Serres C, Lefèvre B, Ziyyat A, Cholesterol Depletion Disorganizes Oocyte Membrane Rafts Altering Mouse Fertilization. PLoS ONE. 8 (2013), doi: 10.1371/journal.pone.0062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silvie O, Charrin S, Billard M, Franetich J-F, Clark KL, van Gemert G-J, Sauerwein RW, Dautry F, Boucheix C, Mazier D, Rubinstein E, Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci 119, 1992–2002 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Israels SJ, McMillan-Ward EM, Platelet tetraspanin complexes and their association with lipid rafts. Thromb. Haemost 98, 1081–1087 (2007). [PubMed] [Google Scholar]
- 46.Vences-Catalán F, Kuo C-C, Rajapaksa R, Duault C, Andor N, Czerwinski DK, Levy R, Levy S, CD81 is a novel immunotherapeutic target for B cell lymphoma. J. Exp. Med 216, 1497–1508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D, Gallagher S, Monson NL, Herbst R, Wang Y, Inebilizumab, a B Cell-Depleting Anti-CD19 Antibody for the Treatment of Autoimmune Neurological Diseases: Insights from Preclinical Studies. J. Clin. Med 5 (2016), doi: 10.3390/jcm5120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill P, Gatsogiannis C, Raunser S, SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SHW, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P, Collaboration gets the most out of software. eLife. 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials will be made available upon request. Accession numbers of the CD19-CD81-Coltuximab Fab model and map are Protein Data Bank (PDB) 7JIC and EMD-22344, respectively.


