Abstract
This study evaluates the impact of physiologically relevant oxygen tensions on the response of HepG2 cells to known inducers and hepatotoxic drugs. We compared transcriptional regulation and CYP1A activity after a 48 h exposure at atmospheric culture conditions (20% O2) with representative periportal (8% O2) and perivenous (3% O2) oxygen tensions. We evaluated cellular responses in 2D and 3D cultures at each oxygen tension in parallel, using monolayers and a paper-based culture platform that supports cells suspended in a collagen-rich environment. Our findings highlight that the toxicity, potency, and mechanism of action of drugs are dependent on both culture format and oxygen tension. HepG2 cells in 3D environments at physiologic oxygen tensions better matched primary human hepatocyte data than HepG2 cells cultured under standard conditions. Despite altered transcriptional regulation with decreasing oxygen tensions, we did not observe the zonation patterns of drug-metabolizing enzymes found in vivo. Our approach demonstrates that oxygen is an important regulator of liver function but it is not the sole regulator. It also highlights the utility of the 3D paper-based culture platform for continued mechanistic studies of microenvironmental influences on cellular responses.
Keywords: 3D-culture, Oxygen, Liver tissue models, Hepatotoxicity, Acetaminophen
1. Introduction
Preclinical in vitro assays cannot accurately predict drug-induced liver injury or hepatotoxicity in patients, two factors that account for a significant number of late-stage drug failures.(Kenna and Uetrecht, 2018; Parasrampuria et al., 2018) These cell-based assays commonly use monolayers of primary human hepatocytes (PHHs) or cell lines presenting hepatocyte-like characteristics. Monolayer cultures lack many aspects of the tissue microenvironment. PHHs on culture-compatible plasticware readily depolarize and lose liver-specific function within days of plating.(Guo et al., 2011; Hewitt et al., 2007; Zeigerer et al., 2017) By simply placing the PHHs in an extracellular matrix-rich environment such as a collagen sandwich, the cells can maintain polarization and metabolic function for prolonged periods.(Dunn et al., 1992) When cultured as spheroids, the HepG2 hepatoma cell line reacquires functions lost to monolayers, including increased expression of phase I and II metabolic enzymes, glycogen storage, and bile transport.(Chang and Hughes-Fulford, 2009; Gaskell et al., 2016; Ramaiahgari et al., 2014) The recovery of these liver-specific functions in a 3D environment suggests including other physiologically relevant culture conditions could increase the utility of HepG2 cells for preliminary evaluations of new drugs or drug libraries in high throughput screens.
While spheroids have clear advantages over monolayer cultures, the analysis of microenvironmental influences on cellular behavior is difficult to assess without histological slicing. The overlapping gradients of oxygen, nutrients, and soluble factors that extend across spheroids also make it difficult to experimentally probe microenvironment-cellular function relationships, individually or in concert.(Hirschhaeuser et al., 2010) Oxygen is of particular interest, as it is a global regulator of cellular function in all tissues.(Prabhakar and Semenza, 2015) Within the liver, changes in oxygen availability correlate with liver zonation, the distribution of metabolic activity in hepatocytes lining each sinusoid. The blood oxygen partial pressures range from 65–60 mmHg O2 (11–8% O2) in the periportal region to 35–30 mmHg O2 (5–3% O2) in the perivenous region.(Jungermann and Kietzmann, 2000; Kietzmann, 2017) Hepatocytes in the periportal region primarily express high densities of sulfotransferases (SULTs).(Jancova et al., 2010) Hepatocytes in the perivenous region primarily express cytochrome P450 enzymes (CYPs) and UGP-glucuronosyltransferases (UGTs).
The hypothesis driving this study was that HepG2 drug-metabolizing enzyme transcript profiles will better match PHHs when placed in physiologically relevant culture environments. Previous works evaluating oxygen effects on HepG2 cells showed substantial changes in cellular responses to drugs when transitioning from standard culture conditions to ones with tumor-relevant oxygen tensions;(Bowyer et al., 2017) these hypoxic conditions initiate global transcriptional reprogramming in HepG2 cells.(Sonna et al., 2003) Few studies have evaluated the oxygen tensions found in healthy livers.
To characterize oxygen’s role in hepatotoxic responses, we probed HepG2 cellular responses at three different oxygen partial pressures in monolayer and 3D culture formats. Cells cultured at ambient conditions (20% O2) served as a reference point; cells cultured at 8% O2 represented the periportal region; cells cultured at 3% O2 represented the perivenous region. To generate the 3D environments, we used the paper-based culture platform first described by Whitesides and further developed in our laboratory.(Cramer et al., 2019; Derda et al., 2009) This setup uses wax-patterned sheets of paper to define 3D culture regions. We demonstrate here that porous structure of the paper can support HepG2 suspended in a collagen matrix, and that the experimental conditions did not cause multicellular aggregates or spheroid-like structures that would result in oxygen- or nutrient-limited environments.
This study highlights the importance of evaluating toxicity in representative in vivo conditions. It also demonstrates the utility of the paper-based culture platform for well-defined mechanistic studies. The datasets we collected show that oxygen tension and the culture format modulate HepG2 responses to known hepatotoxins and aryl hydrocarbon receptor (AhR) inducers. First, we found that muted CYP inducibility in 3D formats is a consequence of the microenvironment and not due to a limited accessibility to AhR inducers in solution. Second, exposing HepG2-laden scaffolds at physiological oxygen tensions to acetaminophen, cyclophosphamide, or aflatoxin B1 yielded responses similar to those observed in PHHs. Third, the altered metabolic profiles caused by decreasing oxygen tension or including a collagen ECM can explain differences in HepG2 responses to drugs.
2. Materials and Methods
2.1. Reagents.
All reagents were used as received unless otherwise noted. Cell culture medium and supplements were purchased from Gibco, except for fetal bovine serum (FBS, VWR) and collagen I (rat tail, Corning). Acetaminophen, aflatoxin B1, cyclophosphamide, 7-ethoxyresorufin (EROD), and 3-methoxycholanthrene (3-MC) were purchased from Sigma Aldrich. Calcein-AM, DMSO, Hoechst 33342, and propidium iodide (PI) were purchased from Fisher Scientific. TCDD (2,3,7,8-tetrachloro-p-dioxin) was purchased from Alfa Chemistry and CellTiter-Glo 2.0 (CTG) from Promega.
2.2. Cell Culture.
HepG2 human hepatoma cells (American Type Culture Collection) were maintained as monolayers at 20% O2, 37 °C, and 5% CO2 in DMEM medium supplemented with 10% FBS, 1% penicillin-streptomycin, and 22.7 mM HEPES. This maintenance medium was exchanged every 2–3 days and the cells were passed at 75% confluency with TrypLE, using standard procedures. All experimental data was obtained from passages 5 – 15 of cryopreserved stocks of HepG2. The HepG2 cells were STR-verified in 2016 and are regularly evaluated for mycoplasma contamination. Experimental studies in standard 96-well plates contained 100 μL of maintenance medium and 40,000 cells/well. The cells were incubated overnight after plating to ensure attachment.
2.3. 3D Culture Preparation.
Sheets of Whatman 105 lens paper were wax-patterned and sterilized as detailed previously by our lab.(Kenney et al., 2019; Lloyd et al., 2017) Briefly, sheets of Whatman 105 lens tissue paper were wax-patterned with a Xerox ColorQube 8750 printer, cut out with a 13” Silver Bullet Professional Series automated cutter, and sterilized overnight under ultraviolet light. Each circular scaffold contained a single cell-containing region, surrounded by a wax border. Small zone scaffolds (6.5 mm in diameter) fit directly into the well of a standard 96-well plate. Large zone scaffolds (18 mm in diameter) fit directly into the well of a standard 6-well plate. Figure S1 contains photographs and detailed schematics of both scaffolds. Prior to use, each scaffold was seeded with either cell-free or cell-laden collagen I (1.2 mg/mL) and incubated overnight before usage. The smaller scaffolds were seeded with 0.5 μL of gel and the larger scaffolds with 12.5 μL. Cell-laden scaffolds had a final density of 80,000 cells/μL (1.81×108 cell/cm3).
2.4. Hypoxia Chamber.
Cells were incubated in a home-built hypoxia chamber (Figure S2) whose gas composition was monitored with a diffusion-based O2 (model 2-BTA, Vernier) and CO2 sensor (model K30, CO2Meter.com). PID controllers (model PXU21A20, Red Lion) maintained the oxygen setpoints in the chamber by monitoring the DC output from each gas sensor in rea-time. The PID controllers used a low-current solid-state relay to actuate solenoid valves (Red Hat model 8262H020, ASCO) connected to supplies of either 100% N2 or 100% CO2 to adjust the gas composition in the chamber as needed. The oxygen controller was set to reverse output mode with the PID parameters P:97, I:150, and D:25. The carbon dioxide controller was set to direct output mode with the PID parameters P:7, I:120, and D:30. The hypoxia chamber was humidified and maintained at 37 °C and 5% CO2 for all experiments. Prior to introducing the cells, the chamber was equilibrated to the appropriate oxygen tension for 18 h.
2.5. Cellular Viability.
Cellular viability was determined with a tri-color live-dead stain. Cell pellets were collected, resuspended in 100 μL of 1X PBS, and stained for 10 min in a 1XPBS solution containing 10 μg/μL of calcein-AM, 5 μg/μL PI, and 10 μg/μL Hoechst. The stained cells were imaged with a Nikon TE-2000i inverted microscope equipped with a QICAM Fast 1394 digital camera (QImaging). Figure S3 contains representative brightfield and fluorescence images at each oxygen tension. Cell counts were determined with a previously published ImageJ method.(Schneider et al., 2012)
2.6. Dose-Response Curves.
Monolayers and small zone scaffolds were dosed for 48 h in 200 μL of drug-containing medium. These solutions were prepared from stocks of acetaminophen (5 M), aflatoxin B1 (200 mM), and cyclophosphamide (5 M) in DMSO. The stock solutions were stored at −20 °C until needed. Prior to analysis, the cells were washed with 1X PBS and lysed for 20 min in a 1:1 (v/v) solution of 1X PBS and CTG. Lysate aliquots (75 μL) were analyzed in an opaque 96-well plate on a Spectramax M5 Multi-Mode Microplate Reader (Molecular Devices) in luminescence mode with a 500 ms integration.
2.7. CYP1A Activity Assays.
Monolayers and small zone scaffolds were incubated for 48 h in 200 μL of induction medium: maintenance medium containing either 3-MC or TCDD. Stock solutions of 3-MC (20 mM) and TCDD (100 μM) were prepared in DMSO and stored at −20 °C until needed. Cells were washed once with 1XPBS and incubated for 1 h in EROD assay solution: 100 μL of maintenance medium containing 10 μM EROD. Aliquots of EROD assay medium (80 μL) were analyzed in an opaque 96-well plate (560/590 ex/em) on a SpectraMax M5 Multi-Mode Microplate Reader. The remainder of the solution was removed from the cell-containing wells, the cells washed with 1X PBS, and viability determined with the CTG assay.
2.8. RT-qPCR Analyses.
Monolayers and large zone scaffolds were dosed for 48 h with maintenance medium containing 3-MC (5 μM), TCDD (1 nM), aflatoxin B1 (10 nM), or acetaminophen (10 mM). Cells were lysed using a TRIzol Plus RNA Purification Kit (Fisher Scientific). The TRIzol reagent was added directly to the monolayer cultures; the cell-containing paper scaffolds were submerged in it. Both culture formats were agitated for 5 minutes at room temperature before RNA isolation. Reverse transcription (RT) was performed immediately after RNA isolation with a High-Capacity cDNA Reverse Transcription Kit (Fisher Scientific) in an Eppendorf Master Cycler.
Table S1 lists the qPCR primer pair sequences, melting temperatures, optimized reaction concentrations, and reaction efficiencies (90–110%) for each primer set. Amplification reactions were performed with PowerUp SYBR Master Mix (Fisher Scientific), in a 384-well plate, on a QuantStudio 6 Flex Real-Time PCR system. Each sample was measured in triplicate, using the following program: 95 °C for 60 sec, followed by 40 cycles of 95 °C for 2 sec and 55 °C for 30 sec. Each transcript was quantified using the ΔΔCt method against ACTB (β-actin).(Schmittgen and Livak, 2008) A fold-change of greater than 2.0 was considered significant.
2.9. Confocal Microscopy.
Images of the HepG2 cells cultured as monolayers and in the paper-based scaffolds were obtained on a Zeiss LSM 710 spectral confocal laser scanning microscope. Prior to mounting the samples with ProLong Gold (Cell Signaling), the cells were fixed with 3.2% paraformaldehyde, permeabilized with a 0.5% (v/v) solution of Triton X-100 in 1XPBS, and stained with Alexa Fluor 488 phalloidin (ThermoFisher). The cells were also counterstained with DRAQ5 (BioLegend). Single plane confocal and z-stack images of the monolayer and 3D cultures are provided in the Supplementary Materials.
2.10. Statistical Analyses.
Datasets were analyzed with GraphPad Prism 7. Values are reported as the average and standard error of the mean (SEM) of at least two separate cell passes (N), with each cell pass containing at least three technical replicates (n). Dose-response datasets were fit to a four-parameter logistics (4PL) fit. Aflatoxin B1 was fit to a three-parameter (3PL) logistics fit due to an ambiguous lower asymptote in the 4PL model, which can be attributed to solubility constraints. EC50 values were compared using an F-test. Statistically significant differences correspond to a p-value of ≤ 0.05.
3. Results
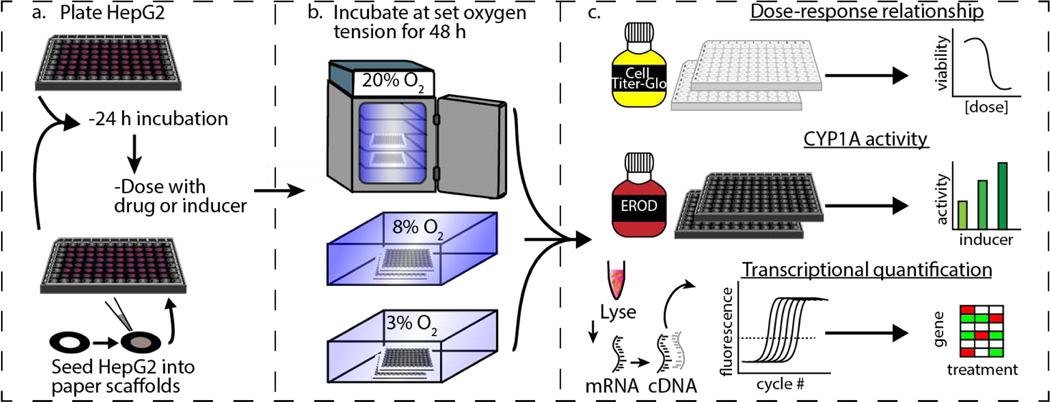
Figure 1 summarizes the experimental workflow used to evaluate hepatotoxicity and induction of phase I and II drug-metabolizing enzymes in HepG2 cells. By simultaneously evaluating monolayer and the paper-based 3D culture setups, we determined the effects of culture dimensionality and oxygen tension in the same passage of cells. We chose the HepG2 cell line for this work because it is readily available and has low genetic drift.(Kitamoto et al., 1993; Zhao et al., 2018) Despite the limited expression of CYP enzymes compared to PHHs, HepG2 cells have a well-characterized liver-representative genotype.(Gerets et al., 2009; Westerink and Schoonen, 2007) When seeded in the paper scaffolds, the suspension of HepG2 cells distribute throughout the wax-defined zone. The collagen attaches to the paper fibers and fills the void spaces, providing the cells a protein-rich region for growth. The Supplementary Materials contain a series of single plane and z-stack images obtained on a confocal microscope. These images show that the cells remain distributed throughout the scaffold after a 48 h culture. The images also show that distribution of cells throughout the scaffolds are not even, with larger cell numbers occurring in regions with higher fiber densities.
Figure 1.
Experimental workflow comparing the responses of HepG2 cells exposed to different oxygen tensions. Cell monolayers were maintained on standard culture plasticware. The 3D cultures were prepared by seeding cells suspended in a collagen I matrix into wax-patterned paper scaffolds. (a) First, cells were placed in the appropriate culture format and incubated for 24 h at atmospheric culture conditions (20% O2, 5% CO2, and 37 °C). (b) Next, the cells were incubated in the presence of a drug or inducer for 48 h at 20%, 8%, or 3% O2. (c) Finally, cellular responses were quantified. Hepatotoxicity was evaluated with the CellTiter-Glo viability assay, CYP1A activity quantified with the EROD assay, and transcriptional regulation was determined with RT-qPCR.
3.1. Cell viability is not impacted by physiological oxygen tensions.
Cell viability was assessed with calcein-AM, Hoechst 33342, and PI staining after a 48 h incubation at each oxygen tension. Figure S3 contains representative brightfield and fluorescence images of the HepG2 cells at each oxygen tension. The fraction of viable cells was determined with Eqn. 1, where nlive is the number of calcein-stained cells, ndead is the number of PI-stained cells, and ntotal is the number of Hoechst-stained cells.
| (Eqn. 1) |
The fraction of viable cells was equivalent at each oxygen tension for the PI staining ratios (0.94 +/− 0.01). The average calcein staining ratios at 20% and 8% O2 were similar (0.90+/− 0.01) and agreed with the PI staining data. The calcein stain at 3% O2 indicated a significant drop in viability (0.42 +/− 0.01). Kang reported a similar observation, which they attributed this difference to decreased esterase availability or an impaired metabolic activity rather than truly reduced viability.(Kang et al., 2020) Table S2 summarizes the viability data for each oxygen tension. We relied on the PI staining data and assumed the 48 h exposure lower oxygen tensions did not significantly affect viability.
3.2. Oxygen tension alters the potency and overall toxicity of known hepatotoxic drugs.
We evaluated three drugs for dose-dependent hepatoxicity at each oxygen tension, using concentration ranges based on previously reported potency (EC50) values: 1–200 mM acetaminophen, 1–100 mM cyclophosphamide, and 0.001–200 nM aflatoxin B1.(Gerets et al., 2012; Wang et al., 2002) Cellular viability was measured after a 48 h incubation and compared to vehicle treatments exposed to the same concentration of DMSO.
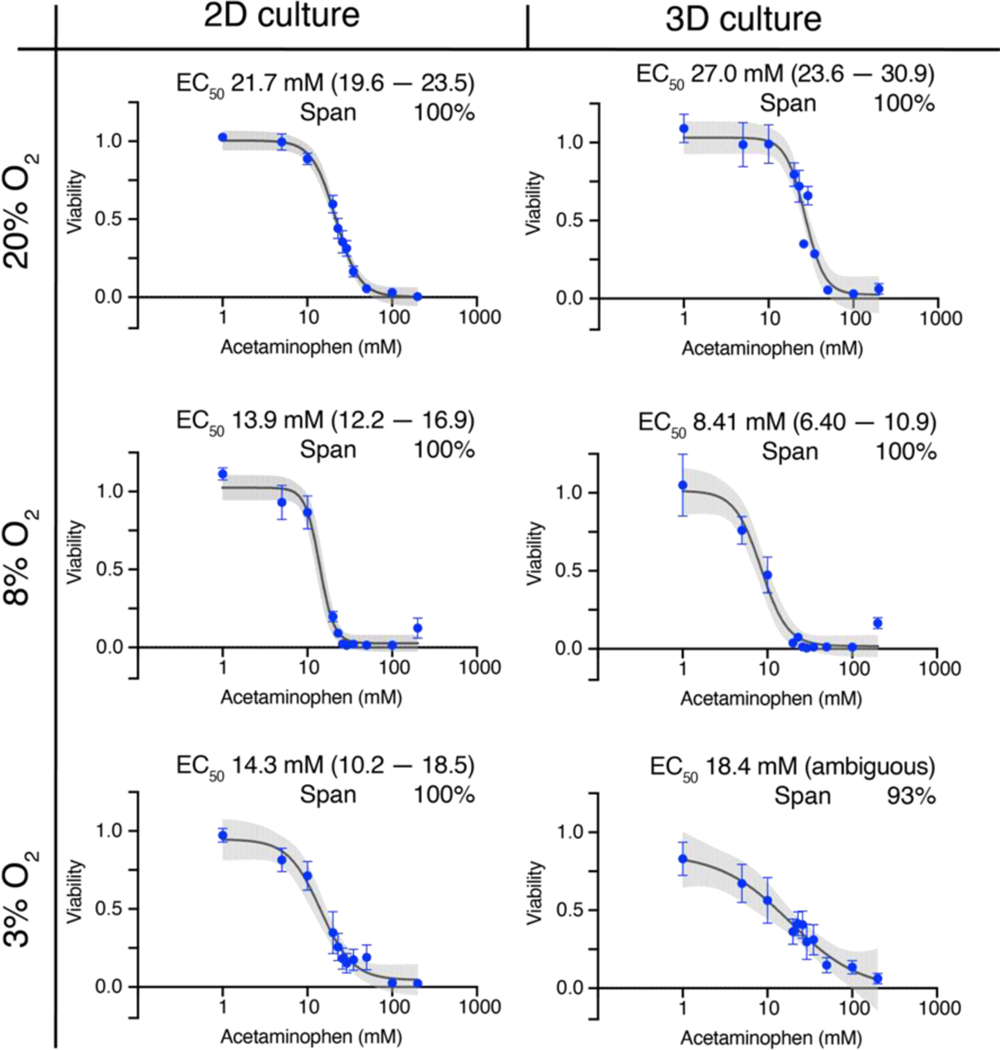
Acetaminophen was equally hepatoxic in the monolayer and 3D culture formats (Figure 2), with 200 mM killing 100% of the cells at each oxygen tension. The potency of acetaminophen increased significantly in both culture formats at physiologically relevant oxygen tensions: in 2D, from 21.7 mM at 20% O2 to 13.9 mM at 8% O2 and 14.3 mM at 3% O2; In 3D, from 27.0 mM at 20% O2 to 8.4 mM at 8% O2 and 18.4 mM at 3% O2. The significant difference in potency compared to 20% O2 suggests an increased rate of accumulation for acetaminophen’s cytotoxic byproduct, N-acetyl-p-benzoquinone imine (NAPQI) at these lower oxygen tensions.
Figure 2.
Dose-response relationships of 40,000 HepG2 cells after a 48 h exposure to acetaminophen at atmospheric (20%), periportal (8%), or perivenous (3%) oxygen tensions. Each point is the average and SEM collected from at least two different cell passages (N=2); each pass contained at least three technical replicates (n=3). The black lines connecting the points represent the best-fit 4PL model; the gray shaded regions represent the 95% confidence intervals of those fits.
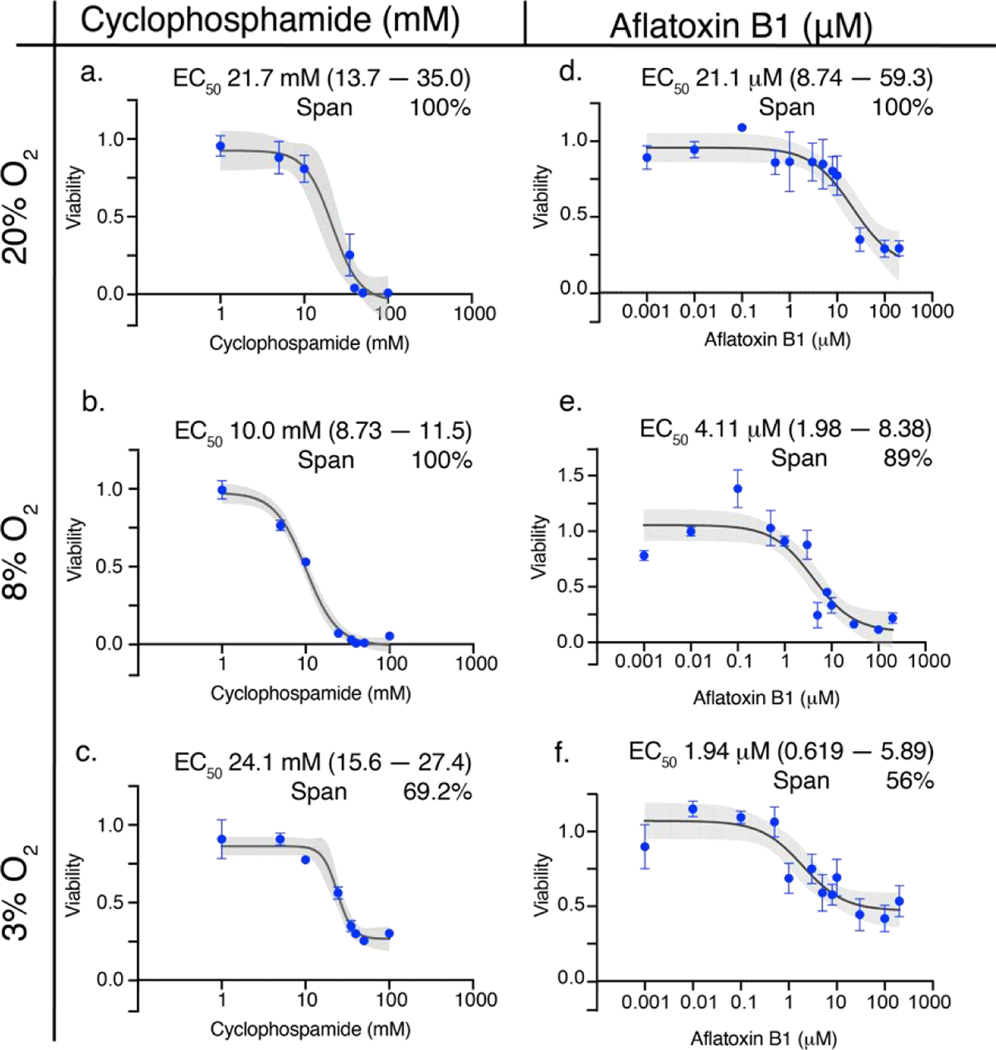
Figure 3 plots the dose-response relationships for aflatoxin B1 and cyclophosphamide in the 3D culture format at each oxygen tension. The overall toxicity of aflatoxin B1 decreased monotonically from 100% at 20% O2 to 56% at 3% O2. Cyclophosphamide killed 100% of the cells at 20% and 8% O2. Its overall toxicity deceased to 69% at 3% O2. The potency of both drugs increased with decreasing oxygen tension. For aflatoxin B1, the EC50 value decreased from 21.1 nM at 20% O2 to 1.9 nM at 3% O2. The potency of cyclophosphamide was decreased from 21.7 mM at 20% to 10.1 mM at 8% O2. Surprisingly the EC50 value at 3% O2 was equivalent to that of 20% O2. Dose-response relationships for both the monolayer and 3D culture formats at each oxygen tension are in the Supplementary Materials.
Figure 3.
Dose-response relationships of 40,000 HepG2 cells in the 3D culture format after a 48 h exposure to either cyclophosphamide (a-c) or aflatoxin B1 (d-f) at atmospheric (20%), periportal (8%), or perivenous (3%) oxygen tensions. Each point is the average and SEM collected from at least two different cell passages (N=2–3); each pass contained at least three technical replicates (n=3). The black lines connecting the points represent the best-fit 4PL model (cyclophosphamide) and 3-PL model (aflatoxin B1); the gray shaded regions represent the 95% confidence intervals of those fits.
3.3. Both oxygen tension and culture alter the drug-metabolizing enzyme transcript profile in HepG2 cells
Transcript-level regulation of eight phase I and phase II drug-metabolizing enzymes was quantified with RT-qPCR. We focused on CYP1A1 and CYP1A2 because they are readily inducible in HepG2 cells; CYP2E1 due to its low inducibility in monolayers at ambient conditions; SULT and UGT genes because of their known contributions to acetaminophen metabolism and their presence in HepG2 cells.(Drahushuk et al., 1998; Gerbal-Chaloin et al., 2014; Ramaiahgari et al., 2014; Sumida et al., 2000; Westerink and Schoonen, 2007) We also quantified the aryl hydrocarbon receptor (AhR), which is known to regulate the expression of CYP1A enzymes.
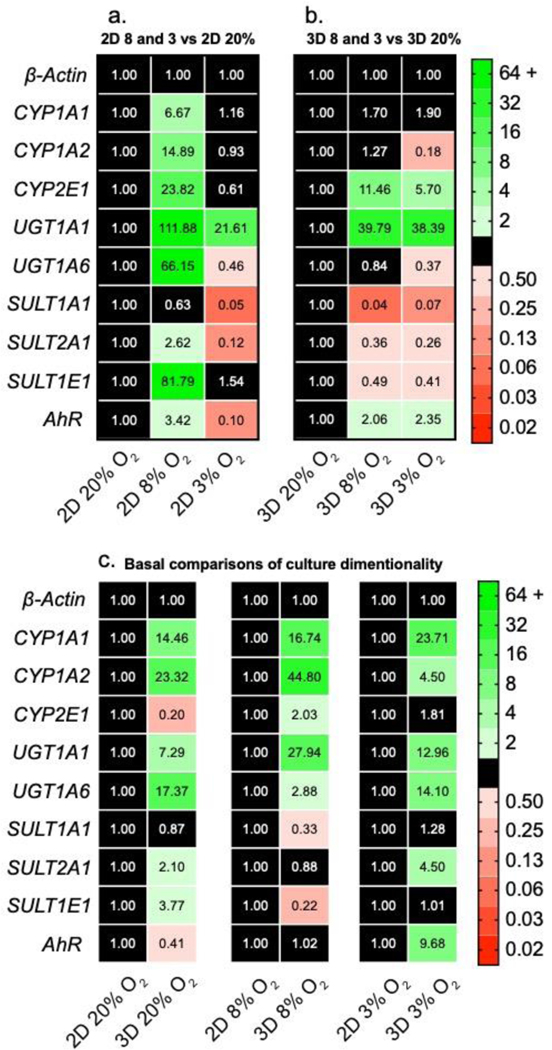
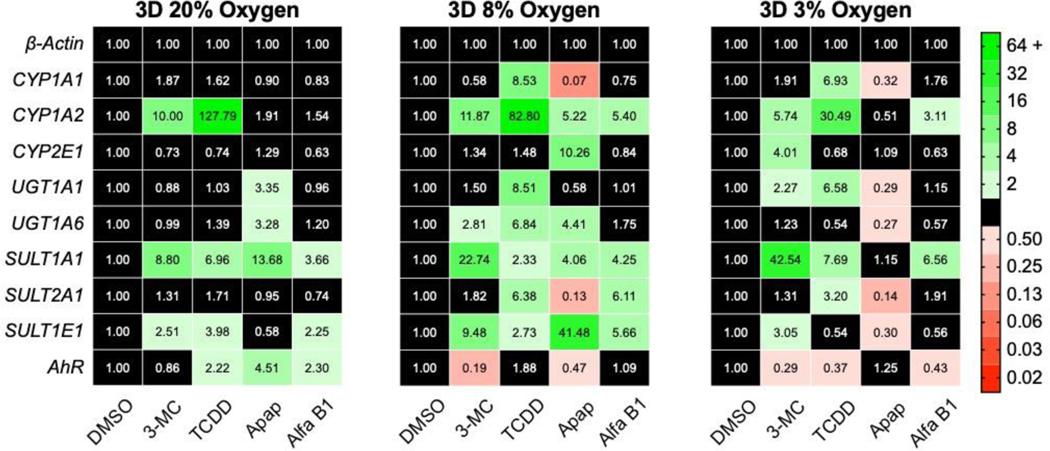
In monolayer cultures (Figure 4a), each drug-metabolizing gene, except for SULT1A1, was upregulated at 8% O2 compared to atmospheric conditions. There was a significant decrease in transcript levels for most genes between 8% and 3% O2; UGT1A6, SULT1A1, SULT1A2, and AhR were also significantly decreased at 3% compared to 20% O2. Interestingly, UGT1A1 was the only gene upregulated at both physiologic oxygen conditions. In the 3D cultures (Figure 4b), the transcript profiles at 8% and 3% O2 were similar, with an overall upregulation of CYP2E1, UGT1A1, and AhR compared to 20% O2. There also was a general downregulation in SULT1A1, SULT1A2, and SULT1E1 in the physiologic oxygen conditions compared to 20% O2.
Figure 4.
Transcript-level regulation of phase I and phase II drug-metabolizing enzymes in HepG2 (a) monolayer and (b) 3D cultures after a 48 h exposure to 20%, 8%, or 3% O2. (c) Transcript-level regulation between monolayer and 3D culture formats at each oxygen tension. Each value is the average of at least two cell passages (N=2–3); each pass contained at least three technical replicates (n=3). A fold-change >2 indicates a significant increase in expression; <0.50 indicates a significant decrease. The numerical values correspond to the average ΔΔCt value of each transcript.
Figure 4c highlights the dramatic changes in transcriptional regulation between the 2D and 3D culture formats. An increased number of CYP1A1, CYP1A2, UGT1A1, and UGT1A6 transcripts in the 3D cultures was conserved across all three oxygen tensions, suggesting the presence of collagen or increased numbers of cell-cell contacts were the primary regulator. Kim and colleagues found that gene regulation in PHHs diverged significantly between monolayers and collagen sandwich cultures.(Kim et al., 2010) In particular, they found that CYP transcripts significantly increased in the collagen sandwich format.
The other genes we probed did not have a consistent trend in regulation that can be attributed to either oxygen or culture format independently. However, there were apparent changes between the 2D and 3D culture environments at each oxygen tension. For example, CYP2E1 was downregulated at 20% O2 but upregulated at 8% O2. The SULT genes show a similar oxygen-dependence with SULT2A1 and SULT1E1 upregulated in 3D at 20% O2; SULT1A1 and SULT1E1 downregulated in 3D at 8% O2; SULT2A1 upregulated in 3D at 3% O2. These results suggest a more nuanced regulation that is dependent on both culture parameters.
3.4. Oxygen tension alters transcriptional regulation in response to known hepatotoxins
Transcription regulation of phase I and phase II drug-metabolizing enzymes in the presence of 10 mM acetaminophen, 10 mM aflatoxin B1, 5 μM 3-MC, 1 nM TCDD were measured with RT-qPCR after a 48 h exposure. At all three oxygen tensions, 3-MC and TCDD upregulated CYP1A2, SULT1A1, and SULT1E1 in the 3D cultures (Figure 5). These molecules differentially induced genes at each oxygen tension, with increased transcripts of phase II enzymes at 8% O2 but increased CYP transcripts at 3% O2. Consistent with previous reports, 3-MC and TCDD upregulated CYP1A1 and CYP1A2 in the monolayer cultures (Figure S6).(Fradette and Du Souich, 2004; Hewitt and Hewitt, 2004; Westerink and Schoonen, 2007)
Figure 5.
Transcript-level regulation of 3D cultures of HepG2 cells after a 48 h incubation with 5 μM 3-MC, 1 nM TCDD, 10 mM acetaminophen (apap), or 10 nM aflatoxin B1 (afla B1). Each value is the fold change of the average ΔΔCt value from at least two cell passages (N=2–3); each pass contained at least three technical replicates (n=3). A fold-change >2 indicates a significant increase in expression; <0.50 indicates a significant decrease. The numerical values correspond to the average ΔΔCt value of each transcript.
The differential gene expression changes with decreasing oxygen tension were most pronounced in the presence of acetaminophen. Acetaminophen upregulated UGT1A1, UGT1A6, SULT1A1, and AhR transcripts in the 3D cultures at 20% O2. At physiologic oxygen tensions, acetaminophen had a much different effect. At 8% O2 nearly every gene measured was affected: CYP1A1, SULT2A1 and AhR were downregulated; CYP1A2, CYP2E1, UGT1A6, SULT1A1, and SULT1E1 were upregulated. At 3% O2, acetaminophen downregulated CYP1A1, UGT1A1, UGT1A6, SULT2A1, and SULT1E1. No one has reported differentially induced genes at various physiological oxygen tensions to our knowledge. Audibert reported reduced drug metabolism in rabbits exposed to hypoxemic conditions.(Audibert et al., 1993) They hypothesized the reduced oxygen tension altered the distribution and expression of CYPs responsible for drug metabolism but did not have molecular-level datasets needed for confirmation.
Transcriptional regulation upon acetaminophen exposure was also dependent on culture format, as shown in Figure S7. At 20% O2, the 2D cultures were more sensitive to acetaminophen with higher expression of CYP and UGT transcripts. At 8% O2, the 3D culture format had significantly more CYP, UGT, and SULT transcripts. At 3% O2, the CYP transcripts were similar in both culture formats, but the 2D cultures had higher expression of UGT and SULT genes.
3.5. CYP1A activity was reduced at physiologic oxygen tensions and in 3D culture formats.
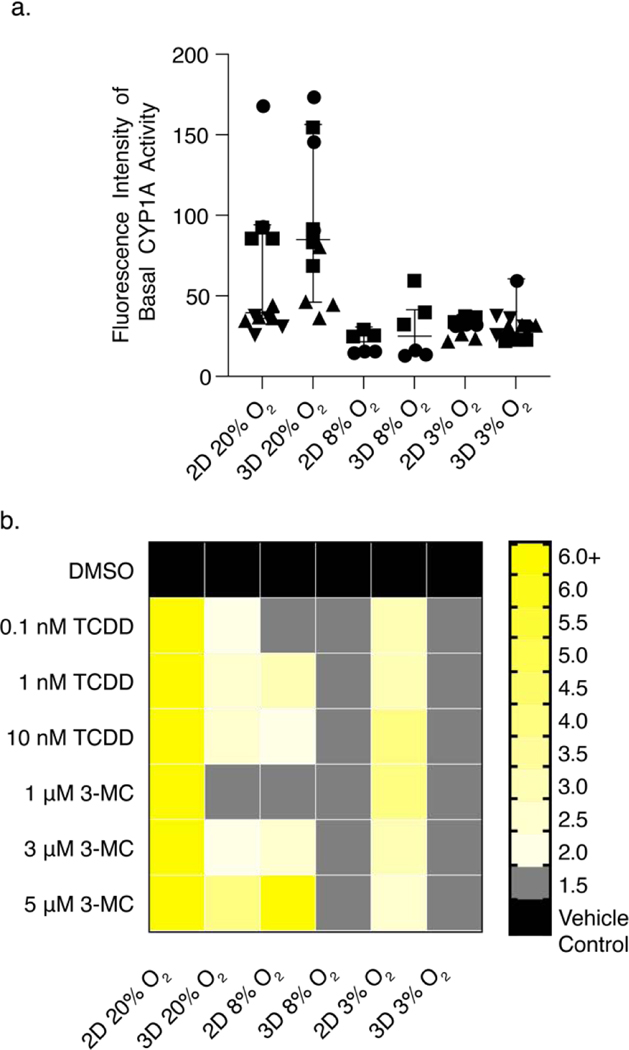
CYP1A activity was determined with an EROD assay. Figure 6a plots the raw fluorescence intensities obtained for the monolayer and 3D cultures after a 48 h incubation at each oxygen tension. This data shows that neither cell culture format nor oxygen tension altered the basal CYP1A activity of the HepG2 cells. We attribute the large variations in the raw fluorescence intensity values to low basal activity levels.
Figure 6.
Average CYP1A activity of HepG2 cells as determined by the EROD assay. (a) Basal CYP1A activity, plotted as the raw fluorescent intensity, after a 48 h incubation. (b) CYP1A activity after a 48 h induction with increasing concentrations of 3-MC or TCDD. Values are the average of at least six data points collected from at least two cell passages (N=2). Each pass contained at least three technical replicates (n=3). A fold-change >2 indicates a significant increase in activity. The numerical values of the heatmap are found in Table S5.
To quantify CYP1A activity in the presence of increasing concentrations of 3-MC or TCDD, we adjusted for potential changes in cellular viability. CYP1A activity per live cell (Eqn. 2) was calculated from the average raw fluorescence intensity collected from replicate EROD assays (IEROD) and the vehicle-normalized luminescence values collected from the CTG assay (ICTG).
| (Eqn. 2) |
Figure 6b is a heatmap of CYP1A activity per live cells at each oxygen tension. These data show that both 3-MC and TCDD significantly induced activity in the 2D cultures at 20% O2. The highest concentrations of 3-MC (10 nM) and TCDD (5 μm) also induced activity at physiologic conditions, with the magnitude of the response decreasing with decreased oxygen tension. We also observed induction by both 3-MC and TCDD in the 3D culture formats. These responses were muted compared to the 2D cultures. At 20% O2, 1 nM TCDD induced activity by 2.05-fold in 3D; the same concentration resulted in a 13.0-fold increase in 2D. 3-MC was able to induce CYP1A activity at 3D 20% O2 only at 5 μM. Both 3-MC and TCDD were unable to induce activity at 8% and 3% O2. Table S5 lists the induction average for each culture condition and format.
4. Discussion
Despite the improved predictability of liver spheroids and organ-on-chip devices there is no 3D culture platform that can quickly assess microenvironmental impacts on the regulation of drug-metabolizing enzymes.(Lauschke et al., 2016; Whitman et al., 2016) Such studies are needed to improve the predictability of current in vitro assays, identifying the structural and chemical components that best approximate the responses of a liver sinusoid or lobule in vivo. In a recent review, Agarwal highlighted the potential of paper-based cultures for generating 3D liver models.(Agarwal et al., 2020) The current work highlights how amenable paper scaffolds are for fundamental cellular studies.
The distinct differences between the two culture reinforces the importance of the tissue microenvironment, but also raises questions about the best way to experimentally control that microenvironment. Both 2D and 3D formats contain oxygen gradients. When cell densities are high, pericellular hypoxia results because oxygen consumption outpaces its diffusion. In monolayers, the differences between the oxygen concentration in an incubator and at the bottom of a medium-containing plate can be substantially different.(Al-Ani et al., 2018) The monolayer cell densities used in this experiment substantially reduce the oxygen concentration at the cell surface for the 8% and 3% O2 conditions, as demonstrated in the Supplementary Materials. Pericellular hypoxia is likely also happening in the paper scaffolds, however it would be less than the monolayer format, as the wax-patterned scaffolds float atop the medium in the plate and reside at the air-water interface throughout the experiment.
4.1. AhR ligand induction is dependent on culture format and oxygen tension.
Quantification of CYP1A transcripts and activity allowed us to assess the responsiveness of HepG2 cells to known AhR inducers as a function of oxygen tension, culture format, and a combination of the two factors. While we focus on TCDD, similar conclusions can be drawn for 3-MC or the hepatotoxic drugs we evaluated in parallel. When exposed to 1 nM TCDD at atmospheric culture conditions, incorporating an ECM reduced the overall CYP1A activity by 7.5-fold compared to monolayers at the same conditions (Figure 6b). Vrba also noted this muted inducibility between 2D and 3D culture formats in an analogous experiment;(Vrba et al., 2014) the CYP1A activity in HepG2 spheroids exposed to 5 nM TCDD was reduced 17-fold compared to monolayers at the same conditions. Transcript changes at 20% O2 correlate with the activity data, with a 1 nM TCDD exposure resulting in a 5-fold lower induction of CYP1A1 in the 3D cultures but a similar induction of CYP1A2 between the two culture formats. These differences could be attributed to altered basal expression levels of AhR, as the 3D culture format contained 2.4-fold lower AhR transcript levels than the monolayer cultures. The basal CYP1A level activity was equivalent between the two formats (Figure 6a). Detailed values of transcript fold-changes are in Tables S6 – S8. A comparison of cellular responses to 1 nM TCDD also showed muted activity in both the 2D and 3D culture formats when transitioning from atmospheric to physiologic oxygen tensions (Figure 6b). In the 2D cultures, the TCDD-induced increase compared to vehicle decreased from 12.9-fold at 20% O2 to 2.6-fold at 8%, and 3.0-fold at 3% O2. In the 3D cultures, the fold-increase compared to the vehicle was only significant at 20% O2 (2.1-fold).
The differences in both transcript numbers and activity suggest the monolayers are more sensitive to inducers, possibly due to differences in cellular energy allocations between the two formats. A second potential reason for this decreased inducibility is a lower effective concentration of TCDD reaching the cells in the 3D formats, due to non-specific adsorption to the collagen matrix or the paper’s cellulose fibers. In previous a previous study, we showed hydrophobic drug molecules distribute evenly throughout a paper-based colon tumor model with 12 sheets of cell-laden thick within six hours of introduction.(Boyce et al., 2017) These results suggest that the single cell-containing scaffolds should see the full inducer concentration in less than an hour of introduction.
4.2. HepG2 responses to acetaminophen at physiologic oxygen tensions are similar to PHHs.
The importance of probing multiple oxygen tensions in drug-induced liver toxicity studies is highlighted by stark differences in the dose-response relationships for acetaminophen, aflatoxin B1, and cyclophosphamide (Figures 2 and 3). The decreased overall toxicity, increased potency, and altered dose-response shapes we observed suggest altered mechanisms of action arise in the presence of an ECM and reduced oxygen tension. Comparing responses to acetaminophen between 2D and the paper-based cultures at atmospheric conditions supports their continued use as a model. Under these conditions, the overall toxicity was consistent with 100% cell loss at the highest tested concentration. The potency of acetaminophen and the shape of its curve was also unchanged, suggesting that the collagen matrix and paper-based scaffolds do not alter its mechanism. These results match previously reported studies with HepG2 monolayers and spheroids.(Parikh et al., 2015; Ramaiahgari et al., 2014)
Direct comparisons of the HepG2 results and in vivo are difficult due to the large variability of the two cellular environments and a disparity of assay readouts. However, direct comparisons to PHHs are an attractive benchmark for determining the utility of HepG2 cells in physiologically relevant conditions as they are the gold standard for in vitro studies of cellular induction, metabolism, and toxicity. When cultured as monolayers, HepG2 cells at 20% O2 have limited expression and induction of drug-metabolizing enzymes. In the collagen-rich environment of the paper scaffolds, HepG2 cells at physiologically relevant oxygen tensions had markedly different responses to inducers and hepatotoxic compounds at the transcript and activity levels. The potency values we obtained for acetaminophen at 8% (8.4 mM) and 3% O2 (18.4) were significantly lower than that at atmospheric oxygen levels (27.0 mM). These values also align with previous reports of 10 mM for PHH collagen sandwiches and spheroids.(Bell et al., 2017; Schyschka et al., 2013)
The altered shape of the dose-response relationship for acetaminophen in 3D cultures exposed to 3% O2 also suggest an oxygen-dependent mechanism of action. Acetaminophen toxicity is due to the accumulation of NAPQI intermediate, which forms protein adducts that lead to glutathione (GSH) depletion and oxidative stress-induced necrosis.(Hinson et al., 2010) The downregulation of UGT and SULT transcripts at 3% O2 suggests acetaminophen is primarily metabolized by CYPs under these conditions, resulting in continued NAPQI-protein adduction at low doses. This proposed mechanism is supported by earlier works, which showed that NAPQI-protein adduct accumulation is localized in the liver’s perivenous region and spreads radially with continued exposure.(Hinson et al., 2010; Roberts et al., 1991)
4.3. HepG2 potency values for cyclophosphamide and aflatoxin B1 at physiologic oxygen tensions are similar to PHHs.
4.3.1. Cyclophosphamide.
The potency values for cyclophosphamide as a function of oxygen tension were markedly different between the 2D and 3D culture formats (Figure S4), supporting the acetaminophen data and changes in the extracellular stimuli can have profound effects on drug sensitivity. In 2D, there was a steady decrease in potency from 37.6 mM at 20% O2 to 7.38 mM at 3%. When placed in the 3D culture format, there are two notable differences. First, the potency values for atmospheric (21.7 mM) and perivenous (24.1 mM) oxygen tensions were indistinguishable; these values were also significantly different from those obtained in 2D. Second, the potency values in the 3D cultures were similar to those recorded for PHHs at 20% O2 (25 mM).(Yokoyama et al., 2018) We note that the decrease in the overall toxicity and altered shape of the dose-response relationships between 20% and 3% O2 also suggest an alternative mechanism of action despite the similar potency values.
The altered shape of the dose-response curves could arise from oxygen-dependent metabolism of cyclophosphamide. Under standard culture conditions cyclophosphamide is converted to 4-hydroxycyclophosphamide, which is further metabolized into the DNA-alkylating agent phosphoramide mustard and acrolein.(Ionescu and Caira, 2005; Rodriguez-Antona and Ingelman-Sundberg, 2006) Decreased oxygen levels could alter the enzyme expression or activity responsible for the detoxification of cyclophosphamide, resulting in the different EC50 values we observed due to an accumulation of either phosphoramide mustard and acrolein. Mechanistic studies of cellular responses to cyclophosphamide in 2D and 3D culture formats should be further explored and compared to PHHs.
4.3.2. Aflatoxin B1.
The potency values of aflatoxin B1 in both culture formats were significantly lower at physiologic oxygen tensions than atmospheric levels. In the 3D cultures, the potency value decreased from 21.1 μM at 20% O2 to less than 4.1 μM. This reduced value agrees with previously reported EC50 values for PHHs on collagen-coated plates at atmospheric oxygen tensions: 10.8 μM (Li et al., 2012) and 4.7– 20 μM.(Li et al., 2012; Yokoyama et al., 2018)
The oxygen-dependent decreases in potency and overall toxicity changes suggest the HepG2 cells are accessing different metabolic pathways. Several mechanisms of action have been reported aflatoxin B1, with a common pathway of CYP3A4 and CYP1A2 metabolism to form aflatoxin B1–8,9-epoxide (AFBO). AFBO forms amine adducts with proteins and DNA. (Deng et al., 2018) It also activates cellular death receptors culminating in apoptosis.(Mughal et al., 2017) Figure 5 shows an increase in CYP1A2 transcripts at both 8% and 3% O2, supporting the increased production at AFBO compared to 20%. The presence of CYP1A enzymes is not the sole contributor, as the percentage of cells killed decreased significantly from 86% at 20% and 8% O2 to 56% at 3% O2.
There also was a notable increase in SULT transcripts at 8% O2. While these enzymes do not metabolize aflatoxin B1, they do activate the nuclear pregnane X receptor.(Ratajewski et al., 2011; Sonoda et al., 2002) These oxygen-dependent changes in response to aflatoxin merits further evaluation, as the activation of the nuclear receptors that orchestrate drug metabolism could prove important in predicting future toxicities or potential drug-drug interactions.
4.4. Is oxygen enough?
Our findings support previous works and agree that an ECM reduces the inducibility of HepG2 cells to AhR ligands while also sensitizing them to certain drugs. The data also highlight the importance of evaluating other physiologically relevant conditions, as HepG2 responses to drugs are oxygen tension-dependent. These oxygen-induced changes in potency or overall toxicity offer insights into potential liver-induced injuries or drug-drug interactions that would be overlooked under standard culture conditions.
Our findings also highlight that oxygen can improve some aspects of hepatic responses. However, it is not the only microenvironmental factor needed to obtain zonal expression of phase I and II drug-metabolizing enzymes. In vivo, zonation is marked by the expression of SULT enzymes at periportal conditions and UGT enzymes at perivenous.(Jancova et al., 2010) Our datasets lacked these basal level expression trends in both 2D and 3D culture formats (Figure 4). The paper-based culture platform is amenable to other studies into the microenvironment-cell function relationships. It is also amenable to other cell lines, such as PHHs, which can improve the platform’s overall a ility to predict drug toxicities. The ability to rapidly prototype and evaluate different paper-based structures, as demonstrated by our lab and others, provides a means to test individual parameters as in this work or generate gradients of oxygen and nutrients similar to those found in vivo. The incorporation of gradients may provide representative responses to change in oxygen tension and perhaps more insight into in vivo happenings.
Supplementary Material
Highlights.
The incorporation of a 3D culture environment and physiologically relevant oxygen tensions lead to more predictive hepatotoxicity models
In a 3D environment, the basal transcript level of phase I and II drug-metabolizing enzymes are elevated at atmospheric and physiological oxygen tensions
Transcriptional regulation of HepG2 cells better match in vivo observations when cultured at physiologically relevant oxygen tensions
CYP1A activity and inducibility decreases in both monolayer and 3D culture formats with decreasing oxygen tensions
Acknowledgements.
This work was supported with funds provided by the National Institute of General Medical Sciences through Grant Award Number R35GM128697. We would like to thank the UNC Physics Machine Shop and the CRiTCL Electronics Core Laboratory for their help in constructing the acrylic-based hypoxia chamber and PID controllers that maintained the gas environments within the chamber. We thank the UNC Microscopy Service Laboratory (MSL) Dr. Pablo Ariel for access to and help with the LSM710 confocal microscope. The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. We would also like to thank Mr. Peter Willard for helpful discussions as we prepared this manuscript and Mrs. Levi Chua, Ms. Pearl Dang, and Mr. Zhi-Wei Lin for their help with preparing paper scaffolds.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal T, Borrelli M, Makvandi P, Ashrafizadeh M, Maiti T, 2020. Paper-based cell culture: Paving the pathway for liver tissue model development on a cellulose paper chip. ACS Appl. Bio Mater 3, 3956–3974. [DOI] [PubMed] [Google Scholar]
- Al-Ani A, Toms D, Kondro D, Thundathil J, Yu T, Ungrin M, 2018. Oxygenation in cell culture: Critical parameters are routinely not reported. PLoS One 13, e0204269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audibert G, Saunier C, du Souich P, 1993. In vivo and in vitro effect of cimetidine, inflammation, and hypoxia on propofol kinetics. Drug Metab. Dispos 21, 7–12. [PubMed] [Google Scholar]
- Bell C, Lauschke V, Vorrink S, Palmgren H, Duffin R, Andersson T, Ingelman-Sundberg M, 2017. Transcriptional, functional, and mechanistic comparisons of stem cell-derived hepatocytes, HepaRG cells, and three-dimensional human hepatocyte spheroids as predictive in vitro systems of drug-induced liver injury. Drug Metab. Dispos 45, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer C, Lewis A, Lloyd A, Phillips G, Macfarlane W, 2017. Hypoxia as a target for drug combination therapy of liver cancer. Anticancer Drugs 28, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce MW, LaBonia GJ, Hummon AB, Lockett MR, 2017. Assessing chemotherapeutic effectiveness using a paper-based tumor model. Analyst 142, 2819–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Hughes-Fulford M, 2009. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng., Part A 15, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer SM, Larson TS, Lockett MR, 2019. Tissue Papers: Leveraging paper-based microfluidics for the next generation of 3D tissue models. Anal. Chem 91, 10916–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derda R, Laromaine A, Mammoto A, Tang SK, Mammoto T, Ingber DE, Whitesides GM, 2009. Paper-supported 3D cell culture for tissue-based bioassays. Proc. Natl. Acad. Sci. USA 106, 18457–18462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahushuk A, McGarrigle B, Larsen K, Stegeman J, Olson J, 1998. Detection of CYP1A1 protein in human liver and induction by TCDD in precision-cut liver slices incubated in dynamic organ culture. Carcinogenesis 19, 1361–1368. [DOI] [PubMed] [Google Scholar]
- Dunn J, Tompkins R, Yarmush M, 1992. Hepatocytes in collagen sandwich: Evidence for transcriptional and translational regulation. J. Cell Biol 116, 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradette C, Du Souich P, 2004. Effect of hypoxia on cytochrome P450 activity and expression. Curr. Drug Metab 5, 257–271. [DOI] [PubMed] [Google Scholar]
- Gaskell H, Sharma P, Colley HE, Murdoch C, Williams DP, Webb SD, 2016. Characterization of a functional C3A liver spheroid model. Toxicol. Res 5, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Dume A, Briolotti P, Kleiber S, Raulet E, Duret C, Fabre J, Ramos J, Maurel P, Daujat-Chavanieu M, 2014. The WNT/β-catenin pathway is a transcriptional regulator of CYP2E1, CYP1A2, and aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol. Pharmacol 86, 624–634. [DOI] [PubMed] [Google Scholar]
- Gerets H, Hanon E, Cornet M, Dhalluin S, Depelchin O, Canning M, Atienzar F, 2009. Selection of cytotoxicity markers for the screening of new chemical entities in a pharmaceutical context: A preliminary study using a multiplexing approach. Toxicol. In Vitro 23, 319–332. [DOI] [PubMed] [Google Scholar]
- Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, Atienzar FA, 2012. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell. Biol. Toxicol 28, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Dial S, Shi L, Branham W, Liu J, Fang J, Green B, Dang H, Kaput J, Ning B, 2011. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos 39, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt N, Gomez-Lechon M, Houston J, Halifax D, Brown H, Maurel P, Kenna J, Gustavsson L, Lohmann C, Skonberg C, Guillouzo A, Tuschl G, Li A, LeCluyse E, Groothuis G, Hangstler J, 2007. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev 39, 159–234. [DOI] [PubMed] [Google Scholar]
- Hewitt N, Hewitt P, 2004. Phase I and II enzyme characterization of two sources of HepG2 cell lines. Xenobiotica 34, 243–256. [DOI] [PubMed] [Google Scholar]
- Hinson J, Roberts D, James L, 2010. Mechanisms of acetaminophen-induced liver necrosis. Handb. Exp. Pharmacol 196, 369–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA, 2010. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol 148, 3–15. [DOI] [PubMed] [Google Scholar]
- Ionescu C, Caira M, 2005. Drug interactions and adverse reactions, Drug Metabolism: Current Concepts. Springer, Dordrecht, pp. 295–358. [Google Scholar]
- Jancova P, Anzenbacher P, Anzenbacher E, 2010. Phase II drug metabolizing enzymes. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc, Czech Repub 154, 103–116. [DOI] [PubMed] [Google Scholar]
- Jungermann K, Kietzmann T, 2000. Oxygen: Modulation of metabolic zonation and disease of the liver. Hepatology 31, 255–260. [DOI] [PubMed] [Google Scholar]
- Kang Y, Eo J, Bulutoglu B, Yarmush M, Usta O, 2020. Progressive hypoxia-on-a-chip: An in vitro oxygen gradient model for capturing the effects of hypoxia on primary hepatocytes in health and disease. Biotechnol. Bioeng 117, 763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna J, Uetrecht J, 2018. Do in vitro assays predict drug candidate idiosyncratic drug-induced liver injury risk? Drug Metab. Dispos 46, 1658–1669. [DOI] [PubMed] [Google Scholar]
- Kietzmann T, 2017. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 11, 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lasher C, Milford L, Murali T, Rajagopalan P, 2010. A comparative study of genome-wide transcriptional profiles of primary hepatocytes in collagen sandwich and monolayer cultures. Tissue Eng., Part C 16, 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto N, Mattion NM, Estes MK, 1993. Alterations in the sequence of the gene 4 from a human rotavirus after multiple passages in HepG2. Arch. Virol 130, 179–185. [DOI] [PubMed] [Google Scholar]
- Lauschke VM, Hendriks DFG, Bell CC, Andersson TB, Ingelman-Sundberg M, 2016. Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Chem. Res. Toxicol 29, 1936–1955. [DOI] [PubMed] [Google Scholar]
- Li A, Uzgare A, LaForge Y, 2012. Definition of metabolism-dependent xenobiotic toxicity with co-cultures of human hepatocytes and mouse 3T3 fibroblasts in the novel integrated discrete multiple organ co-culture (IdMOC) experimental system: Results with model toxicants aflatoxin, cyclophosphamide and tamoxifen. Chem. Biol. Interact 199, 1–8. [DOI] [PubMed] [Google Scholar]
- Mughal M, Xi P, Yi Z, Jing F, 2017. Aflatoxin B1 invokes apoptosis via death receptor pathway in hepatocytes. Oncotarget 8, 8239–8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasrampuria D, Benet L, Sharma A, 2018. Why drugs fail in late stages of development: Case study analyses from the last decade and recommendations. AAPS J. 20, 46. [DOI] [PubMed] [Google Scholar]
- Parikh H, Pandita N, Khana A, 2015. Phytoextract of Indian mustard seeds acts by suppressing the generation of ROS against acetaminophen-induced hepatotoxicity in HepG2 cells. Pharm. Biol 53, 975–984. [DOI] [PubMed] [Google Scholar]
- Prabhakar NR, Semenza GL, 2015. Oxygen sensing and homeostasis. Physiol. 30, 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, den Braver MW, Herpers B, Terpstra V, Commandeur JNM, van de Water B, Price LS, 2014. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol 88, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Ratajewski M, Walczak-Drzewiecka A, Salkowska A, Dastych J, 2011. Aflatoxins upregulate CYP3A4 mRNA expression in a process that involves the PXR transcription factor. Toxicol. Lett 205, 146–153. [DOI] [PubMed] [Google Scholar]
- Roberts D, Bucci T, Benson R, Warbritton A, McRae T, Pumford N, Hinson J, 1991. Immunohistochemical localization and quantification of the 3-(cystein-S-yl)-acetaminophen protein adduct in acetaminophen hepatotoxicity. Am. J. Pathol 138, 359–371. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Antona C, Ingelman-Sundberg M, 2006. Cytochrome P450 pharmacogenetics and cancer. Oncogene 25, 1679–1691. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schyschka L, Martinez Sanchez J, Wang Z, Burkhardt B, Muller-Vieira U, Zeilinger K, Bachmann A, Nadalin S, Damm G, Nussler A, 2013. Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity. Arch. Toxicol 87, 1581–1593. [DOI] [PubMed] [Google Scholar]
- Sonna L, Cullivan M, Sheldon H, Pratt R, Lilly C, 2003. Effect of hypoxia on gene expression by human hepatocytes (HepG2). Physiol. Genomics 12, 195–207. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld J, Barwick J, Guzelian P, Evans R, 2002. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc. Natl. Acad. Sci. USA 99, 13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida A, Fukuen S, Yamamoto I, Matsuda H, Naohara M, Azuma J, 2000. Quantitative analysis of constitutive and inducible CYPs mRNA expression in the HepG2 cell line using reverse transcription-competitive PCR. Biochem. Biophys. Res. Commun 267, 756–760. [DOI] [PubMed] [Google Scholar]
- Vrba J, Havlikova M, Gerhardova D, Ulrichova J, 2014. Palmatine activates AhR and upregulates CYP1A activity in HepG2 cells but not in human hepatocytes. Toxicol. In Vitro 28, 693–699. [DOI] [PubMed] [Google Scholar]
- Wang K, Shindoh H, Inoue T, Horii I, 2002. Advantages of in vitro cytotoxicity testing by using primary rat hepatocytes in comparison with established cell lines. J. Toxicol. Sci 27, 229–237. [DOI] [PubMed] [Google Scholar]
- Westerink W, Schoonen W, 2007. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol. In Vitro 21, 1592–1602. [DOI] [PubMed] [Google Scholar]
- Whitman NA, McIntosh JC, Penley JB, Lockett MR, 2016. Microfabricated devices for studying the metabolism and cytotoxicity of drug candidates. Curr. Pharm. Biotechnol 17, 755–771. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Sasaki Y, Terasaki N, Kawataki T, Takekawa K, Iwase Y, Shimizu T, Sanoh S, Ohta S, 2018. Comparison of drug metabolism and its related hepatotoxic effects in HepaRG, cryopreserved human hepatocytes, and HepG2 cell cultures. Biol. Pharm. Bull 41, 722–732. [DOI] [PubMed] [Google Scholar]
- Zeigerer A, Wuttke A, Marsico G, Seifert S, Kalaidzidis Y, Zerial M, 2017. Functional properties of hepatocytes in vitro are correlated with cell polarity maintenance. Exp. Cell Res 350, 242–252. [DOI] [PubMed] [Google Scholar]
- Zhao YJ, Chen YH, Hu Y, Wang JH, Xie XW, He GX, Chen HS, Shao QX, Zeog H, Zhang HH, 2018. Genomic alterations across six hepatocellular carcinoma cell lines by panel-based sequencing. Transl. Cancer Res 7, 231–239. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.