Abstract
Campylobacter jejuni is a bacterial pathogen that is a common cause of enteritis in humans. We identified a previously uncharacterized type of sensory domain in the periplasmic region of the C. jejuni chemoreceptor Tlp10, termed the DAHL domain, that is predicted to have a bimodular helical architecture. Through two independent ligand-binding sites in this domain, Tlp10 responded to molecular aspartate, isoleucine, fumarate, malate, fucose, and mannose as attractants, and to arginine, galactose and thiamine as repellents. Tlp10 also recognized glycan ligands when present as terminal and intermediate residues of complex structures, such as the fucosylated human ganglioside GM1 and Lewisa antigen. A tlp10 mutant strain lacking the ligand-binding sites was attenuated in its ability to colonize avian caeca and to adhere to cultured human intestinal cells, indicating the potential involvement of the DAHL domain in host colonization and disease. The Tlp10 intracellular signaling domain interacted with the scaffolding proteins CheV and CheW, which couple chemoreceptors to intracellular signaling machinery, and with the signaling domains of other chemoreceptors, suggesting a key role for Tlp10 in signal transduction and incorporation into sensory arrays. We identified the DAHL domain in other bacterial signal transduction proteins, including the essential virulence induction protein VirA from the plant pathogen Agrobacterium tumefaciens. Together, these results suggest a potential link between Tlp10 and C. jejuni virulence.
Introduction
Bacteria have evolved a range of mechanisms to sense chemical changes in their environment and alter their positions accordingly, to avoid harmful substances and unfavorable conditions and move towards nutrient sources and new niches. Motile bacteria sense and move along a chemical gradient using chemosensory receptors that recognize various ligands and trigger an intracellular signal transduction cascade that ultimately directs the cell toward an attractant or away from a repellent (1, 2). One such organism, the intestinal pathogen Campylobacter jejuni, is a leading bacterial cause of gastroenteritis around the globe and reported to affect approximately 10% of the world’s population annually (3–5). The bacterium is commonly found as a commensal bacterium in the gastrointestinal tracts of poultry and other birds. Consumption of undercooked poultry or untreated water can give rise to human infections that lead to severe enteritis with fever, abdominal pain, and diarrhea (4, 6). The mechanisms of molecular pathogenesis and virulence of C. jejuni infections are not well understood; however, motility and chemotaxis are essential for C. jejuni pathogenicity (7, 8).
The mechanism of bacterial chemotaxis is best understood in the model organism Escherichia coli (1, 9), but its principal components are conserved among motile bacteria and archaea (10). Chemoreceptors, also known as the methyl-accepting chemotaxis proteins (MCPs), or transducer-like proteins (Tlps), are the first cellular components of chemosensory signaling pathways. Signaling by transmembrane chemoreceptors is initiated through a periplasmic ligand-binding domain (LBD) and propagated to a cytoplasmic signaling domain, which is connected to the histidine kinase CheA through a docking protein CheW. After undergoing autophosphorylation, CheA transfers the phosphoryl group to the response regulator CheY, which controls the direction of flagellar motor rotation. Phosphorylated CheY interacts with the flagellar switch protein, leading to a change in the direction of flagellar rotation. Chemoattractants inhibit a change in flagellar rotation, thus keeping the bacterium moving along its current trajectory, whereas chemorepellants cause the direction of the motor to switch, thus changing the direction of movement. The phosphatase CheZ, the methyltransferase CheR, and the methylesterase CheB contribute to signal termination and adaptation mechanisms. Despite the core components of the chemotaxis pathway in C. jejuni being similar to those in E. coli, there are some notable differences. The C. jejuni pathway belongs to the evolutionary class F3, whereas the one from E. coli belongs to the F7 class (10), and it contains an additional adaptor protein, CheV, which is missing from E. coli (11). In comparison to E. coli strain K-12, substr. MG1655, which has five chemoreceptors, C. jejuni subsp. jejuni strain NCTC 11168 (ATCC 700819) has eleven chemoreceptors. C. jejuni chemoreceptors can be classified into three groups (A, B and C) based on structural similarities to other organisms (12). The group that senses ligands external to the cell is designated group A and includes Tlp1, Tlp2, Tlp3, Tlp4, Tlp7, Tlp10, and Tlp11. Group A receptors share a common three-domain structure with other external sensory receptors – a periplasmic sensory domain, a cytoplasmic signaling domain, and a transmembrane domain linking the two. Four of the Group A chemoreceptors have been characterized previously: Tlp1 (CcaA) responds to aspartate (13); Tlp3 (CcmL) senses multiple ligands (14); Tlp7 acts as a formic acid sensor (15); and Tlp11 (CcrG) is a galactose chemoreceptor (16). All four of these chemoreceptors contain a dCache_1 (double calcium channels and chemotaxis receptors) domain (17) as their LBD, and crystal structures have been solved for Tlp1 (18) and Tlp3 (19). Although the chemoreceptor repertoire varies among C. jejuni strains, Tlp10 has been found in all strains tested (20). This chemoreceptor has been reported to modulate responses to aspartate and fumarate in C. jejuni strain 81–176; however, the nature of ligand interaction has not been established (21), and the LBD does not match to known domain models (22).
Here, we show that the Tlp10 chemoreceptor contains a previously uncharacterised sensory LBD. This LBD recognized multiple ligands of different classes, including complex glycans, and was found in various bacterial signal transduction proteins, including the histidine kinase VirA from Agrobacterium tumefaciens. We further demonstrated that this chemoreceptor interacted with other chemoreceptors and with the adaptor protein CheV. Finally, we reveal that removal of Tlp10 significantly reduced the adherence of C. jejuni to human cells in vitro and colonization of the avian host in vivo, suggesting a role for this chemoreceptor in host-bacterial interactions.
Results
Tlp10 has an uncharacterized LBD
Protein sequence analysis of Tlp10 (NCBI accession number YP_002343491.1, locus tag Cj0019c) revealed that it has a classical transmembrane topology with two transmembrane regions, a predicted large periplasmic region, and a cytoplasmic signaling domain (Pfam family: MCPsignal, MCP signalling domain, accession number PF00015) (Fig. 1A). Using previously described hidden Markov models (23), we assigned Tlp10 to the 40H chemoreceptor class based on it having 40 helical heptads in the cytoplasmic signaling domain.
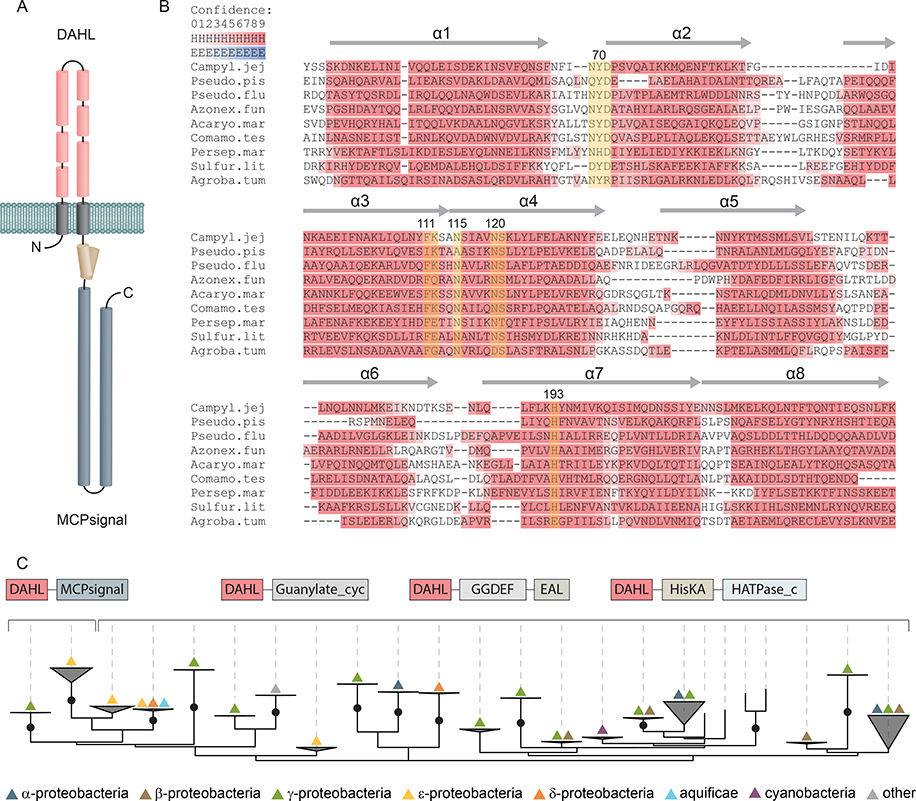
Fig. 1. Secondary structure prediction and phyletic distribution of the DAHL domain.
(A) Schematic representation of Tlp10 topology with the predicted architecture of the DAHL domain as the LBD, the transmembrane region, and the intracellular MCP signaling domain. (B) MSA and secondary structure prediction of periplasmic DAHL domains from various bacterial species. Predicted α-helical regions are labelled and highlighted in red. Conserved amino acid positions are highlighted in yellow. Representative sequences were selected from different clades on the phylogenetic tree. Campyl.jej: C. jejuni, Tlp10 WP_002865311.1; Pseudo.pis: Pseudoalteromonas piscicida, WP_099642651.1; Pseudo.flu: Pseudomonas fluorescens, WP_003172050.1; Azonex.fun: Azonexus fungiphilus, WP_121456889.1; Acaryo.mar: Acaryochloris marina, WP_012165516.1; Comamo.tes: Comamonas testosterone, WP_003076421.1; Persep.mar: Persephonella marina, WP_012676181.1; Sulfur.lit: Sulfurovum lithotrophicum, WP_052746078.1; Agroba.tum: A. tumefaciens, VirA WP_012478065.1. (C) Maximum likelihood phyletic distribution and protein architecture variations of DAHL domain–containing proteins with bootstrap support based on 500 replicates. Neighboring domains are indicated according to Pfam database nomenclature: MCPsignal, MCP signaling domain, PF00015; Guanylate_cyc, adenylate and guanylate cyclase catalytic domain, PF00211; GGDEF, diguanylate cyclase, PF00990; EAL, diguanylate phosphodiesterase, PF00563; HisKA, histidine kinaseA (phospho-acceptor) domain, PF00512; HATPase_c; -, DNA gyrase B-, phytochrome-like ATPases, PF02518. Bootstrap values more than 70% are indicated by black circles.
Searches of Pfam and Conserved Domain Database (CDD) using the Tlp10 periplasmic region (residues 33 to 238), which should contain the LBD, as a query revealed no similarities to any known domain. Fold recognition for this region using Phyre2 also produced hits with low confidence (<42% probability) and low sequence coverage (28% of sequence has been modelled). Profile-profile searches using HHpred (24) returned hits to a partial human Nucleoporin_FG2 (Pfam accession number PF15967.5) domain and a structure for the nuclear pore complex protein Nup155 (PBD accession number 5IJO_T) with low confidence support (<61% probability). We also subjected sequences of predicted periplasmic regions from several Tlp10 LBD-containing proteins from other bacterial species to similar searches that returned the same hits (table S1). Inclusion of transmembrane (TM) regions to the initial sequence query increased the HHpred probability score of homology to Nup155 up to 95%. A common feature of Nup155 and nucleoporin proteins is a preponderance of α-helices connected by short loops. Consequently, we performed secondary structure prediction using Ali2D and Quick2D tools. The periplasmic regions of Tlp10 and homologous sequences were all predicted to have an extensive α-helical character (Fig 1B); however, despite a high amount of helix content, none of the homologues to Tlp10 LBD sequences subjected to a search matched any prokaryotic helical-rich domains, including signal transduction–specific domains, such as four helix bundle (4HB_MCP_1) (17), nitrate and nitrite sensing (NIT) (25), cyclases/histidine kinases associated sensing extracellular (CHASE3) (26), and helical bimodular (HBM) (27).
BLAST search of the NCBI non-redundant database initiated with a predicted periplasmic region of Tlp10 identified more than 200 proteins that contained similar regions – all in a small subset of Campylobacter and Helicobacter strains. To retrieve more homologs, we used the same query in the exhaustive PSI-BLAST search, which identified more than 1700 sequences. We then prepared multiple sequence alignment (MSA) of the entire periplasmic regions from all homologous sequences found by PSI-BLAST, edited it, and used it to construct an HMM model for this newly uncovered domain, which we designated as the DAHL (double all-helical ligand-binding) domain. We deposited the DAHL domain model into the Pfam database (accession number PF19443).
The DAHL domain is found in diverse signal transduction proteins in Proteobacteria
We built a maximum likelihood phylogenetic tree from acquired MSA, with bootstrap support based on 500 replicates (Fig 1C). Examination of the phyletic distribution of the DAHL domain–containing proteins revealed that they are found exclusively in bacterial species, with no detectable homologs in archaea or eukaryotes. The majority of the DAHL domain–containing proteins belonged to alpha-, beta-, gamma- and epsilonproteobacteria. A few DAHLs were found in cyanobacteria (for example, Leptolyngbya, Acaryochloris, Moorea, and Beggiatoa spp.) and even more rarely in Aquificae spp., where its occurrence was restricted to Persephonella species.
Careful analysis of the domain architectures of the DAHL-containing proteins revealed that this domain mainly occurs in two types of sensory proteins: histidine kinases and diguanylate cyclases and phosphodiesterases (Fig 1C, table S1 and data file S1). A few DAHL domains were found in adenylate cyclases. The DAHL domain was found in combination with an MCP domain mostly in epsilonproteobacteria (Campylobacter and Helicobacter spp.), exemplified by the chemoreceptor Tlp10. We also detected the DAHL domain in the histidine kinase VirA from the plant pathogen A. tumefaciens and related histidine kinases from plant-associated alphaproteobacteria (Rhizobium, Sinorhizobium, Mesorhizobium, Ensifer spp., etc.) and Xanthomonas species.
Tlp10 binds specifically to multiple classes of ligands, including amino acids and glycans.
We screened small molecule and glycan arrays containing amino and organic acids, salts, and over 400 glycans ranging from monosaccharides to dodecasaccharides, as well as complex glycans such as mucins (tables S2 and S3), with the purified predicted LBD of Tlp10 (amino acids 33–238, also designated as the DAHL domain) from C. jejuni reference strain NCTC-O 11168-O (Tlp10LBD). Tlp10LBD bound to a diverse range of compounds with a total of 14 interacting ligands, which included nine amino and organic acids and five monomeric glycans (Table 1, fig. S1, A to M). Tlp10LBD also interacted with complex glycans having these same five glycans (galactose, rhamnose, mannose, sialic acids, and fucose) in their terminal structures, such as those present on the human ganglioside GM1, Lewisa antigen, and other mucosal glycans (Table 2, table S4). We used surface plasmon resonance (SPR), which quantifies ligand binding by measuring changes in the refractive index of an immobilized protein when it is exposed to ligand, to measure the KD of Tlp10LBD with the ligands identified from the microarray analyses. SPR analyses showed that the highest affinity Tlp10LBD interaction was with galactose (KD<10 μM; Table 1), but strong interactions (KD<50 μM; Table 1) were also observed for isoleucine, arginine, aspartate, purine, thiamine, mannose, rhamnose, and sialic acid.
Table 1. Summary of monomeric ligand binding and SPR analysis for Tlp10LBD.
The data represents mean values (± SD) of three independent experiments (n = 3).
| Ligands | array | SPR | KD μM) |
|---|---|---|---|
| arginine | + | + | 27.9±3 |
| aspartate | + | + | 32.3±10 |
| isoleucine | + | + | 37.9±7 |
| thiamine | + | + | 59.4±13 |
| purine | + | + | 21±6 |
| fumarate | + | + | 282.1±68 |
| malate | -/+ | + | 168±19 |
| α-ketoglutarate | -/+ | + | 403.4±13 |
| galactose | + | + | 2.9±1.2 |
| mannose | + | + | 39.6±9 |
| rhamnose | + | + | 11.1±2 |
| fucose | + | + | 30±4.1 |
| sialic acid | + | + | 29.3±6.9 |
positive binding
negative binding
weak binding.
Table 2. Effect of substitution mutations in Tlp10LBD on ligand binding affinity.
KDvalues for binding of the listed ligands to WT Tlp10 LBD and the Y70A, N115A, N120A, and H193A substitution mutants.
| Ligand | Tlp10 LBD Wild-type | Tlp10LBDY70A | Tlp10LBDN115A | Tlp10LBDN120A | Tlp10LBDH193A |
|---|---|---|---|---|---|
| arginine | 27.9±3 | 19.1±12 | 41.9±12 | 50.1±8* | 29±8 |
| isoleucine | 37.9±7 | 28.1±6 | 48.13±5 | 51.2±1.2 | 30.8±7 |
| aspartate | 32.3±10 | 38.9 ±11 | 29.5±8 | 48±7 | 30.9±9 |
| purine | 21±6 | 16±1 | 29±8 | 34±3* | 19±2 |
| fumarate | 282.1±68 | 185.1±25 | 242.1±30 | 316.9±34 | 213±59 |
| malate | 168±19 | 184±14 | 176±10 | 215±31 | 161±23 |
| α-ketoglutarate | 403.4±13 | 385±40 | 630±27* | 580±43* | 370±63 |
| thiamine | 59.4±13 | 62±17 | 49.2±10 | 58.27±9 | 50.6±6 |
| galactose | 2.9±1.2 | 3.1±2 | 7±0.34* | 9±0.8* | 2±1.74 |
| mannose | 39.6±9 | 43.3±3 | 52.1±10 | 46.4±12 | 32.1±12 |
| rhamnose | 11.1±2 | 8.2±3 | 18.2±1.9 | 17.9±3.7 | 9.8±0.95 |
| fucose | 30±4.1 | 34±7 | 22±2* | 19.2±4* | 28±3 |
| sialic acid | 29.3±6.9 | 24.1±7 | 34.9±13 | 37.1±6 | 20.4±9 |
The asterisk (*) indicates significant difference in the substitution mutant compared to Tlp10 LBD wild-type (P<0.05). The data represent mean values (± s.d) of three independent experiments (n = 3). Asterisks indicate significant differences (P < 0.05; t-test).
Deletion of the Tlp10 sensory domain alters ligand-directed chemotactic motility of C. jejuni.
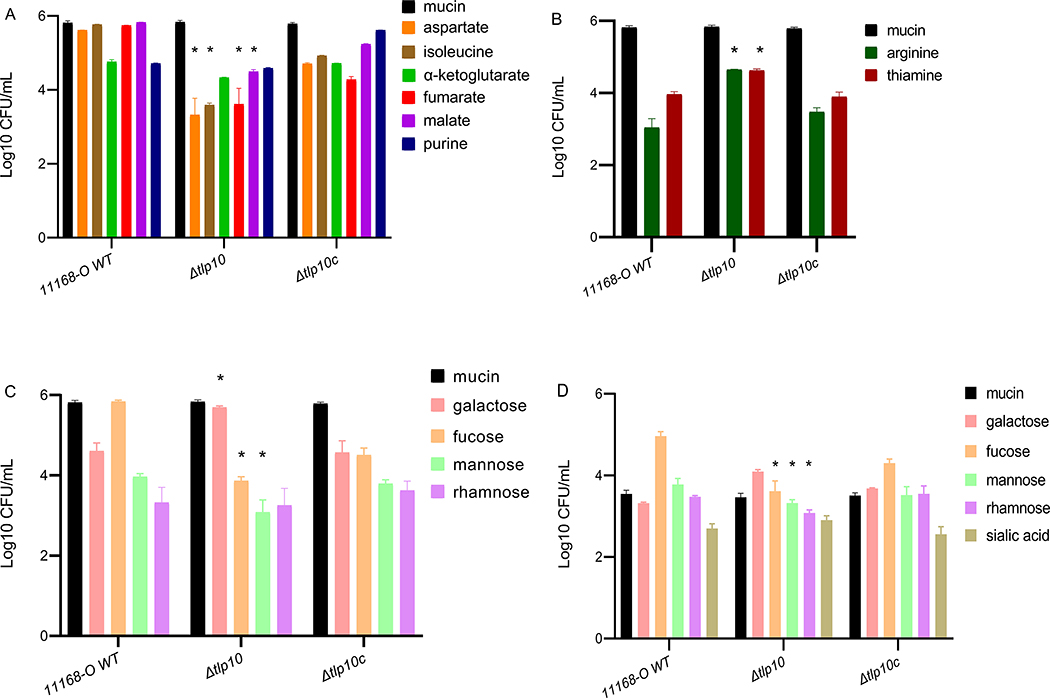
Nutrient-depleted chemotaxis assays (13, 16) indicated that two amino acids (aspartate and isoleucine), two organic acids (fumarate and malate) and two sugars (fucose and mannose) interacted with Tlp10 as attractants (Fig. 2A, table S5). Compared to the wild-type 11168-O strain of C. jejuni, an isogenic strain in which 345 nucleotides encompassing the sensory portion of the LBD of Tlp10 were replaced with a non-polar KmR cassette (Δtlp10LBD) showed a significant reduction in migration towards these ligands. Chemotaxis assays further indicated that Tlp10 mediated a repellent response to arginine and thiamine, because the Δtlp10LBD isogenic strain showed an increase in migration towards these compounds compared to the wild-type strain (Fig. 2B). One-log increase in migration of Δtlp10LBD bacteria towards galactose was also observed (Fig. 2C), indicating that galactose is likely to be sensed as a repellent by Tlp10. Migration towards purine, α-ketoglutarate, or rhamnose was not significantly affected in the Δtlp10LBD strain.
Fig. 2. Chemotaxis assays with wild-type and Tlp10 mutant C. jejuni.
(A and B) Wild-type C. jejuni strain 11168-O (11168-O WT), the isogenic Tlp10LBD mutant (Δtlp10), and the complemented mutant (Δtlp10c) were subjected to chemotaxis assays measuring the movement of starved cells towards test ligands considered attractants (A) and ligands considered repellents (B). (C) Chemotaxis to the monosaccharide ligands galactose, fucose, mannose, and rhamnose. The universal attractant mucin was used a positive control. The C. jejuni non-motile mutant (81116ΔflaA/flaB) and PBS were used as negative controls (Table S5). (D) μ-slide chemotaxis assays testing motility within a gradient of the test ligand were performed using galactose, fucose, mannose, rhamnose, and sialic acid as the test compounds. All data (Log 10 CFU/mL) represent three independent experiments, each performed in triplicate (n = 3). Asterisks indicate significant differences (P < 0.05; t-test).
Considering that galactose has been shown to be a general attractant for wild-type C. jejuni and that C. jejuni strains that have Tlp11, the galactose receptor, display increased chemotaxis toward galactose, we tested wild-type bacteria and the Δtlp10LBD isogenic mutant strain in μ-slide cell enumeration chemotaxis assays to confirm the responses to the glycan chemoeffectors. The μ-slide assay allows enumeration and visualization of bacterial movement in suspension. In this assay, bacteria move from one chamber, through a shallow channel to another chamber, containing a chemoeffector. This assay showed that migration of Δtlp10LBD cells toward galactose was increased (Fig. 2D, table S5), similar to that observed for nutrient depletion chemotaxis assays, confirming that Tlp10 mediates a repellent response to galactose. Migration of Δtlp10LBD cells towards mannose and fucose was decreased, confirming that these ligands also elicit an attractant response through Tlp10 (table S5). No significant differences in cells migration were detected toward rhamnose and sialic acid, compared with wild-type C. jejuni 11168-O. In all cases, wild-type chemotactic motility was restored in a complemented mutant strain (Δtlp10LBDc). The C. jejuni 11168–O wild-type, Δtlp10LBD, and Δtlp10LBDc strains showed the same chemotactic responses toward the universal attractant mucin, demonstrating that deletion of Tlp10LBD did not induce a general motility defect (Fig. 2, A to D).
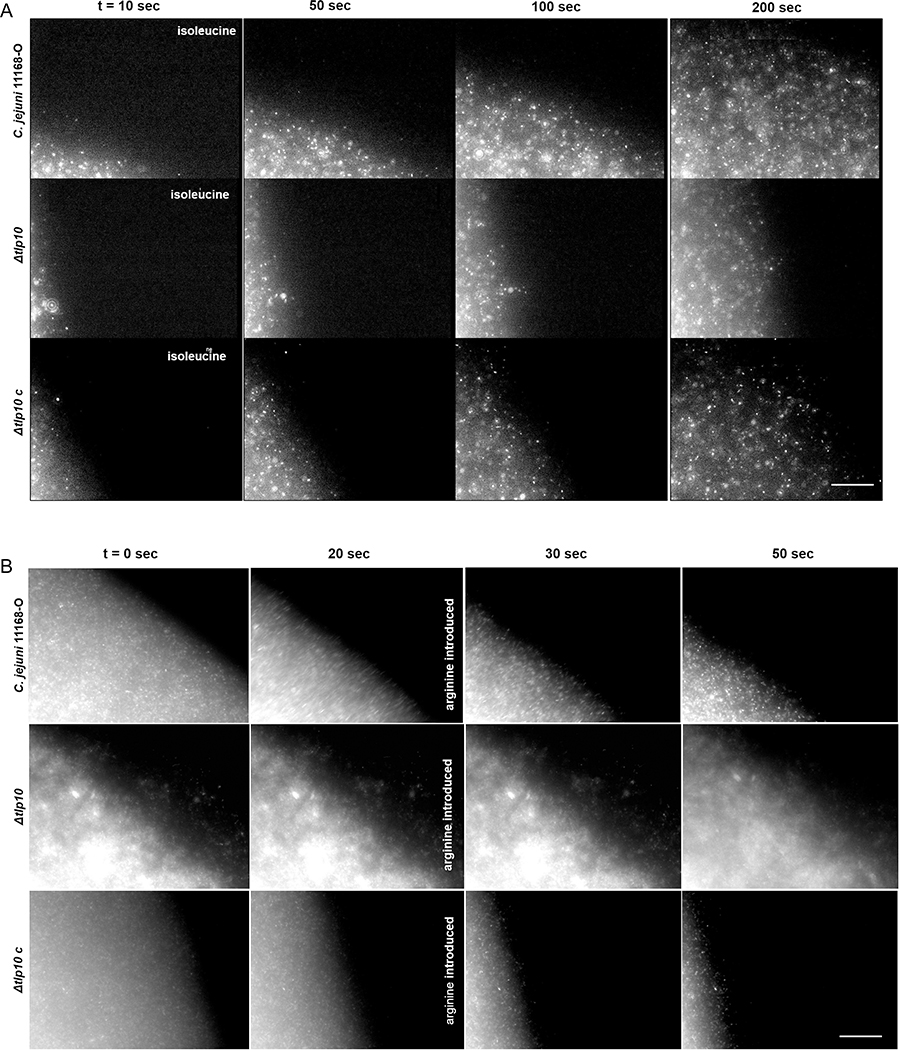
To further elucidate the Tlp10-mediated chemotactic responses of C. jejuni11168-O, we visualized the wild-type, isogenic Δtlp10LBD mutant, and complemented mutant Δtlp10LBDc strains moving up or down gradients of selected chemoeffectors in μ-slide chemotaxis assays. Whereas the migration of wild-type cells toward isoleucine was rapid, the migration of Δtlp10LBD toward isoleucine strain was reduced but restored by complementation (Fig. 3A and movie S1), demonstrating that isoleucine can induce a chemoattractant response in C. jejuni11168-O through Tlp10. To demonstrate that these strains can sense changes in the environment, wild-type cells that had begun to migrate toward a solution of isoleucine were challenged with arginine. The cells rapidly reversed the direction of migration away from the testing solution when arginine was added (Fig. 3B, movie S2). This reversal was not observed when PBS control was added instead of arginine (movie S3). The isogenic Δtlp10LBD mutant cells did not show migratory reversal, demonstrating that Tlp10 mediated a chemorepellent response to arginine in C. jejuni11168-O.
Fig. 3. Migration of 11168-O WT, Δtlp10, and Δtlp10c in response to chemoeffectors.
Time-lapse imaging of fluorescently labelled C. jejuni 11168-O (WT), Δtlp10, and Δtlp10c cells migrating from the left chamber toward the right chamber along a chemoeffector gradient on μ-Slide Chemotaxis chamber. (A) Attractant response toward 10 mM isoleucine. (B) Response of cells that had partially migrated toward 10 mM isoleucine before the introduction of 10 mM arginine into the right chamber at 20 sec. This figure is representative of five independent experiments (n=5). Scale bars, 100 μm.
The Tlp10 LBD has two ligand-binding sites
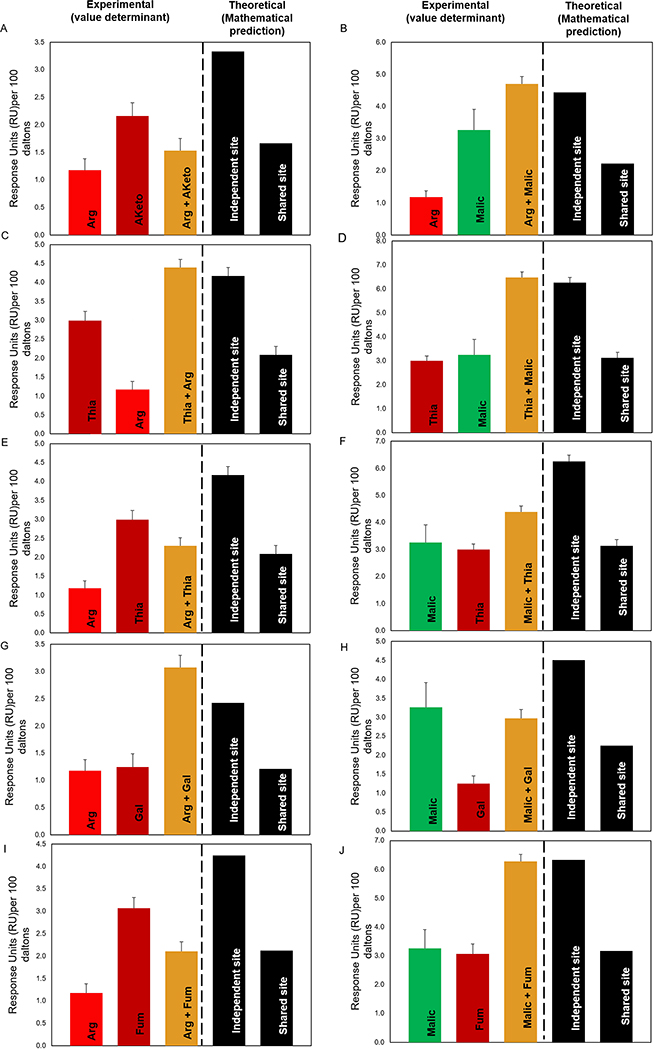
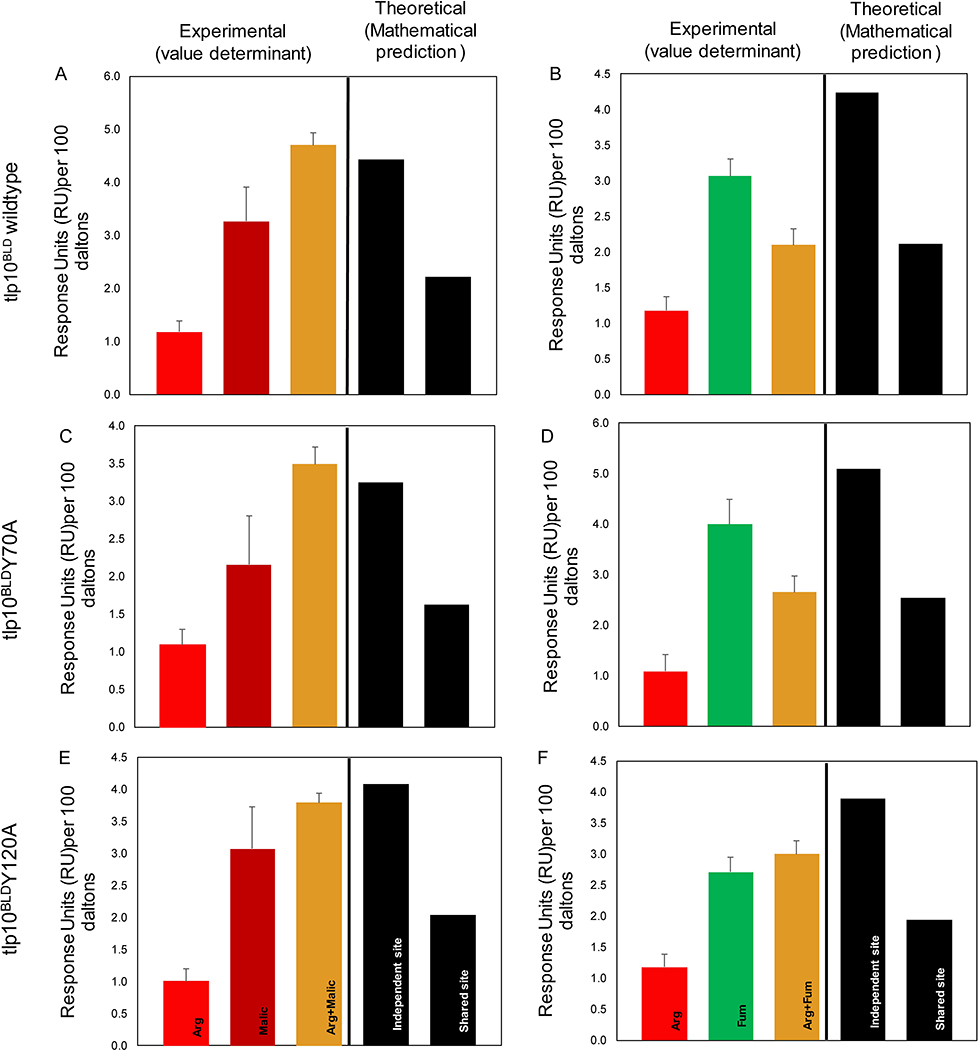
We used competition A-B-A SPR analysis to determine how Tlp10LBD reacted to the presence of multiple ligands, applied in specific sequences, in order to determine whether the ligands interacted with a single or multiple sites on the protein’s sensory domain. The binding status of the ligands to a protein is classified as: i) Independent site (additive, or cumulative effect), where the ligands are not competing for the same binding site and therefore must bind to independent binding sites; ii) Shared site, where the ligands compete for binding to the same binding site; and iii) Preferential Shared site, where the ligands can bind to the same binding site, but one of the ligands binds with higher affinity than the other when in equilibrium. In the A-B-A analysis, the compounds are used at a concentration 10x their respective KD and flowed over the chip with the immobilized protein (Tlp10LBD here) in the following order: analyte 1 (A) (competitor) alone, followed by analyte 1 (competitor) plus analyte 2 (B), followed by analyte 1 (A) (competitor) alone. The assay is designed to detect the presence or absence of a cumulative response when the second analyte (B) is flown across the immobilized protein saturated with the first analyte (A). The experimental value (value determinant) represents actual response units (RU) values, and the theoretical value is determined mathematically.
Competition SPR analysis of the wild-type Tlp10LBD interrogated the binding of arginine (Arg), malate (Malic), thiamine (Thia), galactose (Gal), fumarate (Fum), and α-ketoglutarate (Aketo) to the wild-type Tlp10LBD and identified two clear binding sites, which we referred to as site A and site B, with four types of analyte-LBD interactions (Fig. 4A–4J and table S6). The first type was analyte binding to a shared site, where binding to both analytes existed in equilibrium, resulting in an average of the expected response units (RU) for each analyte. An example of this is the interaction of arginine and α-ketoglutarate (Fig. 4A) with the same site, which we designated site A. The second type of binding interaction was to a single specific site, site A or site B, such as that observed for arginine and malate, where the addition of the two analytes in any combination resulted in a summation of the responses (Fig. 4B). This indicated that each compound was binding independently to one of two separate sites. The third mode was preferential binding to a shared site, where the two analytes bound the same site but when both were present, one of the analytes out-competed the other, resulting in preferential binding of one analyte at that site. An example of this was the binding of galactose and malate to site B (Fig. 4H). A final type of interaction was exemplified by thiamine binding, where it clearly interacted with both sites because its binding was affected by both arginine and malate. The competitive binding results depended on the order of analyte addition (Fig. 4 C, D, E and F), indicating that although thiamine was able to bind to both sites A and B, it preferentially bound to site B (table S6).
Fig. 4. Competition SPR analysis of Tlp10LBD.
Competition analysis of the binding of arginine (Arg), malate (Malic), thiamine (Thia), galactose (Gal), fumarate (Fum), and α-ketoglutarate (Aketo) to WT Tlp10LBD immobilized on a sensor chip. (A) Responses to Arg only, Aketo only, and Arg after saturation with α-ketoglutarate. (B) Responses to Arg only, Malic only, and Arg following saturation with malate. (C) Responses to Thia only, Arg only, and Thia following saturation with arginine. (D) Responses to Thia only, Malic only, and Thia following saturation with malate. (E) Responses to Arg only, Thia only, and Arg following saturation with thiamine. (F) Responses to Malic only, Thia only, and Malic following saturation with Thia. (G) Responses to Arg only, Gal only, and Arg following saturation with Gal. (H) Responses to Malic only, Gal only, and Malic following saturation with Gal. (I) Responses to Arg only, Fum only, and Arg following saturation with Fum. (J) Responses to Malic only, Fum only, and Malic following saturation with Fum. Results are presented for analyte A alone and analyte B alone (red, dark red, and green), and competition assay between analytes A and B (gold). The theoretical values for independent binding of the two analytes to separate sites and for binding to a shared site are shown as response units (RU) values based on mathematical theory (black). All response data were normalized to 100 Da molecular weight for each analyte allowing direct comparison of responses.
These analyses enabled assignment of all Tlp10 chemoeffectors to individual binding sites. Arginine, fumarate, α-ketoglutarate, isoleucine, and purine bound specifically to site A, whereas malate, galactose, fucose, rhamnose, sialic acid, and mannose bound specifically to site B. Aspartate and thiamine bound to both site A and site B with a preference for site B.
To further characterize which residues of Tlp10 were involved in ligand binding, we examined the conservation pattern on the MSA of the Tlp10 periplasmic region homologs, constructed earlier (Fig 1B, data file S1). We identified several highly conserved amino acid residues: Tyr70, Asn115, Asn120, and His193. In-depth sequence analysis revealed that most of the conserved residues were in the N-terminal half of the Tlp10LBD, which implies a key functional role for this region. We individually replaced Tyr70, Asn115, Asn120, and His193 of Tlp10LBD by site-directed mutagenesis with alanine to yield four mutant proteins: Tlp10LBD Y70A, Tlp10LBD N115A, Tlp10LBD N120A, and Tlp10LBD H193A. The mutations did not affect the folding of the protein as determined by the far-UV CD (circular dichroism) spectra (fig. S2) Substitutions N115A and N120A reduced the affinity of Tlp10LBD for arginine, α-ketoglutarate, isoleucine, purine, and galactose, whereas they significantly increased the affinity for fucose compared with wild-type Tlp10LBD. Substitutions Y70A or H193A had little effect on the affinity of any ligand binding (Table 2).
Site-directed mutants also altered the competitive interactions between chemoeffectors (Fig. 5A to 5F). For example, Tlp10LBD N120A shifted the interactions between arginine and fumarate, from a shared equilibrium to an equilibrium that favored the binding of fumarate (Fig. 5F) compared to the equilibrium interactions observed for the wild-type Tlp10LBD (Fig. 5B), whereas no changes occurred with Tlp10LBD Y70A (Fig. 5D).
Fig. 5. Effect of point mutations of Tlp10LBD on ligand binding.
SPR competition analysis of the binding of arginine (Arg), malate (Malic), and fumarate (Fum) to immobilized WT and mutant Tlp10LBD proteins. (A) Responses of Tlp10LBD to Arg only, Malic only, and Arg following saturation with Malic. (B) Responses of Tlp10LBD to Arg only, Fum only, and Arg following saturation with Fum. (C) Responses of Tlp10LBDY70A to Arg only, Malic only, and Arg following saturation with Malic. (D) Responses of Tlp10LBDY70A to Arg only, Fum only, and Arg following saturation with Fum. (E) Responses of Tlp10LBDN120A to Arg only, Malic only, and Arg following saturation with Malic. (F) Responses of Tlp10LBDN120A to Arg only, Fum only, and Arg following saturation with Fum. Results are presented for analyte A alone and analyte B alone (red, dark red, and green), and competition assay between analytes A and B (gold). The theoretical values for independent binding of the two analytes to separate sites and for binding to a shared site are shown as response units (RU) values based on mathematical theory (black). All response data were normalized to 100 Da molecular weight for each analyte allowing direct comparison of responses.
The other site-directed mutants showed similar changes in interactions between competing ligands across both binding sites, providing clear evidence of the existence of two binding sites and specific chemoeffector binding in each site, with Asn115, Asn120 being involved in the first ligand-binding site (tables S7–S10).
The signaling domain of Tlp10 interacts with other proteins and protein domains of the C. jejuni chemotaxis signaling pathway
To further investigate the role of Tlp10 in the chemotaxis signal transduction pathway in C. jejuni11168-O, we analyzed the predicted cytoplasmic signaling domain of Tlp10 (amino acids125 – 197), Tlp10sig, using the yeast two-hybrid protein-protein interaction system (13, 14, 28) (tables S11 and S12). Tlp10sig showed a strong interaction with the full-length CheV protein and with the CheW-like domain of CheV and did not depend on whether Tlp10sig was used as the bait or the prey. A weak interaction was detected between Tlp10sig and CheW only when Tlp10sig was used as the bait.
Analysis of protein-protein interactions among the signaling domains of the group A Tlps showed that Tlp10sig was the only one capable of interacting with the signaling domains of other Group A Tlps, (Tlp1, Tlp2, Tlp3, Tlp4, and Tlp7), which sense the external chemical environment (table S11). Tlp2, Tlp3, and Tlp4 are orthologous proteins with identical signaling domains and differing sensory domains, unique to C. jejuni. Tlp10sig showed interactions with the signaling domain of Tlp1 and the signaling domain common to Tlp2, Tlp3, and Tlp4 in reciprocal combinations of bait and prey, whereas an interaction was detected between the Tlp10 and Tlp7 signaling domains only when Tlp7sig was used as the bait. The signaling domain of Tlp10 was found to interact with itself, suggesting that it forms dimers – and perhaps higher-order structures – through interactions in the cytoplasmic signaling domain.
Mutation of the Tlp10 LBD domain reduces biofilm formation and interaction with human cells
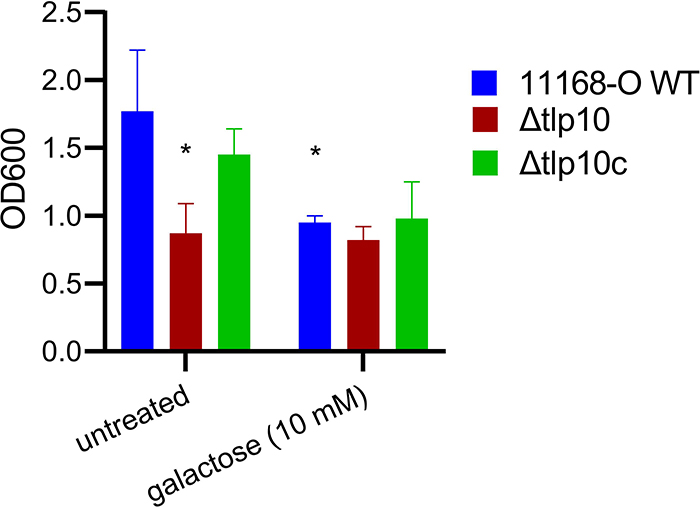
Because mutations in chemoreceptor Tlp3 enhance, whereas mutations in Tlp11 reduce, the ability of C. jejuni11168-O to make biofilms (14, 16), we compared biofilm formation by the C. jejuni11168-O wild-type and Δtlp10LBD isogenic strains using a modified crystal violet assay (29). We observed a 1.5-fold decrease in biofilm formation by Δtlp10LBD isogenic mutant cells compared to wild-type cells, with wild-type biofilm formation restored in the complemented strain, Δtlp10LBDc, (Fig. 6). The addition of free molecular galactose reduced biofilm formation by wild-type C. jejuni 11168–O by up to 46% compared to untreated cells, whereas the Δtlp10LBD mutant showed no significant change in biofilm formation in the presence of galactose, indicating that mutant cells could no longer respond to the presence of galactose in the environment.
Fig. 6. Reduced biofilm formation of Δtlp10 C. jejuni.
Quantification of biofilm formation by wild-type C. jejuni strain 11168-O (11168-O WT), the isogenic Tlp10LBD mutant (Δtlp10), and Δtlp10 complemented \ with full-length Tlp10 (Δtlp10c), with and without 10 mM galactose. The data represents three independent experiments, each performed in triplicate (n = 3). Asterisks indicate significant differences (P < 0.05; t-test).
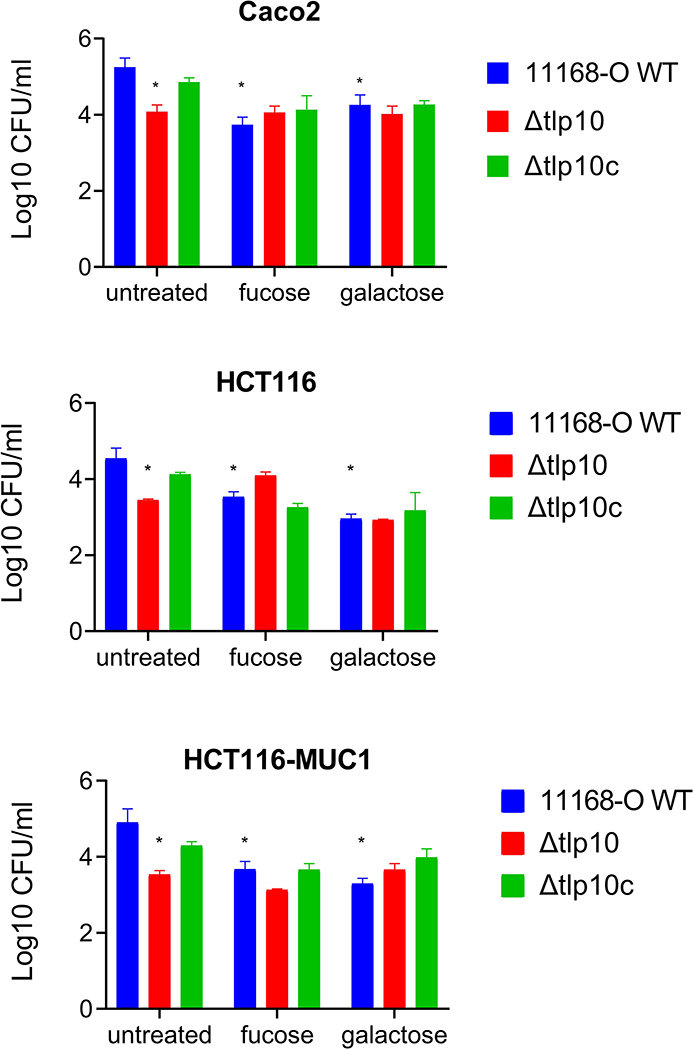
To assess the biological importance of Tlp10 in C. jejuni, we compared adherence and invasion of C. jejuni11168-O wild-type, Δtlp10LBD, and Δtlp10LBDc strains to polarised human colorectal adenocarcinoma Caco-2 cells, the colorectal cancer cell line HCT116, and HCT116 cells expressing the additional surface glycoprotein MUC1 (HCT116MUC1 cells) (30). We observed a significant reduction in adherence of the Δtlp10LBD bacteria to Caco-2 and HCT116 cells compared with 11168-O wild-type bacteria. The adherence of the Δtlp10LBD mutant to HCT116MUC1 cells was reduced to a greater degree than it was to the Caco-2 and HCT cells (Fig. 7). There were no significant differences in invasion between any of the isogenic strains (fig. S3). Molecular galactose and fucose suppressed the adherence of 11168-O wild-type and Δtlp10LBDc isogenic strains to all cell lines tested but had no significant effect on the adherence of Δtlp10LBD mutant strain to any cell line. The inhibition of adherence by fucose was less effective than that by galactose. There was no significant difference between the invasion of HCT116 or HCT116MUC1 cells (fig. S2).
Fig. 7. Reduced adhesion of Δtlp10 C. jejuni to mammalian cells.
Quantification of the adhesion of WT 11168-O C. jejuni (11168-O WT), the isogenic Tlp10LBD mutant (Δtlp10), and Δtlp10 complemented with full-length Tlp10 (Δtlp10c) to Caco2 cells, HCT116 cells, and HCT116 cells overexpressing MUC1 in the presence and absence of 5 mM of fucose or galactose. Adhesion analyses are presented as the mean of adhesion from three independent experiments, each performed in triplicate (n = 3). Asterisks indicate significant differences (P < 0.05; t-test).
Mutation of the Tlp10 LBD domain reduces host colonisation by C. jejuni
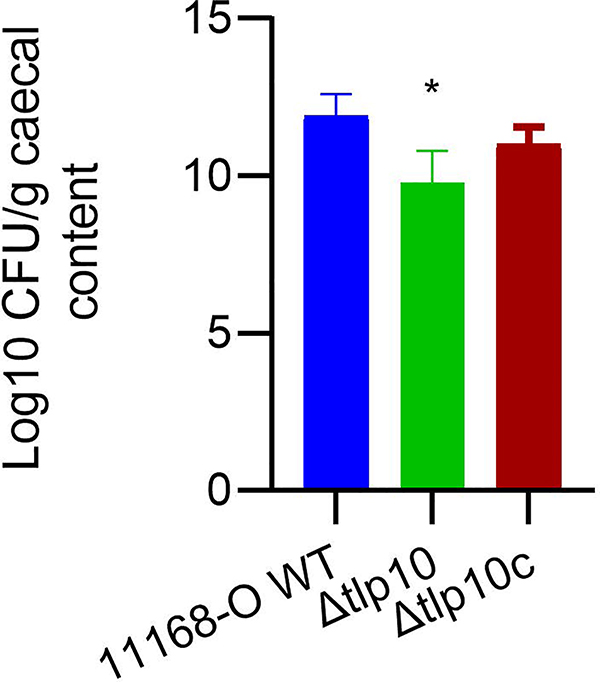
C. jejuni normally lives as a commensal in the avian gut. To test whether Tlp10 was required for colonisation, we orally infected one-day old chicks with 108 wild-type C. jejuni 11168-O, the isogenic mutant Δtlp10LBD, or the complemented isogenic mutant Δtlp10LBDc. The amount of colonisation was assessed 5 days after infection, a time point by which maximal colonisation is reached (14, 31). The Δtlp10LBD isogenic mutant strain displayed a 2-log reduction in colonisation compared to wild-type, and complementation partially restored the wild-type phenotype (Fig. 8).
Fig 8. Reduced colonization of chickens by Δtlp10 C. jejuni.
Chicks were orally infected with 108 CFU of WT C. jejuni (11168-O WT), the Tlp10LBD mutant (Δtlp10), or the complemented mutant (Δtlp10c) at day of hatch. Ceca were recovered at 7 days post-inoculation, homogenized, serially diluted, and plated on selective medium for C. jejuni enumeration. Counts are shown as log CFU/g of caecal contents. n = 7 chicks for each C. jejuni strain. Asterisks indicate significant differences (P < 0.05; t-test).
Discussion
This study reports the characterisation of a C. jejuni reference strain NCTC11168-O chemoreceptor Tlp10, which has a previously unreported type of LBD and specificity for a wide range of effectors, including simple and complex glycans. The Tlp10 chemoreceptor sensed aspartate, mannose, fucose, isoleucine, fumarate, malate, and purine as chemoattractants, and arginine, thiamine, and galactose as chemorepellents. Based on the affinities of the ligands, the binding of repellents appeared to be preferred over the binding of attractants, and there were no chemotactic responses to α-ketoglutarate, rhamnose, purine and sialic acid despite Tlp10 binding to these molecules. It is possible that these ligands elicited a differential chemotaxis response that was below that required for statistical significance or these compounds could act as chemotaxis pathway inhibitors, or antagonists, similar to those reported for PA2652 chemoreceptor in P. aeruginosa (32).
Tlp10 contains a previously undescribed sensor domain that is predicted to consist predominantly of α-helical regions. Although many Pfam models exist for domains of a similar α-helical-rich nature (such as 4HB_MCP clan, PilJ, and NIT), none of them match the LBD in Tlp10. We built a new sequence profile model for the Tlp10 LBD, which we termed the DAHL domain. The DAHL domain was identified not only in chemoreceptors, but also in other bacterial signal transduction proteins. The majority of DAHL domain–containing proteins are histidine kinases and diguanylate cyclases. DAHL-containing chemoreceptors were found only in closely related Campylobacter and Helicobacter spp., and the majority of the DAHL domains in the epsilonproteobacteria were identified in other types of signal transduction proteins. These findings indicate that the Tlp10 LBD likely originated through a recent domain swap in epsilonproteobacteria species. The DAHL domain was also identified in the A. tumefaciens sensor histidine kinase VirA, which is crucial for virulence gene activation in this plant pathogen (33). Genetic evidence indicates that a periplasmic region of VirA corresponding to the DAHL domain interacts with the periplasmic sugar-binding protein ChvE (34–36). However, the exact nature of VirA-ChvE interaction still remains unclear (34, 35, 37, 38).
Sensory domains detect numerous signals and are highly variable in sequence because they evolve to sense specific physicochemical queues (39). DAHL is no exception, and despite high conservation of key amino acid residues Asn115 (conservation is > 80%) and Asn120 (>75%), in some instances they are changed to unrelated amino acids. For example, asparagine in position 115 is substituted with a positively charged lysine in C. fetus or with a negatively charged aspartate in Corallococcus spp. These observations indicate that DAHL domains might bind distinct ligand(s) in different species.
The Tlp10 DAHL domain contains two distinct ligand-binding sites. Site A was specific for arginine, α-ketoglutarate, fumarate, thiamine, isoleucine, purine, and aspartate, and site B was specific for malate, mannose, fucose, rhamnose, sialic acid, aspartate, thiamine, and galactose. It is important to note that the competition SPR assay is not a direct 1:1 binding competition; rather, it is aimed at determining the number of binding sites and which ligands bind each site. Although chemoreceptors with two ligand-binding sites have been previously reported (40), they have limited specificity and respond only to amino acid attractants. At this point, it is unclear whether the two binding sites in the Tlp10 DAHL domain overlap or allosterically effect each other. However, another all-helical domain, HBM, has two non-overlapping, distinct ligand-binding sites: a membrane-proximal module, which binds malate and succinate, and membrane-distal module, which binds acetate (39). Although, the secondary structure prediction pattern of DAHL matched one HBM domain, its sequence could not be recognized by the HBM Pfam model. Tertiary structure of the DAHL domain is yet to be determined.
The ligand-binding associations were confirmed by ligand competition analysis of site-directed mutants wherein substitution N115A significantly decreased the binding affinity for α-ketoglutarate and galactose, and substitution N120A significantly decreased the binding affinity for isoleucine, arginine, purine, galactose, and α-ketoglutarate. Although substitutions Y70A, or H193A did not abolish the ability of Tlp10LBD to bind its ligands, they had a profound effect on the competition between ligands, suggesting that they may be critical to the structural stability of the molecule or the integrity of the LBD folding.
The ability to sense aspartate, isoleucine, and galactose was previously observed for the C. jejuni chemosensors Tlp1, Tlp3, Tlp10, Tlp11, and Tlp12. (13, 20, 41, 42). Ligand-sensing redundancy of C. jejuni chemosensors is an interesting evolutionary phenomenon to explore and allows us to hypothesize that this organism is highly adapted to changing its gene content and gene expression in response to variation in its environment. This would allow the organism to move towards a specific ligand despite the loss of any one receptor, which is consistent with deletion of the Tlp10 LBD resulting in a reduction in, rather than the total loss of, adherence to cultured human cells and colonization of the avian gut. This is similar to C. jejuni strain 520, which bears the rare galactose receptor Tlp11 (CcrG). This strain has an enhanced ability to sense galactose as a chemoattractant and exhibits greater migration rates towards galactose compared to strains that do not have Tlp11 (16). The loss of Tlp11 reduces, but not abrogates, the ability of strain 520 to adhere to human cells and to colonise birds. Strains lacking Tlp11 are still chemotactic towards galactose despite expressing Tlp10, which senses galactose as a repellent (16). This suggests that there is another receptor, or a receptor–periplasmic binding protein pair, that may be responsible for sensing galactose as an attractant, highlighting the complexity of sensory perception and responses directed by C. jejuni chemosensory arrays. Bacteria have long been known to be able to balance between attractant and repellent signals in order to find an optimum environment with sensitivity to repellents being greater than that to attractants (43). C. jejuni appears to follow the same pattern, with the affinity of Tlp11, which senses galactose as an attractant, being lower (17 μM) than that for Tlp10, which senses galactose as a repellent (2.9 μM). It is possible that high concentrations of galactose may be deleterious to C. jejuni in some environments, and a combination of receptors sensing the same molecule as an attractant or a repellent allows the organism to find the best niche possible. The presence of receptors that sense the same chemoeffector as a repellent or an attractant has been reported previously for other organisms, including the McpB and McpC receptors of Salmonella spp., which sense cysteine and are proposed to moderate and orient the response to oxidative stress (44).
The chemoreceptor content redundancy (10, 45) and ability of chemoreceptors to bind to multiple ligands has also been described for many bacterial species (22). However, how the multi-ligand binding affects receptor activity and how the organism balances the signaling output in response to attractants and repellents is still poorly understood. C. jejuni possesses two scaffolding proteins that anchor chemoreceptors within the sensory complex that couples chemosensation to the flagellar motor switch: the canonical CheW, as well as CheV, which contains a response regulator domain. Using yeast two-hybrid assay to test protein-protein interactions, we showed that the signaling domain of Tlp10 interacted with both CheV and CheW as well as with the signaling domains of all other C. jejuni Class A chemoreceptors. In E. coli, homodimeric chemoreceptors of different specificities can form mixed trimers-of-dimers in vivo (46, 47) and then form signaling arrays that enable amplification of the chemotaxis signal (48). Similar sensory arrays in C. jejuni are likely to include all Class A chemoreceptors that could potentially interact with one another either directly or indirectly through Tlp10, which is highly abundant in cells. Assuming that the interactions we detected with the yeast two-hybrid system can occur in C. jeuni cells, it is possible to speculate that C. jejuni Tlp10 may play a role in recruiting mixed ‘teams’ of Tlps into chemosensory arrays, thus enabling a balance of complex multi-ligand interactions and amplification of chemotactic signals.
Considering the niches C. jejuni is able to inhabit, including avian and mammalian intestines, it is likely that the organism recognises and binds multiple glycan structures found within mucus, including glycans featuring terminal fucose, galactose, mannose, or sialic acid (49, 50). Fucosyl residues are typically located in terminal positions on the extensively glycosylated mucin proteins (51–53) that are common in the digestive tract, and the importance of fucose to the intestinal microflora and infectious organisms is well established (54). Tlp10 appears to confer C. jejuni with the ability to sense large glycosylated structures with terminal fucose, galactose, or sialic acid such as the fucosyl GM1 ganglioside [Fucα1–2Galβ1–3GalNAcβ1–4(Neu5Acα2–3) Galβ1–4Glc] and Lewisa (Lea) which has implications for host–pathogen interactions (55, 56). This is consistent with glycans being able to affect colonisation, invasion, and biofilm formation by C. jejuni (57–59) and the fact that differences between mucus composition of humans and chickens may be linked to pathogenicity (30, 60).
The fine-tuning of chemotactic responses is likely to be governed by the interaction of multiple receptors with a wide range of specificities and affinities, allowing the cell to achieve the most beneficial positioning within the host. Addressing the question of how this is accomplished would be a step forward in the deeper understanding of host-pathogen interactions in terms of colonization and disease processes.
Materials and Methods
Microbial strains, growth conditions, and plasmids
Bacterial and yeast strains and plasmids are listed in Table S13. C. jejuni11168-O (12–14) strains were grown at 42°C under microaerobic conditions (5% O2, 10% CO2, 85 % N2) on Muller Hinton agar (Oxoid) with vancomycin 10μg ml−1 (Sigma), and trimethoprim 2.5μg ml−1 (Sigma). Media was supplemented with kanamycin 30μg ml−1 (Roche) or chloramphenicol 20μg ml−1 (Lancaster) when appropriate. The host E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) media (Oxoid) or on LB agar (Oxoid), supplemented with ampicillin 100 μg ml−1 (Sigma) or kanamycin 50 μg ml−1 when appropriate. Yeast strains were grown and prepared according to manufacturer’s instructions (Clontech).
Expression and purification of the periplasmic domain of tlp10
The DNA fragment encoding the periplasmic sensory domain (bp 96–714, representing residues 33–238) of Tlp10 was amplified using primers incorporating start and stop codons (table S14) and ligated into pET-19b to form pGU0708 (table S13). The expression and purification of Tlp10 proteins were performed as described previously (13).
Identification of ligand-binding potential of Tlp10 by glycan and small molecule arrays.
Glycan and small molecule arrays containing amino and organic acids, salts, and over 400 glycans ranging from monosaccharides to dodecasaccharides, as well as complex glycans such as mucins (table S2 and S3), were performed as described previously (49, 61). Briefly, 1 μg of protein precomplexed with anti-His antibody (Cell Signaling Technology) and rabbit anti-mouse/goat anti-rabbit Alexa 555 IgG (Thermo Scientific) in a molar ratio of 1:0.5:0.25 was incubated on the array for 30 min in PBS. The arrays were scanned by a ProScan Array scanner at 488/520 nm and the results analysed by ScanArray Express software program (Perkin Elmer). The binding was defined as a value greater than 1-fold increase above mean background relative fluorescence units (RFU). The mean background was calculated from the average background of empty spots on the array plus three standard deviations. Statistical analysis of the data was performed by a Student’s t-test with a confidence level of 99.99% (p ≤ 0.0001). All arrays were performed in triplicate with a total of 12 data points for each glycan tested and complete dataset and MIRAGE (minimum information regarding a glycomics experiment) are shown in Tables S2 and S15.
Site-directed mutagenesis
Single-point alanine substitutions were introduced into the plasmid pGU0708 (table S13) at positions: Tyr70, Asn115, Asn120 or His193 using oligonucleotide-directed mutagenesis (QuikChange, Stratagene). All the constructed plasmids pG0709 – 0712 (table S13) were sequenced to confirm the desired point mutation in Tlp10 LBD gene.
Surface plasmon resonance (SPR) analysis
Tlp10 wild type, Y70A, N115A, N120A, or H193A mutant LBD proteins were immobilised onto a CM5 series S sensor chip and binding affinities of tested compounds (table 2) were investigated (16, 61). Briefly, using a BIAcoreTM S200 instrument (GE Healthcare), the proteins were captured using amine coupling kit, in which the carboxylmethyl dextran matrix of the sensor chip was activated by a 720 sec injection of a mixture of 0.2M 1-ethyl-3-[(3-dimethylamino) propyl]-carbodiimide (EDC) and 0.05M N-hydroxysuccinimide (NHS) which resulted in the conversion of carboxyl groups to an NHS ester. The proteins were diluted in 10mM sodium acetate buffer pH 4.5 (Tlp10LBD wild type) or pH 4 (Tlp10LBD Y70A, and Tlp10LBD N115A, Tlp10LBD N120A, and Tlp10LBD H193A) at a concentration of 100 μg mL-1. The remaining unreacted NHS ester groups were neutralised by an injection of 1 M ethanolamine-HCl (pH 8.0). Under these conditions, 7948, 7076, 6024 and 7100 response units (RU) of Tlp10LBD Y70A, and Tlp10LBD N115A, Tlp10LBD N120A, and Tlp10LBD H193A were captured on Flow cell 2, 3 and 4 respectively. As a negative control, FC1 was a blank control, which underwent the same treatment as the other flow paths, without the protein injection. This set enabled double reference subtraction of the responses (2–1, 3–1, 4–1). The tested compounds were prepared as a stock concentration of 100–200mM in HBS. The compounds were then diluted between 1 nM-1 mM in a series of 1:10 dilutions in HBS and run over the flow cells at a flow rate of 30 μl/min. Between each sample testing, a series of buffer only injections were run, to enable double blank subtraction for the sensorgram assessment. After the initial run, based on the results, the dilution series ranged from 0.195 μM to 1 mM in 1:4 dilutions in HBS. The samples were run using single-cycle kinetic/affinity methods in triplicate for those compounds that showed sub-millimolar affinity after the initial binding screen. The datasets were analyzed using the BIAcore T200 evaluation software 2.0.2; sensorgrams were double reference subtracted.
SPR competition assays
SPR competition assays were performed by using a BIAcore S200 instrument and the A-B-A inject function (62). Competition A-B-A analyses were used to interrogate specificity of the potential ligand binding site preferences of Tlp10 and to unravel the nature of the ligand-sensor interactions. This assay was designed to show if a cumulative response is observed when a second analyte (B) is flown across the bound protein saturated with the first analyte (A) (Fig. 4 and table S6). As the assay is designed to provide saturation of all analytes tested, this assay does not provide 1:1 competition to indicate which is the preferred analyte for a binding site. The proteins, Tlp10LBD wild type, Tlp10LBD Y70A, and Tlp10LBD N115A, Tlp10LBD N120A, and Tlp10LBD H193A mutants were immobilized as above. The A-B-A was used with combinations of each of the compound (at concentration 10 × KD) and PBS control, with 60 seconds injections of analyte A to ensure saturation or near-saturation was reached prior to competition with analyte B. The results were analyzed using BIAcore S200 evaluation software using the sensorgram mode and data was zeroed to baseline prior to the initial A injection. All response data was normalised to 100Da molecular weight for each analyte allowing direct comparison of responses.
Circular dichroism spectroscopy
Circular dichroism (CD) experiments on Tlp10LBD wild type, Tlp10LBD Y70A, and Tlp10LBD N115A, Tlp10LBD N120A, and Tlp10LBD H193A were carried out with J-1500 CD spectrophotometer (JASCO, United states). The proteins were prepared at a concentration of 1 mg/ml in 100 mM phosphate buffer (pH 7.5). Wavelength scan experiments were performed at 4°C.
Yeast two-hybrid analysis
Yeast two-hybrid analysis of protein interactions was performed as described previously (13). The DNA sequence encoding amino acid residues 377–592 of Tlp10 was amplified with the primers tlp10sigXmaIF (CCCGGGGGAAAATATGCAAGATATGCACA) and tlp10sigBamHIR (GGATCCACTGAAAGCTACTTAATTTTTC) and the DNA sequence encoding amino acid residues 31 – 224 of Tlp7 were amplified with the primers tlp7sigEcoRIF (GAATTCTCAACAGTATCTCAAAATGAAA) and tlp7sigBamHIR (GGATCCAAATTTGAAATTGGTTAAGTCC), designed from the C. jejuni reference strain NCTC11168 genome sequence (63). PCR products were cloned into the ‘target’ vector (pGADT7) to allow for expression of the AD-Tlp10sig and AD-Tlp7sig fusion proteins, and into the ‘bait’ vector (pGBKT7) to allow for expression of the BD-Tlp10sig and BD-Tlp7sig fusion proteins, respectively. Previously constructed ‘target’ and ‘bait’ plasmids (ADD refs 12, 13) expressing the following C. jejuni chemotaxis proteins/domains were also used in yeast two-hybrid analysis and are described in detail in Table S12: CheA, CheW, CheV, the CheW-like domain of CheV (CheVdW), CheY, the signalling domain of Tlp1 (Tlp1sig) and the signalling domains of Tlps 2, 3 and 4 (Tlp2(3, 4)sig). Interactions involving BD-Tlp10sig could only be assessed on intermediate stringency media supplemented with 2.5mM 3-AT, as this media was found to suppress the autonomous activation of reporter gene expression exhibited by this fusion protein. Growth of yeast co-transformants was examined after 5–7 days. Absence of growth indicated no protein-protein interaction, whereas growth of yeast co-transformants on intermediate stringency and/or high stringency indicated a protein-protein interaction. Assay positive controls consisted of pGADT7-T with pGBKT7–53 (Clontech) and pADcheW with pBKcheA. The assay negative control consisted of pGADT7-T with pGBKT7-Lam (Clontech). All control co-transformants were included on each protein-protein interaction assay plate and assays were performed at least twice to verify results.
Mutation and complementation of Tlp10 in C. jejuni 11168-O
C. jejuni reference strain NCTC11168 Tlp10 (Cj0019c) coding region was amplified using forward (TLP10 Mut-F) and reverse (TLP10 Mut-R) primers (table S14). The PCR product was cloned into pGEMT-Easy (Promega) to create pGU0816 (table S13). The inverse PCR mutagenesis strategy was used to mutate the gene. The fragment was amplified by using forward (TLP10 inv-F) and reverse (TLP10 inv-R) primers (table S14) (delete 345bp of tlp10 encoding 115 amino acids) and PCR product was ligated with the chloramphenicol cassette to form pGU0817 (table S13). Then pGU0817 was verified by PCR and sequencing and electro-transformed into competent C. jejuni11168-O to create isogenic Δtlp10LBD mutant (11168Δtlp10LBD::CmR) (table S13) as previously described (16, 64). The 11168Δtlp10LBD::CmR mutant was complemented by the insertion of C. jejuni 11168-O Tlp10 coding region, which was amplified using the forward and reverse primers flanked with BsmBI sites shown in (table S14). The PCR product was cloned into pC46 (65, 66). The tlp10 containing pC46 vector was transformed into 11168 Δtlp10LBD::CmR and plated onto plates containing 50 μg ml−1 kanamycin and 10 μg ml−1 chloramphenicol. The complemented strain 11168Δtlp10LBDΩTlp10 was confirmed by PCR.
Adherence and invasion assays of culture cell lines Caco-2 and HCT116
Adherence rate was calculated against the size of inoculum, (109 bacterial cells per human cell in confluent cell culture) and invasion rate was determined using the gentamicin-kill assays, where adhered, but not invaded cells are removed by this antibiotic for 1 hour, and the rate is calculated against the rate of adhesion. The assays were performed with Caco-2 cells, HCT116 cells, and HCT116 cells overexpressing MUC1 (HCT116MUC1) as previously described (16). Galactose and fucose competition were performed by pre-incubating C. jejuni with 5 mM glycan.
Biofilm assay
Biofilm assay was performed as previously described (13, 14). The effect of free galactose on biofilm was determined by pre-incubating C. jejuni with 10 mM galactose. Briefly, C. jejuni strains were grown microaerobically with shaking overnight at 42°C, diluted to an OD600 of 0.05, and 2 mL of cell suspension were dispersed into 24-wells flat-bottom polystyrene tissue culture plates (Geiner Bio-One, Monroe, NC, USA). The plates incubated at 42 °C under microaerobic conditions for 48 h. Formed biofilm was assessed using modified crystal violet staining assay (29). Three replicates for each strain under each condition were used for each assay; three independent experiments were conducted.
Chemotaxis assay
Chemotaxis assays were performed as previously described (67). Briefly, 6 mm plugs were removed from a 0.5% agar plate and replaced with 0.5% agar with 2 mM of ligands, buffer or mucin. The plates were overlaid with 0.1% agar and left for 2 h to allow for diffusion. A 24 h culture of C. jejuni strains, grown at 42 °C, was sub-cultured in Brucella broth for a further 18 h with shaking at 50 rpm. Cells were collected by centrifugation, resuspended in saline and 108 CFU of C. jejuni in a 100 μl drop was then inoculated in the centre of the petri dish. Viable bacteria that have migrated to amino acid plugs after 4 h at 37 °C, were enumerated by viable count.
μ-Slide Chemotaxis assay
The modified protocol of μ-Slide Chemotaxis (ibidi GmbH, Martinsried, Germany) was carried out as described previously (68). Briefly, μ-Slide is consisting of two separate opposing reservoirs (volume, 60 μl in each), divided by a 1-mm narrow liquid transition zone with a 10 μl volume. A 1% of low melting temperature agarose (LMT) (Sigma) in phosphate buffer saline (PBS, pH 7.5) was applied to the transition zone. The first reservoir was filled with the bacterial cell suspension, overnight C. jejuni cells were harvest off MHA plates using 2 ml of PBS and adjust the OD600 to 1, and mixed with 1% LMT (vol:vol). The second reservoir was filled with 60 μl of chemoeffector (final conc. 10mM) mixed with 0.7% of LMT, to be final concentration 0.35% and incubate for 3 hours at 42°C under microaerobic conditions. After that, 20 μl of samples from the chemoeffectors reservoir was taken and enumerated by viable count. Three biological repeats of all experiments, each in triplicate, including controls, were performed.
μ-Slide chemotactic cell migration visualisation
Chemotactic cell migration was observed using the modified protocol for μ-Slide Chemotaxis (ibidi GmbH, Martinsried, Germany) out. Briefly, the cells were grown and fluorescently labelled with FITC (Fluorescein-5-EX, Succinimidyl Ester, Molecular Probes) as described by Rahman, King, Shewell, Semchenko, Hartley-Tassell, Wilson, Day and Korolik (14). The left and right reservoirs of the μ-Slide were filled, left reservoir with 60 μl of bacterial suspension, and the right reservoir with the same amount of PBS. The transition zone was also filled with 10 μl of PBS. The moving bacteria were then visualized by Olympus Cell^M imaging system (Griffith University) at 40 × magnification lens. Once cells migrated close to the transition zone, 10 μl of 60 mM of isoleucine or arginine directly add into the second reservoir to give a final concentration of 10 mM. Time-lapse video was performed to record the migrating cells toward to isoleucine and away from arginine.
Colonisation potential of C. jejuni wild-type and mutant strains
All Campylobacter strains (table S13) were grown at 42oC in Mueller Hinton broth (MHB) under microaerophilic conditions. Specific pathogen free chicks (n = 7/group) were inoculated orally on the day of hatch with 40 μl of MHB preparations containing 108 CFU of each strain. Each group of birds was housed in separate cages. All birds were further confirmed to be Campylobacter free by periodical testing of cloacal samples. Seven days post-infection birds were euthanized and caecal contents enumerated by viable count. All animal experiments were performed in compliance with Griffith University animal ethics guidelines.
Bioinformatics tools and databases
BLASTp, RPS-BLAST and PSI-BLAST searches (69) against the non-redundant database at NCBI were carried out with default parameters. Protein domain architectures were obtained from the Pfam (70) and Conserved Domain (71) databases. Profile-profile searches for domain identification were carried out TREND (72) and HHpred (24) servers. Fold recognition was performed using Phyre2 (73). Multiple sequence alignments were constructed using LINS-I and FFT-NS-2 methods in MAFFT program (74). Maximum-likelihood phylogenetic trees were constructed using the MEGA X package (75). Sequence alignments were edited in Jalview (76). Secondary structure prediction from amino acid sequences was carried out using Ali2D and Quick2D from the MPI Bioinformatics Toolkit (77).
Hidden Markov Model Construction
We used the sequence of predicted periplasmic and transmembrane regions of Tlp10 as a query in exhaustive PSI-BLAST search against the non-redundant NCBI database in November 2018. The resulted set contain more than 1700 sequences that were verified to contain two transmembrane helices using TREND, aligned using MAFFT program, and then edited in Jalview to discard incomplete sequences and to remove transmembrane regions from the alignment and to reduce redundancy (95% identical sequences). A resulting set of 505 sequences was used to initiate a search against UniprotKB Reference proteomes (78) using the hmmsearch tool from the HMMER server (79). The final set of predicted extracellular regions (non-redundant at 80% identity) consisted of 184 sequences and was used to construct the DAHL HMM using the hmmbuild tool from HMMER.
Statistical Analysis
Statistical significance of data generated in this study was determined using two tailed Student’s t-test, GraphPad Prism (GraphPad Software). P ≤ 0.05 was considered statistically significant. The study uses typical numbers of biological (3+) and technical replicates (3+) for cell/bacterial based laboratory testing. These numbers are sufficient for statistical analysis using Wilcoxon, Mann-Whitney U given a normal distribution post log transformation, the standard Independent Samples t-test. The differences between means are large enough and the level of error within the data is small enough that testing with any of Wilcoxon, Mann-Whitney U or standard Independent Samples t-test yields a significant difference. Any p-values arising from studies with small sample sizes should be treated with caution, with more information taken from the size of the mean difference with respect to the error, rather than making inference on a population. For this reason, we chose to report with standard Independent Sample t-tests in this study.
Supplementary Material
Fig. S1. Representative sensorgrams from SPR analysis of the interaction of Tlp10 LBD with various ligands.
Fig. S2. Mutant Tlp10LBD proteins are correctly folded.
Fig. S3. Invasion of mammalian cells by WT and mutant C. jejuni in the presence and absence of fucose and galactose.
Table S1. HHpred search results for small subset of DAHL domains from various species.
Table S2. Molecules included in the glycan microarray.
Table S3. Molecules included in the small molecules microarray.
Table S4. Complex glycan structures recognized by Tlp10.
Table S5. Chemotaxis responses of WT and mutant C. jejuni strains.
Table S6. Illustration of A-B-A competition SPR analysis of WT Tlp10LBD.
Table S7. Illustration of A-B-A competition SPR analysis of Tlp10LBDY70A.
Table S8. Illustration of A-B-A competition SPR analysis of Tlp10LBDY115A.
Table S9. Illustration of A-B-A competition SPR analysis of Tlp10LBDY120A.
Table S10. Illustration of A-B-A competition SPR analysis of Tlp10LBDH193A.
Table S11. Analysis of protein-protein interactions with Tlp10sig using the yeast two-hybrid system.
Table S12. Fusion proteins for yeast two-hybrid experiments.
Table S13. Microbial strains and plasmids used in this study.
Table S14. DNA Primers used in this study.
Table S15. MIRAGE glycan and small molecule arrays for Tlp10.
Movie S1. C. jejuni migrate toward the attractant isoleucine.
Movie S2. Migration of C. jejuni cells toward the attractant isoleucine is reduced by addition of the repellent arginine.
Movie S3. PBS control for the effect of arginine on the migration of WT C. jejuni toward isoleucine.
Data file S1. Proteins included in DAHL domain MSA.
Acknowledgments:
We acknowledge Dr. Rhys Pritchard, Griffith University, for his help with microscopy preparation.
Funding: This research was supported in part by Griffith University Postgraduate Scholarship for HR, Australian Postgraduate Award (APA) to LH and by National Institutes of Health grant R35GM131760 to IBZ.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The DAHL domain model is deposited into the Pfam database under accession number PF19443. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. An MTA from the Institute for Glycomics, Griffith University, would be required for the transfer of recombinant plasmids. Glycan and small molecule arrays are subject to manufacturing costs.
References and Notes
- 1.Wadhams GH, Armitage JP, Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biology 5, 1024–1037 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Lux R, Shi W, Chemotaxis-guided movements in bacteria. Crit. Rev. Oral Biol. Med. 15, 207–220 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Ketley JM, Pathogenesis of enteric infection by Campylobacter. Microbiology 143, 5–21 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Poly F, Guerry P, Pathogenesis of Campylobacter. Current opinion in gastroenterology 24, 27–31 (2008). [DOI] [PubMed] [Google Scholar]
- 5.W. H. Organization, The global view of campylobacteriosis: report of an expert consultation, Utrecht, Netherlands. World Health Organization, (2012). [Google Scholar]
- 6.Zilbauer M, Dorrell N, Wren BW, Bajaj-Elliott M, Campylobacter jejuni-mediated disease pathogenesis: an update. Trans. R. Soc. Trop. Med. Hyg. 102, 123–129 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Lertsethtakarn P, Ottemann KM, Hendrixson DR, Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korolik V, The role of chemotaxis during Campylobacter jejuni colonisation and pathogenesis. Curr. Opin. Microbiol. 47, 32–37 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Parkinson JS, Hazelbauer GL, Falke JJ, Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuichet K, Zhulin IB, Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50–ra50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortega DR, Zhulin IB, Evolutionary genomics suggests that CheV is an additional adaptor for accommodating specific chemoreceptors within the chemotaxis signaling complex. PLoS computational biology 12, e1004723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchant J, Wren B, Ketley J, Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10, 155–159 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Hartley-Tassell LE, Shewell LK, Day CJ, Wilson JC, Sandhu R, Ketley JM, Korolik V, Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol Microbiol 75, 710–730 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Rahman H, King RM, Shewell LK, Semchenko EA, Hartley-Tassell LE, Wilson JC, Day CJ, Korolik V, Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog 10, e1003822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tareen AM, Dasti JI, Zautner AE, Gross U, Lugert R, Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology 156, 3123–3135 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Day CJ, King RM, Shewell LK, Tram G, Najnin T, Hartley-Tassell LE, Wilson JC, Fleetwood AD, Zhulin IB, Korolik V, A direct-sensing galactose chemoreceptor recently evolved in invasive strains of Campylobacter jejuni. Nature communications 7, 13206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB, Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS computational biology 12, e1004862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machuca MA, Liu YC, Beckham SA, Gunzburg MJ, Roujeinikova A, The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J. Struct. Biol. 194, 205–213 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A, Structural basis for amino-acid recognition and transmembrane signalling by tandem Per–Arnt–Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr. Sect. D. Biol. Crystallogr. 71, 2127–2136 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Day, Hartley-Tassell LE, Shewell LK, King RM, Tram G, Day SK, Semchenko EA, Korolik V, Variation of chemosensory receptor content of Campylobacter jejuni strains and modulation of receptor gene expression under different in vivo and in vitro growth conditions. BMC microbiology 12, 128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekhar K, Gangaiah D, Pina-Mimbela R, Kassem II, Jeon BH, Rajashekara G, Transducer like proteins of Campylobacter jejuni 81–176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front Cell Infect Microbiol 5, 46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega Á, Zhulin IB, Krell T, Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 81, e00033–00017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander RP, Zhulin IB, Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proceedings of the National Academy of Sciences 104, 2885–2890 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Söding J, Biegert A, Lupas AN, The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu CJ, Ulrich LE, Zhulin IB, The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 28, 121–124 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Zhulin IB, Nikolskaya AN, Galperin MY, Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185, 285–294 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega Á, Krell T, The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci. 23, 332–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields S, The two-hybrid system to detect protein-protein interactions. Methods 5, 116–124 (1993). [Google Scholar]
- 29.Tram G, Korolik V, Day CJ, MBDS solvent: an improved method for assessment of biofilms. Advances in Microbiology 3, 5 (2013). [Google Scholar]
- 30.McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA, MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. The Journal of clinical investigation 117, 2313–2324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringoir D, Korolik V, Colonisation phenotype and colonisation potential differences in Campylobacter jejuni strains in chickens before and after passage in vivo. Vet. Microbiol. 92, 225–235 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Martin-Mora D, Ortega A, Perez-Maldonado FJ, Krell T, Matilla MA, The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8, 2102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachel SE, Zambryski PC, virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46, 325–333 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Shimoda N, Toyoda-Yamamoto A, Aoki S, Machida Y, Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J. Biol. Chem. 268, 26552–26558 (1993). [PubMed] [Google Scholar]
- 35.Gao R, Lynn DG, Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 187, 2182–2189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair GR, Lai X, Wise AA, Rhee BW, Jacobs M, Binns AN, The integrity of the periplasmic domain of the VirA sensor kinase is critical for optimal coordination of the virulence signal response in Agrobacterium tumefaciens. J. Bacteriol. 193, 1436–1448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doty SL, Yu MC, Lundin JI, Heath JD, Nester EW, Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J. Bacteriol. 178, 961–970 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyoda-Yamamoto A, Shimoda N, Machida Y, Genetic analysis of the signal-sensing region of the histidine protein kinase VirA of Agrobacterium tumefaciens. Molecular and General Genetics MGG 263, 939–947 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Gavira JA, Gumerov VM, Rico-Jiménez M, Petukh M, Upadhyay AA, Ortega A, Matilla MA, Zhulin IB, Krell T, How bacterial chemoreceptors evolve novel ligand specificities. mBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pineda-Molina E, Reyes-Darias J-A, Lacal J, Ramos JL, García-Ruiz JM, Gavira JA, Krell T, Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proceedings of the National Academy of Sciences 109, 18926–18931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lübke A-L, Minatelli S, Riedel T, Lugert R, Schober I, Spröer C, Overmann J, Groß U, Zautner AE, Bohne W, The transducer-like protein Tlp12 of Campylobacter jejuni is involved in glutamate and pyruvate chemotaxis. BMC microbiology 18, 111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vegge CS, Brøndsted L, Li Y-P, Bang DD, Ingmer H, Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl. Environ. Microbiol. 75, 5308–5314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adler J, Tso W-W, “ Decision”-making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli. Science 184, 1292–1294 (1974). [DOI] [PubMed] [Google Scholar]
- 44.Lazova MD, Butler MT, Shimizu TS, Harshey RM, Salmonella chemoreceptors McpB and McpC mediate a repellent response to L-cystine: a potential mechanism to avoid oxidative conditions. Mol Microbiol 84, 697–711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hida A, Tajima T, Kato J, Two citrate chemoreceptors involved in chemotaxis to citrate and/or citrate-metal complexes in Ralstonia pseudosolanacearum. Journal of bioscience and bioengineering 127, 169–175 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Studdert CA, Parkinson JS, Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A 101, 2117–2122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studdert CA, Parkinson JS, In vivo crosslinking methods for analyzing the assembly and architecture of chemoreceptor arrays. Methods Enzymol 423, 414–431 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Hazelbauer GL, Lai WC, Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol 13, 124–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day CJ, Semchenko EA, Korolik V, Glycoconjugates play a key role in Campylobacter jejuni infection: interactions between host and pathogen. Front Cell Infect Microbiol 2, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day CJ, Tram G, Hartley-Tassell LE, Tiralongo J, Korolik V, Assessment of glycan interactions of clinical and avian isolates of Campylobacter jejuni. BMC microbiology 13, 228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A, L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proceedings of the National Academy of Sciences of the United States of America 108, 7194–7199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coyne MJ, Reinap B, Lee MM, Comstock LE, Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Looft T, Cai G, Choudhury B, Lai LX, Lippolis JD, Reinhardt TA, Sylte MJ, Casey T, Avian intestinal mucus modulates Campylobacter jejuni gene expression in a host-specific manner. Frontiers in microbiology 9, 3215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker DJ, Lowe JB, Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R (2003). [DOI] [PubMed] [Google Scholar]
- 55.Le Pendu J, Marionneau S, Cailleau-Thomas A, Rocher J, Le Moullac-Vaidye B, ClÉment M, ABH and Lewis histo-blood group antigens in cancer. APMIS 109, 9–26 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y, Takeda N, Ishii K, Wakita T, Narimatsu H, Shirato H, Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J. Virol, JVI. 00278–00212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dwivedi R, Nothaft H, Garber J, Xin Kin L, Stahl M, Flint A, van Vliet AH, Stintzi A, Szymanski CM, L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol Microbiol 101, 575–589 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA, Characterization of Campylobacter jejuni biofilms under defined growth conditions. Applied and environmental microbiology 73, 1908–1913 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell R, Blake D, Cell association and invasion of Caco-2 cells by Campylobacter jejuni. Infect. Immun. 62, 3773–3779 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byrne CM, Clyne M, Bourke B, Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 153, 561–569 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Day CJ, Hartley-Tassell LE, Korolik V, in Campylobacter jejuni. (Springer, 2017), pp. 51–63. [Google Scholar]
- 62.Mubaiwa TD, Hartley-Tassell LE, Semchenko EA, Day CJ, Jennings MP, Seib KL, The Bexsero Neisseria meningitidis serogroup B vaccine antigen NHBA is a high-affinity chondroitin sulfate binding protein. Scientific reports 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parkhill J, Wren B, Mungall K, Ketley J, Churcher C, Basham D, Chillingworth T, Davies R, Feltwell T, Holroyd S, The genome sequence of the foodborne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Murphy C, Carroll C, Jordan K, Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 100, 623–632 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Gaskin D, Van Vliet A, Pearson B, in Zoonoses and public health. (BLACKWELL PUBLISHING, 2007), vol. 54, pp. 101–101. [Google Scholar]
- 66.Elgamoudi BA, Ketley JM, Lighting up my life: a LOV-based fluorescent reporter for Campylobacter jejuni. Res. Microbiol. 169, 108–114 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Korolik V, Ottemann KM, in Bacterial Chemosensing. (Springer, 2018), pp. 23–31. [Google Scholar]
- 68.Elgamoudi B, Ketley JM, Determination of the Chemotactic Behavior of Campylobacter jejuni by using the μ-Slide Chemotaxis. User protocols-ibidi. 2016. [Google Scholar]
- 69.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gumerov VM, Zhulin IB, TREND: a platform for exploring protein function in prokaryotes based on phylogenetic, domain architecture and gene neighborhood analyses. Nucleic Acids Res, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 10, 845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K, Rozewicki J, Yamada KD, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ, Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V, A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Consortium U, UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD, HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative sensorgrams from SPR analysis of the interaction of Tlp10 LBD with various ligands.
Fig. S2. Mutant Tlp10LBD proteins are correctly folded.
Fig. S3. Invasion of mammalian cells by WT and mutant C. jejuni in the presence and absence of fucose and galactose.
Table S1. HHpred search results for small subset of DAHL domains from various species.
Table S2. Molecules included in the glycan microarray.
Table S3. Molecules included in the small molecules microarray.
Table S4. Complex glycan structures recognized by Tlp10.
Table S5. Chemotaxis responses of WT and mutant C. jejuni strains.
Table S6. Illustration of A-B-A competition SPR analysis of WT Tlp10LBD.
Table S7. Illustration of A-B-A competition SPR analysis of Tlp10LBDY70A.
Table S8. Illustration of A-B-A competition SPR analysis of Tlp10LBDY115A.
Table S9. Illustration of A-B-A competition SPR analysis of Tlp10LBDY120A.
Table S10. Illustration of A-B-A competition SPR analysis of Tlp10LBDH193A.
Table S11. Analysis of protein-protein interactions with Tlp10sig using the yeast two-hybrid system.
Table S12. Fusion proteins for yeast two-hybrid experiments.
Table S13. Microbial strains and plasmids used in this study.
Table S14. DNA Primers used in this study.
Table S15. MIRAGE glycan and small molecule arrays for Tlp10.
Movie S1. C. jejuni migrate toward the attractant isoleucine.
Movie S2. Migration of C. jejuni cells toward the attractant isoleucine is reduced by addition of the repellent arginine.
Movie S3. PBS control for the effect of arginine on the migration of WT C. jejuni toward isoleucine.
Data file S1. Proteins included in DAHL domain MSA.