SUMMARY
Predators use vision to hunt, and hunting success is one of evolution's main selection pressures. Yet, how viewing strategies and visual systems are adapted to predation is unclear. Tracking predator-prey interactions of mice and crickets in 3D, we find that mice trace crickets with their binocular visual fields and that monocular mice are poor hunters. Mammalian binocular vision requires ipsi- and contralateral projections of retinal ganglion cells (RGCs) to the brain. Large-scale single-cell recordings and morphological reconstructions reveal that only a small subset (9/40+) of RGC types in the ventrotemporal mouse retina innervate ipsilateral brain areas (ipsi-RGCs). Selective ablation of the ipsi-RGCs (< 2% of RGCs) in the adult retina drastically reduces the hunting success of mice. Stimuli based on ethological observations indicate that five ipsi-RGC types reliably signal prey. Thus, viewing strategies align with a spatially restricted and cell-type-specific set of ipsi-RGCs that supports binocular vision to guide predation.
eToc blurb
Johnson et al. show that mice track prey with their binocular visual field and discover that a small subset of retinal ganglion cell types innervates ipsilateral brain targets to support binocular vision and guide predation.
INTRODUCTION
Because hunting success is a central selection pressure in animal evolution (Sillar et al., 2016), understanding how nervous systems mediate hunting promises insights into their functional organization and evolution. Animals use vision to detect, pursue, and capture prey (Ewert et al., 2001; Hoy et al., 2016). In mammals, binocular vision, which combines information from the visual field shared by both eyes, is thought to have arisen for predation (Cartmill, 1974; Pettigrew, 1986). In theory, binocular vision can break a prey's camouflage, estimate the distance between predator and prey more accurately than monocular vision, and improve sensitivity in dim light and low contrast (Nityananda and Read, 2017; Ponce and Born, 2008). Predators tend to have larger binocular visual fields than prey (Heesy, 2008; Walls, 1942). However, whether binocular vision guides mammalian predation and improves hunting success remains to be tested.
Mice use vision to hunt insects (Hoy et al., 2016; Langley, 1989; Whitaker, 1966). Recently, simultaneous head and eye movement measurements revealed that mice track prey primarily by moving their heads (Michaiel et al., 2020), and 2D movies shot from above showed that mice keep crickets within the lateral extent of their binocular visual fields (i.e., the central 40 °) (Hoy et al., 2016; Michaiel et al., 2020). However, at close distances, prey could be obscured from view by the mouse's head, and the role of vision in the final stages of the attack remains uncertain (Hoy et al., 2016). Furthermore, how mice capture crickets is unclear as interactions between predator and prey are only partly visible from above. Thus, the viewing strategies and behavioral repertoire of predation in mice are incompletely understood.
Binocular vision requires the convergence of information from both eyes. In mammals, this convergence is achieved by a partial decussation of retinal ganglion cell (RGC) axons in the optic chiasm (Petros et al., 2008). The primate retina has a strict line of decussation: axons of RGCs on the nasal side of the line cross at the optic chiasm, and RGCs from the temporal side innervate ipsilateral targets (Chalupa and Lia, 1991; Cooper and Pettigrew, 1979). By contrast, cat and mouse retinas lack a clear line of decussation, and only a subset of RGCs in the temporal retina have ipsilateral projections (i.e., ipsi-RGCs) (Dräger and Olsen, 1980; Rompani et al., 2017; Rowe and Dreher, 1982; Stone and Fukuda, 1974). Which RGC types have ipsilateral projections and support binocular vision in mice is unclear.
More than 40 different RGC types send signals from the retina to the brain (Baden et al., 2016; Bae et al., 2018; Rheaume et al., 2018; Tran et al., 2019). This diversity of RGCs is thought to be an adaptation to the diverse behavioral demands on vision (Baden et al., 2020; Turner et al., 2019). The contributions of RGCs to behavior depend on their light responses (i.e., what information they encode) and projections patterns (i.e., where they send this information). To date, few links between the light responses and projection patterns of specific RGC types and particular visual behaviors have been established (Dhande et al., 2015).
Here, we analyze the viewing strategies and hunting behavior of mice in 3D. We combine genetic and projection-specific labeling with large-scale functional recordings and morphological reconstructions to catalog the ipsi-RGC types, which support binocular vision. We analyze the ipsi-RGCs' responses to prey-mimetic stimuli and test their contributions to predation by region- and type-specific cell deletion. Our results reveal that viewing strategies align with region- and cell-type-specific RGC projection patterns and light responses to mediate binocular vision and guide predation.
RESULTS
3D tracking of predator-prey interactions
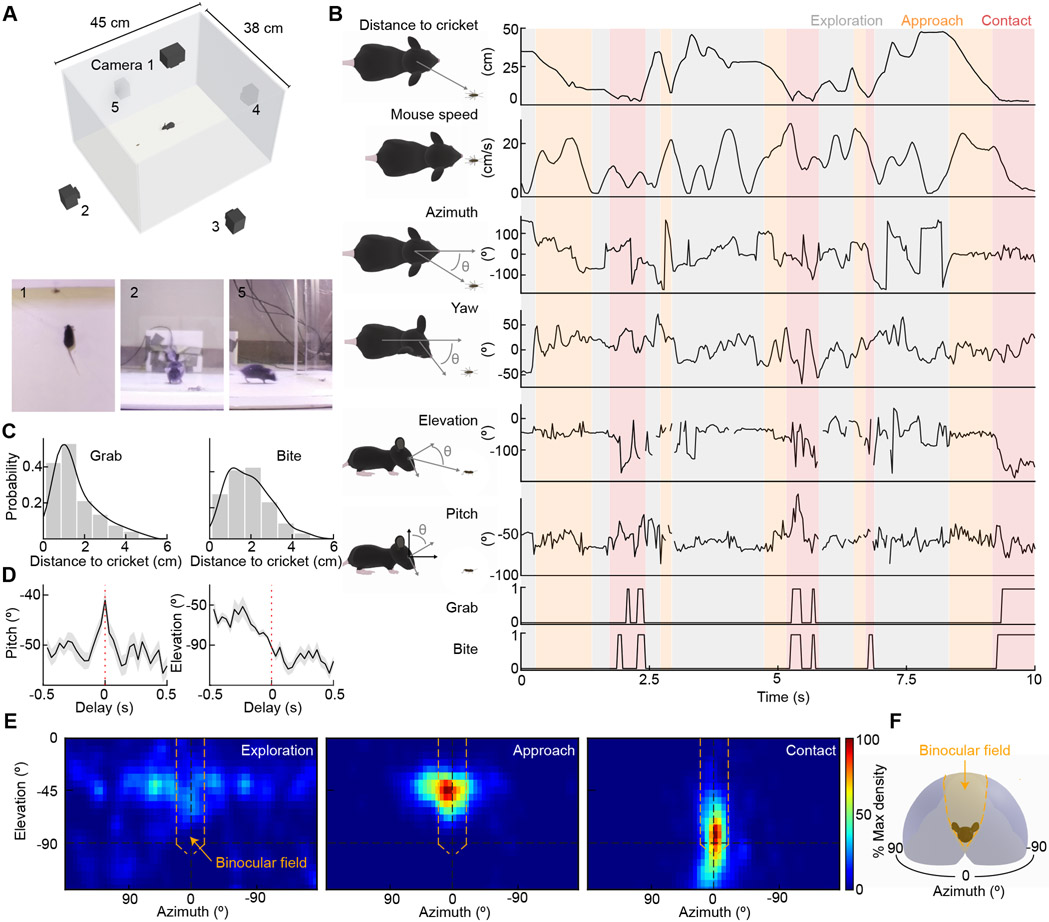
A recent study combined head-mounted sensors and 2D movies shot from above to monitor eye-head coupling and predator-prey interactions of mice hunting crickets (Michaiel et al., 2020). Overhead observations revealed that mice keep crickets within their binocular fields in azimuth (i.e., the lateral angle between the mouse's head and the cricket) (Hoy et al., 2016; Michaiel et al., 2020). However, the visual field elevation of crickets (i.e., the vertical angle between the mouse's head and the cricket) could not be measured from this perspective. Thus, it is unclear whether, at close distances, crickets are obstructed from view by the mouse's head, and the final stages of the attack are independent of vision. Similarly, because predator-prey interactions are only partly visible from above, how mice capture crickets remains unknown. To address these questions, we simultaneously recorded mice hunting crickets on five cameras (one above the arena and four on its sides) and tracked predator-prey interactions in 3D (Figure 1A, see STAR Methods).
Figure 1. 3D tracking of predator-prey interactions.
(A) Schematic diagram of the arena for 3D tracking predator-prey interactions. (1-3) example frames of hunting from each of the three of five synchronized cameras.
(B) Simultaneous measurements of mouse speed and head motion with distance to cricket and position of cricket within the mouse visual field.
(C) Histograms of distance to cricket when mice grab (left) or bite (right) them include all bites and grabs across 13 hunts.
(D) Bite-triggered-average traces (mean ± SEM) of the mice's head pitch (left) and visual field elevation of crickets (right) 0.5 s before to 0.5 s after a bite. Mice keep cricket above the nose until they initiate a bite.
(E) Heatmaps of cricket positions during each of the three hunting phases from 13 hunts.
(F) Schematic of the mouse visual field with monocular (gray) and binocular areas (orange) color-coded.
See also Video S1.
We distinguished three phases of the mouse's hunting behavior: exploration, approach, and contact (Figure 1B, see STAR Methods). First, we defined as 'exploration' periods when mice were neither approaching nor in contact with crickets. Second, we defined as 'approaches' periods when mice were running toward the crickets at speeds > 10 cm/s, decreasing the distance to the crickets by > 7 cm/s, similar to previous definitions (Hoy et al., 2016, 2019). Third, we defined as 'contact' periods when mice were within 4 cm of the crickets. Side-view movies revealed that when mice were within 4 cm of crickets, they started to bite and reach for them with their front paws (i.e., grab, Figure 1C). Bites and grabs occurred in a stereotypic attack sequence (bite-and-grab), in which mice rapidly swung their head up from the nose-down position to bite crickets and, with a slight delay, lifted their front legs to grab crickets (Figure 1C, Video S1). Once mice had secured crickets, they swiftly decapitated them. When we mapped the crickets' positions within the head-centric visual field of mice (Figures 1D and 1E), we found that their distributions were diffuse during exploration but coalesced to a narrow region within the binocular visual field during approach and contact. Mice pitched their heads nose-down to keep crickets visible during approach and contact until a bite-and-grab attack. Thus, our 3D tracking of predator-prey interactions suggests that all phases of the hunt are guided by vision, and that approach and capture may rely on binocular vision.
Monocular mice are poor hunters
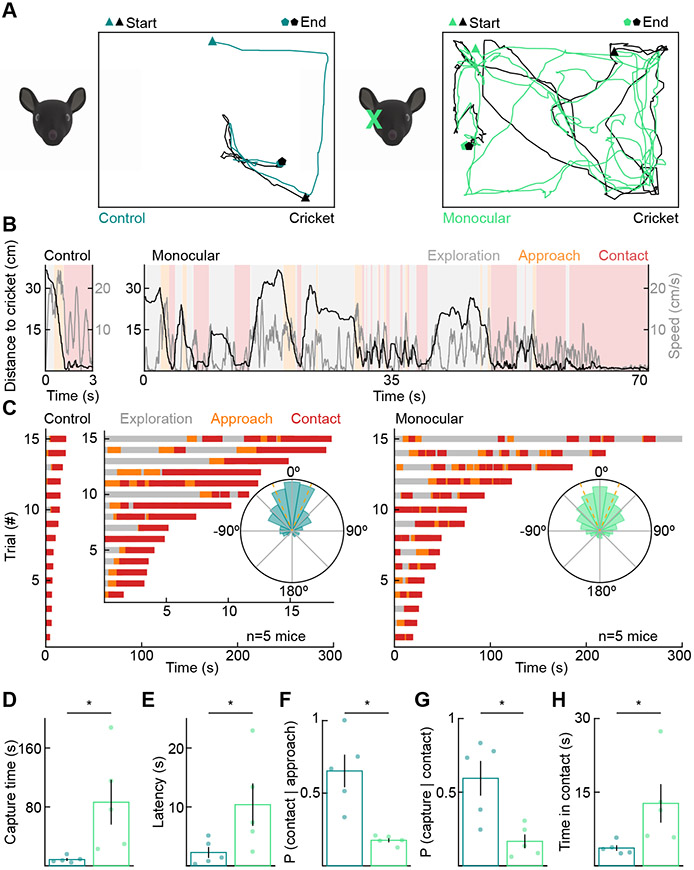
To explore the importance of binocular vision for hunting, we removed one eye from adult mice (postnatal day 30, P30). Compared to control littermates, monocular mice took much longer to capture crickets (Figures 2A-2D, S1, and Video S2). Deficits affected all phases of the hunt. Monocular mice were slower to detect prey and initiate their first approach (Figure 2E). Monocular mice were also less likely to convert approaches into contacts (Figures 2F, S1) and contacts into captures (Figures 2G and 2H). Notably, monocular and control mice did not differ in their maximum running speeds, and the crickets used as prey for both groups were indistinguishable in size (Figure S2). Overhead observations showed that cricket positions were more dispersed in azimuth relative to the mice's heading during approach and contact of monocular compared to control mice (Figure 2C, insets). These results are consistent with the notion that binocular vision guides prey capture. However, because monocular enucleation reduces the overall size of the visual field and generates a large scotoma, they do not prove that the combination rather than the presence of information from both eyes determines hunting success.
Figure 2. Monocular mice are poor hunters.
(A) Representative overhead tracking of control (left) and monocularly enucleated mice (right) hunting crickets.
(B) Same hunts as in (A) showing the distance to the cricket and mouse speed in the three hunting phases.
(C) Exploration, approach, and contact over time for all test-day hunting trials of control (left) and monocularly enucleated (right) mice. We recorded three trials for each mouse. Trials are shown in order of their duration. Insets: circular histograms of the cricket azimuth during approaches across all control (left) and enucleated (right) mice.
(D) Time from the introduction of a cricket to its successful capture (control: 8.55 ± 1.95 s, n = 5, enucleated: 86.37 ± 30.38 s, n = 5, p = 0.008). For D-H, each point is the average of three trials per mouse.
(E) Latency to detect prey and initiate first approach (control: 2.27 ± 0.92 s, enucleated: 10.38 ± 3.61 s, p = 0.032).
(F) Probability that mice successfully convert approaches into contacts (control: 0.65 ± 0.11, enucleated: 0.18 ± 0.01, p = 0.008).
(G) Probability that mice successfully convert contacts into captures (control: 0.59 ± 0.12, enucleated: 0.17 ± 0.05, p = 0.016).
(H) Total time within contact range of the cricket before successful capture (control: 3.63 ± 0.57 s, enucleated: 12.72 ± 3.93 s, p = 0.008).
See also Figures S1, S2, and Video S2.
Distribution of ipsi-RGCs
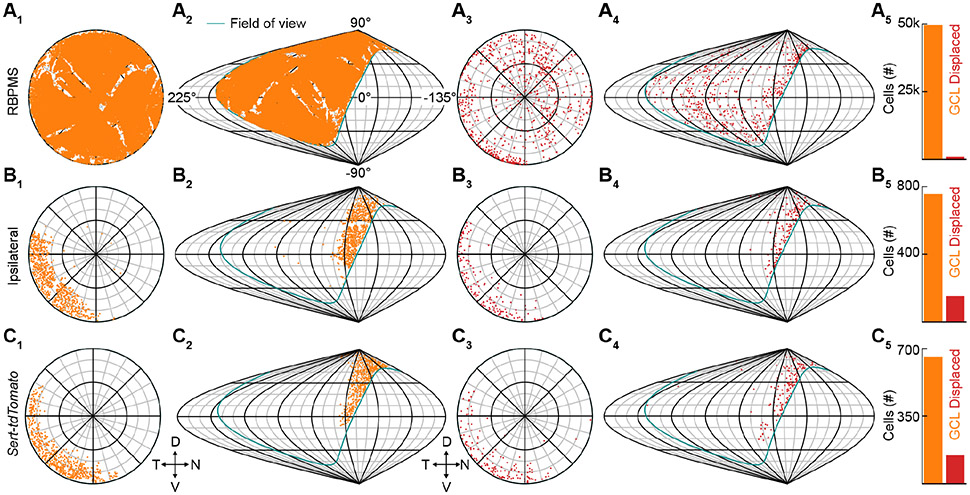
In mammals, binocular vision relies on the partial decussation of RGC axons in the optic chiasm (Petros et al., 2008). In mice, most RGC axons cross at the chiasm, but a subset of RGCs in the ventrotemporal retina innervate ipsilateral targets that combine information from both eyes (Dräger and Olsen, 1980; Rompani et al., 2017). We mapped the distribution of ipsi-RGCs in mice to analyze the cellular substrate of binocular vision. We first stained flat-mounted retinas for RNA-binding protein with multiple splicing (RBPMS), a marker of all RGCs (Tran et al., 2019). We acquired confocal images covering the retina's expanse in the ganglion cell layer (GCL) and inner nuclear layer (INL). We segmented RGCs using a deep learning-based algorithm and generated retinotopic and visuotopic maps of their distributions (Figure 3A) (Stringer et al., 2020). Thus, we found that the mouse retina contains nearly 50,000 RGCs, a small subset of which (~1.4%) is displaced in the INL. The displaced RGCs are distributed unevenly with an area of increased density in the ventrotemporal retina, which covers the contralateral visual field.
Figure 3. Distribution of ipsilaterally projecting retinal ganglion cell.
(A) Distribution of all RBPMS-stained GCL RGCs in a flat-mounted retina (A1) and projection of GCL RGCs onto visual space (A2). The teal outline indicates the edge of the right eye's visual field. Distribution of all RBPMS-stained displaced RGCs in a flat-mounted retina (A3) and projection of displaced RGCs onto visual space (A4). Cell counts of GCL (49,442) and displaced (732) RGCs (A5). Displaced RGCs are ~1.4% of the total RGC population.
(B) Distribution of all CTB-positive GCL RGCs following injection into the ipsilateral dLGN in flat-mounted retina (B1) and projection of ipsilateral GCL RGCs onto visual space (B2). Distribution of all displaced ipsilateral RGCs in a flat-mounted retina (B3) and projection of displaced ipsilateral RGCs onto visual space (B4). Cell counts of GCL (756) and displaced (150) ipsilateral RGCs (B5). Displaced ipsilateral RGCs are ~16% of the total ipsilateral RGC population.
(C) Distribution of Sert-tdTomato GCL RGCs in flat-mount retina (C1) and projection of Sert-tdTomato GCL RGCs onto visual space (C2). Distribution of all displaced Sert-tdTomato RGCs in a flat-mounted retina (C3) and projection of displaced Sert-tdTomato RGCs onto visual space (C4). Cell counts of GCL (655) and displaced (145) Sert-tdTomato RGCs (C5). Displaced Sert-tdTomato RGCs are ~18% of the total ipsilateral RGC population. D, N, V, and T denote dorsal, nasal, ventral, and temporal, respectively, in retinotopic and visuotopic space.
To map the distribution of ipsi-RGCs, we first injected a fluorescent retrograde tracer into one dorsolateral geniculate nucleus (dLGN) or superior colliculus (SC) and charted RGCs labeled in the ipsilateral retina (Figure 3B). Retrogradely labeled ipsi-RGCs localized to the GCL and INL of the ventrotemporal retina. Displaced RGCs accounted for ~16% of the ipsi-RGCs. We next crossed Sert-Cre mice, which target ipsi-RGCs, to a red fluorescent reporter strain (Sert-tdTomato mice, Figure 3C) (Gong et al., 2007; Koch et al., 2011; Madisen et al., 2010). Like retrograde tracing, this genetic approach labeled RGCs in the GCL and INL of the ventrotemporal retina. Furthermore, the total number of RGCs labeled in Sert-tdTomato retinas was close to that obtained from retrograde tracing, as was the fraction of displaced RGCs (~18%). In retinas of Sert-tdTomato mice injected with a green fluorescent tracer into the ipsilateral dLGN, nearly all (~89%) red RGCs were also green. Thus, ipsi-RGCs make up a small fraction of all RGCs in the mouse retina (~1.8%). They are restricted to the ventrotemporal retina, are more likely displaced in the INL than contralaterally projecting RGCs, and can be targeted selectively and comprehensively in Sert-Cre mice.
Few RGC types support binocular vision
The fraction of ipsi-RGCs (~1.8%) is much smaller than the fraction of the retina they occupy (~22%) (Dräger and Olsen, 1980), indicating that ipsi-RGCs are either a small subset of RGC types in the ventrotemporal retina or a subset of RGCs of many types. Despite recent progress in analyzing RGC projection patterns (Martersteck et al., 2017; Rompani et al., 2017), the complement of ipsi-RGC types remains poorly characterized.
Morphology, gene expression patterns, and light responses define RGC types. Large-scale surveys in each category have identified more than 40 RGC types in mice (Baden et al., 2016; Bae et al., 2018; Rheaume et al., 2018; Tran et al., 2019). However, how classifications align across categories remains uncertain as multimodal datasets are scarce (Bae et al., 2018) (rgctypes.org). Therefore, we combined large-scale functional and morphological analyses to classify and comprehensively catalog the ipsi-RGCs of mice.
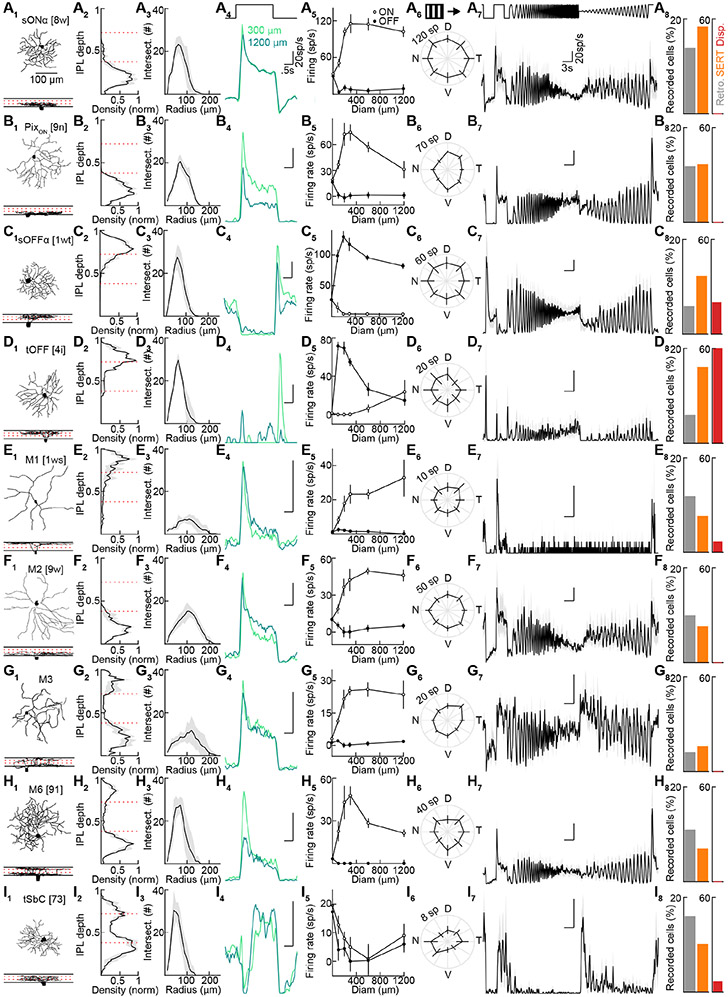
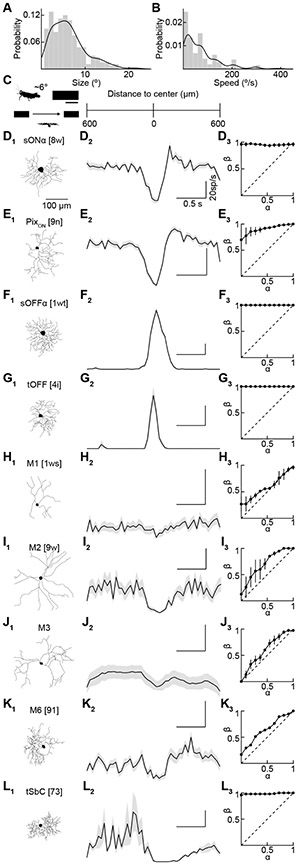
We fluorescently labeled ipsi-RGCs by transgenics (Sert-tdTomato, n = 28 retinas) or retrograde tracing from the dLGN (n = 13 retinas) or SC (n = 5 retinas). All three approaches labeled the same RGC types, and we, therefore, combined their data. We targeted fluorescent ipsi-RGCs for whole-cell patch-clamp recordings under two-photon guidance. We measured their responses to three sets of stimuli: varying size spots, square-wave gratings drifting in different directions, and full-field chirp (Figure 4A). These stimuli allowed us to distinguish key response properties and compare our data to previous functional surveys (Baden et al., 2016; Bae et al., 2018) (rgctypes.org). Simultaneously, we filled RGCs with fluorescent dyes (spectrally separate from the transgenic and retrograde labels) included in the intracellular solution and reconstructed their morphology from two-photon image stacks acquired at the end of each recording. In total, we recorded and reconstructed 232 ipsi-RGCs from 46 retinas. We developed a serial classification approach that reliably divided ipsi-RGCs into nine types (Figure S3). The functional and morphological features of ipsi-RGCs were highly consistent within these types and conspicuously different between them (Figures 4 and S4). The nine ipsi-RGC types fall into three groups. First, ipsi-RGCs include four conventional contrast-coding RGC types (Figures 4A-4D) that differ in their preference for light increments (sONα and PixON) and decrements (sOFFα and tOFF), response transience (tOFF > sOFFα), and surround suppression (PixON > sONα and tOFF > sOFFα) (Johnson et al., 2018; Murphy and Rieke, 2006; Pang et al., 2003; Schwartz et al., 2012). Notably, tOFFα-RGCs are absent from this group (Huberman et al., 2008). Second, ipsi-RGCs encompass the melanopsin-expressing RGC types: M1-, M2-, M3-, M4- (i.e., sONα), M5- (i.e., PixON), and M6-RGCs (Figures 4A and 4E-4H) (Ecker et al., 2010; Johnson et al., 2018; Levine and Schwartz, 2020; Quattrochi et al., 2018; Schmidt et al., 2011; Stabio et al., 2017). M-RGCs signal luminance (particularly M1-, M2, M3, and M4-RGCs) and, to some extent, contrast (particularly M4-, M5-, M6-RGCs) (Do, 2019). Third, transient Suppressed-by-Contrast (tSbC-) RGCs, which have high baseline firing rates transiently suppressed by local or global light increments and decrements, are abundant among ipsi-RGCs (Figure 4I) (Tien et al., 2015).
Figure 4. A catalog of ipsilaterally projecting RGCs.
(A-I) Representative traced cells of each ipsilateral cell type (X1). Stratification profiles for each cell type (X2). Dashed red lines show approximate ChAT band locations. (n = 5 traced cells for each type). (x3) Sholl analyses (X3, n = 5 traced cells for each type). Representative spike responses of individual cells of each type to 300- and 1200-μm spot stimulus (X4). Average firing rates to spots of different size (X5, A: n = 25, B: n = 20, C: n = 13, D: n = 20, E: n = 14, F: n = 14, G: n = 13, H: n = 13, I: n = 20). Spike count responses to 2 s drifting gratings moving in eight directions (X6, A: n = 16, B: n = 18, C: n = 8, D: n = 13, E: n = 12, F: n = 14, G: n = 12, H: n = 13, I: n = 20). Responses to chirp stimuli (X7, A: n = 8, B: n = 3, C: n = 2, D: n = 4, E: n = 3, F: n = 4, G: n = 2, H: n = 6, I: n = 4). Percent of total cells recorded that belonged to each type by targeting retrogradely labeled cells (X8, A: n = 14, B: n = 12, C: n = 6, D: n = 6, E: n = 12, F: n = 10, G: n = 4, H: n = 11, I: n = 16, of 101 total cells), Sert-tdTomato cells (A: n = 24, B: n = 16, C: n = 22, D: n = 21, E: n = 10, F: n = 10, G: n = 7, H: n = 9, I: n = 13, of 131 total cells) and displaced RGCs from either retrogradely or tdTomato-labeled cells (A: n = 0, B: n = 0, C: n = 3, D: n = 9, E: n = 1, F: n = 0, G: n = 0, H: n = 0, I: n = 1, of 15 total cells. See also Figure S3 and S4). Cells that could not be classified as one of these types: retrograde (n = 10), Sert-tdTomato (n = 9), displaced (n = 1).
See also Figures S3-S5.
Our analysis of ipsi-RGC distributions revealed that a significant portion (16-18%) is displaced in the INL. Due in part to their less accessible location, displaced RGCs have not been characterized. We recorded and reconstructed 14 displaced ipsi-RGCs. These overwhelmingly represented tOFF-RGCs (9/14), fewer sOFFα-RGCs (3/14), and rarely M1- (1/14) and tSbC-RGCs (1/14) (Figures S4 and S5). In contrast to this narrow set of displaced ipsi-RGC types, we observed diverse responses among displaced contralaterally projecting RGCs (Figure S5).
Thus, a small subset of RGC types (9/40+) have ipsilateral projections and support binocular vision in mice. The ipsi-RGCs include conventional contrast-encoding types, melanopsin-expressing luminance-encoding types, and a transient Suppressed-by-Contrast type. Large groups of RGCs (e.g., direction-selective RGCs (Wei, 2018), orientation-selective RGCs (Nath and Schwartz, 2016, 2017), and small motion-sensitive RGCs (Jacoby and Schwartz, 2017; Zhang et al., 2012)) are absent from the ipsi-RGC set. Displaced ipsi-RGCs are a subset of the ipsi-RGCs in the GCL, enriched in two types whose dendrites stratify close to the INL.
Ipsi-RGCs guide predation
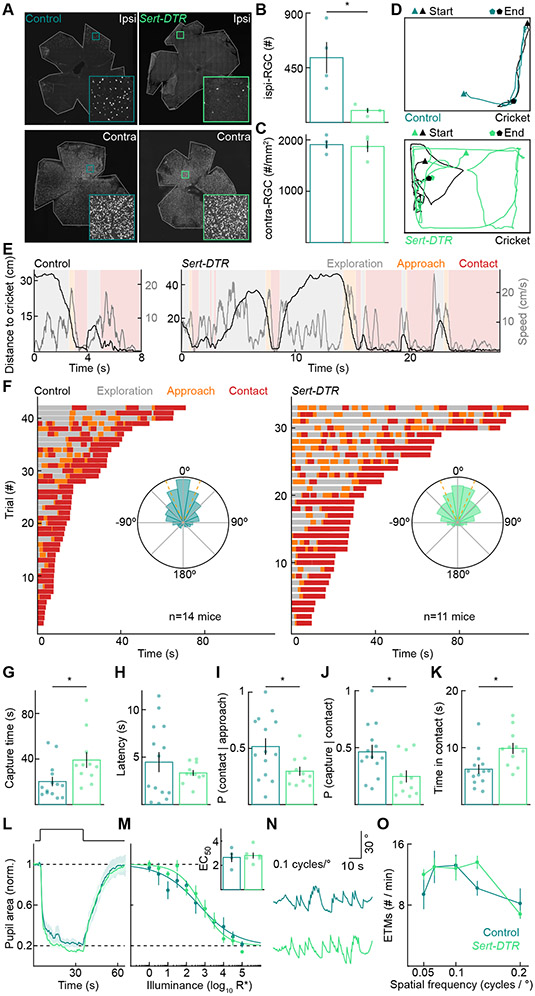
Binocular vision in mammals depends on ipsi-RGCs. To test the importance of binocular vision for predation, we selectively removed ipsi-RGCs from adult mice and examined the impact on cricket hunting. We removed ipsi-RGCs by injecting diphtheria toxin (DT) into both eyes of P30 Sert-DTR mice. Compared to controls (i.e., Cre-negative littermates injected with DT), ipsi-RGC numbers were reduced by 82% in Sert-DTR mice (Figures 5A and 5B). In contrast, the density of contralaterally projecting RGCs, labeled by the same retrograde tracer injections, was unchanged (Figures 5A and 5C). In addition, the density of serotonergic neurons in the brain, which express DTR in Sert-DTR mice, was unaffected by intraocular DT injections (Figure S6). Thus, intraocular DT injections in Sert-DTR mice selectively and nearly completely removed ipsi-RGCs from adult mice. Importantly, this manipulation eliminates binocular vision without creating a scotoma as the binocular visual field is still covered by contralaterally projecting RGCs.
Figure 5. Ipsilaterally projecting RGCs are required for efficient prey capture.
(A) Representative images of control (left) and Sert-DTR (right) ipsilateral (top) and contralateral (bottom) retinas with RGCs retrogradely labeled by CTB injections into the right dLGNs.
(B) Number of ipsi-RGCs retrogradely labeled by dLGN injections in control and Sert-DTR mice (control: 533 ± 129, Sert-DTR: 98 ± 20, p = 0.029).
(C) Peak density of contra-RGCs retrogradely labeled by dLGN injection in the control and Sert-DTR mice shown in (B) (control: 1908 ± 80 RGCs/mm2, Sert-DTR: 1871 ± 109 RGCs/mm2, p = 0.90).
(D) Representative overhead tracking of mouse and cricket positions in control (top) and Sert-DTR mice (bottom).
(E) Same hunts as in (D), showing the distance to cricket and mouse speed with the three hunting phases.
(F) Exploration, approach, and contact over time for all test-day hunting trials of control (left) and Sert-DTR mice (right). We recorded three trials for each mouse. Trials are shown in order of their duration. Insets: circular histograms of the cricket azimuth during approaches across all control (left) and monocularly enucleated (right) mice (p = 1).
(G) Time from the introduction of a cricket to its capture (control: 20.23 ± 4.15 s, Sert-DTR: 39.09 ± 6.89 s, p = 0.011). For G-K, each point is the average of three trials per mouse.
(H) Latency to detect prey and initiate first approach (control: 4.47 ± 1.05 s, Sert-DTR: 3.33 ± 0.31 s, p = 0.94).
(I) Probability that mice successfully convert approaches into contacts (control: 0.52 ± 0.07, Sert-DTR: 0.30 ± 0.04, p = 0.035).
(J) Probability that mice successfully convert contacts into captures (control: 0.46 ± 0.06 s, Sert-DTR: 0.25 ± 0.05 s, p = 0.033).
(K) Total time within contact range of the cricket before successful capture (control: 6.21 ± 0.81 s, Sert-DTR: 9.86 ± 0.97 s, p = 0.001).
(L and M) Representative pupil area traces and summary data for control (n = 4) or Sert-DTR (n = 6) mice to varying illuminance steps. Representative traces (L) illustrate normalized and averaged (± SEM) responses to 5 log10 R*. Summary data (M) plot normalized and averaged (± SEM) pupil area at the point of maximal pupil constriction for each illuminance step and were fitted with a Hill curve for each mouse to derive an EC50 value (inset, p = 0.73). Dashed lines indicate baseline, dark-adapted pupil area, and maximal pupil constriction.
(N and O) Representative eye movement traces and summary data for eye-tracking movements (ETMs) in Control (n = 5) or Sert-DTR (n = 5, p = 0.35) mice. Representative traces (N) illustrate responses to 0.1 cycles/°. Summary data (O) for each spatial frequency are presented as mean ± SEM.
See also Figures S2, S6, S7, and Video S3.
The selective removal of ipsi-RGCs (<2% of all RGCs) caused severe deficits in predation (Figures 5D-5F, S7, Video S3). Mice took, on average, nearly twice as long to capture crickets (Figure 5G). This difference was not due to delays in prey detection and approach initiation (Figure 5H). Instead, Sert-DTR mice failed to convert approaches into contacts (Figure 5I) and contacts into captures (Figure 5J) more frequently than littermate controls. The failure to convert contacts into captures increased overall contact times (Figure 5K). Approaches failed, i.e., crickets escaped, at further distances for Sert-DTR than control mice (Figure S7C). Interestingly, Sert-DTR mice did not alter their viewing strategies during approach and contact (Figures 5F and S7B) and executed their final attack sequence (i.e., bite-and-grab) at similar distances as littermate controls (Figures S7D and S7E). However, they attacked less frequently within contact range (Figures S7D and S7E). Sert-DTR and control mice did not differ in their running speeds (Figure S2) or hunting success in the dark (Figure S6), and the crickets used as prey for both groups were indistinguishable in size (Figure S2).
We found that the pupillary light response and optokinetic nystagmus did not differ significantly between Sert-DTR and control mice (Figures 5L-5O), highlighting the specific importance of ipsi-RGCs to predation and the selectivity of our approach.
Sert-DTR mice tended to catch crickets faster than monocular mice (Sert-DTR: 37.3 ± 6.9 s, monocular: 86.4 ± 30.4 s). Although this trend did not reach statistical significance (p = 0.3), it likely reflects the scotoma generated by enucleation. Consistent with this interpretation, monocular (Figure 2E) but not Sert-DTR mice (Figure 5H) exhibited deficits in prey detection. The normalcy of prey detection in Sert-DTR mice suggests that it is either mediated by members of the ipsi-RGC types outside the ventrotemporal retina or by other RGC types.
To rule out the possibility that predation is sensitive to any RGC loss, we injected TNF-α (1 ng) or saline into both eyes of wild-type mice (Figure S8). TNF-α kills RGCs through microglial activation and an excitotoxic mechanism (Nakazawa et al., 2006; Vargas et al., 2015). Six weeks after injections, TNF-α-injected mice had ~15% fewer RGCs than saline-injected mice (Figures S8A and S8B). This comparatively large RGC loss did not affect the mice's prey capture performance (Figures S8C-S8F). TNF-α-injected mice took as long to detect and approach crickets as saline-injected mice (Figure S8G). They were equally successful in converting approaches into contacts (Figure S8H) and contacts into captures (Figures S8I and S8J).
Thus, the small type- and region-specific ipsi-RGC set, which mediates binocular vision in mice, specifically guides prey pursuit and capture and determines hunting success.
A subset of ipsi-RGCs detects prey-mimetic stimuli
To understand how ipsi-RGCs guide predation and which ipsi-RGC types likely contribute to this behavior, we analyzed their responses to a prey-mimetic stimulus. We used our 3D tracking of predator-prey interactions to estimate stimulus size and speed during predation, taking into account the crickets' and mice's movements relative to each other (Figures 6A and 6B).
Figure 6. A subset of ipsilaterally projecting RGCs reliably signal prey.
(A) Cricket size in ° of visual angle during all approach frames from nine hunts.
(B) Cricket speed in ° of visual angle per second during all approach frames from nine hunts.
(C) Schematic of visual stimulus used to mimic a cricket of size 5.7 x 2.3 ° (195 x 75 μm) moving at 19 °/s (650 μm/s) through the receptive field centered on the RGC soma aligned with the traces in D-L.
(D-L) Representative morphologies of each cell type (X1). Average responses of a representative cell of each type to 24 stimulus repeats (X2, three repeats x eight directions; no cells were direction selective). ROC curves for each ipsi-RGC type illustrating ability to detect cricket stimulus (X3, area under curve D: 0.95 ± 0.024, n = 3, E: 0.91 ± 0.053, n = 4, F: 1.0 ± 0, n = 3, G: 1.0 ± 0, n = 5, H: 0.59 ± 0.017, n = 3, I: 0.72 ± 0.080, n = 2, J: 0.61 ± 0.001, n = 3, K: 0.65, n = 1, L: 0.99 ± 0.015, n = 2).3
Based on the estimated size and speed distributions, we designed a simple prey-mimetic stimulus consisting of a dark bar (width: 5.7 °, height: 2.2 °) moving along its long axis at 19 °/s against a bright background (Figure 6C). We targeted ipsi-RGCs for whole-cell patch-clamp recordings as before, measured their responses to the prey-mimetic stimulus, varying size spots, and gratings drifting in different directions (the latter two for classification), and reconstructed their morphology. Receiver operating characteristics showed that the four contrast-coding RGC types reliably detected the prey-mimetic stimulus. sONα- and PixON-RGC have high baseline firing rates and responded by spike suppression (Figures 6D and 6E), whereas sOFFα- and tOFF-RGCs increased their firing rates to the prey-mimetic stimulus (Figures 6F and 6G). Importantly, M1-, M2-, M3-, and M6-RGCs did not respond consistently to the prey-mimetic stimulus (Figures 6H-6K). Finally, tSbC-RGCs, like sONα- and PixON-RGCs, reliably signaled the prey-mimetic stimulus through spike suppression (Figure 6L). Although natural prey present more complex stimuli than our simple approximation, the failure of M1-, M2, M3, and M6-RGCs to detect this simple stimulus suggests that all or a subset of the five remaining ipsi-RGC types (sONα-, PixON-, sOFFα-, tOFF-, and SbC-RGCs) guide mice in their pursuit and capture of prey.
DISCUSSION
Here, we analyze the predator-prey interactions of mice and crickets in 3D, test the role of binocular vision in predation, and catalog the RGC types that mediate binocular vision and predation in mice. We reach six main conclusions. First, mice move their heads to keep targets within their binocular visual fields as they approach and contact prey. Second, mice engage a stereotyped attack sequence (bite-and-grab) to capture crickets. Third, a small number of RGC types (9/40+, sONα-, PixON-, sOFFα-, tOFF-, M1-, M2-, M3-, M6-, and tSbC-RGCs) in the ventrotemporal retina have ipsilateral projections and support binocular vision. Fourth, two ipsi-RGCs (tOFF- and sOFFα-RGCs) are frequently displaced in the INL. Fifth, ipsi-RGCs are required for efficient prey pursuit and capture. Sixth, a subset of ipsi-RGCs (5/9 types, sONα-, PixON-, sOFFα-, tOFF-, and tSbC-RGCs) signal prey. Thus, viewing strategies align with a small region- and cell-type-specific set of RGCs (<2% of RGCs) to mediate binocular vision and guide predation.
Recent studies revealed two types of eye-head coupling in freely moving mice, including mice on the hunt (Meyer et al., 2020; Michaiel et al., 2020). First, eye movements compensate for head tilt to stabilize the mice's gaze in the horizontal plane (Meyer et al., 2020; Michaiel et al., 2020). Second, eye movements combine with head yaw rotation to generate gaze-shifting saccade-and-fixate sequences (Meyer et al., 2020; Michaiel et al., 2020). We tracked mice (including head movements) and crickets in a shared 3D reference frame (Figure 1). Combined with the recent insights into eye-head coupling, our data demonstrate that mice keep crickets in their binocular visual field during predation and reveal that mice tilt their heads nose-down farther when hunting (pitch: −50 °) than during normal walking (Oommen and Stahl, 2008; Vidal et al., 2004), so that, even at close distances, prey remains visible above their nose.
Our 3D observations provide new insights into the final stages of the attack. We find that mice capture and subdue crickets by biting and grabbing them. In bites, mice quickly swing their heads up from the nose-down position, and in grabs, mice lift their front legs simultaneously and reach for the crickets. Bites and grabs are triggered when mice are within striking distance, < 4 cm from the cricket. Mice can either bite or grab but most frequently deploy these actions in a stereotypic bite-and-grab sequence. Once mice held crickets in their front paws, they swiftly decapitated them. Some mice, like the grasshopper mouse (Onychomys torridus), are specialized predators (Langley, 1989; Mccarty and Southwick, 1977). Insects have been found in the stomachs of wild house mice (Mus musculus) (Whitaker, 1966). It will be interesting to see if the attack sequence we observed is innate or learned through trial and error and whether Mus musculus, like Onychomys torridus (Whitman et al., 1986), displays prey-specific attack behaviors.
Hunting success is one of evolution's main selection pressures (Sillar et al., 2016). It has been hypothesized that binocular vision in mammals arose to support predation (Cartmill, 1974; Pettigrew, 1986). Our finding that binocular vision in mice guides prey pursuit and capture and determines hunting success is consistent with this hypothesis (Figures 2 and 5). Binocular vision could benefit predation through stereopsis. In stereopsis, disparate object positions (i.e., Wheatstone stereopsis) or background occlusions (i.e., da Vinci stereopsis) in the two retinal images support depth perception (Nityananda and Read, 2017; Ponce and Born, 2008). Stereopsis can help track prey against a matching background (i.e., camouflage breaking) and determine the correct distance to release the final attack sequence (i.e., range finding) (Nityananda and Read, 2017; Ponce and Born, 2008). The two retinal images should be consistently aligned to supply reliable depth information, which requires conjugated eye movements (Cumming and DeAngelis, 2001). In humans, small misalignments of retinal images disrupt depth perception and cause double vision (Duwaer and Van Den Brink, 1981; Lyle and Foley, 1955). The eye movements of freely moving mice, including mice on the hunt, are often unconjugated, destabilizing retinal images' alignment and casting doubt on the use of stereopsis in mice (Meyer et al., 2020; Michaiel et al., 2020). However, mice can distinguish stereoscopic surfaces (Samonds et al., 2019), binocular inputs to their dLGN and SC are topographically aligned (Cang and Feldheim, 2013), and their visual cortex contains numerous disparity-tuned neurons (La Chioma et al., 2019; Samonds et al., 2019; Scholl et al., 2013). We speculate that the comparatively low resolution of mouse vision and broad neuronal disparity tuning combine with interocular velocity differences in flow fields (Choi and Priebe, 2020; Nityananda and Read, 2017) to support stereoscopic vision during hunting. Cuttlefish present an evolutionarily distant example of stereopsis without tightly conjugated eye movements (Feord et al., 2020).
Besides stereopsis, binocular vision could improve prey pursuit and capture in dim light and low contrast via the increased convergence of RGCs on binocular neurons in the brain (Ponce and Born, 2008; Rompani et al., 2017). Consistent with this idea, combined input from both eyes enhances responses to weak stimuli in binocular compared to monocular dLGN neurons (Howarth et al., 2014), and mice show increased contrast sensitivity for stimuli presented in their binocular vs. monocular visual fields (Speed et al., 2019). Thus, binocular vision may support predation through stereopsis and improved contrast sensitivity. In addition to mammals (Figures 2 and 5), different forms of binocular vision contribute to the hunting success of evolutionarily distant predators (Bianco et al., 2011; Feord et al., 2020; Gahtan et al., 2005; Gebhardt et al., 2019; Nityananda et al., 2016). This parallel evolution underscores the algorithmic implementation-independent advantages of binocular vision to predation (Marr, 1982).
In mammals, binocular vision relies on the partial decussation of RGC axons in the optic chiasm (Petros et al., 2008). All RGC types in mice have contralateral projections (Dräger and Olsen, 1980; Martersteck et al., 2017). Which RGC types have ipsilateral projections and support binocular vision was unclear. We performed large-scale patch-clamp recordings and single-cell reconstructions (232 cells in 46 retinas) to catalog ipsi-RGCs in mice comprehensively (Figure 4). The combination of light responses and morphological characteristics allowed us to unambiguously identify nine ipsi-RGC types and match them to previous surveys (Bae et al., 2018) (rgctypes.org). This success highlights the benefits of multimodal datasets for robust and reproducible neuron classification (Zeng and Sanes, 2017).
The nine ipsi-RGC types include conventional contrast-encoding types (sONα-, PixON-, sOFFα-, and tOFF-RGCs), melanopsin-expressing types (M1-, M2-, M3-, M4- or sONα-, M5- or PixON, and M6- RGCs), and a transient Suppressed-by-Contrast (tSbC-RGC) type. Notably, the set of mouse ipsi-RGCs excludes feature detector types like the direction-selective RGCs (Wei, 2018), orientation-selective RGCs (Nath and Schwartz, 2016, 2017), and object-motion-sensitive RGCs (Jacoby and Schwartz, 2017; Zhang et al., 2012). This parallels observations in cats, in which most contrast-encoding alpha (or Y) and beta (or X) RGCs in the temporal retina project ipsilaterally, whereas half to two-thirds of the gamma (or W) RGCs in the temporal retina innervate contralateral brain areas (Rowe and Dreher, 1982; Stein and Berson, 1995; Stone and Fukuda, 1974; Wässle and Illing, 1980). Gamma cells encompass multiple types. Although assignments are tenuous, ipsilaterally projecting gamma cells appear to include M-RGCs and SbC-RGCs, whereas direction-selective and object-motion-sensitive gamma cells in the temporal retina innervate contralateral targets (Kirk et al., 1976; Rowe and Stone, 1977; Stein and Berson, 1995). Thus, a similar cast of ipsi-RGCs seems to support predation in cats and mice. This could reflect convergent evolution, homologous RGC types that arose in a common ancestor, or a combination of both.
The ipsilaterally projecting M-RGCs that fail to detect simple prey-mimetic stimuli (i.e., M1-M3 and M6-RGCs) likely contribute to global illumination measurements to other behavioral ends. We speculate that the lack of feature detectors among ipsi-RGCs is related to divergent optic flow fields at visuotopically matched retinal locations in both eyes during translational movements (Sabbah et al., 2017) and the often unconjugated (including rotational) eye movements of mice (Meyer et al., 2018, 2020; Michaiel et al., 2020), which could introduce feature-confusion in downstream neurons receiving input from both eyes.
A retinal specialization with increased cell densities and reduced receptive field sizes (i.e., increased spatial resolution) in the area projecting forward to the binocular field (i.e., area centralis) is a near-universal feature of animals with functional binocular vision (Cartmill, 1974; Pettigrew, 1986). Although no area centralis is evident when looking at all RGCs (Figure 3) (Dräger and Olsen, 1981; Jeon et al., 1998), Bleckert et al. (2014) found that the density of sONα- and sOFFα-RGCs is increased and their dendritic field size reduced in the ventrotemporal mouse retina. Comparisons of our morphological data (Figure S3) to previously published results indicate that dendritic field sizes of the other ipsi-RGC types do not differ between the ventrotemporal and other parts of the retina (Bae et al., 2018; Johnson et al., 2018; Tien et al., 2015). Therefore, the area-centralis-like specialization appears to be specific to sONα- and sOFFα-RGCs, which may play a particularly prominent role in binocular vision and predation. Thus, evolution produced ipsilateral projections of RGC types that can support predation in mice (i.e., the predator hypothesis) (Cartmill, 1974; Pettigrew, 1986), formed a cell-type-specific area centralis for a subset of them, and eliminated ipsilateral projections from RGC types that could cause feature confusion in downstream neurons (i.e., the feature confusion hypothesis).
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for further information, resources, and reagents should be directed to the Lead Contact, Daniel Kerschensteiner (kerschensteinerd@wustl.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw data and analysis routines are available upon request to the Lead Contact, Daniel Kerschensteiner (kerschensteinerd@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Throughout this study, we used ET33 Sert-Cre (Sert-Cre from here on) mice (Gong et al., 2007) to target ipsilaterally projecting RGCs (ipsi-RGCs). To label ipsi-RGCs, we crossed Sert-Cre mice to a tdTomato reporter strain (Ai9 (Madisen et al., 2010), The Jackson Laboratory, stock #007909). To remove ipsi-RGCs, we crossed Sert-Cre mice to a line expressing the diphtheria toxin receptor (DTR) in a Cre-dependent manner (Buch et al., 2005). On postnatal day 30 (P30), we injected 10-15 ng of diphtheria toxin (DT) into both eyes of Cre-positive (Sert-DTR mice) and Cre-negative (control mice) littermates from these crosses. Prey capture experiments began two weeks after injection. In monocular enucleation experiments, P30 Sert-Cre mice either had their right eye removed (monocular mice) or not (control mice). Prey capture experiments began two weeks after the enucleation. DT-injected control mice tended to catch crickets slower than uninjected controls (Figures 2D and 5G). This trend, which did not reach statistical significance (p = 0.07), might reflect differences between litters or visual impairments from binocular injections. For TNF-α experiments, we injected saline or 1 ng of TNF-α intravitreally in P30 wild-type mice (C57BL/6J, The Jackson Laboratory, stock #000664). Prey capture experiments began six weeks after injection. Crickets (Fluker's Farm) aged five to seven weeks (1.2-2.4 cm) were used as prey for all prey capture experiments. We used mice of both sexes. We observed no differences in predation between male and female mice and, therefore, combined their data. All procedures in this study were approved by the Animal Studies Committee of Washington University School of Medicine (Protocols #20170033 and #20-0055) and performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Prey capture training
Forty-eight hours before training began, mice were housed individually, and three crickets were introduced to each cage with food pellets. 16-18 hr before training, food pellets were removed, and three crickets were given to each mouse. On the first day of training, mice were introduced to the behavioral arena (width: 45 cm, length: 38 cm, height: 30 cm, with padded flooring) and given approximately three minutes to acclimate. A cricket was then placed in the center of the arena, and the interactions of mice and crickets were recorded with an overhead camera (30 fps; C310, Logitech). Mice were given up to 5 min to capture prey. Following successful capture or after 5 min, the arena was cleaned, and a new cricket was introduced. Each day, mice had the opportunity to capture three crickets. After three trials, mice were returned to their home cages and given access to food pellets for 6-8 hr. The food pellets were then removed, and mice were given three crickets in their home cages. This sequence was repeated for five days. The first four days were considered training for experiments with manipulations, and the prey capture was tested on the fifth day. All results reported are from the test day unless otherwise noted.
Prey capture recording and analysis
3D recording and analysis.
We filmed mice and crickets' interactions in the arena with five synchronized cameras (30 fps; e3Vision camera; e3Vision hub; White Matter LLC). One camera was mounted overhead, and four cameras were placed on the sides of the arena (Figure 1A, Video S1). Before tracking and analysis, all images were corrected for lens distortions (OpenCV, Python). The cricket and the mouse's nose and ears were tracked in videos from the overhead camera using DeepLabCut (Mathis et al., 2018) and manually edited using custom software (OpenCV, Python). The midpoint between the mouse's ears was defined as its head position and used to measure the distance to the cricket and speed of the mouse. A vector from the head position to the nose defined the heading of the mouse. The azimuth was calculated as the angle between this vector and vector from the head position to the cricket. We tracked the cricket and several points on the mouse's face in videos from the side-view cameras, including the nose and eyes, using DeepLabCut and manually edited this tracking using custom software (OpenCV, Python). Infrared light was used to enhance image contrast and improve tracking. The pitch of the head was approximated as the angle between the vector normal to the plane through the mouse's eyes and nose and the vertical axis. The cricket elevation was approximated as the angle between the same vector perpendicular to the mouse's eyes and a vector from the mouse's eyes to the cricket.
Bites and grabs were scored manually in videos from the side-view cameras. We verified on a subset of videos that three observers independently identified the same bites and grabs. We found biting and grabbing began when mice were within 4 cm of the cricket (Figure 1C). We defined this distance between the mouse and cricket (< 4 cm) as a contact. Contacts were usually preceded by the mice moving towards the cricket at speeds > 10 cm/s. We defined an approach as a period when the mouse was running at speeds > 10 cm/s, and the distance between mice and crickets decreased by > 7 cm/s. An approach ended when these criteria were no longer met or when the mice contacted the cricket. We defined exploration as the time that mice neither approached nor were in contact with crickets. Cricket position heatmaps were made from all video frames in which the elevation and azimuth of the cricket were calculated. The points were binned in 6-° increments, smoothed with a Gaussian window (standard deviation: 6 °), normalized by the total number of frames from that epoch, and the maximum density across the three epochs.
2D recording and analysis.
We calculated speed, azimuth, approach, and contact, as described above. Capture times reported are the average time to capture each of the three crickets on the test day. We calculated latency as the time from the introduction of the cricket to the first approach. The probability of contact given approach was calculated as the number of times contact occurred within 250 ms of an approach ending/the total number of approaches. The probability of capture given contact was calculated as 1/number of contacts. The time in contact was the total amount of time in contact range across all contacts before successful capture. All reported values are the average for three crickets on the test day.
Pupillary light response and optokinetic nystagmus recording and analysis
For pupillometry and optokinetic experiments, mice were headplated one week before behavioral testing. Mice were dark adapted for at least one hour and restrained in a custom head-fixed holder before exposure to visual stimuli. Pupil size and eye movements were tracked and recorded from the left eye using an ETL-200 eye-tracking system (ISCAN) under infrared illumination.
To assess the optokinetic reflex, square wave gratings of varying spatial frequencies (0.05, 0.067, 0.1, 0.13, 0.2 cycles/°) and Michelson contrasts (5%, 7.5%, 10%, 15%, 25%, 100%) moving at 10 °/s in the temporal-to-nasal direction were presented on a monitor 16 cm from the mouse's left eye at a 45 ° angle. Each presentation of the stimulus comprised 10 s of a uniform gray screen, 60 s of drifting gratings, and a final 10 s of a uniform gray screen. Eye-tracking movements were quantified as the number of saccades preceded by a slow tracking motion.
To assess the consensual pupillary light reflex, varying illuminance steps (0.0- 5.0 log10 R* in 0.5 log10 R* increments) were presented to the mouse's right eye using an Arduino-controlled 465 nm LED and a set of ND filters (Thorlabs). Each presentation of the stimulus comprised 5 s of background darkness, 30 s of illumination, and 30 s of post-illumination recovery to baseline, with a 2-min minimum of darkness between presentations. Pupil constriction was normalized to the dark-adapted pupil area, and the relative pupil area for each illuminance was calculated as the 5 s average around the maximum pupil constriction. To derive EC50 values, a Hill equation was fit to the data for each animal.
Tissue preparation
Mice were deeply anesthetized with CO2, killed by cervical dislocation, and enucleated. For patch-clamp recordings, mice were dark-adapted overnight before their retinas were isolated under infrared illumination (> 900 nm) in oxygenated mACSFNaHCO3 containing (in mM) 125 NaCl, 2.5 KCl, 1 MgCl2, 1.25 NaH2PO4, 2 CaCl2, 20 glucose, 26 NaHCO3, and 0.5 L-glutamine equilibrated with 95% O2 5% CO2. For confocal imaging, retinas were isolated in oxygenated mouse artificial cerebrospinal fluid (mACSFHEPES) containing (in mM): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgCl2, 20 HEPES, and 11 glucose (pH adjusted to 7.37 using NaOH), mounted flat on filter paper and fixed for 30 min in 4% paraformaldehyde in mACSFHEPES. Brains were removed and placed in 4% paraformaldehyde overnight. The following day, brains were sectioned in 100-μm thick coronal slices, stained, and mounted for confocal imaging.
Immunohistochemistry
Flat-mount preparations were cryoprotected (1 hr 10% sucrose in PBS at room temperature [RT], 1 hr 20% sucrose in PBS at RT, and overnight 30% sucrose in PBS at 4 °C), frozen and thawed three times, and blocked with 10% normal donkey serum in PBS for 2 hr before incubation with primary antibodies for five days at 4 °C. Flat mounts were washed in PBS (3 × 1 hr) at RT, incubated with secondary antibodies for one day at 4 °C, and washed in PBS (3 × 1 hr) at RT. Brain slices were blocked with 10% normal donkey serum in PBS for 2 hr before incubation with primary antibodies for 3 hr at RT, washed in PBS (3 × 20 min) at RT, incubated with secondary antibodies for 2 hr at RT, and washed in PBS (3 × 20 min) at RT. The following primary antibodies were used in this study: mouse anti-RFP (1:1,000, Abcam), guinea pig anti-RBPMS (1:1000, PhosphoSolutions), rabbit anti-Serotonin (1:200, S5545, Sigma–Aldrich). Secondary antibodies were Alexa 488 and Alexa 568 conjugates (1:1,000, Invitrogen).
Electrophysiology
Whole-cell patch-clamp recordings were obtained from dark-adapted retinas, flat-mounted on transparent membrane discs (13 mm Whatman Anodisc) superfused (~7 mL/min) with warm (30 – 33 °C) mACSFNaHCO3. Fluorescent RGCs were targeted under two-photon guidance (excitation wavelength: 940 nm) in either Sert-Cre Ai9 retinas or RGCs retrogradely labeled with cholera toxin B conjugated to Alexa 488 or Alexa 568 (CTB 488, CTB 568) or fluorescent retrobeads. The intracellular solution for current-clamp recordings contained (in mM) 125 K-gluconate, 10 NaCl, 1 MgCl2, 10 EGTA, 5 HEPES, 5 ATP-Na2, and 0.1 GTP-Na (pH adjusted to 7.2 with KOH). Patch pipettes had resistances of 3–6 MΩ (borosilicate glass). Signals were amplified with a Multiclamp 700B amplifier (Molecular Devices), filtered at 3 kHz (8-pole Bessel low-pass), and sampled at 10 kHz (Digidata 1550, Molecular Devices).
Retrograde labeling
Mice were anesthetized with a cocktail (0.1ml/20g body weight) of ketamine HCl (87 mg/kg) and xylazine (13mg/kg), and CTB 488 or CTB 568 (1 g/L; 300 nL, Thermo Fisher Scientific) were injected into either the right dLGN or SC using a NanojectII (Drummond). SC injections were targeted to 1 mm from the midline, 3.79-3.87 mm caudal from Bregma, and 1.32 mm below the surface of the brain. dLGN injections were targeted to 2-2.18 mm from the midline, 2.15-2.79 mm caudal from Bregma, and 2.75-2.88 mm below the surface of the brain. After completion of the injection, the wound was closed with stainless steel wound clips, and antiseptic ointment (Vetropolycin) applied to the suture. Approximately 72 hr after injection, retinas were prepared for physiological recordings as described above. SC- and dLGN-injections labeled the same ipsi-RGC types, and we, therefore, combined their results.
Visual stimulation
All visual stimuli were written using the Cogent Graphics toolbox (John Romaya, Laboratory of Neurobiology at the Wellcome Department of Imaging Neuroscience, University College London) in MATLAB (The MathWorks). A UV E4500 MKII PLUS II projector illuminated by a 385-nm light-emitting diode (LED, EKB Technologies) was used for stimulus presentation. Stimuli were focused onto the photoreceptors via a substage condenser of an upright two-photon microscope (Scientifica). All stimuli were centered on the soma of the recorded cell. Background luminance of 3,000 rhodopsin isomerizations/rod/s (R*) was used for all visual stimuli unless otherwise noted. Spots of varying diameters (0, 100, 200, 300, 600, 1200 μm) were presented in a pseudo-random sequence and square-wave modulated at 0.25 Hz (Michelson contrast: 100%). To test for direction/orientation-selectivity, full-field square-wave drifting gratings (Bar width: 225 μm; Speed: 900 μm/s; Michelson contrast: 100%) were presented in eight directions. A full-field chirp stimulus of gradually increasing temporal frequency and contrast was presented to a subset of cells (Figure 4) (Baden et al., 2016). To mimic a cricket in a mouse's visual field, we moved a 195 x 75 μm dark bar (5.7 x 2.3 °; Michelson contrast: 100%) through an RGC's receptive field in eight orientations at 650 μm/s (19 °/s). We classified RGCs based on their responses to varying size spots, drifting gratings, and morphological parameters (Figure S2) and matched types to other multimodal RGC classifications (rgctypes.org and museum.eyewire.org) (Bae et al., 2018).
Morphological analysis
We made maximum intensity projections of Alexa 488-filled cells were made in Fiji (Schindelin et al., 2012) to measure dendritic field and soma diameters. Using custom software written in MATLAB, a polygon was drawn around the edges of a cell's dendrites or soma, and the dendritic field or soma diameter was calculated as the longest distance across the polygon. Neurite length was calculated by tracing z stack images of Alexa 488 filled cells using Neurolucida (MBF Bioscience). Dendritic branching patterns were analyzed using the Sholl Analysis function in Neurolucida. To calculate the inner plexiform layer (IPL) depth, IPL borders were detected from transmitted light images. Traced neurites were registered by their relative position within the inner plexiform layer (0%–100% from its border with the INL to its border with the GCL). Dendrite length at each depth was measured using the 3D-wedge analysis function in Neurolucida.
Confocal imaging
Image stacks of whole fixed retinas were acquired through a 20 × 0.85 NA objective (Olympus) on an inverted laser scanning confocal microscope (LSM 800, Zeiss). Image stacks of fixed brains were acquired through a 10X 0.25 NA objective (Olympus) on an upright laser scanning confocal microscope (FV1000, Olympus). All images were processed with Fiji (Schindelin et al., 2012).
Electrophysiology analysis
Total spike counts or average spike rates during 100-200 ms time windows were measured. To construct receiver operator characteristic (ROC) curves, we compared firing rates during pre-stimulus (rpre) and stimulus (rstim) time windows to 12 threshold values (z) spanning the range of rpre values and plotted the hit rate (β) as a function of the false alarm rate (α), with α and β and defined by the following probabilities α(z) = P(rpre ≥ z) and β(z) = P(rstim ≥ z). For RGC types with suppressive responses'≥' was replaced by'≤'. The performance of each cell was then quantified by the area under its ROC curve (0.5 for chance performance to a maximal value of 1). All analyses were performed using custom scripts written in MATLAB.
Cell counting and visual space mapping
TdTomato- and RBPMS-positive RGCs in the GCL were counted in images of whole retinas using Cellpose (Stringer et al., 2020), a deep learning-based algorithm for cell segmentation. CTB-labeled cells and displaced RGCs were counted manually using custom software (OpenCV, Python). Retinas were outlined manually, and cell locations were mapped to retinotopic and visuotopic space using the R package Retistruct (http://davidcsterratt.github.io/retistruct/) (Sterratt et al., 2013).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
No statistical methods were used to predetermine sample sizes. P values were calculated using Mann-Whitney U tests, Kolmogorov–Smirnov tests, or bootstrapping were used to assess the statistical significance of observed differences. Unless otherwise noted, population data are reported as mean ± SEM, and n represents the numbers of animals or cells analyzed.
Supplementary Material
Video S1. Stereotypic bite-and-grab attack. (related to Figure 1)
Representative prey capture sequence simultaneously recorded on five cameras. Videos from the overhead camera and three of the four side cameras are shown.
Video S2. Monocular mice are poor hunters. (related to Figure 2)
Representative hunting examples from control (left) and monocularly enucleated (right) mice recorded on overhead cameras.
Video S3. Ipsilaterally projecting RGCs are required for efficient prey capture. (related to Figure 5)
Representative hunting examples from DT-injected control (left) and Sert-DTR (right) mice recorded on overhead cameras.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-RFP | Abcam | RRID:AB_945213 |
| Guinea pig anti-RBPMS | PhosphoSolutions | RRID:AB_2492226 |
| Rabbit anti-Serotonin (S5545) | Sigma-Aldrich | RRID:AB_477522 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor 488 hydrazide, sodium salt | Thermo Fisher Scientific | Cat# A10436 |
| Alexa Fluor 568 hydrazide, sodium salt | Thermo Fisher Scientific | Cat# A10437 |
| Cholera toxin B conjugated to Alexa Fluor 488 | Thermo Fisher Scientific | Cat# C34775 |
| TNF-α | PeproTech | Cat# 315-01A |
| Experimental models: organisms/strains | ||
| Mouse model: ET33 Sert-Cre | Dr. Erik Ullian | Gong et al., 2007 |
| Mouse model: Ai9 | The Jackson Laboratory | Strain# 007909 |
| Mouse model: DTR | The Jackson Laboratory | Strain# 007900 |
| Mouse model: C57BL76J | The Jackson Laboratory | Strain# 000664 |
| Cricket model: Acheta domesticus | Fluker’s Farm | N/A |
| Software and algorithms | ||
| Python 3 (Anaconda distribution) | Anaconda | https://www.anaconda.com/ |
| OpenCV (Python) | OpenCV | https://opencv.org/ |
| MATLAB | The Mathworks | RRID:SCR_001622 |
| Cogent Graphics Toolbox | LON | http://www.vislab.ucl.ac.uk/cogent_graphics.php |
| R | R Foundation for Statistical Computing | http://www.r-project.org/ |
| Retistruct | Sterratt et al., 2013 | http://davidcsterratt.github.io/retistruct/ |
| DeepLabCut | Mathis et al., 2018 | https://github.com/DeepLabCut/DeepLabCut |
| Neurolucida | MBF Bioscience | RRID:SCR_001775 |
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| Cellpose | Stringer et al., 2020 | https://github.com/MouseLand/cellpose |
Highlights.
Mice track prey with their binocular visual field
Few retinal ganglion cell types (9/40+) innervate ipsilateral brain targets
Ipsilaterally projecting ganglion cells guide predation
A subset of ipsilaterally projecting ganglion cell types (5/9) reliably signal prey
ACKNOWLEDGMENTS
Dr. E. Ullian kindly provided the Sert-Cre mice. We thank all members of the Kerschensteiner lab for helpful discussions throughout this study and Drs. J.L. Morgan, F. Soto, and M. Kerschensteiner for critical reading of the manuscript. This work was supported by the NIH (EY023341, EY026978, EY027411, and EY030623 to D.K., EY029975 to K.J.P., and the Vision Core grant EY0268), the Grace Nelson Lacy Research Fund (director: D.K.), and a Career Development Award to P.R.W. and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Euler T, and Berens P (2020). Understanding the retinal basis of vision across species. Nat. Rev. Neurosci 21, 5–20. [DOI] [PubMed] [Google Scholar]
- Bae JA, Mu S, Kim JS, Turner NL, Tartavull I, Kemnitz N, Jordan CS, Norton AD, Silversmith WM, Prentki R, et al. (2018). Digital Museum of Retinal Ganglion Cells with Dense Anatomy and Physiology. Cell 173, 1293–1306.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Kampff AR, and Engert F (2011). Prey capture behavior evoked by simple visual stimuli in larval zebrafish. Front. Syst. Neurosci 5, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Schwartz GW, Turner MH, Rieke F, and Wong ROL (2014). Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types. Curr. Biol 24, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJAJ, Kremer M, Wunderlich FT, Jung S, and Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426. [DOI] [PubMed] [Google Scholar]
- Cang J, and Feldheim DA (2013). Developmental mechanisms of topographic map formation and alignment. Annu. Rev. Neurosci 36, 51–77. [DOI] [PubMed] [Google Scholar]
- Cartmill M (1974). Rethinking primate origins. Science 184, 436–443. [DOI] [PubMed] [Google Scholar]
- Chalupa LM, and Lia B (1991). The nasotemporal division of retinal ganglion cells with crossed and uncrossed projections in the fetal rhesus monkey. J. Neurosci 11, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi V, and Priebe NJ (2020). Interocular velocity cues elicit vergence eye movements in mice. J. Neurophysiol 124, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper ML, and Pettigrew JD (1979). The decussation of the retinothalamic pathway in the cat, with a note on the major meridians of the cat’s eye. J. Comp. Neurol 187, 285–311. [DOI] [PubMed] [Google Scholar]
- Cumming BG, and DeAngelis GC (2001). The physiology of stereopsis. Annu. Rev. Neurosci 24, 203–238. [DOI] [PubMed] [Google Scholar]
- Dhande OS, Stafford BK, Lim J-HA, and Huberman AD (2015). Contributions of Retinal Ganglion Cells to Subcortical Visual Processing and Behaviors. Annu. Rev. Vis. Sci 1, 291–328. [DOI] [PubMed] [Google Scholar]
- Do MTH (2019). Melanopsin and the Intrinsically Photosensitive Retinal Ganglion Cells: Biophysics to Behavior. Neuron 104, 205–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger UC, and Olsen JF (1980). Origins of crossed and uncrossed retinal projections in pigmented and albino mice. J. Comp. Neurol 191, 383–412. [DOI] [PubMed] [Google Scholar]
- Dräger UC, and Olsen JF (1981). Ganglion cell distribution in the retina of the mouse. Invest. Ophthalmol. Vis. Sci 20, 285–293. [PubMed] [Google Scholar]
- Duwaer AL, and Van Den Brink G (1981). What is the diplopia threshold? Percept. Psychophys 29, 295–309. [DOI] [PubMed] [Google Scholar]
- Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen S-K, LeGates T, Renna JM, Prusky GT, Berson DM, and Hattar S (2010). Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewert JP, Buxbaum-Conradi H, Dreisvogt F, Glagow M, Merkel-Harff C, Röttgen A, Schürg-Pfeiffer E, and Schwippert WW (2001). Neural modulation of visuomotor functions underlying prey-catching behaviour in anurans: perception, attention, motor performance, learning. Comp. Biochem. Physiol. A Mol. Integr. Physiol 128, 417–461. [DOI] [PubMed] [Google Scholar]
- Feord RC, Sumner ME, Pusdekar S, Kalra L, Gonzalez-Bellido PT, and Wardill TJ (2020). Cuttlefish use stereopsis to strike at prey. Sci Adv 6, eaay6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahtan E, Tanger P, and Baier H (2005). Visual prey capture in larval zebrafish is controlled by identified reticulospinal neurons downstream of the tectum. J. Neurosci 25, 9294–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Auer TO, Henriques PM, Rajan G, Duroure K, Bianco IH, and Del Bene F (2019). An interhemispheric neural circuit allowing binocular integration in the optic tectum. Nat. Commun 10, 5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, and Gerfen CR (2007). Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci 27, 9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesy CP (2008). Ecomorphology of orbit orientation and the adaptive significance of binocular vision in primates and other mammals. Brain Behav. Evol 71, 54–67. [DOI] [PubMed] [Google Scholar]
- Howarth M, Walmsley L, and Brown TM (2014). Binocular integration in the mouse lateral geniculate nuclei. Curr. Biol 24, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy JL, Yavorska I, Wehr M, and Niell CM (2016). Vision Drives Accurate Approach Behavior during Prey Capture in Laboratory Mice. Curr. Biol 26, 3046–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy JL, Bishop HI, and Niell CM (2019). Defined cell types in superior colliculus make distinct contributions to prey capture behavior in the mouse. Curr. Biol 29, 4130–4138.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, and Barres BA (2008). Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron 59, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby J, and Schwartz GW (2017). Three Small-Receptive-Field Ganglion Cells in the Mouse Retina Are Distinctly Tuned to Size, Speed, and Object Motion. J. Neurosci 37, 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, and Masland RH (1998). The major cell populations of the mouse retina. J. Neurosci 18, 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Zhao L, and Kerschensteiner D (2018). A Pixel-Encoder Retinal Ganglion Cell with Spatially Offset Excitatory and Inhibitory Receptive Fields. Cell Rep. 22, 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk DL, Levick WR, and Cleland BG (1976). The crossed or uncrossed destination of axons of sluggish-concentric and non-concentric cat retinal ganglion cells, with an overall synthesis of the visual field representation. Vision Research 16, 233–236. [DOI] [PubMed] [Google Scholar]
- Koch SM, Dela Cruz CG, Hnasko TS, Edwards RH, Huberman AD, and Ullian EM (2011). Pathway-specific genetic attenuation of glutamate release alters select features of competition-based visual circuit refinement. Neuron 71, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Chioma A, Bonhoeffer T, and Hübener M (2019). Area-Specific Mapping of Binocular Disparity across Mouse Visual Cortex. Curr. Biol 29, 2954–2960.e5. [DOI] [PubMed] [Google Scholar]
- Langley WM (1989). Grasshopper mouse’s use of visual cues during a predatory attack. Behav. Processes 19, 115–125. [DOI] [PubMed] [Google Scholar]
- Levine JN, and Schwartz GW (2020). The Olivary Pretectal Nucleus Receives Visual Input of High Spatial Resolution. [Google Scholar]
- Lyle TK, and Foley J (1955). Subnormal binocular vision with special reference to peripheral fusion. Br. J. Ophthalmol 39, 474–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D (1982). Vision: A Computational Investigation into the Human Representation and Processing of Visual Information (W. H. Freeman and Company; ). [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, et al. (2017). Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Rep. 18, 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, and Bethge M (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci [DOI] [PubMed] [Google Scholar]
- Mccarty R, and Southwick CH (1977). Paternal care and the development of behavior in the southern grasshopper mouse, Onychomys torridus. Behav. Biol 19, 476–490. [Google Scholar]
- Meyer AF, Poort J, O’Keefe J, Sahani M, and Linden JF (2018). A Head-Mounted Camera System Integrates Detailed Behavioral Monitoring with Multichannel Electrophysiology in Freely Moving Mice. Neuron 100, 46–60.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AF, O’Keefe J, and Poort J (2020). Two Distinct Types of Eye-Head Coupling in Freely Moving Mice. Curr. Biol 30, 2116–2130.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaiel AM, Abe ETT, and Niell CM (2020). Dynamics of gaze control during prey capture in freely moving mice. ELife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GJ, and Rieke F (2006). Network variability limits stimulus-evoked spike timing precision in retinal ganglion cells. Neuron 52, 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, Nakazawa C, and Matsubara A (2006). Tumor necrosis factor-α mediates oligodendrocyte death and delayed retinal ganglion cell loss in a mouse model of glaucoma. Journal Of. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, and Schwartz GW (2016). Cardinal Orientation Selectivity Is Represented by Two Distinct Ganglion Cell Types in Mouse Retina. J. Neurosci 36, 3208–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, and Schwartz GW (2017). Electrical synapses convey orientation selectivity in the mouse retina. Nat. Commun 8, 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nityananda V, and Read JCA (2017). Stereopsis in animals: evolution, function and mechanisms. J. Exp. Biol 220, 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nityananda V, Tarawneh G, Rosner R, Nicolas J, Crichton S, and Read J (2016). Insect stereopsis demonstrated using a 3D insect cinema. Sci. Rep 6, 18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oommen BS, and Stahl JS (2008). Eye orientation during static tilts and its relationship to spontaneous head pitch in the laboratory mouse. Brain Res. 1193, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J-J, Gao F, and Wu SM (2003). Light-Evoked Excitatory and Inhibitory Synaptic Inputs to ON and OFF α Ganglion Cells in the Mouse Retina. J. Neurosci 23, 6063–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros TJ, Rebsam A, and Mason CA (2008). Retinal Axon Growth at the Optic Chiasm: To Cross or Not to Cross. Annu. Rev. Neurosci 31, 295–315. [DOI] [PubMed] [Google Scholar]
- Pettigrew JD (1986). Evolution of binocular vision. Vis. Neurosci 208–222. [Google Scholar]
- Ponce CR, and Born RT (2008). Stereopsis. Curr. Biol 18, R845–50. [DOI] [PubMed] [Google Scholar]
- Quattrochi LE, Stabio ME, Kim I, Ilardi MC, Fogerson PM, Leyrer ML, and Berson DM (2018). The M6 cell: A small-field bistratified photosensitive retinal ganglion cell. J. Comp. Neurol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompani SB, Müllner FE, Wanner A, Zhang C, Roth CN, Yonehara K, and Roska B (2017). Different Modes of Visual Integration in the Lateral Geniculate Nucleus Revealed by Single-Cell-Initiated Transsynaptic Tracing. Neuron 93, 767–776.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe MH, and Dreher B (1982). Retinal W-cell projections to the medial interlaminar nucleus in the cat: implications for ganglion cell classification. J. Comp. Neurol 204, 117–133. [DOI] [PubMed] [Google Scholar]
- Rowe MH, and Stone J (1977). Naming of neurones. Classification and naming of cat retinal ganglion cells. Brain Behav. Evol 14, 185–216. [DOI] [PubMed] [Google Scholar]
- Sabbah S, Gemmer JA, Bhatia-Lin A, Manoff G, Castro G, Siegel JK, Jeffery N, and Berson DM (2017). A retinal code for motion along the gravitational and body axes. Nature 14, 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonds JM, Choi V, and Priebe NJ (2019). Mice Discriminate Stereoscopic Surfaces Without Fixating in Depth. J. Neurosci 39, 8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, Chen S-K, and Hattar S (2011). Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 34, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl B, Burge J, and Priebe NJ (2013). Binocular integration and disparity selectivity in mouse primary visual cortex. J. Neurophysiol 109, 3013–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz GW, Okawa H, Dunn FA, Morgan JL, Kerschensteiner D, Wong RO, and Rieke F (2012). The spatial structure of a nonlinear receptive field. Nat. Neurosci 15, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillar KT, Picton LD, and Heitler WJ (2016). The Neuroethology of Predation and Escape (John Wiley & Sons; ). [Google Scholar]
- Speed A, Del Rosario J, Burgess CP, and Haider B (2019). Cortical State Fluctuations across Layers of V1 during Visual Spatial Perception. Cell Rep. 26, 2868–2874.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabio ME, Sabbah S, Quattrochi LE, Ilardi MC, Fogerson PM, Leyrer ML, Renna JM, Kim MT, Kim I, Schiel M, et al. (2017). The M5 Cell: A Color-Opponent Intrinsically Photosensitive Retinal Ganglion Cell. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JJ, and Berson DM (1995). On the distribution of gamma cells in the cat retina. Vis. Neurosci 12, 687–700. [DOI] [PubMed] [Google Scholar]
- Sterratt DC, Lyngholm D, Willshaw DJ, and Thompson ID (2013). Standard anatomical and visual space for the mouse retina: computational reconstruction and transformation of flattened retinae with the Retistruct package. PLoS Comput. Biol 9, e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, and Fukuda Y (1974). The naso-temporal division of the cat’s retina re-examined in terms of Y-, X- and W-cells. The Journal of Comparative Neurology 155, 377–394. [DOI] [PubMed] [Google Scholar]
- Stringer C, Michaelos M, and Pachitariu M (2020). Cellpose: a generalist algorithm for cellular segmentation. BioRxiv. [DOI] [PubMed] [Google Scholar]
- Tien N-W, Pearson JT, Heller CR, Demas J, and Kerschensteiner D (2015). Genetically Identified Suppressed-by-Contrast Retinal Ganglion Cells Reliably Signal Self-Generated Visual Stimuli. J. Neurosci 35, 10815–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, et al. (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 104, 1039–1055.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MH, Sanchez Giraldo LG, Schwartz O, and Rieke F (2019). Stimulus- and goal-oriented frameworks for understanding natural vision. Nat. Neurosci 22, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JLC, Cueva Vargas JL, Osswald IK, Unsain N, Aurousseau MR, Barker PA, Bowie D, and Di Polo A (2015). Soluble Tumor Necrosis Factor Alpha Promotes Retinal Ganglion Cell Death in Glaucoma via Calcium-Permeable AMPA Receptor Activation. Journal of Neuroscience 35, 12088–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal P-P, Degallaix L, Josset P, Gasc J-P, and Cullen KE (2004). Postural and locomotor control in normal and vestibularly deficient mice. J. Physiol 559, 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls GL (1942). The vertebrate eye and its adaptive radiation (Cranbrook Institute of Science; ). [Google Scholar]
- Wässle H, and Illing RB (1980). The retinal projection to the superior colliculus in the cat: a quantitative study with HRP. J. Comp. Neurol 190, 333–356. [DOI] [PubMed] [Google Scholar]
- Wei W (2018). Neural Mechanisms of Motion Processing in the Mammalian Retina. Annu Rev Vis Sci 4, 165–192. [DOI] [PubMed] [Google Scholar]
- Whitaker JO (1966). Food of Mus Musculus, Peromyscus Maniculatus Bairdi and Peromyscus Leucopus in Vigo County, Indiana. J. Mammal 47, 473–486. [Google Scholar]
- Whitman DW, Blum MS, and Jones CG (1986). Prey-specific attack behaviour in the southern grasshopper mouse, Onychomys torridus (Coues). Anim. Behav 34, 295–297. [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 18, 530–546. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim I-J, Sanes JR, and Meister M (2012). The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proc. Natl. Acad. Sci. U. S. A 109, E2391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Stereotypic bite-and-grab attack. (related to Figure 1)
Representative prey capture sequence simultaneously recorded on five cameras. Videos from the overhead camera and three of the four side cameras are shown.
Video S2. Monocular mice are poor hunters. (related to Figure 2)
Representative hunting examples from control (left) and monocularly enucleated (right) mice recorded on overhead cameras.
Video S3. Ipsilaterally projecting RGCs are required for efficient prey capture. (related to Figure 5)
Representative hunting examples from DT-injected control (left) and Sert-DTR (right) mice recorded on overhead cameras.
Data Availability Statement
Raw data and analysis routines are available upon request to the Lead Contact, Daniel Kerschensteiner (kerschensteinerd@wustl.edu).