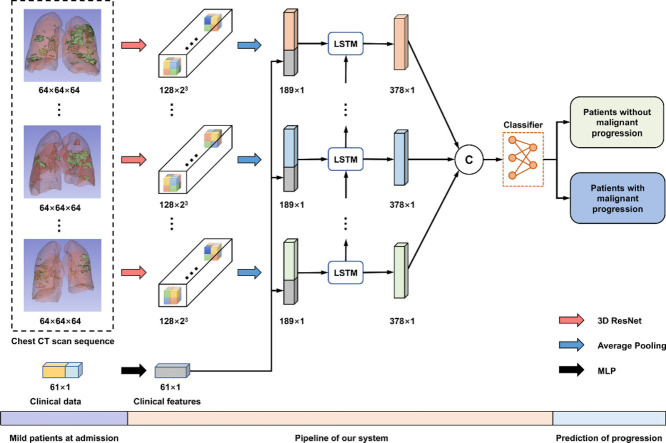
Graphical abstract
Keywords: COVID-19, Domain adaptation, Feature fusion, Malignant progression
Abstract
As COVID-19 is highly infectious, many patients can simultaneously flood into hospitals for diagnosis and treatment, which has greatly challenged public medical systems. Treatment priority is often determined by the symptom severity based on first assessment. However, clinical observation suggests that some patients with mild symptoms may quickly deteriorate. Hence, it is crucial to identify patient early deterioration to optimize treatment strategy. To this end, we develop an early-warning system with deep learning techniques to predict COVID-19 malignant progression. Our method leverages CT scans and the clinical data of outpatients and achieves an AUC of 0.920 in the single-center study. We also propose a domain adaptation approach to improve the generalization of our model and achieve an average AUC of 0.874 in the multicenter study. Moreover, our model automatically identifies crucial indicators that contribute to the malignant progression, including Troponin, Brain natriuretic peptide, White cell count, Aspartate aminotransferase, Creatinine, and Hypersensitive C-reactive protein.
1. Introduction
Since 2020, COVID-19 has had a fundamental effect on people’s lives. As of August 12, 2020, the number of COVID-19 infections in the world has soared to 20.2 million (20,162,474) with a mortality of 3.7 (737,417/20,162,474) (Organization et al., 2020), which greatly challenges public medical systems. France and The United Kingdom have the highest mortality in the world, which is 15.8 (30,227/191,265) and 14.9 (46,526/312,793), respectively. In comparison, the mortality in some other countries is much lower, such as 4.2 (9,207/218,519) in Germany (Organization et al., 2020).
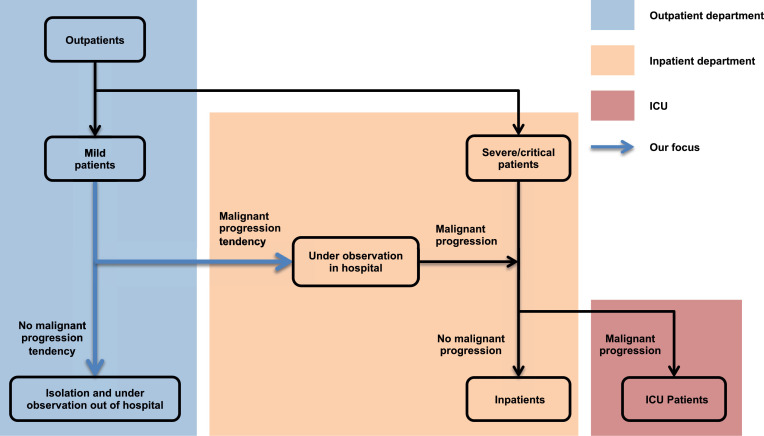
One of the most important causes for such a difference in mortalities is early identification and active intervention of patients with mild symptoms to prevent deterioration (Bennhold, 2020). Clinical observations (Cummings, Baldwin, Abrams, Jacobson, Meyer, Balough, Aaron, Claassen, Rabbani, Hastie, et al., 2020, Wu, McGoogan, 2020) suggest that although around 80 of COVID-19 patients are mild or asymptomatic, some of them may rapidly deteriorate. More importantly, studies (Yang et al., 2020) have shown that over 60 of patients died once they progressed into a severe/critical stage. Thus this group of patients requires special attention and treatment in advance. Therefore, the focus of this study, as illustrated in Fig. 1 , is on an accurate prediction of COVID-19 malignant progression, which is conducive to the timely intervention of clinicians, rational optimization of medical resources, and the effective operation of the entire medical system.
Fig. 1.
Patient stratification from outpatients to ICU. Reasonable hierarchical management of COVID-19 patients is beneficial to optimizing the allocation of medical resources and improving the efficiency of diagnosis and treatment.
Current research mainly focuses on exploiting clinical variables ascertained at hospital admission or quantitative CT parameters for progression prediction via multivariate regression (Gong, Ou, Qiu, Jie, Chen, Yuan, Cao, Tan, Xu, Zheng, et al., 2020, Ji, Zhang, Xu, Chen, Yang, Zhao, Chen, Cheng, Wang, Bi, et al., 2020), Light Gradient Boosting Machine (LightGBM) (Zhang et al., 2020c), or Least Absolute Shrinkage and Selection Operator (LASSO) (Liang et al., 2020a). However, the performance of those methods is still far from that required for practical use, for three reasons: 1) a manual quantization of feature patterns is required, which leads to information loss before analysis; 2) temporal cues are more or less ignored, but crucial to an accurate prediction; 3) chest Computed Tomography (CT) scans and the clinical data capture different characteristics of patients, but the complementarity between them is not fully leveraged.
To address the above issues, we resort to Artificial Intelligence (AI) techniques to deliver an accurate model for predicting COVID-19 malignant progression. Based on deep learning methods (He, Zhang, Ren, Sun, 2016, Hochreiter, Schmidhuber, 1997, Hornik, Stinchcombe, White, et al., 1989), our model effectively mines the complementary information in the static clinical data and the dynamic sequence of chest CT scans. It operates on raw data in an end-to-end manner, which means any manual design of feature patterns or interference of clinicians is not required. Moreover, our model automatically identifies crucial indicators that contribute to the malignant progression, including Troponin, Brain natriuretic peptide, White cell count, Aspartate aminotransferase, Creatinine, and Hypersensitive C-reactive protein.
In summary, our work presents an early warning system and targets early identification of COVID-19 malignant progression for reducing the patient stratification uncertainty, optimizing the diagnosis and treatment, increasing the efficiency of medical resource allocation, improving the emergency response capacity of the medical system, and ultimately decreasing the mortality. Comprehensive experiments on three cohorts demonstrate that our system using both CT scans and the clinical data not only achieves the best performance in the internal validation (Area Under the Receiver Operating Characteristic Curve (AUC): 0.920, 95 Confidence Interval (CI): [0.861, 0.979], cohort one), but more importantly, has robust generalization power in the external validations (AUC: 0.885, 95 CI: [0.847, 0.923], cohort two; AUC: 0.862, 95 CI: [0.789, 0.935], cohort three).
2. Related works
In this section, we first provide a short review of previous studies on the COVID-19 diagnosis and prognosis, then introduce temporal information exploring and domain adaptation on medical images.
2.1. AI-based COVID-19 diagnosis and prognosis
In the past few months, AI-based methods have played an important role in this epidemic. In outbreak areas, COVID-19 patients are in urgent need of diagnosis. Due to fast acquisition, some works perform X-ray (Wong, Lam, Fong, Leung, Chin, Lo, Lui, Lee, Chiu, Chung, Lee, Wan, Hung, Lam, Kuo, Ng, 2019, Sitaula, Hossain, 2020, Minaee, Kafieh, Sonka, Yazdani, Soufi, 2020) and CT scans (Di, Shi, Yan, Xia, Mo, Ding, Shan, Li, Wei, Shao, Han, Gao, Sui, Gao, Shen, 2020, Yang, Xu, Li, Myronenko, Roth, Harmon, Xu, Turkbey, Turkbey, Wang, Zhu, Carrafiello, Patella, Cariati, Obinata, Mori, Tamura, An, Wood, Xu, 2021, Gao, Su, Jiang, Zeng, Feng, Shen, Rong, Xu, Qin, Yang, Wang, Hu, 2020) to identify COVID-19. Besides early screening, the study of malignant progression prediction is also important for treatment planning. Demographic and clinical characteristics (Liang, Liang, Ou, Chen, Chen, Li, Li, Guan, Sang, Lu, et al., 2020, Liang, Yao, Chen, Lv, Zanin, Liu, Wong, Li, Lu, Liang, qiang Chen, Guo, Guo, Zhou, Ou, Zhou, Chen, Yang, Han, Huan, Tang, Guan, Chen, Zhao, Sang, Xu, Wang, Li, Lu, Zhang, Zhong, Huang, He, 2020, Ji, Zhang, Xu, Chen, Yang, Zhao, Chen, Cheng, Wang, Bi, et al., 2020) are the most commonly used input of the prediction model. Simultaneously, quantitative CT features (Zhang et al., 2020b) obtained by radiographic knowledge or deep learning-based method are the alternative input information. Besides, segmentation as the essential step in COVID-19 quantification and diagnosis has been extensively studied. Gao et al. (2020a) develop a dual-branch combination network for COVID-19 diagnosis that can simultaneously achieve individual-level classification and lesion segmentation. Fan et al. (2020) propose a parallel partial decoder with a reverse attention module to model the boundaries and enhance the representations for the semi-supervised framework. Wang et al. (2020) introduce a noise-robust Dice loss with an adaptive self-ensembling framework to learn from noisy labels for the segmentation of COVID-19 Pneumonia Lesions.
2.2. Temporal information exploring on medical images
The Long Short-Term Memory (LSTM) network is the most commonly used sequential information modeling method on medical imaging. Liang et al. (2018) employ multi-phases CT images as sequential data and extract an enhancement pattern via the bi-directional LSTM block from the output of a convolutional neural network (CNN) for the classification of focal liver lesion. Zhang et al. (2020d) extend convolutional LSTM into the spatio-temporal domain by jointly learning the inter-slice 3D contexts and the temporal dynamics from multiple patient studies for tumor growth prediction. Gao et al. (2020b) propose the distanced LSTM by introducing time-distanced gates to handle irregular sampling sequences targeting lung cancer diagnosis. When early predicting Alzheimer’s disease, Zhu et al. (2021) propose a Temporally Structured Support Vector Machine (TS-SVM) model to constrain the partial MR image sequence’s detection score to increase monotonically with AD progression.
2.3. Domain adaptation on medical images
Domain adaptation is a popular learning scenario in medical imaging, referred to as the different domain, same task scenario. In the domain adaptation, we are dealing with, for example, data acquired with different scanners (Opbroek, Ikram, Vernooij, de Bruijne, 2015a, Opbroek, Vernooij, Ikram, de Bruijne, 2015b) or heterogeneous appearances (Bermúdez-Chacón et al., 2016). Some works such as Conjeti et al. (2016), Wachinger and Reuter (2016), Götz et al. (2016), Opbroek et al. (2015a) focus on supervised transfer, with a small amount of labeled data from the target domain. Opbroek et al. (2015b), Cheplygina et al. (2018), Wachinger and Reuter (2016) change the source distribution by weighting training instances to reduce the distribution difference between the source domain and the target domain. On the other hand, another strategy is to align the source and target domains by the feature space transformation (Conjeti, Katouzian, Roy, Peter, Sheet, Carlier, Laine, Navab, 2016, Guerrero, Ledig, Rueckert, 2014, Hofer, Kwitt, Höller, Trinka, Uhl, 2017). In this work, we use a metric-based method to bridge the domain gap between different data centers by a few labeled samples.
3. Material and methods
3.1. Clinical data acquisition and preprocessing
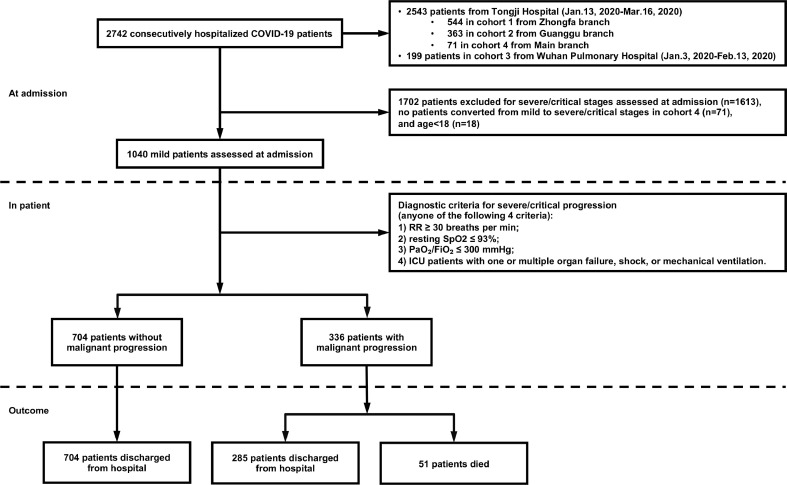
In Wuhan Pulmonary Hospital, data from 199 patients were collected from January 3, 2020, to February 13, 2020. In Tongji Hospital, data from 2,543 patients were collected from January 13, 2020, to March 16, 2020, of which 544 patients came from the Zhongfa branch, 363 patients came from the Guanggu branch, and 71 patients came from the Main branch. All patients were confirmed by a positive viral nucleic acid test. A subset of 1,040 adult patients belonging to the mild type at admission assessments is selected for further investigation. The inclusion criteria are all of the followings: 1) respiratory rate 30 breaths per min; 2) resting blood oxygen saturation 93; 3) the ratio of arterial oxygen partial pressure to fraction of inspiration oxygen 300 mm Hg; 4) non-ICU patients without shock, respiratory failure, mechanical ventilation, and failure of other organs. Anyone who fails to fulfill one of the criteria is considered to progress into a severe/critical stage according to the guidelines for the COVID-19 infection diagnosis and treatment by the National Health Commission of the People’s Republic of China (Version 7). The medical history, physical examination results, and laboratory tests were all collected from the HIS system. The time points of symptoms onset and the beginning of severe/critical stage are recorded for further selections of available CT scans. All the patients in the Main branch have mild prognoses, so our research excludes patients from this branch. Furthermore, considering the age imbalance problem, we exclude patients under 18 years old. We finally obtain 61 clinical indicators for each sample. Fig. 2 illustrates the flowchart of patient selection.
Fig. 2.
Flowchart of patient selection. A total of 1,040 out of 2,742 patients are selected according to the inclusion criteria. All the 1,040 patients have the complete clinical data required for the study and 57.9 of them underwent serial chest CT imaging. Abbreviations: Respiratory rate (RR); Blood oxygen saturation (SpO); Arterial oxygen partial pressure (PaO), Fraction of inspiration oxygen (FiO).
3.2. CT data acquisition and preprocessing
All the patients underwent serial pulmonary CT exams on dedicated CT scanners (GE, SIEMENS, TOSHIBA, and UNITED IMAGING) in two hospitals with the following parameters: slice thickness 1-3 mm, slice gap 0 mm, 130 kV, 50 mAs. All CT scans before the severe/critical stage are included to segment the masks of bilateral lungs and pneumonia on an autonomous system (HUAWEI CLOUD Launches AI-Assisted Diagnosis Platform for COVID-19). Each CT scan is downsampled to a width height slice tensor. Three different resolution settings (64 64 64, 128 128 64, and 256 256 64) are performed. CT image values are clamped to the range [-1250, 250]. Data augmentations including random horizontal flips and random rotations are used.
As traditional machine learning methods cannot handle raw CT scans directly, we design a set of hand-crafted quantitative features for experimental comparison. These quantitative features include the infection pixels proportion and the average CT value of infection pixels for each CT slice. Finally, a 128-dimensional vector can be obtained for each CT scan. We use zero-padding for the missing CT scan tensor.
3.3. Network architecture and training process
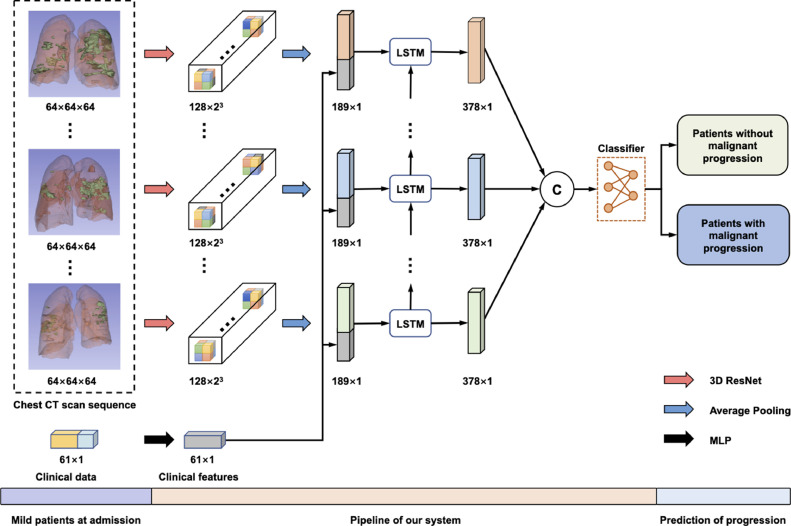
Fig. 3 illustrates the pipeline of our system. As shown, the input of our model includes the clinical data and a sequence of CT scans obtained at different time points. Specifically, the clinical data is a 61-dimensional vector processed by a Multilayer Perceptron (MLP) with identity connections (Supplementary Figure 1). Besides, each CT scan is encoded into a 128-dimensional feature vector by 3D ResNet.
Fig. 3.
The pipeline of our system about the prediction of COVID-19 malignant progression. First, 3D ResNet and MLP encode chest CT scans and the clinical data, respectively. Then, we combine the two features and feed them into an LSTM to model the temporal information. Finally, several fully connected layers are exploited to make the prediction. Abbreviations: Computed Tomography (CT); Long Short-Term Memory (LSTM); Multilayer Perceptron (MLP).
To model the temporal information across the sequence of CT features, we use LSTM for its high capacity in modeling such information (Shi et al., 2017) and densely combine the clinical feature and the CT feature via concatenation at each time step. LSTM employed in this study is a single-layer network with an embedding dimension of 189 and a hidden dimension of 378. The output of LSTM, a 378 7 tensor, is flattened and then fed into several fully connected layers. Finally, we normalize the output with a softmax layer, which can be interpreted as the probability of the patient’s conversion to the severe/critical stage. The whole model is trained with the cross-entropy loss. The detailed architecture of our model is given in Supplementary Table 1.
3.4. Domain adaptation process
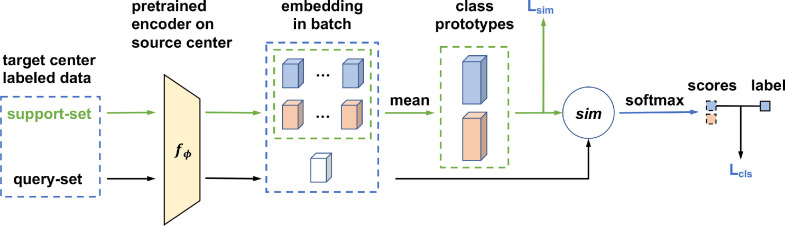
In domain adaptation, we use a metric-based method by using a few labeled samples to bridge the domain gap between the source center (cohort one) and the target center (cohort three). Fig. 4 illustrates the proposed domain adaptation process. Specifically, our method can be decomposed into two stages: a pre-training stage and a domain adaptation stage. For the pre-training stage, we first train a model on the source center then remove the classifier to get the pre-trained encoder . For the domain adaptation stage, which is the core of our method, we adapt the pre-trained model through a metric-based approach, passing the prototype representations learned from the source center to the target center. The details of this stage are elaborated as follows.
Fig. 4.
Multicenter domain adaptation process. First, we pre-train an encoder on the source center, and then, adapt the model through a metric-based approach, passing the prototype representation learned from the source center to the target center.
First, we randomly select N labeled samples from each class in the target center as the support-set to compute prototypes. Simultaneously, we randomly choose one sample per class in the target center as the query-set to compute distances to the prototypes in the embedding space. Specifically, in each class let denotes a small support-set of N labeled samples, where is the feature vector of an example and {0, 1} is the corresponding label. denotes the support-set of examples with class k {0, 1}. We compute the mean vector of the embedded support points as prototypes for the two classes:
| (1) |
Second, we compute the predicted probability distribution for each sample in the query-set based on a softmax over its cosine similarities (denoted by sim(.)) with the prototypes in the embedding space:
| (2) |
Similar to the studies of Gidaris and Komodakis (2018), Oreshkin et al. (2018), Qi et al. (2018), we also use a learnable parameter to control the probability distribution sharpness generated by the softmax function during training.
Finally, we train our framework with the classification and similarity losses jointly. Concretely, the classification loss is computed based on p and the labels in the query-set:
| (3) |
where is the cross-entropy loss. The similarity loss is adopted to increase the distance between the two class prototypes, which is defined as:
| (4) |
Based on the above two losses, our final loss function is formulated as:
| (5) |
where is the coefficient to balance the two loss terms.
In the testing stage, all labeled samples in the support-set and the query-set are used to compute prototypes. A test sample will be classified by the similarity between the feature vector of the sample and the prototype of class k.
3.5. Implementation details
The proposed network is implemented using Python (Version 3.6 with scipy, scikit-learn, and PyTorch). For single-center experiments on the cohort one, the network is trained by Adam optimizer with an initial learning rate of 0.05, and a batch size of 32 on a single NVIDIA Titan X GPU. The learning rate is decayed by a factor of 10 every 30 epochs. We train our model for 100 epochs. Model weights are initialized with the Kaiming method (He et al., 2015), and biases are initialized as 0. For multicenter experiments on the cohort three, we use the same optimizer settings as single-center experiments but with a fixed learning rate of 0.01. We finetune 50 epochs for domain adaptation, and the batch size is the same as the number of labeled samples with a maximum of 20. The cosine scaling parameter is initialized to 5 with a fixed learning rate of 0.1. The loss coefficient is 0.5. The margin of the similarity loss is set to 0.2.
3.6. Performance evaluation criterion
The AUC, accuracy, sensitivity, specificity, and Receiver Operating Characteristic Curve (ROC) are used to evaluate the model performance. The calculation method is shown in Supplementary methods. The 95 bilateral confidence interval is used for all metrics, where the AUC metric uses the Wald-cc interval (Kottas, Kuss, Zapf, 2014, Delong, Delong, Clarke-Pearson, 1988) and the other metrics use the Wilson interval (Brown et al., 2001).
4. Results
4.1. Dataset statistics
All data enrolled in this Institutional Review Board (IRB) approved retrospective study is obtained from two hospitals in Wuhan, including Wuhan Pulmonary Hospital and three Tongji Hospital branches. 1,040 patients with mild COVID-19 pneumonia at admission are considered in our study, including 491 males and 549 females, aged 18 to 95 (57.51 14.75). 32.3 of patients (336/1,040) malignantly progressed to a severe/critical stage during the hospitalization, while the remaining 67.7 (704/1,040) did not. The selected data is divided into three cohorts, of which the cohort one is used for the single-center study, and the cohort two and the cohort three are used for the multicenter study. The clinical data in three cohorts is summarized in Table 1.
Table 1.
Patient and clinical characteristics. Qualitative variables are in number () and quantitative variables are in mean standard deviation, when appropriate. Positive patients: COVID-19 patients with malignant progression. Negative patients: COVID-19 patients without malignant progression. Abbreviations: Hypersensitive C-reactive protein (HCRP); Brain natriuretic peptide (BNP); Alanine aminotransferase (ALT); Aspartate aminotransferase (AST); -Glutamyl transpeptidase (-GT).
| Characteristics | All patients | Cohort one | Cohort two | Cohort three |
|---|---|---|---|---|
| Number | 1040 | 544/1040 (52.3%) | 363/1040 (34.9%) | 133/1040 (12.8%) |
| Age, years | 57.5 14.7 | 58.5 14.7 | 57.7 15.2 | 52.5 12.6 |
| Sex, Male/Female | 491/549 | 259/285 | 166/197 | 66/67 |
| Patients with CTs/Total CT scans | 602/1,601 | 301/852 | 197/498 | 104/251 |
| Positive/Negative patients | 336/704 | 128/416 | 154/209 | 54/79 |
| Days from symptom onset to admission | 17.7 6.1 | 18.8 6.1 | 18.2 5.1 | 11.6 5.1 |
| Hypertension | 314/1,040 (30.2%) | 184/544 (33.8%) | 101/363 (27.8%) | 29/133 (21.8%) |
| Fever | 783/1,040 (75.3%) | 425/544 (78.1%) | 232/363 (63.9%) | 126/133 (94.7%) |
| Respiratory rate, breaths per minute | 19.9 5.1 | 20.1 5.1 | 19.5 5.9 | 20.3 1.6 |
| White cell count, 109/L | 8.2 21.5 | 9.3 28.6 | 7.5 9.3 | 5.5 3.3 |
| Total T lymphocyte count, cell/l | 1980.6 769.9 | 2200.2 570.5 | 2057.0 668.3 | 874.4 805.1 |
| Absolute count of CD3+CD4+T cells, cell/l | 965.4 333.3 | 1060.7 233.1 | 1008.1 272.7 | 459.2 380.7 |
| HCRP, mg/L | 21.9 39.5 | 22.4 40.4 | 14.1 34.5 | 41.2 41.4 |
| Troponin, ng/ml | 15.9 75.1 | 19.7 94.8 | 12.8 51.0 | 8.4 8.5 |
| BNP, pg/ml | 767.5 3820.5 | 866.7 3863.0 | 759.4 4389.8 | 383.9 559.7 |
| ALT, U/L | 29.2 30.2 | 31.3 35.0 | 26.0 22.6 | 29.0 25.9 |
| AST, U/L | 26.3 25.8 | 26.1 17.4 | 24.5 35.5 | 31.8 22.5 |
| Albumin, g/L | 37.7 5.8 | 36.7 6.4 | 39.0 4.9 | 38.5 5.1 |
| -GT, U/L | 41.7 48.3 | 44.6 52.6 | 36.7 35.2 | 43.2 58.7 |
| Urea, mmol/L | 5.2 4.4 | 5.4 4.9 | 5.2 4.4 | 4.6 1.9 |
| Creatinine, mol/L | 80.4 94.3 | 79.8 87.0 | 85.0 117.9 | 70.3 21.4 |
| T4 | 17.0 2.0 | 17.3 2.2 | 16.9 1.5 | 15.9 2.2 |
4.2. Performance evaluation and results
Our work advocates using a sequence of CT scans, captured at different timings after hospitalization, for accurate malignant progression prediction. Unlike Zhang et al. (2020c), Liang et al. (2020a), we do not quantify CT scans to avoid information loss but use a deep learning model to process the raw data directly. In the meantime, effective integration of CT scans and clinical information underpins our system.
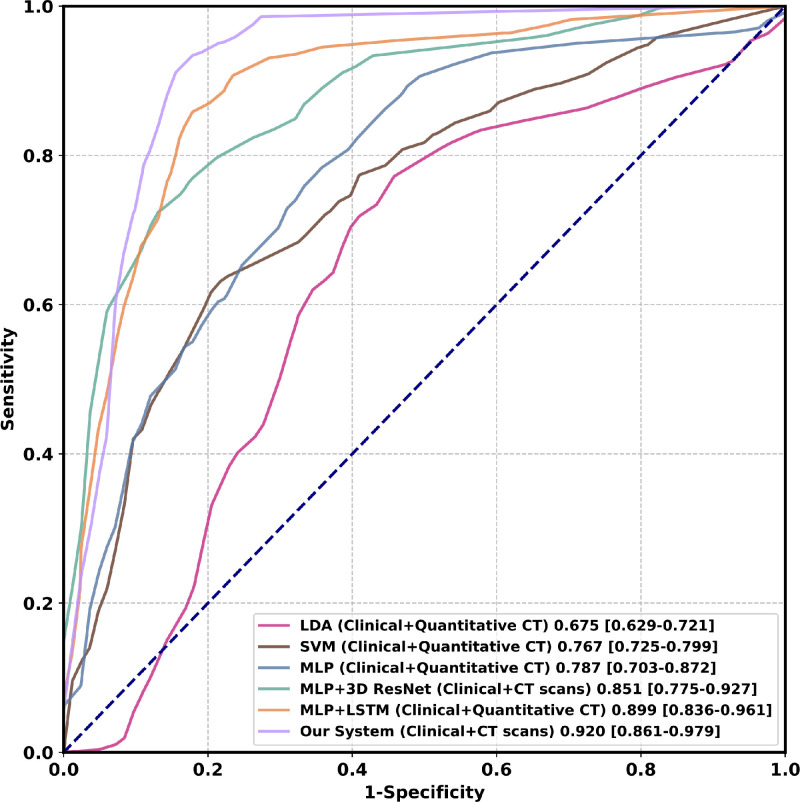
In Table 2, we compare the performance of our model against different methods, including Linear Discriminant Analysis (LDA) (Fisher, 1936), Support Vector Machine (SVM) (Cortes and Vapnik, 1995), and MLP (Hornik et al., 1989). In this experiment, we adopt patients with mild symptoms and without malignant progression as the negative reference standard and divide the cohort one into the training cohort (80) and the validation cohort (20) randomly. We use five-fold cross-validation to evaluate our model. Our system, which fuses sequential CT scans with the clinical information, achieves a mean AUC of 0.920 (95 CI: [0.861, 0.979]) and outperforms the best traditional machine learning methods (mean AUC of 0.767, 95 CI: [0.725, 0.799]) by a large margin.
Table 2.
The performance comparison of different methods. 95% confidence intervals are included in brackets. The best average results are shown in bold. The indicates our method significantly improves the compared method (McNemar’s test) (Dietterich, 1998). Abbreviations: Area Under the Receiver Operating Characteristic Curve (AUC); accuracy (ACC); sensitivity (SEN); specificity (SPEC); Linear Discriminant Analysis (LDA); Support Vector Machine (SVM); Multilayer Perceptron (MLP); Long Short-Term Memory (LSTM); Clinical Data (CD); Quantitative CT features (QCF); CT scans (CS); CT scan resolution 64 64 64 (Our System 64); CT scan resolution 128 128 64 (Our System 128); CT scan resolution 256 256 64 (Our System 256).
| Methods | AUC | ACC (%) | SEN (%) | SPEC (%) | CD | QCF | CS | p-value |
|---|---|---|---|---|---|---|---|---|
| LDA | 0.675[0.629, 0.721] | 73.5[69.8, 77.2] | 20.3[13.3, 27.3] | 89.9[87.0, 92.8] | 0.001 | |||
| LDA | 0.675[0.629, 0.721] | 67.3[63.3, 71.2] | 39.1[30.6, 47.5] | 76.0[71.9, 80.1] | 0.001 | |||
| SVM | 0.652[0.606, 0.699] | 76.3[72.7, 79.9] | 0.80[0.00, 2.30] | 99.5[98.9, 100.0] | 0.001 | |||
| SVM | 0.767[0.725, 0.799] | 76.8[73.3, 80.4] | 19.5[12.7, 26.4] | 94.5[92.3, 96.7] | 0.001 | |||
| MLP | 0.787[0.703, 0.872] | 81.6[78.1, 84.6] | 48.4[40.0, 57.0] | 91.8[88.8, 94.1] | 0.001 | |||
| MLP | 0.823[0.778, 0.853] | 81.1[77.6, 84.1] | 61.7[53.1, 69.7] | 87.0[83.4, 89.9] | 0.001 | |||
| MLP+3D ResNet | 0.851[0.775, 0.927] | 85.3[82.1, 88.0] | 69.5[61.1, 76.8] | 90.1[86.9, 92.7] | 0.021 | |||
| MLP+LSTM | 0.899[0.836, 0.961] | 86.0[82.9, 88.7] | 71.9[63.5, 78.9] | 90.4[87.2, 92.9] | 0.265 | |||
| Our System 64 | 0.920[0.861, 0.979] | 87.7[84.7, 90.2] | 89.1[82.5, 93.4] | 87.3[83.7, 90.1] | ||||
| Our System 128 | 0.923[0.897, 0.949] | 87.7[84.7, 90.2] | 86.7[79.8, 91.5] | 88.0[84.5, 90.8] | 0.151 | |||
| Our System 256 | 0.914[0.883, 0.938] | 88.4[85.5,90.8] | 88.3[81.6,92.8] | 88.5[85.0, 91.2] | 0.231 |
According to the results in Table 2, we can draw the following conclusions: 1) CT scans turns out to be more effective than quantitative CT features. For instance, with LSTM, using CT scans obtain a relative improvement of 2.3 over using quantitative CT features in AUC (0.920 vs. 0.899). Without LSTM, the improvement is more distinctive, reaching 8.1 (0.851 vs. 0.787). 2) Modeling the temporal information brings measurable benefits for boosting the performance of our system. This has been evidenced by an improvement of 14.2 (0.899 vs. 0.787) using LSTM when quantitative CT features are used. Meanwhile, when CT scans are used, the improvement is 8.1 (0.920 vs. 0.851) with a difference of 0.069 in AUC. The corresponding ROCs in Fig. 5 further support our method. 3) Higher resolutions (128 128 64 and 256 256 64) significantly increase the training time and computing resources without improving performance. Considering that the COVID-19 malignant progression prediction is a classification task, we use a relatively small resolution (64 64 64) to extract the global information of the CT scan in the multicenter study.
Fig. 5.
Comparison of ROC curves among different methods on the cohort one. Numbers after parentheses are AUCs. Numbers in brackets are confidence intervals. Figure best viewed in color. Abbreviations: Linear Discriminant Analysis (LDA); Support Vector Machine (SVM); Multilayer Perceptron (MLP); Long Short-Term Memory (LSTM). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.3. Analysis on feature selection algorithms
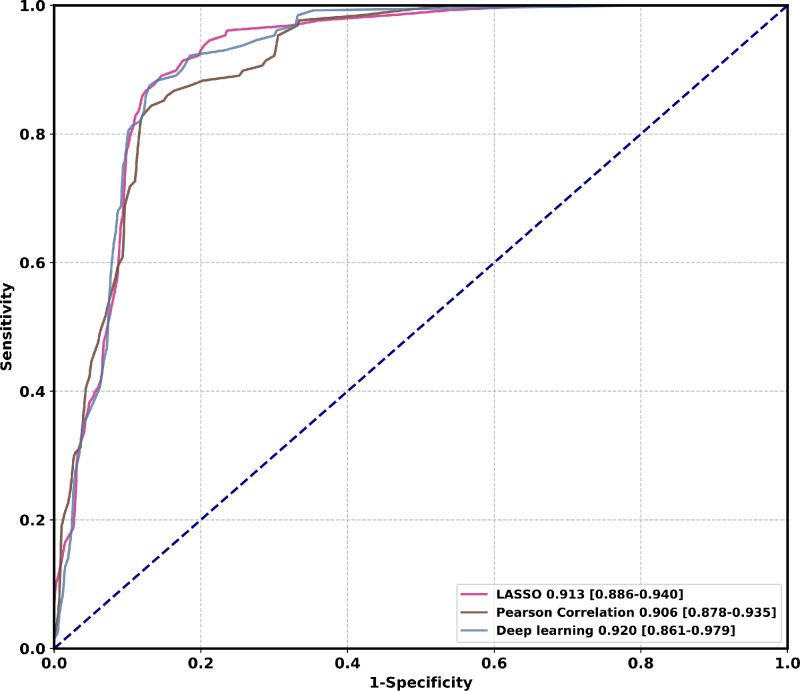
In this study, there are two types of feature selection algorithms for clinical data, i.e., LASSO (Tibshirani, 1996) and Pearson Correlation. Ten features with statistically significant () hazard ratios are identified through LASSO. These are hypertension, age, HCRP, urea, T3, lactate dehydrogenase (LDH), alkaline phosphatase, Total T lymphocyte count, lymphocyte, and alanine aminotransferase (ALT). Another set of features with statistically significant () hazard ratios are identified through Pearson Correlation. These are hypertension, age, expectoration, lymphocyte, alkaline phosphatase, urea, T3, Total T lymphocyte count, CD3+CD4+double-positive T lymphocytes (T cells count), CD3+CD8+T cells count. Among them, hypertension, age, alkaline phosphatase, urea, T3, lymphocyte, and Total T lymphocyte count are features selected by both algorithms. To compare the effectiveness of feature selection algorithms with deep learning features, we replace the clinical features extracted by MLP with the selected clinical features and keep the CT features and the network structure unchanged. Experimental comparison on cohort one is demonstrated in Table 3 and Fig. 6 . Our method using deep learning clinical features is comparable to other feature selection algorithms in AUC, accuracy, specificity, and significantly superior to other algorithms in sensitivity by 3.7% (89.1% vs. 85.9%). This observation demonstrates that deep learning features have a lower missed-detection rate, which is particularly important in COVID-19 epidemic prevention and control.
Table 3.
Experimental comparison on different feature selection algorithms. 95% confidence intervals are included in brackets. The best average results are shown in bold. The indicates our method significantly improves the compared method (McNemar’s test). Abbreviations: Area Under the Receiver Operating Characteristic Curve (AUC); accuracy (ACC); sensitivity (SEN); specificity (SPEC); Least Absolute Shrinkage and Selection Operator (LASSO).
| Feature selection algorithm | AUC | ACC (%) | SEN (%) | SPEC (%) | p-value |
|---|---|---|---|---|---|
| LASSO | 0.913[0.886, 0.940] | 87.5[84.5, 90.0] | 85.9[78.9, 90.9] | 88.0[84.5, 90.8] | 0.004 |
| Pearson Correlation | 0.906[0.877, 0.934] | 86.8[83.7, 89.4] | 85.9[78.9, 90.9] | 87.0[83.4, 89.9] | 0.009 |
| Deep learning | 0.920[0.861, 0.979] | 87.7[84.7, 90.2] | 89.1[82.5, 93.4] | 87.3[83.7, 90.1] |
Fig. 6.
Comparison of ROC curves among different feature selection algorithms on the cohort one. Numbers before brackets are AUCs. Numbers in brackets are confidence intervals. Figure best viewed in color. Abbreviations: Least Absolute Shrinkage and Selection Operator (LASSO). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.4. Performance evaluation in the multicenter study
A high-quality labeling process typically requires time-consuming human effort, which is a prominent drawback during the outbreak of COVID-19, where fast analysis is essential. Hence, how to use a small amount of labeled data to improve the generalization power of the system is of great practical significance. Inspired by metric learning-based methods (Snell et al., 2017), we propose a domain adaptation method to adapt our model to a new domain with only a few labeled samples available. The details are given in the Methods section.
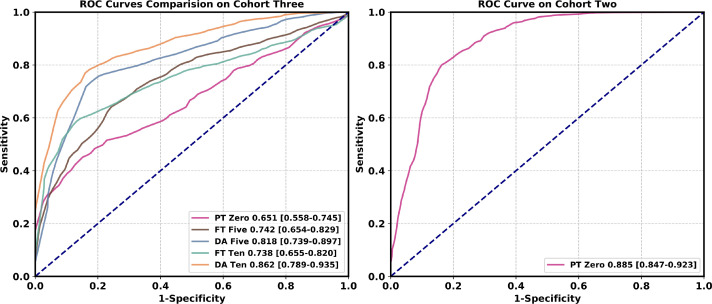
As Table 4 shows, when directly evaluating the model trained from the cohort one on the cohort two, a satisfactory performance is achieved (AUC: 0.885, 95 CI: [0.847, 0.923]). This is because the two cohorts are from different branches of the same hospital, which means that their data distributions are similar to some degree. However, when directly evaluating the model trained from the cohort one on the cohort three, the performance drops a lot. A mean AUC of 0.651 (95 CI: [0.558, 0.745]) is achieved because the two cohorts are from different hospitals. When ten labeled samples in the target domain are used, directly finetuning the model achieves a mean AUC of 0.738 (95 CI: [0.655, 0.820]), still inferior to our system (AUC: 0.862, 95 CI: [0.789, 0.935]). The corresponding ROCs in Fig. 7 further support our domain adaptation method.
Table 4.
Performance comparison in the multicenter study. 95% confidence intervals are included in brackets. indicates the pre-trained model obtained from the source center is directly used on the target center. indicates the number of samples in each class used for fine-tuning. indicates the number of samples in each class used for domain adaptation. Abbreviations: Area Under the Receiver Operating Characteristic Curve (AUC); accuracy (ACC); sensitivity (SEN); specificity (SPEC); pre-trained (PT); fine-tuning (FT); domain adaptation (DA).
| Source domain | Target domain | Methods | AUC | ACC (%) | SEN (%) | SPEC (%) |
|---|---|---|---|---|---|---|
| Cohort one | Cohort two | PT Zero | 0.885 [0.847, 0.923] | 80.6 [78.7, 82.3] | 76.0 [72.8, 78.9] | 83.9 [81.6, 86.0] |
| Cohort one | Cohort three | PT Zero | 0.651 [0.558, 0.745] | 65.6 [61.9, 69.1] | 16.3 [12.4, 21.2] | 99.2 [97.8, 99.7] |
| Cohort one | Cohort three | FT Five | 0.742 [0.654, 0.829] | 70.8 [68.2, 73.3] | 35.1 [31.0, 39.4] | 94.5 [92.6, 95.9] |
| Cohort one | Cohort three | DA Five | 0.818 [0.739, 0.897] | 77.3 [74.9, 79.6] | 74.1 [70.0, 77.8] | 79.5 [76.4, 82.2] |
| Cohort one | Cohort three | FT Ten | 0.738 [0.655, 0.820] | 75.8 [73.3, 78.2] | 52.8 [48.3, 57.3] | 90.9 [88,6, 92.8] |
| Cohort one | Cohort three | DA Ten | 0.862 [0.789, 0.935] | 81.2 [78.9, 83.4] | 74.1 [69.8, 78.0] | 85.8 [83.0, 88.2] |
Fig. 7.
Comparison of ROC curves among different methods in the multicenter study. In each class, the same number of samples are used during the domain adaptation process. Numbers before brackets are AUCs. Numbers in brackets are confidence intervals. Figure best viewed in color. Abbreviations: pre-trained (PT); fine-tuning (FT); domain adaptation (DA). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.5. Prognostic factors of the clinical data
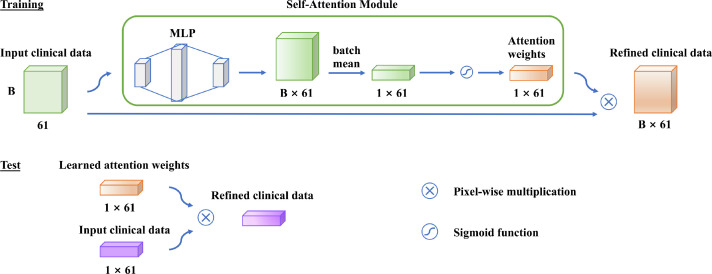
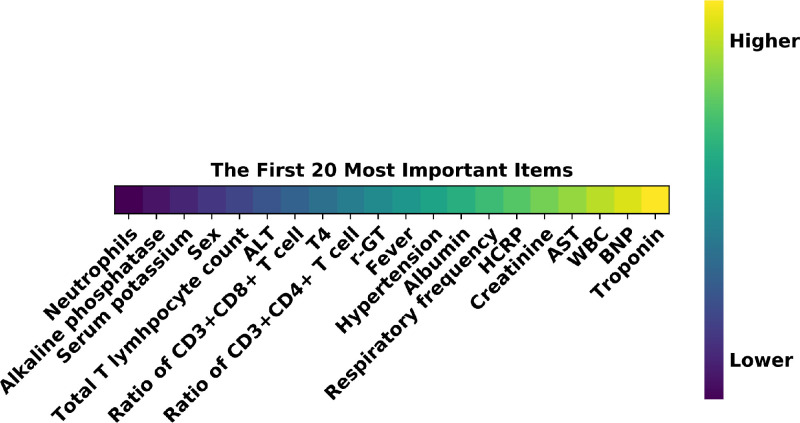
We further investigate the clinical indicators that contribute to predicting the malignant progression by a self-attention layer before the first fully connected layer of the MLP. As shown in Fig. 8 , it automatically learns the attention weight corresponding to each clinical indicator. Each attention weight is normalized to (0, 1) by a sigmoid function to measure the importance of the related clinical indicator for the prediction task. The top 20 clinical indicators with the highest attention weights are listed in Fig. 9 . The most important prognostic clinical indicators are myocardial injury (Troponin and Brain natriuretic peptide), followed by hepatic injury (Aspartate aminotransferase, Albumin, and -Glutamyl transpeptidase), renal failure (Creatinine), and inflammatory status (Hypersensitive C-reactive protein, White cell count, CD3+CD4+T cells count, fever).
Fig. 8.
Self-attention module for prognostic factors. B 61 represents the batch size and the length of the vector. Abbreviations: Multilayer Perceptron (MLP).
Fig. 9.
The top prognostic factors of the clinical data. Figure best viewed in color. Abbreviations: Alanine aminotransferase (ALT); -Glutamyl transpeptidase (r-GT); Hypersensitive C-reactive protein (HCRP); Aspartate aminotransferase (AST); White blood cell (WBC); Brain natriuretic peptide (BNP). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5. Discussion
Coronavirus-induced pneumonia puts tremendous pressure on public medical systems. Such patients without timely and effective treatment will eventually develop multi-organ failure associated with high mortality (Zhou, Yu, Du, Fan, Liu, Liu, Xiang, Wang, Song, Gu, et al., 2020, Wu, McGoogan, 2020). Therefore, early prediction and early aggressive treatment of patients with mild symptoms at a high risk of malignant progression to a severe/critical stage are important ways to reduce mortality.
In this study, we argue that the effective integration of sequential CT scans and the clinical data is important for an accurate prediction of malignant progression. Moreover, the rich temporal information in the sequence of CT scans, which has not been considered by any studies so far, is critical for this specific task. We have conducted extensive experiments to demonstrate that our system, which effectively fuses the two complementary data, achieves much better performance than using either data as input separately (e.g., 0.851 vs. 0.787 in AUC when using the clinical data and quantitative CT features). More importantly, due to the capability of our system in learning temporal information, our system reports a much higher AUC compared with the counterparts that do not consider the temporal information.
Our work is novel because we are among the first attempts to explore ways to fuse sequential CT scans and the clinical data to improve COVID-19 malignant progression predicting in an end-to-end manner. Experimental results show that both CT scans and the clinical data are of paramount importance to this problem. Furthermore, there is little literature concerning the temporal information of CT sequences. However, the temporal cue also contributes significantly to the prediction of malignant progression as it reveals the change of the patient’s health condition.
Traditional machine learning methods heavily rely on domain-specific expertise. Feature patterns to be analyzed are manually designed, leading to information loss before feeding them to the classifier. However, our method attempts to automatically learn complementary and temporal features from raw data and jointly optimizes the feature extractor and classifier in an end-to-end manner.
Deep learning-based methods often encounter performance degradation in the multicenter study, mainly due to the large data distribution discrepancy between different cohorts. This is caused by different CT scanners, different slice thickness, different regions, age distribution discrepancy, and systematic errors during the data collection process. Another notable merit of this work is that we employ domain adaptation to improve the robustness of our system in the multicenter study. From comprehensive experimental results, we observe that inferior performance is achieved when the model trained with a single-center is adapted to a completely different data domain by directly fine-tuning. The proposed domain adaptation process enables our system to transfer the prototype representations learned from the source domain to the target domain with a small number of labeled samples, which greatly improves the generalization power in the multicenter evaluation. A well-trained and mature prediction system in one center, which can be quickly deployed in multiple centers, will greatly reduce the significant demand for diagnostic expertise. It effectively optimizes the treatment strategy, thus improving the emergency response capacity of the medical system.
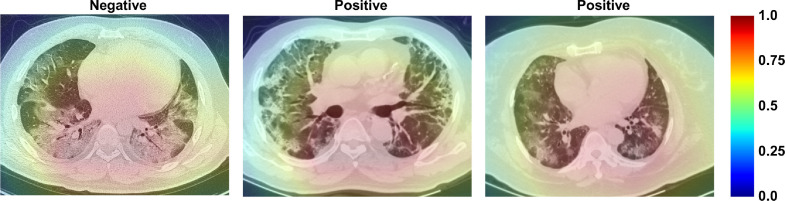
To investigate the interpretability of the CT feature patterns learned by our model, we show the activation maps via using Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2017). Fig. 10 shows that the intra-zone and middle-zone of the pulmonary region have the greatest influence on the prediction task. Hence, they are valuable for malignant progression prediction.
Fig. 10.
Visualization of learned activation maps for COVID-19 patients with mild symptoms. Red regions correspond to high score for class, and our system localizes class-discriminative regions. Figure best viewed in color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Our model could effectively identify valuable indicators for predicting the malignant progression of COVID-19 patients with mild symptoms, which assists in clinical assessment and treatment. According to our results, dysfunction or injuries of multiple organs are essential predictive indicators for COVID-19 malignant progression, of which myocardial injury is the most important one, followed by liver dysfunction and kidney failure. Coronavirus is currently believed to invade host cells through the Angiotensin-Converting Enzyme 2 (ACE2) pathway to cause COVID-19 (Lan, Ge, Yu, Shan, Zhou, Fan, Zhang, Shi, Wang, Zhang, et al., 2020, Zheng, Ma, Zhang, Xie, 2020, Sama, Ravera, Santema, van Goor, Ter Maaten, Cleland, Rienstra, Friedrich, Samani, Ng, et al., 2020, Devaux, Rolain, Raoult, 2020, Bourgonje, Abdulle, Timens, Hillebrands, Navis, Gordijn, Bolling, Dijkstra, Voors, Osterhaus, et al., 2020, Alqahtani, Schattenberg, 2020). Since ACE2 is widely distributed in various human tissues, such as type II alveolar cells, myocardial cells, hepatocytes, cholangiocytes, and proximal tubule cells, multiple organ involvement in COVID-19 is not surprising (Alqahtani, Schattenberg, 2020, Zheng, Ma, Zhang, Xie, 2020, Zhang, Shi, Wang, 2020). The inflammatory storm caused by coronavirus is another essential predictive indicator for COVID-19 malignant progression. Although the exact mechanisms are unclear yet, patients with COVID-19 (Song, Zhang, Fan, Meng, Xu, Xia, Cao, Yang, Dai, Wang, et al., 2020, Ye, Wang, Mao, 2020, Soy, Keser, Atagündüz, Tabak, Atagündüz, Kayhan, 2020) do show high levels of hypersensitive C-reactive protein and high expression of Interleukin-1B (IL-1B), Interferon gamma (IFN-), interferon-inducible protein-10 (IP-10), monocyte chemoattractant protein 1 (MCP-1), etc. These cytokines further activate the T-helper type 1 (Th1) cell response, providing another predictive indicator, CD3+CD4+T cells. The complexity of the clinic and the ambiguity of the pathogenic mechanism significantly increase the difficulty of evaluation and treatment strategy selection for COVID-19 patients.
However, our study still has several limitations. First, samples available for malignant progression prediction are limited. The diverse data in the large-scale dataset will allow deep learning-based methods to gain a more comprehensive understanding of what causes the malignant progression. Second, the data source of our study is limited to three hospital branches of two hospitals in Wuhan. More data needs to be collected from multiple centers, especially from foreign hospitals, to further enhance our model. Third, this study only conducts an interpretable analysis of the relationship between prognostic factors and patients who are easy to deteriorate from the perspective of relevance. Future studies can combine evidence-based medicine to identify the cause and effect of malignant progression.
6. Conclusions
In conclusion, our early warning system, built upon the deep learning techniques and the integration of sequential CT scans and the clinical data, can accurately predict the malignant progression of COVID-19. Compared with traditional machine learning methods, we demonstrate that our deep learning-based method can learn discriminative feature patterns and improve the prediction performance significantly. Furthermore, the generalization power of our method is improved by domain adaptation in the multicenter study. Our method can identify patients with potentially severe/critical COVID-19 outcomes using an inexpensive, widely available point-of-care test. Our system can be potentially deployed on the front line to decrease the mortality of COVID-19.
7. Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
8. Code availability
The code and the pre-trained models are available on GitHub: https://github.com/CongFang/PMP-COVID-19
CRediT authorship contribution statement
Cong Fang: Conceptualization, Methodology, Software, Writing - original draft, Writing - review & editing. Song Bai: Methodology, Writing - review & editing, Supervision. Qianlan Chen: Data curation, Investigation, Validation. Yu Zhou: Methodology, Supervision. Liming Xia: Data curation, Investigation. Lixin Qin: Data curation, Investigation. Shi Gong: Software, Validation. Xudong Xie: Investigation, Validation. Chunhua Zhou: Data curation, Investigation. Dandan Tu: Resources, Supervision. Changzheng Zhang: Resources, Investigation. Xiaowu Liu: Data curation, Investigation. Weiwei Chen: Conceptualization, Supervision, Writing - original draft, Writing - review & editing. Xiang Bai: Conceptualization, Supervision, Resources, Funding acquisition. Philip H.S. Torr: Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Key R&D Program of China (No. 2018YFB1004600), HUST COVID-19 Rapid Response Call (No. 2020kfyXGYJ093, No. 2020kfyXGYJ094), National Natural Science Foundation of China (No. 61703049, No. 81401390), Natural Science Foundation of Hubei Province of China (No. 2020CFB848), the Royal Academy of Engineering under the Research Chair and Senior Research Fellowships scheme, EPSRC/MURI (No. EP/N019474/1) and FiveAI.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.media.2021.102096.
Appendix A. Supplementary materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
References
- Alqahtani S.A., Schattenberg J.M. Liver injury in COVID-19: the current evidence. U. Eur. Gastroenterol. J. 2020;8(5):509–519. doi: 10.1177/2050640620924157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennhold K. A German exception? Why the country’s coronavirus death rate is low. New York Times. 2020;6(4):2020. [Google Scholar]
- Bermúdez-Chacón R., Becker C.J., Salzmann M., Fua P. MICCAI. 2016. Scalable unsupervised domain adaptation for electron microscopy. [DOI] [PubMed] [Google Scholar]
- Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.-L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020 doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L., Cai T.T., DasGupta A. Interval estimation for a binomial proportion. Stat. Sci. 2001;16:101–133. [Google Scholar]
- Cheplygina V., Peña I., Pedersen J.H., Lynch D., Sørensen L., de Bruijne M. Transfer learning for multicenter classification of chronic obstructive pulmonary disease. IEEE J. Biomed. Health Inf. 2018;22:1486–1496. doi: 10.1109/JBHI.2017.2769800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conjeti S., Katouzian A., Roy A.G., Peter L., Sheet D., Carlier S., Laine A., Navab N. Supervised domain adaptation of decision forests: transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization. Med. Image Anal. 2016;32:1–17. doi: 10.1016/j.media.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20(3):273–297. [Google Scholar]
- Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong E.R., Delong D.M., Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44 3:837–845. [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.-M., Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di D., Shi F., Yan F., Xia L., Mo Z., Ding Z., Shan F., Li S., Wei Y., Shao Y., Han M., Gao Y., Sui H., Gao Y., Shen D. Hypergraph learning for identification of COVID-19 with CT imaging. Med. Image Anal. 2020;68:101910. doi: 10.1016/j.media.2020.101910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich T.G. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10:1895–1923. doi: 10.1162/089976698300017197. [DOI] [PubMed] [Google Scholar]
- Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39:2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. The use of multiple measurements in taxonomic problems. Ann. Eugenics. 1936;7(2):179–188. [Google Scholar]
- Gao K., Su J., Jiang Z., Zeng L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y., Wang W., Hu D. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2020;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Tang Y., Xu K., Huo Y., Bao S., Antic S., Epstein E.S., Deppen S., Paulson A.B., Sandler K., Massion P., Landman B. Time-distanced gates in long short-term memory networks. Med. Image Anal. 2020;65:101785. doi: 10.1016/j.media.2020.101785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidaris S., Komodakis N. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Dynamic few-shot visual learning without forgetting; pp. 4367–4375. [Google Scholar]
- Gong J., Ou J., Qiu X., Jie Y., Chen Y., Yuan L., Cao J., Tan M., Xu W., Zheng F., et al. A tool to early predict severe corona virus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M., Weber C., Binczyk F., Polaska J., Tarnawski R., Bobek-Billewicz B., Köthe U., Kleesiek J., Stieltjes B., Maier-Hein K. DALSA: domain adaptation for supervised learning from sparsely annotated mr images. IEEE Trans. Med. Imaging. 2016;35:184–196. doi: 10.1109/TMI.2015.2463078. [DOI] [PubMed] [Google Scholar]
- Guerrero R., Ledig C., Rueckert D. MLMI. 2014. Manifold alignment and transfer learning for classification of Alzheimer’s disease. [Google Scholar]
- He K., Zhang X., Ren S., Sun J. 2015 IEEE International Conference on Computer Vision (ICCV) 2015. Delving deep into rectifiers: surpassing human-level performance on imagenet classification; pp. 1026–1034. [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hofer C., Kwitt R., Höller Y., Trinka E., Uhl A. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 2017. Simple domain adaptation for cross-dataset analyses of brain MRI data; pp. 441–445. [Google Scholar]
- Hornik K., Stinchcombe M., White H., et al. Multilayer feedforward networks are universal approximators. Neural Netw. 1989;2(5):359–366. [Google Scholar]
- Ji D., Zhang D., Xu J., Chen Z., Yang T., Zhao P., Chen G., Cheng G., Wang Y., Bi J., et al. Prediction for progression risk in patients with COVID-19 pneumonia: the call score. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottas M., Kuss O., Zapf A. A modified Wald interval for the area under the ROC curve (AUC) in diagnostic case-control studies. BMC Med. Res. Methodol. 2014;14:26. doi: 10.1186/1471-2288-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liang D., Lin L., Hu H., Zhang Q., Chen Q., Iwamoto Y., Han X., Chen Y. MICCAI. 2018. Combining convolutional and recurrent neural networks for classification of focal liver lesions in multi-phase CT images. [Google Scholar]
- Liang W., Liang H., Ou L., Chen B., Chen A., Li C., Li Y., Guan W., Sang L., Lu J., et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Internal Med. 2020 doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Yao J., Chen A., Lv Q., Zanin M., Liu J., Wong S.-S., Li Y., Lu J., Liang H., qiang Chen G., Guo H., Guo J., Zhou R., Ou L., Zhou N., Chen H., Yang F., Han X., Huan W., Tang W., Guan W., Chen Z., Zhao Y., Sang L., Xu Y., Wang W., Li S., Lu L., Zhang N., Zhong N., Huang J., He J. Early triage of critically ill COVID-19 patients using deep learning. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-COVID: predicting COVID-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opbroek A.V., Ikram M., Vernooij M., de Bruijne M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans. Med. Imaging. 2015;34 5:1018–1030. doi: 10.1109/TMI.2014.2366792. [DOI] [PubMed] [Google Scholar]
- Opbroek A.V., Vernooij M., Ikram M., de Bruijne M. Weighting training images by maximizing distribution similarity for supervised segmentation across scanners. Med. Image Anal. 2015;24 1:245–254. doi: 10.1016/j.media.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Oreshkin B.N., López P.R., Lacoste A. NeurIPS. 2018. TADAM: task dependent adaptive metric for improved few-shot learning. [Google Scholar]
- Organization W.H., et al. situation report 205. 2020. Coronavirus disease 2019 (COVID-19) [Google Scholar]
- Qi H., Brown M., Lowe D. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Low-shot learning with imprinted weights; pp. 5822–5830. [Google Scholar]
- Sama I.E., Ravera A., Santema B.T., van Goor H., Ter Maaten J.M., Cleland J.G., Rienstra M., Friedrich A.W., Samani N.J., Ng L.L., et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur. Heart J. 2020;41(19):1810–1817. doi: 10.1093/eurheartj/ehaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- Shi B., Bai X., Yao C. An end-to-end trainable neural network for image-based sequence recognition and its application to scene text recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:2298–2304. doi: 10.1109/TPAMI.2016.2646371. [DOI] [PubMed] [Google Scholar]
- Sitaula C., Hossain M.B. Attention-based VGG-16 model for COVID-19 chest x-ray image classification. Appl. Intell. 2020:1–14. doi: 10.1007/s10489-020-02055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell J., Swersky K., Zemel R. Advances in Neural Information Processing Systems. 2017. Prototypical networks for few-shot learning; pp. 4077–4087. [Google Scholar]
- Song J.-W., Zhang C., Fan X., Meng F.-P., Xu Z., Xia P., Cao W.-J., Yang T., Dai X.-P., Wang S.-Y., et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020:1. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B. 1996;58:267–288. [Google Scholar]
- Wachinger C., Reuter M. Domain adaptation for Alzheimer’s disease diagnostics. NeuroImage. 2016;139:470–479. doi: 10.1016/j.neuroimage.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from ct images. IEEE Trans. Med. Imaging. 2020;39:2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C., Lui M.M., Lee J.C.Y., Chiu K., Chung T., Lee E., Wan E., Hung F.N., Lam T., Kuo M., Ng M. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2019 doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. Jama. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yang D., Xu Z., Li W., Myronenko A., Roth H.R., Harmon S., Xu S., Turkbey B., Turkbey E., Wang X., Zhu W., Carrafiello G., Patella F., Cariati M., Obinata H., Mori H., Tamura K., An P., Wood B., Xu D. Federated semi-supervised learning for covid region segmentation in chest CT using multi-national data from China, Italy, Japan. Med. Image Anal. 2021;70:101992. doi: 10.1016/j.media.2021.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of thecytokine storm’in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shi L., Wang F.-S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. Hepatol. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Liu X., Shen J., huan Li Z., Sang Y., wang Wu X., Zha Y., Liang W., Wang C., Wang K., Ye L., Gao M., Zhou Z., Li L., Wang J., Yang Z., Cai H., Xu J., Yang L., Cai W., Xu W., Wu S., Zhang W., Jiang S., Zheng L., Zhang X., Wang L., Lu L., Li J., Yin H., Wang W., Li O., Zhang C., Liang L., Wu T., Deng R., Wei K., Zhou Y., Chen T., Lau J.Y.-N., Fok M., He J., Lin T., Li W., Wang G. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433.e11. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020 doi: 10.1016/j.cell.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu L., Zhu R., Bagheri M., Summers R., Yao J. Spatio-temporal convolutional LSTMs for tumor growth prediction by learning 4D longitudinal patient data. IEEE Trans. Med. Imaging. 2020;39:1114–1126. doi: 10.1109/TMI.2019.2943841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-Y., Ma Y.-T., Zhang J.-Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Kim M., Zhu X., Kaufer D., Wu G. Long range early diagnosis of Alzheimer’s disease using longitudinal MR imaging data. Med. Image Anal. 2021;67:101825. doi: 10.1016/j.media.2020.101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/