Abstract
The circadian system of cyanobacteria is built upon a central oscillator consisting of three genes, kaiA, kaiB, and kaiC. The KaiA protein plays a key role in phosphorylation/dephosphorylation cycles of KaiC, which occur over the 24-h period. We conducted a comprehensive evolutionary analysis of the kaiA genes across cyanobacteria. The results show that, in contrast to the previous reports, kaiA has an ancient origin and is as old as cyanobacteria. The kaiA homologs are present in nearly all analyzed cyanobacteria, except Gloeobacter, and have varying domain architecture. Some Prochlorococcales, which were previously reported to lack the kaiA gene, possess a drastically truncated homolog. The existence of the diverse kaiA homologs suggests significant variation of the circadian mechanism, which was described for the model cyanobacterium, Synechococcus elongatus PCC7942. The major structural modifications in the kaiA genes (duplications, acquisition and loss of domains) have apparently been induced by global environmental changes in the different geological periods.
Subject terms: Molecular evolution, Phylogenetics
Introduction
Circadian rhythms or internal biological clock appeared in cells of living organisms as the main tool for adaptation to day–night change caused by the rotation of our planet around its axis1. This mechanism controls timely gene expression of a significant part of a genome.
Adaptation to the daily light cycles makes an important contribution to the ecological plasticity of cyanobacteria and apparently confers a selective advantage2, 3. It seems particularly important for marine cyanobacteria, which are characterized by ecological niche partitioning4.
Cyanobacteria were the first prokaryotes shown to have the circadian system5. The circadian system of cyanobacteria has been comprehensively studied in a model strain Synechococcus elongatus PCC7942. Its key structural and functional element, central oscillator, consists of three genes: kaiA, kaiB, and kaiC6. The corresponding proteins interact with each other: KaiB weakens the phosphorylation of KaiC7, while KaiA inhibits dephosphorylation of KaiC by binding to its respective domains8.
While the role of KaiA in the cyanobacterial circadian mechanism has been extensively studied (see9 for review), the knowledge about its evolution is limited. The first and most comprehensive study so far was published in 200310 and was based on then available GenBank collection of genomic sequences. It suggested the origin of the kaiA gene about 1000 Mya. The growing volume of available genomic data allowed for updating the initially proposed evolutionary scenario for the cyanobacterial circadian system and move the kaiA origin back to 2600–2900 Mya11.
The rapid growth of genomic databases during the last decade prompted for a new, more comprehensive analysis and, respectively, update of the existing evolutionary scenario for kaiA and the other circadian genes. The present study analyzed the occurrence, domain architecture, genetic variation and phylogeny of the kaiA gene homologs. We attempted to reconstruct the evolutionary history and to determine the evolutionary factors that have been operating on this key genetic element of the cyanobacterial circadian oscillator and might contribute to its function. We also updated a timeline for key events in the evolution of both kaiA and the whole circadian system. This study provides new data about the probable functional significance of various residues and motifs in the KaiA protein, and significantly updates our knowledge about the evolution of the cyanobacterial circadian system as a whole.
Results
Occurrence and domain architecture of the kaiA genes and proteins in cyanobacteria
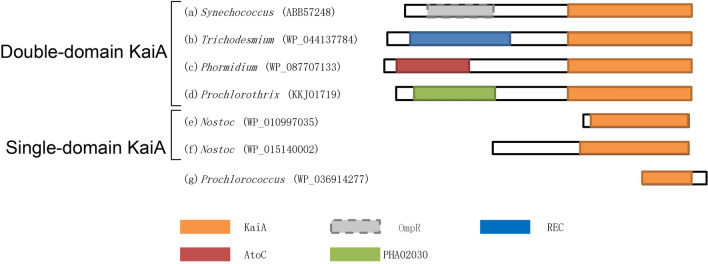
Homologs of kaiA occur in nearly all major cyanobacterial taxa available in GenBank, including Oscillatoriophycideae, Synechococcales, Pleurocapsales, Spirulinales, Chroococcidiopsidales, and Nostocales. All KaiA proteins can be roughly classified by their architecture into two main subfamilies, single-domain and double-domain, respectively (Fig. 1). The double-domain KaiA (ddKaiA) is about 300 aa long and found in Oscillatoriophycideae, Synechococcales, Pleurocapsales and Spirulinales. The KaiA proteins in Chroococcidiopsidales and Nostocales feature a single domain (sdKaiA) and are quite variable in length mostly ranging from 89 to 202 amino acid residues (Table S1). However, in some Nostocales, such as in Richelia intracellularis HH01, sdKaiA experienced even more drastic truncation, up to 45 amino acid residues. Both single-domain and double-domain versions share the conserved KaiA domain (pfam07688), which is the only member of superfamily cl17128.
Figure 1.
The domain architecture of KaiA proteins. (a) Synechococcus (ABB57248); (b) Trichodesmium (WP_044137784); (c) Phormidium (WP_087707133); (d) Prochlorothrix (KKJ01719); (e) Nostoc (WP_015140002); (f) Nostoc (WP_010997035); (g) Prochlorococcus (WP_036914277). Homology to the OmpR domain is weak and denoted by dashed box.
The BLAST search of the GenBank database also returned several short proteins manifesting high homology to other segments of the KaiA domain. For example, the proteins from two cyanobacterial strains annotated as Cyanobacteria bacterium QH_1_48_107 and Cyanobacteria bacterium QS_7_48_42, possess the KaiA homologs of 56 residues long (PSO52447.1 and PSP04869.1, respectively), which match a region between positions 169 and 224 in the bona fide S. elongatus PCC7942 protein (hereinafter the position numbers refer to the bona fide KaiA sequence). Interestingly, both these strains possess KaiC but lack KaiB.
Another example is the KaiA homologs found in some Prochlorococci (Table S1). They vary from 62 to 66 residues in length and, unlike the previous ones, match residues 238–284 in the respective S. elongatus PCC7942 protein (Fig. 1e). In contrast to the above-mentioned two strains, Prochlorococci do possess both KaiB and KaiC.
Several strains of Prochlorococcus sp. (e.g., MIT9303, MIT9313, and MIT1306) possessed a gene located in the genomic region usually occupied by the kaiA gene in the syntenic bona fide kaiABC operon, i.e. between the rplU and kaiB genes. However, unlike kaiA, this gene is located on the reverse complement strand. This gene was previously described as a pseudogene in MIT9303 and MIT931312. However, this is apparently not so: according to the genomic annotations, the gene is apparently translated, because it contains an open reading frame and thus may be functional. The putative respective proteins were about the same length (65–132 aa) as the sdKaiA homologs in other cyanobacteria. However, these proteins showed no apparent homology to either KaiA or any other proteins in the non-redundant NCBI protein database according to the BLAST search. Their function remains unknown.
In addition to cyanobacteria, the KaiA protein was found in other marine and freshwater bacteria, e.g., Propionibacteriaceae bacterium and Planctomycetaceae bacterium TMED241 (Table S1). This finding is unlikely an artefact, because screening of this species’ genome assembly revealed the full syntenic kaiABC operon typically found in cyanobacteria.
According to the Conserved Domain Database13, the N-terminal domain of the bona fide ddKaiA protein of S. elongatus PCC7942 belongs to the OmpR family. However, the observed homology is quite weak and was detected only when a lower E-value was applied. OmpR is a DNA-binding dual transcriptional regulator and is often an element of various two-component regulatory systems. While most ddKaiA proteins share the above architecture, few of them manifest some variability by featuring other domains instead of OmpR, namely REC, AtoC or PHA02030 (Fig. 1). However, regardless of the domain architecture, all KaiA homologs appear to form a homodimer in solution14, 15. On the other hand, this may not be the case for the truncated homologs.
The kaiA genes in some species are annotated as pseudogenes as, for example, in Aphanizomenon ovalisporum (CDHJ01000032, locus tag apha_00336). The functional deficiency of their KaiA might result by lack of the N-terminal fragment.
Conserved residues of possible functional significance
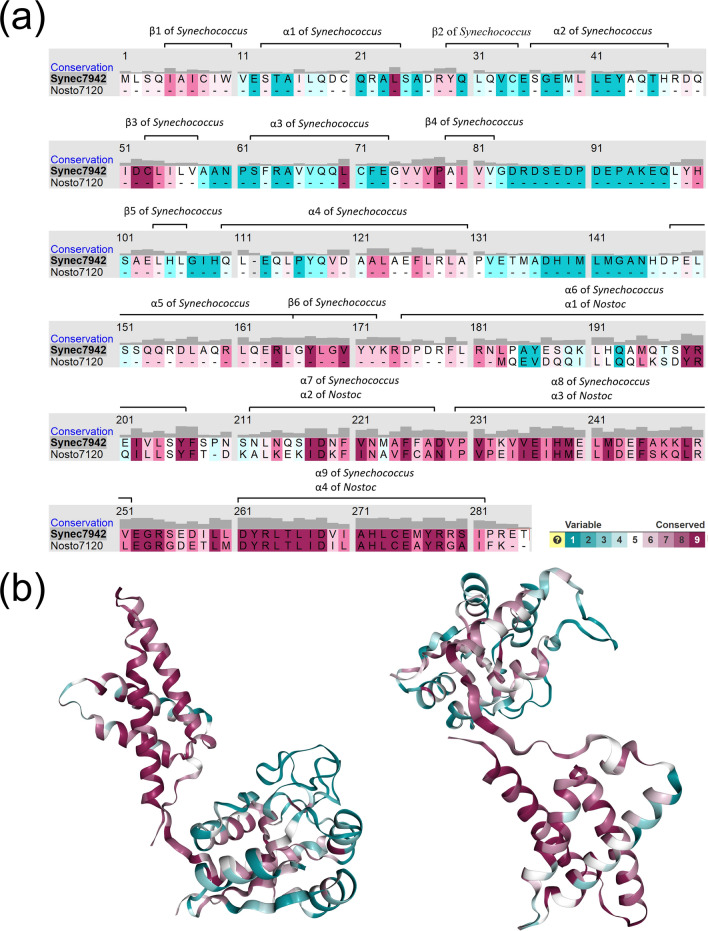
The C-terminal domains of the single-domain and double-domain KaiA homologs have quite similar structure consisting of four conserved helices. The most terminally located helix (α4 of Nostoc and α9 of Synechococcus, sites 259–280) has the highest level of conservation (Fig. 2) and acts as a dimer interface14. However, the KaiA proteins in some species (e.g., Trichormus variabilis ATCC 29413) have a significantly shorter C-terminal region and lack at least two most terminally located KaiC-binding residues. In Synechococcus elongatus PCC 7942, the truncation of C-terminal amino acid residues leads to the shortened circadian periods because the binding between KaiA and KaiC is strengthened16.
Figure 2.
Group-conserved residues identified by ConSurf. Degrees of conservation in subfamilies were visualized by Chimera v.1.10.217. (a) Conserved sites of the KaiA protein. Number the residues is accordant with the Synechococcus KaiA (ABB57248). The black bars above sequence indicate the level of conservation (1–9). (b) Conserved sites labeled (red) in the 3D structure of the Synechococcus elongatus KaiA protein (PDB: 1R8J_A) (left: N-terminal region; right: KaiA domain).
The comparative analysis of the KaiA homologs identified 27 sites in the namesake domain universally conserved in most cyanobacteria according to the BLOSUM62 matrix18. Five sites, M241, D242, E251, L265, and D267, are 100% conserved across all cyanobacteria, including the truncated KaiA homologs in the Prochlorococcus sp. and Trichormus variabilis ATCC 29413 (Table 1). Ten of the 27 conserved sites are fixed, which strongly suggests their high functional significance. However, the respective data are available only about five of them. The known functions of the conserved sites are related to either the maintenance of the KaiA homodimer structure14 or binding the KaiC protein19.
Table 1.
A list of the universally conserved positions in the KaiA homologs of cyanobacteria with the reference to the bona fide protein of S. elongatus PCC7942.
| Position number | Amino acid in S. elongatus PCC7942 | Possible variants in other cyanobacteria | Effect of mutation or putative function | References |
|---|---|---|---|---|
| 198 | Y | None | Unknown | na |
| 201 | I | L, V | Unknown | na |
| 202 | V | L, I | Unknown | na |
| 205 | Y | None | Unknown | na |
| 206 | F | Y | Unknown | na |
| 216 | I | M, L, V | Unknown | na |
| 217 | D | E | Unknown | na |
| 224 | F | Y | Abolishes the rhythm | 18 |
| 234 | V | I, L, M | Unknown | na |
| 237 | H | None | Unknown | na |
| 241a | M | I, V |
Modifies amplitude KaiC binding site |
|
| 242a | D | E |
Modifies amplitude KaiC binding site |
|
| 251a | E | K | Unknown | na |
| 258 | L | I, V | Unknown | na |
| 260 | D | None | Dimer interface site | 14 |
| 261 | Y | None | Unknown | na |
| 262 | R | None | Dimer interface site | 14 |
| 265a | L | I, V | Unknown | na |
| 266 | I | M, L, V | Modifies amplitude, KaiC binding | 19 |
| 267a | D | None | Unknown | na |
| 269 | I | M, L, V | Dimer interface site | 14 |
| 270 | A | S | Dimer interface site | 14 |
| 271 | H | N | Unknown | na |
| 272 | L | M | Dimer interface site | 14 |
| 274 | E | None | Dimer interface site | 14 |
| 276 | Y | None | Dimer interface site | 14 |
| 277 | R | None | Dimer interface site | 14 |
aConserved in the truncated KaiA too.
Functional divergence of the KaiA homologs
The analysis of the functional divergence between the single-domain and double-domain KaiA proteins showed the significantly altered functional constraints (rates of evolution) after duplication of the ancestral gene. On the other hand, no type II functional divergence (radical amino acid changes without a rate shift) was detected. In the analyzed segment of 96 amino acid residues (nearly the full length KaiA domain), the effective number of the type I residues was 33. That means, nearly 1/3 of the domain experienced significant shift in evolutionary rate.
Nucleotide diversity and selection of kaiA
The C-terminal region of ddkaiA is more variable (dN = 0.30 ± 0.03, π = 0.36 ± 0.00) as compared to the single-domain homologs (dN = 0.20 ± 0.02, π = 0.26 ± 0.01) and is much more conserved than the N-terminal one (dN = 0.88 ± 0.06, π = 0.52 ± 0.00). This may be due to the evolutionary younger age of sdKaiAs as compared to ddKaiAs (Nostocales are evolutionary younger than Oscillatoriophycideae, Synechococcales and Pleurocapsales)10 or/and because of the higher functional significance of the C-terminal region of KaiA (binds to KaiB and KaiC)20. Besides, the N-terminal domains of the ddkaiA genes may vary and manifest functional diversity (Fig. 1). None of the applied methods detected positive selection in the kaiA genes.
The phylogeny of the kaiA genes and time estimates of the evolutionary events
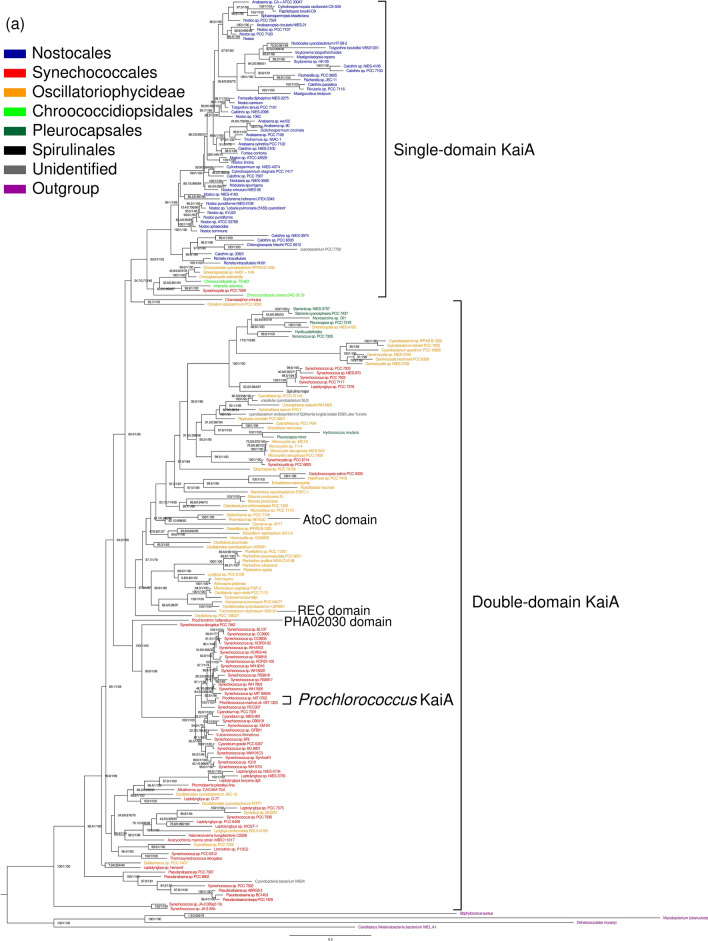
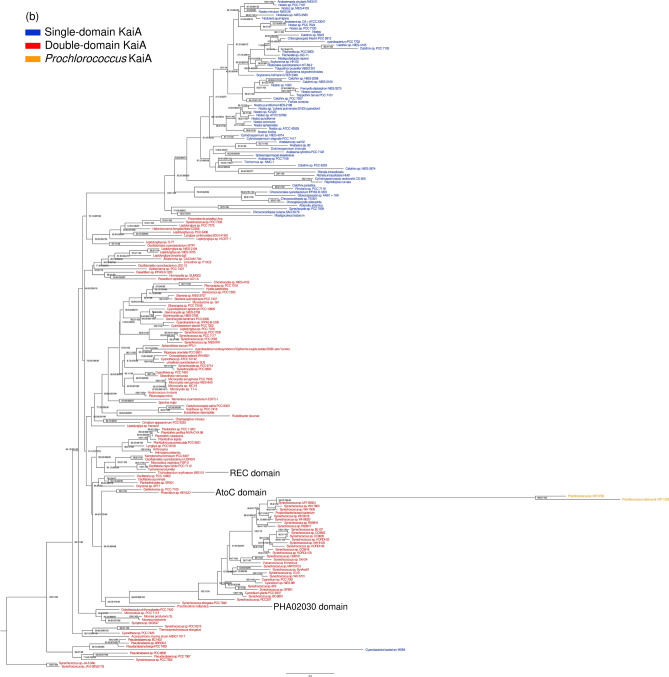
The comparison of the species and gene trees clearly shows they are largely incongruent. Both trees feature several monophyletic clades with a strong statistical support (Figs. 3 and S1). These few clades in both trees match each other by set of taxa, but their positions in the overall tree topologies are quite different, and so are the positions of taxa within the clades. The ML trees manifested better resolution than the Bayesian trees, particularly at deeper nodes. The comparative analysis of the species and gene trees identified 36 lateral gene transfers having occurred in the evolution of the kaiA genes (Supplementary file 1, Fig. S2). However, while the lateral transfers seem to be quite common within the clades, they have been much less frequent or absent between the clades. Furthermore, no HGTs were detected between the taxa with single-domain and double-domain kaiA genes. The results of the HGT analysis suggest that heterocystous cyanobacteria, which are a group possessing the sdkaiA, have experienced the most frequent transfers (Fig. S2).
Figure 3.
The maximum-likelihood phylogenetic trees of: (a) the 16S and 23S rRNA genes (species tree) and (b) KaiA homologs (gene tree). The node support values are ultrafast bootstrap/SH-aLRT branch test/approximate Bayes test.
The time estimates of the major events in the evolution of the kaiA homologs are provided in Table 3. Both Bayesian and ML estimates are similar and suggest three main periods when these events probably occurred: about 30–100, 500–600, and 1000–1500 Mya. The origin of the sdkaiA was apparently associated with the origin of Chroococcidiopsidales that occurred about 1500 Mya.
Table 3.
Bayesian and maximum-likelihood time estimates for the events in the evolution of the kaiA homologs based on the species trees (Mya).
| Evolutionary events | Bayesiana | Maximum likelihoodb |
|---|---|---|
| HGT of kaiA from Synechococcus to Prochlorococcus followed by truncation | 34.2–100 | 31.7–145.6 |
| Loss of ddkaiA in Prochlorococcus | 154.1 (91.1, 222.0) | 202.5 (161.0, 262.2) |
| Domain fusion of AtoC in Phormidium | 508.8 (134.5, 1023.4) | 709.3 (522.2, 971.0) |
| Domain fusion of PHA02030 in Prochlorothrix hollandica | 1032.6 (673.0, 1422.8) | 1363.4 (1151.6, 1605.9) |
| Domain fusion of REC in Trichodesmium erythraeum | 1405.9 (1313.9, 1498.5) | 1513.6 (1314.6, 1752.5) |
| Origin of sdkaiA/Chroococcidiopsidales | 1483.3 (1325.5, 1683.4) | 1650.8 (1530.2, 1808.7) |
| CP1: origin of Nostocales | 1300–1480 | |
| CP2: origin of cyanobacteria | 3000 | |
aPosterior mean (95% HPD).
bMean (95% CI).
The 3D structure of the KaiA homologs
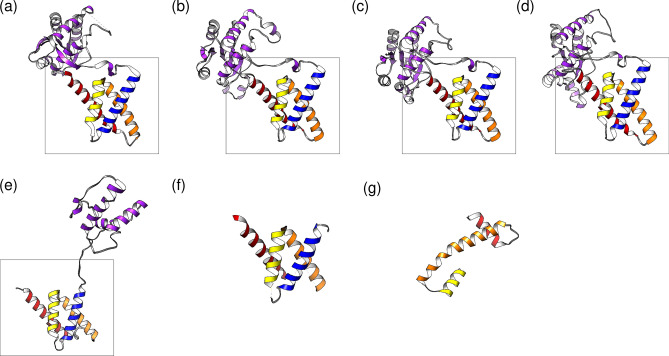
The in silico inferred 3D models of the KaiA homologs have essentially the same structure as those determined experimentally (Fig. 4). They all feature a highly conserved KaiA domain formed by four pairwise paralleled helices. The only exception is a KaiA homolog of Prochlorococcus sp. (Fig. 4g). It is truncated and features three helices, two of which on the termini are short. The long helix, however, is highly homologous to the terminal helix (α9) of the KaiA domain in the bona fide protein of S. elongatus PCC7942 (Fig. 2a).
Figure 4.
Models of the 3D structure of the KaiA homologs from different cyanobacteria. (a) Synechococcus (ABB57248, PDB: 1R8J); (b) Trichodesmium (WP_044137784); (c) Phormidium (WP_087707133); (d) Prochlorothrix (KKJ01719); (e) Nostoc (WP_015140002); (f) Nostoc (WP_010997035, PDB: 1R5Q); (g) Prochlorococcus (WP_036914277). The KaiA domain is boxed. Models (a) and (f) are experimental, the others are computer generated.
Discussion
The occurrence and distribution of kaiA among cyanobacterial taxa suggest an ancient origin of the gene
The kaiA genes were found in all analyzed cyanobacteria except Gloeobacter. The latter is thought to be the most ancient cyanobacterium, which, while being able for photosynthesis, lacks a few structures and genes common for all other cyanobacteria21. Our results on the kaiA occurrence are essentially in agreement with those recently reported by Schmelling et al.22 who performed comprehensive screening of prokaryotes for circadian orthologs. In addition, the present study firstly reports the kaiA homologs and the whole kaiABC operon in prokaryotes other than cyanobacteria. The most probable explanation of this may be a lateral transfer of the operon from cyanobacteria.
The truncated kaiA homologs from Prochlorococcales were not reported by the early evolutionary studies of the circadian system in cyanobacteria (see, e.g.10, 11). This might be due to the much smaller volume of then available genomic data and poor annotations of genomes.
The occurrence of the kaiA homologs across all cyanobacterial taxa suggests that this gene is of ancient origin, probably as old as most cyanobacteria themselves. Indeed, kaiA was found in the thermophilic strains from Yellowstone, Synechococcus sp. JA-2-3B'a(2-13) and Synechococcus sp. JA-3-3Ab, which are located at the root of the cyanobacterial phylogenetic subtree (Figs. 3a and S1a). It might be that the gene was horizontally transferred from the evolutionary younger lineages. However, no such transfers to this clade was detected (Fig. S2).
In the pioneering study about origin and evolution of the cyanobacterial circadian genes, it was hypothesized that kaiA originated about 1000 Mya after two other key circadian genes, kaiB and kaiC10. This hypothesis was later revisited based on the growing available genomic data and much earlier origin of the kaiA gene was suggested23, 24. This revision is further supported by the results of the present study.
The domain architecture underlies evolutionary and functional constraints of the kaiA genes
All kaiA genes can be divided into two large groups according to their domain architecture: single-domain and double-domain, respectively. The occurrence of these two versions of the gene is taxon-specific (Figs. 3 and S2). The sdkaiA occurs exclusively in Chroococcidiopsidales and Nostocales, while ddkaiA was found across all other cyanobacterial taxa. However, the N-terminal domain in ddkaiA varies quite significantly (especially as compared to the kaiA domain) across cyanobacteria and its homology to OmpR is quite weak. This suggests that the ancestral OmpR domain has been under weak selective constraints in the course of evolution that might result in its functional modification or even loss of the original function.
The truncation of the ancient ddkaiA into sdkaiA and the origin of Chroococcidiopsidales were apparently associated with each other (Table 2, Fig. 3). The loss of the N-terminal domain probably conferred evolutionary constraints to the kaiA domain: it is significantly less variable in sdkaiA than in ddkaiA (Table 2). Another evidence comes from the analysis of HGTs: while the transfers have been quite common within the clades of the genes with the same architecture (i.e., either single-domain or double-domain, respectively), no HGTs were determined between these clades (Fig. S2).
Table 2.
Patterns of nucleotide diversity in the kaiA homologs of cyanobacteria.
| dN | π | |||||
|---|---|---|---|---|---|---|
| N-terminal region | kaiA domain | Average over gene | N-terminal region | kaiA domain | Average over gene | |
| ddkaiA, | 0.78 ± 0.05 | 0.35 ± 0.04 | 0.57 ± 0.03 | 0.48 ± 0.00 | 0.38 ± 0.00 | 0.42 ± 0.00 |
| sdkaiA | 0.79 ± 0.05 | 0.26 ± 0.02 | 0.31 ± 0.02 | 0.52 ± 0.02 | 0.27 ± 0.02 | 0.27 ± 0.01 |
| Average over domain | 1.08 ± 0.06 | 0.37 ± 0.03 | 0.54 ± 0.03 | 0.57 ± 0.01 | 0.33 ± 0.01 | 0.33 ± 0.01 |
The rate of synonymous nucleotide substitutions (dS) was not estimated due to saturation.
The KaiA domain of the protein is a key player in its binding to KaiC: several functionally important or critical residues have been identified experimentally in this domain14, 19. However, there are several more highly conserved or invariable residues identified in the present study (Table 1), which are apparently functionally important, but their exact function has yet to be determined.
There are several factors, which likely confer evolutionary constraints to the kaiA genes and limit HGTs even between the clades with the same domain architecture. In particular, this may be related to possible interaction with other elements of the circadian system. For example, some studies showed that KaiA competes with CikA in binding to KaiB and phosphorylation of KaiC25. However, this mechanism is likely not universal, because bona fide CikA is absent in many cyanobacteria11, 22. Therefore, the observed variation in the kaiA domain and, respectively, above mentioned constraints may be related to functional modifications of KaiA to adjust to the circadian input pathway alterations. Wood et al.26 reported that KaiA of S. elongatus PCC7942 binds the quinone by its N-terminal domain (OmpR, Fig. 1). This interaction helps to stabilize KaiA and is important for the mechanism of the KaiC phosphorylation. However, the sdKaiA proteins either lack the N-terminal domain completely or have it truncated (Fig. 1e–g) that means the circadian system in Chroococcidiopsidales and Nostocales should either lack this binding ability completely or have a different one. Furthermore, some cyanobacterial lineages have different N-terminal domains (Fig. 1) that assumes the different (if any) interaction with the quinone.
The variation patterns in the kaiA gene and the encoded protein support the functional diversification of the circadian system in cyanobacteria
There is ample evidence that the cyanobacterial circadian system has experienced extensive evolutionary diversification (see, e.g.11, 24 for review). The results of the present study provide further support for that. Not only did the functional divergence occur between the single-domain and double-domain KaiA proteins, but also it occurred between the clades within these two subfamilies (data not shown).
In its native state, KaiA is a dimer whose only known function is binding to KaiC CII domain and inducing its autophosphorylation27, 28. Therefore, in the circadian system missing KaiA, the timing mechanism may be simplified as it was suggested for Prochlorococcus29. However, it seems that even within the Prochlorococcus lineage, different versions of the simpler circadian system may exist. Indeed, as the results of the present study suggest, some Prochlorococcus strains possess, albeit truncated, but a highly conserved homolog of kaiA (Fig. 1). This extreme conservation, particularly at the functionally important residues common for the KaiA homologs across cyanobacteria, may suggest that the function of this truncated KaiA is somewhat similar to that of the bona fide protein. Strains of Prochlorococcus are known for their niche-specific adaptation, particularly with respect to the different light and temperature regimes, and extensive diversification into many co-existing ecotypes30. The presence/absence of the kaiA homolog or its orphan replacement may be associated with this adaptation. For example, strains MIT9303 and MIT9313, which possess the orphan gene, were reported as adapted to low light30. Importantly, despite the quite significant type I functional divergence (altered evolutionary rate), no type II divergence (radical amino acid changes) was detected in the KaiA domain of the truncated homologs. In these terms, it would be interesting to determine the exact functional significance of the universally conserved residues identified in the present study (Table 1).
Phylogenetic dating supports the hypothesis about the association of the circadian system evolution with the geochronological events
The origin of the kaiA gene was initially estimated about 1000 Mya based on then available genomic data10. Since then, as more data has been accumulated, this estimate has been reconsidered23, 24. The results of the present study suggest that the kaiA gene is evolutionarily much older than it was thought before and its origin can be dated back to that of most cyanobacteria, i.e., about 3000 ± 500 Mya depending on the estimation methods.
The loss of kaiA in Prochlorococcales was apparently associated with the origin of this taxon that occurred about 150–200 Mya (Table 3). This estimate is in broad agreement with the previously reported dating based on the rRNA sequences31. Holtzendorff et al.12 hypothesized that kaiA might experience a stepwise deletion in Prochlorococcales by referring to the “kaiA pseudogene” in strains MIT9303 and MIT9313 as the evidence. However, the results of the present study suggest an alternative scenario: the original kaiA gene was initially either lost in Prochlorococcales or replaced by the orphan gene (the one erroneously referred to as the kaiA pseudogene). This assumption seems quite feasible given that the above two strains as well as others missing the truncated kaiA homologs belong to the earliest branching low-light adapted clades of the Prochlorococcus subtree32, 33.
After that, about 50–100 Mya, the kaiA gene was laterally transferred from the Synechococcus lineage to some Prochlorococcales and underwent a drastic truncation (Fig. 1e, Table 3). One more scenario may be based on the fact that the truncated kaiA homologs are apparently common in various Synechococcales and Nostocales and therefore the truncation might occur prior to the HGT to Prochlorococcales. These HGT and follow-up truncation (if any) might be related to the Cretaceous–Paleogene (K–Pg) extinction, which occurred about 66 MYA due to the asteroid impact having caused global ecological devastation, including rapid acidification of the oceans and light regime change34, 35.
There were several major structural changes in the kaiA genes (Table 3). The origin of sdkaiA and Chroococcidiopsidales falls within the Calymmian Period, the first geologic period in the Mesoproterozoic Era about 1500 MYA. These events might be associated with oxygenation of the Metaproterozoic ocean that occurred about 1570–1600 Mya36. The domain fusion in kaiA of Phormidium occurred about 500–700 Mya, which corresponds to either the Ediacaran Period known for its Avalon explosion37 or the Cambrian explosion38.
Of course, the above interpretation of the obtained estimates has some limitations, one of which is the uncertainty of the fossil calibrations. On the other hand, the dates of the multiple events in the evolution of the kaiA genes inferred by the molecular methods match well the specific events in the Earth geochronology, which indeed might affect this evolution.
Conclusion
The present study provides compelling evidence for the ancient origin of the kaiA gene and thus revises the previously suggested timeline of the cyanobacterial circadian system evolution. It also prompts for further experimental studies to determine the exact functions of the identified universally conserved/fixed residues in the KaiA domain.
Materials and methods
DNA and protein sequences
The sequences of the KaiA proteins and respective genes were retrieved from the GenBank using the KaiA sequence of Synechococcus elongatus PCC7942 (WP_011377921) as a query. We utilized the genomic BLASTP39 to search the database. Only the sequences from the fully sequenced cyanobacterial genomes were used for the analyses. Bit score of 100 was applied as a cutoff value for sequence selection. Finally, the sequences from 226 strains were retained for the analysis. The used sequences are listed in Supplementary Table S1.
Besides, we used the 16S and 23S rRNA genes for the construction of the species tree. The respective DNA sequences of Acaryochloris marina strain MBIC11017 (CP000828) and Nostoc sp. PCC 7107 (CP003548) were used as the probes. In addition to the rRNA genes of cyanobacteria, the respective sequences of Staphylococcus aureus, Dehalococcoides mccartyi, Mycobacterium tuberculosis, and Candidatus Melainabacteria bacterium MEL.A1 were retrieved for the phylogenetic analysis (Table S1). In total 231 sequences were used in the analyses.
Sequence editing and alignment
The full protein sequences were aligned using the combined sequence and structure-based algorithm implemented in the PRALINE server40, 41; the nucleotide sequences were aligned according to the protein alignment by Rev-Trans v.1.4 (http://www.cbs.dtu.dk/services/RevTrans/)42. The rRNA sequences were aligned using MAFFT43. The aligned sequences were inspected visually and trimmed manually to remove poorly aligned regions and thus to improve a phylogenetic signal. The resulting final alignment of the KaiA protein subfamilies included 296 positions; the concatenated 16S-23S rRNA alignment counted 2941 positions.
Identification of conserved residues
The ConSurf server (http://consurf.tau.ac.il/) was utilized to identify group-specific conserved sites in the KaiA proteins44. The analysis was conducted using a Bayesian procedure, the JTT substitution matrix, and Synechococcus elongatus KaiA (SMTL ID: 4G86_A) as a template45.
Analysis of nucleotide diversity and selection
The dN values of the kaiA genes were calculated using the modified Nei-Gojobori method (with Jukes-Cantor correction and 1000 bootstrap replicates)46 as implemented in MEGA X47. To test the saturation of synonymous substitutions, pairwise dS estimates were calculated first. Most pairwise dS values were above 2, thus indicating that synonymous nucleotide substitutions were saturated. Also, the nucleotide diversity of the KaiA was analyzed using DnaSP v. 6.12.0348. The level of variation was estimated by π49.
Positive selection in the kaiA genes was analyzed using several approaches implemented in the Datamonkey server50. Site-specific positive selection was analyzed using FUBAR51; the branch-site positive selection was tested using aBSREL52. A gene-wide test for positive selection was conducted using BUSTED53.
Analysis of the functional divergence
The functional divergence between the KaiA subfamilies at the namesake domain was analyzed using the DIVERGE3 software54. The following parameters were estimated: type I and type II functional divergence55, 56, effective number of sites related to this divergence. The False Discovery Rate (FDR) of the probability cut-off for the predicted sites was set at < 0.05.
Phylogenetic analysis
Using only the KaiA domain for the phylogenetic inference yielded a poorly resolved tree. Therefore, the full KaiA protein alignment of 296 positions was utilized. The maximum-likelihood phylogenetic analysis was conducted using the IQ-TREE software57 with the built-in ModelFinder function58. Based on the ModelFinder analysis results, the JTT model59 with a gamma distribution (α = 0.840) was used for the phylogenetic analysis of the KaiA homologs; the GTR model with a proportion of invariable sites and gamma distribution (GTR + I + 4G, p-inv = 0.106, α = 0.662) was applied to the analysis of the rRNA genes. The node support was inferred according to the ultrafast bootstrap60, SH-aLRT branch test61, and approximate Bayes test62.
The Bayesian relaxed clock as implemented in BEAST v.2.6.263 was used to construct phylogenetic trees. The length of the MCMC chain was set for 100 million with trees sampling every 1,000 steps. The maximum clade credibility tree was determined using TreeAnnotator v.1.7.5 from the BEAST software package.
The horizontal gene transfers were determined using the bipartition dissimilarity algorithm implemented in the HGT-Detection software64.
Time estimates for the evolutionary events
Two internal calibration points (CP1 & CP2) based on cyanobacteria fossil evidence were used for evolutionary time estimates. CP1 indicated the origins of Nostocales (1480–1300 Mya)65, CP2 corresponded to the lower boundary of the estimates for the origin of cyanobacteria and was limited to the mid-Archean, before the Great Oxidation Event (~ 3000 Mya)66. The height of the whole tree was constrained to 4000 Mya. The computations were conducted using BEAST v.2.6.267 and IQ-TREE57 as mentioned above.
Three-dimensional modeling of the KaiA proteins
The predicted 3D models of KaiA proteins with the different domain architecture were constructed and refined using the respective methods implemented in the GalaxyWEB server68. The quality of the models was assessed by the structure assessment tool of the SWISS-MODEL server69.
Supplementary Information
Acknowledgements
The study was supported by Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0408) and National Natural Science Foundation of China (Grant No. 31600341).
Author contributions
V.D. designed the analysis, performed the analysis and wrote the paper. Q.M. collected the data, performed the analysis, prepared the figures and wrote the paper.
Data availability
All data generated or analyzed during this study are included in this published article (and its “Supplementary Information” files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Volodymyr Dvornyk and Qiming Mei.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89345-7.
References
- 1.Pittendrigh CS. Temporal organization: Reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Jacquet S, Partensky F, Marie D, Casotti R, Vaulot D. Cell cycle regulation by light in Prochlorococcus strains. Appl. Environ. Microbiol. 2001;67:782–790. doi: 10.1128/AEM.67.2.782-790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson CH, Golden SS, Kondo T. Adaptive significance of circadian programs in cyanobacteria. Trends Microbiol. 1998;6:407–410. doi: 10.1016/S0966-842X(98)01356-0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 5.Kondo T, et al. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 6.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 7.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swan JA, Golden SS, LiWang A, Partch CL. Structure, function, and mechanism of the core circadian clock in cyanobacteria. J. Biol. Chem. 2018;293:5026–5034. doi: 10.1074/jbc.TM117.001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc. Natl. Acad. Sci. USA. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baca I, Sprockett D, Dvornyk V. Circadian input kinases and their homologs in cyanobacteria: Evolutionary constraints versus architectural diversification. J. Mol. Evol. 2010;70:453–465. doi: 10.1007/s00239-010-9344-0. [DOI] [PubMed] [Google Scholar]
- 12.Holtzendorff J, et al. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms. 2008;23:187–199. doi: 10.1177/0748730408316040. [DOI] [PubMed] [Google Scholar]
- 13.Lu S, et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye S, Vakonakis I, Ioerger TR, LiWang AC, Sacchettini JC. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J. Biol. Chem. 2004;279:20511–20518. doi: 10.1074/jbc.M400077200. [DOI] [PubMed] [Google Scholar]
- 15.Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 2004;23:1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, et al. A novel allele of kaiA shortens the circadian period and strengthens interaction of oscillator components in the cyanobacterium Synechococcus elongatus PCC 7942. J. Bacteriol. 2009;191:4392–4400. doi: 10.1128/JB.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura H, et al. Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology. 2002;148:2903–2909. doi: 10.1099/00221287-148-9-2903. [DOI] [PubMed] [Google Scholar]
- 20.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura Y, et al. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 22.Schmelling NM, et al. Minimal tool set for a prokaryotic circadian clock. BMC Evol. Biol. 2017;17:169. doi: 10.1186/s12862-017-0999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvornyk V. Evolution of the circadian clock system in cyanobacteria: A genomic perspective. Int. J. Algae. 2016;18:5–20. doi: 10.1615/InterJAlgae.v18.i1.10. [DOI] [Google Scholar]
- 24.Dvornyk, V. The circadian clock gear in cyanobacteria: Assembled by evolution. In: Bacterial Circadian Programs (ed. J. Ditty, Mackey, S.R., Johnson, C.H.), Ch. 14, pp. 241–258 (Springer-Verlag Berlin Heidelberg, 2009).
- 25.Kaur M, Ng A, Kim P, Diekman C, Kim YI. CikA modulates the effect of KaiA on the period of the circadian oscillation in KaiC phosphorylation. J. Biol. Rhythms. 2019;34:218–223. doi: 10.1177/0748730419828068. [DOI] [PubMed] [Google Scholar]
- 26.Wood TL, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc. Natl. Acad. Sci. USA. 2010;107:5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: Implications for KaiC regulation. Proc. Natl. Acad. Sci. USA. 2004;101:10925–10930. doi: 10.1073/pnas.0403037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axmann IM, Hertel S, Wiegard A, Dorrich AK, Wilde A. Diversity of KaiC-based timing systems in marine cyanobacteria. Mar. Genom. 2014;14:3–16. doi: 10.1016/j.margen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Zinser ER, et al. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol. Oceanogr. 2007;52:2205–2220. doi: 10.4319/lo.2007.52.5.2205. [DOI] [Google Scholar]
- 31.Sanchez-Baracaldo P. Origin of marine planktonic cyanobacteria. Sci. Rep. 2015;5:17418. doi: 10.1038/srep17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biller SJ, et al. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci. Data. 2014;1:140034–140034. doi: 10.1038/sdata.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doré H, et al. Evolutionary mechanisms of long-term genome diversification associated with niche partitioning in marine picocyanobacteria. Front. Microbiol. 2020;11:567431–567431. doi: 10.3389/fmicb.2020.567431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henehan MJ, et al. Rapid ocean acidification and protracted Earth system recovery followed the end-Cretaceous Chicxulub impact. Proc. Natl. Acad. Sci. USA. 2019;116:22500–22504. doi: 10.1073/pnas.1905989116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez LW, Alvarez W, Asaro F, Michel HV. Extraterrestrial cause for the cretaceous-tertiary extinction. Science. 1980;208:1095–1108. doi: 10.1126/science.208.4448.1095. [DOI] [PubMed] [Google Scholar]
- 36.Zhang K, et al. Oxygenation of the Mesoproterozoic ocean and the evolution of complex eukaryotes. Nat. Geosci. 2018;11:345–350. doi: 10.1038/s41561-018-0111-y. [DOI] [Google Scholar]
- 37.Shen B, Dong L, Xiao S, Kowalewski M. The Avalon explosion: Evolution of Ediacara morphospace. Science. 2008;319:81–84. doi: 10.1126/science.1150279. [DOI] [PubMed] [Google Scholar]
- 38.Butterfield, N. J. Ecology and evolution of Cambrian plankton. In: The Ecology of the Cambrian Radiation (ed. Riding, R. & Zhuravlev, A.), pp. 200–216 (Columbia University Press, 2001).
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Simossis VA, Heringa J. PRALINE: A multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simossis VA, Heringa J. The PRALINE online server: Optimising progressive multiple alignment on the web. Comput. Biol. Chem. 2003;27:511–519. doi: 10.1016/j.compbiolchem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pattanayek R, Sidiqi SK, Egli M. Crystal structure of the redox-active cofactor dibromothymoquinone bound to circadian clock protein KaiA and structural basis for dibromothymoquinone's ability to prevent stimulation of KaiC phosphorylation by KaiA. Biochemistry. 2012;51:8050–8052. doi: 10.1021/bi301222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozas J, et al. DnaSP 6: DNA Sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 49.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver S, et al. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murrell B, et al. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith MD, et al. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015;32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murrell B, et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu X, et al. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013;30:1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- 55.Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- 56.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 57.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 60.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 62.Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouckaert R, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boc A, Philippe H, Makarenkov V. Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst. Biol. 2010;59:195–211. doi: 10.1093/sysbio/syp103. [DOI] [PubMed] [Google Scholar]
- 65.Demoulin CF, et al. Cyanobacteria evolution: Insight from the fossil record. Free Radic. Biol. Med. 2019;140:206–223. doi: 10.1016/j.freeradbiomed.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schirrmeister BE, Gugger M, Donoghue PC. Cyanobacteria and the Great Oxidation Event: Evidence from genes and fossils. Palaeontology. 2015;58:769–785. doi: 10.1111/pala.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ko J, Park H, Heo L, Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterhouse A, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its “Supplementary Information” files).