Abstract
Artemisinin is the frontline fast-acting anti-malarial against P. falciparum. Emergence and spread of resistant parasite in eastern-India poses a threat to national malaria control programs. Therefore, the objective of our study is to evaluate the artesunate-sulfadoxine-pyrimethamine efficacy in Central India. 180 monoclonal P. falciparum-infected patients received standard ASSP therapy during August 2015–January 2017, soon after diagnosis and monitored over next 42-days. Artemisinin-resistance was assessed through in-vivo parasite clearance half-life (PC1/2), ex-vivo ring-stage survivability (RSA), and genome analysis of kelch13 and other candidate gene (pfcrt, pfmdr1, pfatpase 6, pfdhfr and pfdhps). Of 180 P. falciparum positive patients, 9.5% showed increased PC1/2 (> 5.5 h), among them eleven isolates (6.1%) showed reduced sensitivity to RSA. In 4.4% of cases, parasites were not cleared by 72 h and showed prolonged PC1/2(5.6 h) (P < 0.005) along with significantly higher RSA (2.2%) than cured patients (0.4%). None of day-3 positive isolates contained the pfkelch13 mutation implicated in artemisinin resistance. Parasite recrudescence was observed in 5.6% patients, which was associated with triple dhfr–dhps (A16I51R59N108I164–S436G437K540G581T613) combination mutation. Emergence of reduced sensitivity to artesunate-sulfadoxine-pyrimethamine, in central India highlighted the risk toward spread of resistant parasite across different parts of India. Day-3 positive parasite, featuring the phenotype of artemisinin-resistance without pfkelch13 mutation, suggested kelch13-independent artemisinin-resistance.
Subject terms: Antimicrobial resistance, Antiparasitic agents, Haplotypes, Genetic markers, Malaria
Introduction
Drug resistant P. falciparum is one of the major factors for death in malaria. 445,000 deaths and an estimated 216 million confirmed malaria cases—including an increase of about 5 million cases over and above what was recorded in 2015-were reported in 20161. Malaria transmission in India potentially occurs through either P. falciparum or P. vivax infection2,3. In India, 844,558 malaria cases were reported in 2017, of which 529,530 cases were P. falciparum positive4. North-eastern states and central Indian states contributed 80% of the total cases5. Chhattisgarh, one of the states in central India, contributed the second highest malaria incidence in India over the years6. National vector borne disease control program (NVBDCP) had launched artemisinin-based combination therapy (ACT) to wipe-out the burden of chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) resistant malaria in 20097–9. Success of ACT depends on a combination of fast-acting short half-life artemisinin derivatives with late-acting longer half-life 4-aminoquinolines or antifolates10. Global mortality and morbidity associated with malaria were considerably reduced after the introduction of ACT, but emergence and subsequent spread of artemisinin-resistant parasites in the Greater Mekong sub-region had seriously threatened the global malaria control and elimination progress11–14. Artemisinin resistance is characterized by drug failure reflected in slow parasite clearance as assessed by increased in vivo parasite clearance half-life (PC1/2) along with reduced sensitivity of ex-vivo ring-stage parasites to artemisinin15–17. Genome based transfection studies proved the association of artemisinin resistance with kelch13 gene-polymorphism which was directly linked with increased ex-vivo ring-stage survivability and prolonged in-vivo PC1/2 (> 5.5 h)18–21. Previous genome-based reports suggested that polymorphism in pfatpase6, pfmdr1, pfcrt and P. falciparum ferredoxin (pffd) genes also exhibited potential role on artemisinin resistance22–24. In addition to this, presence of day-3 parasite above 10% of day-0 parasitemia indicated a potential treatment failure. The level of PCR adjusted cure rates after 28 days treatment follow-up below 90% against the WHO recommended first line therapy for uncomplicated P. falciparum called for its reassessment25.
Artesunate-sulfadoxine-pyrimethamine (ASSP) is the drug of choice against P. falciparum in India excepting the north-eastern states9. Declining efficacy of ASSP was previously reported from these states and eastern India26–28. The emergence and spread of partial artemisinin resistant parasites were previously reported from West Bengal-an eastern state of India27,28. Partial artemisinin resistance was associated with the failure of partner drugs in combination14,24. Prevalence of mutations in molecular markers (pfdhfr and pfdhps) associated with partner drug resistance (sulfadoxine–pyrimethamine) was previously reported from different parts of India including central India8,29–33. Therefore, the aim of this study was to critically examine the ASSP efficacy through in-vivo, ex-vivo and genome-wide variation studies in central India.
Results
Study population
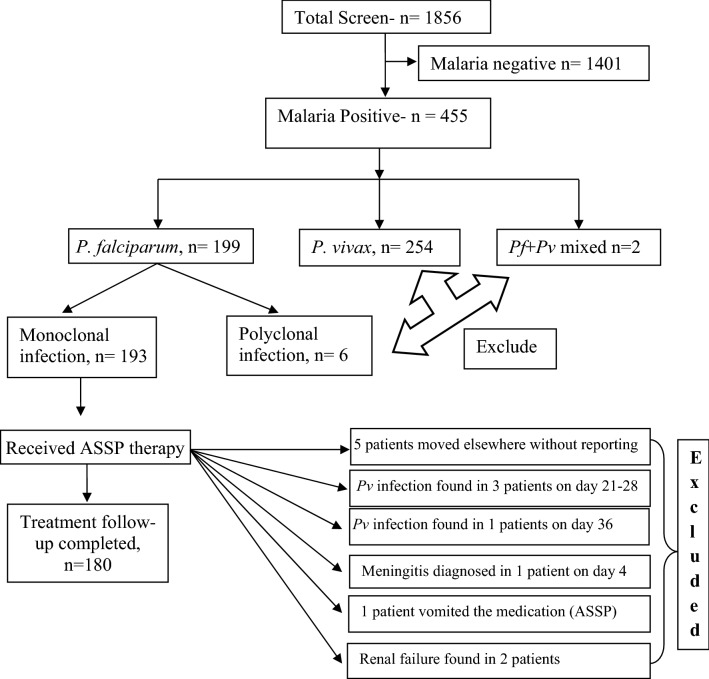
A total of 1856 febrile patients were screened; of them, 199 patients (10.7%) were detected P. falciparum positive. Mean age of P. falciparum-infected persons was 30.7 years (range 8–69 years). 193 patients (193/199) were identified as monoclonal P. falciparum infection and received standard ASSP therapy. Patient characteristics on enrolment of the study were presented in Table 1. 180 patients (180/193) had successfully completed the 42 day’s follow-up. As we have only considered monoclonal P. falciparum infection, therefore patients with P. vivax infection (254/1856) and polyclonal falciparum (6/199) infections were excluded. A detailed information regarding patient selection (inclusion and exclusion criteria) were presented in Fig. 1.
Table 1.
Patient characteristics on enrolment of the study.
| Patient characteristics | Bhilai including durg |
|---|---|
| Age (year) | 30.74 (95% CI 17–58) |
| Sex ratio (women/men) | 73/107 |
| Axillary temperature on day 0 (°C) | 39.22 °C (95% CI 38.16–40.19) |
| Parasite density (parasite/µL) | 44,152 (95% CI 9632–78,810) |
| Mean hemoglobin (g/dL) | 12.3 (95% CI 10.2–14.8) |
| Hematocrit |
Male: 47.1% (95% CI 45.8–49.5) Female: 38.8% (95% CI 37.4–41.7) |
Figure 1.
Schematic presentation of patient selection and entry criteria for randomization with ASSP. Monoclonal P. falciparum infections contained a single allelic form of infection i.e. either of mspI or mspII or glurp. Polyclonal infections along with Pf, Pv mixed infections were excluded. Only P. falciparum monoclonal infections were selected for the study. Patients with additional P. vivax co-infection during the follow-up scheduled were excluded. Patients, who had not completed the 42 days follow-up schedule, were eliminated.
Parasite clearance phenotype
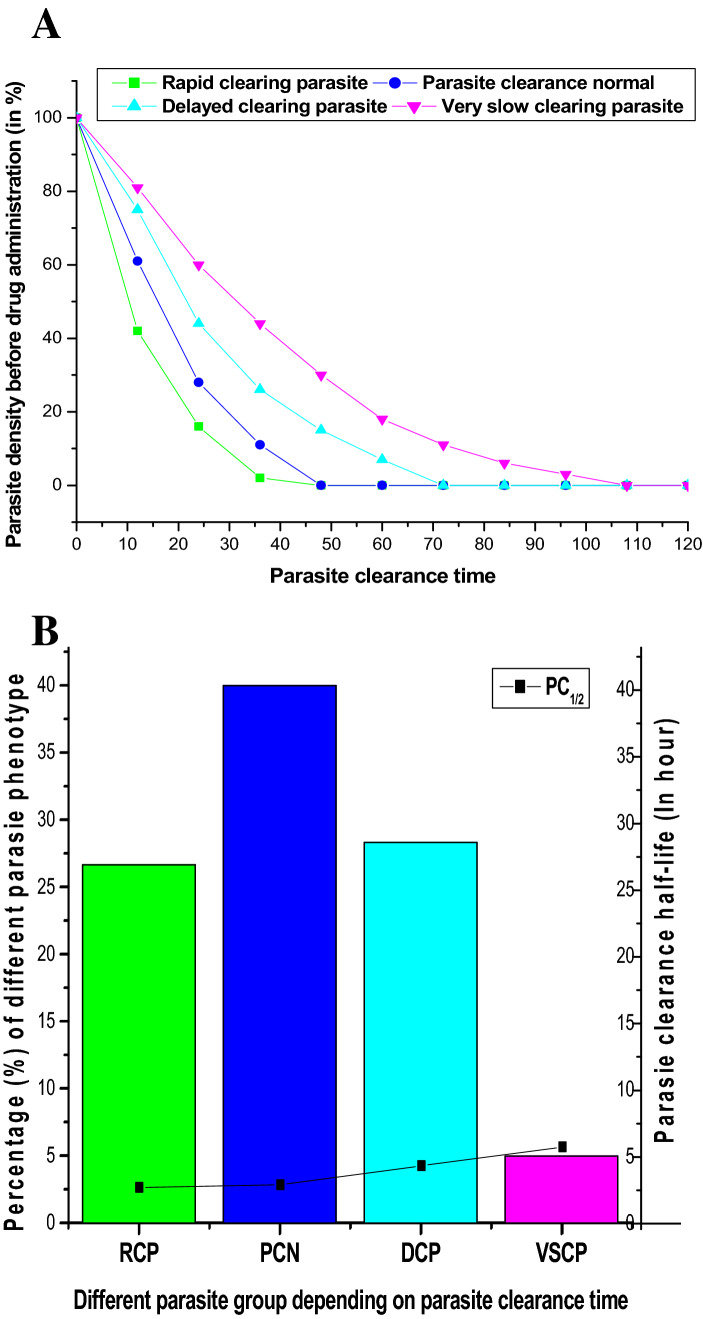
Based on parasite clearance time (PCT), we classified four different parasite clearance phenotypes. We defined, PCT ≤ 36 h, as rapid-clearing parasite (RCP); PCT > 36– ≤ 48 h as parasite-clearance normal (PCN); PCT > 48– ≤ 72 h as delayed-clearing parasite (DCP); and PCT > 72 h as very slow-clearing parasite (VSCP) (Fig. 2A). We found low median PC1/2 in RCP (2.6 h; 95% Cl 2.3–3 h) and PCN phenotype (2.8 h; 95% Cl 2.4–3.2 h). We recorded very high median PC1/2 in VSCP phenotype (5.6 h; 95% Cl 5.5–5.7 h) and higher PC1/2 in DCP phenotype (4.2 h; 95% Cl 3.7–4.8 h) (Fig. 2B). 17 patients (9.5%) showed prolonged PC1/2 (> 5.5 h) (8/52, DCP; and 9/9 VSCP phenotype) (Table 2). We observed a significant statistical difference in PC1/2 among these four parasite phenotypes (Kruskal–Wallis Test, p = 0.0042).
Figure 2.
(A) Proportion of Parasite clearance phenotypes: we had classified four different parasite clearance phenotypes depending on the parasite clearance time. Parasites, those who cleared within 36 h of drug administration were classified as Rapid clearing parasite (RCP) whereas parasites cleared by 48 h of drug exposure were designated as Parasite clearance normal (PCN). In patients, those whose parasites were cleared by > 48 h to ≥ 72, were designated as Delayed clearing parasite (DCP). Parasites were not cleared after 72 h of drug exposure were designated as Very slow clearing parasite (VSCP). (B) Frequency of different Parasite clearance phenotypes in relation to PC1/2: Parasite clearance normal (PCN) phenotype (40%), was most prevalent followed by delayed clearing parasite (DCP; 28.88%) and rapid clearing parasite (RCP; 26.11%). Interestingly, 5% of isolates represented VSCP phenotypes. We found low median PC1/2 in RCP (2.6 h) and PCN phenotype (2.8 h). Higher PC1/2 was observed in DCP phenotype (4.2 h) while very high median PC1/2 was recorded in VSCP phenotype (5.6 h) which proved these phenotypes perhaps less sensitive to ASSP therapy.
Table 2.
Distribution of different candidate genotype in relation to ASSP combination treatment.
| No of isolates | kelch 13 | atpase6 (263 + 431 + 623 + 6 30 + 769) | pfmdr1 (86 + 184 + 1034 + 1042 + 1246) | pfdhfr (16 + 51 + 59 + 108 + 164) | pfdhps (436 + 437 + 540 + 581 + 613) | pfcrt (72–76 + 326 + 356) | PC1/2 | Parasite clearance time | Recrudescence | Ex-vivo AS (RSA) sensitivity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| < 5 h | > 5 h | ≤ 36 h | > 36–≤ 48 h | > 48–≤ 72 h | > 72hETF | (LTF) | S | RS | |||||||
| 21 | Wild | LEAAS | NYSND | ANCSI | SAKAA | CVMNKNI | 21 | – | 10 | 8 | 3 | – | – | 18 | – |
| 11 | Wild | LEAAS | YYSND | ANCNI | SAKAA | CVMNTNI | 11 | – | 7 | 2 | 2 | – | – | 10 | – |
| 6 | Wild | LEAAS | NFSND | AICNI | SAKGA | SVMNTSI | 6 | – | 2 | 4 | – | – | – | 4 | – |
| 16 | Wild | LEAAS | NFSND | AICNI | SGKAA | SVMNTSI | 15 | 1 | 5 | 6 | 5 | – | – | 14 | – |
| 7 | Wild | LEAAS | YYSND | ANRNI | AAKAA | SVMNTNT | 7 | – | 4 | 3 | – | – | – | 6 | |
| 24 | Wild | LEAAS | YYSND | AICNI | AGKAA | SVMNTNT | 23 | 1 | 8 | 11 | 4 | 1 | 2 | 20 | 1 |
| 3 | Wild | LEAAS | NFSND | AICTI | SGKAA | SVMNTSI | 3 | – | 1 | 2 | – | – | – | 3 | – |
| 17 | Wild | LEAAS | YYSND | AICNI | SGKAA | CVIETNT | 17 | – | 4 | 11 | 2 | – | – | 16 | – |
| 6 | N29L | LEAAS | NFSND | AICNI | AGKAA | SVMNTST | 6 | – | 4 | 1 | 1 | – | 1 | 6 | – |
| 10 | Wild | LEAAS | YYSND | ANRNI | SGKGT | SVMNTST | 8 | 2 | – | 2 | 7 | 1a | 2 | 6 | 1 |
| 3 | Wild | LEEAS | NFSND | AICNI | SAKGA | SVMNTNT | 3 | – | – | 3 | – | – | – | 3 | |
| 5 | Wild | LEAAS | NFSND | ANRNI | AGKAA | CVIETNT | 5 | – | – | 3 | 2 | – | 1 | 5 | |
| 5 | Wild | LKAAS | YYSND | ANRNI | AGKAA | SVMNTNT | 4 | 1 | 1 | 3 | 1 | – | – | 3 | – |
| 21 | Wild | LEAAS | YFSND | AIRNI | SGKGT | SVMNTST | 15 | 6 | 1 | 5 | 12 | 3 | 4 | 15 | 3 |
| 8 | Wild | LEEAS | YFSND | AIRNI | AGKAA | SVMNTNT | 6 | 2 | – | 2 | 4 | 2 | 1 | 6 | 1 |
| 3 | A675V | LEAAS | NFSND | ANRNI | AGKAA | CVIETNT | 2 | 1 | 1 | 2 | – | 1 | – | 3 | |
| 14 | Wild | LEAAS | YFSND | ANRNI | SGKGT | CVIETNT | 11 | 3 | – | 5 | 7 | 2 | – | 10 | 2 |
Underlined codons are mutant codons. ETF and LTF stand for early treatment failure and late treatment failure, respectively, whereas S, and RS stand for sensitive, and reduced sensitivity, respectively. Parasite clearance half-life was denoted as PC1/2.
aPatients did not present adequate plasma dihydroartmisinin concentration, therefore not considered as true day-3 positive case.
Plasma availability of dihydroartemisinin
We measured artemisinin exposure in patients through detection of plasma DHA. Mean plasma DHA was recorded as 4052 nM, (95% CI 3925.8–4128.1) at 1.5 h and 2137.5 nM, (95% CI 2069.1–2194.8) at 3 h after ASSP exposure. Among 180 patients, 178 patients (98.8%) attained adequate plasma DHA level.
ACT efficacy
We observed persistence of parasite after 72 h of ASSP exposure in 9 patients (5%), with high median axillary temperature of 38 °C (95% CI 37.8–38.2 °C), corresponding patients also showed prolonged median PC1/2 of 5.6 h (95% Cl 5.5–5.7 h). Among them, 8 patients represented adequate plasma DHA level. Of them, 7 patients showed reduced sensitivity to DHA (in vitro) and designated as early ACT failure cases.
We also observed reappearance of infections in 12 patients (6.7%) during 42-days follow-up. After PCR correction through analysing the msp1, msp2, and glurp genes, we further confirmed the existence of 10 (5.5%) true recrudescence cases among 12 parasite reappearance cases. Crude cure-rate (PCR uncorrected) after ASSP therapy was recorded 89.44% (Kaplan–Meier estimate; 95% CI 83.78–93.35) whereas PCR adjusted cure-rate after day 42 was recorded 90.56% (95% CI 85.07–94.23). We found artemether-lumefantrine (AMLF) rescue therapy was successful without any report of treatment failure (Tables 3, 6).
Table 3.
Summary of treatment After ASSP therapy.
| PCR | Drug | Study population (n) | Day-3 positive parasite | ETF (n) | LTF (n) | ACPR (n) | Recrudescence (n) | Re-infection (n) |
|---|---|---|---|---|---|---|---|---|
| PCR uncorrected | ASSP | 180 | 9 (5%) | 7a (3.9%) | 12 (6.7%) | 159 (88.3%) | – | – |
| PCR corrected | ASSP | 180 | 9 (5%) | 7a (3.9%) | 10b (5.6%) | 161 (89.4%) | 10 (5.6%) | 2 (1.1%) |
ACPR, ETF and LTF stand for adequate clinical parasitological response, early treatment failure and late treatment failure, respectively, whereas ASSP stand for artemisinin-sulfadoxine-pyrimethamine.
aOne patient did not attain the adequate plasma DHA concentration and in another patient, we failed to adapt the in vitro culture for RSA assay.
bInitially reappearance of infection was observed in 12 patients (6.7%), but analyses of msp1, msp2, and glurp gene confirmed that among those 12 patients, 10 (5.5%) were true recrudescence (LTF) case.
Table 6.
Phenotypic and genotypic characteristics of late treatment failure and evaluation of AMLF rescue therapy.
| Case | Drug | Day of recurrence | Parasite load on recurrence | Fever (°C) | PCR correction | pfk13 allele | pfdhfr allele | pfdhps allele | Rescue therapy with AMLF | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACPR | ETF | LTF | |||||||||
| Case 1 | ASSP | 5 | 3642/µL | 38.6 | Recrudescence | Wild | AICNI | AGKAA | √ | – | – |
| Case 2 | ASSP | 7 | 5082/µL | 38 | Recrudescence | Wild | ANRNI | SGKGT | √ | – | – |
| Case 3 | ASSP | 21 | 27,419/µL | 39.1 | Recrudescence | N29L | AICNI | AGKAA | √ | – | – |
| Case 4 | ASSP | 11 | 11,026/µL | 39.4 | Recrudescence | Wild | AICNI | AGKAA | √ | – | – |
| Case 5 | ASSP | 7 | 4957/µL | 38.4 | Recrudescence | A675V | ANRNI | AGKAA | √ | – | – |
| Case 6 | ASSP | 28 | 870/µL | 37.9 | Recrudescence | Wild | AIRNI | SGKGT | √ | – | – |
| Case 7 | ASSP | 35 | 7350/µL | 38.1 | Recrudescence | Wild | ANRNI | SGKGT | √ | – | – |
| Case 8 | ASSP | 32 | 512/µL | 38.3 | Recrudescence | Wild | ANRNI | AGKAA | √ | – | – |
| Case 9 | ASSP | 28 | 51,154/µL | 40.1 | Re-infectiona | Wild | AIRNI | SGKGT | √ | – | – |
| Case10 | ASSP | 21 | 16,822/µL | 39.3 | Recrudescence | Wild | AIRNI | AGKAA | √ | – | – |
| Case11 | ASSP | 35 | 3092/µL | 38 | Re-infectiona | Wild | AIRNI | SGKGT | √ | – | – |
| Case12 | ASSP | 14 | 5168/µL | 38.8 | Recrudescence | Wild | AIRNI | SGKGT | √ | – | – |
Underlined codons are mutant codons. ACPR, ETF and LTF respectively stand for adequate clinical parasitological response, early treatment failure and late treatment failure, whereas ASSP stand for artesunate-sulfadoxine-pyrimethamine and AMLF stands for artemether-lumefantrine. Parasite load was expressed as number of parasite/ micro liter of blood.
aAfter PCR correction, case 9 and case 11 were identified as the case of parasite re-infection.
In vitro drug susceptibility
156 (86.7%) clinical isolates were adapted for their in vitro susceptibility to DHA, sulfadoxine, and pyrimethamine (Table 4). We observed reduced sensitivity to DHA in 11 (7%) isolates. Of those, 7 (77.8%) belonged to VSCP phenotype (mean RSA(0–3 h) = 2.8%; 95% CI 2.4–3.1) and 4 (7.7%) belonged to DCP phenotype (mean RSA(0–3 h) = 1.9%; 95% CI 1.8–2) (Table 4). Lower mean RSA(0–3 h) was recorded in RCP (0.2%; 95% Cl 0.1–0.3) and PCN phenotype (0.2%; 95% CI 0.2–0.3), and were highly sensitive to DHA (p = 0.092).
Table 4.
In vitro drug susceptibility in different parasite phenotype.
| Different parasite phenotype | PC1/2 > 5 h | Culture adaptation | RSA (0–3 h) (mean) | IC50 nMol/L pyrimethamine (mean) | IC50 nMol/L sulfadoxine (mean) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive < 1% | RS ≥ 1% | S < 100 nM | IR 100–2000 nM | R > 2000n M | S < 640 nM | IR 640–3000 nM | R > 3000 nM | |||
| Rapid clearing parasite (PCT ≤ 36 h) | 0/47 | 45/47 | 0.19% (95% CI 0.14–0.25) 45/45 | – | 72.43 (95% CI 55.8–88) 12/45 | 1161.25 (95% CI 523.7–1794.2) 17/45 | 2512.82 (95% CI 2050.2–29.72) 16/45 | 372.24 (95% CI 236.3–510.1) 21/45 | 2271.41 (95% CI 1685.4–2860) 14/45 | 3570.27 (95% CI 3149.2–3987.7) 10/45 |
| Parasite clearance normal (PCT > 36 h ≤ 48 h | 0/72 | 62/72 | 0.23% (95% CI 0.15–0.32) 62/62 | – | 77.21 (95% CI 64.5–91) 8/62 | 1330.62 (95% CI 767.3–1891.4) 25/62 | 4696.70 (95% CI 2675.1–6720) 29/62 | 426.29 (95% CI 291.4–564.3) 12/62 | 2349.60 (95% CI;1821.5–2875.8) 28/62 | 5642.51 (95% CI 4276.2–7012.7) 22/62 |
| Delayed clearing parasite) (PCT > 48- ≤ 72 h) | 8/52 | 41/52 | 0.33% (95% CI 0.22–0.46 37/41 | 1.92% (95% CI 1.83–2.02) 4/41 | 88.9 (95% CI 88–90) 2/41 | 1741.35 (95% CI 1562.8–1920.5) 12/41 | 7442.39 (95% CI 5486–9400.2) 27/41 | 412.54 (95% CI 408.1–417.2) 3/41 | 2718.50 (95% CI 2635.5–2804.1) 6/41 | 8156.17 (95% CI 6480.7–9832.3) 32/41 |
|
Very slow clearing parasite PCT > 72 h |
9/9 | 8/9 |
0.94% 1/8 |
2.76% (95% CI 2.43–3.09) 7/8 | – |
1887.45 1/8 |
8004.17 (95% CI 6780.3–9235) 7/8 | – | – |
8397 (95% CI 7117.2–9680.6) 8/8 |
| 3D7 | 0.17% (95% CI 0.14–0.19 | – | 68.42 ± 2·1 | – | – | – | – | 4320.50 ± 32.65 | ||
Individual ex-vivo ring stage survivability of parasite isolates was represented as RSA. Here S, IR, R and RS stand for Sensitive, Intermittent resistant, Resistant, and Reduced susceptibility, respectively. Parasite clearance time was denoted as PCT and Parasite clearance half-life was denoted as PC1/2.
We observed prevalence of SP resistant parasite. 79 (50.6%) clinical isolates were identified as pyrimethamine resistance whereas 72 (46.1%) isolates were sulfadoxine resistant (Supplementary Fig. 1A, Supplementary Table 1). Only 22 (14.1%) isolates were sensitive to pyrimethamine and 36 (23.1%) were sensitive to sulfadoxine. Two microscopists showed good conformity during assessment of slides (Pearson correlation r = 0.81, p < 0.005).
Parasite genetic architecture in relation to parasite clearance outcome
Only 9 isolates (5%) represented kelch13 mutation (Table 2). Substitution of alanine to valine at codon 675 was observed in 3 (1.7%) isolates (GenBank-N534312-N534314) (Supplementary Fig. 1B), corresponding isolates showed substantially higher RSA(0–3 h) (mean, 2.1%; 95% CI 1.9–2.3) (Table 2). Of those, one patient showed PC1/2 of 5.3 h while the remaining represented PC1/2 of 4.6 h and 4.9 h, respectively. Of whom, 1 was further developed recrudescence (Table 2). Isolates representing kelch13-A675V mutation strongly associated with reduced ex-vivo artemisinin-sensitivity but not showing any relationship in treatment outcome (P = 0.7, Kruskal–Wallis test) Supplementary Fig. 1C). Another 6 (3.3%) patients contained N29L polymorphism, representing lesser PC1/2 (2.6 h, 95% Cl 2.4–2.8 h) along with substantially lower RSA(0–3 h) (0.2%; 95% CI 0.1–0.2). Surprisingly, we observed the prevalence (16/17) of prolonged PC1/2 (> 5 h) related with wild pfkelch13 genotype (Table 2).
We found 11 patients (6.1%) with A623E mutation in pfatpase6 gene (Supplementary Fig. 1B). Of those, two isolates showed PC1/2 > 5.5 h. One of the corresponding isolates presented RSA(0–3 h) of 3.3% while others failed to culture adaptation. The isolates containing A623E mutation were sensitive to DHA (0.7%; 95% CI 0.1–1.2) (Supplementary Table 1). Another 5 isolates (2.8%) contained pfatpase6-E431K polymorphism (L263K431A623A630S769), one of those isolates showed PC1/2 of 5.1 h (Table 2). Among 5 isolates, three were culture adapted and all were found sensitive to DHA (0.4%; 95% CI 0.4–0.5) (Table 2). Isolates contained polymorphism in pfkelch13 (N29L, and A675V) and pfatpase6 gene (E431K, and A623E) represented low to moderate PC1/2 (3.9 h; 95% CI 3.5–4.3) but not associated with early ASSP failure (ETF) (P = 0.74, Kruskal–Wallis test).
We observed prevalence of pfmdr1-N86Y (65%) and Y184F (50%) mutation (Supplementary Fig. 1D). A total of 48 patients represented Y86F184S1034N1042D1246 mutation. Of those, 11 (22.9%) showed PC1/2 > 5.5 h, of whom 6 (12.5%) showed reduced DHA susceptibility. Another 69 patients contained pfmdr1-Y86Y184S1034N1042D1246 mutation. Of those, only four (5.8%) showed PC1/2 > 5.5 h. Isolates containing pfmdr1-Y86F184S1034N1042D1246, and Y86Y184S1034N1042D1246 mutation presented moderate PC1/2 (3.7 h; 95% CI 3.4–4.1), but not associated with ASSP failure (P = 0.56) (Table 2). We also observed prevalence of pfcrt-K76T (88.3%) and I356T (68.3%) mutations (Supplementary Fig. 1D). Among 37 isolates with pfcrt-S72V73M74N75T76S326T356 mutation, 8 (21.6%) showed PC1/2 > 5.5 h. Of those 4 (10.8%) represented day-3 positive parasites. 40 patients had pfcrt-S72V73M74N75T76N326T356 mutation; of those, 4 (10%) had PC1/2 > 5.5 h and 3 (7.5%) had day-3 positive parasite. Another 22 contained pfcrt-C72V73I74E75T76 N326T356 mutation, 4 (18.1%) of those showed PC1/2 > 5.5 h, of whom 2 (9.1%) resulted day-3 positive parasite (Supplementary Table 2). We observed combination of pfmdr1 and pfcrt (Y86F184S1034N1042D1246-S72V73M74N75T76S326T356, Y86Y184S1034N1042D1246-S72V73M74N75T76S326T356, and Y86F184S1034N1042D1246-C72V73I74E75T76N326T356) mutations associated with delayed-clearing parasite (PCT > 48– ≤ 72 h) (P < 0.02), corresponding isolates showed moderate RSA(0–3 h) (0.78%, 95% CI 0.4–1.3) but were not associated with early ASSP failure (P = 0.68, Kruskal–Wallis test).
Molecular cause of day-3 positive parasite
We identified 9 day-3 positive cases (Table 5). Of these, 8 represented adequate plasma DHA (mean 2073.2 nmol/L, 95% CI 1960–2180) after 3 h of ASSP exposure, corresponding isolates showed prolonged PC1/2 (mean 5.6 h, 95% CI 5.5–5.7) which were significantly higher as compared to PC1/2 (3.2 h; 95% CI 2.8–3.6) of cured patients (P < 0.005, Mann–Whitney U-test). Of whom 7 represented significantly higher RSA (mean-2.2%, 95% CI; 1.7–2.8) than the cured patients (mean-0.4%; 95% CI 0.2–0.6). Perhaps these isolates were early ASSP failure cases. The mean axillary temperature [37.9 °C (95% CI 37.8–38.2 °C)] at day three was found very high in those 9 day-3 positive cases. Presence of parasites on day-3 was determined through microscopy as well as PCR-based detection. The mean day-3 parasite load was found 9.1% (95% CI 6.3–11.9%) from that of day-0 parasitemia All day-3 positive isolates had pfcrt-I356T polymorphism, while eight of them represented pfmdr1 N86Y and Y184F double mutations. None of the day-3 positive isolates carried mutation in pfkelch13 gene (the most recognized and validated artemisinin-resistance associated gene), but 2 had mutation in pfatpase6 gene. Day-3 positive parasites showing artemisinin-resistance phenotype without pfkelch13 mutation suggest kelch13-independent artemisinin-resistance.
Table 5.
Distribution of parasite phenotype and genotypes in day-3 positive parasites.
| > 72 h (+) case | Plasma artesunate (nmol/L) | Parasite load on day-0 pd/µL | Parasite load on day-3 pd/µL | Fever on day 0 (°C) | Fever on day 3 (°C) | RSA(0–3 h) (in %) | Treatment response (in h) | Kelch13 genotype | Atpase6 haplotype | Pfmdr1 haplotype | Pfcrt haplotype | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.5 h | 3 h | PCT | PC1/2 | ||||||||||
| Case1 | 4023 | 2361 | 19,368 | 237 | 39.6 | 38.1 | 2.4 | 96 | 6.2 | Wild | LEAAS | YYSND | SVMNTNT |
| Case 2 | 4196 | 2147 | 8432 | 96 | 38.5 | 38 | 3.0 | 102 | 5.6 | Wild | LEAAS | YFSND | CVIETNT |
| Case 3 | 3833 | 2051 | 21,641 | 356 | 38.7 | 38.1 | 2.1 | 108 | 5.6 | Wild | LEAAS | YFSND | SVMNTST |
| Case 4 | 4077 | 1952 | 70,846 | 611 | 38.4 | 37.8 | 2.6 | 108 | 5.8 | Wild | LEAAS | YFSND | SVMNTST |
| Case 5 | 4086 | 2052 | 56,855 | 123 | 38.8 | 37.5 | 1.7 | 96 | 5.3 | Wild | LEAAS | YFSND | SVMNTST |
| Case 6 | 3981 | 2114 | 11,790 | 202 | 40.3 | 38.2 | 3.3 | 120 | 5.9 | Wild | LEEAS | YFSND | SVMNTNT |
| Case 7 | 3591 | 1805 | 44,181 | 728 | 39.8 | 37.9 | – | 102 | 5.7 | Wild | LEEAS | YFSND | SVMNTNT |
| Case 8 | 955 | 548 | 9716 | 455 | 38 | 37.5 | 0.9 | 108 | 5.7 | Wild | LEAAS | YYSND | SVMNTST |
| Case 9 | 4258 | 2104 | 36,847 | 107 | 39.5 | 38 | 2 | 114 | 5.5 | Wild | LEAAS | YFSND | CVIETNT |
PCT represented Parasite clearance time (in h) and parasite clearance half-life was denoted as PC1/2 (in h). We have presented parasite density as PD. Individual ex-vivo ring stage survivability of parasite isolates was represented as RSA(0–3 h). Underline codons were mutant codon. In case 8, RSA(0–3 h) was recorded 0.94% with PC1/2 of 5.7 h while case 7 was failed to culture adaptation. The day-3 parasite load was ranging from 1.8 to 14.2% from that of day-0 parasitaemia.
Molecular characterization of parasite recrudescence
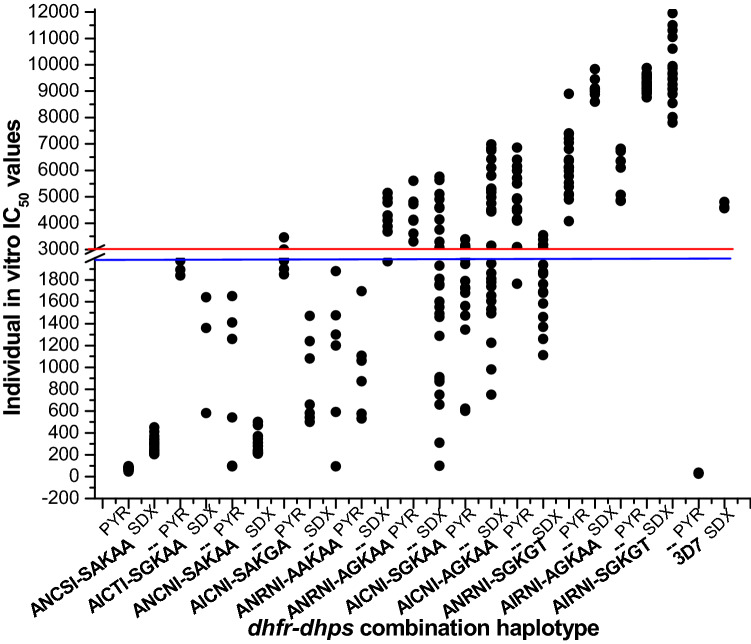
After PCR corrections, we identified 10 cases of recrudescence. The mean parasite load on the day of recrudescence was found 8285/µl (95% CI 2726–13,843). The mean body temperature on the day of recrudescence was very high 38.6 °C (95% CI 38.2–39.0 °C). We observed 3 cases of parasite recrudescence within day-7 and another 7 cases between day-8 to day 42, corresponding isolates representing significantly higher IC50s for sulfadoxine and pyrimethamine. Parasite recrudescence was the true cause of late ASSP failure. We noticed combination of triple dhfr and dhps mutation (A16I51R59N108I164–S436G437K540G581T613) was highly correlated with parasite recrudescence (P < 0.01), while A16N51R59N108I164–S436G437K540G581T613 and A16I51C59N108I164–A436G437K540A581A613 mutations also contributed crucial role in parasite recrudescence (Table 6) (Supplementary Fig. 1E). Isolates contained A16N51R59N108I164–A436G437K540A581A613, A16I51C59N108I164–S436G437K540A581A613, and A16N51R59N108I164–A436A437K540A581A613 mutations exhibited moderate to high IC50 for pyrimethamine and sulfadoxine but never connected with recrudescence (P = 0.73) (Fig. 3). On the day of parasite recurrence, all the LTFs received a standard dose of AMLF and all showed treatment success after 42 days follow-up (Table 6).
Figure 3.
In vitro IC50 of pyrimethamine and sulfadoxine in relation to individual dhfr-dhps genotype: Here PYR and SDX respectively stand for “pyrimethamine, and sulfadoxine.” The blue line (corresponding to 2000 nM of PYR) represented the in vitro PYR resistance, while the red line (corresponding to 3000 nM of SDX) represented the in vitro SDX resistance. We observed prevalence of SP resistant parasites. The isolates presenting triple dhfr and dhps mutation (AIRNI-SGKGT; ANRNI-SGKGT) and double dhfr and dhps combination mutation (AICNI-AGKAA) represented very high IC50 for pyrimethamine and sulfadoxine and proved to be highly resistant to PYR and SDX (P < 0.01). Isolates contained ANRNI-AGKAA, AICNI-SGKAA, and ANRNI-AAKAA mutations exhibited moderate to high IC50 for pyrimethamine and sulfadoxine but never connected with recrudescence (P = 0.73). Pyrimethamine sensitive and sulfadoxine resistant 3D7 strain was used as a control strain.
Discussion
Emergence and spreading of partial artemisinin resistant parasites in eastern India27,28 along with late ACT failures in north-east India34 called for a systematic screening of ASSP in central India, as the second highest number of malarial infections was reported from the state of Chhattisgarh. We found the day-3 positive parasite with prolonged parasite clearance half-life (> 5.5 h), along with recrudescence cases. 9.4% of patients showed prolonged PC1/2 (> 5.5 h) which was an alarming sign. Prolonged parasite clearance was an indicator of decreased efficacy of fast acting artemisinin within ACT which perhaps led to ACT failure12,16,19. We observed the median PC1/2 of very slow-clearing parasite (VSCP) (5.6 h) and delayed-clearing parasite (4.2 h) phenotype were significantly higher from the PC1/2 of Thailand-Myanmar border (3.7 h) and lower from Western Cambodia (5.9 h)16. Identification of these VSCP parasite phenotypes, confirmed the emergence of parasites that became less sensitive to artemisinin in vivo12. In spite of this, we also observed isolates with reduced sensitivity to DHA in vitro. The corresponding isolates of VSCP phenotype (5%) represented higher RSA(0–3 h) (2.2%) than the cured patients (0.46%), which proved those VSCP phenotypes were less sensitive to artemisinin in vitro. Previous reports from Thailand suggested that increased viability of ring-stage parasites (RSA(0–3 h) > 1%) was strongly associated with elevated PC1/216. Recent reports suggested that apart from reduced efficacy of fast acting artemisinin drugs, failure in late-acting combinations also contributes towards the increment of PC1/214. Thus, the reduced artemisinin sensitivity along with elevated PC1/2 of the ring stage parasite resulted in reduced sensitivity to ASSP combination therapy. However, most studies in India reported no evidence of less susceptibility to artemisinin in vivo or in vitro34–36. Some recent studies reported the spreading of parasites with reduced sensitivity to artemisinin in vivo in eastern India27,28. We have identified 9, day-3 positive cases; among them, 7 (3.9%) patients were confirmed as ETF whereas 10 (5.6%) patients were identified as true recrudescence (LTF) cases (Table 2). Given the results of our study, although the numbers of treatment failure cases were not very high, we confirmed the emergence of reduced susceptibility to ASSP combination in this part of India. Nevertheless, findings of reduced efficacy of ASSP in Central India suggested the possibility of emergence of resistant parasites in the near future.
ETF generally occurs due to reduced efficacy of fast-acting artemisinin in combination therapy18. The mechanism behind the reduced susceptibility to artemisinin is not fully clear, but genome wide analyses, ex-vivo RSA, and transfection studies suggested that artemisinin resistance is predominantly related to pfkelch13 gene polymorphism14,18–22. Interestingly, predominance of wild pfkelch13 allele was observed in this parasite population of Central India. Despite the direct correlation with pfkelch13 polymorphism, we had identified 7 (3.9%) ETF cases with prolonged PC1/2 as well as increased ex-vivo RSA(0–3 h) survivability, without pfkelch13 polymorphism. However, the findings of our study were quite uncommon. Our observations from this study suggest several novel aspects of artemisinin-resistance. Firstly, the kelch13-indpendent artemisinin-resistance which was previous reported only in Thailand24,37,38. Although, we identified 3 isolates with pfkelch13-A675V polymorphism which showed positive association with increased ex-vivo RSA(0–3 h), but not with treatment outcome like previous reports from in north-east India and southern Rwanda39,40. The results do not represent outliers, as we studied a statistically valid number of patients. Secondly, the absence of pfkelch13 polymorphism suggests definitive roles for other genetic factors in reduced artemisinin-sensitivity and emergence of artemisinin-resistance. Of note, polymorphism in Ca2+ATPase6 gene (pfatpase6) had some role in reducing susceptibility to artemisinin22. However, the likelihood of this mechanism has been debated41. Likening the observations from Thai-Cambodian border12, we have not so far found any definitive correlation between pfatpase6 mutation and ASSP efficacy In India. Thirdly, pfcrt, pfmdr, and pffd polymorphisms plausibly represent the genetic background required for the onset of pfkelch13 polymorphism, as mutations in those genes showed strong association with the beginning of pfkelch13 mutation during selection of artemisinin-resistance18,23,27,28. Indeed, in our study, day-3 positive cases demonstrated acquired mutations in pfcrt (K76T, I356T) and pfmdr1 (N86Y, Y184F) genes similar to the findings reported in eastern and north-India27,37. A recent report showed P. falciparum strains in South-East Asia having some genetic attenuation to develop novel mutations that caused artemisinin-resistance42. Fourthly, the precise genetic architectures in relation to reduced artemisinin-sensitivity in the East, North-East, South-West, and Central India are plausibly different owing to extensive variations in socio-demographic, environmental, seasonal and parasite-vector relationships26,27,39.
We detected 10 recrudescence cases in our study. The clinical manifestations of recrudescence were related with less susceptibility of longer acting partner drug (SP), as evidence, parasite isolates showed acquired combination mutation in pfdhfr–pfdhps gene8,27,30. Isolates representing sextuple or quintuple dhfr–dhps combination mutations (A16I51R59N108I164–S436G437K540G581T613, A16N51R59N108I164–S436G437K540G581T613 and A16I51C59N108I164–A436G437K540A581A613), exhibited very high IC50s for pyrimethamine and sulfadoxine, proving true resistance towards SP. Quadruple dhfr–dhps combination mutations with reduced SP sensitivity was previously reported from Chhattisgarh29. Despite the high prevalence of molecular markers associated with SP resistance, treatment failure rate especially LTFs were much less in number. The sensitivity of artesunate over this parasite population was still very high. Artemisinin derivatives within the ASSP were therefore able to kill most of the parasite and reduce the burden of partner drugs. That is how all treatment failure cases recovered after AMLF therapy suggesting that a standard six-dose-AMLF could be a potential second-line treatment against three-dose ASSP failure.
In conclusion, emergence of reduced sensitivity to artemisinin and predominance of SP resistant parasites suggested us for the evaluation of national drug policy as artemether-lumefantrine could be successor of ASSP. Finally, a National Malaria Eradication Program requires urgently for centralized and synchronized implementation of new drug-combinations and tracking of genetic mutations that might lead to higher level of resistance to artemisinin and its partner drug.
Methods
Study population
We conducted the study at Bhilai (21.21° N, 81.38° E) and Durg (21.19° N, 81.28° E), districts of Chhattisgarh, India during August 2015–January 2017. Chhattisgarh had contributed the second highest malaria incidence in 20149. The inclusion criteria were age ≥ 3 years, axillary temperature ≥ 37.5 °C, shivering, headache during the past two 2 days or more, and no recent history of anti-malarial medication. All participants were screened for P. falciparum infection by microscopic examination of Giemsa-stained thick and thin blood smears. Quantification of parasitaemia was performed by counting the number of parasites per 8000 WBC. We usually count at least 200 fields using 100× oil immersion objectives. Every field generally comprises 4–5 WBCs. Parasites/µl blood was determined as (parasites/WBCs) × 8000 (WBC count/µl of blood). The minimum detectable parasitemia was 40 parasites/µl of blood. pfmspI (MAD20 and K1) and pfmspII (3D7 and FC27) alleles were screened to detect the clonality of infection. Patients having signs and symptoms of severe malaria, pregnant women, infants and poly-clonal P. falciparum infections were excluded43. The experimental design and protocols were duly approved by Vidyasagar University, Human ethical committee (VU/HEC 15-0051). We strictly followed the WHO and WWARN (WorldWide Anti-malarial Resistance Network) guideline along with the Helsinki protocol. We obtained duly signed informed consents from each patient and patient’s guardian for minor (child patient) (Fig. 1 for patient selection details).
ASSP efficacy
Those patients who fulfilled the inclusion criteria received the quality assured standard ASSP dose (supplied by Ministry of Health and Family-Welfare) of 4 mg/kg body weight artesunate once daily for 3 days and a single dose of 25 mg/kg body weight sulfadoxine along with 1.25 mg/kg bodyweight pyrimethamine on first day, under the supervision of medical officer9. Trained microscopists checked the thin blood smears at an interval of 6 h, until the patients became parasite free. We estimated the PC1/2 by parasite clearance estimator15. Patients who developed renal failure and those who vomited the drug were withdrawn from the study and sent for further care. We performed the follow-up evaluations on day 7, 14, 21, 28, 35, and 42 days after initial therapy, and responses were classified accordingly43. We performed unscheduled follow-up when symptoms of malaria reappear. Patients, not responding to ASSP, received artemether-lumefantrine rescue therapy (6 tablets, each containing 40 mg AM and 240 mg LF) and were further followed-up for next 42 days.
Plasma artemisinin testing
We collected 500 µl of intra-venous blood just before, at 1.5 h and 3 h (± 30 min) after initiation of ASSP therapy. We obtained plasma samples immediately and stored at – 20 °C. We evaluated the plasma dihydro-artemisinin (DHA) to validate the sporadic drug exposure by liquid chromatography as stated previously44.
In vitro drug sensitivity testing
We adapted P. falciparum clinical isolates in vitro as described previously28,45. After culture adaptation parasites were allowed to proliferate for 72 h before doing the anti-malarial drug (sulfadoxine, pyrimethamine, and chloroquine) exposure. We performed sensitivity testing of anti-malarial by hypoxanthine incorporation assay in triplicate, according to our standard laboratory protocol46,47. As following the standard guideline, we defined criteria for sulfadoxine, IC50 < 640 nM, susceptible; IC50 > 640– ≤ 3000 nM, intermediate; and IC50 > 3000 nM, resistant. For pyrimethamine, we defined IC50 < 100 nM, susceptible; IC50 > 100– ≤ 2000 nM, intermediate; and IC50 > 2000 nM, resistant. For chloroquine, we defined IC50 < 100 nM, susceptible; and IC50 > 100 nM, resistant. We used pyrimethamine sensitive and sulfadoxine resistant 3D7 strain as quality-control strain.
Ring-stage survival testing
We performed ring-stage survival (RSA) assay in triplicate after culture adaption of clinical isolates as described earlier17. We treated 0–3 h post-invasive, highly synchronized early ring-stage parasites with 700nMoles of dihydro-artemisinin for 6 h, followed by washing with RPMI-1640 for three times and further cultivated for another 66 h. We measured parasite survival rates by microscopic examination of 10,000 RBCs per treatment replicate in Giemsa-stained thin blood smears.
DNA extraction and sequencing
We extracted parasite DNA from frozen blood aliquots (200 µl) using Mini blood-kit (Qiagen). We performed nested PCR to amplify pfkelch13 gene using Kelch13-F and Kelch13-R primers according to our standard laboratory protocol28. We also performed nested PCRs to amplify pfATPase6, pfdhfr, pfdhps, pfmdr1, and pfcrt gene as described previously46–48. For pfATPase6 gene amplifications, we used primer-pairs (5′TTGGTAATAAAACTCCCGC3′ and 5′TATTCCTCTTAG-CACCACTCC3; for 250–500 codon; 5′AAGAAGGATAAATCACCAAG3′ and 5′AAATACACGTATA-CCAGCC3′; for 520–800 codon). We sequenced the amplicons directly using the BigDye Terminator v3.1 Sequencing Kit (Applied Biosystems), and were run on 3730xl genetic analyzer48. We used online translation tool (http://www.expasy.org) to translate the sequences. We queried single nucleotide polymorphism of individual sequences by using a nucleotide database with BLAST.
Statistical analysis
We expressed our data as a univariate median; mean ± SEM. Fisher’s exact test along with regression analyses were performed to correlate the treatment efficacies with molecular genotyping. We used the Clopper-Pearson method to calculate the 95% confidence intervals. Data were compared between two groups by Mann–Whitney U-test while Kruskal–Wallis-test was used to compare among more than two groups. We considered p < 0.05 statistically significant. All statistical analyses were performed through Graph Pad in-Stat 3.0 and Origin 6.1.
Supplementary Information
Acknowledgements
We are thankful to all patients who took part in this study. The authors express their gratefulness to Vidyasagar University, Midnapore for providing the facilities to execute these studies. We are especially grateful to Goutam Laboratory and their staff for laboratory support. Genome sequencing was done at Indian Institute of Technology, Kharagpur and SCI genome company, Kochi; we thank the staff of IIT KGP for sequencing, and bio-informatics facilities for their contribution. We are also thankful to Hari Kumar, Debandra Sahal, Sundar Bist for processing of the in vivo samples, and coordination.
Author contributions
S.S.D., B.S. and A.K.H. contributed to the designing and conceptualization of the manuscript. A.K.H. and S.D. supervised in vivo ASSP therapy. In vivo data analysis and writing of the article were done by S.D., S.M. (Sayantani Mandal) and A.K.H. A.K., S.M. (Subhankar Manna) and S.R.M. (Samaresh Mandal) analysed the in vitro data and wrote the article. S.S.D. and A.K. contributed to genome analysis and interpretation of the data. S.S.D., A.K.H. and B.S. drafted the manuscript. All authors have approved the final version of the manuscript.
Funding
There is no external research grant. This first author (SSD) has received a personal fellowship of monthly remuneration from Council of Scientific and Industrial Research, India (sanction no: 09/599(0055)2K1-EMR-I).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89295-0.
References
- 1.World malaria report 2017 by World Health Organization (WHO). https://www.who.int/malaria/publications/world-malaria-report-2017/en/. Accessed 29 Nov 2017.
- 2.Zomuanpuii R, et al. Epidemiology of malaria and chloroquine resistance in Mizoram, northeastern India, a malaria-endemic region bordering Myanmar. Malar. J. 2020;19:95. doi: 10.1186/s12936-020-03170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das S, Banik A, Hati AK, et al. Low prevalence of dihydro folate reductase (dhfr) and dihydropteroate synthase (dhps) quadruple and quintuple mutant alleles associated with SP resistance in Plasmodium vivax isolates of West Bengal, India. Malar. J. 2016;15:395. doi: 10.1186/s12936-016-1445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magnitude of the Problem. National vector borne disease control program. https://nvbdcp.gov.in/index4.php?lang=1&level=0&linkid=420&lid=3699. Accessed Mar 2021.
- 5.Disease control programme (NHM). Ministry of health and family welfare, 2018–2019. https://main.mohfw.gov.in/sites/default/files/05%20ChapterAN2018-19.pdf. Accessed Sept 2019.
- 6.Malaria situation in India from 2016, monthly epidemiological situation (mes). https://nvbdcp.gov.in/WriteReadData/l892s/35471409381614152262.pdf. Accessed Feb 2021.
- 7.Das S, et al. Progressive increase in point mutations associates’ chloroquine resistance: Even after withdrawal of chloroquine use in India. Int. J. Parasitol. Drugs Drug Resist. 2017;7:251–261. doi: 10.1016/j.ijpddr.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed A, et al. Prevalence of mutations associated with higher levels of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Car Nicobar Island and Assam, India. Antimicrob. Agents Chemother. 2006;50:3934–3938. doi: 10.1128/AAC.00732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidelines for Diagnosis and Treatment of Malaria in India 2014. New Delhi, India: National Institute of Malaria Research. 2014. http://www.mrcindia.org/Diagnosis%20of%20Malaria%20pdf/Guidelines%202014.pdf. Accessed Mar 2015.
- 10.Li J, Zhou B. Biological actions of artemisinin: Insights from medicinal chemistry studies. Molecules. 2010;15:1378–1393. doi: 10.3390/molecules15031378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noedl H, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 12.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparummalaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ménard D, Khim N, Beghain J, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaratunga C, Andrianaranjaka VH, Ashley E, et al. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments a WWARN individual patient data meta-analysis. BMC Med. 2019 doi: 10.1186/s12916-018-1207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flegg JA, Guerin PJ, White NJ, et al. Standardizing the measurement of parasite clearance in falciparum malaria: The parasite clearance estimator. Malar. J. 2011;10:339. doi: 10.1186/1475-2875-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phyo AP, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: A longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witkowski B, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariey F, et al. A molecular marker of artemisinin resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbengue A, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straimer J, et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckstein-Ludwig U, Webb RJ, Goethem IDA, et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 23.Duraisingh MT, Jones P, Sambou I, et al. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 2000;108:13–23. doi: 10.1016/S0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 24.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO, Guidelines for the treatment of malaria. http://helid.digicollection.org/pdf/s13418e/s13418e.pdf. Accessed Oct 2006.
- 26.Mishra N, Kaitholia K, Srivastava B, et al. Declining efficacy of artesunate plus sulphadoxine-pyrimethamine in northeastern India. Malar. J. 2014;13:284. doi: 10.1186/1475-2875-13-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Saha B, Hati AK, Roy S. Evidence of Artemisinin resistant Plasmodium falciparum malaria in Eastern India. N. Engl. J. Med. 2018;379(20):1962–1964. doi: 10.1056/NEJMc1713777. [DOI] [PubMed] [Google Scholar]
- 28.Das S, Manna S, Saha B, Hati AK, Roy S. Novel pfkelch13 gene polymorphism associates with artemisinin resistance in eastern India. Clin. Infect. Dis. 2019;69:1144–1152. doi: 10.1093/cid/ciy1038. [DOI] [PubMed] [Google Scholar]
- 29.Patel P, et al. Prevalence of mutations linked to anti-malarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: A malaria elimination point of view. Sci. Rep. 2017;7:16690. doi: 10.1038/s41598-017-16866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma YD. Molecular surveillance of drug-resistant malaria in India. Curr. Sci. 2012;102:696–703. [Google Scholar]
- 31.Sharma D, Lather M, Mallick PK, et al. Polymorphism in drug resistance genes dihydrofolate reductase and dihydropteroate synthase in Plasmodium falciparum in some states of India. Parasit. Vectors. 2015;8:471. doi: 10.1186/s13071-015-1080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar AS, Gahlawat K, Singh V. Comparative analysis of Plasmodium falciparum dihydrofolate-reductase gene sequences from different regions of India. Heliyon. 2020;6:e03715. doi: 10.1016/j.heliyon.2020.e03715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S, et al. The novel quadruple mutations in dihydropteroate synthase genes of Plasmodium falciparum in West Bengal, India. Trop. Med. Int. Health. 2012;17:1329–1334. doi: 10.1111/j.1365-3156.2012.03071.x. [DOI] [PubMed] [Google Scholar]
- 34.Mishra N, Bharti RS, Mallick P, et al. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar. J. 2016;15(1):583. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rana R, et al. Sequence analysis of the K13-propeller gene in artemisinin challenging Plasmodium falciparum isolates from malaria endemic areas of Odisha, India: A molecular surveillance study. Biomed. Res. Int. 2020 doi: 10.1155/2020/8475246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra N, Prajapati SK, Kaitholia K, et al. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob Agents Chemother. 2015;59(5):2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee A, et al. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar. J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulle M, et al. Artemisinin-resistant Plasmodium falciparum K13 mutant alleles, Thailand-Myanmar border. Emerg. Infect. Dis. 2016;22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakrabarti R, White J, Babar PH, et al. Decreased in vitro artemisinin sensitivity of Plasmodium falciparum across India. Antimicrob. Agents Chemother. 2019 doi: 10.1128/AAC.00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tacoli C, et al. Artemisinin resistance-associated K13 polymorphisms Plasmodium falciparum in southern Rwanda, 2010–2015. Am. J. Trop. Med. Hyg. 2016;95:1090–1093. doi: 10.4269/ajtmh.16-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haynes RK, et al. Considerations on the mechanism of action of artemisinin anti-malarials: Part 1—the 'carbon radical' and 'heme' hypotheses. Infect. Disord. Drug Targets. 2013;13:217–277. doi: 10.2174/1871526513666131129155708. [DOI] [PubMed] [Google Scholar]
- 42.Beez D, et al. Genetic predisposition favors the acquisition of stable artemisinin resistance in malaria parasites. Antimicrob. Agents Chemother. 2011;55:50–55. doi: 10.1128/AAC.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization (WHO). Assessment and monitoring of anti-malarial drug efficacy for the treatment of uncomplicated falciparum malaria. Geneva. 2003. http://www.cdc.gov/malaria/resources/pdf/drug.../WHO2003_monitoring.pdf.
- 44.Li QG, Peggins JO, Fleckenstein LL, et al. The pharmacokinetics and bioavailability of dihydroartemisinin, arteether, artemether, artesunic acid and artelinic acid in rats. J. Pharm. Pharmacol. 1998;50:173–182. doi: 10.1111/j.2042-7158.1998.tb06173.x. [DOI] [PubMed] [Google Scholar]
- 45.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 46.Das S, Chakraborty SP, Hati AK, Roy S. Association between prevalence of chloroquine resistance and unusual mutation in pfmdr-I and pfcrt gene in India. Am. J. Trop. Med. Hyg. 2013;88(5):828–834. doi: 10.4269/ajtmh.11-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das S, Chakraborty SP, Hati A, Roy S. Malaria treatment failure with novel mutation in the Plasmodium falciparum dihydrofolate reductase (pfdhfr) gene in Kolkata, West Bengal, India. Int. J. Antimicrob. Agents. 2013;41:447–451. doi: 10.1016/j.ijantimicag.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Das S, et al. Double mutation in the pfmdr1 gene is associated with emergence of chloroquine resistant Plasmodium falciparum malaria in eastern India. Antimicrob. Agents Chemother. 2014;58(10):5909–5915. doi: 10.1128/AAC.02762-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.