Graphical abstract
Keywords: Tuberculosis, Genome sequencing, MDR, MycoTRAP-DB, Secondary mutations, SNPs
Highlights
-
•
MycoTRAP-DB, a database of mutations and their impact on normal functionality of protein in M.tb genes.
-
•
Several secondary mutations were identified with significant impact on protein structure and function.
-
•
Comprehensive information gives insight for screening of suspected hotspots in advance to combat drug resistant TB.
Abstract
Tuberculosis (TB) continues to be the leading cause of deaths due to its persistent drug resistance and the consequent ineffectiveness of anti-TB treatment. Recent years witnessed huge amount of sequencing data, revealing mutations responsible for drug resistance. However, the lack of an up-to-date repository remains a barrier towards utilization of these data and identifying major mutations-associated with resistance. Amongst all mutations, non-synonymous mutations alter the amino acid sequence of a protein and have a much greater effect on pathogenicity. Hence, this type of gene mutation is of prime interest of the present study. The purpose of this study is to develop an updated database comprising almost all reported substitutions within the Mycobacterium tuberculosis (M.tb) drug target genes rpoB, inhA, katG, pncA, gyrA and gyrB. Various bioinformatics prediction tools were used to assess the structural and biophysical impacts of the resistance causing non-synonymous single nucleotide polymorphisms (nsSNPs) at the molecular level. This was followed by evaluating the impact of these mutations on binding affinity of the drugs to target proteins. We have developed a comprehensive online resource named MycoTRAP-DB (Mycobacterium tuberculosis Resistance Associated Polymorphisms Database) that connects mutations in genes with their structural, functional and pathogenic implications on protein. This database is accessible at http://139.59.12.92. This integrated platform would enable comprehensive analysis and prioritization of SNPs for the development of improved diagnostics and antimycobacterial medications. Moreover, our study puts forward secondary mutations that can be important for prognostic assessments of drug-resistance mechanism and actionable anti-TB drugs.
1. Introduction
Tuberculosis (TB) has been a deadly disease since 3400BCE with huge social and economic impact worldwide [1]. According to the WHO Global Tuberculosis Report, an estimated 10 million people developed TB in 2019 [2]. Globally, TB is among the top ten causes of death, with multi-drug resistance posing additional challenge in managing this global epidemic [3]. In 2019, it took relatively 1.2 million lives in HIV-negative people and 208,000 deaths amongst HIV-positive people. Mycobacterium tuberculosis (M.tb) pathogen circumvents new strategies by which it can find ways to infect, survive, and disseminate against the barriers set by the host [4]. The evolution of the M.tb genome has been crucial in maintaining its virulence throughout the centuries as the pathogen undergoes reductive evolution and confines its essential functionality to the minimum possible number of genes [5]. The alterations within the genome composition have been a crucial strategy for the pathogen to overcome the stresses and challenges posed by the environment or the host.
The extensive use of antibiotics against M.tb has been posing a threat to the existence of this pathogen, and in response M.tb has been rigorously selecting the strains that could help the pathogen to survive against the antibiotic exposures [6], [7]. With time, the mutations accumulated within the M.tb genome and specifically in the proteins serving as drug targets, have caused the development of resistance against various first and second lines anti-TB drugs and further its self-transformation into pre-extensively drug resistant (pre-XDR), multidrug resistant (MDR) and extensively drug-resistant (XDR) [7], [8]. Drug efflux pumps, with a specific genetic signature could be yet other mechanism for drug resistance [9]. Mutations at the active site of protein precisely affect the binding efficacy of the drug or may leave the protein deprived of its catalytic efficiency. Such mutations are well characterized due to their major role in drug resistance, however there are other mutations present outside the active region, also termed as secondary or accessory mutations which are indirectly responsible for causing resistance [8], [10]. Secondary mutations or accessory mutations cause noticeable rearrangements into the protein structure leading to altered drug interaction and affecting the dynamics of the complex by altering the shape and flexibility of the binding pocket through a complex network of interactions within the structure, defined as “network hypothesis” [11]. These mutations also assist in compensating the fitness cost developed due to the after effects of drug resistance mutations, hence termed as compensatory mutations [12]. It is thus of utmost importance to explore secondary mutations present in the neglected and less focused genomic areas for complete understanding of the mechanism of resistance.
We are also witnessing enormous amounts of genome sequencing data that are being assessed broadly to identify the polymorphisms of relative clinical significance and improve the understanding of drug resistance in M.tb [11], [12], [13], [14]. However, the corresponding information is scattered among the large pool of literature, which renders the researchers to be deprived of the critical information which might be used for identification of novel drug targets and development of anti-TB drugs. Over the years, several databases [15] namely GMTVD [16], TBDreaMDB [17], MUBII-TB-DB [18] have been developed to store and organize data, but they are devoid of the recent records owing to which the conclusions are bound to remain incomplete. Thus, an extensive search of recent literature to acquire all the reported mutations is critical for understanding the evolution of anti-TB drug resistance.
Drug resistance-associated genes rpoB, inhA, katG, pncA, gyrA and gyrB are known to confer resistance against rifampicin (RIF), isoniazid (INH), pyrazinamide (PZA) and fluoroquinolones (ciprofloxacin, CIF; moxifloxacin, MFX; levofloxacin, LFX; and ofloxacin, OFX), respectively [7]. The mutations in these genes are responsible for detrimental effects over the normal functionality of protein serving as a target, disrupting its binding with the drug and subsequently leading to antibiotic resistance [19]. Single nucleotide polymorphisms (SNPs) are the most recurrent genetic variations responsible for disturbing protein functionality and promoting drug resistance in M.tb. Although many of these are benign or neutral and do not contribute much to the phenotypic alteration many others, primarily the non-synonymous mutations, are deleterious which have severe consequences on function of the translated protein and its phenotype. Thus, it is imperative to look for the non-synonymous mutations reported in the drug-resistant isolates as they serve as key leads for explaining the mechanism of resistance development. Apart from SNPs, other variations that include insertions, deletions and frame-shift mutations are also major contributors towards drug resistance, however these were out of the scope of the present study and thus were not included.
In order to study the mechanisms of drug resistance, it is critically important to know the association between mutations in drug targets and their phenotypic profile by establishing minimum inhibitory concentrations (MICs). In-vitro propagation of M.tb is challenging because of its slow growing nature requiring time and resources. Alternative (in-silico) methods can help to prioritize mutations by predicting structural implications, due to the unavailability of a quantitative estimation of phenotypic profiling [20]. In the present study, a rigorous search of the recently reported literature on nearly all the mutations in well recognized drug resistance-associated genes rpoB, inhA, katG, pncA, gyrA and gyrB was performed [21]. Apart from these genes, embC, embA, and embB are also associated with resistance to ethambutol (EMB) [22] which is a first-line drug. However, in 2019, WHO released a new drug classification for the treatment of drug resistant TB, reorganizing second-line drugs into three groups (Group- A, B, and C). Ethambutol has been included in group C [23] and also serves as a key drug in second-line regimens for MDR-TB and thus was not included in this study. Various computational approaches were used to quantify the effects of non-synonymous substitutions over the protein structure, function, and conservation narrowing down our search for the candidates with higher potential of inducing the resistance. Further, molecular docking studies were carried out to examine the impact of the candidate mutations on drug-protein interaction eventually ending up with hampered drug efficacy. This was followed by normal mode analysis (NMA) performed to gain insights into the interaction profiles by taking into account the dynamic aspects of the protein which are requisite parameters for favorable protein-drug interaction, concluding our search for the potential novel active site and secondary mutations responsible for conferring the resistance [24]. The computational analyses and MycoTRAP-DB (Mycobacterium tuberculosis Resistance Associated Polymorphisms Database) platform serving as a high-throughput platform, could provide the scientific basis of an initiation to in-vitro studies that aim for an in-depth characterization of emerging potential resistance conferring mutations.
2. Methodology
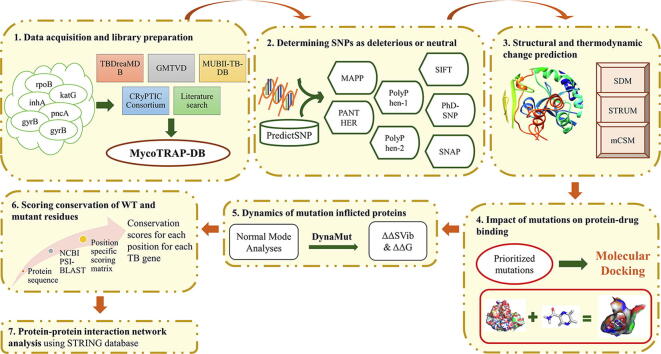
Fig. 1 Illustrates all of the steps of computational methodology followed in the present study. These steps consist of: 1) Acquisition of publicly available data and construction of a library predictive of resistant and susceptible mutations in genes; 2) Determining SNPs as deleterious or neutral using PredictSNP tool; 3) Structural and thermodynamic change prediction showing the impact of mutations on protein stability and conformation using three tools based on different structural features; Site Directed Mutator (SDM), STRUM and mutation Cut-off Scanning Matrix (mCSM); 4) Determining the impact of mutations on protein-drug binding by molecular docking; 5) Identification of the dynamic interaction of mutation inflicted proteins. 6) Scoring conservation of WT and mutant residues; and 7) Protein-protein interaction network analysis. These steps are described in detail below:
Fig. 1.
Illustration of the workflow describing all of the steps of computational methodology consisting of: 1) Acquisition of publicly available data and construction of a library predictive of resistant and susceptible mutations in genes; 2) Determining SNPs as deleterious or neutral using PredictSNP tool; 3) Structural and thermodynamic change prediction showing the impact of mutations on protein stability and conformation using three tools based on different structural features; SDM, STRUM and mCSM; 4) Determining the impact of mutations on protein-drug binding by molecular docking; 5) Identification of the dynamic interaction of mutation inflicted proteins, 6) Scoring conservation of WT and mutant residues; and 7) Protein-protein interaction network analysis.
2.1. Data acquisition and library preparation
A library predictive of resistant and susceptible mutations of rpoB, inhA, katG, pncA, gyrA and gyrB genes was prepared through a rigorous literature search. Mutations from the two publicly available databases, TBDreaMDB [17] and GMTVD [16] were also extracted. Furthermore, mutations reported by web-based tools, MUBII-TB-DB [18] and CRyPTIC Consortium [25] were also incorporated. Based on the intensive data mining of available literature, each SNP was represented as resistance causing, neutral or uncharacterized along with their frequency of occurrence in clinical isolates.
2.2. Determining SNPs as deleterious or neutral
The reference M.tb H37Rv genome used was NC_000962.3. The protein sequences for the M.tb genes investigated in the present study were obtained from UniProt, which is a freely accessible database hosting protein sequence and functional data. The UniProt IDs for rpoB, inhA, katG, pncA, gyrA and gyrB corresponded to P9WGY9, P9WGR1, P9WIE5, I6XD65, P9WG47 and P9WG45. Characterization of non-synonymous (ns) SNPs as deleterious or neutral was performed using PredictSNP tool [26]. PredictSNP is a disease associated mutation classifier and gives a consensus score based on the output of six different amino acid (AA) based function and pathogenicity prediction tools, namely Multivariate Analysis of Protein Polymorphism (MAPP), nsSNPAnalyzer, Protein Analysis through Evolutionary Relationships (PANTHER), PhD-SNP, PolyPhen-1 and PolyPhen-2, Sorting Intolerant From Tolerant (SIFT), and Synonymous Non-synonymous Analysis Program (SNAP) (Table 1). MAPP calculates the difference in physicochemical properties between the wild and mutant AAs. Larger the deviation, higher is the chance that the mutation causes functional disruption of the protein. PANTHER is an evolutionary based tool which anticipates the function of hypothetical genes based on their evolutionary relationship to experimentally characterized genes with known function [27]. PhD-SNP is a support vector machine classifier-based method that predicts deleterious SNPs using sequence and profile information [28]. Polymorphism Phenotyping (PolyPhen) engages a set of empirical rules to determine the functional disruption in the presence of mutation [29]. PolyPhen-2 is different from PolyPhen-1 in terms of features used for prediction and uses eight sequence based and three structure-based AA properties for predicting the functional and structural damage due to AA substitutions [30]. SIFT is an evolutionary based algorithm which measures the impact of AA change on the function of protein based on physical properties and sequence homology [31]. SNAP is based on a set of codon-aligned nucleotide sequences which determine synonymous and non-synonymous substitution rates [32]. Consensus prediction made by PredictSNP represents an accurate and robust alternative to the predictions delivered by individual tools. The AA sequence of a query protein was uploaded in FASTA format on PredictSNP along with the list of mutations in a text format. PredictSNP web server gave consensus prediction along with the predictions made by the individual tools for all selected mutations.
Table 1.
Computational tools used to study the impact of mutations on protein structure and function.
| Software | Information | Description | Value/Range | PMID |
|---|---|---|---|---|
| MAPP | Physicochemical variation | Deleterious/ Neutral | – | 15,965,030 |
| PANTHER | Protein analysis through evolutionary relationship | Pathogenic/Deleterious | – | 20,015,972 |
| PolyPhen-1 | Sequence, phylogenetic and structural information | Deleterious/ Neutral | – | 12,202,775 |
| PolyPhen-2 | Eight sequence based and three structure based amino acid properties | Deleterious/ Neutral | 0–1 (≤0.15 tolerated, 0.15 ≤ 1.0 possibly damaging, 0.85 ≤ 1.0 damaging) | 20,354,512 |
| SIFT | Sequence homology and the physical properties of amino acids | Deleterious/ Neutral | 0 (≤0.05 pathogen; 0–1) | 12,824,425 |
| PhD-SNP | Support vector machine prediction trained on sequence and evolutionary information | Pathogenic/ Benign | 0–1 (≤0.5 benign, >0.5 pathogenic) | 16,895,930 |
| SNAP | Neural network method based on sequence information | Non-Neutral | reliability index measure (range 0–9) | 17,526,529 |
| SDM | Environment-specific substitution tables | Reduced stability/ Increased stability | ΔΔG < 0 reduced stability; ΔΔG > 0 increased stability | 21593128, 28,525,590 |
| STRUM | Sequence profile, structural profile, different energy functions based on I-TASSER model | Destabilizing/ stabilizing | ΔΔG < 0 destabilizing; ΔΔG > 0 stabilizing | 27,318,206 |
| mCSM | Graph-based signatures, pharmacophore properties and experimental conditions | Destabilizing/ stabilizing | ΔΔG < 0 destabilizing; ΔΔG > 0 stabilizing | 24,281,696 |
| mCSM-lig | Protein-ligand affinity change upon mutation | Destabilizing/ stabilizing | ΔΔG < 0 destabilizing; ΔΔG > 0 stabilizing | 27,384,129 |
2.3. Structural and thermodynamic change prediction
To examine the impact of mutations on protein stability and conformation, three tools based on different structural features; SDM, STRUM and mCSM were utilized. SDM calculates a stability score based on environment-specific AA substitution frequencies within homologous protein families [33]. The score is equivalent to the change in free energy (ΔΔG) between the wild-type (WT) and mutant protein. To further enhance the significance of SDM predictions, the nsSNPs that were submitted to SDM were also analyzed by STRUM which combines the WT protein sequence profiles with low-resolution structure models built by I-TASSER to calculate the fold stability change (ΔΔG) of protein molecules upon single-point mutations [34]. Lastly mCSM, a machine learning approach that integrates graph-based signatures, pharmacophore properties and experimental conditions and uses this information to calculate changes in the stability of protein structure [35] was used. The consensus ΔΔG score of all the three tools were considered to denote the impact of mutations on protein stability as highly destabilizing, destabilizing or stabilizing.
Based on the results drawn from various sequence and structure-based tools, consensus mutations, that were predicted to be functionally deleterious and structurally destabilizing, were derived and considered for further analyses.
2.4. Impact of mutations on protein-drug binding
Molecular docking was performed to quantify the influence of mutations on the binding affinity of drugs targeting proteins encoded by rpoB, inhA, katG, pncA, gyrA and gyrB genes. An initial screening was done employing mCSM-lig. This is a freely available online tool that uses graph-based signatures of WT structural environment to assess the structural ramifications on drug binding due to mutations in the protein [36]. The mutations having strong destabilizing effects on protein-drug binding were subjected to docking using Schrodinger suite [37]. The X-ray crystal structures of WT proteins were obtained from Protein Data Bank (PDB) [38] and the structures represented by following PDB IDs: RpoB (5UHB) [39], InhA (1ENY) [40], KatG (1SJ2) [41] (recently a cryo-EM structure of katG was published [42] however it has not yet been released on PDB), PncA (3PL1) [43] and DNA gyrase (5BS8) [44]. The ligand bound crystal structures were available for RpoB, InhA and DNA gyrase (in complex with MFX). In case of mutations in InhA protein, ligand bound crystal structures were available only for I21V (PDB ID: 2AQH) [45] and S94A (PDB ID: 2NV6) [46]. The mutations in WT protein structures were incorporated by employing the Maestro interface available from Schrodinger. Prior to molecular docking, the WT and mutant protein structures were prepared using Schrodinger’s protein preparation wizard [47]. Several tasks were performed during protein preparation which include deleting water molecules, repairing truncated sidechains, adding hydrogens and assigning partial charges. The chemical structures of the first line TB drugs (RIF, INH and PZA) and fluoroquinolones [48] were obtained from PubChem compound database [49] and were used as ligands in the present study. The ligands were prepared using the LigPrep module [47] which generated accurate and energy minimized conformations of compounds. The idea behind generating structurally diverse compounds is to explore all the chemical and structural properties a compound could possess as even small changes can amount to significant variations in computational results. Subsequent to this, a grid box was generated centered on the active site residues of the proteins using the Receptor Grid Generation module. The active site for each of the protein, RpoB, InhA, KatG, PncA and DNA gyrase, was acquired from literature [39], [43], [46], [50], [51]. Lastly, the ligands were docked in the active sites of WT and mutant proteins using the extra precision approach of Schrodinger’s Glide module [52].
2.5. Dynamic interaction of mutation inflicted proteins
Proteins are highly dynamic in nature and their structural fluctuations have an important role in their functions. Therefore, it is quite imperative to assess the impact of mutations on the native conformations of proteins. Post molecular docking of the proteins were run on DynaMut, a web server that takes into account NMA and integrates it with graph-based structural signatures and gives a consensus prediction of the impact of a mutation on the stability of a protein [24]. NMA is a powerful technique for predicting molecular motions and examines the vibrational entropy changes due to harmonic oscillating fluctuations as a consequence of mutations in the protein [53]. Vibrational entropy changes (ΔΔSVib ENCoM in kcal.mol−1.K−1) and NMA predictions (ΔΔG ENCoM in kcal/mol) were recorded to examine the effect of mutations on protein flexibility and stability thus influencing overall protein dynamics.
2.6. Scoring conservation of WT and mutant residues
Conservation analysis of codons helps in understanding the evolutionary conservation of a particular residue in a protein that could be of structural and functional importance. Conservation scores are determined based on the AA frequencies at a certain codon in the alignment. To perform AA conservation analysis in the present study, the sequence for each protein, RpoB, InhA, KatG, PncA, GyrA and GyrB was downloaded from UniProt database [54] followed by a NCBI PSI-BLAST [55] run against the non-redundant protein database for each TB protein. A profile search was generated using five PSI-BLAST iterations with an e-value cut-off of 0.001. Next a position specific substitution matrix (PSSM) was generated, the elements of which were conservation scores for each position for each TB gene. The overall substitution score ranges from −8 to 10 where the positive value represents that the mutation is favorable, fit and evolution can accept this transition over normal WT codon during the time of need. On the other hand, negative values reflect the less likely mutations or mutations that result in altered functionality. For all nsSNPs, the conservation scores of all the substitutions were acquired. Additionally, a score for replacement of WT with mutant AA was analyzed which represents the substitution score. The mutations with high substitution score (>5 in this study) would be less detrimental for the overall integrity of the protein and thus the normal function of the protein would not be hampered.
2.7. Protein-protein interaction network analysis
Protein-protein interaction (PPI) network was analyzed using STRING database version 11.0 for all TB proteins studied in this work. STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is a freely accessible web resource and biological database which harbors information about PPIs curated from numerous sources, including experimental data, computational prediction methods and public text literature collections [56].
3. Results and discussion
3.1. Data acquisition and library preparation
In the present study, we have assembled known mutations and present a database, MycoTRAP-DB for TB-protein coding genes, rpoB, inhA, katG, pncA, gyrA and gyrB from the previously published databases as well as thorough data mining of the available literature till December 2020. A repository comprising a total of 3303 mutations was prepared which included mutations from GMTVD, TBDreaMDB, MUBII-TB-DB and CRyPTIC Consortium. Moreover the remaining mutations, not included in the above databases, were acquired by rigorously searching the published literature related to the drug resistance in M.tb (Supplementary Table 1). The final collective mutations were 701, 56, 504, 476, 822, and 744 for rpoB, inhA, katG, pncA, gyrA and gyrB respectively. By removing the duplicates, synonymous mutations and point mutations leading to insertions and deletions, we obtained 1795 total mutations for further analysis. The final mutations obtained for rpoB, inhA, katG, pncA, gyrA and gyrB corresponded to a total of 406, 37, 446, 402, 258 and 246 mutations, which were further characterized based on their resistance conferring potential (Table 2).
Table 2.
Total number of the mutations gathered from multiple sources and pre-processing for selection of mutations for final analysis.
| Gene | Total mutations in MycoTRAP-DB | Synonymous mutations | INDELS | Non-synonymous mutations | Final analyzed |
|---|---|---|---|---|---|
| rpoB | 701 | 187 | 36 | 478 | 406 |
| inhA | 56 | 11 | 4 | 41 | 37 |
| katG | 504 | 12 | 24 | 468 | 446 |
| pncA | 476 | 5 | 7 | 464 | 402 |
| gyrA | 822 | 552 | 5 | 265 | 258 |
| gyrB | 744 | 442 | 1 | 301 | 246 |
3.2. Amino acid conservation and substitution analysis
Conservation score reflects the significance of a WT AA at a particular codon position in preserving the normal functionality of the protein [57]. On the other hand, the deviation from the normal function caused by a particular substitution based on newly inserted AA is depicted by substitution score. Analyzing these scores for all the AAs at a specific codon presented the information about evolutionarily favorable mutated AAs at that position. The conservation and substitution scores of all the AAs for each gene have been provided as Supplementary Table 2. The positive values for conservation scores showed the high degree of conservation of an AA at that position in retaining the protein function. The overall less substitution scores for mutations than WT in our study indicated that these mutations may deteriorate the protein of its overall original function. This provides an opportunity for evolution to select the advantageous changes bestowing bacteria with mutations that could possibly promote its viability and virulence [58], [59]. M.tb acquires these less favorable mutations which completely or partially eliminate the activity of the associated protein but in turn, make the bacteria more fit to survive against the anti-TB drugs. Similar observations were made for I335T, T262R and T76P mutations of KatG and PncA where the mutations led to substantial deprivation in activity of proteins but in turn, provided resistance to bacteria against two most potent anti-TB drugs namely INH and PZA [58], [59]. Therefore, the secondary mutations identified to be resistance conferring in this work could be declared as suspects that despite being less favorable by evolution hold great importance in the survival of the pathogen in stress conditions.
3.3. Analyzing impact of nsSNPs on protein function
rpoB: A total of 517 nsSNPs were studied for their impact on protein function and virulence. Results were obtained for 406 mutations; the remaining 111 mutations were not used due to irregularity in the WT residues at certain codon positions. Out of 406 nsSNPs, 258 (63.5%) were recorded as deleterious, while 148 (36.5%) were predicted as neutral (Supplementary Table 3). The rpoB gene mutations are predominantly located within RIF-resistance-determining region (RRDR) corresponding to codons 426 to 452 in M.tb [60]. A deeper digging into 258 deleterious nsSNPs brought to light three codon positions, 441, 451 and 456 in RRDR region where four or more AA variants were present. All these mutations, D441F/N/V/Y/A/G/C; H451R/N/Q/C/P/T and S456F/Q/P/W/Y, are known to be associated with RIF drug resistance and were found to be highly deleterious in our analyses.
inhA: A total of 42 nsSNPs were analyzed for their effect on protein function and pathogenicity, of which 37 nsSNPs were obtained by PredictSNP. Among 37 nsSNPs, 13 (35.1%) were predicted as deleterious, while 24 (64.86%) were listed as neutral (Supplementary Table 3). The mutations on the active site of InhA protein through altering its binding with the substrate NADH can disrupt the architecture of the active site and reduce the binding affinity of the drug with InhA. The highly prevalent mutations associated with INH-resistance are found in the active site region (G14, S20, V21, D64, V65, I95, G96, D148, F149, K165, I194, and T196) of InhA [46]. The active site mutations, I21V/T, I95T/P, and I194T were predicted to be deleterious to protein function, the same has been observed in DST [61]. The mutations, I21T, S94A and I95P have been identified to be associated with resistance in both, INH- as well as ETH-resistant clinical isolates [61]. However, S94A and A190S (commonly occurring mutations) were predicted as neutral and did not have any impact on the function of protein.
katG: Among 500 katG nsSNPs analyzed for functional impact and pathogenicity, predictions were obtained for 446 nsSNPs. A total of 351 mutations (78.6%) were recorded as deleterious, while 95 (21%) were predicted as neutral (Supplementary Table 3). Resistance causing nsSNPs in katG are known to be distributed throughout the coding region. The high number of variants present at a single codon position is indicative of its high mutational propensity in unfavorable conditions providing an escape route for bacteria to survive [62]. In the present work, five such codon positions were observed and all the mutations at these positions, N138S/H/D/T, W300G/C/D/I/R, S315L/I/R/T/N/G, W321L/S/R/G/F and W328R/L/C/S/G were predicted to be deleterious with most of them already known to be associated with INH drug resistance [61]. However, the mechanism of resistance in case of W300D, W321G and W328C substitutions has not been explored yet. The deleterious secondary mutations, other than 138, 300, 315, 321 and 328 codon positions, present in the entire coding region of katG were also explored for their effects on the protein.
pncA: The functional and pathogenic analysis in pncA was initiated with a total of 402 mutations. Based on the consensus results from PredictSNP, 281 (70%) of them were identified as deleterious mutations and 121 (30%) as neutral (Supplementary Table 3). The 281 deleterious mutations included mutations at the iron binding site (D49, H51, H57, H71) and catalytic triad (D8, K96, C138). These results were in concordance with the previous reports that demonstrate the mutations within the active site as the highly detrimental or the resistance conferring mutations [43]. Among them, the variants D8H and C138R were less studied thus offering an opportunity to thoroughly explore their impact.
gyrA and gyrB: A total of 271 gyrA and 284 gyrB mutations were analyzed for their functional and pathogenicity impact. Predictions were obtained for 258 and 246 in gyrA and gyrB, respectively. The consensus prediction resulted in 63/258 (24.4%) and 64/246 (26%) as deleterious in gyrA and gyrB, respectively, while 195/258 (75.6%) and 182/246 (74%) were identified as neutral mutations in gyrA and gyrB, respectively (Supplementary Table 3). As evident from the literature, the QRDR of the gyrA (codon 74–113) and gyrB (codon 461–499) are hotspot regions [63]. QRDR mutations A90V, S91P and D94G/N/A/H/Y/V/F in gyrA and D461N/H, D472H/A, N499D/T in gyrB are involved in fluoroquinolone resistance [63], [64] and most of them were identified to be deleterious to protein function by our analyses. However, mutations A90V and D94G of gyrA were predicted to have no impact on protein function despite being resistant in clinical isolates.
3.4. Structural consequences of nsSNPs
The ΔΔG value calculated as the difference in ΔG score of WT and mutant protein structures is considered as the perfect measure for depicting the impact of single point mutation in changing the stability of the protein [65], [66]. Positive and negative values of ΔΔG (kcal mol-1) correspond to increase or decrease in the overall stability of the protein. In the present study, a single point mutation was classified as highly destabilizing (ΔΔG < 0 by all three tools, SDM, STRUM & mCSM), destabilizing (ΔΔG < 0 by any two tools) and stabilizing (ΔΔG > 0 by any two or all three tools). Supplementary Table 4 shows change in protein stability using SDM, STRUM, mCSM web servers in presence of each single point mutation for rpoB, katG, inhA, pncA, gyrA and gyrB.
RpoB: A total of 406 nsSNPs were subjected to stability analyses, and predictions were obtained for 299 nsSNPs. Among these, 4 (1.3%) were highly destabilizing, 182 (60.9%) destabilizing and 113 (37.8%) stabilizing mutations. Few mutations were dropped due to the absence of WT residue in crystal structures (RpoB as well as some other genes). The mutations H451R/N/Q/C/P/T and S456F/Q/P/W/Y of RRDR region were observed to reduce the stability of the protein with the exception of D441F/N/V/A/G/C which did not have a destabilizing effect though affecting the protein at functional level. The mildly destabilizing effect in case of all the variants at D441 position might be associated with lower protein fitness penalty thereby retaining the overall fitness of the bacteria in presence of the drug thus further enriching their frequency in the population [62]. In case of rpoB, lineage defining mutations, T356I and S394L [67] were predicted to stabilize the protein.
InhA: Among 37 nsSNPs analyzed for structural impact on InhA protein, 21 (56.7%), 9 (24.3%), and 7 (19%) were predicted to be highly destabilizing, destabilizing, and stabilizing, respectively. Highly destabilizing effect on structures was noted for certain well known INH-resistance associated mutations, I21V/T, I95P/T and I194T present on the active site of protein. Other than the active site, K8N and S94A mutations, known to cause resistance widely, were also found as destabilizing in our analysis [61]. Lineage defining polymorphism V78A [67] has no role in resistance however, it was predicted to destabilize the protein structure.
KatG: The structural stability prediction tools revealed a total of 434 mutations, out of which 18 (4.1%), 317 (73%) and 99 (22.8%) mutations were classified as highly destabilizing, destabilizing and stabilizing, respectively. Highly destabilizing effect on protein structure was noted for certain well known INH-resistance associated mutations like N138S, W300G, W321G, amongst others with the exception of S315T and S315N. A very low destabilizing effect was observed in terms of ΔΔG value in case of S315T and S315N mutations. This can be attributed to lower protein fitness penalty based on the similar observation in RpoB [62]. The secondary mutations, W90R, A109V, W328G, A614E, R128G, D142G, T275P and W328G were also recorded as destabilizing, pointing towards their possible candidature to effective drug binding. One of the most commonly occurring and lineage defining mutation R463L [67] was predicted to exhibit a stabilizing effect on the protein structure.
PncA: After the completion of functional analysis, we proceeded with all the 402 mutations to examine their role in inducing the structural changes within the protein. A total of 71 highly destabilizing (17.6%), 295 destabilizing (73%) and 36 stabilizing (8.9%) mutations were obtained. As expected, the prominent residues that form the iron binding site (D49, H51, H57, and H71) and the catalytic triad (C138, D8, K96) were predicted to destabilize the protein with H51N, H71D/E/Q and K96E being the most destabilizing ones. All mutations within these sites displayed the deformity in protein structure except D8V. With negligible structural alteration and even displaying the deleterious functionality earlier, D8V hints at the high impact of active site residues over the protein functionality altogether. Being in a catalytic triad and a prime site facilitating the interaction with PZA, a mutation at D8 seems sufficient to compromise the interaction without destabilizing the overall structure [43].
DNA gyrase: Post functional and pathogenicity analyses, the mutations were subjected to structural analysis which showed 26/227 (11.5%) and 13/228 (5.7%) mutations as highly destabilizing; 143/227 (63%) and 128/228 (56.1%) as destabilizing and 58/227 (25.6%) and 87/228 (38.2%) as stabilizing in GyrA and GyrB, respectively. Highly destabilizing effect on protein structure was noted for L96P (GyrA) and D472A (GyrB) which are prevalent in fluoroquinolone resistance associated mutations [64], [68]. The most prevalent QRDR mutations D94G/H/N/A/Y/F/V, G88A or D89N in GyrA and N499D/Y/T/S in GyrB showed very less or no effect on protein stability although these mutations have a remarkable role in fortifying the functionality of the protein. This may be due to the fact that alteration of these key residues does not lead to the deformation of overall structure nonetheless deprives the protein from its function [69]. V125M, a deleterious and highly destabilizing mutation, was observed in the active site region of GyrB pointing towards its possible role in affecting drug binding. Lineage defining mutations, T80A [67] was predicted to increase protein stability and S95T [67] was predicted to have a destabilizing impact on protein.
3.5. Prioritized mutations
For protein-drug interaction studies, mutations were prioritized on the basis of functional and structural effect, prevalence in drug resistant isolates, conservation score, clinically validated, resistance pattern and fold change in drug binding affinity (Supplementary Table 5). Table 3 shows the number of nsSNPs characterized as functionally deleterious or neutral and structurally stabilizing or destabilizing for each gene. The individual tool predictions for the functional and structural impact of prioritized mutations have been provided in Supplementary Tables 6 and 7, respectively. These prioritized mutations were explored for their mechanism of resistance using protein-drug interaction studies.
Table 3.
Depiction of nsSNPs as functionally deleterious or neutral and structurally stabilizing or destabilizing as well as the prioritized mutations for each gene.
| Predict SNP |
Structural analyses |
|||||
|---|---|---|---|---|---|---|
| Gene | Results obtained | Deleterious | Neutral | Highly destabilizing | Destabilizing | Stabilizing |
| rpoB | 406 | 258 | 148 | 4 | 182 | 113 |
| inhA | 37 | 13 | 24 | 21 | 9 | 7 |
| katG | 446 | 351 | 95 | 18 | 317 | 99 |
| pncA | 402 | 281 | 121 | 71 | 295 | 36 |
| gyrA | 258 | 63 | 195 | 26 | 143 | 58 |
| gyrB | 246 | 64 | 182 | 13 | 128 | 87 |
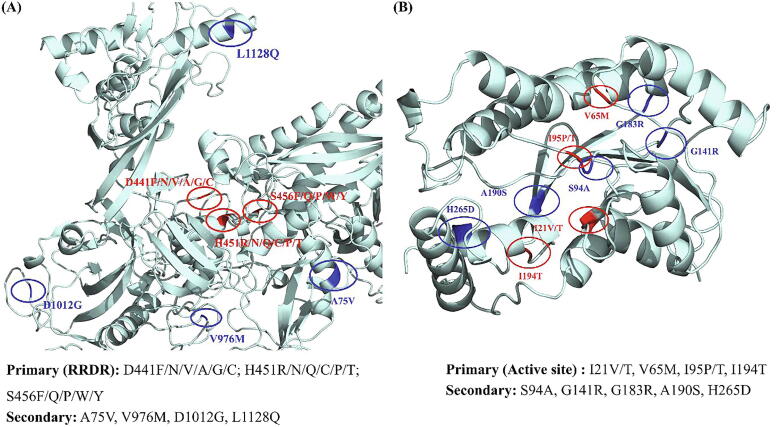
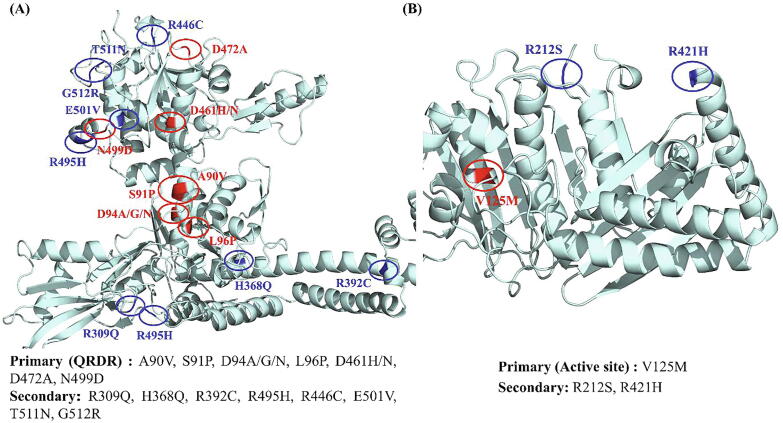
RpoB: The 22 prioritized mutations included the most prevalent and clinically validated RRDR mutations (D441F/N/V/A/G/C, H451R/N/Q/C/P/T, S456F/Q/P/W/Y) and secondary mutations (A75V, V976M, D1012G and L1128Q) (Fig. 2A). Eleven mutations in RRDR and 4 secondary mutations were predicted to be deleterious as well as destabilizing. The remaining 7 RRDR mutations were deleterious for function and stabilizing with respect to structure, but were considered for further analyses due to their high frequency in drug resistance. The H451D and S456L are the most frequently reported mutations among MDR/XDR strains of M.tb across RIF drug resistance spectrum [70], [71]. H451D mutant has been described to provide fitness benefits to the pathogen under hypoxic conditions [70]. Mutations in rpoA, rpoB, and rpoC are also common in clinical isolates carrying S531L mutation in rpoB and play a major role in compensating for the fitness loss of the bacteria [10], [72].
Fig. 2.
Distribution of prioritized primary and secondary mutations in (A) RpoB (B) InhA. Protein is shown in cyan, primary mutations in red and secondary mutations are shown in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
InhA: Among 11 prioritized mutations, 6 mutations, I21V/T, V65M, I95P/T, and I194T were present on the active site of InhA and had a highly destabilizing and deleterious profile (Fig. 2B). The other mutations, S94A, A190S, G141R, G183R and H265D were secondary mutations. Mutations, S94A and A190S did not have any impact on protein function but reduced its stability largely. G141R, G183R and H265D were considered owing to their significant impact and high prevalence [73].
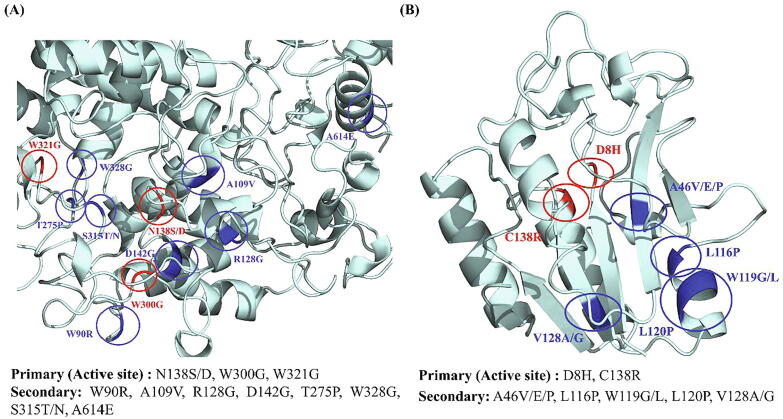
KatG: The 13 prioritized mutations included clinically validated and highly destabilizing mutations (N138S/D, R128G, D142G, T275P, W300G, W328G); frequently occurring yet uncharacterized destabilizing mutation (W321G, less evidence in literature supporting its resistance pattern); most common and stabilizing/less destabilizing mutation (S315T/N), and other secondary mutations (W90R, A109V, A614E) [25], [74], [75]. Fig. 3A shows the prioritized primary and secondary mutations in KatG protein. The INH-resistant M.tb katG (S315T) variant has been studied to carry fully functional catalase peroxidase activity while decreased INH-oxidase activity [61] makes the bacteria more fit to tackle ROS produced by macrophages and also survive in the presence of isoniazid. Mice studies have revealed that M.tb S315T mutation does not affect bacterial fitness, and it remains fully virulent and highly transmissible [76]. Consequently, the S315T variant is more often found in MDR-TB patients than in INH mono-resistant clinical isolates [77]. Our analysis also found that S315T does not confer any highly destabilizing effect on the protein structure due to which its catalase peroxidase activity is retained and bacteria do not lose their overall fitness.
Fig. 3.
Distribution of prioritized primary and secondary mutations in (A) KatG (B) PncA. Protein is shown in cyan, primary mutations in red and secondary mutations are shown in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
PncA: Eleven prioritized mutations include the active site mutations like D8H and C138R and secondary mutations A46V/E/P, L116P, W119G/L, L120P, V128A/G predicted as deleterious and destabilizing by structural and functional analyses (Fig. 3B). Among the secondary mutations, L116P, W119G, and L120P were the commonly occurring mutations [78], [79], [80]. We observed the presence of several mutations D33A, I6L, S179C and E181R in pncA that display a susceptible phenotype against PZA and have also been found in MDR strains of M.tb [81], [82], [83]. Their existence in MDR strains comes with a low fitness cost as these mutations preserve the protein functionality and at the same time can be suspected for their role in the establishment of MDR. This phenomenon justifies the existence and natural selection of those susceptible mutations which contribute to the drug resistance without compromising the overall fitness of the pathogen.
DNA gyrase: A total of 21 DNA gyrase mutations were considered for further analysis, amongst which A90V, S91P, D94A/G/N, L96P, D461H, D461N, D472A and N499D were QRDR mutations. The other mutations, R309Q, H368Q, R392C [84], R495H, V125M, R212S, R421H, R446C [85], E501V [21], T511N [86], [87], [88] and G512R [89], [90], [91], [92] were secondary mutations further analyzed in-depth by virtue of their destabilizing and deleterious profiles. Fig. 4A and B represent the primary and secondary mutations prioritized for further studies in DNA gyrase and GyrB ATPase domain respectively. Development of resistance to fluoroquinolones poses the concern of generating XDR-TB strains [85]. A study showed the presence of mutations in gyrA and gyrB in pre-XDR and XDR M.tb strains where all the strains had at least one mutation in QRDR of gyrA [93] and gyrB [94]. Interestingly, the strains with INH mono-resistance also carried gyrA mutations S91P and D94N/Y [93].
Fig. 4.
Distribution of prioritized primary and secondary mutations in (A) DNA gyrase (B) GyrB (ATPase domain). Protein is shown in cyan, primary mutations in red and secondary mutations are shown in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Protein-drug docking and interatomic interactions analyses
The crystal structures and the protein–ligand complexes for WT RpoB, InhA and DNA gyrase (in complex with MFX) obtained after molecular docking had root mean square deviation (RMSD) value of 0 indicating complete alignment of the structures. In case of InhA, the RMSD values obtained after superimposition of WT to I21V and S94A protein–ligand complexes were 0.45 and 0.29, respectively. Table 4 lists molecular docking scores depicting the affinity of WT and mutant proteins, RpoB, InhA, KatG, PncA and DNA gyrase, with the respective drugs. The vibrational entropy values (ΔΔSVib) depicting molecular flexibility and NMA-based values (ΔΔG) indicating overall stability of WT and mutant proteins mutations are provided in Table 5.
Table 4.
Molecular docking scores depicting the affinity of WT proteins and their mutant forms with the respective drugs.
| Protein | Drug | Mutation | Glide score (kcal/mol) |
|---|---|---|---|
| RpoB | Rifampicin | WT | −6.5 |
| A75V | −4.3 | ||
| D441A | −4.1 | ||
| D441C | −1.6 | ||
| D441F | −1.7 | ||
| D441G | −2.0 | ||
| D441N | −6.1 | ||
| D441V | −3.6 | ||
| D441Y | −6.3 | ||
| H451C | −1.1 | ||
| H451N | −5.4 | ||
| H451P | −4.0 | ||
| H451Q | −1.6 | ||
| H451R | −2.0 | ||
| H451T | −1.5 | ||
| S456F | −3.8 | ||
| S456P | −6.0 | ||
| S456Q | −4.3 | ||
| S456W | −0.6 | ||
| S456Y | −4.1 | ||
| V976M | −4.7 | ||
| D1012G | −4.4 | ||
| L1128Q | −6.2 | ||
| I497F (Benign) | −6.9 | ||
| InhA | INH-NAD | WT | −16.8 |
| I21T | −16.6 | ||
| I21V | −13.5 | ||
| V65M | −10.2 | ||
| S94A | −14.1 | ||
| I95T | −15.9 | ||
| I95P | −14.9 | ||
| G141R | −17.0 | ||
| G183R | −15.7 | ||
| A190S | −8.2 | ||
| I194T | −13.0 | ||
| H265D | −17.0 | ||
| V78A (Benign) | −17.2 | ||
| KatG | Isoniazid | WT | −5.0 |
| W90R | −4.5 | ||
| A109V | −4.1 | ||
| R128G | −4.5 | ||
| N138S | −4.8 | ||
| N138D | −4.9 | ||
| D142G | −4.9 | ||
| T275P | −4.2 | ||
| W300G | −4.5 | ||
| S315T | −4.7 | ||
| S315N | −4.9 | ||
| W321G | −4.4 | ||
| W328G | −4.9 | ||
| A614E | −5.0 | ||
| A110V (Benign) | −5.1 | ||
| PncA | Pyrazinamide | WT | −4.7 |
| D8H | −4.7 | ||
| A46V | −4.4 | ||
| A46E | −4.4 | ||
| A46P | −4.6 | ||
| L116P | −4.6 | ||
| W119G | −4.5 | ||
| W119L | −4.4 | ||
| L120P | −4.4 | ||
| V128A | −4.7 | ||
| V128G | −4.6 | ||
| C138R | −4.7 | ||
| A134D (Benign) | −4.8 | ||
| DNA gyrase | Ofloxacin | WT | −7.3 |
| Moxifloxacin | WT | −6.8 | |
| Ciprofloxacin | WT | −8.2 | |
| Levofloxacin | WT | −4.8 | |
| Ofloxacin | A90V | −5.0 | |
| Moxifloxacin | A90V | −4.8 | |
| Ciprofloxacin | A90V | −4.0 | |
| Levofloxacin | A90V | −3.7 | |
| Ofloxacin | S91P | −4.6 | |
| Moxifloxacin | S91P | −3.3 | |
| Ciprofloxacin | S91P | −4.0 | |
| Levofloxacin | S91P | −2.9 | |
| Ofloxacin | D94A | −6.0 | |
| Moxifloxacin | D94A | −4.8 | |
| Ciprofloxacin | D94A | −3.9 | |
| Levofloxacin | D94A | −3.8 | |
| Ofloxacin | D94G | −4.3 | |
| Moxifloxacin | D94G | −4.9 | |
| Ciprofloxacin | D94G | −4.6 | |
| Levofloxacin | D94G | −3.5 | |
| Ofloxacin | D94N | −3.3 | |
| Moxifloxacin | D94N | −4.3 | |
| Ciprofloxacin | D94N | −6.6 | |
| Levofloxacin | D94N | −3.5 | |
| Ofloxacin | L96P | −3.4 | |
| Moxifloxacin | L96P | −5.2 | |
| Ciprofloxacin | L96P | −4.8 | |
| Levofloxacin | L96P | −3.2 | |
| Ofloxacin | R309Q | −3.3 | |
| Moxifloxacin | R309Q | −6.7 | |
| Ciprofloxacin | R309Q | −4.7 | |
| Levofloxacin | R309Q | −3.6 | |
| Ofloxacin | H368Q | −4.8 | |
| Moxifloxacin | H368Q | −5.8 | |
| Ciprofloxacin | H368Q | −6.0 | |
| Levofloxacin | H368Q | −3.1 | |
| Ofloxacin | R392C | −5.4 | |
| Moxifloxacin | R392C | −5.3 | |
| Ciprofloxacin | R392C | −6.7 | |
| Levofloxacin | R392C | −2.9 | |
| Ofloxacin | R495H | −3.5 | |
| Moxifloxacin | R495H | −3.2 | |
| Ciprofloxacin | R495H | −4.2 | |
| Levofloxacin | R495H | −3.6 | |
| Ofloxacin | R446C | −4.3 | |
| Moxifloxacin | R446C | −4.7 | |
| Ciprofloxacin | R446C | −5.8 | |
| Levofloxacin | R446C | −3.3 | |
| Ofloxacin | D461H | −4.9 | |
| Moxifloxacin | D461H | −3.7 | |
| Ciprofloxacin | D461H | −4.3 | |
| Levofloxacin | D461H | −4.1 | |
| Ofloxacin | D461N | −3.6 | |
| Moxifloxacin | D461N | −2.8 | |
| Ciprofloxacin | D461N | −4.3 | |
| Levofloxacin | D461N | −4.3 | |
| Ofloxacin | D472A | −5.1 | |
| Moxifloxacin | D472A | −5.2 | |
| Ciprofloxacin | D472A | −6.3 | |
| Levofloxacin | D472A | −3.7 | |
| Ofloxacin | N499D | −2.8 | |
| Moxifloxacin | N499D | −5.4 | |
| Ciprofloxacin | N499D | −6.1 | |
| Levofloxacin | N499D | −4.0 | |
| Ofloxacin | E501V | −2.9 | |
| Moxifloxacin | E501V | −3.4 | |
| Ciprofloxacin | E501V | −4.9 | |
| Levofloxacin | E501V | −3.2 | |
| Ofloxacin | T511N | −4.2 | |
| Moxifloxacin | T511N | −4.7 | |
| Ciprofloxacin | T511N | −4.6 | |
| Levofloxacin | T511N | −3.5 | |
| Ofloxacin | G512R | −2.1 | |
| Moxifloxacin | G512R | −2.2 | |
| Ciprofloxacin | G512R | −4.8 | |
| Levofloxacin | G512R | −3.5 | |
| Ofloxacin | S95T (Benign) | −7.3 | |
| Moxifloxacin | S95T (Benign) | −6.8 | |
| Ciprofloxacin | S95T (Benign) | −8.2 | |
| Levofloxacin | S95T (Benign) | −4.8 | |
| GyrB (Nter), ATPase domain | Ofloxacin | WT | −5.5 |
| Moxifloxacin | WT | −5.7 | |
| Ciprofloxacin | WT | −5.0 | |
| Levofloxacin | WT | −3.7 | |
| Ofloxacin | V125M | −5.5 | |
| Moxifloxacin | V125M | −2.7 | |
| Ciprofloxacin | V125M | −7.6 | |
| Levofloxacin | V125M | −3.6 | |
| Ofloxacin | R212S | −4.0 | |
| Moxifloxacin | R212S | −4.1 | |
| Ciprofloxacin | R212S | −4.4 | |
| Levofloxacin | R212S | −1.1 | |
| Ofloxacin | R421H | −6.5 | |
| Moxifloxacin | R421H | −5.6 | |
| Ciprofloxacin | R421H | −6.5 | |
| Levofloxacin | R421H | −2.2 |
Table 5.
Vibrational entropy values depicting molecular flexibility and ΔΔG values indicating overall stability of WT and mutant proteins mutations.
| Protein | Mutation | NMA Based Predictions (ΔΔG ENCoM in kcal/mol) | Outcome | ΔΔSVib ENCoM (kcal.mol-1.K-1) | Molecule flexibility Outcome |
|---|---|---|---|---|---|
| RpoB | A75V | −0.05 | Destabilizing | 0.063 | Increase |
| D441F | 0.039 | Destabilizing | −0.049 | Decrease | |
| D441N | −0.181 | Destabilizing | 0.227 | Increase | |
| D441V | −0.225 | Destabilizing | 0.281 | Increase | |
| D441Y | 0.313 | Destabilizing | −0.391 | Decrease | |
| D441A | −0.252 | Destabilizing | 0.315 | Increase | |
| D441G | −0.469 | Destabilizing | 0.586 | Increase | |
| D441C | −0.197 | Destabilizing | 0.246 | Increase | |
| H451R | −0.031 | Destabilizing | 0.039 | Increase | |
| H451N | −0.563 | Destabilizing | 0.703 | Increase | |
| H451Q | −0.245 | Destabilizing | 0.307 | Increase | |
| H451C | −0.295 | Destabilizing | 0.369 | Increase | |
| H451P | −0.545 | Destabilizing | 0.682 | Increase | |
| H451T | −0.373 | Destabilizing | 0.467 | Increase | |
| S456F | 0.273 | Destabilizing | −0.342 | Decrease | |
| S456Q | 0.098 | Destabilizing | −0.123 | Decrease | |
| S456P | −0.059 | Destabilizing | 0.074 | Increase | |
| S456W | 0.372 | Destabilizing | −0.465 | Decrease | |
| S456Y | 0.278 | Destabilizing | −0.348 | Decrease | |
| V976M | 0.397 | Destabilizing | −0.496 | Decrease | |
| D1012G | −0.161 | Destabilizing | 0.201 | Increase | |
| L1128Q | −0.266 | Destabilizing | 0.332 | Increase | |
| InhA | I21V | −0.344 | Destabilizing | 0.43 | Increase |
| I21T | −0.282 | Destabilizing | 0.352 | Increase | |
| V65M | 0.028 | Destabilizing | −0.035 | Decrease | |
| S94A | −0.242 | Destabilizing | 0.303 | Increase | |
| I95P | −0.498 | Destabilizing | 0.623 | Increase | |
| I95T | −0.272 | Destabilizing | 0.339 | Increase | |
| G141R | 0.613 | Stabilizing | −0.767 | Decrease | |
| G183R | 0.08 | Destabilizing | −0.1 | Decrease | |
| A190S | 0.225 | Destabilizing | −0.281 | Decrease | |
| I194T | −0.085 | Destabilizing | 0.106 | Increase | |
| H265D | −0.073 | Destabilizing | 0.092 | Increase | |
| KatG | W90R | −0.664 | Destabilizing | 0.83 | Increase |
| A109V | 0.216 | Destabilizing | −0.27 | Decrease | |
| R128G | −0.997 | Destabilizing | 1.246 | Increase | |
| N138S | 0.102 | Destabilizing | −0.127 | Decrease | |
| N138D | 0.138 | Destabilizing | −0.172 | Decrease | |
| D142G | −0.577 | Destabilizing | 0.721 | Increase | |
| T275P | −0.03 | Destabilizing | 0.039 | Increase | |
| W300G | −2.195 | Destabilizing | 2.744 | Increase | |
| S315T | 0.345 | Destabilizing | −0.432 | Decrease | |
| S315N | 0.164 | Destabilizing | −0.205 | Decrease | |
| W321G | −1.194 | Destabilizing | 1.492 | Increase | |
| W328G | −1.606 | Destabilizing | 2.008 | Increase | |
| A614E | 0.142 | Destabilizing | −0.178 | Decrease | |
| PncA | D8H | 0.128 | Destabilizing | −0.16 | Decrease |
| A46V | 0.2 | Destabilizing | −0.249 | Decrease | |
| A46P | 0.203 | Destabilizing | −0.254 | Decrease | |
| A46E | 0.148 | Destabilizing | −0.185 | Decrease | |
| L116P | −0.833 | Destabilizing | 1.041 | Increase | |
| L120P | −0.274 | Destabilizing | 0.342 | Increase | |
| W119G | −1.153 | Destabilizing | 1.442 | Increase | |
| W119L | −0.711 | Destabilizing | 0.889 | Increase | |
| V128A | −0.43 | Destabilizing | 0.537 | Increase | |
| V128G | −0.738 | Destabilizing | 0.922 | Increase | |
| C138R | 0.372 | Destabilizing | −0.465 | Decrease | |
| S67L(susceptible) | 0.22 | Destabilizing | −0.274 | Decrease | |
| GyrA | A90V | 0.238 | Destabilizing | −0.298 | Decrease |
| A90S | −0.016 | Destabilizing | 0.019 | Increase | |
| A90T | 0.128 | Destabilizing | −0.16 | Decrease | |
| A90G | 0.034 | Destabilizing | −0.043 | Decrease | |
| A90C | 0.059 | Destabilizing | −0.073 | Decrease | |
| S91P | −0.053 | Destabilizing | 0.066 | Increase | |
| S91A | −0.103 | Destabilizing | 0.129 | Increase | |
| D94G | 0.015 | Destabilizing | −0.019 | Decrease | |
| D94H | 0.237 | Destabilizing | −0.297 | Decrease | |
| D94N | 0.004 | Destabilizing | −0.005 | Decrease | |
| D94V | 0.080 | Destabilizing | −0.100 | Decrease | |
| D94A | −0.013 | Destabilizing | 0.016 | Increase | |
| D94F | 0.230 | Destabilizing | −0.287 | Decrease | |
| D94Y | 0.375 | Destabilizing | −0.469 | Decrease | |
| L96P | −0.358 | Destabilizing | 0.447 | Increase | |
| R309Q | 0.042 | Destabilizing | −0.053 | Decrease | |
| H368Q | −0.287 | Destabilizing | 0.358 | Increase | |
| R392C | −0.149 | Destabilizing | 0.186 | Increase | |
| R392L | −0.086 | Destabilizing | 0.107 | Increase | |
| R495H | −0.115 | Destabilizing | 0.144 | Increase | |
| S95T | −0.034 | Destabilizing | 0.043 | Increase | |
| GyrB | V125M | 0.399 | Destabilizing | −0.499 | Decrease |
| R212S | −0.619 | Destabilizing | 0.773 | Increase | |
| R421H | 0.026 | Destabilizing | −0.032 | Decrease | |
| R446C | −0.557 | Destabilizing | 0.697 | Increase | |
| D461N | −0.037 | Destabilizing | 0.046 | Increase | |
| D461H | −0.045 | Destabilizing | 0.056 | Increase | |
| D472A | −0.126 | Destabilizing | 0.157 | Increase | |
| D472H | −0.07 | Destabilizing | 0.088 | Increase | |
| D472N | −0.107 | Destabilizing | 0.134 | Increase | |
| N499D | −0.048 | Destabilizing | 0.06 | Increase | |
| N499Y | 0.23 | Destabilizing | −0.288 | Decrease | |
| N499T | 0.058 | Destabilizing | −0.072 | Decrease | |
| N499S | 0.021 | Destabilizing | −0.027 | Decrease | |
| T500A | −0.088 | destabilizing | 0.110 | Increase | |
| T500N | −0.085 | destabilizing | 0.106 | Increase | |
| E501V | −0.134 | Destabilizing | 0.168 | Increase | |
| E501D | −0.318 | Destabilizing | 0.397 | Increase | |
| A504V | 0.307 | Destabilizing | −0.384 | Decrease | |
| T511N | −0.247 | Destabilizing | 0.309 | Increase | |
| G512R | 0.597 | Stabilizing | −0.746 | Decrease | |
| M291I | −0.348 | Destabilizing | 0.435 | Increase |
RpoB: Molecular docking of WT and 22 mutant RpoB structures with RIF was performed to study the effect of mutations on RpoB-RIF binding. Of the total 22 mutations, mutations at all codon positions reduced the binding affinity of RIF to RpoB except in the case of D441Y, S456P and L1128Q where glide docking scores (in kcal/mol) were more or less similar to WT (-6.5). Large difference in docking scores was observed for S456W (-0.6) followed by D441C (-1.6), H451C (-1.1), H451Q (-1.6), H451T (-1.5) and H451R (-2.0). Docking scores for D441Y, S456P and L1128Q corresponded to −6.3, −6.0 and −6.2 and did not show any significant reduction in binding affinity. The unexplored secondary mutations A75V (-4.3), V976M (-4.7) and D1012G (-4.4) also had reduced docking scores determining their imperative role in drug resistance. A benign mutation I497F had a slightly higher than WT docking score equivalent to −6.9.
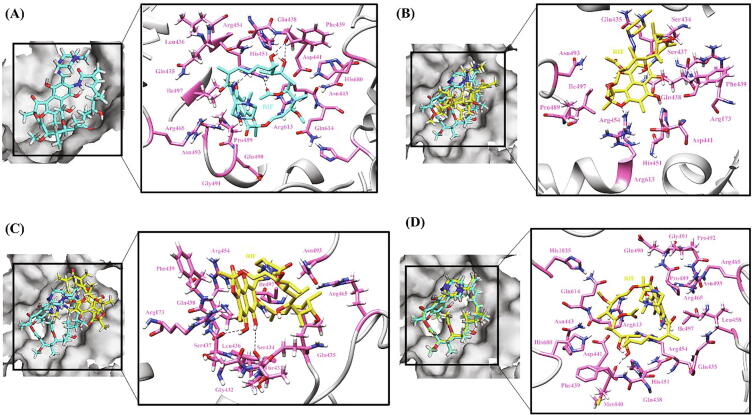
The RIF binding pocket is lined by a group of hydrophobic residues, Leu436, Leu458, Gly459 and Ile497 on one side and a polar Gln435 residue at the other end. In WT RpoB, residues participating in H-bonds with RIF include Phe439 (2 bonds) and Arg454 (2 bonds) along with other key residues Gln438, Asp441, His451, Ser456, Asn493 and Ile497 forming hydrophobic network (Fig. 5A). The influence of RRDR mutations at codon positions 441 (D441F/N/V/A/G/C), 451 (H451R/N/Q/C/P/T) and 456 (S456F/Q/P/W/Y) has been studied in length for their destabilizing role and reduced binding affinity of RIF to RpoB [95].
Fig. 5.
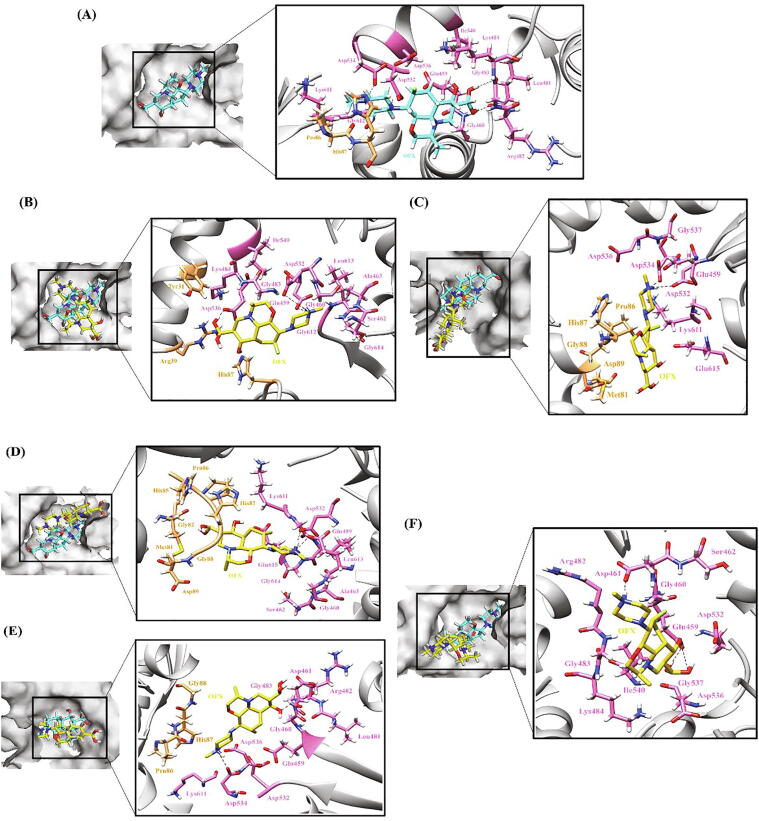
Hydrogen bonding and hydrophobic interactions in RpoB-RIF complex. (A) WT (B) A75V (C) D1012G (D) L1128Q. The left panel is a view of the drug inside the binding pocket of protein and the right panel illustrates protein residues interacting with the drug. RIF is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in pink. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In RIF-resistant isolates, 70 to over 90% of RIF-resistant isolates harbor mutations at RpoB codons 441, 451 and 456 [96]. However, the frequencies of SNPs in these three codons are variable in different geographic regions [96]. In case of mutations D441A/C/F/G/N/V/Y, substitution to F/G/V/C resulted in highly reduced binding affinity and moderately reduced in case of A/N/Y. In case of D441A/N mutation, the loss of H-bond with Arg613 and oxygen of RIF in case of substitution to Ala/Asn led to an altered orientation of ligand and disrupted proper drug binding. No change in ligand position was noticed upon replacement of Asp with Tyr, D441Y, still the resistance induced might be due to the low cell wall permeability since this exclusion barrier is responsible for natural resistance of some strains [97], [98]. In D441C, the drug formed 3H-bonds but with different residues from the WT, which was due to breakage of H-bonds with Phe439 and Arg613 and a drastic reduction in binding affinity for RIF. The D441C substitution changed the position of RIF and exposed it to the solvent in response to a significant reduction in hydrophobic interacting residues further making the binding pocket inaccessible to RIF. In case of D441F mutation, decreased molecular flexibility (ΔΔSVib ENCOM −0.049), alterations in H-bonds as well as decreased hydrophobicity along with large conformational change in the ligand was noticed which restricted the ligand from reaching the catalytic site. The D441V mutation increased the molecular flexibility (ΔΔSVib ENCOM 0.281), the H-bond between carbonyl of RIF and Arg454 shifted to H-bond between oxygen and Arg454 leading to ligand disorientation in the binding pocket and a considerable decrease in ligand binding affinity. All the H-bonds were lost in D441G mutation which did not let the ligand to reach the depth of the binding pocket and moved it towards the surface which is also reflected from the highly reduced docking score as compared to WT.
In RpoB protein, His451 is the most important β-subunit residue, involved in RNAP/RIF complex, through hydrogen bonding with the RIF ring system oxygen atoms as determinants. His451 mutants are considered “affinity mutants” since they interfere with crucial protein polar/hydrophobic RNAP/RIF interactions [99]. The substitutions in codon 451 (H/Y/D/R/L/P) in RpoB are very common in M.tb clinical isolates highly resistant to RIF [100], [101], [102]. Substitution of His to any other AA is quite rare as it is not well physiologically favored due to its chemical properties [103]. The mutation of His451 to D/R/Y has been well studied for the mechanism of resistance [104], [105], [106]. However, the other variants identified to be deleterious to function and destabilizing to structure by our analysis have not been well explored. In the case of H451C/Q/T mutation, the weak interatomic interactions in the mutants lead to an increase in the flexibility of the native protein reaching 0.369, 0.307 and 0.467 (ΔΔSVib ENCOM) for H451C/Q/T, respectively. A considerable change in ligand orientation in the binding pocket of RpoB and loss of H-bonds was seen making RIF exposed to the solvent and leading to much decrease in docking score as compared to WT and other mutations. An overlap in the conformation of ligand to that of WT was found in H451N mutation which is also evident from their almost similar docking scores. The residues in H451P mutation are changing from polar to hydrophobic which resulted in loss of H-bonds with Arg613 and key residue Phe439 and thus change in ligand orientation and an increase in flexibility (ΔΔSVib ENCOM 0.682) of the native protein.
Mutation at codon 456 is another predominant alteration as reported in previous studies [95], [96]. At Ser456, W/F/Q are most frequently occurring substitutions [96] and their mechanisms of resistance have already been explored. Amongst the other variants (S456P/Y), in S456P since Ser is known to mimic Pro, this mutation induced hardly any change in conformation of ligand which is also clear from its similar docking score to WT along with slight increase in molecular flexibility (ΔΔSVib ENCOM 0.074). In the case of S456Y mutation, the single aromatic ring of RIF moved in a direction opposite to WT and formed H-bond with Arg493. All the H-bonds formed amid WT RpoB and RIF were lost leading to a considerable decrease in the docking score as compared to WT.
Amongst the unexplored secondary mutations, in the A75V mutation, only one H-bond, with Arg454, was retained in contrast to 4H-bonds in WT protein–ligand complex (Fig. 5B). Val contains two non-hydrogen substituents i.e., two carbons which increases the bulkiness near the protein backbone resulting in confined movement of nearby residues. So, under the influence of Val, protein flexibility (ΔΔSVib ENCOM 0.063) increased and the position of ligand within the binding site got disturbed further affecting the binding capacity of ligand to the protein. In case of V976M mutation, substitution from Val to Met disturbed the interactions with neighboring protein residues as a result of which the ligand, after entering the pocket, rotated and moved in an opposite direction from the binding pocket and thus low ligand affinity for the protein. As is evident from Fig. 5C, in the case of mutation from Asp (polar) to Gly (non-polar) at position 1012, a H-bond with Arg454, the key residue of binding pocket, is broken which forced the ligand to move away from the hydrophobic binding pocket subsequently leading to reduced affinity of RIF for RpoB thus increasing the flexibility of molecule (ΔΔSVib ENCOM 0.201). The L1128Q mutation did not largely impact ligand binding and all the H-bonds and hydrophobic interactions were retained except that the mutation pushed the ligand marginally inside the binding pocket (Fig. 5D) but an overall increase in randomness (ΔΔSVib ENCOM 0.332) in the protein structure was observed.
InhA: To investigate the impact of mutations on the binding of activated INH with InhA i.e., INH-NAD adduct, molecular docking was performed. All the mutant structures showed reduced docking scores (in kcal/mol) as compared to WT score, −16.8. Mutations I21V (-13.5), S94A (-14.1), I95P (-14.9), I95T (-15.9), G183R (-15.7) and I194T (-13.0) resulted in a low-affinity INH-NAD binding. A large difference in binding affinity was observed for V65M (-10.2) and A190S (-8.2). The score for I21T (-16.6), G141R (-17.0) and H265D (-17.0) was more or less comparable to the WT. The docking score for the benign mutation V78A was more than WT and corresponded to −17.2.
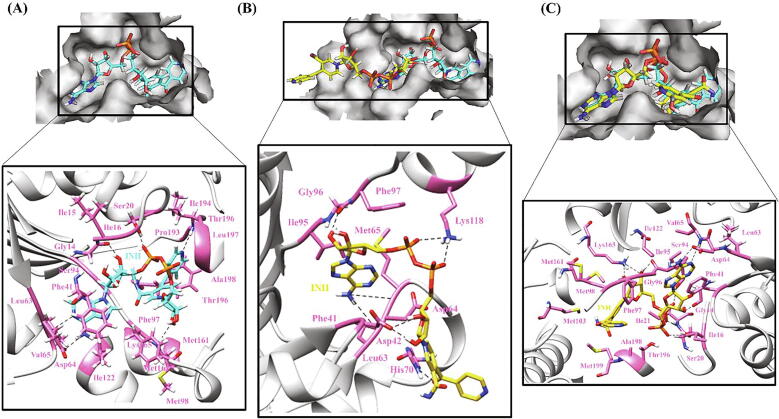
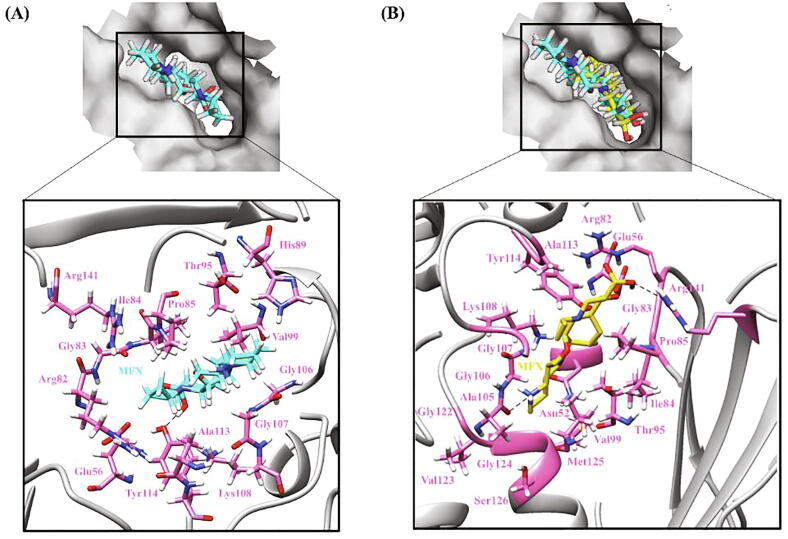
The strong binding in WT is the result of 6H-bonds and 15 hydrophobic interactions formed between WT InhA and INH-NAD (Fig. 6A). INH-NAD is buried in a largely hydrophobic binding pocket in InhA protein thus changes induced in hydrophobicity by certain mutations might have been responsible for decreased binding affinity of activated INH. In I21T, a loss of few hydrophobic residues owing to the weakly polar nature of Thr as well as bulky side chain restricting the movement of neighboring backbone residues was observed. In case of I21V (active site) mutation, though Ile and Val are equally hydrophobic, minor differential changes were observed in terms of interactions with the adjacent residues but an overall increase in randomness (ΔΔSVib ENCOM 0.430) in the protein structure was seen. Ile is known to be highly important in protein–ligand binding. Val at the same time has high frequency of occurrence near the ligand [107] in absence of which INH-NAD ligand changed its coordinates with respect to its conformation in WT leading to low-affinity ligand binding. Major loss of interatomic interactions was observed in I194T (ΔΔSVib ENCOM 0.106) as compared to I95T (ΔΔSVib ENCOM 0.339) depicting its upper hand in modifying the environment inside the binding pocket. In case of I194T, a H-bond formed between Ile194 and INH-NAD was no longer seen and the ligand moved in direction opposite to that in WT on encountering Thr instead of Ile. In case of I95P, replacement of Ile to Pro did not change the H-bonds and hydrophobic interactions however increased the solvent exposure due to which the position of ligand got disturbed reducing its strength of binding to InhA. In another active site mutation V65M, substitution of Val with Met accompanied development of H-bonds with adjoining residues which consequently augmented rigidity (ΔΔSVib ENCOM −0.035) in the affected area and narrowing of the binding pocket. A large reduction in hydrophobicity and breaking of the H-bond with Val and formation with Met in the V65M mutation induced a huge conformational change owing to which the ligand did not reach the hydrophobic pocket and settled on the surface of protein (Fig. 6B).
Fig. 6.
Hydrogen bonding and hydrophobic interactions in InhA-INH-NAD complex. (A) WT (B) V65M (C) A190S. The top panel is a view of the drug inside the binding pocket of protein and the bottom panel illustrates protein residues interacting with the drug. INH-NAD is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in pink. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Mutation S94A increased protein flexibility (ΔΔSVib ENCOM 0.303) by change in interatomic interactions, specifically formation of hydrophobic interactions owing to the hydrophobic nature of Ala. With no Ser at 94 position, the ligand rotated at phosphate linkages and changed its course to reach the binding pocket compromising interactions with the neighboring residues and did not fit well inside the binding pocket. S94A mutation has been shown to reduce the binding of INH-NADH and increase the IC50 and Ki by 17 and 30 times respectively as compared to WT [108]. In secondary mutations, G141R and G183R, substitution from Gly (smallest AA) to Arg (polar) led to the formation of a few new interactions with the neighboring residues which made the affected area quite stable. Also being present away from the binding pocket, the resulting change in flexibility did not influence ligand binding much compared with WT as is reflected from the overlapping coordinates of ligands in WT and mutant protein–ligand complexes. In the A190S mutation, overall molecular flexibility was decreased (ΔΔSVib ENCOM −0.281) due to the change from hydrophobic residue Ala to Ser which is a polar AA with tendency to form H-bonds. A considerable rotation was seen in INH-NAD from its position in WT since the ligand on coming across Ser got exposed to solvent and did not fully reach the binding pocket (Fig. 6C). Exchange of His with any AA is mostly non-favorable as its pKa is very close to that of physiological pH and thus the binding capacity of ligand was not hampered and docking score observed in H265D mutation (-17.0 kcal/mol) was similar to WT.
KatG: Molecular docking of WT KatG and 13 mutant structures with INH was performed to study the effect of mutations on KatG-INH binding. As compared to WT (-5.0), R128G (-4.5), T275P (-4.2), W300G (-4.5) and W321G (-4.4) mutants had lesser docking scores (in kcal/mol) representing their reduced INH binding affinity. The scores for N138S (-4.8), N138D (-4.9), D142G (-4.9), W328G (-4.9), S315T (-4.7), S315N (-4.9) and A614E (-5.0) were more or less comparable to the WT. The docking scores of the unexplored secondary mutations, W90R (-4.5) and A109V (-4.1) determine their possible distinctiveness in minimizing the INH-oxidase activity of KatG and further leading to resistance against INH. Almost similar to WT docking score was obtained in case of benign mutation A110V (-5.10).
The strong binding affinity of INH to WT KatG protein can be explained by 8 hydrophobic network forming residues present around the ligand (INH) along with the 1H-bond between His270 and NH2 group of INH and one pi-pi (Trp107) interaction (Fig. 7A). In the mutations, T275P and D142G, Thr and Asp are changing to non-polar and neutral residues known to induce flexibility (ΔΔSVib ENCOM 0.039) and (ΔΔSVib ENCOM 0.721) in protein, respectively. In the case of T275P, the mutant residues formed an intricate network of interactions with surrounding AAs changing the shape of the binding pocket and leading to a loss in INH interactions which certainly changed the orientation of INH thereby affecting its absolute binding and activation.
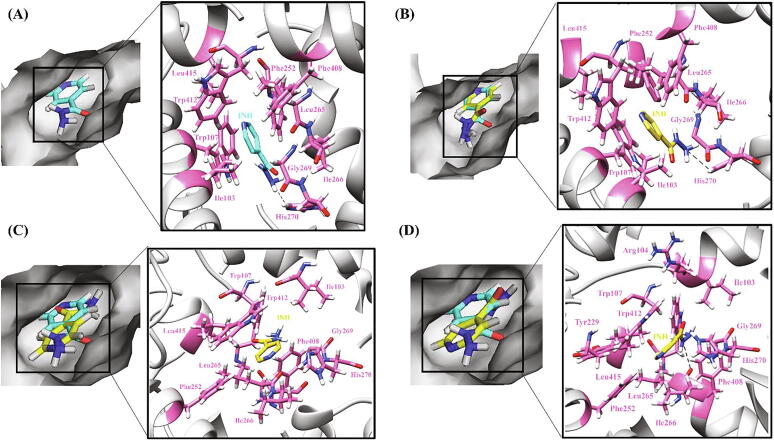
Fig. 7.
Hydrogen bonding and hydrophobic interactions in KatG-INH complex. (A) WT (B) W90R (C) A109V (D) W321G. The left panel is a view of the drug inside the binding pocket of protein and the right panel illustrates protein residues interacting with the drug. INH is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in pink. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Substitution of a nonpolar AA (Trp) with a positively charged residue (Arg) as in matter of W90R, led to a huge loss of interactions with neighboring residues and an increase in local flexibility (ΔΔSVib ENCOM 1.246) although there was no change in orientation of the ligand (Fig. 7B). In the case of A109V mutant structure, more hydrophobic and hydrogen interactions were observed to be made by Val with the surrounding residues along with the increase in rigidity of nearby area (ΔΔSVib ENCOM −0.270) which can result in the constriction or change in the shape of binding pocket [109]. Furthermore, the ligand was observed to adhere away from the hydrophobic core of the binding pocket subsequently leading to its improper binding and activation (Fig. 7C).
In context with the mutations R128G, W300G and W321G, the WT residues are being replaced by small AA Gly which is aliphatic and devoid of any side chains which is thought as a contributory factor for the partial rotation of drug inside the enzyme binding pocket. Moreover, the replacement of an aromatic and high molecular weight AA Trp with Gly as in the case of W300G and W321G (Fig. 7D) mutant models led to the depletion of necessary interactions with the surrounding residues required to stabilize the binding pocket. All these variations collectively contributed to disrupting the ligand binding affinity, by changing its orientation away from the catalytic site to a more solvent exposed area. These mutations are therefore associated with low disturbances in drug binding and less INH resistance (0.5–1 µg/ml) [110], [111]. However, more experimental studies are needed to confirm this hypothesis.
In the mutation A614E, the codon position Ala614 is localized near the C-ter domain of KatG. This conserved codon position appears to mutate and code marginally favorable negatively charged AA Glu to defend bacteria against INH stress. The substitution brings challenges to the protein in terms of reduction in halogen and hydrogen bonds in mutated structure but subsequently the increment in the number of new hydrophobic and weak hydrogen interactions seems to compensate for the onsite loss. This somehow favors the drug to properly adjust inside the binding pocket and thus forms a stable protein-drug complex which explains this substitution as functionally deleterious and overall, structurally destabilizing yet not hampering binding affinity of INH much as compared to the WT. S315T and S315N mutations turned out to be deleterious to protein function but had a stabilizing and mildly destabilizing effect on protein structure respectively. Still, they are most frequently appearing mutations in the drug resistant isolates. Both the mutations increased the interactions of mutated residue with the neighboring AAs which accompanied stability in the enzyme binding pocket. Apart from the minor alterations in the position of nearby residues, a prominent differential change in orientation of the ligand or conformation of the binding pocket was not seen between WT and mutant protein–ligand complexes since this was a conservative substitution. Still the resistance induced by substitution of Ser with Thr at codon position 315 might be due to some other mechanisms like interference in the electron-transfer chain triad and proposed mechanism of threonine steric constraint further helping this mutation in reducing INH-oxidase activity and causing resistance [112]
PncA: Docking studies performed to assess the impact of mutations on protein-drug binding exhibited low ligand binding propensity in the case of all the mutations except D8H, V128A and C138R. The docking score (in kcal/mol) for the WT was found to be −4.7 while the score in case of mutant protein–ligand complexes ranged from −4.4 to −4.7. The benign mutation A134D had higher docking score of −4.8 as compared to the WT.
Hydrophobic interactions are key driving factors in maintaining the conformational integrity of the binding pocket in PncA which facilitates prompt activation of PZA. The residues, Asp8, Lys96 and Cys138 form catalytic triad in PncA where Cys138, upon activation by Asp8, initiates the covalent catalysis by acting as a nucleophile. Lys96 on the other hand stabilizes either the Asp8 or the thiolated form of Cys138 during the catalysis mechanism [113]. The Fe+2 ion has a catalytic role than a structural one and is coordinated inside the active site by residues His57, His71, and His137 [43].
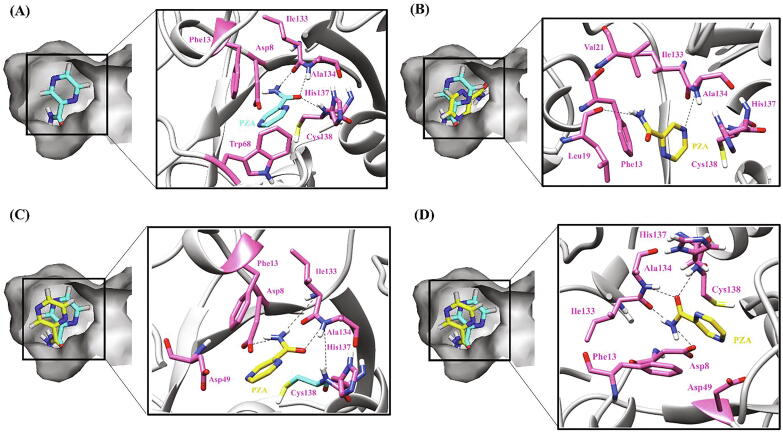
In case of WT PncA, besides the hydrophobic environment, the residues Ala134 and Cys138 form two H-bonds with the carbonyl oxygen atom and Ile 133 forms one with the terminal amino group of PZA thus contributing to the stability of the complex. Furthermore, the polar interactions between Asp8, His71 and His137 of the protein and PZA together with the pi-pi interaction between aromatic Trp68 and aromatic ring of PZA reinforce the integrity of the drug-protein complex and assist the PZA activation (Fig. 8A).
Fig. 8.
Hydrogen bonding and hydrophobic interactions in PncA-PZA complex. (A) WT (B) W119L (C) A46E (D) A46V. The left panel is a view of the drug inside the binding pocket of protein and the right panel illustrates protein residues interacting with the drug. PZA is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in pink. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Substitution of Asp with His as in the case of D8H mutant structure resulted in the formation of more intense inter-atomic interactions (hydrophobic and hydrogen) with the surrounding residues and altered environment of the binding pocket. The sustained rigidity depicts lower flexibility in D8H mutant with ENCOM values for ΔΔSVib reaching down to −0.16. Upon ligand binding, D8H mutation did not interfere much with the hydrophobic structure of the binding pocket and thus all polar interactions and the H-bonds with PZA were retained as found in WT. An increase in docking score can be due to the rotation of the aromatic ring of PZA which caused the ligand to come in close proximity to Trp68 which besides the pi-pi interaction enhanced the hydrophobicity of the pocket, thus enabling Trp68 to firmly stack with nearby hydrophobic residues. In WT, change from Cys138 to Arg enriched the hydrophobic environment of the binding pocket. Moreover, the tendency of Arg to form salt-bridges with Asp placed the Asp49 in the vicinity of PZA, helps in the restoration of key H-bond with its carbonyl oxygen group enabling the mutant C138R to maintain an efficient docking score of −4.7. The stable and compact binding can also be substantiated by the surge in interatomic interactions in the mutant which explains the lower flexibility value, ΔΔSVib ENCOM of −0.465.
Mutations W119G/L presented comparable results by keeping intact most of the interactions visible in WT. In W119G, a change in the position of Asp8 and Phe13 caused a gentle constriction in the binding site thus bringing down the docking score to −4.5 only. On the other hand, a shift of docking score to −4.4 was seen in W119L caused by stacking of interacting hydrophobic residues and loss of the critical H-bond at Cys138 eventually disorienting the drug within the binding site (Fig. 8B). Besides the difference in the stability of the static conformations of W119G/L, the complex of both mutations did not form the multitudes of interactions as the flexibility values were recorded to be 1.442 and 0.889 in W119G and W119L, respectively. Mutations A46E/P/V display variations of docking capacities among each other at the single codon position. Although, due to the rise in hydrophobic interactions and H-bonds within the complex, all the three mutants replicated the results of WT in displaying reduced complex flexibilities with the ΔΔSVib ENCOM values of −0.254, −0.185 and −0.249 in A46E, A46P and A46V, respectively. Compared to the WT, A46P showed the minute shift in docking score, −4.6, due to its presence near the loop forming residues (51–77) that comprise the mouth of the binding site [43]. On the other hand, the docking scores of A46E (-4.4) and A46V (-4.4) were lower than the WT due to fewer hydrophobic residues in these two mutants and also because of the absence of pi-pi interaction at Trp68 compared to the WT. Fig. 8C and 8D illustrate H-bonding and hydrophobic interactions between PZA and A46E and A46V mutant proteins, respectively.
The docking scores in V128A and V128G were close to that of WT and found to be −4.7 and −4.6, respectively. The increased entropies associated with Ala and Gly induced flexibility in the protein with ΔΔSVib ENCOM values being 0.537 and 0.992 each for V128A and V128G. In V128A, the altered positions of hydrophobic Phe13 and Leu19 residues turned out to be favorable for the drug and enhanced its binding with PncA. However, the change in positions of residues, Asp8 and Phe13, in case of V128G resulted in the constriction of the drug binding site but a polar interaction by the nearby His57 slightly restored the binding stability resulting in a very little change from the WT.
The docking score in L120P is −4.4 which shows ineffective binding compared to the WT. The mutation resulted in the loss of crucial Ile133 H-bond and pi-pi interaction with Trp68 and an increase in hydrophobic residues causing deformity in the binding site and thus ligand disorientation. The impact of fewer H-bonds is also reflected from the increased flexibility value of 0.342 in case L120P. The docking score of L116P was recorded to be −4.6 which signals the unaffected interaction of a mutant in comparison to WT except a slight positional movement in Phe13 and Leu19. Being hydrophobic, they tend to remain shifted towards the hydrophobic clusters which causes the slight constriction within the binding site. Moreover, the reduced interatomic interactions soared the flexibility score to 1.041.
DNA gyrase: The docking scores (in kcal/mol) for WT were found to be −7.3, −6.8, −8.2 and −4.8 for OFX, MFX, CIF and LFX, respectively. As compared to WT, ΔG was reduced in A90V (-5.0, −4.8, −4.0, −3.7), S91P (-4.6, −3.3, −4.0, −2.9), D94A (-6.0, −4.8, −3.9, −3.8), D94G (-4.3, −4.9, −4.6, −3.5) and D94N (-3.3, −4.3, −6.6, −3.5) of GyrA and D461N (-3.6, −2.8, −4.3, −4.3), D461H (-4.9, −3.7, −4.3, −4.1), N499D (-2.8, −5.4, −6.1, −4.0) and E501V (-2.9, −3.4, −4.9, −3.2) of GyrB, exhibiting the reduced binding affinity of drugs (in the order OFX, MFX and CIF) in these mutants. In DNA gyrase, for benign mutation S95T the docking scores for OFX, MFX, CIF and LFX were −6.4, −6.4, −7.2 and −3.5, respectively which were lower in comparison to WT.
Fluoroquinolones were docked in the binding pocket of WT and mutant DNA gyrase which is composed of GyrA-NTD (codons 2–500) and GyrB-CTD (codons 426–675) [44]. The strong interaction between the WT DNA gyrase and the ligands are attributed to the formation of 3 (OFX), 3 (MFX), 2 (CIF) and 3 (LFX) H-bonds, salt bridges and hydrophobic interactions and alterations in these interactions was seen in case of mutants leading to disrupted drug binding (Fig. 9A). The impact of DNA gyrase mutations, A90V, S91P, D94G/A/N, D461N/H, N499D and E501V on drug binding has already been reported [84], [85], [114], [115], [116]. Large scale effect of mutations was observed in case of binding of OFX in contrast to CIF and MOX as OFX behaves as H-bond acceptor more than CIF and MFX [117].
Fig. 9.
Hydrogen bonding and hydrophobic interactions in DNA gyrase-OFX complex. (A) WT (B) L96P (C) D472A (D) R392C (E) R495H (F) R446C. The left panel is a view of the drug inside the binding pocket of protein and the right panel illustrates protein residues interacting with the drug. OFX is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in orange for GyrA-NTD and pink for GyrB-CTD. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The QRDR L96P mutation led to a declined score compared to the WT (Fig. 9B) since Proline is a non-favored substitution residue known to introduce kink in the structure and thus disrupting the protein structure and inducing disorientation of ligand as evident from our analysis of the mutation L96P [57].
R309Q showed a drastic reduction in binding affinity for OFX and CIF as compared to WT. CIF forms a H-bond with Glu459 moving towards GyrA-NTD which leads to breakage of H-bonds with Gly614. Substitution of Arg to Gln induced a significant reduction in hydrophobic interacting residues and made the ligand exposed to the solvent further making it inaccessible to the binding pocket. Negligible changes were observed in the case of MFX and LFX.
As already mentioned in the case of RpoB, substitution from His is not favorable, the same was observed in H368Q mutation [57]. The weak interatomic interactions in the mutant lead to an increase in the flexibility (ΔΔSVib ENCOM 0.358) of the native protein. Further, ligand disorientation in the binding pocket and alterations in H-bonds as well as decreased hydrophobicity were noticed. H368Q mutation had maximum impact on binding of OFX followed by CIF, MFX and least by LFX.
In QRDR mutation D472A, change from Asp to a Ala, a comparatively less bulky AA is reported to be a disfavored substitution [103]. This led to the loss of interactions between the ligand and active residues Asp532, Asp534 and Asp536 of protein, which did not let the ligand reach the binding pocket (Fig. 9C).
In R392C, the substitution of polar and positively charged AA by a non-polar, neutral and low molecular weight residue led to the reduced binding affinity of the drugs as evident from their docking scores, which can also correspond to their high MIC value on DST [84]. Substitution of Arg by Cys resulted in the loss of side chain interaction with the surrounding residues leading to the slight increase in protein flexibility (ΔΔSVib ENCOM 0.186). CIF in complex with mutant protein completely overlapped with CIF-WT complex, a slight reduction in hydrophobicity was found accompanied by increase in H-bonds. Similar was the observation in MFX-R392C complex where MFX shifted slightly more towards GyrA-NTD. However, a major difference was seen in case of OFX and LFX. OFX could not enter the binding pocket and moved towards GyrA-NTD where it settled in a horizontal conformation on the surface (Fig. 9D). The orientation of LFX changed from horizontal in WT to vertical in case of mutant.
Although His is generally considered favorable at active sites and thus induces rigidity and stability, the mutant R495H resulted in decreased molecular flexibility. This may be due to the loss of side chain interactions made by the WT Arg with the neighboring residues. In the mutant protein complexed with OFX (Fig. 9E) and MFX, the drugs could not reach the binding pocket as much as in case of WT. CIF and LFX drifted from the binding pocket and moved away from GyrB and largely towards GyrA.
In secondary mutations, the decrease in docking scores in R446C (-4.3, −4.7, −5.8, −3.3 kcal/mol), T511N (-4.2, −4.7, −4.6, −3.5 kcal/mol) and G512R (-2.1, −2.2, −4.8, −3.5 kcal/mol) depicts altered binding of the drugs within the binding site and needs in-depth evaluation. Similar to R392C, Arg substitution to Cys at position 446 increased the overall flexibility of the molecule (ΔΔSVib ENCOM 0.697), and reduced the overall binding affinity of OFX, MFX, CIF and LFX due to a weaker hydrophobic network and a smaller number of H-bonds. Fig. 9F portrays the positioning of OFX in the binding pocket of DNA gyrase where the drug, in case of mutant R446C, did not reach the binding pocket in its entirety and moved away from GyrA-NTD.
In the T511N mutation, Asn introduced flexibility (ΔΔSVib ENCOM 0.309) in the molecule. The low docking score with drugs is evidence of the failure of the ligand to interact with the Asp532 and Asp536 catalytic residues, driving the ligand disorientation.
In the case of G512R, mutation from non-polar to polar AA resulted in an increase in the interacting residues making the mutant protein more rigid (ΔΔSVib ENCOM −0.746). But in the G512R-drug complex, the drugs, MFX and CIF, were unable to reach the depth of the binding pocket as compared to WT due to the loss in all the H-bonds. A huge influence of this mutation was seen in the case of OFX and LFX where OFX placed itself entirely on the surface of GyrB and LFX totally drifted away from the binding pocket.
With the emergence of DR-isolates with varied genetic composition, enumerating the importance of less focused GyrA-CTD and GyrB-NTD has become equally pivotal. GyrA-CTD is involved in binding and wrapping the DNA [118] but the mutations found in this domain had very lesser effects on the stability, functionality and pathogenicity of the protein by our analyses and thus, were not considered for protein–ligand docking studies. GyrB-NTD harbors the ATPase activity and is known to have binding sites for other drugs like coumarins. Mutations in this domain along with GyrA-NTD and GyrB-CTD have been found to be involved in drug resistance in clinical isolates [118]. Thus, highly destabilizing and deleterious mutations from this domain were analyzed further for their effect on drug binding affinity.
V125M which is an active residue of GyrB-NTD resulted in increased molecular rigidity (ΔΔSVib ENCOM −0.499) as a result of the construction of a multibranched network of bonds with the nearby residues. The mutant protein showed similar affinity for OFX (-5.5) and LFX (-3.6), reduced affinity for MFX (-2.7) and increased for CIF (-7.6) as compared to WT OFX (-5.5), LFX (-3.7), MFX (-5.7) and CIF (-5.0). V125 is part of the ATP lid region and is disordered in nature [119] due to which its substitution by Met induced large conformational changes in binding of drugs. The enhanced stability of the V125M-CIF complex is due to the more hydrophobic environment in case of mutant than WT. With OFX, the mutation led to loss of H-bonds but also increasing the number of hydrophobic residues around the ligand and thus exhibiting similar affinity as WT. LFX completely overlapped in case of WT and mutant and a slightly reduced binding affinity was due to loss of hydrophobic residues in mutant protein–ligand complex. In the V125-MFX complex, the ligand rotated and moved inwards towards Sulphur moiety of Met resulting in formation of new hydrogen bonds where in WT protein, the ligand was entirely surrounded by hydrophobic network forming residues (Fig. 10A and 10B).
Fig. 10.
Hydrogen bonding and hydrophobic interactions in DNA gyrase (ATPase)-MFX complex. (A) WT (B) V125M. The top panel is a view of the drug inside the binding pocket of protein and the right panel illustrates protein residues interacting with the drug. MFX is shown in cyan in WT and yellow in mutants. Hydrogen bonding and hydrophobic residues are shown in pink. Black dashed lines represent hydrogen bonds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In mutant R212S, Arg interacts with the surrounding residues using hydrogen, ionic and weak interactions, thus replacement of this residue with a neutral and less bulky Ser enhances the flexibility of the molecule (ΔΔSVib ENCOM 0.773). Presence of Ser at 212 position led to the slight disorientation of the ligands and more exposure to the solvent thus forming new H-bonds. This was due to the presence of a OH group in its side chain which can form H-bonds with the protein backbone thus mimicking Pro [57].
Mutation R421H had binding affinity similar to WT. The mutant protein R421H was observed to have decreased flexibility (ΔΔSVib ENCOM −0.032) which might be due to the introduction of an aromatic ring by His. However, there was not much difference in the interactions with the neighboring residues in the mutant when compared to the WT protein.
3.7. Protein-protein interaction (PPI) analysis
Numerous PPI were obtained/computed for proteins coded by rpoB, katG, inhA, pncA, gyrA and gyrB genes by taking in account their reported co-expressions, fusions, gene neighborhood and co-occurrence. This was done to look into some crucial unexplored interactions which might affect the normal functioning of the proteins mentioned and subsequently impact the drug resistance.
RpoB: The PPI network of RpoB demonstrated, RpoA and RpoC proteins as its highly interacting and co-expressing partners with the confidence score >=0.9. Mutations in RpoA and RpoC are already known for their significant role in RIF drug resistance owing to which various studies are now exploring the role of RpoA and RpoC as compensatory targets against M.tb [120]. Other interacting partners of RpoB included RpsL, RpsK, RpsC, RpsL, RpsG, RpsH and RpsE known for conferring resistance to streptomycin. Moreover, RpsL generates dysfunctional ribosome due to mutations [121], affecting transcription and translation efficiencies ultimately leading to disruption of the machinery, therefore challenging the transcription-translation coupling [122] while the role of remaining interacting partners is not yet studied. Indeed, observations [123] in clinical populations of bacterial pathogens suggest the existence of undiscovered interactions [124]. Thus, there is an urgent need to uncover the spectrum of mutations that suppress the costs arising from epistatic interactions in multidrug-resistant pathogens.
InhA: The highly interacting partners of InhA as analyzed by the STRING PPI network were found to be FabH, KasA, and FabG. Biosynthesis of mycolic acids involves two distinct pathways FAS-I and FAS-II. The InhA, KasA, and FabG work together in the FAS-II system where FabH serves as a connecting link between the two systems determining its relative importance in the mycolic acid synthesis. The involvement of co-expressing InhA and FabG genes in drug resistance is well established [61]. The mutations present in their regulatory region are known to confer INH and ethionamide (ETH) resistance however, the involvement of FabG coding region in ethionamide resistance is not known. Moreover, the higher expression of essential KasA and its compensatory mutations are known to be present in clinical isolates in response to INH stress. Targeting this particular overexpressed protein in INH resistant isolates where InhA is partially or completely deactivated due to mutations, would further deprive the bacteria of its necessary mycolic acid production pathway. The subsequent deactivation of both the essential InhA and KasA proteins would obstruct bacterial replication and survival [125].
KatG: The results of KatG PPI network demonstrated FurA and Rv1907c proteins as its highly interacting and co-expressing partners with the confidence score >=0.9. KatG and FurA are well-studied proteins and mutations in them are known to be highly associated with INH drug resistance. Conversely, their third co-expression partner Rv1907c, already reported for its suspected role in INH resistance by in silico studies [126], is still an uncharacterized conserved hypothetical protein. Other interacting partners of katG included superoxide dismutase (SodA and SodC) and alkylhydroperoxidase (AhpC) proteins which are involved in superoxide degeneration pathway and are known to partially compensate for the loss of KatG protein activity in INH-resistant strains. AhpC protects the pathogen against hydrogen peroxides and its hyper-expression due to the presence of compensatory mutations has also been linked to various INH-resistant strains [127]. Therefore, it would be interesting to elucidate the action of interaction partners like Rv1907c and AhpC which might reveal more players contributing to INH resistance. Further studies on their possible function inside the pathogen and involvement in acquiring drug resistance could project Rv1907c and AhpC as novel therapeutic targets to counter INH-resistant M.tb.
PncA: String analysis of PncA reported interactions with an uncharacterized protein Rv2044c that scores high for the fusion and co-expression. PncA is known to be present downstream of Rv2044c. A frameshift deletion in Rv2044c has been reported that eliminates its stop codon causing its protein to fuse with the downstream PncA. It results in the complex formation of PncA and Rv2044c with a novel and uncharacterized domain DUF2784 [128]. Such a phenomenon is bound to alter the existing pattern of functionality in PncA. It would be interesting to note the consequences of this event in PncA keeping in mind its association with the drug resistance. The newly explored functions and role of such uncharacterized domains could elucidate the alternative pathways that could be exploited to halt the M.tb pathogenesis.
Another high confidence scoring (co-expression, co-occurrence and vicinity) interacting partner for PncA was bifunctional NAD(P)H-hydrate repair enzyme (nnr). PncA is known to have a role in deamination of nicotinamide into nicotinate, required for the salvage pathway of nucleotide biosynthesis. Meanwhile the protein NNR (NAD(P)H-hydrate) is required for the epimerization of the NAD(P)HX, a damaged form of NAD(P)H, thus providing a crucial metabolic factor required for basic cellular function and biosynthetic pathways. The interaction between PncA and NNR within the genome demands an in-depth analysis of their conjoint role keeping in focus their possible contribution in conferring resistance against the drug PZA [129].
DNA gyrase: Interestingly, all the drug resistance associated proteins included in this study (RpoB, KatG, InhA, and PncA) were found to be high confidence interacting partners of GyrA and GyrB. Any mutations leading to malfunction in these proteins would directly affect the properties of cells and other interacting partners by changing their expression profiles. Further subsequent mutations in interacting proteins [130], [131] may also be responsible for the conversion of mono fluoroquinolone resistant isolates to MDR or XDR. Recombinant proteins, RecA and RecF, the high confidence DNA gyrase interacting proteins are involved in the SOS pathway which is known to be upregulated in M.tb persister cells [132]. Targeting these proteins in recalcitrant bacterial subpopulation can be advantageous in improving the efficacy as well as shortening the anti-TB regimen. Another interacting protein is DNA topoisomerase I (TopA) which is emerging as an attractive anti-mycobacterial drug target against which chemotherapeutic agents are being developed to inhibit transcription in persisters [133].
3.8. MycoTRAP-DB database
Our MycoTRAP-DB is an online database that harbors almost all the mutations present in six putative first- and second-line drug resistance associated genes (rpoB, inhA, katG, pncA, gyrA and gyrB). Other than just assembling the mutations, it also provides information about implications of these mutations on the structures and functions of proteins coded by these genes. The database can be used for searching, retrieving, analyzing and downloading the data of mutations present in drug resistant M.tb clinical isolates. MycoTRAP-DB presents information about mutations which have been studied for diagnosis of drug resistance in TB, however, there are other mutations identified to be pathogenic by various sequence and structure-based tools used in the present study. Future experimental studies can focus on these pathogenic mutations to decipher their possible roles and accountability for causing drug resistance and develop more efficient genotypic methods for diagnosis and detection of drug resistant TB. The database is accessible at http://139.59.12.92.
4. Conclusion
An increase in the number of MDR and XDR-TB cases has aggravated the problem of TB worldwide, urging the need to improvise treatment strategies against DR-TB. The breakneck advances in the genomics and computational biology have led to the upgradation in the genomic and transcriptomic data. With the evolution of the microorganisms, mutations have been incorporated into their genomes which may provide them with the evolutionary and ecological advantages. Extensive data for these mutations are available in various widely known databases, but the lack of the updated database places a vacuum among the investigators approaching the real-time reported data. An in-house database, MycoTRAP-DB, of almost all the reported mutations (till December 2020) in resistant and sensitive strains of M.tb was prepared for the genes rpoB, inhA, katG, pncA, gyrA and gyrB. This database brings most of the dispersed information about drug susceptible or resistant polymorphisms at a single place and would also open up the gateway to utilize this dataset to derive a convincing solution against DR-TB. The study is focused on non-synonymous mutations in specific genes encoding drug targets and any variations, primarily non-synonymous being able to alter protein structure, in these genes could affect the function of protein as well as its affinity for the drug resulting in drug resistance. Moreover, from our comprehensive dataset, we identified several secondary mutations which had a similar impact on protein structure and function as compared to known prevalent mutations in target genes: a finding that prompted us to look into the impact of these mutations on the pathogenesis of TB.
Molecular docking studies conducted to explore the impact of mutations on drug-binding revealed a correlation between the docking scores and reported MIC values for WT and mutant protein-drug complexes. The mutations which were found to have scored lower than their respective WT proteins have also been known to have higher MIC. This reflects a relationship between the structural stability of the complex with the reported MIC values. The RpoB mutations, S456W, D441C, H451C, H451Q, H451T and H451R scored contrastingly lower than WT and these results are also supported by the reported higher MIC values [96]. In case of InhA, the I21T and I21V mutants are found to be associated with promoter region mutation mostly with C-15 T. The MIC for only C-15 T was recorded as 8 µg/ml but with co-occurrence of I21T it increased to 32 µg/ml [134]. The MIC for another low scoring mutation, S94A of InhA was 8 µg/ml [134] against WT 0.2 μg/mL [135]. The lesser docking scores for KatG mutations R128G, T275P, W300G and W321G, as compared to WT, correlate with their known resistance profiles and MIC values derived from INH-DST (1–10 µg/ml) [111], [136]. PncA also followed the similar trend as higher MIC value has been reported for its lowest scoring mutation A46P [137]. The drastic reduction in docking scores of mutations reportedly having higher MIC values was also found in case of GyrA and GyrB against OFX, MFX, LFX and CIF. The low scoring mutations, A90V, S91P, D94A, D94G and D94N of GyrA and D461N, D461H, N499D and E501V of GyrB have been stated to have higher MIC values on DST 0.25 to 2 μg/ml (OFX), 0.06 to 0.5 μg/ml (MFX), LFX 0.12–1 μg/ml and 1–8 mg/L (CIF) [138]. The comparison of MIC of these drugs for the WT M.tb H37Rv (≤0.5 mg/L for OFX, ≤0.125 mg/L for MFX and ≤ 1 mg/L for CIF) [125]. Furthermore, high MIC values have been shown for mutation R392C [84] and the secondary mutations, R446C, T511N and G512R of DNA gyrase [84], [85], [89].
The systematic approach used in this study has the ability to expand our understanding related to the mechanism of resistance associated with a particular mutation. However, the results presented in this study are mere predictions and form the foreground for further experimental validations to establish the impact of the identified mutations. The information can be exploited in future by investigating the resistance profile of the M.tb strain from the clinical sample prior to the drug administration process. The most prevalent and evolutionary important mutations can be used for the advancement of drug designing. Drug repurposing can be done to make use of the FDA approved drugs which may bind to the mutant phenotype of the protein with better binding affinity. Also, combination therapy may be incorporated into the drug designing where the combination of the drugs which inhibits two or more common mutant phenotypes of the protein can be developed.
5. Data Availability
The database, MycoTRAP-DB is publicly accessible at http://139.59.12.92. All the other data are available as supplementary data.
CRediT authorship contribution statement
Pooja Singh: Investigation, Data curation, Writing - original draft. Salma Jamal: Methodology, Investigation, Writing - original draft. Faraz Ahmed: Investigation, Writing - original draft. Najumu Saqib: Investigation. Seema Mehra: Investigation, Writing - original draft. Waseem Ali: Investigation, Writing - original draft. Deodutta Roy: . Nasreen Z. Ehtesham: Conceptualization, Writing - original draft. Seyed E. Hasnain: Conceptualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
S.J. and S.M. acknowledge a Young Scientist Fellowship from the Department of Health Research (DHR), India. S.E.H. is a J.C. Bose National Fellow, Department of Science and Technology, Government of India and Robert Koch Fellow, Robert Koch Institute, Berlin, Germany.
Funding
S.E.H. was supported by North East Grants (BT/PR23099/NER/95/632/2017), (BT/PR23155/NER/95/634/2017) from Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.034.
Contributor Information
Nasreen Z. Ehtesham, Email: nzehtesham@gmail.com.
Seyed E. Hasnain, Email: seh@dbeb.iitd.ac.in.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Crubézy E., Legal L., Fabas G., Dabernat H., Ludes B. Pathogeny of archaic mycobacteria at the emergence of urban life in Egypt (3400 BC) Infect Genet Evol. 2006;6(1):13–21. doi: 10.1016/j.meegid.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Organization WH (2020) Global tuberculosis report 2020: executive summary.
- 3.Chakaya J.M., Marais B., du Cros P., Ntoumi F., Mfinanga S., Kapata N. Programmatic versus personalised approaches to managing the global epidemic of multidrug-resistant tuberculosis. Lancet Respir Med. 2020;8(4):334–335. doi: 10.1016/S2213-2600(20)30104-1. [DOI] [PubMed] [Google Scholar]
- 4.Rahman S.A., Singh Y., Kohli S., Ahmad J., Ehtesham N.Z., Tyagi A.K., Hasnain S.E. Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis. mBio. 2014;5 doi: 10.1128/mBio.02020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed N., Dobrindt U., Hacker J., Hasnain S.E. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat Rev Microbiol. 2008;6(5):387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- 6.Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 7.Parsa K., Hasnain S.E. Proteomics of multidrug resistant Mycobacterium tuberculosis clinical isolates: a peep show on mechanism of drug resistance & perhaps more. Indian J Med Res. 2015;141:8–9. doi: 10.4103/0971-5916.154485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eldholm V., Balloux F. Antimicrobial resistance in Mycobacterium tuberculosis: The odd one out. Trends Microbiol. 2016;24(8):637–648. doi: 10.1016/j.tim.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqi N., Pathak N., Banerjee S., Ahmed N., Katoch V.M., Hasnain S.E. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection. 2004;32(2):109–111. doi: 10.1007/s15010-004-3097-x. [DOI] [PubMed] [Google Scholar]
- 10.Comas I., Borrell S., Roetzer A., Rose G., Malla B., Kato-Maeda M. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44(1):106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragland D.A., Nalivaika E.A., Nalam M.N.L., Prachanronarong K.L., Cao H., Bandaranayake R.M. Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease. J Am Chem Soc. 2014;136(34):11956–11963. doi: 10.1021/ja504096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karen van Niekerk R.P., Reva O.N., Korostetskiy I.S., Ilin A.I., Akhmetova G.K. 2018. Basic Biology and Applications of Actinobacteria. 1 - Shymaa Enany Y1–2018-12-05 ed. [Google Scholar]
- 13.Hasnain S.E., O'Toole R.F., Grover S., Ehtesham N.Z. Whole genome sequencing: a new paradigm in the surveillance and control of human tuberculosis. Tuberculosis (Edinb) 2015;95(2):91–94. doi: 10.1016/j.tube.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Miotto P., Tessema B., Tagliani E., Chindelevitch L., Starks A.M. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017;50 doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majeed A.A., Ahmed N., Rao K.R., Ghousunnissa S., Kauser F. AmpliBASE MT: a Mycobacterium tuberculosis diversity knowledgebase. Bioinformatics. 2004;20:989–992. doi: 10.1093/bioinformatics/bth051. [DOI] [PubMed] [Google Scholar]
- 16.Chernyaeva E.N., Shulgina M.V., Rotkevich M.S., Dobrynin P.V., Simonov S.A., Shitikov E.A. Genome-wide Mycobacterium tuberculosis variation (GMTV) database: a new tool for integrating sequence variations and epidemiology. BMC Genomics. 2014;15(1):308. doi: 10.1186/1471-2164-15-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandgren A., Strong M., Muthukrishnan P., Weiner B.K., Church G.M., Murray M.B. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flandrois J.P., Lina G., Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinf. 2014;15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamal S., Khubaib M., Gangwar R., Grover S., Grover A., Hasnain S.E. Author Correction: Artificial Intelligence and Machine learning based prediction of resistant and susceptible mutations in Mycobacterium tuberculosis. Sci Rep. 2020;10:14660. doi: 10.1038/s41598-020-71840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pires D.E., Chen J., Blundell T.L., Ascher D.B. In silico functional dissection of saturation mutagenesis: Interpreting the relationship between phenotypes and changes in protein stability, interactions and activity. Sci Rep. 2016;6:19848. doi: 10.1038/srep19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hameed H.M.A., Islam M.M., Chhotaray C., Wang C., Liu Y., Tan Y. Molecular Targets Related Drug Resistance Mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis Strains. Front Cell Infect Microbiol. 2018;8 doi: 10.3389/fcimb.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L.u., Zhao Y., Gao Y., Wu L., Gao R., Zhang Q.i. Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science. 2020;368(6496):1211–1219. doi: 10.1126/science.aba9102. [DOI] [PubMed] [Google Scholar]
- 23.Iacobino A., Fattorini L., Giannoni F. Drug-Resistant Tuberculosis 2020. Where We Stand. 2020;10(6):2153. doi: 10.3390/app10062153. [DOI] [Google Scholar]
- 24.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium CR, the GP, Allix-Beguec C, Arandjelovic I, Bi L et al. (2018) Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing. N Engl J Med 379: 1403-1415. [DOI] [PMC free article] [PubMed]
- 26.Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, Brezovsky J and Damborsky J (2014) PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol 10: e1003440. [DOI] [PMC free article] [PubMed]
- 27.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S and Thomas PD (2010) PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 38: D204-210. [DOI] [PMC free article] [PubMed]
- 28.Capriotti E., Calabrese R., Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 29.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bromberg Y., Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worth C.L., Preissner R., Blundell T.L. SDM–a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quan L., Lv Q., Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32(19):2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pires DE, Ascher DB and Blundell TL (2014) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30: 335-342. [DOI] [PMC free article] [PubMed]
- 36.Pires D.E., Blundell T.L., Ascher D.B. mCSM-lig: quantifying the effects of mutations on protein-small molecule affinity in genetic disease and emergence of drug resistance. Sci Rep. 2016;6:29575. doi: 10.1038/srep29575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrodinger. (2011). LLC, New York.
- 38.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin W., Mandal S., Degen D., Liu Y.u., Ebright Y.W., Li S. Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition. Mol Cell. 2017;66(2):169–179.e8. doi: 10.1016/j.molcel.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dessen A., Quemard A., Blanchard J., Jacobs W., Sacchettini J. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267(5204):1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 41.Bertrand T., Eady N.A.J., Jones J.N., Jesmin, Nagy J.M., Jamart-Grégoire B. Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem. 2004;279(37):38991–38999. doi: 10.1074/jbc.M402382200. [DOI] [PubMed] [Google Scholar]
- 42.Munir A., Wilson M.T., Hardwick S.W., Chirgadze D.Y., Worrall J.A.R., Blundell T.L. Using cryo-EM to understand antimycobacterial resistance in the catalase-peroxidase (KatG) from Mycobacterium tuberculosis. Structure. 2021 doi: 10.1016/j.str.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrella S., Gelus-Ziental N., Maudry A., Laurans C., Boudjelloul R., Sougakoff W. Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: insights into natural and acquired resistance to pyrazinamide. PLoS ONE. 2011;6(1):e15785. doi: 10.1371/journal.pone.0015785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blower T.R., Williamson B.H., Kerns R.J., Berger J.M. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2016;113(7):1706–1713. doi: 10.1073/pnas.1525047113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliveira J.S., Pereira J.H., Canduri F., Rodrigues N.C., de Souza O.N., de Azevedo W.F. Crystallographic and pre-steady-state kinetics studies on binding of NADH to wild-type and isoniazid-resistant enoyl-ACP(CoA) reductase enzymes from Mycobacterium tuberculosis. J Mol Biol. 2006;359(3):646–666. doi: 10.1016/j.jmb.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 46.Vilcheze C., Wang F., Arai M., Hazbon M.H., Colangeli R., Kremer L. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006;12:1027–1029. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 47.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 48.Arbex M.A., Varella Mde C., Siqueira H.R., Mello F.A. Antituberculosis drugs: drug interactions, adverse effects, and use in special situations. Part 1: first-line drugs. J Bras Pneumol. 2010;36:626–640. doi: 10.1590/s1806-37132010000500016. [DOI] [PubMed] [Google Scholar]
- 49.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metcalfe C., Macdonald I.K., Murphy E.J., Brown K.A., Raven E.L., Moody P.C.E. The tuberculosis prodrug isoniazid bound to activating peroxidases. J Biol Chem. 2008;283(10):6193–6200. doi: 10.1074/jbc.M707412200. [DOI] [PubMed] [Google Scholar]
- 51.Piton J., Petrella S., Delarue M., André-Leroux G., Jarlier V., Aubry A. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS ONE. 2010;5(8):e12245. doi: 10.1371/journal.pone.0012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 53.Frappier V., Najmanovich R.J., MacKerell A.D. A coarse-grained elastic network atom contact model and its use in the simulation of protein dynamics and the prediction of the effect of mutations. PLoS Comput Biol. 2014;10(4):e1003569. doi: 10.1371/journal.pcbi.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.UniProt C. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607-D613. [DOI] [PMC free article] [PubMed]
- 57.Rudnicki W.R., Mroczek T., Cudek P. Amino acid properties conserved in molecular evolution. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0098983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rouse D.A., Li Z., Bai G.H., Morris S.L. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995;39(11):2472–2477. doi: 10.1128/aac.39.11.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheen P., Ferrer P., Gilman R.H., López-Llano J., Fuentes P., Valencia E. Effect of pyrazinamidase activity on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2009;89(2):109–113. doi: 10.1016/j.tube.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siddiqi N., Shamim M., Hussain S., Choudhary R.K., Ahmed N., Prachee Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 2002;46(2):443–450. doi: 10.1128/AAC.46.2.443-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilcheze C and Jacobs WR, Jr. (2014) Resistance to Isoniazid and Ethionamide in Mycobacterium tuberculosis: Genes, Mutations, and Causalities. Microbiol Spectr 2: MGM2-0014-2013. [DOI] [PMC free article] [PubMed]
- 62.Portelli S., Phelan J.E., Ascher D.B., Clark T.G., Furnham N. Understanding molecular consequences of putative drug resistant mutations in Mycobacterium tuberculosis. Sci Rep. 2018;8:15356. doi: 10.1038/s41598-018-33370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E and Aubry A (2012) A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67: 819-831. [DOI] [PMC free article] [PubMed]
- 64.Brossier F., Veziris N., Aubry A., Jarlier V., Sougakoff W. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010;48(5):1683–1689. doi: 10.1128/JCM.01947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park H., Bradley P., Greisen P., Liu Y., Mulligan V.K., Kim D.E. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput. 2016;12(12):6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kellogg E.H., Leaver-Fay A., Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79(3):830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merker M., Kohl T.A., Barilar I., Andres S., Fowler P.W., Chryssanthou E. Phylogenetically informative mutations in genes implicated in antibiotic resistance in Mycobacterium tuberculosis complex. Genome Med. 2020;12(1) doi: 10.1186/s13073-020-00726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singhal R., Reynolds P.R., Marola J.L., Epperson L.E., Arora J., Sarin R. Sequence analysis of fluoroquinolone resistance-associated genes gyrA and gyrB in clinical mycobacterium tuberculosis isolates from patients suspected of having multidrug-resistant tuberculosis in New Delhi, India. J Clin Microbiol. 2016;54(9):2298–2305. doi: 10.1128/JCM.00670-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamaguchi T, Yokoyama K, Nakajima C and Suzuki Y (2017) Quinolone resistance-associated amino acid substitutions affect enzymatic activity of Mycobacterium leprae DNA gyrase. Biosci Biotechnol Biochem 81: 1343-1347. [DOI] [PubMed]
- 70.Rifat D., Campodónico V.L., Tao J., Miller J.A., Alp A., Yao Y. In vitro and in vivo fitness costs associated with Mycobacterium tuberculosis RpoB mutation H526D. Future Microbiol. 2017;12(9):753–765. doi: 10.2217/fmb-2017-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brandis G., Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother. 2013;68(11):2493–2497. doi: 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- 72.Casali N., Nikolayevskyy V., Balabanova Y., Ignatyeva O., Kontsevaya I., Harris S.R. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22(4):735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walker T.M., Kohl T.A., Omar S.V., Hedge J., Del Ojo Elias C., Bradley P. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15(10):1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X., He G., Wang S., Lin S., Chen J., Zhang W. Evaluation of whole-genome sequence method to diagnose resistance of 13 anti-tuberculosis drugs and characterize resistance genes in clinical multi-drug resistance Mycobacterium tuberculosis isolates from China. Front Microbiol. 2019;10:1741. doi: 10.3389/fmicb.2019.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gagneux S., Burgos M.V., DeRiemer K., Enciso A., Muñoz S., Hopewell P.C. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006;2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pym A.S., Saint-Joanis B., Cole S.T. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun. 2002;70(9):4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Soolingen D., de Haas P.W., van Doorn H.R., Kuijper E.d., Rinder H., Borgdorff M. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J Infect Dis. 2000;182(6):1788–1790. doi: 10.1086/317598. [DOI] [PubMed] [Google Scholar]
- 78.Sheen P., Lozano K., Gilman R.H., Valencia H.J., Loli S., Fuentes P. pncA gene expression and prediction factors on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013;93(5):515–522. doi: 10.1016/j.tube.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aung H.L., Tun T., Moradigaravand D., Köser C.U., Nyunt W.W., Aung S.T. Whole-genome sequencing of multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. J Glob Antimicrob Resist. 2016;6:113–117. doi: 10.1016/j.jgar.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuevas-Córdoba B., Xochihua-González S.O., Cuellar A., Fuentes-Domínguez J., Zenteno-Cuevas R. Characterization of pncA gene mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Mexico. Infect Genet Evol. 2013;19:330–334. doi: 10.1016/j.meegid.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 81.Chiu Y.C., Huang S.F., Yu K.W., Lee Y.C., Feng J.Y., Su W.J. Characteristics of pncA mutations in multidrug-resistant tuberculosis in Taiwan. BMC Infect Dis. 2011;11:240. doi: 10.1186/1471-2334-11-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allana S., Shashkina E., Mathema B., Bablishvili N., Tukvadze N., Shah N.S. pncA Gene mutations associated with pyrazinamide resistance in drug-resistant tuberculosis, South Africa and Georgia. Emerg Infect Dis. 2017;23(3):491–495. doi: 10.3201/eid2303.161034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi J., Su R., Zheng D., Zhu Y., Ma X., Wang S. Pyrazinamide resistance and mutation patterns among multidrug-resistant Mycobacterium tuberculosis from Henan Province. Infect Drug Resist. 2020;13:2929–2941. doi: 10.2147/IDR.S260161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farhat M.R., Jacobson K.R., Franke M.F., Kaur D., Sloutsky A., Mitnick C.D. Gyrase Mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2016;54(3):727–733. doi: 10.1128/JCM.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malik S., Willby M., Sikes D., Tsodikov O.V., Posey J.E., Via L.E. New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS ONE. 2012;7(6):e39754. doi: 10.1371/journal.pone.0039754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salah Eldin A., Mostafa N.M., Mostafa S.I. Detection of fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates as determined by gyrA/B gene mutation by using PCR technique. Egypt J Chest Dis Tubercul. 2012;61(4):349–353. [Google Scholar]
- 87.Pantel A., Petrella S., Veziris N., Brossier F., Bastian S., Jarlier V. Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother. 2012;56(4):1990–1996. doi: 10.1128/AAC.06272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yin X., Yu Z. Mutation characterization of gyrA and gyrB genes in levofloxacin-resistant Mycobacterium tuberculosis clinical isolates from Guangdong Province in China. J Infect. 2010;61(2):150–154. doi: 10.1016/j.jinf.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Chien J.-Y., Chiu W.-Y., Chien S.-T., Chiang C.-J., Yu C.-J., Hsueh P.-R. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2016;60(4):2090–2096. doi: 10.1128/AAC.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hameed H.M.A., Tan Y., Islam M.M., Guo L., Chhotaray C., Wang S. Phenotypic and genotypic characterization of levofloxacin- and moxifloxacin-resistant Mycobacterium tuberculosis clinical isolates in southern China. J Thorac Dis. 2019;11(11):4613–4625. doi: 10.21037/jtd.2019.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miotto P., Zhang Y., Cirillo D.M., Yam W.C. Drug resistance mechanisms and drug susceptibility testing for tuberculosis. Respirology. 2018;23(12):1098–1113. doi: 10.1111/resp.13393. [DOI] [PubMed] [Google Scholar]
- 92.Yi L, Aono A, Chikamatsu K, Igarashi Y, Yamada H, Takaki A and Mitarai S (2017) In vitro activity of sitafloxacin against Mycobacterium tuberculosis with gyrA/B mutations isolated in Japan. J Med Microbiol 66: 770-776. [DOI] [PubMed]
- 93.Kabir S., Tahir Z., Mukhtar N., Sohail M., Saqalein M., Rehman A. Fluoroquinolone resistance and mutational profile of gyrA in pulmonary MDR tuberculosis patients. BMC Pulm Med. 2020;20:138. doi: 10.1186/s12890-020-1172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Disratthakit A., Prammananan T., Tribuddharat C., Thaipisuttikul I., Doi N., Leechawengwongs M. Role of gyrB mutations in pre-extensively and extensively drug-resistant tuberculosis in thai clinical isolates. Antimicrob Agents Chemother. 2016;60(9):5189–5197. doi: 10.1128/AAC.00539-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Q., An X., Liu H., Wang S., Xiao T., Liu H. Uncovering the Resistance mechanism of Mycobacterium tuberculosis to rifampicin due to RNA Polymerase H451D/Y/R mutations from computational perspective. Front Chem. 2019;7:819. doi: 10.3389/fchem.2019.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zaw M.T., Emran N.A., Lin Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in Mycobacterium tuberculosis. J Infect Public Health. 2018;11(5):605–610. doi: 10.1016/j.jiph.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 97.Hui J., Gordon N., Kajioka R. Permeability barrier to rifampin in mycobacteria. Antimicrob Agents Chemother. 1977;11(5):773–779. doi: 10.1128/aac.11.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guerrero C., Stockman L., Marchesi F., Bodmer T., ROBERTS G.D., Telenti A. Evaluation of the rpoB gene in rifampicin-susceptible and -resistant Mycobacterium avium and Mycobacterium intracellulare. J Antimicrob Chemother. 1994;33(3):661–663. doi: 10.1093/jac/33.3.661-a. [DOI] [PubMed] [Google Scholar]
- 99.Figueiredo R., Ramos D.F., Moiteiro C., Medeiros M.A., Marcelo Curto M.J., Cardoso de Menezes J. Pharmacophore insights into rpoB gene mutations in Mycobacterium tuberculosis rifampicin resistant isolates. Eur J Med Chem. 2012;47:186–193. doi: 10.1016/j.ejmech.2011.10.041. [DOI] [PubMed] [Google Scholar]
- 100.Williams D.L., Spring L., Collins L., Miller L.P., Heifets L.B., Gangadharam P.R.J. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42(7):1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huitric E., Werngren J., Juréen P., Hoffner S. Resistance levels and rpoB gene mutations among in vitro-selected rifampin-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2006;50(8):2860–2862. doi: 10.1128/AAC.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hwang H.Y., Chang C.Y., Chang L.L., Chang S.F., Chang Y.H., Chen Y.J. Characterization of rifampicin-resistant Mycobacterium tuberculosis in Taiwan. J Med Microbiol. 2003;52:239–245. doi: 10.1099/jmm.0.05045-0. [DOI] [PubMed] [Google Scholar]
- 103.Betts MJ and Russell RB. (2003), Bioinformatics for Geneticists, pp. 289-316.
- 104.Telenti A., Imboden P., Marchesi F., Matter L., Schopfer K., Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341(8846):647–651. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 105.Caws M., Duy P.M., Tho D.Q., Lan N.T.N., Hoa D.V., Farrar J. Mutations prevalent among rifampin- and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J Clin Microbiol. 2006;44(7):2333–2337. doi: 10.1128/JCM.00330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chikaonda T., Ketseoglou I., Nguluwe N., Krysiak R., Thengolose I. Molecular characterisation of rifampicin-resistant Mycobacterium tuberculosis strains from Malawi. Afr J Lab Med. 2017;6:463. doi: 10.4102/ajlm.v6i2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Villar H.O., Kauvar L.M. Amino acid preferences at protein binding sites. FEBS Lett. 1994;349:125–130. doi: 10.1016/0014-5793(94)00648-2. [DOI] [PubMed] [Google Scholar]
- 108.Ramaswamy S., Musser J.M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 109.Pimentel A.L., de Lima Scodro R.B., Caleffi-Ferracioli K.R., Siqueira V.L.D., Campanerut-Sá P.A.Z., Lopes L.D.G. Mutations in catalase-peroxidase KatG from isoniazid resistant Mycobacterium tuberculosis clinical isolates: insights from molecular dynamics simulations. J Mol Model. 2017;23(4) doi: 10.1007/s00894-017-3290-3. [DOI] [PubMed] [Google Scholar]
- 110.Marttila H.J., Soini H., Huovinen P., Viljanen M.K. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996;40(9):2187–2189. doi: 10.1128/aac.40.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jagielski T., Grzeszczuk M., Kaminski M., Roeske K., Napiorkowska A., Stachowiak R. Identification and analysis of mutations in the katG gene in multidrug-resistant Mycobacterium tuberculosis clinical isolates. Pneumonol Alergol Pol. 2013;81:298–307. [PubMed] [Google Scholar]
- 112.Mo L., Zhang W., Wang J., Weng X.H., Chen S., Shao L.Y. Three-dimensional model and molecular mechanism of Mycobacterium tuberculosis catalase-peroxidase (KatG) and isoniazid-resistant KatG mutants. Microb Drug Resist. 2004;10(4):269–279. doi: 10.1089/mdr.2004.10.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du X., Wang W., Kim R., Yakota H., Nguyen H., Kim S.-H. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry. 2001;40(47):14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 114.Pandey B., Grover S., Tyagi C., Goyal S., Jamal S., Singh A. Dynamics of fluoroquinolones induced resistance in DNA gyrase of Mycobacterium tuberculosis. J Biomol Struct Dyn. 2018;36(2):362–375. doi: 10.1080/07391102.2016.1277784. [DOI] [PubMed] [Google Scholar]
- 115.Chen J., Chen Z., Li Y., Xia W., Chen X., Chen T. Characterization of gyrA and gyrB mutations and fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates from Hubei Province, China. Braz J Infect Dis. 2012;16:136–141. doi: 10.1016/s1413-8670(12)70294-5. [DOI] [PubMed] [Google Scholar]
- 116.Kim H., Nakajima C., Yokoyama K., Rahim Z., Kim Y.U., Oguri H. Impact of the E540V amino acid substitution in GyrB of Mycobacterium tuberculosis on quinolone resistance. Antimicrob Agents Chemother. 2011;55(8):3661–3667. doi: 10.1128/AAC.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Daisy P., Vijayalakshmi P., Selvaraj C., Singh S.K., Saipriya K. Targeting Multidrug Resistant Mycobacterium tuberculosis HtrA2 with Identical Chemical Entities of Fluoroquinolones. Indian J Pharm Sci. 2012;74(3):217. doi: 10.4103/0250-474X.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang YY, Deng JY, Gu J, Zhang ZP, Maxwell A, Bi LJ, Chen YY, Zhou YF, Yu ZN and Zhang XE (2006) The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA). Nucleic Acids Res 34: 5650-5659. [DOI] [PMC free article] [PubMed]
- 119.Agrawal A, Roue M, Spitzfaden C, Petrella S, Aubry A, Hann M, Bax B and Mayer C (2013) Mycobacterium tuberculosis DNA gyrase ATPase domain structures suggest a dissociative mechanism that explains how ATP hydrolysis is coupled to domain motion. Biochem J 456: 263-273. [DOI] [PubMed]
- 120.Song T., Park Y., Shamputa I.C., Seo S., Lee S.Y. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the beta' subunit of RNA polymerase. Mol Microbiol. 2014;91:1106–1119. doi: 10.1111/mmi.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ruusala T., Andersson D., Ehrenberg M., Kurland C.G. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984;3(11):2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma C., Yang X., Lewis P.J. Bacterial Transcription as a Target for Antibacterial Drug Development. Microbiol Mol Biol Rev. 2016;80(1):139–160. doi: 10.1128/MMBR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arenz S., Wilson D.N. Blast from the past: reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol Cell. 2016;61(1):3–14. doi: 10.1016/j.molcel.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 124.Casali N., Nikolayevskyy V., Balabanova Y., Harris S.R., Ignatyeva O., Kontsevaya I. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46(3):279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abrahams K.A., Chung C.-w., Ghidelli-Disse S., Rullas J., Rebollo-López M.J., Gurcha S.S. Identification of KasA as the cellular target of an anti-tubercular scaffold. Nat Commun. 2016;7(1) doi: 10.1038/ncomms12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Beg M.A., Shivangi, Thakur S.C., Meena L.S. Structural prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Adv Bioinformatics. 2018;2018:1–12. doi: 10.1155/2018/6152014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sherman D.R., Mdluli K., Hickey M.J., Arain T.M., Morris S.L., Barry C.E. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272(5268):1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 128.Baddam R., Kumar N., Wieler L.H., Lankapalli A.K., Ahmed N., Peacock S.J. Analysis of mutations in pncA reveals non-overlapping patterns among various lineages of Mycobacterium tuberculosis. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-22883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marbaix A.Y., Noël G., Detroux A.M., Vertommen D., Van Schaftingen E., Linster C.L. Extremely conserved ATP- or ADP-dependent enzymatic system for nicotinamide nucleotide repair. J Biol Chem. 2011;286(48):41246–41252. doi: 10.1074/jbc.C111.310847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu Y., Mangan J.A., Dhillon J., Sole K.M., Mitchison D.A., Butcher P.D. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000;182(22):6358–6365. doi: 10.1128/jb.182.22.6358-6365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Torrey H.L., Keren I., Via L.E., Lee J.S., Lewis K., Kaufmann G.F. High Persister Mutants in Mycobacterium tuberculosis. PLoS ONE. 2016;11(5):e0155127. doi: 10.1371/journal.pone.0155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Choudhary E., Sharma R., Kumar Y., Agarwal N. Conditional Silencing by CRISPRi Reveals the Role of DNA Gyrase in Formation of Drug-Tolerant Persister Population in Mycobacterium tuberculosis. Front Cell Infect Microbiol. 2019;9:70. doi: 10.3389/fcimb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ravishankar S., Ambady A., Awasthy D., Mudugal N.V., Menasinakai S., Jatheendranath S. Genetic and chemical validation identifies Mycobacterium tuberculosis topoisomerase I as an attractive anti-tubercular target. Tuberculosis (Edinb) 2015;95(5):589–598. doi: 10.1016/j.tube.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 134.Morlock G.P., Metchock B., Sikes D., Crawford J.T., Cooksey R.C. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2003;47(12):3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jhamb S.S., Goyal A., Singh P.P. Determination of the activity of standard anti-tuberculosis drugs against intramacrophage Mycobacterium tuberculosis, in vitro: MGIT 960 as a viable alternative for BACTEC 460. Braz J Infect Dis. 2014;18(3):336–340. doi: 10.1016/j.bjid.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heym B., Alzari P.M., Honore N., Cole S.T. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15(2):235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 137.Yadon A.N., Maharaj K., Adamson J.H., Lai Y.-P., Sacchettini J.C., Ioerger T.R. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaniga K., Cirillo D.M., Hoffner S., Ismail N.A., Kaur D., Lounis N. A multilaboratory, multicountry study to determine MIC quality control ranges for phenotypic drug susceptibility testing of selected first-line antituberculosis drugs, second-line injectables, fluoroquinolones, clofazimine, and linezolid. J Clin Microbiol. 2016;54(12):2963–2968. doi: 10.1128/JCM.01138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The database, MycoTRAP-DB is publicly accessible at http://139.59.12.92. All the other data are available as supplementary data.