Graphical abstract
Keywords: SHP2, PD-1, NMR spectroscopy, Molecular dynamics, Allosteric coupling
Abstract
SHP2 is a ubiquitous protein tyrosine phosphatase, whose activity is regulated by phosphotyrosine (pY)-containing peptides generated in response to extracellular stimuli. Its crystal structure reveals a closed, auto-inhibited conformation in which the N-terminal Src homology 2 (N-SH2) domain occludes the catalytic site of the phosphatase (PTP) domain. High-affinity mono-phosphorylated peptides promote catalytic activity by binding to N-SH2 and disrupting the interaction with the PTP. The mechanism behind this process is not entirely clear, especially because N-SH2 is incapable of accommodating complete peptide binding when SHP2 is in the auto-inhibited state. Here, we show that pY performs an essential role in this process; in addition to its contribution to overall peptide-binding energy, pY-recognition leads to enhanced dynamics of the N-SH2 EF and BG loops via an allosteric communication network, which destabilizes the N-SH2–PTP interaction surface and simultaneously generates a fully accessible binding pocket for the C-terminal half of the phosphopeptide. Subsequently, full binding of the phosphopeptide is associated with the stabilization of activated SHP2. We demonstrate that this allosteric network exists only in N-SH2, which is directly involved in the regulation of SHP2 activity, while the C-terminal SH2 domain (C-SH2) functions primarily to recruit high-affinity bidentate phosphopeptides.
1. Introduction
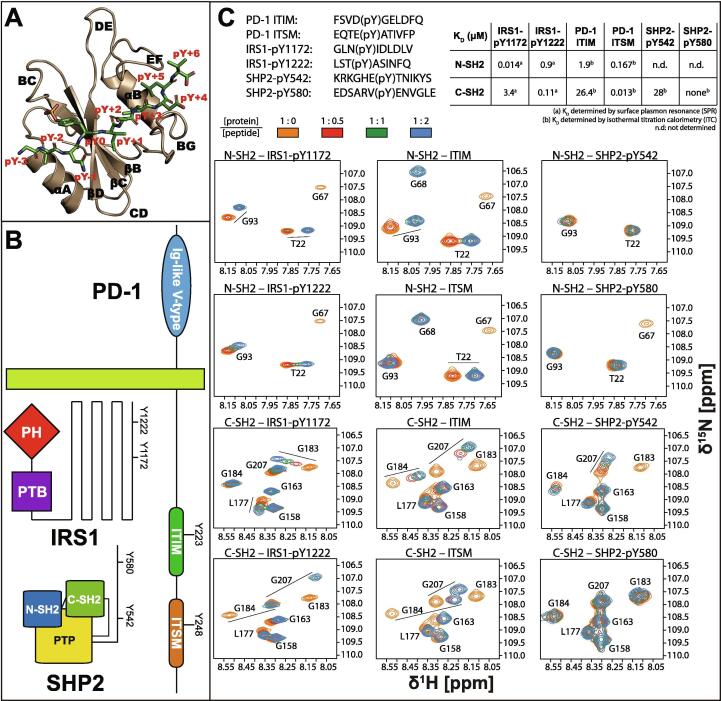
Cellular signaling cascades require precise spatiotemporal control of multiple binding events and thus rely on coordinated tuning of affinities and specificities within the relevant molecular networks [1]. In signaling pathways based on tyrosine phosphorylation, phosphotyrosine (pY) residues, generated by protein tyrosine kinases (PTKs), are recognized by specialized protein modules, of which Src homology 2 (SH2) domains are by far the most prevalent and well-characterized [2], [3]. SH2 domains consist of a globular fold of approximately 100 amino acids, with a three-stranded antiparallel β-sheet core flanked by two α-helices (Fig. 1A) [4]. Peptides that carry a pY mark (phosphopeptides) usually bind to their cognate SH2 domains in an extended conformation, with the peptide backbone arranged perpendicularly to the central β sheet; the pY is held in place by a network of salt bridges, where the invariant residue ArgβB5 (secondary-structure and amino-acid nomenclature according to Eck and coworkers [5], Figure S1A) plays a central role. Further interactions with residues adjacent to pY confer specificity, and thus make SH2 domains moderately selective, despite their structural invariance [6], [7]. In most cases, a hydrophobic patch flanked by the EF and BG loops makes extensive contacts with phosphopeptide residues C-terminal to the pY [8].
Fig. 1.
Binding of SHP-2 SH2 domains to phosphopeptides of different composition. (A) The architecture of SHP2 N-SH2 bound to IRS1-pY1172 highlights the features of a typical SH2 domain–phosphopeptide complex; secondary structure elements (black labels) and peptide residues (red labels) are named according to the scheme introduced by Eck et al., 1993 (see also figure S1A). (B) Schematics of the three proteins involved in this study, showing the sites subject to tyrosine phosphorylation. (C) Upper left panel: sequences of the peptides used in this study. Upper right: KD values determined for PD-1-, IRS1- and SHP2-derived phosphopeptides binding either N-SH2 or C-SH2. The KD values determined by surface plasmon resonance are taken from the literature [19]; those determined by ITC were measured in this work. Lower panel: 1H,15N-HSQC spectra of N-SH2 and C-SH2 in the presence of increasing concentrations of peptide. The species present in each sample are indicated above the individual spectra. The protein concentration was 200 μM and all spectra were measured on a 600 MHz spectrometer at 298 K. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Interestingly, some SH2 domains have evolved the ability to regulate the activity of catalytic domains that are part of the same protein. Protein tyrosine phosphatase SHP2, a 70-kDa protein encoded by the ptpn11 gene on chromosome 12 [9], is comprised of two SH2 domains arranged in tandem (termed N-SH2 and C-SH2), a catalytic PTP domain and a disordered C-terminal tail with hitherto unclear function (Figure S1B) [10]. In its basal state, SHP2 is almost inactive, due to the inhibitory intramolecular interaction of the N-SH2 and PTP domains; the binding of phosphopeptides triggers a conformational change that opposes the binding of the N-SH2 domain to the PTP catalytic site, thereby activating the phosphatase (Figure S1C) [11]. Notably, bidentate peptides, with two properly spaced pY residues, are much stronger activators than mono-phosphorylated peptides [12], [13].
As an almost unique example of a protein tyrosine phosphatase with positive rather than negative signaling function, SHP2 is involved in several signaling pathways, including Ras/MAPK, PI3K and PD-1 (Fig. 1B) [14], [15], [16]. Immune checkpoint receptor PD-1 (Programmed Death-1) recruits and activates SHP2 with two phosphorylated tyrosines, Y223 and Y248, embedded in the Immune Tyrosine Inhibitory Motif (ITIM) and the Immune Tyrosine Switch Motif (ITSM), respectively [17], [18]. This interaction results in a reduction of the functionality of T lymphocytes and may also be exploited by cancer cells to evade immune surveillance [14]. In other cellular pathways, phosphotyrosine sites of the IRS1 (Insulin Receptor Substrate 1) disordered C-terminus, such as pY1172 and pY1222, activate SHP2 [19]. IRS1 is a large scaffolding protein that plays a key role in transmitting signals originating from insulin and insulin-like growth factor (IGF) [20]; nevertheless, the role of the SHP2–IRS1 interaction is poorly characterized. Finally, the disordered C-terminus of SHP2 itself also contains two phosphorylation sites, Y542 and Y580 (Figure S1B) [9], whose role is still under debate: they are thought to recruit other proteins, including Grb2 [21], [22], [23], but their cis interaction with the N-SH2 and C-SH2 domains of SHP2 has also been proposed [24], [25].
The two SH2 domains of SHP2 have a unique feature, shared only with the SH2 domains of the closely related tyrosine phosphatase SHP1, namely the presence of a glycine instead of an arginine at position αA2. As a consequence, the phosphopeptides that bind these SH2 domains carry a hydrophobic residue at position pY–2 (two amino acids before pY) [26], [27]. Apart from this common feature, the N-SH2 and C-SH2 domains are functionally very different and their peptide binders have non-overlapping consensus motifs: C-SH2 is selective for peptides with sequence (T/V/I/y)XpY(A/s/t)X(I/V/L), while N-SH2 is more promiscuous and its binding partners can be divided into four different classes, based on their consensus sequence [28]. Furthermore, N-SH2 is directly responsible for the functional regulation of SHP2: in the basal state, the DE loop of N-SH2 is deeply inserted in the catalytic pocket of the PTP domain, blocking phosphatase activity. Phosphopeptide binding to N-SH2 is associated with a conformational change in the EF and BG loops, which reduces the degree of surface complementarity between the N-SH2 and PTP domains and prevents their intramolecular interaction (Figure S1C). Conversely, C-SH2 appears to play a role in the PTP activation only via bidentate phosphopeptides, which simultaneously engage both SH2 domains to ensure maximum stimulation of SHP2 activity [17]. Unlike N-SH2, the structure of C-SH2 is largely unaffected by peptide binding [11].
Here, we study the SH2 domains of SHP2 to understand how small variations in protein sequence and dynamics influence binding specificity and how binding of pY-containing peptides is coupled to the regulation of enzymatic activity. We select three SHP2-binding motifs, corresponding to phosphopeptides derived from IRS1 (pY1172 and pY1222), PD-1 (ITIM and ITSM) and the disordered C-terminus of SHP2 itself (pY542 and pY580) and rationalize the differences in their binding affinities. By employing a combination of solution-state nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics (MD) simulations, we study the structural and dynamic properties of the SHP2 SH2 domains in isolation and in the presence of either pY or pY-containing peptides PD-1 ITIM and ITSM, and reveal a dynamic network that couples phosphotyrosine binding to SHP2 activation. We find that the central β-strands βC and βD mediate a significant coupling between the pY-binding site and the EF and BG loops of N-SH2. A similar allosteric coupling has previously been found in an analysis of the conformational space of peptide-bound N-SH2 [29], [30]. Here we demonstrate that pY-binding alone is responsible for activating this allosteric network by inducing coupled dynamics of the BC, EF and BG loops, as well as of the tip of βD preceding the DE loop. Some conformational states induced by pY-binding disfavor the interaction of the N-SH2 domain with the PTP domain within SHP2, indicating that the pY-binding component of the phosphopeptide interaction with N-SH2 is principally responsible for increasing the fractional population of open, catalytically active SHP2. The coupling between the pY-binding site and the EF loop is weaker in C-SH2 than in N-SH2, in agreement with their different roles in SHP2 activation.
2. Experimental procedures
2.1. Protein expression and purification
The DNA sequences of N-SH2 (SHP21–105) and C-SH2 (SHP2106–220) were cloned into the pETM22 expression vector (EMBL Collection) and expressed as fusion proteins with a cleavable His6-thioredoxin tag. The “superbinder” N-SH2 construct (N-SH2 N37V/T42A/K55V) was produced with the QuikChange Lightning Site-Directed Mutagenesis protocol (Agilent Technologies). All constructs were confirmed by DNA sequencing.
Recombinant protein expression was performed in Tuner (DE3) cells (Merck): after transformation, the cells were grown at 37 °C in 2-liter flasks (250 rpm) until an optical density (OD600) of 0.6–0.8 was reached. Afterwards, the cultures were quickly cooled in a water–ice mix, and protein expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and continued for 20 h at 20 °C. Preparation of unlabeled samples (for ITC) was achieved by growing the cells in Luria-Bertani broth (LB), whereas the samples that required isotopic enrichment for NMR were prepared by growing the bacteria in minimal medium containing 15NH4Cl (1 g/l, Cambridge Isotope Laboratories) and 13C-D-glucose (4 g/l, Cambridge Isotope Laboratories) as the sole nitrogen and carbon sources, respectively. Once protein expression was complete, the cultures were harvested, pelleted and frozen at –20 °C until further use.
Protein purification began with the lysis of the cells by sonication in lysis buffer (1 M NaCl, 50 mM Tris-HCl, 5% glycerol, 10 mM imidazole, 5 mM β-mercaptoethanol, pH 7.6) supplemented with one tablet of EDTA-free cOmplete™ protease inhibitors (Roche), 100 µg of lysozyme (Roth) and 50 µg of DNAse (NEB). After centrifugation (18000 g for one hour at 4 °C) and filtration through a sterile 0.2-µm syringe filter, the supernatant was loaded on a HisTrap HP column (GE Healthcare), previously equilibrated with wash buffer (1 M NaCl, 50 mM Tris-HCl, 5% glycerol, 10 mM imidazole, 5 mM β-mercaptoethanol, pH 7.6). After loading, the column was washed with 10 column-volumes (CV) of wash buffer and the His-tagged protein was eluted with 5 CV of elution buffer (1 M NaCl, 50 mM Tris-HCl, 5% glycerol, 500 mM imidazole, 5 mM β-mercaptoethanol, pH 7.6). The His6-thioredoxin tag was removed by incubating the eluate with 3C protease (1:100 protease:protein ratio) at 4 °C overnight, while dialyzing against 2 L of wash buffer. The following day, another HisTrap affinity chromatography was performed and the protein of interest was retrieved in the flow-through, which was then concentrated to a final volume of 1–2 ml and loaded on a HiLoad 16/600 Superdex 75 pg column (GE Healthcare), previously equilibrated with NMR buffer (100 mM MES, 150 mM NaCl, 3 mM TCEP, 0.05% NaN3, pH 6.8). The fractions containing the pure protein were pooled, the protein was concentrated to the desired concentration and either used immediately or flash-frozen with liquid nitrogen for long-term storage at –80 °C.
2.2. Ligands
The peptides used in this study (PD-1 ITIM: FSVDpYGELDFQ, PD-1 ITSM: EQTEpYATIVFP, IRS1-pY1172: GLNpYIDLDLV, IRS1-pY1222: LSTpYASINFQ, SHP2-pY542: KRKGHEpYTNIKYS, SHP2-pY580: EDSARVpYENVGLE) were purchased from Caslo ApS (Lyngby, Denmark). All of them were synthesized with N-terminal acetylation and C-terminal amidation. Stock solutions were prepared by dissolving the lyophilized peptide in the appropriate buffer to a nominal concentration of 5 mM. If necessary, the pH was corrected by addition of small volumes of 10 M NaOH. Small aliquots of 20 μL were prepared and stored at –20 °C until further use. In all cases, peptide concentration in the stock solutions was confirmed by integration of appropriate methyl-group proton peaks by NMR. Phosphotyrosine (O-Phospho-L-tyrosine) was purchased from Toronto Research Chemicals (catalog number P365000).
2.3. Isothermal titration calorimetry (ITC)
ITC experiments were carried out on a Nano ITC machine (TA Instruments) in 100 mM MES (pH 6.8), 150 mM NaCl at 25 °C. Peptides (1–1.5 mM) were titrated onto proteins (100–200 μM) in a 300-μL sample cell while stirring at 200 rpm. The data (Figure S2) were analyzed with NanoAnalyze (v3.6.0, TA Instruments) and the KD values were calculated from a single replicate using a single-binding site model.
2.4. NMR spectroscopy
NMR experiments were recorded on Bruker Avance III HD spectrometers at 1H field-strengths of 600 MHz and 850 MHz, equipped with cryogenic inverse HCN probe-heads (N2- and He-cooled, respectively) and running Topspin 3.2 software. All experiments were recorded at a temperature of 298 K. NMR samples were prepared using uniformly-15N-labelled protein at concentrations of 0.3–0.6 mM in NMR buffer (100 mM MES, 150 mM NaCl, 3 mM TCEP, 0.05% NaN3, pH 6.8).
2D 1H-15N correlation maps were recorded as 1H,15N-HSQC spectra, acquired with States-TPPI for frequency-discrimination and with water suppression achieved via a combination of WATERGATE and water flip-back pulses to preserve the water magnetization [31], [32], [33].
In all titration experiments, the protein concentration was 200 µM and the amide chemical shifts of U-15N-labeled N-SH2 or C-SH2 were monitored by acquiring a series of 1H,15N-HSQC spectra in the presence of increasing stoichiometric ratios of peptide. Chemical-shift perturbations (CSPs) were calculated according to:
where ΔδH and ΔδN denote the chemical-shift differences in the 1H and 15N dimensions, respectively.
Backbone-amide 15N relaxation rates (R1 and R1ρ) were measured at a 1H field-strength of 600 MHz using established proton-detected experiments based on a gradient-selected, sensitivity-enhanced, refocused 1H,15N-HSQC sequence [34], [35], [36], [37]. The water signal was preserved using flip-back pulses and weak bipolar gradients during the indirect chemical shift evolution time to maintain the water magnetization along the +z axis. The relaxation delays were varied between 20 ms and 1.2 s for the R1 experiment, and between 3 ms and 100 ms for the R1ρ experiment. In the R1 sequence, N–H cross-relaxation pathways were suppressed by application of 1H amide-selective IBURP-1 inversion pulses [38] at intervals of 10 ms during the relaxation delay. In the R1ρ sequence, cross-relaxation was suppressed by application of between one and four 1H amide-selective inversion pulses during the 15N spin-lock relaxation period [39] and the 15N magnetization was explicitly aligned with the spin-lock field [40], which was applied at a field-strength of 2.5 kHz. Backbone-amide {1H}15N steady-state heteronuclear NOEs were measured using the standard method [41], [42]. Water magnetization was preserved in the reference spectrum as described above. Saturation of the amide proton magnetization was achieved using a 5-s train of high-power 180° pulses applied at 10.9-ms intervals [43]. The reference and saturated spectra were recorded in an interleaved fashion, using a long recycle delay of 15 s to ensure full recovery of the water magnetization at the start of each increment of the reference experiment.
Backbone-amide 15N relaxation-dispersion profiles were measured with the 15N CPMG sequence of Hansen et al. [44] at static 1H field-strengths of 600 MHz and 850 MHz (Figures S3&S4). During the CPMG period, transverse 15N magnetization is maintained as pure in-phase coherence via the application of high-power 1H continuous-wave (CW) decoupling, in which the exact 1H CW field-strength is varied slightly according to the 15N CPMG field-strength so that ν(1H-CW) = 2·k·νCPMG for all νCPMG (k integer). The pulse-sequence was implemented with both 1H and 15N temperature-compensation elements so that RF heating effects were constant for all νCPMG field-strengths and for the reference plane. With the exception of the data for the N-SH2 (apo) sample at 600 MHz, all CPMG data-sets were measured using a constant-time delay of 50 ms, and the CPMG field-strength was varied between 20 and 1200 Hz. For the N-SH2 (apo) sample at 600 MHz, the constant-time delay was 40 ms, and the CPMG field-strength was varied between 25 and 1000 Hz.
All NMR spectra were processed/visualized with a combination of NMRPipe v10.1 [45] and CcpNmr Analysis v2.4 [46]. Relaxation spectra were processed with partial Lorentzian-to-Gaussian apodization in both frequency dimensions and limited linear-prediction in the 15N dimension. Peak intensities were quantified by lineshape-fitting with FuDA [47].
Backbone-amide R1 and R1ρ rates and {1H}15N steady-state heteronuclear NOEs were analysed within the model-free framework of Lipari & Szabo [48], [49] using the program TENSOR2 [50] to fit the relaxation data to the model-free spectral-density and extract the global rotational diffusion tensor, local order-parameters (S2) and, where necessary, local correlation times. The differences in the amide-specific conformational entropies between two states A and B were calculated according to the equation:
which holds either for uncorrelated motions of individual vectors or for order parameters S2 > 0.75 both in the presence and in the absence of correlated motions [51], [52].
CPMG relaxation-dispersion profiles were analysed using the software package ChemEx [53]. Profiles recorded at both static field-strengths were fitted simultaneously on a residue-by-residue basis to a simple two-state exchange model with variable parameters kex (exchange-rate), pb (minor-state population), Δωab (frequency-difference between major and minor states) and (exchange-free transverse relaxation rate) and using an {Îx, Îy, Îz} spin-operator basis-set for integration of the spin-evolution during the CPMG element. The overall exchange contributions, , were extracted from the back-calculated profiles as described in the main text.
2.5. Homology modeling
The models for N-SH2–IRS1-pY1172, N-SH2–IRS1-pY1222, C-SH2–IRS1-pY1172, C-SH2–IRS1-pY1222, N-SH2–SHP2-pY542, N-SH2–SHP2-pY580, C-SH2–SHP2-pY542, C-SH2–SHP2-pY580 and C-SH2–ITIM were generated with Modeller, version 9.23 [54], [55]. The template structures used for the modeling were N-SH2–ITIM (PDB code 6ROY), N-SH2–ITSM (PDB code 6ROZ), C-SH2–ITSM (PDB code 6R5G) and N-SH2–GAB1 (PDB code 4QSY). The root-mean-square deviations (RMSDs) from the template structures are given in Table S1.
2.6. Molecular dynamics
Molecular dynamics (MD) simulations were performed with the GROMACS 2019.3 software package [56], [57], using the AMBER99SB-ILDN parameter set [58]. The initial atomic coordinates were taken from crystallographic (N-SH2: PDB code 1AYD, N-SH2–pY: modified from PDB code 6ROZ) or NMR (C-SH2: PDB code 2SHP, C-SH2–pY: modified from PDB code 6R5G) structures. The “superbinder” N-SH2 domain (N-SH2 N37V/T42A/K55V) and its complex with pY were generated by in-silico mutagenesis with PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC). The topology and parameters for the pY peptide (Ac-GpYG-NH2) were obtained from the GAFF force-field [59] with the ACPYPE server [60]. Each protein or protein complex was placed at the center of a dodecahedral box filled with TIP3P water molecules [61], with at least 1 nm of solvent on all sides. Long-range electrostatic interactions were computed with the particle-mesh Ewald (PME) scheme [62], while short-range non-bonded interactions were truncated at 1 nm. After solvation and neutralization with an appropriate number of counter-ions, each system was minimized once by steepest descent. Afterwards, five independent replicas with different assignments of the initial velocities were prepared from the energy-minimized systems. Each replica underwent two 1-ns rounds of solvent equilibration (NVT and NPT ensembles) to bring it to the target temperature of 310 K and target pressure of 1 bar, while protein and peptide atomic positions were restrained with a harmonic potential. Protein bond-lengths involving H were constrained with LINCS [63], while the bond-lengths and bond-angles of water molecules were constrained with SETTLE [64]. Temperature coupling was achieved by velocity rescaling with a stochastic term [65] and, when necessary, the pressure was fixed with the Parrinello-Rahman barostat [66]. After solvent relaxation, each replica underwent a 250-ns run of productive MD simulation in the NPT ensemble. After removal of the initial 5 ns, the five trajectories were merged to yield an aggregated 1.225-µs trajectory for analysis. Equilibration of the structures was complete after 5 ns (Figure S5).
Free-energy landscapes (FEL) were calculated using the GROMACS tool “sham”, which generates Gibbs free energy landscapes by Boltzmann inversion of multi-dimensional distribution histograms of the quantities of interest.
2.7. Correlation network analysis
The Bio3D v2.3 package [67] was used for the analysis of correlated motions. Correlations of atomic displacement were calculated for each Cα pair as Linear Mutual Information (LMI) values and stored into five different matrices (one per replica, after removal of the initial 50 ns), according to the following equation:
where Ci and Cij are the marginal-covariance matrices for the displacements of Cα atom i and for Cα atoms i & j, respectively. 50 ns were removed instead of only the 5 ns necessary for equilibration due to hardware limitations in handling the long trajectories. A consensus matrix containing average LMI values was built from the five individual LMI matrices, in which zero values were assigned to a Cα pair if their LMI was not greater than 0.5 for all replicas and/or if they were separated by more than 10 Å in at least 70% of cumulative trajectory frames; application of this filter required previous calculation of a contact map that stored all inter-Cα pairwise distances [68], [69]. In the construction of the correlation network, the nodes represent Cα atoms and node-pairs are connected through edges that are weighted by minus the logarithm of their LMI values. Community analysis, node centrality and sub-optimal path calculations were performed with Bio3D. Betweenness centrality (defined as the number of unique shortest paths crossing a node) was used as the measure of node centrality. The parameters for the analysis of sub-optimal paths were the source and sink nodes, as well as the total number of paths to be calculated, which was set to 250.
3. Results
3.1. Binding specificity: A comparison of the residues C-terminal to pY
The sequence determinants of the affinity of phosphopeptides to SH2 domains have been the subject of numerous studies [70], [71], [72], [73], [74]. Here, we use NMR spectroscopy, available structures and homology modelling to rationalize the differences amongst a set of six phosphopeptides that bind with different affinities to SHP2 N-SH2 and C-SH2 domains. Four of these phosphopeptides are known activators of SHP2: PD-1 ITIM and ITSM, and IRS1-pY1172 and -pY1222 (Fig. 1C); the other two comprise the SHP2 C-terminal pY-motifs, containing pY542 and pY580, whose function is still controversial.
The affinities of ITIM, ITSM, IRS1-pY1172 and IRS1-pY1222 for SHP2 N-SH2 and C-SH2 had been determined previously, as had the structures of N-SH2–ITIM, N-SH2–ITSM and C-SH2–ITSM [17], [19]. Here, we generated homology models of the complexes N-SH2–IRS1-pY1172, N-SH2–IRS1-pY1222, C-SH2–IRS1-pY1172, C-SH2–IRS1-pY1222 and C-SH2–ITIM, as described in Methods.
IRS1-pY1172 is the strongest binder of N-SH2, followed by ITSM (with one order-of-magnitude lower affinity) and then ITIM and IRS1-pY1222 (with two orders-of-magnitude lower affinity). NMR spectra indicate that neither SHP2-pY542 nor SHP2-pY580 bind significantly to the N-SH2 domain (Fig. 1C), thus contradicting the hypothesis that SHP2 could be auto-activated by its C-terminal tail.
C-SH2 has the highest affinity for ITSM and IRS1-pY1222, which both match the consensus binding sequence, followed by IRS1-pY1172 and ITIM with affinities in the micromolar range. The NMR spectra demonstrate no binding of C-SH2 to SHP2-pY580, while SHP2-pY542 bound to CSH2 with a dissociation constant KD of 28 µM, as determined by ITC (Fig. 1C and Figure S2).
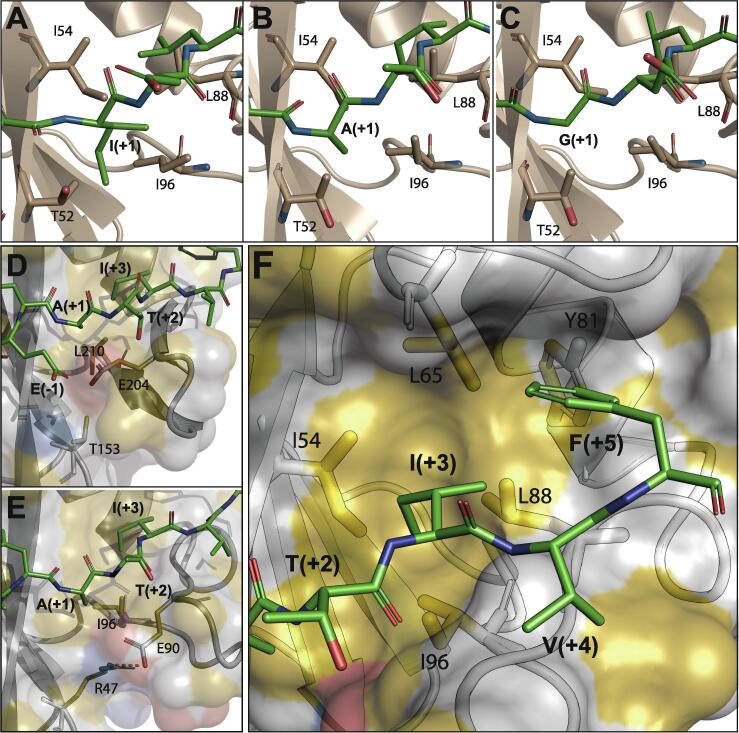
Both N-SH2 and C-SH2 interact with phosphopeptides in a two-pronged manner, with one binding pocket, delimited by βB, βC, βD, αA and the BC loop, hosting the pY residue, and a second hydrophobic pocket, delimited by CD, DE, EF, BG, βD and αB, accommodating residues C-terminal to pY. The affinity of the peptides for N-SH2 correlates strongly with the length of the side-chain in position pY+1, which points towards residues T52, I54 and I96 of the second binding pocket. The homology model of the N-SH2–IRS1-pY1172 complex shows that the long aliphatic side-chain of I(+1) (Fig. 2A) makes a multitude of van der Waals contacts with these residues; fewer contacts can be made by the shorter A(+1) side-chain of ITSM and IRS1-pY1222 (Fig. 2B), while no interactions occur with ITIM G(+1) (Fig. 2C). The T(+1) and E(+1) residues of SHP2-pY542 and SHP2-pY580, respectively, do not complement the hydrophobic character of the pocket. Thus, for N-SH2, the affinity to the phosphopeptides tested here correlates with the nature of the pY+1 amino acid.
Fig. 2.
Recognition of phosphopeptide sidechains at positions pY+1, pY+3 and pY+5. (A, B, C) Interactions between the hydrophobic cluster of N-SH2, formed by I54, L88 and I96, and the pY+1 residue of IRS1-pY1172, ITSM and ITIM, respectively. (D) Same region of the protein as in panels A–C, but for the C-SH2–ITSM complex. E204 in C-SH2 cannot form a salt-bridge with T153 (for which the corresponding residue in N-SH2 is R47) and thus shields L210 from interacting with the peptide residue pY+1. (E) Same as panel B (N-SH2–ITIM complex), highlighting the salt-bridge between E90 and R47, which exposes I96 to the ITSM residue pY+1. (F) The pY+5 binding site of N-SH2 bound to ITSM consists of a pentagonal arrangement of five hydrophobic residues (I54, L65, Y81, L88 & I96), which also contact the side-chain of the pY+3 residue.
Despite the conservation between the two SH2 domains of the hydrophobic residues in the region contacting the pY+1 amino acid (N-SH2/C-SH2: I54/V171; I96/L210), the affinities of the peptides for C-SH2 do not show the same correlation with the length of the pY+1 side-chain. One possible explanation is that the hydrophobicity of this region in C-SH2 is reduced by the presence of E204, whose side-chain partially shields L210 (Fig. 2D). In N-SH2 the same side-chain (E90/E204 in N-SH2/C-SH2) is moved away from I96 by a salt-bridge with R47 (Fig. 2E), which cannot be formed in C-SH2 (R47/T153 in N-SH2/C-SH2). We propose that the reduced hydrophobic character of this protein region lowers the dependency of the binding free-energy on the nature of the pY+1 side-chain.
The side-chain of pY+2 is oriented towards the solvent and would not appear to make significant contributions to the free energy of binding. However, molecular dynamics simulations have shown that an acidic amino acid at this position, as for example the aspartic acid of the strongest N-SH2 binder IRS1-pY1172, can make favorable electrostatic interactions with K89 and K91 of N-SH2 [30]. Both SHP2-pY542 and SHP2-pY580 have an asparagine in position pY+2, which is required for binding to the SH2 domain of Grb2 with a conformation different from the extended one [75]. Thus, both the lack of a strong interaction with either N-SH2 or C-SH2 of SHP-2 and the presence of N(+2) suggest that the phosphorylated motifs of the C-terminal tail of SHP2 function mainly to recruit Grb2.
A hydrophobic aliphatic residue at pY+3 is required for binding to the SH2 domains of SHP2 (and to most SH2 domains in general) [5], [7], [76]. In agreement, all four binding phosphopeptides have an isoleucine (IRS1-pY1222 and ITSM) or leucine (IRS1-pY1172 and ITIM) residue at this position. In all complexes, this residue is encapsulated in a pentagonal-shaped array of hydrophobic residues, namely I54/V170, L65/V181, Y81/Y197, L88/M202 and I96/L210 in N-SH2/C-SH2 (Fig. 2F).
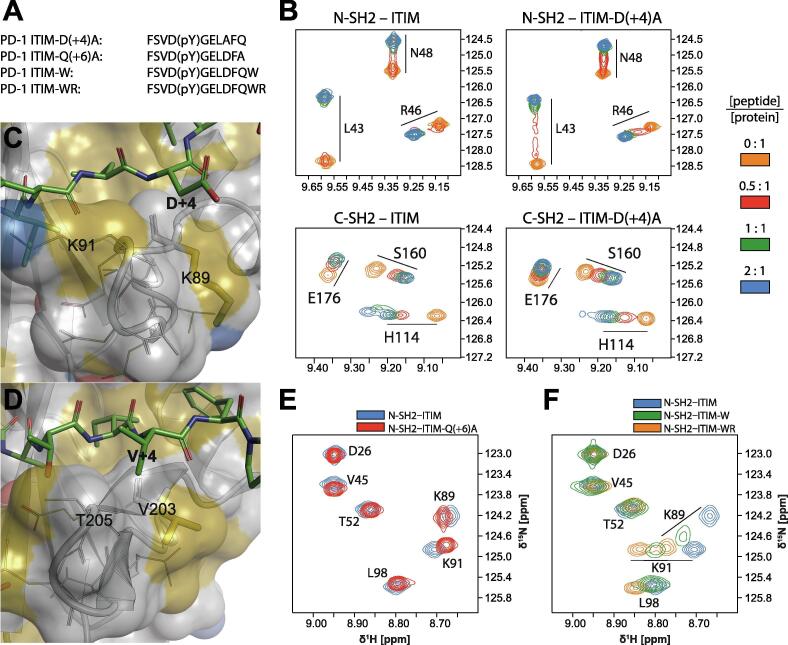
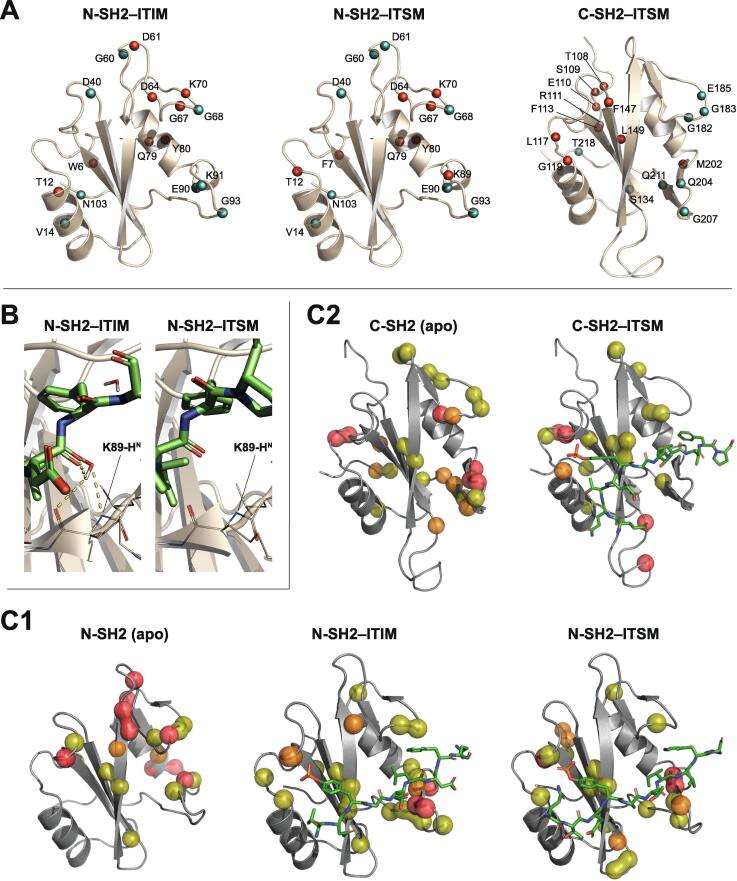
The role of the residue pY+4 is subtle: its side-chain points towards the solvent, but is very close to the BG loop, with which it could interact. The ITIM mutation D(+4)A causes either the association or the dissociation rate of the N-SH2–ITIM complex to increase, as demonstrated by the behavior of the N-SH2 1H-15N NMR peaks upon titration of the peptide (transition from the slow-exchange regime in the presence of ITIM to the intermediate-exchange regime in the presence of the mutant ITIM, Fig. 3B). The same mutation reduces affinity for C-SH2, as demonstrated by the increase in the peptide:protein molar ratio necessary to reach saturation of C-SH2 from 1:1 to 2:1 (Fig. 3B). In complex with N-SH2, D(+4) may participate in transient electrostatic interactions with K89 and K91 of the BG loop (Fig. 3C). However, in complex with C-SH2, these electrostatic interactions are not possible (K89/V203 and K91/T205 in N-SH2/C-SH2) and the detrimental effect of the D(+4)A mutation cannot be rationalized in terms of binding enthalpy. Nevertheless, the hydrophobic character of the BG loop of C-SH2 matches the nature of V(+4) in the strongest binding peptide ITSM (Fig. 3D), while IRS1-pY1222 with N(+4) binds less strongly.
Fig. 3.
Influence of phosphopeptide side-chains at positions pY+4 and pY+6 on binding affinities. (A) Sequences of the ITIM mutants used to study the role of amino acids at position pY+4 and beyond pY+5. (B) 1H,15N-HSQC spectra of N-SH2 and C-SH2 in the presence of increasing concentrations of wild-type and D(+4)A ITIM. (C) Although not evident from the crystal structure, D(+4) may form transient electrostatic interactions with K89 and/or K91 of N-SH2; these interactions would not be possible with the D(+4)A mutation. (D) K89 and K91 in N-SH2 are replaced by V203 and T205 in C-SH2, which makes this area substantially more hydrophobic, in agreement with the high affinity of phosphopeptides possessing a hydrophobic residue at pY+4. (E) Overlay of 1H,15N-HSQC spectra of N-SH2 with a 2:1 peptide:protein molar ratio; the peptide is either wild-type ITIM or Q(+6)A ITIM. (F) Overlay of 1H,15N-HSQC spectra of N-SH2 with a 2:1 peptide:protein molar ratio; the peptide is either wild-type ITIM or mutant ITIM containing one (W) or two (WR) additional amino acids at the C-terminus. The protein concentration was 200 μM and all spectra were measured on a 600 MHz spectrometer at 298 K.
The interaction with residue pY+5 is particular to the SH2 domains of SHP2: this residue is invariably hydrophobic and very often aromatic and interacts with the same hydrophobic patch that surrounds pY+3 (Fig. 2F). The presence of a non-hydrophobic amino acid at this position is strongly unfavorable, as demonstrated by the reduction in affinity by two orders of magnitude when comparing the VSPEPIpYATIDDL peptide with the ASPEPIpYATIDFD peptide [77].
The contribution of residues beyond pY+5 is negligible: both the ITIM mutation Q(+6)A and extension of ITIM by two residues, W(+7) and R(+8), cause only minor local changes in the 1H-15N NMR spectrum of bound N-SH2 in comparison with the spectrum of N-SH2 in complex with wild-type ITIM: the differences are localized to the BG-loop residues K89 and K91 (Fig. 3E and 3F).
3.2. Influence of ITIM and ITSM binding on the dynamics of SHP2 SH2 domains
To establish whether the binding of either ITIM or ITSM affects the dynamics of the SHP2 SH2 domains differently, and thus reveal potential entropic contributions to the binding energy, we measured fast (sub-ns) and slow (200-μs–20-ms) time-scale dynamics by NMR spectroscopy for both the N-SH2 and C-SH2 domains of SHP2 in their apo, ITIM-bound and ITSM-bound forms.
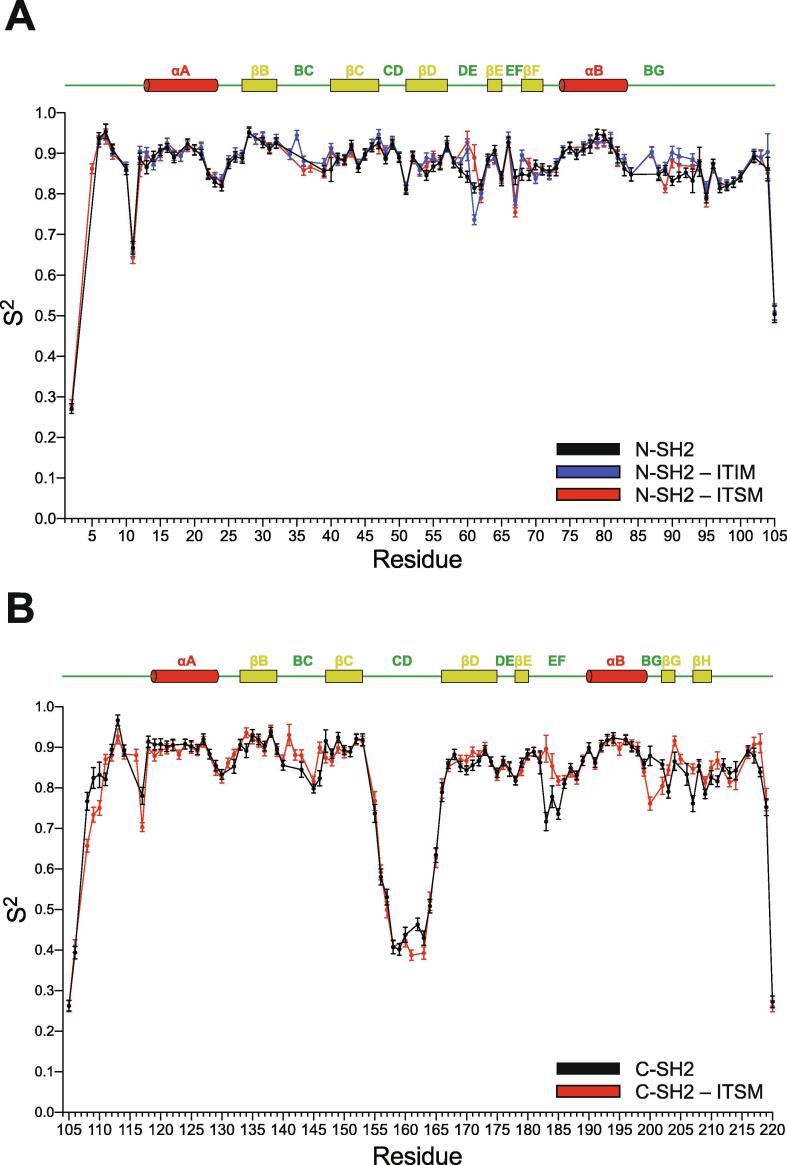
15N R1 and R2 relaxation rates and {1H}15N heteronuclear NOEs were measured for the backbone amide groups of N-SH2, N-SH2–ITIM, N-SH2–ITSM, C-SH2 and C-SH2–ITSM and fitted with the Lipari-Szabo model-free formalism, as described in Methods. From this analysis we extracted the global rotational correlation time τc, the degree of anisotropy of the rotational diffusion tensor, and the S2 order-parameter for each amide group, which is a measure of the magnitude of protein internal motions on the fast time-scale (S2 can vary between 0 and 1; larger internal motions correspond to smaller values of S2). Information on some of the residues of the apo N-SH2 BC and BG loops was missing because the corresponding peaks were very weak (vide infra). The unbound, ITIM-bound and ITSM-bound N-SH2 and C-SH2 domains can be described as globular and approximately spherical entities with rotational correlation times compatible with those predicted from the monomeric state (Table S2). In the absence of phosphopeptide, both N-SH2 and C-SH2 showed only limited internal motion on the fast time-scale, with the exception of the long CD loop of C-SH2 (Fig. 4). The average S2 order-parameter of C-SH2 (0.81) was lower than that of N-SH2 (0.86). Addition of phosphopeptide did not significantly perturb the overall backbone dynamics (Fig. 4), (average values of the S2 order-parameter: N-SH2–ITIM, 0.88; N-SH2–ITSM, 0.87; C-SH2–ITSM, 0.81). Nevertheless, a few subtle differences were observed in functionally relevant areas of the two SH2 domains. To better understand these changes, we converted the differences between the S2 values of the phosphopeptide-bound and unbound states of individual residues to the corresponding changes in conformational entropy [52]. Despite neglecting contributions from fluctuations with correlation time greater than τc and entropy changes due to the ligand or the solvent, this analysis can reveal interesting residue-specific effects. The binding of ITIM and ITSM was associated with an overall negative conformational entropy change on the timescale probed by the NMR measurements (up to 50 ps). This global reduction of fast motions was more pronounced for N-SH2 compared to C-SH2 and for ITIM compared to ITSM: ΔS(N-SH2–ITIM vs N-SH2) = −32.6 J mol–1K−1; ΔS(N-SH2–ITSM vs N-SH2) = −19.5 J mol–1K−1; ΔS(C-SH2–ITIM vs C-SH2) = −15.9 J mol–1K−1; ΔS(C-SH2–ITSM vs C-SH2) = −3.7 J mol–1K−1 (overall entropy-changes were calculated as a sum of the residue-specific entropy-changes, excluding residues with S2 < 0.7). Locally, both ITIM and ITSM increased the conformational entropy of the N-SH2 EF loop, while ITSM decreased that of the same loop in C-SH2 (Fig. 5A). Moreover, binding of both peptides rigidified the BG loop of both domains (Fig. 5A). Upon inspection of individual sites, we observed that K89, in the BG loop of N-SH2, showed an entropy increase in complex with ITSM but not with ITIM. A possible explanation is that in complex with ITSM, the area around K89 is completely dehydrated, presumably because of the high hydrophobicity of the peptide (PDB code 6ROZ); in contrast, in complex with ITIM (PDB code 6ROY), K89 coordinates a water molecule, thus becoming less mobile (Fig. 5B). In conclusion, the fast dynamics of the N-SH2– and C-SH2–phosphopeptide complexes show some peptide- and domain-specific effects, which can contribute to subtle differences in the binding affinities.
Fig. 4.
Fast dynamics of the N-SH2 and C-SH2 domains in their apo states and in complexes with phosphopeptides. S2 order parameters extracted from the fit of the 15N R1, R2 and heteronuclear NOE data to the Lipari-Szabo model-free parametrization of fast motions. A. S2 order parameters of N-SH2, N-SH2–ITIM and N-SH2–ITSM. B. S2 order parameters of C-SH2 and C-SH2–ITSM. The residue numbers are given on the x-axes. Secondary-structure elements are indicated along the sequence.
Fig. 5.
Experimentally-measured dynamics of N-SH2 and C-SH2 in the apo state and in complex with ITIM and ITSM. (A) Amide-specific entropy changes measured between apo N-SH2 and the N-SH2–ITIM complex (left), apo N-SH2 and the N-SH2–ITSM complex (centre), apo C-SH2 and the C-SH2–ITSM complex (right). An entropy increase/decrease is represented by a red/cyan sphere. The entropy changes were calculated from the Lipari-Szabo amide order-parameters and are due to internal dynamics on the ps timescale. (B) Differences in the hydrogen-bond network of the BG loop in the N-SH2–ITIM and N-SH2–ITSM complexes. (C) Transverse-relaxation exchange-broadening contributions due to slow dynamics as measured by CPMG relaxation dispersion for N-SH2 (panel C1: apo state, and in complex with ITIM and ITSM peptides) and C-SH2 (panel C2: apo and in complex with ITSM peptide). Amide nitrogens with significant exchange-broadening are shown as spheres, and are colored yellow, orange and red in order of increasing (according to the thresholds shown in the plots of supplementary figures S3 & S4). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we probed the dynamics in the high-μs–ms timescale, measuring transverse relaxation dispersion experiments on both isolated N-SH2 and C-SH2 and their complexes with ITIM and ITSM. We used a 15N Carl-Purcell-Meiboom-Gill (CPMG) experiment, recorded at both 600 and 850 MHz, with CPMG field-strengths varying between 20 and 1200 Hz applied for a constant-time period of 50 ms. Under these conditions, conformational exchange dynamics can be measured with exchange rates, kex, in the range ~50–5000 Hz. The experimental relaxation dispersion profiles of individual residues were fit to a two-site exchange model using ChemEx [53]. The exchange contributions to the transverse relaxation rates, , were then calculated as the difference between the fitted transverse relaxation rates at the minimum and maximum CPMG field-strengths (). The value of the parameter can be roughly correlated with the extent of slow time-scale dynamics at each amino-acid position.
In apo N-SH2, higher than average values were measured for the DE and EF loops and for the tip of βD before the DE loop. In the BG loop, two amino acids showed enhanced dynamics, together with the tip of the αB helix before the loop (Fig. 5C1 and Supplementary Figure S3). In the 1H-15N correlation spectra, a small number of amide groups of the BC and BG loops were absent, probably due to fast exchange of the amide proton with the solvent, and their relaxation dispersion could not be measured. Overall, apo N-SH2 showed enhanced mobility on the high-μs–ms timescale in regions that bind the peptide residues C-terminal to the phosphotyrosine. Binding of both ITIM and ITSM diminished the dynamics in both the DE and EF loops and the flanking βD. The slow dynamics of the tip of αB was quenched in both the N-SH2–ITIM and N-SH2–ITSM complexes, while slow dynamics was induced by both ITIM and ITSM in the 89KE91K stretch (Fig. 5C1 and Supplementary Figure S3). In addition, ITIM and ITSM had different effects on the exchange-contributions of H85 and L88, which were higher in the N-SH2–ITSM than in the N-SH2–ITIM complex. By contrast, the increase of the slow dynamics of the 89KE91K stretch was more prominent in N-SH2–ITIM than in N-SH2–ITSM. As for the fast dynamics, these variable effects may be due to the different hydrophobicities of the C-terminal halves of ITIM and ITSM.
Similar to the N-SH2 domain, exchange contributions to the 15N transverse relaxation rates of the C-SH2 domain were quite small overall. The peaks for a small number of amide groups of the CD loop were absent, again probably due to fast exchange of the amide proton with the solvent, and their relaxation dispersion could not be measured. In the rest of the protein, the N-terminal tail, the DE and the EF loops and the BG loop together with the C-terminus showed enhanced dynamics (Fig. 5C2 and Supplementary Figure S4). The dynamics of the BG loop and the C-terminal part of the protein were quenched by ITSM binding (Fig. 5C2 and Supplementary Figure S4), in agreement with a stabilization of this region due to the interaction with the C-terminal half of the peptide and in qualitative agreement with the fast dynamics data (Fig. 5A). The dynamics of the DE and EF loops were nearly unchanged in the presence of the peptide, with the exception of V181 and G182, which are in direct proximity to I(+3) in the hydrophobic specificity pocket and were partially rigidified by peptide binding, again in agreement with the fast dynamics data (Fig. 5A).
In conclusion, peptide binding considerably reduces the slow motions of the DE and EF loops of apo N-SH2, while it has a milder effect on the less mobile DE and EF loops of apo C-SH2. By contrast, the slow dynamics of the apo C-SH2 BG loop is quenched by peptide binding, while that of the N-SH2 BG loop is changed in a residue-specific manner. These distinct behaviors of the N-SH2 and C-SH2 domains may contribute to modulate their interactions with phosphopeptides in different manners, according to their different functions.
3.3. A network of structural interactions connects the pY-binding site with remote structural elements
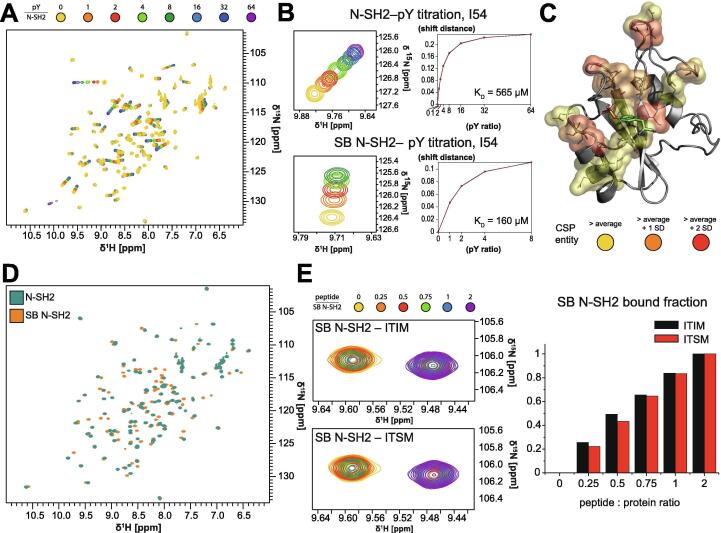
Next, we asked whether pY in the phosphopeptide has a role in inducing the structural changes of N-SH2 that are responsible for its reduced affinity for the PTP domain, or whether it functions solely to increase affinity. To address this question, we titrated pY onto the SHP2 N-SH2 domain and followed the CSPs of the 1H-15N peaks (Fig. 6A). pY binds in the fast-exchange regime with a KD of 565 µM, as calculated by quantification of the NMR CSPs (Fig. 6B). Binding of pY affected the chemical shifts of residues located far from the pY-binding site, such as those in the EF and DE loops (Fig. 6C). This observation suggests the presence of an allosteric network, which propagates the structural changes induced in the BC loop by pY-binding to distant regions of the N-SH2 domain.
Fig. 6.
Effect of pY-binding on wild-type and mutant N-SH2. (A) Overlay of 1H,15N-HSQC spectra of N-SH2 in the presence of increasing concentrations of pY. (B) Left: expansion of the 1H,15N-HSQC spectra, showing the amide peak of I54 of either N-SH2 or the mutant SB N-SH2 in the presence of increasing concentrations of pY. Right: quantification of the CSPs of the spectra shown on the left and corresponding fits to extract the KD values. (C) CSPs produced by addition of pY mapped on the structure of N-SH2. (D) Overlay of 1H,15N-HSQC spectra of N-SH2 and mutant SB N-SH2. (E) Left: overlay of the amide peak of G49 in 1H,15N-HSQC spectra of mutant SB N-SH2 in the presence of increasing concentrations of either ITIM and ITSM. The protein concentration was 200 μM in all spectra, which were measured on a 600 MHz spectrometer at 298 K. Right: quantification of the fraction of bound protein from the peak intensities of residue G49, as measured in the spectra shown on the left.
To further confirm the existence of this allosteric network, we generated the N-SH2 binding-site mutant N37V/T42A/K55V, which was demonstrated to bind pY with increased affinity (pY “superbinder”, SB N-SH2) [78]. NMR titrations confirm that SB N-SH2 binds pY with a lower KD than wild-type N-SH2 (KD = 160 μM, Fig. 4, Fig. 6B). Despite the fact that all the mutations were localized to the pY-binding site, comparison of the NMR spectra of apo SB N-SH2 and apo wild-type N-SH2 showed CSPs for distant residues (Fig. 6D), confirming the presence of an allosteric structural network connecting the pY-binding site with other structural elements of the N-SH2 domain. Interestingly, SB N-SH2 could no longer discriminate between ITIM and ITSM and bound both peptides with similar affinities (Fig. 6E). This finding supports the previous work of Kaneko et al. [78], who proposed that a relatively weak affinity of SH2 domains for pY alone is necessary to discriminate between specific and non-specific binding partners and avoid crosstalk between unrelated pathways.
3.4. Molecular dynamics simulations reveal the nature of the allosteric network
To understand the nature of the allosteric network that connects the pY-binding site with remote structural elements of the N-SH2 domain, we performed molecular dynamics (MD) simulations of N-SH2, N-SH2–pY, SB N-SH2, SB N-SH2–pY, C-SH2 and C-SH2–pY. A similar study has recently been conducted on a pool of phosphopeptide-bound conformations of the SHP2 N-SH2 domain [29], [30]. Here, we focus on understanding the role of pY-binding on the dynamics of the N-SH2 domain, independent of the presence or nature of other interacting amino acids. In addition, we compare the effect of pY-binding on N-SH2, SB N-SH2 and C-SH2 domains.
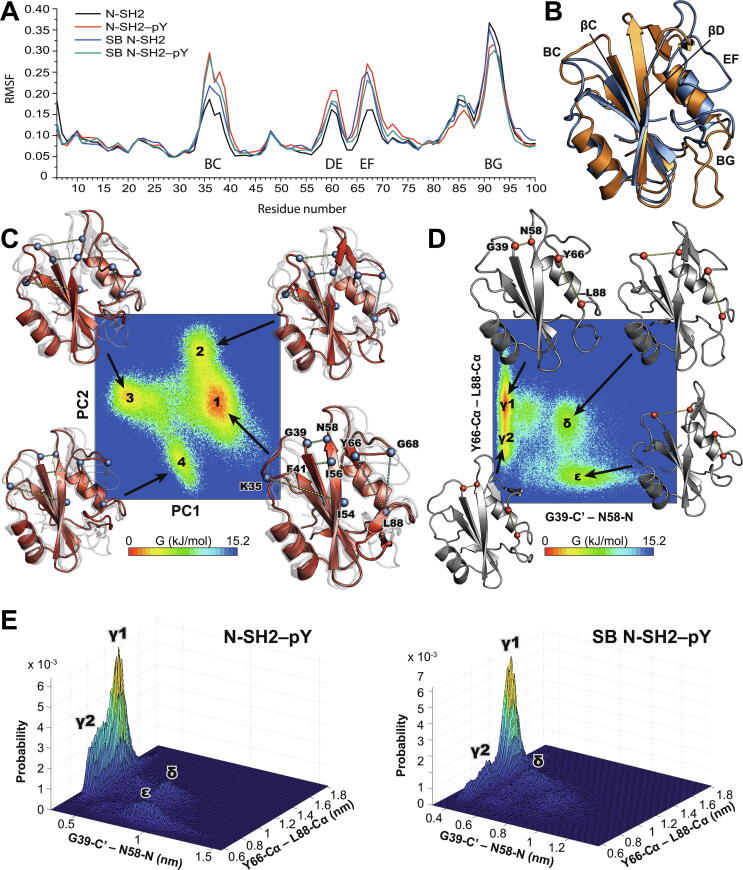
MD trajectories were simulated for a total of 1.225 μs, as described in Methods. N-SH2 showed limited internal mobility, with the highest atomic root-mean-square fluctuations (RMSF) located in the BC, DE, EF and BG loops (Fig. 7A). A few amino-acids acted as hinges intercalated within higher-mobility segments, as for example Y63 and Y64, which separate the DE and the EF loops, and L88, at the start of the BG loop. When a pY moiety was bound, the RMSF of the BC, DE and EF loops increased significantly; conversely, the RMSF of the BG loop decreased slightly.
Fig. 7.
Analysis of the MD simulations of N-SH2–pY and SB N-SH2–pY. (A) Root-mean-square fluctuations (RMSF) calculated over the MD trajectories of N-SH2 and SB N-SH2 in their apo and pY-bound states. (B) Overlay of the two most distant configurations of N-SH2–pY along PC1. The two structures differ mainly in the conformations of the BC, EF and BG loops, as well as in the packing of the β-strands βC and βD. (C) The free-energy landscape of N-SH2–pY mapped onto PC1 and PC2 shows two major and two minor energy minima. The four structures representative of the four minima are shown overlaid in cartoon representation, with the structure representative of each individual minimum in solid red and the other three structures as semi-transparent. The differences among these structures can be characterized by the interatomic distances displayed as dotted lines on the structures. (D) The free-energy landscapes of N-SH2–pY mapped on the distances describing the size of the pY+5 binding site (Y66-Cα–L88-Cα) and the unzipping of the β-strands βC and βD (G39-C’–N58-N). (E) Comparison of the free-energy landscapes of N-SH2–pY and SB N-SH2–pY mapped on the same distances as in panel D. The free-energy landscape panels were generated with the MatLab script felandscape.m [87]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
SB N-SH2 showed increased mobility in the same regions as wild-type N-SH2; however, the extent of motion in the EF loop matched that of N-SH2–pY rather than that of apo N-SH2. Upon addition of pY to SB N-SH2, the dynamics increased in the BC loop, but remained almost unchanged in the DE and EF loops, suggesting that the motions induced by pY binding within the DE and EF loops of wild-type N-SH2 are already present in the apo form of the mutant SB N-SH2 (Fig. 7A).
To study the nature of these motions, we first performed principal component analysis (PCA) of the four trajectories. In the apo states, the first two eigenvectors (PC1 and PC2) comprise only 36% and 33% of the total motions for N-SH2 and SB N-SH2, respectively. The most populated conformational states of this subspace are shown in Figures S6A (N-SH2) and S6B (SB N-SH2). For the pY-bound forms, PC1 and PC2 comprised 50% and 40% of the motions of N-SH2 and SB N-SH2, respectively, suggesting a larger impact of pY-binding on wild-type N-SH2 than on mutant SB N-SH2. For N-SH2–pY, the PC1 eigenvector, which alone accounts for ~32% of the overall motion, connects two states that show substantial differences in the orientation of the BC, EF and BG loops and in the packing of the central β-strands βC and βD (Fig. 7B). PC2, on the other hand, describes the motion of the tip of the BG loop relative to the β-sheet core. Overall, the free-energy landscape (FEL) projected onto PC1 and PC2 is populated by four major states, of which states 1 and 3 are the most frequently sampled (Fig. 7C). These states are characterized by three major structural features: (i) the bending of the BC loop towards the core of the central β-sheet (measured by the distance between K35-Cα and I54-Cα); (ii) the “unzipping” of the βC and βD strands (measured by the length of the G39-C’–N58-N and I56-C’–F41-N hydrogen-bonds); (iii) the size of the pocket hosting the phosphopeptide residue pY+5 (measured by the distance between L88-Cα at the start of the BG loop and either Y66-Cα or G68-Cα in the EF loop), which is largely dictated by the conformation of the EF loop.
To investigate whether the structural parameters distinguishing state 1 from state 3 along PC1 are inter-correlated over the entire trajectory, we mapped FELs on pairwise combinations of the three structural features described above. First, we chose as the first coordinate one of the two distances reporting on the packing of the βC and βD strands and as the second coordinate one of the distances reporting on the size of the pY+5 binding pocket. In this mapping of the FEL, the conformational space of N-SH2–pY is divided into three main configurations: (i) short G39-C’–N58-N/I56-C'–F41-N distances and long Y66-Cα–L88-Cα/G68-Cα–L88Cα distances (denoted as the γ state); (ii) long G39-C’–N58-N/I56-C'–F41-N distances and short Y66-Cα–L88-Cα/G68-Cα–L88Cα distances (ε state); (iii) long G39-C’–N58-N/I56-C'–F41-N distances and medium Y66-Cα–L88-Cα/G68-Cα–L88Cα distances (δ state). The γ state is much more populated than the other two states and can be further sub-divided into a γ1 state with long Y66-Cα–L88-Cα/G68-Cα–L88Cα distances and a γ2 state with medium Y66-Cα–L88-Cα/G68-Cα–L88Cα distances (Fig. 7D). A similarly strong correlation exists between the descriptor of the BC-loop conformation and the packing of βC and βD. In this case, the two major states represent (i) long K35-Cα–I54-Cα distances and short G39-C’–N58-N/I56-C'–F41-N distances; (ii) short K35-Cα–I54-Cα distances and medium G39-C’–N58-N/I56-C'–F41-N distances (Figure S7). This data suggests that pY-binding is communicated to the EF loop through the N- and C-terminal residues of the central β-strands βC and βD, respectively. As a comparison, the conformational space of apo N-SH2 is restricted to short G39-C’–N58-N distances in combination with either medium or long K35-Cα–I54-Cα and Y66-Cα–L88-Cα distances, without any correlation between the last two distances (Figure S6C).
The SB N-SH2–pY complex populates almost exclusively the γ1 state, as shown by the FELs constructed using distances G39-C’–N58-N and Y66-Cα–L88-Cα as the two coordinates (Fig. 7E).
Next, we asked whether the dynamic properties of C-SH2 differ from those of N-SH2. We analyzed the MD simulations of C-SH2 and C-SH2–pY and found that the dynamics are dominated by the long and flexible CD loop. C-SH2 showed a higher RMSF than N-SH2 along the entire sequence, with binding of pY causing an increase in the dynamics of the DE and EF loops but not of the BC and BG loops (Figure S8A). The PCA analysis, conducted with the exclusion of the CD loop residues T153–K166, showed a similar separation of the protein conformations into four clusters, differing in the width of the pY+5 binding pocket and in the packing of the βC and βD elements (Figure S8B). However, when analyzing the correlation between the distances indicative of the opening of the pY+5 binding pocket and those indicative of the unzipping of the βC and βD strands, we saw a more continuous distribution of distances accompanied by the presence of conformations with “closed” pY+5 binding pocket and zipped βC and βD strands (Figure S8C). This observation, together with the absence of significant changes in the dynamics of the BC loop upon pY-binding, suggest a much weaker coupling between the pY-binding site and the conformation of the EF loop than in N-SH2. This is in good agreement with the function of C-SH2, which acts to recruit phosphopeptides rather than to regulate the activity of the phosphatase domain.
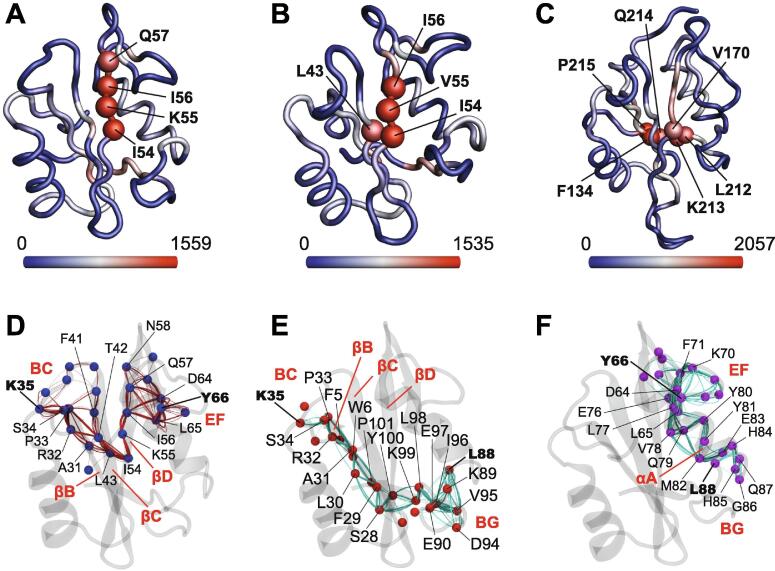
The presence of significant correlation in the motions of different N-SH2 structural elements prompted us to analyze the MD trajectories by Correlation Network Analysis (CNA). In this approach, protein structures are represented by abstract correlation networks, in which the nodes correspond to protein Cα carbons and nodes with motional correlation are connected by “edges”, which are weighted according to the degree of correlation. In this way, CNA reveals the relative importance of each node in the correlated motions and allows identification of the residues with the largest number of edges (greatest degree of centrality) as motional “hubs”. In both N-SH2–pY and SB N-SH2–pY, residues I54, K55/V55 and I56 are the hubs of the network, underlining the importance of the βD C-terminal part in the coordination of the internal dynamics of SH2 domains (Fig. 8A, B). Interestingly, in C-SH2–pY the centrality of this segment is reduced, and the most connected nodes are located very close to the C-terminus of the protein (L212–P215, Fig. 8C).
Fig. 8.
CNA analysis of the MD simulations of N-SH2–pY, SB N-SH2–pY and C-SH2–pY. (A, B, C) Network centrality of the Cα atoms in N-SH2–pY (A), SB N-SH2–pY (B) and C-SH2–pY (C), respectively. The residues with the highest degree of centrality (hubs) are shown as spheres. The color-range from blue to red indicates the degree of betweenness centrality. (D, E, F) Representation of 250 communication pathways between three different regions of N-SH2–pY. Dynamic changes in the BC loop due to pY binding can be relayed to the EF (D) and BG (E) loops via the central β sheet, while αB mediates transfer of dynamic information between the EF and BG loops (F). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Finally, calculation of optimal and sub-optimal paths connecting two selected nodes provides a visualization of how dynamic processes are propagated through the protein. In wild-type N-SH2, changes in the BC loop (for example to K35) are transmitted to the EF loop (for example to Y66) through a series of backbone hydrogen bonds between the BC loop, the C-terminal parts of βB and βD and the N-terminal part of βC, and through another set of hydrogen bonds between βD-I56 and -Q57 and EF-D64 and -L65 (Fig. 8D for N-SH2). Residues 54–56 are crossed by all 250 paths of this network. From the BC (K35) to the BG (L88) loop, dynamics are propagated along βB (up to S28) to BG K99 and Y100 and from there to other residues in the BG loop (Fig. 8E), while communication between the EF and BG loop is almost entirely mediated by the αB helix (Fig. 8F).
In both SB N-SH2–pY and C-SH2, the communication pathways are similar to those described in wild-type N-SH2 (Figure S9). However, the length of the K35–Y66 and Q141–G182 sub-optimal pathways in SB N-SH2 and C-SH2, respectively, was greater than in N-SH2, indicating that the communication between the BC and EF loops is less pronounced.
4. Discussion
Since their discovery, SH2 domains have represented the archetype of a domain with a specific function found as a modular unit in larger proteins [79]. This well-conserved fold binds different phosphopeptides in numerous cellular contexts, where it achieves the necessary level of specificity thanks to a finely tuned network of enthalpic and entropic contributions to the interaction with the cognate phosphopeptide [80]. The regulation of binding specificity, as well as that of the conformational and dynamic changes induced by phosphopeptide binding, becomes even more stringent when SH2 domains modulate the activity of an enzymatic domain within the same protein. In the protein tyrosine phosphatase SHP2, two SH2 domains arranged in tandem couple the enzymatic activity of the catalytic PTP domain with the recognition of upstream partners.
Here, we have characterized the binding to the SHP2 N-SH2 and C-SH2 domains of six phosphorylated peptides from the two SHP2 activators PD-1 and IRS1 and from the disordered C-terminus of SHP2 itself. We find affinities ranging from 10 nM to 20 µM and are able to rationalize many of the differences on the basis of the structures of the complexes. In a first approximation, the binding affinities in-vitro inform on the selectivity and specificity that can be expected in an in-vivo setting. Most SHP2-activator motifs (including those studied here) occur naturally in tandem, which guarantees a far higher degree of binding specificity, mostly because of the spatial constraints imposed by the linker connecting the two pY-motifs [13]. In the case of PD-1, a high-affinity interaction (C-SH2–ITSM) is paired with a two orders-of-magnitude weaker interaction (N-SH2–ITIM). It is likely that this combination is relevant to the physiological role of SHP2 in PD-1 signaling, where ITSM binding to C-SH2 guarantees stable and specific recruitment, while the relatively low-affinity of ITIM for N-SH2 prevents sustained SHP2 activation. ITSM has higher affinity than ITIM for both C-SH2 and N-SH2; this implies that, given a sufficiently high concentration of PD-1, ITSM can displace ITIM from N-SH2, resulting in an arrangement with two PD-1 molecules bound to an individual SHP2 molecule, each via the ITSM. The existence of this 1:2 SHP2:PD-1 complex was originally observed in vitro [17] and later confirmed in live cells [81], but the physiological role of this species is not yet clear. Contrary to PD-1, IRS1 binds SHP2 with two high-affinity motifs. This is likely to ensure retention of the activated phosphatase in the signaling pathway for a longer duration, which might be necessary for SHP2 to dephosphorylate several proximal targets in the Ras/MAPK and PI3K signaling pathways [15], before IRS1 is itself degraded by the ubiquitin–proteasome system [82]. Finally, the very low affinity of the pY motifs derived from the C-terminus of SHP2 for both N-SH2 and C-SH2 make it unlikely that they have a role in regulating phosphatase activity. Instead, the presence within these pY-motifs of the consensus sequence for binding to the Grb2 SH2 domain reinforces the putative role of SHP2 in the recruitment of Grb2 [15].
The traditional view of the activation mechanism of SHP2 posits that a monophosphorylated peptide of sufficiently high affinity inserts its residues at positions pY+1, pY+3 and pY+5 into the specificity pocket of N-SH2; this binding event results in a conformational change of the EF loop, which creates a steric clash with the PTP domain. As a consequence, the interaction between the N-SH2 and PTP domains is disfavored and the active site becomes available for substrate-binding [11]. This view ascribes a “passive” role to phosphotyrosine: it provides free energy of binding (non-phosphorylated peptides do not bind SH2 domains), but it is the residues C-terminal to it that play the crucial role in SHP2 activation. In addition, it remains an open question as to how the peptide C-terminal residues can bind to the N-SH2 domain in the auto-inhibited SHP2 conformation, where the EF and BG loops adopt a conformation that partially closes the specificity pocket. In fact, previous studies demonstrated that the active conformation of SHP2 has higher affinity for phosphopeptides than the inactive conformation [83], [84].
Here we demonstrate that phosphotyrosine binding causes significant NMR chemical-shift perturbations across the entire N-SH2 domain, suggesting that pY alone is capable of inducing global rearrangements of the ground-state protein conformation. Our MD simulations of the N-SH2–pY complex indicate the presence of a correlated network of protein motions that conveys information allosterically from the pY binding-site to the specificity pocket via a few key nodes located in the βC (F41, T42 and L43) and βD (I54, K55, I56 and Q57) strands. In complex with pY, N-SH2 alternates between two major conformations (γ and ε), in only one of which is the specificity pocket accessible to the phosphopeptide residues following pY+3.
The coupling between the pY binding-site and the specificity pocket has been observed previously for simulations of N-SH2 in complex with phosphopeptides [29], [30]. The authors of this study found two distinct conformational states (α and β) for 12 SHP2 N-SH2–phosphopeptide complexes, characterized either by stably zipped βC and βD strands, together with open pY and specificity pockets (β state), or by unzipped βC and βD strands, together with more closed pY and specificity pockets (α state). The N-SH2–phosphopeptide complexes do not transition between the states but adopt either one or the other, depending on the nature and the orientation of the side-chain at position pY+5. A comparison between the α and β states found in this previous work and the γ, δ and ε states found here (Figure S10) shows that our γ and δ/ε states correspond to the β and α states of [29], [30], respectively (Figure S10B and C). This indicates that pY-binding stimulates transitions between the α and β states of Anselmi et al. [29], [30], while the binding of the rest of the phosphopeptide selects one of the two states. In the absence of ligands, N-SH2 is found exclusively in the γ state. In qualitative agreement, the NMR-derived fast-dynamics parameters of the EF and BC loops, as well as those of the C-terminal part of βD in both apo N-SH2 and the N-SH2–ITIM and N-SH2–ITSM complexes do not show enhanced mobility with respect to the rest of the protein (Fig. 5).
The inaccessibility of the pY+5 binding pocket in the ε state leads us to predict that the ε conformation is unable to accommodate residue pY+5 in this pocket (Figure S11). An analysis of the experimentally available conformations of N-SH2–phosphopeptide complexes together with the 12 MD-derived conformations [30] (Figure S12) demonstrates that this indeed the case. When the G39-C’–N58-N distance exceeds 8 Å, the side-chain of the residue at position pY+5 does not occupy the specificity pocket but is rather exposed to the solvent (Figure S12C). In this case, we expect no contribution of this residue to the binding enthalpy. In our MD simulations, the SB N-SH2–pY complex showed less frequent βC-βD unzipping and longer Y66-Cα–L88-Cα/G68-Cα–L88-Cα distances (Fig. 7E and S12D&E). The absence of the ε state in the mutant SB N-SH2 may contribute to the improved affinity of SB N-SH2 to phosphopeptides, over and above that due to the optimized molecular-recognition properties of the pY-binding pocket. In agreement with this hypothesis, the enhanced activity of SHP2 T42A, an overly active variant associated with Noonan syndrome, is thought to be due to increased affinity for upstream binders [83], [85], [86], rather than to modulation of its basal activity.
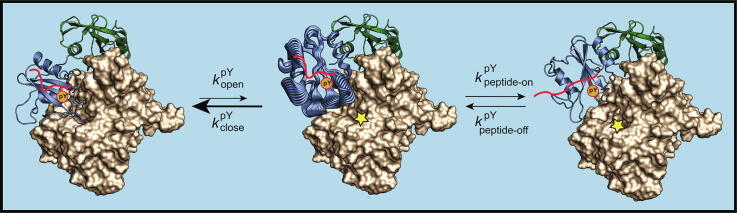
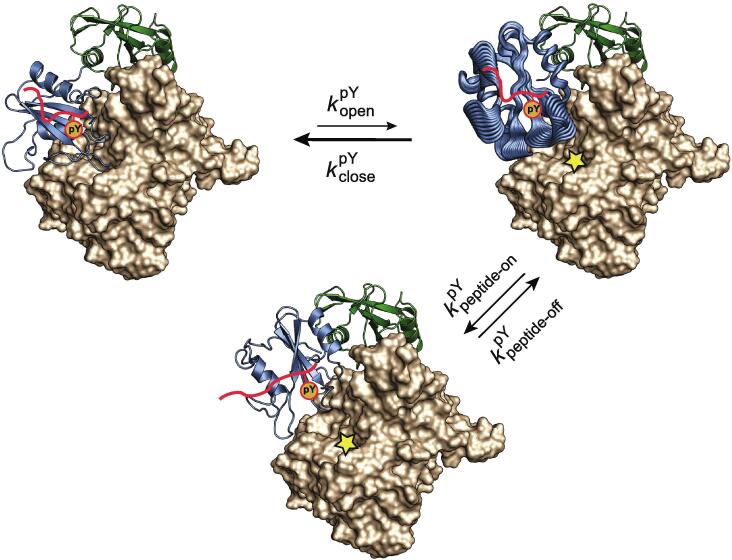
We propose that the correlated dynamics observed in silico for SH2 upon pY-binding acts to destabilize the N-SH2 interface upon binding of the phosphotyrosine residue of activating peptides (Fig. 9). In the SHP2 closed state, the N-SH2 domain of SHP2 occludes the catalytic site of the PTP, inhibiting phosphatase activity. A low level of basal activity remains due to a small percentage of open, active conformation. An activating phosphopeptide with high affinity for N-SH2 cannot occupy its complete binding site in the SHP2 closed conformation due to steric hindrance by the EF loop, but the pY moiety of the peptide can still bind. After spontaneous transition of SHP2–pY to its open conformation, the motions induced by the presence of bound pY perturb the PTP interaction surface (consisting of the EF and DE loop) and thus prevent a re-closing of the N-SH2 domain on the PTP. This results in a slower rate of closure, kclose, in the presence of pY than in its absence. It is also likely that pY-binding increases the rate of transition to the open conformation, kopen; however, our MD simulations carried out with the isolated N-SH2 domain give no information on the dynamics of this domain when it is connected to the PTP domain. In addition to favoring the open SHP2 conformation, the motions induced by pY-binding provide access to different conformations of the specificity pocket that can be selected by the C-terminal half of the peptide for full binding (Fig. 9). Once the C-terminal residues of the peptide are locked into the specificity pocket, our NMR experimental data support a picture where the dynamics of the DE and EF loops are quenched, while the dynamics at longer time-scales is preserved in the BG loop (Fig. 4 and Fig. 5). After the N-SH2–phosphopeptide complex is completely formed, its lifetime (1/kpeptide-off) determines the efficiency of SHP2 activation.
Fig. 9.
A model for the activation of SHP2 by mono-phosphorylated peptides. The N-SH2 domain of SHP2 occludes the catalytic site of the PTP, inhibiting phosphatase activity. A low-level of basal activity remains due to a small percentage of open, active conformation. A high-affinity peptide that carries a phosphotyrosine residue (pY) cannot fully occupy its cognate binding site on N-SH2 due to steric hindrance by the EF loop; however, the pY moiety can still bind. After spontaneous transition of SHP2–pY to its open conformation, the bound pY triggers conformational fluctuations (represented by broad cartoons) in several regions of N-SH2, disfavoring the re-closing of SHP2-pY and simultaneously opening up the N-SH2 specificity pocket to accommodate the phosphopeptide residues C-terminal to pY+3. The full binding of the phosphopeptide then stabilizes the N-SH2 loops in a conformation that precludes interaction with the PTP domain and hence maintains the accessibility of the catalytic site of the PTP domain. The SHP2–phosphopeptide complex is thus kept in its activated state for as long as the phosphopeptide remains bound to the N-SH2 domain. and are the rate constants for the transition between the closed and open conformations of SHP2 in the presence of bound pY. and are the rate constants for binding and unbinding of the C-terminal part of the peptide to the specificity pocket of N-SH2.
The correlated dynamics network appears to be particular to N-SH2, since in C-SH2 the correlation between the motions of the pY and specificity pockets is markedly less pronounced. The stringent selectivity and weakly-correlated dynamics of C-SH2 fit well with the notion that this domain mainly acts to recruit bidentate phosphopeptides to SHP2, while N-SH2, with its more promiscuous binding and correlated dynamics, has evolved to couple phosphopeptide binding with regulation of phosphatase activity [17].
Using MD simulations, Anselmi and Hub find that the state of N-SH2 with unzipped βC- and βD-strands (state α in their work and states ε/δ in our work) is better at inducing the displacement of the N-SH2 domain from PTP [29]. They propose a mechanism of activation of SHP2 where the peptide binds to the closed conformation of the phosphatase, but only peptides binding to N-SH2 in the α state induce opening and activation of SHP2. This hypothesis may seem problematic, as strong activators of the phosphatase, such as PD-1 ITSM and PDGFR-1009, bind N-SH2 without unzipping the βC- and βD-strands (Figure S12). Our model may reconcile this discrepancy by proposing, next to the model of Anselmi and Hub [29], a second activation pathway (Fig. 9), based on the initial recognition of pY by the closed conformation of SHP2 and full peptide binding to the open conformation, whose population is increased by the pY-binding. Once the peptide is bound, the γ state of N-SH2 is also incompatible with the closed conformation of SHP2 because of steric clashes between the EF loop and the PTP domain (Figure S1C). Furthermore, the fact that Anselmi and Hub found that the α conformation induces SHP2 opening supports our hypothesis that pY-binding increases the rate kopen by promoting the transition from the γ to the ε/δ states, which are similar to the α state of Anselmi and Hub [29]. The two mechanisms of SHP2 activation may coexist or be mutually exclusive, depending on the peptide sequence.
5. Conclusions
The N-SH2 domain of SHP2 differs from many other SH2 domains (including SHP2 C-SH2) because its binding to upstream phosphopeptides is coupled to the regulation of enzymatic activity. In this context, phosphotyrosine-recognition not only provides free energy of binding, but also has significant effects on the global structure and internal dynamics of the domain. In the pY-bound state, the pY-binding pocket and the specificity pocket experience correlated fluctuations, which correspond to two principal conformational states of N-SH2–pY; of these two states, only one can accommodate the phosphopeptide residues C-terminal to pY+3. The fluctuations induced by pY-binding disrupt the interaction interface with the PTP domain, thus supporting SHP2 activation. The results described here explain how high-affinity monophosphorylated peptides can bind the N-SH2 domain of auto-inhibited SHP2, despite the inaccessibility of part of the specificity pocket when N-SH2 is engaged with the PTP domain.
In conclusion, we show that, despite their very similar fold, SH2 domains have a range of structural and dynamical differences that support a variety of specificities and functions. These properties cannot be revealed by X-ray structures, as they are related to the allosteric coupling of the dynamics of binding pockets and structural elements. Likely, the concepts introduced here for the SH2 domains of SHP2 also apply to other SH2 domains involved in the regulation of enzymatic activity (as, for example, those of phospholipase Cγ1 or Src-family kinases) and are thus of general relevance for understanding the mechanisms of function of SH2 domains.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the German Science Foundation DFG (grant CA 294/20-1 to TC). MM was supported by a fellowship from the Hannover School for Biomolecular Drug Research (HSBDR) and was a member of the Hannover Biomedical Research School (HBRS) and the MD/PhD program “Molecular Medicine”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.040.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116(2):191-203. Epub 2004/01/28. doi: 10.1016/s0092-8674(03)01077-8. PubMed PMID: 14744431. [DOI] [PubMed]
- 2.Hunter T. Tyrosine phosphorylation: thirty years and counting. Current opinion in cell biology. 2009;21(2):140-6. Epub 2009/03/10. doi: 10.1016/j.ceb.2009.01.028. PubMed PMID: 19269802; PubMed Central PMCID: PMCPMC2670436. [DOI] [PMC free article] [PubMed]
- 3.Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Molecular cell. 2006;22(6):851-68. Epub 2006/06/24. doi: 10.1016/j.molcel.2006.06.001. PubMed PMID: 16793553. [DOI] [PubMed]
- 4.Waksman G, Kumaran S, Lubman O. SH2 domains: role, structure and implications for molecular medicine. Expert reviews in molecular medicine. 2004;6(3):1-18. Epub 2004/02/28. doi: 10.1017/s1462399404007331. PubMed PMID: 14987415. [DOI] [PubMed]
- 5.Eck M.J., Shoelson S.E., Harrison S.C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature. 1993;362(6415):87–91. doi: 10.1038/362087a0. PubMed PMID: 7680435. [DOI] [PubMed] [Google Scholar]
- 6.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767-78. Epub 1993/03/12. doi: 10.1016/0092-8674(93)90404-e. PubMed PMID: 7680959. [DOI] [PubMed]
- 7.Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S. Defining the Specificity Space of the Human Src Homology 2 Domain*. Mol Cell Proteomics. 2008;7(4):768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Ladbury JE, Arold ST. Energetics of Src homology domain interactions in receptor tyrosine kinase-mediated signaling. Methods in enzymology. 2011;488:147-83. Epub 2011/01/05. doi: 10.1016/b978-0-12-381268-1.00007-0. PubMed PMID: 21195228. [DOI] [PubMed]
- 9.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends in biochemical sciences. 2003;28(6):284-93. Epub 2003/06/27. doi: 10.1016/s0968-0004(03)00091-4. PubMed PMID: 12826400. [DOI] [PubMed]
- 10.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92(4):441-50. Epub 1998/03/10. doi: 10.1016/s0092-8674(00)80938-1. PubMed PMID: 9491886. [DOI] [PubMed]
- 11.Barford D, Neel BG. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure (London, England : 1993). 1998;6(3):249-54. Epub 1998/04/29. doi: 10.1016/s0969-2126(98)00027-6. PubMed PMID: 9551546. [DOI] [PubMed]
- 12.Pluskey S., Wandless T.J., Walsh C.T., Shoelson S.E. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. The Journal of biological chemistry. 1995;270(7):2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- 13.Ottinger EA, Botfield MC, Shoelson SE. Tandem SH2 domains confer high specificity in tyrosine kinase signaling. J Biol Chem. 1998;273(2):729-35. Epub 1998/02/14. doi: 10.1074/jbc.273.2.729. PubMed PMID: 9422724. [DOI] [PubMed]
- 14.Longo D.L., Boussiotis V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med. 2016;375(18):1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dance M., Montagner A., Salles J.-P., Yart A., Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell Signal. 2008;20(3):453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Xu E., Schwab M., Marette A. Role of protein tyrosine phosphatases in the modulation of insulin signaling and their implication in the pathogenesis of obesity-linked insulin resistance. Rev. Endocrine Metabolic Disorders. 2014;15(1):79–97. doi: 10.1007/s11154-013-9282-4. [DOI] [PubMed] [Google Scholar]
- 17.Marasco M., Berteotti A., Weyershaeuser J., Thorausch N., Sikorska J., Krausze J. Molecular mechanism of SHP2 activation by PD-1 stimulation. Science. Advances. 2020;6(5):eaay4458. doi: 10.1126/sciadv.aay4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peled M., Tocheva A.S., Sandigursky S., Nayak S., Philips E.A., Nichols K.E. Affinity purification mass spectrometry analysis of PD-1 uncovers SAP as a new checkpoint inhibitor. Proc Natl Acad Sci U S A. 2018;115(3):E468–E477. doi: 10.1073/pnas.1710437115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto S., Wandless T.J., Shoelson S.E., Neel B.G., Walsh C.T. Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. J Biol Chem. 1994;269(18):13614–13622. Epub 1994/05/06 PubMed PMID: 7513703. [PubMed] [Google Scholar]
- 20.Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, et al. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352(6330):73-7. Epub 1991/07/04. doi: 10.1038/352073a0. PubMed PMID: 1648180. [DOI] [PubMed]
- 21.Bennett A.M., Tang T.L., Sugimoto S., Walsh C.T., Neel B.G. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci U S A. 1994;91(15):7335–7339. doi: 10.1073/pnas.91.15.7335. PubMed PMID: 8041791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Nishimura R., Kashishian A., Batzer A.G., Kim W.J., Cooper J.A. A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol Cell Biol. 1994;14(1):509–517. doi: 10.1128/mcb.14.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel W., Ullrich A. Multiple in vivo phosphorylated tyrosine phosphatase SHP-2 engages binding to Grb2 via tyrosine 584. Cell Growth Differ. 1996;7(12):1589–1597. Epub 1996/12/01 PubMed PMID: 8959326. [PubMed] [Google Scholar]
- 24.Lu W, Gong D, Bar-Sagi D, Cole PA. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Molecular cell. 2001;8(4):759-69. Epub 2001/10/31. doi: 10.1016/s1097-2765(01)00369-0. PubMed PMID: 11684012. [DOI] [PubMed]
- 25.Sun J., Lu S., Ouyang M., Lin L.-J., Zhuo Y., Liu B.o. Antagonism between binding site affinity and conformational dynamics tunes alternative cis-interactions within Shp2. Nat Commun. 2013;4(1) doi: 10.1038/ncomms3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huyer G, Ramachandran C. The specificity of the N-terminal SH2 domain of SHP-2 is modified by a single point mutation. Biochemistry. 1998;37(9):2741-7. Epub 1998/04/16. doi: 10.1021/bi9722913. PubMed PMID: 9485424. [DOI] [PubMed]
- 27.Huyer G, Li ZM, Adam M, Huckle WR, Ramachandran C. Direct determination of the sequence recognition requirements of the SH2 domains of SH-PTP2. Biochemistry. 1995;34(3):1040-9. Epub 1995/01/24. doi: 10.1021/bi00003a039. PubMed PMID: 7530043. [DOI] [PubMed]
- 28.Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44(45):14932-47. Epub 2005/11/09. doi: 10.1021/bi051408h. PubMed PMID: 16274240. [DOI] [PubMed]
- 29.Anselmi M., Hub J.S. An allosteric interaction controls the activation mechanism of SHP2 tyrosine phosphatase. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-75409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anselmi M., Calligari P., Hub J.S., Tartaglia M., Bocchinfuso G., Stella L. Structural Determinants of Phosphopeptide Binding to the N-Terminal Src Homology 2 Domain of the SHP2 Phosphatase. J Chem Inf Model. 2020;60(6):3157–3171. doi: 10.1021/acs.jcim.0c00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenhausen G., Ruben D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69(1):185–189. doi: 10.1016/0009-2614(80)80041-8. [DOI] [Google Scholar]
- 32.Sklenar V., Piotto M., Leppik R., Saudek V. Gradient-Tailored Water Suppression for 1H–15N HSQC Experiments Optimized to Retain Full Sensitivity. J Magn Reson, Ser A. 1993;102(2):241–245. doi: 10.1006/jmra.1993.1098. [DOI] [Google Scholar]
- 33.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2(6):661-5. Epub 1992/11/01. doi: 10.1007/BF02192855. PubMed PMID: 1490109. [DOI] [PubMed]
- 34.Dayie K.T., Wagner G. Relaxation-Rate Measurements for 15N–1H Groups with Pulsed-Field Gradients and Preservation of Coherence Pathways. J Magn Reson, Ser A. 1994;111(1):121–126. doi: 10.1006/jmra.1994.1236. [DOI] [Google Scholar]
- 35.Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 36.Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 37.Mulder F.A.A., de Graaf R.A., Kaptein R., Boelens R. An Off-resonance Rotating Frame Relaxation Experiment for the Investigation of Macromolecular Dynamics Using Adiabatic Rotations. J Magn Reson. 1998;131(2):351–357. doi: 10.1006/jmre.1998.1380. [DOI] [PubMed] [Google Scholar]
- 38.Geen H, Freeman R. Band-selective radiofrequency pulses. Journal of Magnetic Resonance (1969). 1991;93(1):93-141. doi: https://doi.org/10.1016/0022-2364(91)90034-Q.
- 39.Korzhnev D.M., Skrynnikov N.R., Millet O., Torchia D.A., Kay L.E. An NMR Experiment for the Accurate Measurement of Heteronuclear Spin-Lock Relaxation Rates. J Am Chem Soc. 2002;124(36):10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- 40.Hansen D.F., Kay L.E. Improved magnetization alignment schemes for spin-lock relaxation experiments. J Biomol NMR. 2007;37(4):245–255. doi: 10.1007/s10858-006-9126-6. [DOI] [PubMed] [Google Scholar]
- 41.Li Y.C., Montelione G.T. Solvent Saturation-Transfer Effects in Pulsed-Field-Gradient Heteronuclear Single-Quantum-Coherence (PFG-HSQC) Spectra of Polypeptides and Proteins. J Magn Reson, Ser B. 1993;101(3):315–319. doi: 10.1006/jmrb.1993.1049. [DOI] [Google Scholar]
- 42.Renner C, Schleicher M, Moroder L, Holak TA. Practical aspects of the 2D 15N-[1h]-NOE experiment. J Biomol NMR. 2002;23(1):23-33. Epub 2002/06/14. doi: 10.1023/a:1015385910220. PubMed PMID: 12061715. [DOI] [PubMed]
- 43.Ferrage F., Cowburn D., Ghose R. Accurate Sampling of High-Frequency Motions in Proteins by Steady-State 15N−{1H} Nuclear Overhauser Effect Measurements in the Presence of Cross-Correlated Relaxation. J Am Chem Soc. 2009;131(17):6048–6049. doi: 10.1021/ja809526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen D.F., Vallurupalli P., Kay L.E. An Improved 15N Relaxation Dispersion Experiment for the Measurement of Millisecond Time-Scale Dynamics in Proteins. J Phys Chem B. 2008;112(19):5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687-96. Epub 2005/04/09. doi: 10.1002/prot.20449. PubMed PMID: 15815974. [DOI] [PubMed]
- 47.Hansen DF. Functional and Data Analysis (FuDA).
- 48.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104(17):4546–4559. doi: 10.1021/ja00381a009. [DOI] [Google Scholar]
- 49.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104(17):4559–4570. doi: 10.1021/ja00381a010. [DOI] [Google Scholar]
- 50.Dosset P., Hus J.-C., Blackledge M., Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16(1):23–28. doi: 10.1023/A:1008305808620. [DOI] [PubMed] [Google Scholar]
- 51.Dhulesia A., Bodenhausen G., Abergel D. Predicting conformational entropy of bond vectors in proteins by networks of coupled rotators. J Chem Phys. 2008;129(9):095107. doi: 10.1063/1.2969809. [DOI] [PubMed] [Google Scholar]
- 52.Li DW, Brueschweiler R. A dictionary for protein side-chain entropies from NMR order parameters. J Am Chem Soc. 2009;131(21):7226-7. Epub 2009/05/09. doi: 10.1021/ja902477s. PubMed PMID: 19422234. [DOI] [PubMed]
- 53.Bouvignies G. ChemEx.
- 54.ASali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779-815. Epub 1993/12/05. doi: 10.1006/jmbi.1993.1626. PubMed PMID: 8254673. [DOI] [PubMed]
- 55.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2016;54:5 6 1-5 6 37. PubMed PMID: 27322406. [DOI] [PMC free article] [PubMed]
- 56.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 57.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701-18. Epub 2005/10/08. doi: 10.1002/jcc.20291. PubMed PMID: 16211538. [DOI] [PubMed]
- 58.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157-74. Epub 2004/04/30. doi: 10.1002/jcc.20035. PubMed PMID: 15116359. [DOI] [PubMed]
- 60.Sousa da Silva A.W., Vranken W.F. ACPYPE-Antechamber python parser interface. BMC Res Notes. 2012;5(1):367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 62.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 63.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 64.Miyamoto S., Kollman P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13(8):952–962. doi: 10.1002/(ISSN)1096-987X10.1002/jcc.v13:810.1002/jcc.540130805. [DOI] [Google Scholar]
- 65.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157-74. Epub 2004/04/30. doi: 10.1002/jcc.20035. PubMed PMID: 15116359. [DOI] [PubMed]
- 66.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 67.Grant B.J., Rodrigues A.P.C., ElSawy K.M., McCammon J.A., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22(21):2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 68.Scarabelli G., Grant B.J. Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule, and neck-linker binding sites. Biophys J. 2014;107(9):2204–2213. doi: 10.1016/j.bpj.2014.09.019. PubMed PMID: 25418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao X.-Q., Malik R.U., Griggs N.W., Skjærven L., Traynor J.R., Sivaramakrishnan S. Dynamic Coupling and Allosteric Networks in the α Subunit of Heterotrimeric G Proteins. J. Biological Chem. 2016;291(9):4742–4753. doi: 10.1074/jbc.M115.702605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou S., Shoelson S.E., Chaudhuri M., Gish G., Pawson T., Haser W.G. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–778. doi: 10.1016/0092-8674(93)90404-E. [DOI] [PubMed] [Google Scholar]
- 71.De Souza D., Fabri L.J., Nash A., Hilton D.J., Nicola N.A., Baca M. SH2 Domains from Suppressor of Cytokine Signaling-3 and Protein Tyrosine Phosphatase SHP-2 Have Similar Binding Specificities. Biochemistry. 2002;41(29):9229–9236. doi: 10.1021/bi0259507. [DOI] [PubMed] [Google Scholar]
- 72.Sweeney M.C., Wavreille A.-S., Park J., Butchar J.P., Tridandapani S., Pei D. Decoding Protein−Protein Interactions through Combinatorial Chemistry: Sequence Specificity of SHP-1, SHP-2, and SHIP SH2 Domains. Biochemistry. 2005;44(45):14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 73.Imhof D., Wavreille A.-S., May A., Zacharias M., Tridandapani S., Pei D. Sequence Specificity of SHP-1 and SHP-2 Src Homology 2 Domains: CRITICAL ROLES OF RESIDUES BEYOND THE pY+3 POSITION *. J Biol Chem. 2006;281(29):20271–20282. doi: 10.1074/jbc.M601047200. [DOI] [PubMed] [Google Scholar]
- 74.Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S. Defining the Specificity Space of the Human Src Homology 2 Domain *. Mol Cell Proteomics. 2008;7(4):768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverría C, Furet P, et al. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. 1996;3(7):586-9. Epub 1996/07/01. doi: 10.1038/nsb0796-586. PubMed PMID: 8673601. [DOI] [PubMed]
- 76.Waksman G, Kominos D, Robertson SC, Pant N, Baltimore D, Birge RB, et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358(6388):646-53. Epub 1992/08/20. doi: 10.1038/358646a0. PubMed PMID: 1379696. [DOI] [PubMed]
- 77.Hayashi T., Senda M., Suzuki N., Nishikawa H., Ben C., Tang C. Differential Mechanisms for SHP2 Binding and Activation Are Exploited by Geographically Distinct Helicobacter pylori CagA Oncoproteins. Cell Reports. 2017;20(12):2876–2890. doi: 10.1016/j.celrep.2017.08.080. [DOI] [PubMed] [Google Scholar]
- 78.Kaneko T., Huang H., Cao X., Li X., Li C., Voss C. Superbinder SH2 domains act as antagonists of cell signaling. Sci Signal. 2012;5(243) doi: 10.1126/scisignal.2003021. [DOI] [PubMed] [Google Scholar]
- 79.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11(12):504-11. Epub 2001/11/24. doi: 10.1016/s0962-8924(01)02154-7. PubMed PMID: 11719057. [DOI] [PubMed]
- 80.Marasco M., Carlomagno T. Specificity and regulation of phosphotyrosine signaling through SH2 domains. J Struct Biol X. 2020;4:100026. doi: 10.1016/j.yjsbx.2020.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patsoukis N, Duke-Cohan JS, Chaudhri A, Aksoylar HI, Wang Q, Council A, et al. Interaction of SHP-2 SH2 domains with PD-1 ITSM induces PD-1 dimerization and SHP-2 activation. Commun Biol. 2020;3(1):128. Epub 2020/03/19. doi: 10.1038/s42003-020-0845-0. PubMed PMID: 32184441; PubMed Central PMCID: PMCPMC7078208. [DOI] [PMC free article] [PubMed]
- 82.Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Molecular cell. 2008;30(4):403-14. Epub 2008/05/24. doi: 10.1016/j.molcel.2008.03.009. PubMed PMID: 18498745; PubMed Central PMCID: PMCPMC2633441. [DOI] [PMC free article] [PubMed]
- 83.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280(35):30984-93. Epub 2005/07/01. doi: 10.1074/jbc.M504699200. PubMed PMID: 15987685. [DOI] [PubMed]
- 84.Bocchinfuso G., Stella L., Martinelli S., Flex E., Carta C., Pantaleoni F. Structural and functional effects of disease-causing amino acid substitutions affecting residues Ala72 and Glu76 of the protein tyrosine phosphatase SHP-2. Proteins Struct Funct Bioinf. 2007;66(4):963–974. doi: 10.1002/prot.21050. [DOI] [PubMed] [Google Scholar]
- 85.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45-68. Epub 2005/08/30. doi: 10.1146/annurev.genom.6.080604.162305. PubMed PMID: 16124853. [DOI] [PubMed]
- 86.Martinelli S, Torreri P, Tinti M, Stella L, Bocchinfuso G, Flex E, et al. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Human Molecular Genetics. 2008;17(13):2018-29. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed]
- 87.Papaleo E, Mereghetti P, Fantucci P, Grandori R, De Gioia L. Free-energy landscape, principal component analysis, and structural clustering to identify representative conformations from molecular dynamics simulations: the myoglobin case. J Mol Graph Model. 2009;27(8):889-99. Epub 2009/03/07. doi: 10.1016/j.jmgm.2009.01.006. PubMed PMID: 19264523. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.