Abstract
Circadian clocks are biochemical time-keeping machines that synchronize animal behavior and physiology with planetary rhythms. In Drosophila, the core components of the clock comprise a transcription/translation feedback loop and are expressed in seven neuronal clusters in the brain. Although it is increasingly evident that the clocks in each of the neuronal clusters are regulated differently, how these clocks communicate with each other across the circadian neuronal network is less clear. Here, we review the latest evidence that describes the physical connectivity of the circadian neuronal network . Using small ventral lateral neurons as a starting point, we summarize how one clock may communicate with another, highlighting the signaling pathways that are both upstream and downstream of these clocks. We propose that additional efforts are required to understand how temporal information generated in each circadian neuron is integrated across a neuronal circuit to regulate rhythmic behavior.
Keywords: locomotion behavior, neuronal network, neurotransmitter, neuropeptide, ionotropic, metabotropic, circadian clock
Circadian rhythms are daily behavioral and physiological changes in bacteria, fungi, plants, and animals that synchronize to daily environmental oscillations. The study of the “clockworks” that underlie circadian rhythms in animals can be separated into three primary approaches: (1) the genetic and molecular, (2) the neuronal, and (3) the behavioral and physiological outputs. For a broader review of the genetic and molecular evidence, we point the reader to other sources (Crane and Young, 2014; Top and Young, 2018; Williams and Sehgal, 2001; Yu and Hardin, 2006). Here, we review how Drosophila circadian clocks communicate with each other across the circadian neuronal network (CNN), and how the information from each circadian clock may be integrated across the network to program circadian rhythms.
Genes, Loops and Regulation of the Circadian Clock
Before exploring the circadian neuronal circuitry, an introduction to the “gears” that comprise the clockworks is necessary. A pioneering forward genetics screen in Drosophila melanogaster revealed three variants of circadian behavior (long, short, null), which were allelic and mapped to a single locus named period (per), which was cloned about a decade later (Bargiello et al., 1984; Konopka and Benzer, 1971; Reddy et al., 1984). Subsequent genetic screens revealed additional components including timeless (tim), doubletime (dbt), cycle (cyc), vrille (vri), clock (clk), clockwork orange (cwo), and PAR domain protein 1ε (pdp1ε) (Allada et al., 1998; Blau and Young, 1999; Kadener et al., 2007; Kloss et al., 1998; Matsumoto et al., 2007; Price et al., 1998; Rutila et al., 1998; Sehgal et al., 1994, 1995; Vosshall et al., 1994; Zheng et al., 2009). These components form a transcription/translation negative feedback loop called the “circadian clock” (Hardin et al., 1990). The primary negative feedback loop is comprised of the CLK/CYC activator complex that initiates the transcription of per and tim, whose protein products later form a transcriptional inhibitor complex that binds CLK/CYC, thereby closing the loop (Glossop et al., 1999). The secondary feedback loop, which itself can be subdivided into two, is comprised of the CLK/CYC activator complex that initiates the transcription of vri and pdp1ε, which inhibit and promote CLK/CYC activity, respectively (Cyran et al., 2003; Glossop et al., 2003). In what can be considered a third feedback loop, CLK/CYC initiates transcription of cwo, which after translation competes for the DNA E-boxes bound by CLK/CYC, inhibiting expression of CLK/CYC target genes (Lim et al., 2007a; Zhou et al., 2016).
Negative feedback loops require built-in delays to create an oscillation. Critically timed and tightly regulated delay mechanisms govern the circadian clock to ensure a transcriptional oscillation of ~24 h. These delays include a delay between transcription and translation of clock genes including per and tim, a delay in nuclear entry of the PER/TIM repressor complex, and a delay in degradation of nuclear PER/TIM. At the protein level, these delays are regulated by post-translational modifications such as phosphorylation, dephosphorylation, ubiquitination and glycosylation (Top and Young, 2018). These mechanisms converge to regulate the oscillating expression of ~10% of Drosophila genes (Abruzzi et al., 2011; Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Meireles-Filho et al., 2014). Rhythmic regulation of post-transcriptional events such as protein translation, membrane localization, and splicing permits the circadian clock to indirectly regulate rhythmic expression of hundreds of additional proteins (Huang et al., 2013; Lear et al., 2005a; Wang et al., 2018).
Neuronal Clocks
The circadian clock is present in numerous tissues throughout the fly (Giebultowicz, 2001; Ito et al., 2008; Kaneko and Hall, 2000; Plautz et al., 1997). Locomotor behavior is primarily used to monitor circadian behavior due to the ease of testing, but it must be noted that other behavioral or physiological circadian outputs are likely to have been unwittingly overlooked due to this focus. For this reason, the focus of understanding the clockworks has been on brain clocks. In the brain, the molecular components of the circadian clock (e.g. VRI, PER, and TIM), are expressed in ~150 neurons organized into 7 neuronal clusters named for their anatomical location (Blau and Young, 1999; Ewer et al., 1992; Helfrich-Förster et al., 2007; Hunter-Ensor et al., 1996; Kaneko, 1998; Kaneko and Hall, 2000; Kaneko et al., 1997; Rothenfluh et al., 2000; Rutila et al., 1996). The clusters include the small ventral lateral neurons (s-LNvs), the large ventral lateral neurons (l-LNvs), the dorsal lateral neurons (LNds), three groups of dorsal neurons (DN1, DN2, and DN3), lateral posterior neurons (LPNs) and the lone 5th s-LNv neuron, often grouped with the LNds (Figure 1) (Helfrich-Förster, 1997; Kaneko, 1998; Kaneko et al., 1997; Schubert et al., 2018). These neuronal clusters can be further subdivided based on their expression of molecular markers, such as neuropeptides, that distinguish the neurons from one another.
Figure 1.
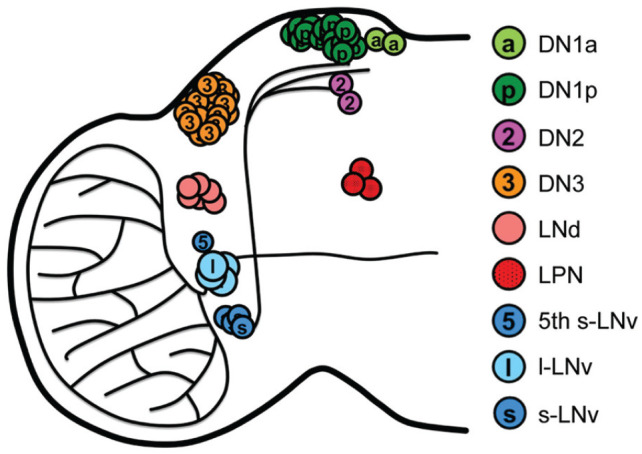
Circadian neuronal network anatomical organization. Schematic of the neuroanatomical locations of the circadian neuronal clusters. Abbreviations: DN1a = Dorsal Neurons 1a; DN1p = Dorsal Neurons 1p; DN2 = Dorsal Neurons 2; DN3 = Dorsal Neurons 3; LNd = Dorsal lateral neurons; LPN = lateral posterior neuron; LNv = ventral lateral neuron; l-LNv = large ventral lateral neuron; s-LNv = small ventral lateral neuron.
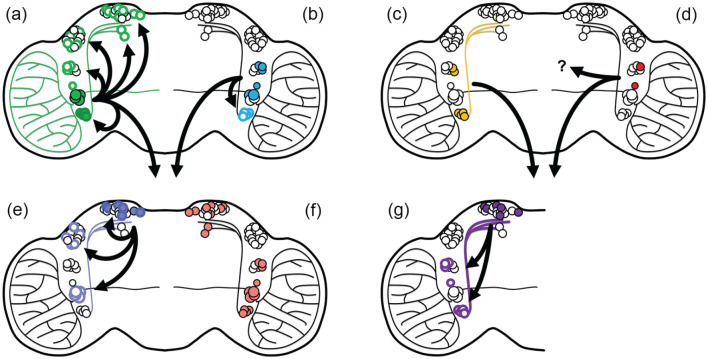
Circadian clocks in the brain communicate with each other across a CNN through neuropeptides and synaptic connections. Disruption of synaptic transmission interferes with locomotor behavior (Kaneko et al., 2000). Effort from a number of labs have revealed a number of neurotransmitters that are used by clocks to communicate across neuronal clusters in the CNN (Guo et al., 2018; Q. He et al., 2017; Schlichting et al., 2016; Schubert et al., 2018). A recent electron microscopy-based synaptic connectivity map of the Drosophila adult central brain, the hemibrain connectome, provides additional information on potential synaptic connectivity in the CNN (Scheffer et al., 2020) (https://neuprint.janelia.org). Interestingly, if taken at face value, this work suggests that there may be synaptic connections across the two hemispheres of the fly brain, mediated by a subset of DN1 posterior (DN1p) neurons, DN1pA, that connect to the contralateral LNds and 5th s-LNv. It is worth noting that this study reveals no synaptic connections between the LPNs or the DN3s with the remainder of the CNN, although others have reported that DN1s may be presynaptic to DN3s (Guo et al., 2018), suggesting that more investigation is required for a complete connectome map. In addition, a lack of synaptic connection does not mean that a synapse does not exist; the hemibrain connectome does not have a time dimension, and changes in synaptic plasticity across the day are well-documented (Cavey et al., 2016; Duhart et al., 2020; Fernandez et al., 2020; Frank, 2016; Frenkel et al., 2017; Gorostiza et al., 2014; Herrero et al., 2020; Krzeptowski et al., 2018; Tang et al., 2017). Although lack of synaptic connections may appear to suggest that the CNN is not a single interconnected network, a number of neuropeptides transmit information by way of diffusion across the CNN, avoiding the need for direct synaptic connections (Figure 2).
Figure 2.
Map of neurotransmitter communication across the circadian neuronal network. The schematic of the brain illustrates the anatomical location of the circadian neurons. The neurons that generate the indicated neurotransmitter are represented by filled circles. The neurons that express the receptor that responds to the indicated neurotransmitter are represented by rings. (a) pigment dispersing factor (PDF; green), (b) neuropeptide F (NPF; cyan), (c) short neuropeptide F (sNPF; yellow), (d) Ion transport peptide (ITP; red), (e) DH31 (lavender), (f) Cryptochrome (pink), (g) glutamate (purple).
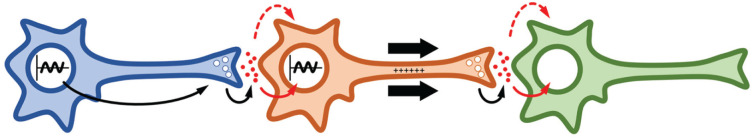
A neurotransmitter released by a neuron can cause ionotropic or metabotropic responses in a downstream neuron that expresses the relevant receptor (Figure 3). When activated, ionotropic receptors flux ions, depolarizing or hyperpolarizing the membrane of the neuron. Voltage-dependent increases in cytosolic calcium (Ca2+) trigger vesicle fusion at presynaptic termini, releasing neurotransmitters. In contrast, metabotropic responses activate second messenger molecules which initiate signaling cascades, leading to activation of ion channels and proteins such as GSK-3/SGG and PKA (Ferkey and Kimelman, 2000; Kaidanovich-Beilin and Woodgett, 2011; L. Kim and Kimmel, 2000; D. Lee, 2015; Mackiewicz et al., 2008), two kinases with prominent regulatory functions in the circadian clock. Ionotropic and metabotropic responses are not mutually exclusive; activation of a metabotropic receptor often initiates intracellular signaling events that cause ion channels to open and depolarize the cell membrane. Conversely, ionotropic receptors can signal to the nucleus through calcium-dependent signals to alter transcriptional programs.
Figure 3.
Models of integrating circadian clock information. In a hypothetical neuronal circuit of three neurons, information from the first neuron is communicated to the third neuron. In metabotropic communication (solid red arrow), the circadian clock from neuron 1 regulates the release of neurotransmitters that communicate to the transcription machinery in neuron 2. In turn, neuron 2 integrates information from its own circadian clock to the signal and communicates to neuron 3. In ionotropic communication (dashed red arrow), the circadian clock from neuron 1 communicates to neuron 3 by bypassing the circadian clock in neuron 2, using neuron 2 as a communications cable. Sinusoidal line in nucleus represents the circadian clock. White circles represent vesicles. Red circles represent neurotransmitters. Thick arrows represent direction of changing action potential (+++++). Small arrows represent steps of communication within and across neurons.
One important convoluting factor is synaptic plasticity across the CNN. It is evident that circadian neurons are remodeled in a rhythmic fashion by small GTPases involved in cytoskeletal rearrangements (Fernández et al., 2008; Petsakou et al., 2015). Assuming a comprehensive communication map of multiple clocks across the CNN, we still face the daunting task of determining how the “circuit board” itself changes with time as it runs the circadian clock “program.” As was the case before the clock genes were arranged into a transcriptional negative feedback loop (Hardin et al., 1990), accumulating components, time-dependent changes to synapses and differential responses to neurotransmitters will help in developing models of neuronal circuitry feedback loops arranged into a coherent mechanism to elicit behavior.
Neurotransmission in the Circadian Neuronal Network
There are two important assumptions made in Drosophila circadian research. The first assumption is that the neuronal clocks are the relevant clocks. The primary output measured as a proxy for circadian clock activity is locomotor activity (i.e. behavior). Thus, the neuron-behavior link has placed emphasis on understanding neuronal clocks. Although we too will focus on behavioral outputs as regulated by neuronal clocks in this review, it is worth remembering that clocks in non-neuronal tissues are likely to have an important effect on the function of neuronal clocks through various feedback mechanisms across these tissues. It is also worth mentioning that limiting clock outputs to primarily one output, locomotor activity, makes an assumption that other behaviors and physiologies are equally changed by the manipulation of clock genes.
The second assumption is that circadian clocks should only be studied in constant darkness conditions. Rhythmic behavior is studied in constant conditions (constant darkness) away from environmental cues that influence and entrain the circadian clock. Under these conditions, the LNvs are the dominant neurons within the CNN (Allada and Chung, 2010; Renn et al., 1999), earning themselves the title “master pacemakers,” which has resulted in a lot of information about these cells. The best-known characteristic of LNvs is their production of the neuropeptide, pigment dispersing fact (PDF) (expanded upon below). Without PDF, flies in constant darkness become arrhythmic in behavior, similar to the phenotype observed in per0 flies, making PDF a key circadian clock neurotransmitter (Renn et al., 1999; R. F. Smith and Konopka, 1981; Wheeler et al., 1993). However, in a light-dark cycle, mutants of these two genes differ, with per0 flies exhibiting loss of morning and evening anticipatory behavior, and pdf 01 flies exhibiting advanced evening anticipatory behavior (expanded below). This difference hints at the complexities that underly circadian behavior regulation.
We will begin by focusing on the LNvs, primarily for historical reasons. However, as we develop our discussion, we will point out how these assumptions do not explain all observed phenomena. We will point out how signaling mechanisms may influence circadian clocks in each neuron differently, and how these signals can be integrated to influence neuronal activity. We begin by pointing out that targeted CRISPR-mediated elimination of PER or TIM in the LNvs knocks out the “master pacemaker clock,” yet permits rhythmic behavior in constant darkness (Delventhal et al., 2019).
Ventral Lateral Neurons and PDF
The LNvs are the most studied circadian neuronal cluster for their inferred role as master pacemakers that dominate the neuronal network in constant dark conditions (DD), and for their expression of the neuropeptide PDF that is critical to maintaining rhythmic behavior (Grima et al., 2004; Stoleru et al., 2004). Among the two LNv clusters, s-LNvs appear to be the “true” master pacemaker (Menegazzi et al., 2017; Shafer and Taghert, 2009). In light-dark conditions (LD), the s-LNvs regulate morning anticipation, described as increased locomotor activity of flies before the lights are turned on (Grima et al., 2004; Stoleru et al., 2004). DN1as are presynaptic to the s-LNvs. GRASP (GFP Reconstitution Across Synaptic Partners) and electron microscopy experiments reveal that the s-LNvs extend to the DN1s to form an active synapse (Guo et al., 2016; Yasuyama and Meinertzhagen, 2010). The hemibrain connectome suggests that s-LNvs may form connections with DN1pAs, DN2s and LNds in the CNN, though a connection with the LNds is disputed (W. J. Kim et al., 2013; Scheffer et al., 2020). Possible lack of synaptic connections between s-LNvs and other CNN neurons may suggest that these connections were missed due to time-dependent synaptic plasticity, due to communication by s-LNvs to the remainder of the CNN by other means, such as neuropeptide signaling, or that communication occurs through the limited clusters s-LNvs form synapses with.
Both s-LNvs and l-LNvs express PDF, with s-LNvs also expressing sNPF, to modulate the amplitude, synchrony, and the pace of rhythmic behavior (Helfrich-Förster, 1995; Johard et al., 2009; Park et al., 2000). Loss of PDF (pdf 01 mutant) causes flies to become arrhythmic in behavior in DD (Renn et al., 1999). PDF acts on other neurons within the CNN, in addition to the s-LNvs, through its receptor, PDFR (Hyun et al., 2005; Lear et al., 2005b; Mertens et al., 2005) (Figure 2a). As with a pdf 01 mutant, pdfr mutants (han5304 and han3369) advance evening anticipation and eliminate morning anticipation in a LD cycle (Hyun et al., 2005; Lear et al., 2005b; Renn et al., 1999). Reintroduction of PDFR to circadian neurons outside of the LNvs restores morning anticipation and timing of evening anticipation, as well as rhythmic behavior in DD (Lear et al., 2009), indicating that morning anticipation behavior is in fact regulated by a PDF signaling response to LNv instruction. This point is underscored by experiments in which tethered PDF, a PDF variant that is anchored to the cellular membrane of the neuron in which it is expressed, is expressed exogenously in non-LNv circadian neurons, which similarly restores morning and evening anticipation behaviors (and rhythmic behavior in DD) (Choi et al., 2012). The LNv PDF autocrine loop appears to reinforce PDF expression, adding robustness to the oscillating clock (Mezan et al., 2016). Later experiments narrowed the behavior-restoring function of PDF-responsive neuronal clusters in DD conditions to the DN1s (Goda et al., 2019). The role that DN1s appear to play in regulating behavioral rhythms in DD is consistent with observed damping of DN1 clock oscillations that correlate with a damping of behavioral rhythms in the first 1 to 3 days of DD, in which PDF signaling is disrupted (Hyun et al., 2005; Lin et al., 2004; Renn et al., 1999; Roberts et al., 2015; Yoshii et al., 2009). Another PDF-responsive cluster, the LNds, synapse with the LNvs and communicate with them through release of acetylcholine (Duhart et al., 2020). The excitatory effect of acetylcholine on the s-LNvs is modulated by circadian changes in synaptic strength (Duhart et al., 2020). Changes in synaptic strength may also be influenced by the s-LNv’s own response to PDF, which results in increased arborization during the day (Herrero et al., 2020). These findings suggest that timing of evening anticipation in LD conditions depends on the response of LNds to PDF, but may also involve LNv response to acetylcholine released by the LNds in the light-to-dark transition. Thus, one possible model is that DN1s establish rhythmic behavior in DD, while an LNv-LNd interaction accurately times evening anticipation. However, this interpretation of a DN1-LNd relationship is likely incomplete; if four of the six LNds and 5th s-LNv are silenced, flies become arrhythmic. This suggests that LNd neuronal activity is important for maintaining rhythmic behavior in DD (Guo et al., 2014), suggesting possible redundancy between the functions of DN1s and LNds, or DN1 reliance on rhythmic release of glutamate from the LNds (Duhart et al., 2020; Lear et al., 2009). Dynamic changes in neuronal partnerships may also explain the interaction between these three clusters. Under light dark conditions, the s-LNvs pair with LNds or DN1s based on the presence of light, underscoring the contribution of multiple oscillators to circadian behavior (Chatterjee et al., 2018; Lamba et al., 2018). Whether the arrhythmic behavior caused by silencing the LNds is due to a loss of cholinergic or glutamatergic signaling from LNds to the CNN, or a break in information transmission across the LNds through its PDF response is unclear.
Pigment Dispersing Factor Receptor and the Cytosolic Response
PDF expressed by the LNvs is presumably released to bind to PDFR, a G-protein coupled receptor. Within the CNN, PDFR is expressed by DN1as, some DN1ps, DN2s, some DN3s, some LNds, the 5th s-LNv, and the s-LNvs (Hyun et al., 2005; Mertens et al., 2005; Shafer et al., 2008) (Figure 2a). Once activated, PDFR elicits an increase in cyclic adenosine monophosphate (cAMP) and regulated calcium oscillations, which serve as signaling molecules to tune circadian clocks (Im and Taghert, 2010; Klose et al., 2016; Liang et al., 2016, 2017, 2019; Mertens et al., 2005; Palacios-Muñoz and Ewer, 2018; Shafer et al., 2008). Using PDF signaling as a model, we explore the different potential neuronal responses to G-protein coupled receptor signaling.
PDF-PDFR-cAMP-PKA Axis
Cyclic AMP has a wide range of functions in cells. Historically, cAMP has been primarily associated with Protein Kinase A (PKA) activity. Adding PKA inhibitor to S2 cells and overexpression of PKA regulatory subunit (PKAR1) suggest that PKA activity stabilizes PER and TIM proteins (Y. Li et al., 2014; Seluzicki et al., 2014). Thus, PKA activity promotes TIM/PER stability likely in response to PDF signaling (Herrero et al., 2020; Y. Li et al., 2014; Seluzicki et al., 2014). Though it is unclear how PKA stabilizes TIM/PER, one possibility is direct phosphorylation; PKA mutant with reduced kinase activity (DC0) increases electrophoretic mobility of PER on an sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel suggesting reduced PER phosphorylation (Majercak et al., 1997). Another possible mechanism by which PKA stabilizes PER may involve regulating PER nuclear localization, where PER is programmed for degradation by DBT (Ko et al., 2002; Price et al., 1998; Seluzicki et al., 2014; Top et al., 2018). This mechanism may involve direct regulation of PER subcellular localization, as with Rel protein (Drier et al., 1999), or indirect regulation through influencing the GSK-3/SGG activity on TIM, which regulates PER subcellular localization (Kaidanovich-Beilin and Woodgett, 2011; Martinek et al., 2001; Saez and Young, 1996; Top et al., 2016). Indeed, in pdf 01 mutants PER subcellular localization is disrupted in time (Yoshii et al., 2009), consistent with a regulatory mechanism downstream of PDF signaling in some circadian neurons. Thus, the coordination between increased morning activity and increased evening activity in the LNvs, LNds and DN1s may be coordinated through PKA-TIM/PER interactions as instructed by the LNvs through PDF signaling.
PDF-PDFR-cAMP-CREB Axis
Another function of cAMP is the activation of cAMP response element-binding protein (CREB). CREB binds to cAMP-responsive elements (CRE) on DNA, recruiting CREB binding protein (CBP) to activate transcription. CBP influences the function of the CLK/CYC transcription activator complex, as well as per expression (Belvin et al., 1999; Hung et al., 2007; Lim et al., 2007b), suggesting one mechanism by which non-clock transcriptional regulatory elements may convey external signals to the circadian clock. These studies demonstrate that CBP overexpression lengthens per transcription oscillation (Hung et al., 2007) and downregulation shortens per/tim transcription oscillation (Lim et al., 2007b), which correlates with an observed advanced evening anticipation in pdfr mutant flies if PDFR were to upregulate CBP activity. Interestingly, another CREB-regulated protein, CREB-regulated transcription co-activator (CRTC) promotes the transcription of tim but not per, suggesting that tim and per are subject to differential regulation (M. Kim et al., 2016).
PDF-PDFR-cAMP-EPAC Axis
Changes in cAMP concentrations within cells also change the activity of exchange proteins activated by cAMP (EPACs) (Bos, 2003; Seino and Shibasaki, 2005). EPACs are guanine nucleotide exchange factors that facilitate swapping of GDP for GTP in GTPases. EPACs appear to aid Rap1, a Ras-related protein 1 GTPase. Although circadian synaptic plasticity is mediated by Rac- and Rho-type GTPases in regulating rhythmic behavior (Petsakou et al., 2015), there is no known role for Rap1 protein in circadian behavior. However, knock down of EPAC protein in the prothoracic gland leads to longer eclosion rhythms in DD conditions (Palacios-Muñoz and Ewer, 2018), suggesting some role in regulating circadian clocks. Since loss of PDF signaling leads to an initial shortening of rhythmic behavior, it is unlikely that PDF signaling through cAMP acts on target neurons through the EPAC pathway. However, given the number of G-protein coupled receptors involved in CNN communication (see below), it is possible that future studies will reveal EPAC-dependent changes to behaviors that have not yet been investigated.
PDF-PDFR-cAMP-HDAC Axis
Histone modifications are cornerstones of transcription regulation. In mammals, CLK has been identified as a histone acetyltransferase and histone acetylation appears to oscillate rhythmically (Doi et al., 2006; Hove et al., 2003). In flies, a hypomorph of a cAMP-dependent histone deacetylase, HDAC4, causes arrhythmic behavior (Fogg et al., 2014). Although the authors of this study did not show whether changes in cytosolic cAMP concentration, activation of a GPCR, or distinct neuronal clusters are responsive to HDAC4 activity, HDAC4 may represent a regulatory element that receives instruction in tissues downstream of the CNN, which do not express a clock themselves. Such possible mechanisms underscore that circadian clocks are not necessary in all tissues for them to exhibit molecular rhythms.
PDF-PDFR-cAMP-Ca2+ Axis
Recent advances in optical methods allow brain-wide in vivo scanning of Ca2+ concentrations across the CNN in real-time across a full day. When Ca2+ activity within the CNN is recorded using genetically encoded Ca2+ sensors, each cluster exhibits a distinct phase of oscillation. Despite distinct phases, these Ca2+ oscillations are circadian clock (period) dependent (Liang et al., 2016). Strikingly, loss of PDF signaling promotes resynchronization of Ca2+ oscillations in the CNN (with a notable exception of the DN1s, see discussion below), suggesting that PDF signaling is necessary for establishing distinct phases (Liang et al., 2016). Thus, despite a presumed synchrony of circadian clock regulated transcription oscillations within the CNN, within the CNN, PDF acts to desynchronize Ca2+ oscillations.
PDF signaling suppresses cytosolic Ca2+ in neurons. Application of synthetic PDF successfully delays the peak of Ca2+ in LNds and DN3s in pdf 01 mutants (Liang et al., 2017). Oddly, DN1s, which express PDFR, are not responsive to this treatment (Guo et al., 2016). PDF signaling also includes an autocrine mechanism of regulation. In the s-LNvs and LNds, active PDFR significantly lowers basal Ca2+ levels (Liang et al., 2017). Thus, PDF can feed back to suppress the Ca2+ wave and terminate PDF signaling, presumably by limiting Ca2 +-dependent vesicle fusion at s-LNv termini. This suggests that the PDF autocrine system in the s-LNvs likely takes the form of a burst of presumed PDF release in the dark-to-light transition stage of an LD cycle, which is supported by a decrease in detectable PDF at the s-LNv termini in the mornings (Park et al., 2000). Indeed, all circadian neuronal clusters in the fly brain exhibit a decrease in cytosolic Ca2+ during subjective or objective day (in LD or DD), except LNds and the l-LNvs (Liang et al., 2016, 2017). The l-LNvs do not express PDFR, offering an explanation for their unresponsiveness to PDF signaling. The LNds, which show a cytosolic Ca2+ peak during the day (Liang et al., 2016, 2017), may be influenced by other neurotransmitters that integrate with PDF signaling to influence peak cytosolic Ca2+. We will revisit this point when discussing DH31.
The differentially timed Ca2+oscillations across the CNN derive from the oscillation of a single peptide, PDF, underscoring how complexity can arise from a simple system. Such differences mediated by PDF signaling propagate to other brain regions, such as the central complex, to modulate locomotor activity (Liang et al., 2019). Although the different neurons in the CNN do not make direct synaptic contacts with central complex neural circuits, they drive the central complex pre-motor centers through the agency of dopaminergic interneurons.
Although Ca2+ levels are often interpreted as changes in neuronal activity, changes in cytosolic Ca2+ does not necessarily reflect changes in membrane potential. For example, despite Ca2+ oscillations in the l-LNvs and DN1ps peaking at different times of day, their resting membrane potential is synchronized. Both clusters exhibit a hyperactive membrane potential late at night/early in the day, similar to what is seen in mammalian circadian neurons (Cao and Nitabach, 2008; Flourakis et al., 2015; Muraro and Ceriani, 2015). One ion channel that regulates the neuronal membrane potential in the DN1ps, Narrow Abdomen, is a Na+ leak channel that is rhythmically transported to the cell membrane by NLF-1 (Flourakis et al., 2015). nlf-1 transcription is regulated by the circadian clock, indicating that transportation of Narrow Abdomen to the cell membrane is under circadian regulation (Flourakis et al., 2015). Similarly, Shaw and Shal potassium channels in the LNvs also oscillate in a circadian manner, though in reverse phase with each other (P. Smith et al., 2019). Such mechanisms may serve as models for discovering how ion channels, ion currents, membrane potential, and basal Ca2+ are integrated through a single circadian clock.
Glycine Cooperation With PDF
Neurotransmitters act in a complex milieu in the brain. Therefore, it is likely and even expected that different neurotransmitters would cooperate to exert an effect on target neurons and tissues. Indeed, Choi et al. (2012) suggest that PDF activity is coupled to a small molecule neurotransmitter. LNv-expressed glycine has recently emerged as a candidate for added inhibition of downstream neurons (Frenkel et al., 2017). Glycine activates glycine receptor, allowing permeation of chloride to lower the membrane potential, thus likely inhibiting downstream LNds and DN1ps (Frenkel et al., 2017). Thus, a cooperative inhibition by both PDF and glycine may reduce the probability of neuronal firing and vesicle release in neurons downstream of LNvs.
Ion Transport Peptide Cooperation With PDF
Ion transport peptide (ITP) is expressed in the 5th s-LNv and one LNd (Hermann-Luibl et al., 2014; Johard et al., 2009) (Figure 2d). Similar to PDF, ITP is rhythmically expressed, targeting the dorsal neurons in the brain. However, ITP differs from PDF in that it regulates evening activity and suppresses nocturnal activity. Although the receptor for ITP is not yet characterized in Drosophila, it is likely to be a GPCR (Nagai et al., 2014), and likely expressed in evening cells such as the DN1s, given its effect on evening anticipatory behavior.
Diuretic Hormone 31 Cooperation With PDF
Although locomotor behavior reveals nearly identical phenotypes for both pdf 01 and pdfr mutants, in which morning anticipation is lost, evening anticipation is advanced, and in DD conditions flies become arrhythmic (Hyun et al., 2005; Lear et al., 2005b; Renn et al., 1999), there are observable differences at the molecular level. When Ca2+ oscillations are monitored in a pdfr mutant background, all neuronal clusters within the CNN show synchrony, with notable exception of the DN1s (Liang et al., 2016). When Ca2+ oscillations are monitored in a pdf 01 mutant background, the LNds (in addition to the DN1s) do not synchronize with the remainder of the CNN neurons (Liang et al., 2017). This suggests that PDFR in the LNds may be involved in mediating a separate signal, and may also explain LNd unresponsiveness in Ca2+ oscillation, in response to loss of PDF.
PDFR has the capacity to respond to a second neurotransmitter. Diuretic hormone 31 (DH31) triggers a PDFR-dependent cAMP response in HEK293 cells, though at half the efficacy of PDF (Mertens et al., 2005). Elimination of DH31 causes a loss of morning anticipation similar to a loss of PDF, but does not alter evening anticipation nor cause arrhythmic behavior in DD conditions (Goda et al., 2019). Indeed, double mutant (DH31 and pdf) experiments measuring locomotion as a behavioral output suggest that both DH31 and PDF cooperate to act through the PDFR, with the DH31 Receptor (also a GPCR) unlikely to play a role in locomotion behavior (Goda et al., 2019). DH31 Receptor instead appears to play a role in temperature preference and is expressed in DN1s, DN3s, and l-LNvs (weakly), but not in DN2s, s-LNvs or LNds (Goda et al., 2016, 2018; Johnson, 2005) (Figure 2e). Temperature cycles that oscillate with day/night cycles can entrain the CNN and the circadian clock (Glaser and Stanewsky, 2007; Matsumoto et al., 1998; Sidote et al., 1998; Yoshii et al., 2005) and appear to act through the DN1ps (posterior DN1s) (Yadlapalli et al., 2018), suggesting DH31 as a candidate for temperature entrainment (Goda et al., 2016). Indeed, DH31 interacts with DN2s through PDFR to guide lower temperature preference by flies at nightfall, though all of the three DN subgroups show calcium responsiveness to temperature changes (Goda et al., 2016; Yadlapalli et al., 2018). Thus, given that DH31 peptide is expressed in the DN1s and also promotes wakefulness (Goda et al., 2016; Kunst et al., 2014), it is tempting to think that a DN1 response to changes in temperature communicate this information to the CNN through DH31 by way of PDFR and/or DH31 Receptor. Indeed, wild type flies that are synchronized to both light-dark and warm-cold cycles exhibit an increase in daytime activity and a decrease in nighttime activity in a sleep analysis when compared to flies synchronized monitored at constant temperature (C. Chen et al., 2018). LNd responsiveness to changes in light regimen through the deep brain photoreceptor Cryptochrome (CRY) or its response to the LNvs, and its glutamatergic communication with the DN1s may provide a mechanism of integrating the CNN response to the two zeitgebers (Duhart et al., 2020). Such cooperation between neuropeptides underscores how combinations of neuronal signaling can fine tune the activity of clock neurons and circadian clocks under diverse environmental conditions.
Glutamatergic Influences on the Ventral Lateral Neurons
Glutamate released from DN1s promotes wakefulness (Figure 2g). LNvs respond to glutamate with a decrease in intracellular calcium, which shortens behavioral period under DD conditions (Guo et al., 2016). Downregulation of glutamate receptor, DmGluRA, in the LNvs lengthens free-running period in DD conditions, and reduces locomotor activity at night in LD conditions (Hamasaka et al., 2007). Expectedly, decreasing glutamate by either blocking neurotransmitter release or decreasing the activity of glutaminergic neurons also promotes sleep at nighttime (Zimmerman et al., 2017). Collectively, this evidence suggests the LNvs are in a circuit feedback loop, placing the LNvs, which often enjoy the pinnacle of CNN hierarchy, into a post-synaptic position within the CNN. Strikingly, loss of glutamate signaling in flies restores rhythmicity in constant light conditions (LL) (de Azevedo et al., 2020), clearly pointing to a critical role for glutamate signaling in light-induced arrhythmia. This is reminiscent of flies carrying a hypomorphic allele or genomic deletion of the deep brain light receptor cryptochrome (cryb and cry01, respectively) in which flies exhibit rhythmic behavior in LL conditions (Dolezelova et al., 2007; Emery et al., 2000; Helfrich-Förster et al., 2001). Whether glutamate signaling and cry function in parallel pathways or cooperate with each other in response to constant light conditions remains to be explored.
A glutamatergic response closes a neuronal feedback loop to PDF neurons. While PDF transmits information to both DN1s and LNds, DN1s release glutamate to silence the s-LNvs and the three LNds/5th s-LNv cluster as indicated by decreased cytosolic Ca2+ (Guo et al., 2016). These studies were conducted using exogenous expression of the P2X2 ATP-responsive cationic channel that can be used to activate neurons. Activation of DN1s by ATP in explanted brains revealed a decrease in s-LNv and LNd Ca2+ levels through glutamate. Hemibrain connectome data support such reciprocal connectivity, with synaptic connections observed between DN1s and s-LNvs, and DN1s and LNds. It is tempting to think of the glutamatergic response to PDF signaling as closing a signaling loop across so-called morning cells (LNvs) and evening cells (DN1s), while signaling to PDF-responsive LNds. However, there are additional complicating factors. Although t-PDF expression in DN1s restores rhythmic locomotor behavior in pdf 01 flies in DD (Goda et al., 2019), DN1s do not appear to be Ca2+ responsive to exogenous PDF added to explanted pdf 01 fly brains in DD, in contrast to other neurons (Liang et al., 2017). Thus, the increase in cytosolic Ca2+ in DN1s that trigger a glutamatergic signal to the s-LNvs and the 3LNds/5th s-LNv cluster may not be caused by PDF signaling alone. It is possible that other neurotransmitters released by the s-LNvs act on the DN1pAs as the means to trigger a glutamatergic response, while a PDF response by DN1a and DN1pAs elicits a separate response. How the glutamate and PDF neurotransmitters integrate into a feedback loop and how they regulate other clusters in the CNN will be an exciting area of investigation.
Light and Dopamine Input to the Ventral Lateral Neurons, and Morning Arousal
There are a number of excellent reviews that describe light input pathways in detail (Helfrich-Förster, 2002, 2020; Schlichting, 2020; Yoshii et al., 2016); therefore, we will focus on some of the neurotransmitter signaling pathways as they relate to communication into the various molecular clocks in the CNN. CRY protein, expressed in a portion of each neuronal cluster, directly communicates blue-light information to the clock transcription machinery by virtue of the semi-translucent nature of the fly cuticle (Figure 2f). However, cry01 flies can be entrained to a new LD phase (Dolezelova et al., 2007), suggesting that the visual system also conveys light to the CNN (Schlichting, 2020). Indeed, cooperative input from CRY and the visual system allow fine tuning of evening anticipation under different photoperiods (Kistenpfennig et al., 2018). The l-LNvs are the target neuronal clusters within the CNN for many arousal promoting neurotransmitters (Mazzotta et al., 2020). The HB-eyelet, a light-sensing body in the retina, and photoreceptors from the eye form synaptic connections with the LNvs, which contributes to morning arousal (Damulewicz et al., 2020; Helfrich-Förster et al., 2002; Hofbauer and Buchner, 1989; Veleri et al., 2007). Light information is communicated to the LNvs through excitatory cholinergic and inhibitory histaminergic signals, though how these two neurotransmitters are coordinated is unclear (Schlichting et al., 2016). Acetylcholine released from HB-eyelets and photoreceptors interacts with nicotinic acetylcholine receptors in both the s- and l-LNvs to trigger increases in cytosolic Ca2+ and cAMP, which causes depolarization of the neuronal membrane (McCarthy et al., 2011; Muraro and Ceriani, 2015; Schlichting et al., 2016; Wegener et al., 2004). Although the HB-eyelet may also synapse with the DN1s as suggested by the hemibrain connectome, there is no evidence of synapses between retinal photoreceptors and DN1s (or any other non-LNv within the CNN) (Scheffer et al., 2020). However, when the LNvs are silenced, the LNds, 5th s-LNv, DN1a and DN3s respond to a hub in the accessory medulla that receives signals from the fly visual system, suggesting that indirect connections between photoreceptors and other clusters within the CNN must exist (M.-T. Li et al., 2018).
Dopamine promotes wakefulness in both insects and mammals, and triggers an increase of cAMP in the LNvs (Andretic et al., 2005; Crocker and Sehgal, 2010; Fernandez-Chiappe et al., 2020; Kume et al., 2005; Lebestky et al., 2009; Riemensperger et al., 2011; Shang et al., 2011). fumin mutations in the dopamine transporter result in an excess of synaptic dopamine and hyperactive fly behavior, underscoring dopamine involvement in arousal (Kume et al., 2005). However, knockdown of dopamine receptors Dop1R1 and Dop1R2 (relevant in l- and s-LNvs respectively), do not affect daytime arousal despite reducing cAMP response, suggesting a more complex regulation of morning arousal than previously assumed (Fernandez-Chiappe et al., 2020). Instead, some dopaminergic neurons respond to PDF (Potdar and Sheeba, 2018), suggesting that the LNvs may be further upstream in the morning arousal process. Underscoring this possibility, DN1s may be involved in morning arousal as discussed above, possibly through instruction received from the LNvs or elsewhere (Guo et al., 2016, 2018; Kunst et al., 2014; Lamaze et al., 2017, 2018; L. Zhang et al., 2010a; Y. Zhang et al., 2010b). Although the LNvs are designated “morning cells” (Grima et al., 2004; Stoleru et al., 2004), it may be more accurate to think of “morningness” as the activity within a circuit, rather than the responsibility of a specific neuronal cluster.
Examples of CNN Output
Coordinated oscillations within the CNN must ultimately synchronize clocks within the rest of the body. Neuropeptides, such as short neuropeptide F (sNPF) and neuropeptide F (NPF), mediate communication within the CNN and to the body. Although these two peptides are similar in name, their sequences are different and they share no homology. sNPF is transcribed rhythmically in the s-LNvs (Abruzzi et al., 2017; Kula-Eversole et al., 2010) (Figure 2c). Knockdown of sNPF leads to increased nighttime activity, suggesting that this neuropeptide promotes sleep at night (Johard et al., 2009; Shang et al., 2013), in contrast to PDF, which promotes wakefulness. One mechanism of action is that sNPF moderately suppresses l-LNv electrical activity, thereby suppressing their arousal function and helping consolidate sleep into the night (Lebestky et al., 2009; Parisky et al., 2008; Shang et al., 2008, 2013; Sheeba et al., 2008). Although we do not yet have a complete picture of where sNPF receptor is expressed to determine how this receptor responds in the fly, application of exogenous sNPF to the BG2-c6 Drosophila neuronal cell line, which expresses sNPFR1, leads to a cAMP response in a dose-dependent manner (W. Chen et al., 2013), analogous to a PDFR response (Mertens et al., 2005). Application of exogenous sNPF to explanted brains reveals a cAMP response in the insulin producing cells (IPCs) in the brain, suggesting they may express sNPF receptor (Nagy et al., 2019). Knockdown of sNPF receptor by RNAi in this part of the brain induces reproductive arrest, indicating that sNPF signaling to the IPCs is critical for reproductive activity (Nagy et al., 2019) and suggesting possible contributions to circadian influences on courtship and mating behavior (Allada and Chung, 2010; Sakai and Kitamoto, 2006). Within the CNN, both PDF and sNPF suppress basal Ca2+ levels in targeted pacemakers with long durations by cell-autonomous actions (Liang et al., 2017). Notably, sNPF released from morning cells appears to be critical for setting the Ca2+ phase of PDF-unresponsive DN1s (Liang et al., 2017).
NPF, the Drosophila homolog of mammalian orexigenic peptide Neuropeptide Y (NPY), regulates diverse behaviors including circadian locomotor activity. The l-LNvs and 3LNds/5th s-LNv express NPF (C. He et al., 2013a; Hermann et al., 2012; W. J. Kim et al., 2013), though expression in the PDF-expressing s-LNvs has also been reported (C. He et al., 2013a) (Figure 2b). The pattern of NPF receptor (NPFR1) expression is not fully resolved. NPFR1 Gal4 drivers suggest that the s-LNvs express the receptor, while immunostaining suggests that instead, DN1s and LNds express NPFR1 (C. He et al., 2013a; W. J. Kim et al., 2013). NPF signaling appears to regulate the phasing and amplitude of evening activity in light-dark cycles and the modulation of evening anticipatory behavior; NPF and NPFR1 loss-of-function mutations lead to an elimination of evening anticipatory behavior (C. He et al., 2013a; Hermann et al., 2012; G. Lee et al., 2006). These phenotypes mirror the effect of PDF and PDFR loss-of-function mutations that lead to an elimination of morning anticipatory behavior, suggesting that PDF and NPF may link morning and evening oscillators. NPF is also involved in sleep regulation (overexpression of NPF increases sleep) and homeostasis of sleep (C. He et al., 2013b); as well as promoting wakefulness and feeding behavior (Chung et al., 2017); alcohol sensitivity (Wen et al., 2005); prolonging mating and courtship (W. J. Kim et al., 2013; W. Liu et al., 2019); and in clock-controlled sexual dimorphism through the LNds (G. Lee et al., 2006). All of these behaviors have a circadian component. It will be exciting to identify the circuits that connect these behaviors to the molecular clock in the CNN.
The Intersection of Molecular Biology, Neuroscience and Circadian Biology
Neurons have discrete genetically programmed oscillators and communicate through circuit connections. Within a network of circadian clocks, all molecular clocks oscillate and encode their own unique molecular timekeeping information. How is the information in these different clocks integrated into various signals that communicate within the CNN and out to the organism? Research of the CNN is unique in its need to assess oscillatory properties of each molecular clock, neuronal output and behavior output, all of which are likely to be distinct in their response to manipulation. If we consider a simplified model of a three-neuron circadian circuit (Figure 3), and if the molecular clock in the first neuron generates temporal information, is this information communicated directly to the third neuron through the second neuron, or does the clock in the second neuron integrate ionotropic, metabotropic, and temporal information encoded by its own clock to this signal as well? In this example, the third neuron is not a clock neuron, yet responds to circadian information. Indeed, two recent examples suggest that leucokinin and DH44 neuropeptides are critical for rhythmic behavior, even though neither peptide is expressed in clock neurons (Cavey et al., 2016; King et al., 2017; Zandawala et al., 2018). As interrogation methods improve in their spatial, temporal, and dynamic precision, and as more neuronal (e.g. Ca2+ and membrane potential) and behavioral outputs can be monitored across circadian time, investigators will be able to assess in increasing detail how clock information is integrated across the brain to regulate behavior.
In prevailing models, LNvs dominate a hierarchy within the CNN, instructing the other neurons how to oscillate within the network. This model is based primarily on changes to locomotor behavior in DD conditions (Grima et al., 2004; Stoleru et al., 2004). In LL conditions, however, there is evidence that the LNd clocks dominate in certain genetic backgrounds (Murad et al., 2007; Picot et al., 2007). This suggests that the CNN may instead function as a “network of equals” where different clusters take control depending on changing environmental cues. Indeed, changes in temperature appear to influence rhythmic behavior through the DN1s (Yadlapalli et al., 2018). Recently, a model in which synaptic plasticity integrates and gates light and temperature input into the CNN was proposed, in agreement with a non-hierarchical model (Fernandez et al., 2020). Here, the authors ablate dorsal medial termini of the s-LNvs and find no effect on behavioral rhythms in DD. Underscoring the link between light and synaptic plasticity, CRY has recently been implicated in circadian rhythmicity in synaptic plasticity (Damulewicz et al., 2020). Another recent report using CRISPR ablation of circadian clocks also undermines a central role for s-LNvs (Delventhal et al., 2019). While restoring per only in the LNvs restores rhythmicity in DD conditions (Grima et al., 2004) and ablating only per in the LNvs permits wild type oscillations (Delventhal et al., 2019). So while LNv clocks are sufficient for rhythmic locomotion (Grima et al., 2004), they may not be necessary (Delventhal et al., 2019). However, restoration of per in LNvs restores the morning anticipatory peak, while CRISPR-editing per removes it, underscoring that the LNvs are involved in morning anticipatory behavior (Stoleru et al., 2005), despite the evidence that DN1s regulate morningness, as discussed above. Thus, the assertion that the LNvs are “master pacemakers” that regulate rhythmic behavior in constant conditions should be revisited.
How is a mechanism in which LNv neuron elimination (Stoleru et al., 2004) creates arrhythmic behavior, but LNv clock elimination does not (Delventhal et al., 2019), possible? Vrille is a protein that is a negative regulator of the CLK/CYC activator complex, and a component of the secondary feedback loop (Blau and Young, 1999). Recently, it was proposed that Vrille rhythmically regulates PDF expression (Gunawardhana and Hardin, 2017). Perhaps LNvs lacking an operational primary loop are able to utilize their secondary loop to signal to downstream neurons. An alternative possibility is that interneuronal communication within the CNN is sufficient to compensate for a loss of a circadian clock in a given cluster (Bulthuis et al., 2019; Schlichting et al., 2019). Regardless, this evidence points to a robust CNN with compensatory mechanisms that do not rely on a single circadian clock.
Emerging data on reciprocal connectivity within the CNN may provide the framework for a cooperative network. The LNvs appear to form a communication loop with both the LNds and the DN1s. The three CRY + LNds and 5th s-LNv express PDFR and are responsive to PDF (Yao et al., 2012), indicating that they take instruction from the LNvs through PDF (Park et al., 2000). The s-LNvs, DN1s and LNds express NPFR1, which is responsive to NPF released by the l-LNvs, CRY + LNds and 5th s-LNv (C. He et al., 2013a; Hermann et al., 2012; Johard et al., 2009; W. J. Kim et al., 2013; G. Lee et al., 2006). This feedback appears to form the foundation for a “dual oscillator” that was predicted to exist decades ago (Pittendrigh and Daan, 1976). Since PDFR promotes cAMP production while NPFR1 inhibits it (Garczynski et al., 2002; W. J. Kim et al., 2013; Yao et al., 2012), it is tempting to think of cAMP oscillations acting in reverse phase as part of the mechanism that underlies this dual oscillator. These same LNvs also form synaptic feedback loops with DN1s and LNds (Guo et al., 2016). Although DN1s do not appear responsive to PDF alone, DN1s are triggered somehow to release glutamate to quiet the s-LNvs, forming a second neuronal feedback loop. Thus, analogous to the genetic feedback loops that comprise the clock, the neuronal clusters that comprise the CNN also appear to form neuronal feedback loops that may be centered around the LNvs. The existence of multiple neuronal feedback loops suggest that communication across the CNN is more complex than a dual oscillator model involving neuronal clusters that regulate morning and evening anticipation (Menegazzi et al., 2020).
Communication pathways that exclude LNvs are also beginning to emerge, underscoring the likelihood of a network model of the CNN, rather than a hierarchical model. Allatostatin C (AstC) is a clock-regulated neuropeptide that peaks in the night-day transition and is expressed in DN1ps and the less well-characterized DN3s and LPNs (Díaz et al., 2019). The AstC receptor AstC-R2 is expressed in the LNds, and ex vivo calcium imaging reveals that one of these LNds is inhibited by the AstC signaling pathway (Díaz et al., 2019). Blocking this pathway results in a delay in evening peak activity in long and short photoperiods (Díaz et al., 2019).
A complete picture of how individual clocks communicate with each other across this neuronal network to regulate various circadian behaviors will require further investigation. Elucidating the logic of circadian circuits and how the molecular programs of circadian transcription are integrated across this network will provide new insights into the basis of circadian behavior.
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-06101). M.A. is supported by the IWK Health Center Project Grant. We would like to thank Nicholas Stavropoulos for comments on the manuscript.
Footnotes
Conflict of Interest Statement: The author(s) have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Deniz Top  https://orcid.org/0000-0002-1042-8460
https://orcid.org/0000-0002-1042-8460
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. (2011) Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25:2374-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, Rosbash M. (2017) RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Gen 13:e1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. (2010) Circadian organization of behavior and physiology in Drosophila. Ann Rev Physiol 72:605-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. (1998) A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93:791-804. [DOI] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. (2005) Dopaminergic modulation of arousal in Drosophila. Curr Biol 15:1165-1175. [DOI] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW. (1984) Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312:752-754. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. (1999) The Drosophila dCREB2 gene affects the circadian clock. Neuron 22:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. (1999) Cycling vrille expression is required for a functional Drosophila clock. Cell 99:661-671. [DOI] [PubMed] [Google Scholar]
- Bos JL. (2003) Epac: a new cAMP target and new avenues in cAMP research. Nat Rev Mol Cell Biol 4:733-738. [DOI] [PubMed] [Google Scholar]
- Bulthuis N, Spontak KR, Kleeman B, Cavanaugh DJ. (2019) Neuronal activity in non-LNv clock cells is required to produce free-running rest: activity rhythms in Drosophila. J Biol Rhythms 34:249-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN. (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci 28:6493-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Collins B, Bertet C, Blau J. (2016) Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci 19:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Lamaze A, De J, Mena W, Chélot E, Martin B, Hardin P, Kadener S, Emery P, Rouyer F. (2018) Reconfiguration of a multi-oscillator network by light in the Drosophila circadian clock. Curr Biol 28:2007-2017.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xu M, Anantaprakorn Y, Rosing M, Stanewsky R. (2018) nocte is required for integrating light and temperature inputs in circadian clock neurons of Drosophila. Curr Biol 28:1595-1605.e3. [DOI] [PubMed] [Google Scholar]
- Chen W, Shi W, Li L, Zheng Z, Li T, Bai W, Zhao Z. (2013) Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem Mol Biol 43:809-819. [DOI] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy EV, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JC, Nitabach MN. (2012) Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in Drosophila. Cell Rep 2:332-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Ro J, Hutter SA, Miller KM, Guduguntla LS, Kondo S, Pletcher SD. (2017) Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep 19:2441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. (2001) Circadian regulation of gene expression systems in the Drosophila head. Neuron 32:657-671. [DOI] [PubMed] [Google Scholar]
- Crane BR, Young MW. (2014) Interactive features of proteins composing eukaryotic circadian clocks. Ann Rev Biochem 83:191-219. [DOI] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. (2010) Genetic analysis of sleep. Genes Dev 24:1220-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J. (2003) vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112:329-341. [DOI] [PubMed] [Google Scholar]
- Damulewicz M, Woźnicka O, Jasińska M, Pyza E. (2020) CRY-dependent plasticity of tetrad presynaptic sites in the visual system of Drosophila at the morning peak of activity and sleep. Sci Rep 10:18161-18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo RVDM, Hansen C, Chen KF, Rosato E, Kyriacou CP. (2020) Disrupted glutamate signaling in Drosophila generates locomotor rhythms in constant light. Front Physiol 11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delventhal R, O’Connor RM, Pantalia MM, Ulgherait M, Kim HX, Basturk MK, Canman JC, Shirasu-Hiza M.(2019) Dissection of central clock function in Drosophila through cell-specific CRISPR-mediated clock gene disruption. eLife 8:e48308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz MM, Schlichting M, Abruzzi KC, Long X, Rosbash M. (2019) Allatostatin-C/AstC-R2 is a novel pathway to modulate the circadian activity pattern in Drosophila. Curr Biol 29:13-22.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. (2006). Circadian regulator CLOCK is a histone acetyltransferase. Cell 125:497-508. 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. (2007) Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177:329-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier EA, Huang LH, Steward R. (1999) Nuclear import of the Drosophila Rel protein Dorsal is regulated by phosphorylation. Genes Dev 13:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhart JM, Herrero A, la Cruz de G, Ispizua JI, Pírez N, Ceriani MF. (2020) Circadian structural plasticity drives remodeling of E cell output. Curr Biol 30:5040-5048.e5. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. (2000) A unique circadian-rhythm photoreceptor. Nature 404:456-457. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. (1992) Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci 12:3321-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkey DM, Kimelman D. (2000) GSK-3: new thoughts on an old enzyme. Dev Biol 225:471-479. [DOI] [PubMed] [Google Scholar]
- Fernández MP, Berni J, Ceriani MF. (2008) Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 6:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MP, Pettibone HL, Bogart JT, Roell CJ, Davey CE, Pranevicius A, Huynh KV, Lennox SM, Kostadinov BS, Shafer OT. (2020) Sites of circadian clock neuron plasticity mediate sensory integration and entrainment. Curr Biol 30:2225-2237.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chiappe F, Hermann-Luibl C, Peteranderl A, Reinhard N, Senthilan PR, Hieke M, Selcho M, Yoshii T, Shafer OT, Muraro NI, et al. (2020) Dopamine signaling in wake promoting clock neurons is not required for the normal regulation of sleep in Drosophila. J Neurosci 40:9617-9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, et al. (2015) A conserved bicycle model for circadian clock control of membrane excitability. Cell 162:836-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg PCM, O’Neill JS, Dobrzycki T, Calvert S, Lord EC, McIntosh RL, Elliott CJ, Sweeney ST, Hastings MH, Chawla S. (2014) Class IIa histone deacetylases are conserved regulators of circadian function. J Biol Chem 289:34341-34348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MG. (2016) Circadian regulation of synaptic plasticity. Biology 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel L, Muraro NI, Beltrán González AN, Marcora MS, Bernabó G, Hermann-Luibl C, Romero JI, Helfrich-Förster C, Castaño EM, Marino-Busjle C. (2017) Organization of circadian behavior relies on glycinergic transmission. Cell Rep 19:72-85. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. (2002) Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides 23:773-780. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM. (2001) Peripheral clocks and their role in circadian timing: insights from insects. Philos Trans R Soc Lond B Biol Sci 356:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. (2007) Synchronization of the Drosophila circadian clock by temperature cycles. Cold Spring Harb Symp Quant Biol 72:233-242. [DOI] [PubMed] [Google Scholar]
- Glossop NRJ, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. (2003) VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37:249-261. [DOI] [PubMed] [Google Scholar]
- Glossop NRJ, Lyons LC, Hardin PE. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286:766-768. [DOI] [PubMed] [Google Scholar]
- Goda T, Doi M, Umezaki Y, Murai I, Shimatani H, Chu ML, Nguyen V, Okamura H, Hamada F. (2018). Calcitonin receptors are ancient modulators for rhythms of preferential temperature in insects and body temperature in mammals. Genes Dev 32:140-155. 10.1101/gad.307884.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda T, Tang X, Umezaki Y, Chu ML, Kunst M, Nitabach MN, Hamada FN. (2016) Drosophila DH31 neuropeptide and PDF receptor regulate night-onset temperature preference. J Neurosci 36:11739-11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda T, Umezaki Y, Alwattari F, Seo HW, Hamada FN. (2019) Neuropeptides PDF and DH31 hierarchically regulate free-running rhythmicity in Drosophila circadian locomotor activity. Sci Rep 9:838-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pírez N, Ceriani MF. (2014) Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431:869-873. [DOI] [PubMed] [Google Scholar]
- Gunawardhana KL, Hardin PE. (2017) VRILLE controls PDF neuropeptide accumulation and arborization rhythms in small ventrolateral neurons to drive rhythmic behavior in Drosophila. Curr Biol 27:3442-3453.e4. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. (2014) PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife 3:e02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Holla M, Díaz MM, Rosbash M. (2018) A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron 100:624-635.e4. [DOI] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M. (2016) Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature 536:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaka Y, Rieger D, Parmentier M-L, Grau Y, Helfrich-Förster C, Nässel DR. (2007) Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J Comp Neurol 505:32-45. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343:536-540. [DOI] [PubMed] [Google Scholar]
- He C, Cong X, Zhang R, Wu D, An C, Zhao Z. (2013. a) Regulation of circadian locomotor rhythm by neuropeptide Y-like system in Drosophila melanogaster. Insect Mol Biol 22:376-388. [DOI] [PubMed] [Google Scholar]
- He C, Yang Y, Zhang M, Price JL, Zhao Z. (2013. b) Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS ONE 8:e74237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Wu B, Price JL, Zhao Z. (2017) Circadian rhythm neuropeptides in Drosophila: signals for normal circadian function and circadian neurodegenerative disease. Int J Mol Sci 18:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. (1995) The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci U S A 92:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. (1997) Drosophila rhythms: from brain to behavior. Semin Cell Dev Biol 7:791-802. [Google Scholar]
- Helfrich-Förster C. (2002) The circadian system of Drosophila melanogaster and its light input pathways. Zoology 105:297-312. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. (2020) Light input pathways to the circadian clock of insects with an emphasis on the fruit fly Drosophila melanogaster. J Comp Physiol A 206:259-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Edwards T, Yasuyama K, Wisotzki B, Schneuwly S, Stanewsky R, Meinertzhagen IA, Hofbauer A. (2002) The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J Neurosci 22:9255-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. (2001) The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30:249-261. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Yoshii T, Wulbeck C, Grieshaber E, Rieger D, Bachleitner W, Cusumano P, Rouyer F.(2007) The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb Symp Quant Biol 72:517-525. [DOI] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Förster C. (2012) Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol 520:970-987. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, Helfrich-Förster C. (2014) The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J Neurosci 34:9522-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Yoshii T, Ispizua JI, Colque C, Veenstra JA, Muraro NI, Ceriani MF. (2020) Coupling neuropeptide levels to structural plasticity in Drosophila clock neurons. Curr Biol 30:3154-3166.e4. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Buchner E. (1989) Does Drosophila have seven eyes? Naturwiss 76:335-336. [Google Scholar]
- Hove JR, Köster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. (2003) Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature 421:172-177. [DOI] [PubMed] [Google Scholar]
- Huang Y, Ainsley JA, Reijmers LG, Jackson FR. (2013) Translational profiling of clock cells reveals circadianly synchronized protein synthesis. PLoS Biol 11:e1001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-C, Maurer C, Kay SA, Weber F. (2007) Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J Biol Chem 282:31349-31357. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A. (1996) Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84:677-685. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. (2005) Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron 48:267-278. [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. (2010) PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518:1925-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito C, Goto SG, Shiga S, Tomioka K, Numata H. (2008) Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc Natl Acad Sci U S A 105:8446-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HAD, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, Nässel DR. (2009) Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol 516:59-73. [DOI] [PubMed] [Google Scholar]
- Johnson EC. (2005) A novel diuretic hormone receptor in Drosophila: evidence for conservation of CGRP signaling. J Exp Biol 208:1239-1246. [DOI] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. (2007) Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21:1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Woodgett JR. (2011) GSK-3: functional insights from cell biology and animal models. Front Mol Neurosci 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M. (1998) Neural substrates of Drosophila rhythms revealed by mutants and molecular manipulations. Curr Opin Neurobiol 8:652-658. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422:66-94. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Förster C, Hall JC. (1997) Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci 17:6745-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. (2000) Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol 43:207-233. [DOI] [PubMed] [Google Scholar]
- Kim L, Kimmel AR. (2000) GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev 10:508-514. [DOI] [PubMed] [Google Scholar]
- Kim M, Lee H, Hur J-H, Choe J, Lim C. (2016) CRTC potentiates light-independent timeless transcription to sustain circadian rhythms in Drosophila. Sci Rep 6:32113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Jan LY, Jan YN. (2013) A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron 80:1190-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AN, Barber AF, Smith AE, Dreyer AP, Sitaraman D, Nitabach MN, Cavanaugh DJ, Sehgal A.(2017) A peptidergic circuit links the circadian clock to locomotor activity. Curr Biol 27:1915-1927.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistenpfennig C, Nakayama M, Nihara R, Tomioka K, Helfrich-Förster C, Yoshii T. (2018) A Tug-of-War between Cryptochrome and the visual system allows the adaptation of evening activity to long photoperiods in Drosophila melanogaster. J Biol Rhythms 33:24-34. [DOI] [PubMed] [Google Scholar]
- Klose M, Duvall LB, Li W, Liang X, Ren C, Steinbach JH, Taghert PH. (2016) Functional PDF signaling in the Drosophila circadian neural circuit is gated by Ral A-dependent modulation. Neuron 90:781-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94:97-107. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. (2002) Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420:673-678. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. (1971) Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeptowski W, Hess G, Pyza E. (2018) Circadian plasticity in the brain of insects and rodents. Front Neural Circuits 12:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. (2010) Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A 107:13497-13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. (2005) Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN. (2014) Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 24:2652-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze A, Krätschmer P, Chen KF, Lowe S, Jepson JEC. (2018) A wake-promoting circadian output circuit in Drosophila. Curr Biol 28:3098-3105.e3. [DOI] [PubMed] [Google Scholar]
- Lamaze A, Öztürk-Çolak A, Fischer R, Peschel N, Koh K, Jepson JEC. (2017) Regulation of sleep plasticity by a thermo-sensitive circuit in Drosophila. Sci Rep 7:40304-40312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P, Foley LE, Emery P. (2018) Neural network interactions modulate CRY-dependent photoresponses in Drosophila. J Neurosci 38:6161-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Lin J-M, Keath JR, McGill JJ, Raman IM, Allada R. (2005. a) The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48:965-976. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin J-M, Schroeder A, Zhang L, Allada R. (2005. b) A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48:221-227. [DOI] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. (2009) The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol 7:e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang J-SC, Dankert H, Zelnik L, Kim Y-C, Han K-A, Wolf FW, Perona P, Anderson DJ. (2009) Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron 64:522-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. (2015) Global and local missions of cAMP signaling in neural plasticity, learning, and memory. Front Pharmacol 6:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH. (2006) Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci U S A 103:12580-12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-T, Cao L-H, Xiao N, Tang M, Deng B, Yang T, Yoshii T, Luo DG. (2018) Hub-organized parallel circuits of central circadian pacemaker neurons for visual photoentrainment in Drosophila. Nat Commun 9:4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo F, Shen J, Rosbash M. (2014) PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc Natl Acad Sci U S A 111:E1284-E1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ho MCW, Zhang Y, Li Y, Wu MN, Holy TE, Taghert PH. (2019) Morning and evening circadian pacemakers independently drive premotor centers via a specific dopamine relay. Neuron 102:843-857.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. (2016) Synchronous Drosophila circadian pacemakers display nonsynchronous Ca²+ rhythms in vivo. Science 351:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. (2017) A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron 94:1173-1189.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. (2007. a) Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol 17:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kim J, Doh E, Choe J. (2007. b) Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol 27:4876-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. (2004) The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci 24:7951-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ganguly A, Huang J, Wang Y, Ni JD, Gurav AS, Aguilar MA, Montell C. (2019) Neuropeptide F regulates courtship in Drosophila through a male-specific neuronal circuit. eLife 8:e49574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Naidoo N, Zimmerman JE, Pack AI. (2008) Molecular mechanisms of sleep and wakefulness. Ann N Y Acad Sci 1129:335-349. [DOI] [PubMed] [Google Scholar]
- Majercak J, Kalderon D, Edery I. (1997) Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Mol Cell Biol 17:5915-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105:769-779. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Matsumoto N, Harui Y, Sakamoto M, Tomioka K. (1998) Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J Insect Physiol 44:587-596. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, et al. (2007) A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 21:1687-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotta GM, Damulewicz M, Cusumano P. (2020) Better sleep at night: how light influences sleep in Drosophila. Front Physiol 11:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EV, Wu Y, Decarvalho T, Brandt C, Cao G, Nitabach MN. (2011) Synchronized bilateral synaptic inputs to Drosophila melanogaster neuropeptidergic rest/arousal neurons. J Neurosci 31:8181-8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107:567-578. [DOI] [PubMed] [Google Scholar]
- Meireles-Filho ACA, Bardet AF, Yáñez-Cuna JO, Stampfel G, Stark A. (2014) Cis-regulatory requirements for tissue-specific programs of the circadian clock. Curr Biol 24:1-10. [DOI] [PubMed] [Google Scholar]
- Menegazzi P, Beer K, Grebler V, Schlichting M, Schubert FK, Helfrich-Förster C. (2020) A functional clock within the main morning and evening neurons of D. melanogaster is not sufficient for wild-type locomotor activity under changing day length. Front Physiol 11:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzi P, Dalla Benetta E, Beauchamp M, Schlichting M, Steffan-Dewenter I, Helfrich-Förster C. (2017) Adaptation of circadian neuronal network to photoperiod in high-latitude European Drosophilids. Curr Biol 27:833-839. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. (2005) PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48:213-219. [DOI] [PubMed] [Google Scholar]
- Mezan S, Feuz JD, Deplancke B, Kadener S. (2016) PDF signaling is an integral part of the Drosophila circadian molecular oscillator. Cell Rep 17:708-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. (2007) A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53:689-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro NI, Ceriani MF. (2015) Acetylcholine from visual circuits modulates the activity of arousal neurons in Drosophila. J Neurosci 35:16315-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai C, Mabashi-Asazuma H, Nagasawa H, Nagata S. (2014) Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J Biol Chem 289:32166-32177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy D, Cusumano P, Andreatta G, Anduaga AM, Hermann-Luibl C, Reinhard N, Gesto J, Wegener C, Mazzotta G, Rosato E, et al. (2019) Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Gen 15:e1008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Muñoz A, Ewer J. (2018) Calcium and cAMP directly modulate the speed of the Drosophila circadian clock. PLoS Gen 14:e1007433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A 97:3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsakou A, Sapsis TP, Blau J. (2015) Circadian rhythms in Rho1 activity regulate neuronal plasticity and network hierarchy. Cell 162:823-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. (2007) Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5:e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. (1976) A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol 106:223-252. [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. (1997) Independent photoreceptive circadian clocks throughout Drosophila. Science 278:1632-1635. [DOI] [PubMed] [Google Scholar]
- Potdar S, Sheeba V. (2018) Wakefulness is promoted during day time by PDFR signalling to dopaminergic neurons in Drosophila melanogaster. eNeuro 5:ENEURO.0129-18.2018. DOI: 10.1523/ENEURO.0129-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94:83-95. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. (1984) Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38:701-710. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99:791-802. [DOI] [PubMed] [Google Scholar]
- Riemensperger T, Isabel G, Coulom H, Neuser K, Seugnet L, Kume K, Iché-Torres M, Cassar M, Strauss R, Preat T, et al. (2011) Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci U S A 108:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, Holmes TC. (2015) Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol 25:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Young MW, Saez L. (2000) A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26:505-514. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. (1998) CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93:805-814. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Zeng H, Le M, Curtin KD, Hall JC, Rosbash M. (1996) The timSL mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron 17:921-929. [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW. (1996) Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17:911-920. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kitamoto T. (2006) Clock, love and memory: circadian and non-circadian regulation of Drosophila mating behavior by clock genes. Sleep Biol Rhythms 4:255-262. [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura S-Y, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. (2020) A connectome and analysis of the adult Drosophila central brain. eLife 9:e57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M. (2020) Entrainment of the Drosophila clock by the visual system. Neurosci Ins 15:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M, Díaz MM, Xin J, Rosbash M. (2019) Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. eLife 8:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M, Menegazzi P, Lelito KR, Yao Z, Buhl E, Dalla Benetta E, Bahle A, Denike J, Hodge JJ, Helfrich-Förster C, et al. (2016) A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J Neurosci 36:9084-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert FK, Hagedorn N, Yoshii T, Helfrich-Förster C, Rieger D. (2018) Neuroanatomical details of the lateral neurons of Drosophila melanogaster support their functional role in the circadian system. J Comp Neurol 526:1209-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. (1994) Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263:1603-1606. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW. (1995) Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 270:808-810. [DOI] [PubMed] [Google Scholar]
- Seino S, Shibasaki T. (2005) PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol Rev 85:1303-1342. [DOI] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. (2014) Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol 12:e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. (2009) RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide’s circadian functions. PLoS ONE 4:e8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. (2008) Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58:223-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. (2013) Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80:171-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. (2008) Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A 105:19587-19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pírez N, Harrington KI, Guo F, Pollack J, Hong P, Griffith LC, Rosbash M. (2011) Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14:889-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou Y-T, Sharma VK, Holmes TC. (2008) Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote D, Majercak J, Parikh V, Edery I. (1998) Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol 18:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Buhl E, Tsaneva Atanasova K, Hodge JJL. (2019) Shaw and Shal voltage-gated potassium channels mediate circadian changes in Drosophila clock neuron excitability. J Physiol 597:5707-5722. [DOI] [PubMed] [Google Scholar]
- Smith RF, Konopka RJ. (1981) Circadian clock phenotypes of chromosome aberrations with a breakpoint at the per locus. Mol Gen Genet 183:243-251. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431:862-868. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. (2005) A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438:238-242. [DOI] [PubMed] [Google Scholar]
- Tang X, Roessingh S, Hayley SE, Chu ML, Tanaka NK, Wolfgang W, Song S, Stanewsky R, Hamada FN. (2017) The role of PDF neurons in setting the preferred temperature before dawn in Drosophila. eLife 6:e23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, Young MW. (2018) Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb Perspect Biol 10:a033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, Harms E, Syed S, Adams EL, Saez L. (2016) GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep 16:357-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, O’Neil JL, Merz GE, Dusad K, Crane BR, Young MW. (2018) CK1/Doubletime activity delays transcription activation in the circadian clock. eLife 7:e32679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Rieger D, Helfrich-Förster C, Stanewsky R. (2007) Hofbauer-Buchner eyelet affects circadian photosensitivity and coordinates TIM and PER expression in Drosophila clock neurons. J Biol Rhythms 22:29-42. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. (1994) Block in nuclear localization of period protein by a second clock mutation, timeless. Science 263:1606-1609. [DOI] [PubMed] [Google Scholar]
- Wang Q, Abruzzi KC, Rosbash M, Rio DC. (2018) Striking circadian neuron diversity and cycling of Drosophila alternative splicing. eLife 7:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener C, Hamasaka Y, Nässel DR. (2004) Acetylcholine increases intracellular Ca2+ via nicotinic receptors in cultured PDF-containing clock neurons of Drosophila. J Neurophysiol 91:912-923. [DOI] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. (2005) Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A 102:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. (1993) Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms 8:67-94. [DOI] [PubMed] [Google Scholar]
- Williams JA, Sehgal A. (2001) Molecular components of the circadian system in Drosophila. Ann Rev Physiol 63:729-755. [DOI] [PubMed] [Google Scholar]
- Yadlapalli S, Jiang C, Bahle A, Reddy P, Meyhofer E, Shafer OT. (2018) Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature 555:98-102. [DOI] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. (2012) Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol 108:684-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA. (2010) Synaptic connections of PDF-immunoreactive lateral neurons projecting to the dorsal protocerebrum of Drosophila melanogaster. J Comp Neurol 518:292-304. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Hermann-Luibl C, Helfrich-Förster C. (2016) Circadian light-input pathways in Drosophila. Commun Integr Biol 9:e1102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Heshiki Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. (2005) Temperature cycles drive Drosophila circadian oscillation in constant light that otherwise induces behavioural arrhythmicity. Eur J Neurosci 22:1176-1184. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. (2009) The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci 29:2597-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. (2006) Circadian oscillators of Drosophila and mammals. J Cell Sci 119:4793-4795. [DOI] [PubMed] [Google Scholar]
- Zandawala M, Marley R, Davies SA, Nässel DR. (2018) Characterization of a set of abdominal neuroendocrine cells that regulate stress physiology using colocalized diuretic peptides in Drosophila. Cell Mol Life Sci 75:1099-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. (2010. a) DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol 20:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. (2010. b) Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol 20:600-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A. (2009) An isoform-specific mutant reveals a role of PDP1 in the circadian oscillator. J Neurosci 29:10920-10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu W, Hardin PE. (2016) CLOCKWORK ORANGE enhances PERIOD mediated rhythms in transcriptional repression by antagonizing E-box binding by CLOCK-CYCLE. PLoS Gen 12:e1006430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Chan MT, Lenz OT, Keenan BT, Maislin G, Pack AI. (2017) Glutamate Is a wake-active neurotransmitter in Drosophila melanogaster. Sleep 40:zsw046. [DOI] [PMC free article] [PubMed] [Google Scholar]