Abstract
The adult mammalian heart is thought to be a terminally differentiated organ given the postmitotic nature of cardiomyocytes. Consequently, the potential for cardiac repair through cardiomyocyte proliferation is extremely limited. Low-density lipoprotein receptor-related protein 6 (LRP6) is a Wnt co-receptor that is required for embryonic heart development. In this study we investigated the role of LRP6 in heart repair through regulation of cardiomyocyte proliferation. Lrp6 deficiency increased cardiomyocyte cell cycle activity in neonatal, juvenile and adult mice. Cardiomyocyte-specific deletion of Lrp6 in the mouse heart induced a robust regenerative response after myocardial infarction (MI), led to reduced MI area and improvement in left ventricular systolic function. In vivo genetic lineage tracing revealed that the newly formed cardiomyocytes in Lrp6-deficient mouse hearts after MI were mainly derived from resident cardiomyocytes. Furthermore, we found that the pro-proliferative effect of Lrp6 deficiency was mediated by the ING5/P21 signaling pathway. Gene therapy using the adeno-associated virus (AAV)9 miRNAi-Lrp6 construct promoted the repair of heart injury in mice. Lrp6 deficiency also induced the proliferation of human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). Our study identifies LRP6 as a critical regulator of cardiomyocyte proliferation, which may lead to the development of a novel molecular strategy to promote myocardial regeneration and repair.
Subject terms: Cell-cycle exit, Cytokinesis
Introduction
Numerous heart diseases are associated with loss or dysfunction of cardiac muscle cells. However, adult cardiomyocytes largely lose their ability to undergo cell division and proliferation after a narrow proliferative window at the neonatal stages.1–3 As a representative organ with poor regeneration capacity, the adult heart is particularly vulnerable to failure following injury responses.4–6 Adult mammalian hearts form fibrotic scars in response to injury, which can lead to heart failure, arrhythmia and death. Current treatments can temporarily improve heart function but do not replace lost cardiomyocytes.7–9
Mammalian tissues exhibit specific rates and mechanisms of cell turnover, and different organs vary widely in repair capacity.10–12 Lineage mapping analyses in neonatal mice and adult zebrafish have suggested that the majority of regenerated cardiomyocytes are derived from pre-existing cardiomyocytes.1,13 In addition, many studies have proposed that the adult mammalian heart is capable of turning over cardiomyocytes following injury, suggesting that cardiomyocyte proliferation could be an effective strategy for heart repair.14–20 Thus, understanding the mechanisms underlying the proliferative capacity of cardiomyocytes and how to control the process of cardiomyocyte proliferation is key for the successful repair of heart injuries.
Low-density lipoprotein receptor-related protein 6 (LRP6), a single-span transmembrane protein that plays a key role in the transduction of the canonical Wnt/β-catenin signaling pathway,21,22 has been demonstrated to be essential for cardiac neural crest and outflow tract development. Ablation of Lrp6 in mice affected the proliferation and survival of cardiac progenitors and caused a range of severe morphogenetic defects during early cardiogenesis.23 In adulthood, LRP6 dysfunction has been linked to coronary artery disease and fatal cardiac arrhythmia.24–26 However, little is known about the potential role of LRP6 in cardiomyocyte cell cycle activity.
In the present study, we found that Lrp6 deficiency increased cardiomyocyte proliferation at different stages in mouse heart and human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). Lrp6 deficiency in the mouse heart reduced the size of the myocardial infarction (MI) and improved heart function. The ING5/P21 signaling pathway was shown to be responsible for the effects of LRP6 on cardiomyocyte proliferation. After injury, administration of an AAV9 construct encoding miRNAi-Lrp6 promoted heart repair in the mice. Thus, our study reveals a novel molecular switch controlling the re-entry of cardiomyocyte cell cycle and a promising intervention target for cardiac regeneration.
Results
Lrp6 deficiency increases neonatal rat cardiomyocyte proliferation in vitro
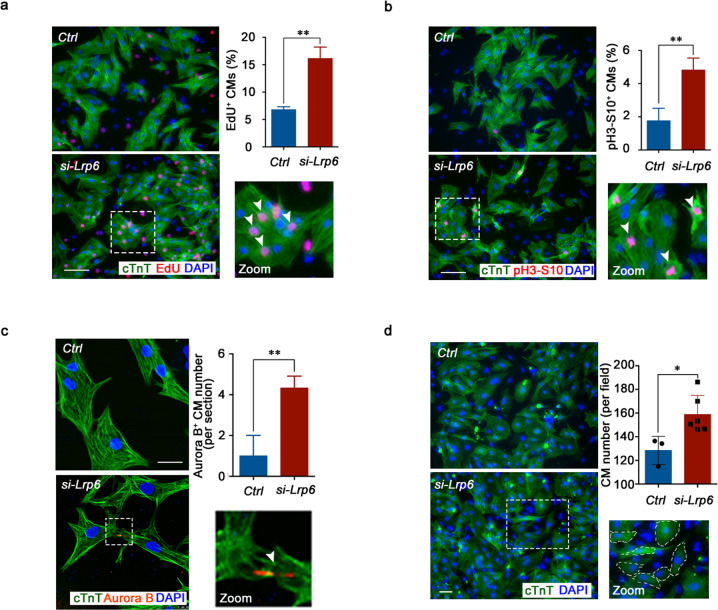
To explore the potential effect of LRP6 on cardiomyocyte proliferation, we first performed in vitro siRNA knockdown using postnatal day 1–3 (P1d–P3d) neonatal rat cardiomyocytes (NRCMs) and found that Lrp6 knockdown enhanced 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Fig. 1a; Supplementary information, Figs. S1a, S2a). To determine whether the increased DNA replication in NRCMs eventually results in increases in karyokinesis and cytokinesis, we stained the cells treated with Lrp6 siRNA (si-Lrp6) for pH3-S10, a marker of late G2 phase/mitosis, and for Aurora B, a kinase localized in midbodies during cytokinesis. Lrp6 knockdown increased both the percentage of cardiomyocytes positive for pH3-S10 and the number of cardiomyocytes presenting midbodies (Fig. 1b, c; Supplementary information, Fig. S2b). Immunofluorescence analysis confirmed that Lrp6 knockdown resulted in a large increase in cardiomyocyte numbers (Fig. 1d). In addition, we found a significant increase in Lrp6 expression from P1d to P7d in mouse hearts (Supplementary information, Fig. S1b), suggesting that LRP6 is potentially involved in the regulation of cell cycle reentry in postnatal cardiomyocytes.
Fig. 1. LRP6 reduction promotes neonatal cardiomyocyte proliferation in vitro.
a–c Representative images of NRCM cultures stained with EdU, pH3-S10, Aurora B and cTnT. White arrows indicate EdU+cTnT+, pH3-S10+cTnT+ and Aurora B+cTnT+ cells. The column shows the percentage or number of proliferating NRCMs in response to Lrp6 downregulation, respectively. In a, c, the results represent three experimental repeats and in b represent four repeats. (a, b) Scale bar = 100 µm. (c) Scale bar = 25 µm. d Immunostaining showing quantification of the number of cTnT+DAPI+ NRCMs in the control (Ctrl) and Lrp6 siRNA-treated (si-Lrp6) cardiomyocytes. Quantitative analysis was performed of counts in multiple fields from three independent samples per group; the results represent three individual experiments. Scale bar = 50 µm. *P < 0.05, **P < 0.01. Data are presented as the means ± SD.
Cardiomyocytes of the Lrp6-CKO mice display activation of the cell cycle at the juvenile and adult stages in vivo
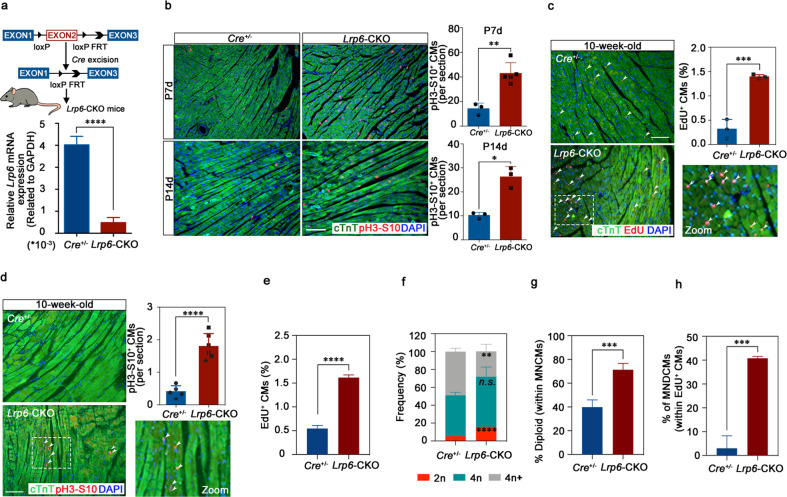
To determine whether LRP6 knockdown prolongs the transient postnatal proliferative window of cardiomyocytes in vivo, we conditionally deleted Lrp6 in cardiomyocytes by crossing the Lrp6flox/flox mice with αMHC/MerCreMer (Cre+/−) mice (Fig. 2a). Analysis of the Lrp6 mRNA expression in Lrp6flox/flox × Cre+/− (Lrp6-CKO) hearts confirmed that the Lrp6flox/flox allele was efficiently recombined in the cardiomyocytes (Fig. 2a). We then induced Lrp6 knockout in the cardiomyocytes at the neonatal stage and examined the cell cycle activity at the juvenile stage (P7d and P14d). Immunofluorescence analysis of the Cre+/− and Lrp6-CKO heart tissue slides showed that the Lrp6-CKO mice had substantial increases in the pH3-S10-positive cardiomyocytes compared to the Cre+/− mice (Fig. 2b; Supplementary information, Fig. S3a, b). No significant differences in heart morphology and cardiac fibrosis were observed between the Cre+/− and Lrp6-CKO mice (Supplementary information, Fig. S4). These results demonstrated that Lrp6 deficiency after birth could override the cardiomyocyte cell cycle exit that occurs during the first postnatal week and broaden the postnatal cardiomyocyte proliferative window.
Fig. 2. Lrp6 deficiency induces cell cycle activation in mouse hearts at the juvenile and adult stages in vivo.
a Generation of the mouse line with Lrp6-CKO. Top, schematic diagram. Bottom, qRT-PCR examination of Lrp6 mRNA levels in the cardiomyocytes (CMs) of the Lrp6-CKO mice. n = 4 mice per group. b Immunostaining showing co-staining of pH3-S10, cTnT and DAPI in the Cre+/− and Lrp6-CKO hearts at P7d and P14d, respectively. The graph shows the number of pH3-S10+cTnT+ nuclei. n = 3–5 mice per group. Scale bar = 100 µm. c, d Immunostaining showing EdU+cTnT+DAPI+ and pH3-S10+cTnT+DAPI+ cardiomyocytes in the Cre+/− and Lrp6-CKO hearts at 10 weeks. The graph shows the percentage of EdU+cTnT+DAPI+ cells and the number of pH3-S10+cTnT+ DAPI+ cells. The results of quantitative analysis represent 3 to 6 independent samples per group (three sections per heart). Scale bar = 100 µm. e Quantifications of proliferating CMs stained positive for EdU in dissociated CMs from adult Cre+/− and Lrp6-CKO mice injected with EdU substrate. Approximately 5000–7000 cardiomyocytes were counted from 3 mice per group. f, g The nucleation and ploidy assessment of dissociated CMs from adult Cre+/− and Lrp6-CKO mice. 2n, diploid, 4n, tetraploid, 4n+, polyploid. MNCMs, mononucleated CMs. n = 4 mice per group. h Quantifications of the percentage of MNDCMs within the EdU+ CMs population from adult Cre+/− and Lrp6-CKO mice. MNDCMs mononucleated diploid CMs. EdU-positive cardiomyocytes were counted from 3 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n.s. not significant. Data are presented as means ± SD.
We next examined several parameters for evaluating cell proliferation in adult Cre+/− and Lrp6-CKO mice. At this stage, cardiomyocyte proliferation was almost undetectable in the control mice. Two weeks after tamoxifen administration, we observed a significant increase in the adult cardiomyocyte proliferation relative to the level in the control mice, as shown by immunostaining for the DNA synthesis marker EdU (Fig. 2c; Supplementary information, Fig. S3c) and the mitosis marker pH3-S10 (Fig. 2d). Considering it is difficult to accurately distinguish EdU-positive cardiomyocytes from EdU-positive non-cardiomyocytes in tissue sections, we further performed the EdU incorporation assays in cardiomyocytes isolated from adult Lrp6-CKO and Cre+/− mice as previously described.27,28 The results showed that the EdU-positive cardiomyocyte was significantly increased in Lrp6-CKO mice compared to Cre+/− mice (Fig. 2e), consistent with our observations on EdU immunostaining in heart sections. We then examined the total numbers of cardiomyocytes and found significantly more cardiomyocytes in the Lrp6-CKO hearts than in the Cre+/− hearts (Supplementary information, Figs. S5a, S6).
To further confirm the cardiomyocyte proliferation induced by LRP6 deficiency, we performed nucleation and ploidy analysis on isolated cardiomyocytes from the adult Lrp6-CKO and Cre+/− mice. The increased ratio of mononucleated cardiomyocytes and decreased ratio of binucleated cardiomyocytes were observed in the Lrp6-CKO mice (Supplementary information, Fig. S5b). We also observed a 2.96-fold increase in diploid cardiomyocytes in Lrp6-CKO mice, while the percentage of polyploid cardiomyocytes was reduced (Fig. 2f). Additionally, the Lrp6-CKO mice had a significant elevation in diploid cardiomyocyte subpopulation in mononuclear cardiomyocyte (Fig. 2g). For the EdU-positive cardiomyocytes, the ratio of mononuclear was 70.4% in Lrp6-CKO mice, which was significantly increased compared to that in Cre+/− mice (Supplementary information, Fig. S7). Furthermore, a significant increase of EdU-positive mononucleated diploid cardiomyocyte was observed in Lrp6-CKO mice (Fig. 2h). As the EdU-positive mononucleated diploid cardiomyocyte indicates a proliferative event,27,28 we suppose the occurrence of cardiomyocyte cytokinesis and proliferation in Lrp6-CKO mice.
The adult Lrp6-CKO and Cre+/− mice had a normal heart size and normal cardiac function, with no change in the heart to body weight ratio or in cardiac fibrosis (Supplementary information, Fig. S8). We found that the LRP6-deficient cardiomyocytes were significantly smaller than the Cre+/− cardiomyocytes; this small size is a feature of proliferative fetal/neonatal cardiomyocytes (Supplementary information, Fig. S9). This finding may explain why the increased cardiomyocyte numbers in the Lrp6-CKO mice were not accompanied by a significant change in ventricular thickness or the heart weight-to-body ratio. Collectively, these results indicate that LRP6 deficiency induced fully differentiated cardiomyocytes to reenter the cell cycle.
Lrp6 deficiency promotes adult cardiac repair after MI through increased cardiomyocyte proliferation
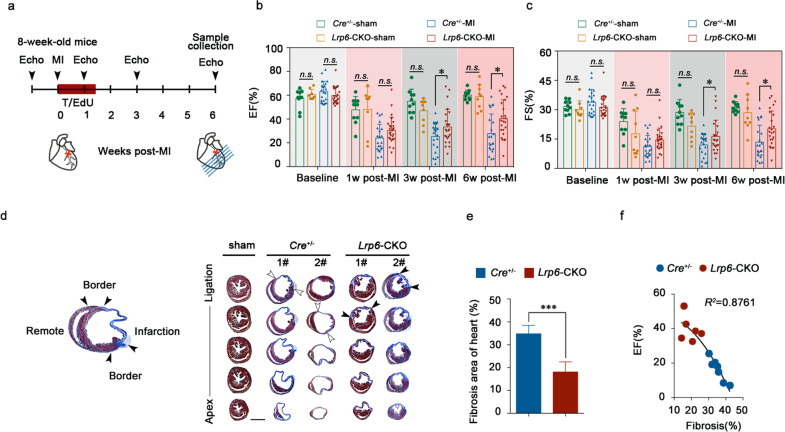
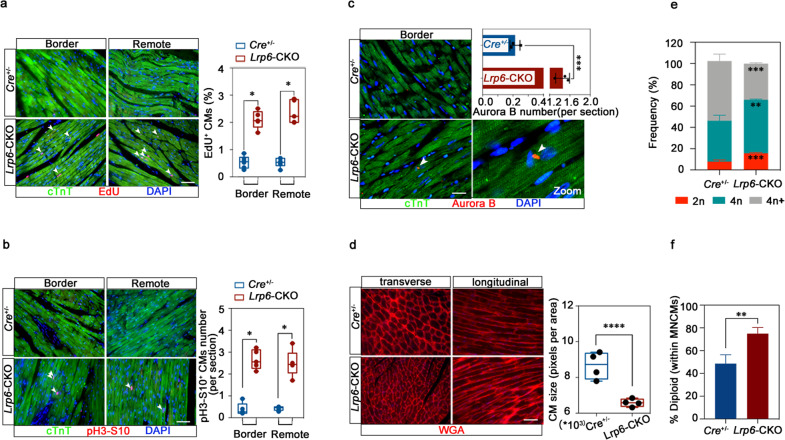
We then examined whether the cardiomyocyte proliferation response mediated by LRP6 plays a role in cardiac repair. We induced MI by permanent ligation of the proximal left anterior descending coronary artery and then induced Lrp6 knockout by tamoxifen administration (Fig. 3a; Supplementary information, Fig. S10a, b). As evidenced by echocardiography at 1 week, 3 weeks and 6 weeks post MI, the left ventricular ejection fraction (LVEF), fractional shortening (FS), and other parameters of cardiac function were significantly improved in the Lrp6-CKO mice relative to the Cre+/− mice from 3 weeks post-MI (Fig. 3b, c; Supplementary information, Fig. S10c). Moreover, analysis of the trichrome-stained heart cross-sections and quantification of the scar size showed that the infarcts were significantly smaller in the Lrp6-CKO mice than in the Cre+/− mice (Fig. 3d, e; Supplementary information, Fig. S10b). Furthermore, correlation analysis showed that the improvement in cardiac function was positively correlated with the reduction in infarct size (Fig. 3f). We observed significant increases in the percentages of EdU-positive cardiomyocytes at both the MI border zone and the remote zone and in the number of cardiomyocytes positive for pH3-S10 and Aurora B kinase (Fig. 4a–c; Supplementary information, Fig. S11). In addition, cardiomyocytes were smaller in the Lrp6-CKO mice than in the Cre+/− mice (Fig. 4d). Considering that increased angiogenesis may contribute to maintaining cardiac function in response to chronic myocardial ischemia, such as MI, we analyzed the density of myocardial capillaries in the Lrp6-CKO and Cre+/− mice after MI. No significant difference was observed between the two groups (Supplementary information, Fig. S12).
Fig. 3. Lrp6 deficiency reduces myocardial infarction size and improves heart function.
a Experimental timeline. MI myocardial infarction, T tamoxifen, Echo echocardiography. Left ventricular systolic function quantified by ejection fraction (EF) (b) and fractional shortening (FS) (c) at 1 week, 3 weeks and 6 weeks post MI. n = 10–25 mice per group. d Masson’s trichrome-stained serial sections 6 weeks after MI. n = 10 per group. Scar boundaries are indicated by arrows. Scale bar = 2 mm. e Left ventricular scar size. Quantification of the fibrotic area relative to the myocardial area in trichrome-stained sections demonstrated a significant decrease in scar formation in Lrp6-CKO hearts relative to Cre+/− hearts. n = 8–10 mice per group. f Correlation analysis of heart fibrosis area and EF value with a second-order polynomial fit. n = 6–7 mice per group. *P < 0.05, ***P < 0.001, n.s. not significant. The data are presented as means ± SD.
Fig. 4. Lrp6 deficiency promotes heart repair after myocardial infarction through cardiomyocyte proliferation.
a Immunostaining revealed EdU+cTnT+ cells in the Cre+/− and Lrp6-CKO hearts at both the infarct border zones and remote areas. n = 5–6 mice per group. Scale bar = 50 μm. b Co-immunostaining for pH3-S10 and cTnT revealed pH3-S10+cTnT+ cells in the Cre+/− and Lrp6-CKO hearts at both the infarct border zones and remote areas. n = 5 mice per group. Scale bar = 50 μm. c Co-immunostaining with Aurora B and cTnT revealed increased cardiomyocyte (CM) mitosis in the Lrp6-CKO hearts. n = 3 mice per group. Scale bar = 25 μm. d WGA staining showed a smaller CM size in Lrp6-CKO post-MI hearts than in the Cre+/− post-MI hearts at the border zone. n = 4 mice per group, 3000 cells were quantified per group. Quantification was performed by analyzing the transverse cell size. Scale bar = 25 μm. e, f The nucleation and ploidy assessment of dissociated CMs from Cre+/− and Lrp6-CKO mice at 4 weeks post MI surgery. 2n, diploid, 4n, tetraploid, 4n+, polyploid. MNCMs, mononucleated CMs. n = 4 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The data are presented as means ± SD.
To distinguish the lineage origin of the newly formed cardiomyocytes in the Lrp6-CKO mice, we used an αMHCMerCreMer; Rosa26-tdTomato genetic lineage tracing system, in which RFP specifically labels cardiomyocytes that have undergone Cre-mediated recombination. Lineage tracing studies were performed after MI and tamoxifen treatment (Supplementary information, Fig. S13a, b). The heart function was improved in the Lrp6-CKO; Rosa26-tdTomato mice relative to the Cre+/−; Rosa26-tdTomato mice (Supplementary information, Fig. S13c, d), but no significant difference was observed in the ratio of EdU-positive cardiomyocytes between Lrp6-CKO; Rosa26-tdTomato and Lrp6-CKO hearts 6 weeks post MI (Supplementary information, Fig. S13e), indicating that the newly formed cardiomyocytes were mainly derived from preexisting cardiomyocytes.
We also performed nucleation and ploidy analysis on isolated cardiomyocytes from adult Lrp6-CKO and Cre+/− mice after MI surgery. An increase in diploid cardiomyocytes and a decrease in polyploid cardiomyocytes were revealed in Lrp6-CKO mice at 4 weeks post MI (Fig. 4e). The significantly increased percentage of diploid cardiomyocyte subpopulation in mononuclear cardiomyocyte was also observed in Lrp6-CKO mice (Fig. 4f). Considering that the diploid cardiomyocyte just appears to be the primary cardiomyocyte subpopulation that is capable of proliferation and of supporting regeneration,29 these data suggested that the increase in mononuclear diploid cardiomyocyte rather than polyploidization, may account for the cardiac regeneration mediated by LRP6 deficiency.
LRP6 regulates cardiomyocyte proliferation through the ING5/P21 pathway
Wnt/β-catenin signaling plays essential roles during embryonic development and organ regeneration.30,31 LRP6 acts as a known coreceptor and conducts Wnt signaling through several cytoplasmic relay components to initiate the canonical β-catenin signaling cascade.32 Previously, we found that LRP6 deficiency did not alter the protein expression and cellular distribution of β-catenin in cardiomyocyte,26 indicating that β-catenin may not be involved in the regulation of cardiomyocyte proliferation mediated by LRP6.
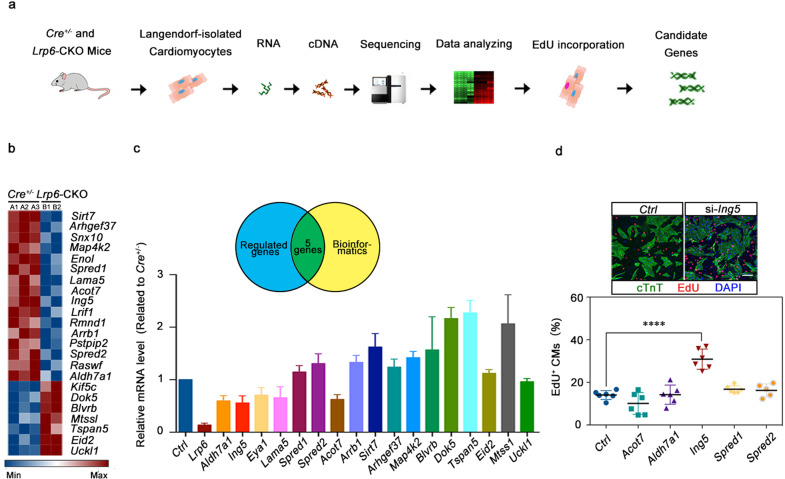
To determine the mechanism by which LRP6 regulates cardiomyocyte proliferation, we collected cardiomyocytes from adult Lrp6-CKO and Cre+/− hearts and performed total RNA sequencing (RNA-seq) (Fig. 5a). Gene Ontology analysis revealed enrichment in the actin cytoskeleton, extracellular matrix and contractile actin filament bundle. Kyoto Encyclopedia of Genes and Genomes mapping suggested that the differentially expressed genes were mainly involved in endothelial cell proliferation, ion transport, the PI3K/Akt pathway and the MAPK signaling pathway (Supplementary information, Fig. S14). Among the differentially expressed genes, 16 downregulated genes and 7 upregulated genes were involved in cell cycle regulation (Fig. 5b). Through PCR validation and bioinformatics analysis, we identified 5 genes (Acot7, Aldh7a1, Ing5, Spred1 and Spred2) that may be downstream of LRP6 (Fig. 5c).
Fig. 5. ING5 is the downstream of LRP6 in cardiomyocyte proliferation regulation.
a Total RNA-seq diagram and b heatmap showing the most differentially regulated genes in the Cre+/− and Lrp6-CKO cardiomyocytes (CMs) as identified by RNA-seq analysis. Red and blue represent up- and down-regulated genes, respectively. n = 2–3 mice per group. c Verification of the expression of genes identified by RNA-seq in the Cre+/− and Lrp6-CKO cardiomyocytes (CMs). The Venn diagram shows five genes that were revealed by RNA-seq and bioinformatics analysis to be highly correlated with cell cycle regulation. n = 4 mice per group. d Immunostaining showing the incorporation of EdU in control (Ctrl) and si-Ing5-treated NRCMs. The results represent 5–6 experiments. Scale bar = 50 μm. The column shows the percentage of proliferating NRCMs after silencing the Acot7, Aldh7a1, Ing5, Spred1, and Spred2 genes in NRCMs. ****P < 0.0001. Data are presented as means ± SD.
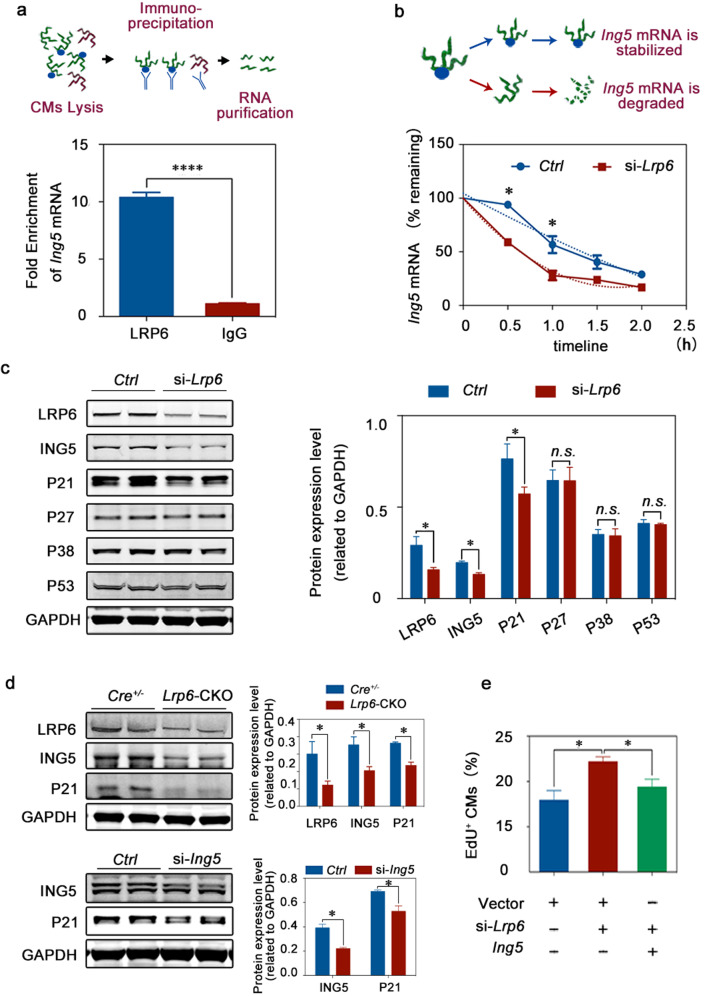
To identify LRP6 target genes, we performed in vitro siRNA knockdown in NRCMs and found that only Ing5 knockdown resulted in a significant increase in cardiomyocyte proliferation, consistent with the effect of LRP6 deficiency (Fig. 5d; Supplementary information, Fig. S15). To determine whether LRP6 can transcriptionally activate Ing5, we generated luciferase reporter constructs that were fused to genomic sequences containing promoter regions of Ing5 (Supplementary information, Fig. S16a). Upon transfecting the Ing5 promoter–reporter gene construct into NRCMs, we found that the reduced Ing5 mRNA level in LRP6 deficient cells was not mediated by the transcription defects (Supplementary information, Fig. S16b). RNA immunoprecipitation-qRT-PCR (RIP-qRT-PCR) with an anti-LRP6 antibody revealed a significant enrichment of Ing5 mRNA, indicating a direct interaction between Ing5 and LRP6 (Fig. 6a). Actinomycin D was used to block de novo RNA synthesis, and the abundance of the existing Ing5 mRNA was measured by qRT-PCR in the presence of Actionmycin D. Lrp6 knockdown accelerated degradation of Ing5 mRNA in the cardiomyocytes (Fig. 6b). Consistent with the mRNA expression findings, LRP6 deficiency reduced the ING5 protein levels in the NRCMs and the Lrp6-CKO heart tissue (Fig. 6c, d). In addition, overexpression of Ing5 attenuated the cardiomyocyte proliferation induced by LRP6 deficiency (Fig. 6e).
Fig. 6. The pro-proliferative effect of LRP6 knockdown is mediated by the ING5/P21 signaling pathway.
a The examination of LRP6 protein binding to Ing5 mRNA in NRCMs was performed by RIP assays as described in the schematic diagram. b Quantitation of the decrease in Ing5 mRNA abundance for the control (Ctrl) and si-Lrp6 NRCMs. The dashed lines illustrate the effects of the two parameters on the shape of the curve. Ing5 mRNA was unstable in the absence of LRP6 protein binding. c The ING5, P21, P27, P38 and P53 protein levels were examined in the control (Ctrl) and si-Lrp6 NRCMs. Left, typical western blots, Right, pooled data. The results represent 3–6 experiments. d The P21 protein levels were reduced in the Lrp6-CKO mouse heart tissues and the Ing5 siRNA-treated NRCMs relative to the control (Ctrl) samples. Top, Cre+/− and Lrp6-CKO tissue samples. Bottom, Ctrl and Ing5 siRNA-treated NRCM samples. Left, typical western blots, Right, pooled data. The results represent 3–6 experiments. e EdU assays revealed that the increased proliferation of CMs induced by LRP6 deficiency was attenuated by the overexpression of Ing5. The results represent three experiments. *P < 0.05, ****P < 0.0001, n.s. not significant. Data are presented as means ± SD.
To assess the potential effect of Ing5 on cardiomyocyte proliferation in vivo, we delivered the AAV9-hTNNT2-Ing5-eGFP or control vector to the Cre+/− or Lrp6-CKO neonatal mice and detected cardiomyocyte proliferation by pH3-S10 and eGFP co-staining (Supplementary information, Fig. S17a–c). We observed that AAV9-mediated Ing5 overexpression significantly decreased the cardiomyocyte proliferation and reversed the increased cardiomyocyte proliferation induced by LRP6 knockdown at P14d (Supplementary information, Fig. S17d).
ING5 is a member of the ING family, which have been identified as an important group of tumor suppressor proteins.33–35 ING5 has been reported to regulate cell proliferation, apoptosis and metastasis in different cancer models mainly through the P53-dependent regulatory pathway.36–39 However, the role of ING5 in the pathophysiology of the heart has not been documented. The current study identified ING5 as a critical regulator of the cardiomyocyte cell cycle downstream of LRP6. We examined the P53 protein expression in cardiomyocytes. LRP6 deficiency did not affect P53 expression but reduced P21 protein levels, whereas the expression of P27 and P38 was not significantly altered by LRP6 deficiency (Fig. 6c). In addition, we observed a reduction in the P21 expression in the Lrp6-CKO hearts and the ING5-deficient cardiomyocytes relative to that in the Cre+/− hearts and the control cardiomyocytes, respectively, suggesting that ING5 regulated cardiomyocyte proliferation in a P53-independent manner (Fig. 6d). Above all, LRP6 served as an RNA-binding protein to regulate the ING5/P21 signaling pathway, controlling the proliferation of cardiomyocytes.
Biological interventions targeting LRP6 promotes heart repair after injury
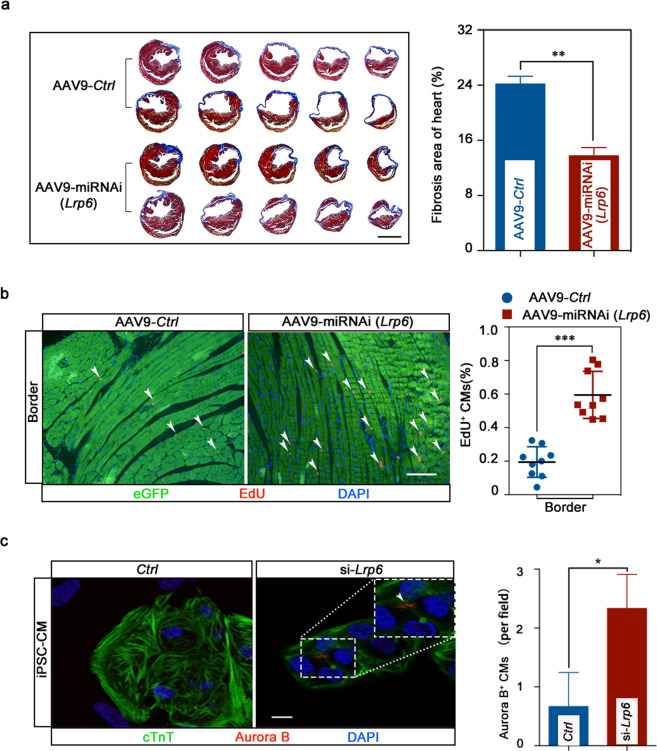
To assess the effect of Lrp6 on cardiac injury repair, we generated an AAV9 construct to knock down Lrp6 in cardiomyocytes (Supplementary information, Fig. S18a, b). The AAV9-miRNAi-Lrp6 construct or a control vector was directly injected into the peri-infarct myocardial area at the time of MI. At 6 weeks after MI and vector delivery, a significantly reduced scar size was detected in the infarcted hearts of animals injected with AAV9-miRNAi-Lrp6 relative to those of the animals treated with the control vector (Fig. 7a). Cardiac function was also preserved in the AAV9-miRNAi-Lrp6 mice (Supplementary information, Fig. S18c, d). Increased numbers of EdU-positive cardiomyocytes were also detected in the infarct border zones in the knockdown mice relative to the control mice (Fig. 7b). Together, these data indicate that Lrp6 knockdown after infarction exerts a notable beneficial effect by stimulating cardiomyocyte proliferation.
Fig. 7. Lrp6 gene therapy reduced scar formation in the hearts of the mice with MI, and LRP6 knockdown increased mitosis in human iPSC-CMs.
a Masson’s trichrome serial sections 6 weeks after MI. Analysis of left ventricle scar size and quantification of fibrotic area relative to myocardium in trichrome-stained sections revealed a significant decrease in scar formation in AAV9-miRNAi-Lrp6 hearts. Scale bar = 2 mm. n = 3 mice per group. b EdU incorporation increased significantly following AAV9-miRNAi-Lrp6 treatment of CMs at the border zone. Scale bar = 100 μm. n = 9 mice. c Confocal images show increased mitosis of human iPSC-CMs following LRP6-knockdown. Graph shows the numbers of Aurora B+cTnT+ CMs. Scale bar = 10 μm. The results represent three experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as means ± SD.
LRP6 inhibition increases mitosis in human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs)
To date, strategies to promote cardiac repair by inducing cardiomyocyte proliferation have been investigated primarily in rodents. Whether these approaches can be applied to restore functional hearts in humans remains to be demonstrated. We were curious whether LRP6 can regulate human cardiomyocyte proliferation; therefore, we investigated the effects of LRP6 on human iPSC-CMs. We found that Lrp6 knockdown significantly increased the number of Aurora B-positive cardiomyocytes (Fig. 7c), indicating that LRP6 could be a potential target for cardiac regeneration in human heart.
Discussion
Our study demonstrated the role of LRP6 in controlling cardiomyocyte proliferation and myocardial repair after injury. Several lines of evidence support this conclusion. First, Lrp6 deficiency induced cardiomyocyte proliferation in neonatal, juvenile and adult mice. Second, cardiomyocyte proliferation by targeting LRP6 expression promoted cardiac repair in both loss-of-function genetic experiments and exogenous virus intervention. Third, Lrp6 knockdown induced differentiated human iPSC-CMs to divide. Finally, ING5/P21 signaling was identified as the molecular basis underlying cardiomyocyte proliferation control downstream of LRP6.
This study revealed a new biological characteristic of LRP6. To date, LRP6 has been investigated in various biological processes, most as a Wnt signaling co-receptor.22,32 For the first time, we demonstrated a novel function of LRP6 in postnatal cardiomyocyte proliferation. The pro-proliferation effects of Lrp6 deficiency were evident in the NRCMs and were detected well beyond the first postnatal week, when most cardiomyocytes normally exit the cell cycle and enter a differentiated phase. We found that the cell cycle was activated in the Lrp6-deficient cardiomyocytes at both the juvenile and adult stages in vivo.
A previous report has shown that Lrp6 deficiency decreases the proliferation and survival of cardiac progenitors in the embryonic stage,23 suggesting the diversity of the biological properties of LRP6 in the life cycle of cardiomyocytes. Such life-cycle-dependent functional difference should have its basis in cell biology. In the present study, immunostaining revealed that LRP6 was localized in cardiac cell nuclei during the embryonic period but became extranuclear during the adult phase (Supplementary information, Fig. S19). Deciphering the localization and molecular interaction network of LRP6 in different periods might provide important insights into cardiac cell proliferation and regeneration.
We previously reported a novel molecular property of LRP6 in regulating gap junction formation in adult mouse hearts in a β-catenin-independent manner.26 In addition, we identified LRP6 as a potential splicing regulator in a muscle development model.40 Here, we identified a critical role of LRP6 as an RNA-binding protein in the postnatal transition of cardiomyocyte proliferation. Lrp6 deficiency accelerated the degradation of Ing5 mRNA by directly binding the Ing5 transcript. Overexpression of Ing5 can attenuate the cardiomyocyte proliferative effect mediated by LRP6 both in vivo and in vitro. Importantly, RNA-binding proteins are important regulators of gene expression and have a wide range of biological effects.41–44 In all of our three studies above, LRP6 acts as an RNA-binding protein performing both biological and pathological functions. However, as an RNA-binding protein, LRP6 and its molecular regulatory network warrant further exploration.
The discovery of the pathologic contributions of LRP6 has led to considerable interest in this protein as a therapeutic target for metabolic syndrome and coronary heart disease.24,45,46 We have reported in the past that the hearts of adult Lrp6-CKO mice were both morphologically and functionally normal under physiological conditions but were particularly susceptible to life-threatening arrhythmias when exposed to stress.26 In addition, we found that the double-knockout of LRP5 and LRP6 in mice led to increased myocardial damage in the infarcted heart at 12 and 24 h post MI.47 The present findings implicate the potential of LRP6-targeting in the clinical applications. We showed that Lrp6 deficiency after MI in mice significantly reduced scar area and improved cardiac function. Importantly, exogenous viral intervention targeting LRP6 also had a significant effect on heart repair. Furthermore, we demonstrated that Lrp6 deficiency could increase the cell cycle activity in human iPSC-CMs. These findings provide insights into the mechanisms of human cardiomyocyte proliferation and reveal a promising molecular basis for therapeutic approaches in human heart injury and congenital heart defects. However, the safety issue of viral intervention in injured hearts remains unclear. Small molecules and synthetic oligonucleotides that specifically target LRP6 might represent a more attractive option than viral vectors and will be a focus of our future work.
Accurate detection of the proliferation of cardiomyocytes, especially in vivo, has always been a challenge.2,48–51 The co-staining of nuclear proliferation markers with cytoplasmic cardiomyocyte markers could not fully distinguish proliferating cardiomyocytes from non-cardiomyocytes. It is also difficult to distinguish between proliferating polyploid mononucleated cardiomyocytes and binucleated cardiomyocytes. Here, in addition to the cytoplasmic cardiomyocyte marker cTNT, we added the nuclear marker PCM1, which is specific to cardiomyocytes, to further distinguish proliferating cardiomyocytes.28,50 Additionally, genetic lineage tracing is a powerful mean to identify different cell lineages and determine cell fate. The Cre-loxP recombination system is the most widely used technology for in vivo cell tracking.52 We used a lineage tracing strategy with cardiomyocytes to precisely determine the source of proliferating cardiomyocytes during regenerative responses by intercrossing αMHC-Cre mice and Rosa26-tdTomato mice. This approach allowed us to ensure that the newly formed cardiomyocytes induced by Lrp6 deficiency after MI were derived from differentiated cardiomyocytes, either pre-existing differentiated cardiomyocytes or cardiomyocytes that differentiated during the tamoxifen induction period.
Unlike the detection of the cell cycle activity using immunostaining, the clonal analysis in vivo appears to be a more direct assay to determine cell division.53 Clonal analysis provides a precise and versatile method to provide direct insights into the proliferative behavior of cardiomyocytes at a single-cell resolution, especially the characteristics of cardiac progenitor cells during embryonic development.54–56 The Brainbow technology and ‘mosaic analysis with double markers’ mouse model have been used in clonal analysis to obtain individual labeled cardiomyocyte in adult mice, and distinguish cell division from just cell cycle activity and/or multinucleation/polyploidization in cardiomyocyte.57–59 However, because of the complexity of the model, few research groups have successfully utilized this system. In addition, it is reported that clonal expansion was primarily detected in non-cardiomyocytes in adult mice in contrast to that in embryonic and neonatal mice. Very rare clones could be detected even after MI in adult mice.54 It seems that this method is more suitable for the qualitative rather than the quantitative analysis at the adult stage.54
Actually, multiple lines of evidence are usually required to prove the induction of cardiomyocyte proliferation. The examination of cardiomyocyte cell cycle activity does not necessarily equate with proliferation as it can have a variety of outcomes such as cell division, endoreduplication, endomitosis and binucleation.60 However, the lack of appropriate unambiguous assays makes it difficult to distinguish cardiomyocyte division from binucleation and polyploidization.59 Recent studies conclude that the mononucleated diploid cardiomyocyte appears to be the primary cardiomyocyte subpopulation that is capable of proliferation and of supporting regeneration, which provides a reliable parameter for determining the proliferation and generation of cardiomyocytes.27,29,61 Thus, the significantly increased ratio of EdU-positive mononucleated diploid cardiomyocytes in Lrp6-CKO mice suggested the cytokinesis and proliferation of cardiomyocyte induced by LRP6 deficiency.
In summary, our study identified that LRP6 regulates cardiomyocyte proliferation through the ING5/P21 pathway other than its well-established role as a Wnt co-receptor. The increased cardiomyocyte proliferation mediated by LRP6 deficiency significantly contributed to adult cardiac repair and improvement of heart function after MI. Our work reveals the novel cellular biological functions and signaling pathways of LRP6 and demonstrates its potential in myocardial regeneration and repair.
Materials and methods
Animals
All animal experiments and procedures were carried out in strict accordance with the Guidelines for the Care and Use of Laboratory Animals set by the U.S. National Institutes of Health (National Academies Press; 2011) and were approved by Tongji University school of medicine. All animal experiments were performed on age-matched mice, and the experimenters were blinded to the genotypes of the mice.
Isolation of NRCMs
Primary cardiomyocytes (CMs) were isolated from Sprague-Dawley rats at the age of P1d–P3d.62 Briefly, the ventricles were separated and cut into pieces (~1–3 mm2), then they were dissociated in calcium-free HBSS (Gibco) containing 0.125 mg/mL trypsin (Gibco), 0.1 mg/mL collagenase type IV (Sigma) and 10 mg/mL DNase II (Sigma). Digestion was performed at 37 °C by continually stirring the heart sections, and the supernatant was collected into HBSS containing 10% FBS every 5 min. The digestion procedure was repeated ~8–10 times. When the tissue was completely digested, the supernatant was centrifuged at 1000 rpm for 10 min and resuspended in DMEM (Gibco) supplemented with 10% FBS and 100 mM 5-bromo-20-deoxyuridine (B5002-250MG, Sigma). The resuspended cells were passed through a cell strainer (100 μm, BD Falcon), seeded onto 10 cm plastic dishes for 2 h at 37 °C to remove fibroblasts, and then plated on 1% gelatin-coated plastic culture dishes at an appropriate density. After 24 h, the medium was changed to low-serum medium, and the cells were cultured for 48 h before use.
Cell transfection
For some gene silencing studies, siRNAs against Lrp6, Ing5, Acot7, Aldh7a1, Spred1, and Spred2 were transfected into NRCMs at a final concentration of 50 nM using Lipofectamine RNAiMAX (13778-150, Invitrogen), respectively. All siRNA sequences are listed in Supplementary information, Table S1. For gene overexpression, plasmids were transfected respectively using Lipofectamine 3000 (L3000-008, Invitrogen). 48–72 h after transfection, the cells were collected for the experiments.
Mouse alleles and transgenic lines
Lrp6-floxed mice were maintained on a mixed C57BL6 genetic background. The mice were genotyped by PCR using genomic DNA isolated from the tails with forward (5′-GGGGTTCTACTTTTGTGTGTGG-3′) and reverse (5′-CCGTCTGTTTGCATAAAGCAACA-3′) primers. On a 2% agarose gel, the wild-type (WT) allele was shown to 343 bp, and the mutant allele generated a 411 bp band. A cardiac-specific deletion Lrp6 knockout (Lrp6-CKO) mouse line was generated by crossing the Lrp6flox/flox mice with αMHC/MerCreMer (Cre+/−) mice. The αMHC-Cre mice were genotyped using the following primers: 5′-TCGATGCAACGAGTGATGAG-3′ and 5′-TCCATGAGTGAACGAACCTG-3′.
To trace cardiomyocyte lineages, we intercrossed αMHC-Cre mice and Rosa26-tdTomato mice.52 The Rosa26-tdTomato mouse line was kindly provided by Bin Zhou, the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences. Rosa26-tdTomato mice were genotyped using the following primers: 5′-AAGGGAGCTGCAGTGGAGTA-3′, 5′-CCGAAAATCTGTGGGAAGTC-3′, 5′-GGCATTAAAGCAGCGTATCC-3′ and 5′-CTGTTCCTGTACGGCATGG-3′. The WT allele was identified as 297 bp, and the mutant allele was determined to be 196 bp. In this line, the mice expressed RFP protein in cardiomyocytes that underwent Cre-mediated recombination. This system allowed us to distinguish whether the proliferating cells were derived from preexisting cardiomyocytes.
Tamoxifen administration
Tamoxifen (T5648, Sigma) was dissolved in 90% peanut oil/10% alcohol. To achieve recombination of the floxed alleles in the experiments with the P7d and P14d mice, tamoxifen was administered on P0d, P1d and P2d at a dose of 40 µg per day using an insulin needle.63 To achieve recombination of floxed alleles in adult mice, tamoxifen was administered via intraperitoneal injection (80 mg/kg, once a day for 5 consecutive days) at 8 months or after MI.64
MI surgery
Eight-week-old αMHC-Cre, Lrp6-CKO, αMHC-Cre; Rosa26-tdTomato, Lrp6-CKO; Rosa26-tdTomato and C57/BL6 mice were anesthetized with 1% isoflurane in a chamber. The mice were placed in a left supine position on a heating pad (37 °C), and the heart was exposed via thoracotomy at the fourth left intercostal space. The pericardium was then opened, and the left coronary artery was permanently ligated with a 7-0 suture. Ligation was considered successful when the left ventricle became pale. For virus-injected mice, the AAV vectors were injected immediately after ligation into the myocardium bordering the infarct zone. The chest was then closed with 6-0 suture. For the αMHC-Cre, Lrp6-CKO, αMHC-Cre; Rosa26-tdTomato, and Lrp6-CKO; Rosa26-tdTomato mice, tamoxifen was administered via intraperitoneal injection (80 mg/kg, once a day for 5 consecutive days) 1 day after surgery. For αMHC-Cre, Lrp6-CKO, αMHC-Cre; Rosa26-tdTomato, Lrp6-CKO; Rosa26-tdTomato mice, echocardiography was performed at 1 week, 3weeks, and 6 weeks after surgery, for AAV vectors-injected mice, echocardiography was performed every week for 6 weeks after surgery. The mice were then euthanized, and the hearts were subjected to histology. Sham controls were used for each genotype.
EdU assay
For NRCMs, the medium was replaced with medium containing 10 µM EdU (c10338, Thermo Fisher Scientific, component A) 48 h after transfection, and cells were fixed 12 h later. For EdU assay of adult heart section, a dose of 500 µg EdU (A10187, Thermo Fisher) per animal was injected intraperitoneally every 2 days for five times. For isolated adult mice cardiomyocytes EdU assay, a dose of 1000 µg EdU (A10187, Thermo Fisher) per animal was injected intraperitoneally once daily for 10 days. For EdU staining, the rehydrated heart sections and cells were incubated with Click-iT EdU 555 Imaging Kit reagents (c10338, Thermo Fisher Scientific) to reveal EdU incorporation according to the manufacturer’s instructions.
Injection of AAV vectors in the neonatal and adult mice
For the experiments using AAV vectors, the αMHC-Cre, Lrp6-CKO neonatal mice at P7d were intraperitoneally injected with AAV vectors (pAAV2-hTNNT2-NC-3Flag-T2A-EGFP, pAAV2-hTNNT2-Ing5-3Flag-T2A-EGFP) at a dose of 8 × 109/g viral genome particles per animal using an insulin syringe with an incorporated 30-gauge needle. Hearts were collected 7 days after AAV injection.
Adult mice underwent an intracardiac injection (bordering infarct zone) of AAV2/9-hTNNT2-eGFP-miRNAi (negative control, NC) or AAV2/9-hTNNT2-eGFP-miRNAi-Lrp6 at a dose of 4 × 1011 viral genome particles per animal underwent MI. Hearts were collected 6 weeks after MI.
Echocardiography
Cardiac function was assessed by serial echocardiography before and after MI. Left ventricular systolic function was evaluated by echocardiography (Visual Sonics Vevo 770) under conditions of 1% isoflurane anesthesia with spontaneous ventilation. Two-dimensional B-mode imaging was used to capture the long-axis projection with guided M-mode images. The LVEF and FS were calculated based on end diastolic and end systolic dimensions obtained from M-mode ultrasound.
Injury region division
The cardiac tissue regions used for image characterization are described as the whole heart, infarct zone (left ventricle free wall), border zone (left ventricle anterior and posterior walls), or distal zone (interventricular septum).
Histology
For histological studies, hearts were collected at the indicated time points. After the blood was removed by retrograde perfusion from the heart apex with cold PBS, the whole hearts were fixed with 4% paraformaldehyde (PFA, Sigma) at 4 °C overnight. Next, the hearts were dehydrated in increasing concentrations of ethanol and embedded in paraffin. For P7d, P14d and P10w baseline mice, the hearts were sectioned longitudinally. Hematoxylin and eosin staining and Masson’s trichrome staining were performed according to previously published methods.65
For analysis of adult cardiac regeneration following the MI procedure, the hearts were sectioned at a thickness of 8 µm, and slides were created with 5 sections per slide. The sections started at the ligation site and ended at the heart apex (~50 slides). The slides were stained with Masson’s trichrome stain to identify areas of fibrosis. The scar size was quantified by examining serial sections from the apex to the ligation site and calculating the average percent fibrotic area of the total area using Image J software based on Masson’s trichrome staining.
Immunofluorescence
Cells were fixed with 4% PFA for 15 min, washed twice with PBS for 5 min, and then permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Then, the nonspecific sites were blocked for 1 h with 4% normal goat serum at room temperature. The cells were then incubated with primary antibodies at 4 °C overnight. Then, the cells were washed with PBST and incubated for 1 h at room temperature with the appropriate secondary antibodies conjugated to Alexa Fluor-488 or Alexa Fluor-555 (1:200, Invitrogen). The cells were washed again in PBST three times and then stained with DAPI (D9542, Sigma) to label the nuclei.
The paraffin-embedded heart slides were de-paraffinized with xylene and then rehydrated in decreasing concentrations of ethanol (100, 100, 95, 90, and 80%) and water. The heart slides underwent antigen retrieval by microwaving in citrate solution for 10 min; then, the slides were blocked with 10% goat serum (Invitrogen) and incubated with primary antibody overnight at 4 °C. The next day, the slides were washed in PBST three times and incubated with the appropriate fluorescent secondary antibody (1:200, Invitrogen) for 1 h at room temperature. The slides were washed in PBST three times and then stained with DAPI (D9542, Sigma) to label the nuclei.
The primary antibodies included mouse anti-cTnT (1:100, Abcam), rabbit anti-histone H3 (1:500, ab32107, Abcam), rabbit anti-Aurora B (1:100, ab2254, Abcam), and an isolectin B4 antibody (Vector Labs), mouse anti-histone H3 (1:500, 9706 S, CST), mouse anti-Aurora B (1:100, ab3609, Abcam), and rabbit anti-PCM1 (1:100, HPA023370, Sigma).
WGA staining
For isolated cardiomyocytes, the cells were collected in 24-well plates for 4 h at 37 °C, stained with WGA conjugated to Alexa Fluor-594 (50 mg/mL, W11262, Thermo Fisher Scientific) in PBS for 30 min, and then washed with PBS. For heart sections, the slides were deparaffinized, rehydrated and then incubated for 1 h at room temperature with WGA conjugated to Alexa Fluor-594 (50 mg/mL, W11262, Thermo Fisher Scientific) in PBS. The slides were then rinsed in PBS.
Image acquisition and analysis
Immunofluorescent images were acquired on a fluorescence microscope (Leica) and analyzed with Image J. For cardiomyocyte proliferation quantification in vitro, data were collected from more than 3 batches (Figs. 1a, c, 6e, 7c; Supplementary information, Figs. S2, S15, n = 3 batches; Fig. 1b, n = 4 batches; Fig. 5d, n = 5–6 batches.) in each group. For cardiomyocyte proliferation quantification in vivo, data were collected from more than 3 mice (Fig. 2b, n = 3–5 mice; Fig. 2c, d, n = 3–6 mice; Fig. 2e, 5000–7000 cells from three mice; Fig. 4a, n = 5–6 mice; Fig. 4b, n = 5 mice; Fig. 4c, n = 3 mice; Fig. 7b, n = 9 mice; Supplementary information, Fig. S3, n = 3 mice; Supplementary information, Fig. S11, n = 3 mice; Supplementary information, Fig. S13e, n = 4–6 mice; Supplementary information, Fig. S17b–d, n = 4–6 mice.) in each group. For cell size analysis, three to eight independent samples per group were analyzed, for heart slides, 4000 cells from different fields and positions of the transverse sections were quantified for each group, for isolated adult cardiomyocytes, 2000–3000 cells from different fields were quantified for each group. For cardiomyocytes nucleation analysis, Fig. S5b, ~7000–9000 cells were counted from 4 mice per group. For cardiomyocytes ploidy analysis, ~800 cells from 4 mice per group were quantified. For nucleation and ploidy analysis of EdU-positive cardiomyocytes, ~300 cells from three Lrp6-CKO mice and 70 cells from 3 Cre+/− mice were quantified. For capillary density analysis, data were collected from three independent animals per group and analyzed with Image J.
Isolation of adult mouse ventricular myocytes
For RNA-seq, cardiomyocytes were isolated as previously described, with a minor modification.66 8- to 10-week-old Cre+/− and Lrp6-CKO mice were injected with 200 μL of heparin (100 IU/mouse) before being anesthetized with pentobarbital (70 mg/kg). The hearts were removed and perfused at 37 °C with a Ca2+-free buffer (in mM: NaCl, 113; KCl, 4.7; KH2PO4, 0.6; Na2HPO4, 0.6; MgSO4, 1.2; HEPES, 10; NaHCO3, 12; KHCO3, 10; taurine, 30; butanedione monoxime, 10; glucose, 5.5) until the effluents became clear. Next, enzymatic digestion was performed with collagenase type II (300 U/mL, Worthington) for 20 min. When the hearts became slightly pale and flaccid, the ventricles were removed, cut into small pieces in stop buffer (Ca2+-free solution, 10% bovine calf serum) and filtered through a cell strainer (100 µm, BD Falcon). The myocytes were centrifuged at 4000 × g for 3 min, and total RNA was isolated for RNA-seq.
For determination of the total number of myocytes and quantify CMs size, cardiomyocytes were isolated as previously described.12 Hearts were fixed with freshly made 4% PFA on a shaker at 120 rpm for 1 h at room temperature. The hearts were rinsed with PBS, cut into pieces, and rinsed with 1× HBSS (1 mM Mg2+ and 4 mM Ca2+) three times. Then, the pieces were digested with collagenase (collagenase type II: 1.8 mg/mL, collagenase type IV: 2.4 mg/mL, Worthington) at 37 °C. On the second day, the supernatant was collected and centrifuged (500 rpm for 2 min) to yield isolated cardiomyocytes. Then, the undigested pieces were re-digested with fresh enzyme. These procedures were repeated until all tissues were digested. All myocytes were collected and stained with DAPI for counting number of cardiomyocytes. For myocyte counts, three different counts/sample and counts for three hearts/group were obtained using a handheld automated cell counter (Millipore) and averaged.
Quantification of cardiomyocyte nucleation and ploidy
Suspended single cells were simply washed with PBS after DAPI staining, then deposited on slides. Pictures slides were captured using a Spot camera holding all exposure settings constant. Cardiomyocyte nucleation was quantified manually.
For ploidy analysis, single suspension cardiomyocytes were stained with DAPI as above. Briefly, nuclei were identified and outlined with a standard threshold requirement using Image J software, and DAPI fluorescence intensity of individual nuclei in cardiomyocytes and in non-cardiomyocytes were calculated in the same picture. The average value of DAPI fluorescence intensity of non-cardiomyocyte nuclei was used as a diploid nucleus standard, cardiomyocyte nuclei were determined as diploid if their normalized intensity values were within a 0.5–1.5 times range of this standard, cardiomyocyte nuclei were determined as tetraploid if their normalized intensity values were within a 1.5–2.5 times range of this standard, cardiomyocyte nuclei were determined as polyploid if their normalized intensity values were greater than 2.5 times range of this standard.28
Luciferase reporter assay
For construction of a luciferase reporter plasmid for the Ing5 promoter, a fragment containing the mouse Ing5 promoter region was amplified from mouse genomic DNA by PCR. The fragment was then inserted into the pGL3-Basic luciferase reporter plasmid between the KpnI and XhoI loci. For determination of the effect of LRP6 deficiency on the activity of the Ing5 promoter, NRCMs were transfected with si-Lrp6 or si-Ctrl. NRCMs were transfected with 0.5 µg of the Ing5 promoter luciferase reporter plasmid and 0.01 µg of the Renilla luciferase plasmids in 24-well plates using Lipofectamine 3000 (Invitrogen). Cells were harvested 24 h after plasmid transfection. The luciferase activity was measured using a dual luciferase reporter assay system (Promega).
RNA immunoprecipitation assay
RIP was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (17–700, Millipore) according to the manufacturer’s instructions. NRCMs in 15 cm dishes were collected, homogenized in ice-cold RIP lysis buffer, and then stored at −80 °C overnight. Magnetic beads were preincubated with 1 µg of LRP6 antibody (c5c7, Cell Signaling Technology) or with 1 µg of normal rabbit IgG (03-241, Millipore), which was used as a negative control. Next, the frozen homogenates were thawed quickly and centrifuged at 14,000 rpm for 10 min, and 10 µl of the supernatant was stored as input. Then, for each RIP reaction, 100 µl of supernatant was incubated overnight with the magnetic bead-antibody complex at 4 °C. On the second day, the RNA/protein immunocomplex was extensively washed with RIP Wash Buffer (provided in the kit). The cross-linking was reversed by incubation with proteinase K. The immunoprecipitated RNA was purified through phenol: chloroform: isoamyl alcohol (125:24:25) isolation. The purified immunoprecipitated RNA was reverse-transcribed into cDNA using an RNA-to-cDNA Kit (RR036A, Takara). qRT-PCR was performed using this cDNA as a template to quantify the Ing5 mRNA.
RNA stability study
NRCMs at 80% confluency were transfected with either anti-Lrp6 siRNA or scrambled Control siRNA in 35-mm culture dishes. After 48 h, the medium was replaced with medium containing 1 µmol/L actinomycin D (A1410, Sigma) to inhibit transcription; every 0.5 h thereafter, total RNA was extracted using TRIzol (Ambion) reagent according to the manufacturer’s instructions. qRT-PCR was performed using specific sets of primers and SYBR Green Master Mix (A25742, Applied Biosystems Life Technologies). GAPDH levels were used to normalize the specific gene expression levels. The primer sequences are listed in Supplementary information, Table S2.
hiPSC-derived CMs culture
hiPSC-derived CMs were purchased from Help Stem Cell Innovations (Nanjing, China). The cells were thawed and seeded on 1% fibronectin pre-coated six-well optical bottom plates (1 × 106 cells/plate). Cells were allowed to adhere for 48–72 h prior to replacement of the medium with cardiac maintenance medium (Help Stem Cell Innovations). The medium was refreshed every 2 days. After 18 days, the cells were digested from the dish, resuspended in fresh cardiac maintenance medium and divided into 48-well plate for immunofluorescence staining.
qRT-PCR
Total RNA was extracted from NRCMs and adult mouse ventricular myocytes (AMVMs) using TRIzol reagent (Invitrogen, USA). cDNA was generated using qScript cDNA SuperMix (RR036A, Takara). qRT-PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) with SYBR Green Supermix (A25742, Applied Biosystems). All procedures were performed according to the manufacturers’ manuals. The PCR primers were listed in Supplementary information, Table S2.
Western blot analysis
Hearts and NRCMs were homogenized and lysed in cold RIPA buffer (Beyotime) containing protease inhibitors (Roche) for protein extraction. Equal amounts of total proteins (70 µg for heart proteins, 50 µg for cell proteins) were mixed and dissolved in 4 × SDS/PAGE sample buffer and heated to 95 °C for 5 min. The proteins were then separated on NuPAGE 10% Bis-Tris Gels (Invitrogen) and electrophoretically transferred onto polyvinylidene fluoride membranes. The membranes were immunoblotted overnight at 4 °C with appropriate primary antibodies, including the following: LRP6 (ab134146, Abcam), ING5 (10665-1-AP, Proteintech), P21 (ab109199, Abcam), P27 (sc-1641, Santa Cruz Biotechnology), P53 (ab26, Abcam), P38 (ab170099, Abcam), and GAPDH (60004-1, Proteintech). On the second day, after washing three times with TBST, the membranes were incubated with conjugated fluorescent secondary antibody (Li-Cor), and the bands were visualized using an Odyssey imager.
Statistical analysis
All statistical data are shown as the means ± standard deviation (SD). Unpaired two-tailed Student’s t-test and one-way ANOVA followed by Tukey’s post hoc test was used for statistical comparison of two and more than two groups, respectively. Statistical analysis were performed using Prism 5.0 (GraphPad Software). A value of P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This work was funded by the Grants from the National Key Research and Development Plan (2019YFA0801501, to Y.-H.C.), the Programs of National Natural Science Foundation of China (81530017, 81930013 and 81770397, to Y.-H.C.; 81770267, to Dandan Liang), the Outstanding Young Talent Training Program of Shanghai Municipal Commission of Health and Family Planning (2017YQ045, to Dandan Liang), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2017-05), Top-level Clinical Discipline Project of Shanghai Pudong District (PWYgf2018-02), Research Unit of Origin and Regulation of Heart Rhythm, Chinese Academy of Medical Sciences (2019RU045), the Fundamental Research Funds for the Central Universities to Y.-H.C. Y.-H.C. is a Fellow at the Collaborative Innovation Center for Cardiovascular Disease Translational Medicine, Nanjing Medical University.
Author contributions
Y.W. and L.Z. performed experiments, analyzed data and wrote the manuscript. H.L. and R.D. performed experiments and analyzed data. H.Z., F.Z., X.H., D.Lu., K.X., M.X., J.Z., Y.L., and L.L. performed experiments. D.Liang. funded the projects, analyzed data and wrote the manuscript. Y.-H.C. designed, led and funded the projects, and wrote the manuscript. All authors have proof-read the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yahan Wu, Liping Zhou, Hongyu Liu, Ran Duan
Contributor Information
Dandan Liang, Email: dandanliang@tongji.edu.cn.
Yi-Han Chen, Email: yihanchen@tongji.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-00411-7.
References
- 1.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soonpaa MH, et al. Cardiomyocyte cell-cycle activity during preadolescence. Cell. 2015;163:781–782. doi: 10.1016/j.cell.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 3.Zhu W, et al. Regenerative potential of neonatal porcine hearts. Circulation. 2018;24:2809–2816. doi: 10.1161/CIRCULATIONAHA.118.034886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 2013;8:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis Stuart SD, De Jesus NM, Lindsey ML, Ripplinger CM. The crossroads of inflammation, fibrosis, and arrhythmia following myocardial infarction. J. Mol. Cell Cardiol. 2016;91:114–122. doi: 10.1016/j.yjmcc.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;25:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 10.Iismaa SE, et al. Comparative regenerative mechanisms across different mammalian tissues. NPJ Regen. Med. 2018;23:3–6. doi: 10.1038/s41536-018-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells JM, Watt FM. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;7705:322–328. doi: 10.1038/s41586-018-0073-7. [DOI] [PubMed] [Google Scholar]
- 12.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 2010;10:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach JP, et al. Hippo pathway deficiency reverse systolic heart failure after infarction. Nature. 2017;7675:260–264. doi: 10.1038/nature24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F, et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat. Commun. 2019;1:1802. doi: 10.1038/s41467-019-09530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Uva G, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 18.Aysu U, Richard TL. Mechanisms of cardiac regeneration. Dev. Cell. 2016;36:362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdos FX, et al. Cardiac regeneration: lessons from development. Circ. Res. 2017;20:941–959. doi: 10.1161/CIRCRESAHA.116.309040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karra R, Poss KD. Redirecting cardiac growth mechanisms for therapeutic regeneration. J. Clin. Investig. 2017;2:427–436. doi: 10.1172/JCI89786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 22.Acebron SP, Niehrs C. β-catenin independent roles of Wnt/LRP6 signaling. Trends Cell Biol. 2016;26:956–967. doi: 10.1016/j.tcb.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Song L, Li Y, Wang K, Zhou CJ. Cardiac neural crest and outflow tract defects in Lrp6 mutant mice. Dev. Dyn. 2010;239:200–210. doi: 10.1002/dvdy.22079. [DOI] [PubMed] [Google Scholar]
- 24.Mani A, et al. LRP6 Mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava R, et al. Impaired LRP6-TCF7L2 activity enhances smooth muscle cell plasticity and causes coronary artery disease. Cell Rep. 2015;13:746–759. doi: 10.1016/j.celrep.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, et al. LRP6 acts as a scaffold protein in cardiac gap junction assembly. Nat. Commun. 2016;7:11775. doi: 10.1038/ncomms11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson M, et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 2017;49:1346–1353. doi: 10.1038/ng.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirose K, et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science. 2019;364:184–188. doi: 10.1126/science.aar2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan PH, Patterson M, Sucov HM. Cardiomyocyte polyploidy and implications for heart regeneration. Annu. Rev. Physiol. 2020;82:45–61. doi: 10.1146/annurev-physiol-021119-034618. [DOI] [PubMed] [Google Scholar]
- 30.Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;11:dev146589. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 31.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;3:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 32.Joiner DM, Ke J, Zhong Z, Xu HE, Williams BO. LRP5 and LRP6 in development and disease. Trends Endocrinol. Metab. 2013;1:31–39. doi: 10.1016/j.tem.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyon Y, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol. Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PubMed] [Google Scholar]
- 35.Gong. W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int. J. Biochem. Cell Biol. 2005;5:1054–1065. doi: 10.1016/j.biocel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Shiseki M, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 37.Liu N, et al. ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 2013;73:3749–3760. doi: 10.1158/0008-5472.CAN-12-3684. [DOI] [PubMed] [Google Scholar]
- 38.Cengiz B, et al. Tumor-specific mutation and downregulation of ING5 detected in oral squamous cell carcinoma. Int. J. Cancer. 2010;127:2088–2094. doi: 10.1002/ijc.25224. [DOI] [PubMed] [Google Scholar]
- 39.Jafarnejad SM, Li G. Regulation of p53 by ING family members in suppression of tumor initiation and progression. Cancer Metastasis Rev. 2012;1–2:55–73. doi: 10.1007/s10555-011-9329-5. [DOI] [PubMed] [Google Scholar]
- 40.Yuan T, et al. Low-density lipoprotein receptor-related protein 6 regulates alternative pre-mRNA splicing. J. Cell Mol. Med. 2018;10:4653–4663. doi: 10.1111/jcmm.13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner M, Díaz-Muñoz MD. RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 2018;2:120–129. doi: 10.1038/s41590-017-0028-4. [DOI] [PubMed] [Google Scholar]
- 42.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;7:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;8:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Conlon EG, Manley JL. RNA-binding proteins in neurodegeration: mechanisms in aggregate. Genes Dev. 2017;15:1509–1528. doi: 10.1101/gad.304055.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Go GW. Low-density lipoprotein receptor-related protein 6 (LRP6) is a novel nutritional therapeutic target for hyperlipidemia, non-alcoholic fatty liver disease, and atherosclerosis. Nutrients. 2015;6:4453–4464. doi: 10.3390/nu7064453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keramati AR, et al. Wild-type LRP6 inhibits, whereas atherosclerosis-lingked LRP6R611C increases PDGF-dependent vascular smooth muscle cell proliferation. Proc. Natl. Acad. Sci. USA. 2011;5:1914–1918. doi: 10.1073/pnas.1019443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wo D, et al. Opposing roles of Wnt inhibitors IGFBP-4 and Dkk1 in cardiac ischemia by differential targeting of LRP5/6 and β-catenin. Circulation. 2016;24:1991–2007. doi: 10.1161/CIRCULATIONAHA.116.024441. [DOI] [PubMed] [Google Scholar]
- 48.Engel FB, Schebesta M, Keating MT. Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell Cardiol. 2006;4:601–612. doi: 10.1016/j.yjmcc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Hesse M, et al. Midbody positioning and distance between daughter nuclei enable unequivocal identification of cardiomyocyte cell division in mice. Circ. Res. 2018;9:1039–1052. doi: 10.1161/CIRCRESAHA.118.312792. [DOI] [PubMed] [Google Scholar]
- 50.Alkass K, et al. No Evidence for cardiomyocyte number expansion in preadolescent mice. Cell. 2015;4:1026–1036. doi: 10.1016/j.cell.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 51.Engel FB, Hsieh PC, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. USA. 2006;42:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L, et al. Enhancing the precision of genetic lineage tracing dual recombinases. Nat. Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leone M, Magadum A, Engel FB. Cardiomyocyte proliferation in cardiac development and regeneration: a guide to methodologies and interpretations. Am. J. Physiol. Heart Circ. Physiol. 2015;309:H1237–H1250. doi: 10.1152/ajpheart.00559.2015. [DOI] [PubMed] [Google Scholar]
- 54.Sereti KI, et al. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat. Commun. 2018;1:754. doi: 10.1038/s41467-018-02891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yelon D. Developmental biology: heart under construction. Nature. 2012;7395:459–460. doi: 10.1038/484459a. [DOI] [PubMed] [Google Scholar]
- 56.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;7395:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin ZQ, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali SR, et al. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl. Acad. Sci. USA. 2014;111:8850–8855. doi: 10.1073/pnas.1408233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leone M, Engel FB. Advances in heart regeneration based on cardiomyocyte proliferation and regenerative potential of binucleated cardiomyocytes and polyploidization. Clin. Sci. 2019;133:1229–1253. doi: 10.1042/CS20180560. [DOI] [PubMed] [Google Scholar]
- 60.Zebrowski DC, Becker R, Engel FB. Towards regenerating the mammalian heart: challenges in evaluating experimentally induced adult mammalian cardiomyocyte proliferation. Am. J. Physiol. Heart Circ. Physiol. 2016;310:H1045–H1054. doi: 10.1152/ajpheart.00697.2015. [DOI] [PubMed] [Google Scholar]
- 61.Shen H, et al. Mononuclear diploid cardiomyocytes support neonatal mouse heart regeneration in response to paracrine IGF2 signaling. eLife. 2020;9:e53071. doi: 10.7554/eLife.53071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collesi C, Zentilin L, Sinagra G, Giacca M. Notch1 signaling stimulates proliferation of immature cardiomyocytes. J. Cell Biol. 2008;183:117–128. doi: 10.1083/jcb.200806091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao G, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534:119–123. doi: 10.1038/nature17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji YX, et al. The ubiquitin E3 ligase TRAF6 exacerbates pathological cardiac hypertrophy via TAK1-dependent signaling. Nat. Commun. 2016;7:11267. doi: 10.1038/ncomms11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heallen T, et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Köhncke C, et al. Isolation and Kv channel recordings in murine atrial and ventricular cardiomyocytes. J. Vis. Exp. 2013;12:e50145. doi: 10.3791/50145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.