Abstract
Bi-allelic loss-of-function (LoF) variants in the base excision repair (BER) gene NTHL1 cause a high-risk hereditary multi-tumor syndrome that includes breast cancer, but the contribution of heterozygous variants to hereditary breast cancer is unknown. An analysis of 4985 women with breast cancer, enriched for familial features, and 4786 cancer-free women revealed significant enrichment for NTHL1 LoF variants. Immunohistochemistry confirmed reduced NTHL1 expression in tumors from heterozygous carriers but the NTHL1 bi-allelic loss characteristic mutational signature (SBS 30) was not present. The analysis was extended to 27,421 breast cancer cases and 19,759 controls from 10 international studies revealing 138 cases and 93 controls with a heterozygous LoF variant (OR 1.06, 95% CI: 0.82–1.39) and 316 cases and 179 controls with a missense variant (OR 1.31, 95% CI: 1.09–1.57). Missense variants selected for deleterious features by a number of in silico bioinformatic prediction tools or located within the endonuclease III functional domain showed a stronger association with breast cancer. Somatic sequencing of breast cancers from carriers indicated that the risk associated with NTHL1 appears to operate through haploinsufficiency, consistent with other described low-penetrance breast cancer genes. Data from this very large international multicenter study suggests that heterozygous pathogenic germline coding variants in NTHL1 may be associated with low- to moderate- increased risk of breast cancer.
Subject terms: Cancer genomics, Mutation, Cancer genetics, Breast cancer, Gene expression
Introduction
NTHL1 encodes a DNA glycosylase that is a critical component of the DNA base excision repair (BER) pathway involved in the repair of oxidatively damaged DNA. It has recently been shown that carriers of bi-allelic loss-of-function (LoF) variants in NTHL1 are predisposed to colorectal adenomatous polyposis and colorectal cancer1, and to a multi-tumor syndrome that includes a high incidence of breast cancer in female carriers2–5. Grolleman et al.4 described the largest set of carriers of bi-allelic germline NTHL1 variants (29 carriers from 17 families), and reported that 9 of 15 female carriers (60%) were diagnosed with breast cancer at an earlier age than observed in the general population (48.5 years compared to 62 years). In contrast to the previously described BER defect caused by MUYTH deficiency, multiple tumor types from carriers of germline bi-allelic NTHL1 LoF variants exhibit a distinctive somatic mutation pattern (Single Base Substitution Signature 30 [SBS30] in the COSMIC database1,6) characterized by an abundance of C > T transitions at non-CpG sites, indicating that an NTHL1-driven BER defect was the predominant mutational process driving the development of these tumors. These data demonstrated that germline bi-allelic inactivation of NTHL1 predisposes to breast cancer, although individuals with two LoF variants are very rare in the population4. In contrast, 0.37% of non-cancer participants in gnomAD are carriers of monoallelic LoF variants (gnomAD V2.1.1, 134,187 participants), but whether carriers are also predisposed to breast cancer has not been evaluated. To address this question, this study sequenced all exons and exon-intron boundaries of NTHL1 in 9,771 subjects in the hereditary BrEAst Case CONtrol (BEACCON) study, comprising index cases from hereditary breast cancer families who tested negative for germline pathogenic variants in BRCA1 and BRCA2 and cancer-free older female controls (average 49.7 years vs. 65.6 years) in the same population. In addition, whole-genome and targeted sequencing was performed on formalin-fixed, paraffin-embedded (FFPE)-derived DNA from the breast cancers of 20 germline NTHL1 LoF variant carriers. Further NTHL1 sequencing data were analyzed from nine additional case–control studies, to give a combined analysis of 47,180 subjects including 27,421 cases and 19,759 controls.
Results
Germline variants in NTHL1 are associated with breast cancer susceptibility in the BEACCON hereditary case–control study
All exons and exon-intron boundaries of the NTHL1 gene were sequenced in the BEACCON study of index cases from 4985 hereditary breast cancer families and 4786 cancer-free female controls from the same Australian population. A total of eight unique LoF variants were identified among 39 cases and 15 controls (0.78% vs. 0.31%, odds ratio [OR] 2.51, 95% CI: 1.35–4.90, P = 0.002) (Table 1). p.(Gln90Ter) was the most frequent variant accounting for 25 (0.50%) cases and 11 (0.23%) controls, followed by p.(Gln287Ter) accounting for 9 (0.18%) cases and 2 (0.04%) controls. The observed frequency of these two variants in the controls were consistent with the carrier frequency reported among 134,187 non-cancer subjects in the gnomAD database (0.29% and 0.03%, respectively; database version v2.1.1). A single individual, homozygous for p.(Gln90Ter) variant, was the only bi-allelic carrier identified; a case subject with a personal history of multiple primary cancers including bilateral breast cancer and colorectal cancer, consistent with the previously reported syndrome for bi-allelic LoF of NTHL11 (Supplementary Table 1, C21552; pedigree data for this individual was published previously4). In contrast, the heterozygous case carriers were predominantly only affected with breast cancer (Supplementary Table 1). A case–case analysis of cancer incidence in the families of individuals harboring heterozygous NTHL1 LoF variants found no statistically significantly elevated incidence of colorectal cancer, female breast cancer, male breast cancer or ovarian cancer when compared to the NTHL1 wild-type families, although the number of available NTHL1 families was small and the statistical power was limited (Table 2).
Table 1.
NTHL1 LoF variants identified in familial breast cancer cases and cancer-free controls in BEACCON study.
| Consequence | Case n = 4985 | Control n = 4786 | Nucleotide changea | Protein changeb | Exon (of 6) | dbSNP ID | GnomADc |
|---|---|---|---|---|---|---|---|
| Frameshift | 1 | 0 | c.64_83delAGCCTGGGACCCGGGGCTGG | p.Ser22AlafsTer5 | 1 | – | 0 |
| Stop Gained | 25d | 11 | c.268 C > T | p.Gln90Ter | 2 | rs150766139 | 1.44 × 10−3 |
| Frameshift | 1 | 0 | c.380_383dupTACG | p.Arg129ThrfsTer42 | 3 | rs566860680 | 4.48 × 10−6 |
| Stop Gained | 0 | 1 | c.390 C > A | p.Tyr130Ter | 3 | rs371328106 | 2.36 × 10−5 |
| Stop Gained | 0 | 1 | c.390 C > G | p.Tyr130Ter | 3 | – | 0 |
| Stop Gained | 2 | 0 | c.457 C > T | p.Arg153Ter | 3 | rs374489979 | 1.28 × 10−5 |
| Stop Gained | 1 | 0 | c.760 A > T | p.Lys254Ter | 5 | – | 0 |
| Stop Gained | 9 | 2 | c.859 C > T | p.Gln287Ter | 6 | rs146347092 | 1.62 × 10−4 |
| Total | 39 | 15 | – | – | – | – | – |
aENST00000219066.1(NM_002528.5).
bENSP00000219066.1(NP_002519.1).
cgnomAD, the minor allele frequency of each variant in 134,187 samples from non-cancer cohorts in GnomAD database V2.1.1.
dIncluding one homozygous carrier.
Table 2.
Incidence of other cancers in NTHL1 families compared to non-NTHL1 families in BEACCON study.
| Family historya | NTHL1 familiesb n = 22 | Non-NTHL1 familiesc n = 3239 | OR | 95% CI | p Value | ||
|---|---|---|---|---|---|---|---|
| Cancer (%) | Non-cancer | Cancer (%) | Non-cancer | ||||
| Breast cancer | 28 (6.93) | 376 | 3929 (6.46) | 56,872 | 1.08 | 0.71–1.59 | 0.68 |
| Male breast cancer | 1 (0.25) | 403 | 77 (0.13) | 60,724 | 1.96 | 0.05–11.29 | 0.40 |
| Colorectal cancer | 9 (2.23) | 395 | 1559 (2.56) | 59,242 | 0.87 | 0.39–1.66 | 0.87 |
| Ovarian cancer | 6 (1.49) | 398 | 541 (0.89) | 60,260 | 1.68 | 0.61–3.71 | 0.18 |
aCancer affected family members in the first and second degree of relatives of index cases.
bNTHL1 families, families in which the index cases carry a germline monoallelic LoF variant in NTHL1.
cNon-NTHL1 families, families in which the index cases do not carry any LoF variants in NTHL1.
Missense variants in NTHL1 were also significantly enriched in the cases compared to the controls (75, 1.50% vs. 47, 0.98%, OR 1.54, 95% CI: 1.05–2.27, P = 0.02). All the variants were individually “rare” with the highest minor allele frequency [MAF] detected in the controls being 0.0015 in BEACCON and 0.0018 in gnomAD (Supplementary Table 2). Consistent with the frequencies reported in gnomAD, p.(Arg100Cys) and p.(IIe176Thr) were the most common missense variants in both the case and control cohorts. The association of NTHL1 missense variants with hereditary breast cancer remained when applying a population rarity filter or in silico prediction tools, Condel, PolyPhen2, SIFT, CADD, and REVEL to enrich for likely pathogenic variants, with the strongest effect observed for the missense variants that were selected for rarity (MAF ≤ 0.001, 39 versus 20, OR 1.88, 95% CI: 1.07–3.41) (Supplementary Table 3).
In contrast to the high penetrance reported for the multi-tumor syndrome associated with bi-allelic LoF in NTHL14, the OR for heterozygous LoF or missense variants suggests only a moderate- to low-penetrance effect. In this context, the background genetic risk contributed by common, low-penetrance single nucleotide polymorphisms (SNPs) may have an important risk-modifying effect as described for other low-moderate penetrance genes7. To assess this possibility, 70 SNPs with well-established, significant associations with breast cancer were used to calculate a polygenic risk score (PRS) for each carrier as described previously8. A multivariable logistic regression model was used to simultaneously evaluate the association of NTHL1 LoF variants, missense variants, and the PRS with breast cancer in the case–control cohort. The ORs observed for LoF variants, missense variants, and the OR per unit standard deviation for the PRS were 2.41 (95% CI: 1.30–4.44, P = 0.005), 1.57 (95% CI: 1.09–2.28, P = 0.02) and 1.56 (95% CI: 1.50–1.63, P < 0.001), respectively. The ORs for LoF and missense variants in NTHL1 were not attenuated by the inclusion of PRS in the model indicating that the risk associated with NTHL1 coding variants is independent of any familial aggregation of polygenic risk that may be present in this cohort due to the ascertainment based on family history. When the effect of the PRS and NTHL1 status was considered in combination the OR for NTHL1 carriers in the highest 20% of PRS was 3.88 (95% CI: 1.25–15.97, P = 0.012) for the LoF variants and 3.03 (95% CI: 1.44–6.90, P = 0.001) for the missense variants. This result indicates that although the measured level of risk associated with potentially pathogenic NTHL1 variants in isolation is below the current threshold for clinical intervention, a proportion of NTHL1 germline variant carriers in the highest PRS quintile reach a clinically actionable level, as has been described for other low-moderate breast cancer genes7–9. The distribution of NTHL1 LoF and missense variant carriers by PRS quintile (relative to the controls in the BEACCON study) is shown in Supplementary Fig. 1.
Co-segregation analysis in families with germline NTHL1 variants
Co-segregation analysis of breast cancer in families was performed in 16 multi-case families from the BEACCON study segregating NTHL1 LoF variants where detailed pedigree information was available. Sanger sequencing was performed to determine the genotype of 38 additional family members. Analysis of co-segregation using a full-likelihood method10 calculated a maximum likelihood ratio of 1.18 at an odds ratio for breast cancer of 1.69—insufficient to either support or reject an association with breast cancer predisposition.
Evaluation of bi-allelic inactivation in NTHL1 associated breast cancers
Histopathologic characteristics are summarized for 22 breast cancers from 20 NTHL1 LoF variant carrier cases in the BEACCON study where pathology information was available (Supplementary Table 4). The individual with a homozygous p.(Gln90Ter) variant had bilateral breast cancer at age 47 (grade 2, invasive lobular carcinoma, ER+, PR+, HER2−) and 53 (grade 2, invasive ductal carcinoma, ER+, PR+, HER2−). The 21 breast cancers from 19 heterozygous LoF variant carrier cases were predominantly high-grade, invasive, ductal carcinomas (19/21) and hormone receptor-positive (16/21) of which three had ERBB2 amplification (HER2+).
To assess the occurrence of somatic bi-allelic inactivation in NTHL1 and characteristic mutational signatures in NTHL1-associated tumors, targeted sequencing was performed on tumor DNA from cases with a germline NTHL1 LoF variant using a custom-designed panel of 259 genes (total targeted region of 1.337 Mb) that included all exons and exon-intron boundaries of NTHL1 and 27 breast-cancer driver genes11. Fourteen breast cancers and ovarian cancer from individuals with heterozygous NTHL1 germline variants, together with breast cancer and colorectal cancer from the individual homozygous for germline LoF variants, were sequenced. The alternative allele frequency of the germline variants, reconstructed copy number profile, and tumor purity estimation was used to determine loss of heterozygosity in the NHTL1-associated tumors as described previously12,13. The bi-allelic mutation was confirmed in the breast and colorectal cancers from the homozygous carrier, however, there was no evidence of loss of the wild type allele or a second point mutation in NTHL1 in any cancers from heterozygous germline variant carriers. Since CpG island methylation in the promoter region is a possible alternative mechanism of gene silencing, bisulfite sequencing was performed across the NTHL1 promoter region for 13 breast cancers with sufficient DNA available. No evidence of hypermethylation was observed in any of the cancers tested (Fig. 1a, Supplementary Fig. 2).
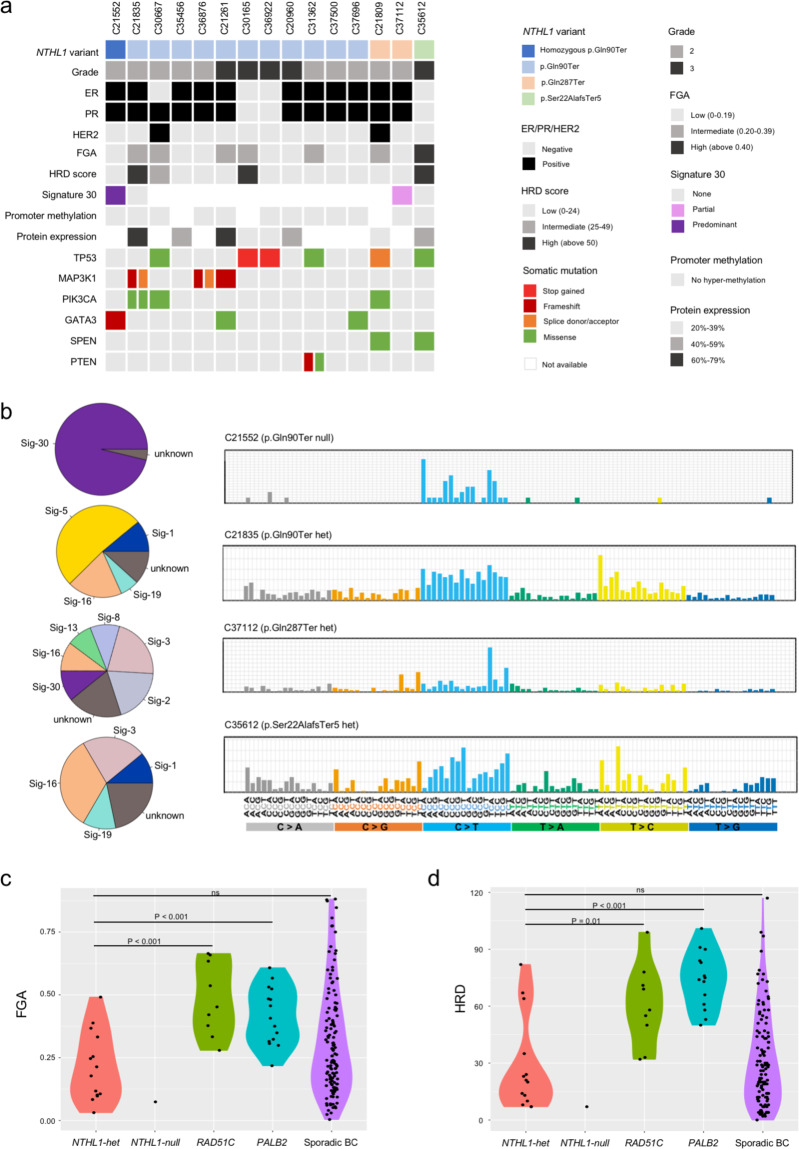
Fig. 1. Genomic characterization of breast and ovarian cancers from carriers of NTHL1 germline loss-of-function variants.
a Germline variants, somatic mutations and HRD score of NTHL1 associated tumors. Germline and somatic variant types are color-coded according to the legend. The phenobar provides information about estrogen receptor (ER), progesterone receptors (PR) and human epidermal growth factor receptor 2 (HER2) status, and somatic mutations in driver genes. b The weighted contribution of mutational signatures from breast cancers of NTHL1 germline variant carriers. c Fraction of genome altered (FGA) and d Homologous recombination deficiency score (HRD) for NTHL1-null (n = 1) and NTHL1-het (n = 14) breast cancers compared to breast cancers from PALB2 (n = 15) and RAD51C (n = 9) germline LoF variant carriers, and sporadic breast cancers (n = 115).
Mutational signatures in NTHL1 associated breast cancers
Targeted sequencing in 14 breast cancers from individuals harboring an NTHL1 heterozygous variant (NTHL1-het) identified somatic mutations in breast cancer driver genes including TP53 (6/14 cases), PIK3CA (3/14 cases), MAP3K1 (3/14 cases), and GATA3 (2/14 cases) (Fig. 1a). This spectrum of somatic mutations in NTHL1-het tumors was similar to that observed in sporadic breast cancer, with mutations in TP53 and PIK3CA the most common, and mutations in MAP3K1 and GATA3 frequently occurring in ER+ cancers14.
To investigate whether NTHL1-associated breast cancers are driven by the same mutagenesis mechanism as the colorectal cancers from carriers of homozygous mutations1,6, whole-genome sequencing was performed on the breast cancers from the NTHL1-null and NTHL1-het carriers of three different germlines LoF variants (p.(Gln90Ter), p.(Gln287Ter), and p.(Ser22AlafsTer5)), together with nine sporadic breast cancers with no known germline cancer predisposition gene mutations as controls. A predominant mutational signature SBS30 was observed in the NTHL1-null breast cancer (Fig. 1b), consistent with the previously reported NTHL1 bi-allelic loss driving tumorigenesis mechanism. In line with the absence of bi-allelic inactivation, the three NTHL1-het breast cancers each exhibited a mixture of mutational signatures, with the top contributing signatures including COSMIC signatures SBS3, SBS5, and SBS16, similar to the sporadic breast cancers. While one of the three NTHL1-het cancers showed a minor proportion of SBS30 (C37112, p.(Gln287Ter), Fig. 1b), this was also observed in one of the nine sporadic cancers, suggesting no major difference in mutational processes between NTHL1-het and sporadic control cancers (BC3, Supplementary Fig. 3).
Fraction of genome alteration and HRD scores in NTHL1 associated breast cancers
Genomic instability and HRD were measured using genome-wide copy number data from the NTHL1 associated cancers. A genomic instability index, a fraction of the genome altered by copy number (FGA)15, and an HRD score16,17 were generated for 14 NTHL1-het cancers and one NTHL1-null cancer. A comparison cohort of breast cancers with expected HR defects from PALB2 (n = 15) and RAD51C (n = 9) germline LoF variant carriers, and 115 sporadic breast cancers sequenced using the same platform, were also evaluated. The NTHL1-het cancers exhibited a broad range of FGA scores (Fig. 1c) that were significantly lower than the PALB2 associated cancers (median 0.20 vs. 0.41, P < 0.001 by Mann-Whitney test) or the RAD51C associated cancers (median 0.20 versus 0.45, P < 0.001) and not statistically significantly different to the FGA scores observed in the sporadic breast cancers (median 0.20 vs. 0.26, P = 0.11). Similarly, the HRD scores for NTHL1-het cancers were significantly lower than those observed in the PALB2 associated cancers (median 21.5 vs. 74.5, P < 0.001) or the RAD51C associated cancers (median 21.5 vs. 58, P = 0.01), and not significantly different to those observed in sporadic cancers (median 21.5 vs. 27, P = 0.79) (Fig. 1d). The one NTHL1-null breast cancer also showed a very low FGA and HRD score (0.07 and 7).
NTHL1 protein expression in NTHL1 associated breast cancers
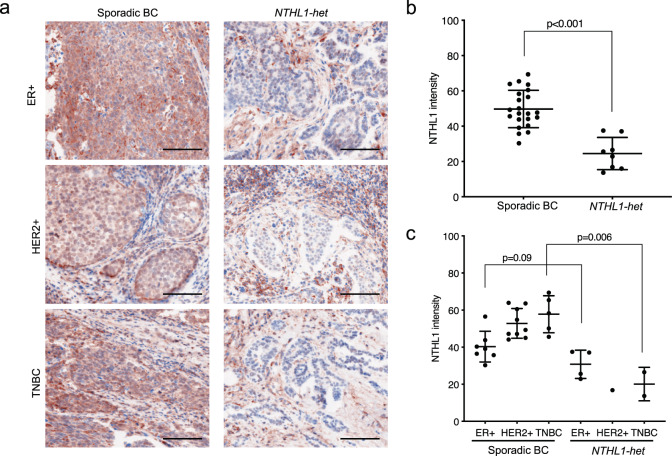
To investigate whether heterozygous NTHL1 LoF variants were associated with reduced protein expression and/or altered cellular location in breast cancers as has been observed in gastric tumors18, fluorescent immunohistochemistry was used to measure NTHL1 protein levels, along with an epithelial cell marker (Cytokeratin AE1/AE3), in 8 NTHL1-het breast cancers and 21 sporadic breast cancers. NTHL1 had a predominantly nuclear localization in both wild-type and NTHL1-het cancers. Compared to sporadic breast cancers, NTHL1-het cancers as a group showed a 51% reduction in NTHL1 staining in the AE1/AE3-positive cancer cells (average staining intensity 24.51 vs. 49.83, P < 0.001 by unpaired t test) (Fig. 2a, b) and a 40% reduction in the AE1/AE3-negative non-cancer cells (35.62 vs. 59.65, P < 0.001). In addition, for 5 of the 8 NTHL1-het cancers, the expression of NTHL1 in the breast cancer cells (AE1/AE3 positive cells) was reduced by >30% compared to the surrounding non-cancer cells (AE1/AE3 negative cells) (Supplementary Fig. 4), while only a small proportion of control cancers showed the same phenomenon (5 of 21), suggesting that NTHL1 expression may be attenuated further specifically in the cancer cells of NTHL1 LoF variant carriers. When considering the pathological subtypes individually the average NTHL1 expression level was lower in NTHL1-het tumors than in the controls in all three major subtypes: ER+, HER2+, and triple-negative cancers, but only reached statistical significance in the triple-negative cancers (Fig. 2c).
Fig. 2. NTHL1 protein expression in sporadic breast cancer (n = 21) and NTHL1-het breast cancer (n = 8).
a NTHL1 expression in sporadic breast cancer and NTHL1-het cancer of ER+, HER2+, and triple-negative types. Multiplex immunofluorescent staining approach was used and the fluorescence signal was displayed in colorimetric pattern for better contrast. NTHL1: brown color; DIPI: blue color. The epithelial marker AE1/AE3 (Supplementary Fig. 4) was used to identify cancer cells in the breast cancer tissue in NTHL1 expression quantitation in (b) and (c). Scale bar = 100 μm. b The average intensity of NTHL1 in sporadic cancer group compared to NTHL1-het group. c The average intensity of NTHL1 in sporadic cancer group compared to NTHL1-het group according to ER, PR, and HER2 status. BC breast cancer, TNBC triple-negative breast cancer.
Analysis of NTHL1 in multi-center international case–control cohorts
To evaluate the role of NTHL1 in breast cancer predisposition in diverse populations and independent studies, a total of 27,421 cases and 19,759 controls were screened for the entire coding region of NTHL1 from ten case–control studies including BEACCON as discovery dataset, and nine additional studies as validation dataset. The validation dataset included SEARCH, UK Population-based Breast Cancer Study; GC-HBOC, German Consortium for Hereditary Breast and Ovarian Cancer; GENESIS, French familial BRCAx study (some data were previously published19); VHIO, familial breast cancer, and control study of the Vall d’Hebron Institute of Oncology of Barcelona; OFBCR, Ontario Familial Breast Cancer Registry; DFBBCS, the Dutch Familial Bilateral Breast Cancer Study; HABC, Hispanic–American Breast Cancer Study; ABCFR, Australian Breast Cancer Family Registry; and CARTaGENE, Québec Population-based Breast Cancer Study. The information of cohorts and subjects, sequencing platform, and coverage are summarized in Supplementary Table 5.
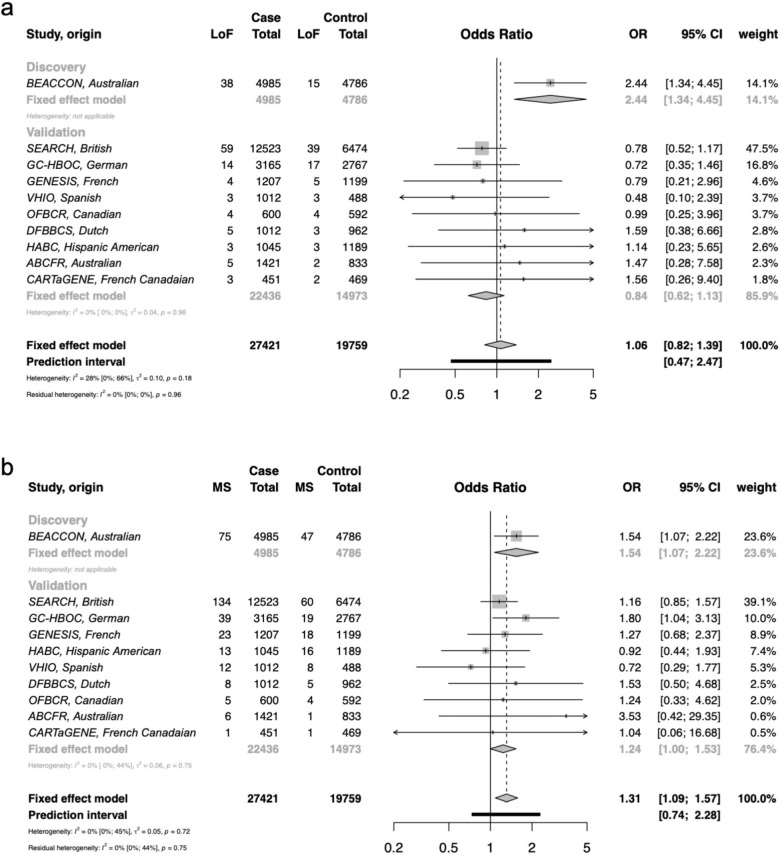
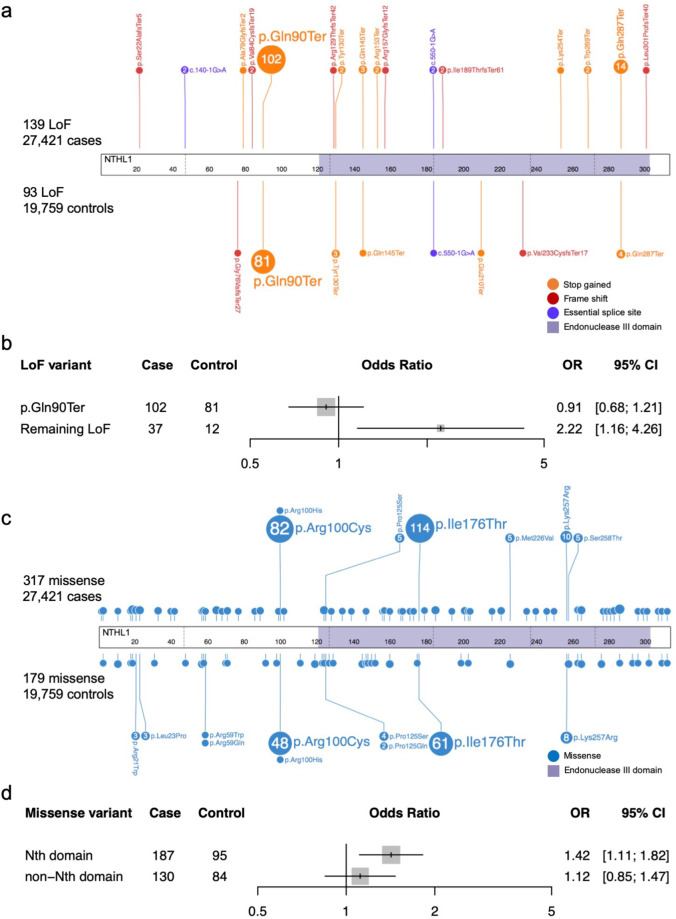
No additional bi-allelic LoF variants were identified in subjects of the validation dataset, indicating germline bi-allelic loss of NTHL1 is extremely rare as a cause of breast cancer (1/27,421 breast cancer cases and 0/19,759 controls). In the heterozygous state, the overall effect observed in the validation dataset (OR = 0.84, 95%CI = 0.62-1.13) did not support the finding in the BEACCON dataset, with 4/9 studies showing a weak positive association while the others showed no effect or a weak negative association, although the sample sizes of most studies were small (Fig. 3a). The frequency of LoF variants observed in each study varied greatly among both the cases (0.29–0.76%) and the controls (0.24–0.68%), largely driven by differences in frequency of the predominant variant p.(Gln90Ter) (Fig. 4a; Supplementary Table 6). The remaining LoF variants identified were all rare in gnomAD (MAF < 0.001), with an overall statistically significant enrichment, observed in the cases in the combined data for these variants from all 10 studies (37 vs. 12, OR 2.22, 95% CI: 1.16–4.26, Fig. 4b).
Fig. 3. Frequency of heterozygous germline variants in NTHL1 and odds ratios for breast cancer observed in case–control data from ten multicenter international cohorts.
a Heterozygous loss-of-function variants in NTHL1 and odds ratios in ten case–control cohorts. b Heterozygous missense variants in NTHL1 and odds ratios in ten case–control cohorts. The overall effect of odds ratios were computed based on a fixed-effect model. BEACCON hereditary BrEAst Case CONtrol study, SEARCH UK Population-based Breast Cancer Study, GC-HBOC German Consortium for Hereditary Breast and Ovarian Cancer, GENESIS French familial BRCAx study (some data were previously published19), VHIO familial breast cancer and control study of the Vall d’Hebron Institute of Oncology of Barcelona, OFBCR Ontario Familial Breast Cancer Registry, DFBBCS the Dutch Familial Bilateral Breast Cancer Study, HABC Hispanic-American Breast Cancer Study, ABCFR Australian Breast Cancer Family Registry, CARTaGENE Québec Population-based Breast Cancer Study.
Fig. 4. Germline NTHL1 variants identified in 27,421 cases and 19,759 controls.
a Lollipop plot of all LoF variants identified in 27,421 cases and 19,759 controls. b Odds ratio (OR) and 95% confidence interval (95% CI) for recurrent LoF variants p.(Gln90Ter) and all the rest of rare LoF variants (MAF < 0.001 according to gnomAD). c Lollipop plot of all missense variants identified in 27,421 cases and 19,759 controls. Nth, Endonuclease III. d OR and 95% CI for missense variants located in functional domain and the rest of the NTHL1 protein.
In contrast to the LoF variants, the frequency of missense variants was higher among the cases in the majority of studies in the validation dataset (6/9 studies), and was statistically significant in the overall analysis of the validation dataset (OR 1.24, 95% CI: 1.00–1.53) and the combined BEACCON and validation datasets (OR 1.31, 95% CI: 1.09–1.57) (Fig. 3b). While the detected missense variants were distributed across the whole gene (Supplementary Table 6), there was an enrichment of missense variants in the cases compared to controls in the Endonuclease III domain of NTHL1 (187 vs. 95, OR 1.42, 95% CI: 1.11–1.82) (Fig. 4c, d). When a number of in silico prediction tools and population rarity filters were applied to enrich for likely pathogenic variants in the combined data from the ten studies, the predicted missense variants were observed in excess in the cases compared to the controls and reached statistical significance in all the in silico prediction groups: Condel, PolyPhen2, SIFT, CADD, and REVEL (Table 3). The strongest enrichment was observed for variants with a high REVEL score of pathogenicity (REVEL > 0.75, OR 1.44, 95% CI: 1.08–1.94), and interestingly these likely pathogenic variants according to REVEL were also predominantly located in the conserved functional domain of NTHL1 protein, Endonuclease III (COG0177, a member of ENDO3c Superfamily) (Supplementary Fig. 5).
Table 3.
Likely pathogenic missense variants of NTHL1 selected by population rarity or deleterious in silico tool predictions in case–control data of ten multi-center international cohorts.
| NTHL1 missense variants | Case n = 27,421 | Control n = 19,759 | OR | 95% CI | p Value | ||
|---|---|---|---|---|---|---|---|
| Carriers | % | Carriers | % | ||||
| Totala | 316 | 1.15 | 179 | 0.91 | 1.28 | 1.06–1.54 | 0.01 |
| Rare (MAF ≤ 0.001)b | 121 | 0.44 | 70 | 0.35 | 1.25 | 0.92–1.7 | 0.16 |
| Rare (MAF ≤ 0.0001)b | 92 | 0.34 | 49 | 0.25 | 1.35 | 0.95–1.96 | 0.09 |
| Condel (deleterious) | 175 | 0.64 | 92 | 0.47 | 1.37 | 1.06–1.79 | 0.02 |
| PolyPhen2 (damaging) | 161 | 0.59 | 83 | 0.42 | 1.4 | 1.07–1.85 | 0.01 |
| SIFT (deleterious) | 268 | 0.98 | 146 | 0.74 | 1.33 | 1.08–1.63 | 0.006 |
| CADD (≥10) | 282 | 1.03 | 155 | 0.78 | 1.31 | 1.08–1.61 | 0.006 |
| REVEL (≥0.75) | 144 | 0.53 | 72 | 0.36 | 1.44 | 1.08–1.94 | 0.01 |
aIncludes all the missense variants identified.
bMAF, minor allele frequency in non-cancer non-Finnish Europeans in gnomAD database.
Discussion
NTHL1 is a DNA glycosylase involved in the earliest steps of the repair of oxidative DNA damage through the BER pathway. Through its endonuclease activity it acts to excise damaged nucleotides, predominantly pyrimidines, and loss of NTHL1 activity would be expected to impact the effectiveness of normal DNA repair. This effect was shown to be clinically significant by the discovery of a specific cancer predisposition syndrome involving complete loss of normal NTHL1 function through bi-allelic pathogenic germline LoF variants. The resulting condition not only demonstrates high risk for a range of different malignancies, but it was possible to show that specific mutational processes arising from the loss of NTHL1-mediated BER dominated the genetic pathology of the resulting cancers, reflected in the distinctive somatic mutational signature, COSMIC signature SBS30. Although the recessive condition has proved to be very rare, the analogy with other tumor suppressor genes raised the possibility that inheritance of a single germline pathogenic variant followed by somatic inactivation of the wildtype allele could achieve the same outcome through a “two-hit” mechanism with the result that heterozygous variants would also be associated with inherited cancer risk. We have examined this possibility specifically in regard to the risk of breast cancer through sequencing of a large number of cases and controls and examination of the somatic landscape of cancers occurring in women carrying a single NTHL1 LoF variant. The results have shown that germline bi-allelic loss is not a major contributor to breast cancer in the populations studied and there is equally no evidence that heterozygous variants act through a traditional two-hit pathway to cause breast cancer. However, the data do support a possible low-risk effect for at least some heterozygous LoF variants and rare, deleterious missense variants. Our data reflects the recent findings that heterozygous LoF variants in NTHL1 do not confer any substantial risk for colorectal cancer and do not undergo bi-allelic inactivation20.
The difficulty in providing clear evidence of a breast cancer risk association for LoF variants in the cases and controls appears to reflect, in part, a higher degree of heterogeneity among the ten analyzed studies compared to little variability between studies for missense variants. The statistically significant two-fold excess in the BEACCON study was not observed in the validation dataset and this discrepancy was largely driven by the variable frequency of LoF variants in control populations across cohorts which ranged from 0.24 to 0.68%. In particular, the frequency of the p.(Gln90Ter) variant, which accounted for the large majority of LoF variants, was highly variable among the 10 studies, as it is across different ethnic groups reported in gnomAD; ranging from ~1 in 280 in the Finnish population to less than 1 in 3000 in African and Asian populations. Differences in the ethnic background and ascertainment methods among the different studies may explain the diverse results in the sub-studies. This variability was, however, limited to the p.(Gln90Ter) variant. If the analysis of the data from the ten studies in combination was restricted to the remaining rarer LoF variants, a statistically significant two-fold excess was observed in cases compared to the controls (37, 0.13% of 27,421 cases compared to 12, 0.06% of 19,759 controls, OR 2.22, 95% CI: 1.16–4.26). With the exclusion of the most common LoF variant, the numbers are small and the result requires validation in further studies. Stronger evidence for a low-risk predisposition effect was found for missense variants, which were more consistently enriched in cases across sub-studies but also showed further enrichment when selected for features most likely to be associated with disruption of normal gene function: rarity, deleterious in silico properties or location in a major functional domain.
Tumor sequencing and promoter methylation analyses demonstrated that NTHL1 does not undergo bi-allelic inactivation in breast cancers, which is supported by the fact that NTHL1-het tumors retain approximately 50% of the wildtype protein expression. Although Drost et al.6 reported loss of the wild-type allele in a single breast cancer carrying an NTHL1 LoF variant (p.(Gln287Ter)), our data suggest this is not a common event in NTHL1-associated breast tumors. The reduced, but not complete loss of NTHL1 expression in NTHL1-het cancers is consistent with the fact that, while the mutational spectrum of the NTHL1-null breast cancer strongly resembled COSMIC mutational signature SBS30, only minor components of SBS30 were observed in one of three NTHL1-het tumors examined. Although the total number of tumors analyzed remains small, the NTHL1-het tumors appeared similar to sporadic breast cancers in regard to the somatic mutational features, and in other respects (HRD scores, histopathology, and FGA).
The absence of a second hit in NTHL1 may be a generic feature of low-moderate penetrance alleles. While many high-risk breast cancer genes BRCA121, BRCA222, PALB223, and RAD51C12 follow the Knudson two-hit model, where a germline mono-allelic variant in the gene is accompanied by somatic loss of the wild-type allele, the limited data currently available for alleles which confer low- (RR 1–2) to moderate- (RR 2–4) risk suggest they do not always undergo a second hit. For example, a second hit rarely occurs in breast cancers from carriers of the lower penetrance CHEK2 p.(Ile157Thr) variant24 or the BRCA2 p.(Lys3326Ter) variant22, while limited reports suggest that it does occur in breast cancers from women carrying germline CHEK2 LoF variants24,25. It is possible that alleles associated with low or moderate risk mediate cancer predisposition through pathways that do not require a second hit, such as haploinsufficiency where reduced protein levels rather than the complete loss are sufficient to increase the risk of disease24–33. Reduced NTHL1 expression has been previously described as a feature of specific malignancies34,35 and recent reports have found that NTHL1 missense variants can induce cellular transformation and genomic instability in vitro while retaining normal cellular location and enzymatic function36,37, raising the possibility that a non-canonical function may be involved. Alternatively, the distinction between high-risk cancer susceptibility variants that undergo a somatic second hit and low-risk alleles that do not—even where bi-allelic loss appears to convey a clear oncogenic advantage, as demonstrated in the case of NTHL1 by the recessive cancer syndrome—directly reflects the fact that these alleles are less prone to obtain a second hit leading to a complete loss of the function, and always retain some activity in the tumor.
In summary, this is the first study to investigate the role of heterozygous NTHL1 LoF and missense variants in breast cancer predisposition, which included 47,180 subjects from ten international case-control studies. The data suggest that NTHL1 may be associated with a modest increase in breast cancer risk that would not be considered clinically actionable in isolation under current clinical guidelines but could be relevant when combined with additional risk factors. Molecular analyses of breast cancers from carriers indicate that NTHL1 may be included in the growing list of low-penetrance breast cancer genes that appear to function via haploinsufficiency rather than the bi-allelic inactivation mechanism almost universally observed for high-risk breast cancer predisposition genes.
Methods
Case–control Subjects
All exons and exon-intron boundaries of NTHL1 were analyzed in the index cases of 4985 hereditary breast cancer families and in 4786 cancer-free women in the hereditary BEACCON study. The cases were female breast and/or ovarian-cancer affected patients (>95% breast cancer affected) in the Variants in Practice (ViP) Study that were ascertained from the combined Victorian and Tasmanian Familial Cancer Centres, and Pathology North, NSW Health Pathology, Newcastle, Australia. The controls were cancer-free women who were greater than or equal to 40 years old from the same population from the Lifepool study as described previously38. A hereditary breast cancer family is defined as those assessed by a specialist Familial Cancer Clinic where most of the affected family members meet family history criteria or have individual risk factors that predict a greater than 10% chance of having a BRCA1 or BRCA2 pathogenic variant (a detailed guide is in https://www.eviq.org.au/cancer-genetics/adult/genetic-testing-for-heritable-mutations/620-brca1-and-brca2-genetic-testing). All cases tested negative for pathogenic variants in BRCA1 and BRCA2 including large scale genome rearrangements. The average breast cancer diagnosis age of the cases was 49.7 years (range 19.0–94.8). The average age of controls was 65.6 years (range 40.0–97.5). The study was approved by the Human Research Ethics Committee at the Peter MacCallum Cancer Centre (Approval # 09/29) and all participating centers. All participants provided informed consent for genetic analysis of their germline DNA.
Nine international case-control studies of diverse countries of origin and sample sizes in which all exons of NTHL1 gene were analyzed were used as validation cohorts. The studies were; UK Population-based Breast Cancer Study (SEARCH), German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC), French familial BRCAx study (GENESIS, some data were previously published19), familial breast cancer and control study of the Vall d’Hebron Institute of Oncology of Barcelona (VHIO), Ontario Familial Breast Cancer Registry (OFBCR), the Dutch Familial Bilateral Breast Cancer Study (DFBBCS), Hispanic–American Breast Cancer Study (HABC), Australian Breast Cancer Family Registry (ABCFR), and Québec Population-based Breast Cancer Study (CARTaGENE). The case subjects in SEARCH, ABCFR, and CARTaGENE were recruited in relevant populations without consideration of enrichment for family history (population-based cohort), while the case subjects in the other studies were ascertained using various standards to enrich for high-risk breast cancer patients (hereditary cohort). The case and control subjects in individual cohorts were sequenced using the same platform and had sufficient and comparable sequencing coverage (Supplementary Table 5).
NTHL1 sequencing using germline DNA
Germline DNA from the cases in the BEACCON study was obtained from blood and was extracted in clinical laboratories, and the DNA from controls were obtained from blood (87%) and saliva (13%) and were extracted by Lifepool researchers. All exons and 10 bp into each exon–intron boundary of the NTHL1 gene were sequenced using a customized targeted HaloPlex HS Targeted Enrichment Assay panel (Agilent Technologies, Santa Clara, CA) which was designed using Agilent’s SureDesign tool at https://earray.chem.agilent.com/suredesign/. A set of 74 Ancestry Informative Markers (AIMs)39–41 was included in the sequencing of 3409 subjects (1747 cases and 1662 controls) to determine the ethnicity background of study subjects. Furthermore, a total of 70 SNPs that were reported to be associated with breast cancer in GWAS studies8 were sequenced in all subjects to calculate breast cancer PRSs. Library preparation was performed using the Agilent NGS Bravo automation system (Agilent Technologies) according to the manufacturer’s protocol (Agilent Technologies, HaloPlex HS Target Enrichment System Automation Protocol For Illumina Sequencing. https://www.agilent.com/cs/library/usermanuals/public/G9931-90010.pdf). Sequencing was performed by the Australian Genome Research Facility (North Melbourne, VIC, Australia) on an Illumina Hiseq2500 sequencer. Library pools of 96 samples were sequenced on a HiSeq2500 Genome Analyzer using 100 bp paired-end reads (Illumina, San Diego, CA) with an average read depth target of >250X. The average sequencing depth yield for NTHL1 gene was 396.13 and 358.64 for the cases and controls, respectively, with 99.32% and 99.19% of the target bases sequenced ≥10 reads.
Germline DNA sequencing alignment and variant calling
Sequencing data from the BEACCON study were processed, aligned, and analyzed through a pipeline constructed using Seqliner v0.1a (http://bioinformatics.petermac.org/seqliner) by Bioinformatic Core Facility of Peter MacCallum Cancer Centre as described in detail elsewhere42. GATK Unified Genotyper v2.4 (Broad Institute, Cambridge, MA)43, Haplotype caller44, and PLATYPUS45 were used for variant calling. ENST00000219066.1(NM_002528.5) and ENSP00000219066.1(NP_002519.1) were used to annotate the variants identified in NTHL1. LoF variants were defined as frameshift, stop gained or essential splice site variants. The MAF reported in ExAC and gnomAD databases were used as a population frequency reference for the variants identified.
Variant filters and validation
Filters were applied to the sequencing data from BEACCON to remove sequencing artifacts and included a quality score over 30, a minimum of 10 reads, and at least 5 reads supporting the alternative alleles and a variant allele proportion of greater than 20%. In addition, all variants included in the analysis had to pass all the internal filters of at least two out of three variant callers GATK, Unified Genotyper, Platypus, and Haplotype caller. Sequencing BAM files were viewed in IGV to manually curate the accuracy of the variant calls, and all the LoF and missense variants detected in NTHL1 were validated using Sanger sequencing (primer sequences are listed in Supplementary Table 7). In addition, any case or control carrying an LoF variant in NTHL1 was reviewed for any pathogenic variants in known hereditary breast cancer genes (CHEK2, PALB2, ATM, TP53, CDH1, PTEN, and STK11) or LoF variants in proposed breast cancer genes (RAD51C, RAD51D, BRIP1, BARD1, MRE11A, RAD50, and NBN). Two case carriers each had a CHEK2 c.1100delC variant, and one case carrier had a splice acceptor variant c.2071-1 G > A in NBN. No pathogenic variants or LoF variants in any proposed breast cancer genes were identified in any of the control carriers.
Co-segregation analysis in families with germline NTHL1 variants
Pedigree information for 16 families with an NTHL1 LoF variant were obtained and Sanger sequencing was performed to determine the genotype for a total of 38 additional family members from these families. A full-likelihood method was used for co-segregation analysis as described previously10. A full pedigree likelihood was calculated as a means of assessing the linkage between the variant and disease based on all available genotype information from the family, including any unaffected individuals who have been tested.
Tumor microdissection and DNA extraction
FFPE breast or ovarian tumor blocks were obtained from carriers of germline variants in the ViP study, and cancer cells were collected through needle microdissection under a dissecting microscope. Hematoxylin and eosin (H&E) stained slide was reviewed for each case to identify cancer cell-enriched regions to guide the microdissection and achieve high tumor purity (aimed to achieve >70% tumor cells). Between 15 and 30 slides per block (depending on the size of the tumor and the proportion of cancer cells) of 8–10 μM thickness was needle microdissected, and the DNA of the collected cancer cells was extracted and purified using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). DNA was quantified using Qubit dsDNA high-sensitivity Assay kit (ThermoFisher Scientific, MA, USA). A multiplex polymerase chain reaction (PCR)-based quality-control method reported by van Beers et al.46, was used to identify DNA samples with sufficient quality for molecular analysis.
Whole-genome sequencing and targeted sequencing of tumor DNA
Whole-genome sequencing (30× coverage, 100 bp paired-end) of four tumor-normal pairs was performed on the BGI-Seq platform using 500 ng of DNA extracted from FFPE tumors or from blood. Targeted sequencing of tumor DNA was performed using an Agilent SureSelect XT Custom Panel that targeted all exons of a total of 259 genes (total targeted region of 1.337 Mb) including all the candidate genes in Halo3 design, and an additional 27 breast cancer driver genes11. Sequencing libraries were generated using 300 ng tumor DNA and were sequenced on an Illumina Next Seq 500 (75 bp paired-end reads).
Tumor DNA sequencing alignment and variant calling
Paired-end sequence reads from tumor DNA were aligned to the GRCh37 human reference genome using BWA-MEM15. PCR and optical duplicate reads were removed using Picard (http://broadinstitute.github.io/picard/) and then local realignment around indels and base quality score recalibration were performed using the Genome Analysis Tool Kit (GATK). SNP and indel variants were called using GATK Unified Genotyper43, Platypus45, and Varscan247.
A quality score over 150, a minimum of 10 reads, and an alternate allele frequency of more than 10% were used to rule out sequencing artifacts. In paired tumor-normal sequencing data, the somatic mutations were identified by removing the germline variants. In tumor sequencing data without matched germline data, somatic mutations were identified by applying stringent filters on the population frequency (minor allele frequency, MAF ≤ 0.0001 in ExAC and gnomAD48), the frequency in the in-house sequencing cohort (<20% of 166 breast tumors with the exception of variants in PIK3CA and TP53), and removing the variants identified in germline sequencing data.
Genome-wide copy number analysis
Genome-wide copy number data were generated from off-target sequencing reads using CopywriteR v2.10 with 50 kb bins49. NEXUS Copy NumberTM (software v8.0 with build version 9169, BioDiscovery Inc.) was used for CN segmentation using the SNPFASST2 algorithm with default parameters. Copy number gains and losses were called with log2 ratio thresholds of >0.2 and < −0.2, respectively.
Fraction of genome altered (FGA) and homologous recombination deficiency (HRD) scores
Using the genome-wide copy number data, the FGA was calculated with adjustment according to chromosome sizes as described50,51. An HRD score, combined from three HRD score components: number of telomeric allelic imbalances52, HRD-loss of heterozygosity53, and large-scale state transitions54 was calculated as described in detail elsewhere13.
Mutational signature analysis
Mutational signatures were performed and plotted using deconstructSigs55 package in R v3.3.256 based on the somatic mutations identified by the whole genome or targeted sequencing of the tumors of interest. For each sample, the somatic substitutions were categorized into six basic base substitutions (C > A, C > G, C > T, T > A, T > C, and T > G) and subcategorized into 96 subcategories according to the trinucleotide context. Mutational signatures were determined referring to the COSMIC mutational signature database.
NTHL1 promoter methylation analysis
The promoter regions of NTHL1 were examined for methylation status by Sanger sequencing the promoter region using bisulfite-treated DNA. Tumor DNA was treated by the EpiTect Fast DNA Bisulfite Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions, followed by PCR amplification and Sanger sequencing of the promoter regions. The primer pairs were designed using the default settings of the Bisulfite Primer Seeker tool (https://www.zymoresearch.eu/bisulfite-primer-seeker, Supplementary Table 7), and/or from previously published study57. CpGenome Human Methylated DNA Standard (Millipore, USA) served as a methylation positive control and Female Genomic Reference DNA (Promega, Madison, WI, USA) as a negative control to assess the methylation status of tumor samples.
NTHL1 protein expression analysis
Opal multiplex fluorescent immunohistochemistry method (PerkinElmer) was used to evaluate the NTHL1 protein expression in cancer and non-cancer cells in FFPE breast cancer tissue. NTHL1 antibody (Abcam, Branford, Connecticut, USA; 1:1000 dilution) and AE1/AE3 antibody (multi-cytokeratin antibody, Leica Biosystems, Wetzlar, Germany; 1:1000 dilution) were used as primary antibodies. HRP-labeled anti-rabbit antibody (PerkinElmer Waltham, Massachusetts, USA; 1:1000 dilution) and anti-mouse antibody (PerkinElmer, Waltham, Massachusetts, USA; 1:1000 dilution) were used as secondary antibodies. Spectral DAPI (PerkinElmer, Waltham, Massachusetts, USA) was used to label the nuclear. The epithelial marker AE1/AE3 was used to identify cancer cells in the breast cancer tissue, and the average intensity of nuclear expression of NTHL1 was quantitated using PerkinElmer Vectra Automated Multispectral Imaging System.
Statistical analysis
OR and two-tailed p value by Fisher’s exact test for the case and control study were calculated in R56. The conditional Maximum Likelihood Estimate was used for OR. Fisher’s exact test (two-sided) was used for the comparisons of case–control data, and a p value of ≤0.05 was considered as statistically significant. PRS was calculated based on 70 low penetrance breast cancer predisposition SNPs following a multiplicative risk model (calculated by the sum of the minor alleles weighted by the per-allele log OR) described by Mavaddat et al.8. Mann–Whitney U test was performed for FGA and HRD score comparisons between groups of tumors in GraphPad Prism version 7.00 (California, USA). Unpaired t test was used for comparing NTHL1 expression in different comparison groups. The meta-analysis for multi-center international cohorts was performed using meta R package58 under a fixed-effect model for analysis within subgroups or analyzing all ten cohorts.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The BEACCON study was supported by the National Breast Cancer Foundation (IF-15-004, I.G.C. and P.A.J.), Cancer Australia/National Breast Cancer Foundation (PdCCRS_1107870, I.G.C. and P.A.J.), the Victorian Cancer Agency (Tumor Stream Grant, P.A.J.) and the National Health and Medical Research Council of Australia (GNT1023698, P.A.J.; GNT1041975, I.G.C.). Na Li is supported by Cancer Council Victoria. The authors thank Simone M. Rowley, Jue Er Amanda Lee, Norah Grewal, Gisela Mir Arnau, Timothy Semple, Jason Li, Kaushalya Amarasinghe and Richard Lupat for helping with the sequencing and bioinformatic analysis. We also thank Lyon Mascarenhas, Rebecca Driessen, for the ViP study site principal investigators Geoffrey Lindeman, Marion Harris, Tom John, and Ingrid Winship, and the staff at the Victorian and Tasmanian Familial Cancer Centers who enrolled participants and provided clinical data. We also thank all the participants of the ViP and Lifepool studies for donating their DNA samples and clinical information. GENESIS (GENE SISters) is a French national study coordinated by D. Stoppa-Lyonnet and N. Andrieu and sponsored by UNICANCER (Sinilnikova et al. BMC Cancer 2016). We wish to thank the genetic epidemiology platform (the PIGE, Plateforme d’Investigation en Génétique et Epidemiologie: Séverine Eon-Marchais, M. Marcou, D. Le Gal, L. Toulemonde, J. Beauvallet, N. Mebirouk, E. Cavaciuti), the biological resource center (S. Mazoyer, F. Damiola, L. Barjhoux, C. Verny-Pierre, V. Sornin) and all the GENESIS collaborating cancer clinics. Financial support for GENESIS was provided by the Ligue Nationale contre le Cancer (grants PRE05/DSL and PRE07/DSL), the French National Institute of Cancer (INCa grant No b2008-029/LL-LC) and the comprehensive cancer center SiRIC (Site de Recherche Intégrée sur le Cancer: Grant INCa-DGOS-4654). Exome sequencing was supported by the France Génomique National infrastructure, funded as part of the « Investissements d’Avenir » program managed by the Agence Nationale pour la Recherche (ANR-10-INBS-09) and the Centre National de Recherche en Génomique Humaine, CEA. The PERSPECTIVE project was supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (GPH-129344), the Ministère de l'Économie, de la Science et de l’Innovation du Québec through Genome Quebec, and the Quebec Breast Cancer Foundation. The PRE3VENTION project was supported by a grant from the Ministère de l'Économie, de la Science et de l’Innovation du Québec through the PSR-SIIRI-949 program. The CARTaGENE study was supported by Genome Canada. The German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) is funded by the German Cancer Aid (#110837, #70111850, coordinator: Rita K. Schmutzler, Cologne). Next-generation sequencing of female control individuals was supported by the Ministry for Innovation, Science, and Research of the State of North Rhine-Westphalia (#323-8.0302.16.02-132142) and LIFE ¬¬- Leipzig Research Center for Civilization Diseases, Universität Leipzig. LIFE is funded by means of the European Union, by the European Regional Development Fund (ERDF), and by means of the Free State of Saxony within the framework of the excellence initiative. This work was also supported by grant UM1 CA164920 from the USA National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. SEARCH is funded by Cancer Research UK [C490/A10124, C490/A16561] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. The sequencing and analysis for this project were funded by the European Union’s Horizon 2020 Research and Innovation Program (BRIDGES: grant number 634935). The work in VHIO was supported by the Spanish Instituto de Salud Carlos III (ISCIII) funding granted to Sara Gutiérrez-Enríquez (PI16/01218 and PI19/01303), an initiative of the Spanish Ministry of Economy and Innovation partially supported by European Regional Development FEDER Funds. Sara Gutiérrez-Enríquez is recipient of an ISCIII Miguel Servet Program contract (CP16/00034). The VHIO authors acknowledge the Cellex Foundation for providing research facilities and thank CERCA Program / Generalitat de Catalunya for institutional support. Tu Nguyen-Dumont is the recipient of a Career Development Fellowship from the National Breast Cancer Foundation (ECF-17-001, Australia).
Author contributions
N.L. contributed to project management, data curation, data analysis and visualization, manuscript writing, and editing; M.Z. and N.T. contributed to bioinformatics analysis and visualization; S.M. and L.D. contributed to the collection of study materials or patients; Y.K.H and D.C. contributed to the curation of protein expression data and analysis; S.G.E., A.M.F., O.D., T.N.D., M.C.S., J.L.H., J.S., M.D., P.S., A.M., R.S., M.K.S., M.A.A., I.L.A., E.H., C.E., F.L., E.G., S.L.N., E.Z., J.A., and D.F.E. contributed to data curation and validation for the case-control data in validation cohorts and manuscript editing. R.J.S. contributed to the provision of patients’ material and data interpretation; K.L.G. contributed to data interpretation, supervision, and manuscript review and editing; P.A.J. and I.G.C. contributed to project conceptualization, supervision, clinical interpretation, manuscript review, and editing; All authors contributed to drafting, editing and final approval of the manuscript.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1420829359. The sequencing data have been deposited in the European Genotype-phenotype Archive under the following accession: https://identifiers.org/ega.dataset:EGAD0000100702560 (study ID: EGAS00001005043). These data include NTHL1 sequencing using germline DNA, Alignment and variant calling, Whole-genome sequencing and targeted sequencing of tumor DNA, Genome-wide copy number analysis, Mutational signature analysis. In addition, the following data are not openly available to protect patient privacy: NTHL1 protein expression analysis, FCC patient database, cohorts summary, NTHL1 promoter methylation analysis. Data requests for these data should be made to the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Paul A. James, Ian G. Campbell.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-021-00255-3.
References
- 1.Weren RD, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 2.Fostira F, et al. Extending the clinical phenotype associated with biallelic NTHL1 germline mutations. Clin. Genet. 2018;94:588–589. doi: 10.1111/cge.13444. [DOI] [PubMed] [Google Scholar]
- 3.Belhadj S, et al. NTHL1 biallelic mutations seldom cause colorectal cancer, serrated polyposis or a multi-tumor phenotype, in absence of colorectal adenomas. Sci. Rep. 2019;9:9020. doi: 10.1038/s41598-019-45281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grolleman JE, et al. Mutational signature analysis reveals NTHL1 deficiency to cause a multi-tumor phenotype. Cancer Cell. 2019;35:256–266 e255. doi: 10.1016/j.ccell.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Groves A, Gleeson M, Spigelman AD. NTHL1-associate polyposis: first Australian case report. Fam. Cancer. 2019;18:179–182. doi: 10.1007/s10689-018-0107-1. [DOI] [PubMed] [Google Scholar]
- 6.Drost J, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher S, et al. Association of a polygenic risk score with breast cancer among women carriers of high- and moderate-risk breast cancer genes. JAMA Netw. Open. 2020;3:e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavaddat, N. et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl Cancer Inst. 10.1093/jnci/djv036 (2015). [DOI] [PMC free article] [PubMed]
- 9.Muranen TA, et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk. Genet. Med. 2017;19:599–603. doi: 10.1038/gim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson D, Easton DF, Goldgar DE. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 2003;73:652–655. doi: 10.1086/378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, N. et al. Combined tumor sequencing and case/control analyses of RAD51C in breast cancer. J. Natl Cancer Inst. 10.1093/jnci/djz045 (2019). [DOI] [PMC free article] [PubMed]
- 13.Lee JEA, et al. Molecular analysis of PALB2-associated breast cancers. J. Pathol. 2018;245:53–60. doi: 10.1002/path.5055. [DOI] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vollan HK, et al. A tumor DNA complex aberration index is an independent predictor of survival in breast and ovarian cancer. Mol. Oncol. 2015;9:115–127. doi: 10.1016/j.molonc.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquard AM, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telli ML, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto M, et al. Altered expression of the human base excision repair gene NTH1 in gastric cancer. Carcinogenesis. 2009;30:1345–1352. doi: 10.1093/carcin/bgp108. [DOI] [PubMed] [Google Scholar]
- 19.Girard, E. et al. Familial breast cancer and DNA repair genes: Insights into known and novel susceptibility genes from the GENESIS study, and implications for multigene panel testing. Int. J. Cancer10.1002/ijc.31921 (2018). [DOI] [PMC free article] [PubMed]
- 20.Elsayed, F. A. et al. Monoallelic NTHL1 loss of function variants and risk of polyposis and colorectal cancer. Gastroenterology10.1053/j.gastro.2020.08.042 (2020). [DOI] [PMC free article] [PubMed]
- 21.Elledge SJ, Amon A. The BRCA1 suppressor hypothesis: an explanation for the tissue-specific tumor development in BRCA1 patients. Cancer Cell. 2002;1:129–132. doi: 10.1016/S1535-6108(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 22.Polak P, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. E. A. et al. Molecular analysis of PALB2-associated breast cancers. J. Pathol.10.1002/path.5055 (2018). [DOI] [PubMed]
- 24.Mandelker D, et al. The landscape of somatic genetic alterations in breast cancers from CHEK2 germline mutation carriers. JNCI Cancer Spectr. 2019;3:pkz027. doi: 10.1093/jncics/pkz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suspitsin EN, et al. Development of breast tumors in CHEK2, NBN/NBS1 and BLM mutation carriers does not commonly involve somatic inactivation of the wild-type allele. Med. Oncol. 2014;31:828. doi: 10.1007/s12032-013-0828-9. [DOI] [PubMed] [Google Scholar]
- 26.Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031–2045. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 27.Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004;23:2330–2338. doi: 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 28.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J. Pathol. 2011;223:116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 29.Papa A, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue K, Fry EA. Haploinsufficient tumor suppressor genes. Adv. Med. Biol. 2017;118:83–122. [PMC free article] [PubMed] [Google Scholar]
- 31.Konishi H, et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl Acad. Sci. USA. 2011;108:17773–17778. doi: 10.1073/pnas.1110969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedic M, Kuperwasser C. BRCA1-hapoinsufficiency: unraveling the molecular and cellular basis for tissue-specific cancer. Cell Cycle. 2016;15:621–627. doi: 10.1080/15384101.2016.1141841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sedic M, et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat. Commun. 2015;6:7505. doi: 10.1038/ncomms8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Z, et al. Expression analyses of 27 DNA repair genes in astrocytoma by TaqMan low-density array. Neurosci. Lett. 2006;409:112–117. doi: 10.1016/j.neulet.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Karger S, et al. Distinct pattern of oxidative DNA damage and DNA repair in follicular thyroid tumours. J. Mol. Endocrinol. 2012;48:193–202. doi: 10.1530/JME-11-0119. [DOI] [PubMed] [Google Scholar]
- 36.Galick HA, et al. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc. Natl Acad. Sci. USA. 2013;110:14314–14319. doi: 10.1073/pnas.1306752110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsden CG, et al. Expression of a germline variant in the N-terminal domain of the human DNA glycosylase NTHL1 induces cellular transformation without impairing enzymatic function or substrate specificity. Oncotarget. 2020;11:2262–2272. doi: 10.18632/oncotarget.27548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li N, et al. Mutations in RECQL are not associated with breast cancer risk in an Australian population. Nat. Genet. 2018;50:1346–1348. doi: 10.1038/s41588-018-0206-9. [DOI] [PubMed] [Google Scholar]
- 39.Kidd JR, et al. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig. Genet. 2011;2:1. doi: 10.1186/2041-2223-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosoy R, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nassir R, et al. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genet. 2009;10:39. doi: 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, et al. Evaluating the breast cancer predisposition role of rare variants in genes associated with low-penetrance breast cancer risk SNPs. Breast Cancer Res. 2018;20:3. doi: 10.1186/s13058-017-0929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Beers EH, et al. A multiplex PCR predictor for aCGH success of FFPE samples. Br. J. Cancer. 2006;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin SF, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birkbak NJ, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popova T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 55.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2016).
- 57.Hansmann T, et al. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum. Mol. Genet. 2012;21:4669–4679. doi: 10.1093/hmg/dds308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li, N. et al. Metadata record for the manuscript: Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: an international multi-center study of 47,180 subjects. figshare. 10.6084/m9.figshare.14208293 (2021). [DOI] [PMC free article] [PubMed]
- 60.European Genotype-phenotype Archive. https://identifiers.org/ega.dataset:EGAD00001007025 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1420829359. The sequencing data have been deposited in the European Genotype-phenotype Archive under the following accession: https://identifiers.org/ega.dataset:EGAD0000100702560 (study ID: EGAS00001005043). These data include NTHL1 sequencing using germline DNA, Alignment and variant calling, Whole-genome sequencing and targeted sequencing of tumor DNA, Genome-wide copy number analysis, Mutational signature analysis. In addition, the following data are not openly available to protect patient privacy: NTHL1 protein expression analysis, FCC patient database, cohorts summary, NTHL1 promoter methylation analysis. Data requests for these data should be made to the corresponding author.