Abstract
We opine on the recent advances in experiments and modeling of modular signaling complexes assembled on mammalian cell membranes (membrane signalosomes) in the context of several applications including intracellular trafficking, cell migration, and immune response. Characterizing the individual components of the membrane assemblies at the nanoscale, ranging from protein-lipid and protein-protein interactions, to membrane morphology, and the energetics of emergent assemblies at the subcellular to cellular scales pose significant challenges. Overcoming these challenges through the iterative coupling of multiscale modeling and experiment can be transformative in terms of addressing the gaps between structural biology and super-resolution microscopy, as it holds the key to the discovery of fundamental mechanisms behind the emergence of function in the membrane signalosome.
Keywords: membrane organization, signaling complex, membrane trafficking, multiscale modeling, emergent function
Graphical Abstract
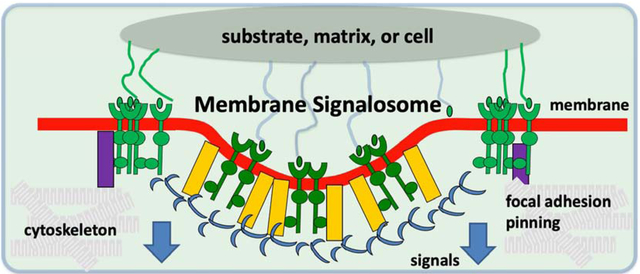
Membrane signalosome paradigm for how physical and biochemical factors impact signals and function
Membrane Signalosomes: Assembly and Function
Intracellular and intercellular signaling and trafficking mechanisms are driven by collective interactions between components of the proteome and the lipidome and are crucial for many cellular processes, including nutrient uptake, cell motility, cell-cell communication, and membrane homeostasis. These cellular transport mechanisms dynamically regulate and impact signaling by modulating receptor activity and protein localization. Spatio-temporal orchestration of functional protein-lipid assemblies transduce and actuate outside-in as well as inside-out signaling, and ultimately link (often govern) signaling, function, and fate in the cellular microenvironment [1].
Formation of protein-lipid assemblies on the membrane interface referred to here as membrane signalosomes (Figure 1) are often linked to the engagement of ligands of the extracellular matrix (ECM) and recruitment and reorganization of the cytoskeleton, which together determine cell morphology and define the forces driving multiple biological processes. Most such processes are coupled to the deformation of the cell membrane. Examples of such membrane-cytoskeleton coupled processes include the formation of filopodia, lamellipodia, and podosomes for cell movement or cancer-cell invasion, endocytosis, phagocytosis, exocytosis, and cytokinesis.
Figure 1:
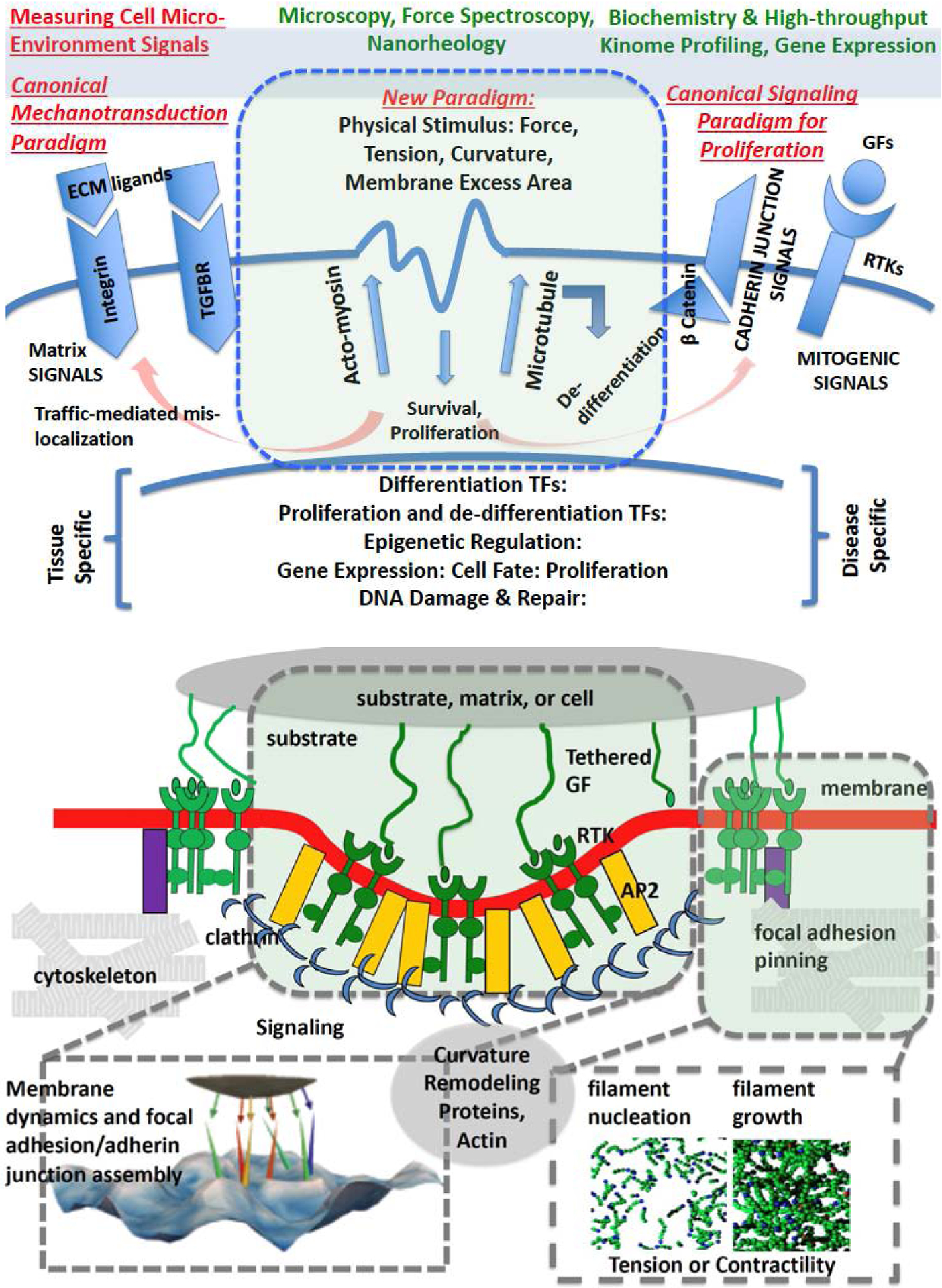
(Top) Membrane signalosome: formation of protein-lipid assemblies resulting in signaling with well-defined emergent functionality; (bottom) the formation of the signalosome is often accompanied by spatial organization of membrane-bound receptors, and can modulate as well as be modulated by external cues such as the extracellular matrix, or internal components of the cytoskeleton. The canonical paradigm revolves around biochemically triggered signals, while the new paradigm revolves around mechanical cues from the microenvironment, and novel molecular mechanisms of mechanosensing, transduction, and actuation. Abbreviations: TF- transcription factor, ECM- extracellular matrix, GF- growth factor, RTK- receptor tyrosine kinase, TGFBR- transforming growth factor beta receptor, AP2- adaptor protein 2.
The recruitment of effector proteins and specific lipids (e.g. phosphoinositides) into functional nanodomains, and their defined spatial organization, are key to the orchestration of trafficking processes [2, 3]. Self-assembly of these protein/lipid complexes and membrane nanodomains is driven by electrostatic interactions, line-tension, restricted diffusion, and membrane curvature effects. One focus in the study of functional domains is lipid clustering (especially for phosphoinositides) and signalosome assembly at the initial step of endocytosis, induced by clathrin, ENTH and BAR domains, and protein domains that recognize specific lipids [4]. Protein domains modify lipid distribution [5], and lipid assembly at the nanoscale and curvature at the mesoscale (10–100 nm) influence protein distribution [6] in processes such as exocytosis, endosomal sorting, and recycling. Moreover, these bidirectional interactions are mechanosensitive and are finetuned to respond physical as well as biochemical changes in the cell microenvironment (Figure 1).
Studies at the mesoscale of the membrane signalosomes are extremely challenging [7]. They are too large for atomic resolution studies yet fall below the diffraction limit, and so elude many optical methods. The dynamics/kinetics of complex assembly in cells are followed using live-cell imaging experiments, including optical microscopy [8], electron microscopy (EM) [9], and super-resolution microscopy [10]. Membrane signalosomes capable of force sensing, transduction, or actuation [11] are investigated through quantitative biophysical techniques including force spectroscopy [12], atomic force microscopy [13, 14], and traction force measurements.
Multiscale Modeling of Membrane Components
Models and simulations at multiple resolutions have provided valuable insight into biophysical interactions of the various components of the membrane signalosome. These studies have been designed to develop and validate spatial, stochastic, temporal computational models of membrane components at the atomic as well as continuum scale; quantify and rationalize the driving forces, specificity, and cooperativity in protein-lipid interactions leading to the assembly of functional nanodomains; quantify and delineate the underlying biophysical and biochemical interactions leading to curvature generation and membrane remodeling at the molecular scale as well as at the mesoscale (Figure 3).
Figure 3:
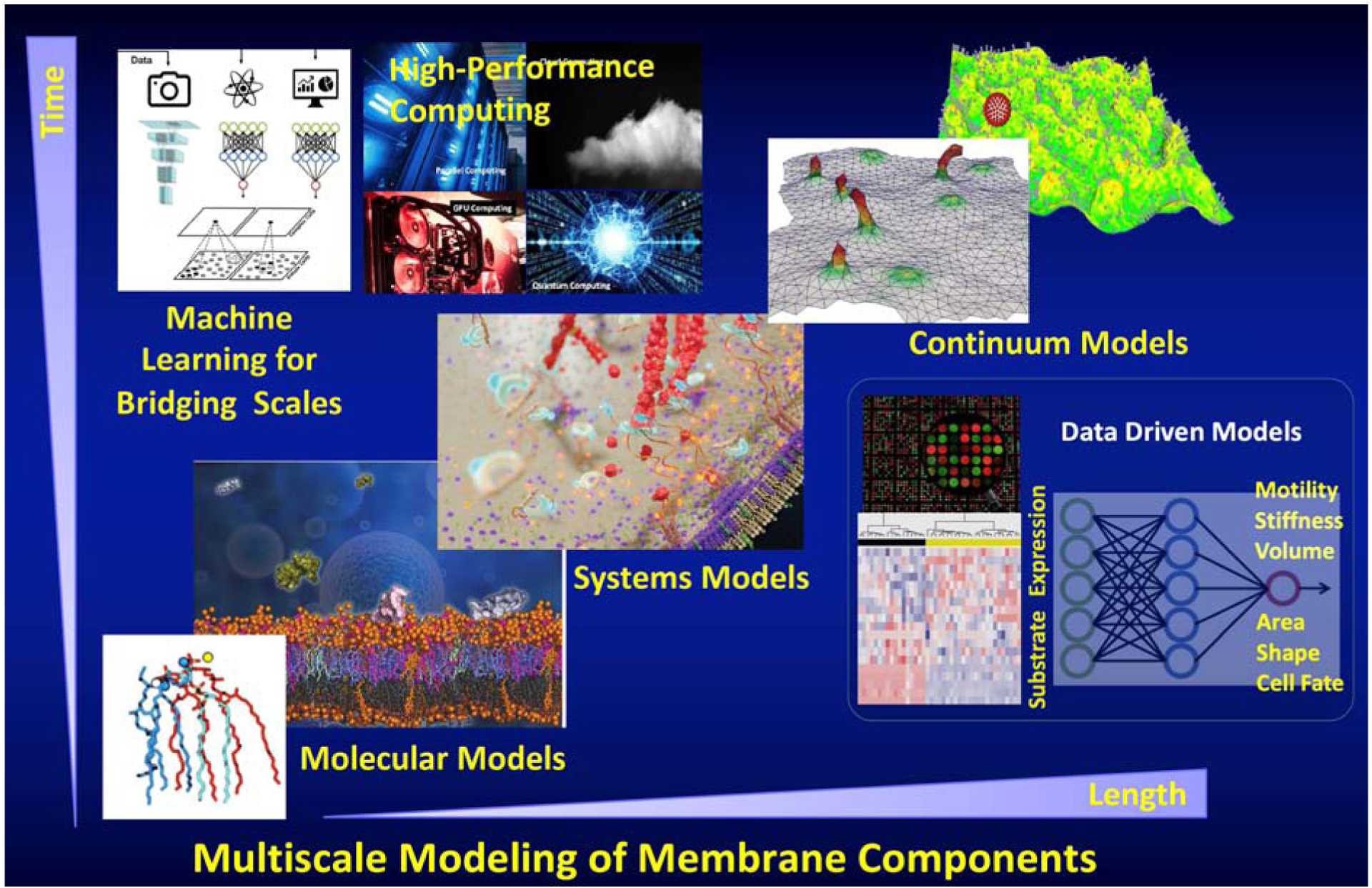
Depiction of modeling at the molecular and mesoscales through atomistic, coarse-grained, and continuum models. Bridging scales to produce a multiscale method is enabled by coupling algorithms based on machine learning. The biophysically based multiscale models often produce explainable and interpretable models with predictive power. This is in contrast to the purely data-driven models produced by machine learning which even with predictive power, are often not interpretable or explainable. However, the two approaches are complementary and can be synergistically leveraged to gain maximal mechanistic insight as well as utilize the rich experimental data.
Molecular dynamics (MD) simulations allow us to investigate the behavior of lipids over a range of time and length scales – from picoseconds to microseconds and from Angstroms to microns – to study the dynamics of biological membranes [15]. Additionally, electronic structure methods and MD simulation methods allow the inspection and manipulation of physicochemical properties of membrane structures – such as their net charge and partial atomic charge distribution – that are intimately related to their biological function by mediating intermolecular contacts with other lipids and ions. Many in vitro and in vivo experimental characterizations of lipids have focused on their localization [16] and structure (neutron scattering) [17], and these measurements generally report static properties. MD simulations are indispensable for testing variable membrane compositions in different ionic environments in the absence [18, 19] and the presence of proteins [2].
Membrane remodeling at mesoscale involves cooperative interactions of multiple proteins at length scales that are several times the thickness of the bilayer, making it prohibitive for a fully atomistic treatment [20]. Particle-based coarse-graining has provided an optimal path forward to model phase behavior in multicomponent lipids, multiple proteins interacting with the bilayer and membrane morphological changes due to cooperative interactions with several proteins [21]. An example of such studies is shown in Figure 2, where an atomic model of a lipid bilayer in which phosphatidylinositol 4,5 bisphosphate (PtdIns(4,5)P2) clusters recruit and activate the actin nucleator mDia2 is superimposed on an electron micrograph of the cytoplasmic face of a cell membrane in which PtdIns(4,5)P2 clusters are seen adjacent to actin filaments and endocytic vesicles [2].
Figure 2.
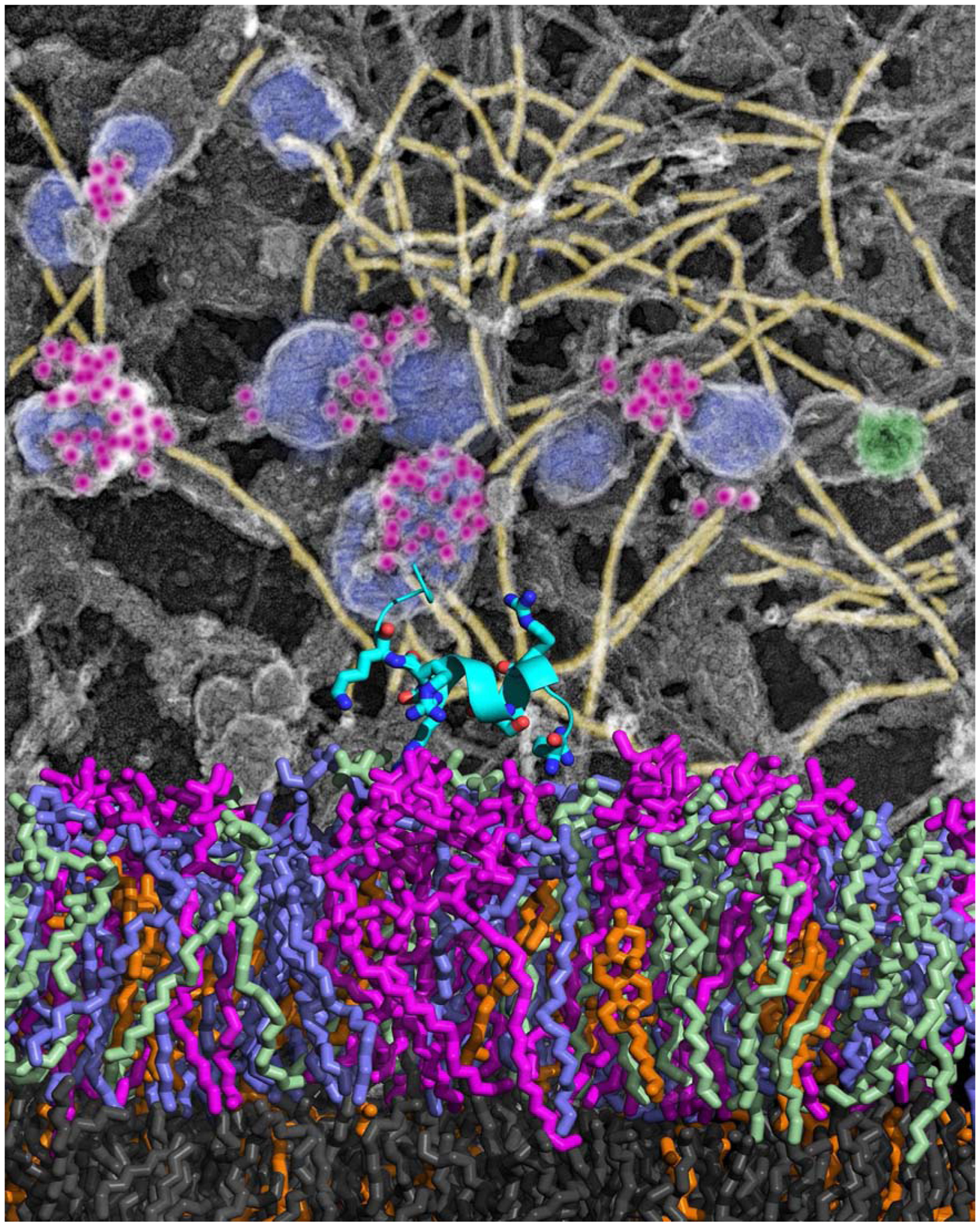
Lateral distribution of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] within plasma membranes alters the capacity of PI(4,5)P2 to nucleate actin assembly. Clusters of PI(4,5)P2 immunogold (Purple) associate with actin filaments (Yellow) at the surface of caveolae (Blue) or clathrin-coated structures (Green) on plasma membranes sheets from electron microscopy (Top). The bottom shows atomistic molecular dynamics simulation of protein mDia2 bound to bilayers containing PI(4,5)P2 (Purple).
Continuum models based on curvature elasticity of thin films have been the method of choice for lengthscales approaching organelles and cells. The literature on the biophysics of thin shells is vast, and we refer to recent reviews written in the context of protein-dependent remodeling of membranes [22]. Of particular interests are studies that have explored the thermodynamics of membrane morphological transitions at the cellular scale and their applications to endocytosis, exocytosis, and endosomal sorting mechanisms [23–26]; these studies have revealed the importance of entropic contributions to membrane remodeling at large lengthscales. An interesting phenomenon in protein remodeling of membranes is the ability to sense curvature at large lengthscales despite the fact that the interactions between the proteins and the lipids occur at the molecular scale. Coarse-grained simulations have revealed possible mechanisms of curvature sensing through scaffolding as well as through curvature-undulation coupling mechanisms [24, 27, 28]. The ability to sense curvature has a profound impact on the spatial localization of these curvature remodeling proteins. The computational models at multiple scales described above have also been extended to include cytoskeletal components to study the effects of the cytoskeleton on membrane morphology [29–31] and to couple the membrane morphological changes to small-world networks of actin assembly [32].
Multiscale Biophysical Models of the Membrane Signalosome
Historically, computational models of signaling have led to paradigm-shifting insights in various subdisciplines of physiology and cell biology. Notable examples include the description of the initiation and the wave propagation of action potentials in neurons through the Hodgkin–Huxley model or the early description of the chemical basis of pattern formation and morphogenesis by Turing. With the evolution of cell biology as a data-rich and quantitative discipline, the physical biology of the cell has emerged to be a powerful approach to construct computational models encoding higher levels of regulation and complexity [33], including those aiming to describe the membrane signalosome. Spatial and non-spatial systems models describing the nonlinear dynamics of chemical reactions have certainly led the way in describing membrane reactions in signaling [34]. However, they often fail to describe or capture a class of emergent phenomena or function where morphology influences signaling and signaling influences morphology simultaneously, which is an essential characteristic of outside-in and inside-out signaling mentioned earlier.
The challenge in modeling the signalosome is to incorporate both biophysical models of material interaction and response at multiple length scales and signaling dynamics at multiple timescales. Although the treatment of larger lengthscale processes in models at a sub-cellular scale necessitates approximations, such models provide a unique opportunity to connect with cellular experiments. The key to such a model is the bridging of length and time scales [35], which provides a platform and a suite of approaches for integrating interactions and dynamics at molecular and cellular scales (Figure 3).
Some of the earliest successes of computational studies of mechanosignaling models and computational models of the membrane signalosome include the synapse assembly model describing the formation of the immunological synapse during T-cell recognition of foreign antigens. Chakraborty et al. [36] described the mechanical coupling of two membrane surfaces representing the antigen-presenting cell and the T-cell and the dynamics of pattern formation of multivalent receptor-ligand complexes involving T-cell receptors (TCRs) and integrins with their respective ligands. This mechanochemical model was further evolved by coupling the model to TCR signaling [37], and incorporating the role of cytoskeleton-generated force exerted on the TCR [38].
Another example of a system that has enjoyed much success in terms of computational models of signaling and membrane assembly impacting the experimental research is endocytosis. A detailed parts-list for the protein-protein interactions in clathrin-mediated endocytosis has been available for some time [39]. However, recent experimental, theoretical, and computational tools have proved to be critical in establishing the sequence of events, cooperative dynamics, and energetics of the intracellular process. On the experimental front, total internal reflection fluorescence microscopy, photo-activated localization microscopy, and spinning-disk confocal microscopy have focused on assembly and patterning of endocytic proteins at the membrane [8]. In contrast, on the theory front, minimal theoretical models for clathrin nucleation, biophysical models for membrane curvature and bending elasticity, as well as methods from computational structural and systems biology of phosphoinositide signaling, have proved insightful in describing membrane topologies, curvature mechanisms, and energetics [39–41].
Models considering the effects of extracellular matrix adhesion of mammalian cells mediated by the binding of cell-surface integrin receptors by specifically probing the effect of the ligand affinity on the assembly of focal adhesions have been reported [42]. Among these, some computational models have shed light on important contributions from membrane-mediated interactions arising from membrane deformations and mechanics on cluster size distributions of focal adhesion complexes [43, 44]. Similar mechanisms involving kinetic traps formed by membrane deformations have been employed to explain the hyperactivation of integrin signaling in cancer cells, which accumulate glycocalyx on their membrane surface [45].
A class of models has investigated how cells control their shapes and the structures that they form on their surface, exposing the building blocks that cells use to deform their membranes. Such models explore quantitatively different possible mechanisms that cells can employ to initiate the spontaneous formation of shapes and patterns on their membranes [46]. One study examines how the actin cortex senses and transmits forces and how cytoskeletal proteins interact in response to the forces to explain mechanosensing from molecular to cellular scales [47]. Contributions of intracellular fluid flow, including those of a viscous active cytoskeletal gel, to cell motility, have also been investigated through mathematical models [48]. Recent works have highlighted the explicit coupling of signaling using calcium dynamics, and the remodeling of cell membrane curvature in the assembly and activation of SNARE complexes in calcium triggered exocytosis [49].
In addition to convection and diffusion-based mechanisms, long-range communication through membrane tension has also been hypothesized for synchronizing actin assembly with membrane morphological changes during cell motility [50]. A unified mathematical model of chemotaxis involving the coordinated action of separable but interrelated processes, namely, motility, gradient sensing, and polarization, has been proposed: here, the central module is an excitable network that accounts for random migration, while the response to gradients is mediated by an excitable network, and a polarization module linked to the excitable network through the cytoskeleton allows for sensing and saturation of signals consistent with cellular behaviors that mirror those of genetically altered cell lines [51].
Incorporating Molecular Specificity in Models of the Membrane Signalosome
An aspirational goal of modeling the membrane signalosome is to encode and explain the emergence of function and behavior. However, given the molecular specificity of signals and with the advent of experimental tools to probe large biomolecular complexes at a near-atomic resolution [52], building bridges between structural biology and network-based system biology is a crucial challenge to be overcome. The integration of structural and network-based systems biology is paramount for the improved understanding of how lipids and proteins interact to modulate the behavior of complex biological systems such as the membrane signalosome [53]. Some of the early efforts in this regard with applications to receptor tyrosine kinase signaling and trafficking have been reported in structural systems biology [54–56]. With the advent of artificial intelligence-based algorithms for structure prediction such as the alpha fold, computational modeling of protein-protein interaction networks at atomic resolution is poised for an exciting revolution in the near future [57]. Emergent features of signaling modules analyzed through nonlinear dynamics have been useful to connect mathematical models to cell fate decisions [58, 59]. New paradigms of cellular regulation and cell fate, which rely on kinetic proofreading [60, 61], have proposed that such signaling networks rely on spatial clustering [62] and mechanisms of microphase separations [63], where the underlying bases for driving forces are not yet known. Development of new membrane signalosome models that correctly encode these principles to correctly predict the effects of ligand agonism [64, 65] and mechanosensitivity on cell fate [66] could represent promising advances in the field.
Acknowledgments:
We thank Changsong Yang and Tatyana Svitkina for providing the image for figure 2. We acknowledge financial support from the National Institutes of Health through grants CA227550, CA244660 (RR) and GM136259 (PAJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: none
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Anitei M and Hoflack B, Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol, 2012. 14(1): p. 11–9. [DOI] [PubMed] [Google Scholar]
- ●2.Bucki R, et al. , Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation. Journal of Biological Chemistry, 2019. 294(12): p. 4704–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides experimental evidence and describes the mechanism of actin assembly initated through phosphoinositide clustering.
- 3.Janmey PA, Bucki R, and Radhakrishnan R, Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms. Biochemical and Biophysical Research Communications, 2018. 506(2): p. 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemmon MA, Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol, 2008. 9(2): p. 99–111. [DOI] [PubMed] [Google Scholar]
- 5.van den Bogaart G, et al. , Membrane protein sequestering by ionic protein-lipid interactions. Nature, 2011. 479(7374): p. 552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aimon S, et al. , Membrane shape modulates transmembrane protein distribution. Dev Cell, 2014. 28(2): p. 212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chierico L, et al. , Live cell imaging of membrane/cytoskeleton interactions and membrane topology. Sci Rep, 2014. 4: p. 6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●8.Picco A and Kaksonen M, Quantitative imaging of clathrin-mediated endocytosis. Current Opinion in Cell Biology, 2018. 53: p. 105–110. [DOI] [PubMed] [Google Scholar]; This article describes the advances and applications or super-resolution microscopy based methods to the study of clathrin mediated endocytosis.
- 9.Svitkina TM, Ultrastructure of the actin cytoskeleton. Current Opinion in Cell Biology, 2018. 54: p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Möckl L and Moerner WE, Super-resolution Microscopy with Single Molecules in Biology and Beyond–Essentials, Current Trends, and Future Challenges. Journal of the American Chemical Society, 2020. 142(42): p. 17828–17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diz-Muñoz A, Fletcher DA, and Weiner OD, Use the force: membrane tension as an organizer of cell shape and motility. Trends in Cell Biology, 2013. 23(2): p. 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Z, et al. , Cell Membranes Resist Flow. Cell, 2018. 175(7): p. 1769–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haase K and Pelling AE, Investigating cell mechanics with atomic force microscopy. Journal of The Royal Society Interface, 2015. 12(104): p. 20140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapus A and Janmey P, Plasma membrane-cortical cytoskeleton interactions: a cell biology approach with biophysical considerations. Compr Physiol, 2013. 3(3): p. 1231–81. [DOI] [PubMed] [Google Scholar]
- 15.Corradi V, et al. , Emerging Diversity in Lipid–Protein Interactions. Chemical Reviews, 2019. 119(9): p. 5775–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●16.Wen Y, Vogt VM, and Feigenson GW, Multivalent Cation-Bridged PI(4,5)P2 Clusters Form at Very Low Concentrations. Biophysical Journal, 2018. 114(11): p. 2630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides direct experimental evidence of ion-initiated nanocluster formation of phosphoinositide clusters.
- 17.Ghosh SK, Aeffner S, and Salditt T, Effect of PIP2 on bilayer structure and phase behavior of DOPC: an X-ray scattering study. Chemphyschem, 2011. 12(14): p. 2633–40. [DOI] [PubMed] [Google Scholar]
- 18.Han K, Gericke A, and Pastor RW, Characterization of Specific Ion Effects on PI(4,5)P2 Clustering: Molecular Dynamics Simulations and Graph-Theoretic Analysis. The Journal of Physical Chemistry B, 2020. 124(7): p. 1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley RP, et al. , Divalent cations bind to phosphoinositides to induce ion and isomer specific propensities for nano-cluster initiation in bilayer membranes. R Soc Open Sci, 2020. 7(5): p. 192208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simunovic M, et al. , Protein-mediated transformation of lipid vesicles into tubular networks. Biophys J, 2013. 105(3): p. 711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingólfsson HI, et al. , Computational ‘microscopy’ of cellular membranes. Journal of Cell Science, 2016. 129(2): p. 257. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan N, Sunil Kumar PB, and Radhakrishnan R, Mesoscale computational studies of membrane bilayer remodeling by curvature-inducing proteins. Physics Reports, 2014. 543(1): p. 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tourdot RW, et al. , Multiscale computational models in physical systems biology of intracellular trafficking. IET Syst Biol, 2014. 8(5): p. 198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●24.Ramakrishnan N, et al. , Biophysics of membrane curvature remodeling at molecular and mesoscopic lengthscales. J Phys Condens Matter, 2018. 30(27): p. 273001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article summarizes molecular and mesoscale approaches to treat cell membrane curvature under the influence of curvature remodeling proteins.
- 25.Bahrami AH, et al. , Scaffolding the cup-shaped double membrane in autophagy. PLOS Computational Biology, 2017. 13(10): p. e1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, et al. , Exo70 generates membrane curvature for morphogenesis and cell migration. Dev Cell, 2013. 26(3): p. 266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmood MI, Noguchi H, and Okazaki K.-i., Curvature induction and sensing of the F-BAR protein Pacsin1 on lipid membranes via molecular dynamics simulations. Scientific Reports, 2019. 9(1): p. 14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradley RP and Radhakrishnan R, Curvature-undulation coupling as a basis for curvature sensing and generation in bilayer membranes. Proc Natl Acad Sci U S A, 2016. 113(35): p. E5117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, et al. , Endocytic vesicle scission by lipid phase boundary forces. PNAS, 2006. 103(27): p. 10277–10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●30.Kandy SK and Radhakrishnan R, Emergent membrane morphologies in relaxed and tense membranes in presence of reversible adhesive pinning interactions. Physical Biology, 2019. 16(6): p. 066011. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides mechanisms for vesicle biogenesis through nonspecific mechanisms not involving curvature remodeling proteins.
- 31.Ramakrishnan N, et al. , Excess area dependent scaling behavior of nano-sized membrane tethers. Phys Biol, 2018. 15(2): p. 026002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatunmbi O, et al. , A multiscale biophysical model for the recruitment of actin nucleating proteins at the membrane interface. Soft Matter, 2020. 16(21): p. 4941–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips R, Kondev J, and Theriot J, Physical biology of the cell. 2009: Garland Science. [Google Scholar]
- 34.Kholodenko BN, Hancock JF, and Kolch W, Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol, 2010. 11(6): p. 414−−426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radhakrishnan R, A survey of multiscale modeling: Foundations, historical milestones, current status, and future prospects. AIChE Journal, 2020. n/a(n/a): p. e17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SJE, et al. , The synapse assembly model. Trends Immunol, 2002. 23(10): p. 500–502. [DOI] [PubMed] [Google Scholar]
- 37.KH L, et al. , The immunological synapse balances T cell receptor signaling and degradation. Science, 2003. 302: p. 1218–22. [DOI] [PubMed] [Google Scholar]
- 38.Kaizuka Y, Regulations of T Cell Activation by Membrane and Cytoskeleton. Membranes, 2020. 10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramanan V, et al. , Systems biology and physical biology of clathrin-mediated endocytosis. Integr Biol (Camb), 2011. 3(8): p. 803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, et al. , The mechanochemistry of endocytosis. PLoS Biol, 2009. 7(9): p. e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chabanon M, Stachowiak JC, and Rangamani P, Systems biology of cellular membranes: a convergence with biophysics. WIREs Systems Biology and Medicine, 2017. 9(5): p. e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng X, et al. , Cell adhesion nucleation regulated by substrate stiffness: A Monte Carlo study. Journal of Biomechanics, 2012. 45(1): p. 116–122. [DOI] [PubMed] [Google Scholar]
- 43.Zhao T, Li Y, and Dinner AR, How focal adhesion size depends on integrin affinity. Langmuir, 2009. 25(3): p. 1540–6. [DOI] [PubMed] [Google Scholar]
- 44.Kato M and Mrksich M, Using model substrates to study the dependence of focal adhesion formation on the affinity of integrin-ligand complexes. Biochemistry, 2004. 43(10): p. 2699–707. [DOI] [PubMed] [Google Scholar]
- 45.Paszek MJ, et al. , The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature Publishing Group, 2014. 511(7509): p. 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●46.Gov NS, Guided by curvature: shaping cells by coupling curved membrane proteins and cytoskeletal forces. Philosophical Transactions of the Royal Society B: Biological Sciences, 2018. 373(1747): p. 20170115. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes approaches to couple membrane curvature with actin assembly.
- 47.Luo T, et al. , Molecular mechanisms of cellular mechanosensing. Nat Mater, 2013. 12(11): p. 1064–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mogilner A and Manhart A, Intracellular Fluid Mechanics: Coupling Cytoplasmic Flow with Active Cytoskeletal Gel. Annual Review of Fluid Mechanics, 2018. 50(1): p. 347–370. [Google Scholar]
- 49.Dhara M, et al. , Synergistic actions of v-SNARE transmembrane domains and membrane-curvature modifying lipids in neurotransmitter release. eLife, 2020. 9: p. e55152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houk AR, et al. , Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell, 2012. 148(1–2): p. 175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C, et al. , Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol, 2013. 9(7): p. e1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callaway E, Revolutionary cryo-EM is taking over structural biology. Nature, 2020. 578(7794): p. 201. [DOI] [PubMed] [Google Scholar]
- 53.Chasapis CT, Building Bridges Between Structural and Network-Based Systems Biology. Molecular Biotechnology, 2019. 61(3): p. 221–229. [DOI] [PubMed] [Google Scholar]
- 54.Aloy P and Russell RB, Structural systems biology: modelling protein interactions. Nat Rev Mol Cell Biol, 2006. 7(3): p. 188–97. [DOI] [PubMed] [Google Scholar]
- 55.Telesco SE and Radhakrishnan R, Structural systems biology and multiscale signaling models. Ann Biomed Eng, 2012. 40(11): p. 2295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shih AJ, Purvis J, and Radhakrishnan R, Molecular systems biology of ErbB1 signaling: bridging the gap through multiscale modeling and high-performance computing. Mol Biosyst, 2008. 4(12): p. 1151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senior AW, et al. , Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins: Structure, Function, and Bioinformatics, 2019. 87(12): p. 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kholodenko BN, Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol, 2006. 7(3): p. 165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chickarmane V, et al. , Transcriptional dynamics of the embryonic stem cell switch. PLoS Comput Biol, 2006. 2(9): p. e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●60.Lin JJY, et al. , Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci Signal, 2019. 12(564). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides experimental evidence connecting the dwell times of ligand binding to T-cell receptor signaling using single molecule approaches.
- 61.Kiyatkin A, et al. , Kinetics of receptor tyrosine kinase activation define ERK signaling dynamics. Sci Signal, 2020. 13(645). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●62.Goyette J and Gaus K, Mechanisms of protein nanoscale clustering. Current Opinion in Cell Biology, 2017. 44: p. 86–92. [DOI] [PubMed] [Google Scholar]; This article summarizes various physical and kinetic mechanisms of nanoscale cluster formation on membrane interfaces.
- 63.Cable J, et al. , Phase separation in biology and disease. Annals NY Acad. Sci, 2019: p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●64.Freed DM, et al. , EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell, 2017. 171(3): p. 683–695.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides structural basis for how multiple ligands can initiate varied signals through the same receptor.
- 65.Ghosh A and Radhakrishnan R, Time-dependent antagonist-agonist switching in receptor tyrosine kinase-mediated signaling. BMC Bioinformatics, 2019. 20(1): p. 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar A, Placone JK, and Engler AJ, Understanding the extracellular forces that determine cell fate and maintenance. Development, 2017. 144(23): p. 4261–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]


