Chlamydia sp. injects numerous effector proteins into host cells to manipulate cellular functions important for bacterial survival. Based on findings in Chlamydia trachomatis, it has been proposed that between 5% and 10% of the Chlamydia pneumoniae genome, a related respiratory pathogen, encodes secreted effectors.
KEYWORDS: effector molecules, Chlamydia pneumoniae, secretion system, GSK3β, phospho-GSK3β
ABSTRACT
Chlamydia pneumoniae is a Gram-negative, obligate intracellular pathogen that causes community-acquired respiratory infections. C. pneumoniae uses a cell contact-dependent type III secretion (T3S) system to translocate pathogen effector proteins that manipulate host cellular functions. While several C. pneumoniae T3S effectors have been proposed, few have been experimentally confirmed in Chlamydia sp. In this study, we expressed 382 C. pneumoniae genes in Chlamydia trachomatis as fusion proteins to an epitope tag derived from glycogen synthase kinase 3β (GSK3β) which is the target of phosphorylation by mammalian kinases. Based on the detection of the tagged C. pneumoniae protein with anti-phospho-GSK3β antibodies, we identified 49 novel C. pneumoniae-specific proteins that are translocated by C. trachomatis into the host cytoplasm and thus likely play a role as modifiers of host cellular functions. In this manner, we identified and characterized a new C. pneumoniae effector, CPj0678, that recruits the host cell protein PACSIN2 to the plasma membrane. These findings indicate that C. trachomatis provides a powerful screening system for detecting candidate effector proteins carried by other pathogenic and endosymbiotic Chlamydia species.
IMPORTANCE Chlamydia sp. injects numerous effector proteins into host cells to manipulate cellular functions important for bacterial survival. Based on findings in Chlamydia trachomatis, it has been proposed that between 5% and 10% of the Chlamydia pneumoniae genome, a related respiratory pathogen, encodes secreted effectors. However, only a few C. pneumoniae effectors have been identified and experimentally validated. With the development of methods for the stable genetic transformation of C. trachomatis, it is now possible to use C. trachomatis shuttle plasmids to express and explore the function of proteins from other Chlamydia spp. in a more relevant bacterial system. In this study, we experimentally determined that 49 C. pneumoniae-specific proteins are translocated into the host cytoplasm by Chlamydia secretion systems and identified a novel effector required to recruit the host factor PACSIN2 to the plasma membrane during infection.
INTRODUCTION
Chlamydia pneumoniae is an obligate intracellular pathogen that causes acute and chronic respiratory infections in humans, with most adults likely contracting C. pneumoniae infections at least once in their lifetime (1). The following two species of Chlamydia are major human pathogens: Chlamydia trachomatis is the most common sexually transmitted bacterial infection worldwide and the leading cause of infectious blindness (2), and C. pneumoniae causes acute and chronic infections of the upper and lower respiratory tracts and has been associated with increased risk for the development of cardiovascular diseases (3, 4). Seroepidemiological studies suggest that 50% to 70% of the human adult population has been exposed to C. pneumoniae at some point during their lifetime (5). Chlamydia infections begin with the attachment of the elementary body (EB), the invasive form of the bacterium, to host cells. The EB is endocytosed into a plasma membrane-derived vesicle, which is quickly modified by the pathogen to establish a replicative vacuole, termed the inclusion (6). Within the nascent inclusion, the EB transitions into the replicative form of the pathogen, the reticulate body (RB) (7). The inclusion escapes lysosomal degradation most likely as a result of Chlamydia effector proteins that are secreted to the inclusion membrane and mammalian cytoplasm to inhibit lysosomal maturation and modulate membrane vesicle trafficking pathways (8).
We previously determined the sequence of the C. pneumoniae J138 genome (9) and found that this strain has 1,069 open reading frames (ORFs) with approximately one-half of the predicted ORFs coding for proteins without any known function. It has been reported that 5% to 10% of Chlamydia proteins may constitute putative type III secretion (T3S) effector molecules (10); however, only a few effectors have been identified and experimentally validated in C. pneumoniae. Genes encoding components of the Chlamydia T3S system are grouped into three major loci, and yet, genes encoding targets of T3S are dispersed throughout the chromosome in all Chlamydia species (11, 12). T3S systems are conserved membrane-embedded nanomachines that deliver virulence effector proteins directly from the bacterial cytoplasm into the host cell. T3S components include >20 proteins that form the basal body that spans the bacterial envelope and membrane-embedded oligomeric rings that are connected by a transperiplasmic rod to a hollow needle, through which unfolded effectors are secreted (13).
Several approaches have been applied to identify Chlamydia virulence factors, including screens for proteins that cause growth defects in yeast (14), lead to abnormal membrane trafficking (15), or are secreted by other bacteria such as members of Yersinia (16) or Shigella (17, 18) via their T3S systems. The identification of putative T3S targets in these bacterial expression systems is based on chimeras containing the first 20 to 30 amino acids of C. trachomatis or C. pneumoniae ORFs fused to reporter proteins, such as a β-lactamase TEM1 or an adenylate cyclase, and an analysis of whether the secretion of the reporter proteins occurred in a T3S-dependent manner (16–18).
While these approaches revealed candidate T3S substrates in C. pneumoniae, experimental validation of the delivery of the intact proteins by Chlamydia sp. into host cells is largely lacking. With the development of methods for the stable genetic transformation of C. trachomatis (19), it is now possible to use C. trachomatis shuttle plasmids as an expression platform for exploring the function of other Chlamydia proteins in the closely related, genetically tractable lymphogranuloma strain LGV-L2. In this manner, we expect that the selectivity and function of the C. trachomatis T3S machinery can be leveraged to identify new C. pneumoniae effectors which may not be readily translocated by evolutionarily distant bacteria (20). In addition to adenylate cyclase and β-lactamase, other reporters of T3S have been generated, including a small peptide derived from glycogen synthase kinase 3β (GSK3β) (21). This 13-residue tag is efficiently phosphorylated by eukaryotic protein kinases in cytoplasmic compartments of host cells and does not interfere with secretion, translocation, or function of effectors. This reporter tag can therefore be used to assess the presence of T3S effectors in the mammalian cytoplasm by monitoring the phosphorylation of the GSK3β tag with phospho-specific antibodies. This approach has been successfully adapted to detect translocated C. trachomatis effector proteins (22).
In this study, we used C. trachomatis as a protein expression platform to screen GSK-tagged full-length C. pneumoniae ORFs and identify proteins translocated during infection. Of 382 C. pneumoniae ORFs tested, 49 novel C. pneumoniae proteins were successfully expressed and determined to be translocated into host cells by C. trachomatis.
RESULTS
Expression of epitope-tagged C. pneumoniae proteins in C. trachomatis.
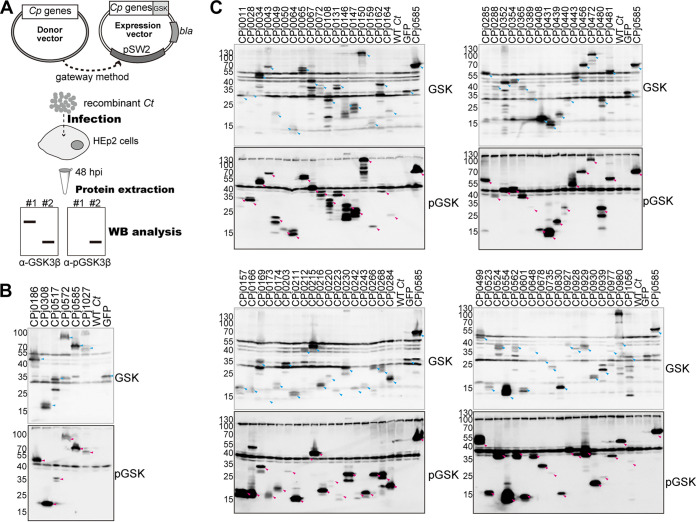
We designed a Gateway-based recombination cloning system to insert C. pneumoniae ORFs into a range of expression vectors (23). DNA inserts present in a C. pneumoniae J138 library of ORFs in Gateway entry vectors (23) were transferred into the C. trachomatis gateway expression shuttle vector pEAS7-Gateway, which drives the expression of recombinant genes from a Neisseria meningitidis promoter. The recombinant gene is C-terminally fused to a myc epitope tag and amino acids 1 to 13 of GSK3β (see Fig. S1 in the supplemental material). Recombinant plasmids were introduced into C. trachomatis LGV-L2 by the calcium transformation method and amplified by selection on penicillin as previously described (24). After 6 to 10 passages in Vero cells, transformed C. trachomatis bacteria were recovered from infected Vero cells and used to infect HEp-2 cells. Protein extracts from the infected HEp-2 cells were analyzed by immunoblotting with anti-GSK3β antibodies to detect the total recombinant proteins expressed and anti-phospho-GSK3β antibodies to assess the efficiency of C. pneumoniae protein translocation to the host cytoplasm (Fig. 1).
FIG 1.
Summary of C. pneumoniae ORFs identified as targets of translocation by C. trachomatis. (A) A total of 382 C. pneumoniae genes in a pDONR221 Gateway vector (entry clone) were transferred to a C. trachomatis expression plasmid (pEAS7) to generate C. pneumoniae protein fusions to a peptide tag derived from glycogen synthase kinase (GSK3β). (B, C) HEp-2 cells were infected for 48 h with C. trachomatis expressing C. pneumoniae proteins, and total proteins were extracted and analyzed by immunoblotting using anti-GSK3β antibodies to identify recombinant C. pneumoniae proteins and anti-phospho-GSK3β antibodies to identify proteins that were exposed to host cytoplasmic GSKs. Six previously characterized C. pneumoniae effectors and GFP-expressing recombinant C. trachomatis were analyzed by immunoblotting (B), and 69 novel C. pneumoniae proteins were detected by both anti-GSK3β and anti-phospho-GSK3β antibodies (C). GFP-GSK was included as a control for a nonsecreted protein. Cyan and magenta arrowheads indicate each protein detected by anti-GSK3β and anti-phospho-GSK3β antibodies, respectively.
C. pneumoniae T3S effectors are translocated by C. trachomatis to the cytoplasm of infected host cells.
To determine if C. trachomatis can translocate C. pneumoniae effector proteins, we first expressed six well-characterized C. pneumoniae effectors tagged with a GSK3β epitope (Fig. 1A). CPj0186, CPj0308, CPj0517, CPj0585, and CPj1027 are predicted inclusion membrane proteins (18). CPj0186 is the orthologue of the C. trachomatis fusogenic factor IncA and has two transmembrane domains in its N-terminal region and a coiled-coil SNARE-like domain at its C-terminal region, which presumably enables the fusion of inclusions (25). CPj0308 and CPj0585 were predicted to be T3S substrates based on the ability of their N-terminal regions to enable T3S-dependent secretion in Shigella flexneri (17, 18). Both CPj0517 (also known as CP0236) and CPj0585 are C. pneumoniae-specific proteins that bind to NF-κB activator 1 (Act1) (26) and a subset of Rab GTPases, respectively (27). CPj0572 is the orthologue of the C. trachomatis protein TarP, which is implicated in the recruitment of actin to the site of internalization of EBs (28). The domain responsible for actin filament recruitment is restricted to the C-terminal half of TarP, placing that region inside host cells upon translocation (29). We used the green fluorescent protein (GFP) as a negative control for secretion, as this protein lacks any signal peptides or predicted T3S export signals. Immunoblot analysis with anti-GSK3β antibodies (Fig. 1B) indicated that all six GSK3β-tagged C. pneumoniae proteins and GSK3β-tagged GFP were expressed in C. trachomatis and detected at the predicted molecular weight as follows: CPj0186-GSK, 46 kDa; CPj0308-GSK, 17 kDa; CPj0517-GSK, 35 kDa; CPj0572-GSK, 82 kDa; CPj0585-GSK, 80 kDa; CPj1027-GSK, 64 kDa; and GFP-GSK, 30 kDa.
The six C. pneumoniae proteins were also detected with anti-phospho-GSK3β antibodies at the expected position on immunoblots (magenta arrowheads), while GFP-GSK3β was not (Fig. 1B). These findings indicate that C. trachomatis can express C. pneumoniae-specific effector proteins and that the molecular mechanisms mediating their secretion/translocation are conserved in C. trachomatis.
A screen of C. pneumoniae ORFs of unknown function in C. trachomatis identifies new translocated effector proteins.
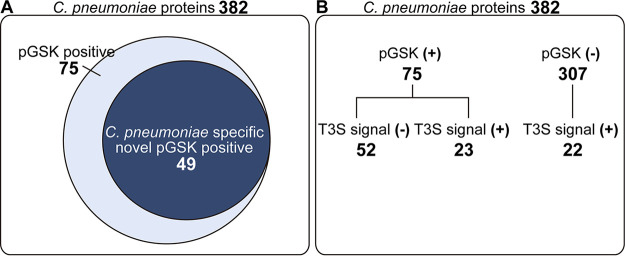
We expressed 382 C. pneumoniae ORFs of unknown function as GSK3β-tagged proteins in C. trachomatis and assessed the degree to which they were expressed and translocated to the host cytoplasm by immunoblotting lysates of infected cells with anti-GSK3β and anti-phospho-GSK3β antibodies, respectively (Fig. 1C). Untransformed C. trachomatis and GFP-GSK3β-expressing C. trachomatis were used as negative controls, and CPj0585-GSK3β-expressing C. trachomatis was used as a positive control for protein secretion. Of the 382 C. pneumoniae proteins expressed in C. trachomatis, 75 proteins were detectable with anti-phospho-GSK3β antibodies and presumed to be translocated into host cells (Fig. 2A; see Table S1 in the supplemental material). Notably, the function of 49 of these C. pneumoniae-specific proteins has not been previously described (Fig. 2A).
FIG 2.
Overlap between predicted and validated C. pneumoniae proteins translocated into the host cell cytoplasm. (A) A total of 382 C. pneumoniae proteins (∼35% of genome) were expressed in C. trachomatis in GSK-tagged forms and tested for phosphorylation of the GSK3β tag with phospho-specific antibodies. Of the 75 phospho-GSK3β-positive clones, 58 proteins were specific to C. pneumoniae, including 49 proteins whose localization during infection had not been previously determined. (B) Bioinformatic analysis of the 382 C. pneumoniae tested proteins indicated that 45 are predicted to have a T3S signal, but only 23 of these proteins were positive for translocation.
In this C. pneumoniae library, some bacterial outer membrane proteins (OMPs) or polymorphic membrane proteins (PMPs) were included, namely, CPj0020, CPj0021, CPj0278, CPj0301, CPj0441, CPj0498, CPj0595, CPj0667, CPj0795, CPj0797, CPj0798, and CPj1033. Those proteins were previously reported to be localized to the bacterial outer membrane or periplasm (30). Those proteins were detected by anti-GSK3β antibodies but not anti-phospho-GSK3β antibodies, indicating these OMPs are unlikely to access the host cytoplasm and reemphasizing the fidelity of our screening system (Table S1).
The Chlamydia genome encodes the following two major secretion systems that could deliver proteins into host cells: the Sec and a T3S system (31). Based on bioinformatic predictions with SecretomeP (http://www.cbs.dtu.dk/services/SecretomeP/) or SignalP (http://www.cbs.dtu.dk/services/SignalP/) (32), 45 of the C. pneumoniae proteins (∼11%) we expressed are predicted to harbor a T3S signal (Table S1). Interestingly, 52 of the proteins expressed in C. trachomatis that were recognized by anti-phospho-GSK3β antibodies do not possess a discernible T3S signal (32). We speculate that these proteins are either secreted through Sec or T2S systems into host cells or that current T3S prediction algorithms fail to accurately identify C. pneumoniae T3S effectors. Conversely, 22 C. pneumoniae proteins with putative T3S signals could not be detected with anti-phospho-GSK3β antibodies (Fig. 2B).
The T3S systems of Yersinia or Shigella members can translocate (16–18) some Chlamydia effectors proteins. In these heterologous secretion systems (16–18), the N-terminal amino acid sequences of 21 C. pneumoniae proteins were sufficient to enable the secretion of reporter proteins. Those 21 C. pneumoniae full-length ORFs were successfully expressed in C. trachomatis and detected with anti-GSK3β antibodies but not with anti-phospho-GSK3β antibodies, which suggested they were not secreted into host cells (Table S1). Overall, our findings suggest that there are significant differences in the recognition of T3S cargo between Yersinia/Shigella and Chlamydia species. A previous study using the N terminus of Chlamydia putative secreted proteins fused to the reporter protein β-lactamase TEM-1 in Yersinia sp. demonstrated that 23 C. trachomatis protein–β-lactamase chimeras were secreted; however, less than one-half of the full-length versions of these proteins were exported (16). It was suggested that nonsecreted proteins with potential T3S signals would fail to be translocated if such signals were masked by other activities within cells (16). Hence, we expected that our system, which uses the full-length proteins, is a more accurate measure of T3S-dependent translocation than those using only fragments of proteins.
C. pneumoniae secreted proteins associate with a range of host organelles when ectopically expressed in HeLa cells.
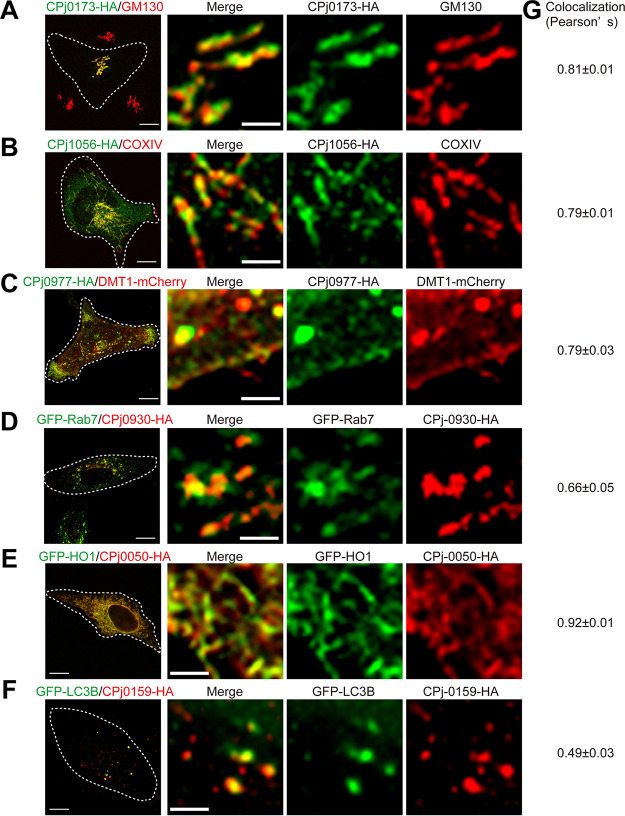
We predicted that C. pneumoniae effector proteins will bind to host factors to exert their functions, including the manipulation of host organelles to recruit lipids, redirection of membrane trafficking, or provision of nutrients to fuel bacterial replication within the inclusion. Alternatively, effectors may mediate protection from cell autonomous innate immune defenses. To gain further insights into the potential functions of translocated C. pneumoniae proteins, we tagged the 49 C. pneumoniae newly secreted proteins we identified in our screens with a C-terminal hemagglutinin (HA) epitope and expressed them in HeLa cells. Next, we assessed the subcellular localization of these proteins in transiently transfected cells. Subcellular localization was assessed with the following markers: calnexin, GFP-HO1 (endoplasmic reticulum [ER]), EEA1, GFP-Rab5, GFP-Rab7 and GFP-Rab11 (endosomes), LAMP1 (lysosomes), Golgin97 and GM130 (Golgi apparatus), COXIV (mitochondria), DMT1-mCherry (plasma membrane), GFP-LC3B (autophagosomes), and GFP-Rab8 and GFP-Rab10 (intracellular vesicles). CPj0173-HA, which has two transmembrane regions typical of inclusion membrane proteins (33), colocalized with the Golgi marker GM130 (Fig. 3A). Similarly, CPj0049, Cpj0067, Cpj0203, CPj0211, CPj0223, CPj0352, and Cpj0439 colocalized with Golgi markers. CPj0028, CPj0162, CPj0242, and Cpj0927 were partially at the Golgi and were also observed as the punctate structures at perinuclear regions (Table S1).
FIG 3.
Subcellular localization of novel C. pneumoniae effector candidates. HeLa cells were transfected with plasmids expressing C-terminally HA-tagged C. pneumoniae proteins. At 48 h posttransfection, cells were fixed and immunostained with mouse anti-HA, the Golgi marker GM130 (A), or the mitochondrial marker COXIV (B). HeLa cells were cotransfected with plasmids expressing C-terminally HA-tagged C. pneumoniae genes and plasmids expressing markers for the plasma membrane (DMT1-mCherry [C]), late endosomes (GFP-Rab7 [D]), smooth ER (GFP-HO1 [E]), or autophagosomes (GFP-LC3B [F]). Images were acquired with a Zeiss LSM880 confocal microscope. Thin scale bars = 10 μm, and thick scale bars = 2.5 μm. (G) Quantitation of colocalization by Pearson’s correlation coefficient. Numbers indicate standard error of the mean from 3 independent experiments with >15 cells per condition.
CPj1056-HA colocalized with COXIV, a component of the mitochondrial inner membrane (Fig. 3B). CPj1056 consists of 234 amino acids and has no predicted transmembrane regions based on hydropathy plots. We speculate that CPj1056 may be secreted into host cells and associate with mitochondrial outer membrane proteins (34).
CPj0977-HA (Fig. 3C) and CPj0678-HA (Fig. 4C) colocalized with the plasma membrane marker DMT1A-I, suggesting that it may perform functions related to early invasion. CPj0930-HA partially colocalized with the late endosomal marker GFP-Rab7 (Fig. 3D). Heme oxygenase (HO) is an enzyme that catalyzes the degradation of heme-producing biliverdin, ferrous iron, and carbon monoxide (35). HO forms a transient complex with NADPH cytochrome P450 reductase at smooth ER membranes (36). CPj0050-HA, CPj0064-HA, and CPj0481-HA colocalized with GFP-HO1, indicating those molecules are localized at the smooth ER (Fig. 3E; Table S1). Some CPj0159-HA-positive puncta were detected in the cytosol and colocalized with GFP-LC3 (Fig. 3F).
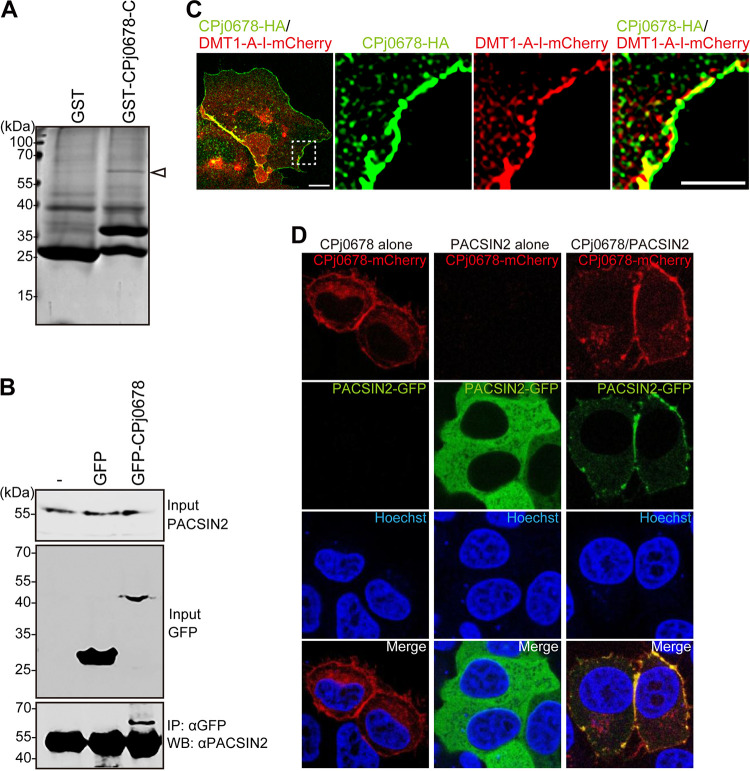
FIG 4.
CPj0678 recruits PACSIN2 on the plasma membrane through its C terminus. (A) The purified GST or GST–CPj0678-C protein and the total protein extracted from HEp-2 cells were mixed and precipitated with glutathione-Sepharose 4B beads. Eluates from the beads were subjected to SDS-PAGE and analyzed by mass spectrometry. Arrowhead indicates a band at a molecular size of approximately 60 kDa. (B) GFP or CPj0678-GFP was transfected into HEp-2 cells. Cells were lysed in lysis buffer, and the expression of PACSIN2, GFP, and GFP-CPj0678 were determined by immunoblotting with specific antibodies. The extracted protein was immunoprecipitated with anti-GFP antibodies conjugated with protein A beads and analyzed by immunoblotting using anti-PACSIN2 antibodies. The experiments were performed in triplicate. (C) CPj0678-HA and DMT1A-I were cotransfected into HeLa cells. Cells were fixed and stained with mouse anti-HA antibodies. Thin scale bar = 10 μm, and thick scale bar = 5 μm. (D) PACSIN2-GFP alone, CPj0678-mCherry alone, or both PACSIN2-GFP and CPj0678 were transfected into HEp-2 cells. Cells were fixed with 4% paraformaldehyde (PFA), and nuclei were stained with Hoechst33342. The representative image is from experiments performed in triplicate.
CPj0499-HA or CPj0268-HA appeared in both the cytoplasm and nucleus, and CPj0268-HA localized to punctate structures in the cytoplasm (see Fig. S2A and B in the supplemental material). CPj0131-HA mainly localized to the nucleus (Fig. S2C), and CPj0980-HA was diffusely distributed throughout the cytoplasm (Fig. S2D). CPj0034-HA partially colocalized with β-tubulin, suggesting that it could impact multiple cytoskeletal activities, including organelle positioning, vesicle trafficking, or centrosome organization (37) (Fig. S2E).
Of the 49 C. pneumoniae effector proteins tested, we could not unambiguously assess the localization of 9 proteins in HeLa cells (data not shown).
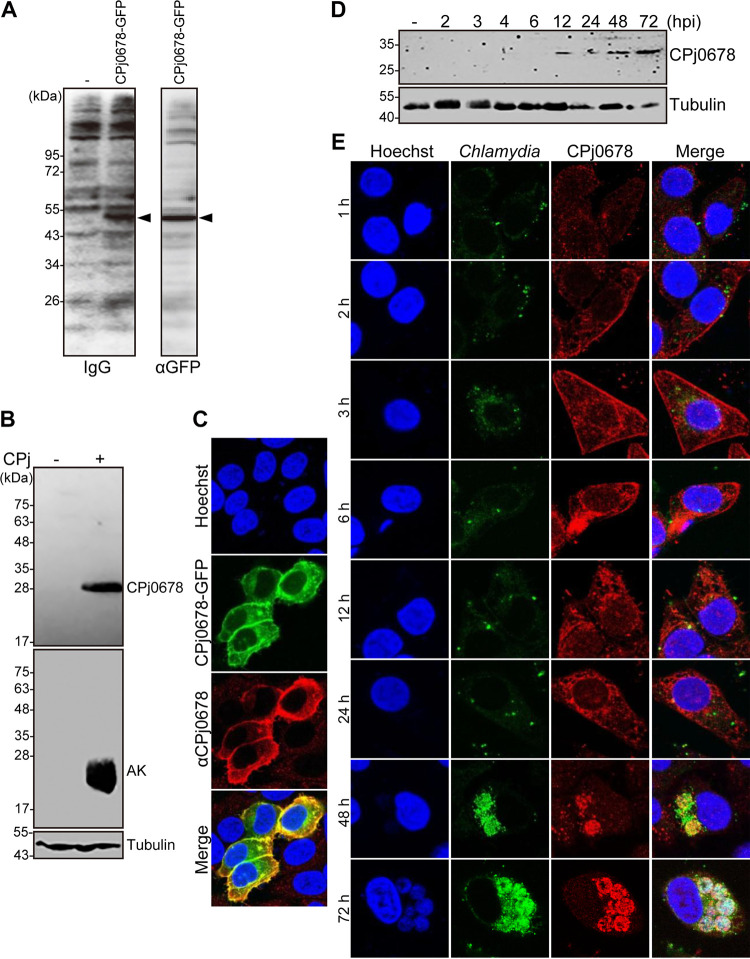
CPj0678 expression in C. pneumoniae-infected cells.
Recombinant CPj0678 expressed in yeast is recognized by antisera derived from patients who had been diagnosed with primary acute C. pneumoniae infection (Fig. 5A) (38), suggesting that CPj0678 is expressed during C. pneumoniae human infections. Because CPj0678 is restricted to C. pneumoniae and a secreted effector based on our screening system (Fig. 1C), we chose to characterize this effector in greater detail by generating specific antisera to recombinant CPj0678.
FIG 5.
The predicted CPj0678 ORF is expressed in C. pneumoniae-infected cells. (A) Serum samples from eight patients who had been clinically diagnosed with a primary acute C. pneumoniae infection specifically recognize CPj0678-GFP expressed in yeast. Total protein lysates were analyzed by immunoblotting with human antisera or anti-GFP antibodies. The predicted molecular size of CPj0678-GFP is 50 kDa (arrowhead). (B) HEp-2 cells were infected with C. pneumoniae and incubated at 37°C for 72 h. Total proteins lysates were resolved by SDS-PAGE and immunoblotted with antibodies against CPj0678, C. pneumoniae adenylate kinase, or tubulin protein. (C) HEp-2 cells were transfected with CPj0678-GFP for 48 h, fixed, and immunostained with anti-CPj0678 antibodies. (D) HEp-2 cells were infected with C. pneumoniae for the indicated times, and CPj0678 or tubulin protein expression was assessed by immunoblotting with specific antibodies. (E) HEp-2 cells were infected with C. pneumoniae for the indicated time. Cells were fixed and stained with anti-CPj0678 antibodies. C. pneumoniae was detected with FITC-labeled mouse anti-Chlamydia antibodies, and nucleic acids were stained with Hoechst 33342. Images are representative of experiments performed in triplicate.
We first tested the expression of CPj0678 in HEp-2 cells infected with C. pneumoniae J138. Anti-CPj0678 antisera recognized an immunoreactive band only in C. pneumoniae-infected cell lysates after 72 hours postinfection (hpi) (Fig. 5B). Moreover, HEp-2 cells transfected with a CPj0678-GFP expression construct and immunostained with anti-CPj0678 antibodies showed complete colocalization of both GFP fluorescence and signals from fluorescently labeled secondary antibodies (Fig. 5C), highlighting the specificity of the antisera. We next investigated at which stage of the C. pneumoniae developmental cycle CPj0678 was expressed. CPj0678 was detected from 12 to 72 hpi by immunoblot analysis (Fig. 5D). By indirect immunofluorescence staining, CPj0678 was detected by 2 to 3 hpi and localized primarily to the plasma membrane of host cells (Fig. 5E). After 6 hpi, CPj0678 shifted its localization from the plasma membrane to the cytoplasm. By 48 to 72 hpi, the CPj0678 signal in the host cytoplasm decreased and became most prominent in association with bacteria within the inclusion.
Overall, the expression of C. pneumoniae proteins in the C. trachomatis system enabled the identification of a novel secreted protein, which we validated with the use of specific antibodies.
CPj0678 recruits PACSIN2 to the plasma membrane.
To identify host protein(s) that interact with CPj0678, we expressed in Escherichia coli the C-terminal region of CPj0678 (amino acids 140 to 213) fused to glutathione S-transferase (GST) for affinity-based purification of associated mammalian proteins. Upon incubation of GST–CPj0678-C with a lysate of HEp-2 cells and affinity purification over glutathione beads, a specific band of 60 kDa was detected (Fig. 4A). Mass spectrometric analysis revealed that this 60-kDa band corresponds to PACSIN2.
To confirm the interaction between CPj0678 and PACSIN2, either GFP-tagged full-length CPj0678 (CPj0678-GFP) or GFP alone was expressed in HEp-2 cells. After cell lysis and immunoprecipitation with anti-GFP antibodies as previously described (15, 39), we determined that PACSIN2 specifically coimmunoprecipitated with CPj0678-GFP but not GFP alone (Fig. 4B). The results demonstrated that CPj0678 can associate with PACSIN2 in mammalian cells.
In our survey for the subcellular localization of C. pneumoniae effectors, we determined that transfected CPj0678-HA localized to the plasma membrane (Fig. 4C) (36). We next expressed a CPj0678-mCherry protein fusion in HeLa cells and also observed prominent localization to the plasma membrane (Fig. 4D). In contrast, C-terminally GFP-tagged PACSIN2 displayed a cytosolic distribution when expressed alone in HeLa cells (Fig. 4D). However, upon cotransfection of CPj0678-mCherry and PACSIN2-GFP, the PACSIN2-GFP signal was redistributed to the plasma membrane (Fig. 4D), further strengthening the conclusion that these proteins interact and that CPj0678 can drive the relocalization of PACSIN2.
CPj0678 is secreted by C. trachomatis and recruits PACSIN2 to the plasma membrane.
We next tested if CPj0678-myc/GSK expressed in C. trachomatis displayed similar activities to what was predicted from transfection experiments and immunolocalization of endogenous CPj0678 in C. pneumoniae-infected cells. HeLa cells were infected with C. trachomatis expressing CPj0678-myc/GSK for 24 h, and the localization of PACSIN2 and CPj0678 was assessed by indirect immunofluorescence (Fig. 6A). In uninfected cells, PACSIN2 was found in scattered puncta in the cytoplasm. In CPj0678-expressing C. trachomatis, PACSIN2 was recruited to CPj0678-myc/GSK-enriched sites at the plasma membrane in infected cells but not in cells infected with C. trachomatis alone (Fig. 6A). We confirmed the interaction between CPj0678-myc/GSK secreted by C. trachomatis and PACSIN2 by performing coimmunoprecipitations (co-IPs) with anti-myc antibodies in lysates of cells infected with C. trachomatis expressing either GFP-myc/GSK or CPj0678-myc/GSK. PACSIN2 coimmunoprecipitated with CPj0678-myc/GSK but not GFP-myc/GSK (Fig. 6B). Overall, these data demonstrate that recombinant C. pneumoniae proteins expressed in C. trachomatis can be secreted and can maintain their relevant biological activities.
FIG 6.
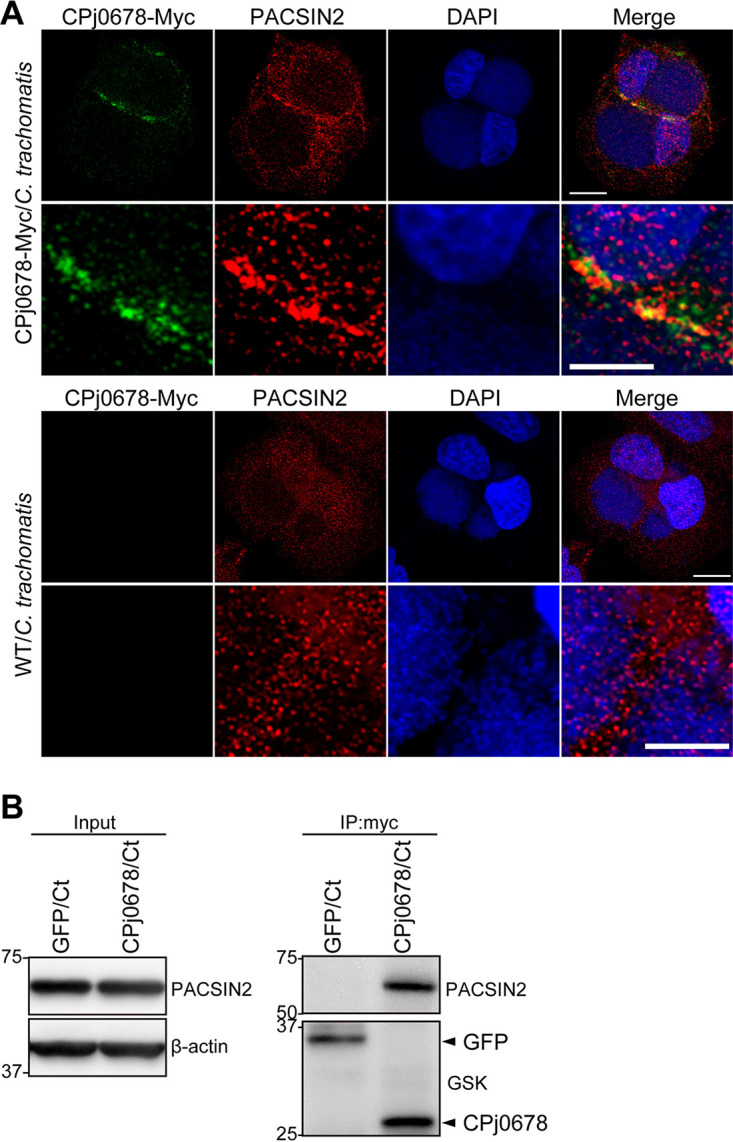
C. trachomatis expressing CPj0678 recruits PACSIN2. (A) HeLa cells were infected with C. trachomatis expressing CPj0678 (top) or wild type (WT; bottom). After a 24-h infection, cells were fixed and stained with mouse anti-Myc (green) and rabbit anti-PACSIN2 (red) antibodies. Nuclei were stained with DAPI (blue). A thin scale bar = 10 μm, and a thick scale bar = 5 μm. (B) HeLa cells were infected with C. trachomatis expressing GSK3β/Myc-tagged GFP or CPj0678. After 24 h of infection, cells were lysed in protein lysates and mixed with mouse anti-Myc antibodies at 4°C for 16 h, and associated proteins were isolated with magnetic protein G. Precipitated samples were analyzed by immunoblotting using anti-PACSIN2 or anti-GSK antibodies. Arrowheads indicate the precipitated GFP or CPj0678 protein.
DISCUSSION
Obligate intracellular bacteria like C. pneumoniae have substantially reduced genomes, with strain J138 having a 1.16-Mb chromosome (9). Approximately two-thirds of the predicted proteins are shared across species, which reflects the genetic conservation and the evolutionary constraints that are imposed by their intracellular lifestyle and shared developmental cycle. In addition, Chlamydia bacteria carry a large number of putative effector proteins, which can comprise 5% to 10% of their coding potential (18, 40). Effectors are translocated to the host cytoplasm and membranes and likely function to recruit host proteins to the inclusion membrane that promote nutrient acquisition, inhibit fusion with degradative compartments, hijack organelles and signaling pathways, and overall promote pathogen replication and dissemination. Chlamydia effectors can participate in numerous processes, including the rearrangement of the host cell cytoskeleton, regulation of membrane dynamics, centrosome tethering, lipid acquisition, and resistance to programmed cell death (41).
Putative C. pneumoniae effector proteins have been identified by leveraging heterologous expression systems (17). Most Chlamydia species, including C. trachomatis or animal isolates of C. pneumoniae, carry plasmids sized approximately 7.5 kb (42). With the generation of C. trachomatis-E. coli shuttle plasmids (24), it became feasible to express C. pneumoniae genes in C. trachomatis. The C. trachomatis and C. pneumoniae genomes are 94% conserved (30), although C. pneumoniae has ∼15% unique proteins, which likely contribute to the differences in how these two species interact with host cells (30, 43). This expanded repertoire of effectors includes a larger number of Inc and Pmp proteins (33). Recently, Shima et al. developed a genetic transformation system in C. pneumoniae which will eventually enable the direct study of virulence factors in C. pneumoniae (44).
The recognition of cargo for secretion by virulence-associated secretion systems is selective. For instance, T3S systems require not only information at the N terminus of the proteins for secretion but also binding sites for chaperones that either stabilize effectors and/or enhance their secretion (32). We reasoned that the C. trachomatis T3S would be more effective at enabling the secretion of effectors than those of Yersinia or Shigella sp.
In this study, we identified 49 C. pneumoniae-specific proteins that are translocated into host cells by C. trachomatis. Of these novel effectors, 32 proteins contained a bilobal hydrophobic domain (e.g., CPj0011 or CPj0034) that is predictive of localization to the inclusion membrane (33). Seven proteins had no predicted transmembrane domains and thus are potentially released into the host cell cytoplasm. An additional 10 proteins were predicted to have 1 transmembrane domain, and CPj0977 was predicted to have 2 by HMMTOP (http://www.enzim.hu/hmmtop/) and TMHMM server V2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
The bioinformatics tools we applied have been optimized for predicting protein secretion in a small number of model bacteria. Based on our findings in C. pneumoniae, it is clear that such predictions need to be confirmed experimentally and that bioinformatic tools can give high levels of false positives and missed many targets of T3S. Previous studies and the functional secretion screen described here indicated that nine C. pneumoniae proteins, including Tarp (CPj0572) and Cap1 (homologous to CPj0648), were secreted despite the absence of predictable T3S signals (29, 45). Moreover, even when signals at the N terminus of Chlamydia proteins permit T3S-dependent translocation of a reporter protein in Yersinia sp., they are not sufficient to enable secretion of the full-length proteins from which they were derived (16). Therefore, while bioinformatic predictions of T3S provided attractive targets for further characterization and Yersinia or Shigella sp. provide a platform for experimental validation of secretion, both approaches do not capture the complexity of secretion requirements for Chlamydia proteins. Indeed, these 49 proteins were not identified as being secreted in previous studies.
We focused our in-depth analysis on CPj0678, a protein that we had identified as immunoreactive to convalescent-phase sera from patients with C. pneumoniae infections (38), gave a robust secretion signal in our secretion screen in C. trachomatis, and did not have a predicted T3S signal bioinformatically (32). By immunofluorescence microscopy, we detected CPj0678 within 6 hpi in association with bacteria and the host plasma membrane. Later during the infection, the CPj0678 signal was exclusively in association with bacteria within inclusions, which lead us to speculate that CPj0678 is packed into EBs at the end of the developmental cycle. CPj0678 in EBs is presumably secreted early in infection and localizes to the plasma membrane of infected host cells, where it interacts with the host protein PACSIN2 (Fig. 4 and 6).
There are three isoforms of PACSIN, as follows: PACSIN1 is neuron specific (46), PACSIN2 is nearly ubiquitously expressed, and PACSIN3 is predominantly expressed in muscle tissues (47). PACSINs contain an N-terminal Bin-amphiphysin-Rvs (F-BAR; also termed extended EFC) domain and a C-terminal Src homology 3 (SH3) domain (48–50). The propensity of the F-BAR domain in PACSINs to bend plasma membranes is regulated by autoinhibition (49, 50). The F-BAR domain also mediates homo- and hetero-oligomerization of PACSIN proteins (51), and this oligomerization is important for their capacity to act as adaptor proteins, linking actin-regulatory proteins with the endocytic machinery (51). Many proteins are known to bind to the SH3 domain of PACSINs, including dynamin (52), neural Wiskott-Aldrich syndrome protein (N-WASP) (52, 53), and synaptojanin (52). The classic SH3 ligand-binding site accommodates a hydrophobic proline-rich PXXP motif and a positively charged specificity-determining residue (51–53). By binding these proteins, PACSIN2 modulates clathrin-mediated endocytosis (54), vesicle budding from the trans-Golgi network (55), the biogenesis of caveolae (56), and the formation/stabilization of microspikes at the cell surface (57) and regulates cell spreading and migration (58). CPj0678 was predicted to have six PXXP motifs which may have an important role for interacting with PACSIN2. We hypothesize that CPj0678 is injected into host cells at the early stage of infection to either regulate actin remodeling or endocytic pathways through its interaction with PACSIN2 and influence C. pneumoniae invasion.
Our data demonstrated that C. pneumoniae-specific proteins can be secreted by other Chlamydia species and provide a platform for the functional characterization of their activities. Based on the localization of expressed proteins in HeLa cells, we can generate hypotheses as to how these translocated Chlamydia proteins may function within infected host cells.
MATERIALS AND METHODS
Cell culture and infection.
HeLa (ATCC CCL-2), Vero (ATCC CCL-81), and HEp-2 (ATCC CCL-23) cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM; Sigma-Aldrich) with 10% fetal bovine serum without antibiotics at 5% CO2 and 37°C in a humidified atmosphere. C. trachomatis strain L2/434/Bu (CTL2; ATCC VR-902B) and derivatives described in this study were propagated in Vero or HeLa cells, and C. pneumoniae J138 was grown in HEp-2 cells. Cells were tested routinely for Mycoplasma contamination using either a MycoALert mycoplasma detection kit (LT07-318; Lonza) or PCR.
Plasmids.
Plasmids were amplified in E. coli JM109, DH5α, or DB3.1 and purified using the polyethylene glycol 8000 (PEG 8000) precipitation method. In brief, each plasmid was extracted from E. coli by the alkaline lysis method (59), and an aqueous nucleic acid sample was purified by precipitation with 6.5% PEG 8000 and 400 nM NaCl (60). DNA sequence verification of all plasmids was performed by using an ABI 3100 automated DNA sequencer.
To generate the Gateway-compatible C. trachomatis expression vector pEAS7-gateway, a Gateway cassette consisting of attR1, Cm, ccdB, and attR2 genes was cloned into NheI and KpnI sites of the C. trachomatis expression vector pEAS7. To reduce the host human genomic DNA contamination, C. pneumoniae genomic DNA was obtained from the EBs of the C. pneumoniae J138-infected HEp-2 cells (23). All 382 C. pneumoniae ORFs were inserted into the Gateway donor vector pDONR221 as described previously (23). To generate pEAS7-Gateway-CPj or pcDNA-gateway-HA, pDONR221-CPj was recombined into pEAS7-Gateway or pcDNA-gateway-HA using the LR clonase II reaction mix (Invitrogen).
To express human DMT1, Rab7, LC3B, and PACSIN2 or C. pneumoniae CPj0678 in mammalian cells, cDNA was cloned into a pRSET-B-mCherry vector, pEGFP-N1, or pEGFP-C1 (Clontech, Palo Alto, CA) vector.
Chlamydia trachomatis transformations.
Chlamydia transformations were performed as described previously (24) with minor modifications. Briefly, C. trachomatis LGV-L2 434/Bu was mixed with 10 to 20 μg of the plasmid of interest in CaCl2 buffer (10 mM Tris [pH 7.4] in 50 mM CaCl2) and incubated for 30 min at room temperature (RT). Then, the mix was added to a confluent monolayer of Vero cells seeded in all wells of a 6-well plate. Twelve hours postinfection, 1 IU/ml penicillin (MP Biomedicals) was added. At 40 to 48 hpi, crude lysates of infected cells were prepared by hypotonic lysis with sterile water (61) (passage 0 [P0]) and immediately used to infect a 6-well plate seeded with fresh Vero cells. At 40 to 48 hpi, the harvesting step was repeated, and the crude lysates (P1) were used to infect half of a 12-well plate seeded with Vero cells. Passages were repeated every 40 to 48 hpi with 10 IU/ml penicillin until the appearance of penicillin-resistant transformants (P6 to P10). Antibiotic was added in all passages for selection, and transformants were kept in −80°C until use. This study was approved by the Institutional Recombinant DNA and Biosafety Committee in Kawasaki Medical School.
C. pneumoniae protein expression in C. trachomatis.
To assess the levels of expression of GSK-tagged proteins, HEp-2 cells were lysed 40 to 48 h after infection in Laemmli buffer and sonicated in a Bioruptor instrument for 10 sec (Wakenyaku). The lysates were centrifuged at 10,000 × g for 10 min, and 10% to 15% of the lysate was analyzed by SDS-PAGE electrophoresis followed by immunoblotting with anti-GSK3β (1:2,000, Cell Signaling Technology) and anti-phospho-GSK3β (1:2,000) antibodies (Cell Signaling Technology).
Immunofluorescence staining and light microscopy.
HeLa or HEp-2 cells were plated onto coverslips, and 300 ng of the indicated plasmids was transfected with polyethyleneimine (PEI) HCl MAX 4000 (Polysciences, Inc.) as previously described (62). After 36 to 48 h posttransfection, cells were fixed with 4% paraformaldehyde for 20 min, permeabilized for 3 min in 0.1% to 0.2% Triton X-100, and blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Cells were incubated with primary antibodies for 2 h at room temperature. Secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 568 were incubated on cells for 60 min at room temperature.
The following antibodies were diluted: mouse anti-HA (1:1,500; MBL), rabbit anti-COXIV (1:500; Cell Signaling Technology), rabbit anti-GM130 (1:500; Cell Signaling Technology), rabbit anti-PACSIN2 (1:400; Abgent), rabbit anti-PACSIN2 (1:400; Proteintech), mouse anti-Myc (9E11) (1:4,000; Cell Signaling Technology), and rabbit anti-CPj0678 (1:50; in this study). Highly cross-absorbed heavy plus light chains (H+L) secondary antibodies (Life Technologies) conjugated to Alexa Fluor 488 or Alexa Fluor 568 were used at 1:2,000. Coverslips were mounted with Vectashield mounting medium H-1200 (Vector Laboratories).
Images were obtained using the Leica SP2 confocal laser-scanning microscope system, an Olympus BX51 fluorescence microscope, a Zeiss LSM880 confocal microscope, and a 63 ×, 1.4-numerical-aperture (NA) oil immersion objective or Olympus SpinSR10 confocal microscope and 100 × 1.4 NA oil immersion objective. Images were analyzed using Fiji (https://fiji.sc/) and presented as maximum intensity projections. Colocalization was quantified using the plugin JaCoP. Pearson’s correlation coefficient was used to quantify colocalization between fluorophores. Numbers indicate standard error of the mean from 3 independent experiments with >15 cells per condition.
Antibody generation.
Antibody generation was performed as previously described (15, 63). Briefly, the recombinant molecule of CPj0678, which was expressed in E. coli as a glutathione S-transferase (GST) fusion protein, was purified on glutathione-Sepharose (GSH) 4B beads and used to immunize Japanese white rabbits. The immunoglobulin fraction was separated by ammonium sulfate precipitation.
CPj0678-GFP expression in Saccharomyces cerevisiae.
CPj0678 ORF was cloned into a tetracycline-inducible pMT830 vector, which was constructed as previously described (23, 38). This vector system allows a protein of interest to be expressed with GFP fused to the C terminus. The vector was transformed into the Saccharomyces cerevisiae strain MTY483. Total protein was extracted in urea-SDS cracking buffer (6 M urea, 1% SDS, and 50 mM Tris HCl [pH 7.5]) buffer and analyzed by immunoblotting as previously described (23, 38).
CPj0678 expression in C. pneumoniae-infected cells.
Inclusion forming units (IFUs) were determined as previously described (64) with minor modifications. Briefly, HEp-2 cell monolayers were infected with serial dilutions of bacteria. At 36 hpi, cells were fixed with 4% formaldehyde, and inclusions were stained with fluorescein isothiocyanate (FITC)-labeled mouse anti-Chlamydia antibody (Denka Seiken) and 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Inclusions were counted using a Cellomics ArrayScan VTI HCS automated imaging system (Thermo Fisher Scientific).
HEp2 cells were infected with C. pneumoniae J138 (10 IFU/cell), and total protein was extracted at the indicated time in lysis buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, and protease inhibitor cocktail). Total protein extracts were analyzed by immunoblotting with mouse anti-human α-tubulin antibodies (1:3,000; BD Transduction Laboratories) and rabbit anti-CPj0678 antibodies (1:1,000; in this study).
For indirect immunofluorescence, HEp-2 cells were plated on coverslips and infected with C. pneumoniae J138 (multiplicity of infection [MOI] of 100 for the period 1 to 12 hpi, MOI of 5 for 24 to 72 hpi). Cells were fixed with 4% paraformaldehyde for 20 min after incubation as indicated, permeabilized for 3 min in 0.1% to 0.2% Triton X-100, and blocked with 2% BSA in PBS. Cells were incubated with anti-CPj0678 antibodies (1:50) for 2 h followed by incubation with secondary antibodies coupled to Alexa Fluor 568 for 60 min at room temperature. C. pneumoniae was stained with FITC-labeled mouse anti-Chlamydia antibody (Denka Seiken).
Glutathione affinity purification and mass spectrometric analysis.
For purification of GST or GST–CPj0678-C (amino acids 140 to 213), E. coli JM109 cells were transformed with protein-expression plasmids and grown in Luria broth (LB) with 100 μg/ml ampicillin at 37°C to an optical density at 600 nm (OD600) of 0.8; then, recombinant GST gene expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 1.0 mM for 2 h at 30°C. Cells were harvested and disrupted by sonication. Recombinant GST or GST–CPj0678-C was purified on glutathione-Sepharose (GSH) beads. HEp-2 cells were lysed in lysis buffer. After incubation for 30 min on ice, the cell suspension was centrifuged at 20,000 × g for 15 min at 4°C. The lysate, purified GST, or GST–CPj0678-C and GSH beads were mixed and incubated at 4°C overnight. GSH beads were washed with lysis buffer three times, and bound proteins were extracted with Laemmli buffer. Samples were analyzed by SDS-PAGE and stained by Coomassie R250. A band was excised from the gel and incubated with trypsin, and then the masses of tryptic fragments were determined with a Bio liquid chromatography system (Thermo Fisher Scientific) coupled to HCT-plus ion trap mass spectrometry (Bruker Daltonics, Bremen, Germany). Masses were compared with possible tryptic fragments of proteins in the GenBank database using Sequest software.
Co-IP assay.
HeLa cells were infected with CPj0678 or GFP-expressing C. trachomatis. After 24 h of incubation, cells were lysed in IP lysis buffer (20 mM HEPES [pH 7.4], 150 mM KCl, 0.5% NP-40, 1 mM EDTA, 1 mM dithiothreitol [DTT], and proteinase inhibitor) (65). One milligram of total protein prelysate was precleared by mixing magnetic Protein G beads (DB10003; Thermo Fisher) for 40 min at RT. Precleared samples were mixed with 1 μg mouse anti-Myc antibodies (9E11; Cell Signaling Technology) for 16 h at 4°C and precipitated with magnetic protein G beads for 40 min at RT. Beads were washed with P lysis buffer 4 times, and precipitated proteins were eluted in Laemmli buffer. Samples were analyzed by immunoblotting with anti-PACSIN2 and anti-GSK antibodies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grant numbers 26860223 and 22790419 to I.Y. and AI100759 to R.H.V.
The pRSET-B-mCherry vector was kindly provided by Roger Y. Tsien (University of California, San Diego, CA).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev 8:451–461. 10.1128/CMR.8.4.451-461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black CM. 1997. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin Microbiol Rev 10:160–184. 10.1128/CMR.10.1.160-184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinonen M, Saikku P. 2002. Evidence for infectious agents in cardiovascular disease and atherosclerosis. Lancet Infect Dis 2:11–17. 10.1016/s1473-3099(01)00168-2. [DOI] [PubMed] [Google Scholar]
- 4.Saikku P, Leinonen M, Tenkanen L, Linnanmäki E, Ekman MR, Manninen V, Mänttäri M, Frick MH, Huttunen JK. 1992. Chronic Chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki Heart Study. Ann Intern Med 116:273–278. 10.7326/0003-4819-116-4-273. [DOI] [PubMed] [Google Scholar]
- 5.Marchello C, Dale AP, Thai TN, Han DS, Ebell MH. 2016. Prevalence of atypical pathogens in patients with cough and community-acquired pneumonia: a meta-analysis. Ann Fam Med 14:552–566. 10.1370/afm.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72:1843–1855. 10.1128/iai.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol 18:221–245. 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- 9.Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, Shiba T, Ishii K, Hattori M, Kuhara S, Nakazawa T. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res 28:2311–2314. 10.1093/nar/28.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes H-W, Horn M, Rattei T. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog 5:e1000376. 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. 2007. Type III secretion à la Chlamydia. Trends Microbiol 15:241–251. 10.1016/j.tim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Hefty PS, Stephens RS. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol 189:198–206. 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, Worrall LJ, Hong C, Vuckovic M, Atkinson CE, Caveney N, Yu Z, Strynadka NCJ. 2018. Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Commun 9:3840. 10.1038/s41467-018-06298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Lesser CF, Lory S. 2008. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456:112–115. 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanatori I, Yasui Y, Ouchi K, Kishi F. 2015. Chlamydia pneumoniae CPj0783 interaction with Huntingtin-protein14. Int Microbiol 18:225–233. 10.2436/20.1501.01.254. [DOI] [PubMed] [Google Scholar]
- 16.da Cunha M, Milho C, Almeida F, Pais SV, Borges V, Maurício R, Borrego MJ, Gomes JP, Mota LJ. 2014. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 14:40. 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subtil A, Parsot C, Dautry-Varsat A. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol Microbiol 39:792–800. 10.1046/j.1365-2958.2001.02272.x. [DOI] [PubMed] [Google Scholar]
- 18.Subtil A, Delevoye C, Balañá M-E, Tastevin L, Perrinet S, Dautry-Varsat A. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 56:1636–1647. 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y-S, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. 2014. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betts-Hampikian HJ, Fields KA. 2010. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front Microbiol 1:114. 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia JT, Ferracci F, Jackson MW, Joseph SS, Pattis I, Plano LRW, Fischer W, Plano GV. 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect Immun 74:5645–5657. 10.1128/IAI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabuchi M, Kawai Y, Nishie-Fujita M, Akada R, Izumi T, Yanatori I, Miyashita N, Ouchi K, Kishi F. 2009. Development of a novel functional high-throughput screening system for pathogen effectors in the yeast saccharomyces cerevisiae. Biosci Biotechnol Biochem 73:2261–2267. 10.1271/bbb.90360. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani G, McCauley M, Lobley A, Bryer AJ, Wesolowski J, Greco DL, Lokareddy RK, Ronzone E, Perilla JR, Paumet F. 2019. Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA. Nat Commun 10:2747. 10.1038/s41467-019-10806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf K, Plano GV, Fields KA. 2009. A protein secreted by the respiratory pathogen Chlamydia pneumoniae impairs IL-17 signalling via interaction with human Act1. Cell Microbiol 11:769–779. 10.1111/j.1462-5822.2009.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes C, Rzomp KA, Tvinnereim A, Scidmore MA, Wizel B. 2007. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun 75:5586–5596. 10.1128/IAI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101:10166–10171. 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiwani S, Alvarado S, Ohr RJ, Romero A, Nguyen B, Jewett TJ. 2013. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains that bundle actin filaments. J Bacteriol 195:708–716. 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens R. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 21:385–389. 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 31.Chen D, Lei L, Lu C, Flores R, DeLisa MP, Roberts TC, Romesberg FE, Zhong G. 2010. Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology (Reading) 156:3031–3040. 10.1099/mic.0.040527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog 5:e1000375. 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. 2011. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics 12:109. 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derré I, Pypaert M, Dautry-Varsat A, Agaisse H. 2007. RNAi screen in Drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog 3:e155. 10.1371/journal.ppat.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maines MD. 1988. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J 2:2557–2568. 10.1096/fasebj.2.10.3290025. [DOI] [PubMed] [Google Scholar]
- 36.Yanatori I, Tabuchi M, Kawai Y, Yasui Y, Akagi R, Kishi F. 2010. Heme and non-heme iron transporters in non-polarized and polarized cells. BMC Cell Biol 11:39. 10.1186/1471-2121-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishnan GK, Splitter GA. 2012. Modulation of host microtubule dynamics by pathogenic bacteria. Biomol Concepts 3:571–580. 10.1515/bmc-2012-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasui Y, Yanatori I, Kawai Y, Miura K, Suminami Y, Hirota T, Tamari M, Ouchi K, Kishi F. 2012. Genomic screening for Chlamydophila pneumoniae-specific antigens using serum samples from patients with primary infection. FEMS Microbiol Lett 329:168–176. 10.1111/j.1574-6968.2012.02520.x. [DOI] [PubMed] [Google Scholar]
- 39.Yanatori I, Richardson DR, Imada K, Kishi F. 2016. Iron export through the transporter ferroportin 1 is modulated by the iron chaperone PCBP2. J Biol Chem 291:17303–17318. 10.1074/jbc.M116.721936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun 76:2746–2757. 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seth-Smith HMB, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 44.Shima K, Wanker M, Skilton RJ, Cutcliffe LT, Schnee C, Kohl TA, Niemann S, Geijo J, Klinger M, Timms P, Rattei T, Sachse K, Clarke IN, Rupp J. 2018. The genetic transformation of Chlamydia pneumoniae. mSphere 3:e00412-18. 10.1128/mSphere.00412-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fling SP, Sutherland RA, Steele LN, Hess B, D'Orazio SE, Maisonneuve J, Lampe MF, Probst P, Starnbach MN. 2001. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci U S A 98:1160–1165. 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plomann M, Lange R, Vopper G, Cremer H, Heinlein UA, Scheff S, Baldwin SA, Leitges M, Cramer M, Paulsson M, Barthels D. 1998. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem 256:201–211. 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- 47.Ritter B, Modregger J, Paulsson M, Plomann M. 1999. PACSIN 2, a novel member of the PACSIN family of cytoplasmic adapter proteins. FEBS Lett 454:356–362. 10.1016/s0014-5793(99)00830-3. [DOI] [PubMed] [Google Scholar]
- 48.Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. 2000. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci 113:4511–4521. [DOI] [PubMed] [Google Scholar]
- 49.Plomann M, Wittmann JG, Rudolph MG. 2010. A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing. J Mol Biol 400:129–136. 10.1016/j.jmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. 2010. Molecular basis for SH3 domain regulation of F-BAR–mediated membrane deformation. Proc Natl Acad Sci U S A 107:8213–8218. 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qualmann B, Kelly RB. 2000. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol 148:1047–1062. 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saksela K, Permi P. 2012. SH3 domain ligand binding: what’s the consensus and where’s the specificity? FEBS Lett 586:2609–2614. 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 53.Kessels MM, Qualmann B. 2002. Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J 21:6083–6094. 10.1093/emboj/cdf604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor MJ, Perrais D, Merrifield CJ. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 9:e1000604. 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kessels MM, Dong J, Leibig W, Westermann P, Qualmann B. 2006. Complexes of syndapin II with dynamin II promote vesicle formation at the trans-Golgi network. J Cell Sci 119:1504–1516. 10.1242/jcs.02877. [DOI] [PubMed] [Google Scholar]
- 56.Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. 2011. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J Cell Sci 124:2032–2040. 10.1242/jcs.086264. [DOI] [PubMed] [Google Scholar]
- 57.Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. 2010. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Lett 584:1111–1118. 10.1016/j.febslet.2010.02.058. [DOI] [PubMed] [Google Scholar]
- 58.de Kreuk B-J, Nethe M, Fernandez-Borja M, Anthony EC, Hensbergen PJ, Deelder AM, Plomann M, Hordijk PL. 2011. The F-BAR domain protein PACSIN2 associates with Rac1 and regulates cell spreading and migration. J Cell Sci 124:2375–2388. 10.1242/jcs.080630. [DOI] [PubMed] [Google Scholar]
- 59.Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira GNM, Cabral JMS, Prazeres DMF. 1999. Development of process flow sheets for the purification of supercoiled plasmids for gene therapy applications. Biotechnol Prog 15:725–731. 10.1021/bp990065. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen BD, Valdivia RH. 2012. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A 109:1263–1268. 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods 138:85–98. 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 63.Miura K, Inouye S, Sakai K, Takaoka H, Kishi F, Tabuchi M, Tanaka T, Matsumoto H, Shirai M, Nakazawa T, Nakazawa A. 2001. Cloning and characterization of adenylate kinase from Chlamydia pneumoniae. J Biol Chem 276:13490–13498. 10.1074/jbc.M009461200. [DOI] [PubMed] [Google Scholar]
- 64.Sixt BS, Bastidas RJ, Finethy R, Baxter RM, Carpenter VK, Kroemer G, Coers J, Valdivia RH. 2017. The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21:113–121. 10.1016/j.chom.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanatori I, Yasui Y, Tabuchi M, Kishi F. 2014. Chaperone protein involved in transmembrane transport of iron. Biochem J 462:25–37. 10.1042/BJ20140225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.