Abstract
The Gram-positive Bacillus methanolicus shows plasmid-dependent methylotrophy. This facultative ribulose monophosphate (RuMP) cycle methylotroph possesses two fructose bisphosphate aldolases (FBA) with distinct kinetic properties. The chromosomally encoded FBAC is the major glycolytic aldolase. The gene for the major gluconeogenic aldolase FBAP is found on the natural plasmid pBM19 and is induced during methylotrophic growth. The crystal structures of both enzymes were solved at 2.2 Å and 2.0 Å, respectively, and they suggested amino acid residue 51 to be crucial for binding fructose-1,6-bisphosphate (FBP) as substrate and amino acid residue 140 for active site zinc atom coordination. As FBAC and FBAP differed at these positions, site-directed mutagenesis (SDM) was performed to exchange one or both amino acid residues of the respective proteins. The aldol cleavage reaction was negatively affected by the amino acid exchanges that led to a complete loss of glycolytic activity of FBAP. However, both FBAC and FBAP maintained gluconeogenic aldol condensation activity, and the amino acid exchanges improved the catalytic efficiency of the major glycolytic aldolase FBAC in gluconeogenic direction at least 3-fold. These results confirmed the importance of the structural differences between FBAC and FBAP concerning their distinct enzymatic properties. In order to investigate the physiological roles of both aldolases, the expression of their genes was repressed individually by CRISPR interference (CRISPRi). The fbaC RNA levels were reduced by CRISPRi, but concomitantly the fbaP RNA levels were increased. Vice versa, a similar compensatory increase of the fbaC RNA levels was observed when fbaP was repressed by CRISPRi. In addition, targeting fbaP decreased tktP RNA levels since both genes are cotranscribed in a bicistronic operon. However, reduced tktP RNA levels were not compensated for by increased RNA levels of the chromosomal transketolase gene tktC.
Keywords: transketolase; methylotrophy; glycolysis; gluconeogenesis; CRISPR interference; fructose-1,6-bisphosphate aldolase; sedoheptulose 1,7-bisphosphate aldolase
Introduction
Bacillus methanolicus MGA3 is a Gram-positive, thermophilic, methylotrophic bacterium originally isolated from freshwater marsh soil (Schendel et al., 1990; Arfman et al., 1992). Methylotrophs such as B. methanolicus utilize reduced one-carbon compounds as their sole sources of carbon and energy (Anthony, 1991; Chistoserdova et al., 2009; Chistoserdova, 2011). MGA3 can utilize methanol via the ribulose monophosphate (RuMP) cycle (Anthony, 1991; Arfman et al., 1992), a trait that makes it a promising candidate for biotechnological applications. Bacillus methanolicus is a methylotroph with a functionally active TCA cycle and glyoxylate shunt (Heggeset et al., 2012; Müller et al., 2015a; Delépine et al., 2020), although this is unusual since some methylotrophs, including some that use the RuMP cycle, do not require a complete TCA to cover their energy needs (Chistoserdova et al., 2009). 13C-labeling experiments revealed that the TCA cycle flux was lower during growth with methanol as substrate than with mannitol or arabitol (Delépine et al., 2020).
As a facultative methylotroph, B. methanolicus MGA3 can grow on a limited substrate spectrum besides methanol: metabolic pathways for the utilization of glucose and mannitol have been described (Heggeset et al., 2012), as well as the recent characterization of its fourth carbon and energy source arabitol (López et al., 2019). The increasing interest lays, however, on the production of fuels and chemicals from methanol, with its attractiveness laying on its low cost, abundant availability, and the reduced risks of microbial contamination during industrial fermentations due to the toxicity of the derivative formaldehyde (Dijkhuizen et al., 1985; Irla et al., 2015; Müller et al., 2015a). Additionally, methanol is not in competition with conventional feedstocks used in biotechnological processes. In order to improve our understanding of B. methanolicus metabolism, and specifically its methylotrophy, full sequencing of the genome (Heggeset et al., 2012; Irla et al., 2014) together with a series of studies at the transcriptomics (Irla et al., 2015; López et al., 2019), proteomics (Müller et al., 2014), and metabolomics (Müller et al., 2015b; Carnicer et al., 2016; Delépine et al., 2020) levels have already been carried out. Furthermore, to enable genetic manipulation for industrial purposes, tools for gene expression of B. methanolicus MGA3 have been developed: rolling circle- and theta-replicating plasmids for controlled gene overexpression (Irla et al., 2016). These advances have contributed to the application of B. methanolicus MGA3 for methanol-based production of l-glutamate and l-lysine (Brautaset et al., 2007), gamma-aminobutyric acid (GABA), the five-carbon diamine cadaverine (Naerdal et al., 2015; Irla et al., 2016), and the platform chemical (R)-acetoin (Drejer et al., 2020). The most recent addition to the restricted genetic toolbox of B. methanolicus has been the application of CRISPR interference (CRISPRi) (Schultenkämper et al., 2019), a relevant tool that opens the possibility for gene knock-down studies in this organism for the first time.
CRISPR interference is a genetic perturbation technique that allows for sequence-specific repression of gene expression in bacteria, archaea and eukaryotes (Qi et al., 2013). The tool has a huge impact on strain development and physiological screening of target genes (Schultenkämper et al., 2020). The CRISPRi system only requires the co-expression of a catalytically deactivated Cas9 protein (dCas9), which has two substitutions in the nuclease domains that render it inactive, and a customizable single guide RNA (sgRNA). The dCas9-sgRNA complex binds to DNA elements complementary to the sgRNA and causes a steric block that halts transcript elongation by RNA polymerase (RNAP), resulting in the repression of the target gene (Larson et al., 2013). If the target DNA sequence belongs to the protein-coding region, the dCas9–sgRNA–DNA complex blocks the movement of the RNAP and subsequent transcription elongation. Furthermore, the CRISPRi-dCas9 technology has already been developed and established in the model organisms Escherichia coli (Elhadi et al., 2016; Cress et al., 2017; Wu et al., 2017; Fontana et al., 2018; Sander et al., 2019), Corynebacterium glutamicum (Cleto et al., 2016; Zhang et al., 2016; Gauttam et al., 2019), Bacillus subtilis (Westbrook et al., 2016, 2018; Wu et al., 2018a; Wang et al., 2019), Paenibacillus sonchi genomovar Riograndensis SBR5 (Brito et al., 2020), and recently in B. methanolicus MGA3 (Schultenkämper et al., 2019).
The genome of B. methanolicus MGA3 is composed of a circular chromosome and the two naturally occurring plasmids pBM19 and pBM69 (Heggeset et al., 2012; Irla et al., 2014). The genes present in the pBM19 plasmid are required for its methylotrophy: sequence analysis of this plasmid showed the presence of one copy of the mdh gene, encoding a methanol dehydrogenase, which was shown to be crucial for methanol consumption in this bacterium (Brautaset et al., 2004). In addition, five additional genes [pfk, encoding phosphofructokinase; rpe, encoding ribulose-5-phosphate 3-epimerase; tkt, encoding transketolase; glpX, encoding fructose-1,6-bisphosphatase; and fba, encoding fructose-1,6-bisphosphate (FBP) aldolase] with deduced roles in methanol assimilation via the RuMP assimilation pathway are present in pBM19 (Brautaset et al., 2004). FBP aldolase (FBA) is an enzyme that catalyzes the reversible reaction of the aldol FBP cleavage into the triose phosphates dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP), thus representing an important enzyme of the glycolysis and gluconeogenesis pathways.
In the genome of B. methanolicus two fba genes encode FBA: fbaC, which is located on the chromosome, and fbaP, located on the natural plasmid pBM19 (Stolzenberger et al., 2013b). Aldolases can be divided into two classes depending on their mechanism of catalysis: class I aldolases, which form a Schiff base as an intermediate, and class II aldolases, which depend on divalent metal ions (Penhoet et al., 1966; Tittmann, 2014). Bacillus methanolicus FBAs belong to the latter group, and among the divalent metal ions that affect their activities it was found that manganese and cobalt acted as activators, while copper and EDTA completely inhibited their activities (Stolzenberger et al., 2013b). Based on the amino acid sequence, class II aldolases can be further divided into type A and type B (Plaumann et al., 1997; Shams et al., 2014). Thereby, type A enzymes are dimeric in contrast with the type B enzymes, which can be dimeric, tetrameric, or octameric (Sauvé and Sygusch, 2001; Nakahara et al., 2003). Bacillus methanolicus FBAs show a tetrameric subunit structure, thus belonging to the latter type B (Nakahara et al., 2003; Izard and Sygusch, 2004; Galkin et al., 2007). Usually the homotetramer structure of aldolases consists of a triosephosphate isomerase (TIM) beta/alpha-beta fold and the fold designation is based upon eight alpha helices and eight parallel beta-strands [(α/β)8] in a closed barrel of each monomeric subunit (Dalby et al., 1999; Wierenga, 2001; Wood, 2008). This typical structure is also called TIM barrel (Figures 1, 2) and is named after the TIM, a highly conserved metabolic enzyme (Banner et al., 1975). Mobility of the active site zinc is necessary to orient the catalytic aspartyl side chain and to polarize the substrate for proton transfer from the substrate FBP (Galkin et al., 2007, 2009). Additionally, kinetic parameters allowed to distinguish the B. methanolicus FBAC as the major glycolytic aldolase and FBAP as the major gluconeogenic one (Stolzenberger et al., 2013b).
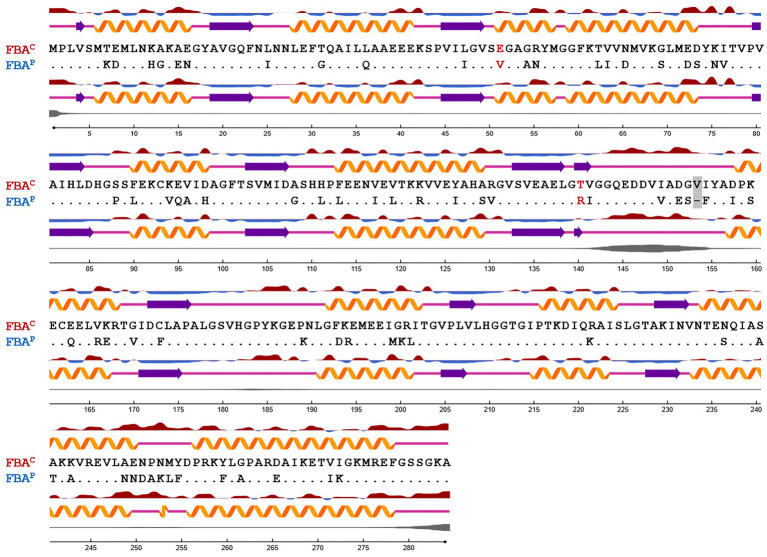
Figure 1.
Sequence alignment based on the secondary structure of FBAP and FBAC from B. methanolicus. Protein secondary structure and surface accessibility predictions were performed with NetSurfP (Petersen et al., 2009; Klausen et al., 2019). The secondary structure is depicted with orange α-helices, purple β-strands, or coils with pink lines, respectively. Protein surface accessibility is shown by red (exposed) or blue (buried) waves, thresholder at 25%. Dots are representing identical amino acids. Amino acid residues exchanged by SDM are highlighted in red. A gray box depicts a gap in the amino acid sequence of FBAP in comparison to FBAC.
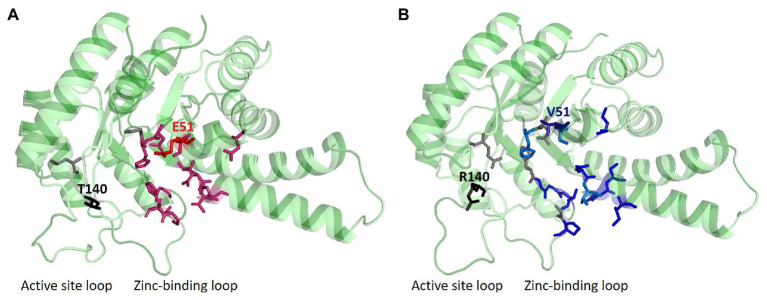
Figure 2.
Ligand and structure prediction of FBAC and FBAP. (A) 3D model of the FBAC showing the protein structure, which was depicted with PyMOL (DeLano, 2002), and predicted ligands via COACH-D (Wu et al., 2018b) analysis. (B) 3D protein model of FBAP. In gray, residues that are important for the zinc-binding are shown; while in red residues that are necessary for binding the substrate FBP are plotted. Blue residues are representing important sites for GAP or DHAP interaction. Sites for SDM are marked in the 3D structures of the protein models (E51V, T140R). The 3D structure of the protein models indicates typical triosephosphate isomerase (TIM) barrels.
In this study, crystal structures of both enzymes were obtained, at resolutions of 2.2 Å and 2.0 Å, respectively, which allowed identifying differences at crucial residues: glutamic acid (E) at position 51 and threonine (T) at position 140 were identified in FBAC while, on the other hand, valine (V) at position 51 and arginine (R) at position 140 were identified in FBAP (Figures 1, 2). The glutamic acid residue at position 51 (E51) is located at the FBP-binding site of FBAC, the FBP-cleaving major glycolytic FBA. Thus, exchanging the amino acid residues at position 51 from glutamic acid to valine (E51V) should affect the binding affinity of FBP (KM value) as well as the glycolytic specific activity (Vmax). Position 140 coordinates zinc-binding, and when no substrate is around (apoenzyme), conformational changes near the zinc atom transform the apoenzyme into a holoenzyme, a crucial step for catalysis. The cofactor zinc coordinated in such a manner is involved in DHAP catalysis (Jacques et al., 2018). It is known that DHAP enters first into the catalytic centrum. Class II aldolases catalyze the aldol reaction by making use of divalent zinc or cobalt metal ions that is suggested to polarize the DHAP (or FBP in glycolysis) carbonyl group and stabilize the carbanion intermediate during catalysis (Belasco and Knowles, 1983; de la Paz Santangelo et al., 2011; Jacques et al., 2018). Hence, changing the position 140 from T to R should affect the gluconeogenic Vmax and the KM of DHAP as well as the glycolytic Vmax and KM of FBP.
To test these hypotheses and to gain further knowledge about the catalytic mechanisms of the B. methanolicus FBAs, the crystal structures of both FBAC and FBAP were solved and site-directed mutagenesis (SDM) was performed in order to exchange identified residues suggested to be crucial to interchange activities of FBAC and FBAP. Both native enzymes and generated SDMs were examined in two different enzyme assays to test glycolysis and gluconeogenesis reactions. Moreover, gene expression analyses of fbaC and fbaP were performed with the help of the recently developed CRISPR interference technique in B. methanolicus and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis.
Materials and Methods
Structure Determination and Analysis
Amino acid sequences from B. methanolicus FBAC (EU83897, AIE61578, and WP_03346852) and FBAP (EIJ77593, AIE61790, and WP_003349819) from the National Center of Biotechnology Information (NCBI) were used for a multiple amino acid sequence alignment with BLASTp (Altschul et al., 1990) and their secondary structures and protein surface accessibility were predicted using NetSurfP (Petersen et al., 2009; Klausen et al., 2019). PyMOL files of the distinct FBAC and FBAP were computationally generated with PHYRE2 (Kelley et al., 2015). Sequences in FASTA format of FBAC and FBAP from B. methanolicus were used as input and the output file was analyzed with COACH-D (Wu et al., 2018b) to gain insights concerning the important residues for ligand binding. Finally, PyMOL (the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC; DeLano, 2002) was used for the depiction of the protein structures and the crucial residues for binding the substrates or zinc as a cofactor.
To obtain the crystal structures from the native B. methanolicus FBAs, the proteins were first purified. Consequently, E. coli strain BL21(DE3) was used for protein production (Studier and Moffatt, 1986). After a 4 h induction with 100 μM isopropyl-β-d-1-thio-galactopyranoside (IPTG), cells were harvested by centrifugation (5,000 × g, 10 min). After sonication (2 × 1 min) on ice and centrifugation (30,000 × g, 20 min, 4°C), the His-tagged enzymes were purified by IMAC column (GE Healthcare) process. Protein samples were stored in Tris-EDTA buffer pH 8. Proteins were crystallized by sitting-drop vapor diffusion. Crystals for FBAC were obtained at 20°C in a reservoir containing 0.1 M sodium acetate trihydrate pH 4.5, 2.0 M ammonium sulfate, and a ratio of protein and reservoir with 25 and 75%, respectively. FBAP crystals grew at 8°C with 50% protein and 50% reservoir containing 20% PEG 4000, 0.2 M imidazole/malate buffer at pH 6. Crystals were flash cooled in liquid nitrogen for storage and data collection. X-ray diffraction data of FBAC were collected in-house at 100 K on a Rigaku MicroMax 007HF rotating anode X-ray generator equipped with amar research mar345dtb image plate detector. Data of FBAP were collected on beamline X06DA with PILATUS 2 M-F detector (Dectris) at Swiss Light Source (Villigen, Switzerland). The FBA structures were solved by molecular replacement with MOLREP (Vagin and Teplyakov, 2010) using aldolase from Bacillus anthracis (PDB ID code 3Q94; Tan et al., 2011) as search model. Improvement of the initial model was carried out in cycles of refinement with REFMAC5 (Murshudov et al., 2011) and phenix.refine (Adams et al., 2010; Afonine et al., 2012), and manual rebuilding was done in COOT (Emsley and Cowtan, 2004). Data collection and refinement statistics are summarized in Supplementary Table S5.
Strains, Media, and Culture Conditions
E. coli DH5α (Hanahan, 1983) was used as the standard cloning host. The strains used in this study are listed in Supplementary Table S1. E. coli strains were routinely cultivated at 37°C and 180 rpm in Lysogeny Broth (LB) medium or on LB agar plates supplemented with antibiotics (100 μg ml−1 ampicillin, 25 μg ml−1 chloramphenicol, and 50 μg ml−1 kanamycin) and 0.5 mM IPTG when necessary. B. methanolicus was used as expression host and cultivated at 50°C and 200 rpm in minimal MVcMY media as described in Brautaset et al. (2003) with 200 mM methanol and with 5 μg ml−1 chloramphenicol. For CRISPR interference, 12.5 mM mannitol was added as an inducer to the media to drive dCas9 gene expression when required. Recombinant B. methanolicus strains were routinely plated on SOB agar plates with 5 μg ml−1 chloramphenicol. Main cultures of all B. methanolicus experiments were inoculated at a start optical density (OD600) of 0.1. Cultivations were performed in 500 ml baffled shake flasks with 50 ml media volume and in biological triplicates.
Recombinant DNA Work
Molecular cloning was performed as described (Sambrook, 2001) using primer sequences listed in Supplementary Table S2. Total DNA isolation from B. methanolicus was performed following the indications of Eikmanns et al. (1994). Inserts were amplified by PCRs with ALLin™ HiFi DNA Polymerase (HighQu, Kraichtal, Germany) and purified using the NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany). CRISPRi Plasmids were constructed using the isothermal DNA assembly method (Gibson et al., 2009; Gibson, 2011) with generated fragments by Annealing Oligo method (using the primer pairs for tfbaC and tfbaP, Supplementary Table S2), and the piCas vector cut with restriction enzymes (New England Biolabs, Ipswich, United Kingdom) as described in Schultenkämper et al. (2019). For plasmid isolation, the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Waltham, United States) was used. Transformation of chemically competent E. coli cells was performed following the procedure described by Mandel and Higa (1970). Plasmids were transformed by polyethylene glycol-mediated procedure into B. methanolicus cells (Cue et al., 1997). FBA mutants were generated by SDM with PfuTurbo™ DNA Polymerase (Agilent, Böblingen, Germany). The generated PCR products were subsequently incubated with DpnI to digest the parental non-mutated template. FBAC; E51V and FBAC; T140R were generated by backbone amplification of pET16b-fbaC using primer pairs SDM1_fwd and SDM1_rev and SDM2_fwd and SDM2_rev, respectively, while FBAP; V51E and FBAP; R140T were generated by backbone amplification of pET16b-fbaP using primer pairs SDM3_fwd and SDM3_rev and SDM4_fwd and SDM4_rev, respectively (Supplementary Table S3). The double mutants FBAC; E51V, T140R and FBAP; V51E, R140T were constructed based on the single mutants FBAC; E51V and FBAP; R140T, respectively. The amino acid sequences are listed in Supplementary Table S4. A detailed description of each performed mutation, the respective plasmid templates and primer pairs used can be found in Supplementary Table S3. All cloned DNA fragments were verified by sequencing (Sequencing Core Facility, Bielefeld University).
CRISPRi Targeting of fbaC and of fbaP
Our recently developed piCas plasmid system (Schultenkämper et al., 2019) was used to target fbaC and fbaP in B. methanolicus. The piCas plasmid was linearized with AvaI and XbaI double restriction and with help of the CRISPy webtool (Blin et al., 2016) the most efficient target sequence (antisense strand) was chosen for fbaC and fbaP (Supplementary Table S2) as described previously (Schultenkämper et al., 2019). The plasmids piCas-tfbaC and piCas-tfbaP were transformed as described above into B. methanolicus, and cells were then cultivated in MVcMY media with 200 mM methanol and 12.5 mM mannitol for the induction of the dCas9 system when needed. When the cells reached an OD600 of 1.0, they were harvested and prepared based on the protocol described by Brautaset et al. (2004) and used for further enzymatic assays and RNA isolation.
Protein Overproduction and Purification
Plasmids for the production of N-terminal decahistidine-tagged FBA proteins using E. coli BL21 (DE3) were previously constructed based on pET16b (Novagen, Madison, WI, United States; Stolzenberger et al., 2013a) and subsequently used in this work for SDM experiments. The already available pET28a-fbp plasmid was used for the production of the N-terminal His-tagged GlpX from C. glutamicum (Rittmann et al., 2003a), which was required as a coupling enzyme in several of the enzymatic assays performed. The plasmids used in this study are listed in Supplementary Table S1. Protein production and purification was performed following the procedure described by Lindner et al. (2007) except for cell lysis, which was performed by sonication (UP 200 S, Dr. Hielscher GmbH, Teltow, Germany) on ice at an amplitude of 55% and a duty cycle of 0.5 for 8 min. Supernatants were subsequently filtered using a 0.2 μm filter and purified by nickel affinity chromatography with nickel-activated nitrilotriacetic acid-agarose (Ni-NTA; Novagen, San Diego, CA, United States). GlpX and the distinct FBA variants eluted with 20 mM Tris, 300 mM NaCl, 5% (vol/vol) glycerol, and 50, 100, 200, or 400 mM imidazole were analyzed by 12% SDS-PAGE (Laemmli, 1970). Fractions showing the highest protein concentrations (i.e., eluted with 100 and 200 mM imidazole) were pooled and buffered in 50 mM Tris-HCl (pH 7.5) using Vivaspin® columns (10,000 MW, Sartorius, Göttingen, Germany). After purification, the His-tag was cleaved using the Factor Xa Cleavage Capture Kit (Novagen, San Diego, CA, United States) following the manufacturer’s recommendations (Supplementary Figure S1). Protein concentration was measured according to the Bradford method (Bradford, 1976) using bovine serum albumin (BSA) as a reference. The purified proteins were subsequently applied for enzymatic assays.
Enzyme Activity Assays
Activity measurements of the different FBA variants in the glycolytic and gluconeogenic directions were performed following the indications of Stolzenberger et al. (2013b). Determination of the FBA activity in the direction of FBP cleavage was performed using an NADH-linked enzyme assay with the coupling enzyme α-glycerol 3-phosphate dehydrogenase (G3PDH) from rabbit muscle (Sigma-Aldrich, St. Louis, MO, United States) and FBP as substrate. The assay mixture contained 50 mM Tris-HCl buffer (pH 7.5), 0.25 mM NADH, 5 U G3PDH, and 0.05–20 mM FBP to initiate the reaction and varying concentrations of the FBA variants, which were pre-warmed for 4 min at 50°C, in a total volume of 1 ml. The oxidation rate of NADH was monitored at 340 nm and 50°C for 3 min using a Shimadzu UV-1202 spectrophotometer (Shimadzu, Duisburg, Germany). Determination of the FBA activity in the direction of FBP synthesis toward 6-phosphogluconolactone (6PGL) was performed by an NADPH-linked enzyme assay with the coupling enzymes phosphoglucoisomerase (PGI) from Saccharomyces cerevisiae (Sigma-Aldrich, St. Louis, MO, United States), glucose-6-phosphate dehydrogenase (G6PDH) from Leuconostoc mesenteroides (Sigma-Aldrich, St. Louis, MO, United States), purified GlpX (FBPase) from C. glutamicum, and DHAP and GAP as substrates. The assay mixture contained 50 mM Tris-HCl buffer (pH 7.5), 0.25 mM NADP+, 5 U PGI, 5 U G6PDH, 1.5 U GlpX, 50 mM KCl, 2 mM MnCl2, and 0.5–5 mM DHAP and GAP to initiate the reaction and varying concentrations of the FBA variants, which were again pre-warmed for 4 min at 50°C, in a total volume of 1 ml. In order to determine the kinetic parameters of GAP and DHAP, one of the substrates’ concentration was left constant while the other substrate concentration was varied (Michaelis et al., 2011). The reduction rate of NADP+ was monitored at 340 nm and 50°C for 3 min using a Shimadzu UV-1202 spectrophotometer (Shimadzu, Duisburg, Germany).
Gene Expression Analysis Using qRT-PCR
Total RNA was isolated from B. methanolicus MGA3(piCas-tfbaC) and MGA3(piCas-tfbaP) cells growing exponentially (at OD600 = 1.0) in MVcMY medium containing 200 mM methanol and 12.5 mM mannitol to induce dcas9 expression. The pellets were stored at −80°C. For the RNA isolation, the pellets were thawed on ice, and the samples were homogenized by resuspending the cells in 100 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8) containing 5 mg ml−1 lysozyme. After an incubation step at 37°C for 30 min, total RNA was extracted using NucleoSpin® RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. Thereafter, RNA samples were treated with DNase digestion using RNase-free DNase Set and RNeasy MinElute kits (Qiagen, Hilden, Germany) to eliminate possible genomic DNA contamination. Furthermore, quality control was performed in order to determine the purity and integrity of isolated RNA. A series of PCRs were performed using Taq polymerase (New England Biolabs) amplifying regions of sizes between 1,000 and 2,000 bp. Additionally, RNA concentration was measured using a spectrophotometer (NanoDrop®, ND-1000). Fifty nanograms of each sample were used to perform cDNA synthesis. All qRT-PCRs were performed according to the manufacturer’s instructions using the SensiFAST™ SYBR® No-ROX One-Step Kit (Bioline, London, United Kingdom) and the CFX96 cycler system (Bio-Rad, Hercules, United States). The temperature profile was (1) 45°C for 10 min (reverse transcription); (2) 95°C for 2 min; (3) 40 cycles of 95°C for 5 s, 55°C for 10 s, and 72°C for 5 s; and (4) melt curve analysis with measures between 65 and 95°C. The ΔΔCt method was used in calculations (Higuchi et al., 1992; Pfaffl, 2001; Bustin et al., 2009) with the reference gene parA (Plasmid partition protein A) and empty vector as control. For each sample, three independent qRT-PCR experiments were performed.
Results
Structural Comparison of Native FBAC and FBAP
The B. methanolicus class II type B enzymes FBAP and FBAC that share 75% identical amino acids differ considerably regarding their kinetic parameters (Stolzenberger et al., 2013b). To gain insight into the structure-function relationship of these enzymes, they were crystallized, and their structures were solved to 2.2 Å and 2.0 Å, respectively. Secondary structure elements are mapped onto the amino acid sequence in Figure 1. Approximately, 46% of the residues in the model are in helical conformations and 16% are in β strands. This can also be observed in the overall fold of the FBA protomer structure in Figure 2. The tetrameric FBA is composed of two pairs of dimers. The core of the structure consists of an eight-stranded parallel β-strand, twisted of strand–helix–strand winds, and building the typical barrel structure. The residues E51 and V51, in FBAC and FBAP respectively, were proposed to play an important role in recognition of the substrate FBP. The crystal structures show that this residue is located at the N-terminus of strand β3 of one subunit, directed into the middle of the TIM barrel with FBP bound at the active site of the partner subunit. It is assumed that the DHAP component will therefore be positioned at the other end of the active site near the catalytic zinc ion (Figure 2, gray and black residues; Cooper et al., 1996). The residues T140 and R140, of FBAC and FBAP, respectively, are in the active center. Since the residues differ at positions 51 and 140 between FBAP and FBAC they may play a role in their different kinetic parameters. Therefore, SDM was performed, and three variants of each enzyme were generated and purified: FBAC; E51V, FBAC; T140R, FBAC; E51V, T140R, FBAP; V51E, FBAP; R140T, and FBAP; V51E, R140T (Supplementary Table S3). His-tagged proteins were purified from recombinant E. coli and used for enzymatic characterization after His-tag cleavage.
Influence of Amino Acid Residues 51 and 140 of FBAP and FBAC on Their Glycolytic Aldol Cleavage Activities
In order to test if changing the amino acid residues relevant for FBP and zinc ion binding negatively affect the glycolytic aldol cleavage activity of the major glycolytic aldolase FBAC, the mutants FBAC; E51V, FBAC; T140R, and FBAC; E51V,T140R were analyzed. The amino acid exchange E51V did not increase the KM value for FBP (i.e., decrease in affinity), while the change T140R and changing both residues increased the KM value for FBP two- to three-fold (Table 1). The kcat was reduced about 2-fold when changing these residues individually, but the effect was not additive (Table 1). In comparison to the native aldolase FBAC, the double mutant FBAC; E51V, T140R showed about 6 to 7-fold reduced catalytic efficiency for FBP cleavage. Compared to the major gluconeogenic aldolase FBAP, the double mutant showed a comparable kcat, but a 3-fold lower KM value for FBP. Thus, exchanging the two amino acid residues E15 and T140 relevant for FBP and zinc ion binding in FBAC were almost sufficient to reduce its glycolytic aldol cleavage activity to that of the native major gluconeogenic aldolase FBAP.
Table 1.
Kinetic parameters [fructose-1,6-bisphosphate (FBP) cleavage] of native FBAs and site-directed mutagenesis (SDMs) in glycolysis.
| Sample | Vmax (U mg−1) | KM (FBP, mM) | kcat (s−1) | Catalytic efficiency (s−1 mM−1) |
|---|---|---|---|---|
| FBAC | 4.2 | 0.3 | 2.1 | 6.9 |
| FBAC; E51V | 2.8 | 0.2 | 1.4 | 7.2 |
| FBAC; T140R | 2.4 | 0.7 | 1.3 | 1.8 |
| FBAC; E51V,T140R | 2.2 | 1.0 | 1.1 | 1.1 |
| FBAP | 1.8 | 3.4 | 1.0 | 0.3 |
| FBAP; V51E | 0.5 | 5.8 | 0.3 | 0.1 |
| FBAP; R140T | 0.4 | 4.7 | 0.2 | <0.1 |
| FBAP; V51E, R140T | 0.01 | <0.01 | <0.01 | <0.01 |
In order to test if changing the amino acid residues relevant for FBP and zinc ion binding increases glycolytic aldolase activity of the major gluconeogenic aldolase FBAP, the mutants FBAP; V51E, FBAP; R140T, and FBAP; V51E, R140T were analyzed. Surprisingly, these changes led to a complete loss of FBP aldol cleavage activity rather than increasing it (Table 1). It has to be noted that the proteins are known to be active since they showed activity in the reverse gluconeogenic direction (see below). Thus, exchanging these amino acids in the FBAP backbone was not sufficient to increase its FBP aldol cleavage activity.
Influence of Amino Acid Residues 51 and 140 of FBAP and FBAC on Their Gluconeogenic Aldol Condensation Activities
Coupled enzyme activity assays were performed with the purified native enzymes and their SDM variants to quantify aldol condensation of GAP and DHAP to FBP. The coupled activity assay to determine gluconeogenic FBP synthesis from GAP and DHAP required three helping enzymes. While PGI from S. cerevisiae and G6PDH from L. mesenteroides are available commercially, GlpX had to be purified from C. glutamicum. It showed a specific GlpX activity of 6.9 ± 0.6 U mg−1 at 50°C and was used to assay FBP synthesis from GAP and DHAP.
In order to test if the gluconeogenic aldol condensation activity of the major gluconeogenic aldolase FBAP is negatively affected upon changing the amino acid residues relevant for FBP and zinc ion binding, the mutants FBAP; V51E, FBAP; R140T, and FBAP; V51E, R140T were analyzed. The amino acid exchange in the FBP binding site (V51E) resulted in an increased KM value for GAP as substrate, while that for the substrate DHAP was in a much lower range (Table 2). The kcat values were reduced about 2-fold for both substrates as a consequence of the R140T exchange in the zinc ion binding site in the active center (Table 2). It has to be noted that the FBAP variant with both amino acid exchanges (FBAP; V51E, R140T) was active in the gluconeogenic direction at a rate similar to the major glycolytic aldolase FBAC (Table 2), while no significant activity in the glycolytic direction could be detected for this mutant (Table 1).
Table 2.
Kinetic Parameters (FBP synthesis) of native FBAs and SDMs in gluconeogenesis.
| GAP as substrate | DHAP as substrate | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Vmax (U mg−1) | KM (mM) | kcat (s−1) | Catalytic efficiency (s−1 mM−1) | Vmax (U mg−1) | KM (mM) | kcat (s−1) | Catalytic efficiency (s−1 mM−1) |
| FBAC | 2.9 | 0.5 | 1.5 | 3.0 | 3.6 | 0.2 | 1.8 | 9.0 |
| FBAC; E51V | 10.1 | 0.2 | 5.2 | 25.8 | 21.8 | 3.3 | 11.1 | 3.4 |
| FBAC; T140R | 8.9 | 0.2 | 4.5 | 22.7 | 6.1 | 0.5 | 3.1 | 6.2 |
| FBAC; E51V, T140R | 9.0 | 0.2 | 4.6 | 23.0 | 5.6 | 0.1 | 2.9 | 28.6 |
| FBAP | 4.8 | 0.3 | 2.6 | 8.6 | 4.0 | 0.5 | 2.1 | 4.3 |
| FBAP; V51E | 2.9 | 1.0 | 1.6 | 1.6 | 4.1 | 0.8 | 2.2 | 2.7 |
| FBAP; R140T | 2.5 | 0.4 | 1.3 | 3.3 | 2.2 | 0.3 | 1.2 | 3.9 |
| FBAP; V51E, R140T | 2.1 | 0.3 | 1.1 | 3.7 | 2.7 | 0.3 | 1.4 | 4.8 |
In order to test if the gluconeogenic aldol condensation activity of the major glycolytic aldolase FBAC can be increased by changing the amino acid residues relevant for FBP and zinc ion binding, the mutants FBAC; E51V, FBAC; T140R, and FBAC; E51V,T140R were analyzed. The amino acid exchange in the FBP binding site (E51V) reduced the KM value for the substrate GAP about 2-fold but increased that for the substrate DHAP more than 10-fold (Table 2). This amino acid exchange also increased the kcat values 3- and 6-fold for GAP and DHAP, respectively (Table 2). The amino acid exchange T140R also increased the kcat values about 2- and 3-fold (Table 2).
The FBAC double mutant FBAC; E51V, T140R showed catalytic efficiencies of 28.6 and 23.0 s−1 mM−1 for GAP and DHAP, respectively. Thus, the combined introduction of the amino acid exchanges E51V and T140R into the major glycolytic aldolase FBAC improved the catalytic efficiencies for the substrates GAP and DHAP about 7- and 3-fold, respectively. Notably, the catalytic efficiencies of FBAC; E51V, T140R also exceeded those of FBAP, the major native gluconeogenic aldolase of MGA3.
Repression of fbaC and fbaP by CRISPR Interference
In order to characterize the roles of FBAC and FBAP for B. methanolicus MGA3, expression of their genes was repressed individually by CRISPRi, and the effects on growth, on aldolase enzyme activity in crude extracts, and fbaC and fbaP RNA levels were determined. Cultivation of MGA3(piCas-tfbaC) and MGA3(piCas-tfbaP) revealed no growth deficit upon dcas9 induction with mannitol (Supplementary Figure S2). A dcas9 specific qRT-PCR analysis showed that dcas9 expression was induced upon mannitol addition (Figures 3A,B).
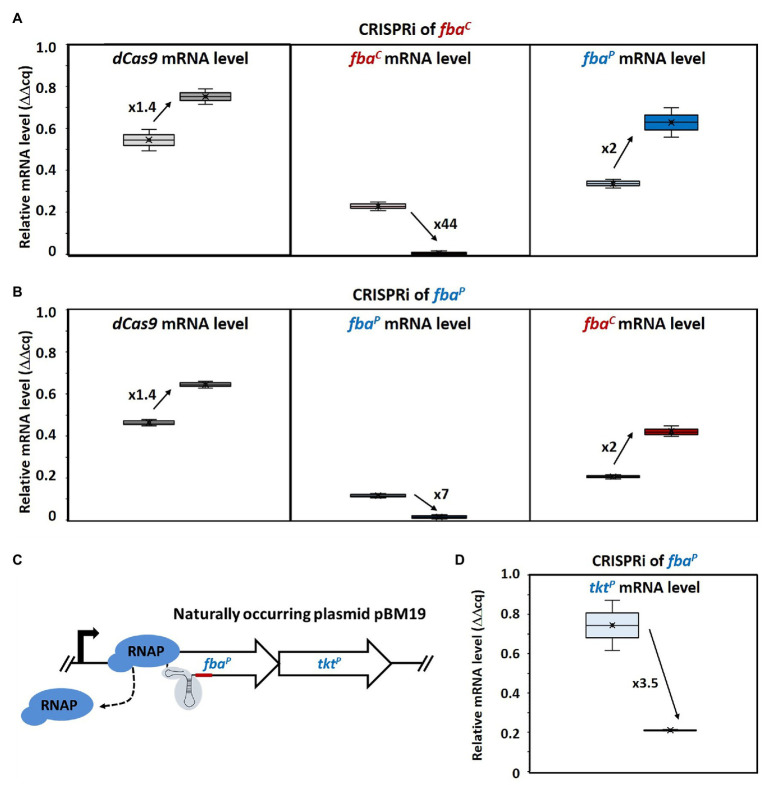
Figure 3.
Changed gene expression levels due to dCas9 targeting fbaC and fbaP and polar effect on tktP by CRISPRi targeting of fbaP. Relative mRNA levels of dCas9 (left), fbaC (middle), and fbaP (right) in MGA3(piCas-tfbaC) (A) and relative mRNA levels of dCas9 (left), fbaP (middle), and fbaC (right) in MGA3(piCas-tfbaP) (B) are shown by Box and Whisker plots as detected by qRT-PCR. Schematic representation of CRISPRi targeting fbaP, which is cotranscribed with tktP as bicistronic operon on the natural plasmid pBM19 of B. methanolicus MGA3 (C). CRISPR interference by blocking transcription elongation by RNA polymerase (RNAP; blue) is shown. qRT-PCR analysis of tktP mRNA levels upon CRISPRi targeting of fbaP in MGA3(piCas-tfbaP) (D). Black arrows with adjacent numbers indicate up- or downregulation without (lighter color, left) and with (darker color, right) CRISPRi induction.
Consequently, mRNA levels of the targeted fbaC gene in MGA3(piCas-tfbaC) were 44-fold lower when mannitol was added as an inducer (Figure 3A). Surprisingly, increased mRNA levels (2-fold) of the plasmid-encoded gene fbaP were also observed in this strain. Vice versa, in strain MGA3(piCas-tfbaP), the mRNA levels of the targeted fbaP were reduced 7-fold upon induction whereas the mRNA level of the chromosomal fbaC was 2-fold higher (Figure 3B). This indicated a compensatory effect at the mRNA level. We aimed then at determining how this compensatory effect at the mRNA level would affect enzyme activity levels. To this end, aldolase enzyme assays with crude extracts of MGA3(piCas-tfbaC) and MGA3(piCas-tfbaP) were performed in the FBP cleavage direction. It has to be noted that assaying crude extracts for fructose bisphosphate aldolase activity will yield the sum of both FBAC and FBAP activities. The aldolase enzyme activity detected in crude extracts did not change for strain MGA3(piCas-tfbaP) under induced as compared to non-induced conditions, while the activity detected was higher upon induction for strain MGA3(piCas-tfbaC) (Table 3). This confirmed a compensatory effect, and in the case of strain MGA3(piCas-tfbaC), an overcompensation was observed. Since FBAC and FBAP differ in their affinity for FBP, the crude extracts were analyzed to derive an estimate of the KM value for FBP (Table 3). The KM value estimated using the crude extracts of strain MGA3(piCas-tfbaC) increased by 20-fold upon induction. This may be explained if repression of fbaC reduced the levels of the low-KM aldolase FBAC (KM value of 0.3 mM for FBP for the purified enzyme) and if compensatory induction of fbaP yielded higher levels of the high-KM aldolase FBAP (KM value of 3.4 mM for FBP for the purified enzyme).
Table 3.
FBA (FBP cleavage) activities in crude extracts of Bacillus methanolicus MGA3(piCas-tfbaC) and MGA3(piCas-tfbaP) without (−) and with (+) induction of CRISPR interference (CRISPRi).
| Strain | Induction | Vmax (U mg−1) | KM (FBP, mM) |
|---|---|---|---|
| MGA3(piCas-tfbaC) | − | 0.1 ± 0.03 | 0.1 ± 0.01 |
| MGA3(piCas-tfbaC) | + | 0.3 ± 0.04 | 2.0 ± 0.05 |
| MGA3(piCas-tfbaP) | − | 0.1 ± 0.02 | 0.2 ± 0.02 |
| MGA3(piCas-tfbaP) | + | 0.1 ± 0.03 | 0.2 ± 0.03 |
CRISPR interference of a gene in an operon may affect the expression of downstream genes. Since fbaP is encoded in a bicistronic operon on the naturally occurring plasmid pBM19 in B. methanolicus and cotranscribed with transketolase gene tktP (Figure 3C), tktP mRNA levels were determined in the MGA3(piCas-tfbaP) strain targeting fbaP. Indeed, a polar effect on tktP was observed when fbaP was targeted by CRISPRi since the tktP mRNA levels were reduced by almost 4-fold (Figure 3D).
B. methanolicus possesses a second transketolase gene encoded on its chromosome by the monocistronic gene tktC. To examine if the expression of the transketolase genes may also be regulated by compensation as observed for the aldolase genes fbaC and fbaP, tktP and tktC were subjected to CRISPRi targeting (Figures 4A,B). CRISPRi targeting tktC repressed tktC almost 2-fold, while qRT-PCR analysis revealed an unchanged mRNA level of tktP (Figure 4A). Similarly, CRISPRi targeting tktP did not affect tktC mRNA levels, but reduced tktP mRNA levels more than 2-fold (Figure 4B). Thus, a compensatory regulation at the mRNA level was not observed for the transketolase genes tktC and tktP.
Figure 4.
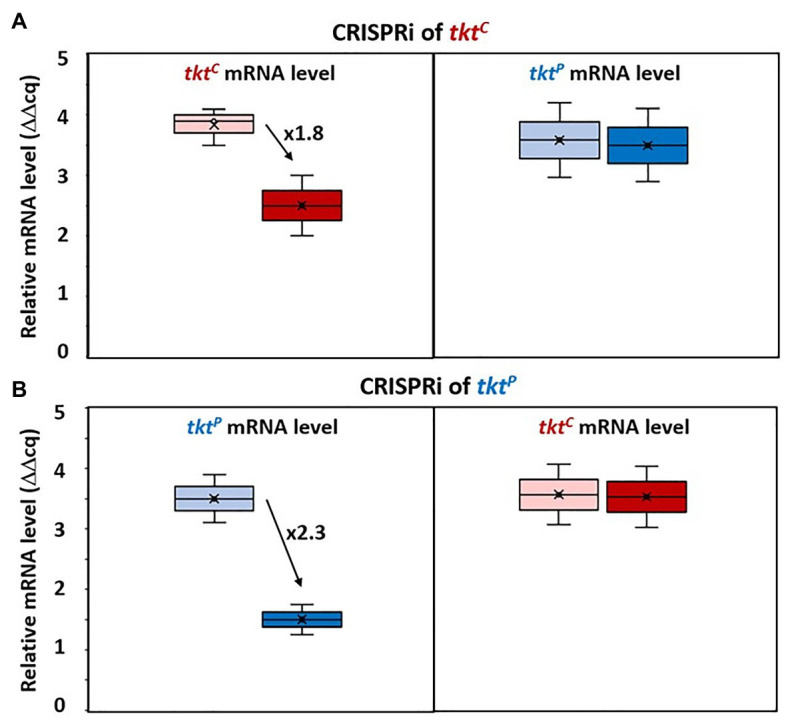
Changed gene expression levels due to dCas9 targeting tktC and tktP. Relative mRNA levels of tktC (left) and tktP (right) in MGA3(piCas-ttktC) (A) and relative mRNA level of tktP (left) and tktC (right) in MGA3(piCas-ttktP) (B) are shown by Box and Whisker plots as detected by qRT-PCR. Black arrows with adjacent numbers indicate up- or downregulation without (lighter color, left) and with (darker color, right) CRISPRi induction.
Discussion
With the help of CRISPRi and SDMs, the characterization of FBAC and FBAP of B. methanolicus was significantly improved in this work. Using the SDM approach, we could show that exchange of the chosen amino acid residues in order to alternate FBA activity supported previous reports on the distinct roles displayed by FBAC and FBAP. Furthermore, CRISPRi revealed a cooperative operation of the two FBAs, since higher gene expression of the untargeted fba could be observed when the other fba gene was repressed, and vice versa. In addition, we could show the effect of CRISPRi-derived repression of gene expression of a gene downstream of the target gene belonging to the same operon for the first time in B. methanolicus.
FBA is a key enzyme in glycolysis and gluconeogenesis in many organisms, and in B. methanolicus it plays a crucial role in the assimilation of methanol through the RuMP cycle. In the evolution of FBA enzymes, it is known that SBPases are of archaeal origin, whereas FBPases are descended from bacteria (Gütle et al., 2016, 2017). Class II FBAs catalyze the second reversible step of the glycolytic pathway, producing GAP and DHAP from the cleavage of the open-chain form of FBP (Marsh and Lebherz, 1992; Pegan et al., 2013). Little to no information has been reported on the interaction of class II or bacterial FBA aldolases with the substrate FBP, thus, there is a clear motivation to study this interaction in more detail. Recently, the X-ray structure of class II FBA from Mycobacterium tuberculosis (MtFBA) was determined with the aim to use it as a new pharmacological target against tuberculosis, since the class II MtFBA differs from the class I FBA in humans (Pegan et al., 2013). In addition, surface labeling and enzyme activity measurements revealed that MtFBA was exported to the cell surface of M. tuberculosis and produced under various axenic growth conditions, including oxygen deficiency (de la Paz Santangelo et al., 2011). However, the exact binding mechanism of class II FBAs to its substrate FBP is not yet fully understood. In this work, it could be shown through an SDM approach that we were able to alternate and significantly improve the catalytic efficiency of FBAC in gluconeogenesis in the double mutant FBAC; E51V,T140R, even surpassing that of native FBAP by at least 3-fold (Table 2). On the other hand, instead of the expected alternation from major gluconeogenic to major glycolytic aldolase (Supplementary Figure S3), a complete loss of glycolytic activity of FBAP could be observed when performing both V51E and R140T amino acid exchanges (FBAP;V51E, R140T). Nevertheless, these results strongly indicate that the chosen amino acid residues are important for glycolytic activity and suggest that the mutant FBAP; V51E, R140T is a strict gluconeogenic enzyme.
As previously reported (Galkin et al., 2007; Stolzenberger et al., 2013b), the presence of zinc decreased the FBAC and FBAP activity significantly. Therefore, the zinc-binding site was additionally chosen for mutagenesis purposes as it represented a strong candidate for the activity alternation. Single mutants FBAC; T140R and FBAP; R140T were constructed and investigated with regard to their enzyme activity. However, the obtained results indicate that the performed mutations at the zinc-binding site are not crucial to change the binding affinity of the substrates, since the observed KM values of mutants in the gluconeogenic direction did not differ from that of the native proteins, and only a slight decrease in the binding affinity of FBAP; R140T for FBP in the glycolytic direction could be observed. Taken together, these results strongly indicate that the amino acid residue divergence at the FBP binding site is crucial for the reactivity change since KM values for FBAP; V51E for the substrates FBP, GAP, and DHAP increased dramatically in comparison to the native protein (Table 1 and Table 2).
Moreover, the Hill coefficients determined (data not shown) indicate that the affinity of binding an FBP molecule is not dependent on whether other FBP molecules are already bound to FBA. This is based on the observation that Hill coefficient values are ≈ 1 for all FBA variants. Furthermore, the Lineweaver Burk Plots generated to determine the kinetic parameters in gluconeogenesis (Supplementary Figures S4, S5) for two different substrates indicate that the FBAs follow the rules of an equilibrium-ordered mechanism (Fromm, 1976), which describes sequential substrate binding to the enzyme. Once all substrates are bound, there is a central complex where the conversion of the substrates to the products takes place, which is subsequently released from the complex (Gates and Northrop, 1988). Since the crossing points to determine the KM and Vmax values for DHAP are in the second quadrant and since the crossing points for GAP as substrates are located close to the ordinate, it can be assumed that DHAP has to bind the FBA enzyme before the substrate GAP is able to bind, as reported by Segel (1975) and Fromm (1976). In addition, Rozovsky and McDermott (2007) used solid-state and solution-state nuclear magnetic resonance (NMR) to analyze the identity and steady-state populations of chemical entities bound to the TIM barrel. It can be assumed that the predominance of DHAP on the enzyme would support a mechanism where the initial proton abstraction in the reaction from DHAP to GAP is significantly slower than the subsequent chemical steps. This is also supported by the fact that most calculated Vmax values for DHAP as substrate were lower than those for GAP (Table 2).
Regarding the physiological role of FBA, it is reported that the FBA has a general “household function,” which is to maintain a rapid equilibrium between FBP, GAP, and DHAP. Further, this ensures the rapid equilibrium of triose phosphates produced by aldolase in glycolysis, which is interconnected to lipid metabolism, the GAP shuttle, and the pentose phosphate pathway (PPP) (Orosz et al., 2009; Malabanan et al., 2012). The major gluconeogenic aldolase FBAP is expected to play an important role during growth of B. methanolicus with carbon sources requiring gluconeogenesis such as acetate or propionate. The glyoxylate shunt genes have not only been annotated, but shown to function as their heterologous expression complemented a glyoxylate deficient C. glutamicum mutant and their overexpression improved (R)-acetoin production by a metabolically engineered strain of B. methanolicus (Drejer et al., 2020). The genome of B. methanolicus also encodes propionyl-CoA carboxylase and methylmalonyl-CoA mutase, thus, providing ability to grow on propionic acid. The two chromosomally encoded and the plasmid encoded NAD+ dependent methanol dehydrogenases have been shown to oxidize other alcohols (ethanol, propanol, butanol, isopropanol, 1,2-propanediol, and 1,3-propanediol) besides methanol (Krog et al., 2013). This may increase the carbon source spectrum to include ethanol besides acetate, propanol, propionate, and possibly more alcohols that require gluconeogenesis to support growth of B. methanolicus. Future work has to address if the amino acid changes introduced here into FBAP affect the ability of B. methanolicus to grow with these C2 and C3 carbon sources.
In B. methanolicus, both aldolase enzymes FBAC and FBAP do not only cleave FBP (glycolysis) and synthesize it from GAP and DHAP (gluconeogenesis), but they have a third role in the so-called SBPase variant of the RuMP cycle that was shown to operate in vivo by 13C-labeling experiments (Delépine et al., 2020). FBAP and FBAC are able to synthesize seduheptolose-1,7-bisphosphate from DHAP and erythrose 4-phosphate, which was revealed by a coupled discontinuous enzyme assay and subsequent LC-MS/MS analysis (Stolzenberger et al., 2013b). Only the plasmid-encoded phosphatase enzyme GlpXP is active as FBPase and SBPase, whereas GlpXC was only found to be active as FBPase (Stolzenberger et al., 2013b). It remains to be studied if the amino acid changes introduced here into FBAP and FBAC affect their activities in the synthesis of seduheptolose-1,7-bisphosphate from DHAP and erythrose 4-phosphate and the consequences for their roles in the SBPase variant of the RuMP cycle.
In several pathogenic bacteria, genetic studies have shown that loss of the fba gene led to a loss of viability of the organism (Gerdes et al., 2003; Kobayashi et al., 2003; Baba et al., 2006; Capodagli et al., 2014). Beyond that, it was recently reported that FBA plays a direct role in the transcriptional regulation of the katG and rpoA genes, which code for a catalase and an RNAP subunit, respectively, in Francisella tularensis (Ziveri et al., 2017). Therefore, FBA is suspected to be involved in the control of host redox homeostasis and inflammatory immune response (Ziveri et al., 2017). Furthermore, in the yeast S. cerevisiae the class II FBA, in addition to its function in glycolysis, physically interacts with RNAP III and plays a role in controlling its transcription (Cieśla et al., 2014; Ziveri et al., 2017). It can therefore be assumed that the FBAs in B. methanolicus will also take on further regulatory tasks that have not been discovered yet. This underlines the importance to study the function of FBAs more deeply, especially in poorly characterized methylotrophs to better understand and, in turn, engineer the metabolism. The present study gives first insights on the activity alternation from FBAC toward FBAP and vice versa by the kinetic parameters shown for the double mutant FBAC; E51V, T140R and FBAP; V51E, R140T both in FBP cleavage and synthesis direction. The expectation of the double mutant FBAP; V51E, R140T leading to a change in reactivity of FBAP toward a more glycolytic role could not be confirmed due to lack of activity detected in said mutant in glycolysis. However, an improved gluconeogenic FBA was engineered with FBAC; E51V, T140R, overcoming the major gluconeogenic FBAP.
CRISPR interference mediated gene repression of fbaC and fbaP revealed compensatory expression of the paralogous gene that was not targeted by CRISPRi (Figure 3). Since an increase in RNA levels of the non-targeted fba could be observed, the mechanism may either involve transcriptional regulation or mRNA degradation control. Up to date, transcriptional regulatory proteins affecting the expression of fbaC or the fbaP-tktP operon have not been described. Similarly, cis-regulatory RNA elements such as riboswitches have not been identified in their 5'UTRs (Irla et al., 2015). However, methylotrophic genes present in the natural pBM19 plasmid including the fbaP-tktP operon, but notably not fbaC, were reported to show increased RNA levels in the presence of methanol (Jakobsen et al., 2006; Heggeset et al., 2012) and it is plausible that the underlying regulatory mechanism may be relevant to explain the compensatory induction of the fbaP-tktP operon upon CRISPRi mediated repression of fbaC. The compensatory induction of fbaC when fbaP is targeted by CRISPRi is even less clear. The observed phenomena in response to genetic perturbation by CRISPRi may be due to physiological robustness that has been described as the persistence of certain characteristics or traits in a biological system under perturbations or conditions of uncertainty (Kitano, 2004; Stelling et al., 2004; Félix and Wagner, 2008). The observed changes in RNA levels of fbaC and fbaP indicate robustness by genetic compensation (Lynch and Conery, 2000; Gu et al., 2003; Plata and Vitkup, 2014; Bunton-Stasyshyn et al., 2019) rather than by alternative signaling or metabolic pathways (Wagner, 2000; Papp et al., 2004; Plata and Vitkup, 2014). Genetic robustness typically involves compensatory genes that are paralogs (Lynch and Conery, 2000; Bunton-Stasyshyn et al., 2019), as is the case of fbaC and fbaP. The CRISPRi approach used here allowed for gene repression of fbaC and fbaP. However, in the absence of CRISPR genome editing or other methods for gene deletion in B. methanolicus, it remains unclear if fbaC and/or fbaP are essential or conditionally essential for methylotrophic growth, and if there are additional aldolase enzymes that may compensate for their absence (μ = 0.36 h−1, 0.35 h−1; Figure 2). Genetic robustness appears not to involve all methylotrophic genes since CRISPRi repression of tktP did neither lead to increased tktC RNA levels nor did CRISPRi targeting of tktC increase tktP RNA levels (Figure 4). As the transketolases TKTP and TKTC share very similar kinetic parameters (Markert et al., 2014) they would be well suited for genetic compensation. It is plausible that the limited CRISPRi mediated repression of tkt (about 2-fold, Figure 4) did not reduce TKT enzyme levels below a threshold that might trigger genetic compensation. Future work with the aim of improving the genetic toolbox of B. methanolicus will have to include making use of stronger and gratuitous inducers in the CRISPRi system and, eventually, to develop tools for genetic modifications in this organism. In this study, the currently available CRISPRi system was successfully employed to demonstrate the first loss of function analysis of a methylotrophic target gene and thus improve the characterization of key RuMP enzymes in B. methanolicus.
Data Availability Statement
Crystal structures have been submitted to PDB with accession codes 7NC7 and 7NCC. The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
KS, ML, and LK carried out the genetics and biochemistry experiments of the present study. OE and LZ performed structure elucidation experiments. All authors analyzed the data. VW coordinated the study. KS and ML drafted the manuscript. All authors revised the manuscript and VW finalized the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the ERA CoBioTech project C1Pro (No. 722361). Support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University was acknowledged. The funding bodies had no role in the design of the study or the collection, analysis, or interpretation of data or in writing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669220/full#supplementary-material
References
- Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., et al. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., et al. (2012). Towards automated crystallographic structure refinement with phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367. 10.1107/S0907444912001308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2, PMID: [DOI] [PubMed] [Google Scholar]
- Anthony C. (1991). Assimilation of carbon by methylotrophs. Biotechnology 18, 79–109. 10.1016/b978-0-7506-9188-8.50011-5, PMID: [DOI] [PubMed] [Google Scholar]
- Arfman N., Dijkhuizen L., Kirchhof G., Ludwig W., Schleifer K. H., Bulygina E. S., et al. (1992). Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int. J. Syst. Bacteriol. 42, 439–445. 10.1099/00207713-42-3-439, PMID: [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., et al. (1975). Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5Å resolution: using amino acid sequence data. Nature 255, 609–614. 10.1038/255609a0, PMID: [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Knowles J. R. (1983). Polarization of substrate carbonyl groups by yeast aldolase: investigation by Fourier transform infrared spectroscopy. Biochemistry 22, 122–129. 10.1021/bi00270a018, PMID: [DOI] [PubMed] [Google Scholar]
- Blin K., Pedersen L. E., Weber T., Lee S. Y. (2016). CRISPy-web: an online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 1, 118–121. 10.1016/j.synbio.2016.01.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3, PMID: [DOI] [PubMed] [Google Scholar]
- Brautaset T., Jakobsen Ø. M., Flickinger M. C., Valla S., Ellingsen T. E. (2004). Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J. Bacteriol. 186, 1229–1238. 10.1128/JB.186.5.1229-1238.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautaset T., Jakobsen Ø. M., Josefsen K. D., Flickinger M. C., Ellingsen T. E. (2007). Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50 degrees C. Appl. Microbiol. Biotechnol. 74, 22–34. 10.1007/s00253-006-0757-z, PMID: [DOI] [PubMed] [Google Scholar]
- Brautaset T., Williams M. D., Dillingham R. D., Kaufmann C., Bennaars A., Crabbe E., et al. (2003). Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in l-lysine-secreting mutants. Appl. Environ. Microbiol. 69, 3986–3995. 10.1128/AEM.69.7.3986-3995.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito L. F., Schultenkämper K., Passaglia L., Wendisch V. F. (2020). CRISPR interference-based gene repression in the plant growth promoter Paenibacillus sonchi genomovar Riograndensis SBR5. Appl. Microbiol. Biotechnol. 104, 5095–5106. 10.1007/s00253-020-10571-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunton-Stasyshyn R. K. A., Wells S., Teboul L. (2019). When all is not lost: considering genetic compensation in laboratory animals. Lab. Anim. 48, 282–284. 10.1038/s41684-019-0397-4, PMID: [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797, PMID: [DOI] [PubMed] [Google Scholar]
- Capodagli G. C., Lee S. A., Boehm K. J., Brady K. M., Pegan S. D. (2014). Structural and functional characterization of methicillin-resistant Staphylococcus aureus’s class IIb fructose 1,6-bisphosphate aldolase. Biochemistry 53, 7604–7614. 10.1021/bi501141t, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicer M., Vieira G., Brautaset T., Portais J.-C., Heux S. (2016). Quantitative metabolomics of the thermophilic methylotroph Bacillus methanolicus. Microb. Cell Factories 15:92. 10.1186/s12934-016-0483-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ. Microbiol. 13, 2603–2622. 10.1111/j.1462-2920.2011.02464.x, PMID: [DOI] [PubMed] [Google Scholar]
- Chistoserdova L., Kalyuzhnaya M. G., Lidstrom M. E. (2009). The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499. 10.1146/annurev.micro.091208.073600, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieśla M., Mierzejewska J., Adamczyk M., Farrants A.-K. Ö., Boguta M. (2014). Fructose bisphosphate aldolase is involved in the control of RNA polymerase III-directed transcription. Biochim. Biophys. Acta 1843, 1103–1110. 10.1016/j.bbamcr.2014.02.007, PMID: [DOI] [PubMed] [Google Scholar]
- Cleto S., Jensen J. V., Wendisch V. F., Lu T. K. (2016). Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth. Biol. 5, 375–385. 10.1021/acssynbio.5b00216, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S. J., Leonard G. A., McSweeney S. M., Thompson A. W., Naismith J. H., Qamar S., et al. (1996). The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. Struct. Lond. Engl. 4, 1303–1315. 10.1016/s0969-2126(96)00138-4, PMID: [DOI] [PubMed] [Google Scholar]
- Cress B. F., Leitz Q. D., Kim D. C., Amore T. D., Suzuki J. Y., Linhardt R. J., et al. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Factories 16:10. 10.1186/s12934-016-0623-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Lam H., Dillingham R. L., Hanson R. S., Flickinger M. C. (1997). Genetic manipulation of Bacillus methanolicus, a gram-positive, thermotolerant methylotroph. Appl. Environ. Microbiol. 63, 1406–1420. 10.1128/AEM.63.4.1406-1420.1997, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby A., Dauter Z., Littlechild J. A. (1999). Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications. Protein Sci. Publ. Protein Soc. 8, 291–297. 10.1110/ps.8.2.291, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsltr. Protein Crystallography 40, 82–92. [Google Scholar]
- de la Paz Santangelo M., Gest P. M., Guerin M. E., Coinçon M., Pham H., Ryan G., et al. (2011). Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli. J. Biol. Chem. 286, 40219–40231. 10.1074/jbc.M111.259440, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delépine B., López M. G., Carnicer M., Vicente C. M., Wendisch V. F., Heux S. (2020). Charting the metabolic landscape of the facultative methylotroph Bacillus methanolicus. mSystems 5, e00745–e00820. 10.1128/mSystems.00745-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkhuizen L., Hansen T. A., Harder W. (1985). Methanol, a potential feedstock for biotechnological processes. Trends Biotechnol. 3, 262–267. 10.1016/0167-7799(85)90026-5 [DOI] [Google Scholar]
- Drejer E. B., Chan D. T. C., Haupka C., Wendisch V. F., Brautaset T., Irla M. (2020). Methanol-based acetoin production by genetically engineered Bacillus methanolicus. Green Chem. 22, 788–802. 10.1039/C9GC03950C [DOI] [Google Scholar]
- Eikmanns B. J., Thum-Schmitz N., Eggeling L., Lüdtke K.-U., Sahm H. (1994). Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140, 1817–1828. 10.1099/13500872-140-8-1817, PMID: [DOI] [PubMed] [Google Scholar]
- Elhadi D., Lv L., Jiang X.-R., Wu H., Chen G.-Q. (2016). CRISPRi engineering E. coli for morphology diversification. Metab. Eng. 38, 358–369. 10.1016/j.ymben.2016.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158, PMID: [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Wagner A. (2008). Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity 100, 132–140. 10.1038/sj.hdy.6800915, PMID: [DOI] [PubMed] [Google Scholar]
- Fontana J., Dong C., Ham J. Y., Zalatan J. G., Carothers J. M. (2018). Regulated expression of sgRNAs tunes CRISPRi in E. coli. Biotechnol. J. 13:e1800069. 10.1002/biot.201800069, PMID: [DOI] [PubMed] [Google Scholar]
- Fromm H. J. (1976). Criteria for distinguishing between the rapid equilibrium ordered and random bi bi kinetic mechanisms. Biochem. Biophys. Res. Commun. 72, 55–60. 10.1016/0006-291X(76)90959-1, PMID: [DOI] [PubMed] [Google Scholar]
- Galkin A., Kulakova L., Melamud E., Li L., Wu C., Mariano P., et al. (2007). Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia. J. Biol. Chem. 282, 4859–4867. 10.1074/jbc.M609534200, PMID: [DOI] [PubMed] [Google Scholar]
- Galkin A., Li Z., Li L., Kulakova L., Pal L. R., Dunaway-Mariano D., et al. (2009). Structural insights into substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase*. Biochemistry 48, 3186–3196. 10.1021/bi9001166, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauttam R., Seibold G. M., Mueller P., Weil T., Weiß T., Handrick R., et al. (2019). A simple dual-inducible CRISPR interference system for multiple gene targeting in Corynebacterium glutamicum. Plasmid 103, 25–35. 10.1016/j.plasmid.2019.04.001, PMID: [DOI] [PubMed] [Google Scholar]
- Gerdes S. Y., Scholle M. D., Campbell J. W., Balázsi G., Ravasz E., Daugherty M. D., et al. (2003). Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684. 10.1128/JB.185.19.5673-5684.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates C. A., Northrop D. B. (1988). Kinetic distinction between rapid-equilibrium random and abortive ordered enzymatic mechanisms using alternative substrates or kinetic isotope effects. Biochem. Biophys. Res. Commun. 152, 406–410. 10.1016/s0006-291x(88)80728-9 [DOI] [PubMed] [Google Scholar]
- Gibson D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361. 10.1016/B978-0-12-385120-8.00015-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. 10.1038/nmeth.1318, PMID: [DOI] [PubMed] [Google Scholar]
- Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., Li W.-H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. 10.1038/nature01198, PMID: [DOI] [PubMed] [Google Scholar]
- Gütle D. D., Roret T., Hecker A., Reski R., Jacquot J.-P. (2017). Dithiol disulphide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci. Int. J. Exp. Plant Biol. 255, 1–11. 10.1016/j.plantsci.2016.11.003, PMID: [DOI] [PubMed] [Google Scholar]
- Gütle D. D., Roret T., Müller S. J., Couturier J., Lemaire S. D., Hecker A., et al. (2016). Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. U. S. A. 113, 6779–6784. 10.1073/pnas.1606241113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. 10.1016/S0022-2836(83)80284-8, PMID: [DOI] [PubMed] [Google Scholar]
- Heggeset T. M. B., Krog A., Balzer S., Wentzel A., Ellingsen T. E., Brautaset T. (2012). Genome sequence of thermotolerant bacillus methanolicus: features and regulation related to methylotrophy and production of l-lysine and l-glutamate from methanol. Appl. Environ. Microbiol. 78, 5170–5181. 10.1128/AEM.00703-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Dollinger G., Walsh P. S., Griffith R. (1992). Simultaneous amplification and detection of specific DNA sequences. Biotechnology 10, 413–417. 10.1038/nbt0492-413, PMID: [DOI] [PubMed] [Google Scholar]
- Irla M., Heggeset T. M. B., Nærdal I., Paul L., Haugen T., Le S. B., et al. (2016). Genome-based genetic tool development for Bacillus methanolicus: theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front. Microbiol. 7:1481. 10.3389/fmicb.2016.01481, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla M., Neshat A., Brautaset T., Rückert C., Kalinowski J., Wendisch V. F. (2015). Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genomics 16:73. 10.1186/s12864-015-1239-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla M., Neshat A., Winkler A., Albersmeier A., Heggeset T. M. B., Brautaset T., et al. (2014). Complete genome sequence of Bacillus methanolicus MGA3, a thermotolerant amino acid producing methylotroph. J. Biotechnol. 188, 110–111. 10.1016/j.jbiotec.2014.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Izard T., Sygusch J. (2004). Induced fit movements and metal cofactor selectivity of class II aldolases: structure of Thermus aquaticus fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 279, 11825–11833. 10.1074/jbc.M311375200, PMID: [DOI] [PubMed] [Google Scholar]
- Jacques B., Coinçon M., Sygusch J. (2018). Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases. J. Biol. Chem. 293, 7737–7753. 10.1074/jbc.RA117.001098, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen Ø. M., Benichou A., Flickinger M. C., Valla S., Ellingsen T. E., Brautaset T. (2006). Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 188, 3063–3072. 10.1128/JB.188.8.3063-3072.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015). The Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc. 10, 845–858. 10.1038/nprot.2015.053, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. (2004). Biological robustness. Nat. Rev. Genet. 5, 826–837. 10.1038/nrg1471, PMID: [DOI] [PubMed] [Google Scholar]
- Klausen M. S., Jespersen M. C., Nielsen H., Jensen K. K., Jurtz V. I., Sønderby C. K., et al. (2019). NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins Struct. Funct. Bioinform. 87, 520–527. 10.1002/prot.25674, PMID: [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., et al. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. 100, 4678–4683. 10.1073/pnas.0730515100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krog A., Heggeset T. M. B., Müller J. E. N., Kupper C. E., Schneider O., Vorholt J. A., et al. (2013). Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS One 8:e59188. 10.1371/journal.pone.0059188, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0, PMID: [DOI] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., Qi L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. 10.1038/nprot.2013.132, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner S. N., Vidaurre D., Willbold S., Schoberth S. M., Wendisch V. F. (2007). NCgl2620 encodes a class II polyphosphate kinase in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73, 5026–5033. 10.1128/AEM.00600-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M. G., Irla M., Brito L. F., Wendisch V. F. (2019). Characterization of d-arabitol as newly discovered carbon source of Bacillus methanolicus. Front. Microbiol. 10:1725. 10.3389/fmicb.2019.01725, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. 10.1126/science.290.5494.1151, PMID: [DOI] [PubMed] [Google Scholar]
- Malabanan M. M., Koudelka A. P., Amyes T. L., Richard J. P. (2012). Mechanism for activation of triosephosphate isomerase by phosphite dianion: the role of a hydrophobic clamp. J. Am. Chem. Soc. 134, 10286–10298. 10.1021/ja303695u, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. (1970). Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53, 159–162. 10.1016/0022-2836(70)90051-3 [DOI] [PubMed] [Google Scholar]
- Markert B., Stolzenberger J., Brautaset T., Wendisch V. F. (2014). Characterization of two transketolases encoded on the chromosome and the plasmid pBM19 of the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. BMC Microbiol. 14:7. 10.1186/1471-2180-14-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. J., Lebherz H. G. (1992). Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem. Sci. 17, 110–113. 10.1016/0968-0004(92)90247-7, PMID: [DOI] [PubMed] [Google Scholar]
- Michaelis L., Menten M. L., Johnson K. A., Goody R. S. (2011). The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269. 10.1021/bi201284u, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J. E. N., Heggeset T. M. B., Wendisch V. F., Vorholt J. A., Brautaset T. (2015a). Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl. Microbiol. Biotechnol. 99, 535–551. 10.1007/s00253-014-6224-3, PMID: [DOI] [PubMed] [Google Scholar]
- Müller J. E. N., Litsanov B., Bortfeld-Miller M., Trachsel C., Grossmann J., Brautaset T., et al. (2014). Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3. Proteomics 14, 725–737. 10.1002/pmic.201300515, PMID: [DOI] [PubMed] [Google Scholar]
- Müller J. E. N., Meyer F., Litsanov B., Kiefer P., Vorholt J. A. (2015b). Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol. Microbiol. 98, 1089–1100. 10.1111/mmi.13200, PMID: [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., et al. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367. 10.1107/S0907444911001314, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naerdal I., Pfeifenschneider J., Brautaset T., Wendisch V. F. (2015). Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microb. Biotechnol. 8, 342–350. 10.1111/1751-7915.12257, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K., Yamamoto H., Miyake C., Yokota A. (2003). Purification and characterization of class-I and class-II fructose-1,6-bisphosphate aldolases from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44, 326–333. 10.1093/pcp/pcg044, PMID: [DOI] [PubMed] [Google Scholar]
- Orosz F., Oláh J., Ovádi J. (2009). Triosephosphate isomerase deficiency: new insights into an enigmatic disease. Biochim. Biophys. Acta 1792, 1168–1174. 10.1016/j.bbadis.2009.09.012, PMID: [DOI] [PubMed] [Google Scholar]
- Papp B., Pál C., Hurst L. D. (2004). Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429, 661–664. 10.1038/nature02636, PMID: [DOI] [PubMed] [Google Scholar]
- Pegan S. D., Rukseree K., Capodagli G. C., Baker E. A., Krasnykh O., Franzblau S. G., et al. (2013). Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis. Biochemistry 52, 912. 10.1021/bi300928u, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penhoet E., Rajkumar T., Rutter W. J. (1966). Multiple forms of fructose diphosphate aldolase in mammalian tissues. Proc. Natl. Acad. Sci. U. S. A. 56, 1275–1282. 10.1073/pnas.56.4.1275, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B., Petersen T. N., Andersen P., Nielsen M., Lundegaard C. (2009). A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9:51. 10.1186/1472-6807-9-51, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plata G., Vitkup D. (2014). Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Res. 42, 2405–2414. 10.1093/nar/gkt1200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaumann M., Pelzer-Reith B., Martin W. F., Schnarrenberger C. (1997). Multiple recruitment of class-I aldolase to chloroplasts and eubacterial origin of eukaryotic class-II aldolases revealed by cDNAs from Euglena gracilis. Curr. Genet. 31, 430–438. 10.1007/s002940050226, PMID: [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. 10.1016/j.cell.2013.02.022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittmann D., Schaffer S., Wendisch V. F., Sahm H. (2003a). Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch. Microbiol. 180, 285–292. 10.1007/s00203-003-0588-6, PMID: [DOI] [PubMed] [Google Scholar]
- Rozovsky S., McDermott A. E. (2007). Substrate product equilibrium on a reversible enzyme, triosephosphate isomerase. Proc. Natl. Acad. Sci. 104, 2080–2085. 10.1073/pnas.0608876104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. (2001). Molecular Cloning: A Laboratory Manual. 3rd Edn. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press, [2001] ©2001. [Google Scholar]
- Sander T., Wang C. Y., Glatter T., Link H. (2019). CRISPRi-based downregulation of transcriptional feedback improves growth and metabolism of arginine overproducing E. coli. ACS Synth. Biol. 8, 1983–1990. 10.1021/acssynbio.9b00183, PMID: [DOI] [PubMed] [Google Scholar]
- Sauvé V., Sygusch J. (2001). Molecular cloning, expression, purification, and characterization of fructose-1,6-bisphosphate aldolase from Thermus aquaticus. Protein Expr. Purif. 21, 293–302. 10.1006/prep.2000.1380, PMID: [DOI] [PubMed] [Google Scholar]
- Schendel F. J., Bremmon C. E., Flickinger M. C., Guettler M., Hanson R. S. (1990). l-lysine production at 50 degrees C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56, 963–970. 10.1128/AEM.56.4.963-970.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultenkämper K., Brito L. F., López M. G., Brautaset T., Wendisch V. F. (2019). Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus. Appl. Microbiol. Biotechnol. 103, 5879–5889. 10.1007/s00253-019-09907-8, PMID: [DOI] [PubMed] [Google Scholar]
- Schultenkämper K., Brito L. F., Wendisch V. F. (2020). Impact of CRISPR interference on strain development in biotechnology. Biotechnol. Appl. Biochem. 67, 7–21. 10.1002/bab.1901, PMID: [DOI] [PubMed] [Google Scholar]
- Segel I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. Vol. 115. ed. Segel I. H. (New York, London: John Wiley & Sons; ), 18–22. [Google Scholar]
- Shams F., Oldfield N. J., Wooldridge K. G., Turner D. P. J. (2014). Fructose-1,6-bisphosphate aldolase (FBA)-a conserved glycolytic enzyme with virulence functions in bacteria: “ill met by moonlight.” Biochem. Soc. Trans. 42, 1792–1795. 10.1042/BST20140203, PMID: [DOI] [PubMed] [Google Scholar]
- Stelling J., Sauer U., Szallasi Z., Doyle F. J., Doyle J. (2004). Robustness of cellular functions. Cell 118, 675–685. 10.1016/j.cell.2004.09.008, PMID: [DOI] [PubMed] [Google Scholar]
- Stolzenberger J., Lindner S. N., Persicke M., Brautaset T., Wendisch V. F. (2013a). Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J. Bacteriol. 195, 5112–5122. 10.1128/JB.00672-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenberger J., Lindner S. N., Wendisch V. F. (2013b). The methylotrophic Bacillus methanolicus MGA3 possesses two distinct fructose 1,6-bisphosphate aldolases. Microbiol. Read. Engl. 159, 1770–1781. 10.1099/mic.0.067314-0, PMID: [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. 10.1016/0022-2836(86)90385-2, PMID: [DOI] [PubMed] [Google Scholar]
- Tan K., Zhou M., Kwon K., Anderson W. F., Joachimiak A. (2011). The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. “Ames Ancestor.” PDB Entry – 3Q94.
- Tittmann K. (2014). Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data. Bioorg. Chem. 57, 263–280. 10.1016/j.bioorg.2014.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25. 10.1107/S0907444909042589, PMID: [DOI] [PubMed] [Google Scholar]
- Wagner A. (2000). Robustness against mutations in genetic networks of yeast. Nat. Genet. 24, 355–361. 10.1038/74174, PMID: [DOI] [PubMed] [Google Scholar]
- Wang C., Cao Y., Wang Y., Sun L., Song H. (2019). Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb. Cell Factories 18:90. 10.1186/s12934-019-1139-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook A. W., Moo-Young M., Chou C. P. (2016). Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl. Environ. Microbiol. 82, 4876–4895. 10.1128/AEM.01159-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook A. W., Ren X., Oh J., Moo-Young M., Chou C. P. (2018). Metabolic engineering to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Metab. Eng. 47, 401–413. 10.1016/j.ymben.2018.04.016, PMID: [DOI] [PubMed] [Google Scholar]
- Wierenga R. K. (2001). The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 492, 193–198. 10.1016/S0014-5793(01)02236-0, PMID: [DOI] [PubMed] [Google Scholar]
- Wood E. J. (2008). Fundamentals of biochemistry: life at the molecular level (third edition) by D. Voet, J. Voet, and C. W. Pratt. Biochem. Mol. Biol. Educ. 36, 319–320. 10.1002/bmb.20198 [DOI] [Google Scholar]
- Wu Y., Chen T., Liu Y., Lv X., Li J., Du G., et al. (2018a). CRISPRi allows optimal temporal control of N-acetylglucosamine bioproduction by a dynamic coordination of glucose and xylose metabolism in Bacillus subtilis. Metab. Eng. 49, 232–241. 10.1016/j.ymben.2018.08.012, PMID: [DOI] [PubMed] [Google Scholar]
- Wu Q., Peng Z., Zhang Y., Yang J. (2018b). COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 46, W438–W442. 10.1093/nar/gky439, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-Y., Sung L.-Y., Li H., Huang C.-H., Hu Y.-C. (2017). Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1,4-BDO biosynthesis. ACS Synth. Biol. 6, 2350–2361. 10.1021/acssynbio.7b00251, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu Z.-Q., Liu C., Zheng Y.-G. (2016). Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production. Biotechnol. Lett. 38, 2153–2161. 10.1007/s10529-016-2207-z, PMID: [DOI] [PubMed] [Google Scholar]
- Ziveri J., Tros F., Guerrera I. C., Chhuon C., Audry M., Dupuis M., et al. (2017). The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun. 8:853. 10.1038/s41467-017-00889-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structures have been submitted to PDB with accession codes 7NC7 and 7NCC. The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.