Abstract
Viruses remain one of the leading causes of animal and human disease. Some animal viral infections spread sporadically to human populations, posing a serious health risk. Particularly the emerging viral zoonotic diseases such as the novel, zoonotic coronavirus represent an actual challenge for the scientific and medical community. Besides human health risks, some animal viral infections, although still not zoonotic, represent important economic loses to the livestock industry. Viral infections pose a genuine concern for which there has been an increasing interest for new antiviral molecules. Among these novel compounds, antiviral peptides have been proposed as promising therapeutic options, not only for the growing body of evidence showing hopeful results but also due to the many adverse effects of chemical-based drugs. Here we review the current progress, key targets and considerations for the development of antiviral peptides (AVPs). The review summarizes the state of the art of the AVPs tested in zoonotic (coronaviruses, Rift Valley fever viruses, Eastern Equine Encephalitis Virus, Dengue and Junín virus) and also non-zoonotic farm animal viruses (avian and cattle viruses). Their molecular target, amino acid sequence and mechanism of action are summarized and reviewed.
Antiviral peptides are currently on the cutting edge since they have been reported to display anti-coronavirus activity. Particularly, the review will discuss the specific mode of action of AVPs that specifically inhibit the fusion of viral and host-cell membranes for SARS-CoV-2, showing in detail some important features of the fusion inhibiting peptides that target the spike protein of these risky viruses.
Keywords: Antiviral peptides, Zoonotic viruses, Animal viruses, Coronavirus
1. Introduction
Emerging and re-emerging viral pathogens that originate in wild or farm animals have become increasingly important throughout the world in recent decades, as they have substantial impacts on human health and agricultural production [1,2]. Infectious diseases that can jump from a non-human animal to humans are called zoonotic diseases. The transmission can be either directly through contact or indirectly through contaminated inanimate objects, intermediate hosts and bites of insect vectors, etc. [3].
In recent years, zoonotic viruses have been responsible for several outbreaks with high mortality rates. Besides human health, there are also serious concerns about viruses that infect animals and cause epidemics, affecting livestock production.
In December 2019 the first reports of a new zoonotic viral outbreak arrived from Wuhan, central China. Since then, the world was hit with a devastating pandemic outbreak: the novel coronavirus SARS-CoV-2, in the form of an acute respiratory illness [4]. Different antiviral agents have been tested or repurposed [5], unfortunately, unlike bacterial infections, there are no "broad-spectrum" antiviral agents, so current therapeutic options are more limited in the case of viruses, some of which represent epidemic risks [6]. Viruses use host cell machinery for replication, so any antiviral compound should specifically avoid interfering with host cell functions. Additionally, differences between RNA and DNA viruses represent another obstacle for broad-spectrum antivirals, actually only a few antivirals have comparable spectrums of activity to penicillin [6].
Generally, we could divide the life cycle of a virus into six steps: 1. Attachment (receptor or co-receptor binding); 2. Fusion and penetration; 3. Uncoating and release of the viral genome; 4. Gene expression and multiplication; 5. Assembly and packaging; 6. Release of viral particle [7,8]
Some of these steps have been specially evaluated as suitable targets for antiviral therapies.
Interaction with functional receptor(s) and subsequent attachment to host cells is the first step in viral infection. Most viral pathogens are membrane-enveloped viruses. For these viruses the fusion of viral and cellular membranes is a key requirement to deliver its genetic material into the target cell. In this type of viruses, the viral proteins located on the outer envelope of the virion are responsible for the recognition of receptors and the attachment to host cells. This very particular process represents a suitable target for the development of broad-spectrum antiviral drugs [9]. The inhibition of this process would block all of the subsequent intracellular steps [10].
1.1. Antiviral peptides (AVPs)
Among the defense arsenal that organisms have developed against bacterial or viral infections are antimicrobial peptides (AMPs) also called host defense peptides (HDPs). AMPs play a vital role in the initial immediate non-specific immune response generated to begin to control infection [[11], [12], [13]]. These small and usually cationic peptides are produced by almost every organism. Also, organisms without an immune system may display an AMP defense against bacteria, viruses or fungi. In general, natural AMPs are non-specific, targeting components of a microbe that are integral or not exclusive, and as members of the innate immune system, they also help to facilitate a larger immune response [14]. Natural and sometimes designed AMPs can have other functions and activities besides antimicrobial, such as apoptosis, wound healing and immune modulation [15,16]
AMPs have been classified according to their structure and function [17,18]. In order to organize our knowledge on AMPs some complete and useful databases have been created [19,20]
While all of the AMPs classes have been shown to possess antimicrobial activity, only a few classes have demonstrated antiviral properties, which are called antiviral peptides (AVPs). In recent years these molecules gained attention as antiviral or virucidal agents [14,[21], [22], [23]].
Some AVPs are capable of inhibiting the fusion of viruses to the cell (interacting with membrane envelope and glycoproteins of the virus), by inhibiting viral entry (e.g. heparan sulphate interaction or by fusing with specific cellular receptors) and others can interfere with proteins or enzymes needed for viral replication [11], in the same way that for bacterial inhibition, these peptides are usually non-specific (Table 1 ).
Table 1.
Some examples of non-specific designed peptides that act as antivirals against enveloped viruses.
| Peptide | Sequence | Target Virus | Mechanism proposed | Reference |
|---|---|---|---|---|
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | VEEV | Entry inhibition, Immunomodulator | [54] |
| Melitin | GIGAVLKVLTTGLPALISWIKRKRQQ | VJ | ND | [58] |
| Cecropin A | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK | VJ | Inhibition at late stages of virus multiplication cycle | [58] |
| Indolicidin | ILPWKWPWWPWRR | VJ | ND | [58] |
| AMLs | Lipopeptides produces from B. subtilis fmbj that included surfactin, fengycin and their isoforms | NDV | Interaction with the viral membrane | [75] |
| SIAMP | Multi peptide preparation from swine small intestines | IBV | block attachment and replication of the virus | [77] |
| Bursin | LHG | ALV | block attachment to the host cells | [81] |
| Bovine lactoferrin | CTISQPEWFKCRRWQWRMKKLGAPSITCVRRAFALECIRAIAEKKADAVTLDGGMVFEACRDPYKLRPVAAEIYGTKESPQTHYYAVAVVKKGSNFQLDQLQGRKSCHTGLGRSAGWIIPMGILRPYLSWTESLEPLQGAVAKFFSASCVPCIDRQAYPNLCQLCKGEGENQCACSSREPYFGYSGAFKCLQDGAGDVAFVKETTVFENLPEKADRDQYELLCLNNSRAPVDAFKECHLAQVPSHAVVARSVDGKEDLIWKLLSKAQEKFGKNKSRSFQLFGSPPGQRDLLFKDSALGFLRIPSKVDSALYLGSRYLTTLKNLRETAEEVKARYTRVVWCAVGPEEQKKCQQWSQQSGQNVTCATASTTDDCIVLVLKGEADALNLDGGYIYTAGKCGLVPVLAENRKSSKHSSLDCVLRPTEGYLAVAVVKKANEGLTWNSLKDKKSCHTAVDRTAGWNIPMGLIVNQTGSCAFDEFFSQSCAPGADPKSRLCALCAGDDQGLDKCVPNSKEKYYGYTGAFRCLAEDVGDVAFVKNDTVWENTNGESTADWAKNLNREDFRLLCLDGTRKPVTEAQSCHLAVAPNHAVVSRSDRAAHVKQVLLHQQALFGKNGKNCPDKFCLFKSETKNLLFNDNTECLAKLGGRPTYEEYLGTEYVTAIANLKKCSTSPLLEACAFLTR | BVDV | Prevents viral infection at its early stage | [96] |
| Chicken egg lysozyme | MRSLLILVLCFLPLAALGKVFGRCELAAAMKRHGLDNYRGYSLGNWVCAAKFESNFNTQATNRNTDGSTDYGILQINSRWWCNDGRTPGSRNLCNIPCSALLSSDITASVNCAKKIVSDGNGMNAWVAWRNRCKGTDVQAWIRGCRL | BVDV | ND | [96] |
| Nisin | ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | BVDV | ND | [96] |
ND: not determined.
Viral life cycles or host response factors have been the target of the novel antiviral peptides’ strategies, (Table 2 ). As previously mentioned, these antiviral peptides can inhibit or interfere with different mechanisms of the viral infection or replication; particularly some AVPs were specifically designed to inhibit or block the fusion of the viral and cellular membranes in enveloped virus, interacting with the pre-fusion conformations of proteins needed for membrane fusion. The interaction of these peptides can destabilize the fusion proteins, preventing the formation of fusion intermediate structures [9] ( Fig. 1 ). Bioinformatics and molecular modeling are a very useful tools to design new antiviral drugs, in fact many AVPs have been obtained with specific bioinformatics tools; these novel sequences are usually called designed AVPs [[24], [25], [26], [27]].
Table 2.
Specifically designed antiviral peptides against enveloped viruses.
| Peptide | Sequence | Target Virus | Mechanism proposed | Reference |
|---|---|---|---|---|
| P9 | NGAICWGPCPTAFRQIGNCGHFKVRCCKIR | MERS-CoV, SARS-CoV, AIV | prevents endosomal acidification, which blocks membrane fusion | [37] |
| SARSWW-I | MWKTPTLKYFGGFNFSQIL | SARS-CoV | Entry inhibition | [29] |
| SARSWW-II | ATAGWTFGAGAALQIPFAMQMAY | SARS-CoV | Entry inhibition | [29] |
| EK1 | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL | SARS-CoV, MARS-CoV | Fusion inhibitor | [34] |
| YKYRYL | YKYRL | SARS-CoV | Block attachment to the host cells | [43] |
| RVFV-6 | WNFFDWFSGLMSWFGGPLK | RVFV, Ebola virus, Andes virus and vesicular stomatitis virus | Fusion inhibitor | [47] |
| DN59 | MAILGDTAWDFGSLGGVFTSIGKALHQVFGAIY | DENV | Viral membrane disruption | [64] |
| AVP-p | LNLFKKTINGLISDSLVIR | JUNV | Fusion inhibitor | [60] |
| NOVEL-1 | NASDMEIKKVNKKIEEYIKKIEEVEKKLEEVNKK | NDV and IBV | Entry inhibition | [69] |
| NOVEL-2 | VNKKIEEIDKKIEELNKKLEELEKKLEEVNKK | NDV and IBV | Entry inhibition | [69] |
| TLTTKLY | TLTTKLY | NDV | Entry inhibition | [67,72] |
| CTLTTKLYC | CTLTTKLYC | NDV | Entry inhibition | [67,72] |
| TP1 | SWLVNRDWFHDLNLPWTGSSAGTWQ | TMUV | Block attachment to the host cells | [84,85] |
| TP2 | MVALGDTAWDFGSVGGVLTSIGKGIHQVFGSAFKSL | TMUV | Block attachment to the host cells and viral membrane disruption | [84,85] |
| NDFRSKT | NDFRSKT | AIV | Block attachment to the host cells | [86,87] |
| CNDFRSKTC | CNDFRSKTC | AIV | Block attachment to the host cells | [85,86] |
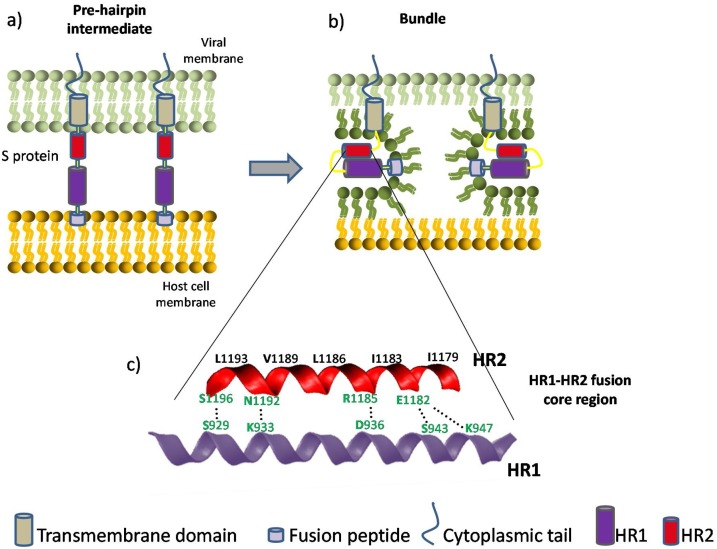
Fig. 1.
Coronavirus viral fusion pathway based on class I fusion protein model. For simplicity, only two stages are depicted. After the S protein achieves a pre-fusion metastable state, relevant proteases enable the fusion peptide to insert in the host membrane and allow the S protein to form the pre-hairpin intermediate (a). The pre-hairpin begins to fold back on itself due to HR1 and HR2 interactions forming the bundle (b). During the S protein foldback, the two membranes will approach each other until the outer leaflets merge and eventually the inner leaflets merge (pore formation). Adapted from [99,100]. (c) Cartoon representation and closer look of the HR1-HR2 fusion core region of SARS-CoV-2. The HR motifs consist of a group of tandemly arranged seven-residue repeats. This sequence feature has the capacity to generate a hydrophobic interface along an α-helix, enabling the formation of a coiled-coil structural module when several HR-containing polypeptide chains are arranged together in parallel [101]. This fusion core is a particularly well studied target for antiviral peptides, as they can be designed in order to competitively inhibit this interaction. Relevant binding residues are labeled. Among the important amino acids responsible for the HR1-HR2 interaction, the residues colored in green may contribute to the enhanced interactions between HR1 and HR2 and stabilize 6-HB conformation of SARS-CoV- 2 compared with those of SARS-CoV (adapted from [34]).
Using these tools or the traditional trial-and-error approach, some AVPs have been evaluated in human or animal viruses, some of them important zoonotic agents. The pursuit of antiviral or virucidal agents has positioned AVPs, natural or designed as possible candidates for future effective drugs for veterinary or human health. Animal viral infections represent not only a risk for human health, considering that some of them may become important zoonotic epidemics, but also for the economic loses that infected farm animals represent.
2. AVPs against zoonotic viruses
2.1. Coronaviruses
Coronaviruses enter target cells by fusing the membranes of the virus and the host cell, which is mediated by the viral spike glycoprotein (S protein) [28], in the case of SARS-CoV and SARS-CoV-2, this type I viral fusion protein consists of two subunits (S1 and S2) on the viral membrane. The S2 subunit characteristically contains two heptad repeat regions (HR1 and HR2), that mediates the viral and host cell fusion.
The receptor binding is mediated by the S1 subunit, while the viral and cell membrane fusion is driven by the S2 subunit [29,30]. Within the S1 subunit, we can find the receptor-binding domain (RBD), which binds to the human angiotensin-converting enzyme 2 (ACE2), the cellular receptor on the cell surface, [31].
In the case of SARS-CoV-2, unlike other coronavirus of the same clade, the S protein contains a furin-like cleavage site [32]. The S2 subunit is composed of fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain (TM), and cytoplasmic domain fusion (CP) [33]
After the RBD-ACE2 binding, HR1 and HR2 domains in the S2 subunit interact with each other to form a six-helix bundle (6-HB) fusion core, this interaction brings the viral and cellular membranes into close to each other for fusion and subsequent infection [34]. The coronavirus fusion pathway is depicted in Fig. 1, showing some details of the HR1-HR2 fusion core region of SARS-CoV-2. Some relevant binding residues are labeled on the alpha-helix conformation. A group of these residues are believed to be responsible for the enhanced interactions and subsequent 6-helix bundle improved stabilization, contributing to the enhanced viral transmissibility or infectivity of SARS-CoV- 2 compared with SARS-CoV [34]
One of the main anti-viral peptide-based strategies employed against this virus has been the inhibition of the S protein-mediated membranes fusion. This mechanism that type I fusion protein viruses use to enter target cells has been well determined, and particularly the HR1-HR2 fusion core emerged as an interesting target for AVP therapies.
It has been demonstrated that the formation of the 6-HB could be competitively inhibited with peptides derived from the HR2 regions of SARS-CoV and MERS-CoV S protein, thereby preventing viral fusion and entry into host cells [35]
Another strategy has been to develop AVPs that target the RBD and inhibit its binding with ACE2, or the use of antimicrobial peptides that target the lipid membrane of this enveloped viruses ( Fig. 2 ), as it will be discussed below.
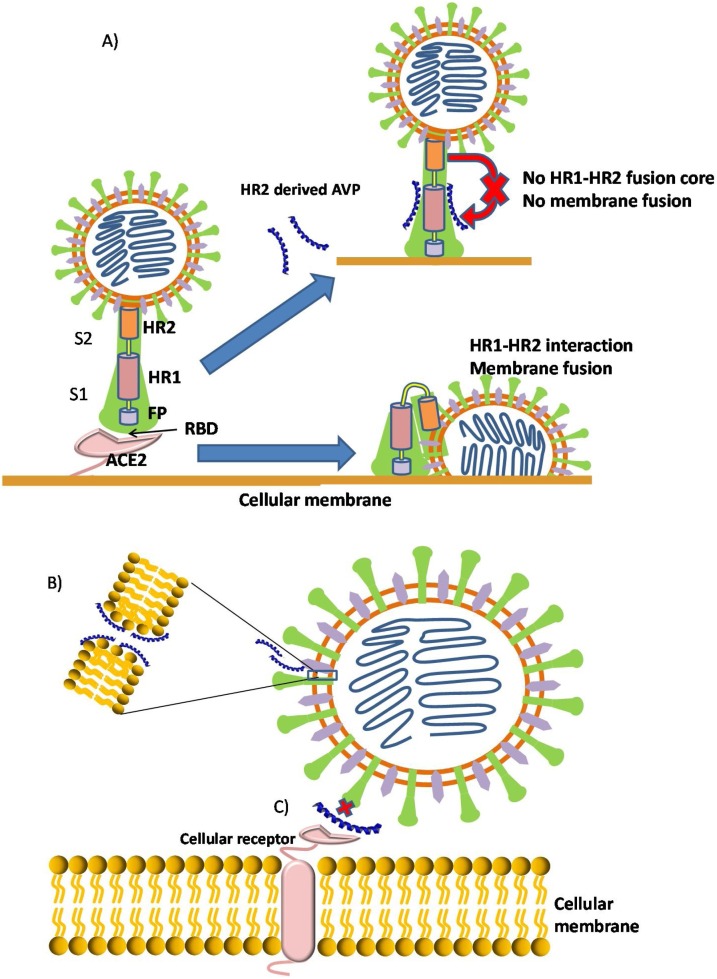
Fig. 2.
Antiviral peptides mechanisms of action against coronaviruses. A) Antiviral peptides blocking the HR1 and HR2 from forming a fusion-active core, thus preventing the viral and cellular membrane fusion. For simplicity, only one monomer is represented. B) Interaction of cationic amphipatic peptide can produce viral membrane disturbance and pore formation. C) Inhibition of viral entry with peptides that interrupt the recpetor binding, like the RBD –ACE2 interaction in coronavirus infection.
2.1.1. MERS-CoV
Middle East Respiratory Syndrome (MERS) is a viral respiratory illness caused by the zoonotic MERS coronavirus (MERS-CoV). MERS-CoV was first reported in 2012 in dromedaries in the Middle East and has since spread to other several countries in Africa and South Asia.
A series of peptides have been designed and tested with variable results against MERS-CoV since its discovery. A complete and comprehensive review together with a detailed list of these sequences and their characteristics have been described by Mustafa et al. [36].
Antimicrobial peptides targeting the lipid membrane of this virus were also evaluated. Mouse β-defensin-4 derived peptides, were designed and evaluated in a group of enveloped viruses. Hanjun Zhao and co-workers tested the antiviral activity of 11 peptides derived from this defensin and found that a short peptide, P9, exhibited potent and broad-spectrum antiviral effects against multiple respiratory viruses in vitro and in vivo, including influenza A virus, H1N1, H3N2, H5N1, H7N7, H7N9, but also in MERS-CoV and SARS-CoV [37]
It is worth to notice that peptides derived from either the virus spike protein or a mouse defensin, both displayed antiviral activity, but the defensin-derived peptide P9, like most AMPs against bacteria, displayed broad-spectrum activity, showing antiviral activity against multiple respiratory viruses. It is interesting that P9 did not prevent the entry of the virus to the cells, but impaired viral uncoating and viral RNA release from late endosomes for subsequent replication.
2.1.2. SARS-CoV
Severe acute respiratory syndrome (SARS) is a viral respiratory disease caused by a SARS-associated coronavirus (SARS-CoV). This virus is the cause of an atypical pneumonia that affected Asia, North America and Europe in 2002–2003 and then hit the whole world in a terrible pandemic in 2019-to date (SARS-CoV-2). Both viruses are genetically closely related. SARS-CoV was more lethal but it disappeared after intense sanitary measures to mitigate the spread of the virus. By contrast, SARS-CoV-2, which came on the scene in December 2019 in Wuhan, China, it spread all around the globe in the lapse time of a few months [38].
Using a computational approach Bruno Sainz Jr. and co-workers identified regions of the SARS-CoV fusion protein that potentially interact with bilayer membranes [29]. The authors identified five regions of high interfacial hydrophobicity within the S2 subunit of SARS-CoV and murine hepatitis virus (MHV). They found that peptides analogous to regions of the N-terminus or the pre-transmembrane domain of the S2 subunit inhibited infection of SARS-CoV by 40–70 % (evaluated as plaque formation counts) at concentrations ranging from 15 to 30 μM.
Shuai Xia and co-workers found that the HR1 binding peptide (HR2-derived peptide) EK1, showed interesting activity inhibiting multiple human coronaviruses. EK1 displayed inhibitory activity against all coronaviruses tested, including MERS-CoV and SARS-CoV, as well as bat SARS-CoVs. In In vivo experiments the authors demonstrated that EK1 administered via the nasal route exhibited protective effects and safety profiles [39,40]. It was also found that EK1C4, one of the modified EK1 peptides, was much more potent inhibiting the membrane fusion mediated by the S protein of SARS-CoV-2. These results lead the authors to suggest that EK1C4 could be a feasible therapeutic candidate against the currently circulating SARS-CoV-2 and other emerging SARS-CoVs [34]
Using molecular dynamics simulation of the fusion core, Ling and co-workers designed and computational-optimized antivirus peptides based on the HR2 region of SARS-CoV-2 S protein [33]. The authors designed a peptide that specifically inhibits the pre-hairpin conformation of SARS-CoV-2 as it binds to the HR1 domain, thus potentially blocking SARS-CoV-2 infection. Although they did not evaluate them in vitro or in vivo, they predicted that the antiviral peptides could prevent SARS-CoV-2 membrane fusion by competitively binding with HR1 to prevent forming of the fusion core. Fig. 3 shows the S protein sequence with its HR regions highlighted and the sequences of the designed candidate peptides.
Fig. 3.
a) Partial amino acid sequence of the surface glycoprotein of SARS-CoV-2 [ID: YP_009724390.1], where the fusion core regions of HR1 and HR2 are underlined and the candidate sequences for antiviral peptides HR1-P, HR2-P and HR2-antiP are colored. It is believed that, as it occurred in SARS-CoV, HR2-like peptide would competitively inhibits the binding of the HR2 domain to the HR1 domain, thus blocking the formation of viral fusion core, while HR1-like peptide would probably be less efficient in preventing HR1 and HR2 binding. Adapted from [33]. b) The SARS-CoV-2 S protein structure simulated by SWISS-MODEL.
Another important target for AVPs, is the receptor binding domain (RBD) on the S protein, which have epitopes that may serve as leads for the design of effective entry inhibitors [41]. Blocking the attachment of the virus to the human cell by specific peptides also demonstrated to be a suitable approach (Fig. 2 A). The binding affinity between ACE2 receptor on the host cell and RBD in S protein it was found to be higher in SARS-CoV-2 compared to SARS-CoV, which may explain the increased infectivity and transmissibility of SARS-CoV-2 [42]
Struck and co-workers identified the key binding motif of SARS-CoV viral protein that is used for its attachment to the human cell [43]. A group of sixteen linear 12mer peptides (RBD-1 to RBD-16) were synthesized which together they comprised residues N318-T509 of the viral RBD. Using surface plasmon resonance (SPR) binding studies with these peptides, the binding motifs in interaction with the human receptor ACE2 could be identified. With this data a hexapeptide was selected that carries a significant portion of the binding affinity of the virus to the human cell. The group demonstrated that the peptide (Tyr–Lys–Tyr–Arg–Tyr–Leu) was capable to reduce viral infection of epithelial cells by a factor of 600. This peptide displayed specificity against coronaviruses that binds to the ACE2 receptor, showing no interference with other viruses that utilize different receptors.
2.2. Rift Valley fever virus
Rift Valley fever (RVF) is a viral infection with significant importance for human health and economy, affecting people and livestock throughout the Arabian Peninsula and Africa [44,45]. The etiological agent is the Rift Valley fever virus (RVFV, family Bunyaviridae). The disease is most commonly seen in domesticated animals such as camels, buffalo, goats, sheep and cattle. People can get RVF through bites from infected mosquitoes but also from contact with body fluids or tissues of infected animals. RVFV infection is severe in animals, resulting in high mortality rates in newborns and near 100 % abortion rates in pregnant animals. According to the WHO, in humans the disease can be lethal, but it ranges from a mild flu-like illness to severe hemorrhagic fever. Besides, in about 1–10 % of infected patients the retinal inflammation can lead to permanent vision loss [44].
RVFV is an enveloped virus that requires the fusion of viral and cellular membranes for the viral genome to enter the cell. This entry into the cells is mediated by two glycoproteins, Gn and Gc, and the process is a pH-dependent activation mechanism following endocytosis [46].
Koehler and co-workers showed that peptides analogous to the RVFV Gc stem region inhibited RVFV infectivity in cell culture by inhibiting the fusion process [47]. Interestingly, one of the peptides (RVFV-6) was capable of inhibiting multiple, diverse viruses in addition to RVFV: Ebola virus, Andes virus or vesicular stomatitis virus, which encode class I, class II and class III fusion proteins, respectively. They could demonstrate that the peptide, although it did not prevent binding to host cells, it interacts with the glycoprotein Gc after acidification, preventing the viral fusion.
The authors propose that RVFV-6 prevents infection by first attaching to the viral and cellular membranes and subsequent internalization together with the virion. After internalization, the low pH environment of the endosome triggers a conformational rearrangement of Gc that exposes a previously hidden stem domain and RVFV-6, analogous to the RVFV stem, may interact with this domain, blocking interactions between the virion and cell membranes [47].
2.3. Eastern equine encephalitis virus
Alphaviruses - including Venezuelan Equine Encephalitis Virus (VEEV), Eastern Equine Encephalitis Virus (EEEV), and Western Equine Encephalitis Virus (WEEV) – are single-stranded positive sense RNA viruses that belong to the Togaviridae family [48,49]. The transmission between species and between humans occurs via mosquitos. Therefore, these viruses are also classified as arboviruses, as the virus replicates in arthropod vectors before being transferred to vertebrates. The virus affects equines and is also important human pathogens. Horses are dead-end hosts for EEE and WEE virus but not for some VEE strains. In this case, horses are the reservoir species and cause clinical disease in both horses and humans [50,51]
Infections with VEEV, after a very acute course, can be lethal in horses, even without any neurologic signs. In spite of that high mortality rate in animals, in humans the disease has low morbidity and mortality [52].
Western equine encephalitis (WEE) is an uncommon viral illness of horses and human, these species are often “dead-end” hosts [53]. Most people infected with WEE virus will have either no symptoms or a very mild illness. The WEEV mortality rate in horses is higher than in humans, ranging between 20%–50% in horses showing clinical signs.
Treatment of these viral infections is supportive, as there are no specific antiviral therapies. In search for new approaches to this viral illness, Ahmed and co-workers [54] have evaluated the antiviral activity of the well-known human cathelicidin LL-37 against different strains of VEEV. The assays were carried out in multiple neuronal cell lines. The authors observed a significant decrease in viral replication after treatment with this peptide, evaluated at early time points (3 h post-infection, hpi) and at 16 hpi (confirmed by genomic quantification of viral particles and by viral titer), demonstrating that the peptide could exert its antiviral activity interacting with the virion as an entry inhibitor, affecting early stages of the infectious process. Authors suggest that LL-37 would probably penetrates the viral membrane or otherwise destroys its integrity, in agreement with a previously published work [55] that demonstrated the LL-37 inhibition of Kaposi sarcoma-associated herpesvirus by viral membrane disruption. Moreover, the authors also determined that LL-37 could induce the expression of immunomodulatory cytokines (modulating interferon expression).
As a result they concluded that this peptide inhibits, in a direct and indirect mode, VEEV replication, establishing an antiviral state in the host cells.
2.4. Junin virus
Junin virus (JUNV, family Arenaviridae) is the etiological agent of Argentine hemorrhagic fever (AHF), an endemo-epidemic disease restricted geographically to the Argentinean most fertile areas. JUNV is a enveloped virus with two ambisense single-stranded RNA molecules. It is generally associated with rodent-transmitted infections in human beings [56,57]. In the USA ribavirin is the only approved antiviral drug for treating arenaviruses; although it has reported undesirable secondary reactions [58,59]. Therefore, there is an urgent demand to develop novel therapeutics against arenaviruses. For this purpose, many antimicrobial peptides have been assayed against JUNV.
As was pointed above, some AMPs can also exhibit antiviral activity, in this sense, Malatani and co-workers tested different cationic AMPs against JUNV. Melittin, the principal constituent in the venom of the European honeybee Apis mellifera, has shown in vitro inhibition of JUNV replication at non-toxic concentration ranges (0.5–3 μM). It was found that 3 μM of this peptide was able to reduce a 99 % of JUNV infectivity, suggesting that this peptide could be a serious candidate for the development of an anti-viral therapy against this virus [58]. Results with cercopin A showed that this peptide effectively inhibited JUNV multiplication with a much higher selectivity index than melittin. Cecropin A treatment reduced viral protein synthesis, but it did not affect host cell protein synthesis. The analysis of the effects of cecropin A as a function of time indicated that the inhibitory action of the compound takes place mainly at the late stages of the viral multiplication cycle. This inhibition is achieved through the prevention of viral morphogenesis and release from infected cells [58]. Finally, indolicidin, another well-known AMP, also showed virucidal activity against JUNV but much less potent than cecropin A. Unlike cecropin A, indolicidin seems to be capable to directly inactivate the virus particles when exerting its antiviral activity [58].
Spence and co-workers have identified AVP-p, a peptide derived from a membrane-anchored fusion protein GP2 from arenaviruses [60]. The arenavirus GP2 is considered a class I viral fusion protein, thus virus-host cell fusion is mediated by the rearrangement of GP2 trimers. The results obtained showed antiviral activity with a 50 % inhibitory dose (IC50) at 7 μM of AVP-p, showing complete inhibition of viral plaque formation at approximately 20 μM, without acute cytotoxicity. AVP-p demonstrated activity against viruses possessing the arenavirus viral glycoprotein complex of Old and New World but not against other enveloped virus families. Regarding its mechanism of action, it was shown by fusion assays with liposomes, that the peptide impairs the fusion machinery of the virions with the host cells by preventing binding to the cell receptor or the subsequent endosomal fusion.
2.5. Dengue virus
Dengue virus (DENV, family Flaviviridae) is a positive-sense RNA virus, encapsulated by a lipid membrane. The four dengue virus serotypes are major mosquito-transmitted human pathogens that infect approximately 100 million people annually with no available therapeutic treatment yet. The viral genome has three structural proteins: capsid, precursor membrane and envelope (E), and seven non-structural proteins also implicated in viral replication [61]. Several peptide-based approaches have been evaluated in order to inhibit DENV infection. These approaches have been directed both at the host cell receptors and at structural and non-structural proteins of the virus; most of these strategies are summarized in the review of Chew and co-workers [62]. Among these different approaches, it is particularly interesting the peptide DN59, a 33 amino acid peptide that mimics a highly conserved region of the stem region of the E protein, and inhibits the infectivity of the Dengue 2 virus and West Nile virus, another flavivirus [63]. Interestingly, in a later study, it was found that DN59 after binding the protein E can reach the viral lipid membrane inducing the formation of holes (Fig. 2B) causing the release of the viral genome, resulting in an irreversibly destroy of the virus infectivity capacity [64].
3. AVPs against non-zoonotic farm animal viruses
3.1. Poultry viruses
Poultry represents the largest domestic animal stock in the world, representing more than 40 % of all animal production (Meat market review: Emerging trends and outlook, FAO December 2020). In this sense, poultry diseases have a great impact on human well-being, especially in rural areas where chickens are a crucial source of income and food. In particular, viral infections represent a great concern especially in countries with industrialized poultry production, which have been spending large budgets to prevent this type of infection or at least reduce economic losses [65]. As an attempt to find an effective treatment for this kind of diseases, some peptide-based approaches were successfully applied.
3.1.1. Newcastle disease (ND) and avian infectious bronchitis (IB)
Among viral infections, Newcastle disease (ND) can have dramatic effects on the global economy of domestic poultry production [66]. However, despite extensive research, there is no effective therapy available against this viral infection. Newcastle disease virus (NDV, family Paramyxoviridae) is an enveloped virus with a genome of negative-sense single-stranded RNA. Two different glycoproteins protrude outside from the lipid membrane, the haemagglutinin-neuraminidase (HN) and the fusion protein (F). The first one is a multifunctional protein that is also a major antigenic determinant of paramyxoviruses [66,67].
Avian infectious bronchitis (IB) is an acute, highly contagious disease caused by infectious bronchitis virus (IBV, family Coronaviridae) an avian Coronavirus of the domestic chicken (Gallus gallus). As for ND, the economic consequences are severe in the poultry industry worldwide [68].
In order to inhibit the fusion of the viral and cell membranes, recombinant peptides based on the heptad repeat regions of NDV fusion glycoproteins were designed, obtaining a new series of peptides called NOVEL [69]. It is worth noticing that both viruses (NDV and IBV) adopt a similar fusion mechanism, which has been described above (type I fusion proteins), in which a key step is the formation of a coiled coil between the trimer HR1 and HR2 of viral fusion proteins. These peptides were tested by plaque formation and chicken embryo infectivity assays and it was found that peptides NOVEL-1 and NOVEL-2 were the most active molecules. Interestingly these peptides completely inhibited both single virus infections as well as mixed infections caused by NDV and IBV. These results point out the feasibility of designing relatively broad-spectrum antiviral peptides capable to reduce the effects of mixed-infections [69]. Later, as a strategy for improving the activity of the NOVEL-2 peptide described above, a Cholesterol Tag or PEG moieties in the N-terminus of the peptide were added [10]. It has been previously reported for other antiviral peptides that the antiviral potency can improve by directing the peptide to the cell compartment where fusion occurs, with the addition of cholesterol as membrane anchor [70,71]. The results obtained for the NOVEL-2 conjugated with cholesterol or polyethylene glycol exhibited even more promising antiviral activity in experiments with animal models. Both peptides (cholesterol- and cholesterol-PEG3-tagged) were protective for chicken embryos against several serotypes of NDV and IBV, when administered 12 h before virus inoculation. On the other hand, for untagged peptides, an intervention near the time of viral inoculation was required to achieve similar protection levels [10].
In another work carried out by Ramanujam and co-workers, the phage display technique was used to select specific ligands that interact with NDV. Using this technique the amino acid sequence Thr-Leu-Thr-Thr-Lys-Leu-Tyr was selected. Afterward, synthetic peptides with the sequence Cys-Thr-Leu-Thr-Thr-Lys-Leu-Tyr-Cys, either in linear or cyclic conformations were obtained and evaluated against NDV. The two flanking cysteine residues added to the sequence increased the inhibition activity of the synthetic peptide. Both peptides showed inhibition of the hemolytic activity of the virus as well as its propagation in embryonated chicken eggs [67]. Afterwards, structure studies on the peptides showed that the peptide displayed better activity as a cyclic arrangement than a linear one, suggesting that cyclization may allow a more stable conformation compared to the linear peptide [72].
Considering that many AMPs have been characterized with dual activity against bacteria and viruses, either directly or by the recruitment of immune cells [73,74], a more general strategy was designed. The antiviral effect of antimicrobial lipopeptides (AMLs) isolated from Bacillus subtilis fmbj that contain the lipopeptides surfactin, fengycin and their isoforms were evaluated against NDV [75]. The results obtained in that work demonstrated that AMLs had a direct inactivation, and successfully inhibited infection and replication of NDV with EC50 values of 7.31 μM. Although the actual antiviral mechanism was not described by the authors, they could demonstrate that surfactin is active against several viruses. The fact that this lipopeptide showed higher activity against enveloped viruses could suggest that the antiviral action of surfactin is primarily by surfactant action of this molecule on the lipids of the virus membrane [76].
In a similar strategy but now against IBV, swine intestine antimicrobial peptides (SIAMP) were evaluated against IBV in chick embryos [77]. SIAMP were obtained from an acetic acid extraction from pig small intestines and contains among others cecropin P1, a well-known antimicrobial peptide [78]. Embryos treated with SIAMP after being inoculated with a lethal dose of IBV showed no evident differences compared with the control; in contrast to non-treated inoculated embryos. However, when embryos were pretreated with SIAMP the mortality induced by IBV dropped dramatically. Although the antiviral mechanism was not characterized, the findings of the study suggested that preincubation with SIAMP may prevent viral fusion to host cells and therefore decrease the IBV infection [77]. Further studies of cecropin P1 against other viruses as Porcine reproductive and respiratory syndrome virus, showed that this peptide blocks the attachment and replication of the virus [79].
3.1.2. Avian leukosis
Avian leukosis (AL) is a tumorigenic disease produced by avian leukosis virus (ALV, family Retroviridade). This virus is also a naturally occurring avian virus that affects poultry worldwide causing enormous economic issues in the poultry industry since its emergence [80]. In this sense, programs for virus eradication should be applied to reduce economic damage caused by ALVs [81]. In chickens, based on their viral envelope glycoproteins, the ALVs are divided into six subgroups: A, B, C, D, E, and J [80]. Zeng et al. (2019) evaluated a novel recombinant hybrid polypeptide based on bursin peptide. It was previously demonstrated that bursin, a tripeptide (Lys-His-Gly-NH2) hormone selectively stimulates avian B cell differentiation [81] and promotes immune-globulin (Ig) switching from IgM to IgG [9]. Furthermore, a novel stabilized bursin mimetics, Gagnon’s tetrapeptide (Lys-Asn-Pro-Tyr), induces the selective expansion of B cells as well as the stimulation of cytotoxic T lymphocytes [10]. In this way, the authors evaluated a hybrid polypeptide (BLP) containing a tandem array of Gagnon’s peptide and albumin-binding peptide, in in vivo studies they found that BLP injection-induced body weight recovery in chickens infected with ALV. To dissect the antiviral action of this synthetic peptide, in vitro studies were carried out showing that the peptide might inhibit virus adsorption onto DF-1 cells. On the other hand, BLP treatment subsequently applied to virus inoculation did not significantly reduce virus infection. Overall the authors stated that peptide BLP showed potential as an antiretroviral drug in chickens diminishing the effects of ALV infection [81].
3.1.3. Tembusu virus
Tembusu virus (TMUV, family Flaviviridae) poses a positive-sense single-stranded RNA genome. TMUV exhibits a wide range of natural host species, including mosquitos, chickens, ducks, geese, pigeons, and sparrows [82]. Particularly, when TMUV infects ducks and geese these animals exhibit a rapid decrease in daily food intake and egg production. In the last outbreak in China, the rapid spread of TMUV resulted in important economic losses in the major waterfowl-breeding.
As was shown above for other viruses, blocking the first step of the viral interaction with the host cell prevents the infection itself. In this sense in the case of TMUV, the envelope (E) protein at the surface of mature virions drives the attaching of the virion to the cell surface receptors. After the virus is internalized by endocytosis, under the low-pH conditions of the endosome, as for other class II type fusion proteins, the dissociation of E homodimer exposed the fusion loop in domain II of the protein (DII) that inserts into the target membrane, then the E protein interacts to form a protein trimer that folds back directing the E stem anchor domain towards the fusion loop [83]. In a recent study, the peptide TP1 derived from domain DII and peptide TP2 derived from the stem region of E protein were synthesized and evaluated against TMUV infection. Both peptides showed antiviral activity by preventing the binding of TMUV to the target cells. This inhibition was achieved by direct binding of TP1 or TP2 to TMUV, but not to target cells. Furthermore, RNase digestion assay showed that virions treated with TP1 are sensitivities, suggesting that the peptide was able to disrupt the viral membrane allowing the escape of genomic viral particles. As it was previously reported for other peptides, after the peptide binds to the target protein it likely inserts itself between the E protein and the lipid membrane, resulting in a disruption of the virion membrane bilayer structure [84]. In the case of TP2, this peptide showed antiviral activity against TMUV as well as Japanese encephalitis virus (another Flaviviridae virus), and due to the highly conserved stem regions of TMUV and other flaviviruses, TP2 may probably inhibit other flaviviruses. Therefore, TP2 and TP2 derivatives or analogous peptides might probably work as broad-spectrum flavivirus inhibitors [85].
3.1.4. Avian influenza
Avian influenza Type A viruses (AIV, family Orthomyxoviridae) is a group of enveloped viruses with a segmented and negative-stranded RNA genome. This pathogen is distributed throughout the world and is responsible for avian flu, one of the most serious avian diseases that cause serious economic losses to the poultry industry [86]. Although AIV is also an important pathogen of humans, responsible of severe and some times deadly respiratory diseases, in this section, we will only discuss antiviral peptides strategies against livestock infections.
By using a phage display library against AIV subtype H9N2 the peptide sequence Asn-Asp-Phe-Arg-Ser-Lys-Thr among other major sequences has emerged. Subsequently, this peptide in its linear and circular form (with the sequence Cys-Asn-Asp-Phe-Arg-Ser-Lys-Thr-Cys) was tested in vitro and in ovo. The antiviral activity was evaluated in embryonated chicken eggs for both synthetic peptides and the fusion phages. All the peptides displayed high levels of anti-viral activity against AIV, demonstrating their value as antiviral molecules at various potential levels, being the cyclic peptide the most active molecule. Due to the peptides showed a strong anti-viral activity inhibiting the hemagglutination when were applied at a pre-infection time but not at post-infection stages, the authors claimed that both peptides prevent the viral replication at the initial step of the life cycle of the virus, either inhibiting the attachment or the entry of the virus into the target cells [86]. In a later study conducted by the same group, they confirmed that the linear version of peptide Cys-Asn-Asp-Phe-Arg-Ser-Lys-Thr-Cys inhibits the replication of AIV, by specifically blocking its attachment to the host cell thus preventing the expression of early viral genes [87]. Interestingly, the short sequence of the peptides implies an advantage for its further commercialization as it can greatly reduce the production costs.
3.2. AVPs against cattle viruses
3.2.1. Bovine viral diarrhea virus
Bovine viral diarrhea virus (BVDV, family Flaviviridae) is an enveloped single-stranded, positive-sense RNA virus of approximately 12.3 Kb. The genome presents an open reading frame (ORF) flanked by untranslated regions (UTRs) [88,89], which encodes a poly-protein of almost 4000 amino acids [90]. BVDV is a pathogen that causes economic losses for the national [91] and the global livestock industry, mainly due to reproductive complications [92,93]. BVDV infection can be acute, fatal (syndrome called "Mucosal Disease") or can cause infertility, abortions and congenital anomalies [94,95]. The infection of pregnant females between 40 and 120 days of gestation is epidemiologically relevant since it leads to the birth of persistently infected (PI) calves. PI animals are immunotolerant to the infecting BVDV strain and they continuously disperse the virus into the herd [91].
Some countries have eradication and control programs for the disease. Immunity induced by commercial vaccines, even attenuated ones, requires more than 2 weeks to develop and more than one dose, leaving a window of vulnerability. The use of antiviral treatments could prevent the spread of BVDV in herds in cases of an outbreak and even protect pregnant females from aborting or producing PI individuals. This procedure could be used in combination with vaccination and would avoid the period of vulnerability to which animals are exposed to until adaptive immunity develops.
For this purpose, natural and synthetic peptides have been tested as potential agents that can inhibit or diminish BVDV infection. It is important to note that naturally occurring substances are characterized generally by greater biocompatibility and safety. Although many compounds have demonstrated potent anti-BVDV activity, the involved mechanisms of action for all of them is still to be elucidated.
Małaczewska and co-workers [96] characterized the antiviral effects of different positively charged natural compounds: two animal proteins, bovine lactoferrin and chicken egg lysozyme, and nisin a bacteriocin from Lactococcus lactis. These substances were in vitro tested, both individually and in combination on BVDV infection. The bacteriocin nisin demonstrated efficacy in reducing the extracellular virus titer and the intracellular BVDV RNA. Bovine lactoferrin showed the strongest anti-BVDV effect and lysozyme the weakest among all the tested compounds. Although none of the natural compounds had virucidal activity, all showed better effects while they were present throughout the course of the infection. Interestingly, the only compound that displayed a protective effect on the cell and was able to inhibit viral attachment was lactoferrin.
When the authors tested combination of peptides they observed that nisin plus lactoferrin and lysozyme plus lactoferrin mixtures were more effective in inhibiting virus replication, compared to the single treatments. These results, obtained after the combined administration of peptides with different modes of action, would make it possible to decrease the therapeutic dose and to reduce possible side effects [97,98].
4. Conclusions
Antiviral peptides are being considered a promising and novel strategy against viruses, principally for enveloped RNA viruses, which represent a plausible pandemic threat. The different strategies that peptides can display range from inhibiting viral entry into the host cell to inactivating the viral particle through peptide-membrane interactions. These characteristics highlight the versatile capabilities that peptides possess as antiviral drugs, probably driving the future research and development in the post−COVID-19 era.
Acknowledgments
The authors acknowledge the financial support of CONICET (PIP 11220130100383CO), ANPCyT-FONCyT (PICT 2016-0478, PICT 2017-2349), Universidad Nacional de Santiago del Estero (PI-UNSE 23A/250), Universidad Nacional de Hurlingham (PIUNAHUR- Art. 11-09/2020) and Universidad Nacional de Quilmes (Programa Microbiología básica y aplicada a Agronomía, Alimentos y Salud). PCM, NPC and AH are members of the Research Career of CONICET. JCE acknowledge a fellowship from CONICET. PCM also wants to acknowledge the CYTED Program (Programa Iberoamericano de Ciencia y Tecnología para el Dearrollo, RED 219RT0573 – Desarrollo de péptidos antivirales y antimicrobianos para cepas multirresistentes)
References
- 1.Bengis R.G., Leighton F.A., Fischer J.R., Artois M., Mörner T., Tate C.M. The role of wildlife in emerging and re-emerging zoonoses. OIE Rev. Sci. Tech. 2004;23:497–511. [PubMed] [Google Scholar]
- 2.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bres P., Seeliger H. Public health action in emergencies caused by epidemics. Mycoses. 2009;30 pp. 613–613. [Google Scholar]
- 4.Rajeev R., Prathiviraj R., Kiran G.S., Selvin J. Zoonotic evolution and implications of microbiome in viral transmission and infection. Virus Res. 2020;290 doi: 10.1016/j.virusres.2020.198175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeraka N.M., Sadhu S.P., Madhunapantula S.R.V., Rao Pragada R., Svistunov A.A., Nikolenko V.N., Mikhaleva L.M., Aliev G. Strategies for targeting SARS CoV-2: small molecule inhibitors—the current status. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.552925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adalja A., Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. Anti. Ther. 2019;17:467–470. doi: 10.1080/14787210.2019.1635009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigant F., Santos N.C., Lee B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C.-G., Tang W., Chi X.-J., Dong Z.-M., Wang X.-X., Wang X.-J. A cholesterol tag at the N terminus of the relatively broad-spectrum fusion inhibitory peptide targets an earlier stage of fusion glycoprotein activation and increases the peptide’s antiviral potency in vivo. J. Virol. 2013;87:9223–9232. doi: 10.1128/JVI.01153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenssen H., Hamill P., Hancock R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez M.L., Martínez M.M.B., Maffia P.C. Natural antimicrobial peptides: pleiotropic molecules in host defense. CellBio. 2013;02:200–210. [Google Scholar]
- 13.van Harten R.M., van Woudenbergh E., van Dijk A., Haagsman H.P. Cathelicidins: immunomodulatory antimicrobials. Vaccines. 2018;6 doi: 10.3390/vaccines6030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shartouny J.R., Jacob J. Mining the tree of life: host defense peptides as antiviral therapeutics. Semin. Cell Dev. Biol. 2019;88:147–155. doi: 10.1016/j.semcdb.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Lai Y., Gallo R.L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez M., Polizzotto A., Flores N., Semorile L., Maffía P.C. Antibacterial, anti-biofilm and in vivo activities of the antimicrobial peptides P5 and P6.2. Microb. Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103886. [DOI] [PubMed] [Google Scholar]
- 17.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollmann A., Martinez M., Maturana P., Semorile L.C., Maffia P.C. Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018;6 doi: 10.3389/fchem.2018.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G., Li X., Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:1087–1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X., Wu H., Lu H., Li G., Huang Q. LAMP: a database linking antimicrobial peptides. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed A., Siman-Tov G., Hall G., Bhalla N., Narayanan A. Human antimicrobial peptides as therapeutics for viral infections. Viruses. 2019;11 doi: 10.3390/v11080704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilas Boas L.C.P., Campos M.L., Berlanda R.L.A., de Carvalho Neves N., Franco O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019;76:3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Cogliano M.E., Hollmann A., Martinez M., Semorile L., Ghiringhelli P.D., Maffía P.C., Bentancor L.V. Cationic antimicrobial peptides inactivate shiga toxin-encoding bacteriophages. Front. Chem. 2017;5 doi: 10.3389/fchem.2017.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney C., Haslam N.J., Pollastri G., Shields D.C. Towards the improved discovery and design of functional peptides: common features of diverse classes permit generalized prediction of Bioactivity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Martínez R., Ramírez-Salinas G.L., Correa-Basurto J., Barrón B.L. Inhibition of influenza a virus infection in vitro by peptides designed in Silico. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang K.Y., Yang J.R. Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakur N., Qureshi A., Kumar M. AVPpred: Collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012;40:199–204. doi: 10.1093/nar/gks450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sainz B., Mossel E.C., Gallaher W.R., Wimley W.C., Peters C.J., Wilson R.B., Garry R.F. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taguchi F., Shimazaki Y.K. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- 31.Huang I.C., Bosch B.J., Li F., Li W., Kyoung H.L., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., Jiang Y. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130 doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F.W., Du L., Yu F., Ma C., Ye S., Yuen K.Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mustafa S., Balkhy H., Gabere M.N. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J. Infect. Public Health. 2018;11:9–17. doi: 10.1016/j.jiph.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K.M., Chan C.C.S., Leung H.C., Fai N., Lin Y.P., Zhang A.J.X., Jin D.Y., Yuen K.Y., Zheng B.J. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:238–244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV - A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loganathan S.K., Schleicher K., Malik A., Quevedo R., Langille E., Teng K., Oh R.H., Rathod B., Tsai R., Samavarchi-Tehrani P., Pugh T.J., Gingras A.C., Schramek D. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science. 2020;367:1264–1269. doi: 10.1126/science.aax0902. [DOI] [PubMed] [Google Scholar]
- 43.Struck A.W., Axmann M., Pfefferle S., Drosten C., Meyer B. A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 2012;94:288–296. doi: 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madani T.A., Al-Mazrou Y.Y., Al-Jeffri M.H., Mishkhas A.A., Al-Rabeah A.M., Turkistani A.M., Al-Sayed M.O., Abodahish A.A., Khan A.S., Ksiazek T.G., Shobokshi O. Rift valley fever epidemic in Saudi Arabia: epidemiological, clinical, and laboratory characteristics. Clin. Infect. Dis. 2003;37:1084–1092. doi: 10.1086/378747. [DOI] [PubMed] [Google Scholar]
- 45.Adam A.A., Karsany M.S., Adam I. Manifestations of severe Rift Valley fever in Sudan. Int. J. Infect. Dis. 2010;14:179–180. doi: 10.1016/j.ijid.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 46.de Boer S.M., Kortekaas J., Spel L., Rottier P.J.M., Moormann R.J.M., Bosch B.J. Acid-activated structural reorganization of the rift valley fever virus gc fusion protein. J. Virol. 2012;86:13642–13652. doi: 10.1128/JVI.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koehler J.W., Smith J.M., Ripoll D.R., Spik K.W., Taylor S.L., Badger C.V., Grant R.J., Ogg M.M., Wallqvist A., Guttieri M.C., Garry R.F., Schmaljohn C.S. A fusion-inhibiting peptide against rift valley fever virus inhibits multiple, diverse viruses. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver S.C., Ferro C., Barrera R., Boshell J., Navarro J.C. Venezuelan equine encephalitis. Annu. Rev. Entomol. 2004;49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- 49.Hsiung G.D., Chang P.W. Handb. Zoonoses. second ed. Sect. B Viral Zoonoses; 2017. Parainfluenza viral infection; pp. 409–421. [Google Scholar]
- 50.Eloit M., Schmitt B. World Organisation for Animal Health; Paris, France: 2017. OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2017. [Google Scholar]
- 51.White G., Ottendorfer C., Graham S., Unnasch T.R. Competency of reptiles and amphibians for eastern equine encephalitis virus. Am. J. Trop. Med. Hyg. 2011;85:421–425. doi: 10.4269/ajtmh.2011.11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Go Y.Y., Balasuriya U.B.R., Lee C. Zoonotic encephalitides caused by arboviruses: transmission and epidemiology of alphaviruses and flaviviruses. Clin. Exp. Vaccine Res. 2014;3 doi: 10.7774/cevr.2014.3.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zacks M.A., Paessler S. Encephalitic alphaviruses. Vet. Microbiol. 2010;140:281–286. doi: 10.1016/j.vetmic.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed A., Siman-Tov G., Keck F., Kortchak S., Bakovic A., Risner K., Lu T.K., Bhalla N., de la Fuente-Nunez C., Narayanan A. Human cathelicidin peptide LL-37 as a therapeutic antiviral targeting Venezuelan equine encephalitis virus infections. Antiviral Res. 2019;164:61–69. doi: 10.1016/j.antiviral.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Brice D.C., Toth Z., Diamond G. LL-37 disrupts the Kaposi’s sarcoma-associated herpesvirus envelope and inhibits infection in oral epithelial cells. Antiviral Res. 2018;158:25–33. doi: 10.1016/j.antiviral.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paweska J.T. Emerg. Infect. Dis. Clin. Case Stud. Elsevier; 2014. Lujo virus hemorrhagic fever; pp. 95–110. [Google Scholar]
- 57.King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B. Vol. 9. Elsevier; 2011. pp. 1–1327. (Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses). [Google Scholar]
- 58.Albiol Matanic V.C., Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int. J. Antimicrob. Agents. 2004;23:382–389. doi: 10.1016/j.ijantimicag.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 59.Grant A., Seregin A., Huang C., Kolokoltsova O., Brasier A., Peters C., Paessler S. Junín virus pathogenesis and virus replication. Viruses. 2012;4:2317–2339. doi: 10.3390/v4102317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spence J.S., Melnik L.I., Badani H., Wimley W.C., Garry R.F. Inhibition of arenavirus infection by a glycoprotein-derived peptide with a novel mechanism. J. Virol. 2014;88:8556–8564. doi: 10.1128/JVI.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murrell S., Wu S.C., Butler M. Review of dengue virus and the development of a vaccine. Biotechnol. Adv. 2011;29:239–247. doi: 10.1016/j.biotechadv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 62.Chew M.F., Poh K.S., Poh C.L. Peptides as therapeutic agents for dengue virus. Int. J. Med. Sci. 2017;14:1342–1359. doi: 10.7150/ijms.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lok S.M., Costin J.M., Hrobowski Y.M., Hoffmann A.R., Rowe D.K., Kukkaro P., Holdaway H., Chipman P., Fontaine K.A., Holbrook M.R., Garry R.F., Kostyuchenko V., Wimley W.C., Isern S., Rossmann M.G., Michael S.F. Release of Dengue Virus Genome Induced by a Peptide Inhibitor. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suarez D.L., Miller P.J., Koch G., Mundt E., Rautenschlein S. In: Diseases of Poultry. 14th edn. Swayne D.E., Martine B., Logue C.M., McDougald L.R., Venugopal N., Suarez D.L., editors. 2020. Newcastle disease, other avian paramyxoviruses, and avian metapneumovirus infections; pp. 109–166. [Google Scholar]
- 66.Brown V.R., Bevins S.N. A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Vet. Res. 2017;48 doi: 10.1186/s13567-017-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramanujam P., Tan W.S., Nathan S., Yusoff K. Novel peptides that inhibit the propagation of Newcastle disease virus. Arch. Virol. 2002;147:981–993. doi: 10.1007/s00705-001-0778-y. [DOI] [PubMed] [Google Scholar]
- 68.Khataby K., Fellahi S., Loutfi C., Mustapha E.M. Avian infectious bronchitis virus in Africa: a review. Vet. Q. 2016;36:71–75. doi: 10.1080/01652176.2015.1126869. [DOI] [PubMed] [Google Scholar]
- 69.Wang X.J., Li C.G., Chi X.J., Wang M. Characterisation and evaluation of antiviral recombinant peptides based on the heptad repeat regions of NDV and IBV fusion glycoproteins. Virology. 2011;416:65–74. doi: 10.1016/j.virol.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Hollmann A., Matos P.M., Augusto M.T., Castanho M.A.R.B., Santos N.C. Conjugation of cholesterol to HIV-1 fusion inhibitor C34 increases peptide-membrane interactions potentiating its action. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pessi A., Langella A., Capitò E., Ghezzi S., Vicenzi E., Poli G., Ketas T., Mathieu C., Cortese R., Horvat B., Moscona A., Porotto M. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chia S.L., Tan W.S., Shaari K., Abdul Rahman N., Yusoff K., Satyanarayanajois S.D. Structural analysis of peptides that interact with Newcastle disease virus. Peptides. 2006;27:1217–1225. doi: 10.1016/j.peptides.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 73.Hsieh I.N., Hartshorn K.L. The role of antimicrobial peptides in influenza virus infection and their potential as antiviral and immunomodulatory therapy. Pharmaceuticals. 2016;9 doi: 10.3390/ph9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomes B., Augusto M.T., Felício M.R., Hollmann A., Franco O.L., Gonçalves S., Santos N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018;36:415–429. doi: 10.1016/j.biotechadv.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 75.Huang X., Lu Z., Zhao H., Bie X., Lü F.X., Yang S. Antiviral activity of antimicrobial lipopeptide from Bacillus subtilis fmbj against pseudorabies virus, porcine parvovirus, Newcastle Disease Virus and infectious bursal disease virus in vitro. Int. J. Pept. Res. Ther. 2006;12:373–377. [Google Scholar]
- 76.Zhao H., Shao D., Jiang C., Shi J., Li Q., Huang Q., Rajoka M.S.R., Yang H., Jin M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017;101:5951–5960. doi: 10.1007/s00253-017-8396-0. [DOI] [PubMed] [Google Scholar]
- 77.Sun Q., Wang K., She R., Ma W., Peng F., Jin H. Swine intestine antimicrobial peptides inhibit infectious bronchitis virus infectivity in chick embryos. Poult. Sci. 2010;89:464–469. doi: 10.3382/ps.2009-00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee J.Y., Boman A., Chuanxin S., Andersson M., Jornvall H., Mutt V., Boman H.G. Antibacterial peptides from pig intestine: isolation of a mammalian cecropin. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9159–9162. doi: 10.1073/pnas.86.23.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo C., Huang Y., Cong P., Liu X., Chen Y., He Z. Cecropin P1 inhibits porcine reproductive and respiratory syndrome virus by blocking attachment. BMC Microbiol. 2014;14:1–11. doi: 10.1186/s12866-014-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Payne L.N., Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 81.Zeng Y., Gong Z., Wu B., Guan W., Yu S., An Y., Lu R., Zhao J., Wu Y., Huang Y., Wu X. A novel Bursin-like peptide as a potential virus inhibitor and immunity regulator in SPF chickens infected with recombinant ALV. BMC Vet. Res. 2019;15 doi: 10.1186/s12917-019-2192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou H., Yan B., Chen S., Wang M., Jia R., Cheng A. Evolutionary characterization of Tembusu virus infection through identification of codon usage patterns. Infect. Genet. Evol. 2015;35:27–33. doi: 10.1016/j.meegid.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 83.Rodenhuis-Zybert I.A., Wilschut J., Smit J.M. Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010;67:2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Y., Deng Y.Q., Zou P., Wang Q., Dai Y., Yu F., Du L., Zhang N.N., Tian M., Hao J.N., Meng Y., Li Y., Zhou X., Chan J.F.W., Yuen K.Y., Qin C.F., Jiang S., Lu L. A peptide-based viral inactivator inhibits Zika virus infection in pregnant mice and fetuses. Nat. Commun. 2017;8:1–12. doi: 10.1038/ncomms15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao D., Zhang L., Han K., Liu Q., Yang J., Huang X., Liu Y., Li Y., Zhao P. Peptide inhibitors of tembusu virus infection derived from the envelope protein. Vet. Microbiol. 2020;245 doi: 10.1016/j.vetmic.2020.108708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rajik M., Jahanshiri F., Omar A.R., Ideris A., Hassan S.S., Yusoff K. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol. J. 2009;6:1–12. doi: 10.1186/1743-422X-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajik M., Omar A.R., Ideris A., Hassan S.S., Yusoff K. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int. J. Biol. Sci. 2009;5:543–548. doi: 10.7150/ijbs.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duffell S.J., Harkness J.W. Bovine virus diarrhoea-mucosal disease infection in cattle. Vet. Rec. 1985;117:240–245. doi: 10.1136/vr.117.10.240. [DOI] [PubMed] [Google Scholar]
- 89.Becher P., Orlich M., Shannon A.D., Horner G., König M., Thiel H.J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 1997;78:1357–1366. doi: 10.1099/0022-1317-78-6-1357. [DOI] [PubMed] [Google Scholar]
- 90.Vilček Š., Nettleton P.F., Paton D.J., Belák S. Molecular characterization of ovine pestiviruses. J. Gen. Virol. 1997;78:725–735. doi: 10.1099/0022-1317-78-4-725. [DOI] [PubMed] [Google Scholar]
- 91.Coria M.F., McClurkin A.W. Specific immune tolerance in an apparently healthy bull persistently infected with bovine viral diarrhea virus. J. Am. Vet. Med. Assoc. 1978;172:449–451. [PubMed] [Google Scholar]
- 92.Kale M., Yavru S., Ata A., Kocamüftüoǧvlu M., Yapici O., Hasircioǧlu S. Bovine viral diarrhea virus (BVDV) infection in relation to fertility in heifers. J. Vet. Med. Sci. 2011;73:331–336. doi: 10.1292/jvms.10-0254. [DOI] [PubMed] [Google Scholar]
- 93.Muñoz-Zanzi C.A., Thurmond M.C., Hietala S.K. Effect of bovine viral diarrhea virus infection on fertility of dairy heifers. Theriogenology. 2004;61:1085–1099. doi: 10.1016/j.theriogenology.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 94.Bolin S.R., Ridpath J.F. Frequency of association of noncytopathic bovine viral diarrhea virus with bovine neutrophils and mononuclear leukocytes before and after treatment with trypsin. Am. J. Vet. Res. 1990;51:1847–1851. [PubMed] [Google Scholar]
- 95.Corapi W.V., Elliott R.D., French T.W., Arthur D.G., Bezek D.M., Dubovi E.J. Thrombocytopenia and hemorrhages in veal calves infected with bovine viral diarrhea virus. J. Am. Vet. Med. Assoc. 1990;196:590–596. [PubMed] [Google Scholar]
- 96.Małaczewska J., Kaczorek-ŁUkowska E., Wójcik R., Krzysztof Siwicki A. Antiviral effects of nisin, lysozyme, lactoferrin and their mixtures against bovine viral diarrhoea virus. BMC Vet. Res. 2019;15 doi: 10.1186/s12917-019-2067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Villa T.G., Feijoo-Siota L., Rama J.L.R., Ageitos J.M. Antivirals against animal viruses. Biochem. Pharmacol. 2017;133:97–116. doi: 10.1016/j.bcp.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luna-Castro S., Samaniego Barron L., Serrano Rubio L.E., Olvera I.C., Avalos Gomez C., de la Garza M. Lactoferrin: a powerful antimicrobial protein present in milk. Adv. Dairy Res. 2017;05 [Google Scholar]
- 99.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y., Geng H., Li H., Wang Q., Xiao H., Tan W., Yan J., Gao G.F. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of middle east respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]