Abstract
The circadian clock is a master regulator of mammalian physiology, regulating daily oscillations of crucial biological processes and behaviors. Notably, circadian disruption has recently been identified as an independent risk factor for cancer and classified as a carcinogen. As such, it is imperative to discern the underpinning mechanisms by which circadian disruption alters cancer risk. Emergent data, reviewed herein, demonstrate that circadian regulatory functions play critical roles in several hallmarks of cancer, including control of cell proliferation, cell death, DNA repair, and metabolic alteration. Developing a deeper understanding of circadian-cancer regulation cross-talk holds promise for developing new strategies for cancer interception, prevention, and management.
Introduction
The circadian clock is an evolutionarily conserved, molecular time-keeping mechanism that regulates daily oscillations of biological processes and behaviors (1–4). The central clock is generated and maintained in the suprachiasmatic nucleus (SCN) of the hypothalamus, but cell-autonomous subordinate clocks also exist in peripheral tissues (e.g., liver, kidney, skin, intestine, lung, pancreas, ovary, and heart), which synchronize with one another by the SCN clock through neural and humoral inputs. The SCN synchronizes to environmental cues, such as ambient light, to coordinate circadian outputs and manage selected molecular and physiologic functions on a 24-hour cycle (5, 6). Although the basic molecular architecture of the SCN and peripheral clocks is the same, the SCN clock is the “master clock,” which signals and synchronizes peripheral clocks and others via circadian output pathways, including the autonomic nervous system and the neuroendocrine system. On balance, the circadian clock is vital to maintaining physiologic homeostasis and normal function of all organisms.
As a result of both population and laboratory-based findings, the World Health Organization designated circadian disruption as a likely carcinogen (7–9), thus raising interest in understanding how disruption of diurnal patterns promotes tumor development. Epidemiologic studies indicate that circadian rhythm disruptions (e.g., via jet lag, shift work, sleep disruption, and exposure to light at night) are associated with increased cancer risk, including cancers of the prostate, breast, colon, liver, pancreas, ovary, and lung (10–13). Furthermore, loss of circadian control is associated with poor efficacy of anticancer treatments and early mortality amongst patients with cancer (14, 15). In addition, visually impaired individuals who are insensitive to light changes in the environment, and thus depend largely on free-running endogenous circadian clocks to synchronize daily physiology, have a lower overall cancer risk (16). While mechanistic understanding of observed increased cancer risk is incompletely understood, recent findings have begun to uncover the molecular impact of circadian disruption on cancer phenotypes. As will be discussed, circadian processes impact key pathways affecting cancer development and progression, including cell-cycle control, apoptosis, metabolic regulation, and the DNA damage response (DDR; Fig. 1).
Figure 1.
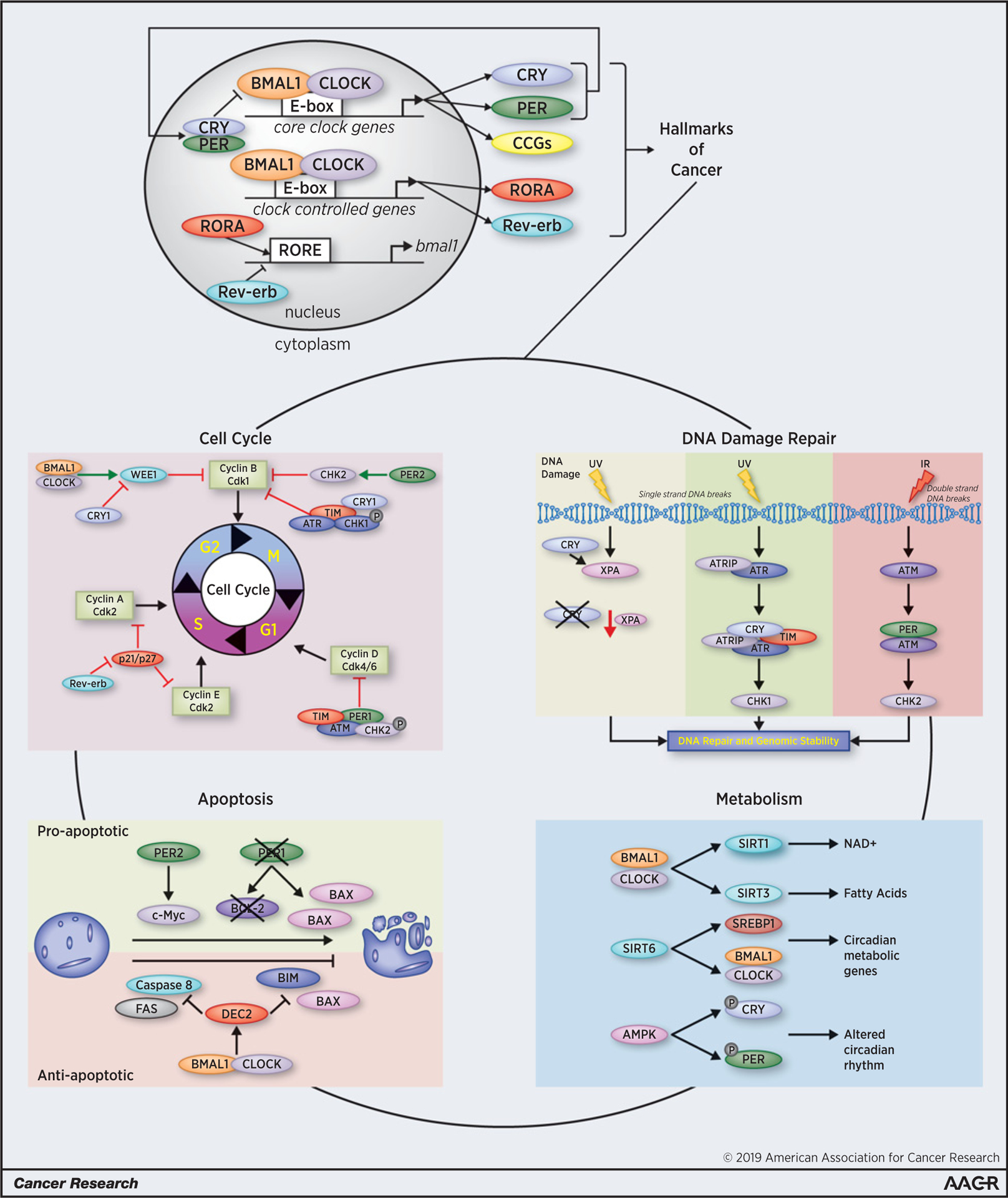
“Hallmarks” of the circadian clock. The core clock machinery consists of positive (CLOCK and BMAL1) and negative [Cryptochrome (CRY) and Period (PER)] regulators that maintain daily rhythmicity throughout an organism, impacting cell cycle, apoptosis, DNA repair, and metabolic regulation. CLOCK/BMAL1 heterodimers bind to E-box sites to regulate expression of core clock genes (CCGs), including CRY1, CRY2, PER1, PER2, and PER3. CLOCK/BMAL1 also regulates expression of additional clock-controlled genes, such as RORA and Rev-erb, which, in turn, regulate expression of BMAL1 through binding to ROR response elements (RORE). Thus, this autoregulatory network consisting of positive and negative transcription-translation feedback loops confer daily rhythmicity for homeostatic maintenance. The circadian clock influences several biological processes impacting tumor development and progression. Circadian-controlled processes are vast, including cell cycle, apoptosis, metabolic regulation, and DDR, which are all crucial for physiologic homeostasis. Disruption of circadian homeostasis through various factors is associated with increased cancer incidence and is an important, independent risk factor of cancer development in humans.
Circadian Dysfunction and Cancer
Circadian clock regulation
The core clock machinery is composed of an autoregulatory network consisting of positive and negative transcription-translation feedback loops (TTFL; Fig. 1; refs. 17–19). Transcriptionally, the clock is driven by positive regulators of the loop. Basic helix–loop–helix heterodimeric transcription factors (CLOCK/BMAL1 or BMAL1/NPAS2) regulate expression of key circadian genes including Cryptochromes (CRY1 and CRY2) and Period (PER1, PER2, and PER3) genes, which are the negative regulators of the circadian loop. CRY and PER form a transcriptional repressor complex that enters the nucleus to repress CLOCK/BMAL1 activity, thus creating a negative feedback loop to control the clock. Bmal1 is also rhythmically controlled by its own transcriptional target. In brief, Rora, Rev-erbα, and Rev-erbβ. RORα stimulate Bmal1 expression, while Rev-erbα, and β suppress Bmal1 expression. The importance of these master clock regulators is underscored by the observation that CLOCK/BMAL1 controls expression of approximately 10% of clock-controlled genes, which regulate molecular, biochemical, and physiologic processes (1–4). Furthermore, posttranslational modifications of CRY and PER regulate protein stability, control nuclear entry of CRY/PER repressors, and impact the autoregulatory clock feedback loops. These overlapping mechanisms conferring daily rhythmicity of cellular, metabolic, and physiologic functions for homeostatic maintenance are depicted in Fig. 1 (17, 20, 21). In sum, the circadian clock is tightly controlled through a discrete set of transcriptional regulatory factors; as will be discussed, recent findings link alteration of clock-regulatory factors as contributing to cancer phenotypes.
Tumor-specific functions of CLOCK and BMAL
Numerous studies have linked disruption of circadian clock function to tumorigenesis. Studies to date indicate that CLOCK and BMAL1 may harbor tumor-suppressive roles that are conserved among humans and rodents (2). In humans, single-nucleotide polymorphisms (SNP) in Clock and/or Bmal1 genes are associated with increased susceptibility to prostate, breast, ovarian, and pancreatic cancers (22–26). SNPs associated with increased cancer susceptibility are located in rs3749474 and rs11943456 (CLOCK) and in rs117104877, rs2290035, rs2278749, and rs969485 (BMAL1). Functional studies will be needed to determine how these alterations may contribute to tissue-specific cancers. Conversely, mice expressing dominant negative Clock mutation (ClockΔ19) incur an expected disruption of circadian rhythm, altering the expression of genes controlling metabolism, chromatin remodeling, DDR, and tumor suppression (27, 28). Moreover, ClockΔ19-mutant mice have significantly decreased survival when compared with wild-type mice, resulting from cardiac dysfunction and metabolic dysregulation. While Clock-mutant mice do not generate spontaneous tumors, this may be attributed to the decreased overall survival time, and transcriptional profiling reveals an induction of protumorigenic signaling. Similarly, there is significant evidence to suggest that Bmal1 also serves to restrict tumor development. Suppression of Bmal1 expression significantly increased the metastasizing potential of prostate, lung, and glioma cancers, both in vitro and in vivo. Mechanistically, tumor-suppressive qualities of BMAL1 are posited to occur through regulation of the PI3K–AKT signaling axis (29, 30). For instance, the AKT pathway component, ribosomal S6 protein kinase 1 (S6K1), phosphorylates BMAL1 allowing for BMAL1 to both associate with translational machinery and to also stimulate protein synthesis to impact tumorigenesis (31). Furthermore, upstream regulators may play a role in modulating tumor-suppressive potential of CLOCK and BMAL1, as studies have shown that the unfolded protein response induces a phase shift in circadian oscillations via direct regulation of miR-211 to suppress Clock and Bmal1 expression impacting tumor progression (32). While these preliminary studies indicate that Clock and BMAL1 may serve tumor suppressor–like functions, exceptions exist in colorectal cancer, wherein Clock and Bmal1 expression is elevated, and modeling studies linked high CLOCK expression to increased proliferation (33, 34). In addition, in acute myeloid leukemia (AML), Clock and Bmal1 are required for growth of AML (35). Taken together, early data suggest that CLOCK/BMAL1 serve tumor-protective roles, but these functions may be context- and disease-specific.
Tumor-suppressive functions of the CRY and PER family
In addition to CLOCK/BMAL1, evidence exists to implicate CRY and PER genes in tumor suppression. SNPs and/or upregulation of Cry1,2 and Per1, 2, or 3 are associated with increased susceptibility to prostate, breast, endometrial, colorectal, and skin cancer (22–26). Similar to CLOCK/BMAL1, PER’s role as pro or antioncogenic need to be further defined; although, current data indicate tumor-suppressive roles. For instance, several mouse models have demonstrated that mice lacking both alleles of Per1 and/or Per2 display an increased risk of spontaneous and radiation-induced tumor development compared with their wild-type counterparts, and this phenotype was further amplified in circadian phase–shifted conditions (36–38). In addition, Per2-inactivating mutations in mice exhibit increased risk of neoplastic growth. Specifically, suppression of Per2 increased cell proliferation in human colon cancer models through regulation of β-catenin and c-Myc, highlighting the tumor-suppressive role of Per2 (39). PER1 directly interacts with ATM to mediate its effect on tumor-suppressive genes, including TP53 and CHK2 to impact tumorigenesis. This interaction is maintained in the presence of DNA damage, where overexpression of PER1 sensitized human cancer cells to DNA damage–induced apoptosis (40). Thus, PER1 and PER2 clearly exhibit tumor-suppressive roles.
Similarly, mice lacking both alleles of Cry1 and/or Cry2 also display an increased risk of spontaneous and radiation-induced tumor development compared with their wild-type counterparts, which was further amplified in circadian phase–shifted conditions (36–38). Interestingly, mice with Cry1 and Cry2 mutations are arrhythmic, further supporting their role in maintaining circadian homeostasis (41). Inhibition of Cry2 expression in breast cancer models leads to dysregulation of genes involved in proliferation, apoptosis, migration, and invasion, again suggestive of a link to tumor development (42). While these collective observations link CRY and PER gene disruption to increased cancer susceptibility, functional dissection of CRY and PER pathways in controlling tumor development remain nascent.
Mechanisms linking clock dysfunction to the hallmarks of cancer
Molecular understanding of Clock, BMAL1, CRY, and PER family functions in circadian regulation are well understood, but data are emergent with regard to the molecular basis of tumor-suppressive functions. As described herein, preliminary findings have identified cross-talk of clock-regulatory proteins with key hallmark pathways controlling cancer development and progression.
Cell-cycle cross-talk.
The mitotic cell cycle and the circadian clock share many similarities as biologic oscillators in dividing cells, as each displays periodic phases of activation and repression. In organisms from cyanobacteria to unicellular eukaryotes (43–46), there is molecular coupling of the processes, wherein the molecular clock serves as an additional “checkpoint” for mitosis that restricts cell division to specific circadian phases (47, 48). However, the relevance of this coupling mechanism is unclear in mammalian cells and is the basis of controversy; whereas a subset of studies reported that most cell divisions occur in specific phases of the circadian cycle, others failed to identify a gating requirement of the cell cycle on circadian clock positioning (49–51). Although the basis of these divergent observations is not known, molecular observations in mammalian cells strongly support the contention that circadian pathways cross-talk with the cell-cycle machinery through transcriptional control or direct protein–protein interactions (Fig. 1).
Studies have demonstrated that circadian clock components can induce or repress cell-cycle progression depending on the time of day, inducing rhythmic transcriptional and posttranscriptional control of the cell cycle. Thus, each phase of the cell cycle has the potential to be influenced by the circadian clock. For example, in G1 phase, Rev-erbα and RORα/γ repress transcription of the cyclin-dependent kinase (CDK) regulator p21cip1, thereby promoting cell-cycle progression (52–56). Conversely, the clock-controlled gene, NONO, regulates the CDK inhibitor p16ink4a expression in a PER-dependent manner at the G1–S transition causing cellular senescence (52–56). In addition, PER1 and the circadian gene Timeless (TIM) inhibit the G1–S transition through interaction with ataxia-telangiectasia–mutated (ATM) and checkpoint 2 (CHK2), causing cell-cycle arrest (40). Similar paradigms exist for the G2–M transition, wherein both positive and negative influence of circadian clock factors has been reported. CRY1 promotes cell-cycle progression by inhibiting WEE1, the G2–M regulatory kinase, and thereby inducing mitotic entry. CLOCK/BMAL1 also regulates WEE1 controlling WEE2 rhythmic transcription in a manner that is sufficient to control cell-cycle arrest (52–56). Conversely, in different analyses it was shown that CRY1 restricts mitosis by modulating the ATM, ATR, CHK1-mediated G2–M transition by interacting with TIM in a circadian-controlled manner (57, 58). Lastly, expression of several known oncogenes (β-catenin, c-Myc, and Mdm2), cyclins (CCND1, CCNB, and CCN1), and additional cell-cycle regulators (Cdk4, Wnt3, and Tcf4) are all clock-controlled (59, 60). Thus, tight circadian control at key checkpoints is necessary to promote proper cell-cycle control at the G2–M checkpoint and maintain physiologic homeostasis. Taken together, these observations indicate circadian factors significantly influence both the G1–S and G2–M transition, but elicit distinct effects, dependent on context and the phase of circadian rhythm.
Conversely, cross-talk exists wherein the cell-cycle regulatory machinery impinges on circadian clock function, especially as related to transcriptional silencing occurring during mitosis. As observed, the pivotal tumor suppressor protein p53 is central to coupling circadian and cell-cycle oscillators. Molecular studies demonstrated that p53 directly binds to Per2 promoter and represses Per 2 expression, thus disrupting CLOCK/BMAL1 regulation and shifting the circadian cycle (61). In addition, a second tumor suppressor, PML, binds to and promotes PER2 nuclear localization, thus altering circadian timing and homeostasis (62). The overall impact of circadian clockcell-cycle cross-talk on tumor risk is complex, and likely contributes to the impact of circadian disruption on tumor development. Future studies are needed to link these molecular observations to cancer risk mediated by circadian disruption.
Regulation of cancer cell signaling.
Sustaining proliferative signaling remains a key factor among hallmarks of cancer (63, 64), and preliminary evidence exists linking circadian factors to regulation of growth factor processes in cancer. For example, upstream components of the JNK and p38 pathways exhibit the high circadian rhythmicity, including the following: ASK2, MKK7, MMK3, MMK6, p38γ, p38α, and JNK3 (65, 66). There is also evidence of cross-talk in that MKK7-mediated JNK activation increases the half-life of PER2 through phosphorylation, resulting in altered circadian timing (67). Furthermore, GADD45 family members that directly interact with JNK/p38 components also respond to circadian clock–regulated signaling (68). Both CRY1 knockdown in vitro and PER-mutant mice demonstrated impaired the circadian expression of GADD45α increasing cellular proliferation (38, 42). Finally, downstream components of the ERK1/2 pathway, ERK2 and MKK2, also show significant circadian rhythmicity, implicating connectivity of multiple growth factor signaling pathways with the circadian clock (65, 66). How these observations connect to specific growth factor responses in cancer remains an open question to be addressed.
At the tissue level, the central clock controls cell proliferation in peripheral tissues via the sympathetic nervous system (SNS), which innervates all peripheral organs to control intracellular signaling (69, 70). Studies showed that deregulated SNS signaling, either through surgical ablation or in jet-lagged mouse models, abolishes circadian oscillation and promotes tumor initiation (36, 71). Deregulated SNS signaling causes loss-of-function in the peripheral clock abolishing CLOCK/BMAL activation and leading to Myc oncogenic activation (36, 72). Taken together, balanced circadian control of cellular proliferation through transcriptional control and kinase regulation is key to cellular homeostasis and preventing tumor development.
Clock influence on cell death.
Recent studies established direct relationship between the core circadian clock and apoptosis. Similar to what was observed with cell-cycle regulation, circadian factors can both promote and restrict apoptosis, dependent on cellular context and clock status. With respect to promoting cell death, CRY1/2 and PER1 influence the extrinsic TNFα-dependent pathway and intrinsic apoptotic pathways, respectively (73). Mechanistically, PER2 sensitizes cancer cells to radiation-induced apoptosis through activation of Myc-mediated proapoptotic pathways. In addition, knockdown of PER1 results in downregulation of antiapoptotic BCL-2 and upregulation of proapoptotic BAX in hepatocellular carcinoma cells, increasing apoptosis. Thus, circadian factors can promote apoptosis through multiple mechanisms. Conversely, Clock can inhibit apoptosis. Mice defective in CLOCK show decreased expression of apoptosis-inducing factors, which contributes to increased tumor growth (74). In addition, the apoptotic regulators DEC1 (proapoptotic) and DEC2 (anti-apoptotic) repress CLOCK/BMAL–induced transactivation of the Per1 promoter, thereby influencing circadian regulation of apoptosis (75–77). These collective observations underscore the need to more fully understand the factors that control circadian-mediated cell death regulation, and the impact of these processes on tumorigenesis.
Clock and the DDR.
Perturbed DDRs contribute to cancer phenotypes (78, 79), and a multitude of evidence links circadian clock to DDR competence. In murine models, clock disruption results in accumulation of DNA damage and increased risk of neoplasia. The link between these processes is likely evolutionarily conserved, as Cryptochromes are structurally related to DNA photolyases that catalyze light-dependent DNA repair in plants and Drosophila (80). The direct role of CRYs in DDR in mammals still has to be fully defined mechanistically; however, several causative studies have shown the importance of CRY1,2 in DDR. UVB irradiation in the epidermis of Cry1−/−; Cry2−/− mice exhibits dampened circadian rhythm in the nucleotide excision repair gene XPA (81, 82). Time-restricted feeding also shifts the phase and amplitude of the epidermis circadian clock, ultimately altering sensitivity to UVB-induced DNA damage and expression of XPA, hindering repair (83). Furthermore, UV-induced DNA damages induce CRY2 interaction with ATR and CHK1 to regulate intra-S checkpoint function (84). The overall involvement of CRYs with DDR regulation underscores the need to fully investigate the contribution of tumor-derived CRY alterations in not only cancer development, but response to therapeutic intervention.
Importantly, other components of the molecular clock including PER1, PER2, and TIM play pivotal roles in multiple DDR processes. PER1 directly interacts with ATM/CHK2 in response to radiation-induced DSBs (40), PER1 overexpression activates Myc-mediated apoptosis in response to radiation-induced DSBs, and conversely, downregulation of PER2 confers resistance to radiation-induced apoptosis due to delayed CHK2 activation (38, 85). Accordingly, Per2−/− mice incur an increased risk of lymphoma (38, 86). TIM also has functions in DDR, including modulation of CHK1 and ATR downstream of single-strand DNA breaks, and activation of CHK2 via ATM modulation downstream of double strand breaks (87). Taken together, distinct circadian components are necessary to elicit canonical DDR for both single-strand and double-strand DNA breaks.
Complementing these findings, the positive circadian component Bmal1 has been preliminarily linked to DDR. Bmal1 knockdown abolishes radiation-induced p53 activation, releasing cells from cell-cycle arrest (88). In addition, in vivo keratinocyte-specific deletion of Bmal1 dampens UVB-induced DDR and increases accumulation of DNA lesions in the epidermis of mice (89). Conversely, the DNA repair factor CCAR2 represses BMAL1 and CLOCK expression and can modulate circadian rhythm (90), providing yet more evidence of cross-talk between DDR and circadian pathways. Understanding the specific timing and pathway regulation by each circadian gene will be pivotal for discerning proper therapeutic regimens to induce sustained responses to genotoxic therapies.
Cancer cell metabolism and the clock.
Coordinated interaction between the molecular circadian clock and the intricate network of metabolic pathways is required for maintenance of physiologic homeostasis in healthy cells. First identified in mammalian red blood cells (91), the metabolic circadian clock is independent of transcriptional activity and is sustained through the redox cycle of peroxiredoxin/thioredoxin/NADPH enzymes (92–94). This complex displays a 24-hour redox fluctuation metabolizing H2O2 in various tissues throughout the body. Multiple redox pairs, including: thiols (glutathione-GSH/GSSG) and coenzymes (FADH2/FAD+, NADH/NAD+, and NADPH/NADP+), dictate the global cellular redox state to influence electron flux and cellular homeostasis (95, 96). In particular, NADPH is able to extend or shorten the 24-hour circadian rhythm in drosophila, mouse tissue, and human cells (97). Thus, NADPH is a critical cofactor that has the potential to function as a circadian-regulated and cancer-promoting metabolite.
Cancer-associated reprogramming of energy metabolism to predominantly utilize glycolytic activity, despite aerobic conditions (Warburg effect), is characterized with higher NADPH formation, decreased TCA cycle activity, and increased fatty acid synthesis (98), and recent studies established a relationship between this process and the circadian clock. For example, melatonin elevation due to light exposure changes leads to decreased growth of prostate and breast cancer xenografts due to disruption of the Warburg effect (99–103). In addition, alterations in the pentose phosphate pathway, which generates NADPH, is tightly controlled in a circadian manner (96). Moreover, sirtuins (SIRT), a family of NAD+-dependent proteins, interact with the circadian clock to control chromatin remodeling and metabolic output (104). SIRT1 expression is regulated by CLOCK/BMAL; conversely, SIRT1 directly interacts with CLOCK and is known to deacetylate PER2 altering the circadian clock (105–107). This SIRT-circadian system also impacts the TCA cycle through SIRT3 and BMAL, and fatty acid metabolism through SIRT6 governing CLOCK/BMAL recruitment of SREBP1 to circadian promoters (108, 109). On the basis of these findings, evidence is strong that circadian disruption influences metabolic adaptions in favor of cancer development.
Consistent with these findings, controlled feeding times improve metabolic disease even when mice are fed a high-fat diet (110, 111). For example, clock-mutant mice exhibit impaired cholesterol metabolism and promotion of atherosclerosis (112). In addition, RORγ−/− mice display reduced hepatic gluconeogenesis and improved insulin sensitivity; while cell-specific deletion of Bmal1 led to hypoglycemia (113, 114). The influence of the circadian clock on metabolism also impacts lipogenesis, bile acid synthesis, cardiovascular disease, and inflammation (115). For instance, chronic jet-lag to disrupt the circadian rhythm induced spontaneous hepatocellular carcinoma (HCC) in wild-type mice due to deregulated nuclear receptor–controlled cholesterol/bile acid and xenobiotic metabolism, in addition to global liver metabolic dysfunction similar to the pathway observed in obese patients (116). Moreover, recent studies showed that 50% of detected metabolites oscillate in a mouse liver, and 18 of those metabolites also oscillate in human cell-autonomous U2OS cells (117). Intriguingly, BMAL1, CRY1, and CRY2 knockdown decouples transcriptional and metabolite rhythms by shortening, lengthening, or diminishing rhythms, respectively. Conversely, metabolic alterations can alter the circadian oscillation as well. For example, AMPK is a key nutrient sensor that can destabilize CRY1 and PER2 through phosphorylation, which alters the circadian rhythm (118, 119). Moreover, UBE3A binds and degrades BMAL1 in a ubiquitin ligase–dependent manner to disrupt circadian oscillation (120). In addition, MYC directly activates negative regulators of CLOCK/BMAL1, leading to disruption of circadian metabolic oscillation (121). The insulin-FOXO3-Clock signaling cascade mediates function in hepatic metabolism and oxidative sensitivity (122, 123). Lastly, a recent study showed that a high-fat diet influenced the circadian transcriptome and metabolome due to impaired BMAL1 and PPARγ recruitment (124). Thus, through transcriptional remodeling and posttranslational modifications, the circadian clock regulates metabolism and integrates nutrient signaling critical to maintaining tumorigenesis.
Leveraging circadian rhythm function in cancer management (chronotherapy)
Given the substantial data linking circadian clock dysfunction of cancer pathways, it is reasonable to consider how this process might be modulated to influence tumor growth and survival. The concept of chronotherapy, which considers the body’s natural rhythms and cycles to treat an illness or disorder, was utilized even before the molecular mechanism of the core circadian clock was defined (125). For example, the chemotherapeutic agent, cisplatin, exhibits significant difference in outcomes for patients with prostate, breast, cervical, and ovarian cancer when morning and evening doses were compared, indicating chronotherapy improves the toxic therapeutic ratio of cisplatin and enhances efficacy (126). Furthermore, additional chemotherapeutic drugs exhibited optimal dosing timings to improve outcome in bladder, colorectal, endometrial, and renal cancer (127–130). The activity of several anticancer drugs may be restricted by their side effects and toxicities to healthy cells. Hence, chronotherapeutics aims to maximize the antitumor effects of cancer chemotherapy by minimizing toxicity and undesirable side effects, while simultaneously increasing tolerability to improve the survival rate for patients with cancer.
The role of the circadian clock in chemotherapy administration has also been evaluated using wild-type and circadian clock–mutant mice (131). Clock-mutant and Bmal1 knockout mice are sensitive to chemotherapy regardless of the time of administration of treatment (132), whereas Cry1−/− Cry2−/− mice are more resistant to chemotherapy compared with wild-type (133). These in vivo studies also revealed that both host tolerability and drug efficacy are affected by circadian timing. Accordingly, it was inferred that response to chemotherapy varies with the time of day, indicating the circadian clock plays an important role in therapeutic outcome (131). In addition, genome-wide circadian gene expression profile studies in mice, nonhuman primates, and human lung and liver tissues indicated that at least 80% of the FDA-approved drugs tested exhibited daily rhythm in their targets and respective downstream functions (134, 135).
Consequently, the successful chronotherapeutic preclinical results were used as justification for several randomized clinical trials for advanced cancers, which revealed that anticancer chronotherapy is most beneficial to patients who maintain their endogenous circadian clock and rhythm (136). This highlights the intricate connection of therapeutic efficacy with individual innate circadian function for ideal outcome. However, a dysregulated circadian clock could alter the efficacy of anticancer drugs, so additional investigation needs to be performed to discern optimal timing for treatment in those disease states. On the basis of these collective data, discerning the mechanisms by which the circadian clock alters cancer therapy may provide insight into refining and augmenting outcomes for the therapeutic intervention.
Conclusions
Circadian disruption is an independent risk factor for cancer and has been classified as a carcinogen. As described herein, perturbations of the circadian clock strongly influence neoplastic transformation and tumor growth through alterations of multiple cancer regulatory pathways including cell cycle, apoptosis, DDR, and metabolism. While the robust link between circadian dysfunction and cancer is well established, mechanistic understanding is nascent. Therefore, it is imperative to continue to elucidate the mechanisms by which the mammalian circadian clock regulates cancer progression.
Key questions remain that must be addressed to delineate the complex roles of the molecular clock in human malignancy. First, is a circadian lifestyle change that incorporates timing of sleep, physical activity, and nutrition enough to diminish disruption of circadian rhythm to reduce cancer incidence? It is intriguing to speculate that cancer risk reduction could be achieved through behavior modification and/or pharmacologic normalization of the circadian cycle. Second, what molecular mechanisms are involved in tissue-specific circadian gene expression and how does that impact tumorigenesis? Peripheral clocks are found in several tissue types, but not all, and the lack of coordinated circadian rhythm leads to differential expression of core circadian genes. The impact of circadian expression on tumorigenesis and other age-related diseases needs to be evaluated. Third, because circadian genes have been associated with high concentrations of sex hormones (137), it would be intriguing to discern the possible role of circadian genes in hormone-related cancers to uncover novel mechanisms of action. Finally, to establish a rational chronotherapeutic strategy, determining the underlying basis of effectivity and optimal implementation strategy will be critical. Future studies will prove instrumental in enriching our understanding of circadian influence on tumor initiation and progression. In conclusion, discerning the intricate workings of the circadian clock on cancer development and progression has the potential for transformative impact with respect to cancer prevention and management.
Acknowledgments
We gratefully thank all the members of the Knudsen laboratory for their invaluable support and input. This work was supported by Prostate Cancer Foundation Young Investigator Award (to A.A. Shafi, 2017) and NIH/NCI grants to K.E. Knudsen (grant nos. R01 CA176401, R01 CA182569, R01 CA217329, P30 CA056036-14).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Sancar G, Brunner M. Circadian clocks and energy metabolism. Cell Mol Life Sci 2014;71:2667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci 2013;119:221–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002;109:307–20. [DOI] [PubMed] [Google Scholar]
- 4.Delaunay F, Laudet V. Circadian clock and microarrays: mammalian genome gets rhythm. Trends Genet 2003;18:595–7. [DOI] [PubMed] [Google Scholar]
- 5.Stevens RG, Blask DE, Brainard GC, Hansen J, Lockley SW, Provencio I, et al. Meeting report: the role of environmental lighting and circadian disruption in cancer and other diseases. Environ Health Perspect 2007; 115:1357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 2010;72:551–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol 2008;8:1065–6. [DOI] [PubMed] [Google Scholar]
- 8.Stevens RG, Hansen J, Costa G, Haus E, Kauppinen T, Aronson KJ, et al. Considerations of circadian impact for defining “shift work” in cancer studies: IARC Working Group report. Occup Environ Med 2011;68: 154–62. [DOI] [PubMed] [Google Scholar]
- 9.Lunn RM, Blask DE, Coogan AN, Figueiro MG, Gorman MR, Hall JE, et al. Health consequences of electric lighting practices in the modern world: a report on the National Toxicology Program’s workshop on shift work at night, artificial light at night, and circadian disruption. Sci Total Environ 2018;607–608:1073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendeu-Foyet MGG, Menegaux F. Circadian disruption and prostate cancer risk: an updated review of epidemiological evidences. Cancer Epidemiol Biomarkers Prev 2017;26:985–91. [DOI] [PubMed] [Google Scholar]
- 11.Dickerman BA, Markt SC, Koskenvuo M, Hublin C, Pukkala E, Mucci LA, et al. Sleep disruption, chronotype, shift work, and prostate cancer risk and mortality: a 30-year prospective cohort study of Finnish twins. Cancer Causes Control 2016;27:1361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markt SC, Flynn-Evans EE, Valdimarsdottir UA, Sigurdardottir LG, Tamimi RM, Batista JL, et al. Sleep duration and disruption and prostate cancer risk: a 23-year prospective study. Cancer Epidemiol Biomarkers Prev 2016;25:302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigurdardottir LG, Valdimarsdottir UA, Fall K, Rider JR, Lockley SW, Schernhammer E, et al. Circadian disruption, sleep loss, and prostate cancer risk: a systematic review of epidemiologic studies. Cancer Epidemiol Biomarkers Prev 2012;21:1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancar A Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture). Angew Chem Int Ed Engl 2016;55:8502–27. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A, Lindsey-Boltz L, Gaddameedhi S, Selby C, Ye R, Chiou Y-Y, et al. Circadian clock, cancer, and chemotherapy. Biochemistry 2015;54: 110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kliukiene J, Tynes T, Andersen A. Risk of breast cancer among Norwegian women with visual impairment. Br J Cancer 2001;84:397–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 2006;15:R271–7. [DOI] [PubMed] [Google Scholar]
- 18.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A 2007;104:3342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2001;2:702–15. [DOI] [PubMed] [Google Scholar]
- 20.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem 2011; 286:25891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker-Weimann S, Wolf J, Herzel H, Kramer A. Modeling feedback loops of the Mammalian circadian oscillator. Biophys J 2004;87:3023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res 2010;69:9315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg 2013;17:443–50. [DOI] [PubMed] [Google Scholar]
- 24.Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J-I, Higashimoto M, Mashiko M, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand 2008;87:1060–70. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi H, Fernández AF, Seti en F, Ropero S, Ballestar E, Villanueva A, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 2009;69:8447–54. [DOI] [PubMed] [Google Scholar]
- 26.Gu F, Zhang H, Hyland PL, Berndt S, Gapstur SM, Wheeler W, et al. Inherited variation in circadian rhythm genes and risks of prostate cancer and three other cancer sites in combined cancer consortia. Int J Cancer 2017;141:1794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009;324:651–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem 2003;278:41519–27. [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z-LL, Wu M-WW, Sun J, Sun Y-LL, Cai Y-CC, Huang Y-JJ, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem 2010;148:319–26. [DOI] [PubMed] [Google Scholar]
- 30.Jung C-HH, Kim EM, Park JK, Hwang S-GG, Moon S-KK, Kim W-JJ, et al. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol Rep 2013;29:2109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 2015;161:1138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu Y, Yoshida A, Chitnis N, Altman BJ, Tameire F, Oran A, et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat Cell Biol 2018;20:104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karantanos T, Theodoropoulos G, Pektasides D, Gazouli M. Clock genes: their role in colorectal cancer. World J Gastroenterol 2014;20:1986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Qian R, Sun N, Lu C, Chen Z, Hua L. Circadian gene hClock enhances proliferation and inhibits apoptosis of human colorectal carcinoma cells in vitro and in vivo. Mol Med Rep 2015;11:4204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell 2016;165:303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 2010;5:e10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003;302:255–9. [DOI] [PubMed] [Google Scholar]
- 38.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002;111:41–50. [DOI] [PubMed] [Google Scholar]
- 39.Wood PA, Yang X, Taber A, Oh E-YY, Ansell C, Ayers SE, et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res 2008;6: 1786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler P. The circadian gene Per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell 2005;22:375–82. [DOI] [PubMed] [Google Scholar]
- 41.Filipski E, Subramanian P, Carrière J, Guettier C, Barbason H, Lévi F. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res 2009;680:95–105. [DOI] [PubMed] [Google Scholar]
- 42.Hoffman AE, Zheng T, Ba Y, Stevens RG, Yi C-HH, Leaderer D, et al. Phenotypic effects of the circadian gene Cryptochrome 2 on cancer-related pathways. BMC Cancer 2010;10:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu B, Dacso CC, O’Malley BW. Unveiling “Musica Universalis” of the cell: a brief history of biological 12-hour rhythms. J Endocr Soc 2018;2: 727–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fung-Uceda J, Lee K, Seo PJ, Polyn S, De Veylder L, Mas P. The circadian clock sets the time of DNA replication licensing to regulate growth in arabidopsis. Dev Cell 2018;45:101–13. [DOI] [PubMed] [Google Scholar]
- 45.Johnson CH. Circadian clocks and cell division: what’s the pacemaker? Cell Cycle 2010;9:3864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaix A, Zarrinpar A, Panda S. The circadian coordination of cell biology. J Cell Biol 2016;215:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altinok A, Gonze D, Lévi F, Goldbeter A. An automaton model for the cell cycle. Interface Focus 2011;1:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clairambault J Physiologically based modelling of circadian control on cell proliferation. Conf Proc IEEE Eng Med Biol Soc 2006;1:173–6. [DOI] [PubMed] [Google Scholar]
- 49.Zámborszky J, Hong CI, Csikász Nagy A. Computational analysis of mammalian cell division gated by a circadian clock: quantized cell cycles and cell size control. J Biol Rhythms 2008;22:542–53. [DOI] [PubMed] [Google Scholar]
- 50.Chauhan A, Lorenzen S, Herzel H, Bernard S. Regulation of mammalian cell cycle progression in the regenerating liver. J Theor Biol 2011;283: 103–12. [DOI] [PubMed] [Google Scholar]
- 51.Gérard C, Goldbeter A. Entrainment of the mammalian cell cycle by the circadian clock: modeling two coupled cellular rhythms. PLoS Comput Biol 2012;8:e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levens D Disentangling the MYC web. Proc Natl Acad Sci U S A 2002;99: 5757–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mir SE, De Witt Hamer PC, Krawczyk PM, Balaj L, Claes A, Niers JM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell 2010;18: 244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharya A, Schmitz U, Wolkenhauer O, Schönherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 2013;32:3175–83. [DOI] [PubMed] [Google Scholar]
- 55.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell 2012;22:438–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang T-HH, Leem S-HH. Modulation of ATR-mediated DNA damage checkpoint response by cryptochrome 1. Nucleic Acids Res 2014;42: 4427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol 2007;17:311–7. [DOI] [PubMed] [Google Scholar]
- 59.Massagué J G1 cell-cycle control and cancer. Nature 2004;432:298–306. [DOI] [PubMed] [Google Scholar]
- 60.Kettner NM, Katchy CA, Fu L. Circadian gene variants in cancer. Ann Med 2014;46:208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates Period2 expression and the circadian clock. Nat Commun 2013;4:2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miki T, Xu Z, Chen-Goodspeed M, Liu M, Van Oort-Jansen A, Rea MA, et al. PML regulates PER2 nuclear localization and circadian function. EMBO J 2012;31:1427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg R. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 64.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res 2017;7:1016–36. [PMC free article] [PubMed] [Google Scholar]
- 65.Munshi A, Ramesh R. Mitogen-activated protein kinases and their role in radiation response. Genes Cancer 2013;4:401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soták M, Sumová A, Pácha J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann Med 2014;46:221–32. [DOI] [PubMed] [Google Scholar]
- 67.Uchida Y, Osaki T, Yamasaki T, Shimomura T, Hata S, Horikawa K, et al. Involvement of stress kinase mitogen-activated protein kinase 7 in regulation of mammalian circadian clock. J Biol Chem 2012;287: 8318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura RE, de Vasconcellos JF, Sarkar D, Libermann TA, Fisher PB, Zerbini LF. GADD45 proteins: central players in tumorigenesis. Curr Mol Med 2012;12:634–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson KL, Marques FZ, Lim K, Davern PJ, Head GA. Circadian differences in the contribution of the brain renin-angiotensin system in genetically hypertensive mice. Front Physiol 2018;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ni Y, Ma L, Wu T, Lim AL, Zhang W, Yang L, et al. The involvement of sympathetic nervous system in essence of chicken-facilitated physiological adaption and circadian resetting. Life Sci 2018;201: 54–62. [DOI] [PubMed] [Google Scholar]
- 71.Adams SL, Roxe DM, Weiss J, Zhang F, Rosenthal JE. Ambulatory blood pressure and Holter monitoring of emergency physicians before, during, and after a night shift. Acad Emerg Med 1998;5:871–7. [DOI] [PubMed] [Google Scholar]
- 72.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell 2005;122:803–15. [DOI] [PubMed] [Google Scholar]
- 73.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci U S A 2009;106:2841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsunaga N, Kohno Y, Kakimoto K, Hayashi A, Koyanagi S, Ohdo S. Influence of CLOCK on cytotoxicity induced by diethylnitrosamine in mouse primary hepatocytes. Toxicology 2011;280:144–51. [DOI] [PubMed] [Google Scholar]
- 75.Wu Y, Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, et al. BHLH transcription factor DEC2 regulates pro-apoptotic factor Bim in human oral cancer HSC-3 cells. Biomed Res 2012;33:75–82. [DOI] [PubMed] [Google Scholar]
- 76.Wu Y, Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, et al. Basic helix-loop-helix transcription factors DEC1 and DEC2 regulate the paclitaxel-induced apoptotic pathway of MCF-7 human breast cancer cells. Int J Mol Med 2011;27:491–5. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Sato F, Kawamoto T, Fujimoto K, Morohashi S, Akasaka H, et al. Anti-apoptotic effect of the basic helix-loop-helix (bHLH) transcription factor DEC2 in human breast cancer cells. Genes Cells 2010;15:315–25. [DOI] [PubMed] [Google Scholar]
- 78.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432: 316–23. [DOI] [PubMed] [Google Scholar]
- 79.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 2004;38: 445–76. [DOI] [PubMed] [Google Scholar]
- 80.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 2011;62:335–64. [DOI] [PubMed] [Google Scholar]
- 81.Kang T-HH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res 2011;39:3176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang T-HH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A 2010;107: 4890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H, van Spyk E, Liu Q, Geyfman M, Salmans ML, Kumar V, et al. Time-restricted feeding shifts the skin circadian clock and alters UVB-induced DNA damage. Cell Rep 2017;20:1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science 2003;302:439–42. [DOI] [PubMed] [Google Scholar]
- 85.Yang X, He X, Yang Z, Jabbari E. Mammalian PER2 regulates AKT activation and DNA damage response. Biochem Cell Biol 2013;90: 675–82. [DOI] [PubMed] [Google Scholar]
- 86.Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol 2008;18:286–91. [DOI] [PubMed] [Google Scholar]
- 87.Yang X, Wood PA, Hrushesky WJ. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J Biol Chem 2010;285:3030–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One 2009;4:e4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A 2012;109:11758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Magni M, Buscemi G, Zannini L. Cell cycle and apoptosis regulator 2 at the interface between DNA damage response and cell physiology. Mutat Res 2018;776:1–9. [DOI] [PubMed] [Google Scholar]
- 91.O’Neill J, Reddy A. Circadian clocks in human red blood cells. Nature 2011;469:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ray S, Reddy A. Crosstalk between circadian clocks, sleepwake cycles, and metabolic networks: dispelling the darkness. Bioessays 2016;38: 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Milev N, Reddy A. Circadian redox oscillations and metabolism. Trends Endocrinol Metab 2015;26:430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robinson I, Reddy AB. Molecular mechanisms of the circadian clockwork in mammals. FEBS Lett 2014;588:2477–83. [DOI] [PubMed] [Google Scholar]
- 95.Ying W NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 2008;10:179–206. [DOI] [PubMed] [Google Scholar]
- 96.Rey G, Valekunja U, Feeney K, Wulund L, Milev N, Stangherlin A, et al. The pentose phosphate pathway regulates the circadian clock. Cell Metab 2016;24:462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun F, Dai C, Xie J, Hu X. Biochemical issues in estimation of cytosolic free NAD/NADH ratio. PLoS One 2012;7:e34525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srivastava A, Mannam P. Warburg revisited: lessons for innate immunity and sepsis. Front Physiol 2015;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mao L, Dauchy R, Blask D, Dauchy E, Slakey L, Brimer S, et al. Melatonin suppression of aerobic glycolysis (Warburg effect), survival signalling and metastasis in human leiomyosarcoma. J Pineal Res 2016;60:167–77. [DOI] [PubMed] [Google Scholar]
- 100.Dauchy RT, Hoffman AE, Wren-Dail MA, Hanifin JP, Warfield B, Brainard GC, et al. Daytime blue light enhances the nighttime circadian melatonin inhibition of human prostate cancer growth. Comp Med 2016;65: 473–85. [PMC free article] [PubMed] [Google Scholar]
- 101.Dauchy RT, Wren-Dail MA, Dupepe LM, Hill SM, Xiang S, Anbalagan M, et al. Effect of daytime blue-enriched LED light on the nighttime circadian melatonin inhibition of hepatoma 7288CTC Warburg effect and progression. Comp Med 2018;68:269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hill SM, Belancio VP, Dauchy RT, Xiang S, Brimer S, Mao L, et al. Melatonin: an inhibitor of breast cancer. Endocr Relat Cancer 2015;22: R183–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blask D, Dauchy R, Dauchy E, Mao L, Hill S, Greene M, et al. Light exposure at night disrupts host/cancer circadian regulatory dynamics: impact on the Warburg effect, lipid signaling and tumor growth prevention. PLoS One 2014;9:e102776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene 2014;33:1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bellet MM, Masri S, Astarita G, Sassone-Corsi P, Della Fazia MA, Servillo G. Histone deacetylase SIRT1 controls proliferation, circadian rhythm, and lipid metabolism during liver regeneration in mice. J Biol Chem 2016;291:23318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bellet M, Nakahata Y, Boudjelal M, Watts E, Mossakowska D, Edwards K, et al. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl Acad Sci U S A 2013;110: 3333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008;134:329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014;158:659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peek C, Affinati A, Ramsey K, Kuo H-Y, Yu W, Sena L, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 2013;342:1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong E, Gill S, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 2012;15: 848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 2014;19: 319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pan X, Jiang X-CC, Hussain MM. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 2013;128: 1758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jetten AM, Takeda Y, Slominski A, Kang HS. Retinoic acid-related Orphan Receptor γ (RORγ): connecting sterol metabolism to regulation of the immune system and autoimmune disease. Curr Opin Toxicol 2018;8: 66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crumbley C, Wang Y, Banerjee S, Burris T. Regulation of expression of citrate synthase by the retinoic acid receptor-related orphan receptor α (RORα). PLoS One 2012;7:e33804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bass J, Takahashi J. Circadian integration of metabolism and energetics. Science 2010;330:1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 2016;30:909–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, et al. Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab 2017;25:1206. [DOI] [PubMed] [Google Scholar]
- 118.Lamia K, Sachdeva U, DiTacchio L, Williams E, Alvarez J, Egan D, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009;326:437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem 2007;282:20794–8. [DOI] [PubMed] [Google Scholar]
- 120.Gossan N, Zhang F, Guo B, Jin D, Yoshitane H, Yao A, et al. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res 2014;42:5765–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab 2015;22:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chaves I, van der Horst G, Schellevis R, Nijman R, Koerkamp M, Holstege F, et al. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol 2014;24:1248–55. [DOI] [PubMed] [Google Scholar]
- 123.Zheng X, Yang Z, Yue Z, Alvarez J, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A 2007;104:15899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eckel-Mahan K, Patel V, de Mateo S, Orozco-Solis R, Ceglia N, Sahar S, et al. Reprogramming of the circadian clock by nutritional challenge. Cell 2013;155:1464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hrushesky WJ. Circadian timing of cancer chemotherapy. Science 1985; 228:73–5. [DOI] [PubMed] [Google Scholar]
- 126.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol Int 2002;19: 237–51. [DOI] [PubMed] [Google Scholar]
- 127.Lévi F Circadian chronotherapy for human cancers. Lancet Oncol 2001;2: 307–15. [DOI] [PubMed] [Google Scholar]
- 128.Lévi F, Giacchetti S, Zidani R, Brezault-Bonnet C, Tigaud JM, Goldwasser F, et al. Chronotherapy of colorectal cancer metastases. Hepatogastroenterology 2001;48:320–2. [PubMed] [Google Scholar]
- 129.Chan S, Rowbottom L, McDonald R, Bjarnason GA, Tsao M, Danjoux C, et al. Does the time of radiotherapy affect treatment outcomes? A review of the literature. Clin Oncol (R Coll Radiol) 2017;29:231–8. [DOI] [PubMed] [Google Scholar]
- 130.Chan S, Zhang L, Rowbottom L, McDonald R, Bjarnason GA, Tsao M, et al. Effects of circadian rhythms and treatment times on the response of radiotherapy for painful bone metastases. Ann Palliat Med 2017;6: 14–25. [DOI] [PubMed] [Google Scholar]
- 131.Sulli G, Manoogian ENCN, Taub PR, Panda S. Training the circadian clock, clocking the drugs, and drugging the clock to prevent, manage, and treat chronic diseases. Trends Pharmacol Sci 2018;39:812–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A 2005;102: 3407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 1999;96: 12114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci U S A 2017;114:5312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018;359:pii: eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lévi F, Focan C, Karaboué A, de la Valette V, Focan-Henrard D, Baron B, et al. Implications of circadian clocks for the rhythmic delivery of cancer therapeutics. Adv Drug Deliv Rev 2007;59:1015–35. [DOI] [PubMed] [Google Scholar]
- 137.Mong JA, Baker FC, Mahoney MM, Paul KN, Schwartz MD, Semba K, et al. Sleep, rhythms, and the endocrine brain: influence of sex and gonadal hormones. J Neurosci 2011;31:16107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


