Summary
S. aureus USA300 isolates utilize the copBL and copAZ gene products to prevent Cu intoxication. We created and examined a ΔcopAZ ΔcopBL mutant strain (cop-). The cop- strain was sensitive to Cu and accumulated intracellular Cu. We screened a transposon (Tn) mutant library in the cop- background and isolated strains with Tn insertions in the mntABC operon that permitted growth in the presence of Cu. The mutations were in mntA and they were recessive. Under the growth conditions utilized, MntABC functioned in manganese (Mn) import. When cultured with Cu, strains containing a mntA::Tn accumulated less Cu than the parent strain. Mn(II) supplementation improved growth when cop- was cultured with Cu and this phenotype was dependent upon the presence of MntR, which is a repressor of mntABC transcription. A ΔmntR strain had an increased Cu load and decreased growth in the presence of Cu, which was abrogated by introduction of mntA::Tn. Over-expression of mntABC increased cellular Cu load and sensitivity to Cu. The presence of a mntA::Tn mutation protected iron-sulfur (FeS) enzymes from inactivation by Cu. The data presented are consistent with a model wherein defective MntABC results in decreased cellular Cu accumulation and protection to FeS enzymes from Cu poisoning.
Keywords: Staphylococcus aureus, manganese, copper, MntABC, iron-sulfur cluster
Abbreviated Summary:
To combat antibiotic resistance, we need to develop new prevention and therapeutic approaches. Copper (Cu) has shown promise as an antimicrobial compound; however, questions remain about how Cu enters cells, kills microorganisms, and how microorganisms prevent intoxication by Cu. Here we present data showing that a dysfunctional MntABC Mn transporter results in decreased cellular Cu accumulation. We also demonstrate that cytosolic Cu accumulation inhibits S. aureus FeS enzymes.
Introduction
Staphylococcus aureus continues to be a serious public health concern as S. aureus infections result in high morbidity and mortality rates (Turner et al., 2019). Although historically known as a nosocomial pathogen, there has been an increase in community-acquired (CA) S. aureus cases among both immunocompetent and immunocompromised groups (Tenover et al., 2006). These CA S. aureus strains often cause skin and soft tissue infections that can develop into invasive and systemic infections (Turner et al., 2019). Treatment of S. aureus infections is complicated due to the ability of this pathogen to evolve and/or acquire resistance to antibiotics (Malachowa & DeLeo, 2010). To combat these problems, we need to develop new prevention and therapeutic approaches including the characterization of new promising antimicrobial targets.
Copper (Cu) is gaining popularity as an antimicrobial but killing or preventing growth of microorganisms using Cu is an age-old technology (Dollwet & Sorenson, 1985, Grass et al., 2011). Copper is increasingly used as an intrinsic antibacterial and in metallic copper or copper-containing alloys on touch surfaces (Grass et al., 2011). Mammals also use Cu to help clear infections by increasing Cu loads at sites of inflammation (Hodgkinson & Petris, 2012, Beveridge et al., 1985). Cu accumulates within macrophage intracellular vesicles (Achard et al., 2012) and ultimately in phagosomes (Wagner et al., 2005). The addition of Cu to macrophages increases killing efficiency and genetic depletion of the Cu transporter ATP7A suppressed macrophage-dependent bacterial killing suggesting that macrophages that are defective in trafficking Cu to the phagolysome have a decreased ability to kill bacterial pathogens (White et al., 2009). Bacterial pathogens, including S. aureus, that are defective in exporting Cu from the cytosol have decreased survival in macrophages and in other models of infection (Purves et al., 2018, White et al., 2009, Zapotoczna et al., 2018, Ladomersky et al., 2017).
Copper is an important micronutrient in some organisms (Andreini et al., 2008) but, as alluded to above, cytosolic Cu overload results in intoxication (Purves et al., 2018). Various mechanisms have been proposed to explain the toxic effects of Cu; however, the molecular mechanism(s) of Cu poisoning is not entirely clear, and it is likely multifaceted. In support of this, Cu intoxication involves perturbations of many stress and non-stress regulons resulting in multi-dimensional responses (Moore et al., 2005, Baker et al., 2010, Chillappagari et al., 2010, Quintana et al., 2017).
Metalloproteins often have an essential requirement for their cognate metal cofactor (Imlay, 2014). Solvent-accessible protein-associated metal ions can be displaced by oxidative stress or by alternate metals (Xu & Imlay, 2012, Gardner & Fridovich, 1991, Anjem & Imlay, 2012). Metalloproteins can also be mismetalated during maturation or oxidative stress (Waldron & Robinson, 2009, Gu & Imlay, 2013). Accumulation of Cu in the cytosol decreases the activities of iron-sulfur (FeS) enzymes (Macomber & Imlay, 2009, Djoko & McEwan, 2013). Biochemical studies found that Cu can poison enzymes with solvent-exposed FeS clusters by disrupting the FeS cluster (Macomber & Imlay, 2009). Cu can also inhibit the assembly of FeS proteins by forming stable complexes with FeS cluster synthesis machineries and/or FeS protein maturation factors (Tan et al., 2017, Chillappagari et al., 2010, Tan et al., 2014, Brancaccio et al., 2017). It was recently shown that glyceraldehyde-3-phosphate dehydrogenase is also poisoned by Cu in S. aureus (Tarrant et al., 2019).
Intoxication of Streptococcus pneumoniae by Cu was alleviated by the addition of manganese (Mn) to the growth medium (Johnson et al., 2015). Accumulation of cytosolic Cu led to increased expression of proteins in the nucleotide synthesis pathway including the anaerobic ribonucleotide reductase system (NrdDG) suggesting that cells were starved for deoxynucleotide triphosphates (dNTPs). The aerobic ribonucleotide reductase system (NrdEF) is Mn-dependent (Martin & Imlay, 2011). A S. pneumoniae ΔnrdD mutant was more sensitive than the parent to Cu intoxication. These data led to the hypothesis that Cu inhibits aerobic dNTP synthesis by mismetalation of NrdF.
All S. aureus contain the copAZ operon (Sitthisak et al., 2007). CopA is a transmembrane P1-type Cu(I) exporting ATPase (Arguello et al., 2007) and CopZ is a cytosolic Cu(I)-binding chaperone that can deliver Cu(I) to CopA for export (Singleton et al., 2009). We and others have recently shown that some S. aureus strains also contain the copBL operon to aid Cu detoxification (Rosario-Cruz et al., 2019, Purves et al., 2018). CopB is a P1-type Cu exporting ATPase and genetic evidence suggests that it has a degree of functional redundancy with CopA (Rosario-Cruz et al., 2019). CopL is an external Cu binding lipoprotein that aids in preventing Cu from entering the cell (Rosario-Cruz et al., 2019). Some strains also encode a multicopper oxidase (Mco) and a S. aureus Δmco mutant was more sensitive to Cu than the parental strain (Zapotoczna et al., 2018). S. aureus contain csoR which encodes for a transcriptional regulator of the copBL and copAZ operons (Grossoehme et al., 2011, Purves et al., 2018). Upon binding to Cu(I), holo-CsoR relieves repression of copBL and copAZ (Grossoehme et al., 2011, Purves et al., 2018).
During infection, the host restricts the bioavailability of metal ions including Mn (Becker & Skaar, 2014). To overcome Mn limitation, S. aureus employ two Mn uptake systems (Horsburgh et al., 2002, Kehl-Fie et al., 2013). The mntABC operon encodes for an ATPase to provide energy for uptake (MntA), a permease (MntB), and an extracellular Mn binding lipoprotein (MntC) (Gribenko et al., 2013). The mntH gene encodes a Mn permease of the NRAMP family (Que & Helmann, 2000, Horsburgh et al., 2002). Cytosolic Mn is sensed by the transcriptional regulator MntR. When associated with Mn, holo-MntR represses mntABC transcription by binding to the promoter of mntA (Horsburgh et al., 2002, Glasfeld et al., 2003, Handke et al., 2013).
Given the inherent differences between bacterial species, the cellular targets of Cu intoxication are likely not universal, and the goal of this study was to investigate how Cu toxifies S. aureus. To this end, we constructed a strain that is defective in copper detoxification (cop- strain). This strain was sensitive to Cu and it accumulated Cu. We screened for cop- strains that had increased tolerance to Cu. Strains containing transposon (Tn) insertions in mntA were protected against Cu intoxication. The data presented herein are consistent with the hypothesis that S. aureus with defective MntABC permease have decreased Cu uptake which protects FeS enzymes from Cu poisoning. These findings highlight the challenges that organisms face when trying to acquire an essential micronutrient and help to explain why having redundant tightly regulated systems such as MntH and MntABC to uptake metals is beneficial.
Results
A S. aureus ΔcopBL ΔcopAZ strain accumulates intracellular Cu.
We created a ΔcopAZ ΔcopBL strain in the USA300_LAC background. We compared the growth of the ΔcopAZ ΔcopBL strain to that of the wild type (WT), ΔcopAZ, and ΔcopBL strains. When compared to the growth of the WT, the ΔcopBL strain was more sensitive to growth on solid medium in the presence of Cu whereas the ΔcopAZ displayed little to no Cu sensitivity phenotype (Figure 1A). The ΔcopAZ ΔcopBL double mutant was more sensitive to Cu than the ΔcopAZ and ΔcopBL strains. We henceforth refer to the ΔcopAZ ΔcopBL strain as cop-.
Figure 1. S. aureus CopBL and CopAZ function in copper detoxification.

Panel A; Spot plate analysis of wild-type (JMB1100), ΔcopAZ (JMB8571), ΔcopBL (JMB7901), ΔcopAZ ΔcopBL (cop-) (JMB8573). Overnight cultures were serial diluted and spot plated on TSB medium containing various concentrations of Cu. Panel B; Spot plate analysis of cop- (JMB8573) and cop- csoR::Tn (JMB8723). Overnight cultures were serial diluted and spot plated on TSB medium with and without 1.25 mM Cu. Panel C; Total cellular 63Cu and 55Mn loads were determined in the WT and cop- strains using inductively coupled plasma mass spectrometry after growth in TSB medium in the presence and absence of 10 μM Cu. The data represent the mean of three biological replicates and errors are presented as standard deviations. Paired student t-tests were performed on the samples and N.S. denotes not significant (p > 0.1) and * denotes p ≤ 0.05. Photos from representative experiments are shown in Panels A and B.
We hypothesized that the copAZ and copBL are the primary genetically encoded elements in LAC specifically utilized for Cu ion homeostasis. CsoR is a transcriptional repressor that relieves repression of genes within its regulon when it is bound to Cu (Ma et al., 2009, Grossoehme et al., 2011). We created a cop- ΔcsoR strain and found that this strain phenocopied the cop- strain (Figure 1B). Although not conclusive, these data are consistent with the hypothesis that copAZ and copBL are the primary Cu detoxification genes transcriptionally controlled by CsoR in USA300_LAC.
We tested the hypothesis that the cop- strain accumulated intracellular Cu. We used inductively coupled plasma mass spectrometry (ICP-MS) to monitor total Cu load in the WT and cop- strains after culturing in the presence and absence of 10 μM Cu(II). After co-culture with Cu in tryptic soy broth (TSB), both strains accumulated Cu; however, the cop- strain accumulated much more Cu (Figure 1C). There was not a noticeable difference in Cu load between the WT and cop- strains after culture in TSB. The overall cellular Mn load of the cultures was not significantly affected. Co-culture with 10 μM Cu(II) did not result in an appreciable decrease in cell pellet weight (Figure S1A) or significant growth defect (Figure S1B) in the cop- strain.
Mutational analyses provide insight into copper homeostasis.
We built a transposon (Tn) library in the cop- strain. We plated cells from the Tn mutant library and ten strains were isolated that grew with 2.5 mM Cu(II). After reconstruction and phenotypic verification, we mapped the transposon mutations to three locations. Two strains had mutations in mntA of the mntABC operon. One strain had a mutation in apt, which encodes a predicted adenine phosphoribosyltransferase. Seven strains had mutations in ispA, which encodes a geranylgeranyl diphosphate synthase II. All mutants provided a similar growth advantage when cultured on Cu containing solid medium (Figure 2A). For the purposes of this study we focused efforts on determining why the mntA::Tn strains permitted growth in the presence of Cu(II). The mntA::Tn insertions were located at the TA/AT sites located at +84 (mntA1::Tn) and +91 (mntA2::Tn). When cultured in the presence of Cu(II) the mntA::Tn mutants phenocopied one another. For simplicity, only the mntA1::Tn mutation was further investigated. The mntA1::Tn allele also improved the growth of the cop- strain in liquid TSB (Figure 2B).
Figure 2. S. aureus strains with individual transposon insertions have increased growth in the presence of Cu.

Panel A; Overnight cultures of the cop- (JMB8573), cop- mntA1::Tn (JMB8914), cop- apt::Tn (JMB 8902), and cop- ispA::Tn (JMB8898) strains were serial diluted and spot plated on TSB medium containing 0 (no addition) or 2 mM Cu(II). Photos from a representative experiment are shown. Panel B; Overnight cultures of the cop- (JMB8573) and cop- mntA1::Tn (JMB8914) were diluted to 0.05 OD (A600) in liquid TSB medium with and without 50 μM Cu and culture density was monitored over time. The data represent the mean of three biological replicates and errors are presented as standard deviations. Error bars are included for all data, but often obscured by the symbol.
The mntA1::Tn mutations are recessive.
The cop- mntA1::Tn displayed increased growth when compared to the cop- strain when cultured in the presence of Cu(II) on solid or liquid TSB medium (Figure 2), as well as in liquid defined medium (Figure 3A). The finding that suppression also occurred in defined medium allows us to expand phenotypic analyses. When we returned the mntABC genes at a secondary chromosomal location, the resulting cop- mntA1::Tn pLL39_mntABC strain grew similar to the cop- pLL39 (empty vector) strain. These data suggested that the mntA1::Tn mutations were recessive.
Figure 3. The mntA1::Tn mutation is recessive and null mutants in mntA, mntB, or mntC promote growth in the presence of Cu(II).
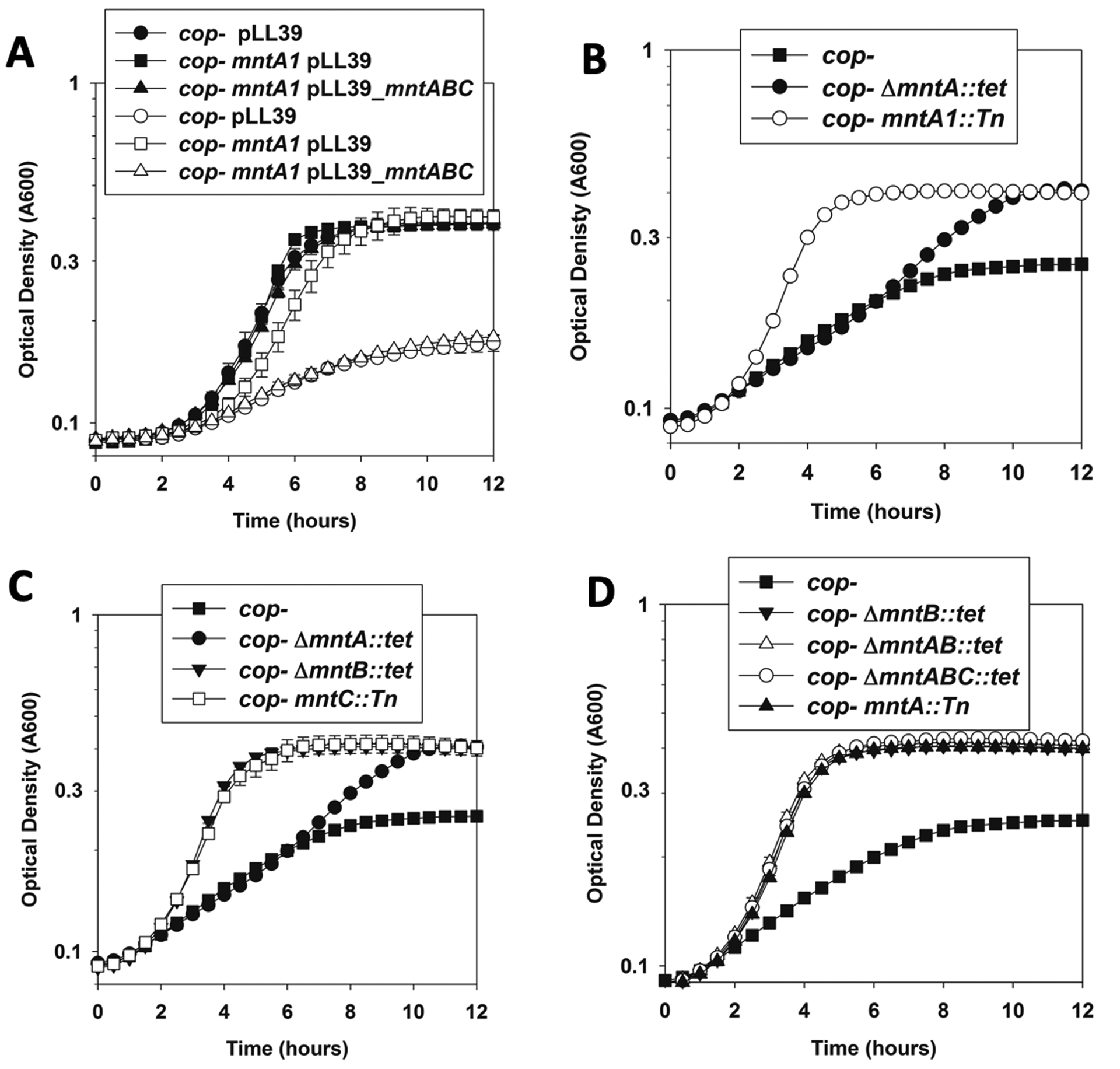
Growth was monitored in chemically defined media containing 20 amino acids (AA) with and without copper. Panel A; Genetic complementation of the Cu(II) resistance phenotype of the cop- mntA1::Tn strain. Growth of the cop- pLL39 (circles; JMB9535), cop- mntA1::Tn pLL39 (squares; JMB9534), and cop- mntA1::Tn pLL39_mntABC (triangles; JMB9469) in the presence (non-filled symbols) and absence (filled symbols) of 15 μM Cu(II) is shown. Panel B; A ΔmntA::tetR allele partially protected against Cu(II) intoxication. Growth of the cop- (squares; JMB8573), ΔmntA::tetR (black circles; JMB 9201), and mntA1::Tn insertion (white circles; JMB8914) in the presence of 20 M μCu(II) is shown. Panel C; non-functional mntA::tetR, mntB::tetR, and mntC::Tn alleles improve the growth of the cop- strain in the presence of Cu(II). Growth of the cop- (filled squares; JMB8573), cop- ΔmntA::tetR (circles; JMB 9201), cop- ΔmntB::tetR (triangles; JMB9208), and cop- mntC::Tn (white squares; JMB 8965) in the presence of 20 M μCu(II) is shown. Panel D; the phenotypes associated with null mutants in mntA, mntB, and mntC are not genetically additive. Growth of the cop- (squares; JMB8573), ΔmntA::tetR (filled circles; JMB 9201), and ΔmntB::tetR (filled triangles; JMB9208), ΔmntAB::tetR (open triangles; JMB9246), ΔmntABC::tetR (open circles; JMB9325) in the presence of 20 μM Cu(II) is shown. The data represent the mean of two biological replicates and errors are presented as standard deviations. Error bars are included for all data, but often obscured by the symbol.
To further explore this idea, we created a cop- ΔmntA::tetR strain. The mntA::tetR mutation provided a growth advantage in medium containing Cu(II), but it did not provide as robust of a growth advantage as the mntA1::Tn mutation (Figure 3B). We hypothesized that this was due to the mntA1::Tn mutation having polar effects on downstream operon components. We compared the growth of the cop- ΔmntA::tetR, cop- ΔmntB::tetR, and cop- mntC::Tn strains in the presence and absence of Cu(II). Whereas all three mutants showed better growth than the cop- strain when cultured with Cu(II), the ΔmntB::tetR and ΔmntC::Tn mutants provided better growth than the ΔmntA::tetR mutation (Figure 3C). The growth of the ΔmntB::Tn strain phenocopied that of the mntA::Tn strain (Figure 4D). None of the mnt mutant strains had a growth defect or growth advantage in the absence of Cu under the growth conditions utilized (data not shown).
Figure 4. The cop- mntA1::Tn strain accumulate less Cu post challenge.

Cultures of the cop- (JMB8573) and cop- mntA1::Tn (JMB8914) strains were cultured in chelex treated TSB and challenged with 1,5, and 10 μM Cu(II) and incubated for an additional 15, 30, or 60 minutes before harvesting and determining Cu load by ICP-MS. Data showing total cellular Cu and total cellular Mn are shown. The data represent the mean of three biological replicates and errors are presented as standard deviations.
We tested the hypothesis that the mntABC gene products work in conjunction with one another to provide enhanced growth in the presence of Cu(II). We created cop- ΔmntAB::tetR, and cop- ΔmntABC::tetR strains and compared them to the growth of the cop- ΔmntB::tetR strain. The strains containing the ΔmntB::tetR, ΔmntAB::tetR, and ΔmntABC::tetR mutations phenocopied one another and all three had enhanced growth in the presence of Cu(II) when compared to the cop- strain (Figure 3D). Taken together, these data suggested that the mntA1::Tn mutation had polar effects on expression of downstream operon components and that mutations in any of the mntABC genes was sufficient to suppress the Cu(II) sensitivity phenotype of the cop- strain.
The small RNA RsaC does not modulate the Cu(II) sensitivity of the cop- strain.
The rsaC gene encodes for a small RNA that is located downstream of mntC and is co-transcribed with mntABC in response to Mn starvation (Lalaouna et al., 2019). We examined whether rsaC has a role in 1) in the Cu(II) sensitivity phenotype of the cop- strain, and 2) in the growth advantage phenotype afforded by the mntA1::Tn insertion mutation. The cop- ΔrsaC::tetM strain phenocopied the cop- strain on TSB medium supplemented with Cu(II) (Figure S2). The cop- mntA1::Tn and cop- mntA1::Tn ΔrsaC::tetM strains also phenocopied one another and both strains grew better than the cop- strain in the presence of Cu(II).
To further explore the role of rsaC in Cu(II) homeostasis, we created vectors that expressed rsaC either in the sense or antisense orientation. The plasmid encoded rsaC variants were under the transcriptional control of an anhydrotetracycline inducible promoter. The cop- strain containing the empty vector (pML100) or the vectors expressing rsaC cloned in the sense (pML100_rsaC) or antisense (pML100_rsaC-antisense) direction behaved identical when plated on TSB medium with or without Cu(II) (Figure S3). As a control for rsaC expression, we also plated the same strains on TSB medium supplemented with the reactive oxygen species generating molecule methyl viologen. Strains that overproduce rsaC are sensitive to methyl viologen as a result of decreased superoxide dismutase (sodA) expression (Lalaouna et al., 2019). The cop- strain containing the pML100_rsaC had decreased growth in the presence of inducer and methyl viologen when compared to the cop- strain containing pML100 or pML100_rsaC-antisense. The results further suggest that RsaC does not impact Cu(II) ion homeostasis in the cop- strain under the conditions examined.
The mntA1::Tn mutation results in decreased Cu accumulation
We tested the hypothesis that the mntA1::Tn mutation provides enhanced growth in the presence of Cu(II) by decreasing cellular Cu accumulation. Cultures were grown for eight hours before 0, 1, 5, or 10 μM Cu(II) was added. Cultures were incubated for an additional 15, 30, or 60 minutes before cells were harvested. Cells were washed to remove any adventitiously bound metal and cellular Cu and Mn were quantified using ICP-MS. Cu accumulated in both strains and it accumulated as a function of both time and the concentration of Cu(II) added. When compared with cop- strain, the cop- mntA1::Tn mutant accumulated less copper across all Cu concentrations of Cu utilized and all sampling times (Figure 4). Co-incubation with Cu(II) had little effect on the cellular Mn content. The cop- mntA1 strain had slightly less Mn than the cop- strain at a few of the time points examined.
mntABC function in Mn homeostasis and are repressed by MntR in a manganese-dependent manner under the growth conditions utilized.
MntR was reported to act as a negative and positive regulator of mntABC and mntH in S. aureus, respectively (Horsburgh et al., 2002). An alternate study found that mntH transcription was not altered as a variable of Mn concentration (Kehl-Fie et al., 2013). We quantified mntABC and mntH transcripts in the cop- and cop- mntR::tetR strains after culturing in the presence and absence of 10 μM Mn(II). The transcription of mntABC was repressed in the presence of Mn(II) and this repression was dependent upon supplementing the growth medium with Mn(II) and the presence of MntR (Figure 5A). Under the growth conditions utilized, neither culturing with Mn(II), or the presence of MntR, had an effect on mntH transcription.
Figure 5. MntR represses transcription of the mntABC in a manganese-dependent manner under the growth conditions utilized.

Panel A; The data show fold-induction of mntA, mntB, mntC, and mntH in the cop- (JMB8573) and cop- ΔmntR::tetR (JMB9151) strains. The strains were cultured in TSB with and without 10 μM Mn(II) before RNA was isolated and transcripts quantified. Panel B; Data represent fold-induction of mntA, mntB, and mntC in the cop- (JMB8573) strain after culture in TSB, TSB with 10 μM Mn(II), Chelex treated TSB, or Chelex treated TSB with 10 μM Mn(II). Panel C; Total 55Mn was quantified in the cop- (JMB8573) and cop- mntA1::Tn (JMB8914) strains using ICP-MS after the strains were cultured for 8 hours in Chelex-treated TSB. The data presented in Panels A, B, and C represent the average of biological triplicates with errors presented as standard deviations. Paired student t-tests were performed on the samples and N.S. denotes not significant (p > 0. 0.05) and * denotes p ≤ 0.05.
Treating TSB with Chelex resin decreases the concentrations of divalent metals. We cultured the cop- strain in TSB, TSB with 10 μM Mn(II), Chelex treated TSB, or Chelex treated TSB supplemented with 10 μM Mn(II) before quantifying mntABC transcripts. The transcription of mntABC was repressed when cells were cultured in TSB medium with Mn(II) when compared to cells cultured in TSB (Figure 5B). These data suggested that our laboratory TSB medium is not replete with Mn. Culturing in Chelex treated medium further increased transcription of mntABC. The addition of 10 μM Mn(II) to the Chelex treated medium returned mntABC transcription to levels noted in the cells grown in TSB medium containing 10 μM Mn(II). These data suggested that we could further remove Mn from the TSB medium by treatment with Chelex.
We sought to verify a role for MntABC in Mn homeostasis in the growth conditions utilized herein. We cultured the cop- and cop- mntA1::Tn strains in Chelex-treated TSB before harvesting cells and determining Mn load using ICP-MS (Figure 5C). The cop- mntA1::Tn mutant had a lower Mn load than the cop- strain confirming a role for MntABC in Mn homeostasis under the growth conditions utilized.
Derepression of mntABC transcription results in increased Cu accumulation and toxicity.
We examined the effect of MntR-dependent regulation of mntABC on cellular Cu loads. The cop-, cop- mntA1::Tn, cop- mntR::tetR, and cop- mntA1::Tn mntR::tetR strains were cultured in Chelex-treated TSB before dosing the cells with and without 5 μM Cu(II). The cells were incubated for an additional 60 minutes before harvesting, washing away adventitiously associated metal, and quantifying cellular Cu and Mn pools. The cop- mntA1::Tn strain accumulated less Cu than the cop- strain (Figure 6A). The cop- mntR::tetR strain accumulated more Cu than the cop- strain; however, this Cu accumulation was dependent upon a functional MntABC. The cop- mntA1::Tn mntR::tetR strain had a Cu load that phenocopied that of the cop- mntA1::Tn strain. In general, the strains had similar Mn pools. The addition of Cu(II) did not have a significant effect on the Mn concentration associated with the cop- strain. The strains containing mntA1::Tn mutation had decreased Mn pools when compared to the cop- parent strain.
Figure 6. Derepression of the mntABC operon results in an increased Cu load and Cu sensitivity.

Panel A; the cop- (JMB8573), cop- mntA1::Tn (JMB8914), cop- ΔmntR::tet (JMB9151), and cop- ΔmntR::tet mntA1::Tn (JMB9244) strains were cultured in Chelex-treated TSB for 8 hours before challenge with 5 μM Cu for 60 minutes before quantifying total Cu and Mn loads using ICP-MS. Panel B; cop- (JMB8573), cop- mntA1::Tn (JMB8914), cop- ΔmntR::tet (JMB9151), and cop- ΔmntR::tet mntA1::Tn (JMB9244) strains were spot plated on solid TSB medium with 1.75 or 2 mM Cu and 0 or 5 μM Mn. The data in Panel A represent the mean of three biological replicates and errors are presented as standard deviations. Paired student t-tests were performed on the samples and N.S. denotes not significant (p > 0.05) and * denotes p ≤ 0.05. Photos from a representative experiment are shown in Panel B.
We tested the hypothesis that derepression of mntABC transcription would increase Cu sensitivity. The cop-, cop- mntA1::Tn, cop- mntR::tetR, and cop- mntA1::Tn mntR::tetR strains were spot-plated on solid TSB containing 1.75 or 2 mM Cu(II). The cop- mntR::tetR strain was more sensitive to Cu(II) than the cop- strain suggesting that removal of MntR-dependent repression of mntABC transcription increased sensitivity to Cu(II) (Figure 6B). The cop- mntA1::Tn mntR::tetR strain phenocopied the cop- mntA1::Tn strain.
The findings that MntR modulates Cu accumulation and homeostasis via MntABC and that our TSB medium is not replete with Mn led us to hypothesize that we could decrease sensitivity to Cu(II) by supplementing the growth medium with Mn(II). We examined the growth of cop-, cop- mntA1::Tn, cop- mntR::tetR, and cop- mntA1::Tn mntR::tetR strains with 1.75 or 2 mM Cu(II) in the presence and absence of 5 μM Mn(II). The presence of Mn(II) improved the growth of the cop- strain when cultured with Cu, but not the growth of the cop- mntR mutant (Figure 6B). The presence of Mn(II) did not alter the Cu(II) sensitivities of the cop- mntA1::Tn and cop- mntA1::Tn mntR::tetR strains.
Increased mntABC expression results in Cu sensitivity and Cu accumulation.
We tested the hypothesis that increased mntABC expression results in increased Cu(II) sensitivity and Cu accumulation. We placed mntABC under the transcriptional control of a xylose inducible promoter. When cultured in the presence of Cu(II), the cop- strain containing the plasmid pEPSA5_mntABC had decreased growth when compared to the cop- strain containing the pEPSA5 (empty vector) (Figure 7A). The WT strain with pEPSA5_mntABC behaved like the WT strain containing pEPSA5.
Figure 7. Over-production of MntABC increases sensitivity to Cu and increases the cellular Cu load.

Panel A; the cop- pEPSA5 (JMB9062) , cop- pEPSA5_mntABC (JMB9397) , WT pEPSA5 (JMB1304) , and WT pEPSA5_mntABC (JMB9478) overnight cultures were serial diluted and spot platted on TSB solid media containing 0.08 % xylose with and without 1.25 mM Cu(II). Panel B; the cop- pEPSA5 (JMB9062) and cop- pEPSA5_mntABC (JMB9397) strains were grown in Chelex-treated TSB for 8 hours before challenge with 5 μM Cu(II) for 15 min. Cells were harvested and Cu quantified using ICP-MS. The data represent the mean of three biological replicates and errors are presented as standard deviations. Paired student t-tests were performed on the samples and N.S. denotes not significant (p > 0.1) and * denotes p ≤ 0.05. Photos from a representative experiment are shown in Panel A.
We compared the total Cu loads of the cop- strain containing either pEPSA5_mntABC or pEPSA5. The cop- strain containing pEPSA5_mntABC had an increased Cu load compared to the cop- strain containing pEPSA5 (Figure 7B). These data are consistent with the hypotheses that overproduction of MntABC in the cop- strain results in increased Cu accumulation and that the WT strain, which contained copAZ and copBL, was able to detoxify Cu even when MntABC was over produced.
The lack of a functional MntABC protects FeS enzymes from Cu poisoning.
Recent work by Johnson et al. reported that the Streptococcus pneumonia aerobic Mn-dependent ribonucleotide reductase (NrdEF) is poisoned by Cu (Johnson et al., 2015). S. aureus utilizes the NrdEF and NrdDG ribonucleotide reductases primarily during aerobic and anaerobic growth, respectively (Cotruvo & Stubbe, 2012, Rabinovitch et al., 2010). We tested the hypothesis that the activity of essential NrdEF is decreased by intracellular Cu accumulation. The cop- and cop- nrdD::Tn strains were spot plated on solid medium with and without Cu and/or the ribonucleotide reductase inhibitor hydroxyurea. Hydroxyurea and Cu(II) individually inhibited the growth of the strain aerobically, but no difference was noted between the cop- and cop- nrdD::Tn strains (Figure S4). However, these compounds showed great synergy in inhibiting growth when provided simultaneously. The addition of Mn did not significantly affect growth in the presence of Cu or hydroxyurea. The cop- nrdG::Tn strain phenocopied the cop- nrdD::Tn strain (data not shown).
During anaerobic growth the cop- nrdD::Tn strain had smaller colony size phenotype and it was more sensitive to hydroxyurea than the cop- strain (Figure S4). Both strains had slow growth phenotype in the presence of Cu(II) and there was not a significant decrease in growth of the cop- nrdD::Tn strain with Cu(II) (Figure S5). There was not a synergistic effect of hydroxyurea and Cu. Taken together, these data suggest that NrdEF is not a primary target of Cu toxicity in S. aureus.
Intracellular Cu accumulation can directly and indirectly decrease the activities of enzymes that require solvent-exposed clusters (Macomber & Imlay, 2009, Tan et al., 2014). S. aureus utilizes the SufCDSUB machinery to synthesize iron–sulfur (FeS) clusters from monoatomic Fe2+, S0, and electrons (Roberts et al., 2017). We previously characterized a strain that contains a Tn insertion between the sufC and sufD genes (sufD*) of the sufCDSUB operon. The sufD* mutation results in decreased transcription of suf genes resulting in a decreased capacity to maturate FeS proteins (Roberts et al., 2017). When compared to the cop- strain, the cop- sufD* strain had a greatly decreased growth when cultured with Cu(II) (Figure 8A). The presence of the mntB::tetR allele rescued the growth of the cop- sufD* strain in the presence of Cu(II) (Figure 8B).
Figure 8. Defective MntABC protects iron-sulfur proteins from Cu poisoning.

Panel A; Growth of the cop- (JMB8573) and cop- sufD* (JMB8625) strains were monitored in defined medium in the presence and absence of 5 μM Cu(II). Panel B; Growth of the cop- sufD* (JMB8625) cop- sufD* ΔmntB::tetR (JMB9604) strains was monitored in defined medium in the presence and absence of 5 μM Cu(II). Growth in panels A and B was monitored in chemically defined media containing 20 amino acids (AA). Panel C; Growth of the cop- (JMB8573) and cop- mntA1::Tn (JMB8914) strains in defined medium containing 18 amino acid medium lacking leucine and isoleucine with and without 5 μM Cu(II). Panel D; AcnA activity was monitored in cell-free lysates from the cop- acnA::tetR and cop- acnA::tetR mntA1::Tn strains containing pEPSA5_mntABC after culture in presence of 0–20 μM Cu(II). Panel E. AcnA activity was monitored in cell-lysates generated from the cop- strain cultured without Cu. The lysates were treated with Cu before assaying AcnA. The data in panels A-C represent the average of biological duplicates with error expressed as standard deviations. The data in panels D and E represent the average of triplicates or biological triplicates (panel D) with error expressed as standard deviations. Paired student t-tests were performed on the samples in D and E and N.S. denotes not significant (p > 0.05) and * denotes p ≤ 0.05.
Previous work found that Cu inhibits Leu and Ile synthesis by inactivation of the FeS cluster-dependent dehydratases necessary to synthesize the amino acids (Macomber & Imlay, 2009). We also noted that the addition of 5 μM Cu(II) to defined medium lacking Leu and Ile inhibited the growth of the cop- strain (Figure 8C). The cop- strain was capable of growth in defined medium containing 5 μM Cu(II) when it was supplemented with Leu and Ile (data not shown). The cop- mntA1::Tn strain was able to grow in defined medium containing 5 μM Cu(II) medium lacking Leu and Ile. These data suggested that the presence of the mntA1::Tn mutation protects FeS cluster containing dehydratases from inactivation in the presence of Cu.
We examined the effect of Cu(II) on aconitase (AcnA) activity, which is an FeS cluster-dependent dehydratase (Beinert et al., 1996). For these experiments, we used a strain that contained a null acnA allele, and a secondary plasmid encoded acnA allele that was under the transcriptional control of a xylose inducible promoter (pacnA). This decreased the possibility that Cu was modulating acnA transcription. Growth in the presence of Cu(II) resulted in a concentration-dependent decrease in AcnA activity in the cop- acnA::tetR strain containing pacnA. AcnA activity was nearly undetectable after culture with 20 μM Cu(II) (Figure 8D). We next examined the effect of introducing the mntA1::Tn mutation on in vivo AcnA activity. The cop- acnA::tetR mntA1::Tn strain containing pacnA had decreased AcnA activity when cultured in the presence of Cu(II), but the decrease in activity was substantially less than that noted for the cop- acnA::tetR strain (Figure 8D).
We sought to determine if the S. aureus AcnA was poisoned by Cu in vitro. Cell lysates from the cop- strain were created anaerobically and then combined with Cu(II) before monitoring AcnA activity. The presence of Cu(II) decreased AcnA activity in a concentration dependent manner (Figure 8E). Ascorbate can reduce Cu(II) to Cu(I) which has a higher affinity for sulfur atoms and is a more avid disrupter of FeS clusters (Macomber & Imlay, 2009). We repeated the in vitro AcnA assays, but this time, when we co-incubated the lysates with Cu(II) we included 100 μM ascorbate in one-half of the samples. The inclusion of ascorbate did not enhance Cu-dependent poisoning of AcnA in vitro (Figure S6).
Discussion
The goal of this study was to investigate how Cu toxifies S. aureus. To this end, we created and examined a S. aureus strains lacking the CopAZ and CopBL Cu detoxification factors. As previously reported, the ΔcopBL and ΔcopAZ mutants had intermediate sensitivities to growth in the presence of Cu(II) whereas the phenotypes of the ΔcopBL and ΔcopAZ deletions displayed genetic synergy. The cop- (ΔcopBL ΔcopAZ) strain also had a high Cu load after dosing with Cu(II). CsoR represses Cu detoxification systems and a cop- ΔcsoR strain had the same sensitivity to Cu(II) as the cop- strain. Although not conclusive, these data are consistent with the hypothesis that copAZ and copBL are the primary Cu detoxification genes in USA300_LAC that are under CsoR transcriptional control.
The data presented herein, in conjunction with our previous work, as well as work from others, has resulted in the following working model for Cu homeostasis in S. aureus (Figure 9). Under Mn deplete conditions, mntABC is expressed and Cu(II) enters S. aureus cells by a MntABC-dependent mechanism. Once inside, cellular reductants reduce Cu(II) to Cu(I) (Scarpa et al., 1996, Rigo et al., 2004, Ngamchuea et al., 2016). The Cu(I) can associate with the CsoR transcriptional repressor resulting in derepression of the copAZ and copBL operons (Grossoehme et al., 2011, Rosario-Cruz et al., 2019). CopA and CopB function as Cu(I) export systems (Grossoehme et al., 2011, Purves et al., 2018). CopZ acts as an intracellular Cu(I) binding protein that buffers the cytosol from Cu toxicity (Banci et al., 2001). Holo-CopZ traffics Cu(I) to CopA for export (Singleton et al., 2009). After export by CopA or CopB, or before Cu enters the cell, CopL binds to Cu(I) and prevents it from (re)entering (Rosario-Cruz et al., 2019). Defective Cu removal results in poisoning of FeS cluster-dependent enzymes.
Figure 9. Working model for copper ion homeostasis in Staphylococcus aureus.

Under Mn deplete conditions, MntR derepresses mntABC transcription and mntABC is expressed. Cu enters S. aureus cells in a MntABC-dependent manner. Once the Cu ions have entered the cell, they are sensed by the CsoR transcriptional regulator. Cu association with CsoR results in derepression of the copAZ and copBL operons. CopA and CopB function as Cu(I) export systems. CopZ acts as an intracellular Cu(I) binding protein that buffers the cytosol from Cu toxicity. Holo-CopZ traffics Cu(I) to CopA for export. After export by CopA or CopB, or before Cu enters the cell, CopL binds to Cu(I) and prevents it from (re)entering.
The S. pneumoniae genome encodes for the MntABC and MntR homologues PsaBCA and PsaR (Dintilhac et al., 1997, McAllister et al., 2004, Johnston et al., 2006, Lisher et al., 2013). PsaB is an ATPase and PsaA is a Mn-specific permease (Claverys, 2001). PsaA is a high-affinity is Mn(II) substrate-binding protein (SBP) that can also bind Zn(II) (Li et al., 2014, McDevitt et al., 2011). PsaA binding to Zn(II) can preclude Mn binding and PsaA association with Mn is thought to be necessary PsaBA-mediated Mn(II) import (Counago et al., 2014). Therefore, Zn(II) association with PsaA can result in decreased Mn(II) influx (McDevitt et al., 2011). Consistent with this, co-culturing S. pneumoniae with high levels of Zn(II) results in growth phenotypes that mimic that of a ΔpsaA mutant. Thermostability and metal binding studies were used to show that PsaA can also associate with Cu(II) (Counago et al., 2014). When co-cultured with Cu(II), the wild-type and ΔpsaA strains had similar Cu accumulation suggesting that PsaBA does not import Cu in S. pneumoniae; however, it should be noted that these strains possessed the CopA Cu(I) exporter and the CupA Cu(I) chaperone possibly masking potential effects that the ΔpsaA mutation had on cellular Cu accumulation (Counago et al., 2014, Shafeeq et al., 2011). The addition of Cu(II) to the growth medium did not significantly impact the concentration of Mn associated with the S. aureus cop- strain under the growth conditions utilized in our studies suggesting the Cu intoxication is not occurring by interfering with Mn uptake.
The mntAB gene products work in conjunction as an ABC transporter (Dintilhac et al., 1997). Results herein show that the ΔmntA::tetR and ΔmntB::tetR mutants both suppressed the Cu sensitivity phenotype of the cop- strain; however, unexpectedly, the ΔmntA::tetR mutation did not suppress to the same extent as the ΔmntB::tetR or mntA1::Tn mutations. The genetic analyses presented show that the effects of the individual mutations were not additive suggesting that MntABC do work in conjunction with one another. Similar to the results presented herein, Novick et al. reported that strains with plasmid insertion mutations in psaA, psaB, or psaC resulted in strains with slightly different transformation efficiencies and adherence properties (Novak et al., 1998). McAllister et al. created and analyzed a library of psa clean deletion mutants and reported that the individual psa mutants did not phenocopy one another (McAllister et al., 2004). Lim et al. found that Neisseria gonorrhoeae mntAB and mntC mutants have different phenotypes with the mntAB mutant having a more severe growth defect and a small colony phenotype (Lim et al., 2008). The authors hypothesized that in the absence of MntC, the permease still functions, but it has a lower affinity for the metals of interest. In this case, the authors hypothesized that MntABC transported both Zn and Mn. TcyABC is an ABC transporter for cystine in Streptococcus mutans. Kim et al. reported differential growth phenotypes of tcyA, tycB, and tycC mutant strains (Kim et al., 2012). The tcyABC and tcyC mutants did not grow in low cysteine containing medium whereas the tcyA and tcyB mutants were capable of growth abet with a slow growth phenotype. TcyA, TcyB, and TcyC are predicted to be an SBP, a permease, and an ATPase, respectively.
RsaC is a sRNA that is co-transcribed with mntABC in S. aureus (Lalaouna et al., 2019). In the FPR3757 genome, the +1 of the rsaC coding region resides 113 base-pairs downstream of mntC. RsaC interacts with a number of mRNAs that function in ROS/RNS metabolism including sodA which encodes for a Mn-dependent superoxide dismutase. Five results are presented herein that suggest that rsaC is not involved in the increased Cu(II) ion resistance phenotype of the cop- mntA1::Tn strain under the growth conditions we utilized. First, we returned the mntABC genes in trans to the cop- mntA1::Tn mutant and found that they could complement the Cu sensitivity phenotype. Both the pLL39_mntABC and pEPSA5_mntABC vectors that we utilized lack the rsaC ORF and both plasmids work for genetic complementation of the Cu sensitivity phenotype of the mntA::Tn strain. Second, a cop- ΔrsaC::tet mutant strain displayed the same sensitivity to Cu(II) as the cop- strain. Third, the ΔrsaC::tet mutation did not alter the ability of the mntA1::Tn mutation to suppress the Cu(II) sensitivity phenotype of the cop- strain. Fourth, overproduction of rsaC did not alter the sensitivity of the cop- strain to Cu(II). Fifth, over-expression of mntABC using either pEPSA5_mntABC or a ΔmntR strain results in increased sensitivity to Cu and increased cellular Cu accumulation. Introduction of the mntA1::Tn allele nullified the effects of the ΔmntR mutation. Taken together, these findings are consistent with the hypothesis that the disruption of something within mntABC by the mntA1::Tn insertion improves growth in the presence of Cu(II) and this phenotype is independent of rsaC expression.
Compared to other transition metals, Cu uptake pathways are the least understood in bacterial pathogens and questions remain about how Cu enters cells (Begg, 2019). Dedicated Cu uptake systems have been described in Rhodobacter capsulatus and Synechocystis which require increased Cu loads for specific metabolic processes (Ekici et al., 2012, Tottey et al., 2001). A dedicated Cu import system has not been described for S. aureus.
A number of metals including lead (Pb), silver (Ag), cadmium (Cd) and mercury (Hg) have no described biological roles and are potent poisons of intracellular enzymes (Xu & Imlay, 2012, Jaroslawiecka & Piotrowska-Seget, 2014). For the most part, dedicated import systems for these metals do not exist. Positively charged metal ions have a difficult time passing through hydrophobic membranes, which has led to the hypothesis that these metals enter cells through import systems that function primarily to import alternate compounds that are required for proper functioning of the cell (Tynecka et al., 1981, Laddaga & Silver, 1985). This is paradoxical because these transporters are expressed to allow entry of essential nutrients while maintaining an electrochemical gradient and preventing the entry of toxic compounds. Cellular systems have evolved to transform and/or efflux these toxic metals thereby decreasing cytosolic concentrations and maintaining homeostasis (Borremans et al., 2001, Gupta et al., 1999, Gaballa & Helmann, 2003, Norambuena et al., 2017).
Metals such as Zn, Fe, Mn, and Cu, often have cellular roles, but cytosolic overload can be toxic (Chandrangsu & Helmann, 2016, Macomber & Imlay, 2009, Huang et al., 2017, Guan et al., 2015). The mechanisms of Mn, Zn, Fe acquisition have been thoroughly examined (reviewed here (Palmer & Skaar, 2016)). Nearly all described ABC transporters only transport in one direction (Wilkens, 2015). In support of this, recent work has found that some bacteria utilize Mn, Zn, and Fe efflux systems to prevent cytosolic accumulation (Guan et al., 2015, Huang et al., 2017, Gaballa & Helmann, 2003, Chandrangsu et al., 2017).
Many studies suggest that metal import is a promiscuous process, but few studies have identified and conclusively shown that metal uptake systems inadvertently uptake alternate metals. In S. aureus, Cd(II) uptake by membrane vesicles was decreased upon the addition of CCCP, valinomycin, or Mn(II) (Perry & Silver, 1982). These data led to the hypothesis that Cd(II) uptake required the PMF and was entering through a Mn(II) transporter. An alternate study found that a S. aureus mntA mutant had increased resistance to Cd(II) (Horsburgh et al., 2002). These data have been interpreted to suggest that Cd(II) is transported into S. aureus via MntABC (Papp-Wallace & Maguire, 2006), but direct evidence for this is lacking.
Studies using other organisms have provided more convincing data that some metal ion transporters can transport more than one metal. Escherichia coli strains over-producing the ZIP family Zn importer ZupT resulted in sensitivity to Fe and Co, as well as cellular accumulation of the metals (Grass et al., 2005). Expression of the Lactobacillus plantarum MntA P-type ATPase Mn importer in E. coli increased cellular Cd(II) and Mn(II) loads (Hao et al., 1999). Expression of the sitABCD in E. coli increased cellular Fe and Mn loads (Sabri et al., 2006) and the Yersinia pestis YfeABCD metal transporter was shown to transport both Mn and Fe.
Unlike Streptococcus pneumoniae, NrdF does not appear to be a primary target of Cu toxicity in S. aureus. However, a S. aureus strain deficient in synthesizing FeS clusters had increased sensitivity to Cu. Moreover, the addition of Cu resulted in an inability to grow without supplementing the growth medium with the amino acids Leu and Ile, which is a phenotype common to S. aureus strains defective in FeS protein maturation (Mashruwala et al., Rosario-Cruz et al., 2015, Mashruwala et al., 2016a). Introduction of the mntB::tetR allele corrected both phenotypes suggesting that decreasing Cu influx protects FeS proteins from Cu poisoning. Furthermore, in vivo and in vitro incubation with Cu(II) inhibited aconitase in a concentration dependent manner. A lower concentration of Cu was necessary to inhibit in vivo than needed for inhibition in cell-free lysates. This may be the result of Cu poisoning both the FeS cluster synthesis machinery and holo-AcnA in vivo resulting in a compounding effect. Intracellular Cu(I) is a more avid disruptor of AcnA than the Cu(II) (Macomber & Imlay, 2009); however, the inclusion of ascorbic acid to reduce Cu(II) to Cu(I) did not increase AcnA poisoning in vitro under the assay conditions utilized. This could be a result of an adequate concentration of molecules in the lysates that can reduce Cu(II). This may result in nullifying the effect of ascorbic acid on Cu(II) reduction. We recently noted a similar effect in the inhibition of AcnA in Thermus thermophilus. The concentration of Hg necessary to inhibit AcnA in vivo was less than that necessary to inhibit the enzyme in cell lysates (Norambuena et al., 2019).
Similar to the results presented herein, Johnson et al. reported the supplementing the medium with Mn protected Streptococcus pneumoniae from Cu intoxication (Johnson et al., 2015). We noted that the addition of Mn, at a >10-fold lower concentration than that used by Johnson et al., protected S. aureus against Cu intoxication. This difference could be a result of differing bacterial uptake rates, but these findings led us to hypothesize that the lower amount of Mn required to protect S. aureus from Cu intoxication was the result of regulatory effects. Protection in S. aureus was dependent upon the presence of the Mn sensing transcriptional regulator MntR. A cop- ΔmntR strain had increased mntABC transcription and decreased growth in the presence of Cu. The decreased growth of the cop- ΔmntR strain was corrected by the introduction of the mntA1::Tn allele. The cop- ΔmntR strain also had a larger Cu load than the cop- strain, and again, this phenotype was mitigated by the introduction of mntA1::Tn. These data have led us to modify our working model (Figure 9) wherein Mn protects S. aureus against Cu intoxication by binding to MntR resulting in MntR-dependent repression of mntABC transcription and ultimately decreasing MntABC-dependent Cu uptake. In light of our findings it would be interesting to examine the effect of the absence of psaR and/or psaBCA on Cu intoxication in S. pneumoniae. We note that there are many variables that may contribute to the differences in the intracellular targets of Cu(II) in S. pneumonia and S. aureus including growth conditions, growth media utilized and physiology.
To summarize, we have demonstrated that a S. aureus strain lacking CopAZ and CopBL is defective in Cu efflux. This strain is protected by loss of function mutations in mntABC. S. aureus lacking MntABC have decreased cellular copper loads whereas increased transcription of mntABC increase Cu loads. We also established that FeS enzymes are a primary target of Cu toxicity. Taken together, these findings expand our understanding of bacterial metal ion homeostasis. Further biochemical studies will be necessary to determine if MntABC can physically import Cu.
Experimental Procedures
Materials and services
Phusion DNA polymerase, deoxynucleoside triphosphates, the quick DNA ligase kit, and restriction enzymes were purchased from New England BioLabs. The plasmid miniprep kit, gel extraction kit, and RNA Protect were purchased from Qiagen. TRIzol and High-Capacity cDNA reverse transcription kits were purchased from Life Technologies. Oligonucleotides, obtained from Integrated DNA Technologies, are listed in Table 2. DNase I was purchased from Ambion. Lysostaphin was purchased from Ambi Products. TSB was purchased from MP Biomedical. Difco BiTek agar was added (15 g L−1) for solid medium. Phosphate buffered saline tablets were purchased from Calbiochem. Distilled and deionized water was used to prepare chemicals, and glassware was often acid washed prior to use. Chelex-100 resin was purchased from Bio-rad. Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich and were of the highest purity obtainable. DNA was sequenced by Genewiz (South Plainfield, NJ).
Table 2.
Primers used in this study
| Primer name | Sequence |
|---|---|
| Ycc Pjb38 forward | AATAGGCGTATCACGAGGCCCTTTCGTCTTCAAGAATTCGGTGGCACT TTTCGGGGAAA |
| Ycc rseA rev | ACCTGATGAATAGCCACCTGAACGGCTAGCGCACATTAGGACCGTTATAGTTACGCTAT |
| Ycc rseA for | ATAGCGTAACTATAACGGTCCTAATGTGCGCTAGCCGTTCAGGTGGCTATTCATCAGGT |
| rseA tetR for | TATTAAAAGGTTGAAATGGTAGAGATAGCACGCGTCGGATTTTATGACCGATGATGAAG |
| rseA tetR rev | CTTCATCATCGGTCATAAAATCCGACGCGTGCTATCTCTACCATTTCAACCTTTTAATA |
| tetR rseA for | TATAAACATTCTCAAAGGGATTTCTAAACGCGTAGATCGATGTGTAACATTACGTTCTA |
| tetR rseA rev | TAGAACGTAATGTTACACATCGATCTACGCGTTTAGAAATCCCTTTGAGAATGTTTATA |
| rseA pJB38 rev | CTAGAGGATCCCCGGGTACCGAGCTCGAATTCCGAAATGCCAAGCTCAAAGTACATCCA |
| YCC CopAZ rev | GGACCT AATTGT GGA TTC TAC ACG CTA GCG CAC ATT AGG ACC GTT ATA GTT ACG CTA T |
| CopAZ Up for | ATA GCG TAA CTA TAA CGG TCC TAA TGT GCG CTA GCG TGT AGA ATC CAC AAT TAG GTC C |
| CopAZ up rev | CGA CAT CGT AAC CTT GAT CTT CAC GCG TGT CAT ACC AGT GAT ATC TAA TGT TG |
| CopAZ dwn for | CAACATTAGATATCACTGGTATGACACGCGTGAAGATCAAGGTTACGATGTCG |
| pJB38 CopAZ rev | CTA GAG GAT CCC CGG GTA CCG AGC TCG AAT TCG ACA ATG TTG AAG GTG TCG CTG G |
| YccmntRfor | AGC GTA ACT ATA ACG GTC CTA ATG TGC GCT AGC AAG ATT CTA ATT CTA AAT CGC TTA AT |
| mntRtetRrev | TTT CTT CAT CATCGG TCA TAA AAT CCG ACG CGT ACT TTC ACC TCA CAT ACA TTG TCT AT |
| tetRmntRfor | GTA TAT AAA CAT TCT CAA AGG GAT TTC TAA AAT AAA GAA GCC ATA AAG ATA TCC ATG AT |
| pJB38mntRrev | TAG AGG ATC CCC GGG TAC CGA GCT CGA ATT CTA TTA TGT TAT TGA TAG GTT TAG CGT CG |
| mntRtetRfor | ATA GAC AAT GTA TGT GAG GTG AAA GTA CGC GTC GGA TTT TAT GAC CGA TGA TGAAGA AA |
| tetRmntRrev | ATC ATG GAT ATC TTT ATG GCT TCT TTA TTT TAG AAA TCC CTT TGA GAA TGT TTA TAT AC |
| YCCmntAfor | ATA GCG TAA CTA TAA CGG TCC TAA TGT GCG CTA GCC CAG CGT CCG AGT CCA CAG TGG GA |
| mntAtetRfor | CTT TTA ATT AGG AGGTAT AAA CGA CGC GTC GGA TTT TAT GAC CGA TGA TGAAGA AAA GA |
| mntAtetRrev | TCT TTT CTT CAT CATCGG TCA TAA AAT CCG ACG CGT CGTTTA TAC CTC CTA ATT AAA AG |
| tetRmntAfor | ATA TAA ACA TTC TCA AAG GGA TTT CTA ACC ACCATG ATC TAT CAA AAG CAA AGC AAT AC |
| tetRmntArev | GTA TTG CTT TGC TTT TGA TAG ATC ATG GTG GTT AGA AAT CCC TTT GAG AAT GTT TAT AT |
| pJB38mntArev | ACT CTA GAG GAT CCC CGG GTA CCG AGC TCG AAT TCT GTG GCC TAA AAT ATT GGA GAT AC |
| tetRmntABrev | GGT ACT AAT TTT TTC ATG TTA AAC TTC CTC GTT AGA AAT CCC TTT GAG AAT GTT TAT AT |
| tetRmntABfor | ATA TAA ACA TTC TCA AAG GGA TTT CTA ACG AGG AAG TTT AAC ATG AAA AAATTA GTA CC |
| YCCmntABCfor | ATA GCG TAA CTA TAA CGG TCC TAA TGT GCG CTA GCC GTT ATT GTT GCA GCT TGT TAT AT |
| mntABCtetRrev | TCT TTT CTT CAT CATCGG TCA TAA AAT CCG ACG CGT TTC TAA CAA ACG TTT ATA CCT CC |
| mntABCtetRfor | GGA GGT ATA AAC GTT TGT TAG AAA CGC GTC GGA TTT TAT GAC CGA TGATGA AGA AAA GA |
| tetRmntABCrev | TCT TAC TTC ATT AAA ACA CAG CGT GTT ATT AGA AAT CCC TTT GAG AAT GTT TAT ATA |
| tetRmntABCfor | TAT ATA AAC ATT CTC AAA GGG ATT TCT AAT AAC ACG CTG TGT TTT AAT GAA GTA AGA |
| pJB38mntABCrev | ACT CTA GAG GAT CCC CGG GTA CCG AGC TCG AAT TCA GTA GGG AGA ACA GTT GTC CAA TC |
| pJB38mntABCrev | ACT CTA GAG GAT CCC CGG GTA CCG AGC TCG AAT TCA GAT GAA GTA TCA GAA GGT ACA GC |
| pEPmntABCfor | CCTCTAGAGTCGACTTAGGAGGATGATTATTT TTGTTAGAAACAAAAGATTTAAATCTG |
| mntABCYCCrev | GCG TAA CTT TTC CCC GAA AAG TGC CAC CACGCG TGG TAA TAT ATA TTT AAC GCA CGA TA |
| pLLYCC5 | CTG TAA TGG GCC CAA TCA CTA GTG AAT TCC CGA AGC GGT GGC ACT TTT CGG GGA AA |
| mntABCYCC3 | AAA GAG GAC TAT TTA AAG GCA ATC CTT ACG CTA TAT TAC CCT GTT ATC CCT A |
| YccmntABC | TAG GGA TAA CAG GGT AAT ATA GCG TAA GGA TTG CCT TTA AAT AGT CCT CTT T |
| mntABCpLL39 | GTG CTA AAG AAGTTG TAG GTA ATA AAA AAG CTT GCC AAT ATT TTA GGT TGC ATC AAC ATC |
| upRsaCtetfwd | CGAAGGCTAATAGTCCCATATCGTGCGCGGATTTTATGACCGATGATGAAGAAAAG |
| tetupRsaCrev | CTTTTCTTCATCATCGGTCATAAAATCCGCGCACGATATGGGACTATTAGCCTTCG |
| downRsaCtetrev | ATAGCCACACTCATATGACATCGGTTAGAAATCCCTTTGAGAATGTTTATATACATTCAAGG |
| tetdownRsaCfwd | CCTTGAATGTATATAAACATTCTCAAAGGGATTTCTAACCGATGTCATATGAGTGTGGCTAT |
| pJB38rsaCrev | ACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCGCCATTATTAACGAAACCTGCACCTAAAACAG |
| YCCrsaCfor | ATAGCGTAACTATAACGGTCCTAATGTGCGCTAGCGACGAATTCAATTTTATATGATATGGCTAAAAATG |
| RsaCSalIantisense | GGGGTCGACGGTGAAGTTAAGTGTATTCACAAAAAATAGCCACACTCATATG |
| RsaCEcoRIantisense | GGGGAATTCGGCCACACATCAACATAACAAAGTCGAAGGCTAATAGTCCCATATC |
| RsaCSalIsense | GGGGTCGACGGCCACACATCAACATAACAAAGTCGAAGGC |
| RsaCEcoRIsense | GGGGAATTCGGTGAAGTTAAGTGTATTCACAAAAAATAGCCACACTCATATG |
| Rt_mntC Fwd | GCA GTG ATA AGT CAA ATG GCA AAT T |
| Rt_mntC rev | TCT CCA CCAACA TTT TTA GCC ATA |
| Rt_mntA fwd | ATA CCA GTA CGC GGC GAA ATA |
| Rt_mntA rev | AGG GAA GAT TTA CCA GCA CCA TT |
| Rt_mntB fwd | TCA CGC AGT ATT ACC TGG TGT TG |
| Rt_mntB rev | ACC AGT TAT AAG TGC GCC TAC AAA |
| Rt_mntH fwd | TTA GTC GCT GTT GGT TAC ATG GA |
| Rt_mntH rev | GCG CCA CCT TGC ATT GAT |
| 16sfwdRT | TGA AAG CCC ACG GCT CAA |
| 16srevRT | TTC TGC ACT CAA GTT TTC CAG TTT |
| gyrAfor | GGACGTCAACGTATTGTTGTCACT |
| gyrARev | CGAGCTCTGCAATTTTTTCAATC |
| SCV8 | GCACATAATTGCTCACAGCCA |
| SCV9 | GCTGATCTAACAATCCAATCCA |
| Tn Buster | GCTTTTTCTAAATGTTTTTTAAGTAAATCAAGTACC |
| Martn-ermR | AAACTGATTTTTAGTAAACAGTTGACGATATTC |
Bacterial strains, media and growth conditions
Unless specified, the S. aureus strains used in this study (Table 1) were isogenic and constructed in the community associated S. aureus MRSA strain USA300_LAC that had cured of the native plasmid pUSA03 that confers erythromycin resistance (Pang et al., 2013).
Table 1.
Microbial strains and plasmids used in this study.
| Microbial strains utilized in this study | ||
|---|---|---|
| Name | Chromosomal Genotype | Allele or strain reference |
| S. aureus USA300 LAC strains | ||
| JMB 1100 | USA300 wild-type strain- LAC | AR Horswill |
| JMB 1432 | fur::tetR | (Horsburgh et al., 2001, Mashruwala & Boyd, 2017) |
| JMB 8571 | ΔcopAZ (SAUSA300_2494–5) | This study |
| JMB 7901 | ΔcopBL (SAUSA300_0078–9) | (Rosario-Cruz et al., 2019) |
| JMB 9621 | ΔcopBL mntA1::Tn (ermB) (SAUSA300_0620) | This study and (103) |
| JMB 8573 | ΔcopBL ΔcopAZ (cop-) | This study |
| JMB 8914 | cop- mntA1::Tn (ermB) (SAUSA300_0620) | This study |
| JMB 8915 | cop- mntA2::Tn (ermB) (SAUSA300_0620) | This study |
| JMB 8898 | cop- ispA::Tn (ermB) (SAUSA300_1470) | This study |
| JMB 8902 | cop- apt::Tn (ermB) (SAUSA300_1591) | This study |
| JMB 9313 | mntA1::Tn (ermB) (SAUSA300_0620) | This study |
| JMB 8965 | cop- mntC::Tn (ermB) (SAUSA300_0618) | This study |
| JMB 9977 | cop- ΔrsaC::tetR | This study |
| JMB 10082 | cop- mntA1::Tn (ermB) ΔrsaC::tetR | This study |
| JMB 9201 | cop- ΔmntA::tetR | This study |
| JMB 9208 | cop- ΔmntB::tetR (SAUSA300_0619) | This study |
| JMB 9325 | cop- ΔmntABC::tetR (SAUSA300_0619–21) | This study |
| JMB 9151 | cop- ΔmntR:tetR (SAUSA300_0621) | This study |
| JMB 9244 | cop- ΔmntR:tetR mntA1::Tn (ermB) | This study |
| JMB 8625 | cop- sufD* | This study and (Roberts et al., 2017) |
| JMB 9604 | cop- sufD* ΔmntB::tetR | This study and (Roberts et al., 2017) |
| JMB 9535 | cop- attP::pLL39 | This study |
| JMB 9534 | cop- mntA1::Tn (ermB) attP::pLL39 | This study |
| JMB 9469 | cop- mntA1::Tn (ermB) attP::pLL39_mntABC | This study |
| JMB 9320 | cop- acnA::tetR | This study and (Somerville et al., 2002) |
| JMB 9517 | cop- mntA1::Tn (ermB) acnA::tetR | This study and (Somerville et al., 2002) |
| JMB 8723 | cop- csoR::Tn | This study and (Fey et al., 2013) |
| Other microbial strains | ||
| S. aureus RN4220 | Restriction minus for transformation | (Kreiswirth et al., 1983) |
| Escherichia coli PX5α | Used for molecular cloning | Protein Express |
| Saccharomyces cerevisiae FY2 | Used for YCC cloning | 13 |
| Plasmids used in this study | ||
| Name | Function | reference |
| pJB38 | Construction of gene deletions | (Bose et al., 2013) |
| pJB38_rsaC::tetR | Construction of rsaC deletion | This study |
| pJB38_ΔmntA::tetR | Construction of mntA deletion | This study |
| pJB38_ΔmntB::tetR | Construction of mntB deletion | This study |
| pJB38_ΔmntABC::tetR | Construction of mntABC deletion | This study |
| pJB38_ΔmntR::tetR | Construction of gene deletion | This study |
| pEPSA5 | XylR dependent transcription | (Forsyth et al., 2002) |
| pEPSA5_mntABC | mntABC under xylose inducible promoter | This study |
| pEPSA5_acnA_FLAG | AcnA expression | (Mashruwala et al.) |
| pLL39 | Genetic complementation | (Luong & Lee, 2007) |
| pLL39_mntABC | mntA::Tn complement | This study |
| pMG020 | Tn library generation | (Grosser et al., 2018) |
| pBursa | Tn library generation | (Bae et al., 2008) |
| pLL2787 | Use to generate attP::pLL39 integrant | (Luong & Lee, 2007) |
| pML100 | TetR dependent transcription | (Lei et al., 2011) |
| pML100_rsaC | Expresses rsaC | This study |
| pML100_rsaC-antisense | Expresses antisense rsaC | This study |
Abbreviations: Tn, transposon
S. aureus strains were cultured in TSB and grown at 37 °C with shaking at 200 rpm. Unless stated otherwise, cells were cultured in 10- or 30-mL capacity culture tubes containing 1 or 7.5 mL of liquid medium, respectively. The chemically defined minimal medium was described previously and was supplemented with 0.5 μg mL−1 lipoic acid (Mashruwala et al., 2016b). Chelex-treated TSB was prepared by incubating liquid TSB overnight at 4 °C with Chelex-100 resin and continuous stirring. The chelated medium was filter sterilized before use. When necessary, antibiotics were added at the final following concentrations: 150 μg mL−1 ampicillin (Amp); 30 μg mL−1 chloramphenicol (Cm); 10 μg mL−1 erythromycin (Erm); 3 μg mL−1 tetracycline (Tet); 100 μg mL−1 spectinomycin; 150 ng mL−1 anhydrotetracycline (Atet). For routine plasmid maintenance, liquid media were supplemented with 10 μg mL−1 or 3.3 μg mL−1 of chloramphenicol or erythromycin, respectively. The CuSO4 and MnCl2 stocks were prepared in deionized and distilled water and filter sterilized. Liquid phenotypic analysis was conducted in 96-well microtiter plates containing 200 μL of medium per well using a BioTek 808E visible absorption spectrophotometer. Plates were continually shaken at shake speed medium. Culture densities were read at 600 nm. The cells used for inoculation were cultured for 18 hours in TSB medium before washing with PBS. The optical densities (OD) of the cell suspensions were adjusted to 2.5 (A600) in PBS. Two microliters of cells were added to 198 μL of medium. For growth analyses using solid media, strains were cultured for 18 hours in TSB medium before harvesting by centrifugation. Cells were washed with PBS, serial diluted in PBS, and 5 μL aliquots were spotted upon solid media.
Aconitase Enzyme assays
Aconitase (AcnA) assay was conducted as previously described (Mashruwala et al., 2016a). Briefly, Strains were cultured overnight in TSB before washing them with PBS and diluting them to an optical density of 0.05 (A600) in 7.5 mL of chemically defined medium supplemented with 1 % xylose and Cm. Strains were cultured in 30 mL culture tubes for 9 hours. After assaying AcnA, protein concentrations were determined using a copper bicinchoninic acid based colorimetric assay modified for a 96-well plate. Cu inhibition of aconitase was conducted by adding 0–100 μM Cu(II) to cell lysates 20 minutes before assaying activity.
RNA isolation and quantification of mRNA transcripts.
Bacterial strains were cultured for 18 hours in TSB and diluted to a final OD of 0.1 (A600) in 30 mL of fresh TSB or Chelex-treated TSB. The cells were cultured in 250 mL flasks and incubated with shaking till growth reached an OD of 0.8 (A600). One mL was transferred to 10 mL capacity tubes in triplicates and 10 μM MnCl2 final concentration was added to Mn(II) treated group. Both Mn(II) treated and non-treated cultures were then incubated for 30 minutes and harvested. Harvested cells were treated with RNAProtect (Qiagen) for 10 minutes at room temperature, pelleted by centrifugation, and stored at −80 °C. Cell pellets were thawed and washed twice with 0.5 mL of lysis buffer (20 mM RNase-free Sodium acetate, 1 mM EDTA, 0.5 % SDS). The cells were lysed by the addition of 4 μg lysostaphin and incubated for 40 min at 37 °C until confluent lysis was observed. RNA was isolated using TRIzol reagent according to the manufacturer’s instructions. DNA was digested with the Turbo DNA-free kit. The cDNA libraries were constructed using isolated RNA as a template and a High Capacity RNA-to-cDNA kit. An Applied Biosystems StepOnePlus thermocycler and Power SYBR green PCR master mix (Applied Biosystems) were used to quantify DNA abundance. The primer pairs were designed using Primer Express 3.0 software (Applied Biosystems).
Whole cell metal quantification
S. aureus were subcultured in triplicate, from 18 hour grown overnights, to an OD of 0.05 (A600) in 7.5 mL of chelexed TSB in a 30 mL capacity culture tubes. Cultures were incubated with shaking for 8 hrs, before 0–10 μM CuSO4 was added. After 15, 30, or 60 minutes of incubation, samples were transferred into pre-weighted metals free propylene tubes and cells were pelleted by centrifugation using a prechilled table top centrifuge (Eppendorf, Hauppauge, NY). Pellets were washed three times with 10 mL of ice-cold PBS. After decanting the PBS, a fourth spin was conducted to help separate the pellets from any remaining liquid, which was removed using metals free pipettes. Tubes were weighed again to quantify the weight of the cell pellets. All samples were kept at −80 °C or on dry ice until processing.
Cell pellets were acid digested with 2 mL Optima grade nitric acid (ThermoFisher, Waltham, MA) and 500 μL hydrogen peroxide (Sigma, St. Louis, MO) for 24 h at 60 °C. After digestion, 10 mL UltraPure water (Invitrogen, Carlsbad, CA) was added to each sample. Elemental quantification on acid-digested liquid samples was performed using an Agilent 7700 inductively coupled plasma mass spectrometer (Agilent, Santa Clara, CA). The following settings were fixed for the analysis Cell Entrance = −40 V, Cell Exit = −60 V, Plate Bias = −60 V, OctP Bias = −18 V, and collision cell Helium Flow = 4.5 mL min−1. Optimal voltages for Extract 2, Omega Bias, Omega Lens, OctP RF, and Deflect were determined empirically before each sample set was analyzed. Element calibration curves were generated using ARISTAR ICP Standard Mix (VWR). Samples were introduced by peristaltic pump with 0.5 mm internal diameter tubing through a MicroMist borosilicate glass nebulizer (Agilent). Samples were initially up taken at 0.5 rps for 30 seconds followed by 30 seconds at 0.1 rps to stabilize the signal. Samples were analyzed in Spectrum mode at 0.1 rps collecting three points across each peak and performing three replicates of 100 sweeps for each element analyzed. Sampling probe and tubing were rinsed for 20 s at 0.5 rps with 2 % nitric acid between each sample. Data were acquired and analyzed using the Agilent Mass Hunter Workstation Software version A.01.02.
Transposon library construction, mutant selection, and Tn location determination.
The transposon library was constructed in the cop- strain as previously described by Grosser et al. (Bae et al., 2004, Grosser et al., 2018). Briefly, pMG020 (harboring transposase) was freshly transformed into RN4220 and plated and incubated on TSA Tet (10 μg/mL) at 30 °C. Single colonies were selected and grown in TSB Tet (10 μg mL−1) at 30 °C and lysates were generated. The cop- strain carrying pBursa was transduced with pMG020 and selected with TSA-Cm-Tet at 30 °C. Individual colonies were struck on Cm-Tet plates. Individual colonies were suspended in 200 μL sterile water and 15 μL aliquots were spread onto TSA plates containing 10 μg mL−1 Erm and incubated at 43 °C for 24 hours to allow for transposition. In total, colonies from 170 Petri plates containing approximately 3000 colonies each were pooled using TSB 10 μg mL−1 Erm with 25 % glycerol. Aliquots were thoroughly mixed by vortexing and combined into a single pool of transposon mutants. This mutant library was cultured for 2 hours with shaking at 43 °C before 1 mL aliquots were frozen and stored at −80 °C. An aliquot of the cop- Tn library was plated on TSA plates containing 2.5 mM CuSO4 to select for Cu resistant mutants. Strains were further studied after reconstruction and phenotypic verification.
The protocol outlined by Fey et al. was used to determine the locations of transposon insertions (Fey et al., 2013). Briefly, chromosomal DNA was digested using AciI and the resulting DNA fragments were ligated using quick ligase kit and PCR reactions were performed using the Tn buster and Martn-ermR primers. The PCR products were gel purified and sequenced to identify the transposon genome junction sites.
Recombinant DNA and genetic techniques
JMB1100 chromosomal DNA was used as a template for PCR reactions. Escherichia coli PX5α (Protein Express) was used as a cloning host and for plasmid propagation. Plasmids were isolated and transformed into S. aureus strain RN4220 using standard protocol (Grosser & Richardson, 2016) and were conducted using phage 80α (Novick, 1991). All strains were verified by PCR and/or sequencing.
Creation of plasmids and mutant strains
Construction of plasmids for mutant generation or genetic complementation was done using yeast homologous recombination cloning as previously outlined (Mashruwala & Boyd, 2015, Joska et al., 2014). The following primer pairs were used to create the amplicons necessary to make pJB38_ΔcopAZ: Ycc Pjb38 for and YCC CopAZ rev; CopAZ Up for and copAZ up rev; copAZ dwn for and pJB38 copAZ rev. The amplified PCR fragments were combined with EcoRI linearized pJB38 plasmid and transformed into competent Saccharomyces cerevisiae FY2. To create pEPSA_mntABC, pEPSA5_citB_FLAG (Mashruwala et al., 2015) was linearized with NheI and MluI. The mntABC insert was amplified using primers pEPmntABCfor and mntABCYCCrev, combined with vector, and transformed into competent Saccharomyces cerevisiae FY2For all tetR insertion mutants, the tetracycline cassette was amplified from strain JMB1432. The pJB38_rsaE::tetR was used as a backbone to create the pJB38_mnt plasmids. To generate the pJB38_rseA::tetR, pJB38 was linearized with EcoRI and combined with PCR amplicons generated using the following primer pairs: pJB39_yCC forward and Ycc rseA rev; Ycc rseA for and rseA tetR rev; rseA tetR for and tetR rseA rev; tetR rseA for and rseA pJB38 rev
To generate all mnt and rsaC pJB38 vectors, pJB38_ΔrsaE::tetR was linearized using NheI and MluI. The gel purified vector was then combined with amplicons and transformed into competent Saccharomyces cerevisiae FY2. The following primer pairs were used to create the amplicons necessary to make pJB38_ΔmntR::tetR: YccmntRfor and mntRtetRrev; tetRmntRfor and pJB38mntRrev. The following primer pairs were used to create the amplicons necessary to make pJB38_ΔmntA::tetR: YCCmntAfor and mntAtetRrev; mntAtetRfor and tetRmntArev; tetRmntAfor and pJB38mntArev. The following primer pairs were used to create the amplicons necessary to make pJB38_ΔmntB::tetR: YCCmntBfor and mntBtetRrev; mntBtetRfor and tetRmntBrev; tetRmntBfor and pJB38mntArev. The following primer pairs were used to create the amplicons necessary to make pJB38_ΔmntAB::tetR: YCCmntAfor and mntAtetRrev; mntAtetRfor and tetRmntABrev; tetRmntABfor and pJB38mntArev. The following primer pairs were used to create the amplicons necessary to make pJB38_ΔmntABC::tetR: YCCmntABCfor and mntABCtetRrev; mntABCtetRfor and tetRmntABCrev; tetRmntABCfor and pJB38mntABCrev. The following primer pairs were used to create pJB38_ΔrsaC::tetR: YCCrsaCfor and tetupRsaCrev; upRsaCtetfwd and downRsaCtetrev; tetdownRsaCfwd and pJB38rsaCrev. The following primer pairs were used to create the amplicons necessary to make pLL39_mntABC: pLLYCC5 and mntABCYCC3; YccmntABC and mntABCpLL39. The pLL39 vector was linearized with Sall, combined with PCR amplicons, and transformed into Saccharomyces cerevisiae FY2. The pLL39_ mntABC construct was transformed into RN4220 containing pLL2787 and integrated onto the chromosome at the Φ11 attB site as previously described (Roberts et al., 2017). Episome integration was verified using the Scv8 and Scv9 primers. The pML vectors were created by linearizing pML using EcoRI and SalI and was ligated with similarly digested amplicons. The following primer pairs were used to generate the amplicons for the pML100_rsaC and pML100_rsaC antisense clones, respectively: RsaCSalIsense and RsaCEcoRIsense; RsaCSalIantisense and RsaCEcoRIantisense.
Mutant strains were constructed using the pJB38 allelic exchange vectors as described previously (Rosario-Cruz et al., 2015). The cop- strain was created using the ΔcopBL mutant strain (JMB7901) (ΔSAUSA300_0078–0079) and pJB38_ΔcopAZ.
Supplementary Material
Acknowledgements:
The Boyd lab is funded by NIAID award 1R01AI139100-01, NSF award MCB-1750624, and USDA MRF project NE-1028.
Footnotes
Data Availability Statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Achard ME, Stafford SL, Bokil NJ, Chartres J, Bernhardt PV, Schembri MA, Sweet MJ, and McEwan AG (2012) Copper redistribution in murine macrophages in response to Salmonella infection. Biochem J 444: 51–57. [DOI] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I, and Rosato A (2008) Occurrence of copper proteins through the three domains of life: a bioinformatic approach. Journal of proteome research 7: 209–216. [DOI] [PubMed] [Google Scholar]
- Anjem A, and Imlay JA (2012) Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem 287: 15544–15556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JM, Eren E, and Gonzalez-Guerrero M (2007) The structure and function of heavy metal transport P1B-ATPases. Biometals 20: 233–248. [DOI] [PubMed] [Google Scholar]
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, and Missiakas DM (2004) Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101: 12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Glass EM, Schneewind O, and Missiakas D (2008) Generating a collection of insertion mutations in the Staphylococcus aureus genome using bursa aurealis. Methods Mol Biol 416: 103–116. [DOI] [PubMed] [Google Scholar]
- Baker J, Sitthisak S, Sengupta M, Johnson M, Jayaswal RK, and Morrissey JA (2010) Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol 76: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Del Conte R, Markey J, and Ruiz-Duenas FJ (2001) Copper trafficking: the solution structure of Bacillus subtilis CopZ. Biochemistry 40: 15660–15668. [DOI] [PubMed] [Google Scholar]
- Becker KW, and Skaar EP (2014) Metal limitation and toxicity at the interface between host and pathogen. FEMS microbiology reviews 38: 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg SL (2019) The role of metal ions in the virulence and viability of bacterial pathogens. Biochem Soc Trans 47: 77–87. [DOI] [PubMed] [Google Scholar]
- Beinert H, Kennedy MC, and Stout CD (1996) Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chemical reviews 96: 2335–2373. [DOI] [PubMed] [Google Scholar]
- Beveridge SJ, Garrett IR, Whitehouse MW, Vernon-Roberts B, and Brooks PM (1985) Biodistribution of 64Cu in inflamed rats following administration of two anti-inflammatory copper complexes. Agents Actions 17: 104–111. [DOI] [PubMed] [Google Scholar]
- Borremans B, Hobman JL, Provoost A, Brown NL, and van Der Lelie D (2001) Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol 183: 5651–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Fey PD, and Bayles KW (2013) Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79: 2218–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio D, Gallo A, Piccioli M, Novellino E, Ciofi-Baffoni S, and Banci L (2017) [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper. J Am Chem Soc 139: 719–730. [DOI] [PubMed] [Google Scholar]
- Chandrangsu P, and Helmann JD (2016) Intracellular Zn(II) Intoxication Leads to Dysregulation of the PerR Regulon Resulting in Heme Toxicity in Bacillus subtilis. PLoS Genet 12: e1006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Rensing C, and Helmann JD (2017) Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15: 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, and Miethke M (2010) Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol 192: 2512–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP (2001) A new family of high-affinity ABC manganese and zinc permeases. Res Microbiol 152: 231–243. [DOI] [PubMed] [Google Scholar]
- Cotruvo JA Jr., and Stubbe J (2012) Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics : integrated biometal science 4: 1020–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counago RM, Ween MP, Begg SL, Bajaj M, Zuegg J, O’Mara ML, Cooper MA, McEwan AG, Paton JC, Kobe B, and McDevitt CA (2014) Imperfect coordination chemistry facilitates metal ion release in the Psa permease. Nat Chem Biol 10: 35–41. [DOI] [PubMed] [Google Scholar]
- Dintilhac A, Alloing G, Granadel C, and Claverys JP (1997) Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol 25: 727–739. [DOI] [PubMed] [Google Scholar]
- Djoko KY, and McEwan AG (2013) Antimicrobial action of copper is amplified via inhibition of heme biosynthesis. ACS Chem Biol 8: 2217–2223. [DOI] [PubMed] [Google Scholar]
- Dollwet HH, and Sorenson JR, (1985) Historic uses of copper compounds in medicine, p. 80–87. Humana Press Inc., Arkansas. [Google Scholar]
- Ekici S, Yang H, Koch HG, and Daldal F (2012) Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, and Bayles KW (2013) A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4: e00537–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C KG, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, and Zyskind JW (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43: 1387–1400. [DOI] [PubMed] [Google Scholar]
- Gaballa A, and Helmann JD (2003) Bacillus subtilis CPx-type ATPases: characterization of Cd, Zn, Co and Cu efflux systems. Biometals 16: 497–505. [DOI] [PubMed] [Google Scholar]
- Gardner PR, and Fridovich I (1991) Superoxide sensitivity of the Escherichia coli aconitase. Journal of Biological Chemistry 266: 19328–19333. [PubMed] [Google Scholar]
- Glasfeld A, Guedon E, Helmann JD, and Brennan RG (2003) Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat Struct Biol 10: 652–657. [DOI] [PubMed] [Google Scholar]
- Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, and Rensing C (2005) The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol 187: 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Rensing C, and Solioz M (2011) Metallic copper as an antimicrobial surface. Appl Environ Microbiol 77: 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribenko A, Mosyak L, Ghosh S, Parris K, Svenson K, Moran J, Chu L, Li S, Liu T, Woods VL Jr., Jansen KU, Green BA, Anderson AS, and Matsuka YV (2013) Three-dimensional structure and biophysical characterization of Staphylococcus aureus cell surface antigen-manganese transporter MntC. J Mol Biol 425: 3429–3445. [DOI] [PubMed] [Google Scholar]
- Grosser MR, Paluscio E, Thurlow LR, Dillon MM, Cooper VS, Kawula TH, and Richardson AR (2018) Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLoS Pathog 14: e1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser MR, and Richardson AR (2016) Method for Preparation and Electroporation of S. aureus and S. epidermidis. Methods Mol Biol 1373: 51–57. [DOI] [PubMed] [Google Scholar]
- Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, and Giedroc DP (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem 286: 13522–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, and Imlay JA (2013) Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol 89: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Arguello JM, and Helmann JD (2015) PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol 98: 787–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Matsui K, Lo JF, and Silver S (1999) Molecular basis for resistance to silver cations in Salmonella. Nature medicine 5: 183–188. [DOI] [PubMed] [Google Scholar]
- Handke LD, Hawkins JC, Miller AA, Jansen KU, and Anderson AS (2013) Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PLoS One 8: e77874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Chen S, and Wilson DB (1999) Cloning, expression, and characterization of cadmium and manganese uptake genes from Lactobacillus plantarum. Appl Environ Microbiol 65: 4746–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson V, and Petris MJ (2012) Copper homeostasis at the host-pathogen interface. J Biol Chem 287: 13549–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Ingham E, and Foster SJ (2001) In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, and Foster SJ (2002) MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44: 1269–1286. [DOI] [PubMed] [Google Scholar]
- Huang X, Shin JH, Pinochet-Barros A, Su TT, and Helmann JD (2017) Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol Microbiol 103: 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA (2014) The mismetallation of enzymes during oxidative stress. J Biol Chem 289: 28121–28128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroslawiecka A, and Piotrowska-Seget Z (2014) Lead resistance in micro-organisms. Microbiology 160: 12–25. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Kehl-Fie TE, and Rosch JW (2015) Copper intoxication inhibits aerobic nucleotide synthesis in Streptococcus pneumoniae. Metallomics : integrated biometal science 7: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JW, Briles DE, Myers LE, and Hollingshead SK (2006) Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect Immun 74: 1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joska TM, Mashruwala A, Boyd JM, and Belden WJ (2014) A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J Microbiol Methods 100: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, and Skaar EP (2013) MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81: 3395–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Senadheera DB, Levesque CM, and Cvitkovitch DG (2012) TcyR regulates L-cystine uptake via the TcyABC transporter in Streptococcus mutans. FEMS Microbiol Lett 328: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, and Novick RP (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305: 709–712. [DOI] [PubMed] [Google Scholar]
- Laddaga RA, and Silver S (1985) Cadmium uptake in Escherichia coli K-12. J Bacteriol 162: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomersky E, Khan A, Shanbhag V, Cavet JS, Chan J, Weisman GA, and Petris MJ (2017) Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Infect Immun 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Baude J, Wu Z, Tomasini A, Chicher J, Marzi S, Vandenesch F, Romby P, Caldelari I, and Moreau K (2019) RsaC sRNA modulates the oxidative stress response of Staphylococcus aureus during manganese starvation. Nucleic Acids Res 47: 9871–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei MG, Cue D, Roux CM, Dunman PM, and Lee CY (2011) Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol 193: 5231–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Yang XY, Guo Z, Zhang J, Cao K, Han J, Zhang G, Liu L, Sun X, and He QY (2014) Varied metal-binding properties of lipoprotein PsaA in Streptococcus pneumoniae. J Biol Inorg Chem 19: 829–838. [DOI] [PubMed] [Google Scholar]
- Lim KH, Jones CE, vanden Hoven RN, Edwards JL, Falsetta ML, Apicella MA, Jennings MP, and McEwan AG (2008) Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect Immun 76: 3569–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisher JP, Higgins KA, Maroney MJ, and Giedroc DP (2013) Physical characterization of the manganese-sensing pneumococcal surface antigen repressor from Streptococcus pneumoniae. Biochemistry 52: 7689–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong TT, and Lee CY (2007) Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods 70: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Cowart DM, Scott RA, and Giedroc DP (2009) Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry 48: 3325–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, and Imlay JA (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106: 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malachowa N, and DeLeo FR (2010) Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67: 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JE, and Imlay JA (2011) The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 80: 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Bhatt S, Poudel S, Boyd ES, and Boyd JM (2016a) The DUF59 Containing Protein SufT Is Involved in the Maturation of Iron-Sulfur (FeS) Proteins during Conditions of High FeS Cofactor Demand in Staphylococcus aureus. PLoS Genet 12: e1006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, and Boyd JM (2015) De Novo Assembly of Plasmids Using Yeast Recombinational Cloning. Methods Mol Biol 1373: 33–41. [DOI] [PubMed] [Google Scholar]
- Mashruwala AA, and Boyd JM (2017) The Staphylococcus aureus SrrAB Regulatory System Modulates Hydrogen Peroxide Resistance Factors, Which Imparts Protection to Aconitase during Aerobic Growth. PLoS One 12: e0170283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, and Boyd JM Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol 95: 383–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, and Boyd JM (2015) Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol 95: 383–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Roberts CA, Bhatt S, May KL, Carroll RK, Shaw LN, and Boyd JM (2016b) Staphylococcus aureus SufT: An essential iron-sulfur cluster assembly factor in cells experiencing a high-demand for lipoic acid. Mol Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, and Paton JC (2004) Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol Microbiol 53: 889–901. [DOI] [PubMed] [Google Scholar]
- McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, and Paton JC (2011) A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog 7: e1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Gaballa A, Hui M, Ye RW, and Helmann JD (2005) Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol 57: 27–40. [DOI] [PubMed] [Google Scholar]
- Ngamchuea K, Batchelor-McAuley C, and Compton RG (2016) The Copper(II)-Catalyzed Oxidation of Glutathione. Chemistry 22: 15937–15944. [DOI] [PubMed] [Google Scholar]
- Norambuena J, Hanson TE, Barkay T, and Boyd JM (2019) Superoxide Dismutase and Pseudocatalase Increase Tolerance to Hg(II) in Thermus thermophilus HB27 by Maintaining the Reduced Bacillithiol Pool. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena J, Wang Y, Hanson T, Boyd JM, and Barkay T (2017) Low molecular weight thiols and thioredoxins are important players in Hg(II) resistance in Thermus thermophilus HB27. Appl Environ Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R, Braun JS, Charpentier E, and Tuomanen E (1998) Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol 29: 1285–1296. [DOI] [PubMed] [Google Scholar]
- Novick RP (1991) Genetic systems in staphylococci. Methods Enzymol 204: 587–636. [DOI] [PubMed] [Google Scholar]
- Palmer LD, and Skaar EP (2016) Transition Metals and Virulence in Bacteria. Annu Rev Genet 50: 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YY, Schwartz J, Bloomberg S, Boyd JM, Horswill AR, and Nauseef WM (2013) Methionine Sulfoxide Reductases Protect against Oxidative Stress in Staphylococcus aureus Encountering Exogenous Oxidants and Human Neutrophils. J Innate Immun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace KM, and Maguire ME (2006) Manganese transport and the role of manganese in virulence. Annual review of microbiology 60: 187–209. [DOI] [PubMed] [Google Scholar]
- Perry RD, and Silver S (1982) Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J Bacteriol 150: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves J, Thomas J, Riboldi GP, Zapotoczna M, Tarrant E, Andrew PW, Londono A, Planet PJ, Geoghegan JA, Waldron KJ, and Morrissey JA (2018) A horizontally gene transferred copper resistance locus confers hyper-resistance to antibacterial copper toxicity and enables survival of community acquired methicillin resistant Staphylococcus aureus USA300 in macrophages. Environmental microbiology 20: 1576–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, and Helmann JD (2000) Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35: 1454–1468. [DOI] [PubMed] [Google Scholar]
- Quintana J, Novoa-Aponte L, and Arguello JM (2017) Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J Biol Chem 292: 15691–15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch I, Yanku M, Yeheskel A, Cohen G, Borovok I, and Aharonowitz Y (2010) Staphylococcus aureus NrdH redoxin is a reductant of the class Ib ribonucleotide reductase. J Bacteriol 192: 4963–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo A, Corazza A, di Paolo ML, Rossetto M, Ugolini R, and Scarpa M (2004) Interaction of copper with cysteine: stability of cuprous complexes and catalytic role of cupric ions in anaerobic thiol oxidation. Journal of inorganic biochemistry 98: 1495–1501. [DOI] [PubMed] [Google Scholar]
- Roberts CA, Al-Tameemi HM, Mashruwala AA, Rosario-Cruz Z, Chauhan U, Sause WE, Torres VJ, Belden WJ, and Boyd JM (2017) The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect Immun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario-Cruz Z, Chahal HK, Mike LA, Skaar EP, and Boyd JM (2015) Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol Microbiol 98: 218–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario-Cruz Z, Eletsky A, Daigham NS, Al-Tameemi H, Swapna GVT, Kahn PC, Szyperski T, Montelione GT, and Boyd JM (2019) The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane-associated copper-binding protein. J Biol Chem 294: 4027–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri M, Leveille S, and Dozois CM (2006) A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152: 745–758. [DOI] [PubMed] [Google Scholar]
- Scarpa M, Momo F, Viglino P, and Rigo A (1996) Activated oxygen species in the oxidation of glutathione A kinetic study. Biophysical Chemistry 60: 53–61. [Google Scholar]
- Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, and Morrissey JA (2011) The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81: 1255–1270. [DOI] [PubMed] [Google Scholar]
- Singleton C, Hearnshaw S, Zhou L, Le Brun NE, and Hemmings AM (2009) Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem J 424: 347–356. [DOI] [PubMed] [Google Scholar]
- Sitthisak S, Knutsson L, Webb JW, and Jayaswal RK (2007) Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology 153: 4274–4283. [DOI] [PubMed] [Google Scholar]
- Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, and Musser JM (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70: 6373–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Cheng Z, Pang Y, Landry AP, Li J, Lu J, and Ding H (2014) Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol Microbiol 93: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Yang J, Li T, Zhao J, Sun S, Li X, Lin C, Li J, Zhou H, Lyu J, and Ding H (2017) Anaerobic Copper Toxicity and Iron-Sulfur Cluster Biogenesis in Escherichia coli. Appl Environ Microbiol 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant E, G PR, McIlvin MR, Stevenson J, Barwinska-Sendra A, Stewart LJ, Saito MA, and Waldron KJ (2019) Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism. Metallomics : integrated biometal science 11: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, and Dunman PM (2006) Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 44: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Rich PR, Rondet SA, and Robinson NJ (2001) Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem 276: 19999–20004. [DOI] [PubMed] [Google Scholar]
- Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, and Fowler VG Jr. (2019) Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17: 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynecka Z, Gos Z, and Zajac J (1981) Reduced cadmium transport determined by a resistance plasmid in Staphylococcus aureus. J Bacteriol 147: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE 3rd, Honer Zu Bentrup K, Russell DG, and Bermudez LE (2005) Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Immunol 174: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Waldron KJ, and Robinson NJ (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 7: 25–35. [DOI] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, and Petris MJ (2009) A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284: 33949–33956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S (2015) Structure and mechanism of ABC transporters. F1000Prime Rep 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu FF, and Imlay JA (2012) Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol 78: 3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapotoczna M, Riboldi GP, Moustafa AM, Dickson E, Narechania A, Morrissey JA, Planet PJ, Holden MTG, Waldron KJ, and Geoghegan JA (2018) Mobile-Genetic-Element-Encoded Hypertolerance to Copper Protects Staphylococcus aureus from Killing by Host Phagocytes. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


