Abstract
Mycobacterium tuberculosis (Mtb) has complex and dynamic interactions with the human host, and subpopulations of Mtb that emerge during infection can influence disease outcomes. This study implicates zinc ion (Zn2+) availability as a likely driver of bacterial phenotypic heterogeneity in vivo. Zn2+ sequestration is part of “nutritional immunity”, where the immune system limits micronutrients to control pathogen growth, but this defense mechanism seems to be ineffective in controlling Mtb infection. Nonetheless, Zn2+-limitation is an environmental cue sensed by Mtb, as calprotectin triggers the zinc uptake regulator (Zur) regulon response in vitro and co-localizes with Zn2+-limited Mtb in vivo. Prolonged Zn2+ limitation leads to numerous physiological changes in vitro, including differential expression of certain antigens, alterations in lipid metabolism and distinct cell surface morphology. Furthermore, Mtb enduring limited Zn2+ employ defensive measures to fight oxidative stress, by increasing expression of proteins involved in DNA repair and antioxidant activity, including well described virulence factors KatG and AhpC, along with altered utilization of redox cofactors. Here, we propose a model in which prolonged Zn2+ limitation defines a population of Mtb with anticipatory adaptations against impending immune attack, based on the evidence that Zn2+-limited Mtb are more resistant to oxidative stress and exhibit increased survival and induce more severe pulmonary granulomas in mice. Considering that extracellular Mtb may transit through the Zn2+-limited caseum before infecting naïve immune cells or upon host-to-host transmission, the resulting phenotypic heterogeneity driven by varied Zn2+ availability likely plays a key role during early interactions with host cells.
Author summary
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), has plagued humanity for millennia and remains the world’s deadliest bacterium today. Bacterial heterogeneity is one of the most important characteristics of Mtb that complicates TB treatment. Access to zinc ion (Zn2+) may influence bacterial heterogeneity, considering microenvironments developed during TB create a perpetual cycle exposing Mtb to high and low concentrations of Zn2+. Here we show that Zn2+ limitation drives changes in gene expression patterns of well described virulence factors in Mtb, and Zn2+-limited Mtb show increased resistance to oxidative stress and increased replication in vivo. Our results suggest that host-pathogen interactions are influenced by pre-exposure of Mtb to Zn2+ and mycobacteria that transit through a Zn2+-depleted microenvironment are primed to withstand impending oxidative stress upon subsequent contact with immune cells in the same, or a naïve host. Considering that the standard mycobacterial media recapitulates a Zn2+-replete environment, the Zn2+-dependent phenotype of the pathogen may confound our fundamental understanding of initial interactions between Mtb and immune cells.
Introduction
The success of Mycobacterium tuberculosis (Mtb) as a human pathogen is enabled by a genetic arsenal that allows it to withstand a myriad of immune defenses, survive in diverse host environments, and establish persistent infection [1]. Disease outcome is influenced by subpopulations of Mtb arising within heterogeneous microenvironments in vivo, but specific cues from the host leading to their development are mostly unknown [2–5].
One cue that triggers physiological adaptation in many pathogens is the limitation of essential micronutrients, e.g., free zinc ion (Zn2+), a strategy known as ‘nutritional immunity’ [6]. Indeed, drastic changes in Zn2+ concentration [Zn2+] are experienced by Mtb throughout infection. Zn2+ rapidly accumulates in phagosomes [7], but Mtb is protected from zinc poisoning through upregulation of heavy metal efflux P1-type ATPase as it colonizes the intracellular niche [8]. Infected macrophages release pro-inflammatory cytokines which recruit immune cells to the site of infection forming the hallmark pathology of TB, the granuloma [1,9]. As TB disease progresses, neutrophils infiltrate granulomas, promoting necrosis of unresolved Mtb-infected immune cells [3,10]. Necrotic cells release their contents, including Mtb, into the extracellular milieu, a microenvironment rich in neutrophil-derived Zn2+ and Mn2+-binding protein calprotectin (CP), part of the ‘nutritional immunity’ host response [10]. Therefore, throughout infection Mtb may transit between high [Zn2+] inside macrophages and low [Zn2+] in CP-rich caseum. The spectrum of diverse immunopathologies of granulomas and necrotic cavities give rise to distinct subpopulations of Mtb that exist simultaneously within the host [3,5], and [Zn2+] may be a contributing factor in their development. Further, [Zn2+]-dependent physiological changes may affect interactions with the immune system, thus influencing disease progression and response to treatment.
As with many other bacteria, Mtb has a Zn2+-responsive transcriptional repressor–zinc uptake regulator (Zur) that controls 21 genes with upregulated expression during Zn2+-limiting conditions [11]. Zn2+ limitation is likely a cue sensed by Mtb during infection, considering upregulation of Zur-regulated genes (e.g., genes in the altRP operon [12]) detected in Mtb from human sputum [13,14]. However, the Zur regulon may be just one aspect of the global response to Zn2+ availability, as suggested by transcriptomic studies in other Zn2+-limited bacteria [15]. Since [Zn2+] is tied to specific microenvironments in vivo, it may cue Mtb to trigger adaptive responses beyond maintaining zinc homeostasis. We hypothesize that Mtb has a dynamic global response enabling endurance through periods of prolonged Zn2+ limitation and reason that Zn2+-limited Mtb, such as those found in sputum or exposed to CP-rich regions in the extracellular microenvironment, are phenotypically distinct from Mtb residing in a Zn2+-replete niche.
In this study, we show that Mtb enduring Zn2+-limited and Zn2+-replete environments have distinct signatures, suggesting that [Zn2+] may likewise delineate subpopulations of Mtb in vivo. We use biochemical and multi-omics approaches to describe the effect of prolonged Zn2+ limitation at a global scale in Mtb grown in vitro. The analysis of Zn2+-limited Mtb revealed a response that goes beyond the Zur regulon, including activation of the oxidative stress response, altered utilization of reducing cofactors, changes in the lipidome and a distinct cell surface morphology. In addition, Zn2+-limited Mtb are more resistant to oxidative stress and more sensitive to the prodrug isoniazid compared to Zn2+-replete Mtb. Finally, in an aerosol mouse model of infection, Zn2+-limited inoculum exhibited greater bacterial burden and pulmonary granulomas than Zn2+-replete inoculum. Together these findings define a novel adaptive mechanism employed by Mtb during Zn2+ limitation that triggers formation of a distinct population that may play a role in TB pathogenesis.
Results
Mtb responds to Zn2+ limitation in vivo and in vitro
Accumulation of CP in the caseum of necrotic granulomas is a marked feature of human pulmonary tuberculosis [16]. Cavitation of necrotic granulomas provides the route for Mtb transmission into the airways and out of the host via sputum [3]. Accordingly, we detected CP in sputum from patients with active TB (S1 Table), suggesting that extracellular Mtb in the caseum and sputum are in contact with CP. CP has been shown to induce altRP expression in vitro [12] and the increased altRP expression detected in Mtb from human sputum suggests this subpopulation is experiencing Zn2+-limitation due to Zur-regulation of the altRP operon [13,14]. However, other environmental stressors may also upregulate altRP expression [17], so to validate the notion that increased altRP expression is in response to Zn2+-limiting conditions sensed by Mtb in contact with Zn2+-binding CP, we further investigated expression of all genes in the Mtb Zur regulon in response to CP in vitro. As expected, we saw significant upregulation of the Zur regulon (except Rv2060 for which we did not detect gene expression), including the altRP operon (Rv2055c-Rv2058c), which was one of the most highly expressed features in response to CP (Fig 1A). In agreement, recent high throughput sequencing of Mtb transcriptomes from human sputum reveal upregulation of Zur regulon just as we have shown for Mtb exposed to CP [18]. The clear evidence that CP induces expression of Zur-regulated genes demonstrates that Mtb in contact with CP experience Zn2+ limitation, and the observation that Mtb from sputum also have increased expression of Zur regulon suggests this subpopulation of bacteria is likely enduring Zn2+-limitation.
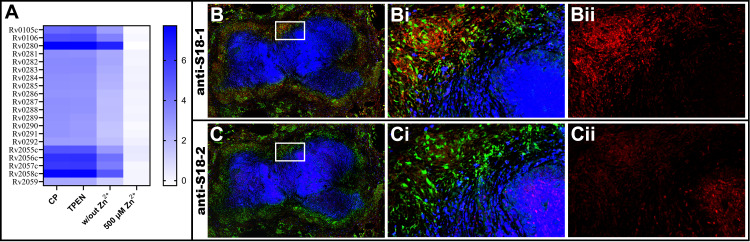
Fig 1. CP upregulates the Zur regulon in vitro and detection of Zur-regulated protein expression in regions containing CP in vivo.
(A) Heatmap showing log2FC values for genes in the Zur regulon after growth in media without added Zn2+ followed by short-term exposure to CP, TPEN, 6 μM Zn2+ and 500 μM Zn2+. In each condition the change in expression (log2FC) is relative to the 6 μM Zn2+ condition. Log2FC expression values are calculated from the normalized expression counts of biological replicates (n = 3). Confocal microscopy images of serial sections of lung granulomas at 20X magnification stained for Mtb ribosomal proteins with polyclonal anti-S18-1 (B) and anti-S18-2 (C) antibodies (red), CD68+ macrophages (green) and CP (blue). The field in B and C bound by the white box is magnified in panels Bi-ii and Ci-ii and corresponds to a region containing both CD68+ epithelioid macrophages and CP-stained neutrophils and caseum, as well as distribution of Mtb S18-1 (Bi) and S18-2 (Ci). Single channel images from staining with antibodies specific to Mtb ribosomal proteins only is shown for S18-1 (Bii) and S18-2 (Cii).
Beyond the Zn2+-limited signature of Mtb from sputum and the observed increase in altRP expression from Mtb in artificial granulomas in mice [19], there is sparse information regarding the existence or localization of Zn2+-limited Mtb in vivo. Because the caseum is a microenvironment rich in CP [10], we predict that extracellular Mtb in necrotic granulomas (caseum) have limited access to Zn2+ and will be delineated from Zn2+-replete bacteria by expression of genes in the Zur regulon. Therefore, we hypothesized that S18-2 protein, one of the products of the Zur-regulated altRP operon, will be observed in the Zn2+-limited, CP-rich cores of necrotic granulomas. To test this hypothesis, we stained serial sections of necrotic granulomas from experimentally infected cynomolgus macaques [10] with polyclonal antibodies targeting either S18-1 or S18-2 ribosomal proteins in combination with antibodies against CD68 and CP to identify macrophages and CP-rich necrotic regions, respectively (Fig 1B and 1C).
The antibody against S18-1 protein is useful as marker for the presence of Mtb antigens, but does not delineate subpopulations of Mtb, considering S18-1 protein is detected in Mtb cultures with and without S18-2 protein expression [12] and S18-1 antibody has non-specific binding to numerous other Mtb antigens (S1 Fig). The signal from S18-1 antibody associated with the macrophage rich cellular ring surrounding the necrotic core of the granuloma, consistent with the description of this microenvironment as one that contains intracellular bacteria [10] (Fig 1B). The signal from S18-2 antibody, which is specific for S18-2 protein and is selectively detected in Zn2+-limited Mtb cultures [12] (S1 Fig), was associated with the signal from CP found within the acellular core (i.e., caseum) of necrotic granulomas, a microenvironment that contains extracellular bacteria [10] (Fig 1C). Although we could not co-stain for individual bacteria because of technical limitations with our staining protocol, extracellular bacilli are detected in the caseum of necrotic macaque granulomas [10] and intact acid-fast bacteria are observed in association with necrotic cells and debris in the caseum of patients with post-primary TB [20], suggesting that the signal detected from S18-2 is from extracellular bacteria in the CP-rich caseum.
Mtb growth during prolonged Zn2+ limitation in vitro
We seek to understand how Mtb responds to Zn2+ limited conditions, such as what may occur during the transition from intracellular to CP-rich extracellular microenvironments. Expression of Zur-regulated genes delineates a subpopulation of Mtb in the CP-rich, Zn2+ limited microenvironment in vivo, so we reasoned that, to analyze the Zn2+-dependent phenotype of Mtb, expression of Zur-regulated genes may be used as a proxy to establish Zn2+-limiting conditions in vitro. Beyond the small-scale study of Zur expression in Mtb exposed to CP, it was not feasible to use recombinant CP to study the response to prolonged Zn2+-limitation, so we also followed expression of Zur regulon in Mtb exposed to N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine (TPEN), a synthetic Zn chelator that has a precedence for being used to study cellular responses to Zn2+ limitation. Recently, it was shown that use of TPEN leads to Zn2+-starved mycobacteria that progress from ribosome remodeling (i.e., expression of AltRPs and their incorporation into ribosomes) to hibernation and growth arrest [21]. Furthermore, although considered a Zn2+-specific chelator, TPEN has been shown to influence the transcriptional profile of bacteria beyond changes relevant to Zn2+ alone [22]. Indeed, we observed induction of the Zur regulon in Mtb exposed to TPEN (Fig 1A), however we also observed a significant increase in the expression of genes involved in mycobactin synthesis (i.e., mbt operon) in TPEN vs. CP conditions (S2 Table), suggesting that use of TPEN intertwines the response to Zn2+ and iron limitation in Mtb. Growth of Mtb in chemically defined Sauton’s medium without addition of Zn2+ also upregulated the Zur regulon compared to Mtb grown in presence of the standard 6 μM Zn2+ supplementation (Fig 1A). Adding Zn2+ to the levels found in phagosomes [7], i.e., 500 μM Zn2+, did suppress the Zur regulon, but not significantly more than when Mtb was grown with 6 μM Zn2+ (Fig 1A). Therefore, we decided to use 6 μM Zn2+ supplemented Sauton’s medium as Zn2+ replete medium (ZRM) and Sauton’s medium without added Zn2+ as Zn2+ limited medium (ZLM) for all our subsequent experiments. This Zn2+-limiting condition (i.e., bacterial growth in ZLM) induces mycobacterial ribosome remodeling, but maintains translational activity of alternative ribosomes [23] and was the ideal condition to study the response to prolonged Zn2+-limitation in Mtb. To detect the onset of Zn2+-limitation in ZLM, we created a transcriptional reporter strain with the Zur-regulated altRP operon promoter fused to mCherry fluorescent protein (PaltRP-mCherry), as fluorescence from this strain has previously been shown to correlate with altRP gene and protein expression in Mtb [12].
We monitored growth of virulent Mtb (strain H37Rv) and the attenuated double auxotroph (strain mc2 6206, a safe and suitable model organism for Mtb research [24]) in parallel with fluorescence from the PaltRP-mCherry reporter in both Mtb strains in ZLM and ZRM. Fluorescence was first detected in Mtb (strains H37Rv and mc2 6206) after 4 days of growth in ZLM, indicating the onset of Zn2+ limitation in ZLM (S2A and S2B Fig). Thus, we determined that the response to persistent Zn2+ limitation in Mtb can be studied using growth in ZLM, and while we do not know the specific [Zn2+] at which Zur-regulated altRP expression is induced, omitting use of Zn2+-chelation enables us to conclude it is below the concentration of [Zn2+] in ZLM which was determined by ICP-MS analysis to be 115 ± 37 nM (average and standard deviation from three independent media preparations). To investigate adaptations employed by Mtb during prolonged Zn2+ limitation, cultures grown in ZLM were compared to cultures grown in ZRM. We recently described a unique morphogenesis of the non-pathogenic mycobacteria Mycobacterium smegmatis (Msm) marked by cell elongation upon Zn2+ limitation [25], however, in contrast to Msm, we did not observe any growth-related impairments due to Zn2+ limitation (S2C and S2D Fig), nor did we observe obvious changes in cell length when Mtb was grown in ZLM (S2E and S2F Fig). Based on observations of PaltRP-mCherry fluorescence in ZLM (S2A and S2B Fig), while potentially maximizing the effect of metabolic and structural remodeling due to Zn2+ limitation to occur at all levels of cellular structures, we chose to further investigate the expression profiles and physiology of Mtb in late log phase (i.e., after 10 days of growth) in ZLM and ZRM.
Global changes in the transcriptome of Zn2+-limited Mtb
To elucidate the global response employed upon prolonged Zn2+ limitation, we analyzed the transcriptomes of Zn2+-replete and Zn2+-limited Mtb H37Rv after 10 days of growth in ZRM and ZLM, respectively, using RNA sequencing (RNAseq). Analyzing the transcriptomes from ZLM and ZRM with RNAseq yielded identification of over 99% of the coding and non-coding RNAs in the Mtb genome, leading to identification of 379 upregulated and 346 downregulated genes in Zn2+ limited condition, i.e., ZLM vs. ZRM (Fig 2A). When visualized on a multidimensional scaling plot, we found that [Zn2+] was the first dimension describing over 90% of variation in the data, with the second dimension representing intra-sample (ZRM n = 3, ZLM n = 3) variation having a much smaller effect (S3A Fig).
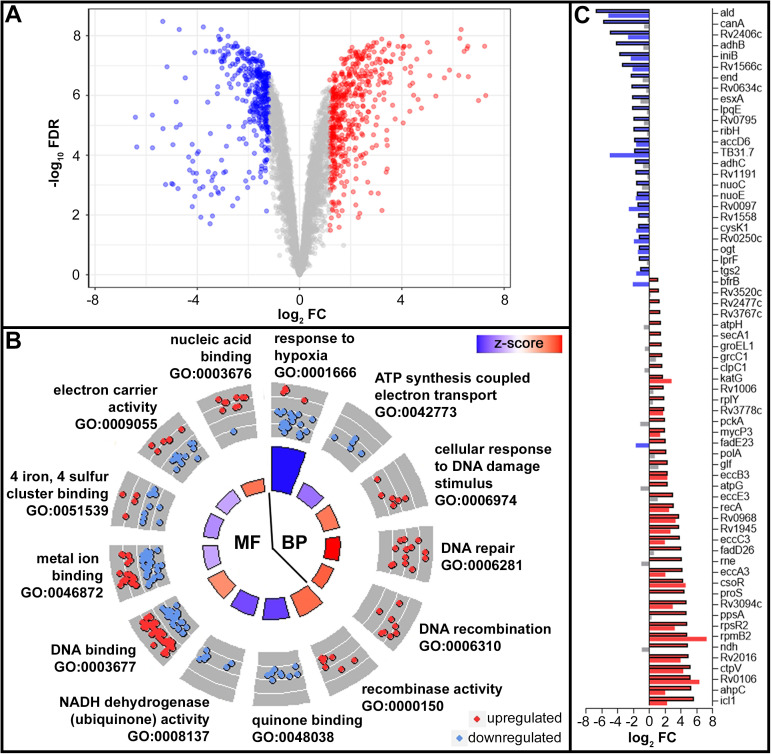
Fig 2. Adaptive response to Zn2+ limitation in Mtb.
(A) Volcano plot of DE genes (absFC >2, FDR <0.05) in Mtb H37Rv in ZLM vs. ZRM. Genes colored in red, blue, and grey are significantly upregulated, downregulated or not significantly regulated in ZLM vs. ZRM, respectively. (B) Circle plot showing GO terms of biological processes (BP) and molecular functions (MF) significantly enriched in the list of DE genes in ZLM. Each pie slice of the circle is labeled with the enriched GO term, the outer circle shows a scatter plot of DE genes with the given GO annotation and their log2FC values where red circles display upregulated and blue circles downregulated genes in ZLM vs. ZRM. The color of bars in the inner circle indicates whether the given biological process is more likely to be increased (red) or decreased (blue) in the dataset and the height represents the -log10(FDR) for the enriched term (larger bars have smaller FDR). (C) Horizontal bar graph showing the overlap between transcript and protein expression for the 65 DE proteins identified in ZLM (absFC >1.5, FDR <0.05). For each gene, the bar on top (black outline) represents the log2FC value of the protein and the bar below it (no outline) represents the log2FC value of its transcript. Red bars show upregulation and blue bars show downregulation of the protein or transcript in ZLM vs. ZRM while grey bars represent no significant differential expression for transcripts. RNA and proteins for DE analysis were isolated from the same cultures of day 10 cells from ZRM (n = 3) and ZLM (n = 3).
While we observed significant upregulation of most Zur-regulated genes (S3 Table), there were numerous other differentially expressed (DE) genes greatly surpassing the induction level of genes in the Zur regulon (S4 Table). Filtering DE genes by those with the greatest level of upregulation in ZLM yielded a surprising observation that most tRNAs are highly upregulated, with tRNA-leuV being the most highly expressed feature in ZLM (S4 Table). In fact, 33/45 tRNAs in the Mtb genome are identified as DE genes upregulated in ZLM whereas only one tRNA was downregulated (S4 Fig).
To better understand the functionality of genes that are differentially regulated during prolonged Zn2+ limitation in Mtb, we conducted an enrichment analysis for gene ontology (GO) terms on the list of DE genes in ZLM. The enrichment analysis showed 65% of the DE genes had at least one functionally enriched GO term which yielded 15 functional categories that were significantly enriched in the list of DE genes (S5A Fig). Fig 2B shows a circle plot of genes that are significantly enriched in the GO analysis. The circle plot shows specific downregulation of genes involved in the hypoxic response, electron transport and quinone binding as well as metal ion binding function during Zn2+ limitation. On the other hand, there was a strong upregulation of genes involved in DNA repair and synthesis, recombination and the general response to DNA damage stimulus during Zn2+ limitation. Similar results are apparent when assigning DE genes to pathways as defined by KEGG (S6 Fig and S6 and S7 Tables). These results suggest that energy production, electron flow and oxygen consumption are reduced while processes involved with DNA repair are increased in Mtb during prolonged periods of [Zn2+] limitation.
Changes in the proteome mirrors transcriptome during Zn2+ limitation in Mtb
The abundance of an mRNA molecule and the protein it codes for are not always correlated; post-transcriptional or post-translational processes affect protein abundance [26,27]. Moreover, the shift in translational machinery from primary to alternative ribosomes upon Zn2+ limitation in Mtb creates a differential population of ribosomes, and this switch could alter ribosomal specificity [12]. Indeed, selective translation from alternative ribosomes in Msm has been recently described [28]. If alternative ribosomes in Mtb also exhibit selective translation, these changes may be undetectable at the transcript level but ultimately affect the proteome. Therefore, we also probed the proteomes of Mtb in ZLM and ZRM to investigate proteins that may be differentially correlated with transcript abundance and to define the core response to Zn2+ limitation conserved between the transcriptome and proteome.
We isolated proteins from the same cultures of Mtb H37Rv used for RNAseq and coordinately analyzed the proteomes of cultures from ZLM and ZRM with label-free quantitation using mass spectrometry. As is typical with shotgun proteomics, coverage of the genome was much lower than with RNAseq; only about half of theoretical proteins were identified using spectral counting (SpC) and removing proteins with low expression resulted in 738 proteins used for differential expression analysis, among which 65 were identified as DE proteins with 25 being upregulated and 40 being downregulated in ZLM (Fig 2C and S5 Table). When visualized on a multidimensional scaling plot, we found that [Zn2+] explains the largest portion of variation in the data (nearly 80%), with intra-sample variation having a much smaller effect (S3B Fig). GO enrichment of DE proteins yielded six significantly enriched categories with “cell wall” being the only category present in the GO analysis of both genes and proteins (S5 Fig). However, there was considerable overlap between DE genes and DE proteins, with half of the DE proteins (33/65) correlating with a DE gene and in only two of these cases (FadE23, BfrB) was the regulation level discordant (Fig 2C). We reasoned that DE genes and DE proteins that follow the same level of regulation (n = 31) represent a robust biological indication of the response to Zn2+ limitation and this list is given in Table 1.
Table 1. Conserved response to Zn2+ limitation in Mtb as defined by the concordance of DE genes and proteins in ZLM vs. ZRM.
| H37Rv Locus | Gene name | Product (Protein name) | logFC RNA | logFC Protein |
|---|---|---|---|---|
| Rv2058c | rpmB2 | 50S ribosomal protein L28 (L28-2) | 7.24 | 4.83 |
| Rv0106 | Rv0106 | Conserved hypothetical protein | 6.32 | 5.26 |
| Rv0967 | csoR | Copper-sensitive operon repressor (CsoR) | 4.58 | 4.32 |
| Rv0969 | ctpV | Probable metal cation transporter P-type ATPase (CtpV) | 4.30 | 5.20 |
| Rv2016 | Rv2016 | Hypothetical protein | 3.95 | 4.96 |
| Rv0968 | Rv0968 | Conserved protein | 3.31 | 3.81 |
| Rv2055c | rpsR2 | 30S ribosomal protein S18 (S18-2) | 3.21 | 4.80 |
| Rv3094c | Rv3094c | Conserved hypothetical protein | 2.95 | 4.70 |
| Rv1908c | katG | Catalase-peroxidase-peroxynitritase (KatG) | 2.81 | 1.73 |
| Rv1945 | Rv1945 | Conserved hypothetical protein | 2.68 | 3.81 |
| Rv2737c | recA | Recombinase A protein (RecA) | 2.51 | 3.07 |
| Rv0283 | eccB3 | ESX-3 type VII secretion system protein (EccB3) | 2.28 | 2.32 |
| Rv0467 | icl1 | Isocitrate lyase (Icl) | 2.24 | 5.67 |
| Rv0282 | eccA3 | ESX-3 type VII secretion system protein (EccA3) | 2.02 | 4.18 |
| Rv2428 | ahpC | Alkyl hydroperoxide reductase C (AhpC) | 2.00 | 5.31 |
| Rv0284 | eccC3 | ESX-3 type VII secretion system protein (EccC3) | 1.96 | 3.82 |
| Rv3778c | Rv3778c | Possible aminotransferase | 1.75 | 1.94 |
| Rv0291 | mycP3 | Probable membrane-anchored mycosin (MycP3) | 1.36 | 2.03 |
| Rv3809c | glf | UDP-galactopyranose mutase (Glf) | 1.16 | 2.29 |
| Rv1316c | ogt | Methylated-DNA—protein-cysteine methyltransferase (Ogt) | -1.45 | -1.36 |
| Rv2334 | cysK1 | Cysteine synthase a (CysK1) | -1.65 | -1.43 |
| Rv3734c | tgs2 | Putative triacylglycerol synthase (Tgs2) | -1.65 | -1.13 |
| Rv2247 | accD6 | Acetyl/propionyl-CoA carboxylase, beta subunit (AccD6) | -1.66 | -1.97 |
| Rv3149 | nuoE | Probable NADH dehydrogenase I, chain E (NuoE) | -1.72 | -1.62 |
| Rv0250c | Rv0250c | Conserved protein | -1.94 | -1.37 |
| Rv1566c | Rv1566c | Possible Inv protein | -2.13 | -3.44 |
| Rv0341 | iniB | Isoniazid inductible gene protein (IniB) | -2.35 | -3.82 |
| Rv0097 | Rv0097 | Possible oxidoreductase | -2.59 | -1.50 |
| Rv2406c | Rv2406c | Conserved protein | -2.68 | -5.02 |
| Rv2623 | TB31.7 | Universal stress protein family protein (TB31.7) | -5.00 | -1.93 |
| Rv2780 | ald | Secreted L-alanine dehydrogenase, 40 kDa antigen (Ald) | -5.19 | -6.80 |
Zn2+-limited Mtb maintain redox homeostasis
Zn2+, a potent antioxidant [29], assists in preservation of cellular redox homeostasis, and Zn2+ deficiency is a condition widely associated with oxidative stress [30,31]. Reactive oxygen species (ROS), produced during oxidative stress, are associated with increased levels of protein and lipid oxidation and DNA damage [32]. As such, mechanisms involved in detoxification of ROS have been reported as an adaptive response during Zn2+ depletion [31,33]. Based on these observations and the results obtained from the multi-omics analysis, we postulated that Zn2+ limited Mtb experience increased exposure to ROS. Specific clues in the data were selective upregulation of some key features in ZLM including the antioxidants catalase (KatG) and alkylhydroperoxide reductase (AhpC) and many genes involved in DNA replication, repair and response to DNA damage (Fig 2 and Table 1), consistent with observations in Mtb treated with oxidizing agents [34].
It was previously shown that Mtb treated with oxidizing agents had a decreased ratio of NADH/NAD+ which was associated with increased sensitivity to oxidation [35,36]. To determine if Zn2+-limited Mtb experienced altered redox homeostasis, we quantified the amount of oxidized and reduced forms of the nicotinamide cofactors in Mtb mc2 6206 cultures grown in ZLM and ZRM. There was no significant difference in the ratio of NADH/NAD+ for bacteria grown with or without added Zn2+ (Fig 3A), however there was a small, but significant increase in the NADPH/NADP+ ratio of Zn2+ limited cultures (Fig 3B). Considering the compartmentalized role of these two cofactors in metabolism, these data suggest that the change in redox equilibrium is not generalized, but specifically affecting NADPH-binding enzymes, i.e., how this factor is utilized.
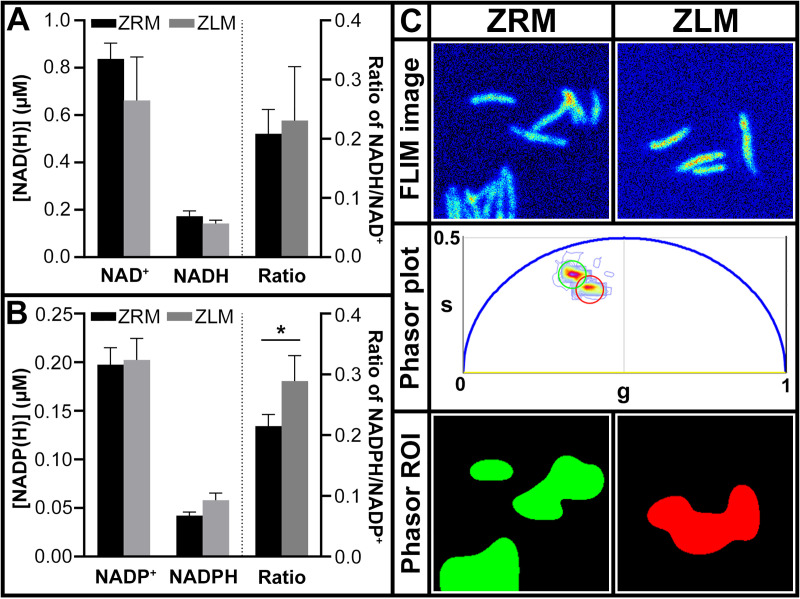
Fig 3. Zn2+-limited Mtb maintain redox homeostasis and exhibit increased reducing power.
Quantification of oxidized (NAD(P)+) and reduced (NAD(P)H) nicotinamide adenine dinucleotide cofactors (A) and its phosphorylated forms (B) in Mtb mc2 6206 cultures after 10 days of growth in ZLM and ZRM. Data, given as the average of biological replicates (ZRM n = 3, ZLM n = 3) with error bars representing standard deviation, are representative of three independent experiments. Asterisks represent a statistically significant difference (t-test, p-value <0.05) in the ratio of oxidized to reduced forms of nicotinamide cofactors in ZRM and ZLM. (C) Fluorescence lifetime imaging (FLIM) of reduced nicotinamide cofactors (NADH and NADPH) for Mtb mc2 6206 cultures after 10 days of growth in ZRM and ZLM. The FLIM micrographs in the top panel are representative fluorescence intensity images for each condition. The phasor plot in the middle shows the phasor positions obtained from the FLIM micrographs where green and red circles encompass the phasor fingerprints of cells grown in ZRM and ZLM respectively. The bottom panels show the region of interest (ROI) encompassed by the encircled areas in the phasor plot projected onto the FLIM micrographs, demonstrating distinct phasor fingerprints between cultures in ZRM and ZLM.
To further assess utilization of redox cofactors, we visualized the biological activity of NAD(P)H from Mtb mc2 6206 in ZLM vs. ZRM using fluorescence lifetime imaging microscopy (FLIM). NAD(P)H FLIM-phasor technique has previously been used to differentiate metabolic states in live bacterial populations at the single-cell level [37]. The phasor positions of cells corresponds to the ratio of free to protein-bound NAD(P)H, and changes in response to environmental factors such as growth phase and history of exposure to antibiotics [37]. Using NAD(P)H FLIM-phasor, we determined that cultures from ZLM and ZRM have distinct phasor fingerprints with Zn2+-limited cultures having shorter fractional intensities, indicating a shift towards a freer state for NAD(P)H than that observed in Zn2+ replete cultures (Fig 3C). This result indicates that the metabolic state of Zn2+-replete and Zn2+-limited Mtb are distinctly different from one another marked by an increased amount of free reduced nicotinamide cofactors available during Zn2+ limitation.
Finally, another coenzyme that has a substantial role in the redox reactions of Mtb is cofactor F420 [38]. Using fluorescence intensity scans, we observed that oxidized coenzyme F420 was detectable in late-log phase cultures (and supernatants) from ZRM while those from ZLM had a loss of peak intensity from the oxidized form of this actinobacterial redox cofactor (S7A and S7B Fig). The loss of peak fluorescence from F420 can be quantified by calculating the ratio of fluorescence intensity at excitation wavelengths of 375 nm and 420 nm (Ex375/Ex420) which offers a simple and robust way to verify the Zn2+-limited phenotype (S7C Fig). Although these data do not provide a detailed picture of the mechanisms involved in maintaining redox homeostasis and utilization of the redox cofactors during Zn2+ limitation, they indicate that [Zn2+] influences the redox signature of Mtb.
Changes in lipidome composition of Zn2+-limited Mtb
During oxidative stress in Msm, in addition to selective upregulation of key detoxifying enzymes (e.g., catalase and alkylhydroperoxidase) along with the global response to DNA repair, a metabolic switch in lipid metabolism is observed [39]. In our RNAseq and proteomic datasets, several enzymes involved in lipid metabolism were differentially regulated, and “cell wall” was an enriched term in the GO analysis from both genes and proteins in Mtb cultures grown in ZLM (S5 Fig and S6 and S7 Tables). Additionally, a clear shift from fatty acid biosynthesis to fatty acid degradation upon Zn2+ limitation was observed when looking at DE genes in the context of a global metabolic network (S8 Fig). Together this led us to investigate whether [Zn2+] may affect lipidome composition.
We analyzed the lipidomes of Mtb H37Rv grown in ZLM and ZRM using LC-MS. Lipid analysis showed remodeling of the lipidome during Zn2+ limitation. Many features had decreased relative abundance in Zn2+-limited condition as seen by the number of compounds downregulated on the cloud plot in Fig 4A. From the precursor ions detected, 307 features were identified as mycobacterial lipids, with 229 of these lipids having decreased relative abundance and 77 having increased relative abundance in ZLM vs. ZRM (S8 Table). In Mtb, de novo synthesis of short-chain fatty acids from acetyl-CoA initiates with the eukaryotic-like fatty acid synthase I (fas) [40]. Fas gene was significantly downregulated in ZLM (S4 Table) and this enzyme is responsible for many of the reactions involved in fatty acid biosynthesis that are downregulated in ZLM (S8 Fig). Additionally, 4/5 genes in the fasII operon (acpM, kasA, kasB and accD6), responsible for chain elongation in the biosynthesis of meroacids [40], were also downregulated in ZLM (S4 Table), with AccD6 being downregulated at both the RNA and protein level (Table 1).
Fig 4. Lipidomic analysis and corresponding surface morphology of Mtb in ZLM vs. ZRM.
(A) Cloud plot of retention time vs. m/z for 7,408 features detected from LC-MS of lipids extracted from Mtb H37Rv cells after 10 days of growth in ZRM (n = 3) and ZLM (n = 3). Each bubble on the graph represents a unique feature where red and blue bubbles indicate upregulated and downregulated features respectively in ZLM vs. ZRM. For each bubble, the p-value is represented by opacity (lower p-values appear darker) and the fold-change is represented by the radius (larger fold-changes have larger radii). Only features with p-value < 0.01 and absFC >2 are shown. (B) Violin plot showing log2FC for each compound identified from selected lipid classes in ZLM vs. ZRM. The horizontal lines in each violin plot represent the interquartile range. Abbreviations of lipid classes can be found in S1 Text. (C) Scanning electron micrographs representative of Mtb mc2 6206 cultures after 10 days of growth in ZRM (n = 3) and ZLM (n = 3). All images are taken at 120kX magnification, the scale bar applies to all images.
Mycolic acids are an integral component of the mycobacterial cell wall and are required for virulence of Mtb [40]. Many forms of mycolic acids had decreased relative abundance in the Zn2+-limited condition, specifically mycolate moieties esterified to carbohydrates in the outer leaflet of the mycomembrane (i.e., trehalose monomycolate -TMM and trehalose dimycolate -TDM) (Fig 4B and S8 Table). Full-length, matured mycolic acids are transferred across the lipid bilayer to be incorporated into the cell wall by inner membrane transporters, upon which antigen 85 complex transfers mycolyl groups from TMM onto cell-wall arabinogalactan forming arabinogalactan-mycolate or onto other TMM moieties forming TDM [41]. In agreement with the decreased relative abundance of TMM and TDM in ZLM, the gene encoding the secreted antigen 85C protein (fbpC) was strongly downregulated in ZLM (S4 Table). In addition, Zn2+-limited Mtb showed a marked decrease in relative abundance of triacyl glycerides (TG) (Fig 4B), a class of lipids stored as energy reserves during stress conditions [42], consistent with the downregulation of TG synthase enzymes tgs1 and tgs2 (S4 Table). Overall, the results show that [Zn2+] affects the lipidome of Mtb with Zn2+-limited bacilli exhibiting decreased relative abundance of specific lipid classes, including triacyl glycerides and mycolic acids.
Next, we investigated whether the global changes observed in the lipidome were reflected in surface features of Mtb mc2 6206 grown in ZRM vs. ZLM. Using scanning electron microscopy (SEM), we observed two features of cells grown in ZLM that differentiated them from those grown in ZRM; the cells were more clumpy (even after enrichment for single cells) (S9 Fig) and most remarkably, there were many finger-like protrusions (fibrils) extending from cells in ZLM (Fig 4C). The protrusions from the cell walls of ZLM cultures observed with SEM are very similar in appearance to those features observed in Mtb mutants deficient in kasA, a key enzyme of mycolic acid biosynthesis [43] that is among the fasII operon enzymes downregulated in ZLM. Therefore, morphological changes observed on the surface of Mtb grown under Zn2+ limited conditions may be caused by differential expression of some key enzymes involved in lipid biosynthesis.
Zn2+-limited Mtb exhibit increased resistance to oxidative stress in vitro and cause higher bacterial burden and pathology in vivo
As presented above, an expected consequence of Zn2+ limitation is exposure to ROS which is supported by increased antioxidant (e.g., KatG and AhpC) expression in ZLM (Table 1). Considering Zn2+-limited bacteria exhibit many features of cells exposed to elevated levels of ROS that correlate with pathogen virulence [44,45], we investigated whether Zn2+ limitation contributes to altered susceptibility of this subpopulation to specific antibiotics, exogenous oxidative stress, and/or affects virulence.
To test the effect of [Zn2+] on antibiotic susceptibility, we used a flow cytometry method to analyze Mtb mc2 6206 grown in ZRM or ZLM and treated with several common antibiotics and oxidizing agents (S10 Fig). This method captures information about viable bacteria not immediately culturable under standard conditions after antibiotic exposure [46]. Using flow cytometry, we also observed increased clumping as seen with SEM, Zn2+-limited cultures (even after enrichment for single cells) had increased abundance of particles at higher forward scatter intensities indicating larger particle sizes (clumps) in ZLM condition (S7 Fig). Results demonstrated that, consistent with upregulation of antioxidant and enzymes involved in DNA repair in ZLM, cells grown in this condition exhibited increased viability when exposed plumbagin, an oxidizing and genotoxic agent (Fig 5A). A similar trend was observed for rifampicin, an antibiotic known to generate ROS in Mtb [47]. A difference in killing with rifampicin was more prominent at the later stages, consistent with the observation that sterilizing activity of rifampicin is time dependent [46] (Fig 5A). On the other hand, cultures from ZLM experienced significantly more rapid killing when treated with isoniazid, consistent with the upregulation of KatG (confirmed in mc2 6206 strain with western blot, S11 Fig), the isoniazid-activating enzyme in Mtb [48] (Fig 5A). No difference in killing was observed for ethambutol or kanamycin, indicating that differences in antibiotic sensitivities are specific (S12 Fig). In conclusion, the Zn2+-limited population exhibited differential response to certain treatments compared to the Zn2+ replete population.
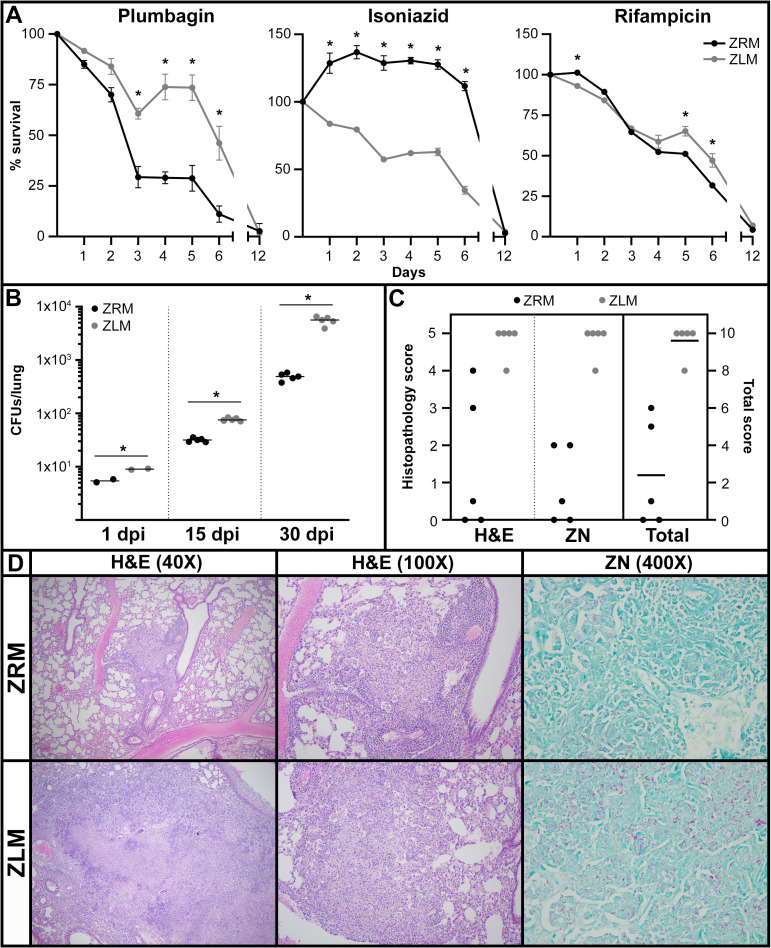
Fig 5. Zn2+-limited Mtb exhibit increased resistance to oxidative stress in vitro and cause higher bacterial burden and pathology in vivo.
(A) Survival of Mtb mc2 6206 after growth in ZRM or ZLM and subsequent exposure to the indicated chemical or antibiotic. Survival was monitored with flow cytometry for twelve days following treatment and was calculated as the percentage of live cells in untreated cultures at the beginning of treatment (S10 Fig). The data are representative of three independent experiments and are given as the average of biological replicates (ZRM n = 3, ZLM n = 2) with error bars representing standard deviation. Asterisks represent a statistically significant difference (t-test, p-value <0.05) between survival of cultures in ZRM vs. ZLM at any given time-point. (B) Bacterial burden in the lungs of C3HeB/FeJ (Kramnik) mice after 1, 15 and 30 days post-infection (dpi) with Mtb H37Rv pre-grown in ZRM or ZLM. Horizontal bars across the data points represent the average CFUs. Asterisks represent a statistically significant difference (t-test, p-value <0.05) between CFUs of mice infected with ZRM vs. ZLM at each timepoint. (C) Blinded histopathology scores from hematoxylin and eosin (H&E) and Ziehl-Neelsen (ZN) staining in panel D. Maximal score of five for H&E and ZN stained cross sections is based on observed changes in lung morphology and immune cell infiltration and bacterial burden respectively and these scores are plotted on the left y-axis. The total score is the sum of the H&E and ZN score for each lung and is given on the right y-axis with a score of 10 representing the maximal level of disease pathology. The horizontal bar through the data for total score represents the average total pathology score. The non-parametric Mann-Whitney U-test for significance was applied to histopathology scores from H&E, ZN and total, all resulting p-values <0.02 indicating a significant difference in pathology scores for mice infected with Mtb pre-grown in ZRM or ZLM. (D) Histopathology micrographs of the lungs of mice infected with Mtb H37Rv pre-grown in ZRM or ZLM at 30 days post-infection. In blinded studies, histology cross-sections of lung tissues were stained with H&E to score changes in lung morphology and immune cell infiltration or ZN stain for mycobacteria (pink) to assess bacterial burden.
In addition to altered survival with exposure to antibiotics, differential expression of key enzymes upregulated in ZLM has been correlated with increased survival and virulence in vivo [44,45]. Considering that certain changes in the transcriptome and proteome of Zn2+-limited Mtb affect virulence, along with the finding that Mtb transits through a Zn2+ limited environment during infection and is likely primed by low [Zn2+] before being transmitted to naïve hosts, it is relevant to determine if [Zn2+] alone can affect virulence of Mtb. To test this, we used C3HeB/FeJ (Kramnik) mice which develop liquefied necrotic granulomas resembling human disease pathology [49]. Mice were infected by aerosol delivery of Mtb pre-grown in ZLM or ZRM (S13 Fig), producing comparable, albeit significantly different (1.7-fold ZLM vs. ZRM, p-value = 0.01265) bacterial numbers in the lungs at 1 day post-infection (dpi) (Fig 5B). Interestingly, bacteria preconditioned by prolonged growth in Zn2+-limiting conditions exhibited significantly higher bacterial burden in the lungs by 15 (2.4-fold, p-value <0.000001) and 30 (11.3-fold, p-value = 0.000004) dpi. (Fig 5B). Accordingly, throughout the infection mice infected with Mtb pre-grown in ZLM provoked increased pulmonary granulomas, with more observable bacteria, considerable tissue destruction, and increased neutrophilic infiltration in the lungs (Figs 5C, 5D and S14A). Although pathology was absent or minimal in livers and spleens (S14 and S15 Figs), by 30 dpi there was a significant increase in the bacterial burden in these organs when mice were infected with inoculum from ZLM vs. ZRM (S15A Fig). These data indicate that exposure to different [Zn2+] may contribute to virulence of Mtb, considering inoculum from ZLM had increased ability to replicate and cause tissue damage in the lungs.
Adaptive response to prolonged Zn2+limitation in Mtb
We have demonstrated that [Zn2+] influences a myriad of changes at the gene level correlating with changes at the protein level and translating into altered physiological characteristics of [Zn2+]-derived populations of Mtb. Many Zn2+ binding proteins have essential biological functions [50], but we found no evidence of a generalized decrease in expression/abundance of these proteins in the response to Zn2+ limitation (S9 Table). This observation indicates that Mtb grown in ZLM were not severely limited for this nutrient, which is also evident from typical growth curves (S2C and S2D Fig). Nonetheless, the response to prolonged Zn2+-limitation in Mtb is robust and complex given the vast global adaptations employed by bacteria grown in ZLM, most of which are not directly related to Zn2+ conservation. In addition to the changes described in the sections above, there were many other genes and pathways affected by [Zn2+] whose significance is unknown and beyond the scope of this paper but support the existence of an adaptive response during Zn2+-limitation. These changes include: 1.) downregulation of many well described antigens in ZLM (e.g., fbpC {Ag85-C}, ald {Ald, 40 Kd-Ag, TB43}, TB31.7, 35 Kd-Ag, hspX {16 Kd-Ag, α-crystallin}, esxA {ESAT-6}) (Table 1 and S4 and S5 Tables) consistent with decreased expression of ABC transporters (S6B Fig and S4 Table), 2.) decreased electron transport, oxidative phosphorylation and ATP production (S6B and S8 Figs) as indicated by decreased cytochrome bc (qcrCAB) and bd (cydB-D) oxidases, NADH-dehydrogenase I (nuoE-N) and ATP-synthase (atpC-E) (S4 Table), and 3.) a significant decrease in genes involved in the hypoxic response (39/49 genes the Mtb H37Rv DosR regulon [51]) (Fig 2B and S10 Table).
In addition to Zur-regulated genes (S3 Table) and the numerous processes involved in DNA replication, mismatch repair and recombination (Figs 2B and S6A and S6 Table) other genes upregulated in ZLM included many PE/PPE proteins, genes involved in biosynthesis pathways and metabolism of diverse substrates (e.g., amino acids) (S6 Table) and isocitrate lyase (Icl) was significantly upregulated at the gene and protein level (Table 1). Another consequence of Zn2+ limitation was altered Cu2+ homeostasis; specifically, there was a strong induction of the copper-sensitive operon repressor CsoR and copper export transporter CtpV (Table 1). Zn2+ limitation altered the expression of numerous genes involved in transcriptional regulation; whiB6 and sigG expression were increased while sigE, sigD, and ten genes involved in two-component signal transduction systems (e.g., devS/devR, mprA/mprB and narG/narI/narX) were decreased in ZLM (S4 and S7 Tables). Altogether, Zn2+ limitation contributes to phenotypic heterogeneity, causing dramatic changes in expression patterns of many genes and pathways that directly influence physiology of Mtb.
Discussion
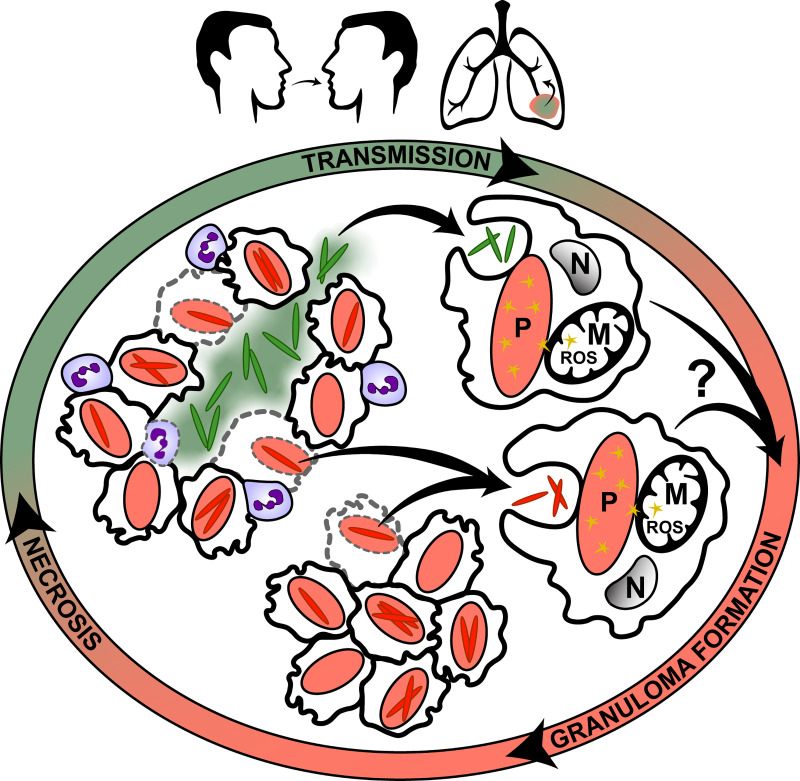
Numerous pathogens experience drastic changes in free [Zn2+] during infection and have adapted elaborate mechanisms to endure Zn2+ toxicity and Zn2+ limitation in vivo [52,53]. Despite the relevance of the Zn2+-depleted niche in the lifecycle of Mtb, the response to prolonged Zn2+ limitation remained undefined in this pathogen. In this study, we suggest that [Zn2+] is likely a physiologically relevant signal experienced by Mtb in vivo, since we were able to demonstrate that this micronutrient triggers the formation of distinct populations in vitro. Zn2+-limited Mtb exhibit a global adaptive response that affects physiology, confers resiliency to oxidative stress and possibly leads to increased virulence. Mtb depends on a cycle of exit and re-entry into host immune cells to perpetuate its lifecycle, and [Zn2+] may be a major cue experienced in this cycle that could potentially affect host-pathogen interactions and disease outcome (Fig 6).
Fig 6. Changes in [Zn2+] throughout TB infection cycle drives formation of Zn2+-limited Mtb with anticipatory adaptations.
The gradient around the outside of the figure represents the cycle of changing [Zn2+] throughout infection; red indicates Zn2+-replete and green indicates Zn2+-limited microenvironments. These [Zn2+]-defined microenvironments drive formation of physiologically distinct subpopulations of Mtb with Zn2+-replete Mtb (red rods) in phagosomes and Zn2+-limited Mtb (green rods) exposed to CP, e.g., in the caseum. After phagocytosis of Mtb, solid granulomas form (bottom) and sustained inflammation leads to the recruitment of neutrophils (purple) which cause necrosis (left). Mtb may be transmitted from necrotic or apoptotic cells (grey dashed margins) in either solid or necrotic granulomas. With the latter, Mtb transit through the Zn2+-limited caseum before being transmitted host-to-host or within a single infected individual (top). The Zn2+-limited Mtb subpopulation has adaptations that could enable this subpopulation to anticipate forthcoming stress and resist host killing (e.g., oxidative stress–yellow stars) and/or affect immune cell activation. The exact mechanisms of how [Zn2+]-derived changes in physiology of Mtb may affect disease outcome are unclear and we highlight the importance of defining the host response to Zn2+-replete and Zn2+-limited Mtb (question mark). Drawings are representative and not to scale and some components of granulomas have been omitted for clarity. Abbreviations: nucleus (N), mitochondria (M), phagosome (P).
After establishing in vitro conditions that induce Zur regulon, we used a multi-omics approach to compare Zn2+-replete and Zn2+-limited Mtb. We detected upregulation of genes in the Zur regulon including those involved in Zn2+ acquisition and replacing Zn2+-dependent ribosomal proteins, as expected [11,12,54]. Interestingly, the ESX-3 gene cluster had the lowest expression levels among Zur-regulated genes, however this finding is explained by the dual regulation of this gene cluster by both Zn2+ (i.e., Zur) [11] and iron (i.e., IdeR) [55], which is not limited in ZLM. The effect of copper (Cu2+) was the opposite, Zn2+-limited Mtb appear to reduce Cu2+ uptake as seen in other Zn2+-limited bacteria [15]. Importantly, the oxidative stress response was a marked feature of Zn2+-limited Mtb including significantly increased expression of antioxidant enzymes KatG and AhpC and genes involved in DNA replication, mismatch repair and recombination, along with accumulation of reducing cofactor NADPH. While there is a precedence for increased antioxidant production and mechanisms to conserve NADPH in Zn2+-limited yeast [31,33], similar protective mechanisms have not been discovered for Zn2+-limited bacteria until now.
In agreement with our observations and consistent with the notion that Zn2+ limitation associates with oxidative stress, multiple different mechanisms leading to elevated oxidative stress in Mtb also modulate expression of antioxidants and enzymes involved in lipid biosynthesis and DNA repair [34,56,57]. Increased expression of sigG in ZLM, the alternative sigma factor upregulated by DNA damaging agents [58], further confirms the response to DNA damage during Zn2+ limitation. Beyond increased capacity to repair damaged DNA, Zn2+ limitation affected other physiological responses that have roles in antioxidant defense or are induced upon oxidative stress, including increased expression of genes involved with iron storage (bfrB) and regulation (furA) [56,59], isocitrate lyase (icl1) [56,60], PE and PPE genes [34], transfer RNAs [61], thioredoxin reductase (trxB2) [62] and WhiB6 gene [45,63]. In this study, Zn2+-limitation is implicated for the first time in induction of the oxidative stress response in Mtb, an adaptive response highly similar to Zn2+-limited pathogenic fungi [64]. The combined effect of an upregulated oxidative stress response, increased reducing power (NADPH), and processes involved in DNA repair likely affords Mtb the ability to adapt to diverse host environments in the context of [Zn2+].
There is a precedence that mycobacteria primed by exposure to oxidative stress have enhanced survival in the host [65,66]. This is, in part, due to two well described virulence factors, catalase (KatG) and alkylhydroperoxide reductase (AhpC), critical elements of the antioxidant defense system, whose expression is induced by oxidative stress and correlates with enhanced survival and hypervirulence in animal models [34,44,45,67]. Owing to inactivation of the oxidation-sensing regulator OxyR in the genome of Mtb, this pathogenic mycobacterium is assumed to have evolved altered regulation of the oxidative stress response [68] and ahpC expression is thought to be silenced under aerobic conditions [69]. Here, we provide evidence that ahpC is not always silenced under aerobic conditions—it is induced upon Zn2+ limitation even in an aerobic environment, a significant finding considering de-repression of ahpC is a mechanism believed to enable survival of bacteria specifically during the transition from latency to active infection [70]. AhpC expression in bacilli exposed to Zn2+ limited environments provides a possible mechanism for sustained survival in the host. Therefore, our results suggest that oxidative stress associated with Zn2+ limitation is a previously unappreciated signal modulating the expression of virulence factors in Mtb.
Hypoxia, not [Zn2+], is the best characterized and appreciated property of the tubercle necrotic granuloma [71]. However, a spectrum of granuloma types exists within a single infected individual [3] and while hypoxia is strongly associated with deeper regions of necrotic granulomas [71], CP-expressing neutrophils are present in both necrotic and non-necrotic granulomas [10]. It is interesting that transcriptomic and phenotypic analysis of Mtb from sputum were found to have a decrease in enzymes involved in PDIM synthesis, protein export systems and a low energy state (decreased ATP synthase genes) [13,14,72], just as we have described to occur upon Zn2+ limitation. The two major differences between transcripts and phenotypes from Zn2+-limited Mtb and those from sputum were decreased accumulation of TGs and expression of genes involved in the hypoxic response (e.g., DosR regulon) in Zn2+-limited condition, another feature shared with Zn2+-limited pathogenic fungi [64]. One possible explanation for this observation is due to decreased oxidative phosphorylation, and presumably a concurrent decrease in oxygen consumption during Zn2+ limitation, which may be an anticipatory response signaled by low [Zn2+] in preparation for transit through the hypoxic caseum. The complete lack of DosR-mediated hypoxic response in Zn2+-limited Mtb demonstrates that [Zn2+] alone does not recapitulate the microenvironment of sputum or the necrotic granuloma. We suggest that, while hypoxia is a relevant cue to bacilli in certain microenvironments during tuberculosis, [Zn2+] may be another cue that delineates bacterial physiology in vivo, possibly even independent of hypoxia, and we emphasize that the combined effects of hypoxia and Zn2+ limitation remains obscure.
[Zn2+] alone dramatically contributes to changes in mycobacterial physiology. We discovered that Zn2+-limited bacteria have increased resistance to exogenous oxidation as predicted by increased antioxidant expression in this condition. While Zn2+-limitation did not have a broad effect on antibiotic susceptibility, Zn2+-limited bacteria were more resistant to ROS-producing rifampicin while increased KatG expression sensitized this subpopulation to the prodrug isoniazid [73]. In addition, changes in the cell wall and other lipid classes in Zn2+-limiting condition can be attributed to a global downregulation in fatty acid biosynthesis at both the gene (fas and fasII operon) and lipidome level, which may be associated with the increased abundance of free NADPH observed during Zn2+ limitation considering NADPH is an essential cofactor for fatty acid biosynthesis. Decreased mycolic acid content of the Zn2+-limited mycomembrane resulted in an observable phenotype marked by finger-like protrusions (fibrils) extending from the surface of cells in ZLM. Cell wall remodeling is utilized by Mtb to interact and persist within the host [74] and the Zn2+-dependent changes in the cell wall may contribute to altered virulence of Zn2+-limited Mtb. Moreover, Zn2+-limited cells showed increased clumping behavior, another cell surface related phenotype that also may play a role in pathogenesis. It has been shown that once a clump of Mtb initiates death of a single macrophage it can lead to serial killing of other macrophages and loss of control over the infection, and it was highlighted that an important next step will be to show how the original clump of Mtb is formed [75]. All in all, Zn2+ limitation drives phenotypic heterogeneity in Mtb in vitro and may be a signal used by Mtb to anticipate forthcoming stress and alter its physiology in ways to promote survival and dissemination.
Classically described as an intracellular parasite, Mtb invades and replicates within host macrophages, and extracellular Mtb in the necrotic milieu face one of two fates for survival; expulsion from the lungs through aerosols (a required step for disease transmission), or re-phagocytosis from competent immune cells at the site of the active lesion. Either way, Mtb must re-enter host immune cells to perpetuate its life-cycle, and, upon phagocytosis, the infected macrophage initiates a respiratory burst producing high levels of ROS to kill the pathogen [76] (Fig 6). However, Mtb possesses resistance mechanisms to evade or counter the phagocytic respiratory burst (e.g., KatG), thus masking the impact of this antimicrobial defense [77]. The fact that Zn2+ limitation primes individual bacilli with the resistance mechanisms needed to evade ROS-derived killing and persist in the host suggests that transit through the Zn2+ limited environment could aid bacterial survival in vivo. Indeed, we show that Zn2+-limited Mtb manifest more severe disease in mice, marked by increased bacterial burden and disease pathology. It is unclear whether the modest difference in bacterial numbers at 1 dpi was due to more efficient initial inoculation that might not reflect altered virulence per se, however this initial difference does not fully explain the large difference in bacterial numbers and disease pathology from Zn2+-limited inoculum observed at later time points. The heightened disease outcome in response to Zn2+-limited inoculum may also be due to increased evasion of killing upon initial contact with macrophages and/or differential priming of the immune system, given the many changes to the lipidome, surface morphology and antigens produced in Zn2+-limited bacteria, their clumping behavior or other unknown mechanisms (Fig 6). Albeit just one study in one mouse strain, our findings suggest that Zn2+-dependent phenotypic heterogeneity in Mtb may have a significant effect on disease progression. We speculate that Zn2+ limitation triggers an anticipatory response against impending phagocytic attack to promote host colonization, as seen in pathogenic fungi exposed to iron limitation [78]. Anticipatory metabolic mechanisms have been described in Mtb [79] and the capacity for adaptive prediction of environmental changes in microorganisms has been shown [80]. Although further in vivo studies are warranted to provide more evidence for the role of [Zn2+] in TB pathogenesis in humans and animal models, our in vitro data suggest that Zn2+ limitation may cue an anticipatory response in extracellular Mtb, stimulating the bacillus to employ protective mechanisms in preparation for the imminent phagocytosis.
Modes of phenotypic heterogeneity in Mtb have been widely recognized as driving forces acting upon individual bacilli which can significantly impact host-pathogen interactions and treatment outcomes [81]. In this study, we demonstrate significant changes in the transcriptome, proteome and lipidome of Mtb depending on [Zn2+], including upregulation of numerous transcription factors and enzymes involved in the oxidative stress response leading to a global adaptive response during Zn2+ limitation. Accordingly, predisposition of individual Mtb bacilli to the Zn2+-limited microenvironment could prime them to interact differentially with the host during infection (Fig 6). Here, for the first time, we suggest that [Zn2+] itself may be a driving factor in the development of phenotypic heterogeneity in Mtb in vivo, and subpopulations of Mtb developing in the Zn2+-depleted niche of necrotic granulomas may affect containment and spread of the bacillus both within the host and the human population.
Materials and methods
Ethics statement
Granulomas from cynomolgus macaques were obtained from animals that were enrolled in completed studies and all work had been previously approved by the University of Pittsburgh’s Institutional Animal Care and Use Committee. All experimental animals (mice) used in this study were approved by the Texas A&M University Institutional Animal Care and Use Committee. Sputum leftovers from TB testing were obtained without identifiers from Hawaii Department of Health and were not considered human subject study. Detailed materials and methods are provided in S1 Text.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Acknowledgments
We sincerely thank Dr. JoAnne L. Flynn for donating non-human primate granuloma material and Dr. Walter J. Chazin for donating recombinant calprotectin and for his insightful discussion on technical limitations of defining free [Zn2+] in necrotic granulomas. We thank Dr. A. Christian Whelen, Diagnostic Laboratory Services, Inc., and Hawaii Department of Health for providing sputum material. We also thank Dr. Robert Husson and Dr. William R. Jacobs Jr. for their contribution of Mtb strains H37Rv and mc2 6206, respectively. We thank the John A. Burns School of Medicine Biocontainment Facility staff for their assistance. This is publication # 115 from the School of Life Sciences, University of Hawaiʻi at Mānoa.
Data Availability
All transcriptomics data are available from the GEO database (accession numbers GSE168513 and GSE168659), proteomics from the PRIDE database (accession number PXD024389), and lipidomics from the MassIVE database (accession number MSV000087030).
Funding Statement
This work was supported by the National Science Foundation (NSF) CAREER Award 1844854 (www.nsf.gov) and Hawaii Community Foundation 17ADVC-86185 (www.hawaiicommunityfoundation.org) to S.P., and the National Institute of Health (NIH) (www.nih.gov) grants: NIAID R21 AI109293 and NIGMS P30 GM114737 to S.P., NIGMS P20 GM113134 and NIGMS R01 GM123048 to N.J. The purchase of the Agilent QTOF LCMS was funded by MRI grant 1532310 from the NSF, awarded to P.G.W. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Philips JA, Ernst JD. Tuberculosis Pathogenesis and Immunity. Annu Rev Pathol Mech Dis. 2012;7: 353–384. 10.1146/annurev-pathol-011811-132458 [DOI] [PubMed] [Google Scholar]
- 2.Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7: 845–855. 10.1038/nrmicro2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenaerts A, Barry CE, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev. 2015;264: 288–307. 10.1111/imr.12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoff DR, Ryan GJ, Driver ER, Ssemakulu CC, De Groote MA, Basaraba RJ, et al. Location of Intra- and Extracellular M. tuberculosis Populations in Lungs of Mice and Guinea Pigs during Disease Progression and after Drug Treatment. Tailleux L, editor. PLoS One. 2011;6: e17550. 10.1371/journal.pone.0017550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadena AM, Fortune SM, Flynn JL. Heterogeneity in tuberculosis. Nat Rev Immunol. 2017;17: 691–702. 10.1038/nri.2017.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zackular JP, Chazin WJ, Skaar EP. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J Biol Chem. 2015;290: 18991–18998. 10.1074/jbc.R115.645085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner D, Maser J, Lai B, Cai Z, Barry CE, Höner zu Bentrup K, et al. Elemental Analysis of Mycobacterium avium -, Mycobacterium tuberculosis -, and Mycobacterium smegmatis -Containing Phagosomes Indicates Pathogen-Induced Microenvironments within the Host Cell’s Endosomal System. J Immunol. 2005;174: 1491–1500. 10.4049/jimmunol.174.3.1491 [DOI] [PubMed] [Google Scholar]
- 8.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, et al. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10: 248–59. 10.1016/j.chom.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. 2012;12: 352–366. 10.1038/nri3211 [DOI] [PubMed] [Google Scholar]
- 10.Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, et al. Microenvironments in Tuberculous Granulomas Are Delineated by Distinct Populations of Macrophage Subsets and Expression of Nitric Oxide Synthase and Arginase Isoforms. J Immunol. 2013;191: 773–784. 10.4049/jimmunol.1300113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maciąg A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, et al. Global Analysis of the Mycobacterium tuberculosis Zur (FurB) Regulon. J Bacteriol. 2007;189: 730–740. 10.1128/JB.01190-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prisic S, Hwang H, Dow A, Barnaby O, Pan TS, Lonzanida JA, et al. Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol Microbiol. 2015;97: 263–280. 10.1111/mmi.13022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garton NJ, Waddell SJ, Sherratt AL, Lee S-M, Smith RJ, Senner C, et al. Cytological and Transcript Analyses Reveal Fat and Lazy Persister-Like Bacilli in Tuberculous Sputum. Neyrolles O, editor. PLoS Med. 2008;5: e75. 10.1371/journal.pmed.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockwood N, Lai RPJ, Seldon R, Young DB, Wilkinson RJ. Variation in pre-therapy levels of selected Mycobacterium tuberculosis transcripts in sputum and their relationship with 2-month culture conversion. Wellcome Open Res. 2019;4: 106. 10.12688/wellcomeopenres.15332.1 [DOI] [Google Scholar]
- 15.Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol. 2013;15: 702–715. 10.1111/j.1462-2920.2012.02849.x [DOI] [PubMed] [Google Scholar]
- 16.Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, et al. S100A8/A9 Proteins Mediate Neutrophilic Inflammation and Lung Pathology during Tuberculosis. Am J Respir Crit Care Med. 2013;188: 1137–1146. 10.1164/rccm.201304-0803OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011;2: e00100–11. 10.1128/mBio.00100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai RP, Cortes T, Marais S, Rockwood N, Burke ML, Garza-Garcia A, et al. Transcriptomic characterization of tuberculous sputum reveals a host Warburg effect and microbial cholesterol catabolism. BioRxiv. 2020. 10.1101/2020.03.09.983163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, et al. Dormancy Phenotype Displayed by Extracellular Mycobacterium tuberculosis within Artificial Granulomas in Mice. J Exp Med. 2004;200: 647–657. 10.1084/jem.20040646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter RL. Pathology of post primary tuberculosis of the lung: An illustrated critical review. Tuberculosis. 2011;91: 497–509. 10.1016/j.tube.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Corro JH, Palmer CD, Ojha AK. Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria. Proc Natl Acad Sci. 2020;117: 19528–19537. 10.1073/pnas.2013409117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigdel TK, Easton JA, Crowder MW. Transcriptional response of Escherichia coli to TPEN. J Bacteriol. 2006;188: 6709–6713. 10.1128/JB.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobiasson V, Dow A, Prisic S, Amunts A. Zinc depletion does not necessarily induce ribosome hibernation in mycobacteria. Proc Natl Acad Sci. 2019;116: 2395–2397. 10.1073/pnas.1817490116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouton JM, Heunis T, Dippenaar A, Gallant JL, Kleynhans L, Sampson SL. Comprehensive Characterization of the Attenuated Double Auxotroph Mycobacterium tuberculosisΔleuDΔpanCD as an Alternative to H37Rv. Front Microbiol. 2019;10: 1–13. 10.3389/fmicb.2019.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dow A, Prisic S. Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc limiting conditions. Pavelka M, editor. PLoS One. 2018;13: e0196300. 10.1371/journal.pone.0196300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halbeisen RE, Gerber AP. Stress-Dependent Coordination of Transcriptome and Translatome in Yeast. Bähler J, editor. PLoS Biol. 2009;7: e1000105. 10.1371/journal.pbio.1000105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerashchenko M V, Lobanov A V, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci. 2012;109: 17394–17399. 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y-X, Xu Z, Ge X, Sanyal S, Lu ZJ, Javid B. Selective translation by alternative bacterial ribosomes. Proc Natl Acad Sci. 2020;117: 19487–19496. 10.1073/pnas.2009607117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell SR. The Antioxidant Properties of Zinc. J Nutr. 2000;130: 1447S–1454S. 10.1093/jn/130.5.1447S [DOI] [PubMed] [Google Scholar]
- 30.Oteiza PI. Zinc and the modulation of redox homeostasis. Free Radic Biol Med. 2012;53: 1748–1759. 10.1016/j.freeradbiomed.2012.08.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C-Y, Bird AJ, Winge DR, Eide DJ. Regulation of the Yeast TSA1 Peroxiredoxin by ZAP1 Is an Adaptive Response to the Oxidative Stress of Zinc Deficiency. J Biol Chem. 2007;282: 2184–2195. 10.1074/jbc.M606639200 [DOI] [PubMed] [Google Scholar]
- 32.Oteiza PL, Olin KL, Fraga CG, Keen CL. Zinc Deficiency Causes Oxidative Damage to Proteins, Lipids and DNA in Rat Testes. Biochem Mol Roles Nutr. 1995;125: 823–829. 10.1093/jn/125.4.823 [DOI] [PubMed] [Google Scholar]
- 33.Wu C-Y, Roje S, Sandoval FJ, Bird AJ, Winge DR, Eide DJ. Repression of Sulfate Assimilation Is an Adaptive Response of Yeast to the Oxidative Stress of Zinc Deficiency. J Biol Chem. 2009;284: 27544–27556. 10.1074/jbc.M109.042036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The Response of Mycobacterium Tuberculosis to Reactive Oxygen and Nitrogen Species. Front Microbiol. 2011;2: 1–12. 10.3389/fmicb.2011.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurumurthy M, Rao M, Mukherjee T, Rao SPS, Boshoff HI, Dick T, et al. A novel F 420 -dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol Microbiol. 2013;87: 744–755. 10.1111/mmi.12127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boshoff HIM, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE. The Transcriptional Responses of Mycobacterium tuberculosis to Inhibitors of Metabolism. J Biol Chem. 2004;279: 40174–40184. 10.1074/jbc.M406796200 [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharjee A, Datta R, Gratton E, Hochbaum AI. Metabolic fingerprinting of bacteria by fluorescence lifetime imaging microscopy. Sci Rep. 2017;7: 3743. 10.1038/s41598-017-04032-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selengut JD, Haft DH. Unexpected Abundance of Coenzyme F420-Dependent Enzymes in Mycobacterium tuberculosis and Other Actinobacteria. J Bacteriol. 2010;192: 5788–5798. 10.1128/JB.00425-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Wu J, Han J, Hu Y, Mi K. Distinct Responses of Mycobacterium smegmatis to Exposure to Low and High Levels of Hydrogen Peroxide. Chatterji D, editor. PLoS One. 2015;10: e0134595. 10.1371/journal.pone.0134595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PaweŁczyk J, Kremer L. The Molecular Genetics of Mycolic Acid Biosynthesis. Microbiol Spectr. 2014;2. 10.1128/microbiolspec.MGM2-0003-2013 [DOI] [PubMed] [Google Scholar]
- 41.Takayama K, Wang C, Besra GS. Pathway to Synthesis and Processing of Mycolic Acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18: 81–101. 10.1128/CMR.18.1.81-101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, et al. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 2006;152: 2717–2725. 10.1099/mic.0.28993-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR. Conditional Depletion of KasA, a Key Enzyme of Mycolic Acid Biosynthesis, Leads to Mycobacterial Cell Lysis. J Bacteriol. 2005;187: 7596–7606. 10.1128/JB.187.22.7596-7606.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Kelley C, Collins F, Rouse D, Morris S. Expression of katG in Mycobacterium tuberculosis Is Associated with Its Growth and Persistence in Mice and Guinea Pigs. J Infect Dis. 1998;177: 1030–1035. 10.1086/515254 [DOI] [PubMed] [Google Scholar]
- 45.Chawla M, Parikh P, Saxena A, Munshi M, Mehta M, Mai D, et al. Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol Microbiol. 2012;85: 1148–1165. 10.1111/j.1365-2958.2012.08165.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendon-Dunn CL, Doris KS, Thomas SR, Allnutt JC, Marriott AAN, Hatch KA, et al. A Flow Cytometry Method for Rapidly Assessing Mycobacterium tuberculosis Responses to Antibiotics with Different Modes of Action. Antimicrob Agents Chemother. 2016;60: 3869–3883. 10.1128/AAC.02712-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piccaro G, Pietraforte D, Giannoni F, Mustazzolu A, Fattorini L. Rifampin Induces Hydroxyl Radical Formation in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58: 7527–7533. 10.1128/AAC.03169-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase—peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358: 591–593. 10.1038/358591a0 [DOI] [PubMed] [Google Scholar]
- 49.Driver ER, Ryan GJ, Hoff DR, Irwin SM, Basaraba RJ, Kramnik I, et al. Evaluation of a Mouse Model of Necrotic Granuloma Formation Using C3HeB/FeJ Mice for Testing of Drugs against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56: 3181–3195. 10.1128/AAC.00217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riccardi G, Milano A, Pasca MR, Nies DH. Genomic analysis of zinc homeostasis in Mycobacterium tuberculosis. FEMS Microbiol Lett. 2008;287: 1–7. 10.1111/j.1574-6968.2008.01320.x [DOI] [PubMed] [Google Scholar]
- 51.Park H-D, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48: 833–843. 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capdevila DA, Wang J, Giedroc DP. Bacterial Strategies to Maintain Zinc Metallostasis at the Host-Pathogen Interface. J Biol Chem. 2016;291: 20858–20868. 10.1074/jbc.R116.742023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ong CY, Berking O, Walker MJ, McEwan AG. New Insights into the Role of Zinc Acquisition and Zinc Tolerance in Group A Streptococcal Infection. Freitag NE, editor. Infect Immun. 2018;86: e00048–18. 10.1128/IAI.00048-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serafini A, Pisu D, Palù G, Rodriguez GM, Manganelli R. The ESX-3 Secretion System Is Necessary for Iron and Zinc Homeostasis in Mycobacterium tuberculosis. Delogu G, editor. PLoS One. 2013;8: e78351. 10.1371/journal.pone.0078351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an Essential Gene in Mycobacterium tuberculosis: Role of IdeR in Iron-Dependent Gene Expression, Iron Metabolism, and Oxidative Stress Response. Infect Immun. 2002;70: 3371–3381. 10.1128/iai.70.7.3371-3381.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyagi P, Dharmaraja AT, Bhaskar A, Chakrapani H, Singh A. Mycobacterium tuberculosis has diminished capacity to counteract redox stress induced by elevated levels of endogenous superoxide. Free Radic Biol Med. 2015;84: 344–354. 10.1016/j.freeradbiomed.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiwari S, van Tonder AJ, Vilchèze C, Mendes V, Thomas SE, Malek A, et al. Arginine-deprivation–induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc Natl Acad Sci. 2018;115: 9779–9784. 10.1073/pnas.1808874115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smollett KL, Dawson LF, Davis EO. SigG Does Not Control Gene Expression in Response to DNA Damage in Mycobacterium tuberculosis H37Rv. J Bacteriol. 2011;193: 1007–1011. 10.1128/JB.01241-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sala C, Forti F, Di Florio E, Canneva F, Milano A, Riccardi G, et al. Mycobacterium tuberculosis FurA Autoregulates Its Own Expression. J Bacteriol. 2003;185: 5357–5362. 10.1128/jb.185.18.5357-5362.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahn S, Jung J, Jang I, Madsen EL, Park W. Role of Glyoxylate Shunt in Oxidative Stress Response. J Biol Chem. 2016;291: 11928–11938. 10.1074/jbc.M115.708149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong J, Xiao C, Gu W, Du G, Sun X, He Q, et al. Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. Ibba M, editor. PLOS Genet. 2015;11: e1005302. 10.1371/journal.pgen.1005302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaeger T, Budde H, Flohé L, Menge U, Singh M, Trujillo M, et al. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;423: 182–191. 10.1016/j.abb.2003.11.021 [DOI] [PubMed] [Google Scholar]
- 63.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Differential Gene Expression in Response to Exposure to Antimycobacterial Agents and Other Stress Conditions among Seven Mycobacterium tuberculosis whiB-Like Genes. Antimicrob Agents Chemother. 2006;50: 2836–2841. 10.1128/AAC.00295-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vicentefranqueira R, Amich J, Marín L, Sánchez C, Leal F, Calera J. The Transcription Factor ZafA Regulates the Homeostatic and Adaptive Response to Zinc Starvation in Aspergillus fumigatus. Genes (Basel). 2018;9: 318. 10.3390/genes9070318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganief N, Sjouerman J, Albeldas C, Nakedi KC, Hermann C, Calder B, et al. Associating H 2 O 2- and NO-related changes in the proteome of Mycobacterium smegmatis with enhanced survival in macrophage. Emerg Microbes Infect. 2018;7: 1–17. 10.1038/s41426-017-0002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Idh J, Andersson B, Lerm M, Raffetseder J, Eklund D, Woksepp H, et al. Reduced susceptibility of clinical strains of Mycobacterium tuberculosis to reactive nitrogen species promotes survival in activated macrophages. Neyrolles O, editor. PLoS One. 2017;12: e0181221. 10.1371/journal.pone.0181221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H-N, Lee N-O, Han SJ, Ko I-J, Oh J-I. Regulation of the ahpC Gene Encoding Alkyl Hydroperoxide Reductase in Mycobacterium smegmatis. Manganelli R, editor. PLoS One. 2014;9: e111680. 10.1371/journal.pone.0111680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deretic V, Philipp W, Dhandayuthapani S, Mudd M, Curcic R, Garbe T, et al. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene:implications for sensitivity to isoniazid. Mol Microbiol. 1995;17: 889–900. 10.1111/j.1365-2958.1995.mmi_17050889.x [DOI] [PubMed] [Google Scholar]
- 69.Springer B, Master S, Sander P, Zahrt T, McFalone M, Song J, et al. Silencing of Oxidative Stress Response in Mycobacterium tuberculosis: Expression Patterns of ahpC in Virulent and Avirulent Strains and Effect ofahpC Inactivation. Kaufmann SHE, editor. Infect Immun. 2001;69: 5967–5973. 10.1128/IAI.69.10.5967-5973.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Master SS, Springer B, Sander P, Boettger EC, Deretic V, Timmins GS. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology. 2002;148: 3139–3144. 10.1099/00221287-148-10-3139 [DOI] [PubMed] [Google Scholar]
- 71.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, et al. Tuberculous Granulomas Are Hypoxic in Guinea Pigs, Rabbits, and Nonhuman Primates. Infect Immun. 2008;76: 2333–2340. 10.1128/IAI.01515-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, Ryndak MB, Aggarwal AN, Yadav R, Sethi S, Masih S, et al. Transcriptome analysis of mycobacteria in sputum samples of pulmonary tuberculosis patients. Neyrolles O, editor. PLoS One. 2017;12: e0173508. 10.1371/journal.pone.0173508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pym AS, Domenech P, Honore N, Song J, Deretic V, Cole ST. Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Mol Microbiol. 2001;40: 879–889. 10.1046/j.1365-2958.2001.02427.x [DOI] [PubMed] [Google Scholar]
- 74.Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol. 2014;12: 550–562. 10.1038/nrmicro3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahamed D, Boulle M, Ganga Y, Mc Arthur C, Skroch S, Oom L, et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife. 2017;6: 1–26. 10.7554/eLife.22028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 2009;11: 1170–1178. 10.1111/j.1462-5822.2009.01335.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52: 1291–1302. 10.1111/j.1365-2958.2004.04078.x [DOI] [PubMed] [Google Scholar]
- 78.Pradhan A, Avelar GM, Bain JM, Childers D, Pelletier C, Larcombe DE, et al. Non-canonical signalling mediates changes in fungal cell wall PAMPs that drive immune evasion. Nat Commun. 2019;10: 5315. 10.1038/s41467-019-13298-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eoh H, Wang Z, Layre E, Rath P, Morris R, Branch Moody D, et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol. 2017;2: 17084. 10.1038/nmicrobiol.2017.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460: 220–224. 10.1038/nature08112 [DOI] [PubMed] [Google Scholar]
- 81.Dhar N, McKinney J, Manina G. Phenotypic Heterogeneity in Mycobacterium tuberculosis. Microbiol Spectr. 2016;4: 1–27. 10.1128/microbiolspec.TBTB2-0021-2016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Data Availability Statement
All transcriptomics data are available from the GEO database (accession numbers GSE168513 and GSE168659), proteomics from the PRIDE database (accession number PXD024389), and lipidomics from the MassIVE database (accession number MSV000087030).