Abstract
Blast crisis (BC) predicts dismal outcomes in patients with chronic myeloid leukaemia (CML). Although additional genetic alterations play a central role in BC, the landscape and prognostic impact of these alterations remain elusive. Here, we comprehensively investigate genetic abnormalities in 136 BC and 148 chronic phase (CP) samples obtained from 216 CML patients using exome and targeted sequencing. One or more genetic abnormalities are found in 126 (92.6%) out of the 136 BC patients, including the RUNX1-ETS2 fusion and NBEAL2 mutations. The number of genetic alterations increase during the transition from CP to BC, which is markedly suppressed by tyrosine kinase inhibitors (TKIs). The lineage of the BC and prior use of TKIs correlate with distinct molecular profiles. Notably, genetic alterations, rather than clinical variables, contribute to a better prediction of BC prognosis. In conclusion, genetic abnormalities can help predict clinical outcomes and can guide clinical decisions in CML.
Subject terms: Cancer genetics, Haematological cancer, Mutation
In chronic myeloid leukaemia (CML), the drivers of blast crisis and resistance to tyrosine kinase inhibitors are not fully characterised. Here, the authors analyse a cohort of CML samples with genomic technologies and find that at least one driver alteration is associated with progression and worse prognosis.
Introduction
Chronic myeloid leukaemia (CML) is a myeloproliferative disorder caused by the BCR-ABL1 gene fusion generated in the Philadelphia (Ph) chromosome, der(22)t(9;22)(q34;q11.2). Recently, the prognosis of CML has been dramatically improved by the development of tyrosine kinase inhibitors (TKIs) targeting the BCR-ABL1 fusion protein. However, a minority of patients in the chronic phase (CP) fail to respond to TKI therapy, progress to blast crisis (BC), and show dismal clinical outcomes1. While a mutation in the BCR-ABL1 kinase domain is known to be one of the major determinants of TKI resistance and a risk for blastic transformation2, additional genetic alterations have been hypothesised to be necessary for the progression to BC. In fact, recent studies have demonstrated several driver mutations acquired during blastic transformation3–17. However, the current understanding of the genetic basis of TKI resistance, and progression of CML-CP to BC remains limited by the small number of patients and/or genes analysed in each study, as well as the paucity of matched CP and BC samples.
Another point of interest is the improved 5-year overall survival (OS) of BC patients from 16% during 2000–2004 to 33% during 2010–2016, which may be attributed to increased use of TKIs18. TKIs may be effective for certain patients with BC and contribute to improved survival; however, the majority of BC patients no longer show response to TKIs. If this is indeed the case, it is of considerable clinical importance to predict which BC patients can respond to TKI, for better management of CML. Unfortunately, only a few clinical factors or biomarkers are currently known to be correlated with clinical outcomes of BC patients treated with TKI-based regimens.
In this study, we investigated a large cohort of CML patients to reveal the landscape of genetic lesions in CML during both CP and BC. We aimed to identify genetic alterations that correlated with TKI-resistance and blastic transformation, as well as those that predicted clinical outcomes. For this, genetic alterations in both CP and BC samples, including paired CP and BC samples, were analysed using unbiased sequencing.
Results
Clonal evolution of CML
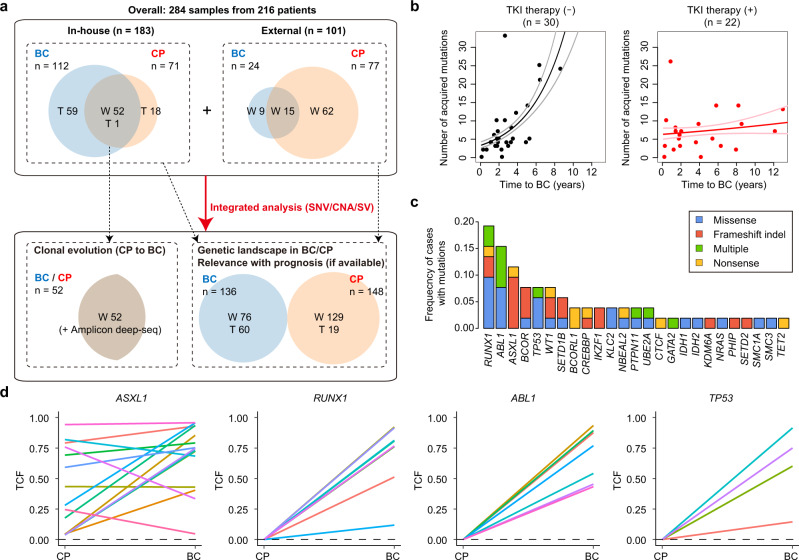
First, we performed whole-exome sequencing (WES) of paired CP and BC samples obtained from 52 patients with CML, to identify genetic alterations that were relevant to the clonal evolution to CML-BC, at a mean depth of 157 (Fig. 1a, Supplementary Fig. 1a, and Supplementary Table 1). On average, 5.3 (0–21) nonsynonymous single-nucleotide variants (SNVs; or 0.088/Mb) were acquired during disease progression from CP to BC, with a median time of 26.7 months (0.7–155.1; Fig. 1b and Supplementary Fig. 1b). Notably, a Poisson regression model revealed that the number of mutations acquired during progression from CP to BC was independently and positively correlated with the interval between the progression (P = 9.4 × 10−12), and negatively correlated with TKI therapy after CP diagnosis (P = 9.3 × 10−3; Fig. 1b). The correlation with the number of recurrent mutations in CML-BC was not clear (Supplementary Fig. 1c). A similar trend was observed in a previous cohort of paired samples13, even though our findings did not show statistical significance owing to the small number of evaluable samples after quality control (n = 13; Supplementary Fig. 1d). These results suggest that transformation from CP to BC is associated with accumulation of somatic mutations with time in the absence of effective therapy, and this accumulation is noticeably suppressed by TKI therapy, which may prevent the transformation from CP to BC.
Fig. 1. Somatic mutations acquired during clonal evolution in CML.
a Scheme demonstrating the study cohort. Numbers indicate analysed samples. Numbers in overlapping regions of CP and BC circles indicate the number of cases analysed for both CP and BC. W whole-exome sequencing, T targeted capture sequencing. b Scatter plots for time to progression (horizontal axis) and number of acquired SNVs (vertical axis) during progression from CP to BC in 52 cases for whom WES was performed, using paired CP and BC samples. Regression lines with 95% confidence intervals were also plotted. Cases with or without TKI therapy after CP diagnosis are indicated separately. c Frequencies of mutations acquired during evolution from CP to BC in 52 cases. Recurrently acquired or known driver genes are described. Categories of mutations are depicted in different colours, and “multiple” indicates ≥2 distinct mutations found in the same gene in the same patient. d TCFs of the indicated mutations in the corresponding CP and BC samples determined by conducting deep amplicon sequencing. Black dashed lines indicate a TCF of 0%. Colours represent individual cases.
In the CML-BC samples, mutations were frequently found in the driver genes implicated in myeloid malignancies, including RUNX1, ABL1, ASXL1, BCOR/BCORL1, TP53, and WT1 (Fig. 1c and Supplementary Fig. 1e). We also identified recurrent mutations in recently reported genes in BC, such as UBE2A13,14 and SETD1B13, as well as in previously unreported genes, such as KLC2 and NBEAL2. Deep amplicon sequencing of these mutations at a mean depth of ×2589 in paired CP and BC samples revealed that ASXL1 mutations were already present in the CP samples, whereas other major drivers, including RUNX1, ABL1, and TP53 mutations were initially absent in CP and emerged during progression to BC (Fig. 1d). In a few patients, mutations in other genes, such as WT1 and IDH2, were also found in the corresponding CP samples with lower tumour cell fractions (TCFs) calculated by variant allele frequencies (VAFs) than those found in the BC samples (Supplementary Fig. 1f). These results suggest distinct roles for the different mutations in the progression of CML. TCFs of ASXL1 mutations were increased in nine, decreased in three, and almost stable (<10% difference) in three patients during disease progression from CP to BC. Almost all patients with ASXL1 mutations showed acquisition of other additional genetic abnormalities during progression to BC (93.3%, 14 out of 15 cases), including mutations (12/15) in RUNX1 (4/15), TP53 (3/15), BCOR (2/15), and SETD1B (2/15) genes. Typically, at least one accompanying mutation had TCFs comparable to ASXL1 mutations and was probably present in the major clones in the BC samples (Supplementary Fig. 2). Therefore, ASXL1-mutated CP clones may be preferentially selected and may evolve by acquiring other drivers during the clonal development to BC.
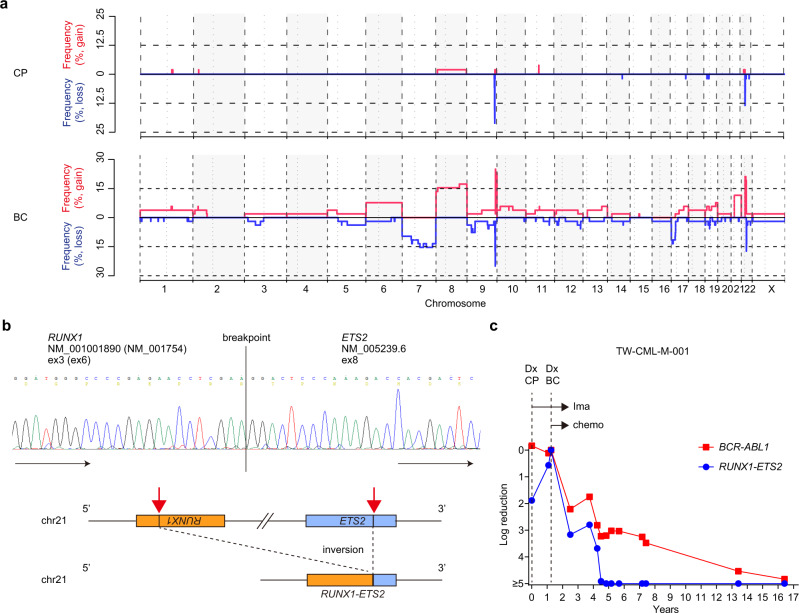
We also performed sequencing-based copy-number analysis19. Copy-number alterations (CNAs) were frequently found in BC, but were rarely present in CP (Fig. 2a), suggesting that CNAs were also acquired during progression from CP to BC. Frequently identified CNAs in BC included −7/del(7p), +8, del(17p), amp(17q), +21, and an extra Ph chromosome (+Ph). We also analysed structural variations (SVs) close to the gene bait regions based on an algorithm, utilising both breakpoint-containing junction read pairs and improperly aligned read pairs20. Although the ability to detect SVs in our pipeline depends on the location of breakpoints and gene baits (see “Methods”), this approach led to the identification of an inversion event, resulting in a RUNX1-ETS2 fusion in a patient with myeloid BC, which was confirmed by performing reverse transcription PCR (RT-PCR) and Sanger sequencing (Fig. 2b and Supplementary Fig. 3). Quantitative RT-PCR (RT-qPCR) at several time points demonstrated that this fusion was already present at the time of CP diagnosis, and the burden of this fusion, which was reduced after successful chemotherapy for CML-BC, was correlated with that of BCR-ABL1 (Fig. 2c). Thus, RUNX1-ETS2 may play a role in the rapid progression of CML-CP to BC.
Fig. 2. CNAs and RUNX1-ETS2 fusion in CML-CP and BC.
a Sequencing-based copy-number profiling in CML-CP and BC samples from 52 cases. Frequencies of copy-number gains or losses are depicted along the chromosomal regions. b RUNX1-ETS2 fusion detected in a patient (TW-CML-M-001) by conducting WES analysis. Sanger sequencing of cDNA obtained from the BC samples was used to validate the fusion breakpoint (top). Simplified scheme of RUNX1-ETS2 fusion (bottom). Inversion 21 involves the RUNX1 and ETS2 genes for generation of the fusion. The karyotyping result of this sample at CP diagnosis was as follows (results at BC diagnosis were not obtained): 47,XY,+8,inv(9)(p11q13),t(9;22)(q34;q11). c Time chart showing dynamic changes in RUNX1-ETS2 and BCR-ABL1 burdens assessed by RT-qPCR at several time points in a patient (TW-CML-M-001). The horizontal and vertical axes represent the time from CP diagnosis (Dx) and levels of the indicated transcripts, respectively. The patient received imatinib (Ima) treatment after CP diagnosis and achieved CHR, while the BCR-ABL1 burden was not reduced significantly. Approximately 15 months after CP diagnosis, there was an abrupt development of BC, which was successfully treated with cytarabine and hydroxyurea in addition to imatinib. Thereafter, BCR-ABL1 transcript levels showed continuous reductions, and were below the threshold of CMR at 15 years after BC diagnosis. The RUNX1-ETS2 transcript was detected at the time of CP diagnosis and increased markedly during BC development, declined following chemotherapy with imatinib, and was undetectable 4 years after BC transformation.
Genetic landscape of CML-BC
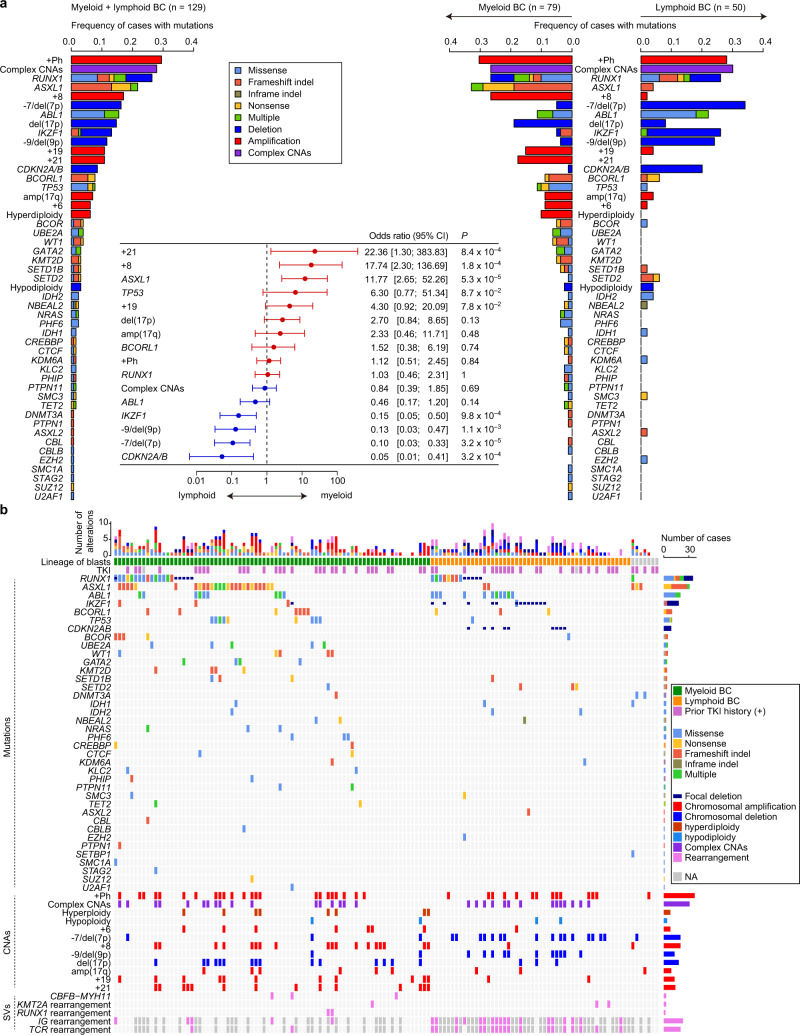
Next, we performed targeted capture sequencing that covered 104 myeloid tumour-associated genes, including candidates of drivers found by WES in an additional 60 BC and 19 CP samples at a mean depth of ×585 (Supplementary Table 2). Combined with the WES data from the 52 paired BC and CP samples, as well as external WES data for 24 BC12,13 and 77 CP13,21,22 patients, we comprehensively analysed a total of 136 BC and 148 CP cases for SNVs, CNAs, and SVs (Fig. 1a and Supplementary Table 1). Clinical information of the external data of patients was typically limited to a few factors, including age, TKI therapy and response, and lineage of BC blasts (Table 1 and Supplementary Table 3). Myeloid and lymphoid crises accounted for 62.8% and 37.2% of the total cohort, respectively. In addition, 42.5% of the cohort presented with a prior history of TKI therapy before BC diagnosis, where in most cases, the initial TKI used was imatinib (94.3%). In total, 126 out of 136 patients with CML-BC harboured at least one mutation or CNA (Fig. 3, Supplementary Figs. 4 and 5, Supplementary Table 4, and Supplementary Data 1). Most mutations (89.7%, 192/214) involved genes implicated in epigenetic regulation and signalling, such as chromatin modification, DNA methylation, transcription factors, the cohesin complex, and signalling pathways. In addition to KLC2 and NBEAL2, we found another recurrent mutational target, PHIP, which was mutated in two patients. Sequencing-based copy-number analysis disclosed the presence of complex CNAs (defined as ≥3 abnormal CNAs) in 26.5% of the BC patients, although complex karyotypes in CML-BC have previously been reported at a lower rate (10–12%) based on conventional karyotyping23, whose resolution is relatively limited.
Table 1.
Characteristics of patients with CML-BC.
| Number of cases | 136 |
| Age at BC diagnosis (y), median (range) (n = 126) | 50 (16–86) |
| Sex, n (%) (n = 132) | |
| Male | 79 (59.8) |
| Female | 53 (40.2) |
| Lineage of blasts, n (%) (n = 129) | |
| Myeloid | 81 (62.8) |
| Lymphoid | 48 (37.2) |
| WBC (×103/uL), median (range) (n = 123) | 40,600 (1700–580,000) |
| Hb (g/dL), median (range) (n = 123) | 9.6 (5.0–15.8) |
| PLT (×103/uL), median (range) (n = 123) | 105,000 (3000–2,740,000) |
| LDH (U/L), median (range) (n = 79) | 696 (75–6332) |
| Blasts in BM (%), median (range) (n = 126) | 59.5 (1.0–98.4) |
| Prior history of CP diagnosis, n (%) (n = 135) | |
| Yes | 103 (76.3) |
| No | 32 (23.7) |
| Time from CP diagnosis (m), median (range) (n = 93) | 34.3 (0.27–363) |
| Age at CP diagnosis (y), median (range) (n = 92) | 45 (14–85) |
| Prior TKI before BC, n (%) (n = 134) | |
| Yes | 57 (42.5) |
| No | 77 (57.5) |
| TKIs used for CP, n (%) (n = 53) | |
| Imatinib | 50 (94.3) |
| Dasatinib | 2 (3.8) |
| Nilotinib | 1 (1.9) |
| TKI-based therapy for BC, n (%) (n = 119) | |
| Yes | 72 (60.5) |
| No | 47 (39.5) |
| TKIs used for BC, n (%) (n = 72) | |
| Imatinib | 36 (50.0) |
| Dasatinib | 32 (44.4) |
| Nilotinib | 2 (2.8) |
| Ponatinib | 2 (2.8) |
| Final status, n (%) (n = 123) | |
| Alive | 26 (21.1) |
| Dead | 97 (78.9) |
| Method, n (%) (n = 136) | |
| Whole-exome sequencing | 76 (55.9) |
| Targeted capture sequencing | 60 (44.1) |
| Median follow-up time (m), median (range) (n = 123) | 38.2 (0.13–365) |
Fig. 3. Genetic landscape of myeloid and lymphoid BC in CML.
a Frequencies of mutations in 129 BC patients evaluable for blast lineage (left panel), and myeloid (n = 79) and lymphoid BC (n = 50) (right panel). Categories of mutations are depicted in different colours, and “multiple” indicates ≥2 distinct alterations found in the same gene in the same patient. The forest plot shows odds ratios with 95% confidence intervals (CI) for enrichment of each genetic lesion in myeloid BC. The dashed line represents an odds ratio of 1. Positive and negative odds ratios are indicated by red and blue colours, respectively. Genetic lesions found in >10 cases were included. P values were calculated using the Fisher’s exact test. b Summary of genetic lesions in all 136 BC patients. Each column indicates one patient. Lineage of blasts and prior history of TKI therapy before BC diagnosis are also shown. Categories of alterations are depicted in different colours, and “multiple” indicates ≥2 distinct mutations found in the same gene in the same patient. The rearrangement of immunoglobulin (IG) and T-cell receptor (TCR) genes is shown in cases analysed by WES. NA not available.
We further explored the relationship between the genetic abnormalities and the lineage of BC (Fig. 3). Certain lesions, such as those attributed to +21, +8, +19, and ASXL1 and TP53 mutations, were enriched in myeloid crisis compared to those in lymphoid crisis, while others were enriched in lymphoid BC, which included CDKN2A/B and IKZF1 deletions, −7/del(7p), and −9/del(9p). In contrast, abnormalities such as RUNX1 mutations and +Ph were almost equally observed in both crises. The rearrangement of immunoglobulin and/or T-cell receptor genes, which was assessed by WES, was confirmed in most evaluable lymphoid samples and in a few samples of myeloid crisis. Analysis of pairwise correlations between the genetic lesions showed the existence of co-occurring patterns depending on the combination of lesions (Supplementary Fig. 6). Among these, the most conspicuous correlations were those observed between +6, +8, +19, and +21, which were highly specific to myeloid crisis. Another visible correlation was between −7/del(7p), −9/del(9p), and CDKN2A/B deletions, which were highly specific to lymphoid crisis. We also found that del(17p) co-occurred with TP53 mutations, +Ph, +8, and amp(17q). Taken together, although substantial overlaps in genetic lesions were observed, myeloid and lymphoid BC cases were distinct in terms of lineage commitment, as well as their molecular profiles.
We next investigated whether a prior history of TKI therapy influenced the genetic profile in BC, whereby we compared genetic alterations in BC patients with and without a prior history of TKI therapy. As expected, ABL1 mutations were almost exclusively found in patients who had received TKIs, representing the most frequent mutation (Supplementary Fig. 7). All ABL1 mutations were accompanied by other genetic alterations, such as mutations in RUNX1 (7/20), ASXL1 (5/20), and BCORL1 (5/20), focal deletions in CDKN2A/B (6/20), and IKZF1 (4/20), and cytogenetic abnormalities, such as −7/del(7p) (6/20), −9/del(9p) (6/20), and +8 (4/20), which was consistent with previous reports24. Moreover, BC patients with a history of TKI treatment were more likely to harbour −7/del(7p), −9/del(9p), and complex CNAs than those without TKI treatment, while BCOR, TP53, and RUNX1 mutations were less frequent in TKI-treated patients. This suggests that genetic profiles in CML-BC differ between TKI-treated and untreated patients, although it remains unclear whether non-ABL1 mutations can affect TKI response/resistance.
Prognostic relevance of genetic abnormalities in CML-BC
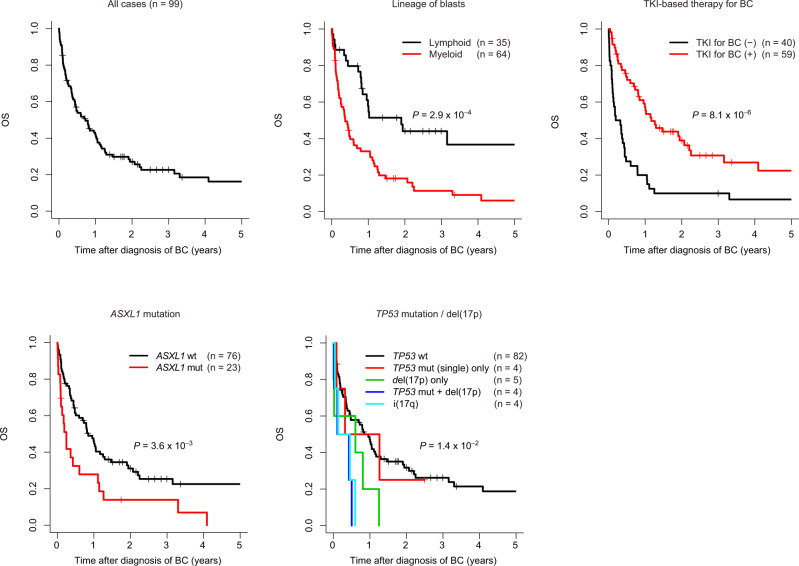
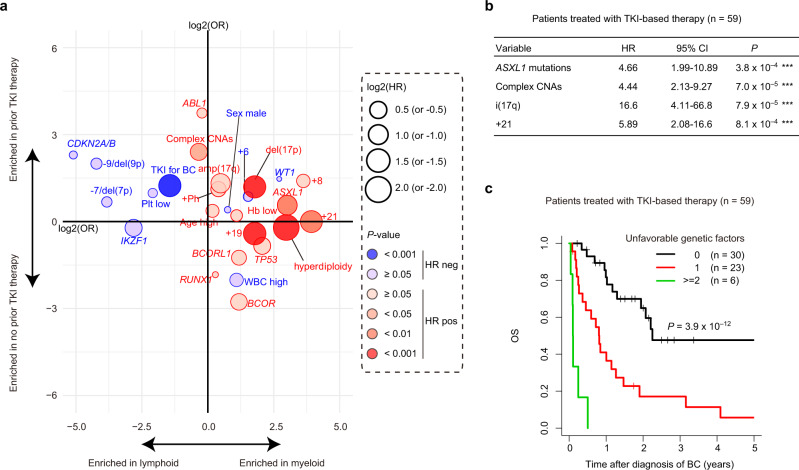
We next analysed the prognostic relevance of the genetic abnormalities, as well as the clinical features in 99 CML-BC patients for whom survival information was available. With a median follow-up of 3.2 years (0.48–30.4), the estimated 2-year OS rate for all patients was 27.1% (95% confidence interval, 19.3–38.0; Fig. 4 and Supplementary Table 5). In the univariate analysis of clinical features for OS, TKI therapy for BC and lineage of blasts were significantly associated with OS, while other parameters, including age, sex, blood counts, and TKI history before BC, did not exhibit significant associations (Figs. 4 and 5a, and Table 2). Univariate analysis of genetic lesions observed in >5% of the patients revealed a negative prognostic impact of ASXL1 mutations, del(17p), i(17q) (isochromosome 17q, resulting in one copy of 17p and three copies of 17q), +19, +21, hyperdiploidy (as defined by presence of ≥48 chromosomes assessed by sequencing-based copy-number profiling), and complex CNAs (Table 2). Conspicuously, patients with concurrent TP53 mutations and del(17p), which were predictive of biallelic targeting of TP53, and i(17q), showed an especially grim outcome (Fig. 4). Consistent with a previous report demonstrating the association of multiple-hit TP53 mutations with complex karyotypes and poor outcomes in myelodysplastic syndromes25, three out of four cases with biallelic TP53 mutations in CML-BC patients revealed the existence of a complex karyotype. After performing adjustment for blast lineage and TKI-based therapy to correct for the possible effects of an association between the clinical and genetic factors, complex CNAs, −7/del(7p), and amp(17q) showed significant association with OS (Table 2 and Supplementary Fig. 8). As shown in Fig. 5a, several genetic lesions were considerably and strongly associated with the lineage of BC, prior TKI-based treatment history, and/or prognosis in CML-BC, compared with clinical features, such as age, sex, white blood cell (WBC) count, haemoglobin levels, and platelet count. Therefore, genetic abnormalities may be good biomarkers for predicting clinical outcomes in BC patients.
Fig. 4. Prognostic relevance of genetic abnormalities in CML-BC.
Kaplan–Meier survival curves for OS in 99 patients with CML-BC according to the indicated clinical or genetic factors. The prognostic impact of each factor on OS was calculated using the log-rank test. For TP53 mutations and/or del(17p), we classified patients according to the presence or absence of TP53 mutations, del(17p), and i(17q), based on which patients were divided into the following five groups: patients (1) without any TP53 mutations/del(17p)/i(17q), (2) with TP53 mutations alone, (3) with del(17p) alone, (4) with concurrent TP53 mutations and del(17p), and (5) with i(17q). One patient harboured two distinct TP53 mutations, as well as del(17p), and was classified into group 4. No patients with i(17q) harboured TP53 mutations.
Fig. 5. Prognostic impact of each clinical or genetic factor in CML-BC.
a Summary of the relationship between individual clinical or genetic factors and lineage of BC, prior TKI history, and OS in 99 BC patients. Odds ratios for the enrichment of individual factors in myeloid lineage of BC and in the history of prior TKI therapy are represented in the horizontal and vertical axes, respectively. The size of the circles corresponds to the hazard ratio (HR) of the effect of each factor on the OS in the univariate analysis. Positive and negative HR values are shown as red and blue circles, respectively. The colour gradient indicates P values calculated by the Cox proportional hazards regression model in the univariate analysis. b Cox proportional hazards regression model with a stepwise variable selection identifying the independent risk factors, predicting OS in 59 patients treated with TKI-based therapy for BC. ***P < 0.001. c Kaplan–Meier survival curves for OS of 59 patients treated with TKI-based therapy for BC according to the number of unfavourable genetic factors listed in b. P values were calculated using the log-rank test.
Table 2.
Univariate analysis for OS of CML-BC.
| Variable | All, n = 99 | TKI treated, n = 59 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Adjusted | Univariate | Adjusted | |||||||||||
| n | HR | 95% CI | P | HR | 95% CI | P | n | HR | 95% CI | P | HR | 95% CI | P | |
| Clinical factors | ||||||||||||||
| Age at BC (>60 y) | 27 | 1.26 | 0.76–2.10 | 0.36 | 1.89 | 1.08–3.31 | 2.5 × 10−2* | 19 | 1.34 | 0.68–2.63 | 0.40 | 1.35 | 0.68–2.67 | 0.39 |
| Sex (male) | 59 | 0.98 | 0.62–1.55 | 0.92 | 1.01 | 0.63–1.62 | 0.97 | 37 | 0.85 | 0.44–1.63 | 0.62 | 0.87 | 0.45–1.68 | 0.68 |
| WBC (>50,000 × 103/uL) | 50 | 0.77 | 0.49–1.23 | 0.27 | 0.73 | 0.46–1.17 | 0.19 | 27 | 0.50 | 0.26–0.96 | 3.6 × 10−2* | — | — | — |
| Hb (<9.3 g/dL) | 49 | 1.19 | 0.75–1.87 | 0.46 | 0.79 | 0.49–1.27 | 0.32 | 25 | 1.20 | 0.63–2.27 | 0.58 | 1.07 | 0.55–2.08 | 0.85 |
| Plt (<96,000 × 103/uL) | 49 | 0.92 | 0.58–1.44 | 0.70 | 1.38 | 0.83–2.27 | 0.21 | 30 | 1.16 | 0.61–2.22 | 0.64 | 1.20 | 0.60–2.41 | 0.61 |
| Lineage of blasts (myeloid) | 64 | 2.59 | 1.53–4.39 | 3.8 × 10−4*** | — | — | — | 33 | 2.08 | 1.06–4.07 | 3.3 × 10−2* | — | — | — |
| Prior history of TKI before BC | 36 | 0.93 | 0.58–1.51 | 0.78 | 1.05 | 0.64–1.73 | 0.84 | 26 | 1.41 | 0.74–2.70 | 0.29 | 0.70 | 0.32–1.56 | 0.39 |
| TKI-based therapy for BC | 59 | 0.36 | 0.23–0.58 | 2.0 × 10−5*** | — | — | — | 59 | — | — | — | — | — | — |
| Mutations | ||||||||||||||
| ABL1 | 13 | 1.11 | 0.58–2.11 | 0.75 | 1.17 | 0.62–2.23 | 0.63 | 8 | 1.55 | 0.67–3.55 | 0.30 | 1.09 | 0.46–2.59 | 0.85 |
| ASXL1 | 23 | 2.09 | 1.26–3.47 | 4.4 × 10−3** | 1.30 | 0.76–2.25 | 0.34 | 11 | 2.31 | 1.08–4.91 | 3.0 × 10−2* | 2.61 | 1.07–6.37 | 3.4 × 10−2* |
| BCOR | 5 | 1.58 | 0.63–3.96 | 0.33 | 1.75 | 0.69–4.42 | 0.24 | 4 | 2.99 | 1.03–8.70 | 4.4 × 10−2* | 2.57 | 0.84–7.85 | 9.7 × 10−2 |
| BCORL1 | 5 | 1.41 | 0.57–3.49 | 0.46 | 1.34 | 0.53–3.41 | 0.54 | 3 | 1.44 | 0.44–4.72 | 0.55 | 0.90 | 0.27–3.07 | 0.87 |
| CDKN2A/B | 7 | 0.95 | 0.41–2.19 | 0.91 | 1.84 | 0.70–4.87 | 0.22 | 5 | 1.97 | 0.75–5.17 | 0.17 | 2.82 | 0.87–9.13 | 8.4 × 10−2 |
| IKZF1 | 12 | 0.61 | 0.27–1.41 | 0.25 | 1.03 | 0.42–2.53 | 0.95 | 10 | 1.09 | 0.42–2.81 | 0.86 | 1.87 | 0.65–5.35 | 0.24 |
| RUNX1 | 28 | 1.02 | 0.63–1.67 | 0.93 | 1.06 | 0.64–1.76 | 0.81 | 21 | 1.45 | 0.76–2.77 | 0.26 | 1.68 | 0.86–3.27 | 0.13 |
| TP53 | 8 | 1.62 | 0.74–3.53 | 0.23 | 1.46 | 0.66–3.22 | 0.35 | 3 | 0.90 | 0.22–3.75 | 0.88 | 0.80 | 0.19–3.36 | 0.76 |
| CNAs | ||||||||||||||
| +Ph | 26 | 1.44 | 0.87–2.36 | 0.16 | 1.28 | 0.78–2.12 | 0.33 | 14 | 1.71 | 0.84–3.46 | 0.14 | 1.46 | 0.72–2.97 | 0.29 |
| Complex CNAs | 28 | 1.65 | 1.02–2.67 | 4.3 × 10−2* | 1.66 | 1.00–2.73 | 4.8 × 10−2* | 16 | 2.99 | 1.55–5.78 | 1.1 × 10−3** | 3.37 | 1.69–6.69 | 5.3 × 10−4*** |
| Hyperdiploidy | 6 | 4.38 | 1.84–10.5 | 8.7 × 10−4*** | 2.28 | 0.93–5.57 | 7.0 × 10−2 | 1 | 10.99 | 1.28–94.1 | 2.9 × 10−2* | 5.90 | 0.67–51.7 | 0.11 |
| +6 | 6 | 0.91 | 0.33–2.52 | 0.86 | 0.75 | 0.27–2.08 | 0.58 | 2 | 0.63 | 0.08–4.65 | 0.65 | 0.21 | 0.03–1.68 | 0.14 |
| −7/del(7p) | 13 | 0.89 | 0.46–1.74 | 0.74 | 2.41 | 1.06–5.45 | 3.5 × 10−2* | 9 | 0.84 | 0.35–2.01 | 0.69 | 1.63 | 0.56–4.72 | 0.37 |
| +8 | 18 | 1.29 | 0.72–2.32 | 0.39 | 0.91 | 0.50–1.65 | 0.76 | 8 | 1.08 | 0.42–2.80 | 0.88 | 0.46 | 0.16–1.32 | 0.15 |
| −9/del(9p) | 9 | 0.87 | 0.40–1.89 | 0.72 | 1.30 | 0.52–3.25 | 0.57 | 5 | 1.97 | 0.75–5.17 | 0.17 | 2.82 | 0.87–9.13 | 8.4 × 10−2 |
| del(17p) | 13 | 2.78 | 1.50–5.18 | 1.2 × 10−3** | 1.78 | 0.94–3.38 | 7.6 × 10−2 | 5 | 4.54 | 1.63–12.6 | 3.8 × 10−3** | 2.86 | 0.97–8.44 | 5.6 × 10−2 |
| amp(17q) | 7 | 1.98 | 0.90–4.34 | 8.8 × 10−2 | 2.44 | 1.10–5.45 | 2.9 × 10−2* | 5 | 2.59 | 1.00–6.73 | 5.0 × 10−2 | 2.62 | 0.98–6.99 | 5.5 × 10−2 |
| i(17q) | 4 | 2.81 | 1.00–7.84 | 4.9 × 10−2* | 2.79 | 0.98–7.91 | 5.4 × 10−2 | 3 | 5.50 | 1.58–19.2 | 7.5 × 10−3** | 2.89 | 0.78–10.7 | 0.11 |
| +19 | 13 | 2.85 | 1.54–5.27 | 8.4 × 10−4*** | 1.66 | 0.87–3.18 | 0.12 | 4 | 4.48 | 1.49–13.5 | 7.5 × 10−3** | 3.09 | 0.98–9.71 | 5.3 × 10−2 |
| +21 | 11 | 2.63 | 1.37–5.05 | 3.6 × 10−3** | 1.69 | 0.87–3.31 | 0.12 | 5 | 3.93 | 1.49–10.4 | 5.7 × 10−3** | 3.23 | 1.16–8.97 | 2.4 × 10−2* |
Univariate analysis for OS by Cox proportional hazard regression model. For all cases (n = 99), values adjusted for lineage of blasts and TKI-containing therapy for BC are also shown. For TKI treated cases (n = 59), values adjusted for WBC and lineage of blasts are also shown. Information for prior history of TKI is missing in two patients treated by TKI for BC.
95% CI 95% confidence interval, HR hazard ratio.
*P < 0.05; **P < 0.01; ***P < 0.001.
We next analysed survival, focusing on 59 TKI-treated patients, because TKI-based therapy has been shown to significantly improve OS and thus, is the current therapy of choice for patients with CML-BC1,18,26. In the univariate analysis, WBC count, lineage of blasts, ASXL1 and BCOR mutations, complex CNAs, del(17p), i(17q), +19, and +21 were significantly associated with OS (Supplementary Fig. 9a). We also evaluated the relative effects of genetic alterations using Cox proportional hazard regression modelling with a standard backward selection of clinical and genetic variables, and identified ASXL1 mutations, complex CNAs, i(17q), and +21 as independent predictors of worse prognosis (Fig. 5b). To internally validate this finding, modelling was performed 100 times by conducting the bootstrap, in which all four variables of the final model were selected at a frequency of >70%, with a mean concordance statistic of 0.74 (Supplementary Fig. 9b). Based on the number of these unfavourable factors, TKI-treated BC patients were classified into three subgroups showing distinct prognosis, where the 2-year OS rate was 65.0%, 17.1%, and 0% for patients with 0, 1, and ≥2 unfavourable genetic risk factors, respectively (P = 3.9 × 10−12; Fig. 5c). We also analysed an independent external cohort reported in a recent publication13, in which 17 CML-BC patients were evaluable for survival with 12 receiving TKI-based therapy. Although the number of cases was limited, several similar associations were observed in the external cohort. TKI-based therapy was associated with better OS, while ASXL1 mutations and i(17q) predicted poor prognosis (Supplementary Fig. 10a). Even though the difference was not statistically significant, owing to the small number of samples analysed, patients with genetic risk factors tended to show a poor prognosis (Supplementary Fig. 10b). Therefore, our results suggest that genetic risk factors may help identify a subset of patients, who may be refractory to TKI therapy.
Genetic landscape and clinical outcome of CML-CP
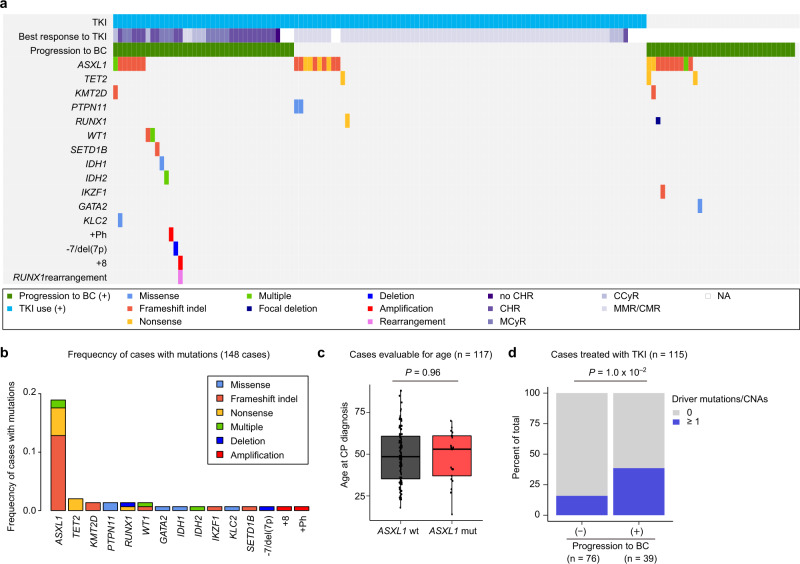
Finally, we explored the effect of genetic abnormalities on the clinical outcomes of CML-CP patients. Our cohort contained more patients who ultimately developed BC (48%, 71/148) compared to other cohorts, because we intentionally included paired CP and BC samples to investigate the molecular pathogenesis of the clonal evolution in CML (Fig. 1, Fig. 6a, and Supplementary Table 3). The median age at diagnosis for patients with CP was 49 (14–88) years and 77.7% of the patients had received TKI therapy, with imatinib being the most commonly used therapeutic agent (80%). Based on the best response, TKI induced complete haematologic response (CHR) in 17.9%, major/complete cytogenetic response (MCyR/CCyR) in 16.9%, and major/complete molecular response (MMR/CMR) in 64.2% of the patients.
Fig. 6. Genetic landscape of CML-CP.
a Summary of genetic lesions in 148 CP patients. Each column indicates one patient. Clinical annotations including TKI usage for CP treatment, best response to TKI, and progression to BC are also shown. NA not available. b Frequencies of mutations in 148 CP patients. Recurrently mutated or known driver genes are described. Categories of mutations are depicted in different colours, and “multiple” indicates ≥2 distinct mutations found in the same gene in the same patient. c Box plot showing age at CP diagnosis according to the presence of ASXL1 mutations. The median, first, and third quartiles are indicated and whiskers extend to the furthest value within 1.5× the interquartile range. The two-sided Wilcoxon rank-sum test was used to calculate the P value. wt wild type, mut mutated. d Proportion of cases harbouring any genetic abnormalities according to progression to BC. P values were evaluated using the Fisher’s exact test.
In total, additional genetic alterations were found in 25.7% of the patients with CP at diagnosis. As expected based on the analysis described above (Fig. 1), only ASXL1 was mutated in CP at a frequency comparable to that in BC, while other mutations, including TET2, KMT2D, PTPN11, RUNX1, and WT1, were found at much lower frequencies compared to those in BC (Fig. 6a, b, Supplementary Table 6, and Supplementary Data 2). Since, only ASXL1 mutations were frequently found in CP and are common in age-related clonal haematopoiesis27, we evaluated the correlation between ASXL1 mutations and patient age. Consistent with previous reports10,13,28, many patients with ASXL1 mutations were younger than 60 years at the time of diagnosis of CP, as opposed to those with age-related clonal haematopoiesis. Moreover, there was no significant impact of age on the frequency of ASXL1 mutations in CML-CP patients (Fig. 6c). We also evaluated whether genetic alterations in CP could predict BC progression in patients treated with TKIs. Interestingly, patients who received TKI and later experienced BC progression often harboured at least one mutation and/or CNA (38.5%, 15/24) than those without TKI treatment (15.8%, 12/76; P = 1.0 × 10−2; Fig. 6d). Thus, even when rarely observed, genetic alterations in CP play a role in driving CML cells to undergo transformation to BC under TKI pressure, which is in agreement with previous reports10,29,30.
Discussion
Based on a comprehensive genetic analysis of a large cohort of CML patients with detailed clinical information and longitudinal sampling, we demonstrated the genetic landscape, clonal evolution, and their relevance to clinical features and outcomes in CML-CP/BC. Although the development of TKIs has substantially improved the outcomes of patients with CML-BC, the prognosis of BC patients remains bleak. To improve the clinical outcome of BC patients, it may be useful to stratify patients according to reliable prognostic markers to enable better clinical decision-making. We demonstrated that certain genetic lesions were better predictors of survival than clinical parameters. For instance, patients with multiple-hit TP53 mutations and i(17q) presented with a particularly dismal outcome, which was consistent with previous studies reporting poor outcomes in TP53-mutated patients with other myeloid neoplasms, such as acute myeloid leukaemia and myelodysplastic syndromes19,25,31–33, and in CML-BC patients with i(17q)34–36. Moreover, ASXL1 mutations, complex CNAs, i(17q), and +21 were independent predictors of poor prognosis in patients receiving TKI-based therapy based on multivariate analysis. Despite the limited number of samples analysed, our results indicate that genetic abnormalities can be effectively used as biomarkers to predict the outcome of BC patients to guide clinical decisions. As the ABL1 mutation itself was not associated with a worse prognosis of CML-BC and was almost always accompanied by other mutations, targeting other mutations in combination with TKIs may be a promising treatment strategy in the future. Of note, sensitivity to various drugs is associated with the mutational profile of patients with acute myeloid leukaemia37, raising the possibility that in the near future, treatment may be personalised depending on individual mutational profiles.
Exome analysis of serial samples revealed that CML cells accumulated somatic mutations over time, which was markedly suppressed by TKI-based therapy. Although the precise reason for this remains unclear, it is possible that the application of TKIs eliminated the rapidly cycling BCR-ABL1-harbouring cells, ignoring the slow-cycling cells, which are less likely to ameliorate the impact of somatic mutations. The TKI-mediated reduction in blastic transformation may be attributable to a drastic reduction in the number of tumour cells at a risk of acquiring driver mutations for BC. However, our results suggest that the suppression of random mutational events may contribute to the reduced risk of BC in TKI-treated patients.
As previously reported3–17, most CML-BC patients in our cohort acquired additional mutations, which not only included mutations previously reported in haematological malignancies and CML-BC, but also included previously unreported recurrent mutations, such as NBEAL2, PHIP, and KLC2 mutations. NBEAL2 encodes a protein with a role in megakaryocyte alpha-granule biogenesis and has been reported to be mutated in patients with Grey platelet syndrome38–40 and factor XIII deficiency41. In contrast, to the best of our knowledge, mutations in PHIP and KLC2 have not been reported in the context of haematological disorders. We also identified a RUNX1-ETS2 fusion. RUNX1 rearrangements are well-known driver events, which were first identified in core-binding factor leukaemia (RUNX1-RUNX1T1) and are present in a variety of myeloid and lymphoid malignancies, which are represented by RUNX1-MECOM in CML-BC, ETV6-RUNX1 in B-precursor acute lymphoblastic leukaemia, and other fusions with unknown partners42. The fusion discovered in our cohort is another RUNX1 rearrangement involving ETS2, which encodes a key haematopoietic transcription factor. Considering that genetic alterations in RUNX1 are not restricted to SNVs, but also appear as CNAs and SVs, application of NGS-based techniques to detect multiple variant types may be important to monitor cases in CML patients. Among the mutations detected in CML, ASXL1 mutations are more likely to be present at the time of CP diagnosis, and showed comparable TCFs between CP and BC samples. Thus, ASXL1 mutations are unique compared to other mutations, as they were rarely detected in CP samples, but expanded with the onset of BC. Since, patients with CP who subsequently developed BC were more likely to harbour mutations, such as ASXL1 mutations, compared to those who did not harbour such mutations, and because BC patients with ASXL1 mutations had a poor prognosis, CP patients harbouring ASXL1 mutations may require careful management. As most patients received imatinib as the first-line TKI for CP in our cohort, second- or third-generation TKIs may be better candidates for therapy to improve the outcomes of these patients, as suggested by a recent study29. As expected, myeloid and lymphoid BC cases exhibited distinct molecular profiles, which was in agreement with a previous report that patients with CML-BC presented with distinct additional chromosomal alterations depending on the lineage of BC11. The genetic profile of CML-BC is also influenced by a prior history of TKI therapy before BC diagnosis, which may explain, at least in part, the differences in the genetic profiles of BC patients. In summary, our study demonstrates the diverse mutational profiles and clonal evolution of CML in a large cohort and bridges genetic abnormalities and clinical features, including outcomes. Our results will hopefully lead to the development of efficacious therapy and strategies for better management of patients with CML.
Methods
Patient samples
We performed WES, targeted capture sequencing, and/or deep amplicon sequencing, using 112 CML-BC and 71 CP samples at diagnosis from 130 patients at ten institutions enroled in this study, according to the protocols approved by the Institutional Review Boards (Fig. 1a and Supplementary Table 1). This study was approved by the institutional ethics committees of Kyoto University, Kobe City Medical Center General Hospital, Tokyo Medical University, Akita University Graduate School of Medicine, Juntendo University School of Medicine, Gifu University Hospital, Kurashiki Central Hospital, Hyogo College of Medicine, Dokkyo Medical University, the University of Tokyo (Japan), and Chang Gung Memorial Hospital-Linkou (Taiwan), and was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants. Clinical information from these patients was collected if available, and cases of 99 patients with CML-BC were assessed for survival. Combined with the external WES data of 24 BC12,13 and 77 CP13,21,22 patients, we comprehensively analysed a total of 136 BC and 148 CP samples obtained from 216 CML patients for SNVs, CNAs, and SVs (Fig. 1a and Supplementary Table 1). Peripheral blood or bone marrow samples and matched buccal samples (if available) were collected from the patients who participated in this study. Genomic DNA from the samples was extracted using the QIAamp DNA mini kit (Qiagen) or Genetra PureGene kit (Qiagen). RNA from the samples was extracted using Trizol (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA).
Detection and quantification of fusion transcripts
RT-PCR for detection of BCR-ABL1 fusion transcripts43 and RT-qPCR with TaqMan assay for BCR-ABL1 (ref. 44) was conducted with our lab-specific conversion factor obtained from the international reference laboratory at Adelaide, Australia for International Scale (IS) calculation. BCR-ABL1 levels were expressed as IS45, along with a log reduction value. SYBR-green RT-qPCR was performed using the ABI 7900HT system (Applied Biosystems, Foster City, CA) for RUNX1-ETS2 determination using freshly frozen samples, identical to those used for quantification of the BCR-ABL1 transcript, according to the manufacturer’s instructions. The primer set for the measurement of the RUNX1-ETS2 transcript comprised RUNX1-ETS2-QF (5′-CTTCACAAACCCACCGCAAG-3′) and RUNX1-ETS2-QR (5′-AGGGAGTCTGAGCTCTCGAAG-3′) with the same ABL1 internal control gene for BCR-ABL1. Each RT-qPCR reaction was performed in duplicate and the relative expression after analysis of follow-up samples was compared with that of the diagnostic sample, using comparative quantification (2−ΔΔCt method) and expressed as log reduction.
Whole-exome sequencing
WES was performed using the SureSelect Human All Exon kit V6 (Agilent Technology, Santa Clara, CA, USA), according to the manufacturer’s instructions. Captured targets were sequenced using the HiSeq 2500, NovaSeq 6000 (Illumina), or DNBSEQ G400 (MGI) instrument with a standard 125- or 150-bp paired-end read protocol46. Sequence alignment and mutation calling were performed using the hg19 reference genome and Genomon pipeline (https://github.com/Genomon-Project). For analysis of recurrent mutations, data on mutations were called for (i) BC samples using either CP or germline samples (if available) as control, and (ii) CP samples using germline samples as controls (if available). Data on putative somatic mutations with (i) Fisher’s exact P value <0.05; (ii) P value for EBCall47 < 0.0001, were filtered by excluding (i) synonymous SNVs; (ii) variants occurring in repetitive genomic regions; and (iii) known single-nucleotide polymorphisms (SNPs) listed in the 1000 Genomes Project (October 2014 release), NCBI dbSNP build 138, National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) 6500, or our in-house dataset. In addition, (i) pathogenic mutations found in >1 haematological malignancies in the COSMIC database (v84); (ii) mutations expected to cause premature termination of protein translation (frameshift or nonsense mutations), were reviewed using a less stringent filter with (i) Fisher’s exact P value <0.1; (ii) P value for EBCall < 0.01, and data were manually curated. Mapping errors were removed by performing visual inspection using the Integrative Genomics Viewer (IGV). For conducting paired analysis of CP and BC samples, data on the putative recurrent mutations were subjected to validation using deep amplicon sequencing in both CP and BC samples, and mutations with VAF ≥ 0.02 were considered to be validated. Primers used for deep amplicon sequencing are summarised in Supplementary Table 7. All variants validated by deep amplicon sequencing were found to possess VAF > 0.03 in the WES data. For cases without germline controls, we also independently analysed the WES data of the CP and BC samples without using controls, and filtered data on the candidate mutations using the criteria used for performing targeted capture sequencing, as described below to rescue potentially overlooked recurrent mutations already present at the time of CP diagnosis. By doing so, we created three mutation lists for cases analysed for both CP and BC without germline controls, i.e., somatic mutations acquired in BC compared to CP, and independently called candidate recurrent mutations in CP and BC. We then merged the mutation lists to generate a single list for each BC sample. To analyse the number of SNVs acquired during disease progression (Fig. 1b and Supplementary Fig. 1b, d), data on the mutations were called for BC samples using CP as control and data on somatic mutations with (i) Fisher’s exact P value <0.01; (ii) VAF > 0.05; (iii) P value for EBCall < 0.001, were filtered by excluding (i) variants occurring in repetitive genomic regions; (ii) SNPs listed in the database as described above. For the external cohort13, WES data of both CP and BC samples were available for 15 patients. Of these, 13 were subjected to analysis after excluding 1 case, which lacked information on progression time from CP to BC and 1 in which a much lower depth was observed compared to the other samples. The estimated TCFs harbouring the relevant mutation were calculated with the total copy number (TCN) of the region and observed VAF values as follows19; TCF = TCN × VAF for deletions, TCF = 2VAF for regions without copy-number changes, and TCF = TCN × VAF for gains.
Targeted capture sequencing
Targeted capture sequencing was performed using the SureSelect custom kit (Agilent Technologies), for which 104 genes were selected from those found to be mutated in the WES data of 52 CML-CP and BC pairs, and/or known oncogenes or tumour suppressor genes in haematological malignancies. Sequencing, alignment, and mutation calling were performed as per the WES analysis, except for the filtering criteria. Relevant somatic mutation data with (i) VAF > 0.05; (ii) depth > 100; (iii) P value for EBCall < 0.0001, were filtered by exclusion based on (i) synonymous SNVs; (ii) variants present only in unidirectional reads; (iii) variants occurring in repetitive genomic regions; (iv) missense SNVs with VAF of 0.4–0.6 or <0.04; and (v) known variants listed in SNP databases (as described in the “Whole-exome sequencing” section). In addition, (i) pathogenic mutations found in >1 haematological malignancies in the COSMIC database (v84) and (ii) mutations expected to cause premature termination of protein translation were reviewed by using a less stringent filter with (i) VAF > 0.02; (ii) depth >50; (iii) P value for EBCall > 0.001, and data were manually curated.
CNA and SV analysis
CNAs and SVs were detected using the CNACS algorithm and Genomon-SV pipeline, respectively19,20. Briefly, CNACS analyses sequencing depth and allele frequencies of heterozygous SNPs to determine genome-wide copy numbers and detect CNAs19. We also used the ExomeDepth package in R48 to detect microdeletion events occurring in the exons of the IKZF1 and RUNX1 genes. CNA data were manually curated using IGV. The Genomon-SV pipeline detects SVs by utilising both breakpoint-containing junction read pairs and improperly aligned read pairs20. Data on putative SVs detected by using the Genomon pipeline were filtered by removing (i) those with Fisher’s exact P value >0.03 and (ii) those present in control normal samples, whose breakpoints were manually inspected using IGV. Since sequencing reads are enriched in gene exons and target genes in WES and targeted capture sequencing, respectively, detection of SV breakpoints is limited to regions close to those covered by gene baits. Nevertheless, a few SVs were detected even when they occurred in intronic regions. For instance, our pipeline could detect a RUNX1-ETS2 fusion as the breakpoint was observed in the intronic region close to the ETS2 gene exon (Supplementary Fig. 3a). Moreover, we were able to identify the BCR-ABL1 fusion gene with the typical breakpoint, in the intronic region, through targeted capture sequencing and WES. Through targeted capture sequencing, we identified BCR-ABL1 fusions in 51 out of 60 cases (85%); however, only 4 out of 76 cases (5.3%) were detected by WES. Therefore, the ability to detect SVs in our pipeline largely depended on the location of breakpoints and gene baits. Rearrangements of immunoglobulin and T-cell receptors were detected as microdeletion events involving loci observed in the WES analysis. Complex CNAs were defined as the presence of ≥3 abnormal CNAs detected by CNACS. We considered the co-amplification of 9q and 22q derived from +Ph abnormality as a single event. Hyperdiploidy and hypodiploidy were defined as two or more gains (≥48) and losses (≤44) of chromosomes assessed by CNACS, respectively, and data on these two aspects were removed from those on complex CNAs.
Statistical analysis
Statistical analyses were performed using R (v3.5.0). Comparisons between groups were based on the two-sided Wilcoxon rank-sum test for continuous data and the Fisher’s exact test for categorical data. The correlation between the number of acquired mutations and time necessary for progression from CP to BC (Fig. 1b) was assessed by using the Poisson regression model and the glm function in R. Pairwise correlation of genetic legions and clinical factors (Supplementary Figs. 6 and 8) was assessed by the Fisher’s exact test with Benjamini–Hochberg correction49. Molecular responses for CML were defined as follows: haematologic remission, IS ≥ 10% for CHR, IS < 10% for MCyR, IS < 1% for CCyR, IS < 0.1% for MMR, and IS < 0.0032% for CMR. Survival analysis was performed for 99 patients with CML-BC for whom survival and treatment data for BC were available, and observations were censored at the last follow-up. The median follow-up was 3.2 years in surviving patients, and 24 (24.2%) patients were alive at the last follow-up. The Kaplan–Meier method was used to estimate the OS, and the differences in OS were assessed using the survival package in R. The effects of genetic lesions on OS were evaluated by using the Cox proportional hazards regression model, and data were adjusted for clinical factors that were significantly associated with OS in the univariate analysis. For blood counts, the following rounded median values were used as the threshold: 50,000 (×103/uL) for WBC counts, 9.3 (g/dL) for haemoglobin levels (Hb), and 96,000 (×103/uL) for platelet counts (PLT). Old age was defined as age of a patient ≥60 years, as per a previous study18. Multivariate analysis was performed for patients treated with TKI-based regimens (n = 59) by performing Cox proportional hazards regression modelling with a stepwise selection of variables using P value to exclude variables, in which genetic or clinical factors with a univariate Cox P value <0.10 were considered. To evaluate the validity of the established model, we also conducted bootstrapping 100 times to construct the test models with the factors subjected to the multivariate modelling using the rms package in R. Each model was assessed for validity by calculating the concordance statistic and frequency of each covariate included in the models. All P values were calculated using two-sided tests, and P < 0.05 was considered to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank Maki Nakamura, Takeshi Shirahari, Kazuhide Onishi, and Satoko Yabuta (Department of Pathology and Tumor Biology, Kyoto University) for providing technical assistance. This work was supported by the Grant-in-Aid for JSPS KAKENHI (26221308, 26253060, JP19H05656: to S.O., JP17J05245: to Y.O.), Grant-in-Aid for Scientific Research on Innovative Areas (15H05909, 15H05912: to S.O. and S.M.), grants from AMED (JP19cm0106501h0004, JP19km0405207h9904: to S.O.), MEXT (hp180198, hp190158, hp190179: to S.O., hp200138: S.M. and S.O.), “Stem Cell Aging and Disease” (14430052: to M.S.), Takeda Science Foundation (to S.O.), and JSPS Core-to-Core Programme (to S.O.), Japan, and grants from the Ministry of Science and Technology, Taiwan (MOST108-2314-B-182-006: to L.-Y.S.), and the Ministry of Health and Welfare, Taiwan (MOHW109-TDU-B-212-134011: to L.-Y.S.). S.B. is supported by the National Health and Medical Research Council of Australia APP1027531 and APP1104425, the Ray and Shirl Norman Cancer Research Trust, and the Royal Adelaide Hospital Research Foundation.
Author contributions
Y.O., K.Y., S.O., and L.-Y.S designed the study. Y.-J.H., M.-C.K., K.S., K.M., N. Hosoya, N. Hiramoto, T.I., S.B., N.S., K.O., N.T., T.T., S.T., N.K., N.N., Y.U., and S.Y. managed the patients and prepared samples. Y.O., Y.-J.H., R.B., and L.Z. performed the experiments. Y.O., K.Y., Y.-J.H., Y.N., Y. Shiozawa, J.T., Y.W., R.O., H.M., M.S., and A.T.-K. analysed the data. S.M., Y. Shiraishi, K.C., and H.T. developed sequence data processing pipelines. Y.O., S.O., and L.-Y.S. wrote the manuscript.
Data availability
The WES data in this study are deposited in the European Genome-phenome Archive under accession code EGAS00001005075. The data is available under restricted access, and access can be obtained by contacting S.O. (sogawa-tky@umin.ac.jp). The public WES data used in this study are available in the European Genome-phenome Archive under accession code EGAS00001003071, and the European Nucleotide Archive under accession code PRJEB20846 (Supplementary Table 8). The remaining data are available within the article, Supplementary Information, or available from the authors upon request.
Competing interests
S.B. is a member of the advisory boards of Qiagen, Novartis, and Cepheid and has received honoraria from Qiagen, Novartis, Bristol-Myers Squibb, and Cepheid, as well as research support from Novartis. The remaining authors declare no competing interests relevant to this study.
Footnotes
Peer review information Nature Communications thanks Susan Branford, David Kent and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Seishi Ogawa, Email: sogawa-tky@umin.ac.jp.
Lee-Yung Shih, Email: sly7012@adm.cgmh.org.tw.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-23097-w.
References
- 1.Hehlmann R. How I treat CML blast crisis. Blood. 2012;120:737–747. doi: 10.1182/blood-2012-03-380147. [DOI] [PubMed] [Google Scholar]
- 2.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 3.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 4.Hosoya N, et al. Genomewide screening of DNA copy number changes in chronic myelogenous leukemia with the use of high-resolution array-based comparative genomic hybridization. Genes Chromosomes Cancer. 2006;45:482–494. doi: 10.1002/gcc.20303. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 6.Nowak D, et al. SNP array analysis of tyrosine kinase inhibitor-resistant chronic myeloid leukemia identifies heterogeneous secondary genomic alterations. Blood. 2010;115:1049–1053. doi: 10.1182/blood-2009-03-210377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makishima H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menezes J, et al. ASXL1, TP53 and IKZF3 mutations are present in the chronic phase and blast crisis of chronic myeloid leukemia. Blood Cancer J. 2013;3:e157. doi: 10.1038/bcj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt M, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28:2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 10.Kim T, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017;129:38–47. doi: 10.1182/blood-2016-04-708560. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, et al. Cytogenetic landscape and impact in blast phase of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Leukemia. 2017;31:585–592. doi: 10.1038/leu.2016.231. [DOI] [PubMed] [Google Scholar]
- 12.Kim T, et al. Exome sequencing reveals DNMT3A and ASXL1 variants associate with progression of chronic myeloid leukemia after tyrosine kinase inhibitor therapy. Leuk. Res. 2017;59:142–148. doi: 10.1016/j.leukres.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Branford S, et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood. 2018;132:948–961. doi: 10.1182/blood-2018-02-832253. [DOI] [PubMed] [Google Scholar]
- 14.Magistroni V, et al. De novo UBE2A mutations are recurrently acquired during chronic myeloid leukemia progression and interfere with myeloid differentiation pathways. Haematologica. 2019;104:1789–1797. doi: 10.3324/haematol.2017.179937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adnan Awad S, et al. Mutation accumulation in cancer genes relates to nonoptimal outcome in chronic myeloid leukemia. Blood Adv. 2020;4:546–559. doi: 10.1182/bloodadvances.2019000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko TK, et al. An integrative model of pathway convergence in genetically heterogeneous blast crisis chronic myeloid leukemia. Blood. 2020;135:2337–2353. doi: 10.1182/blood.2020004834. [DOI] [PubMed] [Google Scholar]
- 17.Branford S, et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia. 2019;33:1835–1850. doi: 10.1038/s41375-019-0512-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain P, et al. Prognostic factors and survival outcomes in patients with chronic myeloid leukemia in blast phase in the tyrosine kinase inhibitor era: cohort study of 477 patients. Cancer. 2017;123:4391–4402. doi: 10.1002/cncr.30864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshizato T, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129:2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kataoka K, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 2015;47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 21.Mitani K, et al. Somatic mosaicism in chronic myeloid leukemia in remission. Blood. 2016;128:2863–2866. doi: 10.1182/blood-2016-06-723494. [DOI] [PubMed] [Google Scholar]
- 22.Togasaki E, et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7:e559. doi: 10.1038/bcj.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asnafi AA, Deris Zayeri Z, Shahrabi S, Zibara K, Vosughi T. Chronic myeloid leukemia with complex karyotypes: Prognosis and therapeutic approaches. J. Cell Physiol. 2019;234:5798–5806. doi: 10.1002/jcp.27505. [DOI] [PubMed] [Google Scholar]
- 24.Grossmann V, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25:557–560. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- 25.Bernard E, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020;26:1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochhaus A, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34:966–984. doi: 10.1038/s41375-020-0776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst T, et al. Frequent ASXL1 mutations in children and young adults with chronic myeloid leukemia. Leukemia. 2018;32:2046–2049. doi: 10.1038/s41375-018-0157-2. [DOI] [PubMed] [Google Scholar]
- 29.Nteliopoulos G, et al. Somatic variants in epigenetic modifiers can predict failure of response to imatinib but not to second-generation tyrosine kinase inhibitors. Haematologica. 2019;104:2400–2409. doi: 10.3324/haematol.2018.200220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mologni L, Piazza R, Khandelwal P, Pirola A, Gambacorti-Passerini C. Somatic mutations identified at diagnosis by exome sequencing can predict response to imatinib in chronic phase chronic myeloid leukemia (CML) patients. Am. J. Hematol. 2017;92:E623–E625. doi: 10.1002/ajh.24865. [DOI] [PubMed] [Google Scholar]
- 31.Lindsley RC, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N. Engl. J. Med. 2017;376:536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porta MGD, et al. Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J. Clin. Oncol. 2016;34:3627–3637. doi: 10.1200/JCO.2016.67.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley TJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fioretos T, et al. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood. 1999;94:225–232. doi: 10.1182/blood.V94.1.225.413k24_225_232. [DOI] [PubMed] [Google Scholar]
- 35.Kanagal-Shamanna R, et al. Myeloid neoplasms with isolated isochromosome 17q represent a clinicopathologic entity associated with myelodysplastic/myeloproliferative features, a high risk of leukemic transformation, and wild-type TP53. Cancer. 2012;118:2879–2888. doi: 10.1002/cncr.26537. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, et al. Risk stratification of chromosomal abnormalities in chronic myelogenous leukemia in the era of tyrosine kinase inhibitor therapy. Blood. 2016;127:2742–2750. doi: 10.1182/blood-2016-01-690230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyner JW, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunay-Aygun M, et al. NBEAL2 is mutated in gray platelet syndrome and is required for biogenesis of platelet α-granules. Nat. Genet. 2011;43:732–734. doi: 10.1038/ng.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahr WH, et al. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat. Genet. 2011;43:738–740. doi: 10.1038/ng.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albers CA, et al. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat. Genet. 2011;43:735–737. doi: 10.1038/ng.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia S, et al. Identification of novel pathogenic F13A1 mutation and novel NBEAL2 gene missense mutation in a pedigree with hereditary congenital factor XIII deficiency. Gene. 2019;702:143–147. doi: 10.1016/j.gene.2019.03.067. [DOI] [PubMed] [Google Scholar]
- 42.Stengel A, et al. Detection of recurrent and of novel fusion transcripts in myeloid malignancies by targeted RNA sequencing. Leukemia. 2018;32:1229–1238. doi: 10.1038/s41375-017-0002-z. [DOI] [PubMed] [Google Scholar]
- 43.van Dongen JJ, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 concerted action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 44.Gabert J, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 45.Shih LY, et al. Emerging kinetics of BCR-ABL1 mutations and their effect on disease outcomes in chronic myeloid leukemia patients with imatinib failure. Leuk. Res. 2013;37:43–49. doi: 10.1016/j.leukres.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 47.Shiraishi Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plagnol V, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki H, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The WES data in this study are deposited in the European Genome-phenome Archive under accession code EGAS00001005075. The data is available under restricted access, and access can be obtained by contacting S.O. (sogawa-tky@umin.ac.jp). The public WES data used in this study are available in the European Genome-phenome Archive under accession code EGAS00001003071, and the European Nucleotide Archive under accession code PRJEB20846 (Supplementary Table 8). The remaining data are available within the article, Supplementary Information, or available from the authors upon request.