Abstract
Salmonids are important sources of protein for a large proportion of the human population. Mycoplasma species are a major constituent of the gut microbiota of salmonids, often representing the majority of microbiota. Despite the frequent reported dominance of salmonid-related Mycoplasma species, little is known about the phylogenomic placement, functions and potential evolutionary relationships with their salmonid hosts. In this study, we utilise 2.9 billion metagenomic reads generated from 12 samples from three different salmonid host species to I) characterise and curate the first metagenome-assembled genomes (MAGs) of Mycoplasma dominating the intestines of three different salmonid species, II) establish the phylogeny of these salmonid candidate Mycoplasma species, III) perform a comprehensive pangenomic analysis of Mycoplasma, IV) decipher the putative functionalities of the salmonid MAGs and reveal specific functions expected to benefit the host. Our data provide a basis for future studies examining the composition and function of the salmonid microbiota.
Subject terms: Bacterial host response, Applied microbiology, Bacterial genomics, Environmental microbiology
Jacob Rasmussen et al. use metagenomic analyses to examine the diversity of Mycoplasma species among three commonly-fished species of salmonids. Their results establish a phylogeny of candidate salmonid related Mycoplasma species and suggest a mutualistic relationship between these microbial species and salmon hosts.
Introduction
The microbial communities that inhabit the vertebrate gastrointestinal tract are tightly connected to many traits displayed by the host1–4. Nonetheless, we are far from understanding the ecological and evolutionary forces that structure these microbial communities. Microbial generalists have been investigated in teleosts to decipher these ecological forces5. Recently, several reports using 16S rRNA gene amplicon surveys of the gut microbiome in salmonids have revealed that the dominant bacterium belongs to the genus Mycoplasma, and that this bacterium was unknown until recently6–8. These salmonid related Mycoplasma species have been shown to be highly dominant in the gastrointestinal microbiota of all salmonids investigated, including rainbow trout (Oncorhynchus mykiss)6,9,10, Chinook salmon (Oncorhynchus tshawytscha)7 and Atlantic salmon (Salmo salar)11–13. Further, phenotypic evidence now points to an advantageous role of the abundant Mycoplasma including increased disease resiliency revealed by a striking inverse correlation between the abundance of Mycoplasma and a potentially pathogenic Vibrio sp.6.
Mycoplasma is one of the smallest independently self-replicating organisms known. The small genome size of Mycoplasma spp. is hypothesised to be a result of close interaction with their host, which has resulted in subsequent gene loss14,15. Mycoplasma species are bacteria lacking a cell wall and are in general characterised by small physical dimensions and a small genome. Most host-associated Mycoplasma genomes currently sequenced are smaller than 1 Mb and contain fewer than 1000 protein-encoding genes16–18. Especially because of the small genome size, this genus of Mollicutes has one of the most extensive compilations of genomic sequences19. Mycoplasmas are recognised as either parasitic or commensals to their host and have undergone a reductive evolution from the Bacillus/Clostridium branch of Gram-positive eubacteria18, which often results in a reduction of the number of genes in the genome. Despite the reduction of genes, Mycoplasma displays a vast variety of phenotypic characteristics, like adaptations to their host, pathogenesis, and mobility18.
Despite the observed dominance of Mycoplasma in salmonid hosts, very little is known about Mycoplasma and its functional potential in salmonids and other teleosts20. In this study (I) we present the first high-quality metagenome-assembled genomes (MAGs) of Mycoplasma from multiple salmonid species including shotgun sequencing of intestinal content from rainbow trout, Atlantic salmon, and European whitefish (Coregonus lavaretus), (II) we establish the phylogeny of the salmonid associated MAGs based on a comparison with 44 known Mycoplasma genomes, leading to the identification of putative novel Mycoplasma species, (III) we present a comparative genomics analysis of Mycoplasma to study their functionality in salmonids in the context of other species within the same genus. Lastly, (IV) we examine functions of the salmonid MAGs that may be related to a potential adaptation to the intestinal host environment.
Results
Genome resolved metagenomics reveals dominance of Mycoplasma in the gut microbiota across three salmonid species
We sampled gut content from three salmonid species from different environments. Two species relevant for aquaculture were chosen, including juvenile rainbow trout and adult Atlantic salmon. Juvenile rainbow trout were sampled in a land-based, freshwater recycled aquaculture system (RAS) in northern Denmark. Atlantic salmon were sampled from a commercial cohort in an open water net pen near Bergen, Norway. Thirdly, wild European whitefish were sampled from a freshwater lake in northern Norway (Fig. 1a) to represent a phylogenetic outlier for comparison (Fig. 1a).
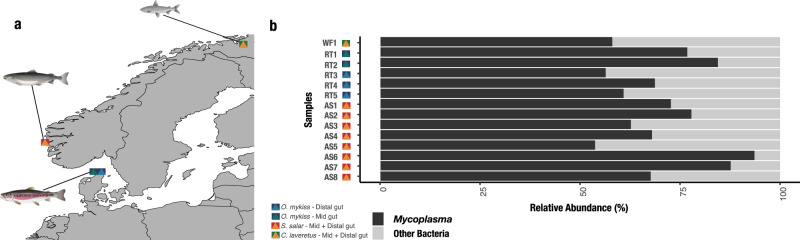
Fig. 1. Sample overview.
a Map of Scandinavia, where sampling points are indicated. Host and sample types are specified in the legend. The European whitefish sample is shown as a yellow triangle with a green square, Atlantic salmon samples are illustrated as a yellow triangle with a red square. Rainbow trout are visualised with two different sample types for the mid and distal gut, with green and light blue triangles, respectively. Both sample types have a dark blue square. b Relative abundance of Mycoplasma across all samples. The black bar plot indicates the presence of Mycoplasma, whereas all other bacteria are concatenated as “other bacteria”.
A total of 2.9 billion metagenomic reads were generated from 12 individuals. Of those, 297 million reads passed quality control and host filtering criteria. Estimates of saturation revealed a sufficient sequencing depth for rainbow trout, but not fully saturated for Atlantic salmon and European whitefish (Supplementary Fig. 1). These filtered reads represent the gut microbiome of the three hosts studied here and were used to establish three metagenomic co-assemblies (Supplementary Data 1). A combination of automatic and manual binning was applied to each co-assembly output, which resulted in one manually curated, low redundant and a near-complete metagenome-assembled genome (MAG), related to Mycoplasma, from each host species (Table 1) (Supplementary Fig. 2). Though sequencing depth was not fully saturated for Atlantic salmon and European whitefish, we do not suspect this to have a major impact on Mycoplasma MAGs, since they all were of a high completion (Table 1).
Table 1.
Summary of metagenomic assembled genomes (MAGs) related to Mycoplasma in three salmonid hosts.
| Host species | No. of contigs | N50 (bp) | Total length (bp) | GC content (%) | CheckM completion (%) | CheckM contamination (%) | No. of genes in the genome |
|---|---|---|---|---|---|---|---|
| Atlantic Salmon | 243 | 3598 | 660,624 | 26.1 | 86.24 | 1.76 | 844 |
| Rainbow Trout | 35 | 88,883 | 697,833 | 24.8 | 95.00 | 0.38 | 627 |
| European Whitefish | 66 | 27,905 | 701,798 | 26.4 | 99.62 | 0.25 | 664 |
Metagenomic data from gut content of juvenile rainbow trout, adult Atlantic salmon, and European whitefish revealed a high relative abundance of Mycoplasma. The salmonid MAGs comprised = 69.44% (SD ± 11.45%) of the metagenomic reads in rainbow trout, = 72.98% (SD ± 13.07%) of the metagenomic reads in Atlantic salmon, and 58.03% of the metagenomic reads in European whitefish (Fig. 1b). Investigation of bacterial load in juvenile rainbow trout, using real-time polymerase chain reaction (PCR) of the 16S rRNA gene, revealed an average cycle threshold (CT) value of 36.3 (SD ± 2.8), where water samples from RAS resulted in an average CT value of 27.6 (SD ± 2.8), clearly indicating a lower bacterial load in the intestinal environment compared to the surrounding water. The amount of sequencing effort to obtain any microbial data and qPCR results of the 16S rRNA gene indicates low bacterial biomass in juvenile rainbow trout gut content samples21 (Supplementary Fig. 1 and Supplementary Table 1).
The GC content of the salmonid MAGs was in the lower end compared to the other Mycoplasma species included (Table 1, Fig. 3). Previous investigations of Tenericutes’ genomes reported GC content varying from 20 to 70%, indicating high plasticity22. Initial analysis of average nucleotide identity (ANI) clustering and gene clusters from each sample to investigate strain variation within each sample revealed identical MAGs within each respective host and clustered each individual salmonid MAGs according to host species (Supplementary Fig. 3a).
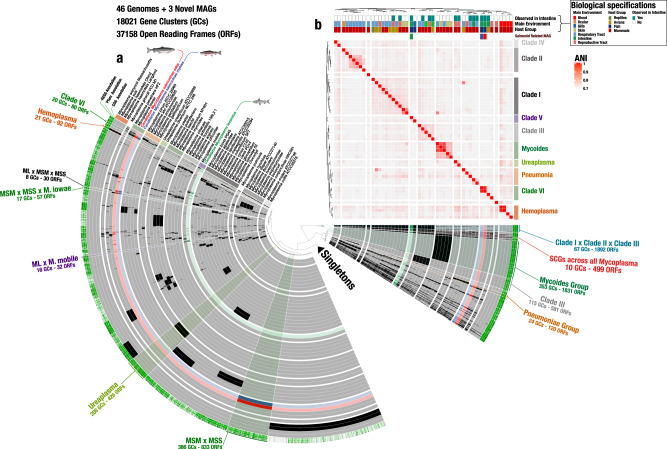
Fig. 3. Comparative genomics of three salmonid co-assembled MAGs with 49 different Mycoplasma genomes.
a Circle diagram based on presence/absence of the 18,021 gene clusters (GCs) containing one or more genes contributed by one or more isolated genomes. Bars in the 49 layers indicate the occurrence of GCs in a given isolated genome and MAGs, black bars indicate presence of GCs, whereas grey indicates absence. Blue, red and green font and layer colour indicate the salmonid MAGs of Candidatus Mycoplasma salmoninae mykiss, Candidatus Mycoplasma salmoninae salar, and Candidatus Mycoplasma lavaretus, respectively. GCs are organised based on their distribution across genomes, and layers of genomes are organised based on their average nucleotide identity (ANI), using Euclidean distance and ward ordination. Selections of co-clustering of genomes were presented in the outer layer and annotations were coloured according to previous clade colours and ANI clusters. Dark red notation indicates single copy core genes (SCGs) between all genomes and MAGs. b Heatmap indicates ANI clusters of Mycoplasma, where red indicates higher similarity between genomes. Colouring of ANI clusters follows the clade definitions shown in the description in Fig. 2. Biological specifications: The three rows describe the biological designation for each genome, including intestinal relation, main environment and host group.
We used fluorescence microscopy to visualise the presence of bacteria on the distal gut epithelial surface as this would further indicate a functional adaptation of the bacteria to the intestinal environment of juvenile rainbow trout. The identity of bacteria could not be established using specific probes, which is likely due to the low bacterial biomass impeding the generation of clear signals. An alternative explanation is the lack of positive control for probe design; unfortunately, no appropriate reference exists since it has yet not been possible to culture these salmonid associated Mycoplasma species20. However, our investigation did reveal a clear DAPI based signal from bacterial cells in close contact with the rainbow trout epithelial surface (Supplementary Fig. 4a–d), which we hypothesise are likely to include Mycoplasma cells, based on the observation that >50% of all microbial reads belonged to Mycoplasma (Fig. 1b).
Phylogenomics reveal candidate species of Mycoplasma in salmonids
We performed a phylogenomic analysis to place the salmonid MAGs within Mycoplasma. To do so, we generated a database of protein clusters across 44 genomes of Mycoplasma isolated from multiple tissue types and host species (Supplementary Data 1). Gene annotation using Hidden Markov Models (HMMs) resulted in 2646 hits, and data filtering for phylogenomics led to a final data set of 55 single-copy core gene alignments from 50 genomes (includes representatives of Ureaplasma and Bacillus).
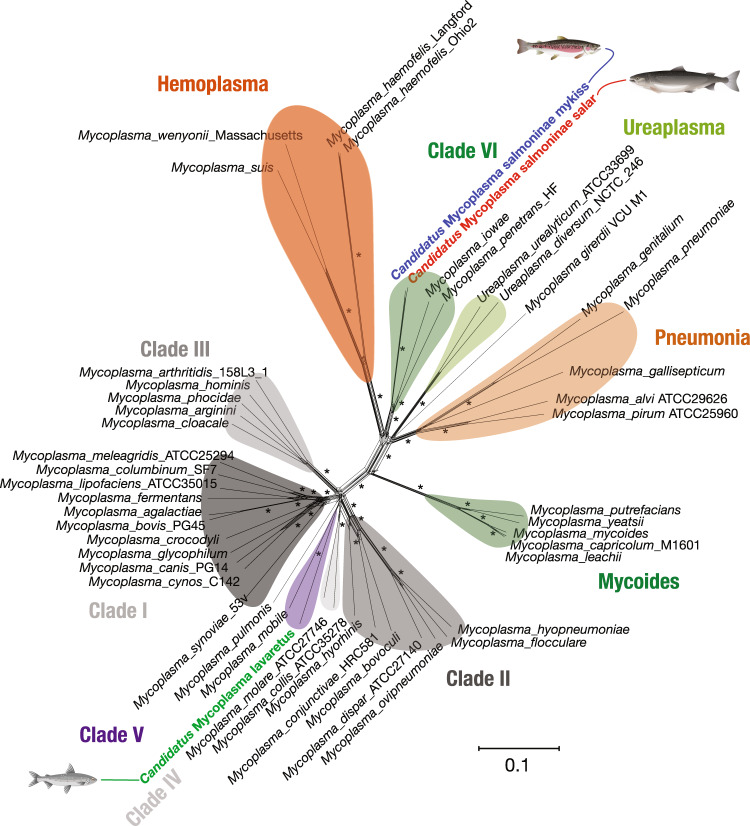
Phylogenetic analyses consistently recovered several highly supported groupings in the genus Mycoplasma and led to a robust placement of our three salmonid MAGs. Two of the salmonid MAGs isolated from the fish subfamily Salmoninae, species rainbow trout and Atlantic salmon, clustered together with an ANI of 96.1% and formed a monophyletic group with the grouping of Mycoplasma penetrans and Mycoplasma iowae (Fig. 2 and Supplementary Fig. 5a), the latter being commonly found in the intestine of turkey (Meleagris gallopavo)23. The two Salmoninae MAGs likely belong to the same species, as suggested by their short terminal branches and uniquely strong branch support according to multiple metrics. The fact that their ANI relative to M. penetrans and M. iowae was <80% further indicated that the two MAGs correspond to, to our knowledge, a new salmonid related Mycoplasma species (Fig. 3). The close relationship of these salmonid MAGs also indicates that they have a close ecological association with Salmoninae, rather than originating from the environment surrounding the host (Fig. 2 and Supplementary Fig. 5a).
Fig. 2. Phylogenomics of salmonid Mycoplasma MAGs.
Phylogenetic network analyses of the bacterial genomes collected. Asterisks represent bootstrap branch support of 100. Branch lengths are measured in the expected number of substitutions per site. Bacillus pumilus ATCC7061 was used as an outgroup for all phylogenomic analysis but is hidden for clarity. Blue, red, and green font indicate the salmonid MAGs of Candidatus Mycoplasma salmoninae mykiss, Candidatus Mycoplasma salmoninae salar, and Candidatus Mycoplasma lavaretus, respectively. Suggested clades of phylogeny are distinguished by colouring to increase clarity.
The third salmonid MAG, characterised by the European whitefish, subfamily Coregoninae, was not identified as a close relative of the isolates from Salmoninae (Fig. 2; Supplementary Fig. 5a). Rather this salmonid MAG appears to be distinct from any of the reference Mycoplasma species, with the closest relative found to be Mycoplasma mobile (ANI < 80%), a pathogen isolated from the gills of tench (Tinca tinca) (Fig. 3).
Overall, our analysis indicated that the salmonid MAGs we characterised represent two salmonid related Mycoplasma species. We tentatively name them according to their respective host species: Candidatus M. salmoninae and Candidatus M. lavaretus. Furthermore, we divided Candidatus M. salmoninae into two biotypes according to their salmonid host, rainbow trout and Atlantic salmon, resulting in Candidatus M. salmoninae mykiss (MSM), Candidatus M. salmoninae salar (MSS) and Candidatus M. lavaretus (ML), respectively.
Beyond the scope of Salmonid MAGs, the phylogenomic analyses also recovered several clades of the genus Mycoplasma with high confidence according to most metrics of phylogenetic branch support (Fig. 2 and Supplementary Fig. 3a, b), confirming the previous phylogenetic placement of Mycoplasma22,24. One metric of branch support was low for most branches, internode certainty for sites (sIC), reflecting the fast accumulation of substitutions in the genus. A fast-evolutionary rate relative to the taxonomic scale of the data is also reflected in the saturation of substitutions found in 12 genes (consequently excluded from phylogenetic analyses), and in the long estimated terminal branches across samples.
An open pangenome with diverse sets of functions is in accordance with niche adaptations of Mycoplasma
We performed a comparative analysis to place the salmonid MAGs within Mycoplasma with respect to their gene content and gene functions. Our Mycoplasma pangenome included 37,158 open reading frames (ORFs) from the 44 different reference Mycoplasma genomes, two Ureaplasma genomes and the three salmonid MAGs, and identified a total of 18,021 gene clusters. Surprisingly, the single-copy core genes which were present among all genomes of Mycoplasma and Ureaplasma genomes only comprised 1.34% of the total ORFs of the pangenome with 10 gene clusters and 499 ORFs (Fig. 3). The amount of singleton gene clusters in the pangenome (i.e., gene clusters only present in a single genome) was 62.8% of all the gene clusters. We investigated the openness of the pangenome using Heaps’ law, resulting in α = 0.281, confirming an open pangenome25, indicating specific adaptations with accessory genes and gene losses, which could derive from specific niche adaptations, like specific host environments and symbiosis (Fig. 3).
Comparison of ANI, environmental relation, and host group of Mycoplasma revealed that Mycoplasma are not only clustering according to phylogeny, but also the origin of host and type of tissue, further emphasising niche-specific adaptations (Fig. 3 and Supplementary Fig. 2a).
Metabolic reconstruction of salmonid MAGs of Mycoplasma suggests adaptation to host environment
Using KEGG we were able to annotate 59.8%, 49.4% and 55.1% of the genes for MSM, MSS and ML, respectively. Especially singletons missed KEGG annotation, indicating that novel functions are yet to be described for many genes in these MAGs (Supplementary Fig. 2a).
Comparison of shared KEGG annotations among MSM, MSS, ML and their nearest relatives M. mobile 163 K and M. iowae 695 revealed 247 shared KEGG functions among the genomes (Fig. 2 and Supplementary Fig. 3b). Investigation of present KEGG annotations revealed that fermentation of sugars through glycolysis appeared to be the main method of ATP production in MSM, MSS and ML. As in many other Mycoplasma species, the genomes are characterised by reduced metabolic functionality as all of the genomes lack general functions, such as the citric acid cycle. Together, these findings are in line with conserved adaptations to, and dependence on, the host gastrointestinal environments across the salmonid related MAGs and their nearest relatives.
Unravelling functions of salmonid MAGs revealed several putatively beneficial functions for their salmonid hosts, including thiamine (B1) biosynthesis26, riboflavin (B2) biosynthesis27 and polyamine metabolism28. Interestingly, we found a complete pathway of isoprenoids biosynthesis by the non-mevalonate (MEP) pathways in two of the salmonid MAGs, including MSM and MSS (Supplementary Fig. 6 and Supplementary Fig. 7). The MEP pathways are rarely found in Mycoplasma, except for the intestinal associated M. iowae and M. penetrans, the sister group to MSM and MSS29,30. We hypothesise this is to reduce the need to obtain isoprenoid precursors from the host and an adaptation towards intestinal environments22.
In brief, our genetic findings are in accordance with a model where Mycoplasma is functionally adapted to the environment in the gut of salmonids. In all three salmonid MAGs, we found uvrABCD, the global genome nucleotide excision repair system (GG-NER). GG-NER is known to protect bacteria against bile salts in gastrointestinal environments31. We found several complete defence systems across the salmonid MAGs (Supplementary Fig. 6), including the stringent response, which is known to react to multiple stress conditions, including amino acid starvation. We found evidence of complete subsystems for lipoic acid metabolism in genomes of clade VI, including MSM, MSS and M. Iowae 695. Lipoic acid metabolism is known to be important for oxidative stress response, in agreement with an adaptation against oxidative stress in the gut (Supplementary Fig. 6 and Supplementary Fig. 7). Furthermore, we found presence of the prtC gene in all genomes of clade VI, including MSM and MSS, which encodes a putative collagenase, responsible for mucus degradation in Helicobacter pylori (Supplementary Data 3). The presence of prtC indicates that MSS and MSM are able to live in gastrointestinal environments by facilitating degradation of mucus in the intestine. Interestingly, we found genetic evidence for a cellobiose and chitobiose degrading complex, known as the cellulosome encoded by celABC, in ML, MSM and MSS. The closest homologues of celABC found in ML, MSM and MSS were found in M. iowae 695 with identities ranging from 58.3 to 64.3%. The cellulosome is responsible for degrading complex polymers, like cellulose, hemicellulose, and chitin32, indicating that intestinal related salmonid MAGs have some putative ability to degrade long chain polymers in the gut, possibly originating from host mucus layers or host diet (Supplementary Data 3).
All three salmonid MAGs lack oligosaccharide ABC transporters, which are otherwise found in other Mycoplasma genomes, indicating that salmonid Mycoplasmas are relying on the phosphotransferase system (PTS), like celABC (Supplementary Fig. 6 and Supplementary Fig. 7). This suggests that the main sources of energy absorbed by the Mycoplasmas from the gastrointestinal tract in its teleost host consist of long-chain polymers, fatty acids, lipoproteins and proteins.
Though the molecular basis of Mycoplasma pathogenicity remains largely elusive, we investigated the presence of Mycoplasma related pathogenicity factors, including the presence of traG/traE33, glpF34, katE35, oppA, mgpA/mgpC36, virulence factor BrkB, toxins, antitoxins, large membrane proteins (LMPs) and adhesion-related proteins. Our investigation revealed a lack of surveyed putative virulence factors in both MSM and MSS. We found a three-gene cluster with virulence factors, including virulence factor BrkB, an anti-toxin and glpF, in ML, indicating that ML still possess some level of pathogenic potential, whereas we found no evidence for pathogenicity of MSS and MSM to its host (Supplementary Fig. 8).
Functional enrichment analysis suggests that intestinal related Mycoplasma species are relying on amino acid synthesis, isoprenoid synthesis and an antioxidative protective system
We performed a functional enrichment analysis by reconstructing metabolic pathways specific for Mycoplasma with RAST37. This analysis revealed 641, 667 and 676 DNA features, including protein-encoding genes and RNA coding genes, in MSM, MSS, and ML, respectively. Our pathway-based comparison among Mycoplasma genomes revealed that Mycoplasma species have a broad range of different functionalities (Supplementary Data 2), which fits the high dissimilarity of the phylogenetic and pangenomic analyses and the hypothesised host adaptation (Fig. 3).
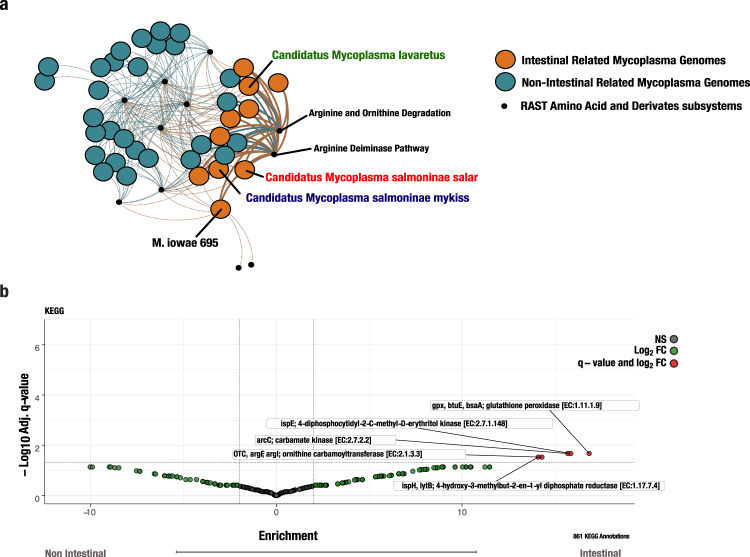
Interestingly, we found a significant enrichment of the subsystems corresponding to arginine biosynthesis in Mycoplasma species and MAGs associated with intestinal environments including the three MAGs described here (87.5%) compared to those in other environments (27.3%) (Fig. 4a, b). Our enrichment analysis also confirmed a higher prevalence of genes encoding the MEP pathway in intestinal environments (Fig. 4b). Lastly, our analysis revealed that glutathione peroxidase was significantly over-represented in Mycoplasma species associated with intestinal environments, indicating that antioxidative protective systems have a putative defensive role in intestinal related Mycoplasma (Fig. 4b) (Supplementary Data 4).
Fig. 4. Functional enrichment analysis between intestinal related Mycoplasma species and non-intestinal related Mycoplasma species.
a Genome clustering network based on the RAST ‘Amino Acid and derivatives’ subsystem. Clustering of genomes was based on RAST ‘Amino Acid and derivatives’ subsystems present among Mycoplasma genomes. The network layout was based on Force Atlas 2 algorithms. Blue colour indicates Mycoplasma genomes not being related to intestinal environments, whereas orange nodes indicate Mycoplasma genomes being related to intestinal environments. Small black nodes indicate RAST subsystems. Dashed edges indicate non-significant relations. Thick solid edges indicated significance between the RAST subsystem and intestinal environment. Blue, red and green font indicate the salmonid MAGs of Candidatus Mycoplasma salmoninae mykiss, Candidatus Mycoplasma salmoninae salar, and Candidatus Mycoplasma lavaretus, respectively. b Volcano plot of 861 KEGG annotated genes in Mycoplasma. Investigated genomes were divided into intestinal and non-intestinal related genomes of Mycoplasma according to Supplementary Data 2. Functional enrichment was inferred using a generalised linear model (GLMs) with the logit linkage function to compute an enrichment score and p value for each function. False detection rate correction to p-values to account for multiple tests was done and only genes with adjusted q-values below 0.05 and an enrichment score above 1 are reported.
Discussion
Our study presents Mycoplasma MAGs characterised from gastrointestinal samples of 12 individuals from 3 different salmonid species. These salmonid related MAGs systematically represented the dominant taxa of the gut microbiota in the three host species. Moving beyond 16S rRNA gene amplicon surveys, we provide the first description of the potential functional importance of Mycoplasma related to the gastrointestinal environment of multiple salmonid host species, including an essential amino acid metabolism of putative advantage to their fish host. We acknowledge that MAG technology comes with limitations, resulting in potential false positives. To minimise these limitations, we used state-of-the-art binning methods, combining CONCOCT and manual curation with anvi’o and applied good practice for the reliable generation of MAGs38. Our Mycoplasma MAG resolved from Atlantic salmon is largely in accordance with another recent assembly20. Together, these findings support a non-neutral evolutionary relationship and warrant further investigations into the use of these Mycoplasma candidate species and their potential to boost gut health and growth performance in commercial production of fish and potentially other animal species. Our 16S rRNA gene-based qPCR investigation of bacterial load in rainbow trout indicated low biomass of bacteria, which we hypothesise reflects the young age of rainbow trout, a relatively sterile RAS environment and that the bacteria present are in a phase of initial colonisation of the gut. The dominance of Mycoplasma in this early life stage further indicates that Mycoplasma is important in young salmonids.
Modifications of the microbiota for a putative gain of beneficial phenotypes, such as increased feed efficiency and host disease resilience, have been exploited for livestock productions. We envisage that our findings will further provide a background for a better understanding of the interaction between Mycoplasma and aquaculturally relevant salmonids and other teleost species to increase the efficiency of aquaculture production. In Atlantic salmon, the abundance of Mycoplasma has been shown to be strongly decreased by the presence of pathogens, which indicates that the abundance of Mycoplasma is positively associated with improved growth8,39, carotenoid utilisation1 and disease resilience of its host40. Together, this suggests that Mycoplasmas may be used as a putative biomarker for monitoring performance and disease status in farmed salmonids6,40.
The phylogeny of salmonid related MAGs of Mycoplasma corresponds to the phylogeny of their hosts, Coregoninae and Salmoninae, indicating putative host-specific adaptations. We deciphered genetic and functional differentiations among Mycoplasma species, using state of the art methods for phylogenomics, comparative genomics, and reconstruction of metabolic subsystems. Despite the fact that Mycoplasma is often reported to be an obligate parasite, our findings revealed that the salmonid-related Mycoplasma species could be specifically adapted for ammonotelic hosts, such as most teleosts, due to the ability to utilise ammonia in the gut. We hypothesise that this might have facilitated an evolutionary beneficial relationship between Mycoplasma and its salmonid hosts. Numerous recent studies have revealed a strong dominance of a salmonid Mycoplasma6,8,11,13; here we add to these intriguing findings by presenting draft genomes, broader phylogenetic relationship and functional potential. A high relative abundance of Mycoplasma has been associated with higher health status in Atlantic salmon indicating a potential adaptive advantage of Mycoplasma to the host fish40. Further, neutral modelling approaches comparing environmental and intestinal frequency distributions of Mycoplasma in Atlantic salmon have previously suggested that Atlantic salmon related Mycoplasmas are well adapted to colonisation of their hosts41. Recent findings of marine Mycoplasma species and uncultured Tenericutes have shed new light over the evolutionary processes and pathogenicity of Mycoplasma, which is corroborated by our findings of symbiotic species of Mycoplasma present in marine environments and marine living vertebrates22,42.
Previous studies have shown the importance of arginine and its derivatives, citrulline and ornithine, in the gastrointestinal tract of farmed fish43–45. Biosynthesis of arginine, citrulline and ornithine has previously been investigated in Mycoplasma, since this pathway is an important energy source in other Mycoplasma species22. We found genetic evidence of MSS, MSM and ML being able to use ammonia as a substrate for biosynthesis of ornithine and citrulline by the presence of the genes encoding carbamate kinase (arcC) and ornithine transcarbamylase (otc). We hypothesise that this is a beneficial trait for salmonids since they lack the ability to de novo synthesise arginine and in addition, this ability of Mycoplasma could increase ammonia detoxification in the gut46, where ammonia are found excessively in salmonids47. Furthermore, ornithine uptake from the gut can lead to increased growth in Atlantic salmon43. These findings indicate that the presence of Mycoplasma in the gut can boost the metabolism of its salmonid host, which we hypothesise could be a result of ammonia detoxification and could result in the increased upper limit of feed utilisation and thereby increased growth of its salmonid host. Further investigations of gene activity related to ammonia detoxification, isoprenoid synthesis and polymer degradation by Mycoplasma would require meta-transcriptomics and metabolomics to further our functional understanding of the role that Mycoplasma plays for its host.
Previous studies investigating the biogeographical dynamics of the intestinal microbiota in Atlantic salmon revealed dominance of Mycoplasma in adults and returning adult salmon, despite the low presence of Mycoplasma in the early life stage of Atlantic salmon13. Combined with our results, we hypothesise that the dominance of Mycoplasma in salmonids is a result of holobiont evolution. Indeed, we show that these salmonid Mycoplasmas harbour genes able to degrade long-chain polymers, such as chitin, which is often abundant in insects and crustaceans that make up an important proportion of the natural diet of juvenile salmonids48. This ability of Mycoplasma could be beneficial for its host since the degradation of long-chain polymers boosts the nutritional value of a chitin rich diet and therefore could be a co-evolutionary driver between the salmonid hosts and Mycoplasma. This hypothesis may also explain the increase of Mycoplasma in aquaculture cohorts, where an increase of Mycoplasma was shown in the intestinal region of rainbow trout reared on an insect-based diet8 and an insect-based diet has subsequently proven beneficial in aquaculture39. We note that our findings could represent the foundation for an optimised production that exploits the beneficial functions of Mycoplasma presence by targeting not only the essential requirements of the fish host but also the requirements of the salmonid specific Mycoplasma.
While several previous studies have demonstrated that Mycoplasma species are commonly found in salmonids using gene marker technologies, our study resolves the phylogeny and functionality of three MAGs of two species of Mycoplasma related to aquaculturally relevant species. Our study facilitates a deeper understanding of the functionality of Mycoplasma in salmonids. Furthermore, our results are in accordance with a mutualistic relationship between Mycoplasma and its salmonid host and imply that Mycoplasmas likely play an important role in nutrition utilisation and possibly health.
Methods
Sample collection and DNA extraction
Sample collections for rainbow trout were carried out at the Research facility of BioMar in Hirtshals, Denmark. Samples from eight individual Atlantic salmon were obtained from a production site in a fjord near Bergen, Norway. The entire gastrointestinal tract was carefully dissected out with sterile tools and gut content was then sampled and preserved in Zymo DNA/RNA shield (Zymo Research). Extraction of DNA from three individuals, including mid and distal intestinal region, of rainbow trouts was carried out using ZymoBiomics DNA miniPrep (Zymo Research), following the protocol of the manufacturer. Extraction of DNA from Atlantic salmon was carried out using MagAttract® PowerSoil® DNA KF Kit, following the protocol of the manufacturer. Samples from a single individual of European whitefish were taken from Lake Suohpatjavri, Norway. The entire gastrointestinal tract was carefully dissected out with sterile tools and gut content was then sampled and preserved in 90% Ethanol. Extraction of DNA from European whitefish was carried out in technical triplicates, using MagAttact Power Soil DNA Kit (Qiagen) with a modified protocol49.
Ethical approval
The methods for rainbow trout were performed in accordance with relevant guidelines and regulations and approved by The Danish Animal Experiments Inspectorate, under license No. 2015-15-0201-00645. The study is thus approved under the Danish law regarding experimental animals. Atlantic salmon and European whitefish included in this study were sacrificed immediately upon catch with a solid hit to the neck region resulting in instant death before tissue and gut content samples were taken. While no particular license is required for such sampling, we stress that all fish handling was supervised by experienced and trained staff in accordance with normal and legal procedures in Norway. We obtained permission for gill net fishing in Lake Suohpatjavri from the County Governor of Finnmark with legal authority through LOV 1992-05-15 nr 47, 113. Fish were euthanized by means of a cerebral concussion prior to sample collection. No ethical permission is required from the Norwegian Animal Research Authority for collection with gill nets and the associated sacrifice of fish (FOR 1996-01-15 nr 23, the Norwegian Ministry of Agriculture and Food).
Biomass monitoring of bacteria in rainbow trout, using real-time PCR of bacterial V3–V4 16S rRNA gene
Real‐time PCR (qPCR) was performed in 20 μl reactions containing either 1 µl 1:1, or 1:10 DNA template, and 1× AccuPrime SuperMix II (Invitrogen), 6.5 µl ddH20, 0.1 µM forward, and reverse primers (341 F: 5′-CCTAYGGGRBGCASCAG-3′, 805R: 5′-GGACTACNNGGGTATCTAAT-3′), and 1 μl of SYBR Green/ROX solution (Invitrogen). qPCR amplifications were performed on an Mx3005 qPCR machine (Agilent Technologies) with the following cycling conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 20 s, and 72 °C for 40 min.
Imaging bacteria in mid- and distal gut sections of rainbow trout
To visualise bacteria in the fish gut sections, formalin-fixed tissue sections were deparaffinised with xylene, dehydrated with 70 and 96% ethanol, respectively, for 3 min each time, and air dried. Vetcashield (Vector Laboratories), an antifade mounting medium with DAPI was added along with a coverslip. Microscopy and image analysis were with an Axioplan II epifluorescence microscope (Carl Zeiss) equipped for epifluorescence with a 100-W mercury lamp. Confocal images were captured on a Leica SPX-5 (Supplementary Fig. 4).
DNA sequencing
Fragmentation of DNA was carried out, using Covaris M220 with microTUBE-50 AFA Fibre Screw-Cap. Samples were normalised prior to library preparation. Library preparation was based on Single-tube library preparation for degraded DNA50. Prior to the indexing of libraries, all libraries were analysed with quantitative PCR to estimate optimal cycle settings on a Mx3005P qPCR System (Agilent Technologies). Indexed libraries were quality controlled with a bioanalyzer 2100 (Agilent Technologies) and sequenced 100PE and 150PE on a BGISEQ-500 or MGI 2000 at BGI Europe.
Genome-resolved metagenomics
Raw sequence reads were quality controlled, using FastQC/v0.11.8 to assess filtering and quality steps. Saturation of sequencing depth to obtain >5X coverage for all reads were estimated for available data, using khmer51. Removal of adapters and low-quality reads was done with AdapterRemoval/v2.2.4, with quality base of 30 and a minimum length of 50 bp. Duplicates were removed, and reads were re-paired to remove singletons, using bbmap/v.38.35. In order to increase assembly efficiency by reducing eukaryotic contaminants, data were filtered for the phiX174 genome, Human (HG19) genome, and the respective host genome (Supplementary Table 2), using minimap252, using default parameters for short accurate genomic reads. Filtered data were initially single assembled to investigate individual variations of Mycoplasma and were also co-assembled to obtain highest completion of MAGs for comparative and phylogenomic analysis, using MegaHIT/v.1.1.1 with a minimal length of 1000 bp per scaffold, using meta-sensitive flag for metagenomic purpose and assembled contigs were quality controlled with Quast/v.5.02. To increase effective binning, we used the anvi’o pipeline53 (available from http://merenlab.org/software/anvio). The subsequent workflow is outlined at http://merenlab.org/2016/06/22/anvio-tutorial-v2/. Briefly, (I) anvi’o was used to profile the scaffolds using Prodigal/v2.6.354 with default parameters to identify genes and HMMER/v.3.355 to identify genes matching archaeal56, Protista (based on http://merenlab.org/delmont-euk-scgs) and bacterial56 single-copy core gene collections. Also, ribosomal RNA based HMMs were identified (based on https://github.com/tseemann/barrnap). The HMMs were used to determine the completeness of metagenome-assembled genomes (MAGs); (II) Kaiju57 was used with NCBI’s non-redundant protein database ‘nr’ to infer the taxonomy of genes (as described in http://merenlab.org/2016/06/18/importing-taxonomy/); (III) we mapped short reads from the metagenomic set to the scaffolds using BWA/v0.7.1596 (minimum identity of 95%) and stored the recruited reads as BAM files using samtools58; (IV) anvi’o profiled each BAM file to estimate the coverage and detection statistics of each scaffold, and combined mapping profiles into a merged profile database for each metagenomic set. Contigs were binned automatically, using CONCOCT59, by constraining the number of clusters per metagenomic set to 10. Each CONCOCT bin was manually curated using the anvi’o interactive interface to ensure high completion and low redundancy. The interface considers the sequence composition, differential coverage, GC-content and taxonomic signal of each scaffold53,60. Identification of Mycoplasma MAGs was initially conducted by three approaches: (I) by short-read mapping with Kaiju, giving the origin of each scaffold, (II) if ribosomal 16S gene were present, we blasted these to nearest relative, (III) we used HMM hits of single-copy core genes to identify bacterial taxa. Relative abundance of each MAG was calculated based on percentage read recruitment across all samples from the specific host. Quality assessment of MAGs was carried out with anvi’o and checkM61.
A summary for each metagenome and Mycoplasma MAGs generated for this study is available at 10.6084/m9.figshare.13019507 and 10.6084/m9.figshare.13019477, respectively.
Phylogenomics of Mycoplasma
Metagenomic co-assemblies from rainbow trout, Atlantic salmon and European whitefish were binned using CONCOCT and each MAG was manually curated using anvi’o v6.1. The MAGs annotated as Mycoplasma were isolated from the dataset for further analysis. To examine the phylogenetic position of these unknown Mycoplasma MAGs, 44 known Mycoplasma species from different hosts and tissues were selected. We also included two samples of the closely related Ureaplasma diversum, and a sample of Bacillus pumilus for use as an outgroup (Supplementary Data 1). We repeated the extraction of HMM hits using HMMER/v.3.3 from each of these additional samples, resulting in 67 orthologous single-copy core genes, using the default anvi’o bacterial profiles, which were aligned using MACSE/v262. Highly divergent taxa were identified using TreeShrink63, followed by the second round of alignment of the raw data excluding the flagged taxa. To further minimise the stochastic error due to substitution saturation, we removed codons in which the most common translated amino acid was different for more than half of the taxa64. We also filtered codons with >50% missing data and assessed overall substitution saturation using PhyloMAd65. The filtering steps resulted in 55 acceptable alignments of core genes.
Phylogenomic analyses were performed using three different models of genome evolution: (I) assuming an identical tree across concatenated genes; (II) assuming free recombination across genes; (III) allowing non-treelike evolution due to recombination, hybridisation or lateral gene transfer.
Phylogenetic analysis was performed on a concatenated data set including the final filtered gene alignments. The substitution models for each gene were selected automatically from the GTR + F + Γ + I + R family of models, allowing gene-models to merge into the best fitting partition scheme66. This step of partition selection was followed by maximum likelihood phylogenetic inference using a model of proportional variation in branch lengths across partition subsets67 implemented in IQ-TREE v1.768. The statistical support for branches was estimated using an Shimodaira–Hasegawa-like approximate likelihood-ratio test (aLRT)69. Branch supports were examined further using internode certainty support metrics, which are based on Shannon’s entropy and indicate the degree of certainty for each branch considering the two most prevalent branches in the data70. Internode certainties for gene trees and for sites were calculated independently, using estimates of site- and gene-concordance factors from IQ-TREE71.
Further phylogenetic analyses were performed for comparison, focusing on estimates of gene trees as a starting point. Individual gene trees were estimated with the best fitting substitution model of the GTR + F + Γ + I + R family in each case, using IQ-TREE v1.7. Species tree inference was then performed assuming free recombination among genes under the multispecies coalescent model, implemented in ASTRAL72. In input trees, all branches with aLRT support below 50 were collapsed, thereby minimising the influence of substantial stochastic error in gene trees arising from having a finite sample of nucleotide sites. Analysis under the multispecies coalescent led to a nearly identical phylogenetic inference to using a concatenated alignment (Supplementary Fig. 2).
Any incongruence in phylogenetic signals across the data was further explored using phylogenetic network analysis. We estimated a network from the concatenated alignment of genes using the Neighbour-Net algorithm73, with pairwise genetic distances calculated under the F81 substitution model and empirically calculated base frequencies, implemented in SplitsTree v4.1674. Statistical supports for network branches were calculated using 1000 bootstrap replicates.
Comparative genomics of Mycoplasma
Mycoplasma genomes from different species were selected based on completion level. Only genomes above 80% completion and below 10% redundancy were selected from Genbank. Selected genomes are referred to as external genomes (Supplementary Data 1).
These external genomes were compared with salmonid MAGs, using anvi’o/v6.1 as the previous studies60. Similarities of each amino acid sequence in every genome were calculated against every other amino acid sequence across all genomes, using blastp. minbit heuristics of 0.5 were implemented to eliminate weak matches between two amino acid sequences75 and a mcl inflation of 10. The MCL algorithms were used to identify gene clusters in amino acid sequence similarity search results76. Euclidean distance and ward linkage were used to organise gene clusters and genomes. ANI was calculated, using PyANI77. A summary of the pan-genome generated for this study is available at 10.6084/m9.figshare.13019543.
Openness estimates calculations of pangenome were calculated, based on Heaps’ law, using R-package micropan25,78, calculations were carried out with 1000 permutations.
Functional description of salmonid MAGs
To increase functional knowledge of the Mycoplasma genomes, we annotated functions, using Pfam79, COG80, KEGG81 and RAST37. RAST was implemented successfully for 39 of the genomes to minimise Mycoplasma specific readthrough of the UGA stop codon82,83.
Functional enrichment analysis of KEGG annotated genes was carried out on all genomes, using generalised linear models (GLMs) through anvi’o/v6.1. GLMs were carried out with the logit linkage function to compute an enrichment score and p value for each function. False detection rate correction to p-values to account for multiple tests was done using the R package qvalue.
Network analysis of genomes based on amino acid metabolism was based on RAST subsystems. Gephi/v0.9.281 was applied to generate a functional network using the Force Atlas 2 algorithm to connect MAGs and RAST functions with 1,000,000 iterations. Only functional nodes related to amino acid metabolism were kept for analysis.
Statistics and reproducibility
Statistics were carried using R/v.4.0.3 and Python/v.3.6, to generate results that are reported in the paper and at https://github.com/JacobAgerbo/Comparative_Mycoplasma.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The research was funded by The Independent Research Fund Denmark (“HappyFish”, grant No. 8022-00005B), GUDP (“Præ-Pro-Fisk”, grant No. 34009-17-1218) and the FHF—Norwegian Seafood Research Fund (“HoloFish”, grant No. 901436). We would like to thank Anni Nielsen, BioMar Trial coordinator, ATC Hirtshals, Denmark and Torunn Forberg, Biomar RD, Trondheim, Norway for assisting with sampling rainbow trout, Cathrine Kalgraff from Lerøy is thanked for assisting with salmon samples.
Author contributions
M.T.L. and J.A.R. conceived the study with input from A.M.B., M.T.P.G. and K.K. Sampling was organised and performed by J.A.R., K.R.V., M.T.L., M.T.P.G., M.D.M., L.C.P., H.S. and K.P. J.A.R., L.C.P., K.R.V., L.V.G.J. and A.M.B. carried out laboratory work. J.A.R., T.O.D and D.A.D. performed the computational analysis. J.A.R. wrote the paper with input from all authors.
Data availability
The salmonid related Mycoplasma MAGs are publicly available at 10.6084/m9.figshare.13019477. Summaries of each metagenome used to generate salmonid related Mycoplasma MAGs are publicly available at 10.6084/m9.figshare.13019507. Summary of pangenome used to analyse salmonid related Mycoplasma MAGs is publicly available at 10.6084/m9.figshare.13019543. Supplementary Data 2 contains source data for Fig. 4. The raw dataset generated during the current study are available in European Nucleotide Archive (ENA) repository with project accession numbers PRJEB40990. Genome accession codes for Candidatus M. salmoninae mykiss (MSM), Candidatus M. salmoninae salar (MSS), and Candidatus M. lavaretus (ML) are GCA_905477455, GCA_90547759 and GCA_905477555, respectively. All other data are available from the corresponding authors upon reasonable request.
Code availability
Any code used for the study used to generate results that are reported in the paper and central to its main claims are available at https://github.com/JacobAgerbo/Comparative_Mycoplasma and on Zenodo at 10.5281/zenodo.4629623.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jacob A. Rasmussen, Email: jacob.rasmussen@bio.ku.dk
Morten T. Limborg, Email: morten.limborg@sund.ku.dk
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02105-1.
References
- 1.Nguyen CDH, Amoroso G, Ventura T, Minich JJ, Elizur A. Atlantic Salmon (Salmo salar L., 1758) gut microbiota profile correlates with flesh pigmentation: cause or effect? Mar. Biotechnol. 2020;22:786–804. doi: 10.1007/s10126-019-09939-1. [DOI] [PubMed] [Google Scholar]
- 2.Huang, Q. et al. Diversity of gut microbiomes in marine fishes is shaped by host-related factors. Mol. Ecol. 10.1111/mec.15699 (2020). [DOI] [PMC free article] [PubMed]
- 3.Perry WB, Lindsay E, Payne CJ, Brodie C, Kazlauskaite R. The role of the gut microbiome in sustainable teleost aquaculture. Proc. Biol. Sci. 2020;287:20200184. doi: 10.1098/rspb.2020.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limborg MT, et al. Applied hologenomics: feasibility and potential in aquaculture. Trends Biotechnol. 2018;36:252–264. doi: 10.1016/j.tibtech.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Kokou F, et al. Core gut microbial communities are maintained by beneficial interactions and strain variability in fish. Nat. Microbiol. 2019;4:2456–2465. doi: 10.1038/s41564-019-0560-0. [DOI] [PubMed] [Google Scholar]
- 6.Brown RM, Wiens GD, Salinas I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss) Fish. Shellfish Immunol. 2019;86:497–506. doi: 10.1016/j.fsi.2018.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciric, M., Waite, D., Draper, J. & Jones, J. B. Characterisation of gut microbiota of farmed Chinook salmon using metabarcoding. bioRxiv10.1101/288761 (2018).
- 8.Rimoldi, S., Gini, E., Iannini, F., Gasco, L. & Terova, G. The effects of dietary insect meal from Hermetia illucens prepupae on autochthonous gut microbiota of rainbow trout (Oncorhynchus mykiss). Animals9, 143 (2019). [DOI] [PMC free article] [PubMed]
- 9.Lowrey L, Woodhams DC, Tacchi L, Salinas I. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl. Environ. Microbiol. 2015;81:6915–6925. doi: 10.1128/AEM.01826-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyons PP, Turnbull JF, Dawson KA, Crumlish M. Phylogenetic and functional characterization of the distal intestinal microbiome of rainbow trout Oncorhynchus mykiss from both farm and aquarium settings. J. Appl. Microbiol. 2017;122:347–363. doi: 10.1111/jam.13347. [DOI] [PubMed] [Google Scholar]
- 11.Holben WE, et al. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 2002;44:175–185. doi: 10.1007/s00248-002-1011-6. [DOI] [PubMed] [Google Scholar]
- 12.Dehler CE, Secombes CJ, Martin SAM. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.) Sci. Rep. 2017;7:13877. doi: 10.1038/s41598-017-13249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llewellyn MS, et al. The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J. 2016;10:1280–1284. doi: 10.1038/ismej.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hottes AK, et al. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helsen J, et al. Gene loss predictably drives evolutionary adaptation. Mol. Biol. Evol. 2020;37:2989–3002. doi: 10.1093/molbev/msaa172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razin S. Peculiar properties of mycoplasmas: the smallest self-replicating prokaryotes. FEMS Microbiol. Lett. 1992;100:423–431. doi: 10.1111/j.1574-6968.1992.tb05735.x. [DOI] [PubMed] [Google Scholar]
- 17.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/MMBR.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dandekar, T. et al. Comparative Genome Analysis of the Mollicutes. in Molecular Biology and Pathogenicity of Mycoplasmas (eds Razin, S. & Herrmann, R.) 255–278 (Springer, USA, 2002).
- 19.Barré A, de Daruvar A, Blanchard A. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 2004;32:D307–D310. doi: 10.1093/nar/gkh114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheaib, B. et al. Genome erosion and evidence for an intracellular niche – exploring the biology of mycoplasmas in Atlantic salmon. Aquaculture, 736772 (2021). [DOI] [PMC free article] [PubMed]
- 21.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli. BMC Genomics. 2020;21:408. doi: 10.1186/s12864-020-06807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei S, et al. Genome sequence of Mycoplasma iowae strain 695, an unusual pathogen causing deaths in turkeys. J. Bacteriol. 2012;194:547–548. doi: 10.1128/JB.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima K, Nishida H. Phylogenetic relationships among mycoplasmas based on the whole genomic information. J. Mol. Evol. 2007;65:249–258. doi: 10.1007/s00239-007-9010-3. [DOI] [PubMed] [Google Scholar]
- 25.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Keinänen M, et al. The thiamine deficiency syndrome M74, a reproductive disorder of Atlantic salmon (Salmo salar) feeding in the Baltic Sea, is related to the fat and thiamine content of prey fish. ICES J. Mar. Sci. 2012;69:516–528. doi: 10.1093/icesjms/fss041. [DOI] [Google Scholar]
- 27.Hemre G-I, et al. Atlantic salmon (Salmo salar) require increased dietary levels of B-vitamins when fed diets with high inclusion of plant based ingredients. PeerJ. 2016;4:e2493. doi: 10.7717/peerj.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn NE, Bird JG, Guthrie AS. Glucocorticoid regulation of amino acid and polyamine metabolism in the small intestine. Amino Acids. 2009;37:123–129. doi: 10.1007/s00726-008-0206-7. [DOI] [PubMed] [Google Scholar]
- 29.Eberl M, et al. Mycoplasma penetrans is capable of activating V gamma 9/V delta 2 T cells while other human pathogenic mycoplasmas fail to do so. Infect. Immun. 2004;72:4881–4883. doi: 10.1128/IAI.72.8.4881-4883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangari FJ, Pérez-Gil J, Carretero-Paulet L, García-Lobo JM, Rodríguez-Concepción M. A new family of enzymes catalyzing the first committed step of the methylerythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in bacteria. Proc. Natl Acad. Sci. USA. 2010;107:14081–14086. doi: 10.1073/pnas.1001962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Begley M, et al. Analysis of the isoprenoid biosynthesis pathways in Listeria monocytogenes reveals a role for the alternative 2-C-methyl-d-erythritol 4-phosphate pathway in murine infection. Infect. Immun. 2008;76:5392–5401. doi: 10.1128/IAI.01376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan, Z. Microbial Energy Conversion. (Walter de Gruyter GmbH & Co. KG, 2018).
- 33.Citti, C., Baranowski, E., Dordet-Frisoni, E., Faucher, M. & Nouvel, L.-X. Genomic islands in mycoplasmas. Genes11, 836 (2020). [DOI] [PMC free article] [PubMed]
- 34.Großhennig S, Schmidl SR, Schmeisky G, Busse J, Stülke J. Implication of glycerol and phospholipid transporters in Mycoplasma pneumoniae growth and virulence. Infect. Immun. 2013;81:896–904. doi: 10.1128/IAI.01212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard RE, Balish MF. Mycoplasma iowae: relationships among oxygen, virulence, and protection from oxidative stress. Vet. Res. 2015;46:36. doi: 10.1186/s13567-015-0170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereyre S, et al. Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 2009;5:e1000677. doi: 10.1371/journal.pgen.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meziti, A. et al. The reliability of metagenome-assembled genomes (MAGs) in representing natural populations: Insights from comparing MAGs against isolate genomes derived from the same fecal sample. Appl. Environ. Microbiol. 87, e02593-20 (2021). [DOI] [PMC free article] [PubMed]
- 39.Rimoldi, S., Antonini, M., Gasco, L., Moroni, F. & Terova, G. Intestinal microbial communities of rainbow trout (Oncorhynchus mykiss) may be improved by feeding a Hermetia illucens meal/low-fishmeal diet. Fish Physiol. Biochem. 10.1007/s10695-020-00918-1 (2021). [DOI] [PMC free article] [PubMed]
- 40.Bozzi, D. et al. Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Animal Microbiome3, 1–17 (2021). [DOI] [PMC free article] [PubMed]
- 41.Heys, C. et al. Neutral processes dominate microbial community assembly in Atlantic Salmon, Salmo salar. Appl. Environ. Microbiol. 86, e02283-19 (2020). [DOI] [PMC free article] [PubMed]
- 42.Lian C-A, et al. Genomic characterization of a novel gut symbiont from the hadal snailfish. Front. Microbiol. 2019;10:2978. doi: 10.3389/fmicb.2019.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berge GE, Sveier H, Lied E. Effects of feeding Atlantic salmon (Salmo salar L.) imbalanced levels of lysine and arginine. Aquacult. Nutr. 2002;8:239–248. doi: 10.1046/j.1365-2095.2002.00211.x. [DOI] [Google Scholar]
- 44.Andersen SM, et al. Dietary arginine affects energy metabolism through polyamine turnover in juvenile Atlantic salmon (Salmo salar) Br. J. Nutr. 2013;110:1968–1977. doi: 10.1017/S0007114513001402. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen TL, et al. Dietary probiotic effect of Lactococcus lactis WFLU12 on low-molecular-weight metabolites and growth of olive flounder (Paralichythys olivaceus) Front. Microbiol. 2018;9:2059. doi: 10.3389/fmicb.2018.02059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li P, Mai K, Trushenski J, Wu G. New developments in fish amino acid nutrition: towards functional and environmentally oriented aquafeeds. Amino Acids. 2009;37:43–53. doi: 10.1007/s00726-008-0171-1. [DOI] [PubMed] [Google Scholar]
- 47.Rubino JG, Zimmer AM, Wood CM. An in vitro analysis of intestinal ammonia handling in fasted and fed freshwater rainbow trout (Oncorhynchus mykiss) J. Comp. Physiol. B. 2014;184:91–105. doi: 10.1007/s00360-013-0781-0. [DOI] [PubMed] [Google Scholar]
- 48.Orlov AV, Gerasimov YV, Lapshin OM. The feeding behaviour of cultured and wild Atlantic salmon, Salmo salar L., in the Louvenga River, Kola Peninsula, Russia. ICES J. Mar. Sci. 2006;63:1297–1303. doi: 10.1016/j.icesjms.2006.05.004. [DOI] [Google Scholar]
- 49.Hildonen M, Kodama M, Puetz LC, Gilbert MTP, Limborg MT. A comparison of storage methods for gut microbiome studies in teleosts: insights from rainbow trout (Oncorhynchus mykiss) J. Microbiol. Methods. 2019;160:42–48. doi: 10.1016/j.mimet.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Carøe C, et al. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 2018;9:410–419. doi: 10.1111/2041-210X.12871. [DOI] [Google Scholar]
- 51.Crusoe MR, et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murat Eren A, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MD. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. 2019;35:4162–4164. doi: 10.1093/bioinformatics/btz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alneberg J, et al. Binning metagenomic contigs by coverage and composition. Nat. Methods. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 60.Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018;6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ranwez V, Harispe S, Delsuc F, Douzery EJP. MACSE: multiple alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE. 2011;6:e22594. doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mai U, Mirarab S. TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics. 2018;19:272. doi: 10.1186/s12864-018-4620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duchêne DA, Duchêne S, Ho SYW. PhyloMAd: efficient assessment of phylogenomic model adequacy. Bioinformatics. 2018;34:2300–2301. doi: 10.1093/bioinformatics/bty103. [DOI] [PubMed] [Google Scholar]
- 66.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duchêne DA, et al. Linking branch lengths across sets of loci provides the highest statistical support for phylogenetic inference. Mol. Biol. Evol. 2020;37:1202–1210. doi: 10.1093/molbev/msz291. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 70.Salichos L, Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- 71.Minh BQ, Hahn MW, Lanfear R. New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 2020;37:2727–2733. doi: 10.1093/molbev/msaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 74.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 75.Benedict MN, Henriksen JR, Metcalf WW, Whitaker RJ, Price ND. ITEP: an integrated toolkit for exploration of microbial pan-genomes. BMC Genomics. 2014;15:8. doi: 10.1186/1471-2164-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012;804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- 77.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods. 2015;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 78.Snipen L, Liland K. H. micropan: an R-package for microbial pan-genomics. BMC Bioinform. 2015;16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Inamine JM, Ho KC, Loechel S, Hu PC. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J. Bacteriol. 1990;172:504–506. doi: 10.1128/JB.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oba T, Andachi Y, Muto A, Osawa S. Translation in vitro of codon UGA as tryptophan in Mycoplasma capricolum. Biochimie. 1991;73:1109–1112. doi: 10.1016/0300-9084(91)90153-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The salmonid related Mycoplasma MAGs are publicly available at 10.6084/m9.figshare.13019477. Summaries of each metagenome used to generate salmonid related Mycoplasma MAGs are publicly available at 10.6084/m9.figshare.13019507. Summary of pangenome used to analyse salmonid related Mycoplasma MAGs is publicly available at 10.6084/m9.figshare.13019543. Supplementary Data 2 contains source data for Fig. 4. The raw dataset generated during the current study are available in European Nucleotide Archive (ENA) repository with project accession numbers PRJEB40990. Genome accession codes for Candidatus M. salmoninae mykiss (MSM), Candidatus M. salmoninae salar (MSS), and Candidatus M. lavaretus (ML) are GCA_905477455, GCA_90547759 and GCA_905477555, respectively. All other data are available from the corresponding authors upon reasonable request.
Any code used for the study used to generate results that are reported in the paper and central to its main claims are available at https://github.com/JacobAgerbo/Comparative_Mycoplasma and on Zenodo at 10.5281/zenodo.4629623.