Abstract
Sertoli–Leydig Cell Tumors (SLCTs) are rare ovarian sex cord-stromal neoplasms, which predominantly affect adolescents and young female adults. The SLCTs clinical diagnosis and treatment remains challenging due to the rarity and the varied presentation. A large majority of SLCTs are unilateral, but also bilateral neoplasms have been reported, sometimes in the context of DICER1 syndrome. In fact, the most significant discovery regarding the molecular genetics basis of SLCTs was the finding of somatic and germline pathogenic variants in the DICER1 gene. The DICER1 protein is a key component of the micro-RNA processing pathway. Germline DICER1 pathogenic variants are typically inherited in an autosomal dominant pattern and are most often loss-of-function variants dispersed along the length of the gene. Contrarily, DICER1-related tumors harbor a characteristic missense “RNase IIIb hotspot” mutation occurring in trans, or, less frequently, loss of heterozygosity (LOH) event involving the wild-type allele. While DICER1 mutations have been identified in approximately 60% of SLCTs, especially in the moderately or poorly differentiated types, there are only a few case reports of ovarian SLCT with underlying germline DICER1 mutations. In this review, we focus on the molecular genetic features of SLCT, performing an extensive survey of all germline pathogenic variants modifying the whole sequence of the DICER1 gene. We point out that DICER1 genetic testing, coupled with an accurate variants classification and timely counseling, is of crucial importance in the clinical management of ovarian SLCT-affected patients.
Keywords: Sertoli–Leydig Cell Tumors, DICER1, molecular diagnosis
1. Introduction
Ovarian Sex Cord-Stromal Tumors (SCSTs) represent approximately 8% of ovarian cancers and comprise a heterogeneous group of malignancies. These tumors are formed by different cell types that arise from the primitive sex cords or stromal cells. Granulosa cell tumors (GCTs) represent 90% of malignant SCSTs, whereas Sertoli–Leydig Cell Tumor (SLCT) account for less than 0.5% of all ovarian cancers and for ≈15% of all ovarian stromal cell tumors [1,2]. The low incidence of SLCTs reflects in part the insufficiency of data regarding the clinical behavior of these tumors and their oncological outcomes. SLCTs are usually predominant in the 2nd and 3rd decades of life and they are characterized by the presence of a testicular structure that produces androgens. Signs and symptoms of virilization appear in two phases, characterized by defeminization followed by the phase of virilization [3]. At premenopausal age, women experience oligomenorrhea or amenorrhea, while virilization can appear with hirsutism, acne, frontal hair thinning, male pattern baldness, deepening of the voice, increased musculature, increased libido, and clitoromegaly [4,5,6,7]. SLCTs can also present with abdominal pain and increased abdominal circumference, frequently with a palpable adrenal mass at physical examination [8]. From a prognostic evaluation of patients, an important feature is the SLCTs degree of differentiation. According to the classification of Female Genital Tumors proposed by World Health Organization (WHO), SLCTs can be subdivided into well-differentiated, moderately differentiated, and poorly differentiated based on the degree of differentiation of the sertoliform component [9]. Well-differentiated tumors show a practically null malignant potential, whereas the risk increases substantially in those with lower degrees of differentiation [10,11]. Most of the ovarian SLCTs are unilateral and diagnosed in stage I, and their treatment is surgical [2,6,11,12]. Moreover, unilateral salpingo-oophorectomy is the appropriate treatment in patients that want to preserve their fertility [5,6,10]. In older patients, as well as in advanced stages, total hysterectomy and bilateral salpingo-oophorectomy or cytoreductive surgery are indicated [4,13]. Lymph node metastasis in ovarian SCSTs is rare, but relapse is not uncommon especially in advanced-stage disease (stage II to IV) [14,15].
Data from Karnezis et al. suggest at least three molecular subtypes of SLCT with distinct clinicopathologic features: DICER1 mutant (younger, more androgenic symptoms, moderately/poorly differentiated, retiform, or heterologous elements), FOXL2 mutant (postmenopausal, abnormal bleeding, moderately/poorly differentiated, no retiform or heterologous elements), and DICER1/FOXL2 wild type (intermediate age, no retiform or heterologous elements, including all well-differentiated tumors) [16]. The DICER1-mutant type is the most common type. In fact, the most significant discovery regarding the molecular genetics of SLCT was the finding of underlying somatic and germline mutations in the DICER1 gene, with the occurrence of SLCT as part of an autosomal dominant disorder named DICER1 syndrome [17,18,19]. DICER1 syndrome, also known as pleuropulmonary blastoma family tumor susceptibility syndrome, shows a large spectrum of clinical phenotypes. Affected individuals are at increased risk of developing pleuropulmonary blastoma, cystic nephroma, rhabdomyosarcoma, multinodular goiter, thyroid cancer, ovarian SLCTs, and other neoplasia. The majority of tumors occur in individuals younger than 40 years [17,18,19,20,21]. As mentioned above, DICER1 variants are typically inherited in an autosomal dominant pattern, but they may also arise de novo in the germline or in a somatic mosaic distribution [21].
Here, we present a literature review of all DICER1 germline pathogenic variants reported in patients with ovarian SLCTs. A spectrum of DICER1 germline alterations was provided, together with their comprehensive molecular and clinical description.
2. Genetics of DICER1
The human DICER1 gene is located on chromosome 14q32.13 and consists of 27 exons encoding for a 1922 amino-acid long protein (200 kDa) [21]. DICER1 is an endoribonuclease protein belonging to the ribonuclease III family and representing a key element of the RNA interference pathway. In particular, DICER1 is a component of the RNA-induced silencing complex (RISC) loading complex (RLC), which is composed of DICER1, Argonaute-2 (AGO2), and trans-activation-responsive RNA binding protein 2 (TARBP2). The DICER1 protein domains include Helicase 1/2, ATP-binding domain, Helicase, C-terminal domain, Dicer dimerization domain (DDD), PAZ domain (PAZ), ribonuclease IIIa domain, and ribonuclease IIIb domain [17]. DICER1 cleaves precursor-microRNA molecules (pre-miRNA) in the citoplasm, producing ≈20–22 nucleotide-long mature regulatory microRNAs (miRNA) [22]. Then, each miRNA is linked to the RISC. The newly formed RISC–miRNA complex binds to specific mRNA, inhibiting ribosomal access and the subsequent translation [23]. Dysregulation of miRNA production is related to a pro-oncogenic effect, as observed in several tumor types. In fact, the overexpression of one miRNA may inhibit the protein translation of a cancer suppressor gene, while the downregulation of another miRNA may increase the protein level of an oncogene [24,25,26].
Some aspects of DICER1 genetics are unique and need to be pointed out. The prevalence of germline DICER1 pathogenic variants has been estimated to be 1/10,600 in the general population [27]. These mutations are most often loss-of-function (LoF) variants dispersed along the length of the protein and are typically inherited in an autosomal dominant pattern, showing an incomplete penetrance [20,28]. The predicted functional effect of all the germline DICER1 LoF variants is essentially equivalent, resulting in a complete or near-complete LoF in miRNA processing. This prediction is based partly on nonsense-mediated decay, but it also reflects the functional domain structure of the DICER1 protein [29]. In addition, the presumed equivalence of DICER1 LoF mutations is also consistent with clinical findings: no correlations are reported between the locations of germline LoF mutations within the DICER1 gene and clinical features, such as age of disease onset, number of disease foci, specific tissue sites involved, or survival [29]. Moreover, DICER1 is not a classical tumor suppressor gene for which “two hits”—loss of function in both alleles—are required to allow tumorigenesis. In fact, in the DICER1 syndrome, the related tumors typically acquire a second missense somatic mutation (second hit) within the RNase IIIb domain that does not fully take out DICER1 ability but causes neomorphic DICER1 function in miRNA processing: the cleavage of mature 5p miRNAs from the 5′ end of pre-miRNA hairpin structures fails, while mature 3p miRNAs continue to be cleaved from the 3′ end normally [29]. This neomorphic function leads to the tumor suppression to falter when the expression of wild-type DICER1 allele is impaired due to an LoF variant or a loss of heterozygosis (LOH) event [28,29]. So, the two mutational events, the RNase IIIb missense and the LoF alterations, are required to promote the initiation of tumorigenesis. However, the occurring of a somatic mutation specifically affecting the RNase IIIb domain is considered a low-probability event, while a LoF mutation along the gene is, relatively, a high-probability event. The consequence of these lopsided probabilities is that the occurrence of an RNase IIIb hotspot mutation becomes the rate-limiting step in the onset of pathogenesis [20,29]. In addition, also the timing in which the second hit occurs is crucial. As proposed, there are apparently windows of risk for neoplastic transformation, coinciding, for example, with specific periods of organ/tissue development. This further lowers the odds of all events coinciding, and conceptually, it could explain the reduced penetrance and the variable expression of familial LoF mutations in DICER1 syndrome [29,30]. In fact, the penetrance of DICER1 pathogenic variants is generally low and is higher in female than in males. Stewart et al. reported that ≈5% of non-proband DICER1 heterozygotes were documented to have developed a neoplasm by age 10 years, which increased to ≈20% by age 50 years [30].
While most individuals with DICER1 syndrome are heterozygous for a germline LoF DICER1 pathogenic variant, other predisposing DICER1 alterations have also been documented as somatic mosaicism [31,32]. In particular, individuals with mosaicism for RNase IIIb hotspot mutations demonstrate an increased penetrance of DICER1 syndrome including earlier onset, greater number of disease foci, and greater range of phenotypes. In these cases, the first hit is the acquisition of the missense RNase IIIb hotspot mutation, while the second loss-of-function mutation can then occur anywhere across the coding region and therefore is stochastically more probable [20,29].
Finally, it was hypothesized that a DICER1 hotspot missense mutation can be sufficient to promote tumorigenesis, even in the presence of an expressed wild-type allele [32,33]. A dominant-negative effect of a RNase IIIb hotspot mutant protein over the wild-type DICER1 could account during the miRNA processing, as demonstrated by Rakheja et al. for DROSHA protein [33].
3. DICER1 Somatic Mutations in Ovarian Sertoli–Leydig Cell Tumor
In the recent years, many studies have investigated the molecular basis of ovarian SCST [34,35,36,37,38,39,40,41,42]. In 2012, using a combination of whole transcriptome and whole exome sequencing analyses, Heravi-Moussavi et al. sequenced 14 non-epithelial ovarian tumors identifying closely clustered missense mutations in the region of DICER1 gene encoding the RNase IIIb domain in a total of four samples [35]. Successively, the authors explored this region by Sanger sequencing, in additional ovarian tumors samples discovering DICER1 mutations in 30 of 102 non-epithelial ovarian tumors (29%). In particular, DICER1 mutations have been identified in approximately 60% of SLCTs (26 of a total of 43) [35]. These mutations were restricted to codons encoding metal-binding sites (p.Glu1705, p.Asp1709, p.Asp1810, p.Glu1813) within the RNase IIIb catalytic centers, which are critical for microRNA interaction and cleavage. Among these, the p.Asp1709Asn (c.5125G>A) was the most common somatic mutation identified in ovarian SLCTs samples (10/26, 38%) [35]. To date, the reason for this clusterization is not fully understood. It was proposed that these mutations specifically affect the processing of miRNA [33,43], potentially conferring a selective growth advantage and thus, they may be selected for by a tumor cell [20].
A subsequent study performed by Conlon et al. confirmed that DICER1 hotspot mutations occur in over half of ovarian SLCTs. The authors subjected 32 ovarian SLCTs to DICER1 hotspot mutation analysis using Sanger sequencing. Twenty of 32 (63%) tumors harbored a DICER1 hotspot mutation, of which 80% had the p.Glu1705Lys variant [40]. In 2017, de Kock et al. reported a very high frequency of DICER1 mutations in SLCTs: of 34 cases diagnosed as SLCTs, a total of 30 (88%) harbored ≥1 DICER1 mutations [42]. Interestingly, all the 30 moderately differentiated or poorly differentiated SLCTs harbored DICER1 mutations, while all four well-differentiated SLCTs were DICER1 wild type [41]. Based on these results, the authors speculated that well-differentiated SLCTs constitute a unique entity with a different pathogenesis from moderately differentiated and poorly differentiated SLCTs [42]. These observations have been confirmed by subsequent studies: all of the SLCTs with DICER1 mutations reported to date have been moderately and poorly differentiated; none of the well-differentiated tumors examined to date have been shown to harbor DICER1 pathogenic variants [16].
4. DICER1 Pathogenic Germline Variants in Ovarian Sertoli–Leydig Cell Tumor
4.1. Literature Review
We have catalogued all DICER1 germline pathogenic variants reported in articles published before February 28, 2012. We choose to consider only papers in which patient’s clinical information was provided. Consequently, we decide to exclude all those variants reported in common public databases (e.g., Exome Aggregation Consortium (ExAC), Genome Aggregation Database (gnomAD), and 1000 Genomes) that are missing that kind of data. Articles were identified via National Center for Biotechnology Information (NCBI) PubMed literature database using the search terms “DICER1 germline mutations and ovarian Sertoli–Leydig Cell Tumor” or “DICER1 germline variants and ovarian Sertoli–Leydig Cell Tumor”. More than 100 papers were read and evaluated for the reliable presence of DICER1 germline variants. The reference list was double checked for additional publications. After the literature review, 32 articles were identified to contain relevant information. We decided to report the identified variants according to Locus Reference Genomic (LRG) DICER1 reference sequence NM_177438.2. Table 1 provides the list of all the 43 DICER1 pathogenic variants related to ovarian SLCT. A full description of each variant was showed, including nucleotide change, amino acid change, DICER1 exon/intron number, and the protein domain involved in the mutation. The germline variant classification was assessed using VarSome tool (https://varsome.com, V6.7, Lausanne, Switzerland; last access date 23 April 2021). If a concomitant DICER1 somatic mutation was described in the same patient, it was reported in brackets below the germinal variant, as for its relative information. In addition, we provided a description of the clinical phenotype of the DICER1-mutated patient (to notice, when more individuals belonging to the same family were affected by SLCT, only the proband was considered; moreover, probands belonging to different families and carrying the same variants were individually described in Table 1). In the reference column, the paper identifying the clinical case was reported.
Table 1.
Spectrum of DICER1 pathogenic variants reported in ovarian SLCT. If described in the corresponding case report, the concomitant somatic mutation is reported in bracket below the germline one.
| Coding DNA Reference Sequence & | Exon/Intron | Codon | Protein Change | Protein Domain Δ | Variant Classification ◊ | Patient Phenotype † |
Ref. |
|---|---|---|---|---|---|---|---|
| c.325C>T | 4 | 109 | p.Gln109Ter | Helicase ATP-binding | Pathogenic | SLCT (7y), MNG (11y) | [44] |
| c.325C>T | 4 | 109 | p.Gln109Ter | Helicase ATP-binding | Pathogenic | MNG, SLCT (38y) | [45] |
| c.443_444delTC* (c.5425G>A) |
5 (25) |
148 (1809) |
p.Leu148HisfsTer22 (p.Gly1809Arg) |
Helicase ATP-binding (RNase IIIb) |
Pathogenic | SCLT (14y) | [46] |
| c.876_879delAAGA | 7 | 293 | p.Arg293IlefsTer4 | TAR RNA binding protein binding domain | Pathogenic | MNG (9y), SLCT (14y), Polyps-GI | [47] |
| c.904-1G>C (c.5439G>A) |
Intron 7 (25) |
- (1813) |
- (p.Glu1813Glu) |
- | Likely Pathogenic | MNG (12y), cERMS (17y), SLCT (18y) | [48] |
| c.1284_1285delGA (c.5437G>C) |
8 (25) |
429 (1813) |
p.Lys429AlafsTer47 (p.Glu1813Gln) |
TAR RNA binding protein binding domain (RNase IIIb) |
Pathogenic | SLCT (54y), Pulmonary Bullae | [42] |
| c.1358_1359delCA (c.5125G>A) |
8 (24) |
453 (1709) |
p.Thr453SerfsTer23 (p.Asp1709Asn) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (12y) | [46] |
| c.1376+1G>T | Intron 8 | - | - | - | Pathogenic | SLCT | [34] |
| c.1376+1G>T | Intron 8 | - | - | - | Pathogenic | PPB type II, MNG, Thyroid-PTC, SLCT | [49] |
| c.1376+1G>T | Intron 8 | - | - | - | Pathogenic | PPB Type II, NCMH, SLCT, Thyroid-DTC, Peritoneal Cyst | [29] |
| c.1376+1G>T | Intron 8 | - | - | - | Pathogenic | PPB (70y), NCMH (13y), Thyroid-PTC, SLCT | [50] |
| c.1376+1G>A (c.5439G>T) |
Intron 8 (25) |
- (1813) |
- (p.Glu1813Asp) |
- (RNase IIIb) |
Pathogenic | PPB-Type-II (5.5y),Thyroid-DTC (8y), SLCT (13.5y), NCMH (13.5y) | [51] |
| c.1376+1G>A (c.5439G>T) |
Intron 8 (25) |
- (1813) |
- (p.Glu1813Asp) |
- (RNase IIIb) |
Pathogenic | SLCT (13y), PPB, Thyroid-Nodule, Nasal Polyp/s | [42] |
| c.1507G>T | 9 | 503 | p.Glu503Ter | Helicase C-terminal | Pathogenic | SLCT | [34] |
| c.1525C>T | 10 | 509 | p.Arg509Ter | Helicase C-terminal | Pathogenic | SLCT-bilateral (8y, 14y), MNG (14y) | [52] |
| c.1525C>T (c.5125G>A) |
10 (24) |
509 (1709) |
p.Arg509Ter (p.Asp1709Asn) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (13y), Thyroid-Nodule | [42] |
| c.1532_1533delAT (c.5429A>T) |
10 (25) |
511 (1810) |
p.His511ArgfsTer16 (p.Asp1810Val) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (16y) | [53] |
| c.1532_1533delAT (c.5438A>C) |
10 (25) |
511 (1813) |
p.His511ArgfsTer16 (p.Glu1813Ala) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (28y), Thyroid-Cyst | [42] |
| c.1532_1533delAT (c.5113G>A) |
10 (24) |
511 (1705) |
p.His511ArgfsTer16 (p.Glu1705Lys) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (15y) | [42] |
| c.1532_1533delAT (c.5429A>T) |
10 (25) |
511 (1810) |
p.His511ArgfsTer16 (p.Asp1810Val) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT (16y)/JGCT | [42] |
| c.1586delC (c.5437G>C) |
10 (25) |
529 (1813) |
p.Pro529GlnfsTer33 (p.Glu1813Gln) |
Helicase C-terminal (RNase IIIb) |
Pathogenic | SLCT-bilateral (20y, 29y) | [54] |
| c.1786dupA (No somatic variant detected) |
11 | 596 | p.Thr596AsnfsTer3 | Helicase C-terminal | Pathogenic | SLCT (59y) | [46] |
| c.1827_1828insAG | 11 | 610 | p.Val610ArgfsTer8 | Between Helicase C-terminal and Dicer dsRNA-binding fold | Pathogenic | PPB Type Ir, CN, SLCT | [29] |
| c.1839delA | 11 | 614 | p.Tyr614MetfsTer3 | Between Helicase C-terminal and Dicer dsRNA-binding fold | Pathogenic | SLCT-bilateral (8y, 14y) | [55] |
| c.1966C>T | 12 | 656 | p.Arg656Ter | Dicer dsRNA-binding fold | Pathogenic | SLCT-bilateral (12y,14y) | [56] |
| c.2246_2247insACTA | 14 | 749 | p.Tyr749Ter | Between Dicer dsRNA-binding fold and PAZ | Pathogenic | SLCT | [34] |
| c.2403C>A (c.5113G>A) |
15 (24) |
801 (1705) |
p. Cys801Ter (p.Glu1705Lys) |
Between Dicer dsRNA-binding fold and PAZ (RNase IIIb) |
Pathogenic | MNG (20y), SCLT (21y) | [57] |
| c.2457C>G | 16 | 819 | p. Tyr819Ter | Between Dicer dsRNA-binding fold and PAZ | Pathogenic/Likely Pathogenic | MNG (9y), SLCT (14y) | [47] |
| c.2457C>G (c.5437G>A) |
16 (25) |
819 (1813) |
p.Tyr819Ter (p.Glu1813Lys) |
Between Dicer dsRNA-binding fold and PAZ (RNase IIIb) |
Pathogenic/Likely Pathogenic | MNG (9y), SLCT (14y) | [35] |
| c.2651-2A>T* (c.5425G > A) |
Intron 16 (25) |
- (1809) |
- (p.Gly1809Arg) |
- (RNase IIIb) |
Pathogenic | SLCT (14y) | [46] |
| c.2825delC (c.5125G>A) |
18 (24) |
942 (1709) |
p.Pro942LysfsTer6 (p.Asp1709Asn) |
PAZ (RNase IIIb) |
Pathogenic | SLCT | [35] |
| c.2903_2904delTT (c.5437G>C) |
18 (25) |
968 (1813) |
p.Phe968CysfsTer5 (p.Glu1813Gln) |
PAZ (RNase IIIb) |
Pathogenic | SLCT (20y) | [42] |
| c.2988-2_2988-1delAGinsCT | Intron 18 | - |
- |
- | Pathogenic | WT (6y), MNG (9y), SLCT (12y) | [56] |
| c.3270-6_4051-1280delinsG | 21 | 1091 | p.Tyr1091SerfsTer28 | Between PAZ and RNase IIIa | Pathogenic | SLCT (6y), MNG (14y) | [58] |
| c.3300dup (c.5437G>A) |
21 (25) |
1101 (1813) |
p.Ser1101IlefsTer3 (p.Glu1813Lys) |
Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | SLCT (53y) | [42] |
| c.3300dup (c.5113G>A) |
21 (24) |
1101 (1705) |
p. Ser1101IlefsTer3 (p.Glu1705Lys) |
Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | PPB-Type-I (1y), LC (1.8y), SLCT (18y) | [42] |
| c.3307_3311delGACAG (c.5439G>T; c.5439G>C) |
21 (25) |
1103 (1813) |
p.Asp1103GlnfsTer8 (p.Glu1813Asp) |
Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | WT-DA (5y), ASK (10y), Thyroid-FA (12y), ERMS-bladder (13y), SLCT-bilateral (15y) | [59] |
| c.3540C>A ** | 21 | 1180 | p.Tyr1180Ter | Between PAZ and RNase IIIa | Pathogenic | SLCT (16y), WDFA (16y) | [60] |
| c.3611_3616delACTACAinsT | 21 | 1204 | p.Tyr1204LeufsTer29 | Between PAZ and RNase IIIa | Pathogenic | MNG (26y), SLCT (29y) | [61] |
| c.3611_3616delACTACAinsT (c.5113G>A) |
21 (24) |
1204 (1705) |
p.Tyr1204LeufsTer29 (p.Glu1705Lys) |
Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | MNG (26y), SLCT (29y) | [35] |
| c.3647C>A *** | 21 | 1216 | p. Ser1216Ter | Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | MNG (13y), SLCT (13y) | [62] |
| c.3726C>A (c.5439G>T) |
21 (25) |
1242 (1813) |
p. Tyr1242Ter (p.Glu1813Asp) |
Between PAZ and RNase IIIa (RNase IIIb) |
Pathogenic | SLCT (13y), NCMH (21y), PPB (27y), MNG | [50] |
| c.4050+1delG | Intron 21 | - | - | - | Pathogenic | SLCT (9y), MNG (20y), cPNET (20y) | [61] |
| c.4050+1G>A | Intron 21 | - | - | - | Pathogenic | PinB (10y), ERMS-cervix, SLCT, FEP-cervix, FEP-vagina, ERMS-brainstem | [63] |
| c.4442G>A ($) | 23 | 1481 | p. Trp1481Ter | Between RNase IIIa and IIIb | Pathogenic | SLCT (6y) | [64] |
| c.4453A>T (c.5428G>C) |
23 (25) |
1485 (1810) |
p.Lys1485Ter (p.Asp1810His) |
Between RNase IIIa and IIIb (RNase IIIb) |
Pathogenic | SLCT-bilateral (12y, 14y) | [46] |
| c.4923_4926delTTTG (c.5438A>G) |
23 (25) |
1641 (1813) |
p. Cys1641Ter (p.Glu1813Gly) |
Between RNase IIIa and IIIb (RNase IIIb) |
Pathogenic | SLCT (17), MNG, Pineal Cyst, Pituitary Cyst | [42] |
| c.5018_5021delTCAA (c.5125G>A) |
23 (24) |
1673 (1709) |
p. Ile1673ThrfsTer31 (p.Asp1709Asn) |
Between RNase IIIa and IIIb (RNase IIIb) |
Pathogenic | SLCT, MNG | [35] |
| c.5122_5128delGGAGATG | 24 | 1708 | p. Gly1708ArgfsTer7 | RNase IIIb | Pathogenic | SLCT-bilateral (17y, 27y) | [56] |
| c.4207-41_5364+1034del (c.5437G>C) |
23-24 (25) |
1403-1788 (1813) |
p. Thr1403_Glu1788del (p.Glu1813Gln) |
RNase IIIb (RNase IIIb) |
Pathogenic | SLCT (13y), MNG (15y) | [65] |
| c.5364+1187T>G | Intron 24 | 1788-1789 | p. Glu1788_ L1789insValTer |
RNase IIIb | Likely Pathogenic | MNG (12y), SLCT (13y), LC (14y) | [66] |
| c.5437G>C (c.4626_4626delG) (Left ovarian) (LOH) (Right ovarian) |
25 (23) |
1813 (1542) |
p. Glu1813Gln (p.Gln1542HisfsTer18) |
RNase IIIb (Between RNase IIIa and IIIb) |
Pathogenic | RC (1.2mo), LC (2.5y),|PinB (7.7y), SLCT-bilateral (13.4y, 15.7y), CBME (17.2y), Nasal-polyps (15.1y), Thyroid-DTC (10.6y) | [31] |
| c.5441C>T | 25 | 1814 | p. Ser1814Leu | RNase IIIb | Pathogenic | Thyroid-Nodule (13y), Thyroid-DTC (18y), SLCT-bilateral (7y, 18y) | [67] |
| c.5441C>T (c.5125G>A) |
25 (24) |
1814 (1709) |
p. Ser1814Leu (p.Asp1709Asn) |
RNase IIIb (RNase IIIb) |
Pathogenic | SLCT (11.5y), MNG (12y) | [68] |
| c.5504_5507delATCC (c.5439G>T) |
25 (25) |
1835 (1813) |
p. Tyr1835SerfsTer2 (p.Glu1813Asp) |
RNase IIIb (RNase IIIb) |
Pathogenic | cERMS (13y), SLCT (13y), Thyroid-PTC (13y), MNG | [69] |
| Full gene deletion | all | - | - | all | Pathogenic | SLCT, WT-NOS | [70] |
| Full gene deletion | all | - | - | all | Pathogenic | cERMS (13y), SLCT (14y), Thyroid-Nodule (13y) | [71] |
& Variants are numbered according to the DICER1 cDNA reference sequence (GenBank accession number NM_177438.2), whereby nucleotide +1 corresponds to the A of the ATG-translation initiation codon. Δ Protein Domains: according to https://www.uniprot.org/uniprot/Q9UPY3. Last access date 23 April 2021. ◊ Classification according to VarSome Software (https://varsome.com/. Last access date 23 April 2021). * Both these variants were detected in the same patient. The allelic phase is not known. ** In the same patient, the c.4206+8_4206+ 9insTT variant was identified. The allelic phase is not known. VarSome classifies this variant as “Benign”. The frequency in the general population is ƒ = 0.00964, according to gnomAD Exomes. *** In the same patient, the c. 3649T>A (p.Tyr1217Asn) variant was identified in cis. VarSome classifies this variant as “Likely Benign”. Frequency is not reported in the general population. $ de novo variant. † Patient’s phenotype: MNG: Multinodular Goiter; Polyps-GI: Bowl Polyps; SLTC: Sertoli–Leydig Cell Tumour; cERMS: Embryonal Rhabdomyosarcoma of uterine cervix; PPB type II: Pleuropulmonary Blastoma Type II; Thyroid-PTC: Papillary Thyroid Carcinoma; NCMH: Nasal Chondromesenchymal Hamartoma; Thyroid-DTC: Differentiated Thyroid Carcinoma; PPB: Pleuropulmonary Blastoma, type not specified; JGCT: Juvenile Granulosa Cell Tumour; PPB Type Ir: Pleuropulmonary Blastoma Type I regressed; CN: Cystic Nephroma; PPB type I: Pleuropulmonary Blastoma Type; LC: Lung Cysts; WT-DA: Wilms Tumour, diffuse anaplasia; ASK: Anaplastic Sarcoma of the Kidney; Thyroid-FA: Thyroid Follicular Adenoma; ERMS-bladder: Embryonal Rhabdomyosarcoma of bladder; WDFA: Well-differentiated Fetal Lung Adenocarcinoma; cPNET: Primitive Neuroectodermal Tumour of cervix; PinB: Pineoblastoma; FEP-cervix: Cervical Fibroepithelial Polyp; FEP-vagina: Vaginal Fibroepithelial Polyp; ERMS-brainstem: Embryonal Rhabdomyosarcoma of brainstem. (y): years at diagnosis.
4.2. The Patients
A total of 57 independent cases were identified. The age at diagnosis of the SLCT was known for 46 patients with a median of 17.6 years. Even if the histological information of some SLCTs reviewed was missed in the corresponding reference papers, it is reasonable to assume that these tumors were all poorly or moderately differentiated. Nine cases of bilateral SLCT were reported. In addition to SLCT, the majority of the patients presented other DICER1 syndrome-associated diseases. In particular, an increased incidence of thyroid lesions was documented: 19 patients with Multinodular Goiter (MNG), 3 with Papillary Thyroid Carcinoma (PTC), 4 with Differentiated Thyroid Carcinoma (DTC), 4 with a thyroid nodule, 1 affected by Thyroid Follicular Adenoma (TFA), and 1 presented a thyroid cyst (Table 1). Based on these findings, some authors suggest that the association between thyroid nodules/thyroid carcinoma and SLCT is highly suggestive for DICER1 syndrome. Interestingly, in the majority of the reported cases, the diagnosis of SLCT has been made on the basis of abdominal pain and/or abdominal/pelvic mass, while data about clinical and/or biochemical hyperandrogenism are available only in few cases. In the study of Yuan et al., eleven patients had androgenic manifestations, three patients presented symptoms related to an inappropriate estrogen secretion, while three patients did not have any manifestation associated with elevated secretion of sex hormones [46]. The patient reported by Luke et al. represents the first case of secondary amenorrhea due to elevated inhibin B levels in a female adolescent with an ovarian SLCT [55]. The two siblings with DICER1 syndrome described by Zhang et al. were diagnosed with ovarian mass and irregular menstruation, but serum testosterone levels were normal in both cases [57]. Elevated testosterone levels associated with signs of hyperandrogenism have been reported in 3 cases [60,62,67].
4.3. Germline DICER1 Alterations
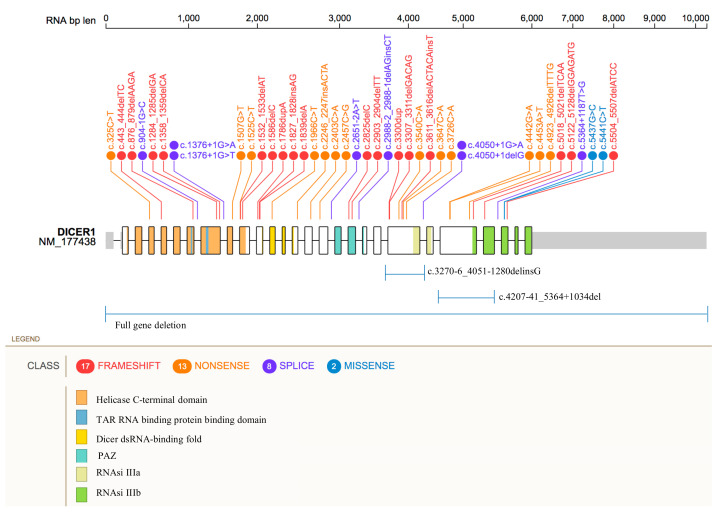
Unlike somatic mutations, which are preferentially located within the RNase IIIb domain at specific hotspot residues, as previously described, pathogenic and likely pathogenic germline variants identified in SLCTs are found to be scattered along the full length of the DICER1 gene (Figure 1).
Figure 1.
Distribution of DICER1 germline pathogenic variants. Figure 1 shows the location of the genetic variants along the DICER1 gene. SNPs and indels mutations are reported as frameshift (red), nonsense (orange), splice-site (purple), and missense (blue) variants. Large deletions are shown below. Protein domains are represented by colored areas.
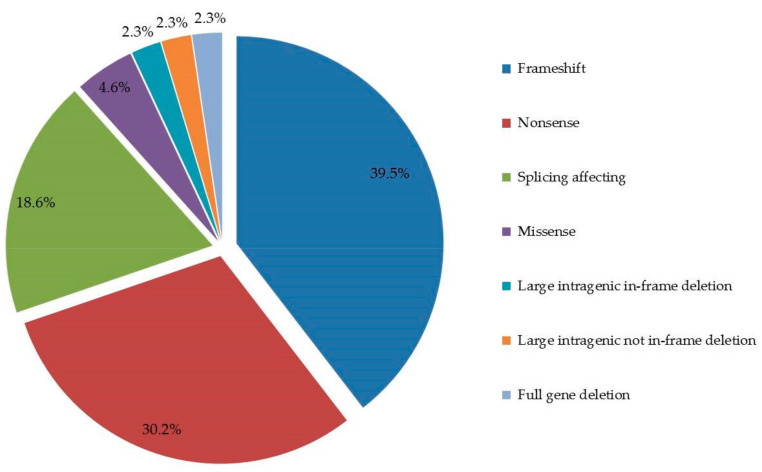
According to our results, the 43 DICER1 germline variants consist of 39.5% (17/43) frameshift mutations, 30.2% (13/43) nonsense mutations, 18.6% (8/43) splicing affecting variants, 4.6% (2/43) missense variants 2.3% (1/43), large intragenic in-frame deletion, 2.3% (1/43) large intragenic not in-frame deletion and, finally, 2.3% (1/43) full gene deletion (Figure 2).
Figure 2.
DICER1 pathogenic variants types. The figure shows the pie chart representative of the numerical proportion of the DICER1 genetic variants types.
Taking into account the distribution of the pathogenic single nucleotide variants (SNVs) and indels accounted in DICER1 gene, the exons carrying the largest number of different mutations were the numbers 21 and 23, harboring a total of six and four pathogenic variants, respectively. To note, these exons are involved also in the two intragenic large deletions, as reported in Table 1. In the remaining exons, only a few mutations (from one to three) were identified, and no pathogenic variants associated to ovarian SLCT were reported within the exons 1–3, 6, 13, 17, 19, 20, and 22. In addition, no different variants, affecting the same codon, have been identified, whereas the donor-splicing sites of introns 8 and 21 were both affected by two different mutations at +1G position (Table 1, Figure 1). The Helicase C-terminal domain, the region between PAZ and RNase IIIa and the RNase IIIb domains resulted as the DICER1 domains harboring the highest number of mutations, with a total of five, seven, and six, respectively.
To note, the two missense variants reported in Table 1 (c.5437G>C; c.5441C>T) are classified as pathogenic variants taking also into account the Varsome bioinformatic tool verdict. In fact, multiple computational evidences support a deleterious effect on the gene product. In addition, the above-mentioned variants are not reported in controls from the Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium.
Among the 43 identified variants, only the c.5437G>C (p.Glu1813Gln) was reported as germline mosaicism [31]. In the corresponding clinical case, the alternative C allele was detected in saliva-derived DNA with a frequency of 0.25%, but it was not found in the patient’s blood DNA. In addition, a second deleterious somatic variant, the c.4626delG (p.Gln1542HisfsTer18), was identified in patient’s left ovarian SLCT, whereas the DICER1 loss of heterozygosity (LOH) was detected in the right ovarian SLCT [31].
Finally, here, we mention the DICER1 c.5096-12G>A variant. This germline intronic substitution was found by Schultz et al. in two adolescents with SLCT [72]. It results in an exact duplication of six bases at the splice site of the intron 23 and exon 24 junction [72]. The predicted improper splicing would lead to the inclusion in the coding sequence of 10 bases of the intronic sequence, with the consequent frameshift and premature truncation of the protein (p.Asp1699AlafsTer8) that disrupts the RNase IIIb domain. To date, the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/. Last access date 23 April 2021) and the VarSome tool classify the c.5096-12G>A substitution as Variant of Uncertain Significance (VUS). In fact, the predicted splicing error has not been confirmed by published transcriptional studies. In conclusion, the currently available evidences are considered insufficient to determine the exact role of this variant in the disease.
5. Discussion
In recent years, many studies have investigated the molecular basis of ovarian SLCT. Consistent literature data support the role of DICER1 in the disease pathogenesis [13,14,34,35,36,37,38,39,40,41,42]. In this article, we present a comprehensive update of the germline DICER1 alterations identified in SLCTs over the past 10 years. A total of 43 pathogenic variants were collected (Table 1). These have been recognized as nonsense or frameshift mutations, leading to stop codons and truncated proteins, or nonsense-mediated RNA degradation. No mutational germline hotspots were identified.
Although rare, DICER1 syndrome can be associated with significant morbidity, often occurring at a young age. For this reason, it is important to emphasize the clinical relevance of DICER1 germline genetic testing, with the aim of detecting precancerous or lower-stage lesions [64,73]. In particular, the prompt diagnosis of SLCTs at an early stage could help avoid chemotherapy and improve survival [74]. Moreover, as recently reported by Yuan et al., the information regarding DICER1 molecular testing could have a prognostic value in SLCT clinical management, where patients harboring germline alterations may be more likely to exhibit a clinical relapse [46]. Therefore, in this context, genetic counseling, molecular sequencing analysis, and variant classification assume a decisive importance.
Full gene Sanger sequencing was the most used approach for the identification of germline DICER1 variants reported in Table 1. However, this approach may fail to detect somatic variants and mosaicism events, if accounted as low frequency alterations. On the contrary, high-throughput techniques such as Next-Generation Sequencing (NGS) performed on blood, tumor tissues, and also non-tumor samples improve the diagnostic confidence of the molecular test [29,46,49,57,59]. In our opinion, an NGS approach could also allow the prediction of Copy Number Variations (CNVs) via bioinformatics analysis of row sequencing data. In this case, the Multiplex Ligation Probe Amplification (MLPA) test could be used as a confirmatory method [58]. Finally, an integrate workflow encompassing NGS, Sanger sequencing, and MLPA is recommended [63].
We want to point out the need of the spread of DICER1 molecular testing in suggestive patients’ management due to the above-mentioned advantages and considering that this is, to date, an uncommon diagnostic practice. During our literature review analysis, we found several SLCT case reports in which the genetic test was not performed. We can speculate that the main reason is to be found above all in the difficulty in testing DICER1 gene status. In this context, the present review intended to be a support for clinicians in terms of diagnosis, management, and familiar counseling of SLCT-affected women, but also in the more comprehensive context of DICER1-syndrome affected patients.
Author Contributions
Conceptualization, P.C.; methodology, E.D.P.; validation, R.M.P.; writing—original draft preparation, P.C.; writing—review and editing, P.C., R.M.P., E.D.P.; visualization, E.D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable. The manuscript is a review of data previously deposited in public databases or sources.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from open access database and publicly archived datasets. Data are however available from the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fuller P.J., Leung D., Chu S. Genetics and genomics of ovarian sex cord-stromal tumors. Clin. Genet. 2017;91:285–291. doi: 10.1111/cge.12917. [DOI] [PubMed] [Google Scholar]
- 2.Sigismondi C., Gadducci A., Lorusso D., Candiani M., Breda E., Raspagliesi F., Cormio G., Marinaccio M., Mangili G. Ovarian Sertoli-Leydig cell tumors. A retrospective MITO study. Gynecol. Oncol. 2012;125:673–676. doi: 10.1016/j.ygyno.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Macut D., Ilić D., Mitrović Jovanović A., Bjekić-Macut J. Androgen-Secreting Ovarian Tumors. Front. Horm. Res. 2019;53:100–107. doi: 10.1159/000494906. [DOI] [PubMed] [Google Scholar]
- 4.Gui T., Cao D., Shen K., Yang J., Zhang Y., Yu Q., Wan X., Xiang Y., Xiao Y., Guo L. A clinicopathological analysis of 40 cases of ovarian Sertoli-Leydig cell tumors. Gynecol. Oncol. 2012;127:384–389. doi: 10.1016/j.ygyno.2012.07.114. [DOI] [PubMed] [Google Scholar]
- 5.Akman L., Ertas I.E., Gokcu M., Terek M.C., Sanci M., Sanli U.A., Zekioglu O., Ozsaran A.A. Ovarian sertoli-leydig cell tumors: A multicenter long-term clinicopathological analysis of 27 patients. J. Cancer Res. Ther. 2016;12:290–294. doi: 10.4103/0973-1482.158037. [DOI] [PubMed] [Google Scholar]
- 6.Bhat R.A., Lim Y.K., Chia Y.N., Yam K.L. Sertoli-Leydig cell tumor of the ovary: Analysis of a single institution database. J. Obstet. Gynaecol. Res. 2013;39:305–310. doi: 10.1111/j.1447-0756.2012.01928.x. [DOI] [PubMed] [Google Scholar]
- 7.Xiao H., Li B., Zuo J., Feng X., Li X., Zhang R., Wu L. Ovarian Sertoli-Leydig cell tumor: A report of seven cases and a review of the literature. Gynecol. Endocrinol. 2013;29:192–195. doi: 10.3109/09513590.2012.738723. [DOI] [PubMed] [Google Scholar]
- 8.Castro B.G.R., Souza C.P., Andrade C.E.M.D.C., Vieira M.A., Andrade D.A.P., Reis R.D. Ovarian Sertoli-Leydig Cell Tumors: Epidemiological, Clinical and Prognostic Factors. Rev. Bras. Ginecol. Obstet. 2019;41:440–448. doi: 10.1055/s-0039-1693056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO Classification of Tumours. [(accessed on 23 April 2021)]; Available online: https://tumourclassification.iarc.who.int/welcome/
- 10.Sahoo T.K., Kar T., Kar A., Panda S. Poorly differentiated Sertoli-Leydig cell tumour of ovary with heterologous elements. J. Clin. Diagn. Res. 2017;11:XD01–XD02. doi: 10.7860/JCDR/2017/25262.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young R.H., Scully R.E. Ovarian Sertoli-Leydig cell tumors. A clinicopathological analysis of 207 cases. Am. J. Surg. Pathol. 1985;9:543–569. doi: 10.1097/00000478-198508000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Nam S.M., Kim J.W., Eoh K.J., Kim H.M., Lee J.Y., Nam E.J., Kim S., Kim S.W., Kim Y.T. A novel clinicopathological analysis of early stage ovarian Sertoli-Leydig cell tumors at a single institution. Obstet. Gynecol. Sci. 2017;60:39–45. doi: 10.5468/ogs.2017.60.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appetecchia M., Cela V., Bernardi F., Burelli A., Cionini R., Pucci E. Sertoli-Leydig cell androgens-estrogens secreting tumor of the ovary: Ultra-conservative surgery. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004;116:113–116. doi: 10.1016/j.ejogrb.2004.01.043. [DOI] [PubMed] [Google Scholar]
- 14.Brown J., Sood A.K., Deavers M.T., Milojevic L., Gershenson D.M. Patterns of metastasis in sex cord-stromal tumors of the ovary: Can routine staging lymphadenectomy be omitted? Gynecol. Oncol. 2009;113:86–90. doi: 10.1016/j.ygyno.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Gouy S., Arfi A., Maulard A., Pautier P., Bentivegna E., Leary A., Chargari C., Genestie C., Morice P. Results from a Monocentric Long-Term Analysis of 23 Patients with Ovarian Sertoli-Leydig Cell Tumors. Oncologist. 2019;24:702–709. doi: 10.1634/theoncologist.2017-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karnezis A.N., Wang Y., Keul J., Tessier-Cloutier B., Magrill J., Kommoss S., Senz J., Yang W., Proctor L., Schmidt D., et al. DICER1 and FOXL2 Mutation Status Correlates with Clinicopathologic Features in Ovarian Sertoli-Leydig Cell Tumors. Am. J. Surg. Pathol. 2019;43:628–638. doi: 10.1097/PAS.0000000000001232. [DOI] [PubMed] [Google Scholar]
- 17.Robertson J.C., Jorcyk C.L., Oxford J.T. DICER1 Syndrome: DICER1 Mutations in Rare Cancers. Cancers. 2018;10:143. doi: 10.3390/cancers10050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz K.A.P., Stewart D.R., Kamihara J., Bauer A.J., Merideth M.A., Stratton P., Huryn L.A., Harris A.K., Doros L., Field A., et al. DICER1 Tumor Predisposition. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews®. University of Washington; Seattle, WA, USA: 2014. pp. 1993–2021. 2014 Apr 24 [Updated 2020 Apr 30] [Google Scholar]
- 19.DICER1 Syndrome Children’s Hospital of Philadelphia. [(accessed on 28 January 2021)]; Available online: http://www.chop.edu/conditions-diseases/dicer1-syndrome.
- 20.De Kock L., Wu M.K., Foulkes W.D. Ten years of DICER1 mutations: Provenance, distribution, and associated phenotypes. Hum. Mutat. 2019;40:1939–1953. doi: 10.1002/humu.23877. [DOI] [PubMed] [Google Scholar]
- 21.Thunders M., Delahunt B. Gene of the month: DICER1: Ruler and controller. J. Clin. Pathol. 2021;74:69–72. doi: 10.1136/jclinpath-2020-207203. [DOI] [PubMed] [Google Scholar]
- 22.Song M.S., Rossi J.J. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity. Biochem. J. 2017;474:1603–1618. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catalanotto C., Cogoni C., Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016;17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Huang J., Yang N., Greshock J., Megraw M.S., Giannakakis A., Liang S., Naylor T.L., Barchetti A., Ward M.R., et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acunzo M., Romano G., Wernicke D., Croce C.M. MicroRNA and cancer-a brief overview. Adv. Biol. Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Kian R., Moradi S., Ghorbian S. Role of components of microRNA machinery in carcinogenesis. Exp. Oncol. 2018;40:2–9. doi: 10.31768/2312-8852.2018.40(1):2-9. [DOI] [PubMed] [Google Scholar]
- 27.Guillerman R.P., Foulkes W.D., Priest J.R. Imaging of DICER1 syndrome. Pediatr. Radiol. 2019;49:1488–1505. doi: 10.1007/s00247-019-04429-x. [DOI] [PubMed] [Google Scholar]
- 28.Foulkes W.D., Priest J.R., Duchaine T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer. 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 29.Brenneman M., Field A., Yang J., Williams G., Doros L., Rossi C., Hill D.A. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000 Res. 2015;4:214. doi: 10.12688/f1000research.6746.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart D.R., Best A.F., Williams G.M., Harney L.A., Carr A.G., Harris A.K., Kratz C.P., Dehner L.P., Messinger Y.H., Rosenberg P.S., et al. Neoplasm Risk Among Individuals with a Pathogenic Germline Variant in DICER1. J. Clin. Oncol. 2019;37:668–676. doi: 10.1200/JCO.2018.78.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Kock L., Wang Y.C., Revil T., Badescu D., Rivera B., Sabbaghian N., Wu M., Weber E., Sandoval C., Hopman S.M., et al. High-sensitivity sequencing reveals multi-organ somatic mosaicism causing DICER1 syndrome. J. Med. Genet. 2016;53:43–52. doi: 10.1136/jmedgenet-2015-103428. [DOI] [PubMed] [Google Scholar]
- 32.Klein S., Lee H., Ghahremani S., Kempert P., Ischander M., Teitell M.A., Nelson S.F., Martinez-Agosto J.A. Expanding the phenotype of mutations in DICER1: Mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J. Med. Genet. 2014;51:294–302. doi: 10.1136/jmedgenet-2013-101943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakheja D., Chen K.S., Liu Y., Shukla A.A., Schmid V., Chang T.C., Khokhar S., Wickiser J.E., Karandikar N.J., Malter J.S., et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz K.A., Pacheco M.C., Yang J., Williams G.M., Messinger Y., Hill D.A., Dehner L.P., Priest J.R. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: A report from the International Pleuropulmonary Blastoma Registry. Gynecol. Oncol. 2011;122:246–250. doi: 10.1016/j.ygyno.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heravi-Moussavi A., Anglesio M.S., Cheng S.W., Senz J., Yang W., Prentice L., Fejes A.P., Chow C., Tone A., Kalloger S.E., et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012;366:234–242. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- 36.Witkowski L., Mattina J., Schönberger S., Murray M.J., Choong C.S., Huntsman D.G., Reis-Filho J.S., McCluggage W.G., Nicholson J.C., Coleman N., et al. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br. J. Cancer. 2013;109:2744–2750. doi: 10.1038/bjc.2013.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M.S., Lee S.H., Yoo NJLee S.H. DICER1 exons 25 and 26 mutations are rare in common human tumours besides Sertoli-Leydig cell tumour. Histopathology. 2013;63:436–438. doi: 10.1111/his.12161. [DOI] [PubMed] [Google Scholar]
- 38.Zou Y., Huang M.Z., Liu F.Y., Yang B.C., Wang L.Q., Wang F., Yu X.H., Wan L., Wan X.D., Xu X.Y., et al. Absence of DICER1, CTCF, RPL22, DNMT3A, TRRAP, IDH1 and IDH2 hotspot mutations in patients with various subtypes of ovarian carcinomas. Biomed. Rep. 2015;3:33–37. doi: 10.3892/br.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulvent T., Ray-Coquard I., Borel S., Haddad V., Devouassoux-Shisheboran M., Vacher-Lavenu M.C., Pujade-Laurraine E., Savina A., Maillet D., Gillet G., et al. DICER1 and FOXL2 mutations in ovarian sex cord-stromal tumours: A GINECO Group study. Histopathology. 2016;68:279–285. doi: 10.1111/his.12747. [DOI] [PubMed] [Google Scholar]
- 40.Conlon N., Schultheis A.M., Piscuoglio S., Silva A., Guerra E., Tornos C., Reuter V.E., Soslow R.A., Young R.H., Oliva E., et al. A survey of DICER1 hotspot mutations in ovarian and testicular sex cord-stromal tumors. Mod. Pathol. 2015;28:1603–1612. doi: 10.1038/modpathol.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato N., Kusumi T., Kamataki A., Tsunoda R., Fukase M., Kurose A. DICER1 hotspot mutations in ovarian Sertoli-Leydig cell tumors: A potential association with androgenic effects. Hum. Pathol. 2017;59:41–47. doi: 10.1016/j.humpath.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 42.de Kock L., Terzic T., McCluggage W.G., Stewart C.J.R., Shaw P., Foulkes W.D., Clarke B.A. DICER1 Mutations Are Consistently Present in Moderately and Poorly Differentiated Sertoli-Leydig Cell Tumors. Am. J. Surg. Pathol. 2017;41:1178–1187. doi: 10.1097/PAS.0000000000000895. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Chen J., Yang W., Mo F., Senz J., Yap D., Anglesio M.S., Gilks B., Morin G.B., Huntsman D.G. The oncogenic roles of DICER1 RNase IIIb domain mutations in ovarian Sertoli-Leydig cell tumors. Neoplasia. 2015;17:650–660. doi: 10.1016/j.neo.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canfarotta M., Riba-Wolman R., Orsey A.D., Balarezo F., Finck C. DICER1 syndrome and thyroid disease. J. Pediatr. Surg. Case Rep. 2016;11:31–34. doi: 10.1016/j.epsc.2016.05.014. [DOI] [Google Scholar]
- 45.Haley M., Bindal P., McAuliffe A., Vredenburgh J. A family with Sertoli-Leydig cell tumour, multinodular goiter, and DICER1 mutation. Curr. Oncol. 2019;26:183–185. doi: 10.3747/co.26.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan Z., Huo X., Jiang D., Yu M., Cao D., Wu H., Shen K., Yang J., Zhang Y., Zhou H., et al. Clinical Characteristics and Mutation Analyses of Ovarian Sertoli-Leydig Cell Tumors. Oncologist. 2020;25:e1396–e1405. doi: 10.1634/theoncologist.2020-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rio Frio T., Bahubeshi A., Kanellopoulou C., Hamel N., Niedziela M., Sabbaghian N., Pouchet C., Gilbert L., O’Brien P.K., Serfas K., et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowan M., Suntum T., Olivas A.D., Perpich M., Applebaum M.A., Lastra M.M., Yamada S.D. Second primary rhabdomyosarcoma of the uterine cervix presenting with synchronous ovarian Sertoli-Leydig cell tumor: An illustrative case of DICER1 syndrome. Gynecol. Oncol. Rep. 2018;25:94–97. doi: 10.1016/j.gore.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pugh T.J., Yu W., Yang J., Field A.L., Ambrogio L., Carter S.L., Cibulskis K., Giannikopoulos P., Kiezun A., Kim J., et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene. 2014;33:5295–5302. doi: 10.1038/onc.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart D.R., Messinger Y., Williams G.M., Yang J., Field A., Schultz K.A., Harney L.A., Doros L.A., Dehner L.P., Hill D.A. Nasal chondromesenchymal hamartomas arise secondary to germline and somatic mutations of DICER1 in the pleuropulmonary blastoma tumor predisposition disorder. Hum. Genet. 2014;133:1443–1450. doi: 10.1007/s00439-014-1474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz K.A., Yang J., Doros L., Williams G.M., Harris A., Stewart D.R., Messinger Y., Field A., Dehner L.P., Hill D.A. DICER1-pleuropulmonary blastoma familial tumor predisposition syndrome: A unique constellation of neoplastic conditions. Pathol. Case Rev. 2014;19:90–100. doi: 10.1097/PCR.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darrat I., Bedoyan J.K., Chen M., Schuette J.L., Lesperance M.M. Novel DICER1 mutation as cause of multinodular goiter in children. Head Neck. 2013;35:E369–E371. doi: 10.1002/hed.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oost E.E., Charles A., Choong C.S., Leung Y.C., Salfinger S., Sonnendecker H., Tan J., Townshend S., Witkowski L., Foulkes W.D., et al. Ovarian sex cord-stromal tumors in patients with probable or confirmed germline DICER1 mutations. Int. J. Gynecol. Pathol. 2015;34:266–274. doi: 10.1097/PGP.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 54.Schrader K.A., Cheng D.T., Joseph V., Prasad M., Walsh M., Zehir A., Ni A., Thomas T., Benayed R., Ashraf A., et al. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol. 2016;2:104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luke A.M., Moroney J.W., Snitchler A., Whiteway S.L. Ovarian Sertoli-Leydig Cell Tumor with Elevated Inhibin B as a Cause of Secondary Amenorrhea in an Adolescent with Germ Line DICER1 Mutation. J. Pediatr. Adolesc. Gynecol. 2017;30:598–600. doi: 10.1016/j.jpag.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Slade I., Bacchelli C., Davies H., Murray A., Abbaszadeh F., Hanks S., Barfoot R., Burke A., Chisholm J., Hewitt M., et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 2011;48:273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y., Ren M., Hong Y., Zhong Y., Cong X., Chen C., Liu Z., Man Y., Yang L. Sertoli-Leydig cell tumor in two siblings with DICER1 syndrome: A case report and literature review. Medicine. 2020;99:e20806. doi: 10.1097/MD.0000000000020806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabbaghian N., Srivastava A., Hamel N., Plourde F., Gajtko-Metera M., Niedziela M., Foulkes W.D. Germ-line deletion in DICER1 revealed by a novel MLPA assay using synthetic oligonucleotides. Eur. J. Hum. Genet. 2014;22:564–567. doi: 10.1038/ejhg.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen K.S., Stuart S.H., Stroup E.K., Shukla A.S., Wang J., Rajaram V., Vujanic G.M., Slone T., Rakheja D., Amatruda J.F. Distinct DICER1 Hotspot Mutations Identify Bilateral Tumors as Separate Events. JCO Precis. Oncol. 2018;2:PO.17.00113. doi: 10.1200/PO.17.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y., Chen D., Li Y., Bian L., Ma T., Xie M. DICER1 mutations in a patient with an ovarian Sertoli-Leydig tumor, well-differentiated fetal adenocarcinoma of the lung, and familial multinodular goiter. Eur. J. Med. Genet. 2014;57:621–625. doi: 10.1016/j.ejmg.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 61.Foulkes W.D., Bahubeshi A., Hamel N., Pasini B., Asioli S., Baynam G., Choong C.S., Charles A., Frieder R.P., Dishop M.K., et al. Extending the phenotypes associated with DICER1 mutations. Hum. Mutat. 2011;32:1381–1384. doi: 10.1002/humu.21600. [DOI] [PubMed] [Google Scholar]
- 62.Rossing M., Gerdes A.M., Juul A., Rechnitzer C., Rudnicki M., Nielsen F.C., Vo Hansen T. A novel DICER1 mutation identified in a female with ovarian Sertoli-Leydig cell tumor and multinodular goiter: A case report. J. Med. Case Rep. 2014;8:112. doi: 10.1186/1752-1947-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Kock L., Sabbaghian N., Druker H., Weber E., Hamel N., Miller S., Choong C.S., Gottardo N.G., Kees U.R., Rednam S.P., et al. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol. 2014;128:583–595. doi: 10.1007/s00401-014-1318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Engelen K., Villani A., Wasserman J.D., Aronoff L., Greer M.C., Tijerin Bueno M., Gallinger B., Kim R.H., Grant R., Meyn M.S., et al. DICER1 syndrome: Approach to testing and management at a large pediatric tertiary care center. Pediatr. Blood Cancer. 2018:65. doi: 10.1002/pbc.26720. [DOI] [PubMed] [Google Scholar]
- 65.Apellaniz-Ruiz M., de Kock L., Sabbaghian N., Guaraldi F., Ghizzoni L., Beccuti G., Foulkes W.D. Familial multinodular goiter and Sertoli-Leydig cell tumors associated with a large intragenic in-frame DICER1 deletion. Eur. J. Endocrinol. 2018;178:K11–K19. doi: 10.1530/EJE-17-0904. [DOI] [PubMed] [Google Scholar]
- 66.Verrier F., Dubois d’Enghien C., Gauthier-Villars M., Bonadona V., Faure-Conter C., Dijoud F., Stoppa-Lyonnet D., Houdayer C., Golmard L. Mutiple DICER1-related lesions associated with a germline deep intronic mutation. Pediatr. Blood Cancer. 2018;65:e27005. doi: 10.1002/pbc.27005. [DOI] [PubMed] [Google Scholar]
- 67.Rutter M.M., Jha P., Schultz K.A., Sheil A., Harris A.K., Bauer A.J., Field A.L., Geller J., Hill D.A. DICER1 Mutations and Differentiated Thyroid Carcinoma: Evidence of a Direct Association. J. Clin. Endocrinol. Metab. 2016;101:1–5. doi: 10.1210/jc.2015-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu M.K., de Kock L., Conwell L.S., Stewart C.J., King B.R., Choong C.S., Hussain K., Sabbaghian N., MacRae I.J., Fabian M.R., et al. Functional characterization of multiple DICER1 mutations in an adolescent. Endocr. Relat. Cancer. 2016;23:L1–5. doi: 10.1530/ERC-15-0460. [DOI] [PubMed] [Google Scholar]
- 69.Moke D.J., Thomas S.M., Hiemenz M.C., Nael A., Wang K., Shillingford N., Biegel J.A., Mascarenhas L. Three synchronous malignancies in a patient with DICER1 syndrome. Eur. J. Cancer. 2018;93:140–143. doi: 10.1016/j.ejca.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herriges J.C., Brown S., Longhurst M., Ozmore J., Moeschler J.B., Janze A., Meck J., South S.T., Andersen E.F. Identification of two 14q32 deletions involving DICER1 associated with the development of DICER1-related tumors. Eur. J. Med. Genet. 2019;62:9–14. doi: 10.1016/j.ejmg.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 71.de Kock L., Geoffrion D., Rivera B., Wagener R., Sabbaghian N., Bens S., Ellezam B., Bouron-Dal Soglio D., Ordóñez J., Sacharow S., et al. Multiple DICER1-related tumors in a child with a large interstitial 14q32 deletion. Genes Chromosomes Cancer. 2018;57:223–230. doi: 10.1002/gcc.22523. [DOI] [PubMed] [Google Scholar]
- 72.Schultz K.A., Harris A., Messinger Y., Sencer S., Baldinger S., Dehner L.P., Hill D.A. Ovarian tumors related to intronic mutations in DICER1: A report from the international ovarian and testicular stromal tumor registry. Fam. Cancer. 2016;15:105–110. doi: 10.1007/s10689-015-9831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schultz K.A.P., Williams G.M., Kamihara J., Stewart D.R., Harris A.K., Bauer A.J., Turner J., Shah R., Schneider K., Schneider K.W., et al. DICER1 and Associated Conditions: Identification of At-risk Individuals and Recommended Surveillance Strategies. Clin. Cancer Res. 2018;24:2251–2261. doi: 10.1158/1078-0432.CCR-17-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J., Field A., Schultz K.A.P., Hill D.A., Stewart D.R. The prevalence of DICER1 pathogenic variation in population databases. Int. J. Cancer. 2017;141:2030–2036. doi: 10.1002/ijc.30907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from open access database and publicly archived datasets. Data are however available from the authors upon reasonable request.