Abstract
Candida auris is a novel and major fungal pathogen that has triggered several outbreaks in the last decade. The few drugs available to treat fungal diseases, the fact that this yeast has a high rate of multidrug resistance and the occurrence of misleading identifications, and the ability of forming biofilms (naturally more resistant to drugs) has made treatments of C. auris infections highly difficult. This review intends to quickly illustrate the main issues in C. auris identification, available treatments and the associated mechanisms of resistance, and the novel and alternative treatment and drugs (natural and synthetic) that have been recently reported.
Keywords: Candida auris, resistance, antifungal, biofilm, infection, novel therapy
1. Introduction
In the last decades, fungal infections have been growing faster, presenting a serious threat to the global population, due to the antimicrobial resistance issues, but also the fact that there are much fewer drug classes available than bacterial diseases [1,2,3]. Despite the fact that the first isolate of Candida auris was reported more than one decade ago, there is still lack of a global identification strategy available, as well as in developing countries, and there is an urgent need for effective therapy against this multidrug-resistant pathogen [4,5]. Indeed, C. auris has been identified in more than 35 countries, most of them documented healthcare-associated person-to-person spread [6]. The high rate of transmission of C. auris is accelerated by its aptitude to colonize skin and other body sites, as well as its ability to persist for weeks on abiotic surfaces and equipment [7]. Importantly, whole-genome sequencing (WGS) has allowed the identification of 4 major populations (clades) in which isolates cluster by geography (isolates from Pakistan, India, South Africa, and Venezuela during 2012–2015, and a specimen from Japan). The clades are, thus, from South Asian, East Asian, African, and South American (I to IV, respectively) [8,9]. Indeed, all infections have clustered in these 4 clades; however, recently, a potential fifth clade was identified, separated from the other clades by >200,000 single-nucleotide polymorphisms, in a clinical isolate from a 14-year-old girl in Iran who had never traveled outside the country. The patient was diagnosed with C. auris otomycosis and her case was the first known C. auris case in Iran [5,10]. Although distanced by thousands of single nucleotide polymorphisms, within each clade, isolates were clonal. Diverse mutations in ERG11 were related directly to azole resistance in all geographic clades. Actually, WGS was proposed nearly immediately, and recently, independent emergence of unlike clonal populations has emerged on 3 continents [9].
The most worrisome aspects associated with infection caused by C. auris can be summarized in three levels: High rate of antifungal drug resistance [9], mode and pace of its easy transmission among healthcare workers and patients in hospitals, and misidentification. This last level is related to difficulties in a correct identification of C. auris strains, which are commonly misidentified as other phylogenetically-related pathogens, but also due to inadequacies of common diagnostic tools to precise identification of C. auris in many countries that result in misidentification and incorrect therapy, which lead to the acquisition of multiple genetic determinants that confer multidrug resistance [11,12,13,14].
This resilient pathogen survives harsh disinfectants, desiccation, high-saline environments (osmotolerance, >10% NaCl), and high temperatures (thermotolerance, >40 °C).
In addition, it rapidly colonizes abiotic surfaces, immunosuppressed individuals, and causes invasive infections that exact a high toll [7,14,15,16,17,18,19]. Studies have demonstrated that the Hog1-related stress-activated protein kinase (SAPK) signaling pathway has a central role in the C. auris reaction to osmotic stress, and it is a crucial virulence factor in several human fungi [1,2]. It is also known that patients can be asymptomatically colonized with C. auris [20].
Sources have been found within the patient’s room/bathroom including beds (e.g., bedding materials and bed side trolley), bathroom doors and walls, faucets, floors, sinks, medical equipment and disposable/reusable equipment (e.g., oxygen mask, axillary temperature probes, intravenous pole), and even on mobile phones, and can endure between one and two weeks [1,2,3,4,5,6,7,8,9]. This yeast has the ability to survive for weeks on different moist and dry abiotic surfaces (e.g., plastic and steel) [1,5,6,10,11]. On biotic surfaces, C. auris can be found on skin in general, and, in particular, on hands and nares of care providers and healthcare workers [4,9,10,11].
The colonization can spread this yeast to other patients, and colonized patients can progress to invasive and/or superficial infections. It is, thus, extremely important to quickly and correctly identify persons with C. auris, in order to contain the spread of C. auris [20].
In terms of infection patterns, a recent report showed that there seem to be no significant differences between C. auris, Candida albicans, Candida glabrata, or Candida haemulonii. The highest in vivo fungal load of the isolates was demonstrated to be detectable in kidney followed by spleen, liver, and lung tested with three different inocula on two different experimental days [1], as similar to previous works [2]. The study indicated no significant difference in the in vivo survival rate between C. auris and either C. albicans or C. glabrata. The tissue fungal burden of C. auris was also similar to C. albicans. In terms of pathogenicity, there was a comparable pathogenic potential in disseminated infection in immunocompetent hosts, which also emphasized a higher pathogenic potential of C. auris, compared to other Candida spp. [1,3].
Until now, a dramatic increase of resistance to antifungal drugs has been reported toward C. auris in different countries, causing either evasion from the efficient therapeutic options or high frequency of mortality [21,22,23,24]. To date, there is a paucity of data to guide the optimal management of C. auris. Based on current ambiguous breakpoints by the CDC, it seems that in the USA about 90, 30, and <5% of isolates have been resistant to fluconazole (Flu), amphotericin B (AmB), and echinocandins, respectively, which has resulted in a low therapeutic success [25]. Echinocandins are currently recommended as first-line therapy for infection in adults and children ≥2 months of age. For neonates and infants <2 months of age, AmB deoxycholate is recommended [26]. In accordance with previous studies, clinical isolates of C. auris represented the highest resistance against azoles globally. C. auris is intrinsically resistant to Flu and can also be resistant to echinocandin and AmB [11,21,22,27]. Both rapid emergence and pan-resistance to available therapeutic options are worrisome in hospital onset [28].
Additionally, with the ongoing coronavirus disease 2019 (COVID-19) pandemic situation concerning millions of cases worldwide, nosocomial infections including C. auris may contribute to worsening of healthcare settings [29,30,31]. Immunodeficient patients are exposed to easy hospital-acquired Candida due to its ability to persist on healthcare facilities’ surfaces notwithstanding the decontamination process [32]. Therefore, hospital measures should be strict in screening upon identification of a new C. auris case and prevent transmission and spreading by contact-tracing, such as when it was reported in Florida [30], to pre-empt outbreaks of infection. New case reports are increasing regarding C. auris infections in patients with COVID-19. Chowdhary (2020) discussed the situation in India, where for four months there was a 60% case-fatality rate of patients with diagnostic coronavirus disease, while two-thirds of them had confirmed C. auris infection [29]. Similar news has been reported in America. Four patients in the specialty care unit in Florida with COVID-19 were infected by C. auris [30]. Another alarming finding from Mexico revealed that mortality in patients with COVID-19-associated C. auris bloodstream infection was enormously high—over 83%, even with antifungal therapy [31].
To address the lack of prevalent awareness, inadequacies of conventional diagnostic systems, and therapeutic challenges, this review aimed to investigate the urgent necessity to perform accurate diagnoses and the search for and application of new therapeutic strategies to fight C. auris. A structured search of bibliographic databases for peer-reviewed research literature was conducted (e.g., PubMed, WOS), using the words “Candida+auris”, “Candida+auris+resistance”, and “Candida+auris+treatment”.
2. Candida auris Identification: Methods and Hitches
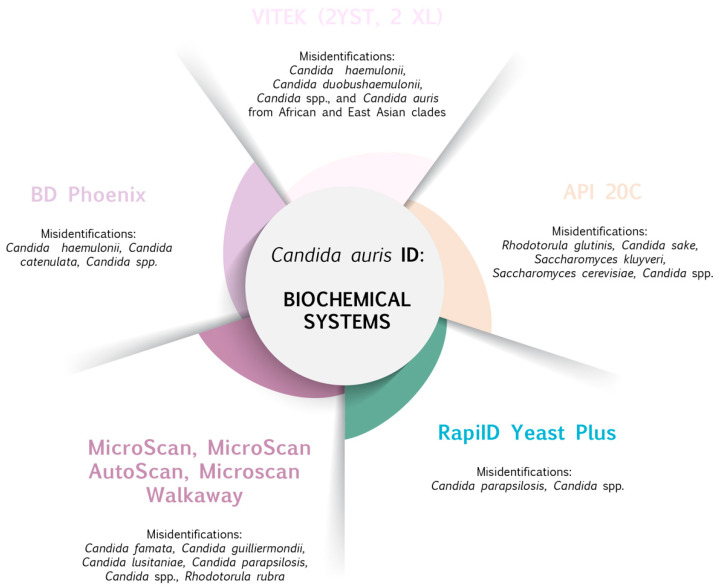
As with other major pathogens, a rapid and correct diagnostic of C. auris is essential [4]. Several methods can be used to detect C. auris from clinical samples; however, there is still the probability of misdiagnosed species/strains (Figure 1 and Figure 2). For instance, C. auris and C. haemulonii are closely related and are not readily distinguishable with phenotypic assays [33] and, taking all this into consideration, C. auris has been systematically incorrectly identified by phenotypic methods in clinical microbiology laboratories [34]. There are developing countries with a lack of available laboratory technology, where C. auris is an unknown pathogen in routine laboratories and most Candida isolates are probably misdiagnosed [5]. Importantly, correct identification of C. auris is necessary to avoid inappropriate therapy [35]. Cultivation on CHROMagar Candida was described by Kumar et al. (2017). The authors supplemented the medium with Pal’s agar for better differentiation. C. auris strains showed confluent growth of white to cream-colored smooth colonies at 37 °C and the C. haemulonii complex showed light-pink colonies at 24 h [35,36]. Nevertheless, the new medium CHROMagar™ Candida Plus (CHROMagar, France) allows for a reliable presumptive identification of C. auris, as a new specific color—light blue with a blue halo—thus obtaining a sensitivity and specificity of 100% at 36 h of incubation [37]. Another biochemical method is identification by API® 20C, though C. auris can be misidentified as Rhodotorula glutinis, Candida sake, and Saccharomyces cerevisiae [38]. Other methods are, for example, BD-Phoenix and Microscan [39]. The VITEK® 2 system is another tool for identifying C. auris strains. Ambaraghassi and colleagues (2019) evaluated a panel of 44 isolates of Candida spp. (C. auris, C. haemulonii, Candida duobushaemulonii) using the VITEK® 2 YST ID card [34]. It was also used to investigate the genomic epidemiology of C. auris isolated in Singapore [40]. Findings of both groups revealed that VITEK® 2 (version 8.01) yeast identification system has a limited ability to correctly identify C. auris and its overall performance probably differs according to the C. auris genetic clade [34].
Figure 1.
Biochemical-based methods commonly used to identify C. auris and species associated with misidentification issues [34,46,47,48].
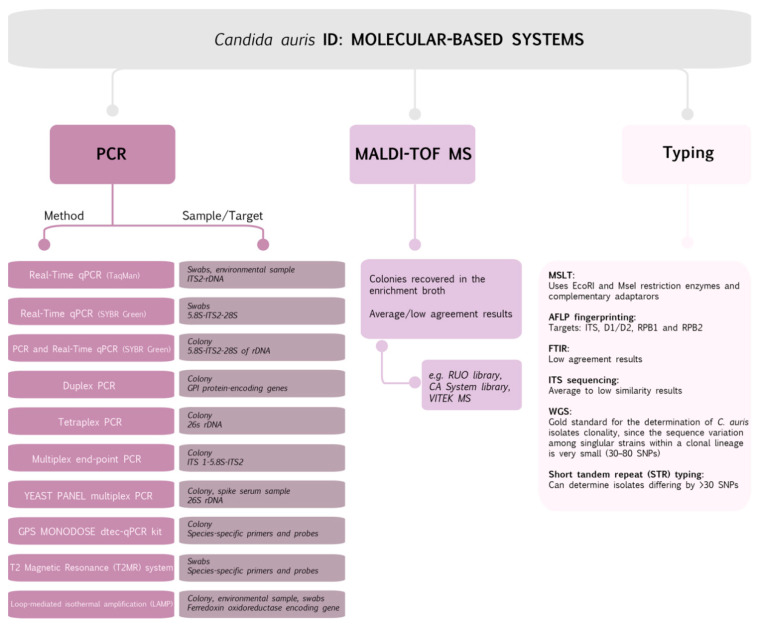
Figure 2.
Molecular-based methods commonly used to identify C. auris [12,49,50,51,52,53,54,55,56,57,58,59,60,61].
The matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based assays offer new perspectives to define the susceptibility pattern of C. auris, by species identification, antifungal susceptibility testing (AFST), and typing [41,42,43]. A study by Kwon and others was focused on two MALDI-TOF MS systems, including Biotyper and VITEK® MS, followed by AFST, sequencing of the ERG11 gene, and genotyping. Using an in vitro diagnostic library of VITEK® MS, 96.7% of the isolates were correctly identified [44]. As another example, isolates collected from blood, urine, ear swab, and groin were confirmed by MALDI-TOF MS and antifungal susceptibility testing was performed using the VITEK® 2 system to detect MICs of six antifungals [45].
It is clear how necessary it is to reliably identify C. auris by molecular methods—polymerase chain reaction (PCR) amplification and/or PCR-sequencing of ribosomal DNA (rDNA). The methods are based on sequencing of the D1–D2 region of the 28s rDNA; however, the internal transcribed spacer (ITS) region can also dependably identify C. auris [32,39,45,49]. Another option is sequences of 18S rRNA gene and phylogenetic analysis for C. auris detection [45]. Sequencing of genetic loci and PCR/qPCR assays have successfully been applied for identification of C. auris. The main advantages of PCR identification concerning GPI protein-encoding genes are its low cost, short time, and specificity because of using species-specific primers for C. auris, C. haemulonii, Candida pseudohaemulonii, C. duobushaemulonii, Candida lusitaniae, and C. albicans. Importantly, it can be used for detection of the isolate of C. auris from spiked blood and serum [4].
3. Candida auris and Treatment Failure of Common Commercial Antifungal Drugs
C andida auris is a multidrug-resistant species associated with high morbidity and mortality in immunocompromised individuals worldwide [12]. As mentioned, the key characteristics of C. auris comprise spreading easily in the environment, transiting quickly among hospitalization patients [62,63], as well as being resilient to common disinfectants in the healthcare setting [64]. The lack of availability of microbiological diagnostic methods and subsequent misidentification [65,66] and high levels of antifungal resistance [36,67] makes C. auris as a potential virulent species, and globally emerging pathogen, causing challenges to control policies against infection.
The number of antifungal drugs targeting the treatment of C. auris infection have been limited. Generally, three main classes of available antifungal including azoles, polyenes, and echinocandins have been used for treatment of infected patients in the clinics [27]. Different countries have reported various levels of resistance to common antifungals used against C. auris infections worldwide, as described by Sekyere [68]. In this study, a systematic review and meta-analysis were carried out on 267 C. auris clinical isolates in 16 countries in 2019, and the analysis of data displayed that resistance to Flu, AmB, voriconazole (Vcz), and caspofungin was 44.29%, 15.46%, 12.67%, and 3.48%, respectively. Two novel drugs, SCY-078 and VT-1598, are currently in the pipeline [68].
In contrast, the first case report in China investigated that the C. auris isolated from bronchoalveolar lavage fluid (BALF) of a hospitalized woman interestingly was sensitive to Flu, AmB, and echinocandins. Additionally, the copper sulfate (CuSO4) also had a potent antifungal effect on C. auris [69]. Likewise, C. auris isolate as a causative agent of urinary tract infection (UTI) showed sensitivity to AmB and echinocandins, and in the case of refractory and persistence of infections and different combination therapy, flucytosine are recommended as an appropriate alternatives therapy [70].
C. auris isolates of patients with candidemia in Russia were susceptible to echinocandins, whereas the high-level of resistance against Flu and AmB was seen in the majority of isolates. More investigation regarding surveillance might be helpful to achieve proper guidelines for the management of candidemia [71].
Response to antifungal agents have also indicated a significant difference between the planktonic and sessile state of C. auris growth. C. auris enable biofilm structures to form on both medical implementation surfaces in immunocompromised patients and bio-surfaces [50,72] that are strongly associated with the type and phenotypic behavior of the isolates [73]. Additionally, biofilms are capable of triggering the antifungal resistance in this species and developing the persistent infection [74]. Romea et al. found that the minimum biofilm eradication concentration (MBEC) was higher than the minimum inhibitory concentration (MIC) against antifungal drugs. In addition, biofilm was resistant to all classes of antifungals drugs; whereas, it was sensitive to echinocandins and polyenes. Therefore, the biofilm feature seems a fundamental factor to antifungal resistance in C. auris [75]. It has been reported that C. auris clinical isolates with a mutation in FKS1 gene were echinocandin resistant, whereas FKS1 wild-type (WT) isolates were sensitive. The findings suggested micafungin represented the most potent echinocandin against C. auris. In addition, all WT isolates showed the Eagle effect (paradoxical growth effect) against the caspofungin susceptibility test performed according to the CLSI microdilution method [76]. However, echinocandins are only available intravenously and are related to increasingly higher rates of resistance by C. auris. Thus, a need exists for novel treatments that reveal potent activity against C. auris. Recently, therapies focused on echinocandins’ synergism with other antifungal drugs were widely explored, representing a novel possibility for the treatment of C. auris infections [77].
4. Facing the Challenges to Fight against Candida auris: Molecular Resistance Mechanisms
The majority of clinical C. auris isolates display resistance to main classes of antifungals (azoles, polyenes, or echinocandins), and there is multi (MDR)- or pan-drug resistance to more than two antifungal classes [13]. Many research groups have studied the molecular mechanisms implicated in resistance development, resulting in limited therapeutic options [13,78]. As previously noted, in order to not fail in treatment of C. auris infections, accurate detection and identification of this pathogen is necessary [35,78]. Setting the MICs of antifungals against C. auris strains has also been requested, since elevated MICs are severe problem [79]. Importantly, it is equally critical to monitor the antifungal resistance in different geographical areas and implement efficient guidelines for treatment [45].
The antifungal resistance in C. auris has been shown to be acquired rather than intrinsic [80]. Currently, the worldwide findings conclude that susceptibility to Flu is the most decreased from C. auris isolates, followed by AmB, and 5-fluorocytosine and echinocandins are also not 100% effective [36,81]. WGS studies revealed that ERG3, ERG11, FKS1, FKS2, and FKS3 homologs were found in the C. auris genome and these loci share around 80% similarity with C. albicans and C. glabrata genes [79,82,83]. In general, preliminary genomic studies showed that the targets of several classes of antifungal drugs are conserved in C. auris, including the azole target lanosterol 14-α-demethylase (ERG11), the echinocandin target 1,3-β-(D)-glucan synthase (FKS1), and the flucytosine target uracil phosphoribosyl-transferase (FUR1) [36,84].
4.1. Mechanisms of Resistance to Azoles
Mechanistically, resistance to azole is acquired through different mechanisms; a number of genes encoding efflux pumps, including ATP-binding cassette (ABC) and overexpressed major facilitator superfamily (MFS) transporters are well known for deliberate major multidrug resistance in C. auris [82,85]. The C. auris genome contains three genes encoding Mrr1 homologs and two genes encoding Tac1 homologs. Mutations in the zinc cluster transcription factors Mrr1 and Tac1 cause intrinsic Flu resistance of C. auris [86]. Deletion of TAC1b decreased the resistance to Flu and Vcz in both mutant Flu-resistant strains from clade III and clade IV. It has been shown that the encoded transcription factor is associated with azole resistance in C. auris strains from different clades [86]. CDR1 expression was not or only minimally affected in the mutants, representing that TAC1b can confer increased azole resistance by a CDR1-independent mechanism. A recent study from Rybak et al. suggested that the Δcdr1 mutation in a resistant isolate was able to increase susceptibility to Flu and itraconazole by 64- and 128-fold, respectively, with notable reductions in MIC also demonstrated in other azoles [87,88]. The function of efflux pumps in azole resistance appears to be predominantly associated with CDR1, as analysis of the Δmdr1 mutant showed minimal effects on increasing azole sensitivity [88].
ERG11 gene mutations are mostly considered to be involved in Flu resistance or higher expression of genes participating in efflux [81,89]. Like other Candida species, specific C. auris ERG11 mutations resulted directly in reduced azole susceptibility [90]. Furthermore, genes encoding efflux pumps may play a role too [33,79,82,83]. Analysis confirmed increased expression of the CDR1 orthologous gene belonging to ABC superfamily transporters and supposed involvement in multidrug resistance in C. auris together with other transporter genes such as CDR4, CDR6, and SNQ2 [91]. Study of C. auris and C. haemulonii clinical isolates from 2 hospitals in central Israel revealed that C. auris exhibited higher thermotolerance, virulence in a mouse infection model, and ATP-dependent drug efflux activity than C. haemulonii [33]. Chawdary et al. found 15 missense mutations when aligned to the C. albicans wild-type ERG11 sequence in C. auris azole-resistant in India, and two variants were found in every resistant strain called Y132F or K143R along 5 mutations that were already identified in azole-resistant C. albicans [32,36,90]. Isolates with the K143R mutation were resistant to Vcz [32]. Research of AlJindan and colleagues showed that all selected C. auris isolates were resistant to Flu and sequencing of ERG11 revealed the same mutation (F132Y and K143R) in ERG11 [45]. Similarly, another only two-hour-lasting assay identified mutations Y132F and K143R in ERG11, and S639F in FKS1 HS1, and results were 100% concordant with DNA sequencing results [78]. From the 350 isolates collected in a hospital in India, 90% of C. auris were Flu resistant, and 2% and 8% were resistant to echinocandins and AmB, respectively [36]. Interestingly, mutations in ERG11 were markedly related with geographic clades (i.e., F126T in South Africa, Y132F in Venezuela, and Y132F or K143R in India/Pakistan) [9,79]. These variants have similarly been reported in the ERG11 gene of C. auris in Columbia [90]. Later, overexpression of ERG11 in resistant isolates of Candida spp. overcame the activity of azole drugs to inhibit the synthesis of lanosterol 14α-demethylase. Molecular experiments have shown that resistant C. auris expressed the increased level of ERG11 genes compared to susceptible species; however, until now, this was not examined on susceptible isolates [36].
Furthermore, multidrug resistance in C. auris is driven by prior antifungal prescription in hospitals and subsequent antifungal treatment failures [92,93]. However, the resistance/sensitivity profile depends on the specific clades. This is the case of C. auris isolates in India, which are almost Flu sensitive, whereas a significant level of Flu resistance has been reported in other geographic areas worldwide [80].
4.2. Mechanisms of Resistance to Polyenes
The mechanisms responsible for polyene resistance are poorly understood in C. auris. Generally, whole-genome sequencing of resistant isolates has identified four novel nonsynonymous mutations, emphasizing a probable correlation with AmB resistance. The reduction in ergosterol content in the cellular membrane [13,25] is due to overexpression of several mutated ERG genes [81]. These mutations included those in genes with homology to the transcription factor FLO8 gene of C. albicans and a membrane transporter [94]. Munoz and colleagues carried out a comparative transcriptional analysis on a resistant isolate and a sensitive isolate after exposure to AmB [84]. Using RNA sequencing (RNA-seq), it was revealed that 106 genes were induced in response to AmB in the resistant isolate. Most notably, genes involved in the ergosterol biosynthesis pathway were highly induced (ERG1, ERG2, ERG6, and ERG13), which logically showed an association with the maintenance of cell membrane stability [84]. Rhodes et al. screened 27 C. auris isolates from the UK for SNPs in ERG genes in strains representing reduced sensitivity to AmB. However, variants lighting these differences in drug susceptibility were not found [95].
4.3. Mechanisms of Resistance to Echinocandins
The development of resistance to echinocandins is rare, and they are the first-line therapy drugs against C. auris infections [13,25]. This resistance is typically associated with hot spot mutations in the FKS1 gene, which encodes the (1,3)-β-D-glucan synthase enzyme, the target of echinocandins, resulting in lower affinity of the enzyme to the drug [76]. Sequencing of the corresponding hot-spot regions of 38 C. auris strains resulted in the discovery that an S639F amino acid substitution was related to pan-echinocandin resistance [36]. Remarkably, this mutation is in the region aligning to the HS1 of C. albicans FKS1 [95]. Indeed, different mutations have also been revealed in the same location in C. auris-resistant isolates including S639Y and S639P, which led to echinocandin resistance in a mouse model in vivo [76]. Identical to other cases of Candida spp., changes in the FKS1 gene leads to caspofungin resistance [81]. On the other side, micafungin-resistant isolates C. auris from patients in Kuwait contained the S639F mutation in hot-spot 1 of FKS1 [32]. Multiple studies have reported the isolation of resistant echinocandins across various geographical regions, with the highest levels of resistance reported in India [76,96].
4.4. Mechanisms of Resistance to Flucytosine (5-Fluorocytosine)
Flucytosine, a nucleoside analog, inhibits nucleic acid synthesis. Since this antifungal compound is less used compared to other drugs, the mechanism of resistance is also less understood because fewer studies have been implemented to discover the resistance of C. auris to this drug. However, a mutation of FUR1 was shown to be associated with flucytosine resistance in non-Candida auris Candida [97], and mutations in the FCY2 and FCY1 genes also seem to be involved in resistance to 5-fluorocytosine [81]. A specific missense mutation of FUR1 causing F211I amino acid substituted in the FUR1 gene was demonstrated in resistant C. auris that had not previously been reported; therefore, more studies are required to confirm the probable mechanisms responsible for the resistance to flucytosine in the tested C. auris strain [95]. Antifungal drugs used for treatment of C. auris infection and mechanism of resistance are summarized in Table 1.
Table 1.
Antifungal drugs commonly used against C. auris infection and mechanisms of resistance already reported.
| Antifungal Drug Class | Mode of Action | Mechanism of Resistance | Reference(s) |
|---|---|---|---|
| Azoles | Inhibit the activity of lanosterol 14-α-demethylase enzyme; prevent converting lanosterol to ergosterol, leading to damaging integrity of cell membrane. | Overexpression of ATP-binding cassette (ABC) and major facilitator superfamily (MFS) transporters. ERG11 point mutation: Y132F and K143R. Mutation in zinc cluster transcription factors Mrr1 and Tac1. |
[9,32,36,45,79,82,85,86,87,88,89,91,92,93] |
| Polyenes | Bind ergosterol molecules in the cytoplasmic membrane; disturb the permeability of cell membrane by formation of pores, causing oxidative damage. | Induction of genes associated with sterol biosynthetic process including ERG1, ERG2, ERG6, and ERG13. SNPs in different genomic loci related to increased resistance. |
[81,84,94,95] |
| Echinocandins | Inhibit β-(1,3)-D-glucan synthase enzyme, leading to defective cell wall formation. | Hot-spot mutation in FKS1 gene associated with S639Y, S639P, and S639Y regions and FKS2. | [32,36,76,81,96] |
| Flucytosine | Inhibit the nucleic acid synthesis (DNA and RNA) of fungi. | Mutation of FUR1 gene, specifically missense mutation of FUR1 causing F211I amino acid substituted in the FUR1 gene in one flucytosine-resistant isolate. Mutations in the FCY2, FCY1 genes. |
[81,95,97] |
5. Combination Therapies and Novel Treatments
To fight C. auris infections, new therapy methods, drugs, and tools are being tested as an urgent demand. One of the promising approaches seems to be synergistic interactions of compounds with antifungals (Table 2). Published data have also focused on the combination therapy as alternative approaches with antifungal drugs open new insights in the management of a global threat of this multidrug-resistant C. auris, as described by Srivastava et al. [98]. In this study, farnesol, a quorum-sensing molecule, suppressed biofilm formation, blocked efflux pumps, and downregulated biofilm and efflux pump-associated genes. Farnesol has been indicated as a promising agent to fight a significant nosocomial multidrug-resistant (MDR) pathogen responsible for causing invasive outbreaks [98]. Similarly, carvacrol, the most active phenolic compound, displayed both antifungal activity with low MIC for clinical isolates of C. auris and a synergistic effect in combination with Flu, AmB, caspofungin, and micafungin, reducing the MIC value of these drugs as well. Interestingly, carvacrol strongly inhibited adherence and enzymatic activity, specifically with proteinase as a potential virulence factor developing pathogenesis [99]. Histatin 5 (Hst 5), a salivary cationic peptide, has attracted much attention due to its substantial antifungal activity against multidrug-resistant fungi. Consistently, Hst 5 at a 7.5-μM concentration killed 90% of Flu-resistant clinical isolates of C. auris, which was highly sensitive to Hst 5. Hence, Hst 5 signifies wide and potent candidacidal activity, leading it to struggle with MDR C. auris strains efficiently [100].
Table 2.
Combination therapies used to treat C. auris infections.
| Combination Therapy | Reference(s) |
|---|---|
| Micafungin and amphotericin B | [107] |
| Sulfamethoxazole, Vcz, and itraconazole | [111] |
| Lopinavir and itraconazole | [112] |
| Suloctidil (sulfur-containing aminoalcohol, vasodilator) and Vcz | [113] |
| Ebselen (synthetic organoselenium molecule) and anidulafungin | [113] |
| Aprepitant and azoles (mainly itraconazole) | [114] |
| Antimicrobial lock therapy: Caspofungin dissolved in low ionic solutions | [115,116] |
To date, researchers have been encouraged to explore newly synthesized compounds with promising anti-fungal approaches regarding C. auris growth in vitro and in vivo. APX001A, a novel agent that inhibits the fungal protein Gwt1 (glycosylphosphatidylinositol-anchored wall transfer protein 1), has been examined against C. auris. The findings considered that APX001A has efficient antifungal activity with lower MIC50 and MIC90 than anidulafungin in vitro and also in an immunocompromised murine model with disseminated infection [101].
Moreover, ibrexafungerp (SCY-078), a novel first-in-class antifungal agent targeting glucan synthase, was used to investigate in vitro activity against C. auris by applying EUCAST. Ibrexafungerp revealed a promising outcome against C. auris, including isolates resistant to echinocandins and/or other agents that were included as comparators [102]. Accordingly, treatment of biofilm with SCY-078 significantly decreased metabolic activity and thickness of sessile cells, indicating that further assessment of this novel bioavailable antifungal is needed [103]. In another study, VT-1598, a tetrazole-based fungal CYP51-specific inhibitor, was assessed in vitro and in vivo against clinical isolates of C. auris. VT-1598 significantly reduced the C. auris burden in the spleen and brain of the infected mouse in a dose-dependent manner compared to caspofungin. These results propose that VT-1598 may be an effective option for the treatment of invasive infections caused by nosocomial outbreak C. auris [104].
Surprisingly, the antifungal activity of crotamine, one of the native polypeptides from the South American rattlesnake, was examined against AmB- and Flu-resistant C. auris. This antimicrobial peptide inhibited the growth of C. auris at 40–80 µM concentration by using the Clinical and Laboratory Standards Institute (CLSI) microdilution assay in vitro. Therefore, it has been suggested that with regard to the limited efficacy of antifungal drugs, new antimicrobial peptides with a lower chance of inducing resistance compared to chemical antimicrobials have the potential power to fight against multidrug-resistant species [105].
The application of probiotic yeasts as a potential alternative/combination therapy against Candida spp., including C. albicans, Candida tropicalis, C. glabrata, Candida parapsilosis, Candida krusei, and C. auris, has been discussed in clinical studies in vitro and in vivo that effectively inhibited Candida spp. growth. The only probiotic yeast commercially available is S. cerevisiae var. boulardii, aggregated to pathogens and able reduce their virulence through inhibiting the adhesion and morphological transition of Candida spp., and could be promising therapeutic options [106].
Based on the findings Zhu’s, Jaggavarapu and colleagues announced that administration of micafungin and AmB might have clinical use against C. auris and seems to be a suitable combination instead of flucytosine [107,108], which is toxic for some groups of patients with bone marrow problems or for pregnant women [107,109,110]. Currently, the screening is focused on molecules with potential inhibitory effect; for example, sulfamethoxazole showed the most potent in vitro synergistic interactions with Vcz and itraconazole [111]. Likewise, lopinavir is an inhibitor of HIV protease but in combination with azole (mainly itraconazole) exhibited a synergistic effect against C. auris [112]. In addition, this agent sensitized C. albicans, C. tropicalis, C. krusei, and C. parapsilosis to azoles, probably by affecting efflux pump activity [112]. Findings of a similar work indicate synergistic interactions between suloctidil (sulfur-containing aminoalcohol, vasodilator) and Vcz, and between ebselen (a synthetic organoselenium molecule) and anidulafungin [113]. Another case of drug repurposing is aprepitant, an agent helping to prevent nausea and vomiting, and on the other hand to enhance antifungal effects of azoles against C. auris by chemosensitizing [114]. Authors reported interference of aprepitant/itraconazole with metal ion homeostasis, which led to losing reactive oxygen species‘ detoxification ability [114].
Biofilms formed by Candida spp. on indwelling catheters are difficult to treat. One of the options to eradicate persistent cells without removal of a catheter is antimicrobial lock therapy [115]. This strategy was selected in the work of Sumiyoshi and colleagues. Caspofungin was dissolved in low ionic solutions and a considerable reduction of candidal (including resistant C. auris) was noticed in the polymicrobial biofilms present in the catheter–lock therapy model [116].
New antifungals have been also explored. This is the case of the triazole PC945, which had a higher inhibitory potential than posaconazole, Vcz, and Flu on C. auris isolates [117]. Similarly, arylamidine T-2307 causes the collapse of fungal mitochondrial membrane potential and was previously successfully when tested in vitro and in vivo against Candida species, also demonstrating activity against C. auris [118].
Regarding novel and alternative treatments (Table 3), C. auris was found to be surprisingly sensitive to translation inhibition by a class of compounds known as rocaglates (also known as flavaglines, natural products found in Meliaceae plants), which activated a cell death program and, therefore, displayed fungicidal activity against this yeast [119].
Table 3.
Novel and alternative (synthetic and natural) therapies to treat C. auris infections.
Another alternative technique could be irradiation of photoactive dyes by a light source with appropriate wavelength, resulting in reactive oxygen species production, called photodynamic therapy. Methylene blue is a commonly used photosensitizer, and after combination with a light-emitting diode reduced the viability of planktonic as well as biofilm C. auris [120]. Finally, a pulsed-xenon ultraviolet mobile device effectively reduced the colony forming unit’s survival of C. auris [121]. Potential of UV-C exposure was investigated to constitute room disinfection. The best results for C. auris killing were obtained after 30 min of UV-C exposure at 2 m [122]. It is important to notice that C. auris has the ability to remain viable on surfaces for at least two weeks [121], which is the main reason that the decontamination processes in hospital surfaces, and on patient medical devices and rooms, is crucial [121,122].
6. Conclusions and Future Perspectives
Candida auris has recently emerged as a serious and major fungal pathogen. Although it was first identified in Japan about a decade ago, presently, several outbreaks have been claimed in several other countries (e.g., Brazil, United States, India, Mexico, Colombia, and in Europe). The main problem is the multi/pan-drug resistance that this yeast presents, which makes the treatment a higher challenge, with, unfortunately, high rates of treatment failure in all antifungals’ classes. Indeed, the fast emergence of antimicrobial resistance traits pose a significant challenge and reduce treatment options all over the world, which urgently calls for new, broadly effective and cost-efficient therapeutic strategies.
As seen, an early and correct identification of patients colonized with C. auris is critical in containing its spread. The elaboration of official guidelines (for instance, from WHO, ECDC, CDC) related to the correct isolation and culture and best methods to avoid misidentification are urgently needed. Additionally, since phenotypic resistance seems to be connected to worse outcomes, this should be taken in perspective with other patient factors (e.g., immunosuppression, previous antibiotherapy). In the occurrence of an undesirable outbreak and having to count the present data on risk factors and transmission mechanisms and the actual pandemic state, contingencies plans should be already prepared in each country, so that all nations could be prepared to act as soon as possible, guiding control measures, to avoid the spread of this pan-resistant pathogen.
Presently, the hope relies on novel and alternative treatments, such as natural compounds (e.g., rocaglates), photodynamic therapy, and novel triazoles (e.g., PC945). Finally, antimicrobial stewardship activities can increase patient results, cut drug adverse effects, and reduce antimicrobial resistance. Collectively, new, precise, and fast diagnostic methods have been established to accelerate effective patient management and progress infection control measures, finally reducing the potential for C. auris transmission. A broad attempt involving clinician, laboratory, and healthcare institutes is required to overcome the challenging of preventing transmission of and treatment for C. auris.
Acknowledgments
L.Č. thanks the Slovak Research and Development Agency under contract No. APVV-15-0347 and the grant of VEGA 1/0537/19 from the Ministry of Education, Science, Research, and the Sport of the Slovak Republic. C.F.R. would like to acknowledge the UID/EQU/00511/2020 Project—Laboratory of Process Engineering, Environment, Biotechnology and Energy (LEPABE)—financed by national funds through FCT/MCTES (PIDDAC).
Author Contributions
Conceptualization, C.F.R. and L.Č.; methodology: L.Č., M.R., S.B., S.T.; C.F.R.; validation, C.F.R., S.T. and S.B.; writing—original draft preparation: All authors; writing—review and editing: L.Č., M.R., S.B., S.T., C.F.R.; supervision, C.F.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rodrigues M.E., Gomes F., Rodrigues C.F. Candida spp./Bacteria Mixed Biofilms. J. Fungi. 2019;6:5. doi: 10.3390/jof6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes R.A., Gow N.A.R., Denning D.W., May R.C., Haynes K. Antifungal resistance: More research needed. Lancet. 2014;384:1427. doi: 10.1016/S0140-6736(14)61861-4. [DOI] [PubMed] [Google Scholar]
- 3.Misseri G., Ippolito M., Cortegiani A. Global warming “heating up” the ICU through Candida auris infections: The climate changes theory. Crit. Care. 2019;23:416. doi: 10.1186/s13054-019-2702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarado M., Bartolomé Álvarez J., Lockhart S.R., Valentín E., Ruiz-Gaitán A.C., Eraso E., de Groot P.W.J. Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses. 2021;64:194–202. doi: 10.1111/myc.13204. [DOI] [PubMed] [Google Scholar]
- 5.Abastabar M., Haghani I., Ahangarkani F., Rezai M.S., Taghizadeh Armaki M., Roodgari S., Kiakojuri K., Al-Hatmi A.M.S., Meis J.F., Badali H. Candida auris otomycosis in Iran and review of recent literature. Mycoses. 2019;62:101–105. doi: 10.1111/myc.12886. [DOI] [PubMed] [Google Scholar]
- 6.Saris K., Meis J.F., Voss A. Candida auris. Curr. Opin. Infect. Dis. 2018;31:334–340. doi: 10.1097/QCO.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 7.Welsh R.M., Bentz M.L., Shams A., Houston H., Lyons A., Rose L.J., Litvintseva A.P. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017;55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Colombo A.L., Calvo B., Cuomo C.A., Desjardins C.A., et al. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernando-Ortiz A., Mateo E., Perez-Rodriguez A., de Groot P.J.W., Quindós G., Erasoa E. Virulence of Candida auris from different clinical origins in Caenorhabditis elegans and Galleria mellonella host models. Virulence. 2021;12:1063–1075. doi: 10.1080/21505594.2021.1908765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow N.A., De Groot T., Badali H., Abastabar M., Chiller T.M., Meis J.F. Potential fifth clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019;25:1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsberg K., Woodworth K., Walters M., Berkow E.L., Jackson B., Chiller T., Vallabhaneni S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019;57:1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 12.Kordalewska M., Perlin D.S. Identification of Drug Resistant Candida auris. Front. Microbiol. 2019;10:1918. doi: 10.3389/fmicb.2019.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirabet V., Salvador C., Valentín A., Escobedo-Lucea C., Navarro L., Gimeno C., Pemán J. Risk assessment of arterial allograft contamination from tissue donors colonized by Candida auris. J. Hosp. Infect. 2021;112:49–53. doi: 10.1016/j.jhin.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Chakrabarti A., Sood P. On the emergence, spread and resistance of Candida auris: Host, pathogen and environmental tipping points. J. Med. Microbiol. 2021;70:001318. doi: 10.1099/jmm.0.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad S., Alfouzan W. Candida auris: Epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms. 2021;9:807. doi: 10.3390/microorganisms9040807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chybowska A.D., Childers D.S., Farrer R.A. Nine Things Genomics Can Tell Us about Candida auris. Front. Genet. 2020;11:351. doi: 10.3389/fgene.2020.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du H., Bing J., Hu T., Ennis C.L., Nobile C.J., Huang G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16:e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz J.F., Welsh R.M., Shea T., Batra D., Gade L., Howard D., Rowe L.A., Meis J.F., Litvintseva A.P., Cuomo C.A. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. 2021 doi: 10.1093/genetics/iyab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kean R., Delaney C., Sherry L., Borman A., Johnson E.M., Richardson M.D., Rautemaa-Richardson R., Williams C., Ramage G. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere. 2018;3 doi: 10.1128/mSphere.00334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidance for Detection of Colonization of Candida Auris|Candida Auris|Fungal Diseases|CDC. [(accessed on 19 April 2021)]; Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-guidance.html.
- 21.Ademe M., Girma F. Candida auris: From multidrug resistance to pan-resistant strains. Infect. Drug Resist. 2020;13:1287–1294. doi: 10.2147/IDR.S249864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrowsky B., Greenko J., Adams E., Quinn M., O’Brien B., Chaturvedi V., Berkow E., Vallabhaneni S., Forsberg K., Chaturvedi S., et al. Candida auris Isolates Resistant to Three Classes of Antifungal Medications—New York, 2019. Morb. Mortal. Wkly. Rep. 2020;69:6–9. doi: 10.15585/mmwr.mm6901a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortegiani A., Misseri G., Fasciana T., Giammanco A., Giarratano A., Chowdhary A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care. 2018;6:1–13. doi: 10.1186/s40560-018-0342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortegiani A., Misseri G., Giarratano A., Bassetti M., Eyre D. The global challenge of Candida auris in the intensive care unit. Crit. Care. 2019;23:150. doi: 10.1186/s13054-019-2449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaabane F., Graf A., Jequier L., Coste A.T. Review on Antifungal Resistance Mechanisms in the Emerging Pathogen Candida auris. Front. Microbiol. 2019;10:2788. doi: 10.3389/fmicb.2019.02788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong C.W., Chen S.C.A., Clark J.E., Halliday C.L., Kidd S.E., Marriott D.J., Marshall C.L., Morris A.J., Morrissey C.O., Roy R., et al. Diagnosis, management and prevention of Candida auris in hospitals: Position statement of the Australasian Society for Infectious Diseases. Intern. Med. J. 2019;49:1229–1243. doi: 10.1111/imj.14612. [DOI] [PubMed] [Google Scholar]
- 27.Pristov K.E., Ghannoum M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019;25:792–798. doi: 10.1016/j.cmi.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 28.Navalkele B.D., Revankar S., Chandrasekar P. Candida auris: A worrisome, globally emerging pathogen. Expert Rev. Anti. Infect. Ther. 2017;15:819–827. doi: 10.1080/14787210.2017.1364992. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhary A., Tarai B., Singh A., Sharma A. Multidrug-resistant candida auris infections in critically Ill Coronavirus disease patients, India, April–July 2020. Emerg. Infect. Dis. 2020;26:2694–2696. doi: 10.3201/eid2611.203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prestel C., Anderson E., Forsberg K., Lyman M., de Perio M.A., Kuhar D., Edwards K., Rivera M., Shugart A., Walters M., et al. Candida auris Outbreak in a COVID-19 Specialty Care Unit—Florida, July–August 2020. Morb. Mortal. Wkly. Rep. 2021;70:56–57. doi: 10.15585/mmwr.mm7002e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villanueva-Lozano H., Treviño-Rangel R.J., González G.M., Ramírez-Elizondo M.T., Lara-Medrano R., Aleman-Bocanegra M.C., Guajardo-Lara C.E., Gaona-Chávez N., Castilleja-Leal F., Torre-Amione G., et al. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad S., Khan Z., Al-Sweih N., Alfouzan W., Joseph L. Candida auris in various hospitals across Kuwait and their susceptibility and molecular basis of resistance to antifungal drugs. Mycoses. 2020;63:104–112. doi: 10.1111/myc.13022. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Ami R., Berman J., Novikov A., Bash E., Shachor-Meyouhas Y., Zakin S., Maor Y., Tarabia J., Schechner V., Adler A., et al. Multidrug-resistant candida haemulonii and C. Auris, tel aviv, Israel. Emerg. Infect. Dis. 2017;23:195. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambaraghassi G., Dufresne P.J., Dufresne S.F., Vallières É., Muñoz J.F., Cuomo C.A., Berkow E.L., Lockhart S.R., Luong M.L. Identification of Candida auris by use of the updated vitek 2 yeast identification system, version 8.01: A multilaboratory evaluation study. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.00884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A., Sachu A., Mohan K., Vinod V., Dinesh K., Karim S. Diferenciación sencilla y de bajo coste de Candida auris del complejo Candida haemulonii con el empleo del medio CHROMagar Candida enriquecido con medio de Pal. Rev. Iberoam. Micol. 2017;34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S., Tarai B., Singh A., Upadhyaya G., Upadhyay S., et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018;73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 37.Mulet Bayona J.V., Salvador García C., Tormo Palop N., Gimeno Cardona C. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagarTM Candida for the detection of Candida auris in surveillance samples. Diagn. Microbiol. Infect. Dis. 2020;98:115168. doi: 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed] [Google Scholar]
- 38.Clancy C.J., Nguyen M.H. Emergence of candida auris: An international call to arms. Clin. Infect. Dis. 2017;64:141–143. doi: 10.1093/cid/ciw696. [DOI] [PubMed] [Google Scholar]
- 39.Tsay S., Kallen A., Jackson B.R., Chiller T.M., Vallabhaneni S. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin. Infect. Dis. 2018;66:306–311. doi: 10.1093/cid/cix744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan Y.E., Teo J.Q.M., Rahman N.B.A., Ng O.T., Kalisvar M., Tan A.L., Koh T.H., Ong R.T.H. Candida auris in Singapore: Genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEK®2 system. Int. J. Antimicrob. Agents. 2019;54:709–715. doi: 10.1016/j.ijantimicag.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Delavy M., Dos Santos A.R., Heiman C.M., Coste A.T. Investigating Antifungal Susceptibility in Candida Species with MALDI-TOF MS-Based Assays. Front. Cell. Infect. Microbiol. 2019;9:19. doi: 10.3389/fcimb.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badali H., Wiederhold N.P. Antifungal Resistance Testing and Implications for Management. Curr. Fungal Infect. Rep. 2019;13:274–283. doi: 10.1007/s12281-019-00354-6. [DOI] [Google Scholar]
- 43.Aslani N., Janbabaei G., Abastabar M., Meis J.F., Babaeian M., Khodavaisy S., Boekhout T., Badali H. Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect. Dis. 2018;18:1–11. doi: 10.1186/s12879-017-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon Y.J., Shin J.H., Byun S.A., Choi M.J., Won E.J., Lee D., Lee S.Y., Chun S., Lee J.H., Choi H.J., et al. Candida auris clinical isolates from South Korea: Identification, antifungal susceptibility, and genotyping. J. Clin. Microbiol. 2018;57 doi: 10.1128/JCM.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aljindan R., Aleraky D.M., Mahmoud N., Abdalhamid B., Almustafa M., Abdulazeez S., Francis Borgio J. Drug resistance-associated mutations in erg11 of multidrug-resistant candida auris in a tertiary care hospital of eastern Saudi Arabia. J. Fungi. 2021;7:1–9. doi: 10.3390/jof7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Identification of Candida Auris|Candida Auris|Fungal Diseases|CDC. [(accessed on 21 April 2021)]; Available online: https://www.cdc.gov/fungal/candida-auris/identification.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Ffungal%2Fcandida-auris%2Frecommendations.html.
- 47.Kordalewska M., Zhao Y., Lockhart S.R., Chowdhary A., Berrio I., Perlin D.S. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J. Clin. Microbiol. 2017;55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fasciana T., Cortegiani A., Ippolito M., Giarratano A., Di Quattro O., Lipari D., Graceffa D., Giammanco A. Candida auris: An Overview of How to Screen, Detect, Test and Control This Emerging Pathogen. Antibiotics. 2020;9:778. doi: 10.3390/antibiotics9110778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arastehfar A., Fang W., Badali H., Vaezi A., Jiang W., Liao W., Pan W., Hagen F., Boekhout T. Low-cost tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front. Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeffery-Smith A., Taori S.K., Schelenz S., Jeffery K., Johnson E.M., Borman A., Manuel R., Browna C.S. Candida auris: A review of the literature. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sexton D.J., Kordalewska M., Bentz M.L., Welsh R.M., Perlin D.S., Litvintseva A.P. Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR green-based quantitative PCR assay. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan Z., Ahmad S., Al-Sweih N., Joseph L., Alfouzan W., Asadzadeh M. Increasing prevalence, molecular characterization and antifungal drug susceptibility of serial Candida auris isolates in Kuwait. PLoS ONE. 2018;13:e0195743. doi: 10.1371/journal.pone.0195743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leach L., Zhu Y., Chaturvedi S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2017;56 doi: 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz-Gaitán A.C., Fernández-Pereira J., Valentin E., Tormo-Mas M.A., Eraso E., Pemán J., de Groot P.W.J. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 2018;308:812–818. doi: 10.1016/j.ijmm.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad A., Spencer J.E., Lockhart S.R., Singleton S., Petway D.J., Bagarozzi D.A., Herzegh O.T. A high-throughput and rapid method for accurate identification of emerging multidrug-resistant Candida auris. Mycoses. 2019;62:513–518. doi: 10.1111/myc.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theill L., Dudiuk C., Morales-Lopez S., Berrio I., Rodríguez J.Y., Marin A., Gamarra S., Garcia-Effron G. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev. Iberoam. Micol. 2018;35:110–112. doi: 10.1016/j.riam.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Arastehfar A., Fang W., Pan W., Lackner M., Liao W., Badiee P., Zomorodian K., Badali H., Hagen F., Lass-Flörl C., et al. YEAST PANEL multiplex PCR for identification of clinically important yeast species: Stepwise diagnostic strategy, useful for developing countries. Diagn. Microbiol. Infect. Dis. 2019;93:112–119. doi: 10.1016/j.diagmicrobio.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Martínez-Murcia A., Navarro A., Bru G., Chowdhary A., Hagen F., Meis J.F. Internal validation of GPS TM MONODOSE CanAur dtec-qPCR kit following the UNE/EN ISO/IEC 17025:2005 for detection of the emerging yeast Candida auris. Mycoses. 2018;61:877–884. doi: 10.1111/myc.12834. [DOI] [PubMed] [Google Scholar]
- 59.Sexton D.J., Bentz M.L., Welsh R.M., Litvintseva A.P. Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses. 2018;61:786–790. doi: 10.1111/myc.12817. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto M., Alshahni M.M., Tamura T., Satoh K., Iguchi S., Kikuchi K., Mimaki M., Makimurab K. Rapid detection of candida auris based on loop-mediated isothermal amplification (LAMP) J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.00591-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lima A., Widen R., Vestal G., Uy D., Silbert S. A TaqMan probe-based real-time PCR assay for the rapid identification of the emerging multidrug-resistant pathogen candida auris on the BD max system. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.01604-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meis J.F., Chowdhary A. Candida auris: A global fungal public health threat. Lancet Infect. Dis. 2018;18:1298–1299. doi: 10.1016/S1473-3099(18)30609-1. [DOI] [PubMed] [Google Scholar]
- 63.Chow N.A., Gade L., Tsay S.V., Forsberg K., Greenko J.A., Southwick K.L., Barrett P.M., Kerins J.L., Lockhart S.R., Chiller T.M., et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: A molecular epidemiological survey. Lancet Infect. Dis. 2018;18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowdhary A., Sharma C., Meis J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durante A.J., Maloney M.H., Leung V.H., Razeq J.H., Banach D.B. Challenges in identifying Candida auris in hospital clinical laboratories: A need for hospital and public health laboratory collaboration in rapid identification of an emerging pathogen. Infect. Control Hosp. Epidemiol. 2018;39:1015–1016. doi: 10.1017/ice.2018.133. [DOI] [PubMed] [Google Scholar]
- 66.Lockhart S.R., Jackson B.R., Vallabhaneni S., Ostrosky-Zeichner L., Pappas P.G., Chiller T. Thinking beyond the common Candida species: Need for species-level identification of Candida due to the emergence of multidrug-resistant Candida auris. J. Clin. Microbiol. 2017;55:3324–3327. doi: 10.1128/JCM.01355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arendrup M.C., Prakash A., Meletiadis J., Sharma C., Chowdhary A. Comparison of EUCAST and CLSI reference microdilution mics of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob. Agents Chemother. 2017;61:e00485-17. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osei Sekyere J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiol. Open. 2018;7:e578. doi: 10.1002/mbo3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., Bing J., Zheng Q., Zhang F., Liu J., Yue H., Tao L., Du H., Wang Y., Wang H., et al. The first isolate of Candida auris in China: Clinical and biological aspects article. Emerg. Microbes Infect. 2018;7:1–9. doi: 10.1038/s41426-018-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Griffith N., Danziger L. Candida auris urinary tract infections and possible treatment. Antibiotics. 2020;9:898. doi: 10.3390/antibiotics9120898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barantsevich N.E., Vetokhina A.V., Ayushinova N.I., Orlova O.E., Barantsevich E.P. Candida auris bloodstream infections in russia. Antibiotics. 2020;9:557. doi: 10.3390/antibiotics9090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horton M.V., Nett J.E. Candida auris Infection and Biofilm Formation: Going beyond the Surface. Curr. Clin. Microbiol. Rep. 2020;7:51–56. doi: 10.1007/s40588-020-00143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh R., Kaur M., Chakrabarti A., Shankarnarayan S.A., Rudramurthy S.M. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses. 2019;62:706–709. doi: 10.1111/myc.12947. [DOI] [PubMed] [Google Scholar]
- 74.Dominguez E.G., Zarnowski R., Choy H.L., Zhao M., Sanchez H., Nett J.E., Andes D.R. Conserved Role for Biofilm Matrix Polysaccharides in Candida auris Drug Resistance. mSphere. 2019;4:e00680-18. doi: 10.1128/mSphereDirect.00680-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romera D., Aguilera-Correa J.J., Gadea I., Viñuela-Sandoval L., García-Rodríguez J., Esteban J. Candida auris: A comparison between planktonic and biofilm susceptibility to antifungal drugs. J. Med. Microbiol. 2019;68:1353–1358. doi: 10.1099/jmm.0.001036. [DOI] [PubMed] [Google Scholar]
- 76.Kordalewska M., Lee A., Park S., Berrio I., Chowdhary A., Zhao Y., Perlin D.S. Understanding echinocandin resistance in the emerging pathogen candida auris. Antimicrob. Agents Chemother. 2018;62:e00238-18. doi: 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cândido E.S., Affonseca F., Cardoso M.H., Franco O.L. Echinocandins as biotechnological tools for treating candida auris infections. J. Fungi. 2020;6:185. doi: 10.3390/jof6030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou X., Lee A., Jiménez-Ortigosa C., Kordalewska M., Perlin D.S., Zhao Y. Rapid detection of ERG11-associated azole resistance and FKS-associated echinocandin resistance in Candida auris. Antimicrob. Agents Chemother. 2019;63:1–7. doi: 10.1128/AAC.01811-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spivak E.S., Hanson K.E. Candida auris: An Emerging Fungal Pathogen. J. Clin. Microbiol. 2018;56:1–10. doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lockhart S.R. Candida auris and multidrug resistance: Defining the new normal. Fungal Genet. Biol. 2019;131:103243. doi: 10.1016/j.fgb.2019.103243. [DOI] [PubMed] [Google Scholar]
- 81.Frías-De-león M.G., Hernández-Castro R., Vite-Garín T., Arenas R., Bonifaz A., Castañón-Olivares L., Acosta-Altamirano G., Martínez-Herrera E. Antifungal resistance in Candida auris: Molecular determinants. Antibiotics. 2020;9:568. doi: 10.3390/antibiotics9090568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharma C., Kumar N., Pandey R., Meis J.F., Chowdhary A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016;13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma C., Kumar N., Meis J.F., Pandey R., Chowdhary A. Draft Genome Sequence of a Fluconazole-Resistant Candida auris Strain from a Candidemia Patient in India. Genome Announc. 2015;3 doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muñoz J.F., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L., Farrer R.A., Litvintseva A.P., Cuomo C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chatterjee S., Alampalli S.V., Nageshan R.K., Chettiar S.T., Joshi S., Tatu U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom. 2015;16:1–16. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mayr E.-M., Ramírez-Zavala B., Krüger I., Morschhäuser J. A Zinc Cluster Transcription Factor Contributes to the Intrinsic Fluconazole Resistance of Candida auris. mSphere. 2020;5:e00279-20. doi: 10.1128/mSphere.00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim S.H., Iyer K.R., Pardeshi L., Muñoz J.F., Robbins N., Cuomo C.A., Wong K.H., Cowen L.E. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. MBio. 2019;10:e02529-18. doi: 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rybak J.M., Doorley L.A., Nishimoto A.T., Barker K.S., Palmer G.E., Rogers P.D. Abrogation of Triazole Resistance upon Deletion of CDR1 in a Clinical Isolate of Candida auris. Antimicrob. Agents Chemother. 2019;63:e00057-19. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanglard D., Ischer F., Koymans L., Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α- demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 1998;42:241–253. doi: 10.1128/AAC.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Healey K.R., Kordalewska M., Ortigosa C.J., Singh A., Berrío I., Chowdhary A., Perlin D.S. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob. Agents Chemother. 2018;62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wasi M., Kumar Khandelwal N., Moorhouse A.J., Nair R., Vishwakarma P., Bravo Ruiz G., Ross Z.K., Lorenz A., Rudramurthy S.M., Chakrabarti A., et al. ABC transporter genes show upregulated expression in drug-resistant clinical isolates of candida auris: A genome-wide characterization of atp-binding cassette (abc) transporter genes. Front. Microbiol. 2019;10:1–16. doi: 10.3389/fmicb.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chowdhary A., Anil Kumar V., Sharma C., Prakash A., Agarwal K., Babu R., Dinesh K.R., Karim S., Singh S.K., Hagen F., et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 93.Ruiz-Gaitán A., Moret A.M., Tasias-Pitarch M., Aleixandre-López A.I., Martínez-Morel H., Calabuig E., Salavert-Lletí M., Ramírez P., López-Hontangas J.L., Hagen F., et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61:498–505. doi: 10.1111/myc.12781. [DOI] [PubMed] [Google Scholar]
- 94.Escandón P., Chow N.A., Caceres D.H., Gade L., Berkow E.L., Armstrong P., Rivera S., Misas E., Duarte C., Moulton-Meissner H., et al. Molecular epidemiology of candida auris in Colombia Reveals a Highly Related, Countrywide Colonization with Regional Patterns in Amphotericin B Resistance. Clin. Infect. Dis. 2019;68:15–21. doi: 10.1093/cid/ciy411. [DOI] [PubMed] [Google Scholar]
- 95.Rhodes J., Abdolrasouli A., Farrer R.A., Cuomo C.A., Aanensen D.M., Armstrong-James D., Fisher M.C., Schelenz S. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris article. Emerg. Microbes Infect. 2018;7:1–12. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rudramurthy S.M., Chakrabarti A., Paul R.A., Sood P., Kaur H., Capoor M.R., Kindo A.J., Marak R.S.K., Arora A., Sardana R., et al. Candida auris candidaemia in Indian ICUs: Analysis of risk factors. J. Antimicrob. Chemother. 2017;72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 97.Vandeputte P., Ferrari S., Coste A.T. Antifungal resistance and new strategies to control fungal infections. Int. J. Microbiol. 2012;68:15–21. doi: 10.1155/2012/713687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srivastava V., Ahmad A. Abrogation of pathogenic attributes in drug resistant Candida auris strains by farnesol. PLoS ONE. 2020;15:e0233102. doi: 10.1371/journal.pone.0233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaban S., Patel M., Ahmad A. Improved efficacy of antifungal drugs in combination with monoterpene phenols against Candida auris. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-58203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pathirana R.U., Friedman J., Norris H.L., Salvatori O., McCall A.D., Kay J., Edgerton M. Fluconazole-resistant Candida auris is susceptible to salivary histatin 5 killing and to intrinsic host defenses. Antimicrob. Agents Chemother. 2018;62:e01872-17. doi: 10.1128/AAC.01872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hager C.L., Larkin E.L., Long L., Abidi F.Z., Shaw K.J., Ghannoum M.A. In vitro and in vivo evaluation of the antifungal activity of APX001A/APX001 against candida auris. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arendrup M.C., Jørgensen K.M., Hare R.K., Chowdhary A. In vitro activity of ibrexafungerp (SCY-078) against Candida auris Isolates as Determined by EUCAST methodology and comparison with activity against C. Albicans and C. Glabrata and with the activities of six comparator agents. Antimicrob. Agents Chemother. 2020;64:e02136-19. doi: 10.1128/AAC.02136-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Larkin E., Hager C., Chandra J., Mukherjee P.K. The Emerging Pathogen Candida auris: Growth Phenotype, Virulence Factors, Activity of Antifungals, and Effect of SCY-078, a Novel Glucan Synthesis Inhibitor, on Growth Morphology and Biofilm Formation Emily. Antimicrob Agents Chemother. 2017;61:1–13. doi: 10.1128/AAC.02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wiederhold N.P., Lockhart S.R., Najvar L.K., Berkow E.L., Jaramillo R., Olivo M., Garvey E.P., Yates C.M., Schotzinger R.J., Catano G., et al. The Fungal Cyp51-Specific Inhibitor VT-1598 Demonstrates In Vitro and In Vivo Activity against Candida auris. Antimicrob. Agents Chemother. 2019;63:e02233-18. doi: 10.1128/AAC.02233-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Colombo A.L., Dal Mas C., Rossato L., Shimizu T., Oliveira E.B., Da Silva Junior P.I., Meis J.F., Hayashi M.A.F. Effects of the natural peptide crotamine from a south american rattlesnake on candida auris, an emergent multidrug antifungal resistant human pathogen. Biomolecules. 2019;9:205. doi: 10.3390/biom9060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kunyeit L., KA A.-A., Rao R.P. Application of Probiotic Yeasts on Candida Species Associated Infection. J. Fungi. 2020;6:189. doi: 10.3390/jof6040189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jaggavarapu S., Burd E.M., Weiss D.S. Micafungin and amphotericin B synergy against Candida auris. The Lancet Microbe. 2020;1:e314–e315. doi: 10.1016/S2666-5247(20)30194-4. [DOI] [PubMed] [Google Scholar]
- 108.Zhu Y.C., Barat S.A., Borroto-Esoda K., Angulo D., Chaturvedi S., Chaturvedi V. Pan-resistant Candida auris isolates from the outbreak in New York are susceptible to ibrexafungerp (a glucan synthase inhibitor) Int. J. Antimicrob. Agents. 2020;55:105922. doi: 10.1016/j.ijantimicag.2020.105922. [DOI] [PubMed] [Google Scholar]
- 109.Fujii S., Yabe K., Kariwano-Kimura Y., Furukawa M., Itoh K., Matsuura M., Horimoto M. Developmental toxicity of flucytosine following administration to pregnant rats at a specific time point of organogenesis. Congenit. Anom. 2019;59:39–42. doi: 10.1111/cga.12282. [DOI] [PubMed] [Google Scholar]
- 110.Vermes A., Guchelaar H.J., Dankert J. Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 2000;46:171–179. doi: 10.1093/jac/46.2.171. [DOI] [PubMed] [Google Scholar]
- 111.Eldesouky H.E., Li X., Abutaleb N.S., Mohammad H., Seleem M.N. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int. J. Antimicrob. Agents. 2018;52:754–761. doi: 10.1016/j.ijantimicag.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 112.Eldesouky H.E., Salama E.A., Lanman N.A., Hazbun T.R., Seleem M.N. Potent synergistic interactions between lopinavir and azole antifungal drugs against emerging multidrug-resistant candida auris. Antimicrob. Agents Chemother. 2021;65:1–5. doi: 10.1128/AAC.00684-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Oliveira H.C., Monteiro M.C., Rossi S.A., Pemán J., Ruiz-Gaitán A., Mendes-Giannini M.J.S., Mellado E., Zaragoza O. Identification of Off-Patent Compounds That Present Antifungal Activity against the Emerging Fungal Pathogen Candida auris. Front. Cell. Infect. Microbiol. 2019;9:1–10. doi: 10.3389/fcimb.2019.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eldesouky H.E., Lanman N.A., Hazbun T.R., Seleem M.N. Aprepitant, an antiemetic agent, interferes with metal ion homeostasis of Candida auris and displays potent synergistic interactions with azole drugs. Virulence. 2020;11:1466–1481. doi: 10.1080/21505594.2020.1838741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walraven C.J.C.J., Lee S.A.S.A. Antifungal lock therapy. Antimicrob. Agents Chemother. 2013;57:1–8. doi: 10.1128/AAC.01351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sumiyoshi M., Miyazaki T., Makau J.N., Mizuta S., Tanaka Y., Ishikawa T., Makimura K., Hirayama T., Takazono T., Saijo T., et al. Novel and potent antimicrobial effects of caspofungin on drug-resistant Candida and bacteria. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-74749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rudramurthy S.M., Colley T., Abdolrasouli A., Ashman J., Dhaliwal M., Kaur H., Armstrong-James D., Strong P., Rapeport G., Schelenz S., et al. In vitro antifungal activity of a novel topical triazole PC945 against emerging yeast Candida auris. J. Antimicrob. Chemother. 2019;74:2943–2949. doi: 10.1093/jac/dkz280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wiederhold N.P., Najvar L.K., Jaramillo R., Olivo M., Patterson H., Connell A., Fukuda Y., Mitsuyama J., Catano G., Patterson T.F. The novel arylamidine T-2307 demonstrates in vitro and in vivo Activity against Candida auris. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02198-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iyer K.R., Whitesell L., Porco J.A., Henkel T., Brown L.E., Robbins N., Cowen L.E. Translation inhibition by rocaglates activates a species-specific cell death program in the emerging fungal pathogen candida auris. MBio. 2020;11:1–17. doi: 10.1128/mBio.03329-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tan J., Liu Z., Sun Y., Yang L., Gao L. Inhibitory Effects of Photodynamic Inactivation on Planktonic Cells and Biofilms of Candida auris. Mycopathologia. 2019;184:525–531. doi: 10.1007/s11046-019-00352-9. [DOI] [PubMed] [Google Scholar]
- 121.Maslo C., Du Plooy M., Coetzee J. The efficacy of pulsed-xenon ultraviolet light technology on Candida auris. BMC Infect. Dis. 2019;19:1–3. doi: 10.1186/s12879-019-4137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.De Groot T., Chowdhary A., Meis J.F., Voss A. Killing of Candida auris by UV-C: Importance of exposure time and distance. Mycoses. 2019;62:408–412. doi: 10.1111/myc.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]