Abstract
Abstract
Specific inhibition of the viral RNA-dependent RNA polymerase (RdRp) of the newly-emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a very promising strategy for developing highly potent medicines for coronavirus disease 2019 (COVID-19). However, almost all of the reported viral RdRp inhibitors (either repurposed drugs or new antiviral agents) lack selectivity against the SARS-CoV-2 RdRp. Herein, I discovered a new favipiravir derivative, (E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide (cyanorona-20), as the first potent SARS-CoV-2 inhibitor with very high selectivity (209- and 45-fold more potent than favipiravir and remdesivir, respectively). Based on the significant reduction in the in vitro SARS-CoV-2 replication/copies, strong computational cyanorona-20 ligand-RdRp protein interactions, and anti-RdRp activity of the parent favipiravir drug, SARS-CoV-2 inhibition is thought to be mediated through the coronaviral-2 RdRp inhibition. This promising selective anti-COVID-19 compound is also, to the best of our knowledge, the first bioactive derivative of favipiravir, the known antiinfluenza and antiviral drug. This new nucleoside analog was designed, synthesized, characterized, computationally studied (through pharmacokinetic calculations along with computational molecular modeling and prediction), and biologically evaluated for its anti-COVID-19 activities (through a validated in vitro anti-COVID-19 assay). The results of the biological assay showed that cyanorona-20 surprisingly exhibited very significant anti-COVID-19 activity (anti-SARS-CoV-2 EC50 = 0.45 μM), and, in addition, it could be also a very promising lead compound for the design of new anti-COVID-19 agents. Cyanorona-20 is a new favipiravir derivative with promise for the treatment of SARS-CoV-2 infection.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11696-021-01640-9.
Keywords: Anti-COVID-19 drug, Coronavirus, SARS-CoV-2, Coronaviral RNA-dependent RNA polymerase (RdRp), Favipiravir, Drug discovery, Cyanorona-20
Introduction
Recently in December 2019, a novel coronavirus (2019-nCoV), officially known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; Fig. 1), suddenly emerged in Wuhan (Wuhan City, Hubei Province, China) (Hui et al. 2020). Despite drastic containment measures, the transmission of this virus is ongoing leading to the spread of the coronavirus disease 2019 (COVID-19) with its major symptoms localizing in the respiratory system of human (i.e., characterized by loss of pulmonary function in humans) (Hui et al. 2020; Li et al. 2020). This outbreak of 2019-nCoV infection has spread across our planet (Hui et al. 2020; Li et al. 2020). Currently, at the end of December 2020, about 85 million COVID-19 cases have been confirmed worldwide, with more than 1.85 millions of lives lost due to this disease (COVID-19 Map 2020). The best efforts of multinational pharmaceutical companies, drug discovery research centers/institutes, international health authorities, and pharmaceutical/medical colleges have focused on the search for effective medications and therapies able to combat the virus (Li et al. 2020; Jiang et al. 2020). No specific antiviral drugs have been officially approved for the treatment of COVID-2019 (Jiang et al. 2020).
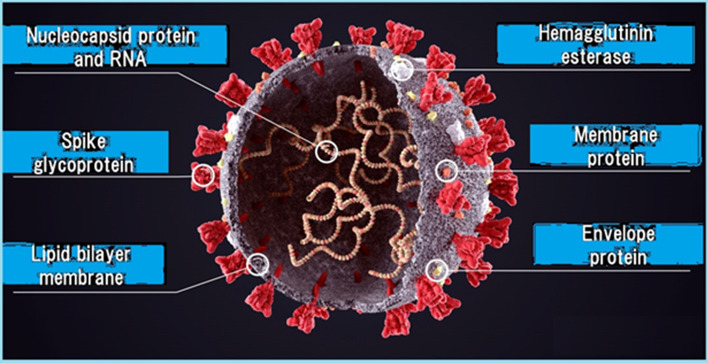
Fig. 1.
A diagrammatic representation of SARS-CoV-2 morphology (its shape when entering the human body carried, mainly, into respiratory droplets) and anatomy (structure)
In this absence of effective anti-COVID-19 therapy, some researchers have suggested the repurposing of the known potent antiinfluenza drug favipiravir (chemically, it is a purine nucleoside analog; approved for medical use in Japan since 2014; Fig. 2) to counteract the novel COVID-19 (Shiraki and Daikoku 2020; Dong et al. 2020; Łagocka et al. 2021; Driouich et al. 2021). Previous studies identified viral RNA-dependent RNA polymerase (RdRp) as a potential drug target in COVID-19 treatment due to its crucial role in SARS-CoV-2 replication and transcription (i.e., in the virus life cycle) (Zhang and Tang 2021). Furthermore, this enzyme has a strategic advantage of being absent in the coronavirus-uninfected human cells (i.e., viral RdRp is a drug target selective for SARS-CoV-2 particles) (Dong et al. 2020; Venkataraman et al. 2018; Wu et al. 2020). Favipiravir, as an antiviral agent, acts by selectively inhibiting viral RdRp (some other researches suggest that favipiravir, in addition of being a potent RdRp inhibitor, induces lethal RNA transversion mutations, thus producing a nonviable viral phenotype) (Shiraki and Daikoku 2020; Furuta et al. 2013). Favipiravir is a prodrug that is metabolized to its active form, favipiravir-ribofuranosyl-5′-triphosphate (favipiravir-RTP), mainly by the enzyme human hypoxanthine-guanine phosphoribosyltransferase (HGPRT) in order to stop the replication process of the viral RNA genome (i.e., of the virus) (Smee et al. 2009). However, limitations have restricted the use of favipiravir as an efficient anti-COVID-19 agent till now, e.g., reliable data regarding in vitro SARS-CoV-2 inhibition are still not available (Shiraki and Daikoku 2020; Dong et al. 2020); broad data regarding in vivo SARS-CoV-2 inhibition and efficacy in preclinical animal studies are still not available (Dong et al. 2020; Cai et al. 2020); the few available animal experiments of favipiravir show the potential for teratogenic effects (Shiraki and Daikoku 2020); favipiravir has not been shown to be effective in primary human airway cells (Yoon et al. 2018); lack of additional virus-toxic functional groups in favipiravir structure to augment its antiviral mechanism of action against the lethal and resistant SARS-CoV-2 (Dong et al. 2020; Abdelnabi et al. 2017); lipophilic/hydrophilic properties of favipiravir are not adequately balanced to achieve maximal bioavailability and distribution (especially to the lungs) in humans (Du and Chen 2020); expected binding affinities of active favipiravir-RTP molecule (as a viral RdRp inhibitor) with SARS-CoV-2 RdRp enzyme protein are not that great (as concluded mainly from the studies of active favipiravir-RTP binding affinities with the viral polymerase, e.g., Abdelnabi et al. 2017); favipiravir/favipiravir-RTP structure is not an ideal hydrogen bond acceptor (Naydenova et al. 2021); data concerning its clinical use in humans are not clear (Shiraki and Daikoku 2020; Cai et al. 2020; Du and Chen 2020); favipiravir as an anti-COVID-19 drug is used primarily off-label (in Japan) as its use as anti-COVID-19 has not been approved (Shiraki and Daikoku 2020; Du and Chen 2020); and many favipiravir published articles and papers have irreproducible data (Dong et al. 2020; Cai et al. 2020).
Fig. 2.
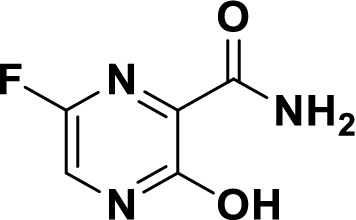
Chemical structure of favipiravir (6-fluoro-3-hydroxypyrazine-2-carboxamide)
These limitations of favipiravir use against COVID-19 prompted us to design a derivative of favipiravir to better inhibit SARS-CoV-2 RdRp. The goal was an improved favipiravir structure with respect to drug-likeness, structure-stabilizing properties, small molecular weight, viral replication inhibition, and biological compatibility. After extensive molecular modeling (of compound libraries with SARS-CoV-2 RdRp), we identified (E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide (cyanorona-20; Fig. 3) as a new derivative of favipiravir expected to have anti-COVID-19 activities.
Fig. 3.
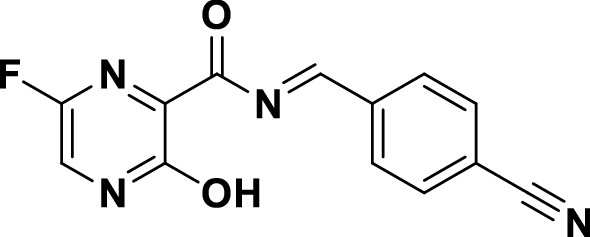
Chemical structure of the newly-designed target compound cyanorona-20 ((E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide)
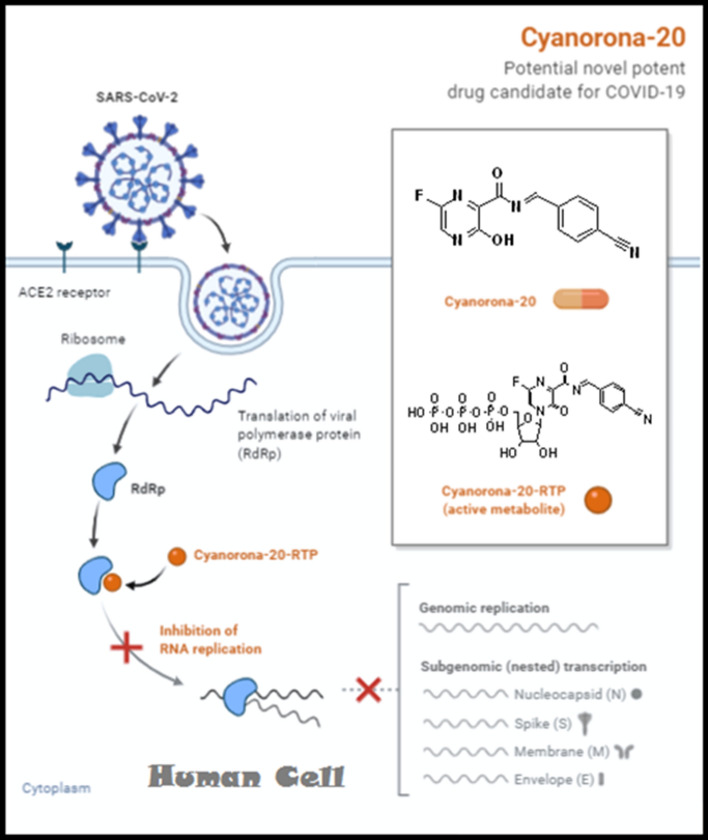
Cyanorona-20 ("cyano" stands for the cyano group, which is the major new moiety added in this derivative of favipiravir; "no" stands for the same word, no; "norona" stands for coronavirus; and "20" stands for the year in which this drug has been discovered, i.e., in 2020) is the 4-cyanobenzylidene derivative of favipiravir at the amino group, and is expected to be a prodrug that is metabolized inside the human body to its active nucleotide triphosphate form, cyanorona-20-ribofuranosyl-5′-triphosphate (cyanorona-20-RTP). Cyanorona-20 molecule has a high degree of drug-likeness (obeys Lipinski's rule of five "Ro5") and fulfills the structural requirements for a potent anti-COVID-19 agent. Figure 4 summarizes the proposed mechanism of anti-COVID-19 action of cyanorona-20.
Fig. 4.
A representation of cyanorona-20 major mechanism of anti-COVID-19 action
One of the interesting features of cyanorona-20 structure is its expected ability to act as a zincophore. Zinc ionophores or zinc ion carriers, e.g., chloroquine (Xue et al. 2014), hydroxychloroquine (Xue et al. 2014), quercetin (Dabbagh-Bazarbachi et al. 2014), and epigallocatechin gallate (Dabbagh-Bazarbachi et al. 2014), transport extracellular Zn2+ ions across the hydrophobic cell membranes to enter the living cells, and have been studied mainly for their antiviral activities, as they have been shown to effectively inhibit the replication of various viruses in vitro (Ishida 2019). Zn2+ inhibits coronavirus RdRp activity (i.e., inhibits coronaviral replication and transcription) in vitro (Zn2+ ion is the only known elemental cofactor and ligand present in the crystal structure of SARS-CoV-2 RdRp, and thus, it has an important role in the activity and performance of this enzyme, i.e., of COVID-19 RNA-synthesizing enzymatic machine) (Hecel et al. 2020). Zinc ionophores evidently block the replication process of coronaviruses intracellularly in cell cultures (Yin et al. 2020; te Velthuis et al. 2010; Derwand and Scholz 2020). Based on this fact, molecules that have good zincophoric properties may be advantageous in inhibiting SARS-CoV-2 RdRp and coronaviral-2 replication. Cyanorona-20 has about six potential zinc-binding centers or moieties (four active nitrogen atoms and two active oxygen atoms), making it an ideal candidate to act as a potent anti-COVID-19 zincophore.
Figure 5 summarizes the key structural features of cyanorona-20. Table 1 shows the structure and nomenclature of both the administered prodrug (or salt) and the active metabolite (or free base) forms of the novel anti-COVID-19 compound cyanorona-20 and the four reference general anti-COVID-19 drugs (favipiravir, remdesivir, arbidol, and hydroxychloroquine) (Dong et al. 2020; Cai et al. 2020; Du and Chen 2020; Shannon et al. 2020; Yin et al. 2020; Derwand and Scholz 2020; Choy et al. 2020; Elfiky 2020; Wang et al. 2020a, b; Kumar et al. 2020). In this research paper, the design, synthesis, characterization, computational studies, and anti-COVID-19 biological activities of the novel compound cyanorona-20 are reported.
Fig. 5.
A detailed presentation of the structural features of the promising anti-COVID-19 ideal model of cyanorona-20
Table 1.
Chemical structures and nomenclatures of both prodrug/salt and active/base forms of cyanorona-20, its parent drug favipiravir, and its three reference antiviral drugs (remdesivir, arbidol, and hydroxychloroquine)
| Compound name | Administered prodrug/salt form | Active metabolite/free base form |
|---|---|---|
| Cyanorona-20 |
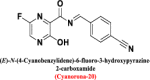
|
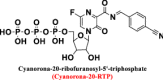
|
| Favipiravir |
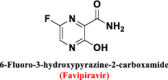
|
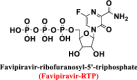
|
| Remdesivir |
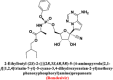
|

|
| Arbidol |

|

|
| Hydroxychloroquine |

|
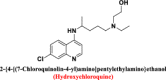
|
Experimental section
Synthesis and stability testing of cyanorona-20
Materials and general data
The conventional and microwave (MW) reactions were performed with commercially available reagents and solvents. Favipiravir (ultrapure) was sourced as a raw material from a representative of Toyama Chemical (Fujifilm group, Japan), 4-cyanobenzaldehyde (98%) was purchased from Merck (Merck KGaA, Darmstadt, Germany), and glacial acetic acid (gla. AcOH; extrapure) was purchased from Sigma-Aldrich. All solvents were of analytical grade, purchased from commercial suppliers, and were used as received without further purification. Microwave irradiation (MWI) for MW reaction was carried out in the laboratory MW synthesizer oven (Samsung type, model M1733N with Triple Distribution System "T.D.S." property, and having a power level of 100–800 W) operated at 2.45 GHz. Thin-layer chromatography (TLC) was used to monitor the progress of both reactions (conventional and MW), and it was carried out on TLC silica gel 60 F254 plates (plates of aluminum sheets precoated with unmodified silica gel 60 F254 to a layer thickness of 0.20 mm, purchased from E. Merck, Merck Millipore Division or Merck Chemicals, Merck KGaA, Darmstadt, Germany) as the stationary phase using n-hexane/ethyl acetate/absolute ethanol (5:2:1, v/v/v) mixture as the mobile phase (the chromatogram spots were visualized and observed under the used ultraviolet "UV" light at a wavelength of 254 nm) for monitoring both reactions. Evaporation/concentration purposes were carried out in a rotavap under reduced pressure. A lyophilizer (freeze dryer, model FD8-8T, SIM international, U.S.A.) was used for the lyophilizing purpose in the MW procedure. Melting point (M.P., °C) of cyanorona-20 was recorded in open glass capillaries using Fisher-Johns melting point apparatus. IR spectrum of cyanorona-20 was recorded on Nicolet™ iS™ 10 Mid-Infrared (Thermo Fisher Scientific) FT-IR spectrometer (υ in cm−1) using potassium bromide disk at the Central Laboratory (Faculty of Pharmacy, Mansoura University, Mansoura, Egypt) (str. = strong; br. = broad; arom. = aromatic; aliph. = aliphatic). 1H-NMR spectrum of cyanorona-20 was recorded on Varian Gemini-300 spectrometer (Mercury-300BB "NMR300") at 300 MHz using tetramethylsilane (TMS) as an internal standard at the Microanalytical Center (Faculty of Science, Cairo University, Cairo, Egypt), and its chemical shifts values (δ) were given in ppm downfield from TMS at a temperature of 30 °C using DMSO-d6 as a solvent. 13C-NMR spectrum of cyanorona-20 was also recorded on Varian Gemini-300 spectrometer (Mercury-300BB "NMR300") at 75 MHz using TMS as an internal standard at the Microanalytical Center (Faculty of Science, Cairo University, Cairo, Egypt), and its chemical shifts values (δ) were given in ppm downfield from TMS at a temperature of 30 °C using DMSO-d6 as a solvent. Mass spectrometry (MS) analysis of cyanorona-20 was performed on Shimadzu Qp-2010 Plus at 70 eV and results were represented by m/z (relative intensity "rel. int." in %) at the Microanalytical Center (Faculty of Science, Cairo University, Cairo, Egypt). Elemental analyses (elem. anal.) of cyanorona-20 were performed at the Microanalytical Center (Faculty of Science, Cairo University, Cairo, Egypt) in order to determine carbon (C), hydrogen (H), and nitrogen (N) atoms contents in %.
Synthetic procedures (conventional and mw)
Favipiravir (15.710 g, 0.1 mol) and 4-cyanobenzaldehyde (13.113 g, 0.1 mol) were gradually dissolved with heating in glacial AcOH (200 mL, 3.5 mol). The resulted reaction mixture was conventionally refluxed for 16 h (or under intermittent MWI at intervals of 30 s for 3 min, i.e., 6 intervals of 30 s, at a power level of 800 W. After MW reaction completion, the reaction mixture paste was cooled to − 20 °C and then it was lyophilized at − 50 °C). Then the reaction mixture (from the conventional step) was concentrated under reduced pressure, cooled to room temperature, and gradually poured onto crushed ice with stirring. The reaction mixture was allowed to stand overnight till the solid was separated and completely settled down. The separated crude solid (or the lyophilized crude solid from the MW step) was filtered and washed thoroughly with cold distilled water (3 × 400 mL), followed by cold absolute ethanol (3 × 350 mL) and then cold hexanes mixture (3 × 300 mL). Finally, the washed solid was dried, then extrapurified by recrystallization from a solvent mixture of absolute ethanol/ethyl acetate/chloroform (300 mL/300 mL/450 mL, i.e., 2:2:3, v/v/v) twice, and left to completely dry to afford the pure cyanorona-20.
Stability testing protocols
Simple short-term stability testing was done to extensively study the stability behavior of cyanorona-20 (mainly testing cyanorona-20 dissolution and hydrolysis profile in aqueous media of different pH ranges using different simulated fluids of the human body fluids by the aid of suitable buffering systems, e.g., simulated gastric and blood fluids of pH ranges of about 1.5–3.5 and 7.35–7.45, respectively, and applying several temperatures in the range of 20–50 °C) using the spectroscopic and chromatographic assays along with monitoring the physicochemical changes (e.g., color, texture, odor, M.P., retention factor, and pH of the aqueous solution). All the observations (including most physicochemical measurements) were done during a period of 3 months (beginning from 0 month interval "just before and just after dissolving the compound in the aqueous solutions", then 1-month interval "after 1 month from dissolving the compound in the aqueous solutions", and finally 3-month interval "after 3 months from dissolving the compound in the aqueous solutions"), and all the results were done in triplicates and compared with the reference favipiravir (which was also exposed to the same conditions for each test). Some stability tests were done on the dried cyanorona-20 which was extracted from its aqueous solution each time interval. The tests included exposure to different degrees of aqueous hydrolysis (the main concern), humidity, heat, light, tight storage, and were done according to the standard international guidelines and methods in stability testing of new compounds or drug substances, e.g., stress testing protocols and procedures, to verify the stability of cyanorona-20 compound (for more details, please see the respective standard guidelines and protocols: Q1A(R2) Stability Testing of New Drug Substances and Products).
Computational molecular studies of cyanorona-20
Pharmacokinetic properties
For the purpose of estimation of the molecular properties of cyanorona-20, Molinspiration web-based software (Molinspiration Cheminformatics 2020 on the Web) was used to calculate the most important molecular properties through using Molinspiration Property Engine (Molinspiration Calculation of Molecular Properties; Molinspiration Web-based Software 2020). Eight different molecular descriptors (parameters) of cyanorona-20 and its four reference repurposed anti-COVID-19 compounds (favipiravir, remdesivir, arbidol, and hydroxychloroquine) were calculated using Molinspiration methodology. The results are shown in Tables S1 and S2, respectively, in the Supplementary Material file.
Predictive anti-COVID-19 pharmacological properties
Prior to its experimental anti-COVID-19 evaluation, molecular docking of cyanorona-20 molecule in the enzyme SARS-CoV-2 RdRp was done using the docking engines of Discovery Studio, GemDock, GOLD, and others. The integration of the predicted pharmacophoric features with the interaction energy analysis revealed important residues in the binding pockets of the expected active/allosteric sites of SARS-CoV-2 RdRp together with in silico predicted common inhibitory binding modes with the highly potent reference compounds. For the purpose of specialized accurate docking of SARS-CoV-2 RdRp and prediction of anti-COVID-19 activities of compounds, both COVID-19 Docking Server and PASS Online web-based software programs (COVID-19 Docking Server Web-based Software 2020; PASS Online Web-based Software 2020) were used.
COVID-19 Docking Server web-based software (AutoDock Vina is used as the docking engine; according to the tutorial of this web server, the Broyden-Fletcher-Goldfarb-Shanno "BFGS" optimization method is used for the optimization purpose, and the Lamarckian genetic algorithm "LGA" is used as the main docking algorithm) is an interactive web server for docking small molecules, peptides, or antibodies against potential protein targets of COVID-19 in order to predict and score the binding modes between COVID-19 targets and the ligands along with screening and evaluating the anti-COVID-19 activities of these ligands. The platform provides a free interactive knowledge-based scoring function to evaluate the candidate binding poses for COVID-19 target-ligand interactions (COVID-19 Docking Server Web-based Software 2020). The structures of all the functional/structural protein targets involved in the SARS-CoV-2 replication life cycle were either collected or constructed based on their known homologs of coronaviruses (by using homology modeling module of Maestro 10, website: www.schrodinger.com), and prepared for direct docking on this web-based software (computational type or module: For docking of only one small molecule, the "Docking" mode box should be specifically selected for every specific target (this is the option used in the present case) (COVID-19 Docking Server Web-based Software 2020). The docked nonstructural enzyme was the SARS-CoV-2 RdRp (simply, the RdRp) and the nonstructural protein 12 (nsp12). Nsp12 is the polymerase which binds to its essential cofactors, nsp7 and nsp8 (the structure of RdRp was constructed based on 6NUR, the RdRp structure of the analogous coronavirus SARS-CoV (Kirchdoerfer and Ward 2019)). Two structures (two nCoV protein targets) were prepared for small molecule docking: One structure was built with RNA from its homolog protein (3H5Y) "RdRp with RNA," while the other one with no RNA in it "RdRp without RNA" (COVID-19 Docking Server Web-based Software 2020). To get significantly accurate results, an average exhaustiveness option of 12 was used. The results of these estimations are shown in Table 2.
Table 2.
Score values of the two computationally-predicted pharmacological anti-COVID-19-related activities (against SARS-CoV-2 RdRp-RNA and against SARS-CoV-2 RdRp) of the target cyanorona-20, the parent favipiravir, and the three references (remdesivir, HCl-arbidol-H2O, and hydroxychloroquine sulfate), along with their five active metabolites/free bases (cyanorona-20-RTP, favipiravir-RTP, GS-441524-TP, arbidol, and hydroxychloroquine), respectively, using COVID-19 Docking Server methodology (the table shows the top docking model score value, i.e., the best binding mode score value or the least predicted binding free energy value, in kcal/mol for each compound with each target)
| Classification | Compound name | Top pose score value for docking of nCoV protein targets (kcal/mol) | |
|---|---|---|---|
| RdRp with RNA | RdRp without RNA | ||
| Prodrugs/salts | Cyanorona-20 | – 10.40 | – 7.80 |
| Favipiravir | – 6.90 | – 6.10 | |
| Remdesivir | – 8.30 | – 7.10 | |
| HCl-Arbidol-H2O | – 7.70 | – 6.00 | |
| Hydroxychloroquine sulfate | – 7.10 | – 5.70 | |
| Active metabolites/free bases | Cyanorona-20-RTP | – 10.50 | – 8.60 |
| Favipiravir-RTP | – 8.40 | – 7.50 | |
| GS-441524-TP | – 9.20 | – 7.90 | |
| Arbidol | – 7.70 | – 6.00 | |
| Hydroxychloroquine | – 7.10 | – 5.70 | |
PASS (Prediction of Activity Spectra for Substances) Online web-based software (PASS Online 2020 on the Web; it is one of the predictive services presented by Way2Drug Predictive Services on the Web) was designed as a software product for evaluating the biological potentials of a drug-like molecule using the Predict New Compound tool (PharmaExpert.ru; PASS Online Prediction of Pharmacological Activities), with an average accuracy of prediction of more than 95% in 2020 (PASS Online Web-based Software 2020; Filimonov et al. 2014). According to PASS Online website, Pa (probability "to be active") estimates the chance that the studied compound is belonging to the subclass of active compounds (actives), while Pi (probability "to be inactive") estimates the chance that the studied compound is belonging to the subclass of inactive compounds (inactives) (PASS Online Web-based Software 2020). The detailed results of these estimations (where, Pa > Pi) are shown in Table 3.
Table 3.
Probability values of the computationally-predicted pharmacological antiviral anti-COVID-19 activities of the target cyanorona-20, the parent favipiravir, and the three references (remdesivir, HCl-arbidol-H2O, and hydroxychloroquine sulfate) along with their five active metabolites/free bases (cyanorona-20-RTP, favipiravir-RTP, GS-441524-TP, arbidol, and hydroxychloroquine), respectively, using PASS Online methodology
| Classification | Compound name | Anti-COVID-19 (nucleotide analog inhibitory or antiviral) activity | |
|---|---|---|---|
| Pa | Pi | ||
| Prodrugs/salts | Cyanorona-20 | 0.651 | 0.009 |
| Favipiravir | 0.498 | 0.014 | |
| Remdesivir | 0.814 | 0.004 | |
| HCl-Arbidol-H2O | 0.740 | 0.004 | |
| Hydroxychloroquine sulfate | 0.520 | 0.028 | |
| Active metabolites/free bases | Cyanorona-20-RTP | 0.741 | 0.009 |
| Favipiravir-RTP | 0.685 | 0.006 | |
| GS-441524-TP | 0.734 | 0.004 | |
| Arbidol | 0.740 | 0.004 | |
| Hydroxychloroquine | 0.520 | 0.028 | |
Antiviral anti-COVID-19 biological activity (in vitro assay) of cyanorona-20
This anti-COVID-19 in vitro assay is based upon the original procedures of Chu and coworkers (Choy et al. 2020; Chu et al. 2020) with very slight modifications (mainly in the prepared stock concentration of the assayed compounds). The complete procedures were carried out in a specialized biosafety level 3 (BSL-3) laboratory (SARS-CoV-2 is classified as a BSL-3 pathogen by the WHO and the FDA) in Hong Kong SAR (China) (the antiviral/cytotoxic evaluation assays were performed as a contract-based collaboration between our laboratory, DARLD, in Egypt and the specialized microbiology laboratory of Prof. Dr. Sahar M.-R. Radwan "professor of microbiology, immunology, and virology" and her coworkers in China, following the guidance of the procedures of Prof. Dr. Chu and coworkers "Choy et al. 2020; Chu et al. 2020"). The assayed SARS-CoV-2 virus, BetaCoV/Hong Kong/VM20001061/2020, was isolated from the fresh nasopharynx aspirate and throat swab of a confirmed middle-aged COVID-19 patient in Hong Kong using Vero E6 cells (ATCC CRL-1586). Stock virus (107.25 TCID50/mL) was prepared after three serial passages in Vero E6 cells in infection media (DMEM supplemented with 4.5 g/L D-glucose, 100 mg/L sodium pyruvate, 2% FBS, 100,000 U/L Penicillin–Streptomycin, and 25 mM HEPES). The four reference compounds were obtained from Toyama Chemical (Fujifilm group, Japan) (favipiravir), MedChemExpress (remdesivir), 3B Scientific (Wuhan) Corporation Limited (HCl-arbidol-H2O), and Sigma-Aldrich (hydroxychloroquine sulfate) and the stocks were accurately prepared with DMSO (100 mM cyanorona-20, 100 mM favipiravir, 100 mM remdesivir, and 100 mM HCl-arbidol-H2O) or with distilled water (100 mM hydroxychloroquine sulfate). To evaluate the in vitro anti-SARS-CoV-2 effect of the target new compound (cyanorona-20) in comparison with the anti-SARS-CoV-2 effects of the standard four reference compounds (mentioned above), Vero E6 cells were pretreated with the five compounds diluted in infection media for 1 h prior to infection by SARS-CoV-2 virus at MOI = 0.02. Antiviral anti-COVID-19 compounds were maintained with the virus inoculum during the 2-h incubation period. The inoculum was removed after incubation, and the cells were overlaid with infection media containing the diluted compounds. After 48-h incubation at 37 °C, supernatants were immediately collected to quantify viral loads by TCID50 assay or quantitative real-time RT-PCR “qRT-PCR” (TaqMan™ Fast Virus 1-Step Master Mix) (Choy et al. 2020; Chu et al. 2020). Note that viral loads in this assay were fitted in logarithm scale (log10 TCID50/mL and log10 viral RNA copies/mL) (Choy et al. 2020; Chu et al. 2020), along with linear scale (Wang et al. 2020b), under increasing concentrations of the tested compounds. Four-parameter logistic regression (GraphPad Prism) was used to fit the dose–response curves and determine the EC50 of the tested compounds that inhibit SARS-CoV-2 viral replication (CPEIC100 was also determined for each compound). Cytotoxicity of each of the five tested compounds was evaluated in Vero E6 cells using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) (Choy et al. 2020; Zhang et al. 2020). The detailed values resulted from the previous assays are shown in Table 4. Final results were represented as the mean ± the standard deviation (SD) from the triplicate biological experiments. Statistical analysis was performed using SkanIt 4.0 Research Edition software (Thermo Fisher Scientific) and Prism V5 software (GraphPad). All reported data were significant at p < 0.05.
Table 4.
Anti-COVID-19/antiviral activities (along with human/mammalian cells toxicities) of cyanorona-20 and the four reference drugs (favipiravir, remdesivir, HCl-arbidol-H2O, and hydroxychloroquine sulfate) against SARS-CoV-2 in Vero E6 cells
| Classification | Compound name | CC50a (μM) |
Inhibition of SARS-CoV-2 in vitro (μM) | ||
|---|---|---|---|---|---|
| 100% CPE inhibitory concentration (CPEIC100)b | 50% reduction in infectious virus (EC50)c |
50% reduction in viral RNA copy (EC50)d | |||
| Target compound | Cyanorona-20 | > 100 | 1.40 ± 0.02 | 0.45 ± 0.03 | 0.48 ± 0.03 |
| Reference compounds | Favipiravir | > 100 | 98.82 ± 1.13 | 94.09 ± 5.01 | > 100 |
| Remdesivir | > 100 | 22.50 ± 0.58 | 20.17 ± 1.99 | 23.88 ± 2.46 | |
| HCl-Arbidol-H2O | > 100 | 81.52 ± 1.12 | 64.20 ± 4.90 | 68.42 ± 6.02 | |
| Hydroxychloroquine sulfate | 93.06 ± 6.92 | > 100 | > 100 | > 100 | |
aCC50 or 50% cytotoxic concentration is the concentration of the tested compound that kills half the cells in an uninfected cell culture. CC50 was determined with serially-diluted compounds in Vero E6 cells at 48 h postincubation using CellTiter-Glow Luminescent Cell Viability Assay (Promega)
bCPEIC100 or 100% CPE inhibitory concentration is the lowest concentration of the tested compound that causes 100% inhibition of the cytopathic effects (CPE) of SARS-CoV-2 virus in Vero E6 cells under increasing concentrations of the tested compound at 48 h postinfection. Compounds were serially twofold or fourfold diluted from 100 μM concentration
cEC50 or 50% effective concentration is the concentration of the tested compound that is required for 50% reduction in infectious SARS-CoV-2 virus particles in vitro. EC50 is determined by infectious virus yield in culture supernatant at 48 h postinfection (log10 TCID50/mL)
dEC50 or 50% effective concentration is the concentration of the tested compound that is required for 50% reduction in SARS-CoV-2 viral RNA copies in vitro. EC50 is determined by viral RNA copies number in culture supernatant at 48 h postinfection (log10 RNA copies/mL)
Results and discussion
Synthesis, structure elucidation (characterization), chemistry, and stability of cyanorona-20
Cyanorona-20 was successfully synthesized, as shown in Scheme 1, in very good yields from its parent favipiravir via direct condensation with 4-cyanobenzaldehyde (equimolar amounts) in the presence of the strong dehydrating agent glacial AcOH. The reaction could proceed either by conventional heating (with 85% yield) or under MWI (with 96% yield). The structure of cyanorona-20 was confirmed through spectroscopic analyses (IR, 1H-NMR, 13C-NMR, and MS) and microanalyses (elem. anal. for the contents of C, H, and N atoms). Spectral data and elemental analyses of this product were in full agreement with the proposed structure of cyanorona-20.
Scheme 1.
Conventional and microwave-assisted synthesis of cyanorona-20 from favipiravir
The pure cyanorona-20 was obtained as a pale white to yellowish beige fine-crystalline powdered solid (22.969 g, 85% conventional yield; 25.941 g, 96% MW yield). M.P.: 294–298 °C (rough); FT-IR (υ in cm−1): Str. and br. 3437 (O–H, arom.), str. 3009 (C–H, aliph.), 2921 (C–H, arom.), 2236 (C≡N, nitrile), 1683 (C=N, aldimine), 1635 (C=O, amide), str. 1606 and 1548 and str. 1500 and 1462 and 1364 (C=C, arom.), str. and br. 1302 (C–F, fluoroheterocyclic), 1266 (C–N, aliph.), 1212 (C-O, phenolic); 1H-NMR (300 MHz, DMSO-d6, δ in ppm): 13.84 (s, 1H, 1 pyrazine phenolic OH), 9.63 (s, 1H, 1 secondary aldimine H), 8.13 (s, 1H, 1 pyrazine H), 7.94–7.69 (m, 4H, 4 benzylidenimine benzene H); 13C-NMR (75 MHz, DMSO-d6, δ in ppm): 168.25 (1C, 1 carbonyl C), 161.42 (1C, 1 pyrazine C–OH), 158.60 (1C, 1 secondary aldimine C), 154.95–152.00 (d, J = 246.2 Hz, 1C, 1 pyrazine C–F), 148.05 (1C, 1 pyrazine C–C=O), 137.65 (1C, 1 benzene C–C=N), 135.85 (2C, 2 similar benzene C attached to C–C≡N), 132.54 (1C, 1 unsubstituted pyrazine C), 128.82 (2C, 2 similar benzene C attached to C–C=N), 118.50 (1C, 1 nitrile C), 113.91 (1C, 1 benzene C–C≡N); GC–MS (EI) (m/z, rel. int. in %, molecular weight "M.Wt." = 270.22): 271.00 ([M + H]+); Elem. Anal. (%, for C13H7FN4O2, calcd (found)): C: 57.78 (57.71), H: 2.61 (2.60), N: 20.73 (20.76).
Cyanorona-20, like favipiravir, is a tautomeric molecule (Guo et al. 2019). According to the computational simulations studies (e.g., Antonov 2020), the molecule favors the enol-like tautomeric structure (the predominant form) in aqueous medium as shown in Scheme 2.
Scheme 2.
Tautomeric forms of cyanorona-20 molecule in aqueous solutions
The stability of the Schiff base-like structure of cyanorona-20 was studied using the analytical and physicochemical methods as demonstrated in the Experimental Section. The results of the aqueous dissolution testing were excellent. Less than 5% (as a maximum) of total cyanorona-20 amount undergoing hydrolysis to minor products and impurities after the period of 3 months, and less than 50% of this 5% amount (i.e., less than 2.5% of total cyanorona-20 amount) was the parent favipiravir (see Chart S4 as a representative analytical chart in the Supplementary Material file), hence proving the practical stability of cyanorona-20.
Computational molecular studies of cyanorona-20
Pharmacokinetic Properties
The values in Tables S1 and S2 (in the Supplementary Material file) reveal that cyanorona-20 has the best balanced predicted molecular properties and pharmacokinetic parameters, among the five evaluated compounds (Molinspiration Web-based Software 2020; Ertl et al. 2000; Lipinski et al. 1997; Ghose et al. 1999; Veber et al. 2002; Yehye et al. 2012). Favipiravir and cyanorona-20 are the smallest molecules among them. Cyanorona-20 has significantly balanced lipophilic/hydrophilic properties ratio and reasonably balanced numbers and types of atoms/bonds (as discussed in details below).
Structurally, cyanorona-20 has a small molecular weight and molecular volume of 270.22 daltons and 219.19 Å3, respectively, which are both less than the value of 500 (the maximum value preferred not to be exceeded for better pharmacokinetic properties), this expectedly helps cyanorona-20 to have extremely excellent biocompatibility and rapid distribution with high bioavailability. Cyanorona-20 has a log P value of 1.29 which is much more moderate and balanced when compared to the other four compounds (all except remdesivir); this interesting balanced value theoretically gives cyanorona-20 the ability to be administered with almost all routes of drug administration (oral, parenteral, nasal, etc.) and to be soluble in all biological fluids (with good predictions to pass through all types of biological membranes with gradual moderate to high rates). The values of nON, nOHNH, nRotB, and TPSA for cyanorona-20 are 6, 1, 2, and 99.24 Å2, respectively, which are balanced moderate values, making cyanorona-20 an ideal candidate drug (better than the other four drugs) to be well fitted into the cavities of the active and allosteric sites of the enzyme SARS-CoV-2 RdRp with the strongest interaction states (contact modes) along with the least possible scores of interaction energies (this is supported in part by the results of the computational docking screening and biological anticopying evaluation). Cyanorona-20 complies with all the preferred values of pharmacokinetic parameters (see Fig. 5), as has no violations from the nine parameters (including those of the Ro5).
Predictive anti-COVID-19 pharmacological properties
The understanding of the COVID-19 target-ligand interactions represents a very important key challenge in drug discovery for COVID-19. The computational simulation prediction of the anti-COVID-19 activities of the new target compound cyanorona-20 along with the up-to-date molecular modeling approaches/studies of the human viruses (e.g., Kumar et al. 2021) greatly helps us to have an overview of the SARS-CoV-2 RdRp-inhibiting properties of this target compound. This prediction mainly gives a detailed idea about the target compound anti-COVID-19 mode of action.
On inspection of the score values (Table 2) of docking RdRp-RNA and RdRp alone using COVID-19 Docking Server, it is noted that cyanorona-20 and its active metabolite cyanorona-20-RTP are generally ranked first in their inhibitory binding affinities and potencies with binding free energies of − 10.40, − 7.80, − 10.50, and − 8.60 kcal/mol, respectively (COVID-19 Docking Server Web-based Software 2020; Kirchdoerfer and Ward 2019). The binding affinities of cyanorona-20-RTP significantly exceed those of all the other four active metabolites of the other four drugs, as this metabolite strongly binds to RdRp (with RNA) in their complex (i.e., cyanorona-20-RTP molecule forms a very stable complex with SARS-CoV-2 RdRp) with a very good binding free energy of − 10.50 kcal/mol which is the lowest among all (i.e., significantly lower than the binding free energies of all the other nine compounds in their complexes with RdRp-RNA). Remdesivir and favipiravir (and their active metabolites) come second in their relative inhibitory potency and efficacy on SARS-CoV-2 RdRp, followed by arbidol and hydroxychloroquine (and their salts). For more illustration, Fig. 6(a–d) shows the COVID-19 Docking Server outputs of the top predicted binding model or mode of docking of SARS-CoV-2 RdRp-RNA and SARS-CoV-2 RdRp with cyanorona-20 and its active metabolite cyanorona-20-RTP, respectively. These results of the predicted binding modes of the two ligands cyanorona-20/cyanorona-20-RTP with the protein SARS-CoV-2 RdRp (either with or without RNA) comply with the suggested mechanism of anti-COVID-19 action of both ligands (see Fig. 4).
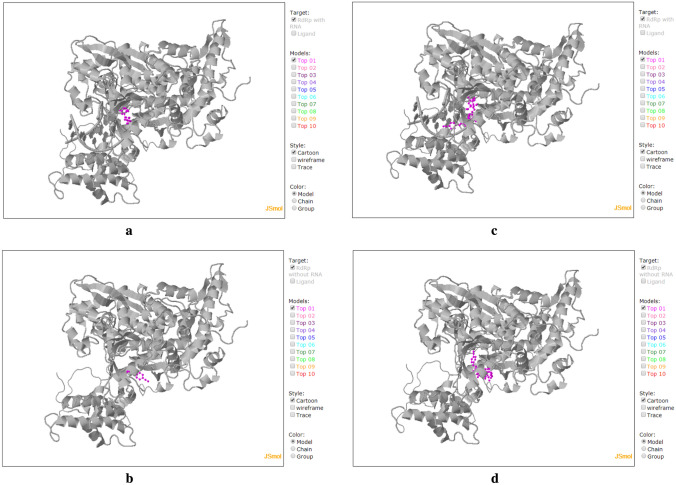
Fig. 6.
Screenshots of COVID-19 Docking Server outputs of the top predicted binding model of docking of: a SARS-CoV-2 RdRp-RNA (colored gray) with cyanorona-20 (colored pink). b SARS-CoV-2 RdRp (colored gray) with cyanorona-20 (colored pink). c SARS-CoV-2 RdRp-RNA (colored gray) with cyanorona-20-RTP (colored pink). d SARS-CoV-2 RdRp (colored gray) with cyanorona-20-RTP (colored pink). PDB code of the docked SARS-CoV-2 RdRp: 7BV2
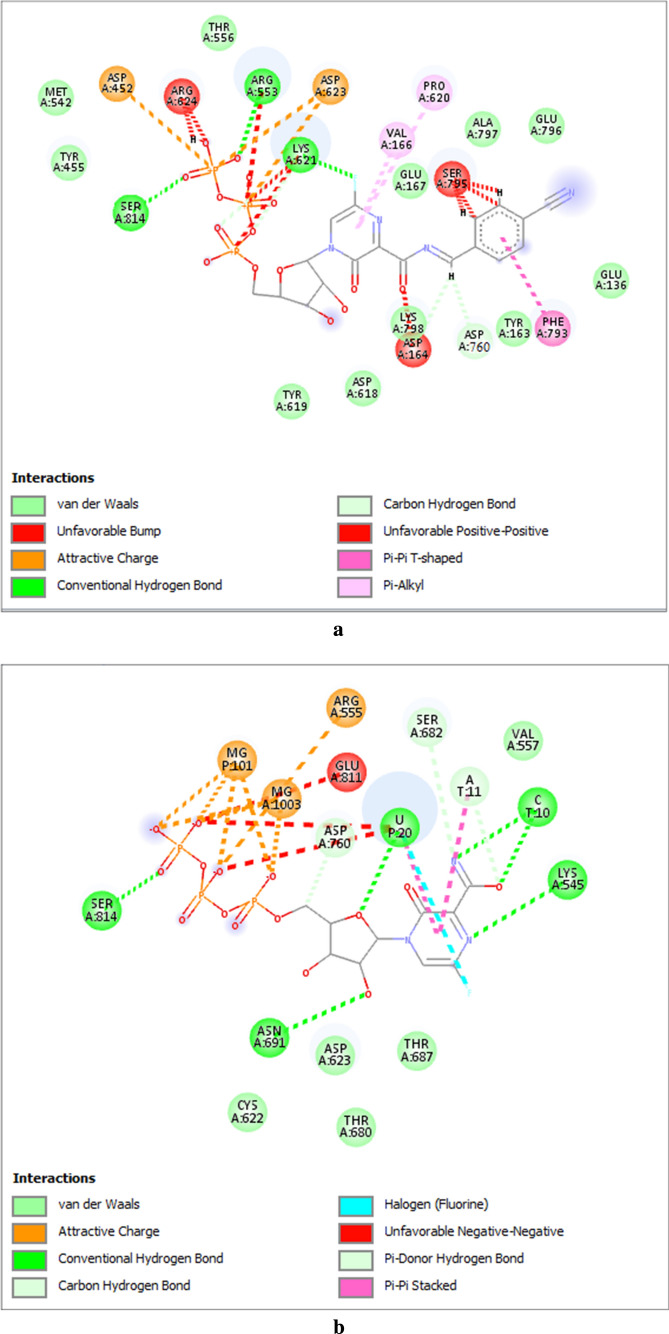
Deep analysis of the computational interaction mode of cyanorona-20-RTP with SARS-CoV-2 RdRp reveals its significant resemblance with that of favipiravir-RTP with the same polymerase, since both ligands form hydrogen bonds and hydrophobic interactions with almost the same or close amino acid residues of the proposed active site of the polymerase (Sada et al. 2020; Picarazzi et al. 2020; Jena 2020) (Fig. 7a,b). Cyanorona-20-RTP molecule, exactly like favipiravir-RTP molecule, strongly binds with the pivotal amino acid residue of the SARS-CoV-2 RdRp active site, Asp760, which is very critical for the initiation and progression of the coronaviral-2 replication processes (Sada et al. 2020; Picarazzi et al. 2020; Jena 2020), thus inhibiting this residue may offer a key role in COVID-19 therapy. Cyanorona-20 and its RTP metabolite mainly depend on the 4-cyanobenzylidene moiety in the binding interaction with the active amino acid Asp760 to effectively inhibit the SARS-CoV-2 RdRp (see Fig. 7a). Importantly, the active metabolite of cyanorona-20 forms slightly higher number of strong interactions with SARS-CoV-2 RdRp than that of favipiravir.
Fig. 7.
The inhibitory binding interactions, of a Cyanorona-20-RTP; b Favipiravir-RTP, with the active amino acids of the SARS-CoV-2 RdRp (2D representations)
An estimate of the probability values (present in Table 3) predicting anti-COVID-19 activities (general antiviral/anti-RNA virus properties and also specific properties such as being nucleos(t)ide analog inhibitor, nucleotide metabolism regulator, and adenosine regulator) of cyanorona-20 and its active metabolite, along with the four references with their metabolites, using PASS Online screening reveals that cyanorona-20, remdesivir, HCl-arbidol-H2O, and their three active metabolites/free bases are generally ranked first in their antiviral and anti-COVID-19 activities and efficacies among the ten ligands (then favipiravir, hydroxychloroquine sulfate, and their active metabolites come second in ranking) (Yehye et al. 2012; PASS Online Web-based Software 2020; Filimonov et al. 2014). Cyanorona-20-RTP has the best probability to be active anti-COVID-19, among all the five screened active nucleotide analog ligands or inhibitors of SARS-CoV-2 RdRp (the five screened active nucleotide analog metabolites, which are cyanorona-20-RTP, favipiravir-RTP, GS-441524-TP, arbidol, and hydroxychloroquine), of more than 74% with a negligible probability to be inactive anti-COVID-19 of less than 1%.
Antiviral anti-COVID-19 biological activity (in vitro assay) of cyanorona-20
The results demonstrated in Table 4 clearly revealed the extremely higher and surprising anti-COVID-19 efficacy of cyanorona-20 (the most potent anti-SARS-CoV-2 compound of the five tested ones). Among the five tested compounds, four compounds (cyanorona-20, remdesivir, HCl-arbidol-H2O, and favipiravir, respectively) inhibit SARS-CoV-2 replication in Vero E6 cells with EC50 under 100 μM, while hydroxychloroquine sulfate was above 100 μM. Surprisingly, cyanorona-20 (EC50 = 0.45 μM, see Chart S5 as a representative curve in the Supplementary Material file) was about 209 and 45 times as potent as favipiravir (EC50 = 94 μM) and remdesivir (EC50 = 20 μM), respectively, in anti-SARS-CoV-2 activity (in vitro). According to the assay, cyanorona-20 is expected to have high clinical selectivity index (SI; SI = CC50/EC50) and safety margin (CC50 is much larger than 100 μM). On the other hand, hydroxychloroquine sulfate is expected to have very narrow clinical therapeutic index (EC50 is just above 100 μM, CC50 = 93 μM). Cyanorona-20 is also having amazingly very small values of the concentration that causes 100% inhibition of the SARS-CoV-2 cytopathic effects in vitro (cyanorona-20 has the best CPEIC100 value, among all the five compounds tested, of 1.4 μM) and of the concentration that is required for 50% reduction in the number of SARS-CoV-2 RNA copies in vitro (cyanorona-20 has the best EC50 value, among all the five compounds tested, of 0.48 μM).
The three nucleoside/nucleotide analogs, cyanorona-20 (guanine analog), favipiravir (guanine analog), and remdesivir (adenosine analog), require intracellular metabolic activation to the triphosphate forms by host cellular enzymes (mainly nucleoside kinases), which may differ among several cell types; thus, evaluation of the actions of nucleos(t)ide analogs in primary human airway epithelial cells would undoubtedly facilitate the interpretation of the results. The metabolic activation would surely add additional anti-COVID-19 activities to the three drugs, and it would also successfully increase the clinical effectiveness of the three drugs. The four reference drugs (favipiravir, remdesivir, HCl-arbidol-H2O, and hydroxychloroquine sulfate) are currently undergoing extensive clinical trials, as anti-SARS-CoV-2/anti-COVID-19 agents, worldwide. The very high value of CC50 of cyanorona-20 indicates that cyanorona-20 may be well tolerated in the human body. The very minute value of anti-SARS-CoV-2 EC50 and the very high value of mammalian cells CC50 (i.e., the desirable high value of SI) of cyanorona-20 indicate that this compound clearly favors resistant RNA virus over DNA virus and mammalian cells, and this, in turn, suggests selective specificity as anti-COVID-19 drug "Corona Antidote or Corona Killer" (see Introduction part). Using a combination formula (a mixture) of cyanorona-20 and remdesivir is a suggested choice, as it may have exceptional combinational synergistic anti-COVID-19 effect in further assays (in vivo) and clinical trials. Almost all the practical results concluded, here, in the antiviral anti-COVID-19 biological evaluation are complying with the previous theoretical results extracted from the computational molecular and pharmacological predictions for the new compound cyanorona-20 and its four reference compounds.
Conclusions
Specific potent blockade of the novel SARS-CoV-2 RdRp is a viable approach for targeted COVID-19 therapy. Our efforts in 2020 focused on designing new drugs (e.g., derivatives of favipiravir) for the more effective inhibition of the viral polymerase. These efforts led to the discovery of a very promising selective specific and potent direct-acting SARS-CoV-2 copying inhibitor, cyanorona-20 ((E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide), which inhibited SARS-CoV-2 replication with significant EC50 values of 0.45 and 0.48 μM, and interestingly presented about 209- and 45-fold anti-SARS-CoV-2 selectivity/potency more than favipiravir and remdesivir, respectively. Cyanorona-20, to the best of our knowledge (up to the date of submitting this discovery article for publication), is the first bioactive derivative of favipiravir. Structural modification at the active amino moiety of the antiinfluenza favipiravir molecule opens the first class of anti-COVID-19 agents (of the type "nucleos(t)ide analogs") which will specifically comprise a series of pyrazine derivatives (Miniyar et al. 2013). Cyanorona-20 fulfills the requirements for an ideal anti-COVID-19 drug (more active than its parent compound favipiravir) (Fig. 5). For example, cyanorona-20 molecule has a virus-toxic cyano group (it may be also called a SARS-CoV-2 RdRp-destabilizing moiety, as it chemically causes major steric clash with the SARS-CoV-2 RdRp molecule at some residues; this greatly helps in RdRp preliminary partial blockade and results in delayed chain termination in RNA synthesis which may speculatively give cyanorona-20 an extrapotency against the major resistance mechanisms that might be emerged by SARS-CoV-2 against favipiravir and other classical potent antiviral nucleos(t)ide analogs (Shannon et al. 2020)) (a strong aliphatic polar group) and a resonance-stabilized benzene ring (in the benzylidene group) (a strong aromatic lipophilic moiety), both groups with a one-carbon-atom linker form a highly stable 4-cyanobenzylidene moiety (a SARS-CoV-2-toxic moiety which is extremely stabilized through strong resonance and inductive effects) which is not present in the parent favipiravir molecule, and predictably adds an exceptional and excellent balanced lipophilic/hydrophilic properties along with electronic extrastability to the molecule (this makes the molecule more bioavailable and more biocompatible).
Extensive computational studies showed that cyanorona-20 has ideal values of the pharmacokinetic and drug-likeness descriptors. Computational modeling analysis of the top inhibitory binding mode of the expected active metabolite of cyanorona-20 inside the human cell, cyanorona-20-RTP, showed that the 4-cyanobenzylidene moiety increases the potency at active and/or allosteric sites of the SARS-CoV-2 RdRp (binding free energy = − 10.50 kcal/mol) when compared to that of the active metabolite of its parent favipiravir inside the human cell, favipiravir-RTP (binding free energy = − 8.40 kcal/mol). Promisingly, cyanorona-20 and its active metabolite surpassed the four moderately to highly potent reference drugs and their active metabolites, respectively, in the values of almost all compared theoretical and practical anti-COVID-19 parameters, scores, and activities. The mechanism of the interaction of cyanorona-20-RTP with SARS-CoV-2 RdRp has not been elucidated, but cyanorona-20 may presumably act through six or more complementary modes of action (i.e., multiaction; Fig. 4 and Fig. 7a) (Smee et al. 2009; Yoon et al. 2018; Jin et al. 2013; Baranovich et al. 2013; Furuta et al. 2009; Shannon et al. 2020), as it may be misincorporated in a nascent SARS-CoV-2 RNA (thus preventing RNA strand elongation and viral proliferation), evade RNA proofreading by viral exoribonuclease (ExoN; thus causing a decrease in SARS-CoV-2 RNA production), competitively bind to conserved polymerase domains (thus preventing incorporation of mainly purine nucleotides for SARS-CoV-2 RNA replication and transcription), cause the SARS-CoV-2 RdRp to pause, induce an irreversible chain termination in the growing SARS-CoV-2 RNA, or induce lethal mutagenesis (thus, mainly, making the virus less effective and reducing its titer "viral titer") during SARS-CoV-2 infection. If cyanorona-20 passes the in vivo bioassays and preclinical/clinical trials with effectively significant results as anti-COVID-19 agent, combination therapy with a second potent antiviral drug, such as remdesivir, may also be advantageous.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
I gratefully thank and deeply acknowledge anyone who gave a hand to make this new discovery and work coming out to light.
Declarations
Conflict of interest
I hereby declare that I totally have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this new research paper.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s11696-022-02315-9
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/10/2022
Editor’s Note: Readers are alerted that the reliability of data presented in this article is currently in question. Appropriate editorial action will be taken once all parties have had the opportunity to respond in full.
Change history
6/4/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s11696-022-02315-9
References
- Abdelnabi R, de Morais ATS, Leyssen P, Imbert I, Beaucourt S, Blanc H, Froeyen M, Vignuzzi M, Canard B, Neyts J, Delang L. Understanding the mechanism of the broad-spectrum antiviral activity of favipiravir (T-705): key role of the F1 motif of the viral polymerase. J Virol. 2017;91:e00487–e517. doi: 10.1128/JVI.00487-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov L. Favipiravir tautomerism: a theoretical insight. Theor Chem Acc. 2020;139:145. doi: 10.1007/s00214-020-02656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovich T, Wong S-S, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol. 2013;87:3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, Liao X, Gu Y, Cai Q, Yang Y, Shen C, Li X, Peng L, Huang D, Zhang J, Zhang S, Wang F, Liu J, Chen L, Chen S, Wang Z, Zhang Z, Cao R, Zhong W, Liu Y, Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K-T, Wong AY-L, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PP-H, Huang X, Peiris M, Yen H-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Docking Server Web-based Software, COVID-19 Docking Server (homepage on the internet), Shan Chang Lab., Institute of Bioinformatics and Medical Engineering, School of Electrical and Information Engineering, Jiangsu University of Technology, Changzhou 213001, China, 2020; available from COVID-19 Docking Server on the web (homepage: http://ncov.schanglab.org.cn); Results were obtained through using the interactive docking tool in COVID-19 Docking Server (Copyright© 2018-2023, Shan Chang; Version 2020) on this website (accessed and cited in 2020, 19–28 May)
- COVID-19 Map (2020), available from Johns Hopkins Coronavirus Research Center. https://coronavirus.jhu.edu/map.html. Accessed in 30 December, 2020
- Dabbagh-Bazarbachi H, Clergeaud G, Quesada IM, Ortiz M, O'Sullivan CK, Fernández-Larrea JB. Zinc ionophore activity of quercetin and epigallocatechin-gallate: from Hepa 1–6 cells to a liposome model. J Agric Food Chem. 2014;62:8085–8093. doi: 10.1021/jf5014633. [DOI] [PubMed] [Google Scholar]
- Derwand R, Scholz M. Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win today’s battle against COVID-19? Med Hypotheses. 2020;142:109815. doi: 10.1016/j.mehy.2020.109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discoveries Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Driouich JS, Cochin M, Lingas G, Moureau G, Touret F, Petit PR, Piorkowski G, Barthélémy K, Laprie C, Coutard B, Guedj J, de Lamballerie X, Solas C, Nougairède A. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat Commun. 2021;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y-X, Chen X-P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin Pharmacol Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- Filimonov DA, Lagunin AA, Gloriozova TA, Rudik AV, Druzhilovskii DS, Pogodin PV, Poroikov VV. Prediction of the biological activity spectra of organic compounds using the Pass Online web resource. Chem Heterocycl Compd. 2014;50:444–457. doi: 10.1007/s10593-014-1496-1. [DOI] [Google Scholar]
- Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- Guo Q, Xu M, Guo S, Zhu F, Xie Y, Shen J. The complete synthesis of favipiravir from 2-aminopyrazine. Chem Pap. 2019;73:1043–1051. doi: 10.1007/s11696-018-0654-9. [DOI] [Google Scholar]
- Hecel A, Ostrowska M, Stokowa-Sołtys K, Wątły J, Dudek D, Miller A, Potocki S, Matera-Witkiewicz A, Dominguez-Martin A, Kozłowski H, Rowińska-Żyrek M. Zinc(II)—The overlooked éminence grise of chloroquine’s fight against COVID-19? Pharmaceuticals. 2020;13:228. doi: 10.3390/ph13090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A, Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T. Review on the role of Zn2+ ions in viral pathogenesis and the effect of Zn2+ ions for host cell-virus growth inhibition. Am J Biomed Sci. 2019;2:28–37. doi: 10.34297/AJBSR.2019.02.000566. [DOI] [Google Scholar]
- Jena NR. Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19. Phys Chem Chem Phys. 2020;22:28115–28122. doi: 10.1039/d0cp05297c. [DOI] [PubMed] [Google Scholar]
- Jiang S, Du L, Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS ONE. 2013;8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Awasthi A, Kumari A, Sood D, Jain P, Singh T, Sharma N, Grover A, Chandra R (2020) Antitussive noscapine and antiviral drug conjugates as arsenal against COVID-19: a comprehensive chemoinformatics analysis. J Biomol Struct Dyn. In Press. 10.1080/07391102.2020.1808072 [DOI] [PMC free article] [PubMed]
- Kumar N, Sood D, Singh S, Kumar S, Chandra R. High bio-recognizing aptamer designing and optimization against human herpes virus-5. Eur J Pharm Sci. 2021;156:105572. doi: 10.1016/j.ejps.2020.105572. [DOI] [PubMed] [Google Scholar]
- Łagocka R, Dziedziejko V, Kłos P, Pawlik A. Favipiravir in therapy of viral infections. J Clin Med. 2021;10:273. doi: 10.3390/jcm10020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-Y, You Z, Wang Q, Zhou Z-J, Qiu Y, Luo R, Ge X-Y. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22:80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Miniyar PB, Murumkar PR, Patil PS, Barmade MA, Bothara KG. Unequivocal role of pyrazine ring in medicinally important compounds: a review. Mini-Rev Med Chem. 2013;13:1607–1625. doi: 10.2174/1389557511313110007. [DOI] [PubMed] [Google Scholar]
- Molinspiration Web-based Software, Molinspiration Cheminformatics (homepage on the internet), Nova ulica, SK-900 26 Slovensky Grob, Slovak Republic, 2020; available from Molinspiration Cheminformatics on the web (homepage: http://www.molinspiration.com); Estimations were calculated through using Molinspiration Property Engine (Version 2018.10; Molinspiration Calculation of Molecular Properties) on this Website (accessed and cited in 2020, 11–22 May)
- Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet MJ, Castro-Hartmann P, Qian P, Sader K, Dent K, Kimanius D, Sutherland JD, Löwe J, Barford D, Russo CJ. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc Natl Acad Sci U S A. 2021;118:e2021946118. doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASS Online Web-based Software, Way2Drug Predictive Services (homepage on the internet), Department for Bioinformatics, Laboratory for Structure-Function Based Drug Design, Institute of Biomedical Chemistry (IBMC), Moscow 119121, Pogodinskaya Str., 10, Russia, 2020; available from PharmaExpert or Way2Drug Predictive Services on the web (homepage: http://www.pharmaexpert.ru or http://www.way2drug.com); Results were obtained through using Predict New Compound tool in PASS software (homepage: http://www.pharmaexpert.ru/passonline or http://www.way2drug.com/PASSOnline; PharmaExpert.ru © or Way2Drug.com ©, 2011-2020, Version 2.0; PASS Online Prediction of Pharmacological Activities) on both websites (accessed and cited in 2020, 20–30 May)
- Picarazzi F, Vicenti I, Saladini F, Zazzi M, Mori M. Targeting the RdRp of emerging RNA viruses: the structure-based drug design challenge. Molecules. 2020;25:5695. doi: 10.3390/molecules25235695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada M, Saraya T, Ishii H, Okayama K, Hayashi Y, Tsugawa T, Nishina A, Murakami K, Kuroda M, Ryo A, Kimura H. Detailed molecular interactions of favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza virus polymerases in silico. Microorganisms. 2020;8:1610. doi: 10.3390/microorganisms8101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A, Le NT-T, Selisko B, Eydoux C, Alvarez K, Guillemot J-C, Decroly E, Peersen O, Ferron F, Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smee DF, Hurst BL, Egawa H, Takahashi K, Kadota T, Furuta Y. Intracellular metabolism of favipiravir (T-705) in uninfected and influenza A (H5N1) virus-infected cells. J Antimicrob Chemother. 2009;64:741–746. doi: 10.1093/jac/dkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis AJW, van den Worm SHE, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6:e1001176. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Prasad BVLS, Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Funct Evolut Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, Yang X, Shi Z, Deng F, Hu Z, Zhong W, Wang M. The antiinfluenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Moyer A, Peng B, Wu J, Hannafon BN, Ding W-Q. Chloroquine is a zinc ionophore. PLoS ONE. 2014;9:e109180. doi: 10.1371/journal.pone.0109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehye WA, Abdul Rahman N, Alhadi AA, Khaledi H, Ng SW, Ariffin A. Butylated hydroxytoluene analogs: synthesis and evaluation of their multipotent antioxidant activities. Molecules. 2012;17:7645–7665. doi: 10.3390/molecules17077645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie Y-C, Tian G, Jiang H-W, Tao S-C, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-J, Toots M, Lee S, Lee M-E, Ludeke B, Luczo JM, Ganti K, Cox RM, Sticher ZM, Edpuganti V, Mitchell DG, Lockwood MA, Kolykhalov AA, Greninger AL, Moore ML, Painter GR, Lowen AC, Tompkins SM, Fearns R, Natchus MG, Plemper RK. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob Agents Chemother. 2018;62:e00766-18. doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J, de Wilde A, Snijder EJ, Liu H, Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tang LV. Overview of targets and potential drugs of SARS-CoV-2 according to the viral replication. J Proteome Res. 2021;20:49–59. doi: 10.1021/acs.jproteome.0c00526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.