Significance
The epithelial-to-mesenchymal transition (EMT) is a critical cell biological process that occurs during normal embryonic development and cancer progression. Our study combines single-cell RNA-sequencing analysis and mathematical modeling to identify critical regulators of EMT. Detailed analyses of TGF-β1–induced EMT by single-cell RNA-sequencing data revealed simultaneous activation of EMT signaling pathways. We created mathematical approaches to identify the master regulatory pathway of EMT and key downstream mediators of this process. This study sheds light on the signaling architecture that governs EMT and informs ongoing efforts to delineate drivers of cancer initiation, progression, and metastasis.
Keywords: EMT, scRNA-seq, signaling cascade, NOTCH, RACIPE
Abstract
The epithelial-to-mesenchymal transition (EMT) plays a critical role during normal development and in cancer progression. EMT is induced by various signaling pathways, including TGF-β, BMP, Wnt–β-catenin, NOTCH, Shh, and receptor tyrosine kinases. In this study, we performed single-cell RNA sequencing on MCF10A cells undergoing EMT by TGF-β1 stimulation. Our comprehensive analysis revealed that cells progress through EMT at different paces. Using pseudotime clustering reconstruction of gene-expression profiles during EMT, we found sequential and parallel activation of EMT signaling pathways. We also observed various transitional cellular states during EMT. We identified regulatory signaling nodes that drive EMT with the expression of important microRNAs and transcription factors. Using a random circuit perturbation methodology, we demonstrate that the NOTCH signaling pathway acts as a key driver of TGF-β–induced EMT. Furthermore, we demonstrate that the gene signatures of pseudotime clusters corresponding to the intermediate hybrid EMT state are associated with poor patient outcome. Overall, this study provides insight into context-specific drivers of cancer progression and highlights the complexities of the EMT process.
During cancer progression, activation of epithelial-to-mesenchymal transition (EMT) results in cancer cells acquiring mesenchymal and stem cell properties. These altered cells can dissociate from the primary tumor mass, invade surrounding tissue, and intravasate into blood vessels (1, 2). Upon extravasation at the distant site, these disseminated cancer cells, depending on the local microenvironment, either begin to proliferate and revert to a more epithelial phenotype or remain in a quiescent phase for an extended period, thereby providing an opportunity for recurrence. In either case, this leads to metastatic disease, the leading cause of death in cancer patients. The stem cell properties that develop during EMT may also impart resistance to various therapies (3, 4).
Cells that have undergone EMT lose adherent junctions due to both transcriptional repression of E-cadherin and the elimination of cell-surface E-cadherin (5). In addition, epithelial markers, such as epithelial cellular adhesion molecule (EpCAM), desmoplakins, and cytokeratins are down-regulated during EMT, and there is an increase of mesenchymal markers, such as Vimentin, N-cadherin, fibronectins, and matrix metalloproteinases (6). These sequential events of EMT are tightly regulated by a set of transcription factors, including Slug, Twist, Snail, Zeb1, and Zeb2, and are highly context-specific (7, 8).
Expression and activation of EMT-inducing transcription factors occur in response to various signaling pathways, including those mediated by TGF-β, BMP, EGF, FGF, PDGF, Wnt, Shh, NOTCH, and integrins (9–14). Signaling pathways have been shown to interact at various levels and a number of feedback activation/repression mechanisms have been demonstrated in different EMT contexts, with each potentially having overlapping/context-specific outputs (15, 16). It is therefore critical to characterize the induction of pathways in various subpopulations of cancer cells over a temporally resolved time course following EMT induction.
Here we used single-cell RNA sequencing (scRNA-seq) to capture the changes in epithelial cells (17) over an extended time course of TGF-β1–induced EMT. The experimental data were analyzed using unsupervised bioinformatic methods to decipher the pseudotime progression. We also utilized a regression-based approach and a circuit randomization procedure to predict couplings between different pathways and the consequences of perturbations to various EMT-related factors. We demonstrate the involvement of several EMT-promoting signaling mechanisms in the cross-talk that integrates the tumor microenvironment with the tumor cells themselves to drive their reprogramming. This study identifies the signaling events and regulators at multiple intermediate stages during EMT and advances our knowledge of tumor progression by elucidating targets for developing novel treatment strategies to combat treatment-resistant and metastatic cancer.
Results
During TGF-β1–Induced EMT, Multiple Signaling Pathways Are Activated Simultaneously.
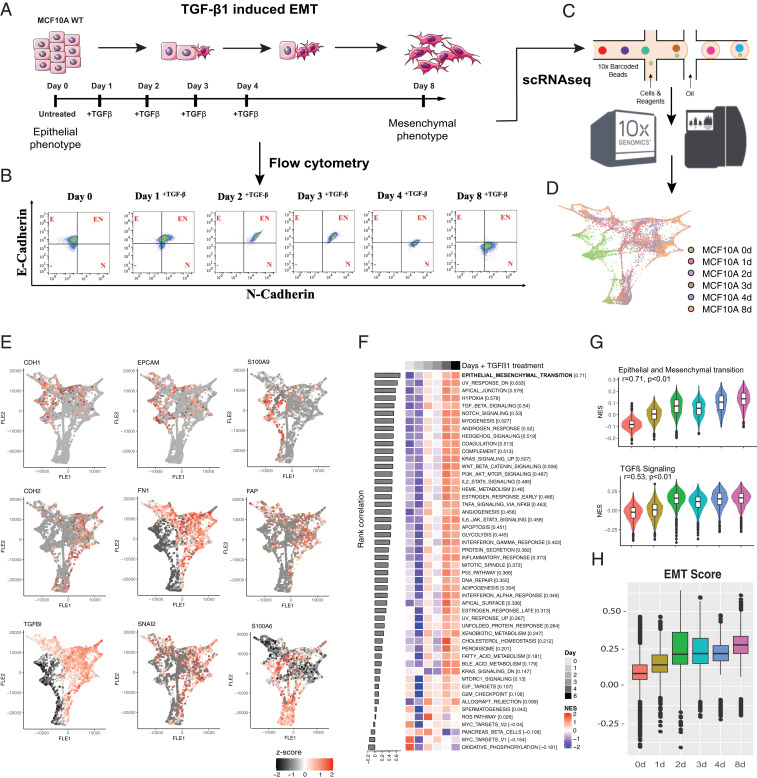
To characterize molecular changes during EMT progression, we used the human immortalized breast cell line MCF10A, which has been well-characterized during TGF- β1–induced EMT. Specifically, when stimulated with TGF- β1, MCF10A cells undergo morphologic and phenotypic EMT-like changes, including cytoskeleton reorganization, mesenchymal marker up-regulation, and cadherin switching (18, 19). MCF10A cells were stimulated with TGF-β1 for 1, 2, 3, 4, or 8 d (Fig. 1A). We initially monitored for changes in the epithelial marker E-cadherin (CDH1) and the mesenchymal marker N-cadherin (CDH2) (Fig. 1B). Almost all uninduced cells expressed E-cadherin without the expression of N-cadherin. N-cadherin was induced in a subset of cells after 1 d of TGF-β1 treatment and within 2 d most of the cells expressed N-cadherin. A decrease in E-cadherin levels began to appear in a subset of cells after 4 d of exposure to TGF-β1 (Fig. 1B), while the complete cadherin switch was observed only after 8 d of TGF-β1 treatment.
Fig. 1.
During TGF-β1–induced EMT multiple signaling pathways are activated simultaneously. (A) Schematic representation of model used to EMT in MCF10A cells. (B) Flow cytometry analyses of MCF10A cells treated over 8 d with TGF-β. (C) scRNA-seq schematic. (D) Force-directed layout embedding (FLE) of the trajectory of TGF-β1–induced EMT in MCF10A. (E) FLE trajectories of epithelial markers (CDH1, EpCAM, and S100A9), mesenchymal markers (CDH2, FN1, and FAP), and EMT transcription factors (TGFB1, SNAI2, and S100A6) across the time course of TGF-β treatment. (F) Rank correlation of Hallmark signaling pathways. The x axis shows the TGF-β1 time course treatment of MCF10A cells and y axis shows the NES. (G) Violin plot of the hallmark EMT and TGF-β signaling pathways NES for each time point during the TGF-β1 treatment. (H) Kolmogorov–Smirnov EMT scores during TGF-β1 time course treatment of MCF10A cells. ANOVA (***P < 0.001).
To identify the signaling pathways that get sequentially induced during EMT in a time-dependent manner, we performed scRNA-seq at the same time points using MCF10A cells untreated (day 0) and treated with TGF-β1 (days 1 to 8) (Fig. 1C). The experiments were performed in two batches; the data were batch-corrected and curated before further analyses (SI Appendix, Fig. S1 and Dataset S1). The force-directed layout (20) embedding of the scRNA-seq data demonstrated a clear shift in the global gene-expression trajectory from epithelial to mesenchymal features, as expected, over the time course (Fig. 1D and SI Appendix, Fig. S2A).
During EMT, E-cadherin and other junctional proteins, including claudins and desmosomes, are repressed, and this facilitates the general dedifferentiation program (5, 6). Therefore, we first analyzed the expression of well-defined epithelial and mesenchymal markers. As previously described, TGF-β1 treatment immediately induced loss of S100 calcium-binding protein A9 (S100A9) expression (21), and also induced the gradual loss of epithelial markers CDH1 and EpCAM (5, 6) (Fig. 1E), as well as other genes linked to the epithelial phenotype (SI Appendix, Fig. S2B). We also observed the gain of mesenchymal markers (CDH2, FN1, FAP) and EMT-associated transcription factors (e.g., SNAI2) and S100 calcium-binding protein A6 (S100A6), a known marker of EMT (22, 23), starting after 2 d of EMT induction (Fig. 1E and SI Appendix, Fig. S2B). Observations of increases in N-cadherin and fibronectin and reduced E-cadherin and EpCAM expression are consistent with previous findings (24, 25).
TGF-β1 signaling is pleiotropic and interacts with various other pathways (26). Not until 2 d after the start of TGF-β1 treatment did we observe increases in specific mRNAs encoding factors involved in NOTCH, Shh and Wnt signaling cascades, with a robust activation after 4 d of stimulation in the total cell population (Fig. 1 F and G) and in the majority of individual cells (SI Appendix, Fig. S2C). We next calculated an EMT score of the cells across different time points using the Kolmogorov–Smirnov metric identified based on epithelial and mesenchymal gene lists, which quantifies the difference between the empirical cumulative distributive functions for the two gene lists. Thus, this EMT score indicates a comparative analysis of the “EMT-ness” of a given sample (27, 28). We observed that the EMT score distribution changed with time, with single cells at each time point showing a range of negative to positive EMT scores indicative of cell-level heterogeneity at each time point. At day 8, the mean EMT score was higher than at other time points, indicating a shift toward a more mesenchymal phenotype at an ensemble level (Fig. 1H). Together, these data show that many EMT regulatory pathways are induced simultaneously, suggesting that EMT may require cross-talk among various signaling pathways in a temporal manner, likely driven by interactions between cells and their microenvironment.
Pseudotime Reconstruction of EMT Reveals Key Regulators.
Global analysis of various time points indicates an overall progression of EMT in response to TGF-β1 stimulation (Fig. 1F). To better understand population heterogeneity and EMT activation at the single-cell level, we performed scRNA-seq analysis. We applied k-nearest neighbor and modularity optimization techniques to cluster the transcriptome data and to characterize the subpopulations of cells that acquire mesenchymal features at various time points during EMT (Fig. 2A). The cluster analysis revealed progression in pseudotime starting from an epithelial to a mesenchymal RNA expression status. When the pseudoprogression was correlated with the time of TGF-β1 treatment, we discovered that the vast majority of the untreated cells (green) start at the epithelial extreme before EMT induction, with only a small fraction of cells falling in clusters that show partial or complete EMT progression (Fig. 2B). An interesting observation was that not all cells progress at the same rate through EMT (Fig. 2B, SI Appendix, Fig. S3A, and Dataset S2). Interestingly, at intermediate time points, single cells with phenotypes across the EMT spectrum were observed. Even after 8 d, about half of the cells analyzed exhibited both epithelial and mesenchymal properties, called hybrid or E/M cells. Most importantly, this analysis revealed 20 pseudotime clusters (Fig. 2B).
Fig. 2.
Reconstruction of EMT using pseudotime clustering reveals key regulators of epithelial-mesenchymal states. (A) t-SNE of cells treated with TGF-β1 (Left) and pseudotime (Right) clusters in principal component analysis space grouped using modularity optimization technique. (B) The fraction of cells in each cluster and time point of origin shown using a Sankey network. The red dots indicate that at least 10% of cells in a cluster mapped to the corresponding time point. (C) Hierarchical network of cells in each cluster with different TGF-β1 treatment time points and ranked in pseudotime. (D) Cluster-specific markers identified using Wilcoxon test (>1.4-fold change, adjusted P < 0.05). Among identified significantly up-regulated genes in each cluster the transcription factors critical for stemness are highlighted. (E) The inferred enrichment for miRNAs based on the miRNA target expression. Before enrichment the average gene expression for each gene in a cluster was calculated. The average expression was scaled across the clusters and used for enrichment analysis. (F) Inferred miRNA enrichment score and relative fraction of target in each cluster. (G) Inferred EMT regulatory miRNA network and putative regulators and model depicting miR217- and miR200a/200b/200c-3p-dependent EMT regulatory checkpoint.
Next, we performed hierarchical clustering based on the 20 identified pseudotime clusters and observed that mRNAs associated with the TGF-β, WNT/β-catenin, NOTCH, Shh, and PI3K/AKT/mTOR pathways were up-regulated after TGF-β1 treatment. (Fig. 2C). We also identified 644 significantly up-regulated mRNAs with TGF-β1 stimulation (SI Appendix, Fig. S3 and Dataset S3), including mRNAs encoding transcription factors with established roles in the regulation of stemness (Fig. 2D). Several stemness factors that we identified, such as myc (29), dnmt1 (30), fos (31), irf6 (32), egr1 (33), and sox4 (34), are known to mediate cancer stem cells and EMT changes in breast cancer cells. We also identified stemness factors, such as hes4 and mxd4, with less clearly defined roles in the EMT process. To get a better understanding of the regulatory pathways involved, particularly to understand the role of microRNA (miRNAs) in regulating differentially expressed transcription factors, we analyzed the miRNA levels in each cluster based on miRNA target gene expression using the mirWalk2.0 database (35) (Fig. 2E). A striking drop of miRNA expression was observed from cluster 8 to cluster 6 in the pseudotime progression. There were large inverse correlations between inferred miRNA enrichment scores and the relative fractions of target genes in each cluster (Fig. 2F). Interestingly, by this analysis we identified several miRNAs previously implicated in EMT regulatory checkpoints (miR217, miR205, and miR200a/200b/200c-3p) (Fig. 2G), as well as novel miRNAs not previously associated with EMT, (miR-30b, miR203A, miR21, miR148-3p and miR192), being suppressed during TGF-β1–induced EMT (Fig. 2F). In addition, several miRNAs were up-regulated in clusters C6 through C17 (miR1268a, miR3140-3p, miR486-5p, miR224, and miR369-5p) with known important roles in regulating EMT. We also identified two miRNAs (miR374-3p and miR613) that had not been previously associated with EMT (Fig. 2F).
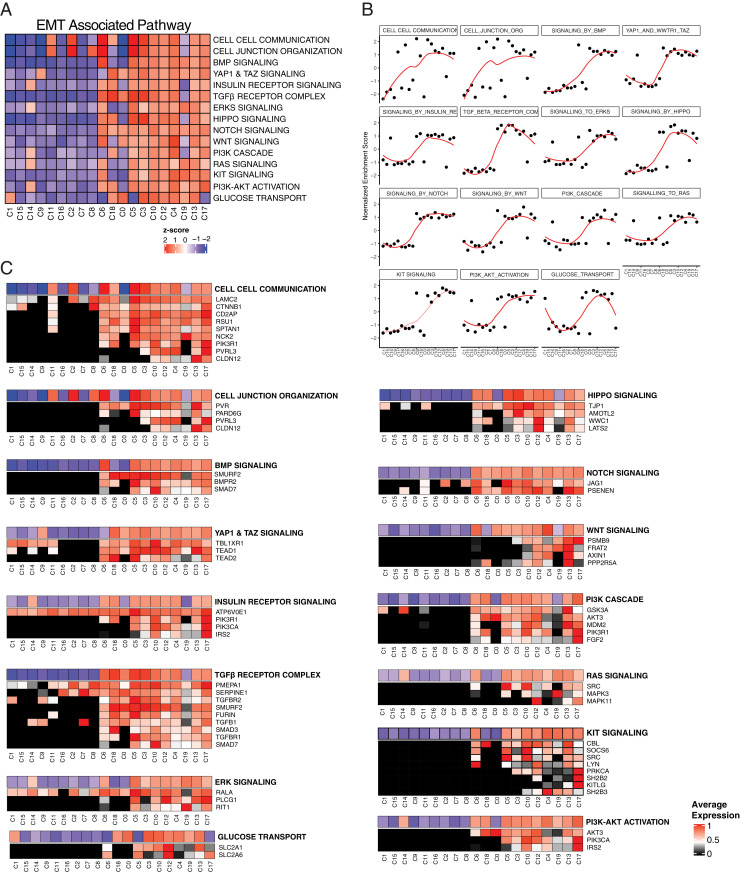
To better understand the parallel and sequential activation of signaling pathways during TGF-β1–induced EMT, we performed gene set enrichment analyses (https://reactome.org/). During early time points of EMT progression, cell–cell communication and cell-junction organization pathways were enriched, suggesting an early role for the contribution of the microenvironment in driving EMT (Fig. 3 A and B). Among known EMT-regulatory genes, CDH1, EPCAM, and several keratins were down-regulated, and CDH2, FN1, VIM, integrin β1, and integrin β5 were up-regulated along with the pseudotime transition (SI Appendix, Fig. S3 C and E). It has been previously reported that during EMT cells switch their metabolism from mitochondrial oxidative phosphorylation to glycolysis (36). In line with this finding, mRNAs encoding proteins involved in mitochondrial oxidative phosphorylation were down-regulated (SI Appendix, Fig. S3 B and G). EMT-associated pathways related to stem cell properties were activated later during EMT, indicated by up-regulation of mRNAs encoding factors involved in BMP, YAP/TAZ, HIPPO, NOTCH, and Wnt pathways (Fig. 3A and SI Appendix, Fig. S3I). Multiple reactome pathways were significantly changed during the pseudotime EMT progression (SI Appendix, Fig. S3D and Dataset S4).
Fig. 3.
scRNA-seq analysis identifies parallel and sequential signaling pathways involved in TGF-β1–driven EMT. (A) Heatmap of enrichment for indicated EMT-associated pathways in each cluster. (B) NES of significantly altered signaling pathways across pseudotime clusters. The red lines are a fit determined by LOESS. (C) Heatmap showing the distributions of mRNA expression of genes that were induced during TGF-β1–induced EMT and maintained increased expression during EMT progression. Genes associated with each EMT-associated pathway and their fraction in each cluster compared to maximum average expression is shown in alignment with EMT-associated pathway activation.
The most striking observation is the binary activation of the EMT-associated pathways (SI Appendix, Fig. S3J). For example, genes implicated in stem cell-related pathways, TGF-β, ERK, PI3K/AKT, and glucose transport pathways, show the most striking binary activation during the transition, which occurs from clusters 8 to 6 (Fig. 3 A and B and SI Appendix, Fig. S3 H and I). Next, we looked at key regulatory genes within the binary EMT-induced pathways that were up-regulated during TGF-β1 induction and stayed up-regulated for the remainder of EMT progression. Many genes within the EMT-associated pathways show a binary increase of expression, and the majority is observed between clusters 8 and 6 (Fig. 3C). Interestingly, binary regulation of mRNAs encoding certain enzymes and transcription factors was also observed (SI Appendix, Fig. S5). This binary activation of EMT-associated pathways during EMT suggests an existence of a checkpoint-like state.
To determine causal relationships between the key EMT-associated pathways (Wnt, Notch, YAP/TAZ, Hippo, and so forth) shown in Fig. 3A, we used a multiple lasso regression-based approach followed by dynamical systems modeling. We used a 15-dimensional vector to represent the state of the population of cells at each pseudotime point. Here, each vector element corresponds to the enrichment score of one of the 15 EMT-associated pathways shown in Fig. 3A. Next, we used multiple regression with lasso regularization to generate a transition matrix that maps the population state at pseudotime point t to the next pseudotime point, t + 1 (37), and created a network representation of the resultant transition matrix (Fig. 4A). To explore the range of dynamic behaviors the inferred network can exhibit, we used the recently proposed random circuit perturbation approach (38). This approach identifies the steady-state behaviors of a network by simulating the network dynamics for an ensemble of parameters (39, 40), and was initially developed to analyze the dynamical behavior of gene regulatory networks with transcription factors as nodes. Here, we directly apply this method to analyze the behavior of a network with different cellular pathways as nodes. In doing so, we assume that the activities of the different genes constituting a “pathway” can be coarse-grained, defining a macroscopic variable which quantifies the overall pathway activity. The assumption is based upon the idea of network coarse-graining as, for example, has previously been described by Drier et al. (41). Similarly, the interactions between the genes in different pathways are functionally represented by a single edge from one pathway node to another, the nature (whether activating or inhibitory) of which is determined in a data-driven manner from the sign of the corresponding entry in the transition matrix (SI Appendix, Methods).
Fig. 4.
Systematic inhibition of EMT signaling reveals key drivers of EMT. (A) Network representation of the transition matrix calculated using multiple regression with lasso regularization mapping the population state at pseudotime t to pseudotime point t + 1. (B) The RACIPE method identifies the steady-state behaviors a network can exhibit by simulating network behavior for an ensemble of parameters. For the constructed network, but not for a set of randomized controls, the variance is dominated by the first principal component (PC1, Left) and there exists a large percentage of parameter values for which the network exhibits more than one stable phenotypic state (Right). (C) Activity of each node was suppressed one-by-one and the change in the distribution of the first principal component was plotted. (D) Effect of suppressing the activity of a given pathway node (shown along the rows) on the activity of the other key network pathways (shown along the columns).
The inferred network has steady states, which mostly vary along with the first principal component (Fig. 4 B, Left, and SI Appendix, Fig. S4A), and exhibits multistability (i.e., two or more steady states for a given parameter set) (Fig. 4 B, Right). Such behavior was not observed in the case of control networks inferred via the same approach but by using randomized data as input instead of the scRNA-seq data. To determine which of the pathway nodes in the network are dominant in driving network behavior, we suppressed each node’s activity individually and determined changes in the distribution of the first principal component of the network steady states (Fig. 4C). The distribution of the first principal component exhibited large deviations from the control case (where the activity of none of the network nodes is suppressed) upon suppression of the TGF-β, NOTCH, and YAP/TAZ signaling pathways, revealing these pathways as the key drivers of EMT in this system (Fig. 4C). This was confirmed by calculating the Kullback–Leibler divergence (42) between the distribution obtained after pathway suppression and that in the control case (SI Appendix, Fig. S4B). The identical three pathways were identified as the key EMT drivers in an alternate approach using a Boolean modeling framework to model the network behavior. This result indicates that our findings are robust and, in particular, not dependent on the choice of the random circuit perturbation approach for network analysis (SI Appendix, Fig. S4 C and D). That the TGF-β signaling pathway is among the identified key pathways serves as a helpful consistency check. Finally, we noted that while suppressing the YAP/TAZ signaling pathway activity did not substantially affect the activity of the NOTCH signaling pathway (Fig. 4 D, Top row), suppression of NOTCH pathway activity suppressed YAP/TAZ signaling activity, including changing the distribution of YAP/TAZ activity from bimodal to unimodal (Fig. 4 D, Middle row). From these analyses, NOTCH signaling appears to be the crucial regulator of TGF-β–driven EMT in the present context.
Gene Signatures of Pseudotime Clusters Correlate with Patient Outcome.
Differentially expressed genes from each cluster were identified using Wilcoxon test (adjusted P < 0.05). We then analyzed whether these pseudotime cluster trajectory-based signatures were correlated with progression-free interval (PFI), disease-free interval (DFI), and overall survival (OS) in the Cancer Genome Atlas (TCGA) breast cancer and pan-cancer cohort, including 32 cancer types (Fig. 5A). We observed increased hazard ratios for PFI, DFI, and OS for patients with C16, C0, C3, C10, and C13 cluster-specific signatures, which lie toward the mid and mesenchymal state of EMT in pseudotime for breast cancer (Fig. 5B and SI Appendix, Fig. S6 A and C) and other cancer types (Fig. 5 C and D, SI Appendix, Figs. S6 and S7, and Dataset S5). Of great interest was that breast cancer and pan-cancer PFI-based survival analysis showed a higher hazard ratio in C16, C0, C10, and C13 clusters, displaying both epithelial and mesenchymal properties, suggesting transient states between epithelial and mesenchymal phenotype on the EMT trajectory, reminiscent of recent in vivo reports (43, 44). Genes within these clusters showed more of an increased hazard ratio than genes in other clusters (Fig. 5E). Together, these data suggest that partial EMT properties identified at the single-cell level are associated with poor prognosis.
Fig. 5.
Gene signatures of pseudotime clusters closer to the mesenchymal state are associated with worse survival. (A) Schematic of identification of differentially expressed (DE) genes from each cluster and signature enrichment in cancer types based on normalized RNA expression data from TCGA. Cancer cohorts (∼11,000 patients) analyzed were adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical and endocervical cancers (CESC), cholangiocarcinoma (CHOL), colon cancer (COAD), lymphoid neoplasm diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS), and uveal melanoma (UVM). (B) Hazard ratios for PFI for TCGA breast cancer cohort (n = 1,092) for patients with each cluster-specific signature. The dashed line indicates hazard ratio 1. (C) Hazard ratios for patients with indicated cancers for each cluster-specific signature. (D) Univariate cox analysis-based hazard ratio observed using PFI data for each cluster in each cancer cohort. (E) Log fold-changes for the cluster C0, C10, and C13 signature genes.
Discussion
A complex network of interconnected pathways mediated by TGF-β, EGF, IGF, Wnt, Shh, and NOTCH regulates EMT (45). Here, we focus on activating EMT via TGF-β, one of the key drivers of EMT in many cancer types (26). TGF-β acts as a tumor suppressor at early stages of tumor development by inhibiting proliferation and inducing apoptosis. Still, at later stages of tumor development, TGF-β acts as a tumor promoter by inducing EMT and suppressing antitumor immune responses (46). Activation of this node leads to an overall activation of additional pathways. These converge on a network of transcription factors and miRNAs that repress epithelial characteristics (47) and induce mesenchymal characteristics (48). Using scRNA-seq, we identified “core” signaling cascades and the critical regulatory network underlying EMT.
The use of single-cell analysis over a time course of TGF-β1 treatment enabled mapping of the signaling cascades that control EMT progression in this context. Our data indicate that EMT-associated signaling pathways are activated sequentially and that stem cell-related pathways are activated relatively quickly, then deactivated, and again, reactivated as a function of pseudotime corresponding to the position along an EMT trajectory. Our single-cell analysis also reconciles conflicting views of activation of signaling pathways in EMT. We show that TGF-β1–induced EMT causes mRNAs encoding certain transcription factors and signaling receptors to accumulate at defined points along the pseudotime trajectory, and that cells fall along a transcriptional continuum during EMT (49). This implies the existence of a cascade of events. Gene variation that activates key signaling pathways could enrich a particular gene-expression profile indicative of a specific EMT intermediate state. Consistent with our findings, cross-talk during EMT has been characterized between TGF-β– and NOTCH-mediated signaling (50), between Wnt and FGF signaling (51, 52), between ERK and TGF-β signaling (53), between PI3K and TGF-β signaling (54), and between hypoxia and NOTCH signaling (55).
Our single-cell analyses also support previous findings that small noncoding RNAs regulate EMT. Expression of the miR200 family is strongly associated with epithelial differentiation, and a reciprocal feedback loop between the miR200 family miRNAs and the ZEB family of transcription factors tightly controls EMT (16). Moreover, additional miRNAs might maintain the epithelial phenotype; an example is miR101, which maintains E-cadherin expression by repressing EZH2 (56)
With our predictive modeling approach, we were able to determine the drivers of EMT network through systematic testing of inhibitory effects of individual signaling pathways on other signaling pathways. Although TGF-β, NOTCH, and YAP/TAZ pathways all regulate EMT, our data indicate that NOTCH signaling is a key driver of EMT, consistent with our previous observations of Notch-Jagged signaling in stabilizing EMT states (57). Activation of EMT is critical for cancer progression and metastasis (58, 59), and there is clinical evidence that cancer cells can disseminate and metastasize early during cancer development (60). Our analysis of gene expression during EMT induced by TGF-β1 demonstrated that although the vast majority of cells during the early induction period have barely entered EMT, rare cells do indeed progress rapidly. These cells may be capable of metastasizing. In support of this, the pseudotime clusters enriched for mesenchymal expression profiles are associated with poor DFI, PFI, and OS of patients with many cancer types. The same clusters have increased hazard ratios among many different cancer types, indicating that the same genes are involved in EMT and cancer progression. Although limitations exist as the signatures were derived from cell-line samples, their overall expression in patients with poor survival helps us nominate important clusters/genes, which can serve as potential targets for the treatment of advanced and metastatic cancers.
Methods
Cell Culture.
MCF10A breast epithelial cells were purchased from ATCC (CRL-10317) and used within 10 passages. Cells were cultured at 37 °C and 5% CO2 in MCF10A complete media (DMEM/F12 [Gibco] supplemented with 5% horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 5 μg/mL insulin, 100 ng/mL cholera toxin, and antibiotic). The cells were treated with 5 ng/mL TGF-β1 to induce EMT. The media was replenished every 2 d.
Flow Cytometry.
MCF10A cells with day 0 and treated with TGF-β1 for 1, 2, 3, 4, and 8 d were harvested using TryplE. Cells were incubated with anti-human CD324 (E-cadherin) Clone 67A4 (BD Biosciences, #562870) conjugated with PE and anti-human CD325 (N-cadherin) Clone 8C11 (Novus, #NBP2-54523APC) conjugated with APC. Antibody incubations were performed in MCF10A complete media. Samples were washed three times with FACS Media (PBS + 10% FBS), resuspended in FACS Media, and analyzed using a BD Accuri C6 Plus. Analysis was performed on FlowJo.
Single-Cell Library Preparation and Sequencing.
The single-cell suspensions of MCF10A cells were prepared as recommended by the 10x Genomics single-cell preparation guide (CG000053 Rev C) and 3′ scRNA-seq libraries were generated according to the instructions for the Chromium Single Cell 3′ Reagent Kits v2 chemistry (CG00052 Rev E, for days 0, 4, and 8 samples, batch 1: marked as MCF10A_0Bd, MCF10A_4d, MCF10A_8d) and v3 chemistry (CG000183 Rev A, for days 0, 1, 2, and 3 samples, batch 2: marked as MCF10A_0d, MCF10A_1d, MCF10A_2d, MCF10A_3d) protocols of the 10x Chromium Single Cell Gene Expression solution (https://www.10xgenomics.com/products/single-cell-gene-expression). Briefly, cells were collected at the day of library preparation, washed twice in 0.04% BSA in PBS, passed through the 40-μm strainer, stained with 0.4% Trypan blue, and quantified and assessed for viability using the cell automated counting machine Cellometer Mini (Nexcelom). Next, the single-cell suspensions with a targeted cell recovery of 3,000 cells per sample were mixed with Master Mix and loaded into the Chromium Chip (A for v2 or B for v3) along with the barcoded single-cell 3′ Gel Beads (v2 or v3) and Partitioning Oil to generate the nanoliter-scale gel beads-in-emulsion (GEMs), in 10x Chromium Controller (10x Genomics). Next, the captured GEMs were incubated to generate cDNA tagged with a cell barcode and unique molecular index (UMI). Then, after breaking the GEMs, the full-length, barcoded cDNA was amplified by PCR to generate sufficient mass for library construction. The quality and quantity of cDNA was assessed using 4200 TapeStation High Sensitivity D5000 reagents (Agilent Technologies). In order to prepare 3ʹ gene-expression libraries, amplified cDNA was first enzymatically fragmented, end-repaired, and A-tailed, followed by fragment-size selection using SPRIselect magnetic beads (Beckman Coulter). Illumina sequencing adapters were subsequently added to the fragments during the ligation step followed up by postligation clean-up (SPRIselect, Beckman Coulter). Finally, the unique 10x sample indices (PN-220103 Chromium i7 Sample Index Plate well ID) were added during PCR amplification to each sample, followed by size selection (SPRIselect, Beckman Coulter). The final libraries were quality control (QC)-checked using 4200 TapeStation High Sensitivity D1000 reagents (Agilent Technologies). All libraries were quantified using Qubit 1X dsDNA HS Assay Kit (Invitrogen), normalized, and pooled based on the chemistry version (v2, v3). The pool of v2 chemistry was sequenced using Illumina NextSeq500 at the MDACC ncRNA core, and a pool of v3 was sequenced on Illumina NovaSeq6000 using S1-100 flow cell type at the MDACC ATGC core, all with 10x Genomics recommended sequencing parameters and targeting 50,000 reads per cell.
scRNA-Seq Data Analysis.
Cell Ranger pipeline (10x Genomics, default settings, v3.1.0) was used to process the raw Illumina sequencing files to generate fastq files and align the sequencing reads to human reference genome (hg19) to generate counts. The downstream analysis, including the quality control steps, normalization, batch correction, and downstream analysis and visualization, were performed in R using Seurat v3.1.0 R package (57).
The aligned sequences resulted in the mapping of 32,738 genes. Between 18,423 and 19,264 genes were expressed at each time point. A UMI count matrix of the number of genes (rows) and unique cells (column) was constructed. The cells with low reads were filtered for greater than 500 features and 0.2 mitochondrial fraction. We obtained data from two batches and combined using the merge function in the Seurat package implemented in R (v3.1.0) (56). The data were normalized and corrected for batch effect using data integration method implemented in Seurat (57). Top variable 15,000 features were selected to identify integrating features and to maximize the number of features in the dataset. Principal component analysis was performed on scaled, log-transformed, library-size-normalized UMI matrices using variable gene sets. Dimensionality reduction and visualization were also performed with the UMAP and t-distributed stochastic neighbor embedding (t-SNE) algorithms. The projections were generated with a perplexity of 30. Graph-based clustering was performed to identify clusters using the first 15 principal components. The redundant day 0 data were removed after batch correction. In total, we used 12,588 of 13,941 cells (>500 features and 20% mitochondrial fraction) for downstream analysis. Transcripts per cluster were identified using a Wilcoxon rank sum test. Significantly differentially expressed transcripts were selected using adjusted P < 0.05 (Benjamini–Hochberg method).
Rank order statistics were calculated using Jonckheere–Terpstra trend test implemented in SAGx (v1.46.0). Before rank order correlation the single-cell normalized count data were averaged over each time point. Using known order of time points (0 to 8 d of TGF-β1 treatment) the rank order correlation was calculated.
The fraction of cells in each cluster attributed to each TGF-β1 time point was calculated using the following equation:
The cells that originated from each time point were visualized using a Sankey network (networkD3, v0.4) with a threshold of 10%.
Rank Correlation.
The rank order statistics was calculated using Jonckheere–Terpstra trend test implemented in SAGx (v1.46.0). Before rank order correlation the single cell-normalized count data were averaged over each time point. Using known order of time point (0 to 8 d) the rank order correlation was calculated.
RNA-Seq to Microarray Conversion.
All previous EMT metrics were identified based on gene expression evaluated using a microarray platform. Therefore, scRNA-seq data were converted to microarray data. The regression parameters used to transform the scRNA-seq data were estimated as described previously (61).
EMT Scoring.
As previously reported by Tan et al. (27), the Kolmogorov–Smirnov EMT scores were calculated. For a given sample, this method compares cumulative distribution functions (CDFs) of epithelial and mesenchymal gene signatures. First, the distance between epithelial and mesenchymal signatures was calculated via the maximum distance between their CDFs. This quantity represents the test statistics used to calculate the EMT score in the subsequent two-sample test. Using hypothesis testing of two alternative hypotheses, the score is determined as follows (with the null hypothesis being that there is no difference in CDF of epithelial and mesenchymal signatures): 1) the CDF of the mesenchymal signature is greater than the CDF of the epithelial signature, and 2) the CDF of epithelial signature is greater than the CDF of the mesenchymal signature. The score range is −1 to +1, where a sample with a positive EMT score has a mesenchymal phenotype and a sample with a negative EMT score has an epithelial phenotype.
Enrichment Analysis.
Single-sample gene-set enrichment analysis was performed using the GSVA package (v1.28.0) using hallmarks from MSigDB (v6.2). The average normalized enrichment score (NES) was calculated for each time point. The NES was compared between samples and scaled values are shown. For miRNA enrichment analysis, miRNAs and their target genes were obtained from the miR database mirWalk2.0 (34). miRNA enrichment was inferred based on the miRNA target expression in scRNA-seq data using the fgsea package (v1.16.0). Before enrichment, the average gene expression for each gene in a cluster was calculated. The average expression was scaled across the clusters and used for enrichment analysis. The NES and P value significance were obtained. For signaling pathway enrichment, the Reactome was downloaded from MSigDB (c2.reactome.v6.2.symbols.gmt, https://reactome.org/).
Inferring the Interplay between the Key Pathways Driving EMT.
The ordering of the different t-SNE clusters was interpreted as a pseudotime trajectory, with each cluster corresponding to one pseudotime point. We represented the population of cells at each such pseudotime point using a 15-dimensional vector where each vector element corresponds to the enrichment score for one of the 15 key pathways. We assumed that the state of the population at a given pseudotime point t depends only on the population state at the previous pseudotime point t − 1. With this set-up, we used multiple linear regression with lasso regularization (1) to obtain a sparse 15 × 15 matrix M that maps the population state from one pseudotime point to the next. The matrix represents a network of regulatory relationships between the different pathways. The range of dynamic behaviors exhibited by this regulatory network was analyzed using Random Circuit Perturbation (RACIPE) (2). Boolean modeling of the regulatory network was carried out using the framework described previously (62). See SI Appendix, Methods for a detailed mathematical description.
Pan-Cancer Survival Analysis.
We downloaded RNA-seq data and clinical characteristics for a cohort of patients with 32 cancer types from the Firehose of the Broad Institute (http://gdac.broadinstitute.org/, January 2016 version). PFI, DFI, and OS were used to perform survival analysis (63). The Kaplan–Meier method was used to determine survival probability. The P values were determined by a log-rank test. The signatures for each cluster were used to retrieve signature enrichment scores. The NES then categorized as high or low based on mean. Univariate Cox proportional hazards models were fitted to calculate the hazard ratios using the coxph function in Survival (v 2.44). P values less than 0.05 were considered to be statistically significant.
Statistical Analysis.
The statistical analyses used are specified in the figure legends.
Supplementary Material
Acknowledgments
The S.A.M. laboratory is supported by NIH/National Cancer Institute Grants (R01CA200970 and 2R01CA155243), Cancer Prevention and Research Institute of Texas Grant (RP170172), and Bowes Foundation. S.A.M. and H.L. are supported by the NSF Grant (PHY-1935762). H.L. is supported by the NSF Grant (PHY-2019745). K.R. is supported by the National Cancer Institute Grant (1R01CA226269). M.K.J. is supported by the Ramanujan Fellowship awarded by the Science and Engineering Research Board, Government of India (SB/S2/RJN-049/2018) and Infosys Young Investigator Award supported by the Infosys Foundation, Bangalore.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102050118/-/DCSupplemental.
Data Availability
All the raw sequencing data have been deposited at National Center for Biotechnology Information Sequence Read Archive (BioProject ID: PRJNA698642). All other study data are included in the article and supporting information.
Change History
May 12, 2021: The license for this article has been updated.
References
- 1.Mani S. A., et al., The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jolly M. K., et al., Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J. R. Soc. Interface 11, 20140962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creighton C. J., et al., Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. U.S.A. 106, 13820–13825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolly M. K., et al., Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 194, 161–184 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Wendt M. K., Taylor M. A., Schiemann B. J., Schiemann W. P., Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol. Biol. Cell 22, 2423–2435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Zhou B. P., Epithelial-mesenchymal transition—A hallmark of breast cancer metastasis. Cancer Hallm. 1, 38–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roche J., The epithelial-to-mesenchymal transition in cancer. Cancers (Basel) 10, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasaikar S. V., et al., EMTome: A resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br. J. Cancer 124, 259–269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taipale J., Beachy P. A., The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Katoh Y., Katoh M., FGFR2-related pathogenesis and FGFR2-targeted therapeutics (Review). Int. J. Mol. Med. 23, 307–311 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Al Moustafa A.-E., Achkhar A., Yasmeen A., EGF-receptor signaling and epithelial-mesenchymal transition in human carcinomas. Front. Biosci. (Schol. Ed.) 4, 671–684 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Espinoza I., Miele L., Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett. 341, 41–45 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Heldin C.-H., Vanlandewijck M., Moustakas A., Regulation of EMT by TGFβ in cancer. FEBS Lett. 586, 1959–1970 (2012). [DOI] [PubMed] [Google Scholar]
- 14.McCormack N., O’Dea S., Regulation of epithelial to mesenchymal transition by bone morphogenetic proteins. Cell. Signal. 25, 2856–2862 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Dong C., et al., G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122, 1469–1486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory P. A., et al., The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., et al., TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 7, ra91 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Maeda M., Johnson K. R., Wheelock M. J., Cadherin switching: Essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J. Cell Sci. 118, 873–887 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Sarrió D., et al., Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68, 989–997 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Jacomy M., Venturini T., Heymann S., Bastian M., ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9, e98679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basso D., et al., Inflammation and pancreatic cancer: Molecular and functional interactions between S100A8, S100A9, NT-S100A8 and TGFβ1. Cell Commun. Signal. 12, 20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Ismaeel Q., et al., ZEB1 and IL-6/11-STAT3 signalling cooperate to define invasive potential of pancreatic cancer cells via differential regulation of the expression of S100 proteins. Br. J. Cancer 121, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li A., et al., S100A6 promotes the proliferation and migration of cervical cancer cells via the PI3K/Akt signaling pathway. Oncol. Lett. 15, 5685–5693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyun K. A., et al., Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 7, 24677–24687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araki K., et al., E/N-cadherin switch mediates cancer progression via TGF-β-induced epithelial-to-mesenchymal transition in extrahepatic cholangiocarcinoma. Br. J. Cancer 105, 1885–1893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J., Lamouille S., Derynck R., TGF-β-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan T. Z., et al., Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 6, 1279–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty P., George J. T., Tripathi S., Levine H., Jolly M. K., Comparative study of transcriptomics-based scoring metrics for the epithelial-hybrid-mesenchymal spectrum. Front. Bioeng. Biotechnol. 8, 220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin S., Cheryan V. T., Xu L., Rishi A. K., Reddy K. B., Myc mediates cancer stem-like cells and EMT changes in triple negative breast cancers cells. PLoS One 12, e0183578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee E., et al., DNMT1 regulates epithelial-mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia 18, 553–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakiri L., et al., Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFβ expression. Cell Death Differ. 22, 336–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke C.-Y., Xiao W.-L., Chen C.-M., Lo L.-J., Wong F.-H., IRF6 is the mediator of TGFβ3 during regulation of the epithelial mesenchymal transition and palatal fusion. Sci. Rep. 5, 12791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W.-S., et al., Snail collaborates with EGR-1 and SP-1 to directly activate transcription of MMP 9 and ZEB1. Sci. Rep. 7, 17753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiwari N., et al., Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Dweep H., Sticht C., Pandey P., Gretz N., miRWalk–database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya D., Scimè A., Metabolic regulation of epithelial to mesenchymal transition: Implications for endocrine cancer. Front. Endocrinol. (Lausanne) 10, 773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibshirani R., Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288 (1996). [Google Scholar]
- 38.Huang B., et al., RACIPE: A computational tool for modeling gene regulatory circuits using randomization. BMC Syst. Biol. 12, 74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang B., et al., Interrogating the topological robustness of gene regulatory circuits by randomization. PLoS Comput. Biol. 13, e1005456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang B., et al., Decoding the mechanisms underlying cell-fate decision-making during stem cell differentiation by random circuit perturbation. J. R. Soc. Interface 17, 20200500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drier Y., Sheffer M., Domany E., Pathway-based personalized analysis of cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 6388–6393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKay D. J. C., Information Theory, Inference, and Learning Algorithms (Cambridge University Press, 2002). [Google Scholar]
- 43.Pastushenko I., et al., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Pastushenko I., et al., Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 589, 448–455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Craene B., Berx G., Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Marcucci F., Stassi G., De Maria R., Epithelial-mesenchymal transition: A new target in anticancer drug discovery. Nat. Rev. Drug Discov. 15, 311–325 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Yang J., Weinberg R. A., Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Mlacki M., Kikulska A., Krzywinska E., Pawlak M., Wilanowski T., Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp. Biol. Med. (Maywood) 240, 1396–1401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McFaline-Figueroa J. L., et al., A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat. Genet. 51, 1389–1398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grego-Bessa J., Díez J., Timmerman L., de la Pompa J. L., Notch and epithelial-mesenchyme transition in development and tumor progression: Another turn of the screw. Cell Cycle 3, 718–721 (2004). [PubMed] [Google Scholar]
- 51.Heisenberg C. P., Solnica-Krezel L., Back and forth between cell fate specification and movement during vertebrate gastrulation. Curr. Opin. Genet. Dev. 18, 311–316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauka-Spengler T., Bronner-Fraser M., A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Zavadil J., et al., Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc. Natl. Acad. Sci. U.S.A. 98, 6686–6691 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakin A. V., Tomlinson A. K., Bhowmick N. A., Moses H. L., Arteaga C. L., Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803–36810 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Peinado H., et al., A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J. 24, 3446–3458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvalho J., et al., Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J. Pathol. 228, 31–44 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Boareto M., et al., Notch-Jagged signalling can give rise to clusters of cells exhibiting a hybrid epithelial/mesenchymal phenotype. J. R. Soc. Interface 13, 20151106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolly M. K., Mani S. A., Levine H., Hybrid epithelial/mesenchymal phenotype(s): The ‘fittest’ for metastasis? Biochim. Biophys. Acta Rev. Cancer 1870, 151–157 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Heerboth S., et al., EMT and tumor metastasis. Clin. Transl. Med. 4, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosseini H., et al., Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao S., Fung-Leung W. P., Bittner A., Ngo K., Liu X., Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9, e78644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tripathi S., Kessler D. A., Levine H., Biological networks regulating cell fate choice are minimally frustrated. Phys. Rev. Lett. 125, 088101 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Liu J.et al.; Cancer Genome Atlas Research Network , An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw sequencing data have been deposited at National Center for Biotechnology Information Sequence Read Archive (BioProject ID: PRJNA698642). All other study data are included in the article and supporting information.