Significance
Calcium-dependent protein kinases (CDPKs) are a family of Ca2+ sensor proteins that contribute to various aspects of plant growth and development. One of these, CPK28, regulates immune homeostasis and reproductive-stage transition in several plant species, including Arabidopsis thaliana. Here, we show that phosphorylation on a single residue, Ser318, results in conformational changes in CPK28 that allow the kinase to be active at low Ca2+ concentrations, enabling it to respond quickly to an immune trigger. Intriguingly, phosphorylation on this residue is not required for its role in development, demonstrating pathway-specific regulation. We further provide evidence that this is a conserved biochemical mechanism in related CDPKs, highlighting the potential for biotechnological applications.
Keywords: phosphorylation, CDPK, Arabidopsis thaliana, plant immunity, calcium signaling
Abstract
Calcium (Ca2+)-dependent protein kinases (CDPKs or CPKs) are a unique family of Ca2+ sensor/kinase-effector proteins with diverse functions in plants. In Arabidopsis thaliana, CPK28 contributes to immune homeostasis by promoting degradation of the key immune signaling receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE 1 (BIK1) and additionally functions in vegetative-to-reproductive stage transition. How CPK28 controls these seemingly disparate pathways is unknown. Here, we identify a single phosphorylation site in the kinase domain of CPK28 (Ser318) that is differentially required for its function in immune homeostasis and stem elongation. We show that CPK28 undergoes intermolecular autophosphorylation on Ser318 and can additionally be transphosphorylated on this residue by BIK1. Analysis of several other phosphorylation sites demonstrates that Ser318 phosphorylation is uniquely required to prime CPK28 for Ca2+ activation at physiological concentrations of Ca2+, possibly through stabilization of the Ca2+-bound active state as indicated by intrinsic fluorescence experiments. Together, our data indicate that phosphorylation of Ser318 is required for the activation of CPK28 at low intracellular [Ca2+] to prevent initiation of an immune response in the absence of infection. By comparison, phosphorylation of Ser318 is not required for stem elongation, indicating pathway-specific requirements for phosphorylation-based Ca2+-sensitivity priming. We additionally provide evidence for a conserved function for Ser318 phosphorylation in related group IV CDPKs, which holds promise for biotechnological applications by generating CDPK alleles that enhance resistance to microbial pathogens without consequences to yield.
Protein kinases represent one of the largest eukaryotic protein superfamilies. While roughly 500 protein kinases have been identified in humans (1), the genomes of Arabidopsis thaliana (hereafter, Arabidopsis) (2) and Oryza sativa (3) encode more than 1,000 and 1,500 protein kinases, respectively, including several families unique to plants. Among these protein kinases are the receptor-like kinases (RLKs), receptor-like cytoplasmic kinases (RLCKs), and calcium-dependent protein kinases (CDPKs or CPKs) that have emerged as key regulators of plant immunity (4–6). Despite encompassing only 2% of most eukaryotic genomes, protein kinases phosphorylate more than 40% of cellular proteins (7, 8), reflecting their diverse roles in coordinating intracellular signaling events. Reversible phosphorylation of serine (Ser), threonine (Thr), and tyrosine (Tyr) residues can serve an array of functions including changes in protein conformation and activation state (9, 10), protein stability and degradation (11, 12), subcellular localization (13–15), and interaction with protein substrates (16–18).
Calcium (Ca2+) is a ubiquitous secondary messenger that acts cooperatively with protein phosphorylation to propagate intracellular signals. Spatial and temporal changes in intracellular Ca2+ levels occur in response to environmental and developmental cues (19–23). In plants, Ca2+ transients are decoded by four major groups of calcium sensor proteins, which possess one or more Ca2+-binding EF-hand motifs (24, 25): calmodulins (CaM), CaM-like proteins, calcineurin B–like proteins, CDPKs, and Ca2+/CaM-dependent protein kinases.
At the intersection of phosphorylation cascades and Ca2+ signaling are CDPKs, a unique family of Ca2+ sensor/kinase-effector proteins. CDPKs have been identified in all land plants and green algae, as well as certain protozoan ciliates and apicomplexan parasites (26, 27). CDPKs have a conserved domain architecture, consisting of a canonical Ser/Thr protein kinase domain and an EF-hand containing Ca2+-binding CaM-like domain (CLD), linked together by an autoinhibitory junction (AIJ) and flanked by variable regions on both the amino (N) and carboxyl (C) termini (28, 29). As their name implies, most CDPKs require Ca2+ for their activation (30). Upon Ca2+ binding to all EF-hands in the CaM-like domain, a dramatic conformational change occurs, freeing the AIJ from the catalytic site of the kinase, rendering the enzyme active (31–33). CDPKs vary in their sensitivity to Ca2+ (30), presumably allowing proteins to perceive distinct stimuli through differences in Ca2+-binding affinity. For example, Arabidopsis CPK4 displays half maximal kinase activity in the presence of ∼3 μM free Ca2+ (30) while CPK5 only requires ∼100 nM (34). Importantly, CDPKs are signaling hubs with documented roles in multiple distinct pathways (4, 24, 35–38) and are therefore likely regulated beyond Ca2+ activation.
Subfunctionalization is at least partially mediated by protein localization and interaction with pathway-specific binding partners, as is well documented for Arabidopsis CPK3 which functions in response to biotic and abiotic stimuli in distinct cellular compartments (39). Recent attention has been drawn to site-specific phosphorylation as a mechanism to regulate the activity of multifunctional kinases. For example, phosphorylation sites on the RLK BRASSINOSTEROID INSENSITIVE 1–ASSOCIATED KINASE 1 (BAK1) are differentially required for its function as a coreceptor with a subset of leucine-rich repeat –RLKs (40). Phosphoproteomic analyses indicate that CDPKs are differentially phosphorylated following exposure to distinct stimuli (41–48); however, the biochemical mechanisms by which site-specific phosphorylation regulates multifunctional CDPKs is still poorly understood.
Arabidopsis CPK28 is a plasma membrane–localized protein kinase with dual roles in plant immune homeostasis (49–51) and phytohormone-mediated reproductive growth (52, 53). In vegetative plants, CPK28 serves as a negative regulator of immune signal amplitude by phosphorylating and activating two PLANT U-BOX–type E3 ubiquitin ligases, PUB25 and PUB26, which target the key immune RLCK BOTRYTIS-INDUCED KINASE 1 (BIK1) for proteasomal degradation (50). Owing to elevated levels of BIK1, CPK28 null plants (cpk28-1) have heightened immune responses and enhanced resistance to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) (51). Upon transition to the reproductive stage, cpk28-1 plants additionally present shorter leaf petioles, enhanced anthocyanin production, and a reduction in stem elongation (52, 53). The molecular basis for developmental phenotypes in the cpk28-1 knockout mutant, beyond hormonal imbalance (52, 53), are comparatively unknown.
Our recent work demonstrated that autophosphorylation status dictates Ca2+-sensitivity of CPK28 peptide kinase activity in vitro (54). While dephosphorylated CPK28 is stimulated by the addition of 100 μM CaCl2 compared to untreated protein, hyperphosphorylated CPK28 displayed similar levels of activity at basal Ca2+ concentrations (54). These results highlight the interesting possibility that phosphorylation status may control the activation of multifunctional kinases in distinct pathways by allowing CDPKs to respond to stimulus-specific Ca2+ signatures.
In the present study, we identify a single autophosphorylation site, Ser318, that decouples the activity of CPK28 in immune signaling from its role in reproductive growth. We show that expression of a nonphosphorylatable Ser-to-Ala variant (CPK28S318A) is unable to complement the immune phenotypes of cpk28-1 mutants but is able to complement defects in stem growth. Further, we uncover a functional role for phosphorylation of Ser318 in priming CPK28 for activation at low free [Ca2+]. Together, we demonstrate that site-specific phosphorylation can direct the activity of a multifunctional kinase in distinct pathways and provide evidence for a conserved mechanism in orthologous group IV CDPKs.
Results
Phosphorylation on Ser318 Is Required for CPK28-Mediated Immune Homeostasis.
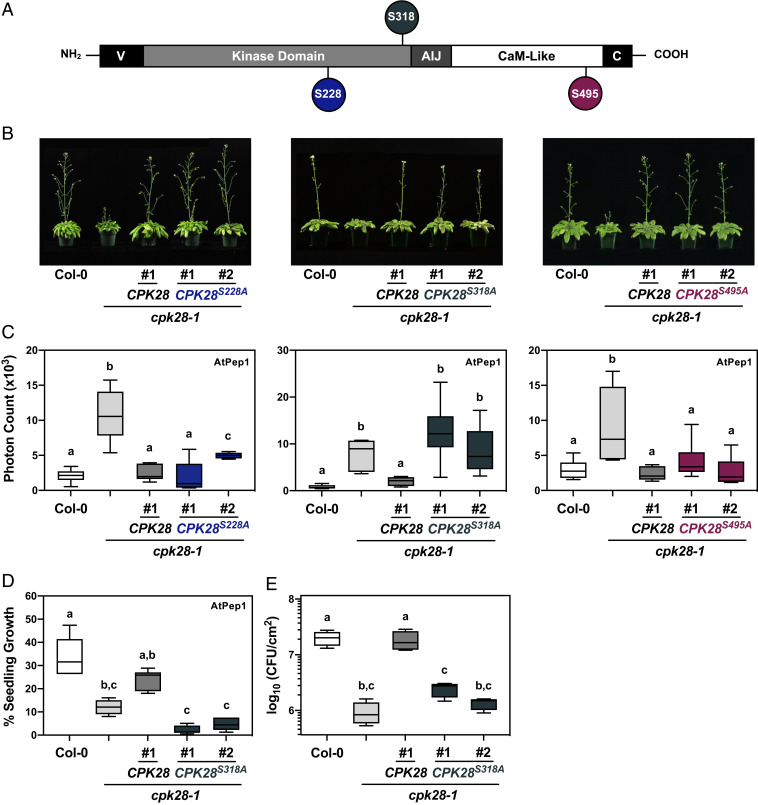
CPK28 is phosphorylated on multiple sites in vitro and in vivo (52, 54–56). To determine whether the function of CPK28 in reproductive stage transition and immunity is bifurcated by site-specific phosphorylation, we generated phosphoablative (Ser-to-Ala) mutations in three known in vivo autophosphorylation sites conserved in CPK28 orthologs across land plants (SI Appendix, Fig. S1): Ser228, Ser318, and Ser495 (52) (Fig. 1A). When driven by the cauliflower mosaic virus 35S promoter, CPK28S228A, CPK28S318A, and CPK28S495A functionally complement the stem elongation phenotype observed in cpk28-1 (52); however, it is unknown if these phosphosites regulate CPK28 function in immune homeostasis. To mitigate the possible effects of ectopic overexpression, we chose to stably express CPK28 mutants under the control of the native pCPK28 promoter in the cpk28-1 background (SI Appendix, Fig. S2) and assessed functional complementation of defects in both stem elongation and immune signaling. Interestingly, we found that while all three mutations were able to complement the stem elongation phenotype of cpk28-1 (Fig. 1B), only CPK28S228A and CPK28S495A complemented the enhanced oxidative burst in cpk28-1 following treatment with the endogenous immune elicitor peptide AtPep1 (Fig. 1C and SI Appendix, Fig. S3). In addition, cpk28-1/pCPK28:CPK28S318A-FLAG lines did not complement cpk28-1 in oxidative burst assays following treatment with the bacterial elicitor peptide elf18 (SI Appendix, Fig. S4) and remained hyperresponsive to AtPep1 in seedling growth inhibition assays (Fig. 1D) and more resistant than wild-type Col-0 to infection with the virulent bacterial pathogen Pto DC3000 (Fig. 1E). Thus, phosphorylation on Ser318 is uniquely required for the function of CPK28 in immune homeostasis.
Fig. 1.
Ser318 phosphorylation differentially regulates CPK28 function in immune homeostasis. (A) Domain structure of CPK28 and position of tested autophosphorylation sites. V= amino-terminal variable domain; AIJ= autoinhibitory junction; and C = carboxyl-terminal variable domain. (B) Stem elongation of 6-wk-old Arabidopsis plants and (C) AtPep1 (500 nM)-triggered oxidative burst in 5-wk-old plants (n = 6) of the indicated genotypes. Data are presented as boxplots indicating first and third quartiles, split by a median line. Whiskers represent maximum and minimum values. (D) Seedling growth inhibition of Col-0, cpk28-1, cpk28-1/pCPK28:CPK28-FLAG, and cpk28-1/pCPK28:CPK28S318A-FLAG lines (n = 6) resulting from continual growth in AtPep1 (500 nM) for 12 d. Values are normalized to untreated seedlings and presented as boxplots indicating first and third quartiles, split by a median line. Whiskers represent maximum and minimum values. (E) Growth of virulent Pseudomonas syringae pv. tomato (DC3000) in Col-0, cpk28-1, cpk28-1/pCPK28:CPK28-FLAG, and cpk28-1/pCPK28:CPK28S318A-FLAG lines (n = 4). Samples were collected 3 d post infection and serially diluted. Values are presented as log transformed colony forming units (CFU) per cm2 and displayed as boxplots indicating first and third quartiles, split by a median line. Whiskers represent maximum and minimum values. At least three independent biological replicates were conducted for all experiments with similar results. Statistically different groups (P < 0.005) are indicated with lowercase letters, as determined by a one-way ANOVA followed by Tukey’s post hoc test.
Mutating a Ser or Thr residue to an Asp mimics the negative charge of phosphorylation and can sometimes be used to determine the effect of constitutive phosphorylation on protein function. We therefore also generated cpk28-1 plants expressing native promoter-driven CPK28S318D and assessed genetic complementation. Although CPK28S318D fully complemented cpk28-1 in both stem elongation (SI Appendix, Fig. S5A) and AtPep1-induced oxidative species production (SI Appendix, Fig. S5B), we did not observe any additional phenotypes that would indicate heightened protein function. This raises the possibility that in vegetative plants, Ser318 may already be phosphorylated prior to immune activation. To test this, we immunoprecipitated 35S promoter–driven CPK28 C-terminally tagged with yellow flourescent protein (YFP) stably expressed in cpk28-1 and monitored Ser318 phosphorylation in plants treated with or without 100 nM elf18 using a phosphorylation and site-specific antibody raised against phosphorylated Ser318 (SI Appendix, Fig. S6). Indeed, we did not observe a detectable increase in Ser318 phosphorylation in elf18-treated plants (SI Appendix, Fig. S7). These results suggest that Ser318 is phosphorylated in plants in a resting state.
Confocal imaging confirmed that CPK28S318A-YFP localizes to the plasma membrane in Arabidopsis stably expressing 35S:CPK28S318A-YFP (SI Appendix, Fig. S8A), suggesting that phosphorylation of Ser318 does not affect the subcellular localization of CPK28. Catalytically inactive CPK28D188A-YFP was also observed at the plasma membrane (SI Appendix, Fig. S8A), indicating that CPK28 autophosphorylation is not required for appropriate localization. Furthermore, ablation of Ser318 phosphorylation did not compromise catalytic activity toward biological substrates PUB25 and PUB26 in in vitro kinase assays (SI Appendix, Fig. S8B), leading us to conclude that Ser318 phosphorylation does not regulate the function of CPK28 in immunity through altered subcellular localization or substrate specificity.
CPK28-Ser318 Undergoes Intermolecular Autophosphorylation.
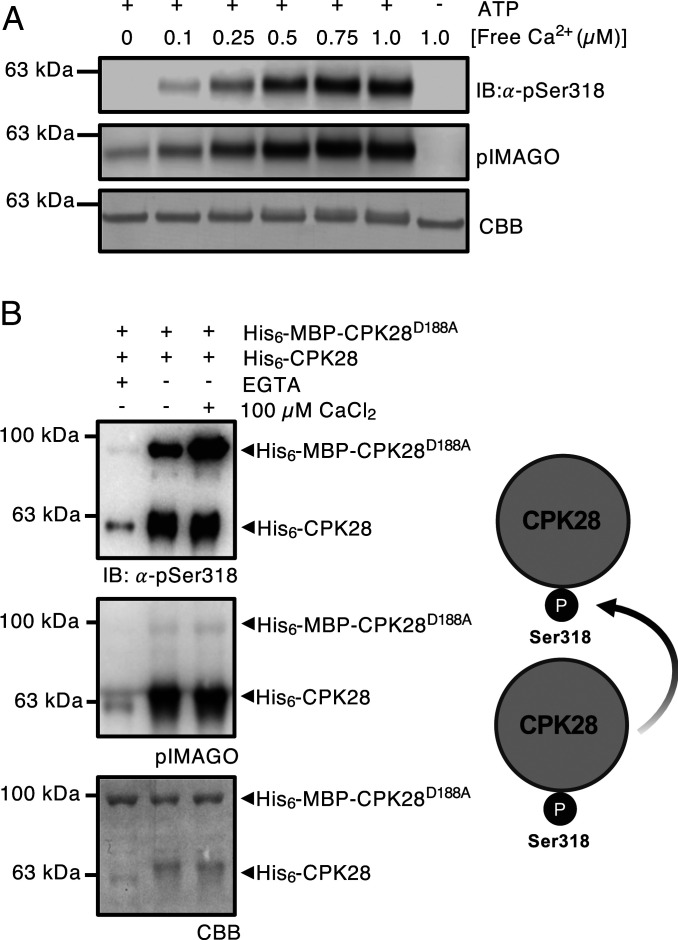
Most protein kinases autophosphorylate in vitro (57). CPK28 peptides containing a phosphorylated Ser318 have been observed in mass spectra from several studies (52, 54–56). We validated that Ser318 is a Ca2+-dependent autophosphorylation site by conducting autophosphorylation assays with increasing levels of CaCl2 and probing with α-pSer318 antibody (Fig. 2A). We also used the pIMAGO phosphoprotein detection reagent to observe total autophosphorylation levels of CPK28. Prediction-based modeling of CPK28 indicated that Ser318 is located in the kinase domain, directed away from the catalytic site (SI Appendix, Fig. S10), suggesting that autophosphorylation in cis may not be the primary mode of action. To determine whether CPK28 autophosphorylates Ser318 in trans, we additionally conducted in vitro kinase assays using recombinantly produced MBP-His6-CPK28D188A as a substrate for His6-CPK28 (Fig. 2B). His6-CPK28 could transphosphorylate MBP-His6-CPK28D188A, indicating that Ser318 autophosphorylation can indeed occur in trans (Fig. 2B). Importantly, these results indicate that CPK28 autophosphorylation, including on Ser318, can occur at levels of free Ca2+ expected to occur under resting conditions in vivo.
Fig. 2.
CPK28 undergoes intermolecular autophosphorylation at Ser318. (A) His6-CPK28 autophosphorylation under increasing Ca2+ concentrations. (B) In vitro kinase assays using recombinant His6-CPK28, His6-CPK28S318A, or His6-CPK28D188A and His6-MBP-CPK28D188A. Blots were probed with pIMAGO for the detection of phosphoproteins or α-pSer318 (1:5,000) antibody. Polyvinylidene difluoride (PVDF) membranes were stained with Coomassie Brilliant Blue (CBB) to assess protein loading. Experiments were conducted three times with similar results.
BIK1 Can Phosphorylate CPK28 on Ser318.
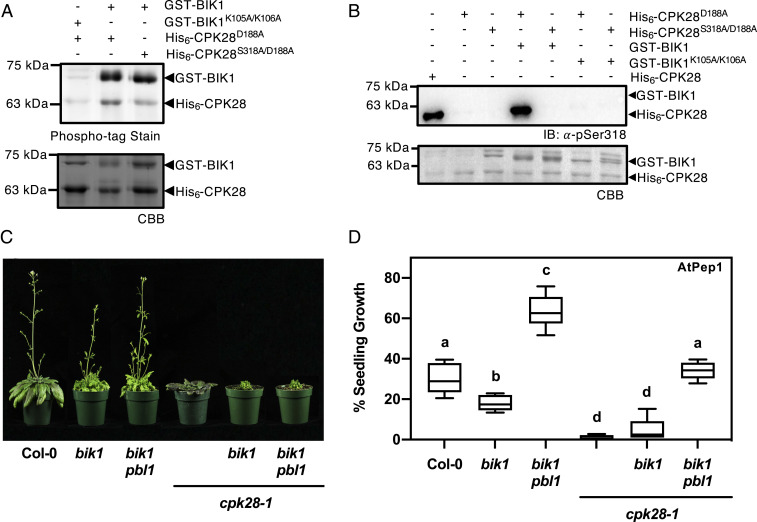
CPK28 is a highly active kinase even at basal cellular levels of Ca2+ (50, 54). Increasing [Ca2+], either by adding CaCl2 to in vitro kinase assays (54) or via immune treatment in vivo (50), increases overall phosphorylation on CPK28, including on Ser318 (Fig. 2A). Although CPK28 can autophosphorylate on Ser318, it is possible that this site is phosphorylated by additional protein kinases. BIK1 is a critical convergent substrate of multiple immune receptors whose activity and abundance are tightly regulated by layers of dynamic posttranslational modifications including phosphorylation/dephosphorylation (58–62) and mono- (63) and polyubiquitination (50, 51, 64). CPK28 phosphorylates both BIK1 (51) and the E3 ubiquitin ligases PUB25 and PUB26 (50). A recent study demonstrated reciprocal phosphorylation between rice orthologs of CPK28 and BIK1, OsCPK4 and OsRLCK176 (65), leading us to hypothesize that a similar mechanism may exist in Arabidopsis. To test whether CPK28 is a substrate of BIK1, we conducted in vitro kinase assays using recombinantly produced GST-BIK1 and catalytically inactive His6-CPK28D188A. Phospho-tag gel stain for the detection of phosphorylated proteins indicated that BIK1 is indeed able to phosphorylate CPK28 in vitro (Fig. 3A). Next, we tested whether BIK1 can phosphorylate Ser318 by conducting in vitro kinase assays between GST-BIK1 and His6-CPK28D188A/S318A compared to His6-CPK28D188A. We observed comparably less phosphorylation when CPK28D188A/S318A was used as a substrate (Fig. 3A), suggesting that Ser318 can be phosphorylated by BIK1. Furthermore, immunoblot analysis using α-pSer318 confirmed that BIK1 is capable of phosphorylating Ser318 in vitro (Fig. 3B). As we still observed some level of transphosphorylation by BIK1 on CPK28D188A/S318A (Fig. 3A), Ser318 is likely not the only site BIK1 phosphorylates on CPK28. Additional BIK1-mediated phosphorylation sites on CPK28 await to be discovered.
Fig. 3.
BIK1 transphosphorylates CPK28 at Ser318 and is required for CPK28-dependent immune signaling. (A) Phospho-tag gel stain and (B) immunoblot analysis (α-pSer318) of in vitro kinase assay using recombinant GST-BIK1 or GST-BIK1K105A/K106A and His6-CPK28D188A or His6-CPK28S318A/D188A. In order to visualize both Ser318 autophosphorylation and GST-BIK1 transphosphorylation, 500 ng of His6-CPK28 and and 2 µg of His6-CPK28D188A were loaded for auto- and transphosphorylation reactions, respectively. Gels and PVDF membranes were stained with CBB to assess protein loading. (C) Stem elongation of 6-wk-old plants and (D) immune-elicited seedling growth following continual treatment with 500 nM AtPep1 for 12 d (n = 6). Values were compared to plants grown without AtPep1 and presented as boxplots indicating first and third quartiles, median values, and whiskers representing maximum and minimum values. Statistically different (P < 0.005) values are denoted by lowercase letters according to a one-way ANOVA followed by Tukey’s post hoc test. All experiments were conducted at least three times with similar results.
BIK1 is part of a large gene family in Arabidopsis and shares biological function with its closest homolog PBL1 (58, 66–68). To investigate the genetic requirement of BIK1 and PBL1 on CPK28-mediated signaling, we generated both bik1 cpk28-1 double and bik1 pbl1 cpk28-1 triple mutants and assessed whether loss of BIK1/PBL1 was able to suppress cpk28-1 phenotypes. In congruence with our finding that Ser318 differentially regulates CPK28 function in immune homeostasis, we found that delayed stem elongation was not suppressed in bik1 cpk28-1 or bik1 pbl1 cpk28-1 (Fig. 3C) but that AtPep1-triggered seedling growth inhibition was partially or fully restored in bik1 cpk28-1 and bik1 pbl1 cpk28-1, respectively (Fig. 3D). These data suggest that the function of CPK28 in immune signaling but not in stem elongation is dependent on BIK1/PBL1 and provides further evidence for complex regulatory feedback between BIK1/PBL1 and CPK28.
Phosphorylation of Ser318 Primes CPK28 Ca2+-Responsiveness.
We previously reported on the phosphorylation-dependent Ca2+-sensitivity priming of CPK28 peptide kinase activity (54) and were interested to understand which phosphorylation site or sites mediate this priming function. We suspected that a phosphorylation site in either the protein kinase domain, the AIJ, or the CLD would be responsible for Ca2+-priming and therefore generated individual nonphosphorylatable (Ser-/Thr-to-Ala) substitutions for autophosphorylation sites within these domains of CPK28 that we identified from in situ phosphorylated recombinant protein (SI Appendix, Fig. S9A) (54). Wild-type hyperphosphorylated CPK28 is insensitive to the addition of excess Ca2+ in peptide kinase assays (SI Appendix, Fig. S9B) (54). We thus hypothesized that nonphosphorylatable substitutions of the site(s) responsible for Ca2+-sensitivity priming would restore CPK28 activation by excess Ca2+. To test this hypothesis, we expressed hyperphosphorylated forms of each phosphoablative mutant and compared protein kinase activity toward a peptide substrate (ACSM+1) in untreated samples versus samples supplemented with 100 µM CaCl2. Much to our surprise, our biochemical analysis converged on Ser318 as a critical regulatory phosphorylation site of CPK28. Of all the phosphoablative mutants tested, only the S318A mutant had enhanced peptide kinase activity upon addition of excess Ca2+. We therefore further characterized the CPK28S318A mutant for Ca2+-dependent autophosphorylation and for peptide kinase activity at different concentrations of Ca2+.
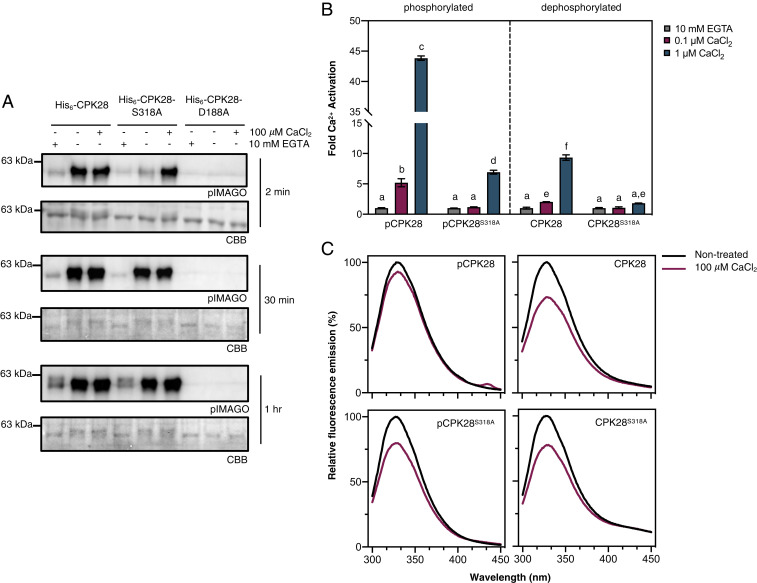
To confirm that phosphorylation of Ser318 plays a role in Ca2+-responsiveness, we used Escherichia coli Lambda phosphatase (LamP)–expressing cells to produce in situ dephosphorylated His6-CPK28 and His6-CPK28S318A and conducted comparative in vitro autophosphorylation assays either in the complete absence of Ca2+ (10 mM EGTA), in the presence of background Ca2+ (no treatment), or with the addition of excess Ca2+ (100 μM CaCl2). Autophosphorylation levels were detected using pIMAGO. Both His6-CPK28 and His6-CPK28S318A showed low levels of autophosphorylation in chelation experiments (Fig. 4A), confirming their Ca2+-dependence. His6-CPK28 was highly active at background Ca2+ and was not substantially stimulated by additional CaCl2 (Fig. 4A). In contrast, at background Ca2+ levels, His6-CPK28S318A displayed dramatically reduced autophosphorylation activity compared to wild-type protein (Fig. 4A). However, when assays were conducted in the presence of excess Ca2+, His6-CPK28 and His6-CPK28S318A exhibited comparable levels of autophosphorylation (Fig. 4A). Together, these data suggest that phosphorylation of Ser318 is required for full activity when Ca2+ levels are not saturating. Interestingly, when autophosphorylation assays were allowed to proceed for longer time intervals (30 min and 1 h), His6-CPK28S318A displayed similar levels of activity as wild-type protein at basal [Ca2+] (Fig. 4A). Collectively, these data suggest that phosphorylation of Ser318 is important for the rapid autophosphorylation of CPK28 under limiting [Ca2+].
Fig. 4.
Phosphorylation at Ser318 primes CPK28 Ca2+-responsiveness. (A) Autophosphorylation of recombinant His6-CPK28 or His6-CPK28S318A without Ca2+ (10 mM EGTA), at background Ca2+ (-EGTA and -CaCl2), or with the addition of 100 µM CaCl2 for 1 to 30 min as indicated. Kinase dead His6-CPK28D188A was used as a negative control. Phosphorylated proteins were detected using pIMAGO, and PVDF membranes were stained using CBB to assess protein loading. (B) Peptide kinase activity of phosphorylated (pCPK28 and pS318A) and dephosphorylated (CPK28 and S318A) purified recombinant proteins at physiological Ca2+ concentrations. Free Ca2+ concentrations were established as described in the Materials and Methods. Means with SD of three replicate reactions are shown. Statistically different (P < 0.01) groups are denoted by lowercase letters according to a one-way ANOVA followed by Tukey’s post hoc test. The experiment was repeated twice with independent preparations of all recombinant proteins with similar results observed both times. (C) Intrinsic Trp fluorescence of hyperphosphorylated (pCPK28 and pS318A) or dephosphorylated (CPK28 and S318A) His-tagged recombinant proteins before (“nontreated”) and after the addition of 100 µM CaCl2. Curves are averages of two scans following appropriate background subtraction. Experiments were performed twice using independent preparations of recombinant proteins with similar results observed in both experiments.
To better understand how phosphorylation of Ser318 affects activation of CPK28 by Ca2+, we assessed CPK28 peptide kinase activity using the ACSM+1 synthetic peptide as substrate (54) at Ca2+ concentrations that would be expected at the lower (0.1 μM) and upper (1.0 μM) range of intracellular physiological conditions. Phosphorylated and dephosphorylated forms of both His6-CPK28 and His6-CPK28S318A were used to determine whether overall phosphorylation status could supersede the requirement for site-specific phosphorylation at Ser318. At free Ca2+ concentrations of 0.1 and 1.0 μM, dephosphorylated His6-CPK28 (Fig. 4B) displayed approximately two- and ninefold activation by Ca2+ relative to EGTA-treated protein, respectively (Fig. 4B). Similarly, phosphorylated CPK28 (Fig. 4B) had approximately fivefold activation with the addition of 0.1 μM free Ca2+ and ∼43-fold activation with 1.0 μM free Ca2+ (Fig. 4B), indicating that phosphorylation enhances CPK28 peptide kinase activity at physiological [Ca2+]. By comparison, His6-CPK38S318A was substantially less active at physiological [Ca2+], having negligible activity at 0.1 µM Ca2+ (relative to EGTA-treated protein) regardless of its overall phosphorylation status, and only approximately two- and sevenfold activation for dephosphorylated and phosphorylated His6-CPK28S318A, respectively, at 1.0 µM Ca2+. Collectively, these results indicate that phosphorylation of Ser318 is prerequisite for substrate phosphorylation by CPK28 at low free Ca2+ and that phosphorylation of Ser318 is required for full responsiveness to Ca2+ elevations within the physiological range. Taken together, these data support the hypothesis that phosphorylation of Ser318 is uniquely important in priming CPK28 for activation by Ca2+ at concentrations that would be expected under cellular conditions.
Ser318 Phosphorylation Promotes a Ca2+-Bound Conformation.
Ser318 is located at the C-terminal end of the canonical protein kinase domain of CPK28, in close proximity to the AIJ (Fig. 1A). We recently demonstrated that CaM binds to an area of the CPK28 AIJ (Leu341–Leu362) to inhibit both in vitro auto- and transphosphorylation activity and that CPK28 autophosphorylation relieves this inhibition (54). Modeling of CPK28 indicates that Ser318 is not in close proximity to the CaM binding region (SI Appendix, Fig. S10) and is therefore unlikely to affect interaction with CaM. We hypothesized that phosphorylation of Ser318 might affect Ca2+-dependent conformational changes of the AIJ–CLD fragment of CPK28. To better understand the biochemical function of Ser318 phosphorylation, we measured Ca2+-induced conformational changes in hyper- and hypophosphorylated CPK28 and CPK28S318A by intrinsic Tryptophan (Trp) fluorescence. Trp fluorescence emission properties depend on the local environment of Trp residues and can thus be used to assess protein conformational changes (69). For dephosphorylated His6-CPK28 and His6-CPK28S318A, Trp fluorescence emission decreased following the addition of 100 µM CaCl2 (relative to nontreated protein), indicating that CPK28 undergoes a Ca2+-dependent conformational change (Fig. 4C). By comparison, Trp fluorescence of hyperphosphorylated His6-CPK28 showed only a marginal decrease after addition of excess Ca2+ (Fig. 4C), suggesting that CPK28 phosphorylation promotes a Ca2+-bound conformation at low levels of Ca2+. In agreement with our kinase activity assays, Trp fluorescence of hyperphosphorylated His6-CPK28S318A decreased similar to the wild-type dephosphorylated protein, suggesting that phosphorylation of Ser318 is responsible for the conformation-based Ca2+-sensitivity changes observed with hyperphosphorylated CPK28.
Ser318 Is a Conserved and Unique Feature of Subgroup IV CDPKs.
CDPK gene families are highly conserved across land plants and form four major subgroups (26, 27). To determine the level of conservation of Ser318, the amino acid sequences of CPK28 and other subgroup IV orthologs from all genomes available on Phytozome 12 were compared. Among the 114 sequences included in our analysis, a Ser residue was strictly conserved at the position orthologous to Ser318 of AtCPK28 (Fig. 5A and SI Appendix, Fig. S1). Comparison of all subgroup I through III CDPKs from 12 representative species spanning all major taxonomic groups indicated that although several flanking residues are highly conserved in all subgroups, conservation of Ser318 is a unique and specific feature of subgroup IV CDPKs (Fig. 5A).
Fig. 5.
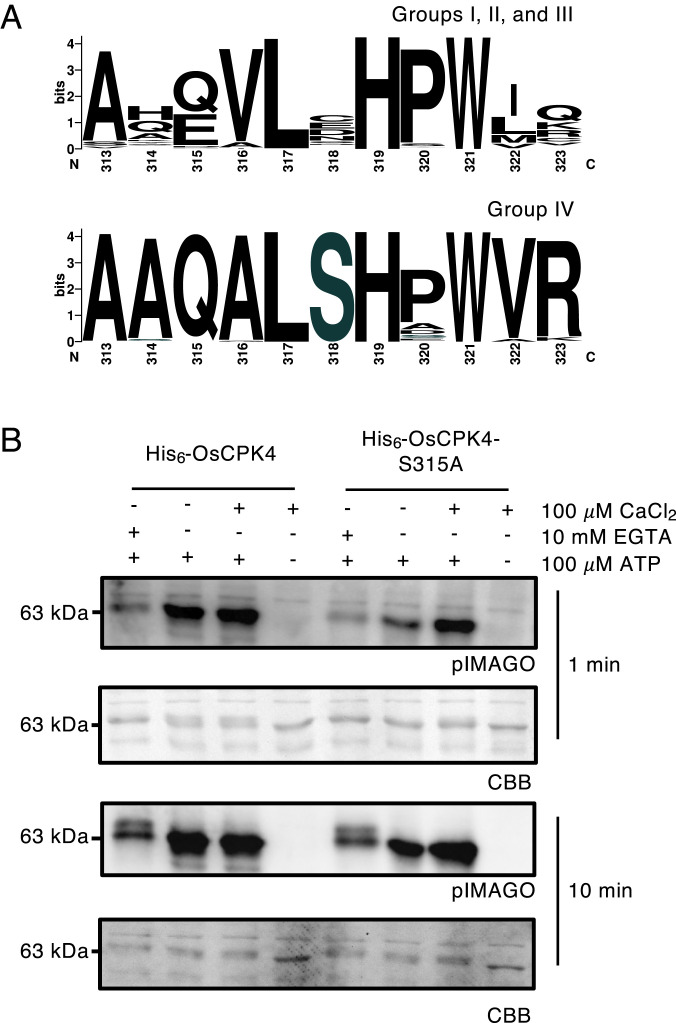
Ser318 is a conserved feature of group IV CDPKs. (A) Amino acid consensus at position 318 (AtCPK28) across CDPK subgroups. Sequences were retrieved from Phytozome 12 and aligned as described in the Materials and Methods. Logos were generated using WebLogo (97). (B) Autophosphorylation of recombinant His6-OsCPK4 and His6-OsCPK4S315A. Reactions were carried out in the absence of Ca2+ (10 mM EGTA), in the presence of background Ca2+(-EGTA and -CaCl2), or with the addition of 100 µM CaCl2 for 1 or 10 min as indicated in figures. Phosphorylation was detected using pIMAGO. PVDF membranes were stained with CBB to assess protein loading. All experiments were conducted at least three times with similar results.
To determine if the Ca2+-priming function of Ser318 autophosphorylation is conserved, we generated a phosphoablative variant of the rice CPK28 ortholog (OsCPK4S315A). Short 1 min autophosphorylation assays were conducted using recombinant His6-OsCPK4 and His6-OsCPK4S315A produced in LamP-expressing E. coli cells, as described above. His6-OsCPK4 displayed a clear requirement for Ca2+ with a marked decrease in overall phosphorylation in the presence of 10 mM EGTA (Fig. 5B). While wild-type His6-OsCPK4 displayed high levels of autophosphorylation at basal [Ca2+], His6-OsCPK4S315A was comparatively less active (Fig. 5B). The addition of 100 μM CaCl2 did not further activate His6-OsCPK4 but did restore His6-OsCPK4S315A autophosphorylation levels to that observed with wild-type protein (Fig. 5B). Together, these data provide evidence of a conserved biochemical function for phosphorylation of this residue in orthologous CDPKs across the plant lineage.
Discussion
Expansion of CDPKs in plants is predicted to have occurred under selective adaptation for kinases with varying Ca2+ sensitivities (27); however, the biophysical properties underlying Ca2+-sensitivity are not fully understood. Analysis of protein sequences from Arabidopsis indicates that CDPKs with little or no requirement for Ca2+ possess one or more degenerated EF-hand motifs (37, 70). However, some CDPKs with the ability to bind Ca2+ do not require it for their activation (30, 71–73), pointing to additional mechanisms regulating CDPK function. Previous work has shown that in situ autophosphorylation “primes” Arabidopsis CPK28 for Ca2+ activation in vitro (54). Here, we demonstrate that phosphorylation at one site, CPK28-Ser318, is responsible for autophosphorylation-based priming when Ca2+ concentrations are limiting (Fig. 4 A and B). Additionally, in vivo phosphorylation at Ser318 is required for CPK28 function in immune homeostasis (Fig. 1 C–E) but not stem elongation (Fig. 1B), suggesting a role in stimulus-specific activation of a multifunctional protein kinase through Ca2+-sensitivity priming.
Autophosphorylation has been correlated with the activation (15, 54, 74–76) or inhibition (73, 76, 77) of several CDPKs, although the mechanisms of regulation remain largely unknown. Phosphorylation could influence interactions with protein substrates (76, 78) or induce changes in secondary protein structure, causing transitions between functional enzyme states (79, 80). A complete crystal structure for a plant CDPK has not yet been resolved; however, a mechanism for Ca2+ activation has been proposed based on the structures of apicomplexan CDPKs (31–33). Experiments using Toxoplasma gondii TgCDPK1/2 and Cytosporidium parvum CpCDPK1 demonstrate that Ca2+ activation is reversible with contact sites between the CaM-like domain and kinase domain stabilizing both active and inactive forms (31). Many of the residues that stabilize these conformations are conserved between plants and protists (32), suggesting similar contact sites may exist in plants.
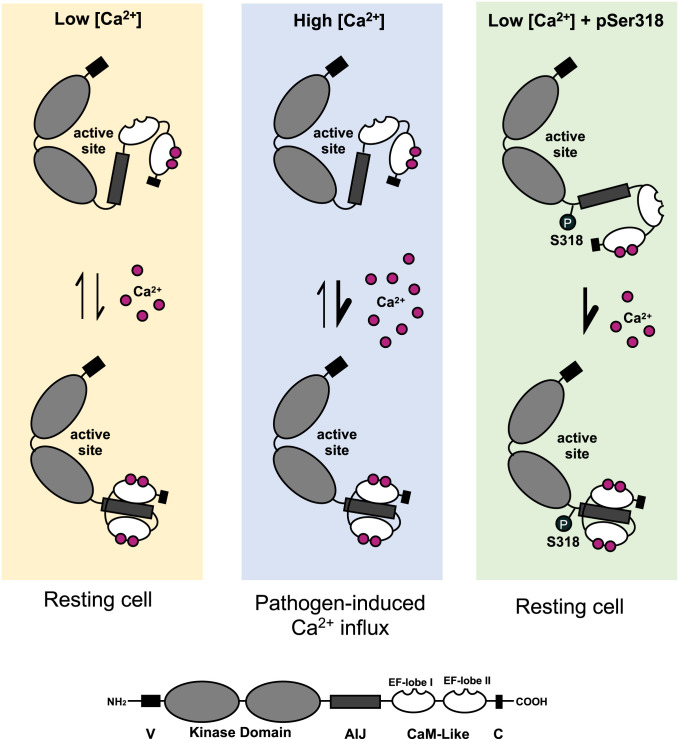
CPK28-Ser318 resides in the C-terminal portion of the kinase domain in close proximity to the AIJ (Fig. 1A). Although we could not generate a high confidence structural model of CPK28 using the crystal structures of Ca2+-bound CDPKs, modeling of CPK28 using inactive TgCDPK1 (31) indicated that Ser318 is likely surface localized, directed away from the active site of the kinase domain (SI Appendix, Fig. S10). In this position, phosphorylated Ser318 would not interact with established contact sites, such as the autoinhibitory triad (31), or other interactions between the pseudosubstrate region and the active site of the kinase domain. We rather propose that phosphorylation of Ser318 could induce a structural change in the AIJ that prohibits stabilization of the inactive conformation. This could cause the protein to adopt an “intermediate” conformation that can more readily move to the active state upon Ca2+ binding (Fig. 6). In support of this idea, our analysis of intrinsic Trp fluorescence of CPK28 and CPK28S318A suggests that Ca2+-dependent conformational changes can occur at a lower concentration of Ca2+ in the hyper- compared to dephosphorylated protein and that the conformational change at low Ca2+ requires Ser318 phosphorylation (Fig. 4C). Phosphorylation of Ser318 could also stabilize the active conformation, serving a similar function to autophosphorylation of a residue in the autoinhibitory region (Thr286) of CaMKII from rat brain, which renders CaMKII substrate phosphorylation independent of both Ca2+ and CaM (81, 82).
Fig. 6.
Model for CPK28 activation. Proposed mechanism of Ca2+-sensitivity priming of CPK28 by phosphorylation of Ser318. In a resting cell, CPK28 transitions between active and inactive conformations, binding and releasing Ca2+ from the amino-terminal EF-hand lobe of the calmodulin (CaM)-like domain. Ca2+ elevation during signaling events shifts this equilibrium toward the activated state via Ca2+-induced conformational changes. When Ser318 is phosphorylated, the transition of CPK28 from an inactive to an active state occurs at lower Ca2+ concentrations possible via stabilizing a conformation in which the AIJ is excluded from the active site.
Overall, our analysis of CPK28 autophosphorylation, peptide kinase activity, and conformational changes collectively suggest that phosphorylation of Ser318 near the AIJ promotes an open, active conformation of CPK28 at low Ca2+ concentrations (Fig. 6). Currently, it is not clear whether this conformational state results in enhanced affinity of the CaM-like domain for Ca2+ or whether pSer318 exerts allosteric effects promoting release of the AIJ from the kinase domain. Resolving an intact crystal structure for CPK28 and other plant CDPKs will provide valuable insight into the activation of these kinases and allow for investigation into the possible structural roles of phosphorylation.
From a physiological perspective, phosphorylation of Ser318 would allow CPK28 to be active in a cell under resting conditions. CPK28 promotes the degradation of BIK1 prior to the activation of immune signaling (50, 51). Limiting BIK1 accumulation is critical to prevent mounting an immune response in the absence of pathogen invasion. Indeed, mass spectrometry analysis identified Ser318 as an in vivo phosphorylation site in unstimulated cells (52), and we did not observe a detectable increase in Ser318 phosphorylation following immune elicitation (SI Appendix, Fig. S7), indicating no requirement for immune activation. A 25-fold increase in phosphorylated peptides corresponding to pSer318 was identified in cpk28-1 protoplasts expressing CPK28-YFP compared to the kinase inactive variant (CPK28D188A-YFP), suggesting that Ser318 is primarily an autophosphorylation site but can be phosphorylated by additional kinases in vivo (52). Our in vitro data indicates that CPK28 undergoes intermolecular autophosphorylation at Ser318 (Fig. 2) and can also be transphosphorylated by BIK1 (Fig. 3 A and B), although a higher level of phosphorylation was detected on autophosphorylated CPK28 in our assays (Fig. 3B). Whether Ser318 phosphorylation occurs by autophosphorylation and/or BIK1-mediated transphosphorylation in vivo is unknown, but our in vitro analyses clearly establish the potential for both mechanisms as drivers of Ser318 phosphorylation in vivo. How these distinct events would contribute to CPK28-mediated BIK1 proteostasis remains an open and challenging question.
Nevertheless, it is tempting to speculate that BIK1 transphosphorylation could contribute to CPK28 regulation. Our epistasis analysis indicates that CPK28-mediated immune signaling is dependent on BIK1 and its homolog, PBL1 (Fig. 3D). However, loss of bik1 pbl1 in cpk28-1 mutants restored cpk28-1 growth inhibition to levels observed in Col-0 seedlings (Fig. 3D), suggesting the involvement of additional RLCKs in response to endogenous immune peptide elicitation. In rice, OsRLCK176 phosphorylates OsCPK4 at three sites: Thr73, Ser210, and Ser381 (65), corresponding to CPK28-Thr76, -Ser213, and -Val384. Phosphorylation at these sites activates OsCPK4 as part of a regulatory feedback loop to control immune output through continual degradation of OsRLCK176 (65). Our in vitro kinase assays suggest that BIK1 phosphorylates CPK28 at Ser318 and additional, currently unknown, residues (Fig. 3A). Given the high conservation of this immune signaling network (83), Thr76 and Ser213 are likely candidates for future analysis. In Arabidopsis, BIK1 turnover is mediated by CPK28-dependent phosphorylation and activation of the E3 ubiquitin ligases PUB25 and PUB26, which target BIK1 for 26S proteasomal degradation (50). Previous work indicates that CPK28 is also capable of phosphorylating BIK1 in vitro (51), although the biochemical and biological consequences of this transphosphorylation are not known. It is therefore plausible that BIK1 accumulation could be modulated by an interplay of BIK1 ubiquitination and transphosphorylation events between CPK28 and BIK1, which is a current area of investigation.
Our cumulative data indicate that phosphorylation of Ser318 would render CPK28 highly responsive to slight increases in Ca2+ following immune activation, preventing initiation of a robust immune response. Ser318 phosphorylation could additionally slow CPK28 deactivation following a primary pathogen attack, making plants more susceptible to secondary infection. In accordance with its role in BIK1 turnover, overexpression of CPK28 dampens the immune-triggered Ca2+ (49) and oxidative (51) bursts, which are dependent on BIK1 and PBL1 (49, 51). This implies that CPK28 overactivation could additionally prevent the establishment of systemic acquired resistance which is reliant on signal propogation by reactive oxygen species and Ca2+ (34, 84, 85). In order to buffer these attenuation mechanisms, the plant would need to adopt a safeguard against sustained activation of CPK28. Recent work has demonstrated that following cell stimulation with AtPep1 (86) or the bacterial flagellin immunogenic epitope flg22 (87), alternative CPK28 transcripts are produced that generate a truncated isoform that lacks the C-terminal EF-hand lobe. This CPK28 variant could act to outcompete Ca2+ “competent” proteins for downstream substrates, alleviating immune attenuation (88). Thus, CPK28 signaling appears to be intricately regulated at both the posttranscriptional and posttranslational levels.
Despite the clear requirement for Ser318 phosphorylation in immune signaling (Fig. 1 C–E), cpk28-1 plants expressing CPK28S318A displayed normal stem elongation (Fig. 1B). The catalytic activity of CPK28 is indispensable for all known biological functions (51, 52); however, the lower kinase activity of CPK28S318A at physiologically relevant Ca2+ concentrations (Fig. 4 A and B) did not impair stem elongation (Fig. 1B), suggesting pathway-specific requirements for full CPK28 catalytic activity. We speculate that more processive genetic programs, such as those that occur in developmental and reproductive processes, would not require CDPKs to be as responsive to Ca2+ as stress-induced signals. In support of this hypothesis, prolonged exposure to low levels of Ca2+ relieved the requirement of Ser318 phosphorylation for full kinase activity in autophosphorylation assays (Fig. 4A). In a physiological context, the phosphorylation status of CPK28 is likely dictated by a combination of kinase and phosphatase activities to precisely control CPK28 function. Although pathway-specific CPK28 binding partners have not yet been identified in the reproductive phase transition, it is possible that such associations could increase kinase activity or lower Ca2+ requirements. How individual Ca2+ sensor proteins distinguish stimulus-specific Ca2+ signatures has remained an outstanding question (89). Our work indicates that phosphorylation could play a role in altering Ca2+-sensitivity, allowing multifunctional CDPKs to decode unique Ca2+ signatures.
The development of pathogen-resistant crops can be complicated by growth trade-offs associated with the overactivation of immune signaling (90, 91). For example, prolonged activation of immune receptors causes seedling growth inhibition (92, 93) and the formation of lesions (94, 95). Ablation of Ser318 phosphorylation allowed us to generate a CPK28 allele that displays enhanced resistance to Pto DC3000 (Fig. 1E) with no consequences to plant growth (Fig. 1B). Group IV CDPKs are highly conserved across all land plants (26) and fulfill conserved roles as regulators of immune signaling (83) and reproductive development (52, 96–98) in multiple plant species. Accordingly, our in vitro analysis of OsCPK4 indicated that, on a biochemical level, phosphorylation of Ser315 (analogous to CPK28-Ser318) has a conserved function in rice (Fig. 4D). Additionally, a Ser residue at this site was identified as a unique feature of group IV CDPKs across all surveyed land plants (Fig. 4C). Cumulatively, our data suggest that ablation of “Ser318” phosphorylation in species with agricultural value could serve as an effective tool for the development of disease resistance without associated costs to fitness or yield.
Materials and Methods
Plant Growth Conditions.
A. thaliana plants were grown either in soil or sterile media depending on the assay. For soil assays, seeds were stratified for 2 to 3 d at 4 °C, sown on soil, and transplanted as one plant per pot in Sunshine Mix 1 soil (Sungro) for 5 wk in temperature-controlled growth chambers in the Queen’s University Phytotron at 22 °C with 10 h light (150 to 160 μE · m2 · s−1) and no humidity control. Oxidative burst and pathogen infection assays were conducted on soil-grown plants 3 to 4 wk post germination (wpg), prior to reproductive stage transition. For assessment of stem elongation, plants were subsequently transferred to a growth chamber maintained at 22 °C, with a 16 h photoperiod (150 to 160 μE · m2 · s−1) and 30% relative humidity for 1 to 2 wk or until all plants had produced reproductive bolts. For sterile assays, seeds were surface sterilized using 50% bleach, stratified for 2 to 3 d at 4 °C, and germinated on 0.8% agar plates with 0.5× Murashige and Skoog (MS) media for 3 to 4 d and then transplanted into liquid 0.5× MS supplemented with 1% sucrose. Sterile seedlings were grown at ambient temperature with 10 h light 150 to 160 μE · m2 · s−1, and assays were conducted at 2 wpg. Soil-grown A. thaliana plants were fertilized biweekly with 1.5 g L−1 20:20:20 N:P:K. Predatory Amblyseius swirskii mites (Koppert Biological Systems) were released into growth chambers biweekly as a precautionary measure against greenhouse pests, according to manufacturer’s instructions.
Plant Materials.
Stable A. thaliana transgenics were generated via Agrobacterium tumefaciens (GV3101)–mediated floral dip transformation (99). T1 plants were selected on MS agar plates containing 50 μg/mL hygromycin. Only lines displaying a 3:1 segregation ratio on selective media in the T2 generation were bred to homozygosity and used in complementation experiments. Stable cpk28-1/35S:CPK28-YFP, cpk28-1/35S:CPK28S318A-YFP, and cpk28-1/35S:CPK28D188A-YFP Arabidopsis lines were previously described (52). Higher-order mutants were generated by crossing bik1 or bik1 pbl1 (66) mutants with cpk28-1 (52) and bred to homozygosity using PCR-based genotyping. SI Appendix, Table S1 includes a list of all germplasm used and generated in this study.
Molecular Cloning.
The pCPK28:CPK28-FLAG construct was cloned by fusing the coding sequence of CPK28 downstream of its native promoter (1,742 bp upstream of the start codon) and in frame with a C-terminal FLAG peptide using digestion–ligation cloning into a pGREENII-based binary vector carrying the aminoglycoside phosphotransferase gene from E. coli for hygromycin B resistance in plants (100). Both the pT7:His6-CPK28 construct in pET28a+ (EMD Biosciences) and the pT7:MBP-His6-CPK28 construct in pOPINM (Novagen) have been described previously (51, 54). Site-directed mutagenesis was used to generate CPK28 mutant constructs using overlapping complementary primers as described previously (51), using either pGREENII-based binary plasmids or pET28a+ clones as the template. The coding region of BIK1 was PCR amplified from previously described pENTR-BIK1 or pENTR-BIK1K105A/K106A vectors (66) and cloned into bacterial expression vector pGex6.1 (GE Healthcare) by Gibson Assembly (NEB) to generate pT7:GST-BIK1 and catalytically inactive pT7:GST-BIK1K105A/K106A constructs. pET100:His6-OsCPK4 and pET100:His6-OsCPK4S315A constructs were synthesized by GeneArt (Thermo Fisher Scientific). The coding region of PUB25 was PCR amplified from previously described pET28a:PUB25 vectors (50) and cloned into bacterial expression vector pMAL-c2× (GE Healthcare) by digestion and ligation cloning using XbaI and PstI-HF (NEB) to produce pMAL-c2×:MBP-PUB25. PUB26 was PCR amplified from pCAMBIA1300-35S:PUB26-FLAG (50) and cloned into pMAL-c2× using XbaI and PstI-HF to produce pMAL-c2×:MBP-PUB26. All clones were confirmed by Sanger Sequencing using plasmid- and/or gene-specific primers (Center for Applied Genomics, Hospital for Sick Children, Toronto, Canada, or Eurofins Genomics, Ebersberg, Germany). All primers used for cloning are listed in SI Appendix, Table S1.
Confocal Microscopy.
Leaf discs were sampled from 4- to 6-wk-old Arabidopsis soil grown plants using a 4 mm biopsy punch (Integra Miltex) and were wet mounted in water with the abaxial surface facing upwards prior to confocal imaging. Imaging was performed using a LSM 710 (Zeiss) confocal microscope with excitation at 488 nM for YFP and a range of 510 to 540 nM for measuring emission. To detect chlorophyll autofluorescence, an excitation wavelength of 543 nM and a range of 680 to 760 nM for detecting emission was used.
Immune Assays.
AtPep1 (101) and elf18 (102) used for immune assays were synthesized by EZBiolab. Seedling growth inhibition assays were performed exactly as described previously (103). Oxidative burst assays were performed on leaf discs from 4- to 5-wk-old soil-grown plants in 96-well plates exactly as described in ref. 103, with an integration time of 1,000 ms per well over 40 min. Infection assays were conducted using virulent Pseudomonas syringae pv. tomato (Pto DC3000) on soil-grown A. thaliana plants using a needless syringe as outlined previously (51).
Generation of the anti-pSer318 Antibody.
The CPK28 pSer318 antibody was generated and purified by LifeTein (New Jersey). Briefly, rabbits were immunized with KLH-coupled synthetic peptide corresponding to the region of CPK28 surrounding Ser318 (NH2-CKDPRARLTAAQALpSHAWV-COO-). KLH coupling was facilitated by the addition of Cys to the N terminus of the peptide. Antibody was purified first by enrichment against the pSer318 phosphopeptide and then by negative enrichment against an unphosphorylated version of the peptide to remove non-phospho–specific IgGs. Purified antibodies were validated by immunoblotting against wild-type, kinase-dead (K91E), or S318A recombinant CPK28 proteins on a polyvinylidene difluoride (PVDF) membrane. Purified antibodies were determined to be phosphorylation and site specific.
Protein Extraction and Immunoprecipitation.
Plant tissues were ground under liquid N2, and proteins were extracted as described previously (50). Protein concentration across samples was normalized using Pierce Coomassie Protein Assay Kit (Thermo Fisher Scientific), and CPK28-YFP was immunoprecipitated from cpk28-1/35S:CPK28-YFP plants using μMACS microbeads conjugated to a monoclonal antibody against green flourescent protein (anti-GFP) (Miltenyi Biotec).
Recombinant Protein Expression and Purification.
All recombinant CPK28 clones were transformed into Lambda phosphatase–expressing BL21 (DE3) E. coli cells for the production of dephosphorylated proteins or into T7 Express cells (New England Biolabs) for production of hyperphosphorylated protein (54). Cultures were grown in Luria–Burtani (LB) broth at 37 °C to an OD600 of ∼0.6 to 0.8. Expression was induced using 1 mM of β-D-1-thiogalactopyranoside (IPTG) for 16 to 18 h at room temperature with gentle shaking. Bacterial cells were harvested at 3,500 x g for 25 min at 4 °C and resuspended in 50 mL of extraction buffer containing 50 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, and 1 protease inhibitor mixture tablet (Sigma-Aldrich). Cells were lysed by passing the resuspended culture through a French press (Glen MillsⓇ High Pressure Cell Disruption) three times. Lysates were clarified by centrifugation at 35,000 × g for 40 min at 4 °C. His6-CPK28 proteins were immobilized on a nickel-nitrilotriacetic acid gravity flow column (Thermo Fisher) as described previously (54). Elution fractions were dialyzed against two exchanges of 2,500 volumes of 25 mM Tris⋅HCl (pH7.5), 50 mM NaCl, and 1 mM DTT at overnight 4 °C. Recombinant production of GST-BIK1 and GST-BIK1K105A/K106A in BL21(DE3)-VR2-pACYC-LamP E. coli cells was conducted as described above in a phosphate-buffered saline (PBS) solution (Thermo Fisher) containing 1 mM DTT, 1 mM PMSF, and 6 mM MgCl2. Proteins were immobilized on a glutathione agarose gravity flow column (Qiagen) washed five times with PBS solution and eluted in 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, and 10 mM reduced glutathione. All proteins were concentrated using Pierce 3000 MWCO concentration columns (Thermo Fisher) to a final concentration of ∼1.5 mg/mL, as determined by Bradford analysis (Bio-Rad) against bovine serum albumin standards. Recombinant production of MBP-PUB25 and MBP-PUB26 was conducted as described above with the following modifications. pMAL-c2×:MBP-PUB25/26 were transformed into BL21(DE3) E. coli cells. Cultures were grown in a baffled flask with LB broth at 37 °C to an OD600 of ∼0.6 to 0.8, then expression was induced using 0.1 mM IPTG for 3 h at room temperature with gentle shaking. Pelleted cells were resuspended in MBP column buffer containing 20 mM Tris HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF. MBP-PUB25/26 were purified using Amylose Resin (NEB) by batch purification according to manufacturer’s instructions. Protein was eluted using 100 µL elution buffer and used for assays on the same day as the purification. Purity was assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis followed by staining with CBB total protein stain. Protein aliquots were flash frozen in liquid N2 and stored at −80 °C until use.
In Vitro Autophosphorylation Assays.
Autophosphorylation assays were conducted by incubating 5 μg of purified His6-CPK28/His6-OsCPK4 or mutant variants in a 50 μL reaction containing 25 mM Tris⋅HCl (pH 7.5), 10 mM DTT, 100 μM ATP, and 100 μM CaCl2 or 10 mM EGTA, where specified. Proteins were allowed to autophosphorylate at room temperature for 1 to 60 min, as specified in figures. Reactions were stopped by the addition of 6× Laemmli sample buffer (LSB) and heating at 80 °C for 5 min. Reactions were analyzed immediately or stored at −20 °C before SDS-PAGE and immunoblotting. Autophosphorylation was detected using the pIMAGO kit (Tymora Analytical) according to manufacturer’s instructions.
In Vitro Transphosphorylation Assays.
Trans-autophosphorylation assays were performed using 2 μg of His6-MBP-CPK28 or His6-MBP-CPK28D188A and 4 μg of His6-CPK28D188A in the same reaction buffer as in the autophosphorylation assays at 30 °C for 30 min. Transphosphorylation assays were conducted using 2 μg of purified GST-BIK1 or GST-BIK1K105A/K106A and 4 μg of His6-CPK28D188A or His6-CPK28D188A/S318A in a 20 μL total reaction volume of 25 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 100 μM ATP at 30 °C for 30 min. Reactions were stopped by adding 6× LSB buffer and heating at 80 °C for 5 min. Reactions were analyzed immediately or stored at −20 °C before visualizing phosphorylated proteins using Phospho-Tag gel stain (APB Bio) according to the manufacturer’s instructions.
Ca2+-Activation Assays.
Analysis of CPK28 kinase activity toward a peptide substrate was carried out as exactly as previously described (54) using the ACSM+1 peptide (NH2-NNLRLSMGKR-COO−) as substrate. Briefly, reactions contained 40 mM Tris⋅HCl, pH 7.5, 1 mM DTT, 10 mM MgCl2, 100 μM ATP, 0.1 μCi/μL [γ-32P]ATP (150 cpm/pmol), 500 ng purified of His6-CPK28 or the S318A site-directed mutant as indicated in the figures, and 10 μM peptide substrate. Reactions were initiated by addition of an ATP/[γ-32P]ATP mixture. Reactions contained combinations of CaCl2 or EGTA as indicated in the appropriate figures. For experiments at physiological Ca2+, final free Ca2+ concentrations were achieved by buffering CaCl2 with EGTA, calculated using the online WEBMAXC Extended calculator (104). Final reaction volumes were 40 μL. After addition of the ATP/[γ-32P]ATP mixture, reactions were allowed to proceed for 10 min at room temperature and were stopped by spotting 35 μL of each reaction onto P81 phosphocellulose cation exchange paper followed by washing three times for 5 min each in 0.45% (vol/vol) o-phosphoric acid. Incorporation of 32P was assessed by liquid scintillation counting.
Intrinsic Trp Fluorescence Measurements.
Intrinsic Trp fluorescence of recombinant purified dephosphorylated or hyperphosphorylated His6-CPK28 or His6-CPK28S318A was performed in a PTI QuantaMaster steady-state spectrofluorimeter (Horiba Scientific) in a quartz cuvette with a 1 cm pathlength (Hellma USA). Data acquisition and background subtraction were performed using the FeliXGX software package (Horiba Scientific). Measurements were carried out on samples of CPK28 at a concentration of 200 nM in a buffer containing 20 mM Hepes-NaOH pH 7.2, 100 mM KCl, and 1 mM DTT. Trp fluorescence was measured at background Ca2+ levels (“nontreated”) and after the titration of 100 µM CaCl2 into the same sample. Samples were measured at an excitation wavelength of 288 nm, and fluorescence was collected between 300 and 450 nm in 1 nm steps with a dwell time of 1 s at each step. Each curve represents an average of two replicate scans of the same sample after appropriate background subtraction of buffer alone or buffer titrated with 100 µM CaCl2. Experiments were performed twice on independent preparations of all recombinant proteins.
Phylogenetic Analysis.
To determine the conservation of Ser318 in group IV CDPKs, the full-length AtCPK28 protein sequence was used as a query in the Phytozome 12 BLAST tool, which identified a total of 114 amino acid sequences from 53 species (Dataset S1). Similarly, the full-length sequences of all group I, II, and III AtCPKs were queried in Phytozome 12 BLAST limited to twelve species spanning the plant lineage (Marchantia polymorpha, Physcomitrium patens, Sphagnum fallax, Selaginella moellendorffi, Amborella trichopoda, O. sativa, A. thaliana, Vitus vinifera, Ricinus comunis, Brassica rapa, Theobroma cacao, and Medicago truncatula), and a total of 327 amino acid sequences were retrieved (Dataset S2). FASTA sequences were aligned using MUSCLE in MEGAX, and the 11–amino acid window spanning Ser318 was extracted for visualization of conservation using WebLogo (105).
Protein Modeling.
CPK28 was modeled using PHYRE Protein Fold Recognition Server (106) on Intensive Mode and visualized using PyMol Molecular Graphics System Version 2.4.0. The crystal structure of inactive TgCDPK1 (PDB:3KU2) was used as a template.
Statistical Analysis.
Statistical significance was determined by a Student’s t test or one-way ANOVA followed by Tukey’s post hoc test using GraphPad Prism version 8, as indicated.
Supplementary Material
Acknowledgments
We gratefully acknowledge Ray Zielinski for thoughtful guidance on aspects of this work related to Ca2+ signaling and for assistance with intrinsic fluorescence experiments. We also extend heartfelt thanks to Cyril Zipfel (The Sainsbury Laboratory and the University of Zurich) for critical discussions regarding early observations of the transphosphorylation between BIK1 and CPK28 and for critically reading this work prior to submission. We thank Tony Papanicolaou, Ruxandra Bogdan, and Andrew Ji (Queen’s University) for their technical assistance and Darrel Desveaux (University of Toronto) for providing Pseudomonas syringae pv. tomato DC3000. We additionally thank all members of the Monaghan laboratory for their helpful and critical feedback of this work. Research in the Monaghan laboratory is funded by Discovery and Accelerator Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chairs Program, as well as infrastructure support through the John R Evans Leaders Fund from the Canadian Foundation for Innovation and the Ontario Ministry of Research and Innovation. Research in the Huber laboratory was funded by NSF-IOS Grant 1354094. Research in the Trujillo laboratory is funded by German Research Foundation. M.B. was supported by an NSERC Postdoctoral Fellowship, A.J.D. was supported by the Queen’s University Summer Work Experience Program, D.C. was the recipient of an NSERC Undergraduate Summer Research Award, and K.E.D. was the recipient of an NSERC Canada Graduate Scholarship for Master’s Students, an Ontario Graduate Scholarship, and an NSERC Michael Smith Foreign Study Supplement to train in the Trujillo laboratory.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024272118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S., The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Adams M. D.et al.; Arabidopsis Genome Initiative , Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Dardick C., Chen J., Richter T., Ouyang S., Ronald P., The Rice Kinase Database , The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 143, 579–586 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredow M., Monaghan J., Regulation of plant immune signaling by calcium-dependent protein kinases. Mol. Plant Microbe Interact. 32, 6–19 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Sun L., Zhang J., Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS Microbiol. Rev. 44, 845–856 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Tang D., Wang G., Zhou J.-M., Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29, 618–637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ubersax J. A., Ferrell J. E. Jr, Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Mergner J., et al., Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579, 409–414 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Khan S. H., McLaughlin W. A., Kumar R., Site-specific phosphorylation regulates the structure and function of an intrinsically disordered domain of the glucocorticoid receptor. Sci. Rep. 7, 15440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobb L., Koyama S., Cohen P., Site-specific phosphorylation by intracellular kinases determines the apoptotic activity of IGFBP-3 in prostate cancer. Cancer Res. 67, 4393 (2007). [Google Scholar]
- 11.Chen Z., et al., Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Ko Y. U., et al., Site-specific phosphorylation of Fbxw7 by Cdk5/p25 and its resulting decreased stability are linked to glutamate-induced excitotoxicity. Cell Death Dis. 10, 579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marceaux C., Petit D., Bertoglio J., David M. D., Phosphorylation of ARHGAP19 by CDK1 and ROCK regulates its subcellular localization and function during mitosis. J. Cell Sci. 131, jcs208397 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Balta E.-A., et al., Phosphorylation modulates the subcellular localization of SOX11. Front. Mol. Neurosci. 11, 211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying S., et al., Regulatory phosphorylation of bacterial-type PEP carboxylase by the Ca2+-dependent protein kinase RcCDPK1 in developing castor oil seeds. Plant Physiol. 174, 1012–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eng G. W. L., Edison D. M., Virshup D. M., Site-specific phosphorylation of casein kinase 1 δ (CK1δ) regulates its activity towards the circadian regulator PER2. PLoS One 12, e0177834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen S. W., et al., Site-specific phosphorylation of PSD-95 PDZ domains reveals fine-tuned regulation of protein-protein interactions. ACS Chem. Biol. 12, 2313–2323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arif A., Jia J., Willard B., Li X., Fox P. L., Multisite phosphorylation of S6K1 directs a kinase phospho-code that determines substrate selection. Mol. Cell 73, 446–457.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thor K., Calcium-nutrient and messenger. Front. Plant Sci. 10, 440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldon D., Mbengue M., Mazars C., Galaud J.-P., Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19, 665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batistič O., Kudla J., Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820, 1283–1293 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Medvedev S. S., Principles of calcium signal generation and transduction in plant cells. Russ. J. Plant Physiol. 65, 771–783 (2018). [Google Scholar]
- 23.Edel K. H., Marchadier E., Brownlee C., Kudla J., Hetherington A. M., The evolution of calcium-based signalling in plants. Curr. Biol. 27, R667–R679 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Ranty B., et al., Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7, 327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender K. W., Zielinski R. E., Huber S. C., Revisiting paradigms of Ca2+ signaling protein kinase regulation in plants. Biochem. J. 475, 207–223 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Hamel L.-P., Sheen J., Séguin A., Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valmonte G. R., Arthur K., Higgins C. M., MacDiarmid R. M., Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 55, 551–569 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Hrabak E. M., et al., The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klimecka M., Muszyńska G., Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. 54, 219–233 (2007). [PubMed] [Google Scholar]
- 30.Boudsocq M., Droillard M. J., Regad L., Laurière C., Characterization of Arabidopsis calcium-dependent protein kinases: Activated or not by calcium? Biochem. J. 447, 291–299 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Wernimont A. K., et al., Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 17, 596–601 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wernimont A. K., et al., Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins 79, 803–820 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Ingram J. R., et al., Allosteric activation of apicomplexan calcium-dependent protein kinases. Proc. Natl. Acad. Sci. U.S.A. 112, E4975–E4984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerra T., et al., Calcium-dependent protein kinase 5 links calcium signaling with N-hydroxy-l-pipecolic acid- and SARD1-dependent immune memory in systemic acquired resistance. New Phytol. 225, 310–325 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Schulz P., Herde M., Romeis T., Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 163, 523–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boudsocq M., Sheen J., CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip Delormel T., Boudsocq M., Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Simeunovic A., Mair A., Wurzinger B., Teige M., Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 67, 3855–3872 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Arimura G., Sawasaki T., Arabidopsis CPK3 plays extensive roles in various biological and environmental responses. Plant Signal. Behav. 5, 1263–1265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perraki A., et al., Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561, 248–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadota Y., et al., Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 221, 2160–2175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benschop J. J., et al., Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Mattei B., Spinelli F., Pontiggia D., De Lorenzo G., Comprehensive analysis of the membrane phosphoproteome regulated by oligogalacturonides in Arabidopsis thaliana. Front. Plant Sci. 7, 1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayapuram N., et al., Quantitative phosphoproteomic analysis reveals shared and specific targets of Arabidopsis mitogen-activated protein kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteomics 17, 61–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rayapuram N., et al., Identification of novel PAMP-triggered phosphorylation and dephosphorylation events in Arabidopsis thaliana by quantitative phosphoproteomic analysis. J. Proteome Res. 13, 2137–2151 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Witte C.-P., et al., Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J. Biol. Chem. 285, 9740–9748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nühse T. S., Stensballe A., Jensen O. N., Peck S. C., Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Fuente van Bentem S., et al., Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 7, 2458–2470 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Monaghan J., Matschi S., Romeis T., Zipfel C., The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal. Behav. 10, e1018497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., et al., A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Monaghan J., et al., The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Matschi S., et al., Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Matschi S., Hake K., Herde M., Hause B., Romeis T., The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27, 591–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bender K. W., et al., Autophosphorylation-based calcium (Ca2+) sensitivity priming and Ca2+/calmodulin inhibition of Arabidopsis thaliana Ca2+-dependent protein kinase 28 (CPK28). J. Biol. Chem. 292, 3988–4002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hegeman A. D., et al., A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics 6, 3649–3664 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Swatek K. N., Wilson R. S., Ahsan N., Tritz R. L., Thelen J. J., Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem. J. 459, 15–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beenstock J., Mooshayef N., Engelberg D., How do protein kinases take a selfie (autophosphorylate)? Trends Biochem. Sci. 41, 938–953 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Lu D., et al., A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couto D., et al., The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog. 12, e1005811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin W., et al., Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 3632–3637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lal N. K., et al., The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23, 485–497.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J., et al., Identification and functional analysis of phosphorylation residues of the Arabidopsis BOTRYTIS-INDUCED KINASE1. Protein Cell 4, 771–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma X., et al., Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581, 199–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang X., et al., Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5, e13568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J., et al., The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol. 176, 1835–1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J., et al., Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Ranf S., et al., Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14, 374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z., et al., BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 6205–6210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vivian J. T., Callis P. R., Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liese A., Romeis T., Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 1833, 1582–1589 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Geiger D., et al., Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 107, 8023–8028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanchiswamy C. N., et al., Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 10, 97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saha P., Singh M., Characterization of a winged bean (psophocarpus tetragonolobus) protein kinase with calmodulin-like domain: Regulation by autophosphorylation. Biochem. J. 305, 205–210 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaudhuri S., Seal A., Gupta M. D., Autophosphorylation-dependent activation of a calcium-dependent protein kinase from groundnut. Plant Physiol. 120, 859–866 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chehab E. W., Patharkar O. R., Hegeman A. D., Taybi T., Cushman J. C., Autophosphorylation and subcellular localization dynamics of a salt- and water deficit-induced calcium-dependent protein kinase from ice plant. Plant Physiol. 135, 1430–1446 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ito T., Ishida S., Oe S., Fukazawa J., Takahashi Y., Autophosphorylation affects substrate-binding affinity of tobacco Ca2+-dependent protein Kinase1. Plant Physiol. 174, 2457–2468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh M.-H., et al., CDPKs are dual-specificity protein kinases and tyrosine autophosphorylation attenuates kinase activity. FEBS Lett. 586, 4070–4075 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Asai S., et al., The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288, 14332–14340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sente A., et al., Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat. Struct. Mol. Biol. 25, 538–545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jonniya N. A., Sk M. F., Kar P., Investigating phosphorylation-induced conformational changes in WNK1 kinase by molecular dynamics simulations. ACS Omega 4, 17404–17416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schworer C. M., Colbran R. J., Keefer J. R., Soderling T. R., Ca2+/calmodulin-dependent protein kinase II. Identification of a regulatory autophosphorylation site adjacent to the inhibitory and calmodulin-binding domains. J. Biol. Chem. 263, 13486–13489 (1988). [PubMed] [Google Scholar]
- 82.Ohsako S., Nakazawa H., Sekihara S., Ikai A., Yamauchi T., Role of threonine-286 as autophosphorylation site for appearance of Ca2(+)-independent activity of calmodulin-dependent protein kinase II α subunit. J. Biochem. 109, 137–143 (1991). [DOI] [PubMed] [Google Scholar]
- 83.Monaghan J., Conserved degradation of orthologous RLCKs regulates immune homeostasis. Trends Plant Sci. 23, 554–557 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Dubiella U., et al., Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marcec M. J., Gilroy S., Poovaiah B. W., Tanaka K., Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 283, 343–354 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Dressano K., et al., Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing. Nat. Plants 6, 1008–1019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bazin J., et al., Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 16, e1008401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wirthmueller L., Romeis T., Sp(l)icing up PepR signalling. Nat. Plants 6, 912–913 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Whalley H. J., Knight M. R., Calcium signatures are decoded by plants to give specific gene responses. New Phytol. 197, 690–693 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Wang J., Long X., Chern M., Chen X., Understanding the molecular mechanisms of trade-offs between plant growth and immunity. Sci. China Life Sci. 64, 234–241 (2020). [DOI] [PubMed] [Google Scholar]
- 91.Karasov T. L., Chae E., Herman J. J., Bergelson J., Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29, 666–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gómez-Gómez L., Felix G., Boller T., A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284 (1999). [DOI] [PubMed] [Google Scholar]
- 93.van Wersch R., Li X., Zhang Y., Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front. Plant Sci. 7, 1717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li X., Clarke J. D., Zhang Y., Dong X., Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14, 1131–1139 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Morita-Yamamuro C., et al., The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 46, 902–912 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Campo S., et al., Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bundó M., Coca M., Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 68, 2963–2975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hettenhausen C., Yang D.-H., Baldwin I. T., Wu J., Calcium-dependent protein kinases, CDPK4 and CDPK5, affect early steps of jasmonic acid biosynthesis in Nicotiana attenuata. Plant Signal. Behav. 8, e22784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M., pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 (2000). [DOI] [PubMed] [Google Scholar]
- 101.Huffaker A., Pearce G., Ryan C. A., An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kunze G., et al., The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bredow M., Sementchoukova I., Siegel K., Monaghan J., Pattern-triggered oxidative burst and seedling growth inhibition assays in Arabidopsis thaliana. J. Vis. Exp. (2019). [DOI] [PubMed] [Google Scholar]
- 104.Bers D. M., Patton C. W., Nuccitelli R., A practical guide to the preparation of Ca(2+) buffers. Methods Cell Biol. 99, 1–26 (2010). [DOI] [PubMed] [Google Scholar]
- 105.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.