Abstract
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is related to new coronavirus disease (COVID-19) has mobilized several scientifics to explore clinical data using soft-computing approaches. In the context of machine learning, previous studies have explored supervised algorithms to predict and support diagnosis based on several clinical parameters from patients diagnosed with and without COVID-19. However, in most of them the decision is based on a “black-box” method, making it impossible to discover the variable relevance in decision making. Hence, in this study, we introduce a non-supervised clustering analysis with neural network self-organizing maps (SOM) as a strategy of decision-making. We propose to identify potential variables in routine blood tests that can support clinician decision-making during COVID-19 diagnosis at hospital admission, facilitating rapid medical intervention. Based on SOM features (visual relationships between clusters and identification of patterns and behaviors), and using linear discriminant analysis , it was possible to detect a group of units of the map with a discrimination power around 83% to SARS-CoV-2-positive patients. In addition, we identified some variables in admission blood tests (Leukocytes, Basophils, Eosinophils, and Red cell Distribution Width) that, in combination had strong influence in the clustering performance, which could assist a possible clinical decision. Thus, although with limitations, we believe that SOM can be used as a soft-computing approach to support clinician decision-making in the context of COVID-19.
Keywords: Covid-19 diagnostic, SARS-CoV-2, Self-organizing maps
Introduction
The global pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has mobilized several scientific areas to explore diagnostic solutions and alternative therapies for this new betacoronavirus disease (COVID-19) (Ciotti et al. (2019)). Considering the absence of efficient prevention and therapies, efforts to control COVID-19 are limited to personal actions (i.e., hand washing, face covering, environmental cleaning and physical distancing), place restrictions adopted at local levels (schools, workplaces, restaurants, malls and other closures), and tracing of infected people. However, it is time-consuming and useful at an early period of infection (Liu et al. (2020)). Although some potential treatments were reported to avoid disease complications and decrease hospitalization period (dexamethasone, remdesivir, and heparin) (Tomazini et al. (2020); Spinner et al. (2020); Wilkinson et al. (2020)), the hope to face COVID-19 is indeed in the development of safe and effective vaccines (Raja et al. (2020)). This scenario is especially critical in this pandemic crisis principally due to the exponential growth of infected people and the number of deaths. In this sense, new strategies to support rapid identification of SARS-CoV-2-positive patients became extremely necessary.
The increasing rate of hospital admission of COVID-19 patients provided an extensive clinical and demographic database that is now available. This dataset permits more ambitious computational approaches to be used in order to find “the needle in the haystack” that could support the clinician decision-making for diagnosis and prognosis of COVID-19 cases. Thus, data mining approaches can be performed with a supervised machine learning algorithm to predict future values by classification and/or regression, or since by unsupervised learning to clustering data (Tan et al. (2016)). Currently, some studies have employed supervised machine learning approaches to predict COVID-19 diagnosis based on recognition/analysis of images (Lalmuanawma et al. (2020)) and hemogram tests (Cabitza et al. (2020)). In our summary review (see in 2), we identified that most of classificatory algorithms utilized strategies with “black box” approaches. The major problem regarding this methodology is associated with the models of decision that are created directly by algorithm when it is applied to a database. Thus, it is not possible to accurately recognize or interpret the variable that had influence in the decision-making process during the prediction. Hence, the black boxes-derived algorithms normally result in complex models that are intrinsically not interpretable for human users (Rai (2020)).
When Considering medical studies both precision and interpretability should be contemplated (Wu et al. (2020)). Consequently, precisely identifying the variables that have an influence on the algorithm analysis allows understand some data patterns and behaviors for better classification. In addition, when considering a human interpretation, data presentation is also important. This attention towards in the decision-making mechanism facilitates human analysis and is designed as explainable artificial intelligence (XAI). Hence, the possibility to explain and interpret data may improve the final result by three reasons: (1) supporting impartiality in decision-making, detecting and correcting bias in the analyzed dataset; (2) improving the robustness of the process, highlighting possible adverse situations that may impact the decision-making process; and (3) assuring that only significant variables have an influence on the outcome (Arrieta et al. (2020)). In fact, data mining of medical studies, it is preferable to identify all the information behind the decision-making than considering a single outcome as part of the prediction process (Holzinger et al. (2017); London (2019); Tjoa and Guan (2019); Arrieta et al. (2020); Samek et al. (2017); Holzinger et al. (2017); Ahmad et al. (2018); Adadi and Berrada (2018); Goebel et al. (2018); Wu et al. (2020)).
In this context, clustering data analysis has an exploratory nature that aims to identify patterns, correlations, trends, trajectories, groups and anomalies that better describe the dataset. As such, it is important to consider that the quality of the clustering method process depends on the fulfillment of specific criteria. For instance, the clustering approach must search for data subsets in samples that share common and, at same time, distinct features with other data subsets.
On the light of these evidence, this study aims at introducing an exploratory research involving clustering analysis to investigate clinical data from blood tests at hospital admission . The data analysis is supported by use of the unsupervised neural network of self-organizing maps (SOM) (Kohonen (2013)). This network allows the visual identification of relationships between clusters and the knowledge produced in the clustering process, as well as the discovery of patterns and behavior of each variable in the database. Besides, with SOM training, other features related to the dataset used in the training process can be mapped on the grid to observe, for example, class or sub-class information distribution. Thus, through visual exploration of relationships between groups, we observed by SOM analysis patterns and behaviors in the blood test dataset which indicated predicted variables to assist clinician decision-making in the context of COVID-19 diagnosis.
For a better organization, this present article is divided as follows: Section 2 summarizes some co-related works; Section 3 shows the novelty of the proposal, describes the methodology and the data mining process applied in the study and also the theoretical foundations of clustering analysis and SOM features; Section 4 represents the materials and methods; Section 5 presents and discusses the main findings, and finally, Sect. 6 indicates the main conclusions and future perspectives are indicated.
Related works
The global pandemic caused by coronavirus 2 (SARS-CoV-2) has mobilized several scientific areas to explore diagnostic solutions, including studies involving data mining approaches in attempt to support a rapid decision-making to COVID-19 diagnosis. According to this, several initiatives have emerged using machine learning methodologies to detect and differentiate COVID-19 positive patients using X-ray and computed tomography (CT) images (Lalmuanawma et al. (2020); Syeda et al. (2020); Duran-Lopez et al. (2020); Civit-Masot et al. (2020); Ohata et al. (2020)). Furthermore, other reports utilized supervised machine learning applications for image recognition and diagnosis in health sciences (Omara et al. (2017); Mei et al. (2017); Mote et al. (2017); Rodrigues et al. (2018); Matsuda et al. (2017); De Souza et al. (2019); O’Driscoll et al. (2017); Chunyan et al. (2016); Mapari et al. (2017); Olivares et al. (2020); Selvachandran et al. (2019); Chouhan et al. (2020); Dourado et al. (2020); Minonzio et al. (2020); Muhammad et al. (2020); Santos et al. (2020); Ding et al. (2020); Bernardo et al. (2019)). However, images-based diagnostics are considered very costly considering that routine blood tests are relatively easy to run and widely used at admission in several clinical centers (Ferrari et al. (2020)).
In this sense, other studies investigated hemogram data from hospitalized patients using different classification algorithms approaches for identification of COVID-19 patients which included: support vector machines and SMOTE Boost algorithms, Naive Bayes classifiers, and others (neural networks, gradient boosted trees, random forests, logistic regression and support vector machine) (Cabitza et al. (2020); Soares et al. (2020); de Moraes Batista et al. (2020); Avila et al. (2020)). Although these findings have indicated an accuracy rate around 80%, the relevant variables to support “decision making” were not precisely identified.
Thus, by precisely comparing the strategies described in the mentioned articles, it was possible detected that in most of them the variable influence in the outcome was not identified. These studies also could not determine the data interpretability essentially comprising of strategies with “black box” approaches, which, as previously mentioned, has several disadvantages in datamining of medical studies (Holzinger et al. (2017); London (2019); Tjoa and Guan (2019); Arrieta et al. (2020); Samek et al. (2017); Holzinger et al. (2017); Ahmad et al. (2018); Adadi and Berrada (2018); Goebel et al. (2018); Wu et al. (2020)). Hence, understanding the features behind the variables influence diagnosis/prognosis prediction of a disease is similarly important to the prediction model, per se, and can be applied in the COVID-19 context, basically exploring unsupervised methods, as, for example, the neural network of self-organizing maps (SOM) was designed to this study.
In Health Sciences, SOM already was designed to cluster clinical and pathological data related to dermatological diseases (Fidan et al. (2016)). The technique was also applied for classification of breast cancer (Omara et al. (2017)). Moreover, SOM was utilized as part of a proposed system to segment magnetic resonance images of brain tumors (Mote et al. (2017)). In other report, the authors found relevant relationships between variables in a database and risk groups for tuberculosis (Orjuela-Cañón et al. (2018)). Finally, SOM was used for choosing the most suitable features to be extracted from electroencephalogram signals in order to discriminate seizures from non-seizures, specifically from patients with nocturnal frontal lobe epilepsy (Pisano et al. (2019)).
Novelty of the proposal
This study uses an adaptation of a data mining framework proposed by De Souza et al. (2019) and based on the Practical Software Systems Measurement (PSM) guide (McGarry (2002)). In De Souza et al. (2019), the authors propose a conceptual framework for data mining, structured in different levels of abstraction which were denominated as visions. In the defined framework, the data mining process is guided by the questions (guidelines) that must be answered at the end of its execution. From the De Souza et al. (2019) the Cluster Analysis View, the Process View and the guideline Information Needs are adapted to this paper. These items from the framework will be used in this study at the context of COVID-19 with the following goals:
Information needs To address the selection of attributes for the training process and procedures for data analysis that allow knowledge discovery, pointing out patterns and features to support identification of patients positive for SARS-CoV-2;
Cluster analysis view To guide the cluster analysis model, the configuration and application of the SOM algorithm, as well as the types of maps that must be generated and their visual interpretations in order to meet the established Information Needs;
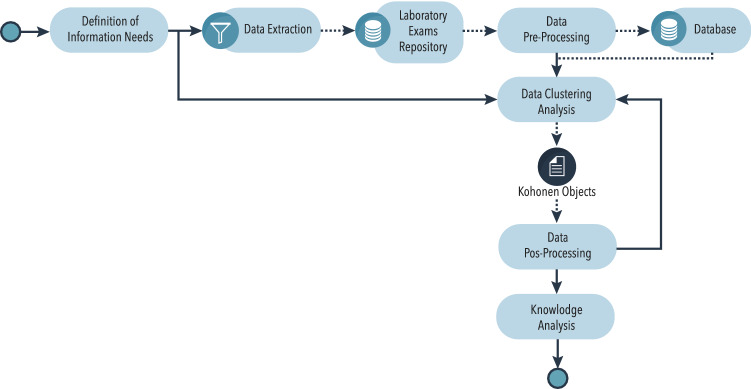
Process view To organize activities that must be performed for data mining: data gathering, pre-processing, analysis and interpretation of the obtained results, in order to meet the established Information Needs. Figure 1 shows all adapted processes of this study.
Fig. 1.
Flowchart plot of process view. Adaptation of the process view structure proposed by De Souza et al. (2019). The Kohonen objects have a set of information resulting from application SOM algorithm such as: dataset, grid of the map and your units, intra-distance of each unit, among others that will be detailed in subsection 3.3
Definition of information needs
In this stage, the Information Needs that should guide the data mining process are defined. Specifically, at this study, the Information Needs are those that may support identification of patterns and behaviors in the blood test dataset that may indicate possible patients positive for SARS-CoV-2. Hence, the main Information Need is then defined as: “Identify which variables in routine laboratory tests in hospitalization present significant patterns for SARS-CoV-2 detection in positive patients.”
Data extraction and pre-processing
These stages are aimed at generating the database with information extracted from laboratory tests to meet the established Information Need. In the data extraction stage, the laboratory tests for data extraction are defined, as well as the information that will be collected from each patient and the procedures to anonymize patient data. In the pre-processing stage the data are cleaned, extracting relevant data and eliminating inconsistencies that may hinder the meeting of the Information Need. The final result is exported in CSV (Comma-Separated Value) files with raw data and a structure composed by lines and columns.
As described in Sect. 4, in this study, it was utilized public data information by Albert Einstein Hospital in São Paulo, Brazil. With the intention to meet the Information Need established for this study, 599 registers were chosen for analysis. These selected registers contained consistent data for the following variables: Hematocrit, Hemoglobin, Platelets, Mean Platelet Volume, Red blood Cells, Lymphocytes, Mean Corpuscular Hemoglobin Concentration (MCHC), Leukocytes, Basophils, Mean Corpuscular Hemoglobin (MCH), Eosinophils, Mean Corpuscular Volume (MCV), Monocytes and Red blood Cell Distribution Width (RDW). These variables were selected due to their frequent presence in routine automated blood test worldwide (George-Gay and Parker (2003); Barnes et al. (2005)) . In the remaining registers, many samples with empty data were found in one or more selected variables, and for this reason they were excluded from the pre-processing step. From the 599 included registers, only 81 of them presented a positive result for SARS-CoV-2 RT-PCR.
Data clustering analysis
Before introducing the cluster analysis model, it is important to contextualize SOM methodology for COVID-19 dataset.
Self-organized maps
Consider that a generic dataset is available for analysis. This dataset is represented by the set , and its samples are such that = . The ith sample is formed by M attributes: = .
The clustering algorithms perform the analysis by comparing the M attributes that describe the N samples. This is done using some measure of similarity, which allows to infer how close are two samples or how similar are the samples attributes (Haykin (2007)).
The outcome of the analysis process is the allocation of each sample in a cluster by maximizing the similarity between elements in the same cluster. That is a problem that can be modeled as an intra-cluster distance minimization problem. In the taxonomy of machine learning techniques, this process is classified as inductive and unsupervised (Tan et al. (2016)).
Among the algorithms in this category, there is the self-organizing maps proposed by Kohonen (1982). SOM is an artificial neural network, interconnected and unsupervised, which allows a self-organizing mapping of samples with multidimensional attributes into a bidimensional space (grid), solving problems with tasks of data clustering, visualization and abstraction. The technique can also be applied to a wider study of the correlation between multiple attributes in the dataset, with no restrictions concerning the quantity of attributes to study. This is due to its property of allowing to map samples with a high number of dimensions in reduced dimensions, converting complex nonlinear statistical relationships into geometric relationships. The mapping is done preserving the original topological relationship of the samples since their physical location on the map shows the similarity between the attributes in the original multidimensional space (Kohonen (2013)).
The similarity between samples is calculated by Euclidean distance; this value is used in the first stage, designed as “training” step. In this stage, samples are used to train the SOM network, and during the mapping of samples to units of the map.
In the training stage, the SOM network is parametrized with U units of map , and a weight is associated with each unit. has the same dimensionality of the samples, that is, . Weights are usually initialized with random values and are adjusted during the training stage. For a detailed description of this process, see Kohonen (2013).
In this way, the Euclidean distance between the ith sample and any unit u in the map is defined as:
| 1 |
The winning unit (nearest) for a given sample is chosen among the U units of map as the one with the smallest distance to the sample. The adjustment of weights is performed to preserve the topology of samples in the dataset; hence, samples with similar attributes will be mapped to the same unit of map or to neighboring units of map and, as the attributes differ, they are represented by adjacent units of map as defined in Kohonen (2013).
The second moment when the distance is used is after the training of the network, when weights are already adjusted. Hence, the operation defined in Eq. (1) is applied to map each sample of the dataset to the map. This is done by attributing an identification number to each unit of the map and, on the other hand, the information of such sample is mapped on the map and allowing at the same time to explore the original sample data.
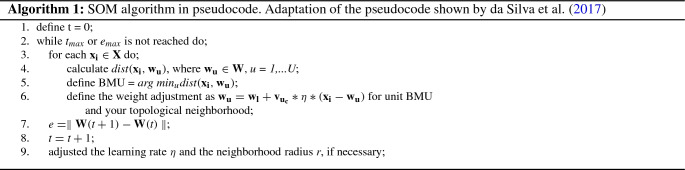
At the end of the training stage, each unit of the map represents a subset of samples used in training which are similar between themselves, but also having similar attributes to closer units of map and distinct in relation to more distant units of map (Moreira and Silva (2017)). In the pseudocode described in Table 1 and Algorithm 1 is shown SOM algorithm.
Table 1.
SOM algorithm input and output
| Input | |
|---|---|
| : an training dataset not labeled, =; | |
| : set of synaptic weights, =; | |
| U: number of units on the map; | |
| : map settings (number of units in the grid, map dimension); | |
| : a vector distance measure. This pseudocode uses Euclidean distance; | |
| da: a matrix distance measure; | |
| : initial learning rate; | |
| : topological neighborhood function between unit u and the unit closest to the sample c; | |
| r: neighborhood radius; | |
| : number of iterations; | |
| : maximum expected error value for weight changes; | |
| Output | |
| : set of adjusted synaptic weights |
Adaptation of the pseudocode shown by da Silva et al. (2017)
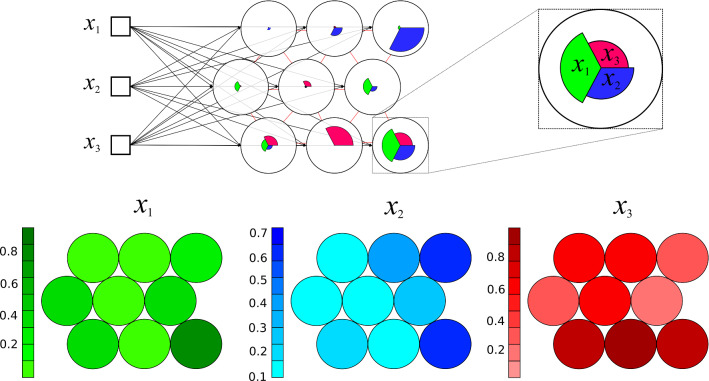
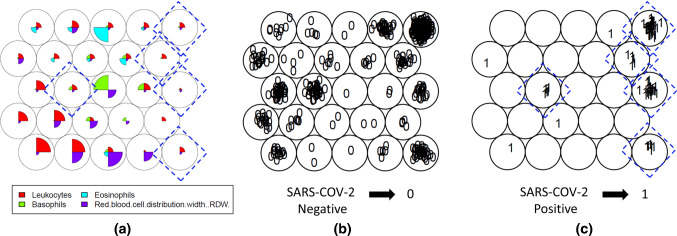
According to Haykin (2007), SOM maps are suitable to the task of selecting features to identify patterns in essential information present in a database. Besides that, other relevant advantage is the visualization of relationships between clusters and the knowledge produced by the clustering process itself as, for instance, the weight of each attribute in the data clustering for each unit, and the consequent distribution of samples on the map. In Fig. 2, a sample of weight values visualization is shown. The weight is displayed for three attributes of the dataset whose elements are mapped onto nine units of map produced by a SOM network training process.
Fig. 2.
Weight of each attribute in the composition of each unit of the map. Overview map of training stage in SOM process different colors represents distinct variables in the dataset
Data post-processing
In this stage the knowledge discovery process was conducted through generation of three types of plots: (i) Feature Vector Visualization Map, (ii) Feature Vector Heatmap and (iii) Topological Distribution Map. This stage is executed every time a different visualization of data is necessary in order to identify tendencies to meet the established Information Need. Data from all samples and the density of features present in the clustering process were used to generate the mentioned plots. The SOM maps generated in this study are presented in Sect. 5.
Knowledge analysis
This final stage is responsible for understanding the behavior, identification patterns, and generation of intrinsic knowledge from findings derived from the laboratory tests present in the database. To support knowledge gain in SOM maps and obtain the “Information Need”, other view data plots as scatter diagrams and others classification tools such as linear discriminant analysis (LDA) can be performed. The processes of analysis execution and results obtainment are fully described in Sect. 5.
Material and methods
In this research, the clustering analysis was performed aimed at discovering patterns that may support the identification of positive patients for SARS-CoV-2, based on data processed in the previous stages (Sect. 3.2). First, the training attributes were defined; furthermore, the training procedures were executed and finally, the quality of the training is evaluated. In the first step, to meet the Information Need, a subset of data from the database was selected for the SOM unsupervised network training. The second activity is related to the analysis of clustering and correlation between variables. This activity is performed using the R language with the Kohonen package (Wehrens and Buydens (2007)) and its SOM function (unsupervised mapping) parameterized as follows: data = number of registers on the database after the pre-processing stage (CSV file with laboratory test data); grid=somgrid() = the dimension of the (xdim,ydim) grid must be set by the square root of the number of registers on the database; topo = hexagonal; toroidal = must be set as false; rlen = quantity of epochs for training (uses the guideline proposed by Haykin (2007): where is the grid dimension and is the number of units on the map; alpha = learning rate, with range of values is established with initial and final values less than 1. dist.fcts = method: Euclidean.
The quality of the training process was assessed by the cohesion of similarity between samples that was grouped into the same unit of map. In addition, the dispersion or separability among units was also assessed, in the sense that different units of map should group elements as distinct as possible. Also, the number of samples in each unit of the map is another metric used to assess the quality.
In attempt to find the Information Need, several training sessions of the SOM unsupervised network were conducted with various configuration parameters. Thus, the best quality clustering configuration was found following the criteria established above. The final configuration for training step is described in Table 2.
Table 2.
SOM function parametrization
| Parameter | Configuration |
|---|---|
| Data | 599 samples |
| grid | somgrid(5,5) |
| topo | Hexagonal |
| toroidal | False |
| rlen | 14.000 |
| alpha | For 14 variables: 0.05 with linear reduction up to 0.01 |
| For 4 variables: 0.08 with linear reduction up to 0.03 | |
| dist.fcts | Euclidean |
Dataset
In this study was utilized public dataset1 which was available by Albert Einstein Hospital in São Paulo, Brazil, in the Kaggle platform at 2020-03-27. A total of 108 different kinds of tests from 5644 patients allocated at the Hospital were enrolled in the analysis, including data of RT-PCR for SARS-CoV-2, patient identification, patient age quantile (range 1 to 19), patient admitted (yes or no) to: regular ward, semi-intensive unit and intensive care unit. The process of the data collection, elimination of patients’ personal information, data cleaning and database generation were performed by the above-mentioned institution. Then, we did not have access to raw clinical data and the data available were standardized by z-score normalization (normalization method made by centering the average of data around zero and scaling by the standard deviation) made by Albert Einstein Hospital institution computational group.
Experimental results
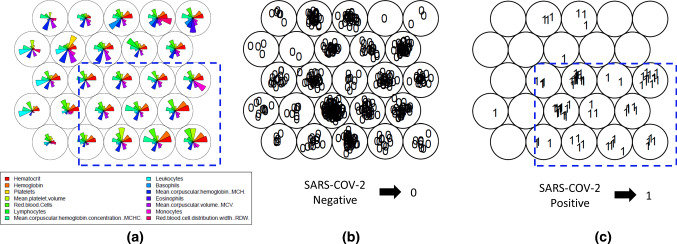
To access the main objective, first it was established the “Information Need”, that in this case, was defined as ”Identify which variables in routine laboratory tests in hospitalization present significant patterns for SARS-CoV-2 detection in positive patients.” Then, the training step of the SOM unsupervised network was performed with parameters described in Sect. 3.3 and blood tests for each patient were used as data input. During training process, 14 variables present in the blood test [Hematocrit, Hemoglobin, Platelets, Mean Platelet Volume, Red blood Cells, Lymphocytes, MCHC, Leukocytes, Basophils, MCH, Eosinophils, MCV, Monocytes and RDW] were investigated to identify patients positive or negative for SARS-CoV-2 with previous confirmation by the gold standard RT-PCR assay. In Fig. 3, three maps resulting from SOM training are shown. The first, in the Feature Vector Visualization Map (a), it is possible to observe vectors of each attribute that represents these above-mentioned variables. The two secondary Topological Distribution Maps represent the topological distribution with both negative (b) and positive (c) SARS-CoV-2 patients.
Fig. 3.
Establishment of Information Needs. It can be observed the general feature of “Vector Visualization color Map” with 14 variables (a) and the Maps comparing between SARS-CoV-2 Negatives (b) and Positives (c) patients with clustering distinction
The maps in Fig. 3 revealed a concentration of positive SARS-CoV-2 patients in the lower region of the map (highlighted in blue in map (c)), although this area also contains SARS-CoV-2 negative patients (map (b)). Considering the cluster analysis produced by the SOM unsupervised network, this aspect indicated that similar weights were attributed for each attribute that composed these units of map and the distinction to weights applied to the other units of map. Therefore, it is possible to determine that the SOM unsupervised network found features that distinguish positive SARS-CoV-2 patients in the set of laboratory tests analyzed. After investigating this mentioned region in the map, it was verified that some units of map have a low weight applied to one or more of the following variables: Leukocytes, Basophils, Eosinophils and Red Blood Cell Distribution Width (RDW).
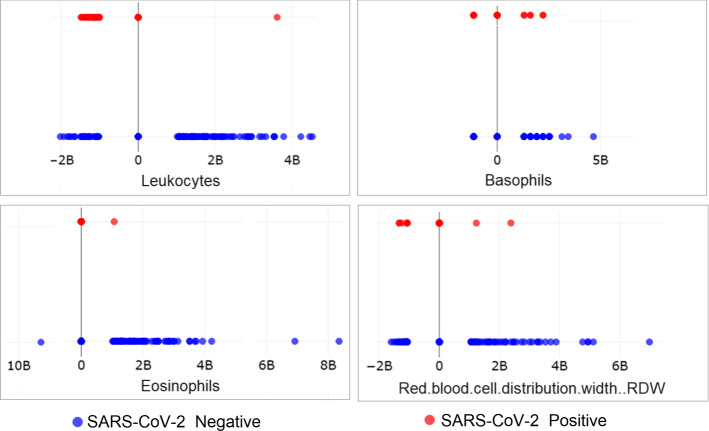
To validate this analysis, a scatter plot of the variables in the blood test, that were previously included in the training set, was prepared. These variables were distributed on the Cartesian plane plot in two levels: upper line with negative SARS-CoV-2 patients and lower line with positive SARS-CoV-2 patients. This plot showed that the variables Leukocytes, Basophils, Eosinophils and Red blood cell distribution width (RDW) had different behavior in positive and negative SARS-CoV-2 patients, as can be seen in Fig. 4.
Fig. 4.
Scatter plot distribution of variables in the blood test applied in SOM analysis: Leukocytes, Basophils, Eosinophils e Red blood Cell Distribution Width (RDW) presented different behavior in positive and negative SARS-CoV-2 groups
Considering this new set of variables, a new training of SOM unsupervised network was carried out using only those pre-selected variables as attributes to verify the more accurate segregation between SARS-CoV-2 negative and positive groups. In Fig. 5, the weights resulting from this training and the topological distribution of patients on the map are shown.
Fig. 5.
Establishment of Information Needs II. It can be observed the general feature of “Vector Visualization color Map” with 4 variables a pre-selected in training I and the Maps comparing between SARS-CoV-2 Negatives b and Positives c patients with evident segregation
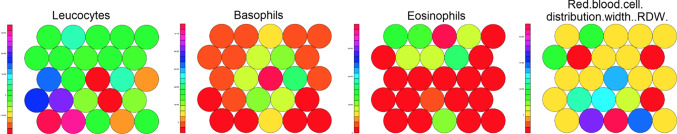
In this new SOM analysis with 4 variables in the blood test, a large concentration of SARS-CoV-2-positive patients was verified in the map units located on right side of the Map (c). In this region, the set of attribute weights for variables: Basophils, Eosinophils, Leukocytes, and Red Blood Cell Distribution Width—RDW—are similar to each other when compared with the other units on the map (a). This aspect can be observed in Fig. 6, in which the profile of each attribute (that represented the 4 variables in the blood test) is separately shown in a color heatmaps distribution.
Fig. 6.
Color heatmaps distribution. It is observed at different colors in the heatmaps the distribution of the attributes (that represented the 4 variables in the blood test) in SOM training II
Although the map units keep overlapping between SARS-CoV-2-positive and SARS-CoV-2-negative patients in this new training using only four variables in the blood test as attributes, it was possible to identify better data segregation when compared with the previous SOM experiment in which 14 variables were used as attributes. To better understand the proportion of positive and negative patients, each unit of the map was identified with more than one positive result in a new topological ordinal distribution (Fig. 7). In this distribution, four map units (5, 15, 20, and 25) were clustered, indicating a strong tendency to discriminate positive patients to SARS-CoV-2 (Fig. 7).
Fig. 7.
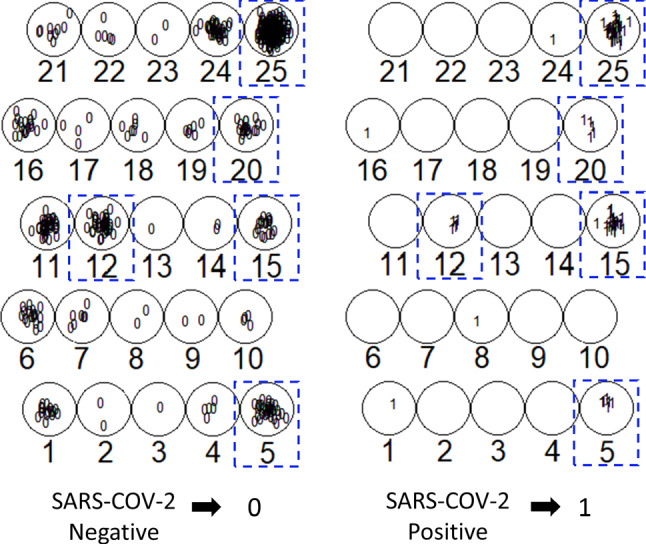
Topological ordinal distribution of units of map. Each units of the map were separately identified and it is observed the distribution of SARS-CoV-2 negative and positive patients at each unit in the map
Then, all map units that were set to positive patients were now plotted and their discriminative potential was analyzed with respect to PCR tests (Table 3). In general, it is possible to identify that 178 negative results were segregated in 20 units of map. Further, it was detected that units of map “5”, “12” and “25” were weak to discriminate positive patients (around 12, 5–15%), and units of map “15” and “20” had the highest percentage of correct positive tests, respectively, 60% and 20% (Table 3).
Table 3.
Positive x negative tests on units of map
| Unit map | Negative tests | Positive tests | Positive tests (%) |
|---|---|---|---|
| 5 | 33 | 6 | 15 |
| 12 | 35 | 5 | 12.5 |
| 15 | 17 | 24 | 60 |
| 20 | 19 | 5 | 20 |
| 25 | 236 | 37 | 13 |
| Others | 178 | 4 | 1.5 |
| Total | 518 | 81 | 13.5 |
To analyze the discriminatory power of units of map (15, 20, and 25) considering the four variables selected in secondary SOM analysis, a linear discriminant analysis (LDA) was performed. The LDA was run using the language R and its package MVar.pt (Ossani et al. (2020)) with the function DA parameterized as follows: prior = 50% for each class (SARS-CoV-2 positive and SARS-CoV-2 negative); type = “lda”; validation = learning; method = “mle”; testing = NA.
The result of the discriminant analysis is shown in the confusion matrix in Table 4. The confusion matrix showed that units of map 15, 20 and 25 were more predicted to correctly classify positive patients, in a unitary and grouped way. They have attributes with similar weights (they are ‘neighbors’ on the map) and can distinguish SARS-CoV-2-negative and SARS-CoV-2-positive patients with more accuracy. For instance, the unit of map “15” has low weight in the Leukocytes attribute and higher weight in the Red Blood Cell Distribution Width (RDW) (Fig. 5).
Table 4.
Confusion matrix LDA units of map 15, 20, 25
| Units of map | ||||||||
|---|---|---|---|---|---|---|---|---|
| 15, 20, 25 | 15 | 20 | 25 | |||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |
| Negative | 250 | 22 | 10 | 7 | 16 | 3 | 167 | 69 |
| Positive | 38 | 28 | 11 | 13 | 2 | 3 | 8 | 29 |
Based on the values presented in Table 4, the group comprised of three map units (15, 20, 25) has 82,24% of accuracy, 86,8% of specificity and presented a sensitivity of 56%. Then, the accuracy rate, the specificity and sensitivity for each selected map unit were individually calculated: The unit “15” had the accuracy rate 56%, specificity 47.62% and sensitivity 65%; the unit “20” had the accuracy rate 79%, specificity 88.89% and sensitivity 50%; and the unit “25” had the accuracy rate 71%, specificity 95.43% and sensitivity 29.59%.
Conclusions and further work
In consideration to global impact of COVID-19 pandemic, the exploration of data repository with infected and non infected patients, by machine learning process, is fundamental to promote new tools for diagnosis or prognosis. This study presented the application of SOM unsupervised network for identification of patterns and distinct behaviors in routine blood tests with the purpose of supporting the diagnosis of patients with COVID-19. Data from a public and anonymized database were analyzed covering 5,644 patients and 108 laboratory tests, which were available by Albert Einstein Hospital Institution. Although with important limitations such as: i) lack of data knowledge in the collection process, z-score normalization which produced negative values and (ii) the nonexistence of raw data, the approach was effective to discriminate positive and negative patients in some specific conditions. The SOM used here grouped 67 of the 81 positive SARS-CoV-2 patients into three principal map units (15, 20, 25) that had common patterns and behaviors. Thus, a significant portion of the negative results (246 from 518) was separated.
Additionally, during the process of finding and establishing the “Information Need,” which in this case was determined as “what variables in routine laboratory blood test at present significant patterns for detection of SARS-CoV-2 positive patients,” the SOM analyses demonstrated some attributes weights (that represented the variables in the blood test) in each unit of the map, generating human explainability and interpretability of the decision performed in the training process. The main variables found in SOM experiments were: Leukocytes, Basophils, Eosinophils, and Red Blood Cell Distribution Width (RDW) which presented a differentiated behavior for patients diagnosed with COVID-19. This was only possible because SOM is oriented toward visualization of the relationships between the clusters and by its ability to generate knowledge, especially, in units of map 15, 20 and 25, where 83% of SARS-CoV-2-positive patients were correctly classified.In fact, it has already been reported that RDW is a significant predictor of severe illness in hospitalized COVID-19 patients (Lippi et al. (2020); Henry et al. (2020)). In line with our findings other group related that subtle differences in white blood cell pattern verified by complete blood count in routine test with differential fluorescence scattergrams can be used to rapid screening of COVID-19 patients (Osman et al. (2020)). Moreover, corroborating our hypothesis to use admission blood test for support decision-making during COVID-19 discrimination, an encouraging work also demonstrated that hematological parameters may be used to assist the diagnosis of COVID-19 at emergency room (Usul et al. (2020)).
Although these evidences show the importance of blood variables found in our SOM analysis (Leukocytes, Basophils, Eosinophils, and RDW), some additional validation steps should be performed to investigate the predictive value of this parameters in facilitating COVID-19 diagnosis. Hence, considering the potential and limitations of this present work, the following topics can be proposed as next steps in future studies: (i) applying new normalization process and incorporation of new analysis aiming to identify the range of values and, if possible, with more clustering organization; and (ii) proposing a conceptual data mining framework for diagnosing diseases using more accurate classificatory analysis. In this sense, we consider for further studies choose or work with balanced dataset, principally to researches that attempt to explore classificatory algorithms with hemogram row data from hospitalized patients.
In summary, in this present work, it was demonstrated that clusters analysis with neural network SOM may be useful to support the decision-making for clinician discrimination of COVID-19. In addition, we point out some relevant variables in routine blood test that could help in the prognostic evaluation and rapid intervention for SARS-CoV-2-positive patients. Thus, it is possible to suggest briefly here that the behavior and pattern identified in the admission blood tests, more particularly for Leukocytes, Basophils, Eosinophils, and Red Blood Cell Distribution Width (RDW) signatures, can altogether assist the emergency sector in the diagnosis and prognosis, facilitating the clinical decision in the context of COVID-19.
Acknowledgements
The authors acknowledge the support of Fapesp Proc. 2015/24341-7, Hospital do Rim, Fundação Oswaldo Ramos (São Paulo, Brazil) and the Albert Einstein Hospital (São Paulo, Brazil) for providing the public database in the digital platform Kaggle
Funding
This work was partially supported by Fapesp Proc. 2015/24341-7, Brazil, and by Hospital do Rim, Fundação Oswaldo Ramos (São Paulo, Brazil).
Availability of data and material
Public data information (https://www.kaggle.com/einsteindata4u/covid19) in Kaggle platform which was available by Albert Einstein Hospital in Sao Paulo, Brazil.
Declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Code availability
The R package with SOM and LDA algorithms are available in https://github.com/Alexandra-Souza/
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s00500-024-10009-5"
Change history
8/8/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s00500-024-10009-5
References
- Adadi A, Berrada M (2018) Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 6:52138 [Google Scholar]
- Ahmad MA, Eckert C, Teredesai A (2018) Interpretable machine learning in healthcare. In: Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 559–560
- Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, García S, Gil-López S, Molina D, Benjamins R et al (2020) Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion 58:82 [Google Scholar]
- Avila E, Dorn M, Alho CS, Kahmann A (2020) Hemogram data as a tool for decision-making in COVID-19 management: applications to resource scarcity scenarios. arXiv preprint arXiv:2005.10227
- Barnes P, McFadden S, Machin S, Simson E et al (2005) The international consensus group for hematology review: suggested criteria for action following automated CBC and WBC differential analysis. Lab Hematol Off Publ Int Soc Lab Hematol 11(2):83 [Google Scholar]
- Bernardo LS, Quezada A, Munoz R, Maia FM, Pereira CR, Wu W, de Albuquerque VHC (2019) Handwritten pattern recognition for early Parkinson’s disease diagnosis. Pattern Recognit Lett 125:78. 10.1016/j.patrec.2019.04.003 [Google Scholar]
- Cabitza F, Campagner A, Ferrari D, Di Resta C, Ceriotti D, Sabetta E, Colombini A, De Vecchi E, Banfi G, Locatelli M et al (2020) Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin Chem Lab Med (CCLM) 1(ahead-of-print)
- Chouhan V, Singh SK, Khamparia A, Gupta D, Tiwari P, Moreira C, Damaševičius R, De Albuquerque VHC (2020) A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl Sci 10(2):559 [Google Scholar]
- Chunyan Z, Xin H, Jing L, Jia Z (2016) SOM neural network for clustering plant and plant stress based on kinetic imaging of chlorophyll fluorescence. In: 2016 Chinese Control and Decision Conference (CCDC), pp. 2085–2088. 10.1109/CCDC.2016.7531328
- Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, Sagnelli C, Bianchi M, Bernardini S, Ciccozzi M (2019) COVID-19 outbreak: an overview. Chemotherapy 64(5–6):215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civit-Masot J, Luna-Perejón F, Domínguez Morales M, Civit A (2020) Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci 10(13):4640 [Google Scholar]
- da Silva LA, Peres SM, Boscarioli C (2017) Introdução à mineração de dados: com aplicações em R. Elsevier, Brasil [Google Scholar]
- De Souza RW, De Oliveira JVC, Passos LA, Ding W, Papa JP, Albuquerque V (2019) A novel approach for optimum-path forest classification using fuzzy logic. IEEE Trans Fuzzy Syst 28(12):3076–3086 [Google Scholar]
- De Souza AA, Barcelos TS, Munoz R, Villarroel R, Silva LA (2019) Data mining framework to analyze the evolution of computational thinking skills in game building workshops. IEEE Access 7:82848 [Google Scholar]
- de Moraes Batista AF, Miraglia JL, Donato THR, Chiavegatto Filho ADP (2020) COVID-19 diagnosis prediction in emergency care patients: a machine learning approach. medRxiv
- Ding W, Abdel-Basset M, Eldrandaly KA, Abdel-Fatah L, de Albuquerque VHC (2020) Smart supervision of cardiomyopathy based on fuzzy Harris Hawks optimizer and wearable sensing data optimization: a new model. IEEE Trans Cybern
- Dourado CM, Da Silva SPP, Da Nóbrega RVM, Rebouças Filho PP, Muhammad K, De Albuquerque VHC (2020) An open IoHT-based deep learning framework for online medical image recognition. IEEE J Sel Areas Commun 39(2):541–548 [Google Scholar]
- Duran-Lopez L, Dominguez-Morales JP, Corral-Jaime J, Vicente-Diaz S, Linares-Barranco A (2020) COVID-XNet: a custom deep learning system to diagnose and locate COVID-19 in chest X-ray images. Appl Sci 10(16):5683 [Google Scholar]
- Ferrari D, Motta A, Strollo M, Banfi G, Locatelli M (2020) Routine blood tests as a potential diagnostic tool for COVID-19. Clinical chemistry and laboratory medicine (CCLM) 1(ahead-of-print)
- Fidan U, Ozkan N, Calikusu I (2016) Clustering and classification of dermatologic data with self organization map (SOM) method. In 2016 Medical Technologies National Congress (TIPTEKNO), pp. 1–4. 10.1109/TIPTEKNO.2016.7863075
- George-Gay B, Parker K (2003) Understanding the complete blood count with differential. J PeriAnesthesia Nurs 18(2):96 [Google Scholar]
- Goebel R, Chander A, Holzinger K, Lecue F, Akata Z, Stumpf S, Kieseberg P, Holzinger A (2018) Explainable AI: the new 42?. In: International Cross-Domain Conference for Machine Learning and Knowledge Extraction (Springer), pp. 295–303
- Haykin S (2007) Neural networks: a comprehensive foundation. Prentice-Hall Inc, Hoboken [Google Scholar]
- Henry BM, Benoit JL, Benoit S, Pulvino C, Berger BA, Olivera MHSd, Crutchfield CA, Lippi G (2020) Red blood cell distribution width (RDW) predicts COVID-19 severity: a prospective, observational study from the cincinnati SARS-CoV-2 emergency department cohort. Diagnostics 10(9):618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Biemann C, Pattichis CS, Kell DB (2017) What do we need to build explainable AI systems for the medical domain?. arXiv preprint arXiv:1712.09923
- Holzinger A, Malle B, Kieseberg P, Roth PM, Müller H, Reihs R, Zatloukal K (2017) Towards the augmented pathologist: Challenges of explainable-ai in digital pathology. arXiv preprint arXiv:1712.06657
- Kohonen T (1982) Self-organized formation of topologically correct feature maps. Biol Cybern 43(1):59. 10.1007/BF00337288 [Google Scholar]
- Kohonen T (2013) Essentials of the self-organizing map. Neural Netw 37:52. 10.1016/j.neunet.2012.09.018 [DOI] [PubMed] [Google Scholar]
- Lalmuanawma S, Hussain J, Chhakchhuak L (2020) Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos, Solitons & Fractals, p 110059
- Lippi G, Henry BM, Sanchis-Gomar F (2020) Red blood cell distribution is a significant predictor of severe illness in coronavirus disease 2019. Acta Haematol pp. 1–5
- Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C (2020) Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta 505:172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London AJ (2019) Artificial intelligence and black-box medical decisions: accuracy versus explainability. Hastings Center Rep 49(1):15 [Google Scholar]
- Mapari S, Chaudhary N, Naik S, Metkewar P (2017) Usage of fuzzy rule and SOM based model to identify a handwritten chemical symbol or structures. In: 2017 Second International Conference on Electrical, Computer and Communication Technologies (ICECCT), pp. 1–4. 10.1109/ICECCT.2017.8117842
- Matsuda N, Tokutaka H, Sato H, Tajima F, Kawata R (2017) Applying the significance degree by SOM to image analysis of fundus using the filter bank. In: 12th International Workshop on Self-Organizing Maps and Learning Vector Quantization. Clustering and Data Visualization (WSOM) 2017:1–6. 10.1109/WSOM.2017.8020011
- McGarry J (2002) Practical software measurement: objective information for decision makers. Addison-Wesley Professional, Boston [Google Scholar]
- Mei PA, de Carvalho Carneiro C, Kuroda MC, Fraser SJ, Min LL, Reis F (2017) Self-organizing maps as a tool for segmentation of magnetic resonance imaging (mri) of relapsing-remitting multiple sclerosis. In: 2017 12th International Workshop on Self-Organizing Maps and Learning Vector Quantization, Clustering and Data Visualization (WSOM) (IEEE), pp. 1–7
- Minonzio JG, Cataldo B, Olivares R, Ramiandrisoa D, Soto R, Crawford B, De Albuquerque VHC, Munoz R (2020) Automatic classifying of patients with non-traumatic fractures based on ultrasonic guided wave spectrum image using a dynamic support vector machine. IEEE Access. 10.1109/ACCESS.2020.3033480 [Google Scholar]
- Moreira LJ, Silva LA (2017) Prototype generation using self-organizing maps for informativeness-based classifier. Comput Intell Neurosci
- Mote SR, Baid UR, Talbar SN (2017) Non-negative matrix factorization and self-organizing map for brain tumor segmentation. In: 2017 International Conference on Wireless Communications, Signal Processing and Networking (WiSPNET) (IEEE), pp. 1133–1137
- Muhammad K, Khan S, Del Ser J, de Albuquerque VHC (2020) Deep learning for multigrade brain tumor classification in smart healthcare systems: a prospective survey. IEEE Trans Neural Netw Learn Syst
- O’Driscoll P, Merenyi E, Grossman R (2017) Using spatial characteristics to aid automation of SOM segmentation of functional image data. In: 12th International Workshop on Self-Organizing Maps and Learning Vector Quantization. Clustering and Data Visualization (WSOM) 2017:1–8. 10.1109/WSOM.2017.8020012
- Ohata EF, Bezerra GM, de Albuquerque VHC, Reboucas Filho PP (2020) Automatic detection of COVID-19 infection using chest X-ray images through transfer learning. IEEE/CAA J Autom Sin 8(1):239–248 [Google Scholar]
- Olivares R, Munoz R, Soto R, Crawford B, Cárdenas D, Ponce A, Taramasco C (2020) An optimized brain-based algorithm for classifying Parkinson’s disease. Appl Sci 10(5):1827. 10.3390/app10051827 [Google Scholar]
- Omara H, Lazaar M, Tabii Y (2017) Classification of breast cancer with improved self-organizing maps. In: Proceedings of the 2nd international Conference on Big Data, Cloud and Applications, pp. 1–6
- Orjuela-Cañón AD, Mendoza JEC, García CEA, Vela EPV (2018) Tuberculosis diagnosis support analysis for precarious health information systems. Comput Methods Programs Biomed 157:11 [DOI] [PubMed] [Google Scholar]
- Osman J, Lambert J, Templé M, Devaux F, Favre R, Flaujac C, Bridoux D, Marque-Juillet S, Bruneel F, Mignon F et al (2020) Rapid screening of COVID-19 patients using white blood cell scattergrams, a study on 381 patients. Br J Haematol 190(5):718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossani PC, Cirillo MA, Ossani MPC (2020) Package MVar. pt., J Stat Softw
- Pisano B, Teixeira CA, Dourado A, Fanni A (2019) Application of self-organizing map to identify nocturnal epileptic seizures. Neural Comput Appl pp. 1–17
- Rai A (2020) Explainable AI: from black box to glass box. J Acad Mark Sci 48(1):137 [Google Scholar]
- Raja AT, Alshamsan A, Al-Jedai A (2020) Status of the current COVID-19 vaccine candidates: implications in the Saudi population. Saudi Pharm J
- Rodrigues MB, Da Nóbrega RVM, Alves SSA, Rebouças Filho PP, Duarte JBF, Sangaiah AK, De Albuquerque VHC (2018) Health of things algorithms for malignancy level classification of lung nodules. IEEE Access 6:18592
- Samek W, Wiegand T, Müller KR (2017) Explainable artificial intelligence: understanding, visualizing and interpreting deep learning models. arXiv preprint arXiv:1708.08296
- Santos MA, Munoz R, Olivares R, Rebouças Filho PP, Del Ser J, de Albuquerque VHC (2020) Online heart monitoring systems on the internet of health things environments: a survey, a reference model and an outlook. Inf Fusion 53:222 [Google Scholar]
- Selvachandran G, Quek SG, Lan LTH, Giang NL, Ding W, Abdel-Basset M, Albuquerque VHC et al (2019) A new design of Mamdani complex fuzzy inference system for multi-attribute decision making problems. IEEE Trans Fuzzy Syst
- Soares F, Villavicencio A, Fogliatto FS, Rigatto MHP, Anzanello MJ, Idiart M, Stevenson M (2020) A novel specific artificial intelligence-based method to identify COVID-19 cases using simple blood exams. medRxiv
- Spinner CD, Gottlieb RL, Criner GJ, López JRA, Cattelan AM, Viladomiu AS, Ogbuagu O, Malhotra P, Mullane KM, Castagna A et al (2020) Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. Jama 324(11):1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syeda HB, Syed M, Sexton KW, Syed S, Begum S, Syed F, Yu F (2020) The role of machine learning techniques to tackle COVID-19 crisis: a systematic review. JMIR Med Inf 9(1): [Google Scholar]
- Tan PN, Steinbach M, Kumar V (2016) Introduction to data mining. Pearson Education India, New York [Google Scholar]
- Tjoa E, Guan C (2019) A survey on explainable artificial intelligence (xai): towards medical xai. arXiv preprint arXiv:1907.07374
- Tomazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, Avezum A, Lopes RD, Bueno FR, Silva MVA et al (2020) Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. Jama 324(13):1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usul E, Şan İ, Bekgöz B, Şahin A (2020) Role of hematological parameters in COVID-19 patients in the emergency room. Biomark Med 14(13):1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens L (2007) Self- and super-organising maps in R: the kohonen package. J Stat Softw 21(5):1–19 [Google Scholar]
- Wilkinson T, Dixon R, Page C, Carroll M, Griffiths G, Ho LP, De Soyza A, Felton T, Lewis KE, Phekoo K et al (2020) ACCORD: a multicentre, seamless, phase 2 adaptive randomisation platform study to assess the efficacy and safety of multiple candidate agents for the treatment of COVID-19 in hospitalised patients: a structured summary of a study protocol for a randomised controlled trial. Trials 21(1):1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ruan W, Wang J, Zheng D, Li S, Chen J, Li K, Chai X, Helal S (2020) Interpretable machine learning for covid-19: an empirical study on severity prediction task
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Public data information (https://www.kaggle.com/einsteindata4u/covid19) in Kaggle platform which was available by Albert Einstein Hospital in Sao Paulo, Brazil.