Abstract
Purpose
Time is a critical factor in drug action. The duration of inhibition of the target or residence time of the drug molecule on the target often guides drug scheduling.
Methods
The effects of time on the concentration-dependent cytotoxicity of approved and investigational agents [300 compounds] were examined in the NCI60 cell line panel in 2D at 2, 3, 7 and in 3D 11 days.
Results
There was a moderate positive linear relationship between data from the 2-day NCI60 screen and the 3-, 7- and 11-day and a strong positive linear relationship between 3-, 7- and 11-day luminescence screen IC50s by Pearson correlation analysis. Cell growth inhibition by agents selective for a specific cell cycle phase plateaued when susceptible cells were growth inhibited or killed. As time increased the depth of cell growth inhibition increased without change in the IC50. DNA interactive agents had decreasing IC50s with increasing exposure time. Epigenetic agents required longer exposure times; several were only cytotoxic after 11 days’ exposure. For HDAC inhibitors, time had little or no effect on concentration response. There were potency differences amongst the three BET bromodomain inhibitors tested, and an exposure duration effect. The PARP inhibitors, rucaparib, niraparib, and veliparib reached IC50s < 10 μM in some cell lines after 11 days.
Conclusions
The results suggest that variations in compound exposure time may reflect either mechanism of action or com-pound chemical half-life. The activity of slow-acting compounds may optimally be assessed in spheroid models that can be monitored over prolonged incubation times.
Keywords: CxT, NCI60, Concentration times time, Epigenetic agents
Introduction
Frequently, the cytotoxicity of anticancer drugs can be estimated as the product of concentration and time for direct cytotoxins such as DNA-damaging agents. However, for cell cycle-specific agents, maximal cytotoxicity is dependent on cellular generation/doubling time [1, 2]. Since anticancer therapeutics have moved away from cytotoxic agents to epigenetic modifiers and targeted drugs, the effect of time on cellular response needs to be re-examined. Cell cycle selectivity is an important factor in understanding the effect of time on cytotoxicity. Drugs such as methotrexate and cytosine arabinoside are specific for cells in S-phase; while drugs such as vincristine and paclitaxel are specifically cytotoxic toward cells in M-phase. Most drugs are more cytotoxic toward cells in exponential growth than toward plateau or stationary phase cells.
Many approved drugs are relatively stable in cell culture medium but have variable half-lives in human circulation (Supplemental Table 1; [3, 4]). Doxorubicin, actinomycin D, bleomycin, and vinblastine were stable in cell culture media for up to 10 days; however, these drugs have circulating half-lives in humans of 14.2, 14, 4, and 2.9 h, respectively. In cell culture media, etoposide lost 60% activity in 10 days; by contrast, the circulating half-life for etoposide in humans is 3.62 h [5]. By HPLC, mitomycin C was stable for 7 days at 37 °C in cell culture media but doxorubicin had a half-life of 10–20 h under the same conditions [6]. In human circulation, mitomycin has a half-life of 0.81 h and doxorubicin of 14.2 h. Cisplatin had a half-life of 48 h in cell culture media due to chemical reactivity and has a half-life of 0.44 h in human circulation [7, 8]. Decitabine, on the other hand, was relatively stable in cell culture media but has a plasma half-life of 20 min in humans’ due to high levels of liver cytidine deaminase which metabolizes the drug to an inactive species [9]. The hydroxamic acid class of histone deacetylase inhibitors including vorinostat, belinostat, and panobinostat had very limited stability (1 day) in aqueous solution, were not stable in human plasma but had better stability in human serum (Supplemental Table 1; [10–12]). The mercaptoacetamide class of histone deacetylase inhibitors exemplified by romidepsin was, in general, more stable in aqueous solution, and plasma than hydroxamic acids [13].
The current study explores the effect of exposure time (2–11 days) on the cytotoxicity and growth inhibition of several classes of anticancer agents.
Material and methods
Cell lines
NCI60 cell lines were obtained from the NCI Developmental Therapeutics Program Tumor Repository. For each lot of cells, the Repository performed Applied Biosystems Amp-FLSTR Identifiler testing with PCR amplification to confirm consistency with the published.
Identifiler STR profile for the given cell line [14, 15]. Each cell line was tested for mycoplasma when it was accepted into the repository; routine mycoplasma testing of lots was not performed. Cells were kept in continuous culture for no more than 20 passages. The optimal seeding densities for each of the cell lines for each time point assessed were determined prior to performing the concentration response studies [16].
Compounds
Three hundred compounds were selected for the assays from the FDA-approved anticancer drugs (available from NCI at: https://dtp.nci.nih.gov/branches/dscb/oncology_drugset_explanation.html) and an investigational agent library, acquired by synthesis or purchase.
NCI60 screen
The cell lines were grown in RPMI 1640 medium (Life Technologies, Gibco) containing 5% fetal bovine serum (Life Technologies, HyClone, Logan, UT) and 2 mM L-glutamine. For experiments, cells were plated in 96-well plates in 100 μl at cell numbers ranging from 5 × 103 to 4 × 104 cells/well. The plates were incubated at 37 °C in humid 5% CO2, for 24 h prior to compound addition. Compounds in DMSO were added at five concentrations from 100 to 0.01 μM. Following compound addition, the plates were incubated for 48 h at 37 °C, in humidified incubators with 5% CO2. For adherent cells, the assay was terminated by addition of cold TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The plates were washed with water and air-dried. Sulforhodamine B (SRB) solution (100 μl) at 0.4% (w/v) in 1% acetic acid was added, and plates were incubated for 10 min at room temperature. After staining, unbound dye was removed by washing with 1% acetic acid and the plates were air dried. Bound stain was solubilized with 10 mM trizma base, and the absorbance read at 515 nm. For suspension cells, the methodology was the same except that the assay was terminated by fixing settled cells at the bottom of the wells by gently adding 50 μl of 80% TCA (final concentration, 16% TCA).
3- and 7-day monolayer screen
12 cell lines (11 lines and A549/ATCC) were screened per run. Day 1, the cells were collected and suspended in 300 ml of media (RPMI 1640 supplemented with 5% FBS and 2 mM L-glutamine), then plated in 42 μl in 384-well plates (CulturPlates, PE, Waltham, MA) using a Tecan Freedom Evo. After incubation overnight, the Tecan Evo was used for compound addition. Each compound was added at 9 concentrations (1.5 nM–10 μM; DMSO concentration 0.25%), then the plates were incubated for 3 or 7 days. The incubation was terminated by adding Cell Titer Glo (Promega, Madison, WI), and luminescence was determined. All assays were performed in triplicate.
11-day spheroid screen
All steps in the assay were performed using an automated high-throughput screening system consisting of two robotic arms interfaced with liquid handlers, pipettors, automated CO2 incubators, dispensers, shakers and plate readers. The screen was controlled via Momentum scheduling software (Thermo, Waltham, MA). Day 0: cells were plated in 384-well Ultra Low Attachment (ULA) plates (Corning, NY) at densities ranging from 1 × 103 to 2 × 104 in RPMI media containing 10% fetal bovine serum in a volume of 42 μL per well [16]. Spheroid formation and morphology were monitored using brightfield imaging on a Cytation 3 high-content imaging system (BioTek, Winooski, VT) equipped with a 4 × objective. Images were captured digitally. 72 h after plating, compounds were added to each well at final concentrations below the Cmax and at half log10 dilutions from the high concentration. Control wells were included on each plate for the concentration response for each single agent being evaluated. The plates were incubated at 37 °C in a humidified atmosphere with 5% CO2 for 11 days. Then, Cell Titer Glo 3D (Promega, Madison, WI) was added to each well, the plates were shaken, incubated at room temperature for 20 min and luminescence (measuring ATP) was determined using Enspire plate readers (PE, Waltham, MA).
Data analysis
The concentration response data were analyzed using a 4-parameter sigmoidal curve fit and IC50 concentrations in micromolar were determined for each assay condition. Initially, treated/control (T/C) was calculated for each compound in each cell line from absorbance or luminescence measurements. The T/C % vs. Log(C) curves were fitted and plotted using the drc R package [17]. When multiple experiments were available for a compound/cell line pair, all data from these experiments were combined to fit a single curve. The lower and upper limits of curves were constrained to ≥ 0 and ≤ 120, respectively. The IC50s were calculated from the fitted curves where the T/C was equal to 50% and only interpolated IC50s were considered in the study (i.e., no extrapolated IC50).
Data reliability was assessed using Pearson correlation comparisons at IC50 values determined under each of the four experimental conditions. Stepwise refinements in the comparisons were: step 1, include only compounds which had IC50s under each of the 4 conditions; step 2, include compounds which had IC50s between 1 nM and 10 μM under each of the 4 conditions (2-day, 3-day and 7-day monolayer and 11-day spheroid screens); step 3, include only compounds for which the fitted response curves had an upper–lower range of ≥ 70% under each of the 4 conditions; and step 4, include only compounds for which the p-values of the EC50 were ≤ 0.2. EC50 is determined as the 50% point on a concentration response curve generated even if the data do not cover the range from 0 to 100%. IC50 determination requires a full concentration response data set [17]. All IC50s and other relevant data are available online at: https://brb.nci.nih.gov/ETvsCT/.
Results
The NCI60 screen is the best known and most widely used compound cell-based screen to determine potential anticancer activity [18–20]. The screen includes human tumor cell lines from nine major tumor types and uses a sulforhodamine B visible absorption endpoint. The NCI60 screen reflects the state-of-the art at its inception when fast-acting DNA cross-linking agents and DNA strand-breaking agents were prominent and therefore, a 2-day exposure to the test compounds was appropriate. The current study was undertaken using the NCI60 panel using longer exposure times (3, 7, and 11 days) and a luminescence readout. There are several caveats including that the optimal cell densities for each time point and culture condition were determined prior to compound testing. In the spheroid condition, spheroid morphologies ranged from densely packed to more loosely clustered and to maintain viability, 10% FBS was used [16]. Of the 320 compounds assayed under four conditions, 295 had complete data sets including concentration response curves in at least 55 cell lines at all time points. Thus, the initial data analysis set was comprised of 16,225 concentration response curves. Pairwise comparisons were made at the IC50 concentrations for each compound at varied exposure times. There was a moderate positive linear relationship between IC50s derived after 2-day exposure and after 3-day exposure at the step 1 stringency (r = 0.527) (Table 1). As the stringency of the analysis was increased, the Pearson correlation increased and at the highest stringency (step 4), there was a strong positive linear relationship between the log IC50s from the 2-day exposure screen and the 3-day exposure screen (r = 0.752). The IC50 correlations decreased but remained moderate as the 2-day screen log IC50s were compared with 7-day and 11-day exposure screen IC50s. Strong positive linear relationships were found when the 3-day IC50s were compared with the 7-day and 11-day log IC50s (r = 0.751–0.875) (Table 1). Finally, there was a strong positive linear relationship between the 7-day screen log IC50s and the 11-day screen log IC50s (r = 0.820–0.869).
Table 1.
IC50 pairwise Pearson correlations for each of the compound exposure times
| Exposure times, days vs days | Step 1 correlation (compound-cell line pairs) | Step 2 correlation (compound-cell line pairs) | Step 3 correlation (compound-cell line pairs) | Step 4 correlation (compound-cell line pairs) |
|---|---|---|---|---|
| 2 vs 3 | 0.527 (5996) | 0.687 (4728) | 0.706 (3185) | 0.752 (1526) |
| 2 vs 7 | 0.421 (5180) | 0.614 (4059) | 0.623 (2801) | 0.645 (1232) |
| 2 vs 11 | 0.385 (5602) | 0.511 (4028) | 0.484 (2877) | 0.562 (1150) |
| 3 vs 7 | 0.838 (5557) | 0.838 (5557) | 0.870 (3945) | 0.875 (2720) |
| 3 vs 11 | 0.751 (5189) | 0.751 (5189) | 0.773 (3667) | 0.790 (2170) |
| 7 vs 11 | 0.820 (5199) | 0.820 (5199) | 0.849 (3982) | 0.869 (2484) |
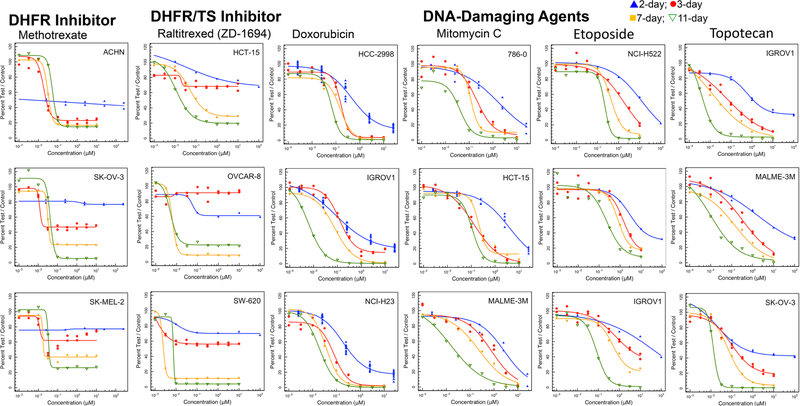
Methotrexate is actively transported into cells and inhibits the enzyme dihydrofolate reductase which is present as a single copy in most cells. Therapeutic benefit arises, in part, because cells in S-phase (i.e., dividing cells), are most vulnerable to dihydrofolate reductase inhibition [21]. In the NCI60 cell panel, methotrexate has a concentration response pattern characteristic of a cytotoxic agent selective for a specific phase of the cell cycle with a plateau when susceptible cells have been growth inhibited (Fig. 1). In some cell lines this occurs at less than 50% growth inhibition and in some lines an IC50 is reached. As the exposure time increased from 2 to 3 to 7 to 11 days the depth of cell growth inhibition of the concentration response curves increased without change in the IC50. A similar pattern was observed with the thymidylate synthase inhibitor raltitrexed. Examining the NCI60 panel median and mean log IC50 values for methotrexate and raltitrexed indicated that after 2-days or 3-days exposure to the drug, the mean log IC50 data were skewed to the left by low IC50 values causing the mean to be 2-logs lower than the median (Supplemental Fig. 1). By 7-days exposure the median and mean log IC50 values were the same. This pattern is consistent with the S-phase specificity of cell growth inhibition by antifolate these drugs.
Fig. 1.
Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the DHFR inhibitor methotrexate, TS inhibitor raltitrexed, and DNA-damaging agents doxorubicin, mitomycin C, etoposide and topotecan
Doxorubicin, a DNA-intercalating topoisomerase 2 inhibitor, produced increasing cytotoxicity with increasing exposure time with a shift in the IC50 to greater potency as time was increased. A similar pattern was observed with the DNA alkylating agent mitomycin C. The IC50 for the topoisomerase 2 inhibitor, etoposide in the NCI60 panel decreased with increasing duration of exposure up to 11 days. A similar pattern was observed with the topoisomerase 1 inhibitor topotecan (Fig. 1). There was good coincidence between the NCI60 panel median and mean log IC50 concentrations over the time course of etoposide and topotecan exposure examined.
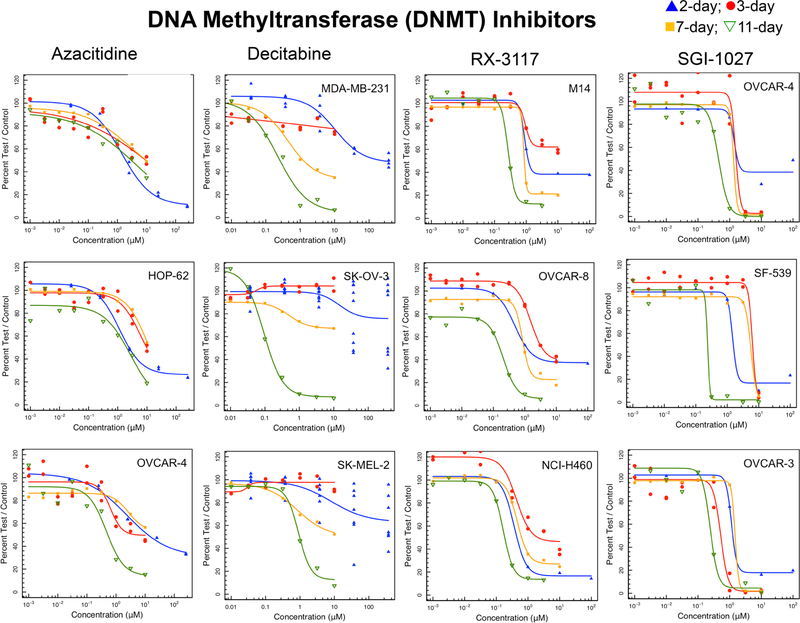
Inhibition of DNA methyltransferases (DNMT1, 3A and 3B) resulting in DNA methylation changes is a major epigenetic target [22–24]. The cytidine nucleoside analogs azacytidine and decitabine cause DNA hypomethylation and DNA damage [25]. With short exposure times (2 or 3 days) decitabine either did not reach an IC50 or only reached an IC50 at concentrations > 10 μM (Fig. 2). Decitabine was only reliably cytotoxic after 11 days exposure. Azacitidine was a more potent cytotoxic agent, reaching greatest potency at the 11-day exposure time. SGI-1027 is a quinoline-based DNA hypomethylating agent which inhibits DNMTs by competing with the cofactor S-adenosyl-methionine [26]. SGI-1027 had a very steep concentration response curve pattern and like the other DNMT inhibitors, demonstrated increased potency after 11 days of exposure. RX-3117 is a cytidine analog, which produces DNA hypomethylation and is also incorporated into RNA and DNA [27]. The IC50s for RX-3117 decreased with increasing duration of exposure demonstrating a pattern somewhat different from the other DNA methyl transferase inhibitors. The NCI60 panel median and mean log IC50 concentrations were in good agreement over the time course of drug exposure.
Fig. 2.
Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the DNA methyltransferase inhibitors RX-3117, SGI-1027, azacytidine and decitabine
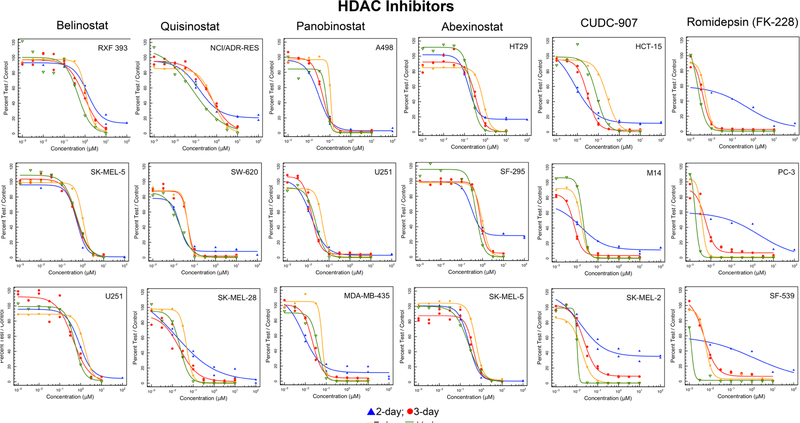
Histone deacetylases are a family of enzymes which remove acetyl groups from histone proteins modifying chromatin between relaxed and condensed forms [28, 29]. Concentration response data for five hydroxamic acid histone deacetylase inhibitors and one mercaptoacetamide histone deacetylase inhibitor are shown in Fig. 3. Hydroxamic acid drugs/investigational agents are not stable in aqueous milieu, therefore, it is not surprising that extending the duration of exposure to the compounds had little effect on concentration response. Romidepsin (FK-228; depsipeptide) had a pattern of response different from the hydoxamic acids (Fig. 3). Results from the 3-, 7- and 11-day screens which had an ATP-content endpoint demonstrated decreasing IC50 with increasing duration of drug exposure. Curve shape and IC50s were distinguished from the 2-day data generated with a sulforhodamine B readout which quantifies cellular protein.
Fig. 3.
Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the histone deacetylase (HDAC) inhibitors belinostat, quisinostat, panobinostat, abexinostat, JNJ-16241199, and romidepsin
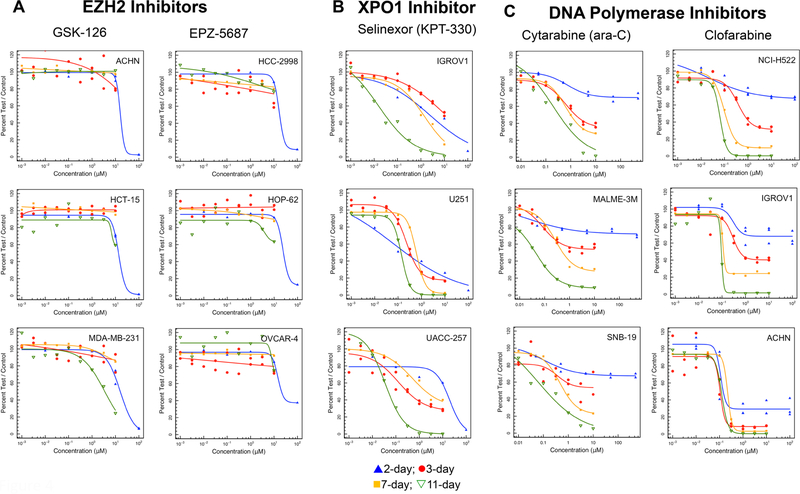
Histone methyltransferases are enzymes involved in covalent histone modification and consequently in gene expression and cell fate [30]. Of interest in this family is EZH2, a histone methyl transferase component of Polycomb repressive complex 2 (PRC2). EZH2 is mutated in some cancers and is often over-expressed in cancer [31]). There are no EZH2 mutated cell lines in the NCI60 panel. The highest gene expression (log2) occurs in hematologic cancer lines. In the 2-day exposure screen of EZH2 inhibitors, IC50s for both GSK-126 and EPZ-5687 were > 10 μM (Fig. 4a). Many lines did not reach an IC50 even after an 11-day exposure to either EZH2 inhibitor.
Fig. 4.
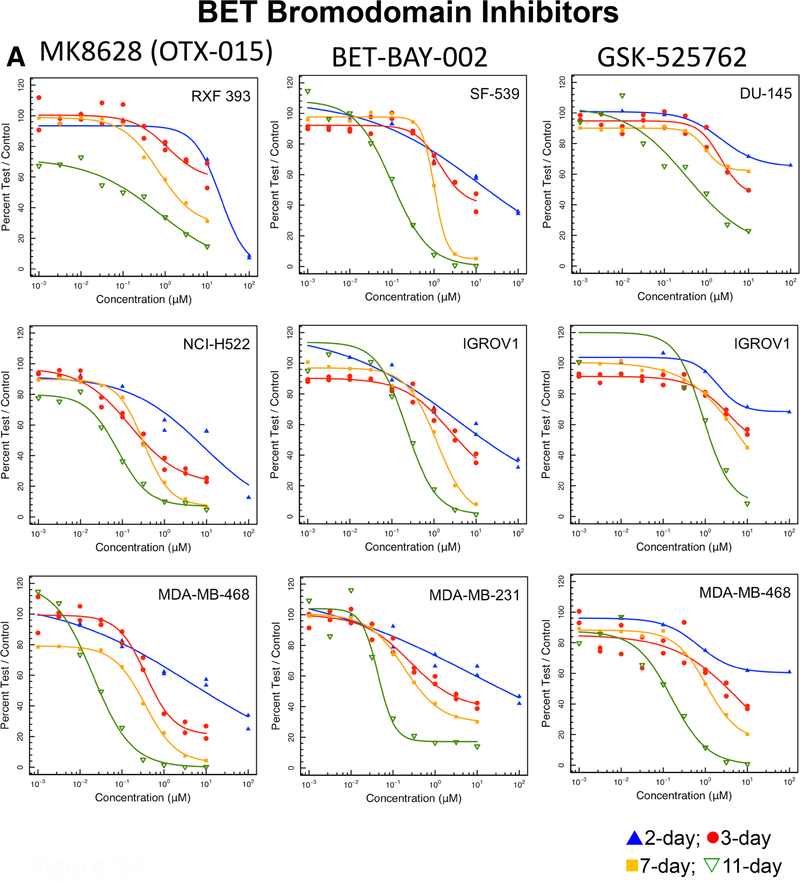
a Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the EZH2 inhibitors GSK-126 and EPZ-5687, the XPO1 inhibitor selinexor, and the DNA polymerase inhibitors cytarabine and clofarabine. b Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the BET bromodomain inhibitors MK8628, BET-BAY-002 and GSK525762 and the gamma-secretase inhibitors MK-0752 and LY-450139
The nuclear pore proteins which facilitate transport of nuclear export signal containing proteins from the nucleus into the cytoplasm are called exportins [32]. Chromosome Region Maintenance 1 [CRM1; also called exportin1 (XPO1)] is one of these proteins [33]. Blocking the transport of proteins from the nucleus to the cytoplasm by inhibiting CRM1/XPO1 activity is beneficial in the treatment of cancer [34]. Selinexor (KPT-330) cytotoxicity was time dependent. As the duration of exposure increased the IC50 of selinexor decreased (Fig. 4b). There was good agreement between the NCI60 panel median and mean log IC50s by selinexor over the time course studied.
Cytarabine (ara-C), a derivative of cytosine and an arabinose sugar which inhibits both DNA and RNA polymerases, intracellularly is rapidly converted to the triphosphate form which damages DNA during S-phase [35]. Cytarabine was increasingly cytotoxic as the duration of drug exposure increased. In most NCI60 cell lines, IC50s were reliably reached after 7 days of drug exposure (Fig. 4c). The activity of clofarabine involves inhibition of DNA synthesis, inhibition of ribonucleotide reductase, and direct induction of apoptosis. Clofarabine competes with dATP for binding to DNA polymerase-α and −ε [36]. Clofarabine action is mainly at terminal sites inhibiting DNA elongation. Clofarabine exposure produced an S-phase growth-inhibition pattern with increased cytotoxicity and decreased IC50 as the exposure duration increased (Fig. 4c).
BET bromodomain (BRD2, 3, 4) inhibitors, target a family of transcriptional coactivators and disrupt the activity of transcription factors. BRD4 cooperates with MYC making it a desirable drug target; however, current inhibitors are non-selective and block all 3 BET proteins [37]. Each BET protein controls distinct transcription factors for functions including insulin production, T cell differentiation, and repression of viruses such as HIV. BET inhibition can reactivate HIV in human cells. Several BET bromodomain inhibitors are in clinical trial. These agents will likely be used in drug combinations [37–40]. Although there were potency differences amongst the three BET bromodomain inhibitors tested, each compound had a clear exposure duration effect (Fig. 5a). The NCI60 panel IC50 values for BET bromodomain inhibitors follow similar trends over a range of potency.
Fig. 5.
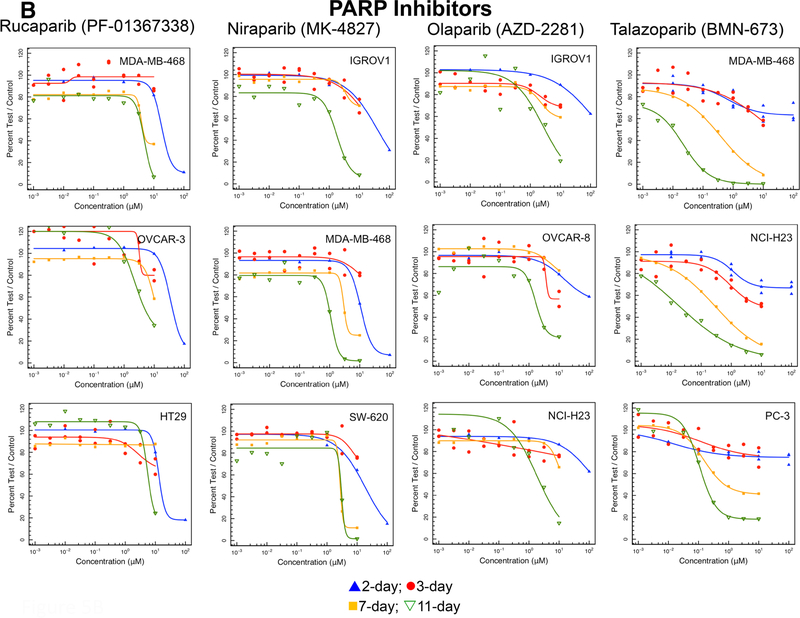
a Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the PARP inhibitors rucaparib, niraparib, olaparib, talazoparib, and veliparib. b Concentration response curves in three representative NCI60 panel cell lines after compound exposure for 2 days (blue), 3 days (red), 7 days (yellow) or 11 days (green) for the SMO inhibitors saridegib and LEQ-506 and the IAP inhibitors birinapant and LCL-161
PARP inhibitors, are NAD-mimetic compounds that prevent the PARylation that occurs in response to DNA damage. PARP inhibitors trap the PARP protein on damaged DNA, preventing binding of DNA repair proteins. The longer the duration that PARP is trapped on damaged DNA, the greater the cytotoxicity. PARP inhibition also disrupts DNA-dependent protein kinase function which is critical in the non-homologous end-joining (NHEJ) process. In patients with BRCA1 mutations, and HR-deficient tumors PARP inhibitors can be synthetically lethal. None of the NCI60 cell lines have a BRCA1/2 mutation or deficiency. There are now four FDA-approved PARP inhibitors [41, 42]. Rucaparib, niraparib, and olaparib reached IC50s at concentrations < 10 μM in some lines with a 11-day exposure (Fig. 5b). Only olaparib did not reach an IC50 after an 11-day exposure. Talazoparib was the most potent and the most time-dependent PARP inhibitor.
The human inhibitor-of-apoptosis (IAP) family has 8 members. c-IAP1, c-IAP2, and XIAP inhibit caspase-mediated apoptosis and RIP-mediated necroptosis. XIAP binds directly to caspases inhibiting their function while c-IAP1, and c-IAP2 act indirectly to prevent cell death [43–45]. The SMAC protein binds to the BIR3 and BIR2 domains of IAPs. Smac mimetics induce apoptosis in a manner dependent upon both XIAP neutralization and cancer cell autocrine tumor necrosis factor-α (TNF-α) production [43]. SMAC mimetics such as birinapant and LCL-161, bind to the BIR domains of SMAC, thus, preventing SMAC from binding to XIAP allowing apoptosis to proceed [46–48]. A longer exposure duration (11 days) markedly increased the cytotoxicity of both birinapant and LCL-161 (Supplemental Fig. 3a). Overall, the NCI60 panel mean log IC50 concentrations for birinapant and LCL-161 over time follow similar patterns with birinapant being more potent (Supplemental Fig. 2).
Kinase inhibitors, multi-kinase inhibitors and highly selective kinase inhibitors, have moved the cancer therapy field into the ‘targeted therapy’ era [49–51]. Sorafenib is a multi-kinase inhibitor, erlotinib is an EGFR inhibitor, crizotinib is an ALK, ROS1, MET, and EML4-ALK fusion inhibitor and nintedanib is a VEGFR, FGFR, and PDGFR inhibitor (Supplemental Fig. 3b). Erlotinib is the least potent among these compounds. In cell culture, exposure duration has little, if any, effect on IC50 or cell growth inhibition by these agents which would be expected to be generally stable in aqueous media. There are no lines in the NCI60 panel with EGFR mutations or amplifications. With serine–threonine intracellular kinases, there was increasing cell growth inhibition over the time course of the study (Supplemental Fig. 3c). Among these agents, the monopolar spindle 1 kinase inhibitor, BAY-1217389, has had a pronounced cytotoxicity time effect. Inhibiting the activity of MPS-1 blocks the spindle-assembly checkpoint, accelerating the mitotic process resulting in chromosome misalignment and destabilizing the mitosis [52, 53]. Comparing the NCI60 panel mean log IC50 concentrations over the time course of the study, the MPS-1 inhibitor was a more potent cytotoxic agent than the EGFR inhibitor, the FAK inhibitor or the AKT inhibitor (Supplemental Fig. 2).
Discussion
The response of cells in culture to drugs and compounds is dependent upon many factors. Most cell culture experiments are performed with cells in exponential growth and, thus, may overestimate the cytotoxicity that some compounds may be expected to achieve in in vivo tumor models. Solid tumors have low growth fractions and are likely to contain large populations of noncycling cells. In cell culture, these noncycling cells can be modeled using stationary phase cultures containing a large fraction of noncycling but potentially clonogenic cells [54, 55]. In 96-well or 384-well cell culture plates, monolayer cultures tend to move toward stationary phase with increasing incubation times which may provide a better estimate of compound cytotoxicity.
However, another important factor is drug/compound stability in cell culture medium and biological fluids. Supplemental Table 1 provides the clinical T1/2 (h) and the clinical Cmax concentration (μM) for 25 FDA-approved drugs [3]. The shortest circulating half-life is that of azacytidine with a T1/2 of 21 min and the longest is that of niraparib with a T1/2 of 36 h. The topoisomerase I inhibitor, topotecan has the lowest clinical Cmax concentration of 15 nM which may limit the clinical effectiveness of this drug. By contrast, another topoisomerase I inhibitor irinotecan has a clinical Cmax of 5.78 micromolar and its active metabolite SN-38 has a clinical Cmax of 143 nM. The DNA polymerase inhibitor cytarabine (araC) has the highest clinical Cmax concentration at 54 micromolar. Stability and solubility of the compound in aqueous solution are important for performance in cell culture. FDA-approved drugs range from an aqueous solution half-life of minutes to complete stability in aqueous solution, therefore, stability in aqueous solution is not a drug requirement. The presence (and percent) or absence of serum and DMSO often have a major impact on the response of cells in culture to compounds especially compounds with limited aqueous solubility. Cell culture likely over-estimates the cytotoxicity of compounds that undergo metabolism in the liver to inactive metabolites and under-estimates the cytotoxicity of compounds which are prodrugs requiring liver metabolism to generate an active species [56, 57].
For more than 20 years cancer drug discovery has focused on molecularly targeted anticancer drugs, the largest class being tyrosine kinase inhibitors, which have greater cytotoxicity toward cells, tumor and normal, that express the molecular target of the drug. More recently, discovery of targeted agents has focused on mutated molecular targets; thus, focusing cytotoxicity further with the expectation that only malignant cells would express the mutated molecular target and be susceptible to the drug. In addition, there has been renewed interest in epigenetic targets that are expressed by all cells. Some classes of epigenetics agents require long exposure times to manifest cytotoxicity. These agents indirectly target cellular DNA as opposed to classic cytotoxic agents that directly target cellular DNA. DNA methyltransferases maintain DNA methylation which represses the expression of the target genes. The classic DNMT inhibitors azacytidine and decitabine have a long history as successful epigenetic drugs. These compounds are incorporated into replicating DNA and bind covalently to DNMTs thus depleting these enzymes in the cell [58]. Both azacytidine and decitabine have pronounced time effects in achieving cytotoxicity in cells (Fig. 2). The newer DNMT inhibitors, RX-3117 and SGI-1027, have steep sigmoidal concentration response curves tending to indicate that these compounds have specific molecular targets in cells (Fig. 2). Histone acetylation is involved in the control of gene expression. Histone deacetylase inhibitors have limited single-agent activity in most cancers and are approved for the treatment of cutaneous or peripheral T-cell lymphomas and multiple myeloma [59]. The histone deacetylase inhibitors tested are chemically unstable in aqueous media, and there is no effect of time on the cytotoxicity of these compounds. The histone deacetylase inhibitors tested were potent cytotoxic agents with panobinostat and romidepsin being the most potent (Fig. 3). Bromodomain-contacting proteins bind with acetylated histones in the chromatin and are involved in transcriptional activation [60]. The bromodomain-containing BRD proteins are important for the expression of genes with highly acetylated promoters. These sites are the targets of the BET bromodomain inhibitors. The three BET bromodomain inhibitors tested had exposure duration effects that corresponded to decreasing IC50s over time. The concentration response curves for these compounds were broad tending to indicate the potential for more than one molecular target for the compounds (Fig. 5a). The two EZH2 histone methyltransferase inhibitors studied, were only effective at concentrations greater than 10 μM even after 11-days exposure (Fig. 4a). None of the cell lines tested had mutant EZH2.
There are three FDA-approved PARP inhibitors, rucaparib, niraparib, and olaparib. PARP protein is involved in the repair of nicks in DNA. Cells with mutations in the homologous recombination repair proteins including BRCA1, BRCA2 or PALB2 are susceptible to PARP inhibitors as single agents [61]. Olaparib was approved for the treatment of patients with germline BRCA-mutated advanced ovarian cancer in 2014. Rucaparib was granted accelerated approval for previously treated BRCA-mutant ovarian cancer in 2016, and in 2017, niraparib was approved for treatment of epithelial ovarian cancer, fallopian tube cancer, and primary peritoneal cancer. Talazoparib and veliparib remain under active clinical investigation. Although none of the cell lines tested in the current study harbor BRCA or PALB2 mutations, the three approved PARP inhibitors were modestly cytotoxic upon long duration exposure. Veliparib was the least cytotoxic PARP inhibitor and talazoparib was the most potent PARP inhibitor (Fig. 5b).
Cancer genomes have many genetic alterations that impact protein kinase signaling networks [62, 63]. Some of these alter protein kinase structure and/or expression. Erlotinib (an EGFR kinase inhibitor) was approved in 2004 for maintenance treatment of patients with locally advanced or metastatic non-small cell lung cancer whose disease had not progressed after first-line chemotherapy, treatment of locally advanced or metastatic NSCLC after failure of one prior chemotherapy regimen, and treatment of patients with locally advanced, unresectable or metastatic pancreatic cancer, along with gemcitabine. Approved in 2005, sorafenib is a broad-spectrum kinase inhibitor indicated for the treatment of impairment of TSH suppression in differentiated thyroid cancer, unresectable hepatocellular carcinoma and advanced renal cell carcinoma. Crizotinib, approved in 2011, is a kinase inhibitor indicated for the treatment of metastatic NSCLC with anaplastic lymphoma kinase (ALK) or ROS1-activation. Nintedanib is a broad-spectrum kinase inhibitor that was approved in 2014 for the treatment of idiopathic pulmonary fibrosis. Nintedanib is actively being studied in clinical trials in major solid tumors. Despite having different kinase targets and approvals in a variety of indications, these four tyrosine kinase inhibitors performed similarly in the NCI60 panel time course screen. The least potent agent, erlotinib, was the only one of these four agents that had an effect of exposure duration (Supplemental Fig. 3b). Potency was similar for the other three tyrosine kinase inhibitors. There was a clear duration of exposure effect with the three-investigational serine–threonine kinase inhibitors (Supplemental Fig. 3c). The most potent kinase inhibitor tested was BAY-1217389, a selective monopolar spindle 1 (MPS1) kinase activity inhibitor. MPS1 is expressed in actively dividing cells and overexpressed in some tumors. MPS1 is a core component of the spindle-assembly checkpoint that during metaphase monitors spindle microtubules and prevents transitions to anaphase until the chromosomes are correctly oriented. MPS1 inhibition causes chromosomal segregation errors and cell death. BAY-1217389 is in phase 1 clinical trial in combination with paclitaxel.
Time is a critical factor in drug action. The duration of inhibition of the target or residence time of the drug molecule on the target often guides drug scheduling. Some targets can be intermittently blocked and still be effective therapeutic targets. S-phase selective agents, DNA-damaging agents, DNA methyltransferase inhibitors, DNA polymerase inhibitors and intracellular serine-threonine kinase inhibitors had clear duration of exposure effects in manifesting growth inhibition. Histone deacetylase inhibitors, receptor tyrosine kinase inhibitors, EZH2 inhibitors, gamma-secretase inhibitors, and SMO inhibitors had no exposure time dependency in manifesting growth inhibition or had no effective on cell viability even after 11-day exposure. The XPO1 inhibitor, BET bromodomain inhibitors, PARP inhibitor and IAP inhibitors had moderate duration of exposure effects on cytotoxicity with the 11-day exposure, generally, showing greater growth inhibition than the 2-, 3- and 7-day exposures.
Cell culture is an important preclinical tool. Like all models, it is important to understand the limitations of cell culture and how that relates to the potential expectations from the testing compounds [1]. The NCI60 is the most highly valued and most highly understood cell culture model systems. In this report, extended exposure time was tested on a selected collection of compounds. One impetus for these studies was wide-spread interest in the use of epigenetic agents which require long exposure times, alone and combination in cancer therapeutic regimens [64]. The results varied with compound target and were found to be similar for compounds directed toward the same target. With many compounds growth inhibition increased with increasing exposure and/or with increasing compound concentration; a caveat is that these effects were observed in the absence of active metabolism by normal tissues and in the absence of disparities caused by heterogenous biodistribution; however, these studies can help to inform in vivo dosing regimens and the selection of appropriate tumor models for further testing.
Supplementary Material
Acknowledgements
This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
Footnotes
Conflicts of interest The authors state that they have no conflicts of interest to report. The study does not involve human subjects or use of animals in research.
Compliance with ethical standards
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00280-019-03863-w) contains supplementary material, which is available to authorized users.
References
- 1.Eastman A (2017) Improving anticancer drug development begins with cell culture: misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 8:8854–8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perego P, Hempel G, Linder S, Bradshaw TD, Larsen AK, Peters GJ, Phillips RM, on behalf of the EORTC PAMM Group (2018) Cellular pharmacology studies of anticancer agents: recommendations from the EORTC-PAMM group. Cancer Chemother Pharmacol 81:427–41 [DOI] [PubMed] [Google Scholar]
- 3.Liston DR, Davis M (2017) Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin Cancer Res 23:3489–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson ES, Adamson RH, Denham C, Oliverio VT (1965) The metabolic fate of tritiated methotrexate I. absorption, excretion and distribution in mice, rats, dogs and monkeys. Cancer Res 25:1008–1017 [PubMed] [Google Scholar]
- 5.Ludwig R, Alberts DS (1984) Chemical and biological stability of anticancer drugs used in a human tumor clonogenic assay. Cancer Chemother Pharmacol 12:142–145 [DOI] [PubMed] [Google Scholar]
- 6.Beijnen JH, Van der Nat JM, Labadie RP, Underberg WJ (1986) Decomposition of mitomycin and anthracycline cytostatics in cell culture media. Anticancer Res 6:39–43 [PubMed] [Google Scholar]
- 7.Niell HD, Webster KC, Smith EE (1985) Anticancer drug activity in platin in human bladder tumor cell lines. Cancer 56:1039–1044 [DOI] [PubMed] [Google Scholar]
- 8.Schuldes H, Bade S, Knobloch J, Jonas D (1997) Loss of in vitro cytotoxicity of cisplatin after storage as stock solution in cell culture medium at various temperatures. Cancer 79:1723–1728 [PubMed] [Google Scholar]
- 9.Karahoca M, Momparler RL (2013) Pharmacokinetic and pharmacodynamic analysis of 5-aza-2’-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin Epigen 5:3–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L, Musson DG, Wang AQ (2006) Stability studies of vorinostat and its two metabolites in human plasma, serum and urine. J Pharmaceut Biomed Anal 42:556–564 [DOI] [PubMed] [Google Scholar]
- 11.Plumb JA, Finn PW, Williams RJ, Bandara MJ, La Thangue NB, Brown R (2003) Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol Cancer Therap 2:721–728 [PubMed] [Google Scholar]
- 12.Wang H, Yu N, Chen D, Lee KCL, Lye PL, Chang JWW, Deng W, Ng MCY, Lu T et al. (2011) Discovery of (2E)-3-{2-butyl-1-[2-(diethylamino)ethyl]-1H-benzimidazol-5-yl}-N-hydroxyacrylamide (SB939), an orally active histone deacetylase inhibitor with a superior preclinical profile. J Med Chem 54:4694–4720 [DOI] [PubMed] [Google Scholar]
- 13.Konsoula R, Jung M (2008) In vitro plasma stability, permeability and solubility of mercaptoacetamide histone deacetylase inhibitors. Int J Pharm 361:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holbeck SL, Camalier R, Crowell JA, Govinharajulu JP, Hollingshead M, Anderson LW, Polley E, Rubenstein L, Srivastava A, Wilsker D, Collins JM, Doroshow JH (2017) The national cancer institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res 77:3564–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, Weinstein J (2009) DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther 8:713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selby M, Delosh R, Laudeman J, Ogle C, Reinhart R, Silvers T, Lawrence S, Kinders R, Parchment R, Teicher BA, Evans DM (2017) 3D models of the NCI60 cell lines for screening oncology compounds. SLAS Discovery 22:473–483 [DOI] [PubMed] [Google Scholar]
- 17.Ritz C, Baty F, Streibig JC, Gerhard D (2015) Dose-response analysis using R. PLoS ONE 10(12):e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker RH (2006) The NCI60 human tumor cell line anticancer drug screen. Nat Rev Cancer 6:813–823 [DOI] [PubMed] [Google Scholar]
- 19.Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, Gray-Goodrich M, Campbell P, Mayo J, Boyd M (1991) Feasibility of a High-flux anticancer drug screen using a diverse panel of cultured tumor cell lines. J Natl Cancer Inst 83:757–766 [DOI] [PubMed] [Google Scholar]
- 20.Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Kohn KW, Fojo T, Bates SE, Rubinstein LV, van Osdol WW, Monks AP et al. (1997) An informative-intensive approach to the molecular pharmacology of cancer. Science 275:343–349 [DOI] [PubMed] [Google Scholar]
- 21.Bertino JR (2009) Cancer research: from folate antagonism to molecular targets. Best Prac Res Clin Hematol 22:577–582 [DOI] [PubMed] [Google Scholar]
- 22.Li H, Li W, Liu S, Zong S, Wang W, Ren J, Li Q, Hou F, Shi Q (2016) DNMT1, DNMT3A and DNMT3B polymorphisms associated with gastric cancer risk: a systemic review and meta-analysis. EBioMed 13:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uysal F, Akkoyunlu G, Ozturk S (2015) Dynamic expression of DNA methyltransferase (DNMTs) in oocytes and early embryos. Biochimie 116:103–113 [DOI] [PubMed] [Google Scholar]
- 24.Fahy J, Jeltsch A, Arimondo PB (2012) DNA methyltransferase inhibitors in cancer: a chemical and therapeutic patent overview and selected clinical studies. Exp Opin Ther Patents 22:1427–1442 [DOI] [PubMed] [Google Scholar]
- 25.Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ (2010) A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS ONE 5:e9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo J, Choi S, Medina-Franco JL (2013) Molecular modeling studies of the novel inhibitors of DNA methyltransferases SGI-1027 and CBC12: implications for the mechanism of inhibition of DNMTs. PLoS ONE 8:e62152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters GJ, Smid K, Vecchi L, Kathmann I, Sarksjan D, Honeywell RJ, Losekoot N, Ohne O, Orbach A, Blaugrund E, Jeong LS, Lee YB, Ahn CH, Kim DJ (2013) Metabolism, mechanism of action and sensitivity profile of fluorocyclopentenylcytosine (RX-3117; TV-1360). Invest New Drugs 31:1444–1457 [DOI] [PubMed] [Google Scholar]
- 28.Eckschlager T, Plch J, Stiborova M, Hrabeta J (2017) Histone deacetylase inhibitors as anticancer drugs. Int J Molec Sci 18:1414–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen L, Orillion A, Pili R (2016) Histone deacetylase inhibitors as immunomodulators in cancer therapeutics. Epigenomics 8:415–428 [DOI] [PubMed] [Google Scholar]
- 30.Kim KH, Roberts CWM (2016) Targeting EZH2 in cancer. Nat Med 22:128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Italiano A (2016) Role of the EZH2 histone methyltransferase as a therapeutic target in cancer. Pharm Therap 165:26–31 [DOI] [PubMed] [Google Scholar]
- 32.Lu C, Figueroa JA, Liu Z, Konala V, Aulakhy A, Verma R, Cobos E, Chiriva-Internati M, Gao W (2015) Nuclear export as a novel therapeutic target: the CRM1 connection. Curr Drug Targets 15:575–592 [DOI] [PubMed] [Google Scholar]
- 33.El-Tanani M, Dakir E, Raynor B, Morgan R (2016) Mechanisms of nuclear export in cancer and resistance to chemotherapy. Cancers 8:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garg M, Kanojia D, Mayakonda A, Said JW, Doan NB, Chien W, Ganesan TS, Chuang LSH et al. (2017) Molecular mechanism and therapeutic implications of selinexor (KPT-330) in liposarcoma. Oncotarget 8:7521–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magina KN, Pregartner G, Zebisch A, Wolfler A, Neumeister P, Greinix HT, Berghold A, Sill H (2017) Cytarabine dose in the consolidation of AML: a systematic review and meta-analysis. Blood 130:946–948 [DOI] [PubMed] [Google Scholar]
- 36.Zhenchuk A, Lotfi K, Juliusson G, Albertioni F (2009) Mechanisms of anti-cancer action and pharmacology of clofarabine. Biochem Pharmacol 78:1351–1359 [DOI] [PubMed] [Google Scholar]
- 37.Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B (2015) Targeting BET bromodomains for cancer treatment. Epigenomics 7:487–501 [DOI] [PubMed] [Google Scholar]
- 38.Andrieu G, Belkina AC, Denis GV (2016) Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today 19:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahai V, Redig AJ, Collier KA, Eckerdt FD, Munshi HG (2016) Targeting BET bromodomain proteins in solid tumors. Oncotarget 7:53997–54009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Vakoc CR (2017) Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harbor Perspect Med 7:a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JS, O’Carrigan B, Jackson SP, Yap TA (2017) Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov 7:20–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord CJ, Ashworth A (2017) PARP inhibitors: synthetic lethality in the clinic. Science 355:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Probst BL, Liu L, Ramesh V, Li L, Sun H, Minna JD, Wang L (2010) Smac mimetics increase cancer cell response to chemotherapeutics in a TNF-α-dependent manner. Cell Death Differ 17:1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonnissen A, Isebaert S, Haustermans K (2015) Targeting the hedgehog signaling pathway in cancer: beyond smoothened. Oncotarget 6:13899–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rimkus TK, Carpenter RI, Qasem S, Chan M, Lo HW (2016) Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers 8:22–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derakhshan A, Chen Z, Van Waes C (2016) Therapeutic small molecules target inhibitor of apoptosis proteins in cancers with deregulation of extrinsic and intrinsic cell death pathways. Clin Cancer Res 23:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peery RC, Liu JY, Zhang JT (2017) Targeting survivin for therapeutic discovery: past, present, and future promises. Drug Discov Today 22:1466–1477 [DOI] [PubMed] [Google Scholar]
- 48.Altieri DC (2010) Survivin and IAP proteins in cell-death mechanisms. Biochem J 430:199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H (2016) Three generations of epidermal growth factor receptor tyrosine kinase inhibitors developed to revolutionize the therapy of lung cancer. Drug Design Dev Therapy 10:3867–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santarpia M, Ligori A, Karachaliou N, Gonzalez-Cao M, Daffina MG, D’Aveni A, Marabello G, Altavilla G, Rosell R (2017) Osimertinib in the treatment of non-small cell lung cancer: design, development and place in therapy. Lung Cancer Targets Therapy 8:109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaumann AKA, Kiefer F, Alfer J, Lang SA, Geissler EK, Breier G (2016) Receptor tyrosine kinase inhibitors: are they real tumor killers? Int J Cancer 138(540–54):52. [DOI] [PubMed] [Google Scholar]
- 52.Wengner AM, Siemeister G, Koppitz M, Schulze V, Kosemund D, Klar U, Stoeckigt D, Neuhaus R, Lienau P et al. (2016) Novel MPS1 kinase inhibitors with potent antitumor activity. Molec Cancer Therap 15:583–592 [DOI] [PubMed] [Google Scholar]
- 53.Ma WW (2011) Development of focal adhesion kinase inhibitors in cancer therapy. Anti-cancer Agents Med Chem 11:638–642 [DOI] [PubMed] [Google Scholar]
- 54.Ray GR, Hahn GM, Bagshaw MA, Kurkjian S (1973) Cell survival and repair of plateau phase cultures after chemotherapy: relevance to tumor therapy and to the in vitro screening of new agents. Cancer Chemother Rep 57:473–475 [PubMed] [Google Scholar]
- 55.Keyes K, Cox K, Treadway P, Mann L, Shih C, Faul MM, Teicher BA (2002) An in vitro tumor model: analysis of angiogenic factor expression after chemotherapy. Cancer Res 62:5597–5602 [PubMed] [Google Scholar]
- 56.Bale SS, Moore L, Yarmush M, Jindal R (2016) Emerging in vitro liver technologies for drug metabolism and inter-organ interactions. Tissue Engineer B 22:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pelkonen O, Turpeinen M, Hakkola J, Abass K, Pasanen M, Raunio H, Vahakangas K (2013) Preservation, induction or incorporation of metabolism into the in vitro cellular system—views to current opportunities and limitations. Toxicol In Vitro 27:1578–1583 [DOI] [PubMed] [Google Scholar]
- 58.Barrero ML (2017) Epigenetic strategies to boost cancer immunotherapies. Int J Mol Sci 18:1108–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halsall JA, Turner BM (2016) Histone deacetylase inhibitors for cancer therapy. An evolutionarily ancient resistance response may explain their limited success. Bioessays 38:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou MM, Dhalliun C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496 [DOI] [PubMed] [Google Scholar]
- 61.McGlynn O, Lloyd B (2002) Recombinational repair and restart of damaged replication forks. Nat Rev 3:859–870 [DOI] [PubMed] [Google Scholar]
- 62.Fleuren EDG, Zhang L, Wu J, Daly RJ (2016) The kinome ‘at large’ in cancer. Nat Rev Cancer 16:83–98 [DOI] [PubMed] [Google Scholar]
- 63.Rotow J, Bivona TG (2017) Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 17:637–658 [DOI] [PubMed] [Google Scholar]
- 64.Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S (2014) Use of epigenetic drugs in disease: an overview. Genet Epigenet 6:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.