Abstract
Neuronal circuits damaged or lost following injury can be regenerated in some adult organisms, but mechanisms enabling this process are largely unknown. We used the planarian Schmidtea mediterranea to study visual system regeneration after injury. We identify a rare population of muscle cells tightly associated with photoreceptor axons at stereotyped positions in both uninjured and regenerating animals. Together with a neuronal population, these cells promote de novo assembly of the visual system in diverse injury and eye transplantation contexts. These muscle guidepost-like cells are specified independently of eyes, and their position is defined by an extrinsic array of positional information cues. These findings provide a mechanism, involving adult formation of guidepost-like cells typically observed in embryos, for axon pattern restoration in regeneration.
One sentence summary:
Adult regeneration requires guidepost-like cells for precise wiring of the visual system in planarians.
Formation of neural circuits during development requires the orchestration of multiple events, such as the specification of neurons, the precise navigation of axons through the extracellular environment towards targets, and the generation of specific synapses. The first axons to extend in developing nervous systems, known as pioneer axons, grow along stereotyped routes and fasciculate with each other to form tracts that are followed by subsequent axons. Ablation of pioneer neurons can perturb the guidance of subsequent axons, causing delay or misrouting, although in most cases these axons still locate their final targets (1–4).
Growing axons navigate with the assistance of cell-extrinsic guidance cues, such as Netrins and their DCC/UNC-40 or UNC-5 receptors; Slits and their Robo receptors; Semaphorins and their Plexin receptors; and Ephrins and their Ephrin receptors (5–8). In addition, transient cell-cell interactions have important roles in neural circuit assembly. Such interactions involve cells referred to as guideposts. Guidepost cells, which were originally identified in the grasshopper limb (9, 10), are discrete, early born, and transient specialized cell populations located at decision or choice points along axonal trajectories (6, 11, 12). Most identified guidepost cells are either glia or neurons (11, 13–16). Upon encountering guidepost cells, axon growth cones can change their responsiveness to extracellular guidance cues and modify their trajectory (6). In most systems, positioning of guidepost cells at choice points along the axonal path is instructed by canonical axon guidance cues themselves (5–7) and proper location of guidepost cells is fundamental for precise axonal tract development (17–20). Since the discovery of guidepost cells in the grasshopper limb, guidepost cells in several other organisms were identified, including midline glia cells in Drosophila, floor plate cells in the vertebrate spinal cord, CD44+ neurons and retinal glial cells found at the developing optic chiasm in mammals (21–24).
The transient developmental programs that guide pioneer axons to their ultimate targets become dispensable once the neuronal circuit is assembled. In regenerative species, however, neuronal circuits that are lost to injury must re-establish themselves in the adult animal. This presents a puzzle: How can an adult neuronal circuit be formed de novo by regeneration in the absence of embryonic and potentially transient guidance mechanisms, such as guidepost cells? In some instances, damage to the nervous system can involve repair by new neuron production, but is limited by the inability of new neurons to wire correctly (25). However, in some other animals, regeneration of the nervous system seemingly restores normal function, although study of the regeneration of wiring patterns in highly regenerative models is sparse. Animals that have the ability to regenerate their neuronal architecture must possess adult mechanism(s) for repairing circuit patterns in the context of diverse injuries. To address how this can occur, we studied these mechanisms in the planarian Schmidtea mediterranea.
Planarians are freshwater flatworms that belong to the Spiralia superphylum (26). They are capable of whole-body regeneration and undergo constant tissue turnover. These processes involve neoblasts, a proliferating cell population containing pluripotent stem cells. In addition to dividing cells, proper regeneration requires positional information. Genes proposed to encode positional information in planarians are predominantly expressed in muscle cells in a regionally restricted manner across body axes (27). Whereas the cells and molecules guiding the fate choices of stem cells into new differentiated cells in planarian regeneration have received substantial attention, the mechanisms by which the architecture of the different planarian tissues is restored remains little studied.
We identified a set of muscle cells and neurons tightly associated with the planarian photoreceptor axons with guidepost-like attributes. These cells were constitutively present and capable of being regenerated following injury. The positioning of these muscle cells was guided by a constitutive system of positional information enabling their replacement at suitable positions after injury to promote precision in the regeneration of visual system architecture. Our results describe a novel strategy for de novo formation of precise axonal projection pattern following injury in the adult.
notum+; frizzled 5/8-4+ muscle cells are tightly associated with the visual system
Many planarian species have a pair of true cerebral eyes that are symmetrically located dorsal to the cephalic ganglia (28). Planarian eyes contain rhabdomeric photoreceptor neurons that extend their axons ipsi- and contralaterally forming an axon bundle, and pigmented cells that build an optic cup (Fig. 1A). The visual axons follow a stereotypic path that can be visualized using an anti-Arrestin monoclonal antibody (28). Contralateral axons defasciculate from ipsilateral axons at choice points and project towards the midline, forming the optic chiasm. Both contralateral and ipsilateral axons project ventrally and posteriorly to the visual targets in the brain (29, 30).
Figure 1. notum+ fz5/8-4+ muscle cells are tightly associated with the planarian visual system.

(A) Schematic diagram of planarian visual system. (B) notum+ cells associated with visual axons. (C) Co-expression and (D) density map (top) of notum and fz5/8-4 in cells associated with visual axons. Dark pink, cells near the eye; light pink, cells at choice points. Bottom: Graph shows positive correlation between number of cells associated with visual circuit and animal size (length). Red line, best fit for linear regression. (E) Transverse cut (red line) shows nuclei position of three notum+ cell subsets. Yellow dotted line, notum+ fz5/8-4+ cells. Dorsal, top; ventral, bottom. (F) SMEDWI-1 expression in a cell associated with visual circuit indicates recent specification. (G-J) notum+ cells associated with visual axons do not express eye-specific markers (G), neuron-specific markers (H), or glia-specific markers (I), but express muscle-specific markers (J; pool: troponin, tropomyosin, colF-2, colF-10). (K) Muscle notum+ cells are associated with visual axons in Schmidtea polychroa embryos. Cartoon shows embryonic stages. White arrows point to notum+ cells associated with visual circuit. Cartoon red box shows location of image taken.
Scale bars, 50μm (B, C, E, G, H, I, J, K) and 10μm for all zoom-ins (C, F, H, I, J).
Using fluorescence in situ hybridizations (FISH) in combination with immunostainings with the anti-Arrestin antibody, we noticed a previously unknown and small population of cells that were in strikingly close association with the visual system, marked by expression of the genes notum and frizzled 5/8-4 (Fig. 1B and C). We found that these cells were mostly concentrated in two regions: 1 – near the eye, where the photoreceptor axons form a bundle projecting out from the eye and 2 – near the axon choice points, located more ventrally (Fig. 1B–D). The number of these notum+; frizzled 5/8-4+ cells found in each animal was variable, but scaled with animal size (Fig. 1D). In a transverse cross section at the level of the eye, these cells were clearly distinguishable from other notum+ cells known to exist in the planarian head, such as anterior pole cells (31) and notum+ neurons (32) located at the anterior commissure of the brain (Fig. 1E; fig. S1A). The nuclei of the notum+; frizzled 5/8-4+ cells found near the eye were located dorsal to the nuclei of the notum+; frizzled 5/8-4+ cells found at the choice points, which were observed immediately dorsal to the cephalic ganglia (Movie S1). To make this DV-location distinction clear, we have colored these two populations differently in illustrations throughout this article. The clear association of these cells at two discrete and important locations near axons of the visual system raised the possibility of a role for these previously unidentified cells in visual system wiring.
Planarian eye cells, like all other cell types assessed in this organism, are replaced during normal tissue turnover. This process requires neoblasts, the only proliferating somatic cells in the animal (33). Neoblasts express the piwi homolog smedwi-1. smedwi-1 transcription ceases as cells leave the neoblast state and begin to differentiate, but the SMEDWI-1 protein transiently perdures as cells differentiate. An antibody that recognizes SMEDWI-1 therefore allows visualization of newly generated cells (34, 35). We observed a small number of notum+; frizzled 5/8-4+ cells positive for SMEDWI-1 protein, indicating that these cells are constantly specified and replaced in uninjured adult animals (Fig. 1F).
To determine the identity of notum+; frizzled 5/8-4+ cells, we examined them for co-expression of different cell-specific markers. These cells did not express the photoreceptor neuron marker opsin or the optic cup marker tyrosinase (Fig. 1G; figs. S1B and C); the neuronal markers pc2, synapsin, or ChAT(Fig. 1H; fig. S1D); or the planarian glia markers if-1 and cali (Fig. 1I). Some fz5/8-4+ ChAT+ cells were found near the eye but did not express notum (fig. S1E). Instead, notum+; frizzled 5/8-4+ cells expressed muscle markers (Fig. 1J) and could occasionally be detected in cross sections co-labeled with an antibody for planarian muscle (6G10; fig. S2A). Muscle cells are broadly distributed within the animal (figs. S2A and B); however, notum+; frizzled 5/8-4+ muscle cells were a rare subpopulation present in association with visual axons (Movie S2). notum+; frizzled 5/8-4+ cells expressed low levels of muscle markers (troponin, tropomyosin, colF-2, mp-1; figs. S2C–E) compared to other muscle cell subsets such as body wall, intestinal, and other dorsal-ventral muscle cells (36, 37) suggesting that these cells could be specialized muscle cells serving a different, e. g., secretory, role. Muscle cells in general are known to secrete signaling factors in planarians (27). Despite lower levels of expression of muscle markers per cell, signal from pooled probes for canonical muscle markers, such as troponin and tropomyosin, was detected in essentially all of the notum+; frizzled 5/8-4+ cells (Fig. 1J; fig. S2F). Whether these cells retain contractile function is unknown. notum+ muscle cells associated with the visual axons were also found during planarian embryonic development. Such notum+ cells were first observed in pre-hatchling stage 7 S. polychroa embryos at the time of optic chiasm formation, but not before (Fig. 1K). After hatching, all free-swimming juveniles displayed a similar array of notum+ muscle cells and visual axons as did their adult counterparts (Fig. 1K). The tight association of notum+; frizzled 5/8-4+ muscle cells with visual axons raised the possibility of a role for these cells in the wiring of the planarian photoreceptor axons.
Regenerating visual axons project towards notum+; frizzled 5/8-4+ muscle cells
We defined notum+; frizzled 5/8-4+ muscle cells near the eye as NMEs: notum+ muscle cells near the eye and those at axon choice points NMCs: notum+ muscle cells at the c point (Fig. 2A). If NMEs and/or NMCs facilitate wiring of photoreceptor axons, they should be present before or concurrently with key axon-pathfinding processes and should also be tightly spatially associated with such events. To examine this possibility, we studied the regeneration of these cells and the dynamics of photoreceptor axons following different regenerative challenges.
Figure 2. notum+ fz5/8-4+ muscle cells are associated with regenerating visual axons.
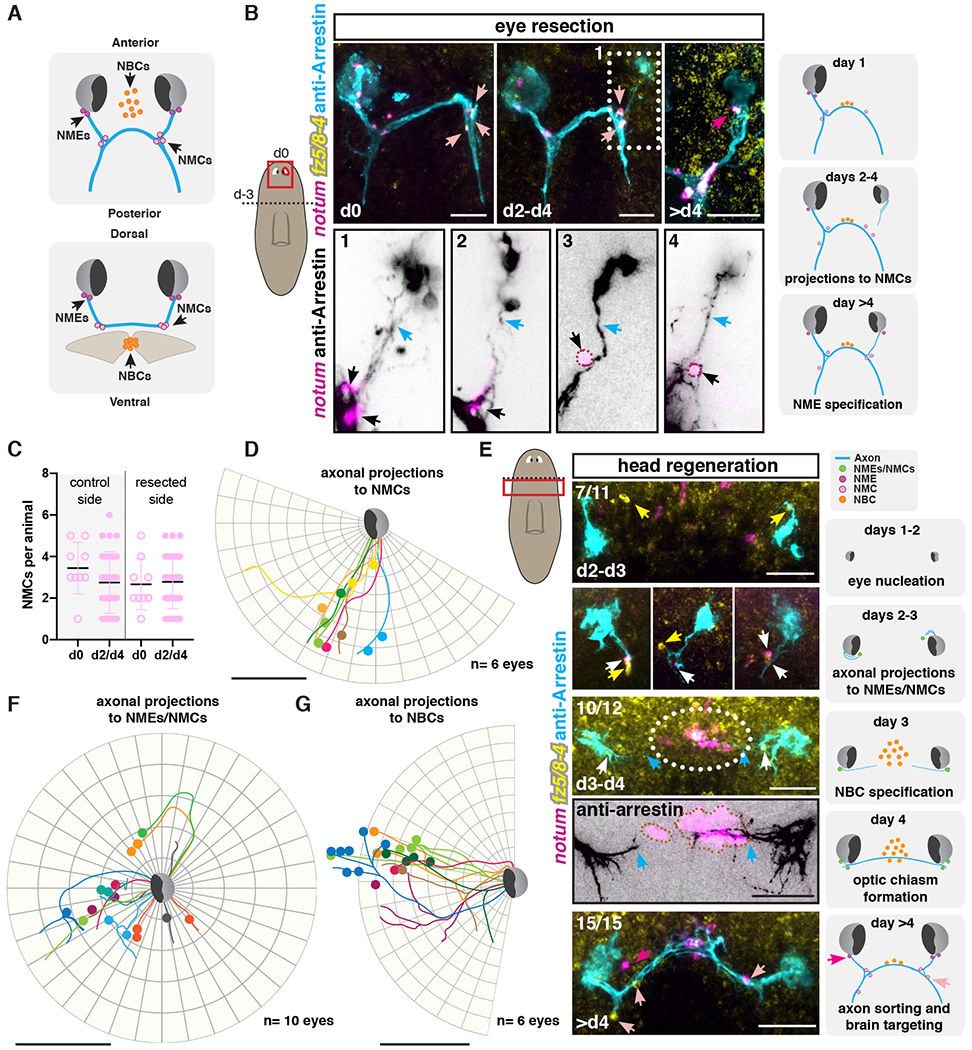
(A) Illustrations show NME, NMC, and NBC positions across different axes. (B) Regeneration of visual system at different timepoints following unilateral eye resection. Bottom: zoom-ins of visual axonal projection examples (1-4) 2-4 days after eye resection. Dotted outline, NMEs/NMCs. Left cartoon shows surgical procedure: head decapitation (dotted line; day −3) and unilateral eye resection three days later (d0; red line). Red box, location of image taken. Right cartoons summarize events observed following eye resection. (C) Graph shows no change in NMC numbers between resected and control sides. (D, F, G) Circular plots show tracing of photoreceptor axonal trajectories (lines) from independent right eyes during regeneration (d2 to d4) of a resected eye (D) or decapitation (F, G). Colored dots represent NMCs (D), NMEs/NMCs (F) or NBCs (G). (E) Regenerating visual system following decapitation. Dotted line in left cartoon indicates amputation line, red box shows location of image taken. Illustrations (right) summarize events observed after decapitation. Blue arrows, axons; white arrows, NMEs and/or NMCs, dark pink arrows or dots, NMEs; light pink arrows or dots and black arrows, NMCs. Orange dots or dotted outline, NBCs; white arrows, NMEs/NMCs; yellow arrows NMEs/NMCs expressing only frizzled 5/8-4.
Scale bars, 50μm (B, D-G).
First, we examined these cells in the context of a unilateral eye resection, which does not perturb the preexisting optic chiasm (Fig. 2B; (38)). This injury removed NMEs, but did not remove NMCs (Fig. 2B). Within two to four days after eye resection, new photoreceptor neurons nucleated dorsally (regenerating the resected eye) and projected their axons ventrally and posteriorly. NMC numbers stayed constant during this time (Fig. 2C). Despite variance in the relative positions of NMCs with respect to the new photoreceptor neurons and in the initial paths of projecting axons, the trajectories of photoreceptor axons essentially always coincided with the NMCs (Fig. 2B and D; figs. S3A and B). This tight association between NMC position and axon path is consistent with the possibility that NMCs have an attractive influence on photoreceptor axons. NMEs regenerated four days after eye resection, suggesting that NMEs are not necessary for early projections during eye regeneration following resection, a scenario where NMCs remained present throughout the repair process.
We next studied the behavior of photoreceptor axons, NMEs, and NMCs following head amputation. In this context, the animals need to regenerate the entire visual system, including NMEs and NMCs. Between days two and three after decapitation, we observed nucleation of new eyes and the appearance of notum+; frizzled 5/8-4+ cells in close proximity to the regenerating eyes (Fig. 2E; fig. S3C). At this early timepoint, the regenerating blastema is small, making distinction between notum+; frizzled 5/8-4+ NMEs and NMCs not possible. These cells displayed detectable fz5/8-4 transcripts first, followed by notum expression (Fig. 2E). At this time, photoreceptor axons from the regenerating eyes projected towards where notum+; frizzled 5/8-4+ cells were observed (Figs. 2E and F; figs. S3A and C). The pattern of axonal projections and notum+; frizzled 5/8-4+ cell location was variable, with some axon tracts projecting anterior-medially and others posteriorly, suggesting the initial steps in axonal pattern formation is noisy, but the spatial association of axons and notum+; frizzled 5/8-4+ cells was apparent nonetheless. This is consistent with the possibility that in the context of de novo visual system formation, these cells have an attractive influence on early axonal projections. It is also possible that photoreceptor axons and notum+; frizzled 5/8-4+ cells might stabilize each other upon interaction.
We noticed that the population of notum+ neurons in the medial brain, normally located ventral to the optic chiasm in a fully formed brain, was in close association with photoreceptor axons during early head regeneration. We refer to this third population of cells associated with photoreceptor axons during regeneration as notum+ brain cells or NBCs (Fig. 2A). NBCs were apparent within three to four days post-amputation at the regenerating anterior brain commissure (Fig. 2E; (32)). Visual axons only projected to the midline, forming the optic chiasm, after NBCs appeared and despite variation in their paths, they were typically associated with NBCs (Fig. 2E and G; figs. S3A and C). This process is reminiscent of optic chiasm formation in mouse embryonic development, in which the site of the future optic chiasm is first populated by a subset of neurons expressing L1 and CD44. Subsequently, retinal ganglion axons grow and project towards the midline using these neurons as a landmark or a scaffold (24). Other cell types, such as radial glial cells at the optic chiasm, or midline cells in insects have a similar scaffolding role for axons crossing the midline during development (39).
In planarian head regeneration, early photoreceptor axonal projections cross the midline (Fig. 2E and G; (40)). At the time of optic chiasm formation, the pattern of axonal projections with choice points and notum+; frizzled 5/8-4+ cells near them became apparent (Fig. 2E). We also observed axonal projections to the brain targets at this time. By day 4 of regeneration, the pattern of the photoreceptor axons was similar across animals, indicating that initial patterning during de novo regeneration resolves with time into a stereotypical pattern. The events that occur can be approximated into the following stages: 1- eye nucleation; 2- NME/NMC regeneration, axon bundle formation and projection towards NME/NMCs; 3- NBC regeneration; 4- axonal projections towards NBCs and optic chiasm formation; 5- axonal projections towards the visual areas in the brain. The close association between photoreceptor axons and NMEs, NMCs, and NBCs in this process is consistent with the possibility that these cells have a function in facilitating visual system wiring.
NMEs and NMCs are specified independently of eyes
If NMEs and NMCs have a guidepost-like function, their formation should at least in part be independent of the photoreceptor neurons themselves. To examine this possibility, we transplanted wild-type eyes (41) at different positions in pre-pharyngeal and post-pharyngeal animal regions. We did not observe any cells co-expressing notum and fz5/8-4 near the transplanted eyes ten days post-transplantation (Fig. 3A; fig. S4A). Interestingly, we observed that axon bundles from transplanted eyes were commonly defasciculated (Fig. 3A; fig. S4A). These findings suggest eyes in ectopic locations are insufficient to induce notum+; frizzled 5/8-4+ muscle cells.
Figure 3. Visual axons are not required for NME and NMC specification but maintenance.
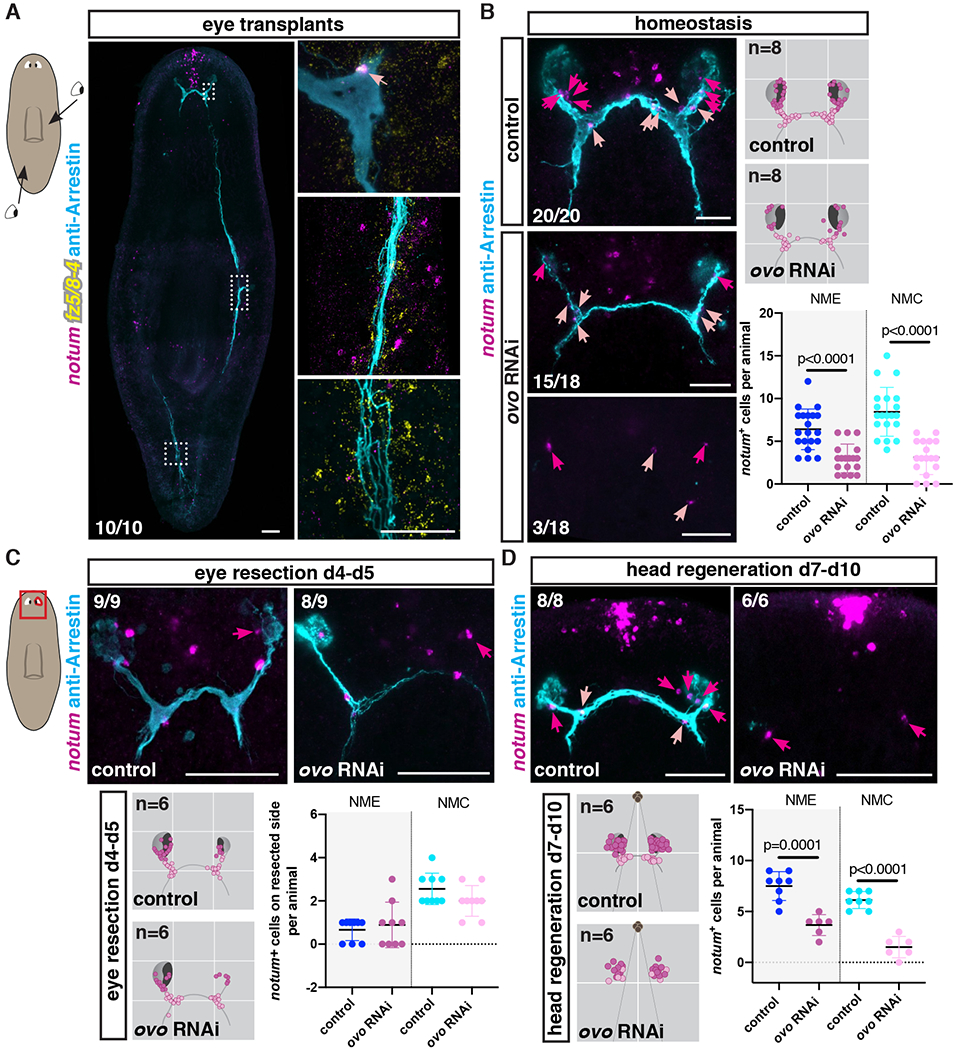
(A) NMCs are observed associated with original visual axons but not with axons from transplanted eyes. Cartoon on left shows location of transplanted eyes. (B) NMEs/NMCs are present in uninjured animals with few or no visual axons (ovo RNAi). Right top: Density map shows NME/NMC distributions in an idealized visual system cartoon. n indicates number of animals mapped. Right bottom: Graph shows NME/NMC numbers in uninjured RNAi animals. Regeneration of NMEs (C) and NMEs/NMCs (D) in ovo RNAi animals after eye resection (C) or head amputation (D). Bottom left: Mapping shows NME/NMC distributions in an idealized visual system cartoon. n indicates number of animals mapped. Bottom right: Graph shows NME/NMC numbers. Red box shows location of image taken. Dark pink arrows or dots, NMEs; light pink arrows or dots, NMCs.
Scale bars, 100μm (A) and 50μm (B-D).
Planarian eye cells (both photoreceptor neurons and optic cup cells) are specified by the transcription factor ovo (42). ovo RNAi animals cannot replace eye cells during normal tissue turnover or regenerate eyes after eye resection or decapitation. Nonetheless, NMEs and NMCs were both present in uninjured ovo RNAi animals, including in animals completely lacking photoreceptor neurons (Fig. 3B). However, their numbers were significantly reduced compared to control animals (Fig. 3B), suggesting a potential role for photoreceptor axons in the maintenance of normal NME and NMC numbers. Next, to determine whether NMEs and NMCs could regenerate independently of eye cells, we performed different injuries in ovo RNAi animals. We found that NMEs were able to regenerate after unilateral eye resection in ovo RNAi animals, indicating that eye cells were not required for NME specification in this context (Fig. 3C). Furthermore, NMEs were still observed 30 days after double-eye resection in ovo RNAi animals, suggesting that these cells were still specified even when no axons were left (fig. S4B). Moreover, NMEs and NMCs also regenerated after decapitation of ovo RNAi animals (Fig. 3D; figs. S4C and D), although the total number of NMEs and NMCs in this context was significantly lower compared to controls (Fig. 3D). In conclusion, both NMEs and NMCs were observed in uninjured or regenerating animals that completely lacked photoreceptor neurons and associated axons. Together, these data suggest that NMEs and NMCs can be formed in the absence of eyes, but their homeostatic numbers are positively influenced by photoreceptor axons.
Axonal projections from transplanted eyes associate with NMEs and NMCs
If visual circuit-associated muscle cells (NMEs and NMCs) have guidepost-like activity, we predicted that visual axonal projections should be correlated to their location not just during regeneration, but also following eye transplantation in the head. We performed unilateral eye transplantations at approximately the original eye position in double-eye-resected ovo RNAi recipients (fig. S5A). As shown above (Fig. 3B), some NMEs and NMCs perdured in intact ovo RNAi animals allowing assessment of correlation between the location of eye projections and NMEs and NMCs. One-to-three days after double-eye resection was not sufficient time to eliminate all preexisting photoreceptor axons in ovo RNAi recipients (figs. S5B and C). Axonal projections from all transplanted eyes at this time point were able to navigate and follow the stereotypical path (figs. S5B and C). To eliminate a potential role for remaining axons in serving as a scaffold for transplanted axons, we transplanted eyes ten to twelve days after double-eye resection of ovo RNAi recipients. Within this time window, 87.5% of the ovo RNAi recipients had no remnant axons left, although some NMEs and NMCs were present at the time of transplantation (d0, Fig. 4A). Axons from all transplanted eyes were prone to more defasciculation and errors, but projected ipsilaterally and crossed the midline in a stereotypical manner (Fig. 4A; fig. S5D). We observed that some NMEs were co-transplanted with the wild-type eye, explaining, at least in part, the increased number observed in NMEs at day seven following transplantation (Fig. 4B). Importantly, in all instances, even when transplanted eyes were slightly mispositioned, the visual axon tracts extended towards the NMCs present in recipient animals, and bifurcated at these choice points (zoom-ins, Fig. 4A; fig. S5D). Moreover, in some cases, we observed that axons locally deviated from the stereotypical route of the main axon bundle towards NMEs or NMCs that were present in the recipient animals (zoom-ins, Fig. 4A; fig. S5D). Trace mappings of axonal trajectories showed that nearly all axons passed close by NME/NMCs (within two cell diameters), and many directly interacted with NMEs/NMCs (Fig. 4C; figs. S6A and B). The locations of eyes, axonal projections, and NMEs/NMCs were variable in these experiments but tight association of projections with NMEs/NMCs was observed nonetheless. These results are consistent with the possibility that NMEs and NMCs might attract and/or stabilize visual axons. Moreover, our observations show that formation of an optic chiasm in adult planarians does not depend on interactions between the axons coming from each eye, as previously shown in vertebrates (23, 39).
Figure 4. NMEs and NMCs facilitate visual system patterning after eye transplantation.
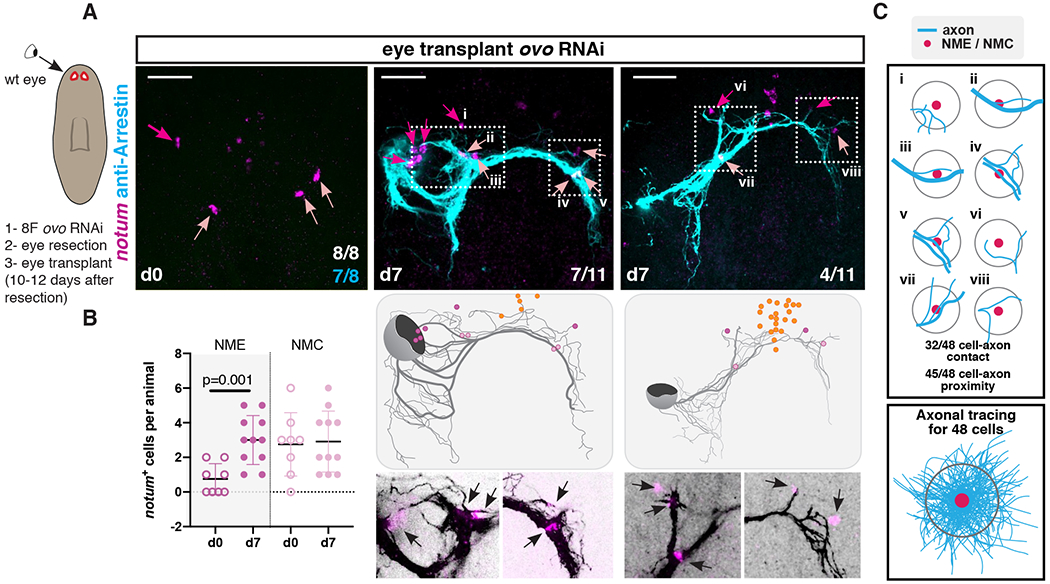
(A) Top: NMEs/NMCs are still present 10-12 days after double-eye resection in ovo RNAi animals before a wild-type eye is transplanted (left). Complete (middle) or incomplete (right) recapitulation of stereotypical visual axonal projections after eye transplantation in double-eye resected ovo RNAi animals. Cartoon on left shows summarized surgical procedure: animals were RNAi fed 8 times (8F), double-eye resection performed, and wild-type eye transplanted 10 to 12 days after eye resection. Animals were fixed just before (d0) or 7 days after (d7) transplantation. Cartoons show axonal tracings of the image shown above and position of NMEs, NMCs, and NBCs. Below: zoom-ins (white dotted box) showing axonal projections from transplanted eyes in close association with NMEs/NMCs (black arrows). (B) Graph shows NME/NMC numbers before and after transplantation. (C) Cartoons show examples from A (i-viii) of NME/NMC associations with visual axons. Numbers of cells that contact an axonal tract or found within two cell-diameter distance to the axonal tract are shown. Bottom: axonal tracing observed near 48 NMEs/NMCs after transplantation. Dark pink arrows or dots, NMEs; light pink arrows or dots, NMCs.
Scale bars, 50μm (A).
Extrinsic axial patterning cues control NME/NMC positioning
If NMEs and NMCs facilitate photoreceptor axonal projection pattern in regeneration, then the positioning of these muscle cells should be regulated, at least in part, independently from the visual system itself. The fact that NMEs and NMCs can be regenerated independently of photoreceptor neurons is consistent with this possibility and suggests that an extrinsic mechanism should exist to position these cells. Correct positioning of guidepost cells is required for the precise wiring of neural circuits in other organisms. In most examined cases, canonical axon guidance cues (Netrins/DCC or Unc-5; Slits/Robo; Semaphorins/Plexins; and Ephrins/Ephrin Receptors) also affect guidepost cell positioning (12). To examine the role of conserved guidance cues in NME/NMC positioning, we inhibited previously reported and newly identified planarian genes encoding guidance cue homologs with RNAi (figs. S7A and B). Inhibition of these genes perturbed photoreceptor axonal projections (figs. S8A and B, (43–46)) but did not alter overall numbers or position of NMEs/NMCs (figs. S8A–C), except when planarian axial patterning was disrupted (see below).
Positional information is essential during both planarian regeneration and normal tissue turnover to inform neoblasts and/or their progeny about the location of tissues that need to be replaced. In planarians, a variety of signaling molecules known as position control genes or PCGs have important roles in axial patterning and constitute adult positional information. PCGs are predominantly expressed in muscle cells along different planarian axes (27, 47), and inhibition of some PCGs results in striking patterning defects, including hypercephalized, animals with duplication of eyes, or pharynges (33, 47–49). Inhibition of PCGs affected the pattern of NMEs and NMCs during normal tissue turnover (Fig. 5A) and regeneration (fig. S8A).
Figure 5. Axial patterning is required for positioning of NMEs and NMCs.
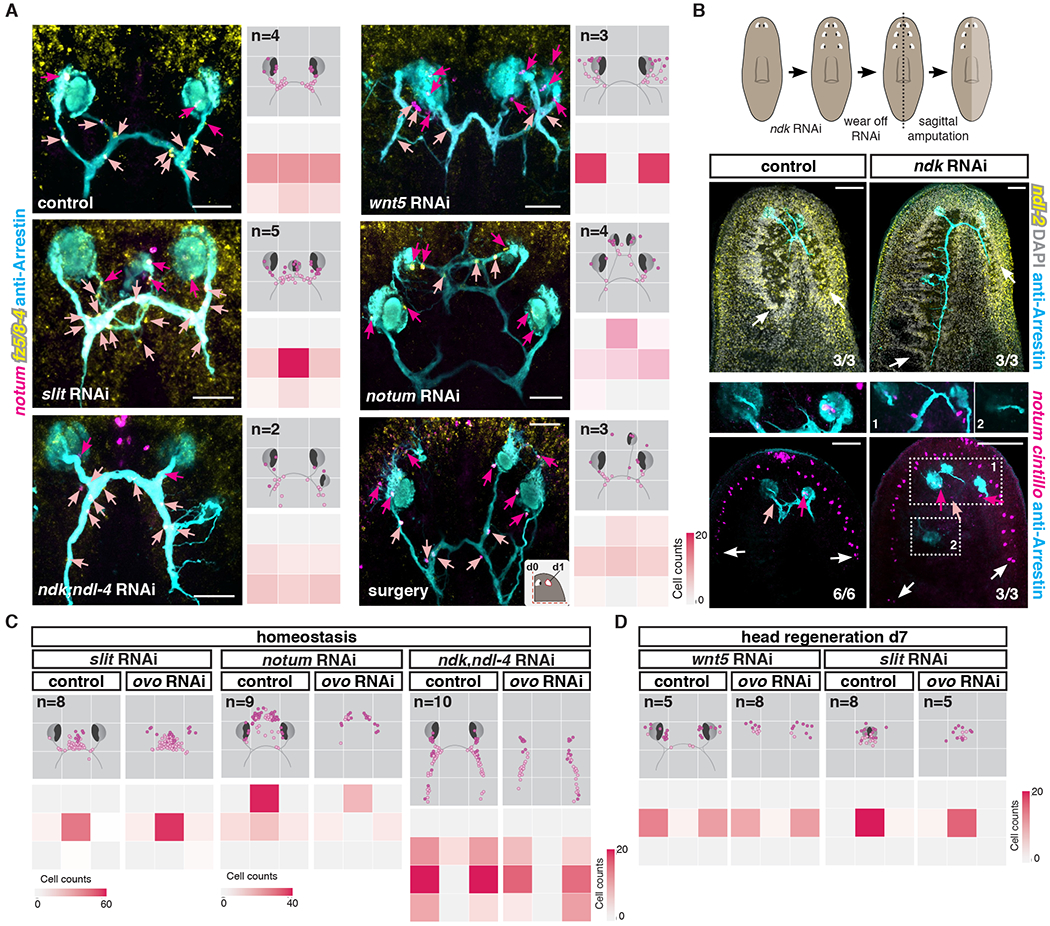
(A) Left: Visual system and NMEs/NMCs in different uninjured RNAi conditions or following surgery. Right top: Density map shows NME/NMC distributions in an idealized visual system illustration. n indicates number of animals mapped. Right bottom: Heatmap shows numbers of NMEs/NMCs in each quadrant. (B) Top: Illustration summarizing experimental procedure. Middle: Rescaling of ndl-2 PCG expression, maintenance of posteriorized brain lobe in old tissue, and regeneration of normal size brain lobe in blastema. Bottom: NMEs/NMCs are present near eyes located at the correct position following PCG rescaling (1) but not near ectopic posterior eyes (2) in an ndk RNAi animal. White arrows point to brain lobes or neurons (cintillo+). (C) Mapping shows NME/NMC distributions in an idealized visual system cartoon of uninjured PCG RNAi animals in the presence (control) or absence (ovo RNAi) of eyes. n indicates number of animals mapped. Heatmap shows total number of NMEs/NMCs located in each quadrant. slit RNAi leads to medialization, notum RNAi leads to anteriorization, and ndk; ndl-4 RNAi leads to posteriorization of NME and NMCs – in each case independently of eyes. (D) wnt5 RNAi animals show lateralized a NME/NMC distribution, whereas slit RNAi animals show medialized cell distribution independently of visual axons during regeneration. Dotted lines in cartoons show amputation planes. Dark pink arrows or dots, NMEs; light pink arrows or dots, NMCs; blue arrows, visual axons.
Scale bars, 50μm(A), 100μm (B).
The planarian medial-lateral (ML) axis is regulated by medial slit and lateral wnt5, with wnt5 restricting slit expression to the midline (50). Inhibition of slit or wnt5 impacted the ML positioning of NMEs and NMCs in both uninjured and regenerating animals (Fig. 5A; fig. S8A). slit RNAi animals showed a shift of these cells towards the midline, where an ectopic or cyclopic eye was observed (Fig. 5A; fig. S8A). roboC RNAi resulted in a similar phenotype to that of slit RNAi, suggesting roboC encodes a major Slit receptor (fig. S8A and D). wnt5 RNAi animals showed increased numbers of NMEs and NMCs laterally, in conjunction with the appearance of ectopic lateral eyes (Fig. 5A). Regenerating wnt5 RNAi animals did not form an optic chiasm and NMEs/NMCs were displaced more laterally (fig. S8A). ror-1 RNAi resulted in a phenotype similar to that of wnt5 RNAi animals during both regeneration and homeostasis (figs. S8A and D), suggesting that ror1 encodes the Wnt5 receptor in this process.
RNAi of genes involved in the patterning of the anterior-posterior (AP) axis affected the AP positioning of NMEs and NMCs (Fig. 5A). notum RNAi causes anteriorization of brain and ectopic anterior eye appearance (32), and these animals showed ectopic anterior NMEs and NMCs. Because of the axial patterning role of notum, it was not possible to readily determine the function of notum within NME and NMCs. nou darake (ndk) RNAi animals develop ectopic posterior eyes (47, 48) and shift anterior PCG expression domains posteriorly (47). ndk RNAi caused ectopic posterior appearance of NMEs and NMCs. Major surgery leading to extensive PCGs shifts in the head fragments (41) also resulted in the appearance of ectopic NMEs and NMCs at positions consistent with the locations of ectopic de novo eyes (Fig. 5A). In PCG RNAi experiments, NMEs and NMCs were sometimes observed in ectopic locations before the nucleation of a new ectopic eye or the projection of a new axon bundle (fig. S9A). This is consistent with the possibility that PCG alteration itself leads to ectopic positioning of NMEs/NMCs as opposed to these cells being induced in ectopic locations only after ectopic eyes or axonal projections appeared.
To further test this hypothesis, we generated posteriorized ndk RNAi animals. After wearing off the RNAi, these animals maintained all ectopic posterior eyes and posteriorly expanded brain lobes, even though ndk function returns and the PCG map scales back to its normal proportions (41). Sagittal amputations at this time result in animals regenerating a side with a new eye and brain lobe at the correct scale and anterior position, generating an asymmetrical body plan (Fig. 5B and (41)). NMEs and NMCs, under these conditions, only regenerated at the correct AP position with respect to the rescaled PCG domains, near the regenerating eye within the blastema. Within the uninjured old tissue side, which maintained a posteriorly expanded cephalic ganglion and ectopic eyes, NMEs and NMCs were also only observed near the eye that was found at the correct position based on the rescaled PCG map. i.e., posterior ectopic eyes did not maintain NMEs and NMCs (Fig. 5B; fig. S9B), unlike animals that were kept under ndk RNAi conditions (Fig. 5A). These results further indicate a role for PCGs, rather than ectopic eyes themselves, in instructing the positioning of newly specified NMEs and NMCs.
Because NMEs and NMCs can be formed in the absence of eyes (Fig. 3D), we inhibited both ovo and PCGs at the same time to ask whether the ectopic positioning of NMEs/NMCs was still observed in PCG RNAi conditions in the absence of eye formation (Figs. 5C and D; figs. S9C and D). In both uninjured or regenerating RNAi animals, NMEs and NMCs were mispositioned, regardless of the presence (in control) or absence (in ovo RNAi) of new photoreceptor neurons. Taken together, our results indicate that PCGs constitutes a positional information system that instructs the positioning of NMEs and NMCs independently of eye cells, providing a mechanism to place cells with candidate guidepost-like function in precise locations.
NMCs are absent in tolloid RNAi animals
Members of the Bmp/TGFβ family have been shown to guide commissural axons of the spinal cord in vertebrates ventrally, through repulsion (51, 52). Inhibition of the planarian bmp4 homolog did not grossly affect the pathfinding of visual axons during regeneration (53). However, inhibition of tolloid, a metalloproteinase which can cleave the Bmp inhibitor Chordin in vertebrates and Drosophila (54, 55), resulted in severe defects in the trajectory of photoreceptor axons (53). In regenerating tolloid RNAi animals, axons were more defasciculated and were not able to cross the midline, resulting in disorganized posterior projections (Figs. 6A and B; fig. S10A). The midline cues slit and netrin-2 were normally expressed in these animals (fig. S10B). NMEs and NBCs were still present, but NMCs were completely absent in regenerating tolloid RNA animals (Fig. 6A). NMCs were also severely reduced in tolloid RNAi animals during eye regeneration following double-eye resection (fig. S10C) and during homeostasis (Fig. 6C). Under all of these conditions, photoreceptor axons showed higher levels of defasciculation.
Figure 6. tolloid, arrowhead, and soxP-5 are required for specification of guidepost-like cells and precise wiring of the visual system.
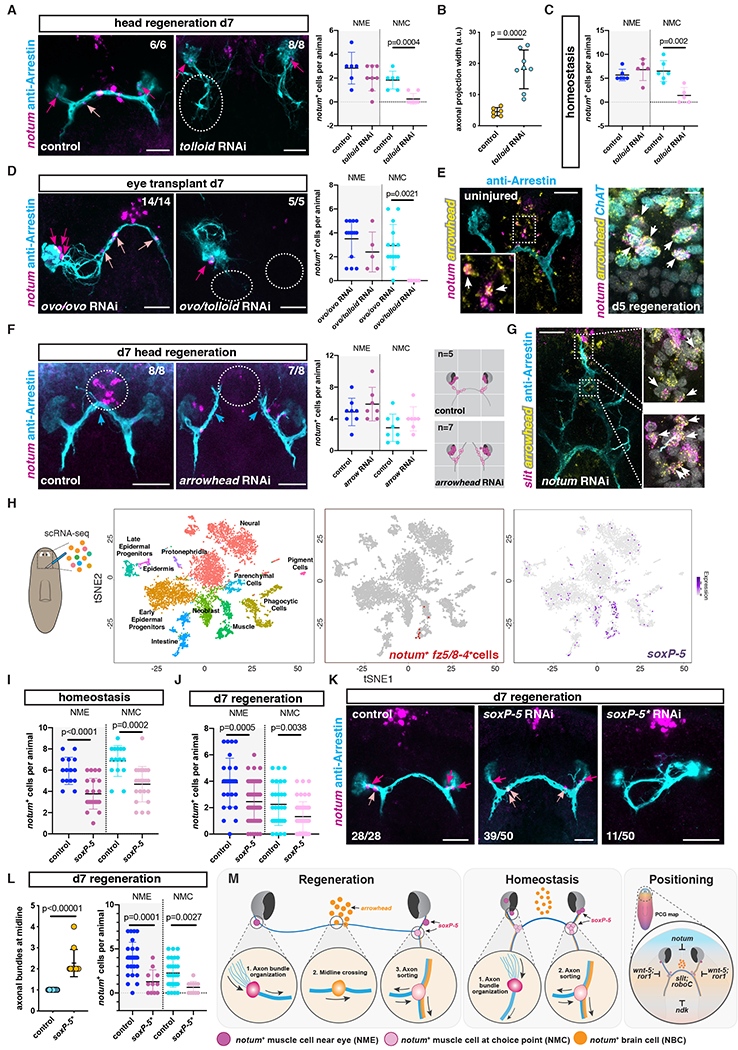
(A) Defasciculation of visual axons (white dotted circle) and absence of NMCs in a regenerating tolloid RNAi animal. Right graph shows NME/NMC numbers after head regeneration. (B) Graph shows axonal projection width in RNAi animals. (C) Graph shows NME/NMC numbers in uninjured animals. (D) Lack of NMCs and inability of transplanted eyes to cross the midline in tolloid RNAi animals. Right graph shows NME/NMC numbers after transplantation. (E) NBCs expressed the transcription factor arrowhead in intact (left) or regenerating (right) animals. (F) Left: Absence of NBCs and lack of optic chiasm (dotted circles) in a regenerating arrowhead RNAi animal. Middle: Graph shows normal NME/NMC numbers in regenerating arrowhead RNAi animals. Right: Mappings show distribution of NMEs/NMCs in an idealized visual system, n indicates total number of animals mapped. (G) slit+; arrowhead+ cells (white arrows) in ectopic anterior brain commissures in a notum RNAi animal coinciding with ectopic optic chiasm. (H) scRNA-sequencing analysis of cells from region depicted in left cartoon. Left: t-SNE representation of clustered cells (dots) colored based on planarian cell types. Middle: t-SNE representation of clustered muscle cells expressing notum; fz5/8-4 (red). Right: t-SNE plot colored by expression of the transcription factor soxP-5(I, L) Graph shows total NME/NMC numbers in uninjured (I) or regenerating (J) RNAi animals. (K) Visual system in presence of normal (middle) or severely reduced (right, soxP-5*) numbers of NMEs/NMCs. (L) Graphs show number of axon bundles at the midline (left), number of NMEs/NMCs (right) in soxP-5* RNAi animals. (M) Model summarizing findings (see text for details). Dark pink arrows or dots, NMEs; light pink arrows or dots, NMCs.
Scale bars, 50μm (A, D, E, F, G, K).
We next transplanted eyes into tolloid; ovo RNAi recipients, which lack NMCs. Independently of the presence or absence of remaining axons (transplantation after three or ten days following double-eye resection), wild-type photoreceptor axons were not able to robustly project in a fasciculated bundle or cross the midline in tolloid; ovo RNAi recipients (Fig. 6D; fig. S10D). These results are consistent with the possibility that NMCs can attract and/or stabilize axons to the choice points and facilitate axon bundle formation. Similar results were observed when transplanting a wild-type eye into a netrin-1; netrin-2; ovo RNAi recipient, or when transplanting an eye from a DCC RNAi animal into an ovo RNAi recipient (fig. S10E). Metalloproteinases have been shown to cleave the DCC receptor (56), raising the possibility of direct or indirect interaction between Tolloid and DCC in planarians. The midline defect in tolloid RNAi animals indicates that additional tolloid-dependent processes likely exist and affect midline axonal crossing.
arrowhead is required for NBC specification and optic chiasm formation
Specific ablation of CD44-expressing chiasm neurons in mice demonstrated a role for these cells in the formation of optic chiasm during embryonic development (57). Cell-specific ablation strategies are not currently available in planarians. However, in some instances, RNAi of transcription-factor-encoding genes involved in cell-fate specification can be used to deplete animals of specific cell populations (34, 42, 58, 59). Inhibition of arrowhead, which is expressed medially to the cephalic ganglia and encodes a Lim homeodomain protein, by RNAi resulted in failed regeneration of the optic chiasm and defective axonal crossing at the midline (60). arrowhead RNAi did not affect the expression of normal midline guidance cues, such as slit (60) and netrin-2 (fig. S11A). Visual axonal projections did not overtly follow other axon bundles at the anterior commissure in regenerating arrowhead RNAi animals (fig. S11B). We hypothesized that arrowhead might be required for the regeneration of NBCs at the anterior commissure, and thereby affect midline crossing of visual axons. Indeed, NBCs expressed arrowhead both in intact and regenerating animals (Fig. 6E; fig. S11C) and arrowhead RNAi animals failed to regenerate NBCs (Fig. 6F). However, we cannot rule out the possibility that other cells specified by arrowhead also contribute to optic chiasm formation. Numbers of NMEs and NMCs were not affected in arrowhead RNAi animals during head regeneration (Fig. 6F) or after double-eye resection (fig. S11D). Intact arrowhead RNAi animals that lost all NBCs during normal tissue turnover were still able to properly rewire the visual system after double eye resection (fig. S11D), suggesting that NBCs might be dispensable for maintenance of the optic chiasm. Importantly, visual axons after double-eye resection perdure for around three days, which gives sufficient time for new photoreceptors to regenerate and project their axons to the brain targets following the pre-existing axonal tract. Alternatively, the photoreceptor axons could also be influenced by the existing brain commissure. In addition, RNAi animals with duplicated optic chiasms (notum RNAi animals; (32)) also had ectopic anterior brain commissures with a duplication of NBCs (Fig. 6G).
soxP-5 is required for NME/NMC specification
To examine the transcriptomic signature of NMEs and NMCs, we performed single-cell RNA sequencing (scRNA-seq) using Drop-seq (61) on cells isolated from the region containing the visual system in intact animals (Fig. 6H). Cells were clustered using Seurat into 28 clusters, each of which could be assigned to one of the eleven broad tissue classes on the basis of tissue-specific gene expression ((62); Fig. 6H). We recovered eight notum+; frizzled5/8-4+ cells that were negative for brain markers (pc2 and ChAT) and anterior pole markers (foxD), and clustered together with muscle cells (Fig. 6H; fig. S12A; Table S1). We performed single-cell-differential expression analysis (SCDE) with these cells, comparing their transcriptomes with the transcriptomes of other muscle cells, and identified enriched transcripts (Table S2). A similar analysis was also performed for NBCs (Table S3). One of the few transcription factors expressed in NMEs and NMCs in the scRNA-seq data was soxP-5 (soxP-5 expression was difficult to detect by FISH). soxP-5 was also broadly expressed in muscle cells, including muscle progenitors, but not in photoreceptor neurons or other cell types (Fig. 6H; fig. S12A; Table S1).
RNAi of soxP-5 led to a significant decrease in the numbers of both NMEs and NMCs in uninjured (Fig. 6I) or double eye-resected animals (fig. S12B), indicating it is required for production of normal numbers of these specialized muscle cells. After head amputation, soxP-5 RNAi animals regenerated head blastemas that were normal in macroscopic size, appearance, and brain architecture (fig. S12C). However, these blastemas displayed lower numbers of NMEs and NMCs – in most soxP-5 RNAi animals multiple NMEs and NMCs did regenerate, but a minority of animals had very few or no detectable NMEs and NMCs (Fig. 6J). In eye-resected soxP-5 RNAi animals, the axonal projection pattern remained mostly unaffected, as expected, because new axons could follow existing axonal tracts in their development (fig. S12B). In regeneration following amputation, the visual system must form de novo, allowing assessment of pattern in animals with reduced NMEs and NMCs. Whereas most soxP-5 RNAi animals regenerated the stereotypical axonal projection pattern, around 20% of the soxP-5 RNAi animals exhibited defects in this pattern (labeled as soxP-5*; Figs. 6K and L; figs. S12D–F). soxP-5* RNAi animals showed ectopic axon bundles at the optic chiasm, abnormal fasciculation, and misrouting of the axon bundles at the choice points (Figs. 6K and L; figs. S12D and E). We found that the soxP-5* animals were those that had a severe reduction in NMEs and NMCs: soxP-5* RNAi animals had significantly fewer NMEs (p=0.0029) and NMCs (p=0.02) than did soxP-5 RNAi animals with a relatively normal axonal projection pattern (fig. S12F). Normal numbers of muscle cells were observed in intact (fig. S12B) and regenerating soxP-5* RNAi animals (figs. S13A and B), indicating that soxP-5 is not essential for general muscle regeneration or maintenance per se. Furthermore, NBCs, as well as other neuronal subsets, normally regenerated in soxP-5* RNAi animals, indicating a specific effect on the muscle cells associated with the photoreceptor axons (figs. S13C and D). The correspondence between low numbers or absence of NMEs/NMCs and the quality of the regenerated axonal projections provides further evidence for a guidepost-like role for these muscle cells. Our results suggest that NME, NMC, and NBC guidepost-like cells, together with constitutive guidance cues expressed broadly in the animal, contribute to the overall assembly and pattern of the precise visual system.
Discussion
Orchestration of precisely wired neural circuits relies on multiple developmental processes. Axon pathfinding is a highly precise process and conserved mechanisms of guidance are used by different organisms. Guidance mechanisms can act in concert, and include long-range or local chemoattractants and chemorepellents (guidance cues), transient cell-cell interactions (guidepost cells), and fasciculation of axons with pioneer or preceding axon tracts that form an initial scaffold. Once the circuit is established, some of these mechanisms become dispensable for the maintenance of pattern. The transient nature of these developmental mechanisms creates a problem for repair after injury: if an injury removes a patterned circuit, the information for bringing the circuit back would be lost, and might result in irreparable damage to the nervous system, even if the mechanisms for new neuron production existed. However, functional regeneration of the nervous system in some animals suggests mechanisms for de novo restoration of the precise pattern of neuronal circuits in adults must exist.
We identified a population of muscle cells expressing notum and fz5/8-4 located in close proximity to axon tracts at key locations in the planarian visual system. The tight association and precise positioning of these muscle cells suggest they have a role in the assembly of the planarian visual system. This finding led us to investigate the possible existence of guidepost-like cells in nervous system regeneration. Guidepost cells in other organisms are often transient neurons or glia. These planarian guidepost-like cells were not only present in regeneration, but constitutively maintained in the adult. It is atypical for a muscle cell to exert a guidepost-like role. However, planarian muscle cells are not only contractile, but they also secrete extracellular matrix proteins (63) and harbor positional information required for regeneration and tissue turnover (27, 47). In addition, specialized subsets of muscle cells serve as anterior (64) and posterior organizers (65, 66). Our findings suggest that non-neuronal connective tissue-like cells can promote neuronal circuit formation in regeneration.
Our results suggest that notum+; frizzled 5/8-4+ muscle cells can locally attract and/or stabilize photoreceptor axons, facilitating formation of visual axon bundles, and possibly helping with axon sorting at choice points (Fig. 6M). Axon-axon interactions are critical for organizing axons into bundles and are required for proper topographic pathfinding. The mechanisms involved in axon bundle formation described in other organisms include either interactions of adhesion molecules expressed on the axons, or repulsive interactions between axons and surrounding tissues (reviewed in (67)). The molecules required in planarians to facilitate this process remain unknown, although members of the Immunoglobulin or Cadherin superfamilies are appealing candidates to play roles in this process. Guidepost cells in other systems exert their function in guiding pioneer axon tracts by secreting attractive or repulsive guidance cues. Inhibition of several genes encoding homologs of canonical guidance cues did not result in defective attraction of the visual axons towards the planarian guidepost-like cells, suggesting that different molecules are at play or that there exists some redundancy within the system.
Our findings suggest a model in which guidepost-like cells act in concert with constitutively active global axon guidance cues to facilitate precision in the regeneration of the visual system.
Our study also identified two different transcription factors required for the specification of distinct subsets of cells with guidepost-like attributes. arrowhead, a Lim domain homeobox gene, is involved in the specification of NBCs, whereas soxP-5, is required for the specification of both NMEs and NMCs. NMEs and NMCs show differences regarding not only their position, but also, based on our observations from tolloid RNAi animals, in the expression of different programs to exert their function. NMCs were sensitive to tolloid inhibition, whereas NMEs remained unaffected. Some of these genes likely regulate systems beyond the NMEs/NMCs. Previously identified targets of the Tolloid protease include the BMP inhibitor, Chordin, a homolog for which has not been found in planarians. However, increasing evidence suggests that members of the Astacin-like family of metalloproteases can also process extracellular matrix components, other TGFβ family ligands, and are likely candidates for modulating the activity of guidance cues or their receptors (56).
How intermediate target or guidepost cells in different organisms are specified at precise locations has not been fully elucidated. Our observations suggest a model in which muscle guidepost-like cells are coarsely specified in a permissive region proximal to the eyes, which is, at least in part, defined by an axial PCG expression map (Fig. 6M). Density maps for the muscle guidepost-like cells suggest that these cells might be maintained only at the positions where photoreceptor axons are present, as indicated by their tight association with photoreceptor axons. This feature may allow dynamic maintenance of muscle guidepost-like cells in precise locations, and allow regeneration of these cells in the face of an unlimited array of regenerative challenges.
Strategies for neuronal patterning in the absence of embryo-specific contexts likely will also be crucial to overcome challenges in regenerative medicine. By studying a naturally occurring context of de novo formation of a neural system in regeneration, we found that small populations of cells, extrinsic to the system itself, provide information to help precisely pattern and assemble axonal projections in regeneration. Our results expand the diversity of strategies utilized by different organisms to organize and properly build the nervous system, especially in the context of injury repair.
Supplementary Material
Acknowledgements
We thank C. McQuestion for help in Drop-seq surgeries and members of the Reddien lab for discussions and comments on the manuscript.
Funding: PWR is an investigator of HHMI and an associate member of the Broad Institute. We thank the Eleanor Schwartz Charitable Foundation for support.
Footnotes
Competing interests: The authors have no competing interests.
Data and materials availability: All data is available in the manuscript or the Supplementary Materials.
References
- 1.Edwards JS, Chen SW, Berns MW, Cercal sensory development following laser microlesions of embryonic apical cells in Acheta domesticus. J Neurosci 1, 250–258 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutter H, Extracellular cues and pioneers act together to guide axons in the ventral cord of C. elegans. Development 130, 5307–5318 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Klose M, Bentley D, Transient pioneer neurons are essential for formation of an embryonic peripheral nerve. Science 245, 982–984 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Pike SH, Melancon EF, Eisen JS, Pathfinding by zebrafish motoneurons in the absence of normal pioneer axons. Development 114, 825–831 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Kolodkin AL, Tessier-Lavigne M, Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol 3, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoeckli ET, Understanding axon guidance: are we nearly there yet? Development 145, (2018). [DOI] [PubMed] [Google Scholar]
- 7.Tessier-Lavigne M, Goodman CS, The molecular biology of axon guidance. Science 274, 1123–1133 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Yu TW, Bargmann CI, Dynamic regulation of axon guidance. Nat Neurosci 4 Suppl, 1169–1176 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Bentley D, Caudy M, Pioneer axons lose directed growth after selective killing of guidepost cells. Nature 304, 62–65 (1983). [DOI] [PubMed] [Google Scholar]
- 10.Bentley D, Keshishian H, Pathfinding by peripheral pioneer neurons in grasshoppers. Science 218, 1082–1088 (1982). [DOI] [PubMed] [Google Scholar]
- 11.Chao DL, Ma L, Shen K, Transient cell-cell interactions in neural circuit formation. Nat Rev Neurosci 10, 262–271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squarzoni P, Thion MS, Garel S, Neuronal and microglial regulators of cortical wiring: usual and novel guideposts. Front Neurosci 9, 248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirata T et al. Guidepost neurons for the lateral olfactory tract: expression of metabotropic glutamate receptor 1 and innervation by glutamatergic olfactory bulb axons. Dev Neurobiol 72, 1559–1576 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Bendito G et al. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell 125, 127–142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niquille M et al. Transient neuronal populations are required to guide callosal axons: a role for semaphorin 3C. PLoS Biol 7, e1000230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, Hirata T, Ogawa M, Fujisawa H, Requirement for early-generated neurons recognized by monoclonal antibody lot1 in the formation of lateral olfactory tract. J Neurosci 18, 7800–7810 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bielle F et al. Slit2 activity in the migration of guidepost neurons shapes thalamic projections during development and evolution. Neuron 69, 1085–1098 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Ito K et al. Semaphorin 3F confines ventral tangential migration of lateral olfactory tract neurons onto the telencephalon surface. J Neurosci 28, 4414–4422 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki T, Ito K, Hirata T, Netrin 1 regulates ventral tangential migration of guidepost neurons in the lateral olfactory tract. Development 133, 845–853 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Nomura T, Holmberg J, Frisen J, Osumi N, Pax6-dependent boundary defines alignment of migrating olfactory cortex neurons via the repulsive activity of ephrin A5. Development 133, 1335–1345 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Bernhardt RR, Patel CK, Wilson SW, Kuwada JY, Axonal trajectories and distribution of GABAergic spinal neurons in wildtype and mutant zebrafish lacking floor plate cells. J Comp Neurol 326, 263–272 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Klambt C, Jacobs JR, Goodman CS, The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell 64, 801–815 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Marcus RC, Blazeski R, Godement P, Mason CA, Retinal axon divergence in the optic chiasm: uncrossed axons diverge from crossed axons within a midline glial specialization. J Neurosci 15, 3716–3729 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sretavan DW, Feng L, Pure E, Reichardt LF, Embryonic neurons of the developing optic chiasm express L1 and CD44, cell surface molecules with opposing effects on retinal axon growth. Neuron 12, 957–975 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amamoto R et al. Adult axolotls can regenerate original neuronal diversity in response to brain injury. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laumer CE et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr Biol 25, 2000–2006 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW, Muscle cells provide instructions for planarian regeneration. Cell Reports 4, 633–641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agata K et al. Structure of the Planarian Central Nervous System (CNS) Revealed by Neuronal Cell Markers. Zool. Sci 15, 433–440 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Okamoto K, Takeuchi K, Agata K, Neural projections in planarian brain revealed by fluorescent dye tracing. Zoolog Sci 22, 535–546 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Sakai F, Agata K, Orii H, Watanabe K, Organization and regeneration ability of spontaneous supernumerary eyes in planarians-eye regeneration field and pathway selection by optic nerves. Zool. Sci 17, 375–381 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Petersen CP, Reddien PW, Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill EM, Petersen CP, Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217–4229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddien PW, The Cellular and Molecular Basis for Planarian Regeneration. Cell 175, 327–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapan SW, Reddien PW, dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet 7, e1002226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scimone ML, Srivastava M, Bell GW, Reddien PW, A regulatory program for excretory system regeneration in planarians. Development 138, 4387–4398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scimone ML, Cote LE, Reddien PW, Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551, 623–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scimone ML et al. , foxF-1 Controls Specification of Non-body Wall Muscle and Phagocytic Cells in Planarians. Curr Biol 28, 3787–3801 e3786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LoCascio SA, Lapan SW, Reddien PW, Eye Absence Does Not Regulate Planarian Stem Cells during Eye Regeneration. Dev Cell 40, 381–391 e383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petros TJ, Rebsam A, Mason CA, Retinal axon growth at the optic chiasm: to cross or not to cross. Annu Rev Neurosci 31, 295–315 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Inoue T et al. Morphological and functional recovery of the planarian photosensing system during head regeneration. Zool. Sci 21, 275–283 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Atabay KD, LoCascio SA, de Hoog T, Reddien PW, Self-organization and progenitor targeting generate stable patterns in planarian regeneration. Science, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapan SW, Reddien PW, Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. Cell Reports 2, 294–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cebria F, Newmark PA, Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132, 3691–3703 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Newmark PA, Reddien PW, Cebria F, Sànchez Alvarado A, Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci 100, 11861–11865 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cebria F, Guo T, Jopek J, Newmark PA, Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol 307, 394–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cebria F, Newmark PA, Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 134, 833–837 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Scimone ML, Cote LE, Rogers T, Reddien PW, Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cebria F et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620–624 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Lander R, Petersen CP, Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurley KA et al. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347, 24–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Augsburger A, Schuchardt A, Hoskins S, Dodd J, Butler S, BMPs as mediators of roof plate repulsion of commissural neurons. Neuron 24, 127–141 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Butler SJ, Dodd J, A role for BMP heterodimers in roof plate-mediated repulsion of commissural axons. Neuron 38, 389–401 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Reddien PW, Bermange AL, Kicza AM, Sànchez Alvarado A, BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043–4051 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Blader P, Rastegar S, Fischer N, Strahle U, Cleavage of the BMP-4 antagonist chordin by zebrafish tolloid. Science 278, 1937–1940 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Piccolo S et al. Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell 91, 407–416 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galko MJ, Tessier-Lavigne M, Function of an axonal chemoattractant modulated by metalloprotease activity. Science 289, 1365–1367 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Sretavan DW, Pure E, Siegel MW, Reichardt LF, Disruption of retinal axon ingrowth by ablation of embryonic mouse optic chiasm neurons. Science 269, 98–101 (1995). [DOI] [PubMed] [Google Scholar]
- 58.Cowles MW et al. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140, 4691–4702 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Scimone ML, Kravarik KM, Lapan SW, Reddien PW, Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem cell reports 3, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts-Galbraith RH, Brubacher JL, Newmark PA, A functional genomics screen in planarians reveals regulators of whole-brain regeneration. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW, Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cote LE, Simental E, Reddien PW, Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nat Commun 10, 1592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oderberg IM, Li DJ, Scimone ML, Gavino MA, Reddien PW, Landmarks in Existing Tissue at Wounds Are Utilized to Generate Pattern in Regenerating Tissue. Curr Biol 27, 733–742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayashi T et al. A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679–3688 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Petersen CP, Reddien PW, Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Jaworski A, Tessier-Lavigne M, Autocrine/juxtaparacrine regulation of axon fasciculation by Slit-Robo signaling. Nat Neurosci 15, 367–369 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


