Abstract
Background
Diet quality is a risk factor for chronic disease and mortality. Differential DNA methylation across the epigenome has been associated with chronic disease risk. Whether diet quality is associated with differential methylation is unknown. This study assessed whether diet quality was associated with differential DNA methylation measured across 445 548 loci in the Women’s Health Initiative (WHI) and the TwinsUK cohort.
Design
The discovery cohort consisted of 4355 women from the WHI. The replication cohort consisted of 571 mono- and dizygotic twins from the TwinsUK cohort. DNA methylation was measured in whole blood using the Illumina Infinium HumanMethylation450 Beadchip. Diet quality was assessed using the Alternative Healthy Eating Index 2010 (AHEI-2010). A meta-analysis, stratified by study cohort, was performed using generalized linear models that regressed methylation on AHEI-2010, adjusting for cell composition, chip number and location, study characteristics, principal components of genetic relatedness, age, smoking status, race/ethnicity and body mass index (BMI). Statistical significance was defined as a false discovery rate < 0.05. Significant sites were tested for replication in the TwinsUK cohort, with significant replication defined by P < 0.05 and a consistent direction.
Results
Diet quality was significantly associated with differential DNA methylation at 428 cytosine-phosphate-guanine (CpG) sites in the discovery cohort. A total of 24 CpG sites were consistent with replication in the TwinsUK cohort, more than would be expected by chance (P = 2.7x10-4), with one site replicated in both the blood and adipose tissue (cg16379999 located in the body of SEL1L).
Conclusions
Diet quality was associated with methylation at 24 CpG sites, several of which have been associated with adiposity, inflammation and dysglycaemia. These findings may provide insight into pathways through which diet influences chronic disease.
Keywords: Epigenome, diet quality, dietary epigenetics, EWAS, Women’s Health Initiative
Key Messages
Given the significant role of diet quality in non-communicable disease (NCD) progression and the more recent evidence establishing the relationship between differential DNA methylation and NCDs, this study examined whether diet quality is associated with differential DNA methylation and the potential functional effects of these differentially methylated sites.
Using genome-wide DNA methylation data, we found 24 CpG sites were associated with diet quality in both the discovery cohort (WHI) and the replication cohort (TwinsUK), with nearly all of the replicated sites (23 of 24) negatively associated with diet quality (poorer diet quality associated with higher methylation).
In one site, cg16379999 in the body of SEL1L, diet quality associated with blood and adipose tissue methylation in the same direction.
These findings may elucidate molecular pathways through which diet influences chronic disease risk.
Background
Poor diet quality is estimated to account for nearly half of the deaths attributable to coronary heart disease (CHD) and type 2 diabetes (T2DM) in the USA.1 Diet influences metabolic conditions, independent of energy balance and adiposity, through effects on glucose–insulin homeostasis, satiety, liver fat synthesis, adipocyte function and metabolic expenditure.2 Exposure to established non-communicable disease (NCD) risk factors, such as smoking,3 particulate matter exposure4 and physical activity,5 has been associated with differential DNA methylation (DNAm) patterns that contribute to regulation of gene expression. The impact of diet quality on the DNA methylome is not well understood. Given the significant influence of diet on NCD risk, diet could plausibly induce changes to DNAm on a causal disease pathway. Assessing the relationship between diet and the methylome, particularly independent of obesity, may reveal pathways linking diet and metabolic conditions.
Few studies have evaluated the association between diet and the methylome among adults, particularly in the context of diet quality and dietary factors causally associated with NCDs. Three studies have examined methylation changes in dietary clinical trials, including a high fat overfeeding trial and the Mediterranean diet.6–8 These studies found some differences in either mean gene methylation or cytosine-phosphate-guanine (CpG) site-specific methylation. Two cross-sectional studies conducted epigenome-wide association studies (EWAS) of dietary fat and fiber. These studies found differential methylation among genes potentially related to metabolism, though neither study validated findings in independent samples.9,10 While all of these studies report associations between dietary factors and the methylome, limitations in sample size and lack of replication support further investigation into the association of diet with the adult methylome. This study therefore evaluated the association between diet quality as measured by the Alternative Healthy Eating Index-2010 (AHEI-2010) and the methylome using cross-sectional data from the Women’s Health Initiative (WHI) and the TwinsUK cohort.
Methods
Study populations
The discovery cohort derives from the WHI, a large, US-based cohort study of post-menopausal women, aged 50–79 years at the time of enrollment, consisting of two study arms: the clinical trial (CT) and the observational study (OS). DNAm data from three ancillary studies in WHI were included: Epigenetic Mechanisms of Particulate Matter-Mediated Cardiovascular Disease (EMPC, n = 2200), the Integrative Genomics for Risk of Coronary Heart Disease and Related Phenotypes in the WHI cohort (BAA23, n = 2107), and Bladder Cancer and Leukocyte Methylation (AS311, n = 882). The replication cohort was derived from the TwinsUK cohort, a large registry of male and female twins between the ages of 19 and 82 years in the UK.11 DNAm derives from a sub-study of female twins (n = 571). Further description of both discovery and replication cohorts have been included in the Supplementary Methods and Results, available as Supplementary data at IJE online.
Inclusion/exclusion criteria
Participants from the WHI cohorts were included if they completed their food frequency questionnaire (FFQ) in the same year as the blood draw on which DNAm was measured. Participants were excluded if they did not have dietary information or if they reported implausible dietary intake (<600 kcal/day or ≥4000 kcal/day).12 These criteria were only used in the discovery cohort. In the replication cohort, TwinsUK, some of the DNAm and diet quality measurements were not obtained from the same time-point.13 The replication analyses included female monozygotic (MZ) and dizygotic (DZ) twins from the TwinsUK from all years of blood sampling for DNAm profiling. In sensitivity analyses, we restricted the replication sample further to those individuals with diet measured within 2 years and 1 year of 2007 (the year of methylation measurement).
Methylation data
Methylation was measured in DNA derived from whole blood samples using the Illumina Infinium HumanMethylation450 Beadchip. The methylation protocols and quality control (QC) procedures are described in the Supplementary Methods and Results, available as Supplementary data at IJE online. After QC, 445 548 CpG sites were available for analysis in the discovery cohort.
Dietary quality assessment
Diet quality was assessed on a scale of 0–100 (lower score indicates poor diet) using the AHEI-2010, which evaluates foods and nutrients strongly predictive of chronic disease.14 AHEI-2010 was assessed through participant FFQ and is composed of dietary and nutritional factors including: linolenic:linoleic fatty acid ratio, vegetable servings, fruit servings, whole grain servings, nuts and legumes servings, sugar-sweetened beverage servings, red/processed meat servings, sodium intake, trans fat intake and alcohol servings. The AHEI-2010 has been extensively evaluated and shown to associate prospectively with CHD and T2DM within the WHI15 and in other settings.14,16
Data analysis
We used R software to conduct all analyses. The discovery analysis flow chart is included as Supplementary Figure S1, available as Supplementary data at IJE online. Overall, 834 women were excluded due to missing dietary intake, implausible dietary intake or overlapping samples. The final discovery cohort included 4355 women. EWAS meta-analysis was conducted by separately regressing methylation β-values for each CpG site on continuous AHEI-2010 score for each ancillary study and combining through inverse-variance weighted meta-analysis. Models were adjusted for study-specific covariates including case/control status (BAA23 and AS311), study year (EMPC), randomization arm (OS vs CT) and CT participant type and randomization assignment (dietary modification, calcium/vitamin D trial or hormone replacement therapy trial). Covariates in all analyses included chip location, estimated cell type proportions, the top three principal components of genetic relatedness (when available), body mass index (BMI), smoking status, age and race/ethnicity, with a random effect for chip number. Significant sites were tested for replication in the TwinsUK cohort using generalized linear regression adjusting for cell composition, age, smoking and BMI as fixed effects, with random effects for chip number and location, genetic relatedness and zygosity. Significant sites were also explored for association between AHEI-2010 and adipose tissue DNAm in 400 female twins from the TwinsUK cohort.17 In the discovery analysis, significance was defined as a false discovery rate (FDR) < 0.05. In replication analyses, significance was defined as a P < 0.05 and a consistent direction of effect. Further description of these analyses is included in the Supplementary Methods and Results, available as Supplementary data at IJE online.
Additional post hoc analyses
We conducted additional post hoc analyses, including evaluation of methylation associated with gene expression in two external cohorts, enrichment testing, and several sensitivity analyses. Further description of these analyses is included in the Supplementary Methods and Results, available as Supplementary data at IJE online.
Results
Demographic characteristics are described by quartile of AHEI-2010 (Table 1). Older women had a higher AHEI-2010 score (indicating a healthier diet) compared with younger women. Those with higher BMI and obesity had a lower AHEI-2010 score. White women had a higher AHEI-2010 score compared with African-American and Hispanic women. Smoking status did not differ by quartile of diet quality.
Table 1.
Demographic and study characteristics by quartile of the AHEI-2010. Counts and means (SD) are presented for categorical and continuous variables, respectively. T-test and chi-square tests were used to examine differences by AHEI-2010 quartile. Quartiles defined as follows: Q1 is <42.7, Q2 is 42.7–49.2, Q3 is 49.3–56.7 and Q4 is >56.7
| n | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | P-value | |
|---|---|---|---|---|---|---|
| WHI Ancillary Study | ||||||
| EMPC | 1613 | 421 | 419 | 400 | 373 | <0.0001 |
| BAA23 | 1914 | 524 | 483 | 459 | 448 | |
| AS311 | 828 | 155 | 174 | 204 | 295 | |
| Clinical trial participant | ||||||
| Yes | 3536 | 926 | 918 | 861 | 831 | <0.0001 |
| No | 819 | 163 | 170 | 228 | 258 | |
| Case/control status (BAA23) | ||||||
| Case | 948 | 267 | 234 | 239 | 208 | 0.01 |
| Control | 966 | 254 | 254 | 225 | 233 | |
| Case/control status (AS311) | ||||||
| Case | 416 | 78 | 84 | 106 | 148 | <0.0001 |
| Control | 412 | 77 | 91 | 109 | 135 | |
| Study year | ||||||
| Baseline | 4097 | 1039 | 1028 | 1019 | 1011 | 0.29 |
| 3 years | 163 | 31 | 37 | 46 | 49 | |
| 6 years | 95 | 19 | 23 | 24 | 29 | |
| Age (years), mean (SD) | 64.0 (7.11) | 62.4 (7.1) | 64.1 (7.1) | 64.3 (7.0) | 65.1 (7.0) | <0.0001 |
| Race/Ethnicity | ||||||
| White | 2495 | 501 | 639 | 628 | 727 | <0.0001 |
| African-American | 1076 | 369 | 261 | 252 | 194 | |
| Hispanic/Latino | 610 | 190 | 153 | 157 | 110 | |
| Asian or Pacific Islander | 105 | 8 | 18 | 27 | 52 | |
| American Indian or Alaskan Native | 38 | 13 | 11 | 12 | 2 | |
| Other | 30 | 8 | 6 | 12 | 4 | |
| BMI (kg/m2) mean (SD) | 29.3 (6.1) | 30.8 (6.4) | 29.7 (6.2) | 29.0 (5.9) | 27.9 (5.5) | <0.0001 |
| BMI categories | ||||||
| Underweight | 23 | 2 | 5 | 10 | 6 | <0.0001 |
| Normal | 1060 | 177 | 246 | 281 | 356 | |
| Overweight | 1506 | 356 | 380 | 379 | 391 | |
| Obese | 1735 | 544 | 449 | 411 | 331 | |
| Smoking status | ||||||
| Former and current | 2108 | 507 | 508 | 549 | 544 | 0.15 |
| No | 2204 | 569 | 570 | 530 | 535 | |
| Income | ||||||
| <$20 000 | 1007 | 326 | 276 | 222 | 183 | <0.0001 |
| $20 000–$49 999 | 1888 | 481 | 464 | 488 | 455 | |
| >$50 000 | 1196 | 215 | 275 | 310 | 396 |
In the discovery analysis (n = 4355), AHEI-2010 was significantly associated with methylation levels of 428 CpG sites (Figure 1, Supplementary Table S1, available as Supplementary data at IJE online). On average, for every 1 SD increase in AHEI-2010 (9.9 units), the β-values (estimated methylation proportions) decreased by 0.0003 at the significant sites. In the WHI population, women in quartile 4 (best diet quality) had AHEI-2010 scores >56.7 and women in quartile 1 (worst diet quality) had AHEI-2010 scores <42.7. Women consuming the best diet had an average difference in methylation of 0.001 at the significant sites compared with those consuming the worst diet (Figure 2, Supplementary Table S2, available as Supplementary data at IJE online). Results of sensitivity analyses are included in the Supplementary Methods and Results, available as Supplementary data at IJE online.
Figure 1.
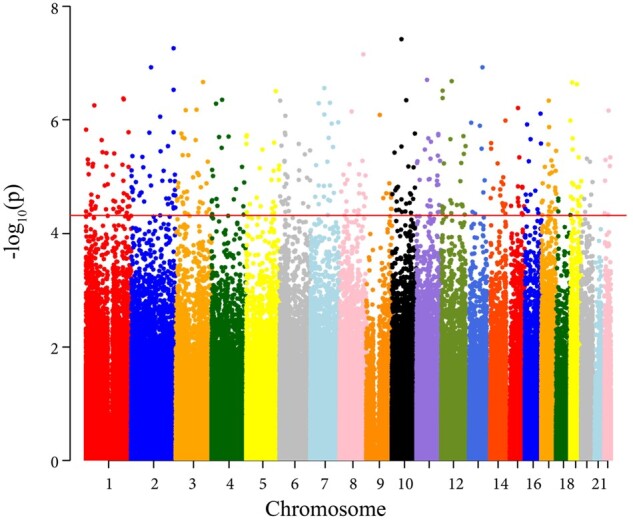
Manhattan Plot of the EWAS of diet quality. The x-axis represents chromosomal position and the y-axis represents P-values on the –log10 scale for each CpG site. The line denotes the threshold for significance P = 4.8 x 10–5.
Figure 2.
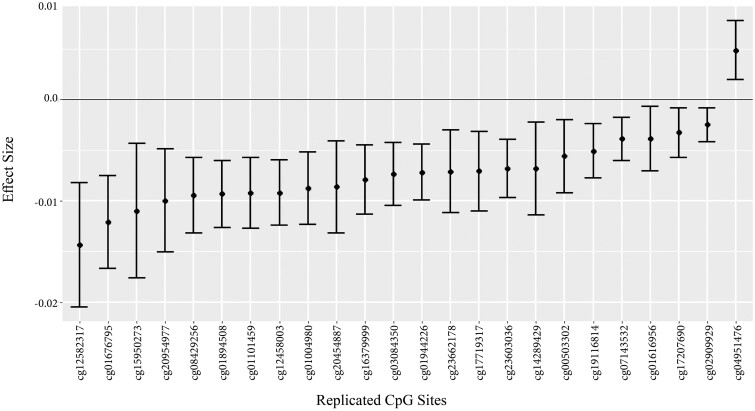
Difference in β-value of replicated CpG sites comparing the best diet score (AHEI-2010 > 56.7) to the worst diet score (AHEI-2010 < 42.7).
Replication in whole blood
A total of 419 of the 428 significant sites passed QC in the TwinsUK cohort and were tested for replication in whole blood samples from 571 women (Supplementary Table S3, available as Supplementary data at IJE online). AHEI-2010 score was significantly associated with methylation at 24 sites with a P < 0.05 and a consistent direction of effect (Table 2), more sites than would be expected by chance (binomial test P = 2.7x10−4). None of the sites was significant after FDR adjustment.
Table 2.
Replicated CpG sites associated with diet quality in the WHI and TwinsUK. Models were adjusted for age, ethnicity (WHI), smoking status, BMI, cell composition, top three principal components of genetic relatedness, study specific covariates (WHI), zygosity (TwinsUK) and batch effects
| WHI |
TwinsUK |
||||||
|---|---|---|---|---|---|---|---|
| CpG Site | Effect size | Standard error | P-value | Effect size | Standard error | P-value | Reference gene |
| cg00503302 | −2.60E−04 | 6.12E−05 | 1.82E−05 | −4.58E−03 | 2.34E−03 | 4.78E−02 | |
| cg01004980 | −2.60E−04 | 5.46E−05 | 2.10E−06 | −6.26E−03 | 2.53E−03 | 1.38E−02 | PRKAR2A |
| cg01101459 | −2.50E−04 | 5.50E−05 | 6.78E−06 | −7.16E−03 | 2.63E−03 | 6.62E−03 | |
| cg01616956 | −2.50E−04 | 5.14E−05 | 1.64E−06 | −5.90E−03 | 2.26E−03 | 8.56E−03 | NMUR1 |
| cg01676795 | −3.60E−04 | 6.92E−05 | 2.75E−07 | −7.75E−03 | 2.46E−03 | 1.47E−03 | POR |
| cg01894508 | −2.10E−04 | 4.94E−05 | 2.76E−05 | −5.48E−03 | 2.54E−03 | 3.03E−02 | ASPRV1 |
| cg01944226 | −1.80E−04 | 4.11E−05 | 2.00E−05 | −7.63E−03 | 3.31E−03 | 2.02E−02 | SLC16A3 |
| cg02909929 | −1.10E−04 | 2.58E−05 | 4.13E−05 | −5.08E−03 | 2.44E−03 | 3.53E−02 | PRF1 |
| cg03084350 | −2.20E−04 | 4.56E−05 | 2.03E−06 | −7.46E−03 | 2.91E−03 | 1.08E−02 | PLCD1 |
| cg04951476 | 2.25E−04 | 4.46E−05 | 4.59E−07 | 6.46E−03 | 2.51E−03 | 1.01E−02 | FAM50B |
| cg07143532 | −1.70E−04 | 3.57E−05 | 2.25E−06 | −6.41E−03 | 2.96E−03 | 3.11E−02 | COL24A1 |
| cg08429256 | −2.50E−04 | 5.49E−05 | 3.50E−06 | −1.10E−02 | 3.12E−03 | 3.68E−04 | SLC16A3 |
| cg12458003 | −2.50E−04 | 5.01E−05 | 4.16E−07 | −1.15E−02 | 3.53E−03 | 1.10E−03 | NFASC |
| cg12582317 | −4.10E−04 | 1.01E−04 | 4.22E−05 | −7.36E−03 | 2.76E−03 | 8.66E−03 | |
| cg14289429 | −3.00E−04 | 7.09E−05 | 2.01E−05 | −5.15E−03 | 2.17E−03 | 1.74E−02 | FAM78A |
| cg15950273 | −4.40E−04 | 1.08E−04 | 4.48E−05 | −6.69E−03 | 3.24E−03 | 3.67E−02 | TRAF3 |
| cg16379999 | −2.20E−04 | 5.07E−05 | 1.24E−05 | −4.43E−03 | 2.19E−03 | 4.55E−02 | SEL1L |
| cg17207690 | −1.50E−04 | 3.61E−05 | 2.11E−05 | −8.05E−03 | 2.57E−03 | 1.62E−03 | NMUR1 |
| cg17719317 | −2.90E−04 | 6.59E−05 | 9.76E−06 | −5.18E−03 | 2.64E−03 | 4.94E−02 | |
| cg19116814 | −1.80E−04 | 4.52E−05 | 4.65E−05 | −9.47E−03 | 3.95E−03 | 1.68E−02 | GPM6A |
| cg20454887 | −3.30E−04 | 7.79E−05 | 1.90E−05 | −5.18E−03 | 2.55E−03 | 4.07E−02 | |
| cg20954977 | −3.40E−04 | 8.16E−05 | 3.64E−05 | −6.99E−03 | 2.85E−03 | 1.43E−02 | B3GNT7 |
| cg23603036 | −2.20E−04 | 4.82E−05 | 5.92E−06 | −5.75E−03 | 2.24E−03 | 9.66E−03 | DHRS3 |
| cg23662178 | −2.60E−04 | 6.30E−05 | 2.89E−05 | −5.13E−03 | 2.61E−03 | 4.79E−02 | |
Replication in adipose tissue
A total of 421 of 428 CpG sites were examined in the adipose tissue of 400 female twins in the TwinsUK cohort (Supplementary Table S4, available as Supplementary data at IJE online). Diet quality was associated with 4 sites with a P < 0.05 and a consistent direction of effect, one of which was also replicated in the blood: cg16379999 (Supplementary Table S5, available as Supplementary data at IJE online). None of the sites was significant after FDR adjustment.
Enrichment
We examined whether the sites identified in the primary analysis were expression quantitative trait methylation loci (eQTMs) found in a previous study of the Grady Trauma Project (GTP) and the Multi-Ethnic Study of Atherosclerosis (MESA).18 The 428 CpGs identified in the discovery analysis were associated with expression of 412 genes in the eQTM database (P < 1x10−5, Supplementary Table S6, available as Supplementary data at IJE online),18 for a total of 1842 CpG-transcript associations. Gene ontology analysis of these 412 genes revealed enrichment for 342 ontologies (FDR < 0.05, Supplementary Table S7, available as Supplementary data at IJE online), which were primarily immune response pathways with several pathways related to metabolism, including regulation of proteins and protein transport, response to fatty acid and cellular response to low-density lipoprotein particle stimulus. We next examined whether cg16379999 associated with expression of specific genes. In the MESA study, cg16379999 (on chromosome 14) positively associated with increased expression in ABHD3 gene (on chromosome 18) representing a trans association between methylation and expression (P = 9.02 x 10–6).
Discussion
Diet quality was associated with 428 CpG sites in the discovery cohort of post-menopausal women from the WHI, with 24 sites consistent with replication, one of which was associated with blood and adipose tissue in a consistent direction.
Among the 24 sites, several have been previously associated with diet-related outcomes. BMI has been associated with cg01101459 in an unannotated gene,19,20 cg12458003 in the body of NFASC,21 cg20954977 in the transcription start site of B3GNT722 and cg01676795 in the body of POR.23 In all the above sites, methylation was negatively associated with diet quality (poorer diets had the highest methylation), and in previous studies these sites were positively associated with BMI. These findings align with our study since poor diet is associated with higher BMI. cg01101459 has also been associated with chronic low-grade inflammation,24 with a positive association between methylation and C-reactive protein (CRP). CRP is another cardiometabolic risk factor playing a direct role in disease progression,25 which has been found to associate with diet patterns,26–29 such that poorer diets can lead to elevated CRP. cg01676795 has been found to associate with dysglycaemia in several studies.23,30 In these studies, higher methylation was positively associated with fasting insulin23 and haemoglobin A1c,23,30 which corroborate our findings, as individuals with the poorest diet quality had the highest methylation.
cg16379999 was found to negatively associate with diet quality in both the blood and adipose tissue. cg16379999 is located in the body of SEL1L. This site has been previously found to associate with obesity,31 air pollution,32 smoking33 and vitamin B12 supplementation.34SEL1L has been shown to play a significant role in lipid metabolism as a regulator of lipoprotein lipase (LPL) secretion.35,36SEL1L knock-out mouse models have elevated fibroblast growth factor 21 (FGF21), a critical metabolic hormone regulating growth, nutrient metabolism and insulin,37 and elevated levels have been associated with obesity38 and have predicted myocardial infarction.39,40 In our study, diet quality was negatively associated with methylation at this site. As this site is located in the gene body, the implications may be difficult to infer as mixed evidence has been reported on the effects of gene body methylation on gene expression.41 However, a large EWAS of mRNA transcripts from the MESA and GTP cohorts found that gene body methylation correlated with reduced gene expression 61% and 72% of the time, respectively,18 which would align with our study. As this gene may play a protective role against metabolic disturbances, the higher methylation patterns associated with poor diet would be deleterious. Methylation was also shown to associate with expression in the ABHD3 gene in the MESA study. ABHD3 has been shown to play a catabolic role in medium-chain and oxidatively-truncated phospholipids.42,43
The 428 CpG sites identified in the discovery cohort were also found to associate with differential expression of 412 genes in the blood. According to gene ontology analysis, this set of genes was enriched for primarily immune response pathways. This finding supports the role of diet quality in the immune response and potentially in an upstream effect of diet on cardiometabolic diseases. Although we adjusted for differences in cell composition,44 there are potentially systemic differences in rarer cell types that would not be captured using this method. Thus the methylation differences we identified may be due to differential inflammatory profiles associated with poor diet. Indeed diet quality was shown to be significantly correlated with natural killer cells, granulocytes and CD8 lymphocytes, even when adjusted for BMI (Supplementary Table S8, available as Supplementary data at IJE online). Improving diet quality has been shown to improve inflammatory profiles and decrease inflammatory markers such as CRP and tumor necrosis factor α.28,29 Moreover, one replicated site was previously associated with CRP levels.24
We conducted several sensitivity analyses in the discovery analysis (exclusion of individual ancillary studies, exclusion of bladder cancer cases, and additional adjustments for BMI and socio-economic status). Although all of these analyses resulted in a change in the number of significant sites (ranging from 0 to 1851 CpG sites), any change in significance was likely due to a change in power as there was very little variation in the effect size (correlation of effect sizes between analyses was >0.98 for all analyses, see Supplementary Methods and Results, available as Supplementary data at IJE online).
Several studies have evaluated the association between various aspects of diet quality and the methylome longitudinally6–8 and cross-sectionally.9,10 One study evaluated adipose methylation following overfeeding of saturated or polyunsaturated fats in 31 participants, finding increased and decreased methylation at 4795 and 138 CpG sites, respectively, and changes in gene expression with saturated fat overfeeding.6 Two studies examined methylation changes following a long-term Mediterranean diet in 40 participants. As neither study observed significant differences when applying a genome-wide significance level, they subsequently filtered CpG sites based on change in methylation for an ingenuity pathway analysis, and reported enrichment in inflammatory pathways.7,8 Two cross-sectional studies have examined metrics of diet quality via EWAS. An EWAS of dietary fat quality conducted in pre-adolescents identified a number of CpG sites and pathways associated with dietary fat quality.10 An EWAS of dietary fiber in African-American adolescents reported three differentially methylated sites in genes associated with adiposity and inflammation.9 However results from these studies have not been replicated, and these CpGs were not significant in our study. Finally, a recent EWAS examined the AHEI-2010 score and the Mediterranean-style diet score (MDS) in 5 population based cohorts. They found significant associations with DNAm and diet quality in 30 sites.45No overlapping sites were identified in our study.
Because the discovery analysis found small effect sizes (±0.0003 per 1 SD diet quality), the biological implications are difficult to infer. A recent review found that most environmental studies resulted in a 2–8% difference in methylation between exposed and unexposed.46 In our study, the best diet had as much as a ∼2% difference in β-value compared with the worst diet (Supplementary Table S2, available as Supplementary data at IJE online, Figure 2). Thus our findings are slightly below the average effect. In terms of functional implications, we do not know what impact this may have on gene expression. However, studies have found differences in expression associated with methylation effect sizes as low as 0.02.47–49
Some limitations in our study are also important to note. There may be epigenetic differences that we were unable to discover due to a narrow distribution of diet quality in the discovery study population and competing effects of nutrients on the epigenome. In the replication analysis, we had the power to detect associations explaining >1% of variation in methylation; however, the partial r2 contribution of diet observed in our discovery analysis was only above this in 76 of the 428 sites in more than one individual ancillary study model (Supplementary Table S1, available as Supplementary data at IJE online). We included women from the TwinsUK cohort with methylation measured within 3 years of diet quality, which may have influenced our replication results. However, the direction of association did not differ in the replicated sites when we restricted the analysis to individuals with methylation and diet quality measured within 2 years or 1 year. The TwinsUK cohort also differed from the WHI cohorts as they were younger and racially homogenous (Supplementary Table S9, available as Supplementary data at IJE online), nevertheless we were able to replicate 24 sites. Additionally, given that the WHI was conducted in post-menopausal women and the TwinsUK cohort was only in women, generalizability to other populations may be limited.
Another potential limitation is the use of blood-based methylation in the context of diet quality. To examine the biological impact of diet on the methylome, the diet-associated blood methylation would correlate with the tissue of interest that is most impacted by diet. We examined adipose tissue methylation and were able to replicate one significant site. Other relevant tissues might include the liver and gastrointestinal cells. However, few studies have examined methylation in these tissues.
In summary, diet quality was significantly associated with methylation at 24 CpG sites in the blood and one site in the adipose tissue among adult women. These sites may mark molecular pathways underlying diet and chronic disease, especially given the previous identification of associations between several of these sites and cardiometabolic risk factors in previous studies.19–24,30,31 Future research should utilize more precise and unbiased estimates of diet quality through use of dietary biomarkers and metabolomic indices to fully elucidate the effect of diet quality on the epigenome.
Supplementary data
Supplementary data are available at IJE online
Funding
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The TwinsUK study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007–2013); National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. The TwinsUK methylation study received support from the ESRC (ES/N000404/1 to J.T.B.) and the Joint Programming Initiative HDHL DIMENSION (administered by the BBSRC UK, BB/S020845/1 to J.T.B.). K.M.V.N., L.S. and W.L.D. were supported in part by the National Institute of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under award number P30DK111024 and the Hubert Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. W.L.D. was supported in part by the Nalini and Ravi Saligram Scholarship. This work was supported in part by National Institute of Environmental Health Sciences grant R01-ES020836 (L.H., E.A.W.) and National Heart, Lung, and Blood Institute contract HHSN268201100046C (K.N.C.). P.B. and K.J. were supported by the American Cancer Society (125299-RSG-13–100-01-CCE) with additional support for K.J. through National Cancer Institute training grants (R25 CA094880 and T32 CA094880). S.H. acknowledges support by U01 AG060908/AG/NIA NIH HHS/US. Y.L. was supported through R01 HL129132.
Supplementary Material
Acknowledgements
The authors thank the WHI investigators and staff for their dedication, and the study participants for making the programme possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 10/15/2019.
Author contributions
W.L.D., K.M.V.N. and K.N.C. conceived of the study. The methodology was developed by W.L.D., K.M.V.N., E.A.W. and K.N.C. The data for WHI were curated by E.A.W., S.H., T.L.A., Y.L., L.H., P.B. and K.J. The TwinsUK data were curated by J.T.B. The formal analysis was completed by W.L.D. with support from K.N.C. Replication analyses were completed by R.C., O.M.M., C.I.L.R. and J.T.B. The manuscript was written by W.L.D. Editorial and content expertise was provided by all authors.
Data availability
The WHI methylation and clinical data are available through the Women’s Health Initiative website. The majority of the TwinsUK methylation datasets analysed in the current study are available through GEO GSE62992 and GSE121633 (blood methylation) and ArrayExpress E-MTAB-1866 (adipose methylation).
Conflict of interest
None declared.
References
- 1. GBD 2015 Risk Factors Collaborators. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1659–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity: a comprehensive review. Circulation 2016;133:187–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prince C, Hammerton G, Taylor AE. et al. Investigating the impact of cigarette smoking behaviours on DNA methylation patterns in adolescence. Hum Mol Genet 2019;28:155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Madrigano J, Baccarelli A, Mittleman Murray A. et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect 2011;119:977–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nitert MD, Dayeh T, Volkov P. et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 2012;61:3322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perfilyev A, Dahlman I, Gillberg L. et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr 2017;105:991–1000. [DOI] [PubMed] [Google Scholar]
- 7. Arpón A, Milagro FI, Razquin C. et al. Impact of consuming extra-virgin olive oil or nuts within a Mediterranean diet on DNA methylation in peripheral white blood cells within the PREDIMED-Navarra randomized controlled trial: a role for dietary lipids. Nutrients 2017;10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arpon A, Riezu-Boj JI, Milagro FI. et al. Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J Physiol Biochem 2016;73:445–55. [DOI] [PubMed] [Google Scholar]
- 9. Chen L, Dong Y, Wang X. et al. Epigenome-wide association study of dietary fiber intake in African American Adolescents. Mol Nutr Food Res 2018;62:e1800155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Voisin S, Almén MS, Moschonis G, Chrousos GP, Manios Y, Schiöth HB.. Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of Greek preadolescents. Eur J Hum Genet 2015;23:654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moayyeri A, Hammond CJ, Valdes AM, Spector TD.. Cohort Profile: TwinsUK and healthy ageing twin study. Int J Epidemiol 2013;42:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patterson RE, Kristal AR, Tinker LF, Carter RA, Bolton MP, Agurs-Collins T.. Measurement characteristics of the Women's Health Initiative food frequency questionnaire. Ann Epidemiol 1999;9:178–87. [DOI] [PubMed] [Google Scholar]
- 13. Bowyer RCE, Jackson MA, Pallister T. et al. Use of dietary indices to control for diet in human gut microbiota studies. Microbiome 2018;6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiuve SE, Fung TT, Rimm EB. et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr 2012;142:1009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. George SM, Ballard-Barbash R, Manson JE. et al. Comparing indices of diet quality with chronic disease mortality risk in postmenopausal women in the Women's Health Initiative Observational Study: Evidence to Inform National Dietary Guidance. Am J Epidemiol 2014;180:616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwingshackl L, Bogensberger B, Hoffmann G.. Diet quality as assessed by the healthy eating index, alternate healthy eating index, dietary approaches to stop hypertension score, and health outcomes: an updated systematic review and meta-analysis of cohort studies. J Acad Nutr Diet 2018;118:74–100.e11. [DOI] [PubMed] [Google Scholar]
- 17. Grundberg E, Meduri E, Sandling JK. et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet 2013;93:876–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kennedy EM, Goehring GN, Nichols MH. et al. An integrated -omics analysis of the epigenetic landscape of gene expression in human blood cells. BMC Genomics 2018;19:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hannon E, Knox O, Sugden K. et al. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet 2018;14:e1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wahl S, Drong A, Lehne B. et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017;541:81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mendelson MM, Marioni RE, Joehanes R. et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: A Mendelian randomization approach. PLoS Med 2017;14:e1002215-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demerath EW, Guan W, Grove ML. et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015;24:4464–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cardona A, Day FR, Perry JRB. et al. Epigenome-wide association study of incident type 2 diabetes in a British population: EPIC-Norfolk Study. Diabetes 2019;68:2315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ligthart S, Marzi C, Aslibekyan S. et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol 2016;17:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Minihane AM, Vinoy S, Russell WR. et al. Low-grade inflammation, diet composition and health: current research evidence and its translation. Br J Nutr 2015;114:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dias JA, Wirfält E, Drake I. et al. A high quality diet is associated with reduced systemic inflammation in middle-aged individuals. Atherosclerosis 2015;238:38–44. [DOI] [PubMed] [Google Scholar]
- 27. Barbaresko J, Koch M, Schulze MB, Nöthlings U.. Dietary pattern analysis and biomarkers of low-grade inflammation: a systematic literature review. Nutr Rev 2013;71:511–27. [DOI] [PubMed] [Google Scholar]
- 28. Arnold K, Weinhold KR, Andridge R, Johnson K, Orchard TS.. Improving diet quality is associated with decreased inflammation: findings from a pilot intervention in postmenopausal women with obesity. J Acad Nutr Diet 2018;118:2135–43. [DOI] [PubMed] [Google Scholar]
- 29. Piccand E, Vollenweider P, Guessous I, Marques-Vidal P.. Association between dietary intake and inflammatory markers: results from the CoLaus study. Public Health Nutr 2019;22:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E. et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet 2016;25:609–19. [DOI] [PubMed] [Google Scholar]
- 31. Cheng Y, Monteiro C, Matos A. et al. Epigenome-wide DNA methylation profiling of periprostatic adipose tissue in prostate cancer patients with excess adiposity-a pilot study. Clin Epigenet 2018;10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de F C Lichtenfels AJ, van der Plaat DA, de Jong K. et al. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the LifeLines Cohort Study. Environ Health Perspect 2018;126:027004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joehanes R, Just AC, Marioni RE. et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 2016;9:436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yadav DK, Shrestha S, Lillycrop KA. et al. Vitamin B12 supplementation influences methylation of genes associated with Type 2 diabetes and its intermediate traits. Epigenomics 2018;10:71–90. [DOI] [PubMed] [Google Scholar]
- 35.SEL1L Gene. [cited; Available from: https://www.genecards.org/cgi-bin/carddisp.pl? gene=SEL1L.
- 36. Sha H, Sun S, Francisco AB. et al. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab 2014;20:458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhattacharya A, Sun S, Wang H. et al. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. Embo J 2018;37:e99277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang X, Yeung DCY, Karpisek M. et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes 2008;57:1246–53. [DOI] [PubMed] [Google Scholar]
- 39. Zhang W, Chu S, Ding W, Wang F.. Serum level of fibroblast growth factor 21 Is independently associated with acute myocardial infarction. PloS One 2015;10:e0129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen Y, Zhang X, Xu Y. et al. Serum FGF21 is associated with future cardiovascular events in patients with coronary artery disease. Cardiology 2018;139:212–28. [DOI] [PubMed] [Google Scholar]
- 41. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- 42. Long JZ, Cisar JS, Milliken D. et al. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol 2011;7:763–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lord CC, Thomas G, Brown JM.. Mammalian alpha beta hydrolase domain (ABHD) proteins: lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim Biophys Acta 2013;1831:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Houseman EA, Molitor J, Marsit CJ.. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 2014;30:1431–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma J, , Rebholz CM, , Braun K et al . Whole Blood DNA Methylation Signatures of Diet Are Associated With Cardiovascular Disease Risk Factors and All-Cause Mortality. Circ Genom Precis Med 2020;13:e002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Breton CV, Marsit CJ, Faustman E. et al. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: The Children's Environmental Health and Disease Prevention Research Center's Epigenetics Working Group. Environ Health Perspect 2017;125:511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Argos M, Chen L, Jasmine F. et al. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect 2015;123:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Everson TM, Punshon T, Jackson BP. et al. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of Two U.S. Birth Cohorts. Environ Health Perspect 2018;126:017010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee MK, Hong Y, Kim S-Y, London SJ, Kim WJ.. DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin Epigenet 2016;8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WHI methylation and clinical data are available through the Women’s Health Initiative website. The majority of the TwinsUK methylation datasets analysed in the current study are available through GEO GSE62992 and GSE121633 (blood methylation) and ArrayExpress E-MTAB-1866 (adipose methylation).