Abstract
Treatment of tuberculosis requires a multi-drug regimen administered for at least 6 months. The long-term chemotherapy is attributed in part to a minor subpopulation of nonreplicating Mycobacterium tuberculosis cells that exhibit phenotypic tolerance to antibiotics. The origins of these cells in infected hosts remain unclear. Here we discuss some recent evidence supporting the hypothesis that hibernation of ribosomes in M. tuberculosis, induced by zinc starvation, could be one of the primary mechanisms driving the development of nonreplicating persisters in hosts. We further analyse inconsistencies in previously reported studies to clarify the molecular principles underlying mycobacterial ribosome hibernation.
Keywords: Mycobacterium tuberculosis, nonreplicating persisters, drug tolerance, ribosome hibernation, zinc starvation, cryo-EM structure analysis
Introduction
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), surpassed HIV in 2019 as the leading cause of mortality in human population by a single infectious agent [1], until it was replaced by SARS-CoV-2 in 2020. The failure frequency of standard treatment regimen for TB is on the rise with emergence of multi-drug-resistant (MDR-TB) strains [1, 2]. While the recent implementation of a 6 month all-oral regimen of new drugs for MDR-TB is a landmark achievement in our long battle against TB [2, 3], a new drug that shortens the treatment remains an unmet challenge. The challenges of daily administration of antibiotics for months under the DOTS (directly observed treatment, short-course) strategy have never been more apparent than during the ongoing COVID-19 pandemic [4, 5].
A lengthy treatment for TB is partly attributed to a specialized subpopulation of Mtb cells with phenotypic tolerance to antibiotics [6–9]. Moreover, these cells, which are also called persisters, are considered to be nonreplicating and metabolically slow, presumably residing in bacteriostatic microenvironments within host tissues [10–13]. A strict requirement of oxygen for growth of Mtb has led to a long-standing hypothesis that hypoxia is an important feature of TB lesions harbouring nonreplicating bacilli, thereby rationalizing in vitro hypoxic cultures as the suitable model to understand dormancy and persistence of the pathogen in vivo [14–22]. However, an antibiotic such as metronidazole that can sterilize nonreplicating Mtb cells in hypoxic cultures in vitro has inconclusive sterilizing activity in various animal models, including those with hypoxic TB lesions [23–27]. Together, these studies suggest that in vivo persisters of Mtb likely have multiple origins, including hypoxia. Following the landmark study by Balaban et al. [28] that provided a visual evidence for the correlation between growth rate and phenotypic drug tolerance in Escherichia coli , studies published over the last decade have expanded our understanding of drug tolerance in mycobacteria [6, 29–31]. Phenotypic drug tolerance in mycobacteria has been broadly categorized into two distinct classes [6, 32]. Class I phenotypic tolerance is proposed to be exhibited by rare cells that emerge from metabolic alterations of diverse origins that may or may not change the growth rate. Examples of such metabolic alterations include asymmetric cell division [33], mistranslation of RpoB [34], stochastic fluctuation in KatG [35] or DnaK [36] expression. Class I tolerance is antibiotic-specific and combination therapy is argued as an effective strategy to counter this phenomenon. By contrast, class II phenotypic tolerance is expressed by the entire population that are rendered nonreplicating by lack of conducive growth conditions such as hypoxia, carbon starvation, low pH [6]. Class II phenotypic tolerance is proposed to be effective against multiple antibiotics, but can be reversed by lifting the underlying condition for growth restriction [6]. Two recent studies offer yet another example of phenotypic drug tolerance in Mtb emerging from a phase variation through hypermutable region in the glycerol kinase gene glpK [37, 38]. As a result of these studies, more inclusive screening approaches have been proposed for discovering small molecules that target phenotypic drug tolerance in mycobacteria [32, 38–42]. However, a significant knowledge gap remains in our understanding of the host microenvironments that give rise to drug-tolerant subpopulation of Mtb in vivo. Addressing this gap is important for maximizing the therapeutic potential of compounds discovered using in vitro models of drug tolerant persisters.
Depletion of zinc to a growth-restrictive level induces hibernation of 70S ribosomes by recruitment of a mycobacterial protein Y (Mpy) [43]. The 70S-Mpy complexes are stabilized as translationally inert particles in a conformational state that resists the binding of kanamycin and streptomycin by hindering the dynamics of the ribosomal RNA (rRNA) bases and protein–RNA interfaces necessary for stable drug binding [43]. Moreover, evidence of zinc-limiting host conditions and Mpy-dependent streptomycin resistance in Mtb during infection [44] together suggest that ribosome hibernation is likely induced in the bacterial population during infection. Here, we review these findings in the context of a hypothesis that ribosome hibernation could be one of the primary survival mechanisms of nonreplicating Mtb during infection. Structure-based design of small-molecule modulators of ribosome hibernation could lead to new therapeutic approaches against persister cells of Mtb.
Ribosome hibernation: a conserved mechanism of persistence in bacteria
The 70S ribosome in bacteria is a dynamic macromolecular machine that performs the essential function of decoding messenger RNAs into polypeptides. Its two unequal-sized subunits – each of which is assembled independently with ribosomal RNA (16S RNA for the smaller 30S subunit, and 23S and 5S RNA for the larger 50S subunit) and multiple ribosomal proteins (over 20 proteins in the 30S subunit and over 30 proteins in the 50S subunit) – interact with the canonical ligands of protein synthesis (mRNA, tRNA as well as initiation, elongation, release and recycling factors) in a spatiotemporally orchestrated manner to synthesize a polypeptide at an average speed of 20 amino acids/second [45]. Over 40 % of the total energy in a growing bacterial cell is consumed in the process of protein synthesis [46]. Not surprisingly, mechanisms regulating the biogenesis of ribosomes and the global translation activity are intricately linked to the cellular growth rate. Unfavourable growth conditions trigger a range of responses in bacterial cells to induce programmed slowdown of metabolism and reduced synthesis of ribosomes. Downregulation of ribosome biosynthesis in many bacterial species is achieved through the stringent response, characterized by accumulation of guanosine (penta)tetraphosphate, (p)ppGpp [47–49]. (p)ppGpp downregulates ribosomal RNA (rRNA) transcription, and thus the global protein synthesis, by directly modulating the interaction between rRNA promoters and RNA polymerase [47–49]. Growth stasis would also generate a sizable pool of nontranslating ribosomes, the fate of which remains a long-standing subject of interest. Degradation of ribosomes during starvation was noted by Jacobson and Gillespie [50], although a later study clarified that degradation is limited to free 30S and 50S subunits and that 70S ribosomes remain largely stable [51]. The intactness of the 70S ribosome in stationary-phase cells is consistent with a growing body of evidence indicating hibernation of ribosomes in growth-arrested bacteria. Ribosome hibernation can be broadly defined as transient and reversible inactivation and stabilization of nontranslating ribosomes by specialized proteins [52]. The presence of stabilized ribosomes in stationary-phase Escherichia coli cells was speculated more than 60 years ago upon visualization of larger-than-usual sized ribonucleoparticles [53, 54]. These particles were later characterized by Ishihama and colleagues as 100S disomes (dimers of the 70S ribosome), and found to be produced predominantly in stationary-phase cells [55]. Moreover, the study identified a protein called ribosome modulating factor (Rmf) associated with the 100S disome [55]. Agafonov et al. later identified another protein, called protein Y (encoded by yfiA, also called raiA), which stabilized and inactivated the ribosome in stationary-phase cells as 70S monosomes [56]. Soon after, Wada and colleagues discovered YhbH (later called hibernation promoting factor, or Hpf) as the second protein associated with 100S ribosomes, and proposed that protein Y and YhbH bind to mutually exclusive populations of ribosomes [57]. Structural analysis of in vitro reconstituted complexes of hibernating ribosomes by Steitz and coworkers elucidated the mechanistic differences between the formation of protein Y bound hibernating 70S and Rmf-Hpf bound 100S ribosomes [58]. Ribosome hibernation is a widely conserved phenomenon in bacteria and eukaryotes [59–66], although unlike E. coli that harbours both hibernating 70S and 100S particles, a majority of characterized bacterial species appear to exclusively harbour 100S disomes. Moreover, the formation of 100S disomes in these species also appears to be different from that in E. coli . While E. coli and other γ-proteobacteria utilize two proteins (Rmf and Hpf) to dimerize 70 ribosomes, other bacterial species forming 100S disomes do not encode Rmf; these species instead encode a longer variant of Hpf, called Hpflong, as a necessary and sufficient factor for 70S dimerization [64] (Fig. 1). Compared to the Hpfshort in E. coli and other γ-proteobacteria, the Hpflong variants have an extended C-terminal region, which forms the dimer interface on the 30S subunit of the respective 70S monomer [67–71] (Fig. 1). Unlike the C-terminal domain of Hpflong, no direct interaction between Rmf of the two monomers could be detected [58, 72, 73]. Instead, Rmf induces conformational changes in the 30S subunit that has been proposed to increase its affinity towards the 30S subunit of the other monomer [58, 72, 73].
Fig. 1.
Comparing various forms of ribosome hibernation factors. (a) Schematic representation of domain organization in short and long variants of Hpf (named Hpfshort and Hpflong, respectively), YfiA and PSRP1. It should be noted that despite an extended C-terminal region in PSRP1 (also referred to as protein Y), it forms a hibernating 70S ribosome. This is also observed for the ribosome hibernation factor in mycobacteria (MSMEG_1878), which is therefore named mycobacterial protein Y (Mpy). (b) Alignment of the protein sequences of representative ribosome hibernation factors from each of the categories shown in (a). An additional N-terminal extension of variable length in PSRP1 and Mpy is notable.
The process regulating the formation of hibernating ribosomes has been investigated in many species (recently reviewed in [52, 74]). These investigations together appear to suggest that each species has evolved to sense distinct signals for inducing ribosome hibernation. For example, (p)ppGpp induces Rmf In E. coli and Hpflong in Bacillus subtilis under nutrient starvation [61, 75], while darkness appears to be the signal for (p)ppGpp-dependent upregulation of Hpflong in Synechococcus elongatus [76]. (p)ppGpp however has no direct role in regulating the levels of Hpflong in Staphylococcus aureus [77]. Moreover, downregulation of yfiA expression in E. coli cells during growth arresting condition of anaerobiosis [78, 79] suggests that regulation of ribosome hibernation is unlikely a default consequence of slowed translation, instead requires specific environmental signals.
Ribosome hibernation is hypothesized to confer stability to nontranslating ribosomes in a state that can revert back to yield translating particles upon growth resumption of cells [55]. Resuspension of stationary-phase E. coli cells in fresh medium results in rapid dissociation of Rmf from 100S disomes and subsequent regeneration of active 70S monosomes [80]. The molecular mechanism of reactivation of hibernating ribosome is better understood in S. aureus, in which the 100S ribosome is split into the constituent 70S monosomes by HflX [77], a conserved GTPase known to split stalled 70S ribosomes during translation [81, 82]. The requirement of GTP as the energy source by Hflx during disassembly of 100S fits well with the argument that reactivation of hibernating ribosomes and cellular growth must be coordinated at molecular levels. Although the physiological consequences of ribosome hibernation remain to be fully understood, prolonged survival of growth-arrested bacterial cells is a common observation reported for P. aeruginosa [83], Vibrio cholerae [84], B. subtilis [85], L. lactis [62], L. monocytogenes [63] and S. aureus [86]. Moreover, ribosome hibernation in L. monocytogenes also confers aminoglycoside resistance [87]. Taken together, ribosome hibernation is a highly conserved and tightly regulated process in bacteria that promotes persistence of growth-arrested cells in a drug tolerant state.
Ribosome hibernation in mycobacteria
The genome of Mtb encodes two homologues of Hpf/YfiA, Rv0079 and Rv3241c, encoding proteins of 273 and 219 amino acid residues, respectively (Fig. 1). While Rv3241c orthologues are universally present in all mycobacterial genomes sequenced so far, an orthologue of Rv0079 is absent in Mycobacterium abscessus , Mycobacterium avium and Mycobacterium leprae [88]. The physiological significance of these proteins was first reported by Williams and coworkers, who observed that Rv0079 and its orthologue in Mycobacterium smegmatis (MSMEG_3935) are induced under hypoxia through DosRS-dependent mechanism [89]. Expression of MSMEG_3935 was associated with stabilization of the 30S subunit, promoting cellular viability in growth-arrested state under hypoxia [89]. Low-level association of ribosomes with either MSMEG_3935 or the Rv3241c orthologue (MSMEG_1878) could also be found in stationary-phase cells but not in actively growing cells [89], suggesting that perhaps other conditions also induce ribosome hibernation by these proteins. Later, studies in our laboratories discovered that depletion of zinc was a condition for inducing hibernation of 70S ribosomes by MSMEG_1878 [43]. We further reported the first high-resolution structure of hibernating ribosomes from mycobacteria [43], which allowed characterization of several important features of MSMEG_1878. Besides the characteristic βαβββα topology of its N-terminal domain, which occupies the space between the two ribosomal subunits and blocks translation, the protein has unstructured C-terminal region of similar length as Hpflong in other bacteria (Fig. 1) [43]. However, unlike Hpflong, MSMEG_1878 forms a hibernating 70S ribosome [43] – a feature similar to protein Y of chloroplast (also called plastid-specific ribosomal protein 1 or PSRP1, (Fig. 1) [66]. MSMEG_1878 therefore was named mycobacterial protein Y (Mpy) [43].
The N-terminal region of Mpy binds to the 30S subunit encompassing the decoding region and making contacts with multiple 16S rRNA helices, including helix 18 (h18, shoulder), h24 (platform), h28 (neck), h31 and h34 (head) and h44 (body), as well as with proteins S3 and S9 [43]. These interactions essentially preclude the binding of canonical protein synthesis ligands (mRNA and tRNAs), thereby arresting the 70S ribosomes in a nontranslating state. Mpy-bound ribosomes, however, are more stable in growth-arrested mycobacterial cells than their Mpy-free counterparts, conferring viability to these cells for an extended period of time in zinc-depleted phosphate-buffered saline [43]. Besides stabilizing the nontranslating ribosome in a growth-arrested mycobacteria, Mpy directly protects the ribosome from multiple antibiotics. Interaction of Mpy with helix 44 directly alters the rRNA dynamics of the ribosome necessary for stable binding of kanamycin and streptomycin – both classes of antibiotics also bind to h44 [43, 90]. Specifically, flipping of the universally conserved rRNA bases A1476 and A1477 (A1492 and A1493 in E. coli ), which occurs upon kanamycin binding and subsequent stabilization of the mini-helix formed by the mRNA codon and the near-cognate anticodon of tRNA [91], is sterically hindered by residue Y107 of Mpy [43]. Streptomycin binding is likely destabilized by additional opening of the streptomycin binding pocket by about 3 Å in the Mpy-bound ribosome [43]. These structural insights of hibernating ribosomes could also explain the Hpf-dependent aminoglycoside resistance observed in stationary-phase L. monocytogenes [87].
Zinc depletion: a specific signal for ribosome hibernation in mycobacteria
Discovery of hibernating ribosomes in zinc-starved mycobacteria was rather unexpected: our original goal was to determine the structural and functional changes resulting from remodelling of ribosomes induced by zinc starvation. Ribosome remodelling involves replacement of multiple ribosomal (r-) proteins containing zinc binding CXXC motif (called C+r-proteins) by their motif-free C- paralogues, which have significant differences in their primary sequences and secondary structures [43, 92, 93]. Ribosome remodelling is a highly conserved process in bacterial species and transcriptionally controlled by a zinc uptake regulator, Zur [93–98]. An apparent purpose of ribosome remodelling is to restore zinc homeostasis in cells by reducing the future demand of zinc through utilization of zinc-free C- ribosomes, while transiently increasing the zinc supply through degradation of pre-existing C+ r-proteins [43, 95, 99].
Zinc being an essential micronutrient raised an ambiguity in our discovery as to whether induced ribosome hibernation is a nonspecific consequence of slowed metabolism in zinc-starved cells, or a specific response to low zinc. The ambiguity was further underscored by the evidence of active C- ribosomes [100, 101], and the structure of hibernating C+ ribosomes purified from zinc-rich cultures [102]. A more recent study from our laboratory clarified this ambiguity by demonstrating that Mpy dissociates from the ribosome upon reintroduction of zinc to a suspension of mycobacterial cells in zinc-depleted phosphate-buffered saline [44]. Moreover, our findings also uncoupled ribosome remodelling from hibernation by showing that Mpy binding is induced at a lower zinc concentration than that inducing assembly of C- ribosomes [44]. The zinc-responsive Mpy binding to the ribosome requires Mpy-recruitment factor (Mrf), which likely binds to the 30S subunit of the C- ribosome upstream from Mpy binding and promotes Mpy recruitment to the ribosome [44]. Mrf protein is stabilized at a zinc level lower than that required for ribosome remodelling [43, 44], and its expression is regulated at both transcriptional and post-translational levels. While transcription of Mrf mRNA is co-induced with C- r-proteins through the Zur-dependent derepression mechanism, Mrf protein is post-translationally degraded by Clp protease in a zinc-dependent manner [44]. The Clp protease system in mycobacteria is a barrel-shaped hetero-tetradecameric peptidase complex of ClpP1P2 that partners with an ATPase (ClpA/ClpC or ClpX) and an adaptor protein (SspB or ClpS) to perform the essential function of degrading proteins with specific sequence determinants, called degrons [103–113]. Zinc binding to Mrf appears to be necessary for its interaction with ClpS, which delivers the protein to ClpCP1P2 complex for degradation [44, 114]. Derepression of Mrf transcription – an event likely preceded by loss of zinc stoichiometry in Zur – at a zinc concentration that is sufficient for its degradation by Clp suggests that Mrf may have greater zinc affinity than Zur, and that this difference may be crucial in separating the ribosome remodelling step from the hibernation process. The broader implication of the two-step regulation of Mrf is a biphasic adaptation of mycobacteria to zinc starvation: an initial phase of efficient zinc utilization through ribosome remodelling is likely followed by gradual slowdown of metabolism through Mrf stabilization and subsequent increase in the fraction of hibernating ribosomes proportionate to decreasing levels of zinc. Furthermore, the underlying mechanism of Mpy dissociation from the ribosome upon zinc replenishment also hinges on restoration of Mrf degradation, because a stable mutant of Mrf with reduced affinity to zinc slows down the Mpy dissociation upon zinc replenishment [44]. These findings together affirm Mpy-dependent ribosome hibernation as a specific response to zinc depletion, while also raising questions about the conditions in which Mishra and coworkers observed binding of Mpy (called Hpf by the authors) to C+ ribosomes [102].
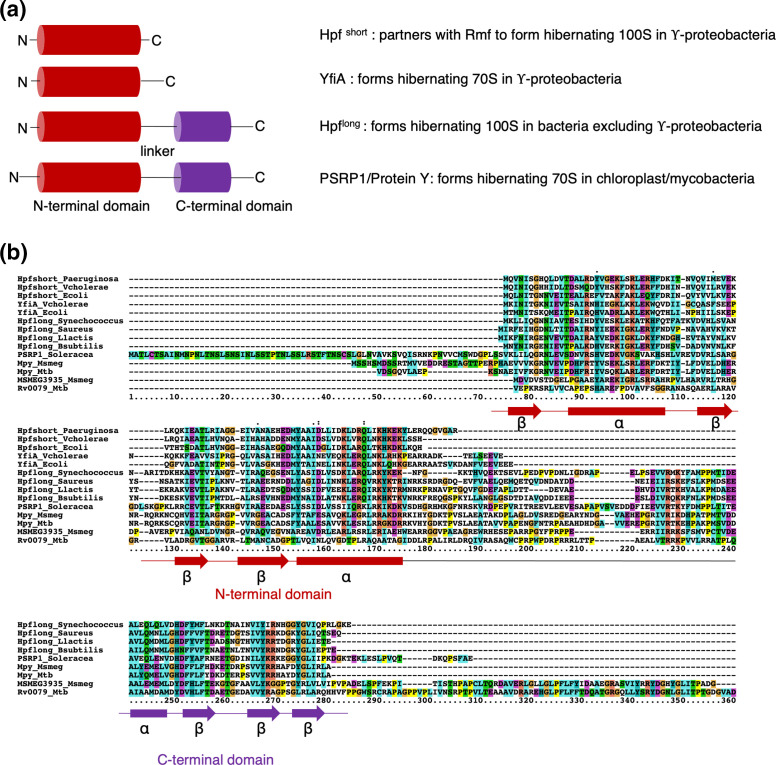
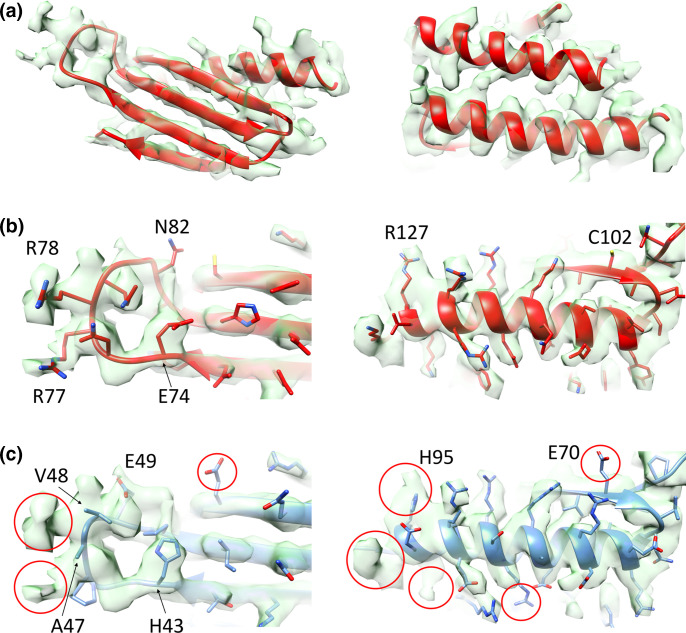
Multiple investigations of stationary-phase ribosomes from zinc-rich cultures in our laboratories failed to observe Mpy-bound C+ ribosomes to the level found in zinc-depleted culture [115]. While preparing this review, we further tested if purified C+ ribosomes can be recognized by recombinant Mpy (rMpy) in vitro to the same level as Mrf-rich C- ribosomes purified from low-zinc cultures of a Δmpy strain. Using the previously standardized assay condition [44], we found that the C+ ribosome forms a relatively poor substrate for rMpy under the tested in vitro condition (Fig. 2). To provide further clarity to our discussion in this review, we compared the cryo-EM density corresponding to Mpy in our structure to that published by Mishra and coworkers. The local density corresponding to Mpy in the refined 3.11 Å resolution cryo-EM map (EMD-23076, PDB ID 6DZI) [116] could be unequivocally attributed to the amino acid side chains of Mpy, while ruling out MSMEG_3935 (Fig. 3). For example, we could identify the densities corresponding to large side chains of amino acid residues (R77 and R78) in the loop region of Mpy that could not be explained by the smaller side chains of amino acid residues (A47 and V48) in the analogous loop region of MSMEG_3935. By contrast, the atomic model of Mpy and a homology model of MSMEG_3935 can be fitted equally well into the corresponding cryo-EM density of Mishra and coworkers (Fig. 4). This analysis suggests that the less resolved protein density found in their structure could represent MSMEG_3935, or perhaps a mixture of both Mpy and MSMEG_3935. Further investigations at higher resolution would be necessary to identify Mpy-like protein(s) in C+ ribosomes and determine the binding mechanism. Interestingly, while Mpy is universally conserved in mycobacteria, species like Mycobacterium kansasii and M. leprae neither encode C- r-proteins nor an orthologue of Mrf, raising the possibility of alternative adaptative responses to low-zinc in these species. Moreover, these species can serve as a suitable model to explore hibernation of C+ ribosomes.
Fig. 2.
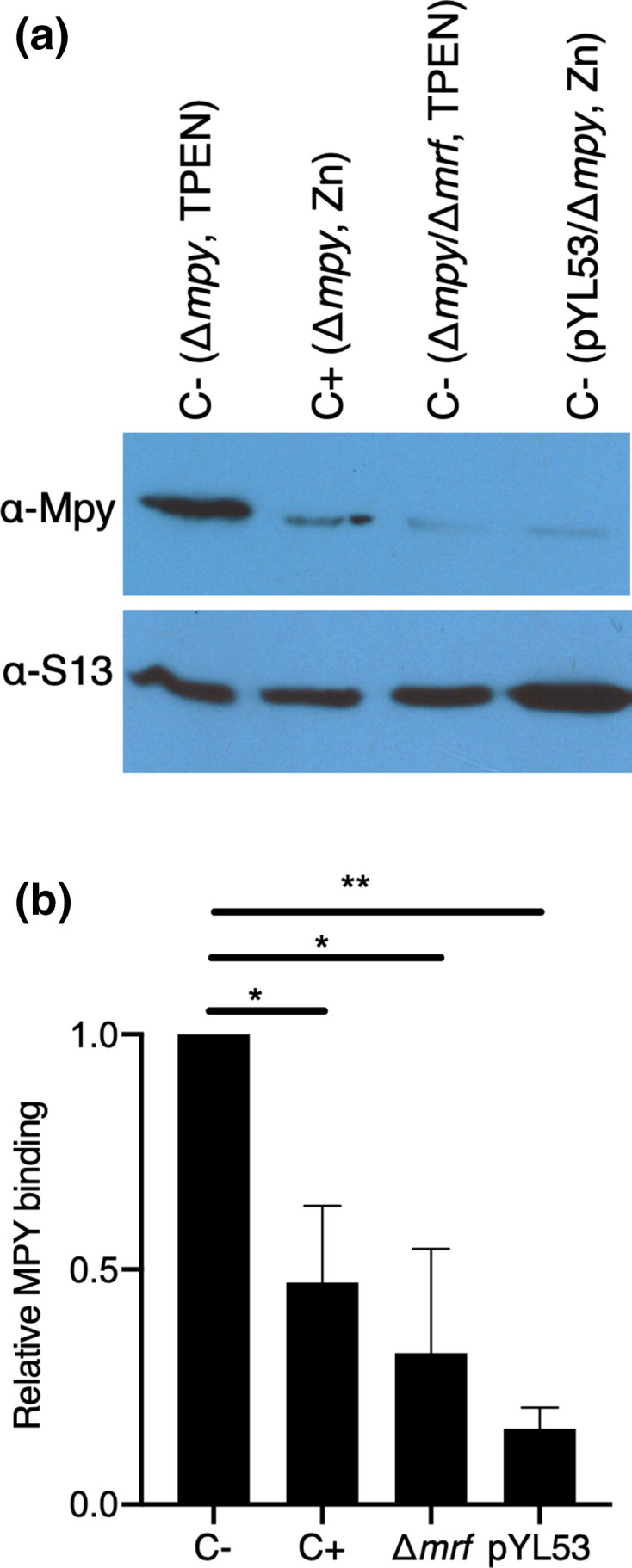
Binding of Mpy requires Mrf-associated ribosomes. (a) Immunoblot analysis of the presence of Mpy in ribosomes purified from a mixture containing recombinant Mpy (rMpy) and crude ribosomes from indicated strains (in parenthesis) grown under either low-zinc (1 µM TPEN) or high-zinc (1 mM ZnSO4) Sauton’s medium for 4 days. The type of ribosomes obtained from each strain under each growth condition is indicated above the lane as C+ or C-. The strain pYL53 constitutively expressed C- ribosomes, even under high-zinc conditions. Crude ribosomes equivalent to OD 0.1 were mixed with 2.4 picomoles of rMpy in a 50 µl reaction and the mixture was incubated at 37 ∘C for 1 h. Then the ribosomes were separated from the unbound rMPY by ultracentrifugation of the mixture on a 32 % sucrose cushion at 100 000 r.p.m. for 3 h in Beckman TLA 100.3 rotor. The pellets containing the ribosomes were analysed for the presence of rMpy. S13 was probed as the loading control. (b) Quantitative difference between the level of rMpy in the indicated ribosome samples, relative to low-zinc C- ribosomes (normalized as 1) obtained from immunoblots including the one shown in (a). Data represent the average of two independent binding experiments from biologically independent preparations of crude ribosomes and rMPy. * and ** represent P-value (t-test) <0.05 and 0.01, respectively.
Fig. 3.
Comparison of the density corresponding to Mpy within the 3.11 Å resolution cryo-EM map of the 70S-Mpy complex (EMD-23076) with atomic models of Mpy and MSMEG_3935. (a) Overall fitting of the atomic model (red) of the N-terminal domain of Mpy into the corresponding cryo-EM density (semitransparent green) shown from two opposite sides of the molecule. (b) Magnified views of some of the modelled segments of Mpy (red) (E74 – N82 and C102 - R127) based on the secondary structural elements and side-chain information inferred from the cryo-EM map (EMD-23076). (c) Magnified views of the analogous segments from the MSMEG_3935 model (blue) docked into the same cryo-EM map (semitransparent green). Red circles highlight the regions of incompatibility between the atomic model of MSMEG_3935 and the cryo-EM density corresponding to Mpy in our 70S-Mpy complex.
Fig. 4.
Comparison of atomic models of Mpy and MSMEG_3935 with the corresponding cryo-EM density of Mishra and coworkers (EMD 6921: PDBID 5ZEP) [102]. Atomic models of (a) Mpy (red) or (b) MSMEG_3935 (blue) could be docked equally well into the cryo-EM density (semitransparent orange).
Although the precise function of Mrf in facilitating Mpy recruitment remains to be determined, its binding to the 30S subunit raises a distinct possibility that Mrf modulates the 30S conformation to favour Mpy binding. This possibility is somewhat comparable to the function of Rmf in the recruitment of Hpfshort to E. coli ribosomes [58], but with clear distinctions. Unlike the Rmf/Hpfshort partnership, which yields 100S ribosomes, the Mrf/Mpy partnership forms hibernating 70S ribosomes. Moreover, the presence of an extended C-terminal region in Mpy (Fig. 1), a feature that is generally considered to be crucial for ribosome dimerization, is intriguing and raises questions about its function. Overall, Mpy binding to the ribosome presents a unique example of ribosome hibernation, many details of which remain to be uncovered. For example, can Mpy bind to C+ ribosomes with the help of a Mrf-like specialized chaperone expressed under yet to be determined conditions?
Ribosome hibernation in Mtb during infection
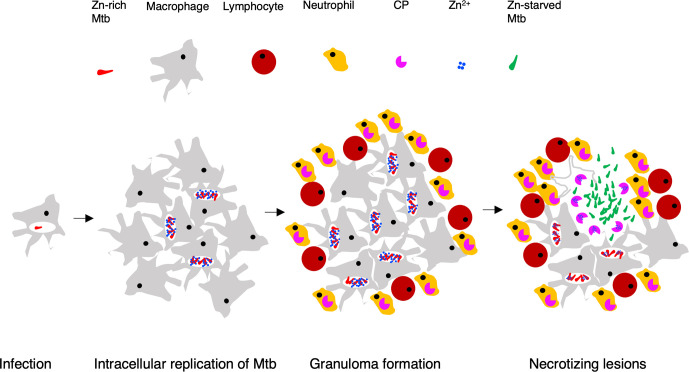
The evidence of zinc depletion as an important condition for Mpy-dependent ribosome hibernation raises the question of whether TB lesions harbouring Mtb are sufficiently zinc starved to induce ribosome hibernation in the pathogen. Mouse infections with a reporter strain of Mtb expressing a fluorescent protein from the promoter of C- r-proteins indicated that more bacilli expressed C- ribosomes during the chronic phase of infection than during the acute phase, which is marked by exponential growth of bacilli inside macrophages [43]. In some mouse models, such as C3HeB/FeJ, nearly all bacilli express C- ribosomes during the chronic phase of infection [43]. The increased frequency of bacilli expressing C- ribosomes highlights a change from a zinc-rich to zinc-depleted microenvironment as a function of the disease progression. A zinc-rich growth condition for Mtb during the acute phase is consistent with active influx of zinc in the phagosomes, which harbour 15-fold more zinc than plasma levels [117, 118]. Mtb induces its own zinc-efflux transporters to survive and grow in macrophages [119], suggesting that the increased influx of zinc in phagosomes is conceivably a host defence mechanism to induce metal toxicity in the pathogen. The origins of zinc starvation in Mtb during chronic infection remain unclear, although zinc chelation by neutrophil-secreted calprotectin is considered an important innate defence mechanism against several bacterial infections [120, 121]. The extent of neutrophil recruitment at the site of Mtb infection correlates with tissue damage and disease pathology [122, 123]. It is therefore reasonable to hypothesize that the extracellular Mtb bacilli in the necrotizing lesions, primarily developed by recruitment of granulocytes, experience zinc starvation from increased calprotectin abundance (Fig. 5). Alternative zinc-deprived host niches such as calprotectin-rich cytosolic environment of neutrophils, which are known to harbour Mtb, cannot be ruled out.
Fig. 5.
A working hypothesis for changing zinc availability to Mtb during progressive TB. Following initial infection, intracellular growth of Mtb is accompanied by enrichment of zinc in the phagosomes [117–119], leading to the expression of C+ribosomes in the pathogen. Infiltrating neutrophils during granuloma formation employ various anti-bacterial mechanisms, including the secretion of zinc-chelating calprotectin (CP) as a component of nutritional immunity [121]. Subsequent lysis of infected macrophages by Mtb-derived factors and Mtb-specific cytotoxic T-cells would cause release of bacilli in extracellular space, where free zinc concentration is expected to be reduced by CP. As a result, Mtb cells will sense zinc starvation and induce C- ribosome expression to conserve intracellular zinc. Induced expression of the C- ribosome is perhaps followed by its hibernation upon further zinc deprivation during exuberant extracellular growth of bacilli.
Because ribosome hibernation is induced at lower zinc concentration than that required for inducing C- r-proteins [44], it is highly likely that differences in local zinc concentrations in host tissues can produce a range of physiological states in Mtb bacilli depending on the ratio of active to Mpy-bound C- ribosomes in each cell. Arguably, Mtb cells harbouring peak-level of hibernating C- ribosomes are expected to have the slowest metabolism, while those with the least level would be most active. Such cell-to-cell variation can potentially lead to extensive phenotypic heterogeneity in the population in terms of antibiotic sensitivity. Interestingly, streptomycin treatment of infection with Δmpy mutant produced ~20 fold fewer bacterial burden than those with wild-type Mtb [44], implying that over 90 % of Mtb population during chronic infection induce some levels of Mpy-dependent ribosome hibernation that are sufficient for the cells to tolerate the drug. However, cells with peak-level of hibernating ribosomes are likely to be only a small fraction of this population, and may represent the nonreplicating persisters.
Future scope
The emerging body of evidence suggest ribosome hibernation as an underlying cause for the development of drug-tolerant, nonreplicating subpopulation of Mtb during infection. This hypothesis, while promising, is still nascent and its maturity depends on addressing crucial gaps in our understanding of the mechanism underlying Mpy recruitment to ribosomes and the physiological consequences of the process. For example, structural studies on how Mrf modulates 30S to facilitate Mpy binding will likely identify intermediates that can be exploited to develop pharmacological modulators of ribosome hibernation. Understanding the physiological consequences of ribosome hibernation however would require carefully designed experimental approaches to address the likely cell-to-cell heterogeneity in terms of the proportion of hibernating to total ribosomes. A method of visualizing mycobacterial cells with saturating levels of hibernating ribosomes will enable tracking of such cells at the single-cell level, thereby facilitating their comparison with the rest of the population. In summary, Mrf/Mpy-dependent ribosome hibernation is mechanistically unique in many ways, and further investigation will likely offer new insights into its significance in mycobacterial persistence. Additional conditions promoting ribosome hibernation in mycobacteria should also be explored to develop an inclusive strategy to counter the process.
Funding information
Authors acknowledge support from NIH to AKO (NIH: AI132422, NIH: AI144474) and RKA (NIH: GM061576).
Acknowledgements
A.K.O. thanks Douglas Young for sharing his unpublished materials on the topic.
Author contributions
Y.L., M.R.S., R.K.K., N.K.B., R.K.A. and A.K.O. designed and performed experiments, analysed data and wrote the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Ignatius EH, Dooley KE. New drugs for the treatment of tuberculosis. Clin Chest Med. 2019;40:811–827. doi: 10.1016/j.ccm.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furin J, Cox H, Pai M. Tuberculosis. The Lancet. 2019;393:1642–1656. doi: 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- 3.WHO Rapid Communication: key changes to the treatment of multidrug- and rifampicin-resistant tuberculosis (MDR/RR-TB). http://wwwwhoint/tb/publications/2018/WHO_RapidCommunicationMDRTBpdf . 2018.
- 4.Adepoju P. Tuberculosis and HIV responses threatened by COVID-19. The Lancet HIV. 2020;7:e319–e320. doi: 10.1016/S2352-3018(20)30109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amimo F, Lambert B, Magit A. What does the COVID-19 pandemic mean for HIV, tuberculosis, and malaria control? Trop Med Health. 2020;48:32. doi: 10.1186/s41182-020-00219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gold B, Nathan C. Targeting phenotypically tolerant Mycobacterium tuberculosis . Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.TBTB2-0031-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCune RM, Feldmann FM, Lambert HP, McDermott W. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med. 1966;123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallis RS, Patil S, Cheon S-H, Edmonds K, Phillips M, et al. Drug tolerance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 1999;43:2600–2606. doi: 10.1128/AAC.43.11.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jindani A, Aber VR, Edwards EA, Mitchison DA. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis. 1980;121:939–949. doi: 10.1164/arrd.1980.121.6.939. [DOI] [PubMed] [Google Scholar]
- 10.Manabe YC, Bishai WR. Latent Mycobacterium tuberculosis–persistence, patience and winning by waiting. Nat Med. 2000;6:1327–1329. doi: 10.1038/82139. [DOI] [PubMed] [Google Scholar]
- 11.Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Ehrt S, Schnappinger D, Rhee KY. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis . Nat Rev Microbiol. 2018;16:496–507. doi: 10.1038/s41579-018-0013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobby GL, Lenert TF. The in vitro action of antituberculous agents against multiplying and non-multiplying microbial cells. Am Rev Tuberc. 1957;76:1031–1048. doi: 10.1164/artpd.1957.76.6.1031. [DOI] [PubMed] [Google Scholar]
- 14.Sever JL, Youmans GP. The relation of oxygen tension to virulence of tubercle bacilli and to acquired resistance in tuberculosis. J Infect Dis. 1957;101:193–202. doi: 10.1093/infdis/101.2.193. [DOI] [PubMed] [Google Scholar]
- 15.Sever JL, Youmans GP. Enumeration of viable tubercle bacilli from the organs of nonimmunized and immunized mice. Am Rev Tuberc. 1957;76:616–635. doi: 10.1164/artpd.1957.76.4.616. [DOI] [PubMed] [Google Scholar]
- 16.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/IAI.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H-D, Guinn KM, Harrell MI, Liao R, Voskuil MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis . Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayuri G, Das TK, Tyagi JS. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR–DevS two-component system, Rv3134c and chaperone α-crystallin homologues. FEMS Microbiol Lett. 2002;211:231–237. doi: 10.1016/S0378-1097(02)00684-5. [DOI] [PubMed] [Google Scholar]
- 21.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim A, Eleuterio M, Hutter B, Murugasu-Oei B, Dick T. Oxygen depletion-induced dormancy in Mycobacterium bovis BCG. J Bacteriol. 1999;181:2252–2256. doi: 10.1128/JB.181.7.2252-2256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis . Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/AAC.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoff DR, Caraway ML, Brooks EJ, Driver ER, Ryan GJ, et al. Metronidazole lacks antibacterial activity in guinea pigs infected with Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2008;52:4137–4140. doi: 10.1128/AAC.00196-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll MW, Jeon D, Mountz JM, Lee JD, Jeong YJ, et al. Efficacy and safety of metronidazole for pulmonary multidrug-resistant tuberculosis. Antimicrob Agents Chemother. 2013;57:3903–3909. doi: 10.1128/AAC.00753-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks JV, Furney SK, Orme IM. Metronidazole therapy in mice infected with tuberculosis. Antimicrob Agents Chemother. 1999;43:1285–1288. doi: 10.1128/AAC.43.5.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J Infect Dis. 2008;198:275–283. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 29.Mandal S, Njikan S, Kumar A, Early JV, Parish T. The relevance of persisters in tuberculosis drug discovery. Microbiology. 2019;165:492–499. doi: 10.1099/mic.0.000760. [DOI] [PubMed] [Google Scholar]
- 30.Vilchèze C, Jacobs WR. The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J Mol Biol. 2019;431:3450–3461. doi: 10.1016/j.jmb.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011;2:e00100–00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryk R, Gold B, Venugopal A, Singh J, Samy R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Javid B, Sorrentino F, Toosky M, Zheng W, Pinkham JT, et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci U S A. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 36.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, et al. Dual-Reporter mycobacteriophages (Φ2 DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. MBio. 2016;7 doi: 10.1128/mBio.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safi H, Gopal P, Lingaraju S, Ma S, Levine C, et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc Natl Acad Sci U S A. 2019;116:19665–19674. doi: 10.1073/pnas.1907631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellerose MM, Baek S-H, Huang C-C, Moss CE, Koh E-I, et al. Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. mBio. 2019;10 doi: 10.1128/mBio.00663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quigley J, Peoples A, Sarybaeva A, Hughes D, Ghiglieri M, et al. Novel antimicrobials from uncultured bacteria acting against Mycobacterium tuberculosis . mBio. 2020;11 doi: 10.1128/mBio.01516-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keren I, Mulcahy LR, Lewis K. Persister eradication: lessons from the world of natural products. Methods Enzymol. 2012;517:387–406. doi: 10.1016/B978-0-12-404634-4.00019-X. [DOI] [PubMed] [Google Scholar]
- 41.Mak PA, Rao SPS, Ping Tan M, Lin X, Chyba J, et al. A high-throughput screen to identify inhibitors of ATP homeostasis in non-replicating Mycobacterium tuberculosis . ACS Chem Biol. 2012;7:1190–1197. doi: 10.1021/cb2004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma S, Morrison R, Hobbs SJ, Soni V, Farrow-Johnson J, et al. Transcriptional regulator-induced phenotype screen reveals drug potentiators in Mycobacterium tuberculosis . Nat Microbiol. 2020 doi: 10.1038/s41564-020-00810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Sharma MR, Koripella RK, Yang Y, Kaushal PS, et al. Zinc depletion induces ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A. 2018;115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Corro J, Palmer C, Ojha AK. Progression from remodelling to hibernation of ribosomes in zinc starved mycobacteria. PNAS. 2020:In press. doi: 10.1073/pnas.2013409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 47.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 48.Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 49.Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A, et al. Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol. 2018;72:163–184. doi: 10.1146/annurev-micro-090817-062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobson A, Gillespie D. Metabolic events occurring during recovery from prolonged glucose starvation in Escherichia coli . J Bacteriol. 1968;95:1030–1039. doi: 10.1128/JB.95.3.1030-1039.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli . RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prossliner T, Skovbo Winther K, Sørensen MA, Gerdes K. Ribosome hibernation. Annu Rev Genet. 2018;52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 53.Tissieres A, Watson JD. Ribonucleoprotein particles from Escherichia coli . Nature. 1958;182:778–780. doi: 10.1038/182778b0. [DOI] [PubMed] [Google Scholar]
- 54.McCarthy BJ. Variations in bacterial ribosomes. Biochim Biophys Acta. 1960;39:563–564. doi: 10.1016/0006-3002(60)90221-3. [DOI] [Google Scholar]
- 55.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a "ribosome modulation factor" associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agafonov DE, Kolb VA, Nazimov IV, Spirin AS. A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci U S A. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueta M, Yoshida H, Wada C, Baba T, Mori H, et al. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli . Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 58.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and Yfia turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Bari H, Berry EA. Structure of Vibrio cholerae ribosome hibernation promoting factor. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:228–236. doi: 10.1107/S1744309113000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueta M, Wada C, Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog Sa HPF. Genes Cells. 2010;15:43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 61.Tagami K, Nanamiya H, Kazo Y, Maehashi M, Suzuki S, et al. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp mutant of Bacillus subtilis triggers YvyD‐dependent dimerization of ribosome. Microbiologyopen. 2012;1:115–134. doi: 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puri P, Eckhardt TH, Franken LE, Fusetti F, Stuart MCA, et al. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol Microbiol. 2014;91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- 63.Kline BC, McKay SL, Tang WW, Portnoy DA. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol. 2015;197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, et al. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells. 2013;18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 65.Krokowski D, Gaccioli F, Majumder M, Mullins MR, Yuan CL, et al. Characterization of hibernating ribosomes in mammalian cells. Cell Cycle. 2011;10:2691–2702. doi: 10.4161/cc.10.16.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, et al. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci U S A. 2007;104:19315–19320. doi: 10.1073/pnas.0709856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beckert B, Abdelshahid M, Schäfer H, Steinchen W, Arenz S, et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. Embo J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flygaard RK, Boegholm N, Yusupov M, Jenner LB. Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat Commun. 2018;9:4179. doi: 10.1038/s41467-018-06724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franken LE, Oostergetel GT, Pijning T, Puri P, Arkhipova V, et al. A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat Commun. 2017;8:722. doi: 10.1038/s41467-017-00718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khusainov I, Vicens Q, Ayupov R, Usachev K, Myasnikov A, et al. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. Embo J. 2017;36:2073–2087. doi: 10.15252/embj.201696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matzov D, Aibara S, Basu A, Zimmerman E, Bashan A, et al. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun. 2017;8:723. doi: 10.1038/s41467-017-00753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, et al. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010;18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 73.Ortiz JO, Brandt F, Matias VRF, Sennels L, Rappsilber J, et al. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trösch R, Willmund F. The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol Chem. 2019;400:879–893. doi: 10.1515/hsz-2018-0436. [DOI] [PubMed] [Google Scholar]
- 75.Izutsu K, Wada A, Wada C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- 76.Hood RD, Higgins SA, Flamholz A, Nichols RJ, Savage DF. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus . Proc Natl Acad Sci U S A. 2016;113:E4867–E4876. doi: 10.1073/pnas.1524915113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basu A, Yap M-NF. Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc Natl Acad Sci U S A. 2017;114:E8165–E8173. doi: 10.1073/pnas.1709588114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, et al. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- 79.Stolper DA, Revsbech NP, Canfield DE. Aerobic growth at nanomolar oxygen concentrations. Proc Natl Acad Sci U S A. 2010;107:18755–18760. doi: 10.1073/pnas.1013435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes to Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Mandava CS, Cao W, Li X, Zhang D, et al. Hflx is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol. 2015;22:906–913. doi: 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
- 82.Rudra P, Hurst-Hess KR, Cotten KL, Partida-Miranda A, Ghosh P. Mycobacterial HflX is a ribosome splitting factor that mediates antibiotic resistance. Proc Natl Acad Sci U S A. 2020;117:629–634. doi: 10.1073/pnas.1906748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akiyama T, Williamson KS, Schaefer R, Pratt S, Chang CB, et al. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci U S A. 2017;114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sabharwal D, Song T, Papenfort K, Wai SN. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae . RNA Biol. 2015;12:186–196. doi: 10.1080/15476286.2015.1017211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akanuma G, Kazo Y, Tagami K, Hiraoka H, Yano K, et al. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis . Microbiology. 2016;162:448–458. doi: 10.1099/mic.0.000234. [DOI] [PubMed] [Google Scholar]
- 86.Basu A, Yap M-NF. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016;44:4881–4893. doi: 10.1093/nar/gkw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKay SL, Portnoy DA. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother. 2015;59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trauner A, Lougheed KEA, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem. 2012;287:24053–24063. doi: 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 91.Francois B, et al. Crystal structures of complexes between aminoglycosides and decoding a site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prisic S, Hwang H, Dow A, Barnaby O, Pan TS, et al. Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis . Mol Microbiol. 2015;97:263–280. doi: 10.1111/mmi.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maciąg A, Dainese E, Rodriguez GM, Milano A, Provvedi R, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Owen GA, Pascoe B, Kallifidas D, Paget MSB. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves Zur and σR. J Bacteriol. 2007;189:4078–4086. doi: 10.1128/JB.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, et al. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol. 2004;52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- 99.Dow A, Prisic S. Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc limiting conditions. PLoS One. 2018;13:e0196300. doi: 10.1371/journal.pone.0196300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Y-X, Xu Z-yu, Ge X, Hong J-Y, Sanyal S, ZY X, ZJ L, et al. Selective translation by alternative bacterial ribosomes. Proc Natl Acad Sci U S A. 2020;117:19487–19496. doi: 10.1073/pnas.2009607117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tobiasson V, Dow A, Prisic S, Amunts A. Zinc depletion does not necessarily induce ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A. 2019;116:2395–2397. doi: 10.1073/pnas.1817490116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mishra S, Ahmed T, Tyagi A, Shi J, Bhushan S. Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA. Sci Rep. 2018;8:13587. doi: 10.1038/s41598-018-31850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmitz KR, Carney DW, Sello JK, Sauer RT. Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery. Proc Natl Acad Sci U S A. 2014;111:E4587–E4595. doi: 10.1073/pnas.1417120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sauer RT, Baker TA. Aaa+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 105.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lo JH, Baker TA, Sauer RT. Characterization of the N-terminal repeat domain of Escherichia coli ClpA-A class I Clp/HSP100 ATPase. Protein Sci. 2001;10:551–559. doi: 10.1110/ps.41401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Varshavsky A. N-degron and C-degron pathways of protein degradation. Proc Natl Acad Sci U S A. 2019;116:358–366. doi: 10.1073/pnas.1816596116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- 109.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli . Embo J. 2009;28:1732–1744. doi: 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao X, Yeom J, Groisman EA. The expanded specificity and physiological role of a widespread N-degron recognin. Proc Natl Acad Sci U S A. 2019;116:18629–18637. doi: 10.1073/pnas.1821060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raju RM, Unnikrishnan M, Rubin DHF, Krishnamoorthy V, Kandror O, et al. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012;8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akopian T, Kandror O, Raju RM, Unnikrishnan M, Rubin EJ, et al. The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. Embo J. 2012;31:1529–1541. doi: 10.1038/emboj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ollinger J, O'Malley T, Kesicki EA, Odingo J, Parish T. Validation of the essential ClpP protease in Mycobacterium tuberculosis as a novel drug target. J Bacteriol. 2012;194:663–668. doi: 10.1128/JB.06142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ziemski M, Leodolter J, Taylor G, Kerschenmeyer A, Weber-Ban E. Genome-wide interaction screen for Mycobacterium tuberculosis ClpCP protease reveals toxin-antitoxin systems as a major substrate class. Febs J. 2021;288:111–126. doi: 10.1111/febs.15335. [DOI] [PubMed] [Google Scholar]
- 115.Li Y, Sharma MR, Koripella RK, Wade JT, Gray TA, et al. Reply to Tobiasson et al.: zinc depletion is a specific signal for induction of ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A. 2019;116:2398–2399. doi: 10.1073/pnas.1821103116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y, Koripella RK, Sharma MR, Lee RE, Agrawal RK, et al. Replacement of S14 protein in ribosomes of zinc-starved mycobacteria reduces spectinamide sensitivity. Antimicrob Agents Chemother. 2020 doi: 10.1128/AAC.01833-20. in press. 23 Dec 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pyle CJ, Azad AK, Papp AC, Sadee W, Knoell DL, et al. Elemental ingredients in the macrophage cocktail: role of ZIP8 in host response to Mycobacterium tuberculosis. Int J Mol Sci. 2017;18:2375. doi: 10.3390/ijms18112375. 09 Nov 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wagner D, Maser J, Lai B, Cai Z, Barry CE, et al. Elemental analysis of Mycobacterium avium, Mycobacterium tuberculosis, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 119.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, et al. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 121.Lonergan ZR, Skaar EP. Nutrient zinc at the host-pathogen interface. Trends Biochem Sci. 2019;44:1041–1056. doi: 10.1016/j.tibs.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lyadova IV. Neutrophils in tuberculosis: heterogeneity shapes the way? Mediators Inflamm. 2017;2017:1–11. doi: 10.1155/2017/8619307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Niazi MKK, Dhulekar N, Schmidt D, Major S, Cooper R, et al. Lung necrosis and neutrophils reflect common pathways of susceptibility to Mycobacterium tuberculosis in genetically diverse, immune-competent mice. Dis Model Mech. 2015;8:1141–1153. doi: 10.1242/dmm.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]