Abstract
Members of Plectosphaerella inhabit different substrates, including plants, soil and insects, and most species are pathogens causing large losses in agriculture. During a survey of endophytic fungi in aquatic plants in southwest China, 112 strains of Plectosphaerella were isolated, representing two new species, P. endophyticasp. nov. and P. sichuanensissp. nov., as well as two known species, P. cucumerina and P. pauciseptata. The novel taxa are described and illustrated here using combined morphological and multi-locus phylogenetic (LSU-ITS-TEF-1α-TUB2) analyses. Our result revealed Plectosphaerella species inhabiting within aquatic plants in southwest China, and the separation frequency of each species was presented.
Keywords: Endophytic fungi, multi-locus phylogeny, new species, Plectosphaerellaceae , Sordariomycetes
Introduction
The genus Plectosphaerella Kleb. was established to accommodate P. cucumeris Kleb. from young cucumber plants (Klebahn 1929). Previously, it was always placed in Hypocreaceae (Sordariomycetes, Hypocreales) and Sordariaceae (Sordariomycetes, Sordariales) (Domsch and Gams 1972; Uecker 1993), until Zare et al. (2007) established Plectosphaerellaceae W. Gams, Summerb. & Zare (Glomerellales) to accommodate it (Réblová et al. 2011). The genus Plectosporium Palm, Gams & Nirenberg was described as the asexual morph of Plectosphaerella (Palm et al. 1995). Given its priority, Plectosphaerella was recommended as the accepted generic name (Réblová et al. 2016), and all species of Plectosporium were transferred into Plectosphaerella (Carlucci et al. 2012). At present, there are 23 species records for Plectosphaerella as listed in Index Fungorum (2021), two species, P. himantia (Pers.) Kirschst. and P. melaena (Fr.) Kirschst. are not recognized for Plectosphaerella.
Species of Plectosphaerella are distributed in a variety of habitats and have wide geographic distribution (Carlucci et al. 2012; Zhang et al. 2019). Some species are pathogens on the fruit, root or collar in Cucurbitaceae plant, causing large losses of melon, pumpkin and zucchini crops (Alfaro-García et al. 1996; Toyozo et al. 2005; Raimondo and Carlucci 2018). Several species harm other plants such as tomato, pepper, bamboo, and asparagus. And the symptoms of hosts are decomposition, breakdown and death (Antignani et al. 2008; Carlucci et al. 2012; Arzanlou et al. 2013). Besides diseased plants, P. sinensis Lei Su & Y.C. Niu was reported as endophytic fungus without causing obvious symptoms (Su et al. 2017). In addition, most species causing plant diseases were also isolated from soil, except for P. oratosquillae (P.M. Duc et al.) A.J.L. Phillips et al. originating from arthropod Oratosquilla oratoria (Duc et al. 2009).
Plectosphaerella cucumerina is the most widely distributed species, thriving on more than nine plant genera: Arabidopsis, Cucumis, Galium, Hydrilla, Nicotiana, Pyrus, Solanum, Viola and Austropotamobius etc., and also occurring in soil and paper (Alderman and Polglase 1985; Palm et al. 1995; Smith-Kopperl et al. 1999; Domsch et al. 2007; Giraldo and Crous 2019). Meanwhile, this species has a wide geographic distribution, and it has been reported from England, Italy, Canada, the Netherlands, Egypt etc. (Raimondo and Carlucci 2018, Giraldo et al. 2019). The second species with wide geographic distribution, P. plurivora A.J.L. Phillips et al., was found in Australia, the Netherlands, Germany, USA etc. Its hosts include Lolium, Solanum, Nicotiana, Asparagus and it also occurs in soil (Carlucci et al. 2012; Giraldo and Crous 2019). However, other species did not show obvious habitats diversity and wide geographic distribution.
During our investigation of endophytic fungal diversity of aquatic plants in southwest China, among 1697 acquired strains, 112 strains belonging to Plectosphaerella were isolated. Based on morphological characteristics and phylogenetic analysis, two known species, P. cucumerina and P. pauciseptata, were described, and two new species, P. endophytica and P. sichuanensis, were proposed and illustrated. The geographic distribution and habitat diversity of Plectosphaerella in this study were also discussed.
Materials and methods
Isolates and Morphology
Samples were collected from Yunnan, Guizhou, Sichuan provinces, Chongqing and Tibet from 2014 to 2017. The dominant hosts are Myriophyllum, Potamogeton, Hydrilla and Hippuris. Samples were placed in plastic bags, labeled and transported to the laboratory. Each leaf and stem was cut into segments 30–40 mm in length and washed thoroughly with tap water, then a surface-disinfection was carried out according to Su et al. (2016). The segments were cut into smaller sections of about 5×5 mm under the aseptic operating table. Firstly, washed 30 s in sterile water and soaked 2 min in 0.5% hypochlorite solution, then 30 s in sterile water and 2 min in 75% ethanol, finally 30 s in sterile water. Each ten sections were randomly isolated on rose bengal agar (RBA, Guangdong Huankai Microbial Sci and Tech), the antibiotics chloramphenicol (0.1 g l–1) was added to restrain bacterial growth. When a fungus grew up from the segments, some hyphae were picked up and transferred to potato dextrose agar (PDA, 200 g potato, 20 g dextrose, 18 g agar, 1000 ml distilled water) plates for incubation at 28 °C. After 10 days, colonies were transferred to different plates, including corn meal agar (CMA, 20 g cornmeal, 18 g agar, 1000 ml distilled water) and oatmeal agar (OA, 30 g filtered oat flakes, 20 g agar, 1000 ml distilled water). Colony characteristics, growth speed and other macrostructure from PDA plates were observed after 10 days. Microscopic characteristics such as mycelium, conidiophores and conidia were examined and measured after 3 days on CMA using BX51 microscope (Olympus); the sterile water was used as a mounting medium.
Pure cultures were deposited in the Herbarium of the Laboratory for Conservation and Utilization of Bio resources, Yunnan University, Kunming, Yunnan, P.R. China (YMF, formerly Key Laboratory of Industrial Microbiology and Fermentation Technology of Yunnan) and at the China Center for Type Culture Collection (CCTCC).
DNA extraction, PCR amplification and sequencing
Actively growing mycelium was scraped off from the surface of the culture and transferred to 2 ml Eppendorf micro-centrifuge tubes. Total genomic DNA was extracted follow the protocol of Guo et al. (2000). The internal transcribed spacer (ITS) and the 28S large subunit nuclear ribosomal RNA (LSU rRNA) were amplified using the primer pairs ITS1/ITS4 (White et al. 1990) and LROR/LR5 (Vilgalys and Hester 1990), respectively. Translation elongation factor 1-alpha (TEF-1α) and partial β-tubulin (TUB2) were amplified using the primer pairs EF-1251R/EF-688F (Alves et al. 2008) and Bt2a/Bt2b (Glass and Donaldson 1995), respectively. The PCR amplifications were conducted in 25 µl final volumes which consisted of 1.0 µl DNA template, 1.0 µl of each forward and reverse primers, 12.5 µl 2 × Master Mix and 9.5 µl ddH2O. The PCR reaction cycles were as follows: initial denaturation at 94 °C for 3 min; followed by 35 cycles of denaturation at 94 °C for 40 sec; the annealing extension dependent on the amplified loci (48 °C for LSU, 54 °C for ITS, 55 °C for TEF-1α and 58 °C for TUB2) for 1 min and extension at 72 °C for 2 min; a final extension at 72 °C for 10 min. PCR products were sequenced by TSINGKE Biological Technology in Kunming, China. The sequences are deposited in GenBank database and the accession numbers are listed in Table 1.
Table 1.
Plectosphaerella species used in phylogenetic analyses.
Strains and sequences generated in this study are emphasized in bold face. Tex-type cultures.
Phylogenetic analyses
The ITS sequences generated in this study were used as a query to search similar DNA sequences using BLASTn. All published DNA sequences were obtained from the GenBank from relevant studies (Su et al. 2017; Phookamsak et al. 2019; Zhang et al. 2019), Monilochaetes infuscans Harter (Australiascaceae) was selected as an outgroup. The generated sequences were manually aligned with CLUSTAL_X v. 1.83 (Thompson et al. 1997) with default parameters. Aligned sequences of multiple loci were concatenated and manually adjusted through BioEdit version v. 7.0.4.1 (Hall 1999), and ambiguously aligned regions were excluded. The combined sequence was converted to a NEXUS file using MEGA6 (Tamura 2013). The alignment was deposited at TreeBase http://purl.org/phylo/treebase/phylows/study/TB2:S27363.
Maximum-likelihood (ML) analysis was conducted by using RAxML (Stamatakis 2006) with the PHY files generated with CLUSTAL_X v. 1.83 (Thompson et al. 1997), using the GTR+GAMMA model. ML bootstrap proportions (MLBPs) were computed with 1000 replicates. Bayesian inference (BI) analysis was executed with MrBayes v. 3.2.2 (Ronquist and Huelsenbeck 2003). The Akaike information criterion (AIC) implemented in jModelTest version 2.0 was used to select the best fit models after likelihood score calculations (Posada 2008). HKY+I+G was estimated as the best-fit model under the output strategy of AIC, Lsetnst=6, rates=gamma. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities. Two runs were executed simultaneously for 6,000,000 generations and sampled every 500th generations, four chains containing one cold and three heated were run until the average standard deviation of the split frequencies dropped below 0.01, the stationarity of the analyses was confirmed in line with standards described by Sun and Guo (2010). The initial 25% of the generations of MCMC sampling were discarded as burn-in. The refinement of the phylogenetic tree was used for estimating BI posterior probability (BIPP) values. The tree was viewed in FigTree version 1.4 (Rambaut 2012).
Results
Phylogenetic analyses
A total of 112 strains were identified as members of Plectosphaerella according to the BLASTn search results using the ITS sequences. At first, we carried out individual phylogenetic analyses with ITS sequences to resolve the taxonomic position of our strains using the sequences of the accepted species into Plectosphaerella. This tree is shown in Figure 1. The result indicated that there are 77 strains clustered together with P. pauciseptata A.J.L. Phillips et al. with 0.92 Bayesian posterior probability, 27 strains clustered together with P. cucumerina with 0.96 Bayesian posterior probability, whereas other strains formed two individual groups. Therefore, three representative strains of P. pauciseptata, a representative strain of P. cucumerina, and three representative strains of two novel taxa were chosen among the 112 strains for single-gene and combined phylogenetic analysis.
Figure 1.
Phylogenetic tree inferred from a Bayesian analysis based on ITS sequences of 112 Plectosphaerella strains obtained in this study. BIPP over 80% are shown on the respective branches. The scale bar shows the expected changes per site. Two new species are given in boldface. Monilochaetes infuscans CBS 379.77 serves as an outgroup.
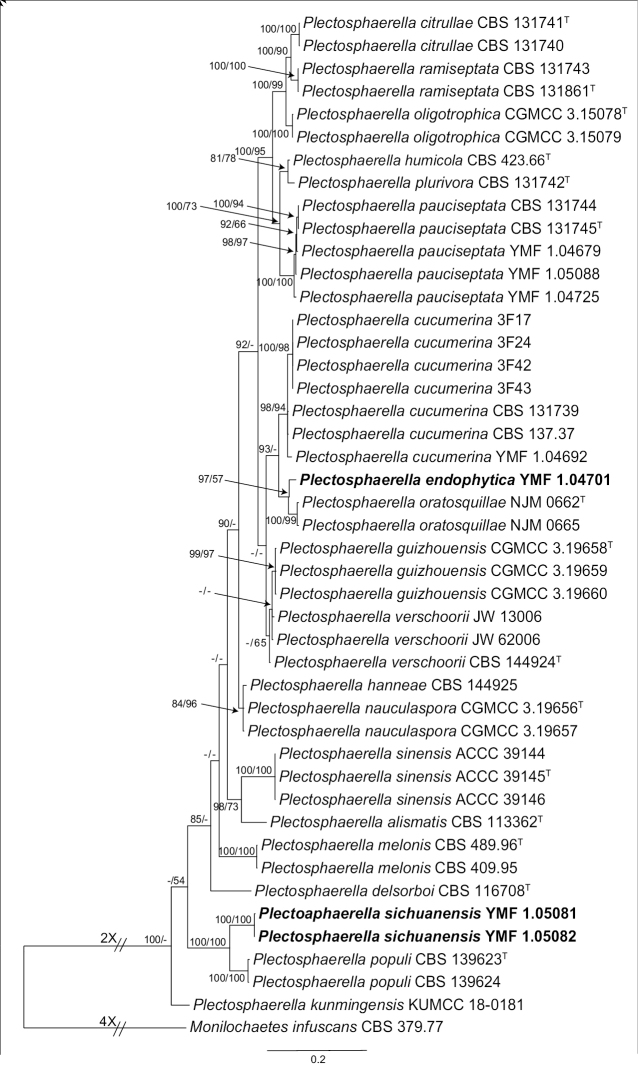
The four Bayesian trees derived from the single-gene sequence alignments (LSU, ITS, TEF-1α, TUB2) confirmed that the novel taxa were distant from other known species in Plectosphaerella. The Bayesian trees are available in the Suppl. material 1. The resulting combined sequence matrix included 2136 nucleotide positions (804 from LSU, 545 from ITS, 413 from TEF-1α, 374 from TUB2), with M. infuscans CBS 379.77 as the outgroup. The tree topology is shown in Figure 2, with the Bayesian posterior probabilities over 80% and ML bootstrap support over 50% indicated for respective clades. In this phylogenetic tree, P. endophytica represented by strain YMF 1.04701 was close to P. oratosquillae (NJM 0662 and NJM 0665) and formed a single clade with 0.97 Bayesian posterior probability and 57% ML bootstrap proportions. Similarly, P. sichuanensis represented by strains YMF 1.05081 and YMF 1.05082 were close to P. populi Ullah et al. (CBS 139623) with 1.00 Bayesian posterior probability and 100% ML bootstrap proportions. Considering distinct morphological differences, we propose to describe our isolates as two new species in Plectosphaerella.
Figure 2.
Phylogenetic tree of Plectosphaerella based on Bayesian analyses and Maximum Likelihood analyses of the combined sequences dataset of LSU, ITS, TEF-1α and TUB2. The numbers above branches represent BIPP (left) and MLBPs (right). BIPP over 80% and MLBPs over 50% are shown on the respective branches. The scale bar shows the expected changes per site. Two new species are given in boldface. Monilochaetes infuscans CBS 379.77 serves as an outgroup.
Taxonomy
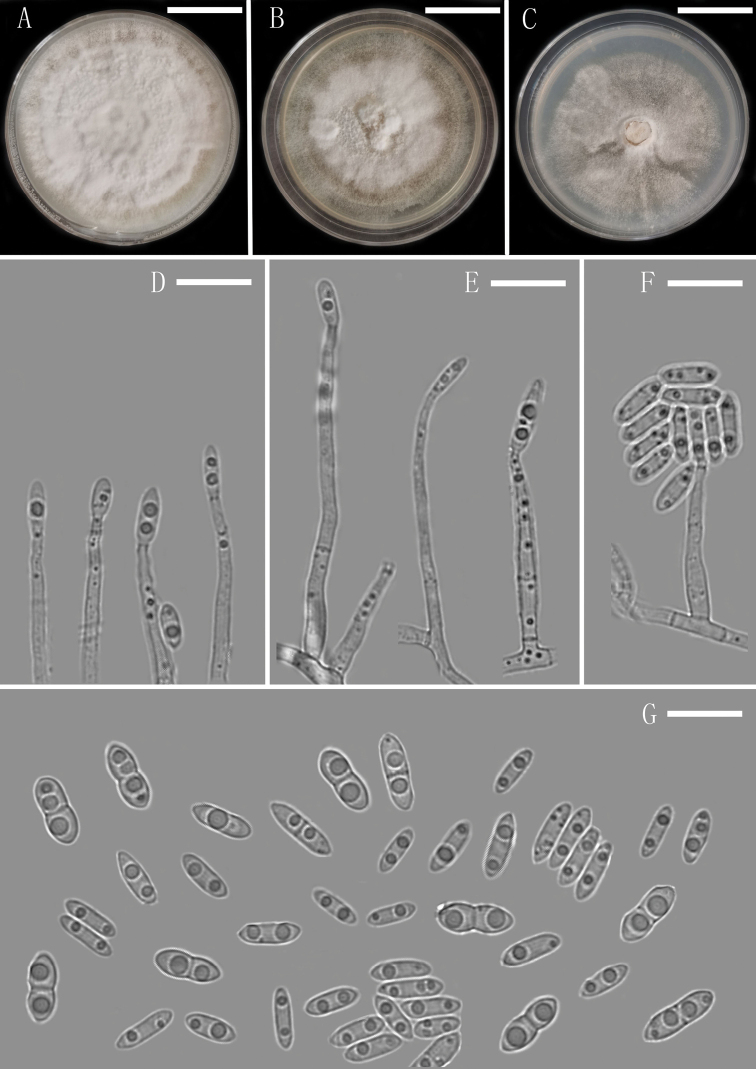
Plectosphaerella endophytica
Z.F. Yu & X.Q. Yang sp. nov.
86C49DCC-9026-5E0C-87F5-050F5A2F27E8
838656
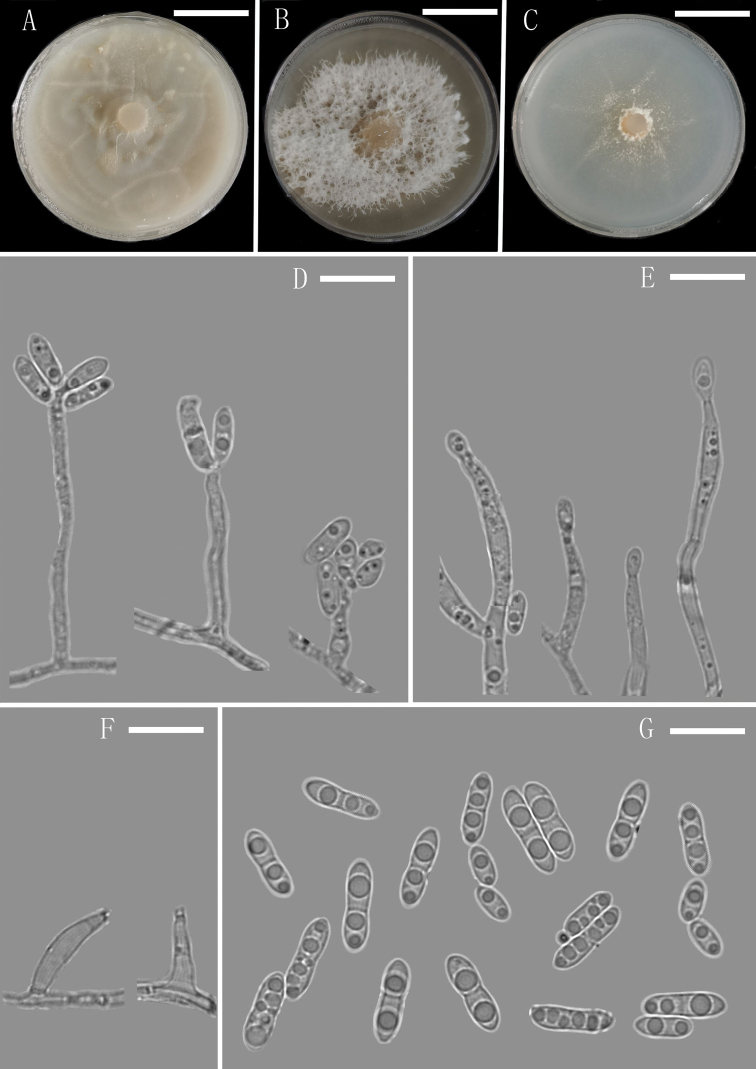
Figure 3.
Plectosphaerella endophytica (YMF 1.04701, holotype) A–C colony on OA, PDA and CMA after 14 d D–F conidiophores and Phialides G conidia. Scale bars: 1.35 cm (A–C), 10 µm (D–G).
Etymology.
Latin, endophytica meaning endophytic, growing within plant tissue.
Description.
Colony on CMA after 3 d, hyphae hyaline, smooth, septate, thin-walled, branched, 1.9–3.3 µm (x̄ = 2.6 μm, n = 10) wide. Conidiophores macronematous, mononematous, erect, straight or flexuous, smooth-walled, hyaline, unbranched or occasionally irregular branched, sometimes 1–2-septate. Conidiogenous cells phialides, subulate, integrated, terminal, determinate, hyaline, smooth-walled. Conidia solitary, acrogenous, broadly navicular to broadly fusiform, suboblong or ellipsoidal, 0–1-septate, usually constricted at septum, bi-guttulate, hyaline, smooth-walled, aseptate conidia abundant, 5–9.1 × 2.5–3.5 µm (x̄ = 7.8 × 3.1 µm, n = 30); septate conidia scarce, 8.8–10.1 × 3.7–4.6 µm (x̄ = 9.4 × 4.1 µm, n = 30), forming hyaline to white mucilaginous masses. Sexual morph and chlamydospores absent.
Culture characteristics.
Colonies on OA reaching 52 mm diameter, on PDA reaching 48 mm diameter and on CMA reaching 43 mm diameter in 14 d at 25 °C. On PDA, colonies white, dense, fluffy hyphae growth in the medium surface, outermost mycelia formed an annule, margin smooth and entire, sporulation abundant, reverse pale yellow to white.
Typification.
China, Yunnan Province, Kunming, The Dian Lake, 24°96'N, 102°66'E, 1886 m alt., isolated from Hydrilla verticillata (L.f.) Royle as an endophyte, 20 Jul. 2014, Z.F. Yu, YMF 1.04701 (Holotype), ex-type CCTCC AF 2021053.
Notes.
Although the phylogenetic analyses showed that our isolate Plectosphaerella endophytica is close to P. oratosquillae, the conidia of P. oratosquillae are aseptate, multi-guttulate (Duc et al. 2009). Furthermore, P. endophytica is most similar to P. verschoorii Hern.-Restr. & Giraldo López in the septa of conidia; both species produce 0–1-septate conidia, and septate conidia are larger than aseptate conidia (P. verschoorii: 1-septate conidia, 8–11.5 × 2–3 μm; aseptate conidia, 3–8.5 × 2–3 μm), but there are obvious difference in the shape of conidia, P. endophytica was deeply constricted at septa. Besides, the phialides of P. verschoorii are shorter (up to 14 μm) (Giraldo et al. 2019).
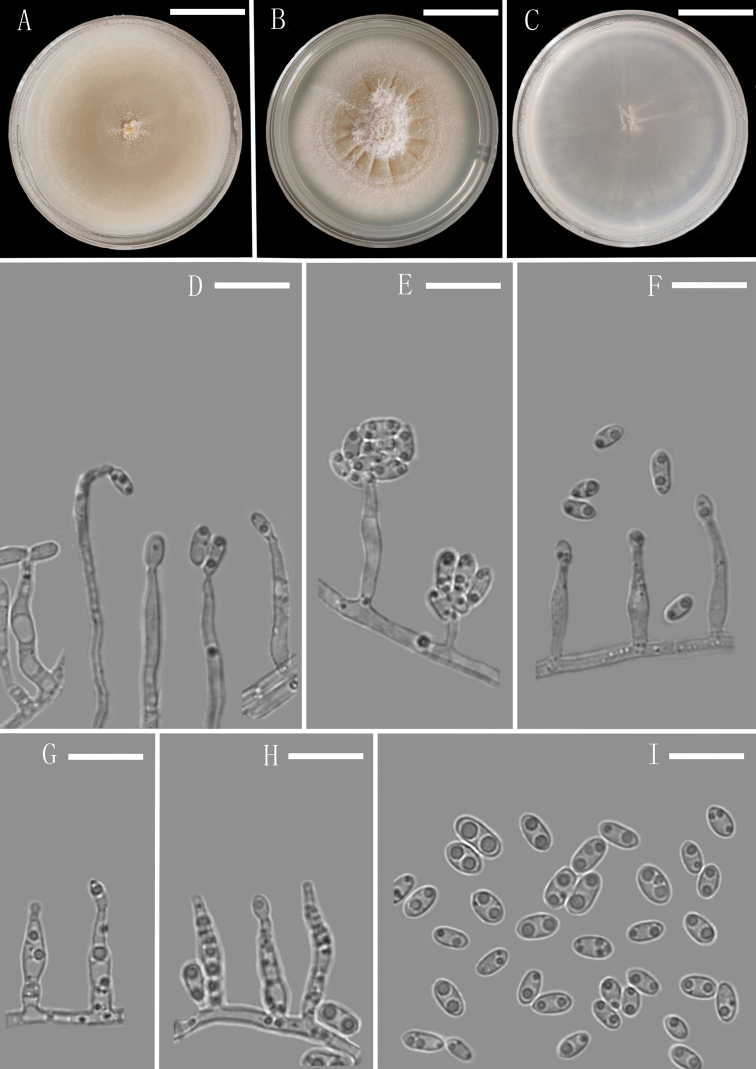
Plectosphaerella sichuanensis
Z.F. Yu & X.Q. Yang sp. nov.
7769D6F4-C3DD-5A3F-A015-9C8B88EE9B56
838657
Figure 4.
Plectosphaerella sichuanensis (YMF 1.05081, holotype) A–C colony on OA, PDA and CMA after 14 d D–H conidiophores and Phialides I conidia. Scale bars: 1.35 cm (A–C), 10 µm (D–I).
Etymology.
Latin, sichuanensis, referred to Sichuan Province, the locality where the fungus was found.
Description.
Colony on CMA after 3 d, vegetative hyphae hyaline, septate, commonly branched, smooth, thin-walled, mostly 2.5–3.5 μm (x̄ = 2.9 μm, n = 10) wide. Conidiophores macronematous, mononematous, erect, straight or flexuous to sinuate, hyaline, smooth, unbranched or rarely branched, aseptate. Conidiogenous cells phialides, integrated, terminal, determinate, subulate, hyaline, smooth. Conidia acrogenous, ellipsoidal, unicellular, smooth-walled, hyaline, 1–3 guttulate, 4.2–6.8 × 2.5–3.7 μm (x̄ = 5.2 × 3.3 µm, n = 30), forming hyaline to white mucilaginous masses. Sexual morph and chlamydospores absent.
Culture characteristics.
Colonies on OA reaching 50 mm diameter, on PDA reaching 47 mm diameter and on CMA reaching 42 mm diameter in 14 d at 25 °C. On PDA, colonies pale brown to white, flat, repressed, plicated, partly immersed, a few white aerial hyphae grew in the middle of the medium, margin regular, frontier distinct, reverse pale brown to white.
Typification.
China, Sichuan Province, Daofu, 30°98'N, 101°13'E, 2960 m alt., isolated from Potamogeton pectinatus as an endophyte, 20 Jul. 2015, Z.F. Yu, YMF 1.05081 (Holotype), ex-type CCTCC AF 2021054, another strain checked: YMF 1.05082.
Notes.
In the phylogenetic tree, the closest species to Plectosphaerella sichuanensis is P. populi, but P. populi can be distinguished from P. sichuanensis by its smaller aseptate conidia (Crous et al. 2015). The size and shape of conidia of P. sichuanensis is more similar to P. cucumerina, expect that P. cucumerina presents longer phialides (up to 69 μm) (Carlucci et al. 2012). In addition, P. sichuanensis resembles P. citrullae Carlucci et al., P. pauciseptata and P. oratosquillae in lacking septate conidia. However, it can be distinguished from P. citrullae by polyphialides conidiogenous cells; from P. pauciseptata and P. oratosquillae by bi-guttulate conidia (Duc et al. 2009; Carlucci et al. 2012).
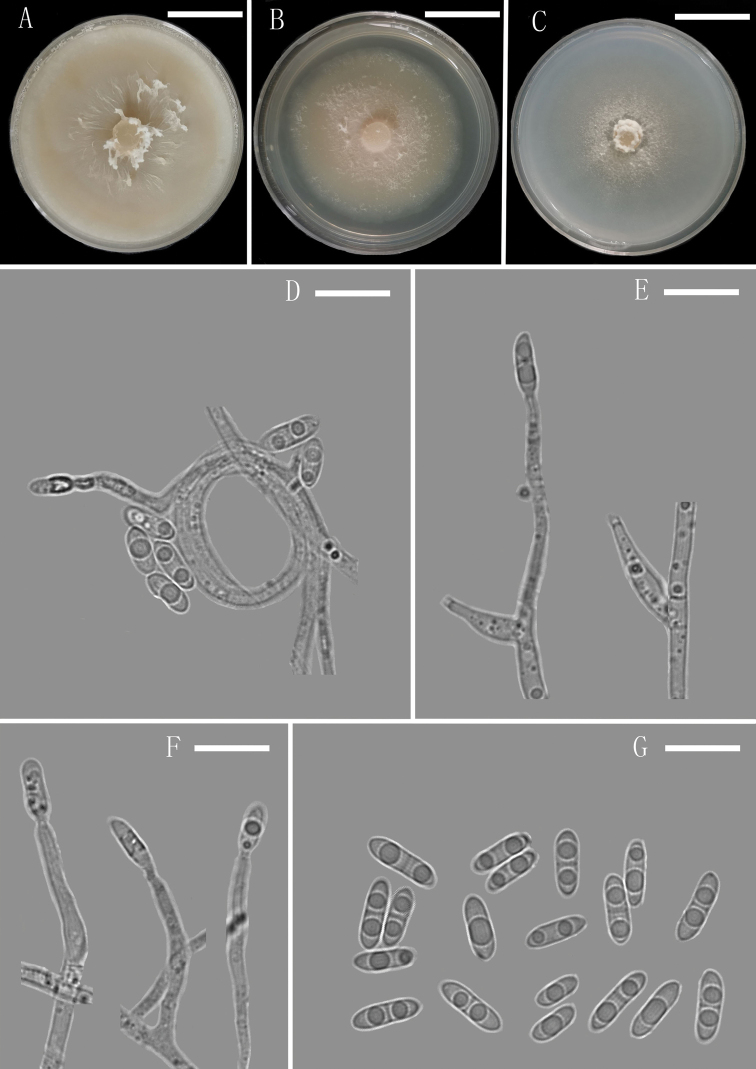
Plectosphaerella cucumerina
(Lindf.) W. Gams, in Domsch & Gams
35CC9E68-95F5-593E-972A-ABCCACA61098
Figure 5.
Plectosphaerella cucumerina (YMF 1.04692) A–C colony on OA, PDA and CMA after 14 d D hyphal coils E, F conidiophores and Phialides G conidia. Scale bars: 1.35 cm (A–C), 10 µm (D–G).
Description.
Colony on CMA after 3 d, hyphae hyaline, septate, smooth, thin-walled, branched, 2.5–3.5 μm (x̄ = 3.2 μm, n = 10) wide. Conidiophores macronematous, mononematous, erect or flexuous to sinuate, hyaline, smooth, branched, occasionally forming hyphal coils. Conidiogenous cells phialides, terminal, determinate, subulate. Conidia acrogenous, hyaline, unicellular, smooth-walled, oblong-ellipsoidal, 1–2 guttulate, 6.6–10.7 × 2.3–3.6 μm (x̄ = 8.7 × 3.1 µm, n = 30), forming hyaline to white mucilaginous masses. Sexual morph and chlamydospores absent.
Culture characteristics.
Colonies on OA reaching 55 mm diameter, on PDA reaching 48 mm diameter and on CMA reaching 44 mm diameter in 14 d at 25 °C. On PDA, colonies pale brown, repressed, flat, partly immersed, some aerial hyphae grew in the middle and margin of the medium, margin regular, reverse pale brown.
Strain examined.
China, Sichuan Province, Baiyu, 31°00'N, 99°41'E, 4013 m alt., isolated from Myriophyllum spicatum as an endophyte, 20 Aug. 2015, Z.F. Yu, YMF 1.04692.
Plectosphaerella pauciseptata
A.J.L. Phillips, A. Carlucci & M.L. Raimondo
DF5B5F92-F840-5A2D-B482-CE2198EBE0DA
Figure 6.
Plectosphaerella pauciseptata (YMF 1.05088) A–C colony on OA, PDA and CMA after 14 d D–F conidiophores and Phialides G conidia. Scale bars: 1.35 cm (A–C), 10 µm (D–G).
Description.
Colony on CMA after 3 d, hyphae hyaline, septate, commonly branched, thin-walled, smooth, 2.5–3.0 μm (x̄ = 2.6 μm, n = 10) wide. Conidiophores macronematous, mononematous, erect, straight or flexuous, hyaline, smooth, aseptate, occasionally branched. Conidiogenous cells phialides, terminal, determinate, subulate, hyaline, smooth, thin-walled. Conidia acrogenous, hyaline, oblong-ellipsoidal, unicellular, smooth-walled, multi-guttulate, 5.5–12.5 × 2.5–3.5 μm (x̄ = 9.7 × 3.3 µm, n = 30), forming hyaline to white mucilaginous masses. Sexual morph and chlamydospores absent.
Culture characteristics.
Colonies on OA reaching 55 mm diameter, on PDA reaching 49 mm diameter and on CMA reaching 45 mm diameter in 14 d at 25 °C. On PDA, colonies white, dense, raised, aerial hyphae growth in the medium surface, margin regular, frontier distinct, reverse pale brown to white.
Strain examined.
China, Yunnan Province, Erhai, 25°43'N, 100°11'E, 1964 m alt., isolated from Myriophyllum spicatum as an endophyte, 31 Jul. 2014, Z.F. Yu, YMF 1.05088, YMF 1.04679, YMF 1.04725.
Discussion
Sexual reproduction of Plectosphaerella has only been reported for three species (Giraldo and Crous 2019; Phookamsak et al. 2019). Most strains, by contrast, show asexual morphs in substrate, so the main distinguishing characteristics of this genus are based on the ratio of septate conidia, conidial dimensions and shape, absence or presence of chlamydospores (Carlucci et al. 2012; Arzanlou et al. 2013). These simple features and similar lifestyle make identification more difficult, so the classical way is no longer a valid and reliable marker (Palm et al. 1995; Zare et al. 2007; Carlucci et al. 2012). The ITS and LSU regions were considered to be a necessary condition for species identification of Plectosphaerella (Duc et al. 2009; Carlucci et al. 2012; Liu et al. 2013; Crous et al. 2015). However, Plectosphaerella species exhibit a relatively low degree of ITS or LSU molecular diversity, showing more molecular markers are needed to distinguish species (Carlucci et al. 2012). Recently, fragments of several protein-coding genes were selected to efficiently elucidate the taxonomy of this genus, including CaM, TEF-1α, TUB2 and RPB2 (Su et al. 2017; Giraldo et al. 2019; Giraldo and Crous 2019; Phookamsak et al. 2019; Zhang et al. 2019). In this study, a multi-locus analysis improved the diagnostic level of the whole genus.
In this study, among 129 sample sites in Yunnan and Sichuan province, Plectosphaerella species were isolated from 13 sampling sites. Regions are arranged in order of isolation frequency: Eehai (39.29%), Dian Lake (20.54%), Fuxian Lake (13.39%), Daofu (8.04%) and other places were below 5%. It seems that distribution of Plectosphaerella is impacted by human activities, because there are more human activities around the first three sites. In addition, Plectosphaerella species only occur in six aquatic plants including Batrachium, Halerpestes, Hippuris, Hydrilla, Myriophyllum, and Potamogeton, although we investigated 30 aquatic plants. So Plectosphaerella species exhibit medium host diversity and relatively narrow geographic distribution when aquatic plants serve as hosts.
Secondly, the separation frequency of Plectosphaerella pauciseptata was the highest at 68.78%, followed by P. cucumerina at 24.11%, and two new species were the lowest at 3.57%. According to this data and previous studies, P. cucumerina and P. pauciseptata are still the most widely distributed species either on land or within aquatic plants, although P. pauciseptata is more common in aquatic plants. On the other hand, only four species were identified among 112 strains. One reason may be that species diversity of Plectosphaerella is low within aquatic plants. Other reasons may be Plectosphaerella species exhibit a relatively low degree of ITS molecular diversity. Based on this, perhaps we treat some different species as two known species, which reduce the diversity of Plectosphaerella. Regrettably, we did not preserve all strains except for representative strains. Or else, we should sequence other loci of all strains, especially for two species, P. pauciseptata, P. cucumerina, more loci are needed to distinguish them.
This study revealed Plectosphaerella species within aquatic plants from the southwest of China, although only 112 strains and four species were reported, which also enrich the ecology and diversity of the genus. However, our survey was limited, hence it is necessary to know if our result is concordant with those of other regions. So in-depth work is required to obtain a more integrated knowledge of their biodiversity and distribution in water bodies.
Supplementary Material
Acknowledgements
This work was financed by the National Natural Science Foundation Program of P.R. China (31770026, 31760012). We are grateful to two reviewers for critically reviewing the manuscript and for providing helpful suggestions to improve this paper.
Citation
Yang XQ, Ma SY, Peng ZX, Wang ZQ, Qiao M, Yu Z (2021) Diversity of Plectosphaerella within aquatic plants from southwest China, with P. endophytica and P. sichuanensis spp. nov.. MycoKeys 80: 57–75. https://doi.org/10.3897/mycokeys.80.64624
Funding Statement
Laboratory for Conservation and Utilization of Bio-resources, Key Laboratory for Microbial Resources of the Ministry of Education, Yunnan University, Kunming, Yunnan, P. R. China. Tianma development office of Yiliang county, Yunnan province, P. R. China
Supplementary materials
Figures S1–S4
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Xiao Qian Yang, Shi Yun Ma, Ze Xiang Peng, Zhong Qiao Wang, Min Qiao, Zefen Yu
Data type
phylogenetic tree
Explanation note
Figure S1. Phylogenetic tree generated by Bayesian inference based on sequences of the ITS. Monilochaetes infuscans CBS 379.77 serves as an outgroup. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S2. Phylogenetic tree generated by Bayesian inference based on sequences of the LSU. Monilochaetes infuscans CBS 379.77 serves as an outgroup. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S3. Phylogenetic tree generated by Bayesian inference based on sequences of the TEF-1α. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S4. Phylogenetic tree generated by Bayesian inference based on sequences of the TUB2. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface.
References
- Alderman DJ, Polglase JL. (1985) Fusarium tabacinum (Beyma) Gams as a gill parasite in the crayfish, Austropotamobius pallipes Lereboullet. Journal of fish disease 8: 249–252. 10.1111/j.1365-2761.1985.tb01222.x [DOI] [Google Scholar]
- Alfaro-García A, Armengol J, Bruton BD, Gams W, García-Jiménez J, Martínez-Ferrer G. (1996) The taxonomic position of the causal agent of Acremonium collapse of muskmelon. Mycologia 88: 804–808. 10.2307/3760975 [DOI] [Google Scholar]
- Alves A, Crous PW, Correia A, Phillips AJL. (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers 28: 1–13. 10.1002/yea.1554 [DOI] [Google Scholar]
- Antignani V, Gams W, Marziano F. (2008) Plectosporium delsorboi sp., a pathogen of Curcuma, Zingiberaceae. Nova Hedwigia 86: 209–214. 10.1127/0029-5035/2008/0086-0209 [DOI] [Google Scholar]
- Arzanlou M, Torbati M, Khodaei S. (2013) Plectosphaerella cucumerina on bamboo in Iran. Mycosphere 4: 627–631. 10.5943/mycosphere/4/3/14 [DOI] [Google Scholar]
- Carlucci A, Raimondo ML, Santos J, Phillips AJL. (2012) Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 28: 34–48. 10.3767/003158512X638251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA, Acharya K, Barber PA, Boekhout T, Dimitrov RA, Dueñas M, Dutta AK, Gené J, Gouliamova DE, Groenewald M, Lombard L, Morozova OV, Sarkar J, Smith MTH, Stchigel AM, Wiederhold NP, Alexandrova AV, Antelmi I, Armengol J, Barnes I, Cano-Lira JF, Castañeda Ruiz RF, Contu M, Courtecuisse PrR, da Silveira AL, Decock CA, de Goes A, Edathodu J, Ercole E, Firmino AC, Fourie A, Fournier J, Furtado EL, Geering ADW, Gershenzon J, Giraldo A, Gramaje D, Hammerbacher A, He X-L, Haryadi D, Khemmuk W, Kovalenko AE, Krawczynski R, Laich F, Lechat C, Lopes UP, Madrid H, Malysheva EF, Marín-Felix Y, Martín MP, Mostert L, Nigro F, Pereira OL, Picillo B, Pinho DB, Popov ES, Rodas Peláez CA, Rooney-Latham S, Sandoval-Denis M, Shivas RG, Silva V, Stoilova-Disheva MM, Telleria MT, Ullah C, Unsicker SB, van der Merwe NA, Vizzini A, Wagner H-G, Wong PTW, Wood AR, Groenewald JZ. (2015) Funsgal planet description sheets: 320–370. Persoonia 34: 167–266. 10.3767/003158515X688433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch KH, Gams W. (1972) Fungi in agricultural soils. Journal of Applied Ecology 10: e897. 10.2307/2401880 [DOI]
- Domsch KH, Gams W, Anderson TH. (2007) Compendium of soil fungi (2nd edn.). IHW Verlag, Germany.
- Duc PM, Hatai K, Kurata O, Tensha K, Yoshitaka U, Yaguchi T, Udagawa SI. (2009) Fungal infection of mantis shrimp (Oratosquilla oratoria) caused by two anamorphic fungi found in Japan. Mycopathologia 167: 229–247. 10.1007/s11046-008-9174-4 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. 10.1128/AEM.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo A, Crous PW. (2019) Inside Plectosphaerellaceae. Studies in mycology 92: 227–286. 10.1016/j.simyco.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo A, Hernández-Restrepo M, Crous PW. (2019) New plectosphaerellaceous species from Dutch garden soil. Mycological Progress 18: 1135–1154. 10.1007/s11557-019-01511-4 [DOI] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147: 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Index Fungorum (2021) http://www.indexfungorum.org/names/Names.asp [accessed 23.1.2021]
- Klebahn H. (1929) Vergilbende junge Treibgurken, ein darauf gefundenes Cephalosporium und dessen Schlauchfrüchte. Phytopathologische Zeitschrift 1: 31–44. [Google Scholar]
- Liu TT, Hu DM, Liu F, Cai L. (2013) Polyphasic characterization of Plectosphaerella oligotrophica, a new oligotrophic species from China. Mycoscience 54: 387–393. 10.1016/j.myc.2013.01.003 [DOI] [Google Scholar]
- Palm ME, Gams W, Nirenberg HI. (1995) Plectosporium, a new genus for Fusarium tabacinum, the anamorph of Plectosphaerella cucumerina. Mycologia 87: 397–406. 10.2307/3760837 [DOI] [Google Scholar]
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN. (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Raimondo ML, Carlucci A. (2018) Characterization and pathogenicity assessment of Plectosphaerella species associated with stunting disease on tomato and pepper crops in Italy. Plant pathology 67: 626–641. 10.1111/ppa.12766 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree v1.4.2. program distributed by the author. http://tree.bio.ed.ac.uk/software/figtree/2012
- Réblová M, Gams W, Seifert KA. (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191. 10.3114/sim.2011.68.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang S-K, Hyde KD, Jayawardena R, Jaklitsch W, Jones EBG, Ju Y-M, Judith C, Maharachchikumbura SSN, Pang K-L, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). Ima Fungus 7: 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Smith-Kopperl ML, Charudattan R, Berher RD. (1999) Plectosporium tabacinum, a pathogen of the invasive aquatic weed Hydrilla verticillate in Flordia. Plant disease 83: 24–28. 10.1094/PDIS.1999.83.1.24 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Su L, Deng H, Niu YC. (2016) Phialemoniopsis endophytica sp. nov., a new species of endophytic fungi from Luffa cylindrica in Henan, China. Mycological Progress 15: e48. 10.1007/s11557-016-1189-5 [DOI]
- Su L, Deng H, Niu YC. (2017) Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycological Progress 16: 823–829. 10.1007/s11557-017-1319-8 [DOI] [Google Scholar]
- Sun X, Guo LD. (2010) Micronematobotrys, a new genus and its phylogenetic placement based on rDNA sequence analyses. Mycological Progress 9: 567–574. 10.1007/s11557-010-0664-7 [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–488. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyozo S, Tadaoki I, Mitsutaka M, Ken W, Keisuke T, Etsuji H. (2005) Plectosporium blight of pumpkin and ranunculus caused by Plectosporium tabacinum. Journal of General Plant Pahtology 71: 127–132. 10.1007/s10327-004-0173-0 [DOI] [Google Scholar]
- Uecker FA. (1993) Development and cytology of Plectosphaerella cucumerina. Mycologia 85: 470–479. 10.2307/3760707 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zare R, Gams W, Starink-Willemse M, Summerbell RC. (2007) Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwigia 85: 463–489. 10.1127/0029-5035/2007/0085-0463 [DOI] [Google Scholar]
- Zhang ZY, Hao W Chen. (2019) Phylogeny and taxonomy of two new Plectosphaerella (Plectosphaerellaceae, Glomerellales) species from China. MycoKeys 57: 47–60. 10.3897/mycokeys.57.36628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S4
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Xiao Qian Yang, Shi Yun Ma, Ze Xiang Peng, Zhong Qiao Wang, Min Qiao, Zefen Yu
Data type
phylogenetic tree
Explanation note
Figure S1. Phylogenetic tree generated by Bayesian inference based on sequences of the ITS. Monilochaetes infuscans CBS 379.77 serves as an outgroup. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S2. Phylogenetic tree generated by Bayesian inference based on sequences of the LSU. Monilochaetes infuscans CBS 379.77 serves as an outgroup. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S3. Phylogenetic tree generated by Bayesian inference based on sequences of the TEF-1α. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface. Figure S4. Phylogenetic tree generated by Bayesian inference based on sequences of the TUB2. Bayesian posterior probability over 80% are shown at the nodes. Two new species are given in boldface.