Abstract
Fused in sarcoma (FUS) is a predominantly nuclear RNA-binding protein with key functions in RNA processing and DNA damage repair. Defects in nuclear import of FUS have been linked to severe neurodegenerative diseases; hence, it is of great interest to understand this process and how it is dysregulated in disease. Transportin-1 (TNPO1) and the closely related transportin-2 have been identified as major nuclear import receptors of FUS. They bind to the C-terminal nuclear localization signal of FUS and mediate the protein's nuclear import and at the same time also suppress aberrant phase transitions of FUS in the cytoplasm. Whether FUS can utilize other nuclear transport receptors for the purpose of import and chaperoning has not been examined so far. Here, we show that FUS directly binds to different import receptors in vitro. FUS formed stable complexes not only with TNPO1 but also with transportin-3, importin β, importin 7, or the importin β/7 heterodimer. Binding of these alternative import receptors required arginine residues within FUS-RG/RGG motifs and was weakened by arginine methylation. Interaction with these importins suppressed FUS phase separation and reduced its sequestration into stress granules. In a permeabilized cell system, we further showed that transportin-3 had the capacity to import FUS into the nucleus, albeit with lower efficiency than TNPO1. Our data suggest that aggregation-prone RNA-binding proteins such as FUS may utilize a network of importins for chaperoning and import, similar to histones and ribosomal proteins.
Keywords: RNA-binding protein, fused in sarcoma (FUS), nuclear transport, transportin, importins, phase separation, RG/RGG motifs
Abbreviations: BSA, bovine serum albumin; CIRBP, cold-inducible RNA-binding protein; CRM1, chromosome region maintenance 1; DAPI, 4′,6-diamidino-2-phenylindole; EGFP, enhanced green fluorescent protein; FTD, frontotemporal dementia; FUS, fused in sarcoma; LCD, low-complexity domain; LLPS, liquid–liquid phase separation; MBP, maltose binding protein; NLS, nuclear localization signal; NTR, nuclear transport receptor; PRMT1, protein arginine N-methyltransferase 1; PY, proline–tyrosine; R, arginine; RBP, RNA-binding protein; RGG3, arginine–glycine–rich; rps, ribosomal proteins; RS, arginine–serine; S, supernatant; SEC, size-exclusion chromatography; SG, stress granule; T, tyrosine; TB, transport buffer; TDP-43, TAR DNA-binding protein 43; TNPO, transportin; TNPO1, transportin-1; TNPO2, transportin-2; TNPO3, transportin-3; unme, unmethylated; Xpo 4, exportin 4
Nuclear depletion and cytoplasmic aggregation of RNA-binding proteins (RBPs) is a key feature of two devastating neurodegenerative diseases, ALS and frontotemporal dementia (FTD) (1, 2, 3). The pathological aggregates are found in neurons and glial cells of the postmortem brain and spinal cord of patients with ALS/FTD. In the majority of cases, these aggregates contain the ubiquitously expressed RBP TAR DNA-binding protein 43 (TDP-43) and, more rarely, fused in sarcoma (FUS) or members of the heterogeneous nuclear ribonucleoprotein A family (3, 4, 5). Both gain-of-function and loss-of-function mechanisms caused by these dysfunctional RBPs have been suggested to contribute to defective RNA metabolism and neurodegeneration (4).
Transport of proteins into the nucleus and back into the cytoplasm is mediated by members of the importin β-family of nuclear transport receptors (NTRs), also referred to as importins and exportins or karyopherins (6). Nuclear import defects have been suggested to contribute substantially to the above-described RBP pathology and ALS/FTD pathogenesis (7, 8, 9). For one, ALS-causing mutations in the C terminus of FUS that also cause motor neuron degeneration in mice (10, 11, 12) were found to disrupt the major nuclear localization signal (NLS) of FUS, thus reducing its nuclear import (13). This NLS consists of a C-terminal proline–tyrosine (PY) consensus sequence preceded by an arginine–glycine–rich (termed RGG3) region and is of low structural complexity (14, 15). Second, the main import receptor for FUS, transportin-1 (TNPO1, also known as karyopherin β2) (16), was found to coaggregate with cytoplasmic FUS inclusions in patients with FTD-FUS and might be functionally impaired in the affected brain regions (17, 18, 19). Moreover, protein arginine N-methyltransferase 1 (PRMT1)-mediated R methylation of the FUS–RGG3 region, which is known to regulate TNPO1 binding and hence FUS nuclear import, is defective in patients with FUS-FTD (14, 20).
In addition to defective nuclear import, enhanced phase separation of FUS has been suggested to contribute to the formation of pathological FUS aggregates (15, 21, 22, 23). Like other RBPs, FUS undergoes liquid–liquid phase separation (LLPS) driven by weak multivalent cation–π interactions between tyrosines in the N-terminal low-complexity domain (LCD) and arginines in C-terminal disordered arginine- and glycine-rich RG/RGG regions (24, 25). We and others have recently shown that TNPO1 can act as a molecular chaperone of FUS: by binding to and shielding of RG/RGG motifs, TNPO1 suppresses phase separation of FUS in vitro and its recruitment into stress granules (SGs) (15, 26, 27, 28). Likewise, other nuclear import receptors have been described to suppress aberrant phase separation of other RBPs, such as TNPO1 for Ewing sarcoma breakpoint region 1 protein, TATA box-binding protein (TBP)-associated factor 15, heterogeneous nuclear ribonucleoprotein A1, and heterogeneous nuclear ribonucleoprotein A2 (26), TNPO1 and transportin-3 (TNPO3) for the cold-inducible RNA-binding protein (CIRBP) (29), or importin α/β for TDP-43 (26). These findings suggest a general function of nuclear import receptors as molecular chaperones for aggregation-prone RBPs (30). Based thereon, elevating nuclear import receptor levels has been proposed as a potential therapeutic strategy to mitigate RBP aggregation and neurodegeneration (31).
Interestingly, a number of nuclear proteins including transcription factors, histones, ribosomal proteins (rps), and RBPs, such as HIV-1 Rev, c-fos, c-jun, and CIRBP are bound and imported by multiple different import receptors (29, 32, 33, 34, 35, 36, 37, 38). Moreover, highly positively charged and aggregation-prone proteins, such as histones and ribosomal proteins, can be chaperoned by multiple import receptors, at least in vitro (39). Whether neurodegeneration-linked aggregation-prone RBPs, such as FUS, are also bound by multiple import receptors for the purpose of import and/or chaperoning has not been investigated.
Here, we demonstrate that FUS directly interacts not only with TNPO1 but also with TNPO3, importin β, importin 7, and the importin β/7 dimer under physiological buffer conditions. Binding of the alternative import receptors depends on arginines in RG/RGG motifs and is weakened by arginine methylation. All interacting importins are able to suppress FUS phase separation in vitro and to reduce its partitioning into SGs in digitonin-permeabilized cells. Import studies in permeabilized cells demonstrate that TNPO3 and, to a lesser extent, importin β, importin 7, and the importin β/7 heterodimer can also import FUS into the nucleus, albeit with lower efficiency than TNPO1. Our data suggest that, similar to histones and ribosomal proteins, FUS may also utilize a network of importins for efficient chaperoning and import.
Results
FUS interacts with different nuclear import receptors in a RanGTP-dependent manner
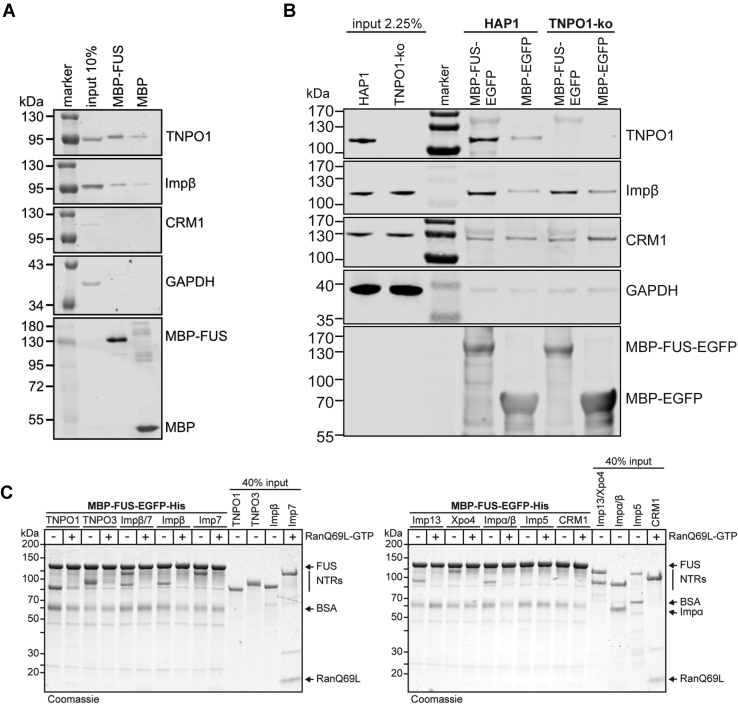
Previously, we and others have shown that FUS is bound and imported into the nucleus by TNPO1 and the highly related transportin-2 (TNPO2) (13, 40, 41). To test whether other NTRs interact with FUS, we first performed pull-down experiments using proteins from a HeLa cell lysate. As shown in Figure 1A, not only TNPO1 but also importin β bound to immobilized FUS fused to the maltose binding protein (MBP–FUS), but only weakly to MBP alone.
Figure 1.
Several nuclear transport receptors interact directly with FUS in a RanGTP-dependent manner.A, MBP or MBP–FUS was immobilized on amylose beads and incubated with HeLa cell lysate. B, MBP–FUS–EGFP or MBP–EGFP was immobilized on MBP-selector beads and incubated with lysates from HAP1 cells (control) or TNPO1-KO cells. A and B, bound proteins were analyzed by SDS-PAGE followed by Western blotting, detecting TNPO1, importin β, CRM1, and GAPDH with specific antibodies. The immobilized fusion proteins were detected using anti-MBP (A) or anti-GFP (B) antibodies. C, MBP–FUS–EGFP was immobilized on GFP–Trap magnetic agarose beads and incubated with His-tagged nuclear transport receptors (TNPO1, TNPO3, importin β/7, importin β, importin 7, importin 13, exportin 4 (Xpo4), importin α, importin 5, and CRM1) in the absence or presence of RanQ69L(aa1-180)–GTP. Bound proteins were analyzed by SDS-PAGE, followed by Coomassie staining. Input: 40%. FUS binds to TNPO1, TNPO3, importin β/7, importin β, importin 7, importin 13, and exportin 4 but not importin α (in the presence of importin β), importin 5, and CRM1 in a RanGTP-sensitive manner. Similar results were obtained when MBP–FUS–EGFP was immobilized on amylose beads (data not shown).
We next addressed the possibility that in the absence of TNPO1, binding of alternative NTRs could be favored and obtained lysates from either the nearly haploid control cells HAP1 or HAP1 cells lacking TNPO1 entirely. As observed for the HeLa cell lysate, TNPO1 and also importin β bound specifically to immobilized MBP–FUS–EGFP (enhanced green fluorescent protein) when the control lysate was used for the reaction (Fig. 1B). Chromosome region maintenance 1 (CRM1), which is not involved in nucleocytoplasmic transport of FUS (42), and GAPDH did not interact with the immobilized protein. In the absence of TNPO1, the amount of importin β bound to the beads did not increase, suggesting that TNPO1 did not prevent binding of the alternative NTR under our experimental conditions.
For a more systematic and direct analysis, we next performed binding experiments with recombinant NTRs. Again, MBP–FUS–EGFP was immobilized on beads and incubated with different His-tagged NTRs in the absence or presence of a truncated version of RanQ69L(aa1-180)–GTP, a Ran mutant deficient in GTP hydrolysis (43). TNPO1, our positive control, showed clear binding to MBP–FUS–EGFP, as expected, and the interaction was reduced when RanQ69L–GTP was added to the reaction (Fig. 1C). In addition, direct binding to FUS was also observed for TNPO3, importin β, importin 7, importin 13, and exportin 4 (Xpo 4). Binding of these NTRs to FUS was either abolished or reduced by RanQ69L, suggesting specific interactions. No binding was detected for importin 5, the export receptor CRM1, or for importin α added to the reaction in addition to importin β. Importin β and importin 7 have previously been shown to dimerize and to function as an import receptor for histone H1 (44). When importin β and importin 7 were added together to immobilized MBP–FUS–EGFP, binding of both NTRs was observed. Again, the addition of RanGTP abolished the interaction.
Together, our pull-down experiments using cell lysates or purified proteins show that several NTRs can engage in specific interactions with FUS.
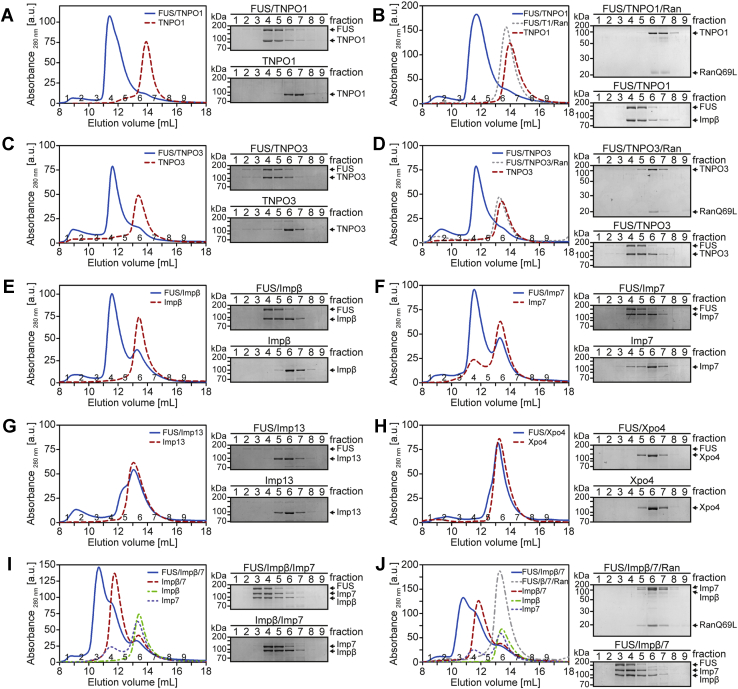
FUS forms a stable complex with TNPO1, TNPO3, importin β, importin 7, and importin β/7
To further address the composition and the stability of FUS–NTR complexes, we subjected them to size-exclusion chromatography (SEC). When analyzed alone the His-tagged NTRs (TNPO1 (Fig. 2A), TNPO3 (2C), importin β (2E), importin 7 (2F), importin 13 (2G), and Xpo 4 (2H); dashed red curves) eluted from the gel-filtration column with peaks in fractions 6 to 7, reflecting the similar sizes of the individual proteins. Upon incubation with MBP–FUS–EGFP, TNPO1, the established import receptor of FUS, eluted earlier from the column (with a peak in fractions 4–5) (Fig. 2A, blue curve), demonstrating that the TNPO1–FUS complex is stable under the conditions of gel filtration. When RanQ69L–GTP was added to the reaction, no complex was observed and TNPO1, together with Ran, was found in fractions 6 to 7 (Fig. 2B, dotted gray curve), confirming the specificity of the interaction. Similar results were obtained for complexes containing FUS and TNPO3 (Fig. 2, C and D), importin β (Fig. 2E), or importin 7 (Fig. 2F). In contrast, importin 13 (Fig. 2G) and Xpo 4 (Fig. 2H) did not form stable complexes with MBP–FUS–EGFP, although binding was observed in the pull-down experiments (Fig. 1C). Next, we analyzed the potential trimeric complex comprising MBP–FUS–EGFP, importin β, and importin 7 in more detail. As expected, the two NTRs alone formed a stable heterodimer, eluting in fractions 4 to 5 from the gel-filtration column (Fig. 2I). When MBP–FUS–EGFP was added to the assembly reaction, the resulting complex eluted as a heterotrimer, peaking in fractions 3 and 4. Importantly, stoichiometric amounts of the three proteins were detected, suggesting a 1:1:1 complex. As for the individual heterodimers described above, the addition of RanQ69L–GTP to the assembly reaction prevented the formation of a stable heterotrimer (Fig. 2J). MBP–FUS–EGFP alone was not detected in gel-filtration experiments with RanQ69L–GTP, as binding to NTRs was prevented by Ran, leading to largely insoluble FUS proteins.
Figure 2.
FUS forms complexes with TNPO3, importin β, importin 7, and importin β/7. MBP–FUS–EGFP was incubated with different His-tagged nuclear transport receptors in the absence (A, C, and E–I) or presence (B, D, and J) of RanQ69L(aa1-180)–GTP and subjected to size-exclusion chromatography, monitoring UV absorbance at 280 nm. Eluate fractions were analyzed by SDS-PAGE, followed by Coomassie staining. Note that FUS alone was not soluble after a 100,000g centrifugation step and was therefore not detected by size-exclusion chromatography.
In summary, just like TNPO1/2, the import receptors TNPO3, importin β, importin 7, and importin β/7 can form stable complexes with FUS.
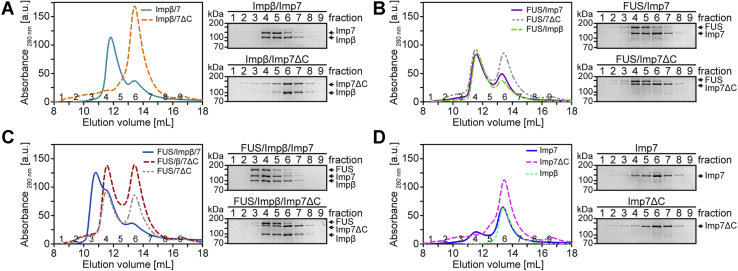
Heterodimerization of importin β and importin 7 is required for the formation of a trimeric complex with FUS
To characterize the trimeric FUS–importin β/importin 7 complex in more detail, we used a mutant version of importin 7 (Imp7ΔC) lacking amino acids 1002 to 1038, that is, the C-terminal end. This deletion was previously shown to impair the interaction of importin 7 with importin β (45). Indeed, full-length importin 7, but not the mutant, formed a complex with importin β, as shown in Figure 3A. The truncated version of importin 7 did, however, interact with FUS (Fig. 3B), showing that cargo-binding is still possible. Finally, importin 7ΔC is unable to engage in a trimeric complex with FUS and importin β (Fig. 3C), suggesting that the importin β–importin 7 interaction is required for trimeric complex assembly. The elution profile of individual proteins is shown in Figure 3D.
Figure 3.
The C terminus of importin 7 is required for the formation of a trimeric FUS–importin β–importin 7 complex.A–D, different combinations of MBP–FUS–EGFP, His–importin β, His–importin 7wt, and His–importin 7ΔC (aa1-1001) were incubated and subjected to size-exclusion chromatography, monitoring UV absorbance at 280 nm. Eluate fractions were analyzed by SDS-PAGE, followed by Coomassie staining. Note that FUS alone was not soluble after a 100,000g centrifugation step and was therefore not detected by size-exclusion chromatography.
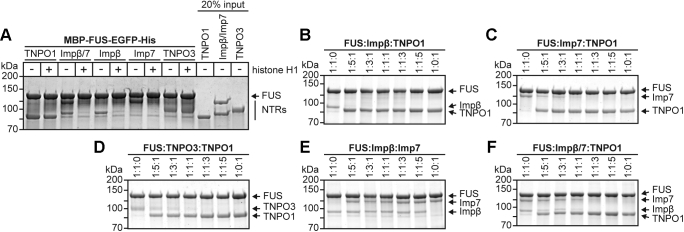
TNPO1 has a high affinity for FUS compared with other NTRs
The importin β/7 dimer has previously been shown to function as an import receptor for histone H1 (44). As another specificity criterion, we therefore performed competition experiments with purified histone H1. As shown in Figure 4A, the addition of a 3-fold excess of the soluble histone over immobilized FUS prevented the interaction of FUS with importin β or importin 7 alone, as well as with the heterodimer. The interaction of FUS with TNPO1 and TNPO3, by contrast, was not affected by the histone.
Figure 4.
TNPO1 has a higher affinity for FUS than TNPO3, importin β, importin 7, and importin β/7. MBP–FUS–EGFP was immobilized on GFP–Trap magnetic agarose beads and incubated with one or two His-tagged nuclear transport receptors in the presence or absence of histone H1 (A) or with two (B–E) or three (F; importin β/importin 7/TNPO1) different His-tagged nuclear transport receptors at different molar ratios, as indicated. Bound proteins were analyzed by SDS-PAGE, followed by Coomassie staining.
We next addressed the question whether different NTRs compete for FUS binding or bind simultaneously. Again, MBP–FUS–EGFP was immobilized on beads and incubated with different ratios of importin β and TNPO1. Clearly, TNPO1, the established import receptor for FUS, prevented binding of importin β to FUS at all ratios (Fig. 4B). Even a 5-fold molar excess of importin β over TNPO1 did not result in significant binding of importin β to the immobilized protein. Likewise, TNPO1 efficiently competed with importin 7 (Fig. 4C) and TNPO3 (Fig. 4D) for binding to immobilized FUS. At a 3- or 5-fold molar excess of importin 7 or TNPO3, however, small amounts of both NTRs bound to FUS, in addition to TNPO1. Thus, importin 7 and TNPO3 appear to have a somewhat higher affinity for FUS than importin β. These results suggest that different NTRs compete for FUS binding, because of either overlapping binding sites or allosteric changes preventing the interaction of FUS with other NTRs. Next, we analyzed the FUS–importin β–importin 7 interaction in more detail. As shown in Figure 4E, a 5-fold molar excess of importin β did not prevent binding of importin 7 to FUS. Likewise, an excess of importin 7 did not reduce the interaction between FUS and importin β, confirming the SEC results (Fig. 2I), showing that the three proteins form a trimeric complex. Finally, we tested the ability of TNPO1 to compete with a combination of importin β and importin 7 for binding to immobilized FUS (Fig. 4F). Again, at a 5-fold molar excess of TNPO1, no other NTR interacted with FUS. When all proteins were present at equimolar concentrations, however, both importin 7 and importin β were found to bind to FUS, besides TNPO1. Similarly, TNPO1 was able to compete with TNPO3, importin β, importin 7, or importin β/7 when these NTRs were allowed to bind to FUS before addition of TNPO1 (Fig. S1, A–D).
Together, these results clearly show that several NTRs are able to directly interact with FUS, with TNPO3, importin β, importin 7, and importin β/7 showing weaker binding than TNPO1. Although TNPO1 occurs at relatively high concentrations (1 to 2 μM in HeLa cells, i.e., similar to importin β and importin 7 (46)), alternative NTRs may thus be used in certain cell types and/or under certain growth conditions.
TNPO3, importin β, importin 7, and importin β/7 bind to FUS RGG regions
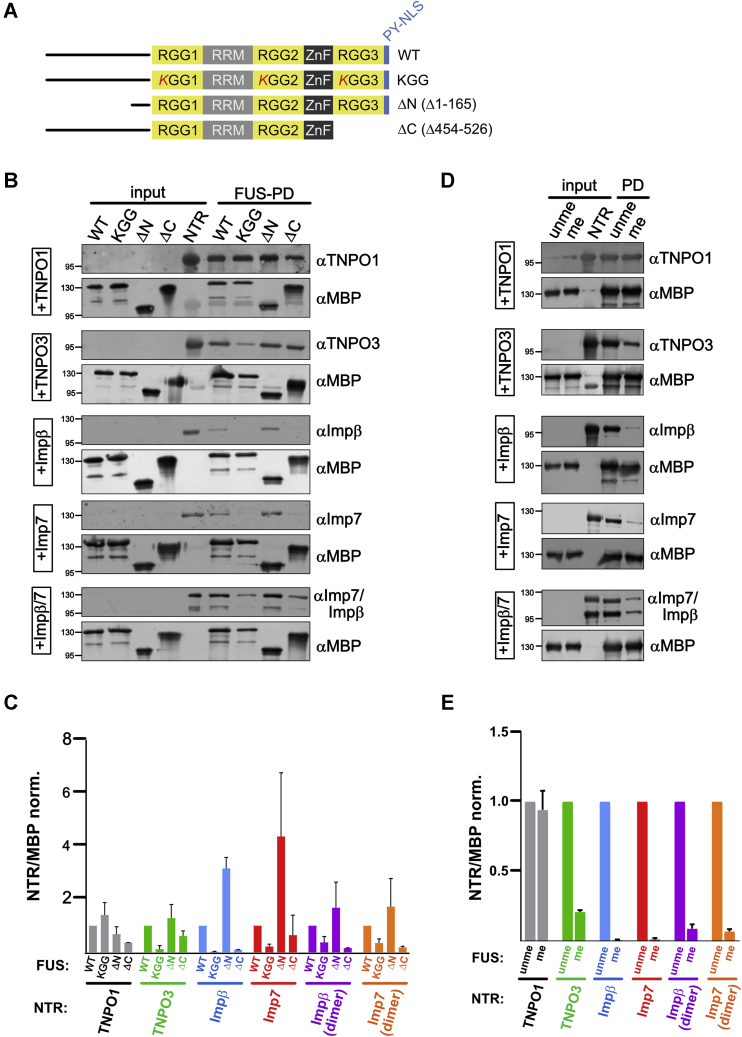
TNPO1 is known to bind with high affinity to the C-terminal RGG3–PY domain of FUS (14, 15). Whether TNPO3, importin β, importin 7, or importin β/7 bind to the same NLS in FUS is unclear. To address this question, we performed binding assays utilizing a series of FUS mutants immobilized to amylose resin: MBP–FUS lacking the N-terminal LCD (FUS Δaa 1–165; FUS ΔN), MBP–FUS lacking the C-terminal RGG3–PY domain (FUS Δaa 454–526; ΔC), or FUS with all RG/RGG motifs mutated to KG/KGG (KGG) (Fig. 5A). Under the physiological buffer conditions used in our assay, TNPO1 showed slightly diminished binding to the C-terminal deletion mutant of FUS (Fig. 5B, quantification shown in Fig. 5C), consistent with the FUS RGG3–PY domain being the main, but not the only binding site for TNPO1 (28). Interestingly, binding of TNPO3 appeared unaffected by deletion of the RGG3–PY domain (ΔC) but was greatly reduced in the KG/KGG mutant in line with our previous results showing binding of TNPO3 to the RG/RGG regions in CIRBP (29). Importin β, importin 7, and importin β/7 all showed reduced binding upon mutation of the RG/RGG motifs (KG/KGG mutant) and upon deletion of the C-terminal RGG3–PY domain (ΔC). In contrast, no impact of importin binding was detected upon deletion of the N-terminal LCD (ΔN). These findings suggest that TNPO3, importin β, importin 7, and importin β/7, in contrast to TNPO1, bind FUS mostly via the RG/RGG motifs, independently of the PY motif, with the C-terminal RGG3 domain being the main interaction site for importin β, importin 7, and importin β/7.
Figure 5.
TNPO3, importin β, importin 7, and importin β/7 bind to RGG motifs in FUS and are affected by arginine methylation.A, scheme illustrating FUS constructs used for pull-down experiments. B, MBP–FUS WT, a FUS mutant with all RG/RGG motifs mutated to KG/KGG (KGG) or FUS deletion mutants lacking either the N-terminal low-complexity domain (aa1-165; ΔN) or the RGG3–PY domain (aa454–526; ΔC) were immobilized on amylose resin and incubated with TNPO1, TNPO3, importin β, importin 7, or importin β/7 as indicated. Binding of importins to the different FUS proteins was analyzed by Western blotting. Successful immobilization of MBP–FUS proteins was verified by MBP Western blot. Input, 15%. C, quantification of the amount of NTR bound to MBP–FUS wt or mutant as the mean of two independent replicates ± SD normalized to TNPO1. D, binding of TNPO1, TNPO3, importin β, importin 7, or importin β/7 to either unmethylated (unme) or methylated (me) MBP–FUS as detected by Western blotting. Immobilization of MBP–FUS proteins was verified by Western blotting. Input, 15%. E, quantification of the amount of NTR bound to unmethylated (unme) or methylated (me) MBP–FUS shown as ratio of NTR/MBP as the mean of three independent replicates ± SEM normalized to unme FUS. NTR, nuclear transport receptor; PY, proline–tyrosine; RGG3, arginine–glycine–rich.
As asymmetric dimethylation of arginine is known to reduce binding of TNPO1 to FUS (14, 15, 20), we sought to address whether methylation of the FUS–RG/RGG regions also affects binding of the alternative importins. Both unmethylated MBP–FUS (unme; incubated with the methyltransferase PRMT1 in the absence of the methyl donor SAM) and methylated (me) MBP–FUS were immobilized on MBP–Trap agarose and tested for binding to TNPO1, TNPO3, importin β, importin 7, and importin β/7. In line with our findings that mutation of all RG/RGGs abolished binding of TNPO3, importin β, importin 7, and importin β/7, binding of these importins was greatly diminished upon R methylation (Fig. 5D, quantification shown in Fig. 5E). Owing to the presence of the PY motif as high-affinity binding site for TNPO1 in the FUS C-terminal domain, TNPO1 binding was only weakly reduced upon R methylation, in line with our previous findings (14, 15).
Together, we demonstrate that TNPO3, importin β, importin 7, and importin β/7 primarily interact with FUS RG/RGG regions and that their binding is strongly reduced upon R methylation. While the presence of the RGG1 and RGG2 domains is sufficient for TNPO3 binding, importin β, importin 7, and the heterodimer importin β/7 appear to require the RGG3 region of FUS, suggesting similar but nonredundant binding modes of different NTRs to the RGG regions of FUS.
TNPO3, importin β, importin 7, and importin β/7 function as chaperones for FUS
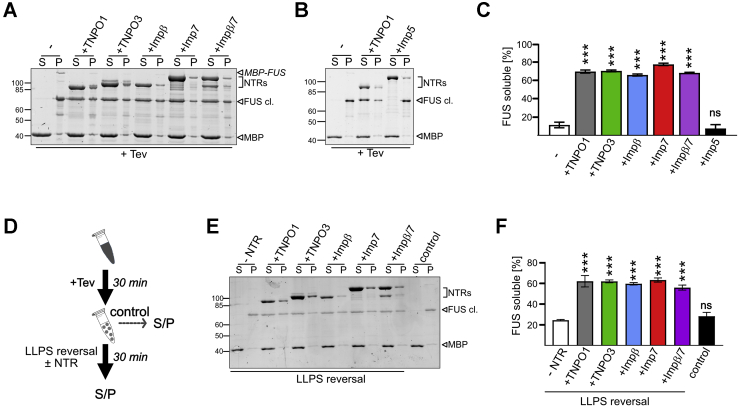
FUS is known to undergo reversible LLPS in a concentration-dependent manner (22, 23, 47, 48, 49, 50). This process is mainly driven by π−π and cation–π interactions of tyrosines in the N-terminal LCD with arginines in the RGG regions (15, 24, 27). We and others have shown that TNPO1 not only mediates nuclear import of FUS but also acts as a chaperone by binding to the RGG regions, thereby preventing LLPS of FUS (15, 26, 27, 28). We therefore hypothesized that the other importins that bind to RG/RGG regions of FUS are also able to suppress FUS phase separation and addressed this hypothesis in a sedimentation assay, where phase-separated condensates are pelleted by centrifugation. Here, FUS was predominantly in the pellet fraction (P) upon Tev protease–mediated cleavage of the MBP-solubility tag (Fig. 6A, see Fig. 6C for quantification). Similar to TNPO1, equimolar levels of TNPO3, importin β, importin 7, or importin β/7 increased FUS levels in the supernatant (S), indicating increased FUS solubility, that is, “chaperoning” activity of all importins tested. Importin 5, which does not bind to FUS (Fig. 1C), could not prevent sedimentation of FUS (Fig. 6, B and C).
Figure 6.
TNPO3, importin β, importin 7, and importin β/7 can suppress and reverse phase separation and sedimentation of FUS.A, equimolar amounts of TNPO3, importin β, importin 7, or importin β/7 can suppress precipitation of MBP–FUS upon Tev cleavage of the MBP-solubility tag in a sedimentation assay similarly to TNPO1. Equal amounts of the supernatant (S) and pellet (P) fraction were visualized by SYPRO Ruby stain. B, importin 5 is not able to suppress FUS precipitation upon Tev cleavage. C, the relative amount of FUS being soluble as the percent of the total (sum of S and P). Values represent the mean of three (TNPO3, importin β, importin β/7, importin 5) or six (untreated, TNPO1) independent experiments ± SEM, ∗∗∗p < 0.0002 defined by one-way ANOVA with Dunnett's multiple comparison test to untreated (“-”). D, experimental setup to assess reversal of FUS phase separation by NTRs. E, reversal of FUS phase separation by TNPO1, TNPO3, importin β, importin 7, or importin β/7 in a sedimentation assay. Equal amounts of the S and P fraction were visualized by SYPRO Ruby stain. F, the relative amount of soluble FUS as percent of the total (sum of S and P). Values represent the mean of three independent experiments ± SEM, ∗∗∗p < 0.0002 defined by one-way ANOVA with Dunnett's multiple comparison test to control lacking NTR (“-NTR”). ns, not significant.
TNPO1 has not only been shown to prevent but also to reverse phase separation of FUS (26). Therefore, we next tested whether TNPO3, importin β, importin 7, or importin β/7 is also able to mediate solubilization of FUS after it has undergone phase separation. To this end, we first induced phase separation of FUS by Tev protease–mediated cleavage of the MBP tag, before adding equimolar amounts of the different import receptors and analyzing FUS solubility in a sedimentation assay (Fig. 6D). As before, the solubility of FUS was greatly reduced upon cleavage of its solubility tag (Fig. 6E, quantification shown in Fig. 6F, “control”). Subsequent addition of TNPO3, importin β, importin 7, or importin β/7 restored solubility, similarly to TNPO1, whereas FUS remained insoluble in a control lacking an NTR.
TNPO3, importin β, importin 7, and importin β/7 suppress SG association of FUS
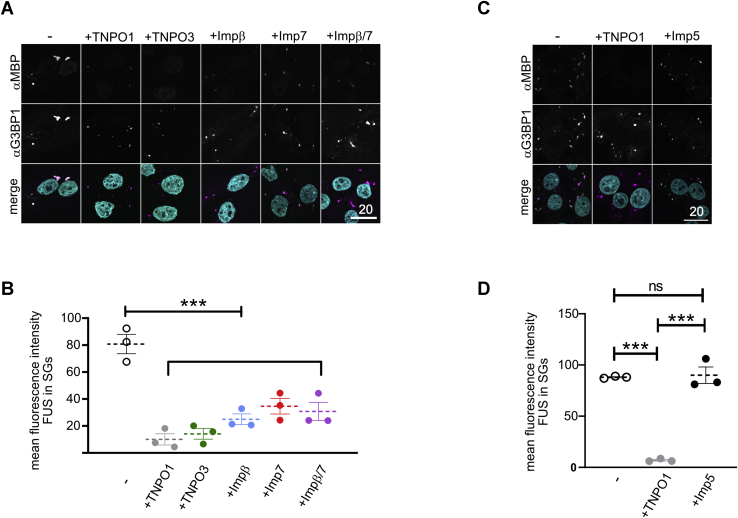
TNPO1 suppresses LLPS of FUS not only in vitro but also in a cellular system, as it reduces the partitioning of FUS into SGs in digitonin-permeabilized cells (15, 51). In this assay, formation of SGs is induced by the proteasome inhibitor MG132. The plasma membrane is then selectively permeabilized using digitonin, and all soluble components are washed out. To analyze association of FUS with SGs in absence of nuclear import, nuclear pores are blocked using wheat germ agglutinin before recombinant MBP–FUS is added in the absence or presence of nuclear import receptors. In the absence of any import receptors, MBP–FUS partitions into SGs, as published previously (15). We used this assay to address whether TNPO3, importin β, importin 7, and importin β/7 are also able to prevent SG association of FUS in a cellular environment. Indeed, all importins were able to suppress SG association of MBP–FUS, with importin β, importin 7, and the heterodimer importin β/7 being slightly less efficient compared with TNPO1 and TNPO3 (Fig. 7, A and B). In contrast, addition of importin 5 could not prevent association of FUS with SGs (Fig. 7, C and D). These results demonstrate that not only TNPO1 but several importins can act as molecular chaperones to prevent LLPS and partitioning of FUS into membrane-less organelles.
Figure 7.
TNPO3, importin β, importin 7, and importin β/7 suppress stress granule association of FUS.A, association of MBP–FUS to stress granules (SGs) in semipermeabilized cells is suppressed by TNPO1, TNPO3, importin β, importin 7, and importin β/7. SGs and MBP–FUS were visualized by staining for the SG marker protein G3BP1 and the MBP-tag. For clarity, signals were converted to gray values in the individual channels (upper two rows). In the merge (lower row), G3BP1 alone is shown in magenta. White pixels indicate colocalization with MBP–FUS (green). DAPI (turquoise) was included in the merge. The scale bar represents 20 μm. B, quantification of the mean fluorescence intensity of MBP–FUS in SGs for three independent replicates ± SEM, ∗∗∗p < 0.0002 defined by one-way ANOVA with Dunnett's multiple comparison test (≥10 cells; ≥ 32 SGs per condition) in the absence or presence of 5-fold excess of TNPO1, TNPO3, importin β, importin 7, or importin β/7. C, importin 5 cannot suppress association of MBP–FUS to stress granules (SGs) in semipermeabilized cells. SGs and MBP–FUS were visualized by staining for the SG marker protein G3BP1 and the MBP tag as before. The scale bar represents 20 μm. D, quantification of the mean fluorescence intensity of MBP–FUS in SGs for three independent replicates ± SEM, ∗∗∗p < 0.0002 defined by one-way ANOVA with Dunnett's multiple comparison test (≥10 cells; ≥ 30 SGs per condition) in the absence or presence of either TNPO1 or importin 5. G3BP1, Ras GTPase-activating protein-binding 1; SGs, stress granules.
FUS can be imported into the nucleus by TNPO1 and TNPO3
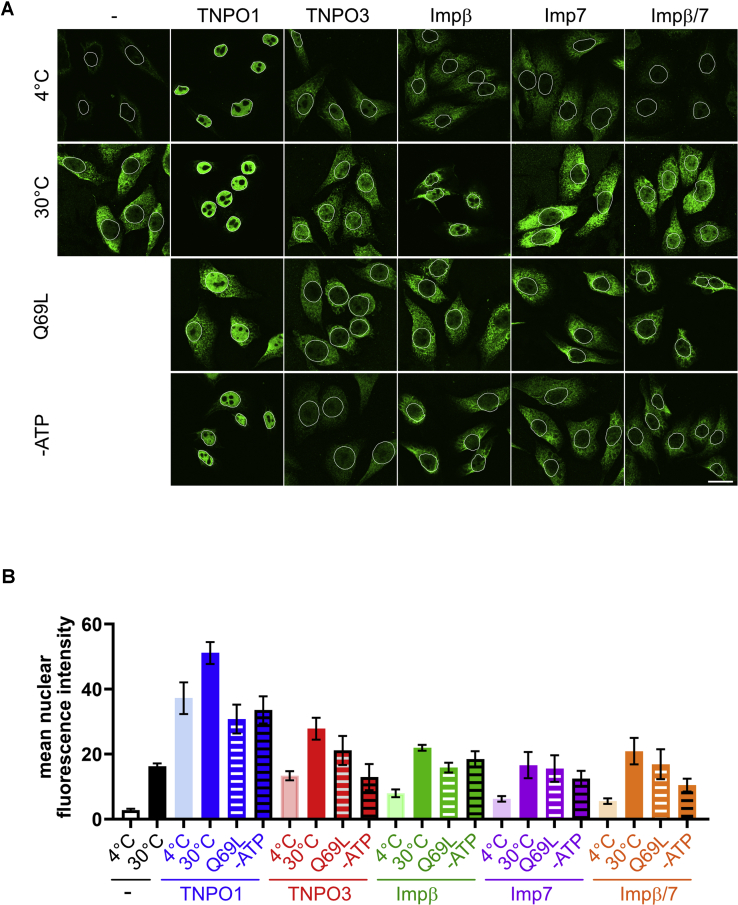
Our results described so far show that FUS can interact with multiple importins, forming complexes that are stable under physiological conditions. Furthermore, not only TNPO1 but also other importin receptors can function as molecular chaperones, increasing the solubility of FUS and preventing its association with SGs. We next asked whether FUS can also be imported into the nucleus by these importins. To this end, we used the established nuclear import assay in digitonin-permeabilized cells with MBP–FUS as the import substrate. As a positive control, we used the main nuclear import factor of FUS, TNPO1, and compared it to TNPO3, importin β, importin 7, and the combination of the latter two. As shown in Figure 8, A and B, in the presence of TNPO1, nuclear import of FUS was observed even at 4 °C in the presence of Ran and an ATP-regenerating system, pointing to the high efficiency of this pathway. Import was further stimulated at 30 °C, whereas RanQ69L, which prevents the interaction of FUS with TNPO1, reduced the import efficiency and resulted in a strong cytoplasmic FUS signal. This observation suggests that without sufficient chaperoning by NTRs, the import substrate is prone to unspecific binding to cellular structures, particularly at 30 °C. Compared with TNPO1, TNPO3 was less efficient in importing FUS into the nucleus, although temperature-dependent transport could be observed in a RanQ69L- and ATP-dependent manner. Addition of importin β, importin 7, or importin β/7 promoted import only slightly.
Figure 8.
Nuclear import of FUS in digitonin-permeabilized cells by TNPO1 and alternative NTRs. Digitonin-permeabilized HeLa cells were incubated with MBP–FUS in the absence or presence of different His-tagged nuclear transport receptors, in the absence or presence of an ATP-regenerating system, with either WT Ran or RanQ69L (Q69L) at 30 °C or 4 °C as indicated. A, cells were subjected to indirect immunofluorescence detecting the MBP tag and analyzed by confocal microscopy. The scale bar represents 20 μm. B, quantification of the mean nuclear import efficiencies of FUS as in panel A for four or three (“-”, 30 °C, importin β, 30 °C) independent experiments ± SEM (25–70 cells per condition).
Together, these data confirm TNPO1 as the major import receptor for FUS but also show that additional importins, in particular TNPO3, can carry out nuclear import of FUS, at least in permeabilized cells. It remains to be investigated, under which conditions these alternative importins, which are functional in vitro, support nuclear import of FUS in intact cells.
Discussion
TNPO1 and TNPO2 are known as major import receptors of FUS (14, 41) and have also been shown to chaperone the FUS protein in the cytoplasm, preventing and reversing its aberrant LLPS (15, 26, 28). Our results now show that additional NTRs can perform similar functions. Of note, a subset of alternative NTRs has also been reported to interact with FUS during the review process of our article (52). Apart from TNPO1/2, TNPO3 (also known as transportin-SR2, Trn-SR2, Trn-3) and its splice variant transportin-SR (53, 54, 55, 56, 57) have been implicated in the nuclear import of several RBPs by binding to their arginine–serine (RS)-rich regions, both in a phosphorylation-dependent and phosphorylation-independent manner (29, 57). RS domains hence have been suggested to act as an NLS for TNPO3. This NLS motif has recently been extended by Bourgeois et al. (29), demonstrating the relevance for tyrosine residues in RSY-rich regions for TNPO3 binding. TNPO3 has also been reported to mediate nuclear import of proteins in the absence of any RS domain (56, 58, 59). Recently, TNPO3 was found to bind to an RGG/RG-rich region in the RBP CIRBP in an arginine-methylation dependent manner, similarly to TNPO1, yet with weaker affinity than TNPO1 (29), suggesting an overlapping cargo spectrum of TNPO1/2 and TNPO3. Our results showing that TNPO3 can also function as a chaperone/NTR for FUS in an RGG-dependent and arginine methylation–dependent manner are very much in line with these findings. It will be interesting to determine which other RGG/RG region containing cargoes can utilize both TNPO1/2 and TNPO3 as import receptors and which features in the “RGG–NLS” determine promiscuous receptor binding.
Similar to TNPO1 and TNPO3, the heterodimeric import receptor importin β/7 acts as a chaperone for various highly basic proteins and mediates their nuclear import, including ribosomal proteins L4 and rpL6, the RNA methyltransferase EMG1, and the linker histone H1 (39, 44, 60). Histone H1 can bind individually to importin β and importin 7 as well. The formation of a functional import complex, however, appears to require the cooperative assembly of an importin β/7-H1-trimer (44, 45, 61). In a large-scale proteomics study for potential cargoes of different importins using reconstituted nuclear import in permeabilized cells (59), FUS was identified as a putative import substrate of TNPO1, TNPO2 importin β, importin 7, and importin 11. Our data show that FUS can also form a heterotrimeric complex with importin β/7, requiring importin β–importin 7 interaction via the importin 7 C-terminal region.
TNPO3, importin β, importin 7, and importin β/7 bind FUS with lower affinity than TNPO1, perhaps because their binding is mediated by RG/RGG motifs or other regions, independently of the PY–NLS. In contrast, the FUS PY–NLS provides a high-affinity docking site for TNPO1, which additionally interacts with various regions throughout the entire length of FUS (28). Under which conditions the weaker-binding importins are utilized by FUS in vivo as chaperones and/or import receptors remains to be determined. We speculate that they may compensate for TNPO1 under conditions where TNPO1–FUS binding is either impaired or TNPO1 is nonfunctional. This is the case in patients with ALS-FUS, where a genetic mutation in the NLS of FUS impairs TNPO1 binding and thereby hampers nuclear import of FUS. It seems possible that the alternative importins partially compensate the reduced TNPO1 binding and restore nuclear import and chaperoning at least to some degree. In addition, TNPO1 in patients with FTD-FUS is found to coaggregate with unmethylated FUS in cytoplasmic inclusions (17, 18, 19). Whether TNPO1 itself is dysfunctional in FTD–FUS and why it is codeposited with FUS are currently unknown. It can be speculated that increased expression levels of TNPO3, importin β, importin 7, or importin β/7 can, at least partially, rescue FUS mislocalization and/or aggregation in these patients. Alternatively, these importins might also be codeposited in postmortem brains of patients with FTD, along with TNPO1, leading to the observed severe mislocalization and aggregation of FUS and other RBPs (62, 63, 64).
In the HeLa cell proteome, importin β, TNPO1, and importin 7 are equally abundant (1–2 μM), whereas TNPO3 has lower levels (∼0.3 μM) (46). Expression levels of NTRs and usage of specific importins might, however, be cell type dependent. Interestingly, FUS inclusions in ALS/FTD are only found in neurons and sometimes glia cells in certain brain regions (3). Based on our data, it seems possible that limiting importin levels might contribute to the RBP pathology in these cell types, possibly in an age-dependent manner. Whether indeed TNPO3, importin β, or importin 7 have higher levels in other cell types and therefore are able to prevent cytosolic aggregation and mediate nuclear import of FUS is currently unclear.
Our data clearly provide further support for a general role of importins as molecular chaperones for aggregation-prone RBPs, as suggested previously (15, 26, 28, 30, 39). Which domains of importins are involved in mediating molecular chaperoning is currently unclear and requires further investigation. Similarly, it is currently unknown whether this mechanism is limited to histones or RBPs rich in basic residues/RGG motifs, such as FUS or ribosomal proteins, or also applies to other disease-linked RBPs that are not characterized by a large number of basic residues, such as TDP-43.
Experimental procedures
Plasmids
Bacterial expression constructs coding for MPB–FUS KG/KGG were generated by inserting a codon-optimized gBlock double-stranded gene fragment (IDT) replacing all codons for RG/RGG with codons for KG/KGG in the original pMal-Tev-Flag-FUS-Tev-His using Bsu36I and HindIII sites. pMal-Tev-Flag FUS (Δaa1-165)-Tev-His (ΔN) was a gift from Marc-David Ruepp. pMal-c2-MBP-EGFP was generated by PCR amplification of the coding sequence of EGFP and cloning into pMal-c2 (New England Biolabs) via EcoRI and HindIII.
Antibodies
Antibodies against importin β (65), CRM1 (66), and importin 7 (67) were described previously. The following antibodies are commercially available: anti-GAPDH (#10494-1-AP, Proteintech); mouse anti-MBP (#66003-1-Ig, Proteintech); rabbit anti-MBP (NEB); rabbit anti-G3BP1 (13057-2-AP, Proteintech); mouse anti-TNPO1 (# T0825, Sigma clone D45); mouse anti-TNPO3 (# ab54353, Abcam). For Western blotting, IRDye 800CW, IRDye 680CW, or IRDye 680LT (LI-COR) was used as the secondary antibody.
In import and SG association assays, recombinant MBP–FUS was detected using mouse anti-MBP and SGs were stained by using rabbit anti-G3BP1. As fluorescent secondary antibodies for microscopy, donkey anti-mouse Alexa 488 (Thermo Fisher) and donkey anti-rabbit Alexa 555 (Thermo Fisher) antibodies were used.
Protein expression and purification
MBP–FUS–EGFP–His6 wt (15), GST–M9 (33), RanWT and RanQ69L (68), RanQ69L(aa1–180) (69), His–importin α (70), His–importin β (71), and His–CRM1 (72, 73) were expressed and purified as described previously. RanQ69L was loaded with GTP as described (74).
His–TNPO3 (29), His–PRMT1 (75), His–Tev (plasmid gift from A. Geerlof), MBP–FUS wt, and FUS ΔRGG3-PY (15) were purified as previously described. MBP–FUS KG/KGG and Δaa1-165 (ΔN) were purified as described for MBP–FUS wt. pMal-c2-MBP-EGFP was transformed into BL21 (DE3) codon+, and bacteria were grown in an LB medium to an absorbance at 600 nm of 0.7. Expression was induced with 0.5 mM IPTG for 4 h at 16 °C. MBP–GFP was purified in MBP buffer (50 mM Hepes, pH 8.0, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and 1 μg/ml each of leupeptin, pepstatin, and aprotinin) using amylose resin (New England Biolabs), followed by SEC (HiLoad 16/60 Superdex 200 prep grade; GE Healthcare) in transport buffer (TB; 20 mM Hepes, 110 mM KOAc, 2 mM Mg(OAc)2, 1 mM EGTA, pH 7.3, 2 mM DTT).
pQE32-His-TNPO1 (34), pQE30-His-importin 5 (34), pQE80-His-importin 7 (Xenopus laevis, (45)), pQE80-His-importin 7-aa1-1001 (X. laevis, (45)), pQE80-His-importin 13 (76), and pQE30-His-Xpo 4 (Mus musculus, gift from Dirk Görlich) were transformed in Escherichia coli JM109 and grown in 2× YT medium supplemented with 2% glycerol and 30 mM K2HPO4 to an absorbance at 600 nm of ∼0.6. The cultures were cooled down for 1 to 2 h at 4 °C, and expression of His–importin 5, His–importin 7, His–importin 13, and His–Xpo 4 was induced with 0.5 mM IPTG overnight at 16 °C. Expression of His–TNPO1 was induced with 0.5 mM IPTG for 3 to 4 h at 25 °C. The proteins were purified in buffer A (50 mM Tris, pH 7.5, 500 mM NaCl, 10 mM Mg(OAc)2, 5% glycerol, 10 mM β-mercaptoethanol, 0.1 mM PMSF, and 1 μg/ml each of leupeptin, pepstatin, and aprotinin) over Ni-NTA Sepharose (# 30230, Qiagen) and separated over a HiLoad 26/60 Superdex 200 prep grade column connected to a ÄKTA purifier system (GE Healthcare) using buffer B (50 mM Tris, pH 7.4, 200 mM NaCl, 5% glycerol, 2 mM DTT).
For LLPS assays and studies using FUS mutants, NTRs were expressed and purified as follows: His–TNPO1 was basically purified as described before (15). Expression of His–importin β and His–importin 7 in Rosetta 2 was induced in the presence of 30 mM K2HPO4 and accompanied by ethanol shock and cold shock. The proteins immobilized on beads were washed with lysis buffer containing 1 M NaCl. After elution, proteins were subjected to SEC and stored in 20 mM Na2HPO4/NaH2PO4, pH 7.5, 75 mM NaCl, 5% (v/v) glycerol, 2 mM DTT (His-TNPO1 and His-importin β), or 50 mM Tris, pH 7.5, 250 mM NaCl, 2 mM MgCl2, 10% (v/v) glycerol, and 2 mM DTT (His–importin 7). Importin 5 was a gift from Dirk Görlich.
In vitro methylation of MBP–FUS
In vitro arginine methylation of MBP–FUS by 2-fold molar excess of PRMT1 was performed using 1 mM SAM overnight at room temperature (RT) and confirmed using methylation-specific antibodies, as previously described (15).
Sedimentation assay
For sedimentation analysis of FUS, the MBP tag of 1 μM purified MBP-Flag-FUS protein in the absence or presence of equimolar amounts of His–TNPO1, His–TNPO3, His–S–importin β, His–importin 7, or His–S–importin β/His–importin 7 was cleaved using 1.6 μM His–Tev in a total volume of 50 μl TP for 30 min at RT, followed by centrifugation at 21,000g and 4 °C for 15 min. Equal volumes of the S and pellet fraction were analyzed by SDS-PAGE and SYPRO Ruby stain (Sigma). Densitometry measurements of band intensities of S and P fractions was performed using implemented plugins of the Image Lab software (Bio-Rad Laboratories), and statistical analysis was performed in GraphPad Prism 8.
SG association assay
HeLa cells were grown on high-precision (No. 1.5), poly-L-lysine–coated 12-mm coverslips, permeabilized with 0.005% digitonin in KPB (20 mM potassium phosphate, pH 7.4, 5 mM Mg(OAc)2, 200 mM KOAc, 1 mM EGTA, 2 mM DTT, and 1 mg/ml each aprotinin, pepstatin and leupeptin). Soluble proteins were removed by several washes (4× 4 min in KPB on ice), and nuclear pores were blocked by 15-min incubation with 0.2 mg/ml wheat germ agglutinin on ice. Cells were then incubated for 30 min at RT with 100 nM MBP–FUS in the absence or presence of 500 nM His–TNPO1, His–TNPO3, His–S–importin β, His–importin 7, or His–S-importin β/His–importin 7 in KPB. Note that a 5-fold excess of importins was required for efficient shielding of FUS, possibly because of other RBPs present in SGs that sequester importins. Subsequently, cells were washed (3× 5 min in KPB on ice) to remove unbound MBP–FUS and processed by immunostaining as described (15, 51) to visualize SGs (by G3BP1 immunostaining) and FUS (by MBP antibody). Cells were analyzed by using confocal microscopy with an inverted Leica SP8 microscope and the LAS X imaging program (Leica), using lasers for 405-nm, 488-nm, and 552-nm excitation. Images were acquired using two-fold frame averaging with a 63× 1.4 oil objective and an image pixel size of 59 nm. The following fluorescence settings were used for detection: 4′,6-diamidino-2-phenylindole (DAPI): 419 to 442 nm, GFP: 498 to 533 nm, Alexa 555: 562 to 598 nm. Recording was performed sequentially to avoid bleed-through using a conventional photomultiplier tube. Images were processed and analyzed using ImageJ/Fiji software (77), applying linear enhancement for brightness and contrast and implemented plugins for measurement of pixel intensities in SGs. Statistical analyses were performed in GraphPad Prism 8.
Cell culture and cell lysate
HeLa cells were grown in Dulbecco's modified Eagle's medium high-glucose GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum and 10 μg/ml gentamicin at 37 °C, 5% CO2 in a humidified incubator.
HAP1 cells and TNPO1 KO cells were obtained from PerkinElmer (Horizon; HZGHC003548c007) and grown in Iscove's Modified Dulbecco's Medium (GIBCO) supplemented with 10% fetal calf serum, 6 mM L-glutamine and 100 U/ml penicillin, 100 μg/ml streptomycin at 37 °C, and 5% CO2.
Cell lysates were obtained from digitonin-permeabilized cells as described previously (51, 78).
Binding assays
For binding assays with HeLa cell lysates, MBP or MBP–FUS (6.25 μg each) was immobilized on amylose resin (NEB) in the TB containing 1 μg/ml each of leupeptin, pepstatin, and aprotinin. The beads were incubated with 300 μl of the cell lysate (2 mg/ml) overnight at 4 °C. After three washing steps with the TB, proteins were eluted with Laemmli buffer and analyzed by SDS-PAGE (10% gel).
For binding assays with HAP1-cell lysates, MBP–GFP or MBP–FUS–EGFP (1 nmol each) were immobilized on 62.5 μl MBP-selector beads (Nanotag Biotechnologies) in the TB containing 1 μg/ml each of leupeptin, pepstatin, and aprotinin and 2 mg/ml hemoglobin (Sigma) as a blocking reagent. The beads were incubated with 300 μl of cell lysate (∼4 mg/ml) and 0.2 μg/ml RNAse A (Qiagen) in a total volume of 0.5 ml overnight at 4 °C. After three washings steps with the TB, proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE (4–12% NuPAGE gels, Invitrogen), followed by Western blotting using the Odyssey system (LI-COR).
For binding assays with purified NTRs, MBP–FUS–EGFP–His (100 pmol) was immobilized on 10 μl GFP–Trap Magnetic Agarose (#gtma-100, Chromotek, Planegg, Germany), equilibrated in the TB supplemented with 20 mg/ml bovine serum albumin (BSA) for 1 h. The immobilized MBP–FUS–EGFP–His was incubated with 100 pmol of His–TNPO1, His–TNPO3, His–importin β/7, His–importin β, His–importin 7, His–importin 13, or His–Xpo 4 in the presence or absence of 300-pmol RanQ69L(aa1-180)–GTP in a total volume of 500 μl for 2 h at 4 °C. After washing 3× with 500 μl TB lacking BSA, bound proteins were eluted in 4× SDS sample buffer, analyzed by SDS-PAGE and Coomassie-stained. Competition experiments were performed as described above, but instead of RanGTP, increasing amounts (100 pmol, 300 pmol, 500 pmol) of two different His-tagged NTRs or 300 pmol of histone H1 (Roche) was added. For sequential competition experiments, 100 pmol MBP–FUS–EGFP was immobilized on amylose HF resin for 1.5 h at 4 °C before allowing 100 pmol of TNPO3, importin β, importin 7, or importin β/7 to bind overnight. Subsequently, increasing amounts of TNPO1 (50–200 pmol) were added for 3 h before samples were processed as described above. Binding of NTRs was visualized by SYPRO Ruby staining.
For binding assays using unmethylated or methylated MBP–FUS WT or MBP–FUS mutants (KG/KKGG, ΔN or ΔC), 5-μg recombinant MBP–FUS protein per reaction was immobilized on preblocked (20 mg/ml BSA) and preequilibrated MBP–Trap agarose (# mbta-20, Chromotek) or amylose resin (# E8021, New England Biolabs), respectively, in the TB supplemented with 100 mM NaCl and 2 mg/ml BSA. Recombinant import receptors (5 μg per reaction) were added for 3 to 4 h at 4 °C. Beads were subsequently washed 3× and then bound proteins were eluted using 2x SDS sample buffer and analyzed by Western blotting.
SEC
To assess complex formation, 1 nmol MBP–FUS–EGFP–His was incubated in the presence or absence of 1 nmol His–TNPO1, His–TNPO3, His–importin β/7, His–importin β/7-aa1-1001, His–importin β, His–importin 7, His–importin 7-aa1-1001, His–importin 13, or His–Xpo 4 in TB in a total volume of 500 μl for 1 h at 4 °C. Some reactions also contained a 3-fold molar excess of RanQ69L(aa1-180)–GTP. After centrifugation at 100,000g for 15 min at 4 °C, 450 μl of protein samples was loaded onto a Superdex 200 Increase 10/300GL column (#28-9909-44, GE Healthcare), equilibrated in TB. Eluted fractions were analyzed by SDS-PAGE, followed by Coomassie staining.
Import assay
For nuclear import assays, HeLa cells were grown on poly-L-lysine–coated 12-mm coverslips, permeabilized with 0.005% digitonin (# CAS11024-24-1, Calbiochem) in TB and washed 4× for 4 min with the TB. Cells were incubated in a total volume of 40 μl with 2 mg/ml BSA, 100 nM MBP–FUS, and 500 nM His–TNPO1, His–TNPO3, His–importin β/7, His–importin β, or His–importin 7 in the absence or presence of 4 μM RanWT, 4 μM RanQ69L–GTP, and an ATP-regenerating system (1 mM ATP, 5 mM creatine phosphate, 20 U/ml creatine phosphokinase) in TB in a humidity chamber for 30 min at 30 °C or 4 °C. The coverslips were washed in cold TB, fixed with 3.7% formaldehyde, and processed by immunostaining for MBP. Nuclei were stained using DAPI, and cells were mounted in ProLong Diamond (Thermo Fisher). Cells were analyzed by confocal microscopy with an inverted Leica SP8 microscope and the LAS X imaging program (Leica), using lasers for 405 nm and 488 nm excitation. Images were acquired using 2-fold frame averaging with a 63× 1.4 oil objective and an image pixel size of 90 nm. The following fluorescence settings were used for detection: DAPI: 419 to 442 nm and GFP: 498 to 533 nm. Images were processed using Fiji, applying linear enhancement for brightness and contrast (77). The area of the nucleus was defined by the DAPI staining using the magic wand tool in Fiji.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Tobias Madl for critically reading the manuscript and Christiane Spillner for technical assistance. We are grateful to Drs. Marina Blenski, Mohamed Hamed, Dirk Görlich, Detlef Doenecke, Achim Dickmanns, Marc-David Ruepp, Elmar Wahle, and Ari Geerlof for the generous gift of constructs or recombinant proteins. We thank Drs Peter Becker and Michael Kiebler for infrastructure and acknowledge the Biomedical Center Core Facility Bioimaging for support.
Author contributions
I. B., S. H., E. S., M. P., M. H. designed and performed experiments; I. B., S. H., D. D., and R. H. K. wrote original draft; I. B., S. H., E. S., D. D., and R. H. K. revision and editing; D. D. and R. H. K. supervision, conceptualization, and funding acquisition.
Funding and additional information
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) within Emmy Noether Grants DO 1804/1-1 and DO 1804/1-2 as well as research grant DO1804/3-1 (to D. D.). D. D. is additionally supported by the Munich Cluster for Systems Neurology (EXC2145 SyNergy—ID 390857198). R. H. K. was supported by the DFG (KE 660/14-1 and SFB860).
Edited by Phyllis Hanson
Footnotes
Present address for Saskia Hutten, Erin Sternburg, and Dorothee Dormann: Johannes Gutenberg-Universität (JGU), Biocenter, Institute of Molecular Physiology & Institute for Molecular Biology (IMB) Mainz, Germany.
Contributor Information
Dorothee Dormann, Email: ddormann@uni-mainz.de.
Ralph H. Kehlenbach, Email: rkehlen@gwdg.de.
Supporting information
References
- 1.Dormann D., Haass C. TDP-43 and FUS: A nuclear affair. Trends Neurosci. 2011;34:339–348. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Ito D., Hatano M., Suzuki N. RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci. Transl. Med. 2017;9:eaah5436. doi: 10.1126/scitranslmed.aah5436. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie I.R.A., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 4.Ling S.-C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor J.P., Brown R.H., Jr., Cleveland D.W. Decoding ALS: From genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried H., Kutay U. Nucleocytoplasmic transport: Taking an inventory. Cell Mol. Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallini C., Khalil B., Smith C.L., Rossoll W. Traffic jam at the nuclear pore: All roads lead to nucleocytoplasmic transport defects in ALS/FTD. Neurobiol. Dis. 2020;140 doi: 10.1016/j.nbd.2020.104835. 104835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutten S., Dormann D. Nucleocytoplasmic transport defects in neurodegeneration — cause or consequence? Semin. Cell Dev. Biol. 2020;99:151–162. doi: 10.1016/j.semcdb.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Kim H.J., Taylor J.P. Lost in transportation: Nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron. 2017;96:285–297. doi: 10.1016/j.neuron.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devoy A., Kalmar B., Stewart M., Park H., Burke B., Noy S.J., Redhead Y., Humphrey J., Lo K., Jaeger J., Mejia Maza A., Sivakumar P., Bertolin C., Soraru G., Plagnol V. Humanized mutant FUS drives progressive motor neuron degeneration without aggregation in 'FUSDelta14' knockin mice. Brain. 2017;140:2797–2805. doi: 10.1093/brain/awx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scekic-Zahirovic J., Oussini H.E., Mersmann S., Drenner K., Wagner M., Sun Y., Allmeroth K., Dieterlé S., Sinniger J., Dirrig-Grosch S., René F., Dormann D., Haass C., Ludolph A.C., Lagier-Tourenne C. Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol. 2017;133:887–906. doi: 10.1007/s00401-017-1687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., Lyashchenko A.K., Lu L., Nasrabady S.E., Elmaleh M., Mendelsohn M., Nemes A., Tapia J.C., Mentis G.Z., Shneider N.A. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 2016;7 doi: 10.1038/ncomms10465. 10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R.A., Capell A., Schmid B., Neumann M., Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormann D., Madl T., Valori C.F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I.R.A., Neumann M., Haass C. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–4275. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.-D., Simons M., Niessing D., Madl T., Dormann D. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719.e713. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Lee B.J., Cansizoglu A.E., Suel K.E., Louis T.H., Zhang Z., Chook Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brelstaff J., Lashley T., Holton J.L., Lees A.J., Rossor M.N., Bandopadhyay R., Revesz T. Transportin1: A marker of FTLD-FUS. Acta Neuropathol. 2011;122:591–600. doi: 10.1007/s00401-011-0863-6. [DOI] [PubMed] [Google Scholar]
- 18.Davidson Y.S., Robinson A.C., Hu Q., Mishra M., Baborie A., Jaros E., Perry R.H., Cairns N.J., Richardson A., Gerhard A., Neary D., Snowden J.S., Bigio E.H., Mann D.M.A. Nuclear carrier and RNA-binding proteins in frontotemporal lobar degeneration associated with fused in sarcoma (FUS) pathological changes. Neuropathol. Appl. Neurobiol. 2013;39:157–165. doi: 10.1111/j.1365-2990.2012.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann M., Valori C.F., Ansorge O., Kretzschmar H.A., Munoz D.G., Kusaka H., Yokota O., Ishihara K., Ang L.-C., Bilbao J.M., Mackenzie I.R.A. Transportin 1 accumulates specifically with FET proteins but no other transportin cargos in FTLD-FUS and is absent in FUS inclusions in ALS with FUS mutations. Acta Neuropathol. 2012;124:705–716. doi: 10.1007/s00401-012-1020-6. [DOI] [PubMed] [Google Scholar]
- 20.Suárez-Calvet M., Neumann M., Arzberger T., Abou-Ajram C., Funk E., Hartmann H., Edbauer D., Kremmer E., Göbl C., Resch M., Bourgeois B., Madl T., Reber S., Jutzi D., Ruepp M.-D. Monomethylated and unmethylated FUS exhibit increased binding to Transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol. 2016;131:587–604. doi: 10.1007/s00401-016-1544-2. [DOI] [PubMed] [Google Scholar]
- 21.Alberti S., Hyman A.A. Are aberrant phase transitions a driver of cellular aging? Bioessays. 2016;38:959–968. doi: 10.1002/bies.201600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami T., Qamar S., Lin J.Q., Schierle G.S.K., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T.S., Michel C.H., Kronenberg-Versteeg D., Li Y., Yang S.-P., Wakutani Y., Meadows W. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., Pozniakovski A., Poser I., Maghelli N., Royer L.A., Weigert M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 24.Bogaert E., Boeynaems S., Kato M., Guo L., Caulfield T.R., Steyaert J., Scheveneels W., Wilmans N., Haeck W., Hersmus N., Schymkowitz J., Rousseau F., Shorter J., Callaerts P., Robberecht W. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 2018;24:529–537.e524. doi: 10.1016/j.celrep.2018.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Choi J.-M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R.V., Alberti S., Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L., Kim H.J., Wang H., Monaghan J., Freyermuth F., Sung J.C., O'Donovan K., Fare C.M., Diaz Z., Singh N., Zhang Z.C., Coughlin M., Sweeny E.A., DeSantis M.E., Jackrel M.E. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173:677–692.e620. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Ströhl F., Meadows W., Ferry R., Dardov V.J., Tartaglia G.G., Farrer L.A. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e715. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshizawa T., Ali R., Jiou J., Fung H.Y.J., Burke K.A., Kim S.J., Lin Y., Peeples W.B., Saltzberg D., Soniat M., Baumhardt J.M., Oldenbourg R., Sali A., Fawzi N.L., Rosen M.K. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018;173:693–705.e622. doi: 10.1016/j.cell.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourgeois B., Hutten S., Gottschalk B., Hofweber M., Richter G., Sternat J., Abou-Ajram C., Göbl C., Leitinger G., Graier W.F., Dormann D., Madl T. Nonclassical nuclear localization signals mediate nuclear import of CIRBP. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8503–8514. doi: 10.1073/pnas.1918944117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Springhower C.E., Rosen M.K., Chook Y.M. Karyopherins and condensates. Curr. Opin. Cell Biol. 2020;64:112–123. doi: 10.1016/j.ceb.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L., Fare C.M., Shorter J. Therapeutic dissolution of aberrant phases by nuclear-import receptors. Trends Cell Biol. 2019;29:308–322. doi: 10.1016/j.tcb.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold M., Nath A., Hauber J., Kehlenbach R.H. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006;281:20883–20890. doi: 10.1074/jbc.M602189200. [DOI] [PubMed] [Google Scholar]
- 33.Arnold M., Nath A., Wohlwend D., Kehlenbach R.H. Transportin is a major nuclear import receptor for c-fos: A novel mode of cargo interaction. J. Biol. Chem. 2006;281:5492–5499. doi: 10.1074/jbc.M513281200. [DOI] [PubMed] [Google Scholar]
- 34.Baake M., Bauerle M., Doenecke D., Albig W. Core histones and linker histones are imported into the nucleus by different pathways. Eur. J. Cell Biol. 2001;80:669–677. doi: 10.1078/0171-9335-00208. [DOI] [PubMed] [Google Scholar]
- 35.Gu L., Tsuji T., Jarboui M.A., Yeo G.P., Sheehy N., Hall W.W., Gautier V.W. Intermolecular masking of the HIV-1 Rev NLS by the cellular protein HIC: Novel insights into the regulation of Rev nuclear import. Retrovirology. 2011;8:17. doi: 10.1186/1742-4690-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jäkel S., Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muhlhausser P., Muller E.C., Otto A., Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldmann I., Wälde S., Kehlenbach R.H. Nuclear import of c-Jun is mediated by multiple transport receptors. J. Biol. Chem. 2007;282:27685–27692. doi: 10.1074/jbc.M703301200. [DOI] [PubMed] [Google Scholar]
- 39.Jakel S., Mingot J.M., Schwarzmaier P., Hartmann E., Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu C., Zhang J., Gao F., Yang L., Jia M., Zhu H., Gong W. FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z.C., Chook Y.M. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proc. Natl. Acad. Sci. U. S. A. 2012;109:12017–12021. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ederle H., Funk C., Abou-Ajram C., Hutten S., Funk E.B.E., Kehlenbach R.H., Bailer S.M., Dormann D. Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci. Rep. 2018;8:7084. doi: 10.1038/s41598-018-25007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klebe C., Bischoff F.R., Ponstingl H., Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- 44.Jäkel S., Albig W., Kutay U., Bischoff F.R., Schwamborn K., Doenecke D., Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlwend D., Strasser A., Dickmanns A., Doenecke D., Ficner R. Thermodynamic analysis of H1 nuclear import: Receptor tuning of Importinbeta/Importin7. J. Biol. Chem. 2007;282:10707–10719. doi: 10.1074/jbc.M610409200. [DOI] [PubMed] [Google Scholar]
- 46.Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., Hyman A.A., Mann M. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 47.Burke K.A., Janke A.M., Rhine C.L., Fawzi N.L. Residue-by-Residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., Grishin N.V., Frantz D.E., Schneider J.W., Chen S., Li L. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z., Diaz Z., Fang X., Hart M.P., Chesi A., Shorter J., Gitler A.D. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutten S., Dormann D. A quantitative assay to measure stress granule association of proteins and peptides in semi-permeabilized human cells. Bio-protocol. 2020;10 doi: 10.21769/BioProtoc.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzalez A., Mannen T., Cagatay T., Fujiwara A., Matsumura H., Niesman A.B., Brautigam C.A., Chook Y.M., Yoshizawa T. Mechanism of karyopherin-beta2 binding and nuclear import of ALS variants FUS(P525L) and FUS(R495X) Sci. Rep. 2021;11:3754. doi: 10.1038/s41598-021-83196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kataoka N., Bachorik J.L., Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai M.C., Lin R.I., Huang S.Y., Tsai C.W., Tarn W.Y. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 55.Lai M.C., Lin R.I., Tarn W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maertens G.N., Cook N.J., Wang W., Hare S., Gupta S.S., Oztop I., Lee K., Pye V.E., Cosnefroy O., Snijders A.P., KewalRamani V.N., Fassati A., Engelman A., Cherepanov P. Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2728–2733. doi: 10.1073/pnas.1320755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun C.Y., Velazquez-Dones A.L., Lyman S.K., Fu X.D. Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J. Biol. Chem. 2003;278:18050–18055. doi: 10.1074/jbc.M211714200. [DOI] [PubMed] [Google Scholar]
- 58.Chook Y.M., Suel K.E. Nuclear import by karyopherin-betas: Recognition and inhibition. Biochim. Biophys. Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M., Morinaka Y., Imai K., Kose S., Horton P., Imamoto N. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. Elife. 2017;6 doi: 10.7554/eLife.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warda A.S., Freytag B., Haag S., Sloan K.E., Görlich D., Bohnsack M.T. Effects of the Bowen-Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum. Mol. Genet. 2016;25:5353–5364. doi: 10.1093/hmg/ddw351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauerle M., Doenecke D., Albig W. The requirement of H1 histones for a heterodimeric nuclear import receptor. J. Biol. Chem. 2002;277:32480–32489. doi: 10.1074/jbc.M202765200. [DOI] [PubMed] [Google Scholar]
- 62.Gami-Patel P., Bandopadhyay R., Brelstaff J., Revesz T., Lashley T. The presence of heterogeneous nuclear ribonucleoproteins in frontotemporal lobar degeneration with FUS-positive inclusions. Neurobiol. Aging. 2016;46:192–203. doi: 10.1016/j.neurobiolaging.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Gittings L.M., Foti S.C., Benson B.C., Gami-Patel P., Isaacs A.M., Lashley T. Heterogeneous nuclear ribonucleoproteins R and Q accumulate in pathological inclusions in FTLD-FUS. Acta Neuropathol. Commun. 2019;7:18. doi: 10.1186/s40478-019-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neumann M., Bentmann E., Dormann D., Jawaid A., DeJesus-Hernandez M., Ansorge O., Roeber S., Kretzschmar H.A., Munoz D.G., Kusaka H., Yokota O., Ang L.C., Bilbao J., Rademakers R., Haass C. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wlde S., Thakar K., Hutten S., Spillner C., Nath A., Rothbauer U., Wiemann S., Kehlenbach R.H. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic. 2012;13:218–233. doi: 10.1111/j.1600-0854.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 66.Roloff S., Spillner C., Kehlenbach R.H. Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1. J. Biol. Chem. 2013;288:3952–3963. doi: 10.1074/jbc.M112.433243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frohnert C., Hutten S., Wälde S., Nath A., Kehlenbach R.H. Importin 7 and Nup358 promote nuclear import of the protein component of human telomerase. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melchior F., Sweet D.J., Gerace L. Analysis of Ran/TC4 function in nuclear protein import. Methods Enzymol. 1995;257:279–291. doi: 10.1016/s0076-6879(95)57032-2. [DOI] [PubMed] [Google Scholar]
- 69.Port S.A., Monecke T., Dickmanns A., Spillner C., Hofele R., Urlaub H., Ficner R., Kehlenbach R.H. Structural and functional characterization of CRM1-Nup214 interactions reveals multiple FG-binding sites involved in nuclear export. Cell Rep. 2015;13:690–702. doi: 10.1016/j.celrep.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 70.Weis K., Mattaj I.W., Lamond A.I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 71.Hu T., Guan T., Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dölker N., Blanchet C.E., Voss B., Haselbach D., Kappel C., Monecke T., Svergun D.I., Stark H., Ficner R., Zachariae U., Grubmüller H., Dickmanns A. Structural determinants and mechanism of mammalian CRM1 Allostery. Structure. 2013;21:1350–1360. doi: 10.1016/j.str.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 73.Guan T., Kehlenbach R.H., Schirmer E.C., Kehlenbach A., Fan F., Clurman B.E., Arnheim N., Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kehlenbach R.H., Dickmanns A., Kehlenbach A., Guan T., Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X., Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mingot J.M., Kostka S., Kraft R., Hartmann E., Görlich D. Importin 13: A novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baade I., Spillner C., Schmitt K., Valerius O., Kehlenbach R.H. Extensive identification and in-depth validation of importin 13 cargoes. Mol. Cell Proteomics. 2018;17:1337–1353. doi: 10.1074/mcp.RA118.000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.