PURPOSE AND APPROPRIATE SAMPLE TYPES
This 40‐color flow cytometry‐based panel was developed for in‐depth immunophenotyping of the major cell subsets present in human peripheral blood. Sample availability can often be limited, especially in cases of clinical trial material, when multiple types of testing are required from a single sample or timepoint. Maximizing the amount of information that can be obtained from a single sample not only provides more in‐depth characterization of the immune system but also serves to address the issue of limited sample availability. The panel presented here identifies CD4 T cells, CD8 T cells, regulatory T cells, γδ T cells, NKT‐like cells, B cells, NK cells, monocytes and dendritic cells. For each specific cell type, the panel includes markers for further characterization by including a selection of activation and differentiation markers, as well as chemokine receptors. Moreover, the combination of multiple markers in one tube might lead to the discovery of new immune phenotypes and their relevance in certain diseases. Of note, this panel was designed to include only surface markers to avoid the need for fixation and permeabilization steps. The panel can be used for studies aimed at characterizing the immune response in the context of infectious or autoimmune diseases, monitoring cancer patients on immuno‐ or chemotherapy, and discovery of unique and targetable biomarkers. Different from all previously published OMIPs, this panel was developed using a full spectrum flow cytometer, a technology that has allowed the effective use of 40 fluorescent markers in a single panel. The panel was developed using cryopreserved human peripheral blood mononuclear cells (PBMC) from healthy adults (Table 1). Although we have not tested the panel on fresh PBMCs or whole blood, it is anticipated that the panel could be used in those sample preparations without further optimization. @ 2020 Cytek Biosciences, Inc. Cytometry Part A published by Wiley Periodicals LLC on behalf of International Society for Advancement of Cytometry.
Keywords: Aurora, broad immunophenotyping, full spectrum, high‐dimensional flow cytometry, OMIP, spectral, PBMCs
This OMIP describes the first 40‐color fluorescent panel using full spectrum flow cytometry to broadly phenotype much of the cellular composition of the human peripheral immune system. The panel has been thoroughly optimized to ensure high‐quality data and well‐resolved populations, enabling the description of most canonical subsets of T cells, B cells, NK cells, monocytes, and dendritic cells. This panel will be particularly useful for studies in which sample availability is limited or unique biomarker signatures are sought.
Background
The need to understand the mechanisms and pathways of immune evasion seen either post immunotherapy or during natural immune responses to cancer, autoimmunity, and infectious diseases, requires methods and protocols which will enable a deeper profiling of the immune system. Greater characterization of immune subpopulations allows for more informed decisions regarding the identification of targetable biomarkers and the development of new therapeutic approaches. (1, 2, 3, 4)
Unraveling the complexity of the human immune response requires the ability to perform high‐throughput, in‐depth analysis, at the single cell and population levels. Flow cytometry has sought to address this need by allowing the characterization of single‐cell protein expression, through the binding of fluorochrome‐labeled antibodies to specific markers of interest. Over the years, manufacturers have increased the capabilities of flow cytometers through the incorporation of additional lasers and detectors, allowing detection of greater numbers of markers per cell. Concurrently, reagent manufacturers have worked to provide additional fluorophores to meet the demands of this rapidly expanding field. This has led to panel expansion over the last two decades, with a 17‐color assay reported in 2004 (5) and up to 28 colors in more recent years (6, 7, 8, 9, 10, 11). With the arrival of mass cytometry in 2009 (12), the number of markers assessed was expanded to 32, using metal‐conjugated antibodies (13), and most recently a panel using 43 markers has been published (14).
In contrast to conventional flow cytometry, which primarily measures the peak emission of each fluorochrome, full spectrum flow cytometry measures the entire emission spectra for every fluorochrome, across all laser lines. As a result of collecting substantially more information about each cell, full spectrum flow cytometry is well suited to the development of highly multiparametric panels. Reports of applying the concepts of measuring fluorescence spectra by flow cytometry can be found as early as the 1970s (15), which was followed by a number of subsequent publications in later years (16, 17, 18, 19, 20). In order to expand the number of fluorochromes beyond the 28‐color mark, a very high level of detail is needed to distinguish fluorochromes whose spectral signatures, particularly their peak emissions, are similar. This level of detail requires high‐quality signals, low noise, and excitation specific full‐emission profiles. It also requires extremely careful panel design and optimization. Here, we define full spectrum flow cytometry as measuring the entire fluorochrome emission, from ultraviolet to near‐infrared, across multiple lasers using many more detectors, when compared to a conventional flow cytometer. This produces very specific spectral fingerprints that are used to mathematically distinguish one fluorophore from another, even when their maximum emissions are very similar. Leveraging this full spectrum technology in a five‐laser system, the ability to combine 30–40 fluorescently labeled antibodies becomes possible using a fluorescence‐based flow cytometer.
As mentioned previously, mass cytometry is also capable of assessing similarly high numbers of parameters. Currently, this technology has the advantage of additional detection channels to accommodate bar coding schemes for sample pooling, and as a more mature technology, high complexity panels using mass cytometry have been previously published and are widely available, including the publication of multiple OMIPs (21, 22, 23, 24). However, limitations such as sample throughput, cell transmission efficiency, and overall cost of ownership have impacted the practicality, and broader adoption, of this technology in some laboratory environments (25, 26, 27). Spectral flow cytometers share a very common workflow with conventional flow cytometers and are therefore not hindered by these limitations. However, there are no previously published reports of panels beyond 28 fluorescent parameters, a fact which further supports the need for a fluorescent OMIP panel of this complexity.
The panel presented in this OMIP examines the frequency of CD4 and CD8 T cells, regulatory T cells (Tregs), γδ T cells, NKT‐like cells, B cells, NK cells, monocytes, basophils, innate lymphoid cells (ILCs), and dendritic cells. Additional markers allow for the characterization of the main B and T cell subsets—naïve, memory, and effector—as well as putative T helper subsets. Dead cells were excluded using a viability dye (LIVE/Dead Fixable Blue). The following markers were used to characterize the indicated cell types: CD45 for all leukocytes; pan‐γδ TCR for γδ T cells; CD3, CD4, and CD8 for the main T cell populations; CD19 and CD20 for B cells; CD16 and CD56 for NK cells; CD123 and HLA‐DR for basophils; lineage markers and CD127 for total ILCs; and CD14 and CD16 for monocytes.
NK cells were identified by expression of CD56 (neural cell adhesion molecule [NCAM]) and CD16 (FcγRIII) (28), while CD2 was used to identify NK cells most likely to be involved in cytotoxicity (29). We included two natural cytotoxicity receptors (NCRs), NKG2D (natural killer group 2, member D [CD314]) and NKp30 (CD337); as well as the inhibitory receptor NKG2A (CD159a), identified as a possible target for immunotherapy (30), and the activation receptor NKG2C (CD159c) known to be relevant for NK cells in infectious diseases and vaccine effector response (31). CD57 was included as a maturation marker for NK cells, identifying cells with potent cytotoxic and reduced replicative potential (32). CD56 was further used for the identification of NKT‐like cells, defined as CD56+CD3+, as cells with this phenotype might play a role in infectious diseases (33).
γδ T cells are regarded as an important bridge between the innate and adaptive immune systems because their response precedes adaptive immunity, making γδ T cells a unique component of the immune system (34, 35, 36). They are also associated with major autoimmune rheumatic diseases, such as rheumatoid arthritis, juvenile idiopathic arthritis, ankylosing spondylitis, systemic lupus erythematosus, and scleroderma (37). Although we included CD16 and CD56 to define NK cell subsets, CD16 and CD56 are also associated with an activated phenotype in γδ T cells (6, 38). γδ T cells have also been shown to have effector/memory subsets based on expression of CD45RA, CD27, and CCR7 (39).
To characterize T cells, the memory and differentiation markers CD45RA, CCR7, CD27, and CD28 were used. Using CD45RA and CCR7, T cells can be classified into naïve (CD45RA+CCR7+), central memory (CD45RA−CCR7+), effector memory (CD45RA−CCR7−), and terminal effector memory (CD45RA+CCR7−) subsets (40, 41). The addition of CD27 and CD28 allows further refinement of those subsets, identifying early effector (CD45RA−CCR7−CD28+CD27+), early‐like effector (CD45RA−CCR7−CD28+CD27−), intermediate effector (CD45RA−CCR7−CD28−CD27+), terminal effector (CD45RA−CCR7−CD28−CD27−), and RA‐terminal effector (CD45RA+CCR7−CD28−CD27−) (42, 43).
Other surface markers, including CD127, CD95, PD‐1, CD57, CD38, and HLA‐DR, were included to further characterize T cell subsets (1, 44, 45). The IL‐7 receptor (CD127) is involved in homeostatic proliferation and survival of memory T cell precursors (46), while CD57 is indicative of cell senescence, failure to proliferate, and susceptibility to activation‐induced cell death (47, 48). CD38 and HLA‐DR were included as T cell activation markers (49). Viral infections such as HIV‐1, dengue virus, or influenza lead to increased frequencies of CD38+HLA‐DR+ activated T cells (50, 51). PD‐1 and CD95 are both upregulated in activated T cells (52). The inhibitory receptor PD‐1 is crucial for the regulation of immune responses and to avoid excessive immune activation (53).
In order to identify Tregs, we used CD25 (IL‐2Rα) and CD127 (IL‐7Rα) markers without the inclusion of FoxP3, which requires intracellular staining. Previous studies have shown that CD25hiCD127lo/‐CD4+ T cells are a good correlate of Tregs (54, 55), although this strategy may over‐ or underestimate their frequency. It has been reported that Tregs can be further subsetted based on CD39 expression (7, 56, 57) and that CD39+ Tregs might play a role in certain autoimmune diseases like multiple sclerosis (58).
CD27 and IgD were chosen for identification of naïve (IgD+CD27−), marginal zone‐like (IgD+CD27+), and memory (IgD−CD27+) B cells as previously described (59). Marginal zone‐like B cells can be further divided into IgM+ marginal zone and IgD only memory B cells (8). When used in combination, CD24 and CD38 distinguish memory (CD24+CD38lo/neg), naïve (CD24intCD38int), and transitional (CD24hiCD38hi) B cells, and have been used for regulatory B cell identification (60, 61). Plasmablasts can be identified based on expression of CD19, CD20, CD27, and CD38 (62). Memory B cells express different B cell receptor isotypes as a result of class switch recombination. This panel includes IgG and IgM, which are the most prevalent subsets of memory B cells found in blood. IgA was not included as this subpopulation of memory B cells is predominantly expressed in mucosa‐associated lymphatic tissues, such as the intestine and mesenteric lymph nodes. IgE was excluded from this panel, since this subset of memory B cells is hardly detectable in human blood (63).
Chemokine receptors are important for the migration and positioning of immune cells (64). This panel includes CCR5, CCR6, CXCR3, and CXCR5. CCR5 is expressed by activated and memory T cells (65), γδ T cells (66), and Tregs (67). On human T cells, CCR6 is attributed to a Th17 (RORγt) phenotype (68). On B cells, CCR6 expression is restricted to functionally mature cells capable of responding to antigen challenge (69). CXCR3 has been reported to be necessary for T cell clustering around antigen presenting cells and T cell bystander activation (70) and also to be expressed on subsets of γδ T cells (66). CXCR5 interacts with CXCL13, which promotes T cell trafficking to B cell follicles and germinal centers. These are crucial sites for the generation of high‐affinity antibody responses (71). Moreover, it has been shown that chronic inflammation leads to modulation of chemokine receptor expression on peripheral blood B cells. In patients with rheumatoid arthritis, B cells show decreased expression of CXCR5 and CCR6 and increased levels of CXCR3 (72).
Monocyte subsets were identified using CD14 (lipopolysaccharide binding protein) and CD16 (FcγRIII). These two markers allow the identification of classical monocytes (CD14++CD16−), non‐classical monocytes (CD14+CD16++), and an intermediate monocyte population (CD14+CD16+) (73). Intermediate monocytes expand in the presence of cytokines and inflammation. Non‐classical monocytes have also been shown to expand in inflammatory diseases. It has been demonstrated that, over the course of infection, there is first an increase in intermediate monocytes followed by an increase in non‐classical monocytes (74).
With the markers present in this panel, basophils were identified as CD45dimCD123+HLA‐DR− (75). The phenotype of these cells can then be further characterized by evaluating expression of CD38, CD95, and CD25.
Total ILCs were identified using a similar strategy as presented in OMIP‐055 (76). This subset was identified as CD45+CD127+Lin‐. For lineage markers, we used CD14, CD19, CD20, and CD3. However, to fully identify these cells other markers should be excluded like CD1a, CD34, CD303, and FCεR1a. Despite lacking these markers, the cells identified as ILCs could be further classified based on expression of CD2, CD4, CCR6, CXCR3, CD27, CD28, CXCR5, and CCR7 as previously reported (77).
Finally, to identify the main dendritic cell subsets, CD11c, HLA‐DR, CD141 (BDCA‐3), CD1c (BDCA‐1), and CD123 were used as previously described (9). pDCs were identified as lineage negative (CD3−CD19−CD56−CD14−) HLA‐DR+CD123+ cells; conventional DCs were identified as CD123−CD11c+HLA‐DR+, and they were further subset based on CD141 (78) and CD1c expression (79).
The manual gating strategy used to identify the main cell subsets, based on the descriptions provided above, is shown in Figure 1A. As it is more likely that a complex data set such as this would be analyzed using a pipeline containing both dimensionality reduction and clustering algorithms, we present in Figure 1B the computationally derived analogs of manually gated canonical subsets using such an unsupervised approach. When preparing for any kind of automated analysis, it is imperative that the data be of the highest quality, as any undesirable events or processing artifacts will negatively affect the downstream results. In this case, the scaling of the data was first checked to ensure the arcsinh transformation was unimodal around 0 and then the data were cleaned by manual gating to remove doublets, debris, and dead cells. The data were then run through flowCut (80) to check for aberrant signal patterns or events, and with none found. UMAP (81) was run to group phenotypically similar events into “islands” to illustrate differences both between and inside each population. FlowSOM (82) was subsequently used to cluster the events based on UMAP parameters and selected surface markers in order to emphasize differences between hard to resolve populations and then the resulting clusters were overlaid on the initial UMAP parameters. A traditional clustered heatmap analysis then followed to aid in the identification and labeling of the FlowSOM clusters (See Supporting Information Fig. S11A).
Figure 1.
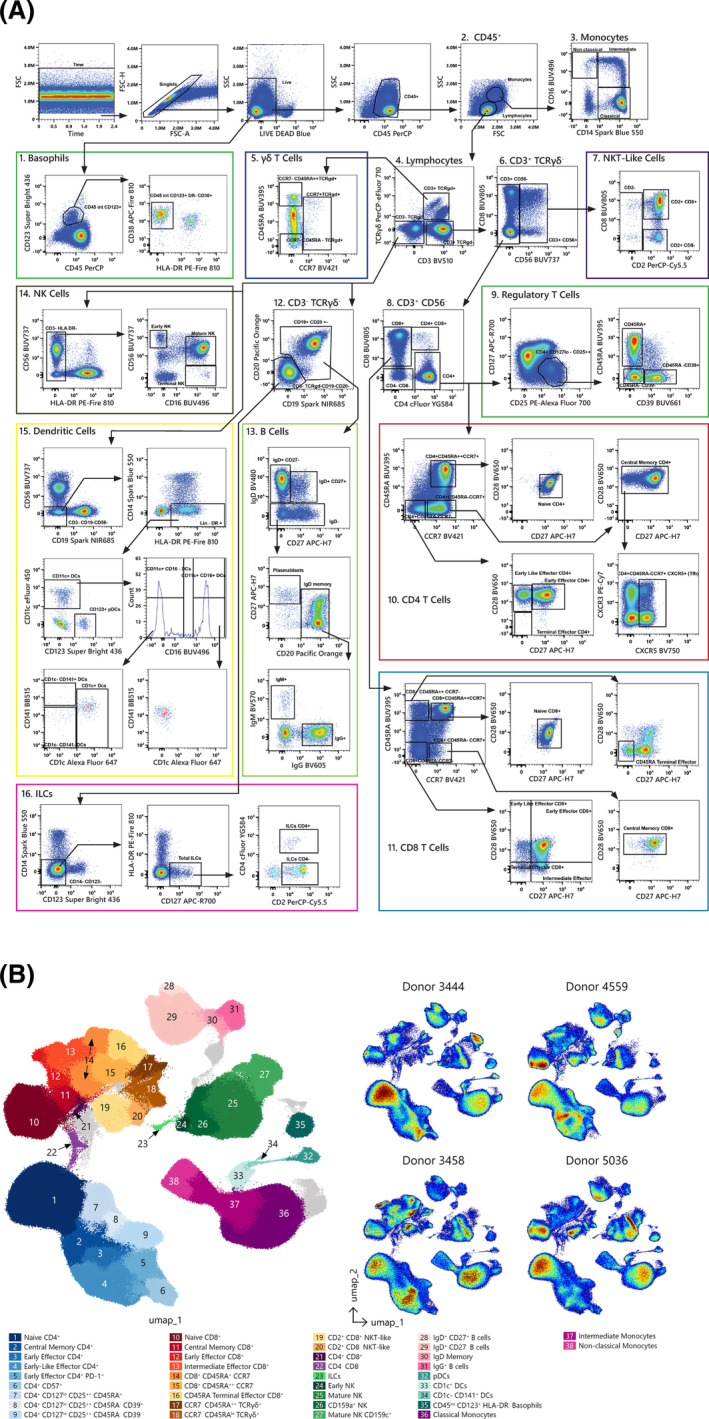
A. Manual gating strategy. The gating strategy used to identify the main cellular subsets is presented. Arrows are used to visualize the relationships across plots, and numbers are used to call attention to populations described here. After doublets and dead cells were excluded, basophils (1) were delineated as CD45+CD123+HLA‐DR‐. Lymphocytes and monocytes (2) were gated based on FSC‐A/SSC‐A properties. Monocytes (3) were then classified by CD14 and CD16 expression as non‐classical (CD14−CD16+), intermediate (CD14+CD16+/low), and classical (CD14+CD16‐). From the lymphocyte gate (2), the following populations were identified: CD3−TCRγδ−, CD3+TCRγδ+, and CD3+TCRγδ− (4). The CD3+TCRγδ+ population (5) was characterized based on CD45RA and CCR7 expression. The CD3+TCRγδ− population was divided in CD3+CD56+ (NKT‐like) and CD3+CD56− subsets (6). The inclusion of CD2 and CD8 enables further classification of the NKT‐like cells (7). CD4+, CD8+, CD4+CD8+ and CD4−CD8− T cells were identified from the CD3+CD56− gate (8). Tregs were identified from the CD4+ population using CD127 and CD25 expression (CD127lo/‐CD25hi) and CD39 and CD45RA were used to further classify these cells (9). CCR7, CD45RA, CD27, and CD28 allowed for further classification of memory/effector CD4 and CD8 T cell subsets (10, 11). CD19+ and/or CD20+ cells (B cells) were gated out of the CD3−TCRγδ− population (12). CD19+CD20+/− cells were further gated as IgD+CD27−, IgD+CD27+, or IgD−CD27+/−; the IgD−CD27+/− subset was divided into plasmablasts or IgD− memory B cells based on CD20 expression and IgG and IgM expression were assessed within the IgD‐ memory B cells (13). NK cells were defined as CD3−TCRγδ−HLA‐DR− and classified as early NK (CD56+CD16−), mature NK (CD56+CD16+), and terminal NK (CD56−CD16+) cells (14). Dendritic cells (DCs, 15) were identified first by gating on CD3−CD19−CD56−CD14−HLA‐DR+ and from there CD123+ (pDCs) and CD11c+ DCs were identified. CD11c+ DCs were further divided into CD16− and CD16+. CD1c and CD141 were then used to further classify the CD11c+CD16− and CD11c+CD16+ DCs. Finally, innate lymphoid cells (ILCs, 16) were identified as CD3−CD19−CD20−CD14−CD123−CD127+ and subsetted based on CD2 and CD4 expression. All data presented is derived from frozen PBMCs of one healthy donor (donor ID 4559). B. High‐dimensional data analysis on PBMCs from four donors displaying FlowSOM clusters projected on to two UMAP dimensions to show concordance between manual and automated analysis techniques. the overlay plot shows concatenated events from all four samples, while the density plots show differences in population distribution between the individual samples. As expected with a combination of high‐resolution and high‐dimensional data, several clusters contain events that evade a canonical definition. These populations are displayed in gray.
This 40‐color panel (Table 2) presents a powerful tool for in‐depth characterization of lymphocytes, monocytes, and dendritic cells present in human peripheral blood. It covers almost the entire cellular composition of the human peripheral immune system and will be particularly useful for studies in which sample availability is limited or unique biomarker signatures are sought. Taking advantage of full spectrum cytometry, we present a panel that highlights the first published OMIP to go beyond 28‐color fluorescence flow cytometry with excellent population resolution.
Table 2.
Reagents used for OMIP‐069
| Specificity | Fluorochrome | Clone | Purpose |
|---|---|---|---|
| Viability | Live Dead UV Blue | — | Viability |
| CD45 | PerCP | HI30 | Leukocytes |
| CD3 | BV510 | SK7 | Pan T cell, NKT‐Like cells |
| CD4 | cFluor YG584 | SK3 | CD4 T and NKT‐Like cells |
| CD8 | BUV805 | SK1 | CD8 T, NK, and NKT‐Like cells |
| CD25 | PE‐Alexa Fluor700 | CD25‐3G10 | Regulatory T cells |
| TCRγδ | PerCP‐eFluor 710 | B1.1 | Pan γδ T cell |
| CD14 | Spark Blue 550 | 63D3 | Monocyte differentiation |
| CD16 | BUV496 | 3G8 | Monocyte, NK cell, and dendritic cell differentiation |
| CD11c | eFluor 450 | 3.9 | Dendritic Cell differentiation |
| CD19 | Spark NIR 685 | HIB19 | B cells |
| CD20 | Pacific Orange | HI47 | B cells |
| CD24 | PE‐Alexa Fluor 610 | SN3 | B cell differentiation |
| CD39 | BUV661 | TU66 | B cell, Tregs, and monocyte differentiation |
| IgD | BV480 | IA6‐2 | B cell differentiation |
| IgG | BV605 | G18‐145 | B cell differentiation |
| IgM | BV570 | MHM‐88 | B cell differentiation |
| CD141 | BB515 | 1A4 | Dendritic cell differentiation |
| CD1c | Alexa Fluor 647 | L161 | Dendritic cells, NKT‐Like cells |
| CD123 | Super Bright 436 | 6H6 | Plasmacytoid dendritic cells |
| CD2 | PerCP‐Cy5.5 | TS1/8 | NK cell differentiation |
| CD56 | BUV737 | NCAM16.2 | Pan NK cell, γδ T cell activation |
| CCR7 | BV421 | G043H7 | T cell differentiation |
| CD27 | APC‐H7 | M‐T271 | T and B cell differentiation |
| CD28 | BV650 | CD28.2 | T cell and NK cell differentiation |
| CD45RA | BUV395 | 5H9 | T cell and dendritic cell differentiation |
| CD95 | PE‐Cy5 | DX2 | T cell and B cell differentiation |
| CD127 | APC‐R700 | HIL‐7R‐M21 | Cytokine receptor; T cell differentiation |
| CD337 | PE‐Dazzle594 | P30‐15 | NK cell differentiation |
| CCR6 | BV711 | G034E3 | Chemokine receptor; T cell and B cell differentiation |
| CCR5 | BUV563 | 2D7/CCR5 | Chemokine receptor; Monocyte, dendritic cell, T cell, and B cell differentiation |
| CXCR5 | BV750 | RF8B2 | Chemokine receptor; T cell differentiation |
| CXCR3 | PE‐Cy7 | G025H7 | Chemokine receptor; Dendritic cell, T cell, and B cell differentiation |
| HLA‐DR | PE‐Fire810 | L243 | T cell and monocyte activation, NK cell lineage discrimination, dendritic cell lineage marker |
| CD38 | APC‐Fire810 | HIT2 | Monocyte, dendritic cell, T cell, and B cell activation/differentiation |
| CD57 | FITC | HNK‐1 | NK and CD8+ T cell immune senescence |
| PD‐1 | BV785 | EH12.2H7 | T cell inhibitory receptor |
| CD159a | APC | REA110 | NK, NKT‐Like, and γδ T cell activation/differentiation |
| CD159c | PE | REA205 | NK cell differentiation |
| CD314 | BUV615 | 1D11 | NK cell differentiation |
Table 1.
Summary table for application of OMIP‐069
| Purpose | Deep subset profiling of immune cells to include subsets of T, B, NK, NKT, Monocyte, and Dendritic cells |
|---|---|
| Species | Human |
| Cell type | PBMCs |
| Cross references | OMIP‐003, OMIP‐004, OMIP‐006, OMIP‐013, OMIP‐015, OMIP‐017, OMIP‐021, OMIP‐023, OMIP‐024, OMIP‐029, OMIP‐030, OMIP‐033, OMIP‐034, OMIP‐039, OMIP‐042, OMIP‐044, OMIP‐050, OMIP‐051, OMIP‐058, OMIP‐060, OMIP‐063 |
Similarity to published OMIPs
This panel is similar to OMIPs ‐015, ‐023, ‐024, ‐030, ‐033, ‐034, ‐042, ‐50, ‐058, ‐063, which are all aimed at identifying the main leukocyte subsets in human blood. It partially overlaps with OMIPs ‐013, ‐017, ‐021, ‐030, and ‐060 for characterization of T cells; OMIPs ‐004, ‐006, and ‐015 for Treg immunophenotyping; OMIP ‐044 for dendritic cells; OMIPs ‐003, ‐033, and ‐051 for B cells; and OMIPs −029, and ‐039 for NK cells (6, 7, 8, 9, 10, 11, 22, 29, 44, 45, 55, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96).
Statement of ethical use of human samples
All human PBMCs used in this study were obtained from AllCells Alameda. Ethical review and regulatory compliance were conducted by Alpha Independent Review Board under Protocol number: 7000‐SOP‐045 (effective through April 26, 2021).
Author contributions
Lily M. Park: Conceptualization; data curation; formal analysis; methodology; validation; visualization. Joanne Lannigan: Data curation; formal analysis; project administration; resources; visualization; writing ‐ original draft; writing ‐ review and editing. Maria C. Jaimes: Conceptualization; data curation; formal analysis; methodology; project administration; resources; software; supervision; validation; visualization; writing ‐ original draft; writing ‐ review and editing.
Conflict of Interest
Lily Park and Maria C. Jaimes are employees of Cytek Biosciences, Inc., the manufacturer of the Aurora full spectrum flow cytometer used in these studies. Joanne Lannigan is a paid consultant for Cytek Biosciences, Inc.
Supporting information
Appendix S1 Supporting Information.
MIFlowCyt MIFlowCyt‐Compliant Items
Acknowledgments
The authors would like to thank BioLegend® for kindly providing the custom HLA‐DR PE‐Fire810 and CD38 APC‐Fire810 (now commercially available). In addition, the authors would like to thank Janelle Shook and James Wei for their assistance with the manuscript and figure preparation, Geoff Kraker for doing the unsupervised analysis of the data, Patrick Duncker for critical reading of the manuscript, and Huimin Gu for her extensive work at testing different fluorochrome combinations in preparation for this panel development.
Literature Cited
- 1. Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol 2012;12:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aghaeepour N, Chattopadhyay PK, Ganesan A, O'Neill K, Zare H, Jalali A, Hoos HH, Roederer M, Brinkman RR. Early immunologic correlates of HIV protection can be identified from computational analysis of complex multivariate T‐cell flow cytometry assays. Bioinformatics 2012;28:1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, Scriba TJ, Mahomed H, Hanekom W, Bart PA, et al. COMPASS identifies T‐cell subsets correlated with clinical outcomes. Nat Biotechnol 2015;33:610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chattopadhyay PK, Roederer M. A mine is a terrible thing to waste: High content, single cell technologies for comprehensive immune analysis. Am J Transplant 2015;15:1155–1161. [DOI] [PubMed] [Google Scholar]
- 5. Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen‐colour flow cytometry: Unravelling the immune system. Nat Rev Immunol 2004;4:648–655. [DOI] [PubMed] [Google Scholar]
- 6. Liechti T, Roederer M. OMIP‐058: 30‐parameter flow cytometry panel to characterize iNKT, NK, unconventional and conventional T cells. Cytom Part A 2019;95A:946–951. [DOI] [PubMed] [Google Scholar]
- 7. Liechti T, Roederer M. OMIP‐060: 30‐parameter flow cytometry panel to assess T cell effector functions and regulatory T cells. Cytom Part A 2019;95A:1129–1134. [DOI] [PubMed] [Google Scholar]
- 8. Liechti T, Roederer M. OMIP‐051 ‐ 28‐color flow cytometry panel to characterize B cells and myeloid cells. Cytom Part A 2019;95A:150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mair F, Prlic M. OMIP‐044: 28‐color immunophenotyping of the human dendritic cell compartment. Cytometry Part A 2018;93A:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nettey L, Giles AJ, Chattopadhyay PK. OMIP‐050: A 28‐color/30‐parameter fluorescence flow cytometry panel to enumerate and characterize cells expressing a wide Array of immune checkpoint molecules. Cytom Part A 2018;93A:1094–1096. [DOI] [PubMed] [Google Scholar]
- 11. Payne K, Li W, Salomon R, Ma CS. OMIP‐063: 28‐color flow cytometry panel for broad human Immunophenotyping. Cytom Part A 2020;97A:777–781. [DOI] [PubMed] [Google Scholar]
- 12. Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time‐of‐flight mass spectrometry. Anal Chem 2009;81:6813–6822. [DOI] [PubMed] [Google Scholar]
- 13. Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single‐cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–696.21551058 [Google Scholar]
- 14. Tsai AG, Glass DR, Juntilla M, Hartmann FJ, Oak JS, Fernandez‐Pol S, Ohgami RS, Bendall SC. Multiplexed single‐cell morphometry for hematopathology diagnostics. Nat Med 2020;26:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wade CG, Rhyne RH Jr, Woodruff WH, Bloch DP, Bartholomew JC. Spectra of cells in flow cytometry using a vidicon detector. J Histochem Cytochem 1979;27:1049–1052. [DOI] [PubMed] [Google Scholar]
- 16. Futamura K, Sekino M, Hata A, Ikebuchi R, Nakanishi Y, Egawa G, Kabashima K, Watanabe T, Furuki M, Tomura M. Novel full‐spectral flow cytometry with multiple spectrally‐adjacent fluorescent proteins and fluorochromes and visualization of in vivo cellular movement. Cytom Part A 2015;87A:830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gauci MR, Vesey G, Narai J, Veal D, Williams KL, Piper JA. Observation of single‐cell fluorescence spectra in laser flow cytometry. Cytometry 1996;25:388–393. [DOI] [PubMed] [Google Scholar]
- 18. Nolan JP, Condello D. Spectral flow cytometry. Curr Protoc Cytom 2013;63(1):1.27.1–1.27.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanders CK, Mourant JR. Advantages of full spectrum flow cytometry. J Biomed Opt 2013;18:037004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gregori G, Patsekin V, Rajwa B, Jones J, Ragheb K, Holdman C, Robinson JP. Hyperspectral cytometry at the single‐cell level using a 32‐channel photodetector. Cytom Part A 2012;81A:35–44. [DOI] [PubMed] [Google Scholar]
- 21. Brodie TM, Tosevski V, Medova M. OMIP‐045: Characterizing human head and neck tumors and cancer cell lines with mass cytometry. Cytom Part A 2018;93A:406–410. [DOI] [PubMed] [Google Scholar]
- 22. Baumgart S, Peddinghaus A, Schulte‐Wrede U, Mei HE, Grutzkau A. OMIP‐034: Comprehensive immune phenotyping of human peripheral leukocytes by mass cytometry for monitoring immunomodulatory therapies. Cytom Part A 2017;91A:34–38. [DOI] [PubMed] [Google Scholar]
- 23. Dusoswa SA, Verhoeff J, Garcia‐Vallejo JJ. OMIP‐054: Broad immune phenotyping of innate and adaptive leukocytes in the brain, spleen, and bone marrow of an Orthotopic murine glioblastoma model by mass cytometry. Cytom Part A 2019;95A:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jaracz‐Ros A, Hemon P, Krzysiek R, Bachelerie F, Schlecht‐Louf G, Gary‐Gouy H. OMIP‐048 MC: Quantification of calcium sensors and channels expression in lymphocyte subsets by mass cytometry. Cytom Part A 2018;93A:681–684. [DOI] [PubMed] [Google Scholar]
- 25. Maecker HT, Harari A. Immune monitoring technology primer: Flow and mass cytometry. J Immunother Cancer 2015;3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nassar AF, Wisnewski AV, Raddassi K. Automation of sample preparation for mass cytometry barcoding in support of clinical research: Protocol optimization. Anal Bioanal Chem 2017;409:2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olsen LR, Leipold MD, Pedersen CB, Maecker HT. The anatomy of single cell mass cytometry data. Cytom Part A 2019;95A:156–172. [DOI] [PubMed] [Google Scholar]
- 28. Montaldo E, Del Zotto G, Della Chiesa M, Mingari MC, Moretta A, De Maria A, Moretta L. Human NK cell receptors/markers: A tool to analyze NK cell development, subsets and function. Cytom Part A 2013;83A:702–713. [DOI] [PubMed] [Google Scholar]
- 29. Mahnke YD, Beddall MH, Roederer M. OMIP‐029: Human NK‐cell phenotypization. Cytom Part A 2015;87A:986–988. [DOI] [PubMed] [Google Scholar]
- 30. Creelan BC, Antonia SJ. The NKG2A immune checkpoint ‐ a new direction in cancer immunotherapy. Nat Rev Clin Oncol 2019;16:277–278. [DOI] [PubMed] [Google Scholar]
- 31. Ma M, Wang Z, Chen X, Tao A, He L, Fu S, Zhang Z, Fu Y, Guo C, Liu J, et al. NKG2C(+)NKG2A(−) natural killer cells are associated with a lower viral set point and may predict disease progression in individuals with primary HIV infection. Front Immunol 2017;8:1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielsen CM, White MJ, Goodier MR, Riley EM. Functional significance of CD57 expression on human NK cells and relevance to disease. Front Immunol 2013;4:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang Y, Cui X, Cui C, Zhang J, Zhou F, Zhang Z, Fu Y, Xu J, Chu Z, Liu J, et al. The function of CD3+CD56+ NKT‐like cells in HIV‐infected individuals. Biomed Res Int 2014;2014:863625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wan F, Hu CB, Ma JX, Gao K, Xiang LX, Shao JZ. Characterization of γδ T cells from zebrafish provides insights into their important role in adaptive humoral immunity. Front Immunol 2016;7:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davey MS, Willcox CR, Joyce SP, Ladell K, Kasatskaya SA, McLaren JE, Hunter S, Salim M, Mohammed F, Price DA, et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR‐dependent adaptive immune surveillance. Nat Commun 2017;8:14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holtmeier W, Kabelitz D. Gammadelta T cells link innate and adaptive immune responses. Chem Immunol Allergy 2005;86:151–183. [DOI] [PubMed] [Google Scholar]
- 37. Bank I . The role of Gamma Delta T cells in autoimmune rheumatic diseases. Cell 2020;9(2):462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryan PL, Sumaria N, Holland CJ, Bradford CM, Izotova N, Grandjean CL, Jawad AS, Bergmeier LA, Pennington DJ. Heterogeneous yet stable Vdelta2(+) T‐cell profiles define distinct cytotoxic effector potentials in healthy human individuals. Proc Natl Acad Sci U S A 2016;113:14378–14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dieli F, Poccia F, Lipp M, Sireci G, Caccamo N, Di Sano C, Salerno A. Differentiation of effector/memory Vdelta2 T cells and migratory routes in lymph nodes or inflammatory sites. J Exp Med 2003;198:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who's who of T‐cell differentiation: Human memory T‐cell subsets. Eur J Immunol 2013;43:2797–2809. [DOI] [PubMed] [Google Scholar]
- 41. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–712. [DOI] [PubMed] [Google Scholar]
- 42. Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: Consensus and issues. Cytom Part A 2008;73A:975–983. [DOI] [PubMed] [Google Scholar]
- 43. Oja AE, Piet B, van der Zwan D, Blaauwgeers H, Mensink M, de Kivit S, Borst J, Nolte MA, van Lier RAW, Stark R, et al. Functional heterogeneity of CD4(+) tumor‐infiltrating lymphocytes with a resident memory phenotype in NSCLC. Front Immunol 2018;9:2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Staser KW, Eades W, Choi J, Karpova D, DiPersio JF. OMIP‐042: 21‐color flow cytometry to comprehensively immunophenotype major lymphocyte and myeloid subsets in human peripheral blood. Cytom Part A 2018;93A:186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wingender G, Kronenberg M. OMIP‐030: Characterization of human T cell subsets via surface markers. Cytom Part A 2015;87A:1067–1069. [DOI] [PubMed] [Google Scholar]
- 46. Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long‐lived memory cells. Nat Immunol 2003;4:1191–1198. [DOI] [PubMed] [Google Scholar]
- 47. Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, et al. Expression of CD57 defines replicative senescence and antigen‐induced apoptotic death of CD8+ T cells. Blood 2003;101:2711–2720. [DOI] [PubMed] [Google Scholar]
- 48. Focosi D, Bestagno M, Burrone O, Petrini M. CD57+ T lymphocytes and functional immune deficiency. J Leukoc Biol 2010;87:107–116. [DOI] [PubMed] [Google Scholar]
- 49. Meditz AL, Haas MK, Folkvord JM, Melander K, Young R, McCarter M, Mawhinney S, Campbell TB, Lie Y, Coakley E, et al. HLA‐DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5‐tropic HIV‐1 RNA in vivo. J Virol 2011;85:10189–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ndhlovu ZM, Kamya P, Mewalal N, Kloverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, et al. Magnitude and kinetics of CD8+ T cell activation during Hyperacute HIV infection impact viral set point. Immunity 2015;43:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z, Zhu L, Nguyen THO, Wan Y, Sant S, Quinones‐Parra SM, Crawford JC, Eltahla AA, Rizzetto S, Bull RA, et al. Clonally diverse CD38(+)HLA‐DR(+)CD8(+) T cells persist during fatal H7N9 disease. Nat Commun 2018;9:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duraiswamy J, Ibegbu CC, Masopust D, Miller JD, Araki K, Doho GH, Tata P, Gupta S, Zilliox MJ, Nakaya HI, et al. Phenotype, function, and gene expression profiles of programmed death‐1(hi) CD8 T cells in healthy human adults. J Immunol 2011;186:4200–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD‐L1 checkpoint. Immunity 2018;48:434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu W, Putnam AL, Xu‐Yu Z, Szot GL, Lee MR, Zhu S, Gottlieb PA, Kapranov P, Gingeras TR, Fazekas de St Groth B, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med 2006;203:1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mahnke YD, Beddall MH, Roederer M. OMIP‐015: Human regulatory and activated T‐cells without intracellular staining. Cytom Part A 2013;83A:179–181. [DOI] [PubMed] [Google Scholar]
- 56. Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 2007;204:1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dwyer KM, Hanidziar D, Putheti P, Hill PA, Pommey S, McRae JL, Winterhalter A, Doherty G, Deaglio S, Koulmanda M, et al. Expression of CD39 by human peripheral blood CD4+ CD25+ T cells denotes a regulatory memory phenotype. Am J Transplant 2010;10:2410–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, Hopner S, Centonze D, Bernardi G, Dell'Acqua ML, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: Hydrolysis of extracellular ATP and immune suppression. Blood 2007;110:1225–1232. [DOI] [PubMed] [Google Scholar]
- 59. Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med 1998;188:1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. von Borstel A, Lintermans LL, Heeringa P, Rutgers A, Stegeman CA, Sanders JS, Abdulahad WH. Circulating CD24hiCD38hi regulatory B cells correlate inversely with the ThEM17 cell frequency in granulomatosis with polyangiitis patients. Rheumatology (Oxford) 2019;58(8):1361–1366. [DOI] [PubMed] [Google Scholar]
- 61. Rosser EC, Mauri C. Regulatory B cells: Origin, phenotype, and function. Immunity 2015;42:607–612. [DOI] [PubMed] [Google Scholar]
- 62. Avery DT, Ellyard JI, Mackay F, Corcoran LM, Hodgkin PD, Tangye SG. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. J Immunol 2005;174:4034–4042. [DOI] [PubMed] [Google Scholar]
- 63. Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol 2008;20:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu Rev Immunol 2014;32:659–702. [DOI] [PubMed] [Google Scholar]
- 65. Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, et al. CCR5 levels and expression pattern correlate with infectability by macrophage‐tropic HIV‐1, in vitro. J Exp Med 1997;185:1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Glatzel A, Wesch D, Schiemann F, Brandt E, Janssen O, Kabelitz D. Patterns of chemokine receptor expression on peripheral blood gamma delta T lymphocytes: Strong expression of CCR5 is a selective feature of V delta 2/V gamma 9 gamma delta T cells. J Immunol 2002;168:4920–4929. [DOI] [PubMed] [Google Scholar]
- 67. Lim HW, Broxmeyer HE, Kim CH. Regulation of trafficking receptor expression in human forkhead box P3+ regulatory T cells. J Immunol 2006;177:840–851. [DOI] [PubMed] [Google Scholar]
- 68. Geginat J, Paroni M, Facciotti F, Gruarin P, Kastirr I, Caprioli F, Pagani M, Abrignani S. The CD4‐centered universe of human T cell subsets. Semin Immunol 2013;25:252–262. [DOI] [PubMed] [Google Scholar]
- 69. Elgueta R, Marks E, Nowak E, Menezes S, Benson M, Raman VS, Ortiz C, O'Connell S, Hess H, Lord GM, et al. CCR6‐dependent positioning of memory B cells is essential for their ability to mount a recall response to antigen. J Immunol 2015;194:505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maurice NJ, McElrath MJ, Andersen‐Nissen E, Frahm N, Prlic M. CXCR3 enables recruitment and site‐specific bystander activation of memory CD8(+) T cells. Nat Commun 2019;10:4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chevalier N, Jarrossay D, Ho E, Avery DT, Ma CS, Yu D, Sallusto F, Tangye SG, Mackay CR. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J Immunol 2011;186:5556–5568. [DOI] [PubMed] [Google Scholar]
- 72. Henneken M, Dorner T, Burmester GR, Berek C. Differential expression of chemokine receptors on peripheral blood B cells from patients with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther 2005;7:R1001–R1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ziegler‐Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ, et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010;116:e74–e80. [DOI] [PubMed] [Google Scholar]
- 74. Strauss‐Ayali D, Conrad SM, Mosser DM. Monocyte subpopulations and their differentiation patterns during infection. J Leukoc Biol 2007;82:244–252. [DOI] [PubMed] [Google Scholar]
- 75. Han X, Jorgensen JL, Brahmandam A, Schlette E, Huh YO, Shi Y, Awagu S, Chen W. Immunophenotypic study of basophils by multiparameter flow cytometry. Arch Pathol Lab Med 2008;132:813–819. [DOI] [PubMed] [Google Scholar]
- 76. Bianca Bennstein S, Riccarda Manser A, Weinhold S, Scherenschlich N, Uhrberg M. OMIP‐055: Characterization of human innate lymphoid cells from neonatal and peripheral blood. Cytom Part A 2019;95A:427–430. [DOI] [PubMed] [Google Scholar]
- 77. Roan F, Stoklasek TA, Whalen E, Molitor JA, Bluestone JA, Buckner JH, Ziegler SF. CD4+ group 1 innate lymphoid cells (ILC) form a functionally distinct ILC subset that is increased in systemic sclerosis. J Immunol 2016;196:2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, Chen CJ, Dunbar PR, Wadley RB, Jeet V, et al. Human CD141+ (BDCA‐3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross‐presents necrotic cell antigens. J Exp Med 2010;207:1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology 2013;140:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meskas J, Wang S, Brinkman R. flowCut — An R package for precise and accurate automated removal of outlier events and flagging of files based on time versus fluorescence analysis. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McInnes L, Healy J, Melville J. UMAP: Uniform manifold approximation and projection for dimension rReduction. ArXiv e‐prints 2018;1802.03426. [Google Scholar]
- 82. Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y. FlowSOM: Using self‐organizing maps for visualization and interpretation of cytometry data. Cytom Part A 2015;87A:636–645. [DOI] [PubMed] [Google Scholar]
- 83. Wei C, Jung J, Sanz I. OMIP‐003: Phenotypic analysis of human memory B cells. Cytom Part A 2011;79A:894–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Biancotto A, Dagur PK, Fuchs JC, Langweiler M, McCoy JP Jr. OMIP‐004: In‐depth characterization of human T regulatory cells. Cytom Part A 2012;81A:15–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Murdoch DM, Staats JS, Weinhold KJ. OMIP‐006: Phenotypic subset analysis of human T regulatory cells via polychromatic flow cytometry. Cytom Part A 2012;81A:281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mahnke YD, Beddall MH, Roederer M. OMIP‐013: Differentiation of human T‐cells. Cytom Part A 2012;81A:935–936. [DOI] [PubMed] [Google Scholar]
- 87. Mahnke YD, Beddall MH, Roederer M. OMIP‐017: Human CD4(+) helper T‐cell subsets including follicular helper cells. Cytom Part A 2013;83A:439–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brodie T, Brenna E, Sallusto F. OMIP‐018: Chemokine receptor expression on human T helper cells. Cytom Part A 2013;83A:530–532. [DOI] [PubMed] [Google Scholar]
- 89. Mahnke YD, Beddall MH, Roederer M. OMIP‐019: Quantification of human gammadeltaT‐cells, iNKT‐cells, and hematopoietic precursors. Cytom Part A 2013;83A:676–678. [DOI] [PubMed] [Google Scholar]
- 90. Wistuba‐Hamprecht K, Pawelec G, Derhovanessian E. OMIP‐020: Phenotypic characterization of human gammadelta T‐cells by multicolor flow cytometry. Cytom Part A 2014;85A:522–524. [DOI] [PubMed] [Google Scholar]
- 91. Gherardin NA, Ritchie DS, Godfrey DI, Neeson PJ. OMIP‐021: Simultaneous quantification of human conventional and innate‐like T‐cell subsets. Cytom Part A 2014;85A:573–575. [DOI] [PubMed] [Google Scholar]
- 92. Brodie T, Rothaeusler K, Sospedra M. OMIP‐033: A comprehensive single step staining protocol for human T‐ and B‐cell subsets. Cytom Part A 2016;89A:629–632. [DOI] [PubMed] [Google Scholar]
- 93. Hammer Q, Romagnani C. OMIP‐039: Detection and analysis of human adaptive NKG2C(+) natural killer cells. Cytom Part A 2017;91A:997–1000. [DOI] [PubMed] [Google Scholar]
- 94. Nowatzky J, Stagnar C, Manches O. OMIP‐053: Identification, classification, and isolation of major FoxP3 expressing human CD4(+) Treg subsets. Cytom Part A 2019;95A:264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bocsi J, Melzer S, Dahnert I, Tarnok A. OMIP‐023: 10‐color, 13 antibody panel for in‐depth phenotyping of human peripheral blood leukocytes. Cytom Part A 2014;85A:781–784. [DOI] [PubMed] [Google Scholar]
- 96. Moncunill G, Han H, Dobano C, McElrath MJ, De Rosa SC. OMIP‐024: Pan‐leukocyte immunophenotypic characterization of PBMC subsets in human samples. Cytom Part A 2014;85A:995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information.
MIFlowCyt MIFlowCyt‐Compliant Items


